
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 September 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01333
This article is part of the Research TopicActivation and Suppression of Plant ImmunityView all 27 articles
The cytosolic RNA exosome, a 3′→5′ exoribonuclease complex, contributes to mRNA degradation in eukaryotes, limiting the accumulation of poorly-translated, improperly translated, or aberrant mRNA species. Disruption of cytosolic RNA exosome activity allows aberrant RNA species to accumulate, which can then be detected by host antiviral immune systems as a signature of pathogen infection, activating antiviral defenses. SKI7 is a critical component of the cytosolic RNA exosome in yeast, bridging the catalytic exoribonuclease core with the SKI2/SKI3/SKI8 adaptor complex that guides aberrant RNA substrates into the exosome. The ortholog of SKI7 was only recently identified in humans as an alternative splice form of the HBS1 gene, which encodes a decoding factor translational GTPase that rescues stalled ribosomes. Here, we identify the plant orthologs of HBS1/SKI7. We found that HBS1 and SKI7 are typically encoded by alternative splice forms of a single locus, although some plant lineages have evolved subfunctionalized genes that apparently encode only HBS1 or only SKI7. In all plant lineages examined, the SKI7 gene is subject to regulation by alternative splicing that can yield unproductive transcripts, either by removing deeply conserved SKI7 coding sequences, or by introducing premature stop codons that render SKI7 susceptible to nonsense-mediated decay. Taking a comparative, evolutionary approach, we define crucial features of the SKI7 protein shared by all eukaryotes, and use these deeply conserved features to identify SKI7 proteins in invertebrate lineages. We conclude that SKI7 is a conserved cytosolic RNA exosome subunit across eukaryotic lineages, and that SKI7 is consistently regulated by alternative splicing, suggesting broad coordination of nuclear and cytosolic RNA metabolism.
Viruses dominate the biosphere, massively outnumbering cellular organisms (Koonin, 2017). Unlike cellular organisms, however, all viruses are obligate parasites that depend on organismal ribosomes for translation and replication (Walsh et al., 2013; Wang, 2015). Viral hosts are under strong selective pressure to recognize and limit viral parasitism, and in parallel, viruses are under strong selective pressure to evade host surveillance mechanisms and coopt the host translation machinery (Walsh et al., 2013; Molleston and Cherry, 2017). Host organisms have evolved various defense and immune systems to protect against viruses, including several approaches to detect non-host (viral) nucleic acids and then trigger defenses and guided destruction of viral genomes (Barrangou et al., 2007; Narayanan and Makino, 2013; Szittya and Burgyán, 2013; Abernathy and Glaunsinger, 2015; Li et al., 2015; Molleston and Cherry, 2017). A famous example is the evolution of bacterial “CRISPR-Cas” systems that incorporate viral dsDNA into short palindromic repeats in the host genome, and then transcribe these incorporated DNA sequences into guides that target DNases to specifically cleave complementary viral DNA (Brouns et al., 2008). Eukaryotes have evolved two major approaches to detect viral nucleic acids and mount immune responses. One of these, the interferon system, evolved in the Gnathostomata lineage (vertebrates with jaws) (Zou et al., 2009). In these organisms, pattern recognition receptors detect aberrant nucleic acids (such as dsRNA) and then rapidly activate transcription of a class of cytokines, called interferons, that are secreted, detected by cell surface receptors (of both infected and uninfected cells), and ultimately trigger transcriptional reprogramming that limits viral replication and spread (Li et al., 2015; Rigby and Rehwinkel, 2015). Outside the Gnathostomata lineage, most eukaryotes employ RNA interference (RNAi) systems to combat viral infections (Shabalina and Koonin, 2008).
RNA interference relies on endoribonucleases of the RNase III family that recognize and cleave double-stranded RNA (dsRNA) as basal defense against viral infection (Seo et al., 2013; Nicholson, 2014). RNase III may recognize dsRNA synthesized by a viral replicase, or the host may synthesize dsRNA from viral RNA templates using an endogenous RNA-dependent RNA polymerase; in either case, the dsRNA is processed by RNase III (Blevins et al., 2006; Szittya and Burgyán, 2013). In plants, two RNase III enzymes, called DICER-LIKE 4 (DCL4) and DCL2, are primarily responsible for defense against RNA viruses (Szittya and Burgyán, 2013; Andika et al., 2015). Cleavage of viral dsRNA by DCL4/DCL2 generates viral short interfering RNAs (vsiRNAs). vsiRNAs are loaded into ARGONAUTE (AGO) proteins in RNAi complexes (RISCs) that search for further, complementary viral RNAs using the 21- or 22-nt siRNA sequence. Any RNA recognized by the RISC, presumably a viral genome, is then cleaved and degraded. In a poorly understood process, RDR6 can be recruited to the cleaved viral RNA to synthesize another dsRNA template for processing by DCL4/DCL2 (Qu et al., 2005, 2008; Qi et al., 2009). This RDR6-dependent process can amplify the number of antiviral siRNAs available for the immune system (Garcia-Ruiz et al., 2010).
The DCL4/DCL2/RDR6 surveillance system has since been coopted in the plant lineage to regulate endogenous gene expression (Peragine et al., 2004; Allen et al., 2005; Cuperus et al., 2010). After cleavage by a 22-nt miRNA (or, in exceptional cases, some 21-nt miRNAs), endogenous transcripts can become templates for copying by RDR6 and cleavage by DCL4/DCL2 into siRNAs (Allen et al., 2005). These siRNAs can then amplify silencing of the miRNA target by guiding RISCs to multiple sites in transcripts with the original miRNA target, or may act in trans by guiding the RISC to other transcripts with complementary (or nearly complementary) ∼21-nt sequences. The DCL4/DCL2/RDR6 system is used by plants to regulate diverse biological processes, including auxin-mediated developmental patterning (Peragine et al., 2004; Vazquez et al., 2004; Fahlgren et al., 2006) and suppression of disease resistance R genes (Li et al., 2012; Deng et al., 2018), whose overexpression can cause autoimmune syndromes (Yi and Richards, 2009).
After cleavage by RISC endonucleases, viral RNA must be degraded by host RNA exoribonucleases (Abernathy and Glaunsinger, 2015). Two major exoribonuclease mechanisms are conserved across eukaryotes: 5′→3′ RNA degradation by the EXORIBONUCLEASEs (XRNs) (Nagarajan et al., 2013) and 3′→5′ RNA degradation by the RNA exosome (Zinder and Lima, 2017). These enzymes are involved in housekeeping degradation of host mRNAs in the cytosol and in processing RNA transcripts in the nucleus. The XRNs have evolved two distinct gene families to handle these processes: the XRN1 family encodes a cytosolic enzyme, and the XRN2 family encodes a nuclear enzyme. In an ancestor of plants, an XRN2 paralog evolved cytosolic localization and the XRN1 gene lineage was lost; the plant cytosolic XRN is therefore called XRN4, but is functionally equivalent to eukaryotic XRN1 (Nagarajan et al., 2013). The RNA exosome forms a large, multiprotein complex, and localizes to both the cytosol and nucleus. Adaptor complexes then guide RNA substrates to the RNA exosome; for example, the nuclear TRAMP complex guides ribosomal RNA (rRNA) and small nucleolar RNA (snoRNA) to the RNA exosome for processing (Tollervey, 2015). In the cytosol, the RNA exosome is chaperoned to substrates by the SKI2/SKI3/SKI8 complex (Schmidt et al., 2016), which facilitates degradation of cleaved RNA (including deadenylated mRNAs) by guiding these RNAs into the RNA exosome catalytic core.
During RNAi, the SKI2/SKI3/SKI8 complex is recruited to siRNA-cleaved mRNAs by stalled ribosomes that reach the cleavage site (Orban and Izaurralde, 2005; Branscheid et al., 2015; Zhang et al., 2015; Szadeczky-Kardoss et al., 2018). Stalled ribosomes at the cleavage site are recognized by PELOTA/DOM34 (a translational decoding factor) and HBS1 (a translational GTPase), which recruit SKI2/SKI3/SKI8 and the RNA exosome to degrade the 5′ fragment of the cleaved mRNA (Orban and Izaurralde, 2005). Supporting this hypothesis, loss of PELOTA, HBS1, or SKI2 stabilizes the 5′ fragment of transcripts cleaved in their open reading frames by miRNA-guided RISCs, in both Drosophila and plants (Orban and Izaurralde, 2005; Szittya and Burgyán, 2013; Branscheid et al., 2015; Hashimoto et al., 2017; Szadeczky-Kardoss et al., 2018). Extending this model, we propose that PELOTA, HBS1, SKI2/SKI3/SKI8, and SKI7 are likely necessary for degradation of messenger viral RNAs after cleavage by vsiRNAs. Moreover, PELOTA, HBS1, SKI2/SKI3/SKI8, and SKI7 are all proposed to contribute to degradation of transcripts with premature termination codons via nonsense-mediated decay (NMD) (Mitchell and Tollervey, 2003; Takahashi et al., 2003; Arribere and Fire, 2018). Structural features of viral RNA can be recognized as nonsense transcripts by eukaryotic cells, leading to viral RNA degradation by NMD. Thus, PELOTA, HBS1, and SKI7 could have antiviral roles by participating in NMD. In tomato, a natural loss-of-function variant of pelota, called ty-5, confers resistance to Tomato yellow curly leaf virus (TYCLV), highlighting the importance of the PELOTA/HBS1 RNA degradation machinery in plant-virus interactions (Lapidot et al., 2015).
In humans and in plants, the SKI2 has been implicated in preventing endogenous RNA from triggering immune responses. Depletion of the human SKI2 ortholog, HsSKIV2L (Kalisiak et al., 2017), allows accumulation of aberrant RNA species that are sensed by nucleic acid pattern recognition receptors, which in turn activate type I interferon expression and trigger autoimmune/autoinflammatory responses (Eckard et al., 2014; Rigby and Rehwinkel, 2015). Loss-of-function mutations in SKIV2L have been genetically linked to autoimmune syndromes (Crow et al., 2006), including systemic lupus erythematosus (Crow et al., 2006), and this association with autoimmune syndromes may be related to its role in limiting autoinflammatory responses to endogenous RNAs (Eckard et al., 2014). In Arabidopsis thaliana, disruption of the SKI2 gene has a similar effect: deadenylated RNA species accumulate in the cytosol (e.g., transcripts cleaved by miRNAs), and become available as templates for RDR6 to generate dsRNA (Branscheid et al., 2015), a process that is comparable to RDR6 copying of cleaved viral RNA. These RDR6-dependent dsRNA molecules are subsequently processed by DCL4/DCL2, generating siRNAs that silence host gene expression. Thus, in both humans and plants, SKI2 is required to limit the accumulation of aberrant RNA species that are otherwise detected by the cell as potential viruses, triggering antiviral immune responses in the absence of pathogen attack.
Recently, structural studies of the cytoplasmic RNA exosome in yeast and humans have revealed the crucial importance of SKI7 in bridging the RNase exosome complex with the SKI2/3/8 adaptor complex that feeds cytosolic RNA substrates into the exosome (Kalisiak et al., 2017). SKI7 was first identified in the same genetic screen as the other cytoplasmic RNA exosome components, but unlike SKI2, SKI3, and SKI8, orthologs of SKI7 were not readily identifiable in other eukaryotic genomes. A genomic investigation of Lachancea kluyveri, a fungus closely related to S. cerevisiae, revealed that SKI7 is encoded by an alternative splice form of the HBS1 locus in that species (Marshall et al., 2013). In S. cerevisiae, HBS1 and SKI7 are functionally distinct homeologs that derive from a whole-genome duplication in a recent ancestor of S. cerevisiae; L. kluyveri diverged from this lineage shortly before the whole-genome duplication. The authors of this study noted briefly that the HBS1 locus is potentially alternatively spliced in other eukaryotic lineages, but did not systematically identify HBS1/SKI7 orthologs in metazoans or plants. Subsequent studies revealed that the vertebrate HBS1 locus is also alternatively spliced, and that one of these splice forms, HBS1Lv3, encodes a protein that serves the same function as SKI7 in S. cerevisiae (Kalisiak et al., 2017).
