
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 September 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01297
This article is part of the Research TopicActivation and Suppression of Plant ImmunityView all 27 articles
A correction has been applied to this article in:
Corrigendum: The Tryptophan decarboxylase 1 Gene From Aegilops variabilis No.1 Regulate the Resistance Against Cereal Cyst Nematode by Altering the Downstream Secondary Metabolite Contents Rather Than Auxin Synthesis
 Qiulan Huang1,2,3
Qiulan Huang1,2,3 Lin Li4
Lin Li4 Minghui Zheng4
Minghui Zheng4 Fang Chen2
Fang Chen2 Hai Long1
Hai Long1 Guangbing Deng1
Guangbing Deng1 Zhifen Pan1
Zhifen Pan1 Junjun Liang1
Junjun Liang1 Qiao Li1
Qiao Li1 Maoqun Yu1*
Maoqun Yu1* Haili Zhang1*
Haili Zhang1*Cereal cyst nematode (CCN, Heterodera avenae) is a most important pathogen of wheat and causes tremendous yield loss annually over the world. Since the lack of resistance materials among wheat cultivars, identification and characterization of the resistance-related genes from the relatives of wheat is a necessary and efficient way. As a close relative of wheat with high resistance against CCN, Aegilops variabilis No.1 is believed to be a valuable source for wheat breeding against this devastating disease. However so far, very few resistance-associated genes have been characterized from this species. In this study, we present that the tryptophan decarboxylase genes from Ae. variabilis No.1 (AeVTDC1 and AeVTDC2) were both induced by CCN juveniles at the early stage of resistance response (30 h post-inoculation), with AeVTDC1 more sensitive to CCN infection than AeVTDC2. Silencing of AeVTDC1 led to compromised immunity to CCN with more CCN intrusion into roots; while overexpression AeVTDC1 in Nicotiana tabacum dramatically enhanced the resistance of plants by reducing the knots formed on roots. Metabolism analysis showed that the contents of secondary metabolites with activity of resistance to varied pathogens correlated with the expression level of AeVTDC1 in both Ae. variabilis No.1 and the transgenic tobacco plants. In addition, the content of IAA was not affected by either silencing or overexpressing of AeVTDC1. Hence, our research provided AeVTDC1 a valuable target that mediates resistance to CCN and root knot nematode (RKN, Meloidogyne naasi) without influencing the auxin biosynthesis.
The cereal cyst nematode (CCN, Heterodera avenae) is a vital pathogen of graminaceous crops, such as wheat and barley. CCN is widely distributed and causes great production losses (Williamson and Kumar, 2006; Hajihasani et al., 2010; Zheng et al., 2015; Li et al., 2016). Many efforts have been made to identify CCN resistance (Cre) genes. However, gene resource resistant to CCN is scarce in wheat but abundant in its relatives (Montes et al., 2008). Rha genes were mapped in barley. Through changing the transcript abundance and composition of cell wall, Rha2-mediated CCN resistance drives rapid deterioration of CCN feeding sites (Aditya et al., 2015). CreX and CreY were identified in Aegilops variabilis (Barloy et al., 2006). Some resistance lines were bred through hybridization. Although several loci related to CCN resistance have been reported, few of them has been cloned and their biological functions hadn’t been clarified (de Majnik et al., 2003; Safari et al., 2005; Zhang et al., 2016). Ae. variabilis No.1 (2n = 4x = 28, UUSvSv), belonging to the genus Aegilops of the Triticeae tribe, is known as a well-resistant material, which confers strong resistance against CCN and root knot nematode (RKN, Meloidogyne naasi) (Barloy et al., 2006; Coriton et al., 2009; Xu et al., 2012; Zheng et al., 2015; Wu et al., 2016).
Plant tryptophan decarboxylase (TDC, 4.1.1.28) catalyzes the formation of tryptamine from tryptophan (Trp) (Giebel and Jackowiak, 1976; Hallard et al., 1997; Canel et al., 1998). Tryptamine is a precursor for the biosynthesis of serotonin, indole alkaloids, and indole acetic acid (IAA) (Bartel, 1997; Valletta et al., 2010; Liu et al., 2012; Dubouzet et al., 2013). Transformation from tryptamine to serotonin is catalyzed by Tryptamine-5-hydroxylase (T5H) (Kang and Back, 2009). TDCs are key enzymes in the biosynthetic pathway of terpenoid indole alkaloids (TIAs), since they link primary to secondary metabolism by converting Trp into tryptamine. Conversion of Trp into tryptamine is a common backbone for many secondary metabolites, which have divergent biological activities regulated by developmental and environmental factors (Mehrotra et al., 2013; Verma et al., 2015). Plant TDC cDNA was firstly isolated from Catharanthus roseus by screening a cDNA expression library (Hallard et al., 1997; Geerlings et al., 1999). Gradually, a few TDC genes had been cloned and characterized from other species, such as Nicotiana tabacum, Mitragyna speciosa, and Withania coagulans (Berlin et al., 1993; Goddijn et al., 1994; Charoonratana et al., 2013; Jadaun et al., 2017). The biological functions of TDCs have been reported in several plant–pathogen interactions. It was reported that TDC gene played a role in resistance against Malacosoma disstria Hub and Manduca sexta L. through tryptamine, which had adverse effects on their feeding behaviors and physiology (Gill and Ellis, 2006). Serotonin defended against Magnaporthe oryzae infection in rice leaves (Gill et al., 2003; Hayashi et al., 2016). Ectopic expression of TDC1 significantly suppressed the growth of insect pests by sufficient tryptamine accumulation in poplar and tobacco leaf tissue (Gill et al., 2003). The inhibition of TDC enzyme activity with S-αFMT resulted in susceptibility of Ae. variabilis No.1 to CCN and RKN, which indicated AeVTDCs play important roles in resistance to nematodes (Li et al., 2016). However, it remains to be clarified which AeVTDC involved in resistance to CCN and RKN and its mechanism of function.
Previous RNA-Seq analysis indicated that the TDC genes of Ae. variabilis No.1 (AeVTDCs) showed different expression pattern between control and CCN invaded roots at different time points (Zheng et al., 2015). AeVTDC1 gene was cloned and its protein had the ability of catalyzing the formation of tryptamine from tryptophan. In this study, we reported AeVTDC1 played a positive role at the early stage of plant resistance to CCN infection and overexpression of AeVTDC1 in tobacco led to reduced susceptibility to RKN. Silencing or overexpression of AeVTDC1 didn’t affect accumulation of IAA, but changed the downstream secondary metabolites of AeVTDC1.
Seeds of Ae. variabilis No.1 and wheat (Fielder) were surface-cleaned by the sterilized water and kept at 4°C for 24 h. Then the seeds were germinated in Petri dishes (5-cm diameter) on wet paper at 20°C under a 16-h light/8-h dark photoperiod. After 2 days, these little seedlings plants were cultured with water or sterilized soil for later use.
AeVTDC1 transgenic tobacco and wild type (WT) were cultured in the sterilized soil in greenhouse under 25°C and 60% humidity.
Second stage juveniles (J2s) of CCN (H. avenae) were hatched with cereal cysts as described previously (Li et al., 2016). For resistance assay and expression inducing assay, about 300 J2s of CCN per pot were inoculated around the root-tips under 19°C. According to the Li’s description, the eggs and second stage juveniles (J2s) of RKN were obtained (Li et al., 2016). For root nematode staining, entire root systems were collected and washed cleanly at 3 days after inoculation (DAI), and immersed in the solution (5% NaClO) for 5 min. Then roots were soaked in tap water for 15 min to remove residual NaClO. The roots were rinsed in boiling stain solution (acid fuchsin, 0.5 g/L) for 30 s, washed with tap water, then placed in 30 mL glycerin acidified with few drops of 5 mol/L HCl for water bath heating 30 s. The dyed roots were kept in glycerin for storage at 4°C. The number of visible pink-stained nematodes present within the roots was counted under a light microscope (Leica, DM3000 LED). At least nine replicate samples (individual plants) were counted for each treatment (Aditya et al., 2015).
To detect gene expression after CCN infection, two-leaf stage seedlings were cultured in sterilized soil (four seedlings each pot), and inoculated with nematodes (300 J2s/pot). The root tissues were respectively collected at 0, 30 h and 3, 9 days. All these samples were stored at -80°C for RNA extraction.
Total RNA was extracted using the TRIzol-A+ reagent (Tiangen, Beijing, China) according to the manufacturer’s instructions. The cDNA synthesis was carried out using the ReverTra Ace qPCR RT Kit (TOYOBO). QPCR was conducted as described (Wang et al., 2013). Gene-specific primers were designed to amplify PCR products about 100–200 bp, and listed in Supplementary Table 1. Elongation factor1-α (EF1α) mRNA was employed as an internal control for normalization (Livak and Schmittgen, 2001). Each sample or treatment was tested in three biological repeats and experiment was performed for three times. The differences were analyzed by t-test and data were presented by software Origin 8.6.