The discovery that SKI7 and HBS1 are encoded by the same locus in many fungi and vertebrates is perhaps surprising because of their apparently unrelated functions. HBS1 is a translational GTPase that is required for the release of stalled ribosomes from mRNA, along with its interacting partner, PELOTA/DOM34 (Figure 1; Shao et al., 2016). SKI7 is instead a bridge between the RNA exosome and the SKI2 complex, and while the S. cerevisiae SKI7 has a C-terminal HBS1-like GTPase domain, this terminus is dispensable for its functions (Horikawa et al., 2016). Highlighting their distinct functions, the two protein isoforms encoded by the HBS1/SKI7 locus in Lachancea kluyveri can complement only one of the S. cerevisiae Δhbs1 or Δski7 mutant strains: the long, SKI7-like isoform only complements Δski7, and the shorter, HBS1-like isoform only complements Δhbs1 (Marshall et al., 2013). In humans, the isoform encoding the functional ortholog of SKI7, HBS1Lv3, loses exons that encode the entire C-terminal HBS1 GTPase domain (which is essential for HBS1 functions), and instead gains an exon that encodes an RNA exosome-interacting surface (Figure 2A). It remains unclear why vertebrate and most fungal genomes would retain a single locus to encode both HBS1 and SKI7, whereas S. cerevisiae has successfully evolved two distinct loci to separate these functions. Here, we take advantage of these recent insights into the gene and protein structures of HBS1/SKI7 in other eukaryotes to identify and characterize HBS1/SKI7 orthologs in the plant lineage.
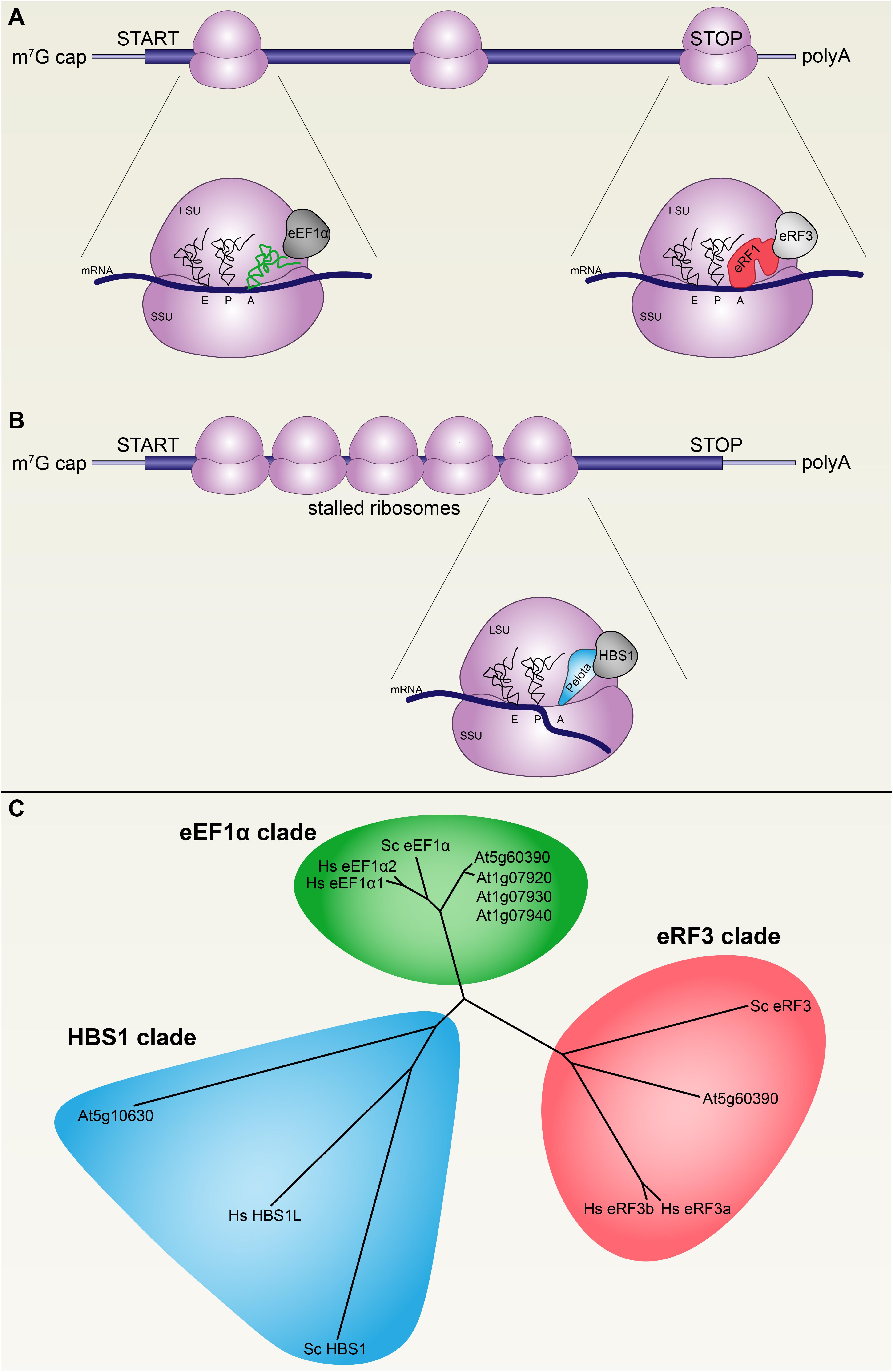
FIGURE 1. Decoding translational GTPases are conserved across eukaryotes. (A) During active translation, mRNA codons are decoded at the aminoacyl site (A site) of 80S ribosomes (purple) by cognate tRNAs (green) in complex with the GTPase eukaryotic translation elongation factor 1 alpha (eEF1α, gray). Stop codons are recognized at the A site by eukaryotic release factor 1 (eRF1) in complex with the GTPase eukaryotic release factor 3 (eRF3), which terminates translation and releases ribosomes. (B) Stalled ribosomes are recognized by PELOTA in complex with the GTPase HBS1, which triggers premature ribosome release and degradation of the mRNA and the nascent polypeptide. Ribosome may stall on mRNAs with complex secondary structures or after endonucleolytic cleavage by RNAi silencing complexes, among other possible causes. (C) The three decoding factor translational GTPases are conserved across the plant, fungal, and metazoan lineages, as represented in this phylogeny by protein sequences from Arabidopsis thaliana, Saccharomyces cerevisiae, and Homo sapiens, respectively.
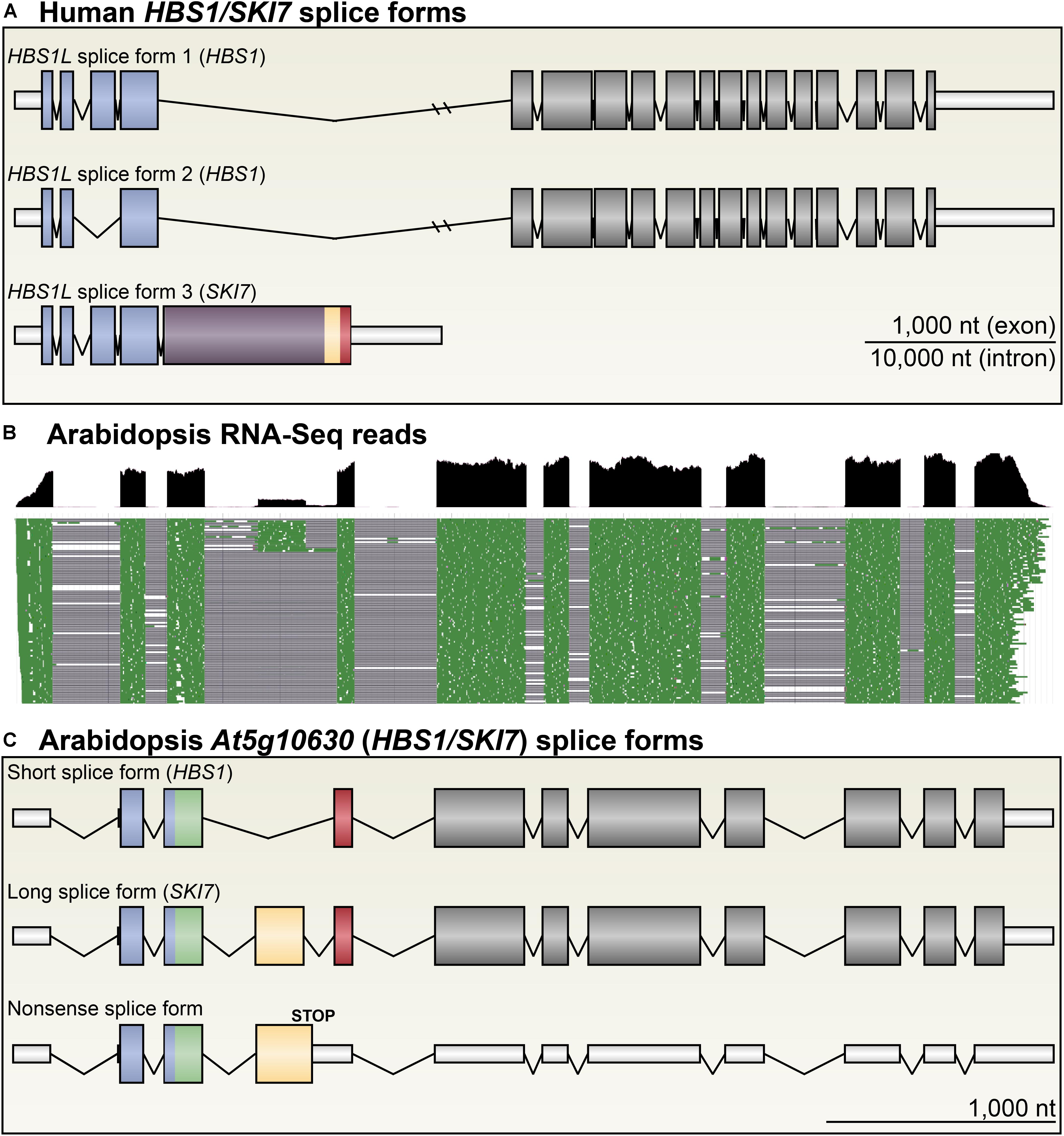
FIGURE 2. Arabidopsis HBS1/SKI7 is alternatively spliced. (A) The human HBS1/SKI7 locus, called HBS1L, produces three primary splice forms: HBS1L splice form 1, a long splice form with 18 exons; HBS1L splice form 2, a slightly shorter splice form skipping exon 3; and HBS1L splice form 3, which selects an alternative exon 5 followed by transcription termination. HBS1Lv3 includes sequences that promote interaction of the protein with the RNA exosome (yellow and red; see Figure 4A). (B) RNA-Seq of light-grown seedlings shows that At5g10630 is alternatively spliced into three major splice forms (Cheng et al., 2017). Cumulative RNA-Seq reads are shown in black (top panel), and select individual aligned reads are shown in green (bottom panel), with spliced sequences indicated by a black line. Note that reads for the fourth exon are ∼20% of the level of reads for the other coding sequence exons. (C) At5g10630 forms three major splice forms. A short splice form (top) skips exon 4 (yellow), yielding a transcript that encodes HBS1 (A). A long splice form (middle) includes exon 4 (yellow), yielding a transcript that encodes SKI7 (A). Rarely, an alternative acceptor site is selected for exon 4, adding five amino acids with no apparently functional consequence. A nonsense splice form (bottom) retains intron 4, which includes two codons, yielding a transcript that is likely subject to NMD. Exons are colored to match protein models in subsequent figures; UTRs are indicated with narrow, white bars.
Transcripts are decoded during translation by duplexes composed of a translational GTPase (trGTPase) and either an aminoacyl-tRNA or a ribosome release factor (Dever and Green, 2012; Shao et al., 2016). There are three major classes of these decoding trGTPases: eEF1α (eukaryotic Elongation Factor 1 alpha), which mediates delivery of aminoacyl-tRNAs to the 80S ribosome; eRF3 (eukaryotic Release Factor 3), which mediates delivery of eRF1 (eukaryotic Release Factor 1) to stop codons to terminate translation and facilitate ribosome dissociation from transcripts; and HBS1 (Hsp80 subfamily B Suppressor 1), which mediates delivery of PELOTA (a.k.a. Dom34 in yeast, Duplication Of Multilocus region) to stalled ribosomes to terminate translation and facilitate ribosome dissociation (Figures 1A,B) (Carr-Schmid et al., 2002; Becker et al., 2012; Shao et al., 2016; Hashimoto et al., 2017). Since all decoding trGTPases are similar to each other, we began by identifying orthologs of each of the three decoding factor trGTPases (eEF1α, eRF3, and HBS1) in Arabidopsis thaliana in order to confidently distinguish plant HBS1 from the other trGTPases (Figure 1C and Supplementary Data File 1). Separate loci encode orthologs of these proteins that localize to mitochondria and/or plastids; these were removed from our analysis to focus only on cytoplasmic proteins. Arabidopsis encodes one copy of eRF3 (At1g18070), four copies of eEF1α (At5g60390 and three tandem paralogs, At1g07920, At1g07930, and At1g07940), and one copy of HBS1 (At5g10630). The three tandem eEF1α paralogs evolved recently (this complex locus is not conserved across Brassicaceae). All of the decoding trGTPases are expressed throughout Arabidopsis development, although it should be noted that eRF3 transcripts are about one order of magnitude more abundant than HBS1 transcripts, and eEF1α transcripts are at least two orders of magnitude more abundant than HBS1 transcripts, consistent with their distinct roles in translation (Cheng et al., 2017).
The Arabidopsis At5g10630 (HBS1) locus is annotated with several different possible transcripts, but there are only two major protein isoforms predicted to be encoded by these transcripts: a long splice form (Figure 2C) encodes a protein that is 738 amino acids long (Figures 3, 4A), and a short splice form (Figure 2C) skips an exon to encode a shorter protein that is 668 amino acids long (Figures 3, 4A). High-throughput sequencing of RNA from light-grown seedlings shows that the alternative cassette exon is included in approximately 20% of At5g10630 transcripts (Figure 2B) (Cheng et al., 2017). A very minor splice form uses a weak 3′ splice site that adds 15nt to the 5′ end of the cassette exon; this is included in at most 4% of At5g10630 transcripts in light-grown seedlings. Because this minor splice form is rare and does not cause significant changes in the protein sequence (it neither induces a frame-shift nor includes a stop codon, and only adds 5 amino acids to the protein), it was not investigated any further. Another very minor splice form (also at most 4% of At5g10630 transcripts in light-grown seedlings, Figure 2B) retains the intron at the 3′ end of the cassette exon (Figures 2C, 3), which introduces two in-frame stop codons (this retained intron can thus be considered a “poison intron”). Since these stop codons are upstream of several exon-exon junctions, this splice form is presumably subject to NMD, and is any case unproductive. We confirmed the RNA sequences of these four splice forms of At5g10630 by RT-PCR using primers surrounding the alternatively spliced exons, followed by TOPO cloning and Sanger sequencing.
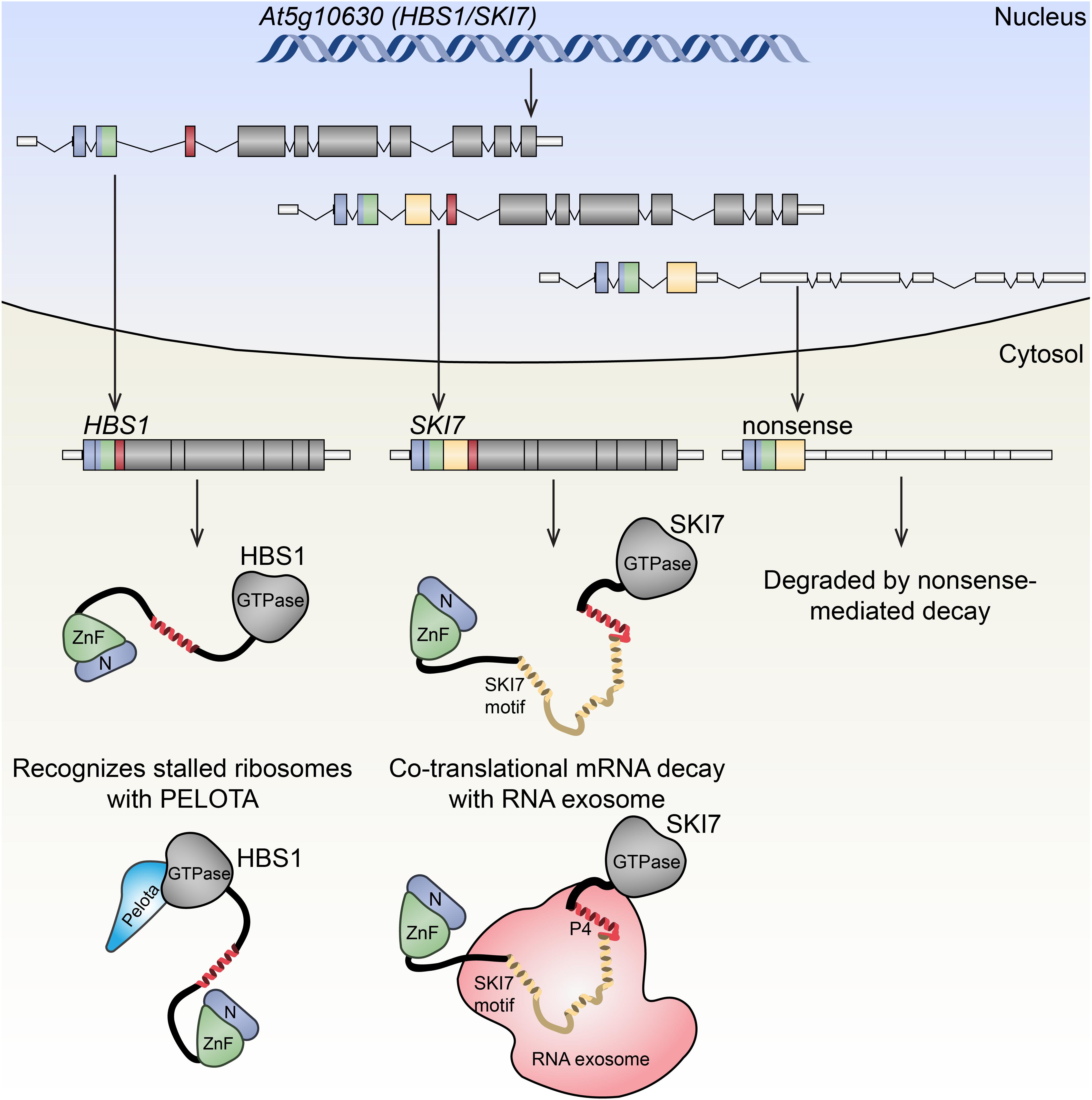
FIGURE 3. Alternative splicing of Arabidopsis HBS1/SKI7 has functional consequences. Arabidopsis HBS1/SKI7 (At5g10630) is alternatively spliced in the nucleus, generating at least three distinct transcripts that are exported to the nucleus. The HBS1 transcript (left) putatitively encodes a translational GTPase, HBS1, that interacts with the decoding factor PELOTA to recognize stalled ribosomes. The SKI7 transcript (center) putatitively encodes SKI7, a protein that associates with the cytosolic RNA exosome, a 3′→5′ exoribonuclease complex that degrades aberrant transcripts. A third splice form (right) encodes a premature translation codon in the fourth exon, presumably triggering NMD after a pioneer round of translation.
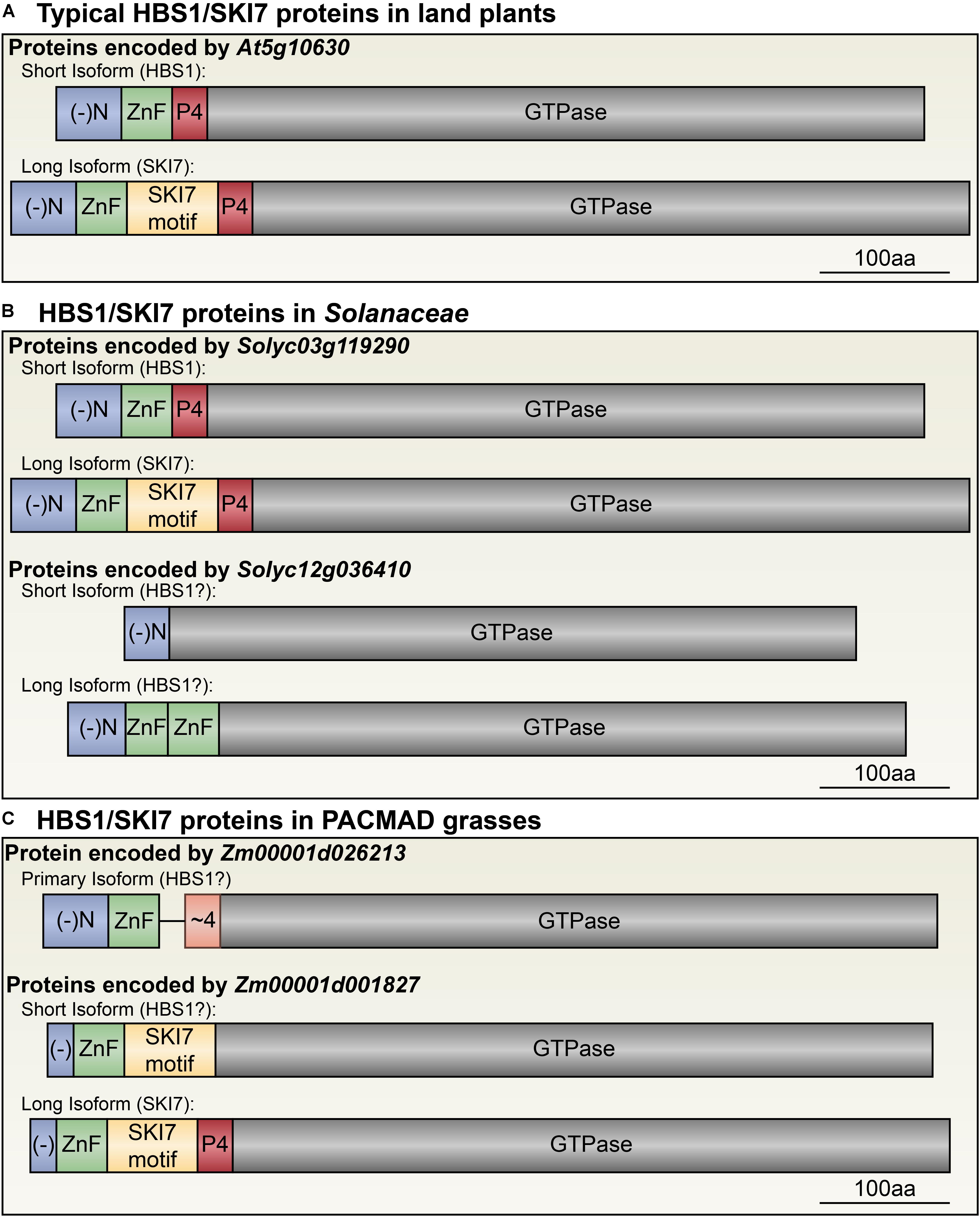
FIGURE 4. Diversity of HBS1/SKI7 isoforms among plant lineages. (A) The typical plant HBS1/SKI7 locus (here exemplified by At5g10630, but also found in P. patens, A. trichocarpa, T. cacao, M. truncata, N. benthamiana, and S. lycopersicum, among other species) encodes two protein isoforms, HBS1 (top) and SKI7 (bottom), defined by the inclusion or exclusion of the SKI7-like motif (yellow). (B) The Solanaceae have two distinct HBS1/SKI7-like loci: one is similar to the typical plant HBS1/SKI7 locus in panel a (exemplified by Solyc03g119290), and the second encodes only an HBS1-like protein, and is alternatively spliced to include or exclude two Zn finger domains (exemplified by Solyc12g036410). (C) In the PACMAD grasses, one HBS1/SKI7-like locus encodes an HBS1-like protein lacking the SKI7-like motif and with a poorly-aligned Patch 4-like motif (exemplified by Zm0001d026213). A second HBS1/SKI7-like locus (exemplified by Zm0001d001827) can encode two proteins: a long SKI7 isoform and a short isoform that excludes the Patch 4-like motif, and thus may function as an HBS1-like protein or be a loss-of-function isoform of SKI7. (-)N, blue, negatively charged N-terminus; ZnF, green, RAN2-type Zinc finger domain; SKI7 motif, yellow, the SKI7-like motif often encoded by a cassette exon; P4, red, the Patch 4-like motif; ∼4, orange, a poorly-aligned Patch 4-like motif unique to grass HBS1 isoforms; GTPase, gray, the HBS1/SKI7 decoding factor translational GTPase domain.