The plasmids used for gene silencing were constructed as previously described (Holzberg et al., 2002). 751∼951 bp of AeVTDC1 coding sequence (ORF) was cloned and ligated to BSMV γ plasmid through NheI to construct plasmid for silencing AeVTDC1. Primers used were listed in Supplementary Table 1.
Infectious BSMV RNA was prepared from each linearized plasmid (α and γ digested with MluI, β digested with SpeI) by in vitro transcription using a Large Scale RNA Production System (T7 RiboMAXTM Express Large Scale RNA Production System). The BSMV inoculum was made by combining an equimolar ratio of α, β, and γ transcripts with excess inoculation buffer containing a wounding agent (GKP buffer: 50 mM glycine, 30 mM dipotassium phosphate, pH 9.2, 1% bentonite, 1% celite) as previously described (Holzberg et al., 2002). The second leaves of two-leaf seedlings were inoculated with BSMV inoculum. BSMV γ empty vector were used as negative controls.
Barley stripe mosaic virus-treated plants were kept in a cultivation chamber at 25°C with 60% humidity. When the virus phenotype was observed (about 10 days after BSMV inoculation), new roots of these plants were sampled and used for RNA isolation. The silencing efficiency for the target gene and expression levels of other related genes compared with control were examined by QPCR. The primers for QPCR were listed in Supplementary Table 1.
PCAMBIA1300-based T-DNA vector was chosen as the skeleton and hygromycin was replaced with bar gene. CaMV35S promoter and NOS terminator were amplified using pJG045 as a template to drive and terminate gene expression (Zhao et al., 2013). For generation of the overexpression construct, the ORF of AeVTDC1 was PCR amplified using template with primers (Supplementary Table 1) (Li et al., 2016). AeVTDC1 ORF was fused to N terminal of yellow fluorescent protein (YFP) sequence and together inserted into the modified binary vector to express AeVTDC1-YFP fusion protein. The insertion sequences were confirmed by nucleotide sequencing. Overexpression construct was introduced into Agrobacterium tumefaciens strain EHA105 for tobacco transformation (Horsch et al., 1985). Cultivar tobacco (Mammoth Gold) was used for transformation. Positive transformants and their offsprings were screened by PCR with specific primers of AeVTDC1. Stable lines (L120 and L133) were selected for QPCR analysis of expression of genes and RKN resistance assay.
In resistance assay, L120, L133 and WT seeds were simultaneously germinated and cultured with sterilized soil under 25°C. Six-leaf seedlings were transplanted into soil containing RKN. The root knots were calculated and analyzed after 8 weeks. No less than 15 individuals each line were used for counting. The photographs were taken by camera (Canon).
The IAA in the roots were measured by UPLC–MS/MS as previously described (Fu et al., 2012).
All fresh roots were collected and freeze-dried for metabolites profiling. Three replicates were used in each treatment. The method was slightly modified according to the description (Chen et al., 2013). Each 0.1 g sample was powdered in liquid nitrogen using a mortar and pestle before 1 mL of extraction solution (70% aqueous methanol) was added, and the mixture was stored overnight in the dark at 4°C. The mixture was then centrifuged at 4°C at 10,000 g for 10 min, and each supernatant was filtered through a 0.22-μm Millipore filter before HPLC–MS/MS analysis.
The sample extracts were analyzed using an LC–ESI–MS/MS system (HPLC, Shim-pack UFLC SHIMADZU CBM30A system1; MS, Applied Biosystems 6500 Q TRAP2). The analytical conditions were as follows, HPLC: column, Waters ACQUITY UPLC HSS T3 C18 (1.8 μm, 2.1 mm × 100 mm); solvent system, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 100:0 V/V at 0 min, 5:95 V/V at 11.0 min, 5:95 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 15.0 min; flow rate, 0.4 ml /min; temperature, 40°C; injection volume: 2 μl. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)–MS.
LIT and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Q TRAP), API 6500 Q TRAP LC/MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in a positive ion mode and controlled by Analyst 1.6 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 500°C; ion spray voltage (IS) 5500 V; ion source gas I (GSI), gas II (GSII), curtain gas (CUR) were set at 55, 60, and 25.0 psi, respectively; the collision gas (CAD) was high. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with collision gas (nitrogen) set to 5 psi. DP and CE for individual MRM transitions were done with further DP and CE optimization. A specific set of MRM transitions were monitored for each period according to the metabolites eluted within this period.
Previous RNA-seq results indicated transcripts of several AeVTDCs were much higher in CCN-treated roots than that in non-treated roots. And primary results revealed inhibition of AeVTDC enzyme activity suppressed resistance to CCN (Li et al., 2016). To determine which member of AeVTDC family mainly contribute to CCN resistance, expression pattern of AeVTDC1 and AeVTDC2 were tested and compared at 0 hour (h), 30 h, 3 day (d), 9 day post-inoculation (dpi) of CCN. At 0 hpi, the expression level of the AeVTDC1 and AeVTDC2 were similar in the CCN-treated and control roots. At 30 hpi, the expression of AeVTDC1 was strikingly induced in CCN-treated roots, which is almost sixfold of that in control sample. Expression of AeVTDC2 was also induced and about twofold to that in control root. At 3 and 9 dpi, expressions of AeVTDC1 remained induced and were nearly threefold of that in control sample. And expressions of AeVTDC2 were still higher but less than two times of that in control sample (Figure 1). These results indicated that expression of AeVTDC1 was much more sensitive to CCN infection than AeVTDC2. Hence, AeVTDC1 was chosen as the candidate gene for the resistance assay.
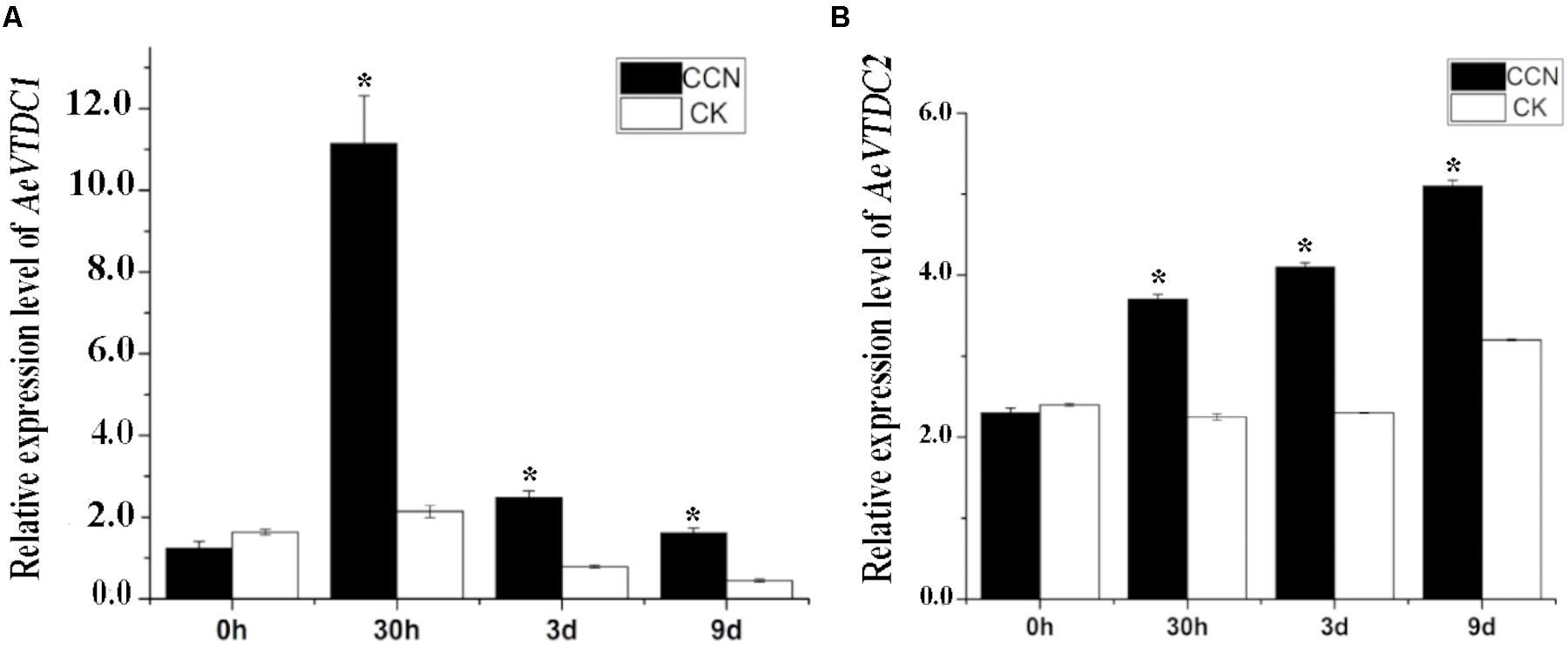
FIGURE 1. Expression of AeVTDC1 and AeVTDC2 gene after CCN inoculation in Aegilops variabilis No.1 roots. Two-leaf stage seedlings were used for CCN J2 inoculation or mock inoculation. CCN treatment group was inoculated with nematodes (300/pot, 4 seedlings/pot), and the control group was inoculated with same volume water. The roots tissues were respectively collected at 0, 30 h and 3, 9 days post-inoculation for RNA extraction and QPCR. The relative expressions were normalized with the AeVEF1α. Data represent the mean ± standard deviation of three replicate samples. The asterisk above the bars indicate values that are significantly different (P < 0.05).