Comparative analysis of the two proteins predicted to be encoded by Arabidopsis HBS1/SKI7 with the functionally characterized orthologs of HBS1 (human HBS1Lv1, baker’s yeast Hbs1, and budding yeast Lachancea kluyveri HBS1) and SKI7 (human HBS1Lv3, baker’s yeast Ski7, and budding yeast L. kluyveri SKI7) allowed us to identify several conserved regions in the Arabidopsis proteins (Figures 4A, 5A). The N-terminus (aa 1–50) begins with a stretch of mostly negatively charged amino acids (40% of the first 50 amino acids are aspartic acid or glutamic acid, Figure 3). This is followed by a single Zinc finger domain of the RanBP2 superfamily (aa 51–75, Figure 5A), putatively involved in protein-protein interactions. If the transcript is alternatively spliced to include a cassette exon, the next region encodes an amino acid sequence with some similarity to the polypeptide encoded by the L. kluyveri alternative exon that determines SKI7 functionality (Marshall et al., 2013). This region includes a motif that corresponds to the HsSKI7 RxxxFxxxL motif required for recruiting HsSKI7 to the RNA exosome (Kalisiak et al., 2017). We have named this the “SKI7-like motif” (Figure 5A). Immediately after the sequence encoded by the cassette exon is the “Patch 4-like” motif (Figure 5A), named after the homologous yeast sequence, which was dubbed the “patch 4” motif (Kowalinski et al., 2016). In the human SKI7 protein, this is called the “PFDFxxxSPDDIVKxNQ motif” (Kalisiak et al., 2017). The Patch 4-like motif is found in all SKI7 proteins, but is not conserved in HBS1 proteins, and is proposed to mediate interactions between SKI7 and the RNA exosome subunit Csl4. Finally, the remaining C-terminus of the At5g10630 protein is an HBS1-like translational GTPase (Figure 4A). Thus, we defined the archetypical plant HBS1/SKI7 protein with five regions: a negatively charged N-terminus, a Zn-finger domain, a SKI7-like motif, a Patch 4-like motif, and the C-terminal GTPase (Figure 4A).
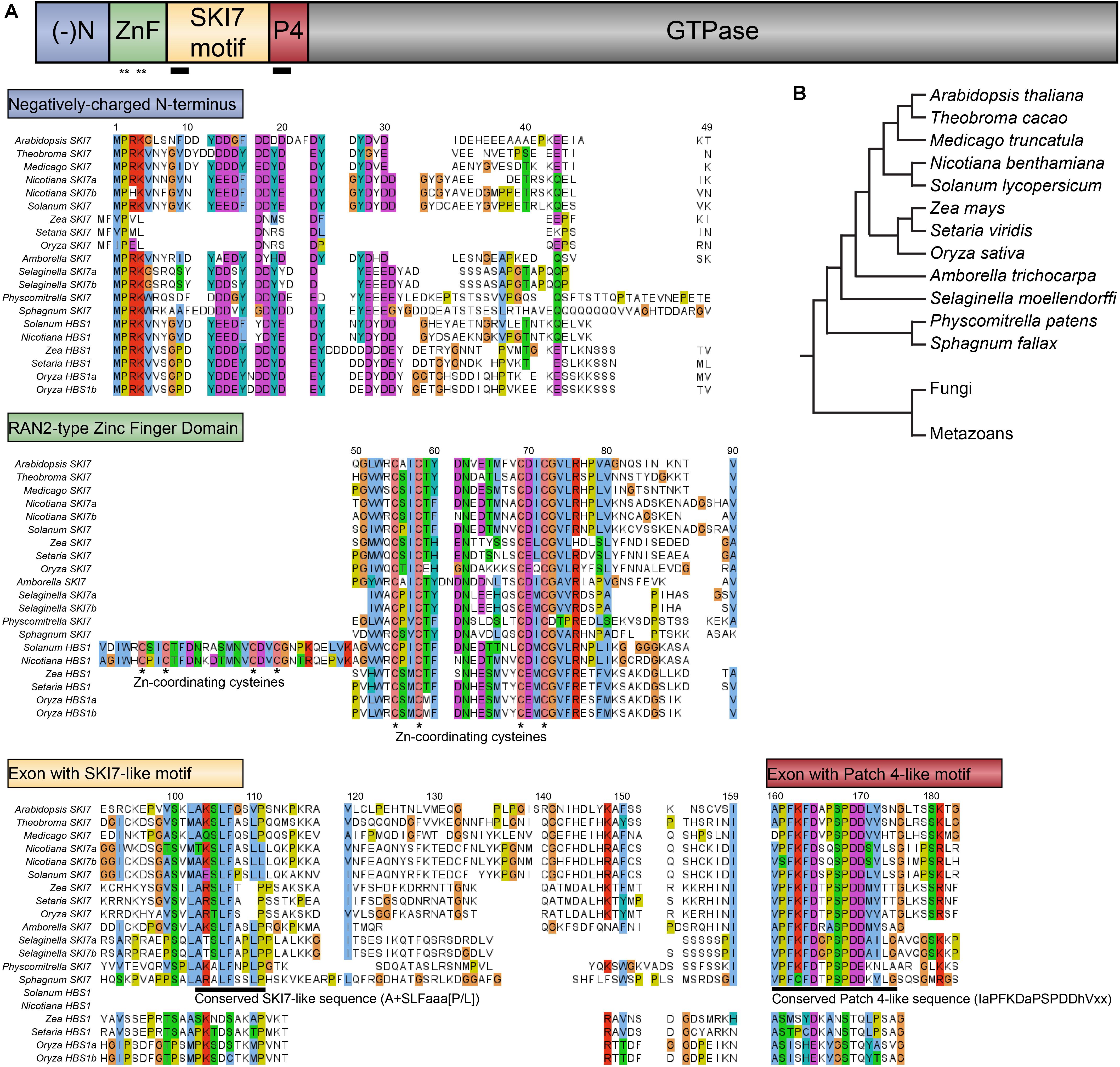
FIGURE 5. Evolution of SKI7-like amino acid sequences illustrated in select plant species. (A) Select SKI7-like proteins encoded by SKI7/HBS1 loci and HBS1-like proteins encoded by exclusively HBS1 loci are aligned to illustrate conserved and divergent sequences. Amino acids are colored following the Clustal standard (cyan – hydrophobic; red – positively charged; magenta – negatively charged; green – polar; cyan – aromatic; coral – cysteines; orange – glycines; yellow – prolines), with poorly conserved amino acids left uncolored. The negatively charged N-terminus (top) is highly similar across all proteins, although the grass SKI7 orthologs encode a much shorter N-terminus. The RAN2-type Zinc finger domain (middle) is highly similar in all plant species, including universal conservation of the Zn-coordinating cysteines (indicated with asterisks). This domain has been duplicated in the Solanaceae HBS1 loci. The cassette exons that encode the SKI7-like motif and the Patch 4-like motif (bottom) are largely well-conserved in SKI7 proteins (deeply conserved eukaryotic sequences indicated with a black line; see Figure 4). Solanaceae HBS1-like proteins have lost both of these motifs. Grass HBS1-like proteins have lost the SKI7-like motif and much of the Patch 4-like motif. (B) Evolutionary relationships among the species included in this figure are depicted by this cladogram.
We next used the Arabidopsis HBS1/SKI7 locus to search for orthologs in the genomes of land plants. We included the following species (Figure 5B): Sphagnum fallax and Physcomitrella patens (two distantly related species representing the moss lineage, which diverged early during land plant evolution), Selaginella moellendorffi (representing the lycophyte lineage, which diverged early during tracheophyte evolution), Amborella trichopoda (an ancient lineage of earliest-diverging flowering plants), Solanum lycopersicum and Nicotiana benthamiana (asterid eudicots in the Solanaceae family), Medicago truncatula (a rosid eudicot in the fabid order), Theobroma cacao (a close relative of Arabidopsis in the malvid order of rosid eudicots), Oryza sativa (a species of the BOP clade of grasses), and Zea mays and Setaria viridis (panicoid grasses). We focused on this limited set of taxa because of their excellent genome and transcriptome sequences, which allowed us to confidently identify potential alternative splice forms of the HBS1/SKI7 locus.
In the genomes of S. fallax, P. patens, A. trichopoda, M. truncatula, and T. cacao, there are single-copy HBS1/SKI7 orthologs that are each very similar to At5g10630 (Figures 2C, 4A, 5A). Like in Arabidopsis, alternative splicing of transcripts from these loci can include or exclude a cassette exon that encodes the SKI7-like region (Figures 2C, 4A). We therefore tentatively propose that this is the ancestral form of HBS1/SKI7 in land plants, although as more deeply annotated transcriptomes of early-diverging land plant species become available, this proposal may require revision. S. moellendorffi encodes two HBS1/SKI7 orthologs that generate several transcript permutations by alternative splicing (Figure 6A). Again, like Arabidopsis, alternative splicing of transcripts from these loci can include or exclude the SKI7-like motif. LOC9640201 can also be alternatively spliced to remove both the SKI7-like and the Patch 4-like motifs. LOC9652039 is even more complex: it can be alternatively spliced to remove only the SKI7-like motif (variant 4), only the Patch 4-like motif (variant 3), or both the SKI7-like and Patch 4-like motifs (variant 5) (Figure 6A). All of these alternatively spliced variants of S. moellendorffi HBS1/SKI7 genes are predicted to encode proteins that function as HBS1, but not as SKI7.