Virus induced gene silencing is an efficient and fast technology in gene function analysis (Holzberg et al., 2002; Liu et al., 2002). BSMV based silencing system has been widely applied in monocots (Pacak et al., 2010). In our study, we found the expression of AeVTDC1 was strikingly induced at 30 hpi, which is almost sixfold of that in control sample. To directly investigate whether AeVTDC1 gene participates in resistance to CCN in Ae. variabilis No.1, we utilized BSMV-mediated gene silencing to knock down expression of AeVTDC1 in roots and observed CCN infection. Expression of AeVTDC1 in roots of AeVTDC1-silenced plants was about 30% of that in vector control (Figure 2C), which indicated VIGS worked well in the roots. Furthermore, the expression of AeVTDC2 in roots of AeVTDC1-silenced plants was similar with that in vector control (Supplementary Figure 1). The number of dyed CCN J2 in the roots of AeVTDC1-silenced plants increased more than 45% compared with that of vector control plants at 3 dpi (Figure 2D). The data was collected from at least nine replicates each time, and the experiment was operated three times. This result showed silencing AeVTDC1 in Ae. variabilis No.1 plants compromised resistance to CCN at the early stage and indicated AeVTDC1 regulates early immune responses to CCN.
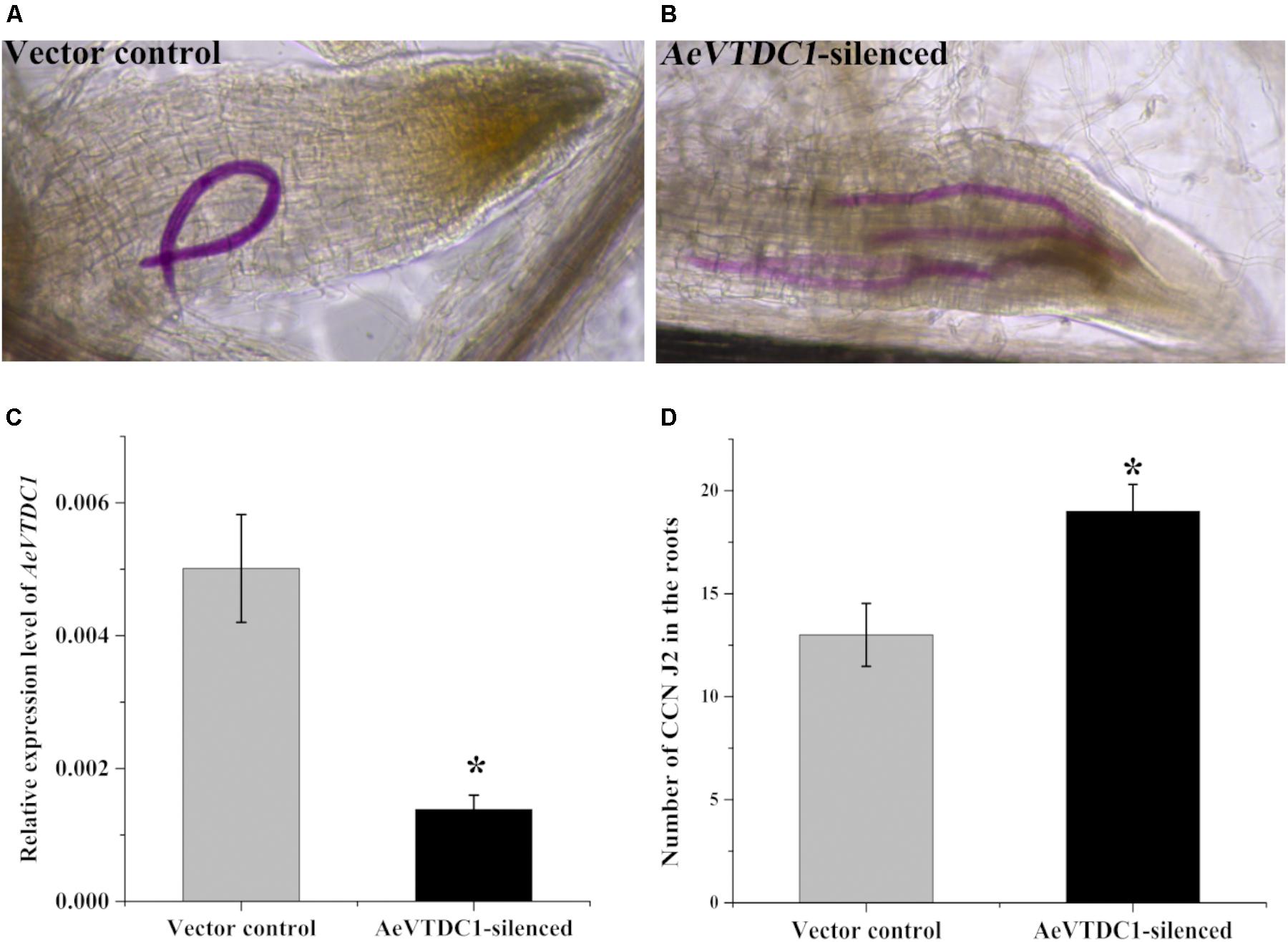
FIGURE 2. Silencing of AeVTDC1 gene compromised the resistance to CCN infection in Ae. variabilis No.1. (A,B) Roots were stained with acid fuchsin to dye nematodes. The visible pink nematodes were counted and photographed. (A) One vision of vector control root under microscope; (B) one vision of the AeVTDC1-silenced root. (C) The relative expression of AeVTDC1 in roots of vector control and AeVTDC1-silenced plants. VIGS inoculation was operated at the two-leaf stage of plant. About 2 weeks after inoculation, the roots of plants displayed BSMV infection symptoms were individually collected to affirm the VIGS effect by QPCR. The results were normalized with the AeVEF1α. (D) Number of CCN in AeVTDC1-silenced roots and vector control plants. The successful silencing plants were used for CCN J2 inoculation. Roots were collected and dyed with acid fuchsin 3 days after CCN inoculation. The number of visible pink-stained nematodes present within the roots was counted under a light microscope. The data were means ± SE. No less than 15 plants were used and calculated in each treatment. The asterisk represented significant differences (P < 0.05).
To analyze in more details of how AeVTDC1 regulated resistance to CCN in Ae. variabilis No.1, the changes of secondary metabolite and expression of related genes were detected in the roots of AeVTDC1-silenced plants.
Specific primers were designed to analyze the expression of genes downstream of TDC. Tryptamine-5-hydroxylase (T5H) catalyzes tryptamine transform into serotonin. Acetylserotonin O-methyltransferase (ASMT) catalyzes the synthesis of melatonin (Back et al., 2016). Methyl easterase (MES) is a key gene in the synthesis pathway of indole alkaloids. In the AeVTDC1-silenced plant roots, the expression of T5H, ASMT, and MES gene markedly declined comparing with vector control (Figure 3A). However, the content of tryptamine and melatonin had no significant changes in the AeVTDC1-silenced plant roots, and the content of serotonin might be too low to be detected both in AeVTDC1-silenced and vector control plants (Figure 3B). Indole and 3-Indolebutyric acid content slightly reduced. Caffeic acid 3-O-methyltransferase (COMT) methylates caffeic acid and 5-hydroxyferulic acid respectively to form ferulic acid and sinapic acid (Doorsselaere et al., 2010; Back et al., 2016). QPCR results displayed that the expression of COMT gene was unchanged (Figure 3A), but the content of caffeic acid, ferulic acid, and sinapic acid obviously decreased in the roots of AeVTDC1-silenced plants compared with vector control plants. Furthermore, the derivatives of serotonin, derivatives of tryptamine, and ferulic acid derivatives sharply declined in AeVTDC1-silenced plants. N-Feruloylserotonin went down 17 times. N-Feruloyltryptamine, N-Feruloylputrescine, and 3-O-Feruloylquinic acid also had significant reduction (Figure 3B). The results showed that silencing of AeVTDC1 gene altered the profile of downstream metabolin. The changes of metabolite (tryptamine derivatives, serotonin derivatives, ferulic acid and its derivatives) might be an important aspect for AeVTDC1 to regulate resistance to CCN in Ae. variabilis No.1.
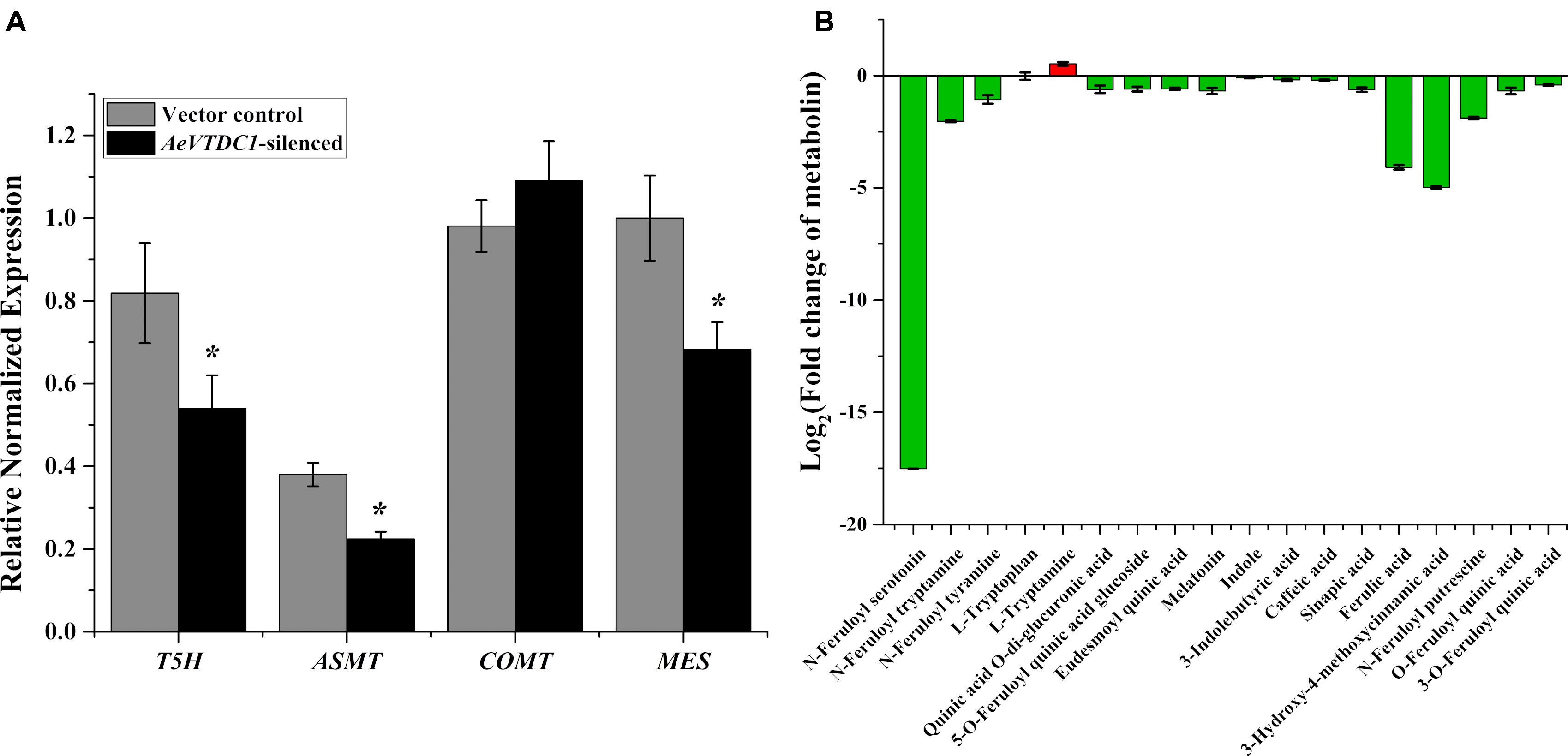
FIGURE 3. Silencing of AeVTDC1 affected expression of downstream genes and contents of relative secondary metabolites in Ae. variabilis No.1 plants. (A) Relative expressions of AeVTDC1 downstream genes (T5H, ASMT, COMT, and MES) were detected in AeVTDC1-silenced and vector control roots. (B) Fold changes of secondary metabolites in AeVTDC1-silenced compared to that in vector control roots. The content of secondary metabolite was detected by UPLC-ESI-MS/MS. Data represent the mean ± standard deviation of three replicate samples. Asterisk above the bars indicate values that were significantly different (P < 0.05).
TDC catalyzes conversion from tryptophan to tryptamine, and tryptamine is a precursor for the biosynthesis of IAA (Dubouzet et al., 2013). Except for regulation of plant development, IAA also involves in plant defense to biotic stress and abiotic stress (Mathesius, 2010; Navarro, 2016).
To understand whether IAA biosynthesis and signaling pathway were altered, we analyzed the transcripts changes of biosynthetic genes and signaling genes of IAA when AeVTDC1 was silenced. QPCR results showed there were no significant differences in the expression levels of biosynthesis genes (indole-3-acetaldehyde oxidase, AAO2; indole-3-pyruvate monooxygenase, YUCCA; nitrilase 2, NIT2; aldehyde dehydrogenase, ALDH2B) and signaling genes (small auxin-upregulated RNA, SAUR15) in AeVTDC1-silenced roots and control vector roots (Figures 4A,B). Liquid chromatography–mass spectrometry (LC–MS) data further attested the level of IAA in the roots of AeVTDC1-silenced plants was similar with that in vector control plants (Figure 4C). These results demonstrated silencing of AeVTDC1 had no effect on expression of biosynthetic and signaling genes of IAA and IAA accumulation in Ae. variabilis plants. There might be other factors related to the resistance reduction in the AeVTDC1-silencing plants.
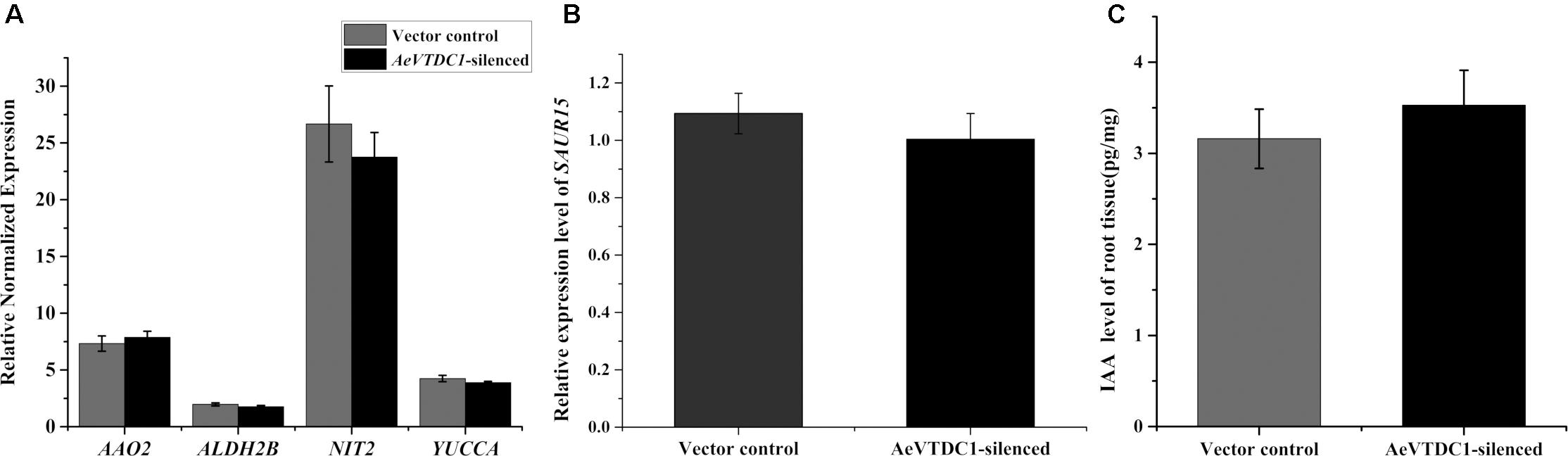
FIGURE 4. Expression of IAA biosynthetic and signaling genes and IAA contents in roots of AeVTDC1-silenced and control plants. (A) Relative expression of IAA biosynthetic genes AAO2, ALDNH2B, NIT2, YUCCA were detected in AeVTDC1-silenced and vector control roots. (B) Expression of IAA signaling gene SAUR15 in AeVTDC1-silenced and vector control roots. The results were normalized with the AeVEF1α. (C) Level of IAA in AeVTDC1-silenced and vector control roots. The content of IAA was detected by UPLC–MS. Data represent the mean ± standard deviation of three replicate samples.