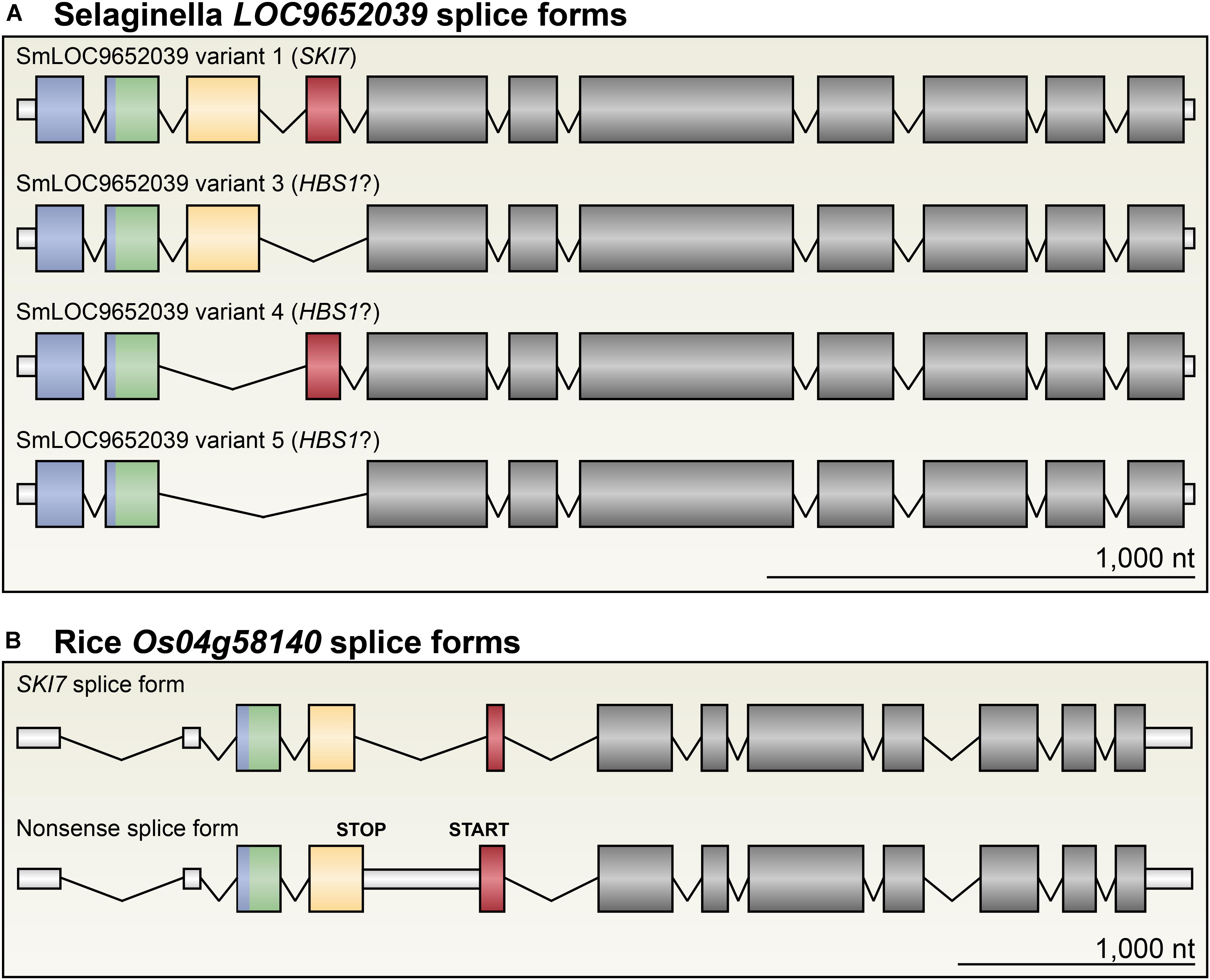
FIGURE 6. SKI7 alternative splicing has distinct functional consequences in different plant linages. (A) LOC9652039 in S. moellendorffi is alternatively spliced to either encode a complete SKI7 protein (variant 1), a putative HBS1 protein lacking the Patch 4-like motif (variant 3), the SKI7-like motif (variant 4), or both (variant 5). (B) Os04g58140 is the only SKI7-encoding locus in rice. Os04g58140 is alternatively spliced to retain intron 4, which includes several premature stop codons. Thus, this alternative splice form (bottom) does not encode a functional HBS1 or SKI7 protein, and is likely subject to NMD. As a result, unlike most other plant lineages examined, no single gene in rice can putatively encode both HBS1 and SKI7.
In the Solanaceae, an ancestral gene duplication resulted in two distinct sets of orthologs (Figure 4B). In S. lycopersicum, one gene, Solyc03g119290, is similar to the typical land plant HBS1/SKI7 orthologs, and is alternatively spliced to include or exclude the SKI7-like region (Figure 4B, upper panel). A second gene, Solyc12g036410, encodes regions that are highly homologous to HBS1, including the negatively charged N-terminus and C-terminal GTPase (Figure 4B, lower panel). The Zn-finger domain has tandemly duplicated, and both Zn-finger domains are on a cassette exon that is alternatively spliced (Figure 4B, lower panel; Figure 5A). The highly conserved Patch 4-like motif is absent in this gene (Figure 4B, lower panel; Figure 5A). A similar situation is found in a distantly related species of Solanaceae, N. benthamiana, which has two homeologous copies of the At5g10630-/Solyc03g119290-like gene, and one ortholog of the Solyc12g036410 gene that has a duplicated Zn-finger domain and has lost the sequences to encode the SKI7-like and Patch 4 motifs (Figure 5A). Thus, duplication of the HBS1/SKI7 locus in an ancestor of Solanaceae led to evolution of two distinct genes: one can encode HBS1 or SKI7-like orthologs, while the other has subfunctionalized to encode only an HBS1-like protein, and not SKI7 (Figure 4B).
The HBS1/SKI7 orthologs are more diverse in the grasses, which have evolved distinct, subfunctionalized HBS1 and SKI7 loci. The grasses are divided into two major lineages: the PACMAD clade, which includes Z. mays (corn) and S. viridis (millet), and the BOP clade, which includes O. sativa (rice). Rice has three HBS1/SKI7 orthologs: Os04g50870, Os04g58140, and Os01g02720 (Figures 5A, 6B and Supplementary Data Sheet 2). Os04g50870 and Os01g02720 are nearly identical paralogs (they encode proteins with 97% amino acid identity) that lack the SKI7-like and Patch 4-like motifs (Figure 5, “Oryza HBS1a” and “Oryza HBS1b”; Supplementary Data Sheet 2). Os04g50870 and Os01g02720 therefore most likely encode functional HBS1 proteins, but not functional SKI7 proteins. Os04g58140 has two splice forms (Figure 6B). Os04g58140.1 encodes a protein that includes the SKI7-like and Patch 4-like motifs (Figures 5A, 6B). Os04g58140.2 retains an intron that includes a premature stop codon and a downstream alternative start codon (Figure 6B and Supplementary Data Sheet 2). If the alternative start codon is selected (either by skipping the upstream open reading frame (uORF) or by reinitiating translation after the uORF), the Os04g58140.2 protein contains only the Patch 4-like motif and the HBS1-like translational GTPase. The Os04g58140.2 transcript may also be subject to NMD, if the first start codon is selected and translation does not reinitiate at the alternative start codon. In either case, retention of the poison intron yields an unproductive transcript of SKI7.
In the PACMAD clade of grasses, S. viridis has two HBS1/SKI7 orthologs, which have apparently subfunctionalized: Sevir3g016200 encodes a complete SKI7-like protein, with the SKI7-like and Patch 4-like motifs, and Sevir9g199100 encodes a protein with only the negatively charged N-terminus, the Zn-finger domain, a poorly-aligned/non-consensus Patch 4-like motif, and the C-terminal HBS1-like GTPase (Figures 4C, 5A and Supplementary Data Sheet 2). RNA-Seq analysis suggests that neither of these transcripts is alternatively spliced, although there are relatively limited data for S. viridis compared to well-established model systems, like tomato and Arabidopsis. Z. mays has orthologs of both S. viridis genes (Zm00001d026213 is orthologous to Sevir9g199100, and Zm00001d001827 is orthologous to Sevir3g016200, Figure 4C), but its SKI7-like gene makes many distinct transcripts. Alternative 5′ and 3′ splice sites near the beginning of the coding sequence select between two different start codons, but only alter the N-terminus by eight amino acids. More importantly, some of these transcripts (such as Zm00001d001827.2) encode an entire SKI7-like protein, while others (such as Zm00001d001827.9) skip an exon that encodes the patch 4 motif (Figure 4C, lower panel), similar to S. moellendorffi LOC9652039 variant 3 (Figure 6A).
To assemble all reliable SKI7-like amino acid sequences, we queried the NCBI RefSeq_protein database1 for proteins with SKI7-like sequences in land plants and metazoans (Supplementary Data Sheets 2, 3, respectively). Using this approach, we found a number of previously unidentified SKI7-like proteins in divergent invertebrate lineages, including orthologs in cnidarians, echinoderms, cephalochordates, mollusks, brachiopods, and priapulids (Supplementary Data Sheet 3). These findings suggest that SKI7-like orthologs are probably ubiquitous in eukaryotes, although deeper sequencing of transcriptomes from diverse phylogenetic clades will be needed to fully support this hypothesis, as well as to determine when the metazoan SKI7-like proteins lost the C-terminal HBS1-like GTPase and how the HBS1/SKI7 gene/transcript structures evolved in the metazoans (Figure 2A).
We aligned SKI7-like sequences from metazoans and land plants to identify any conserved regions that could illuminate how sequences specific to SKI7, but not HBS1, determine its distinct functions (Figure 7B and Supplementary Data Sheets 2, 3). There are two clearly conserved motifs across all SKI7 orthologs, which were previously named RxxxFxxxL and PFDFxxxSPDDIVKxNQ, based on the human SKI7-like protein sequence, and which we have named the SKI7-like motif and the Patch 4-like motif, respectively (Figure 7). Homology modeling of HsSKI7 onto the well-studied structure of the Rrp6/Rrp43 interaction revealed that the SKI7-like motif likely docks HsSKI7 with Rrp43, a core RNA exosome subunit (Kalisiak et al., 2017). The critical yeast SKI7-like motif residues, RxxxFxxxL, are not conserved across all eukaryotes, however; the consensus sequence at this site in metazoans is A+PShFAahL (where + is a positively charged residue, h is a hydrophobic residue, and a is an aliphatic residue; Figure 7B). In plants, this consensus sequence is slightly different: A+SLFaaa[P/L] (where + is a positively charged residue, a is an aliphatic residue, and [P/L] is usually P or L; Figure 7B). The Patch 4-like motif is fairly similar across all metazoans and plants. In metazoans, we found that the consensus Patch 4-like sequence is IaPF[D/R]F[K/D][S/T]aSPDDIV+A (Figure 7B). In plants, the consensus Patch 4-like sequence is IaPFKFDaPSPDDhVxx (Figure 7B). According to the resolved cryo-EM structure of yeast SKI7 in complex with the exosome (Kowalinski et al., 2016), the Patch 4-like motif mediates interactions with the Csl4 RNA exosome subunit. It should be noted that co-immunoprecipitation experiments in humans suggest that the Patch 4-like motif is neither necessary nor sufficient to recruit HsSKI7 to the RNA exosome, but the SKI7-like motif (A+PShFAahL) and neighboring residues were necessary and sufficient to recruit HsSKI7 or GFP to the RNA exosome (Kalisiak et al., 2017). Nonetheless, loss of the Patch 4-like motif did apparently weaken the interaction between SKI7 and the RNA exosome.
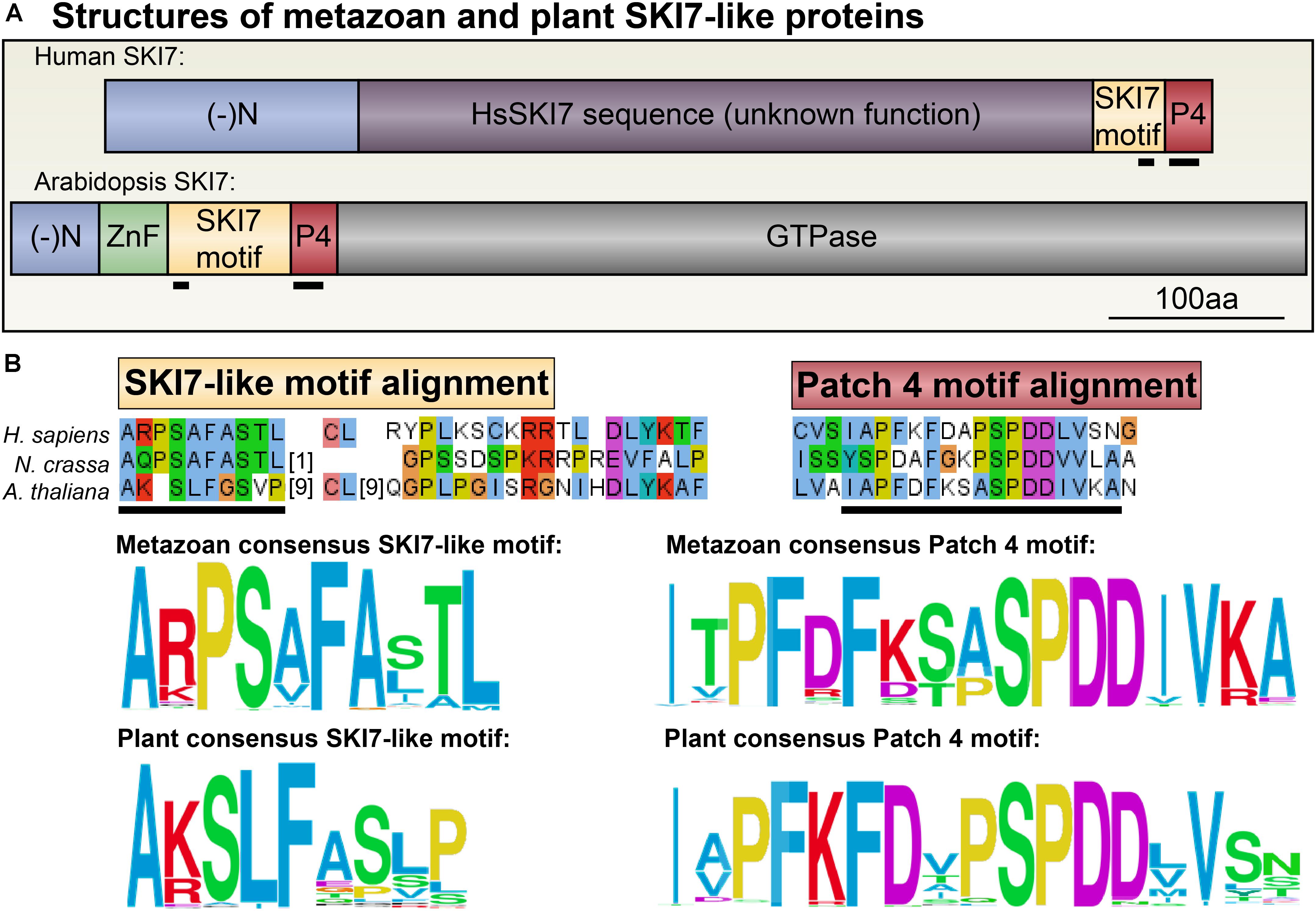
FIGURE 7. Consensus amino acid sequences that define eukaryotic SKI7. (A) Human and Arabidopsis SKI7 proteins do not globally align. Human SKI7 includes a long insert of unknown function (pink) between the negatively charged N-terminus (blue) and the SKI7- and Patch 4-like motifs (red). Arabidopsis SKI7 does not include this insert, but does include a C-terminal trGTPase domain of unclear function for SKI7, but which is required for HBS1 decoding activity. (B) Alignments of NCBI RefSeq SKI7 proteins from metazoans (Supplementary Data Sheet 2) and plants (Supplementary Data Sheet 1) were analyzed to determine consensus sequences for the SKI7-like motifs and the Patch 4-like motifs in these two major eukaryotic lineages. The sequences are largely similar, although the metazoan SKI7-like motif includes a widely conserved proline insertion (at position 3 in the consensus sequence). Alignments of the human, Neurospora crassa (fungus), and Arabidopsis SKI7-like motif and Patch 4-like motifs, as well as surrounding residues, are shown above the consensus sequences as examples. Amino acids are colored following Clustal standards, as above (Figure 5A).
We used homology modeling to predict the structure of the N-terminus of the Arabidopsis SKI7 protein (excluding the C-terminal trGTPase, Figure 8). The negatively charged N-terminus is predicted to form several α helices, which may mediate interactions with the SKI2/SKI3/SKI8 adaptor complex, and the ZnF is predicted to form two β-sheet-like structures (β1 and β2, Figure 5A) followed by an α-helix, as expected for a ZnF domain. The exons encoding the SKI7-like motif and Patch 4-like motif fold into four α-helices (α1 through α4, Figure 5A), very comparable to the resolved yeast SKI7 structure (Kowalinski et al., 2016). α1 overlaps with the deeply conserved SKI7-like motif (in Arabidopsis, AKSLFGSVP, Figure 8A). α3 is highly similar between humans and Arabidopsis (Figure 8A), including residues DLYKAF (Arabidopsis) or DLYKTF (human), and this α-helix has been labeled in the predicted Arabidopsis SKI7 structure (Figure 8B). The Patch 4-like motif is predicted to be highly structured, and forms α4. Immediately after α4, the last amino acids of this N-terminal region of Arabidopsis SKI7 are highly disordered (this pattern continues into the N-terminal residues of the trGTPase region of the protein), allowing the trGTPase to adopt a flexible position relative to the highly structured N-terminus that interacts with the RNA exosome (Figure 8B).
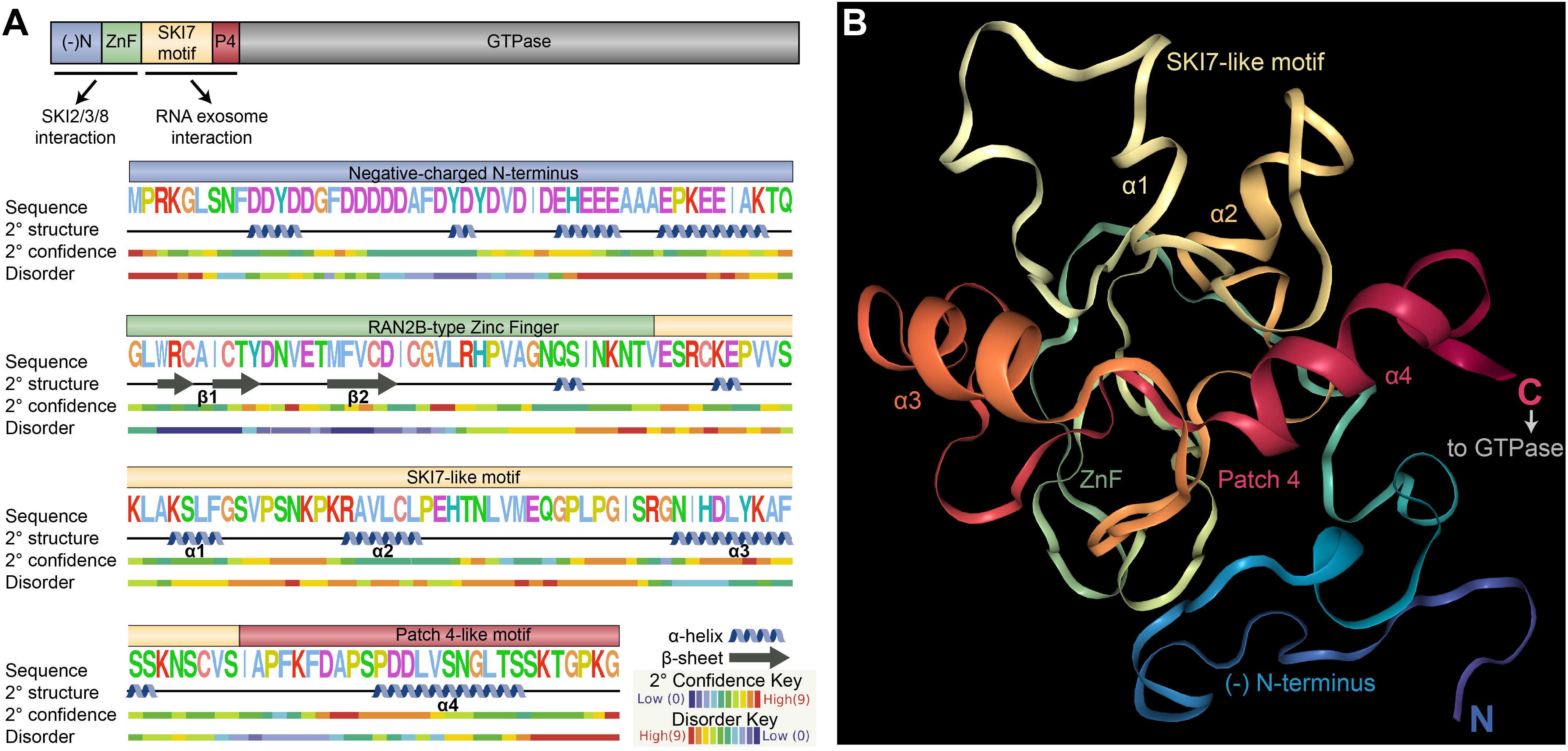
FIGURE 8. Predicted structure of the Arabidopsis SKI7 N-terminus. (A) Phyre2 prediction of the secondary structure and disorder of the N-terminal 183 amino acids of Arabidopsis SKI7. The negatively charged N-terminus (blue, (-)N) and RAN2B-type Zinc Finger (green, ZnF) are predicted to interact with the SKI2/SKI3/SKI8 cytosolic exosome adaptor complex, and the SKI7-like motif (yellow) and Patch 4-like motif (red, P4) are predicted to interact with the surface of the RNA exosome catalytic core. The negatively charged N-terminus includes moderate-confidence α-helices that may promote interaction with SKI2/SKI3/SKI8. The ZnF domain includes two β-sheets (β1 and β2) and an α-helix that coordinate with the Zn ion. The SKI7-like and Patch 4-like motif include four α-helices (α1 through α4), which is structurally comparable to the yeast SKI7 protein. (B) Phyre2 prediction of the structure of the N-terminal 183 amino acids of Arabidopsis SKI7, based on homology modeling against several resolved protein structures (as described in the methods). Residues are colored by position, from blue (N-terminus) to red (C-terminus), closely matching the colors used in panel a and other figures. The defined regions of the protein are labeled, including α1, α2, α3, and α4, which are predicted to mediate interactions with the RNA exosome core. The C-terminus of this structure is predicted to be highly disordered, forming a flexible linker to the C-terminal trGTPase of Arabidopsis SKI7.
Here, we have shown that HBS1/SKI7 is a well-conserved locus in eukaryotes that encodes two proteins with distinct molecular functions. In plants and fungi, HBS1 and SKI7 are nearly identical proteins, with an N-terminus that interacts with the cytosolic RNA exosome SKI2/SKI3/SKI8 complex and a C-terminal translational GTPase. The SKI7 isoform differs from HBS1 by as few as ∼25 amino acids that we propose promote its interaction with the RNA exosome instead of with the ribosome decoding factor, PELOTA (Figure 9).