To test whether AeVTDC1 involved resistance to other pathogen, AeVTDC1 was overexpressed in tobacco. Stable AeVTDC1 transgenic tobacco plants (L120 and L133) were obtained to monitor the susceptibility to RKN. AeVTDC1 transgenic plants and WT plants with similar growth state were planted in the homogenous soil containing RKN. Two months later, all root tissues of plants were taken out to count the number of root knots and photographed (Figure 5A). The knots formed on transgenic plants were much smaller than that on WT. Statistical data revealed that the number of knots formed on the transgenic plants was much less than that on WT (Figure 5B). The results demonstrated ectopic expression of AeVTDC1 in tobacco enhanced defense to RKN and led to reduction of root knots.
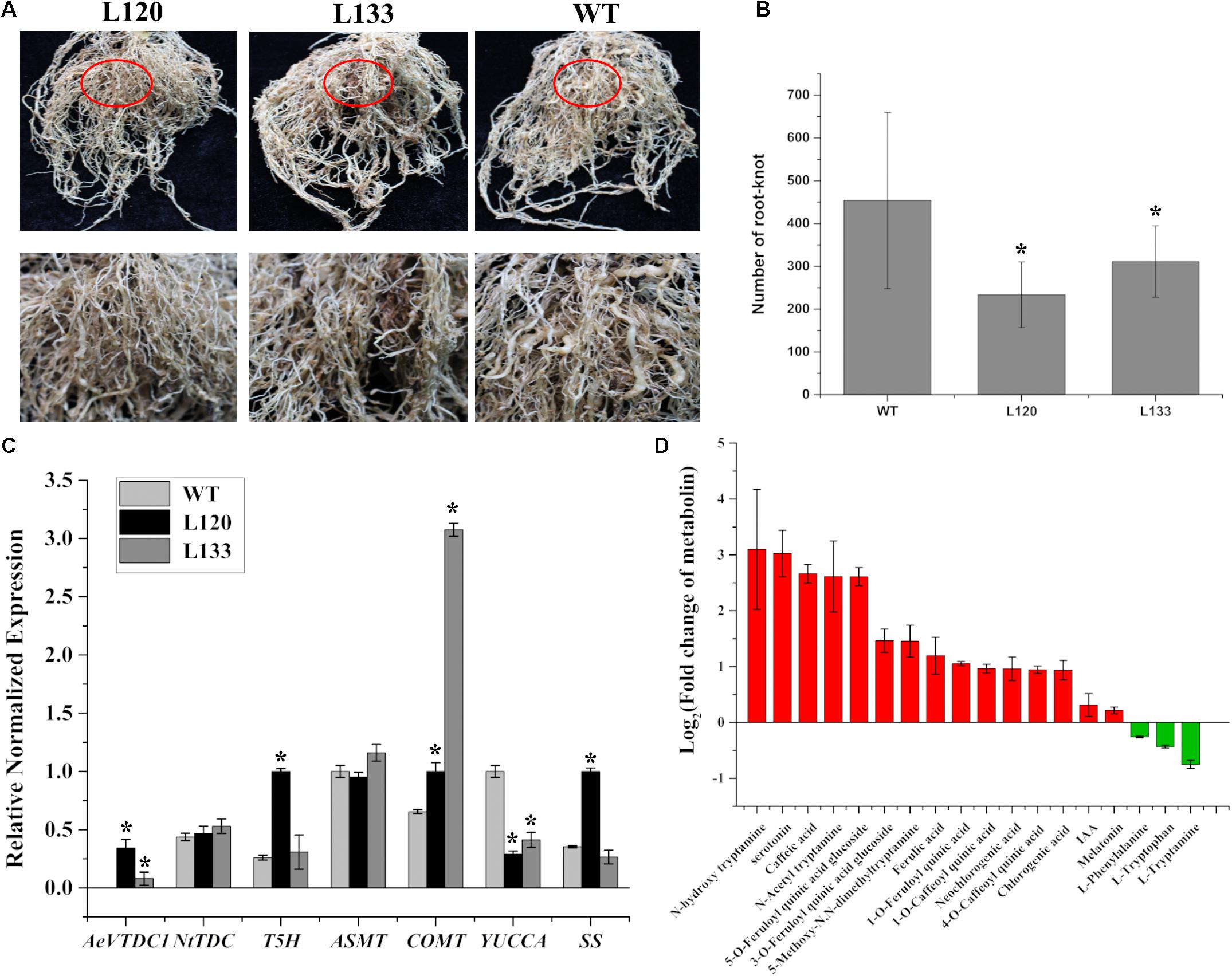
FIGURE 5. Number of root knots formed on roots of AeVTDC1 overexpressing tobacco plants and non-transgenic plants, and contents of downstream metabolites in roots were respectively analyzed. (A) Phenotypes of the root tissues of non-transgenic and AeVTDC1 transgenic tobacco. L120 and L133 were two independent lines of AeVTDC1 transgenic tobacco. The first row showed the overall view of root tissues, and the second row displayed the partial tissue circled with red oval in the first row. Photos were taken 2 months after planting in soil containing RKN. (B) Statistic data of root knots numbers formed on AeVTDC1 overexpressing tobacco and non-transgenic tobaccos. The mean number of knots was calculated and presented. More than 15 replicates were used for counting. (C) Relative expressions of AeVTDC1, NtTDC, TDC downstream genes T5H, ASMT, COMT, SS in tobacco, and IAA biosynthetic related genes YUCCA were detected in root tissues of L120, L133, and WT. (D) Fold change of secondary metabolite in AeVTDC1 overexpressing tobacco plants (L120) and WT roots. The content of secondary metabolite was detected by UPLC-ESI-MS/MS. The error bar represented the standard error. ∗P-value < 0.05.
To further analyze which downstream products of TDC improved resistance to RKN in transgenic tobacco, the roots of the most resistant line (L120) and WT were respectively collected for basic metabolite profile and related gene expression analysis. We found that the expression of NtTDC was similar between L120 and WT (Figure 5C). And downstream genes and substances in tryptophan metabolism were also detected. The content of tryptamine showed no difference in the transgenic tobacco comparative with WT (Figure 5D). The tryptamine may be easily changed to tryptamine derivatives (N-hydroxytryptamine, N-Acetyltryptamine, and 5-Methoxy-N,N-dimethyltryptamine) in the tobacco, which showed obvious increase in L120. Serotonin accumulation strikingly increased in the roots of L120, almost eightfold greater than that of WT plants (Figure 5D), and expression of T5H was also obviously upregulated in the roots of L120 (Figure 5C). The content of IAA and melatonin had no obvious difference (Figure 5D), and the biosynthesis gene ASMT of melatonin similarly had no change, while IAA biosynthesis gene YUCCA was down-regulated compared to the WT (Figure 5C). Moreover, we discovered that the expression of COMT and the content of ferulic acid and ferulic acid derivatives (1-O-Feruloylquinic acid, 3-O-Feruloylquinic acid glucoside, 5-O-Feruloylquinic acid glucoside) were higher in transgenic tobacco than in WT (Figures 5C,D). Furthermore, we also found the level of strictosidine synthase (SS) expression was improved in AeVTDC1 in tobacco. The contents of caffeic acid, chlorogenic acid and some quinine (such as 1-O-Caffeoylquinic acid, 4-O-Caffeoylquinic acid) increased by overexpression AeVTDC1 in tobacco (Figure 5D). These results revealed that the content changes of metabolite (serotonin, tryptamine derivatives, ferulic acid and its derivatives) in transgenic tobacco might contribute to the enhanced resistance to RKN in tobacco. Moreover, the related genes expressions were also detected in L133. Expression of AeVTDC1 was higher in L120 than that in L133. Expression of NtTDC, COMT, ASMT, and YUCCA was similar in the roots of L133 and L120. However, Expression of T5H and SS showed no obvious difference in the roots of L133 and WT (Figure 5C). The expression differences might account for the different resistances by AeVTDC1 overexpression.