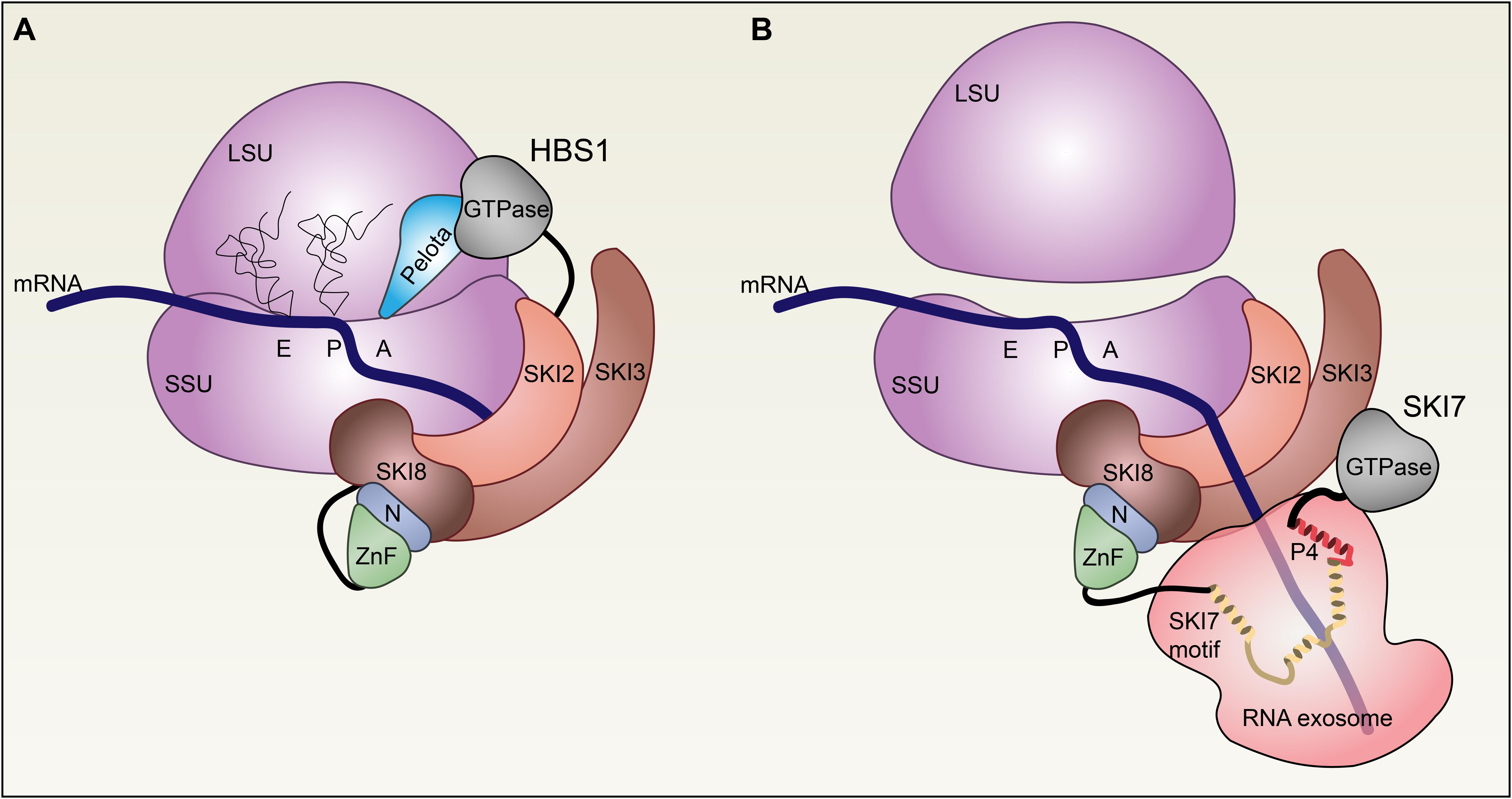
FIGURE 9. Model of the roles of HBS1 and SKI7 in mRNA surveillance and degradation. (A) HBS1 associates with PELOTA (light blue) to recognize stalled ribosomes (purple). The SKI2/SKI3/SKI8 complex (browns) interacts directly with the small subunit (SSU) of the 80S ribosome, perhaps facilitated by the HBS1 N-terminal domains (green and blue), which are known to interact with SKI3 and SKI8. Structural models suggest that PELOTA displaces mRNA from the aminoacyl A site, allowing the mRNA to interact with the SKI2 RNA helicase. (B) PELOTA and HBS1 promote 80S ribosome dissociation from the transcript and splitting into the 60S (large subunit, LSU) and 40S (SSU) subunits. SKI7 recruits the RNA exosome 3′→5′ exoribonuclease complex (light red) to SKI2/SKI3/SKI8 via interactions between the SKI7 N-terminus and SKI3/SKI8. α helices in the SKI7-like motif (yellow) and Patch 4 (P4)-like motif (red) form an interaction surface with the RNA exosome complex.
Although we have shown that many plant genomes encode both HBS1 and SKI7 from a single locus by alternative splicing of an exon encoding the SKI7-like motif, other lineages have evolved distinct HBS1/SKI7 gene structures. In the early-diverging tracheophyte S. moellendorffi, HBS1/SKI7 can be alternatively spliced to exclude or include exons encoding either the SKI7-like motif or the Patch 4-like motif, or to exclude or include both of these exons. In the Solanaceae, a second HBS1 locus has lost the SKI7-like and Patch 4-like motifs, and thus encodes only an HBS1-like protein. In rice, HBS1 and SKI7 are each encoded by their own locus, reminiscent of the subfunctionalization of Hbs1 and Ski7 loci in S. cerevisiae. Panicoid grasses (maize and millet), like rice, have two distinct HBS1 and SKI7 loci: one encodes only HBS1, and the second can encode SKI7. The panicoid grass SKI7 locus is alternatively spliced to include or exclude the Patch 4-like motif, however, which may impact its function. The Patch 4-like motif is universally conserved in all known SKI7 orthologs, but is not conserved in HBS1 orthologs that have lost other SKI7-like features (e.g., the HBS1-specific loci in Solanaceae, rice, and yeast), suggesting that the Patch 4-like motif is critical for SKI7’s functions. In humans, however, Patch 4-like is neither necessary nor sufficient to recruit SKI7 to the RNA exosome (Kalisiak et al., 2017). Thus, while it seems most likely that the panicoid grass SKI7-like protein loses functionality when the exon encoding the Patch 4-like motif is excluded by alternative splicing, this will need to be determined experimentally.
Consideration of the evolutionary history of HBS1/SKI7 loci in eukaryotes reveals an important distinction: subfunctionalized HBS1 paralogs that cannot encode SKI7 have evolved repeatedly, and are often no longer regulated by alternative splicing, but in almost all eukaryotes (except for S. cerevisiae and a handful of other fungi), SKI7 orthologs are alternatively spliced (Marshall et al., 2013; Lambert et al., 2015). Moreover, alternative splicing of SKI7 consistently has strong effects, either yielding an unproductive splice form that is likely degraded by NMD, a splice form that encodes a loss-of-function protein, or a transcript that instead encodes HBS1. This consistent regulation of SKI7 levels by alternative splicing suggests that the activity of the cytosolic RNA exosome is tightly coordinated with nuclear RNA processing, especially conditions that shift alternative splicing dynamics, such as oxidative stress (Staiger and Brown, 2013; Berner et al., 2017). Further characterization of the developmental or physiological conditions that influence alternative splicing of SKI7, as well as assays to determine whether SKI7 protein levels are limiting factors in cytosolic RNA exosome activity, will be needed to unravel how this mechanism influences cytosolic RNA exosome activity. Recently, the Pelota/HBS1 decoding factors and the cytosolic RNA exosome have been implicated in promoting NMD (Arribere and Fire, 2018), and NMD is known to regulate expression of the splicing and translation machinery (Lareau and Brenner, 2015). Our finding that alternative splicing of SKI7 potentially regulates SKI7 levels to limit assembly of the SKI2/SKI3/SKI8-RNA exosome complex (by either excluding an alternative exon of SKI7 to encode HBS1, or by generating a nonsense SKI7 transcript) invites speculation that SKI7 regulates its own splicing and transcript stability via its role in NMD and NMD-mediated regulation of splicing machinery gene expression.
PELOTA and the cytoplasmic RNA exosome are emerging as crucial components of plant immune systems, although their necessary interacting partners, HBS1 and SKI7, have not been comprehensively defined in plants until now. Loss of PELOTA, the decoding factor that recruits HBS1 to stalled ribosomes, confers resistance to TYCLV infection in tomato; with our identification of tomato HBS1 and HBS1/SKI7 genes, it is now possible to test whether loss of HBS1, SKI7, and/or SKI2/SKI3/SKI8 also confer resistance to TYCLV, and how this RNA degradation machinery interacts with other viruses. In rice, a recessive pelota mutant triggers a salicylic acid-associated autoimmune response, including spontaneous lesions and dwarfism, through unclear mechanisms (Ding et al., 2018; Qin et al., 2018; Zhang et al., 2018). The rice pelota defects could be related to hyperaccumulation of aberrant RNA species, similar to the tricohepatoenteric autoimmune syndrome in skiv2l human cells (Eckard et al., 2014), or due to specific dysregulation of transcripts in rice that regulate immunity. For instance, the expression and activity of the disease resistance Toll- and Interleukin-like Receptor (TIR) family of Nucleotide-binding, Leucine-rich repeat Receptors (TIR-NLRs or TLRs) is regulated by NMD in some instances; loss of PELOTA, which contributes to NMD, may therefore deregulate TLR expression, triggering autoimmune defects (Dinesh-Kumar and Baker, 2000; Riehs-Kearnan et al., 2012; Gloggnitzer et al., 2014). Whether pelota mutants can trigger autoimmune defects in other plant species remains to be determined. More broadly, NMD is proposed as a general antiviral mechanism, and so HBS1 and SKI7 may contribute to broad-spectrum antiviral defense via their roles in NMD (Balistreri et al., 2014; Garcia et al., 2014; Rigby and Rehwinkel, 2015; Hamid and Makeyev, 2016).
We have identified the plant orthologs of HBS1 and SKI7, key regulators of RNA metabolism in eukaryotic cells. As a component of the cytosolic RNA exosome, SKI7 not only participates in co-translational RNA surveillance, but is also presumably required to clear 5′ fragments of mRNAs cleaved by RISCs. RNA exosomal degradation of these 5′ fragments prevents copying of host transcripts by RDR6, which can otherwise trigger post-transcriptional silencing of endogenous genes. In diverse eukaryotic lineages, SKI7 levels are controlled by alternative splicing of transcripts; alternative splice forms can either encode the functionally distinct HBS1 protein, or can be unproductive, either by removing critical residues for SKI7 function, or by introducing premature stop codons that likely subject the splice form to NMD. Co-translational RNA decay mechanisms, including HBS1/SKI7-dependent RNA degradation, are becoming more prominent to investigations of eukaryotic immune systems and defenses against viral infection. Our discovery of the alternative splicing of HBS1/SKI7 expression across anciently diverging eukaryotic lineages, including plants and invertebrate clades, implies that co-translational RNA decay mechanisms are under complex regulation to coordinate host gene expression with environmental cues, stress responses, and antiviral defense.
The Landsberg erecta (Ler) ecotype of Arabidopsis was grown under 16 h light (100 μmol photons m-2 s-1 of photosynthetically active radiation)/8 h dark cycles. Shoots were harvested 4 weeks after germination.
RNA was isolated from Ler Arabidopsis plants with the Spectrum Plant Total RNA (Sigma-Aldrich) kit with on-column DNase I digestion (New England Biolabs). cDNA was synthesized from RNA using oligo (dT)18 primers and SuperScript III reverse transcriptase (Fisher Scientific). Splice forms were amplified with Phusion DNA polymerase (New England Biolabs), adding a CACC 5′ overhang to facilitate pENTR/D-TOPO cloning. RT-PCR amplified DNA was gel purified in a 1% agarose gel and extracted using a gel extraction kit (Bioneer). Purified DNA was used for TOPO reactions with pENTR/D-TOPO (Thermo Fisher), transformed into XL1-Blue E. coli chemically competent cells, and screened for resistance to kanamycin on LB agar. Plasmid was purified from positive colonies using a miniprep kit (Bioneer) and sequenced using Sanger technology with the M13F primer.
Oligonucleotides used for cloning were: JB1058: 5′-CACC ATG CCT CGT AAA GGA TTA TCC AAT TTC G-3′, JB1061: 5′-CACC ACA GTT GAG AGCAG ATG CAA AGA AC-3′, and JB1063: 5′-GCC TTT TGG ACC AGT TTT TGAGG ATG-3′. JB1058 + JB1063 surround the alternative exon, and amplified three majors products: the short splice form, the long splice form, and a small amount of the longer, minor splice form. JB1061 is specific to the 5′ end of the alternative exon, and in combination with JB1063, amplified both the long splice form and the longer, minor splice form.
Decoding trGTPases were identified using human protein sequences as queries for a BLASTp search against the Arabidopsis thaliana refseq protein database. Putative trGTPases were filtered to include only cytosol-localized proteins, based on proteomic data and consensus predictions curated by the Subcellular Localization Database for Arabidopsis Proteins 32.