CCN (H. avenae) is soil-borne and invades plants from roots. Production losses caused by CCN gradually become bigger in recent years. However, it is difficult to control CCN disease because CCN infection is not easy to be observed. Bringing resistance-related genes from relatives into wheat has been known as an efficient strategy to enhance wheat resistance to CCN. However, few resistance-related genes were identified. High-throughput methods like RNA-seq and microarray analysis are widely used to find differential expression genes, which provide a mass of candidate genes for following work.
In our previous work, RNA-sequencing was operated to find out differential expression genes before and after CCN infection in Ae. variabilis No.1 (Xu et al., 2012). There were a large number of differential expression genes at 30 hpi, when the early response was conferred to CCN infection. Whereas there were fewer differential expression genes at 3 and 9 dpi, when CCN J2 had migrated in vascular tissues and developed into J2∼J3 stage. It was reported that most of differential expression genes gathered at 3 and 8 dpi in RNA-sequencing analysis of incompatible wheat and a compatible control cultivar infected with H. avenae at 24, 3, and 8 dpi (Kong et al., 2015). The different responses might indicate a different resistance mechanism in Ae. variabilis No.1.
Previous analysis showed transcripts of several genes in tryptophan metabolism were induced by CCN, and AeVTDCs as the key gene were also induced at the 30 hpi early response. In this study, AeVTDC1 and AeVTDC2 were further verified to be induced by CCN at 30 hpi, which was accordant with the transcriptome data (Xu et al., 2012).
In the wheat genome, there are more than 15 copies of TaTDCs which showed various tissue expression patterns (Choulet et al., 2014). Expression patterns of two TaTDCs (TaTDC1 and TaTDC2), highly expressed in roots and with highest homology to AeVTDC1, were tested after CCN inoculation. Expression of TaTDC1 was induced after CCN inoculation, while expression of TaTDC2 showed no obvious changes after CCN infection (Supplementary Figure 2). The different expression pattern of the various isoforms might indicate that not all TaTDCs isoforms respond to CCN. Biological functions of several plant TDCs were gradually disclosed. Overexpression the Catharanthus roseus TDC gene in plants (tobacco, poplar, canola, Petunia hybrida) gave rise to tryptamine accumulation in transgenic plants, which improved tobacco resistance to Manduca sexta and poplar resistance to Malacosoma disstria (Thomas et al., 1999; Leech et al., 2000; Gill et al., 2003). In Ae. variabilis No.1 genomes, there were at least three TDC genes that may have different functions in downstream metabolism process from tryptamine (Facchini et al., 2000; Byeon et al., 2014). However, which AeVTDC participated in CCN resistance remained unclear. In this study, expression of AeVTDC1 showed greater changes than AeVTDC2 after CCN infection (Figures 1A,B). Here, the role of AeVTDC1 in CCN resistance was directly studied. Silencing of AeVTDC1 weakened resistance to CCN and led more CCN invading into roots (Figure 2). This indicated the positive regulation of AeVTDC1 in resistance to CCN. It needs further study whether there was function redundancy among AeVTDC1 and its homologs. Moreover, the role of AeVTDC1 in RKN resistance was also tested by overexpression in tobacco. AeVTDC1 overexpression reduced knots on roots and it revealed AeVTDC1 plays a positive role in RKN resistance as in CCN resistance (Figures 5A,B). The different resistance on L120 and L132 might be related to the expression level of AeVTDC1 in tobacco (Zhang et al., 2013). Hence, we suggested the broad spectrum resistance of AeVTDC1 to crucial nematodes.
In the plant kingdom, diverse metabolites derived from Trp are found and play an important role in the plant immunity and rice (Bohlmann et al., 1995; Zhao and Last, 1996; Matsukawa et al., 2002; Kang and Back, 2009; Ueno et al., 2010; Ishihara et al., 2011; Dharmawardhana, 2013; Dubouzet et al., 2013; Hayashi et al., 2016; Lu et al., 2018). Metabolites, liking tryptamine, serotonin or their derivatives, have strong antioxidant activities and regulate resistance to avoid damage from pathogen attacks (Kang and Back, 2009; Dubouzet et al., 2013). The dramatic increased serotonin suppress leaf damage outside the halo, block expansion of the browning area and attenuate symptom of plant growth inhibition (Hayashi et al., 2016). Ectopic expression of Camptotheca acuminata TDC1 gene allowed sufficient tryptamine to accumulate in poplar and tobacco leaf tissue to significantly suppress the growth of insect pests (Gill et al., 2003). Melatonin-rich rice plants exhibit resistance to herbicide-induced oxidative stress (Park et al., 2013). Recently, it was reported that CYP71A1 mutants with less serotonin content were more susceptible to rice blast Magnaporthe grisea, but more resistant to rice brown spot disease Bipolaris oryzae1, rice brown planthopper and striped stem borer (Lu et al., 2018).
In this study, serotonin concentration in AeVTDC1 transgenic tobacco roots was eightfold greater than in roots from WT plants, while tryptamine and melatonin remained unchanged (Figure 5D). No change of tryptamine was reasonable since it might be easily transformed to its derivatives (N-hydroxytryptamine, N-Acetyltryptamine, and 5-Methoxy-N,N-dimethyltryptamine), which increased in the transgenic tobacco. Our results were similar to that in overexpression of Catharanthus TDC in cell cultures of Peganum harmala (Leech et al., 2000). Serotonin and tryptamine derivatives, caffeic acid, chlorogenic acid and several quinines remarkably increased in transgenic tobacco (Figure 5D). Except for function in plant immunity, serotonin, feruloylserotonin, and 4-coumaroylserotonin were reported contributed to delay senescence of rice (Kang and Back, 2009). High level of serotonin accumulation in rice caused stunt phenotype (Kanjanaphachoat et al., 2012). Even though elevated level of serotonin accumulated in AeVTDC1 transgenic tobacco, the plants didn’t show stunted growth (not shown) as overexpression of TDC in rice (Kanjanaphachoat et al., 2012). In the VIGS assay, the serotonin wasn’t either detected in AeVTDC1-silencing root or in the control roots. That’s probably because of too low level of serotonin in roots of Ae. variabilis No.1. Contents of N-Feruloylserotonin and N-Feruloyltryptamine markedly decreased in the AeVTDC1-silencing plants comparing with the control (Figure 4B). Ferulic acid is a well-known phytoalexins, which inhibited the growth of necrotrophic bacteria Dickeya dadantii (Pérez-Bueno et al., 2016). Furthermore, the decrease of soluble free and soluble conjugated phenolic acids, such as soluble hydrolyzable ferulic acid and sinapic acid, reduced the attraction of Diabrotica virgifera virgifera to the root of maize (Erb et al., 2015). In our study, ferulic acid significantly increased in the AeVTDC1 transgenic tobacco, and decreased in AeVTDC1-silencing plants (Figures 4B, 5D). Here, we found altered expression of AeVTDC1 changed secondary metabolites and nematode resistance. It still needs further studies of direct effects of serotonin, ferulic acid, indole alkaloids, quinines and their derivatives on nematodes.
Plant hormone IAA biosynthesis has several main pathways from Trp, respectively catalyzed by TDC, YUCCA, and NIT (Sugawara et al., 2009; Zhao, 2010). It is well-known that auxin always regulates plant defense as a negative regulator in plant immune system (Mathesius, 2010; Navarro, 2016). A series of evidence has demonstrated that auxin plays roles in balancing plant defense responses and growth in plants. Lionel Navarro reported the repression of auxin signaling made for the improving of bacterial resistance in Arabidopsis (Navarro et al., 2006). Overexpression of OsGH3.1 and OsGH3.8 in rice reduced the IAA content, influence cell growth, and enhanced disease resistance to both fungal and bacterial pathogens (Domingo et al., 2009). In this work, the content of IAA was not changed by silencing AeVTDC1 in Ae. variabilis No.1 (Figure 3C). There was also no change about the content of IAA in the AeVTDC1 transgenic tobacco comparing with WT (Figure 5D). Silencing or overexpression of AeVTDC1 gene also had no effects on the expression of IAA biosynthesis and signaling genes (Figures 3A,B, 5C). The results indicate AeVTDC1 might not function to regulate IAA biosynthesis. Moreover, we used two concentrations (100 and 200 μM) of 2, 4-D to pretreat the roots of Ae. variabilis No.1 to test CCN resistance. Statistical results revealed that there was no difference of CCN number in the roots between IAA pretreat and control (Supplementary Figure 3). These results indicate that IAA has no impact on the interaction between CCN and Ae. variabilis No.1. In addition, IAA is likely subjected to strict monitoring during the interactions of plants and nematodes.
HZ and MY designed the experiments. HZ and QH carried out sample collection, expression analysis, gene silencing, disease resistance assessment, data analysis, and wrote the manuscript. LL transformed transgenic tobacco. MZ analyzed transcriptome data. HL, GD, ZP, JL, QL, and FC assisted the experiments. All authors approved the final manuscript.
This work was supported by National Natural Science Foundation of China (Grant Nos. 31501614 and 31470097), Major transgenic special project (Grant No. 2016ZX08009003-001), and West Light Foundation of Chinese Academy of Sciences (Grant No. 2015XBZG_XBQNXZ_B_010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful of Professor Feng Liu (Shan Dong Agricultural University) for providing soil containing cereal cysts. We are grateful of Professor Deliang Peng and Shujie Luo (Institute of Plant Protection, Chinese Academy of Agricultural Sciences) for providing methods of dying CCN. We thank the expertise of Miss. Shuang Fang and Dr. Jinfang Chu [National Centre for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China] in determining the IAA contents of roots.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01297/full#supplementary-material
Aditya, J., Lewis, J., Shirley, N. J., Tan, H. T., Henderson, M., Fincher, G. B., et al. (2015). The dynamics of cereal cyst nematode infection differ between susceptible and resistant barley cultivars and lead to changes in (1,3;1,4)-beta-glucan levels and HvCslF gene transcript abundance. New Phytol. 207, 135–147. doi: 10.1111/nph.13349
Back, K., Tan, D. X., and Reiter, R. J. (2016). Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437. doi: 10.1111/jpi.12364
Barloy, D., Lemoine, J., Abelard, P., Tanguy, A. M., Rivoal, R., and Jahier, J. (2006). Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol. Breed. 20, 31–40. doi: 10.1007/s11032-006-9070-x
Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 49–64. doi: 10.1146/annurev.arplant.48.1.51
Berlin, J., Rugenhagen, C., Dietze, P., Fecker, L. F., Goddijn, O. J. M., and Hoge, J. H. C. (1993). Increased production of serotonin by suspension and root cultures of Peganum-harmala transformed with a tryptophan decarboxylase cDNA clone from Catharanthus-roseus. Transgenic Res. 2, 336–344. doi: 10.1007/bf01976175
Bohlmann, J., Deluca, V., Eilert, U., and Martin, W. (1995). Purification and cDNA cloning of anthranilate synthase from Ruta-graveolens - modes of expression and properties of native and recombinant enzymes. Plant J. 7, 491–501. doi: 10.1046/j.1365-313X.1995.7030491.x
Byeon, Y., Park, S., Lee, H. Y., Kim, Y. S., and Back, K. (2014). Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J. Pineal Res. 56, 275–282. doi: 10.1111/jpi.12120
Canel, C., Lopes-Cardoso, M. I., Whitmer, S., van der Fits, L., Pasquali, G., van der Heijden, R., et al. (1998). Effects of over-expression of strictosidine synthase and tryptophan decarboxylase on alkaloid production by cell cultures of Catharanthus roseus. Planta 205, 414–419. doi: 10.1007/s004250050338
Charoonratana, T., Wungsintaweekul, J., Keawpradub, N., and Verpoorte, R. (2013). Molecular cloning and expression of tryptophan decarboxylase from Mitragyna speciosa. Acta Physiol. Plant. 35, 2611–2621. doi: 10.1007/s11738-013-1296-8
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Choulet, F., Alberti, A., Theil, S., Glover, N., Barbe, V., Daron, J., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Science 345:1249721. doi: 10.1126/science.1249721
Coriton, O., Barloy, D., Huteau, V., Lemoine, J., Tanguy, A. M., and Jahier, J. (2009). Assignment of Aegilops variabilis Eig chromosomes and translocations carrying resistance to nematodes in wheat. Genome 52, 338–346. doi: 10.1139/g09-011
de Majnik, J., Ogbonnaya, F. C., Moullet, O., and Lagudah, E. S. (2003). The Cre1 and Cre3 nematode resistance genes are located at homeologous loci in the wheat genome. Mol. Plant Microbe Interact. 16, 1129–1134. doi: 10.1094/mpmi.2003.16.12.1129
Dharmawardhana, P. (2013). A genome scale metabolic network for rice and accompanying analysis of tryptophan, auxin and serotonin biosynthesis regulation under biotic stress. Rice 6, 1–15. doi: 10.1186/1939-8433-6-15
Domingo, C., Andrés, F., Tharreau, D., Iglesias, D. J., and Talón, M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 22, 201–210. doi: 10.1094/MPMI-22-2-0201
Doorsselaere, J. V., Baucher, M., Chognot, E., Chabbert, B., Tollier, M. T., Petit-Conil, M., et al. (2010). A novel lignin in poplar trees with a reduced caffeic acid/5-hydroxyferulic acid O-methyltransferase activity. Plant J. 8, 855–864. doi: 10.1046/j.1365-313X.1995.8060855.x
Dubouzet, J. G., Matsuda, F., Ishihara, A., Miyagawa, H., and Wakasa, K. (2013). Production of indole alkaloids by metabolic engineering of the tryptophan pathway in rice. Plant Biotechnol. J. 11, 1103–1111. doi: 10.1111/pbi.12105
Erb, M., Robert, C. A., Marti, G., Lu, J., Doyen, G., Villard, N., et al. (2015). A physiological and behavioral mechanism for leaf-herbivore induced systemic root resistance. Plant Physiol. 169, 2884–2894. doi: 10.1104/pp.15.00759
Facchini, P. J., Huber-Allanach, K. L., and Tari, L. W. (2000). Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54, 121–138. doi: 10.1016/S0031-9422(00)00050-9
Fu, J., Chu, J., Sun, X., Wang, J., and Yan, C. (2012). Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28, 1081–1087. doi: 10.2116/analsci.28.1081
Geerlings, A., Hallard, D., Caballero, A. M., Cardoso, I. L., van der Heijden, R., and Verpoorte, R. (1999). Alkaloid production by a Cinchona officinalis ’Ledgeriana’ hairy root culture containing constitutive expression constructs of tryptophan decarboxylase and strictosidine synthase cDNAs from Catharanthus roseus. Plant Cell Rep. 19, 191–196. doi: 10.1007/s002990050732
Giebel, J., and Jackowiak, N. (1976). Tryptophan decarboxylase in resistant and susceptible potato roots infected with Heterodera-rostochiensis. Nematologica 22, 462–466. doi: 10.1163/187529276x00463
Gill, R. I., Ellis, B. E., and Isman, M. B. (2003). Tryptamine-induced resistance in tryptophan decarboxylase transgenic poplar and tobacco plants against their specific herbivores. J. Chem. Ecol. 29, 779–793. doi: 10.1023/A:1022983529555
Gill, R. I. S., and Ellis, B. E. (2006). Over-expression of tryptophan decarboxylase gene in poplar and its possible role in resistance against Malacosoma disstria. New For. 31, 195–209. doi: 10.1007/s11056-005-2719-1
Goddijn, O. J. M., Lohman, F. P., Dekam, R. J., Schilperoort, R. A., and Hoge, J. H. C. (1994). Nucleotide-sequence of the tryptophan decarboxylase gene of Catharanthus-roseus and expression of tdc-gusA gene fusions in Nicotiana-tabacum. Mol. Gen. Genet. 242, 217–225. doi: 10.1007/bf00391016
Hajihasani, A., Maafi, Z. T., Nicol, J. M., and Seraji, A. (2010). Relationships between population densities of the cereal cyst nematode, Heterodera latipons and yield losses of winter wheat in microplots. Aust. Plant Pathol. 39, 530–535. doi: 10.1071/AP10132
Hallard, D., vanderHeijden, R., Verpoorte, R., Cardoso, M. I. L., Pasquali, G., Memelink, J., et al. (1997). Suspension cultured transgenic cells of Nicotiana tabacum expressing tryptophan decarboxylase and strictosidine synthase cDNAs from Catharanthus roseus produce strictosidine upon secologanin feeding. Plant Cell Rep. 17, 50–54. doi: 10.1007/s002990050350
Hayashi, K., Fujita, Y., Ashizawa, T., Suzuki, F., Nagamura, Y., and Hayano-Saito, Y. (2016). Serotonin attenuates biotic stress and leads to lesion browning caused by a hypersensitive response to Magnaporthe oryzae penetration in rice. Plant J. 85, 46–56. doi: 10.1111/tpj.13083
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general-method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Ishihara, A., Nakao, T., Mashimo, Y., Murai, M., Ichimaru, N., Tanaka, C., et al. (2011). Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry 72, 7–13. doi: 10.1016/j.phytochem.2010.11.001
Jadaun, J. S., Sangwan, N. S., Narnoliya, L. K., Tripathi, S., and Sangwan, R. S. (2017). Withania coagulans tryptophan decarboxylase gene cloning, heterologous expression, and catalytic characteristics of the recombinant enzyme. Protoplasma 254, 181–192. doi: 10.1007/s00709-015-0929-8
Kang, K., and Back, K. (2009). Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 150, 1380. doi: 10.1104/pp.109.138552
Kanjanaphachoat, P., Wei, B. Y., Lo, S. F., Wang, I. W., Wang, C. S., Yu, S. M., et al. (2012). Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxylase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 78, 525–543. doi: 10.1007/s11103-012-9882-5
Kong, L. A., Wu, D. Q., Huang, W. K., Peng, H., Wang, G. F., Cui, J. K., et al. (2015). Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics 16:801. doi: 10.1186/s12864-015-2037-8
Leech, M. J., Burtin, D., Hallard, D., Hilliou, F., Kemp, B., Palacios, N., et al. (2000). Particle Gun Methodology as a Tool in Metabolic Engineering. Dordrecht: Springer. doi: 10.1007/978-94-015-9423-3_4
Li, L., Zheng, M. H., Long, H., Deng, G. B., Ishihara, A., Liu, F., et al. (2016). Molecular cloning and characterization of two genes encoding tryptophan decarboxylase from Aegilops variabilis with resistance to the cereal cyst nematode (Heterodera avenae) and root-knot nematode (Meloidogyne naasi). Plant Mol. Biol. Rep. 34, 273–282. doi: 10.1007/s11105-015-0909-3
Liu, W., Chen, R., Chen, M., Zhang, H., Peng, M., Yang, C., et al. (2012). Tryptophan decarboxylase plays an important role in ajmalicine biosynthesis in Rauvolfia verticillata. Planta 236, 239–250. doi: 10.1007/s00425-012-1608-z
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Luo, T., Fu, H., Wang, L., Tan, Y., Huang, J., et al. (2018). Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 4, 338–344. doi: 10.1038/s41477-018-0152-7
Mathesius, U. (2010). The Role of Auxin in Root-Symbiont and Root-Pathogen Interactions: From Development to Defense. Berlin: Springer.