SKI7 orthologs in the NCBI protein refseq databases were identified using a tBLASTx search with the Arabidopsis SKI7-like and patch 4-like motifs as a query for land plants, and the human SKI7-like and patch 4-like motifs as a query for metazoans. BLASTp results were then filtered to remove identical protein sequences. Protein sequences were aligned using MAFFT via JalView. Uncommon insertions were trimmed from the final alignments for clarity. Transcript structures were obtained for the select plant species described in the text from relevant databases (TAIR10, from arabidopsis.org and araport.org; MaizeGDB.org; Phytozome.jgi.doe.gov; SolGenomics.net; and CosMoss.org), and then confirmed with RNA-Seq evidence from the same databases (or by direct cloning of alternative splice forms, as described above). Consensus sequence logos were generated with WebLogo3.
The N-terminus of SKI7 (through the patch 4-like motif) structure was modeled by Phyre24, which used structures of YY1-associated factor 2 (PDB 2D9G), HBV-associated factor (PDB 2CRC), Rubredoxin B (PDB 2KN9), NEMO CoZi (PDB 4OWF), and TAB3-NZF (PDB 3A9K). The resulting model was visualized by NGL5.
JB designed the project, conducted the experiments, and drafted the manuscript. BB contributed to the experimental design and manuscript.
This project was supported by Innovative Genomics Institute 2017 Award to BB and JB, USDA CRIS 2030-22000-009-00D to BB, and NIH grant 5-DP5-OD023072-02 to JB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Feng Li for stimulating discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01333/full#supplementary-material
Abernathy, E., and Glaunsinger, B. (2015). Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 47, 600–608. doi: 10.1016/j.virol.2015.02.007
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Andika, I. B., Maruyama, K., Sun, L., Kondo, H., Tamada, T., and Suzuki, N. (2015). Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J. 81, 781–793. doi: 10.1111/tpj.12770
Arribere, J. A., and Fire, A. Z. (2018). Nonsense mRNA suppression via nonstop decay. eLife 7:e33292. doi: 10.7554/eLife.33292
Balistreri, G., Horvath, P., Schweingruber, C., Zünd, D., Mcinerney, G., Merits, A., et al. (2014). The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 16, 403–411. doi: 10.1016/j.chom.2014.08.007
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Becker, T., Franckenberg, S., Wickles, S., Shoemaker, C. J., Anger, A. M., Armache, J. P., et al. (2012). Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506. doi: 10.1038/nature10829
Berner, D., Zenkel, M., Pasutto, F., Hoja, U., Liravi, P., Gusek-Schneider, G. C., et al. (2017). Posttranscriptional regulation of LOXL1 expression via alternative splicing and nonsense-mediated mRNA decay as an adaptive stress response. Invest. Ophthalmol. Vis. Sci. 58, 5930–5940. doi: 10.1167/iovs.17-22963
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H. S., et al. (2006). Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Branscheid, A., Marchais, A., Schott, G., Lange, H., Gagliardi, D., Andersen, S. U., et al. (2015). SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43, 10975–10988. doi: 10.1093/nar/gkv1014
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Carr-Schmid, A., Pfund, C., Craig, E. A., and Goss Kinzy, T. (2002). Novel G-Protein complex whose requirement is linked to the translational status of the cell. Mol. Cell. Biol. 22, 2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002
Cheng, C. Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., and Town, C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. doi: 10.1111/tpj.13415
Crow, Y. J., Hayward, B. E., Parmar, R., Robins, P., Leitch, A., Ali, M., et al. (2006). Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38, 917–920. doi: 10.1038/ng1845
Cuperus, J. T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R. T., Takeda, A., et al. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003. doi: 10.1038/nsmb.1866
Deng, Y., Wang, J., Tung, J., Liu, D., Zhou, Y., He, S., et al. (2018). A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 14:e1006756. doi: 10.1371/journal.ppat.1006756
Dever, T. E., and Green, R. (2012). The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 4:a013706. doi: 10.1101/cshperspect.a013706
Dinesh-Kumar, S. P., and Baker, B. J. (2000). Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. U.S.A. 97, 1908–1913. doi: 10.1073/pnas.020367497
Ding, W., Wu, J., Ye, J., Zheng, W., Wang, S., Zhu, X., et al. (2018). A pelota-like gene regulates root development and defence responses in rice. Ann. Bot. doi: 10.1093/aob/mcy075 [Epub ahead of print].
Eckard, S. C., Rice, G. I., Fabre, A., Badens, C., Gray, E. E., Hartley, J. L., et al. (2014). The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat. Immunol. 15, 839–845. doi: 10.1038/ni.2948
Fahlgren, N., Montgomery, T. A., Howell, M. D., Allen, E., Dvorak, S. K., Alexander, A. L., et al. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16, 939–944. doi: 10.1016/j.cub.2006.03.065
Garcia, D., Garcia, S., and Voinnet, O. (2014). Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16, 391–402. doi: 10.1016/j.chom.2014.08.001
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Gloggnitzer, J., Akimcheva, S., Srinivasan, A., Kusenda, B., Riehs, N., Stampfl, H., et al. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16, 376–390. doi: 10.1016/j.chom.2014.08.010
Hamid, F. M., and Makeyev, E. V. (2016). Exaptive origins of regulated mRNA decay in eukaryotes. Bioessays 38, 830–838. doi: 10.1002/bies.201600100
Hashimoto, Y., Takahashi, M., Sakota, E., and Nakamura, Y. (2017). Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem. Biophys. Res. Commun. 484, 1–7. doi: 10.1016/j.bbrc.2017.01.092
Horikawa, W., Endo, K., Wada, M., and Ito, K. (2016). Mutations in the G-domain of Ski7 cause specific dysfunction in non-stop decay. Sci. Rep. 6:29295. doi: 10.1038/srep29295
Kalisiak, K., Kuliński, T. M., Tomecki, R., Cysewski, D., Pietras, Z., Chlebowski, A., et al. (2017). A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic Acids Res. 45, 2068–2080. doi: 10.1093/nar/gkw862
Koonin, E. V. (2017). Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol. Direct 12:5. doi: 10.1186/s13062-017-0177-2
Kowalinski, E., Kögel, A., Ebert, J., Reichelt, P., Stegmann, E., Habermann, B., et al. (2016). Structure of a cytoplasmic 11-subunit RNA exosome complex. Mol. Cell. 63, 125–134. doi: 10.1016/j.molcel.2016.05.028
Lambert, M. J., Cochran, W. O., Wilde, B. M., Olsen, K. G., and Cooper, C. D. (2015). Evidence for widespread subfunctionalization of splice forms in vertebrate genomes. Genome Res. 25, 624–632. doi: 10.1101/gr.184473.114
Lapidot, M., Karniel, U., Gelbart, D., Fogel, D., Evenor, D., Kutsher, Y., et al. (2015). A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genetics 11:e1005538. doi: 10.1371/journal.pgen.1005538
Lareau, L. F., and Brenner, S. E. (2015). Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 32, 1072–1079. doi: 10.1093/molbev/msv002
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, M. M. H., Macdonald, M. R., and Rice, C. M. (2015). To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 25, 320–329. doi: 10.1016/j.tcb.2015.02.001
Marshall, A. N., Montealegre, M. C., Jiménez-López, C., Lorenz, M. C., and Van Hoof, A. (2013). Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genetics 9:e1003376. doi: 10.1371/journal.pgen.1003376
Mitchell, P., and Tollervey, D. (2003). An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′– > 5′ degradation. Mol. Cell. 11, 1405–1413. doi: 10.1016/S1097-2765(03)00190-4
Molleston, J. M., and Cherry, S. (2017). Attacked from all sides: RNA decay in antiviral defense. Viruses 9:2. doi: 10.3390/v9010002
Nagarajan, V. K., Jones, C. I., Newbury, S. F., and Green, P. J. (2013). XRN 5′– > 3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta 1829, 590–603. doi: 10.1016/j.bbagrm.2013.03.005
Narayanan, K., and Makino, S. (2013). Interplay between viruses and host mRNA degradation. Biochim. Biophys. Acta 1829, 732–741. doi: 10.1016/j.bbagrm.2012.12.003
Nicholson, A. W. (2014). Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA 5, 31–48. doi: 10.1002/wrna.1195
Orban, T. I., and Izaurralde, E. (2005). Decay of mRNAs targeted by RISC requires XRN1, the ski complex, and the exosome. RNA 11, 459–469. doi: 10.1261/rna.7231505
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Qi, X., Bao, F. S., and Xie, Z. (2009). Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One 4:e4971. doi: 10.1371/journal.pone.0004971
Qin, P., Fan, S., Deng, L., Zhong, G., Zhang, S., Li, M., et al. (2018). LML1, encoding a conserved eukaryotic release factor 1 protein, regulates cell death and pathogen resistance by forming a conserved complex with SPL33 in rice. Plant Cell Physiol. 59, 887–902. doi: 10.1093/pcp/pcy056
Qu, F., Ye, X., Hou, G., Sato, S., Clemente, T. E., and Morris, T. J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005
Qu, F., Ye, X., and Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Riehs-Kearnan, N., Gloggnitzer, J., Dekrout, B., Jonak, C., and Riha, K. (2012). Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 40, 5615–5624. doi: 10.1093/nar/gks195
Rigby, R. E., and Rehwinkel, J. (2015). RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 36, 179–188. doi: 10.1016/j.it.2015.02.001
Schmidt, C., Kowalinski, E., Shanmuganathan, V., Defenouillere, Q., Braunger, K., Heuer, A., et al. (2016). The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science 354, 1431–1433. doi: 10.1126/science.aaf7520
Seo, J. K., Wu, J., Lii, Y., Li, Y., and Jin, H. (2013). Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 26, 617–625. doi: 10.1094/MPMI-10-12-0255-IA
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587. doi: 10.1016/j.tree.2008.06.005
Shao, S., Murray, J., Brown, A., Taunton, J., Ramakrishnan, V., and Hegde, R. S. (2016). Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167, 1229.e15–1240.e15. doi: 10.1016/j.cell.2016.10.046
Staiger, D., and Brown, J. W. S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656. doi: 10.1105/tpc.113.113803
Szadeczky-Kardoss, I., Csorba, T., Auber, A., Schamberger, A., Nyiko, T., Taller, J., et al. (2018). The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 46, 4632–4648. doi: 10.1093/nar/gky279
Szittya, G., and Burgyán, J. (2013). “RNA interference-mediated intrinsic antiviral immunity in plants,” in Current Topics in Microbiology and Immunology, ed. R. Cullen (Berlin: Springer).
Takahashi, S., Araki, Y., Sakuno, T., and Katada, T. (2003). Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 22, 3951–3959. doi: 10.1093/emboj/cdg374
Tollervey, D. (2015). RNA surveillance and the exosome. RNA 21, 492–493. doi: 10.1261/rna.050989.115
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Walsh, D., Mathews, M. B., and Mohr, I. (2013). Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 5:a012351. doi: 10.1101/cshperspect.a012351
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Yi, H., and Richards, E. J. (2009). Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant. Genetics 183, 1227–1234. doi: 10.1534/genetics.109.105569
Zhang, X., Zhu, Y., Liu, X., Hong, X., Xu, Y., Zhu, P., et al. (2015). Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348, 120–123. doi: 10.1126/science.aaa2618
Zhang, X. B., Feng, B. H., Wang, H. M., Xu, X., Shi, Y. F., He, Y., et al. (2018). A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 60, 160–172. doi: 10.1111/jipb.12613
Zinder, J. C., and Lima, C. D. (2017). Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100. doi: 10.1101/gad.294769.116
Keywords: RNA exosome, innate immunity, antiviral defense, RNA interference, alternative splicing, HBS1, SKI7
Citation: Brunkard JO and Baker B (2018) A Two-Headed Monster to Avert Disaster: HBS1/SKI7 Is Alternatively Spliced to Build Eukaryotic RNA Surveillance Complexes. Front. Plant Sci. 9:1333. doi: 10.3389/fpls.2018.01333
Received: 14 April 2018; Accepted: 24 August 2018;
Published: 12 September 2018.
Edited by:
Feng Qu, The Ohio State University, United StatesReviewed by:
Aardra Kachroo, University of Kentucky, United StatesCopyright © 2018 Brunkard and Baker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacob O. Brunkard, YnJ1bmthcmRAYmVya2VsZXkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.