Matsukawa, T., Ishihara, A., and Iwamura, H. (2002). Induction of anthranilate synthase activity by elicitors in oats. Z. Naturforsch. C 57, 121–128. doi: 10.1515/znc-2002-1-221
Mehrotra, S., Srivastava, V., Rahman, L. U., and Kukreja, A. K. (2013). Overexpression of a Catharanthus tryptophan decarboxylase (tdc) gene leads to enhanced terpenoid indole alkaloid (TIA) production in transgenic hairy root lines of Rauwolfia serpentina. Plant Cell Tissue Organ Cult. 115, 377–384. doi: 10.1007/s11240-013-0369-0
Montes, M. J., Andrés, M. F., Sin, E., Lópezbraña, I., Martínsánchez, J. A., Romero, M. D., et al. (2008). Cereal cyst nematode resistance conferred by the Cre7 gene from Aegilops triuncialis and its relationship with Cre genes from Australian wheat cultivars. Genome 51, 315–319. doi: 10.1139/G08-015
Navarro, L. (2016). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 352:1286.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Pacak, A., Geisler, K., Jorgensen, B., Barciszewska-Pacak, M., Nilsson, L., Nielsen, T. H., et al. (2010). Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 6:26. doi: 10.1186/1746-4811-6-26
Park, S., Lee, D. E., Jang, H., Byeon, Y., Kim, Y. S., and Back, K. (2013). Melatonin-rich transgenic rice plants exhibit resistance to herbicide-induced oxidative stress. J. Pineal Res. 54, 258–263. doi: 10.1111/j.1600-079X.2012.01029.x
Pérez-Bueno, M. L., Granum, E., Pineda, M., Flors, V., Rodriguez-Palenzuela, P., López-Solanilla, E., et al. (2016). Temporal and spatial resolution of activated plant defense responses in leaves of Nicotiana benthamiana infected with Dickeya dadantii. Front. Plant Sci. 6:1209. doi: 10.3389/fpls.2015.01209
Safari, E., Gororo, N. N., Eastwood, R. F., Lewis, J., Eagles, H. A., and Ogbonnaya, F. C. (2005). Impact of Cre 1, Cre8 and Cre3 genes on cereal cyst nematode resistance in wheat. Theor. Appl. Genet. 110, 567–572. doi: 10.1007/s00122-004-1873-8
Sugawara, S., Hishiyama, S., Jikumaru, Y., Hanada, A., Nishimura, T., Koshiba, T., et al. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5430–5435. doi: 10.1073/pnas.0811226106
Thomas, J. C., Akroush, A. M., and Adamus, G. (1999). The indole alkaloid tryptamine produced in transgenic Petunia hybrida. Plant Physiol. Biochem. 37, 665–670. doi: 10.1016/S0981-9428(00)80096-0
Ueno, M., Shibata, H., Kihara, J., Honda, Y., and Arase, S. (2010). Increased tryptophan decarboxylase and monoamine oxidase activities induce Sekiguchi lesion formation in rice infected with Magnaporthe grisea. Plant J. 36, 215–228. doi: 10.1046/j.1365-313X.2003.01875.x
Valletta, A., Trainotti, L., Santamaria, A. R., and Pasqua, G. (2010). Cell-specific expression of tryptophan decarboxylase and 10-hydroxygeraniol oxidoreductase, key genes involved in camptothecin biosynthesis in Camptotheca acuminata Decne (Nyssaceae). BMC Plant Biol. 10:69. doi: 10.1186/1471-2229-10-69
Verma, P., Mathur, A. K., Khan, S. A., Verma, N., and Sharma, A. (2015). Transgenic studies for modulating terpenoid indole alkaloids pathway in Catharanthus roseus: present status and future options. Phytochem. Rev. 16, 19–54. doi: 10.1007/s11101-015-9447-8
Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S., et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383–1399. doi: 10.1105/tpc.112.108993
Williamson, V. M., and Kumar, A. (2006). Nematode resistance in plants: the battle underground. Trends Genet. 22, 396–403. doi: 10.1016/j.tig.2006.05.003
Wu, L., Cui, L., Li, H., Sun, L., Gao, X., Qiu, D., et al. (2016). Characterization of resistance to the cereal cyst nematode in the soft white winter wheat ‘Madsen’. Plant Dis. 100, 679–685. doi: 10.1094/PDIS-06-15-0714-RE
Xu, D. L., Long, H., Liang, J. J., Zhang, J., Chen, X., Li, J. L., et al. (2012). De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genomics 13:133. doi: 10.1186/1471-2164-13-133
Zhang, H., Zhao, J., Liu, S., Zhang, D. P., and Liu, Y. (2013). Tm-22 confers different resistance responses against tobacco mosaic virus dependent on its expression level. Mol Plant 6, 971–974. doi: 10.1093/mp/sss153
Zhang, R., Feng, Y., Li, H., Yuan, H., Dai, J., Cao, A., et al. (2016). Cereal cyst nematode resistance gene CreV effective against Heterodera filipjevi transferred from chromosome 6VL of Dasypyrum villosum to bread wheat. Mol. Breed. 36:122. doi: 10.1007/s11032-016-0549-9
Zhao, J., and Last, R. L. (1996). Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 8, 2235–2244. doi: 10.1105/tpc.8.12.2235
Zhao, J., Liu, Q., Zhang, H., Jia, Q., Hong, Y., and Liu, Y. (2013). The rubisco small subunit is involved in tobamovirus movement and Tm-22-mediated extreme resistance. Plant Physiol. 161, 374–383. doi: 10.1104/pp.112.209213
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
Keywords: cereal cyst nematode, Aegilops variabilis No.1, tryptophan decarboxylase, secondary metabolite, indole acetic acid
Citation: Huang Q, Li L, Zheng M, Chen F, Long H, Deng G, Pan Z, Liang J, Li Q, Yu M and Zhang H (2018) The Tryptophan decarboxylase 1 Gene From Aegilops variabilis No.1 Regulate the Resistance Against Cereal Cyst Nematode by Altering the Downstream Secondary Metabolite Contents Rather Than Auxin Synthesis. Front. Plant Sci. 9:1297. doi: 10.3389/fpls.2018.01297
Received: 13 June 2018; Accepted: 17 August 2018;
Published: 04 September 2018.
Edited by:
Yi Li, Peking University, ChinaReviewed by:
Laurence Veronique Bindschedler, Royal Holloway, University of London, United KingdomCopyright © 2018 Huang, Li, Zheng, Chen, Long, Deng, Pan, Liang, Li, Yu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maoqun Yu, eXVtYW9xdW5AY2liLmFjLmNu; eXVtcUBjaWIuYWMuY24= Haili Zhang, emhhbmdobEBjaWIuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.