- 1Soybean Department, Institute of Field and Vegetable Crops, Novi Sad, Serbia
- 2Industrial Crops Department, Institute of Field and Vegetable Crops, Novi Sad, Serbia
Soybean time of flowering and maturity are genetically controlled by E genes. Different allelic combinations of these genes determine soybean adaptation to a specific latitude. The paper describes the first attempt to assess adaptation of soybean genotypes developed and realized at Institute of Field and Vegetable Crops, Novi Sad, Serbia [Novi Sad (NS) varieties and breeding lines] based on E gene variation, as well as to comparatively assess E gene variation in North-American (NA), Chinese, and European genotypes, as most of the studies published so far deal with North-American and Chinese cultivars and breeding material. Allelic variation and distribution of the major maturity genes (E1, E2, E3, and E4) has been determined in 445 genotypes from soybean collections of NA ancestral lines, Chinese germplasm, and European varieties, as well as NS varieties and breeding lines. The study showed that allelic combinations of E1–E4 genes significantly determined the adaptation of varieties to different geographical regions, although they have different impacts on maturity. In general, each collection had one major E genotype haplogroup, comprising over 50% of the lines. The exceptions were European varieties that had two predominant haplogroups and NA ancestral lines distributed almost evenly among several haplogroups. As e1-as/e2/E3/E4 was the most common genotype in NS population, present in the best-performing genotypes in terms of yield, this specific allele combination was proposed as the optimal combination for the environments of Central-Eastern Europe.
Introduction
Photoperiodism is reaction of a plant to the length of day and night. The discovery of photoperiodism in soybean and tobacco (Garner and Allard, 1930), and subsequently in numerous other plant species, paved the way for investigation of mechanisms which control seasonal rhythm in plants, such as flowering, growth and reproduction (Putterill et al., 2009). Besides plant growth, photoperiod also affects other aspects of plant development, including changes of developmental phases and overall length of vegetation period. Knowledge of photoperiodic response of cultivated crops is one of the vital aspects of modern-day plant production, as shifting seasonal timing of reproduction became a major goal of plant breeding programs in their effort to produce novel varieties better adapted to local environments and variable climate conditions. This also stands for soybean [Glycine max (L.) Merr.] since it is a photoperiod-sensitive short-day plant, i.e., soybean transition from vegetative to reproductive stage directly depends on the length of day (Miladinović and Đorđević, 2011).
Soybean and Photoperiodicity
Estimation of plant phenological stage of development, as a function of specific environment, is the key factor in any attempt of modifying plant growth, adaptation, or productivity (Jones and Laing, 1978; Davidson and Campbell, 1983; Hodges and French, 1985; Wang et al., 1987; Colson et al., 1995; Miladinović and Đorđević, 2011). Soybean phenology is difficult to predict, as it depends on photoperiod (Garner and Allard, 1930; Seddigh et al., 1989), temperature (Egli and Wardlaw, 1980; Board and Hall, 1984), and amount of plant-available water (Meckel et al., 1984; Miladinović et al., 2006).
Dependence of soybean on the length of the day resulted in its geographic distribution into 13 maturity group (MG) zones, from MG000 comprised of varieties which can thrive in higher geographic latitudes, to MGX which includes varieties grown in lower latitudes (Hartwig, 1973). By definition, MGs are the result of classification of soybeans based on their growth and development. The difference between MGs in a certain area are caused by photoperiodic requirements of a variety, while the difference in maturity date between two adjacent MGs ranges from 10 to 18 days. In soybeans, the critical photoperiod, or the definite day length light period above or below which the plant never blooms, decreases progressively from higher to lower geographic latitudes. Photoperiodic requirements thus reduce soybean growing area to a narrow latitude range of around 200 km (Scott and Aldrich, 1983). If grown in higher geographic latitudes compared to its optimal growing area, a soybean variety will flower and mature later, while a variety grown in lower geographic latitudes will flower and mature earlier, resulting in lower vegetative mass and lower yields. Hence, there is an optimal MG for each soybean growing region (Miladinović and Đorđević, 2011).
Soybean and E Genes
Time of flowering and maturity in soybean are genetically controlled by E genes that have different roles in maturity and photoperiod sensitivity. Their allelic combinations contribute to fine adaptation of genotype to certain latitude and climate conditions.
Up to date, 11 major loci (E1–E10 and J) affecting time of flowering and maturity have been identified in soybean (Samanfar et al., 2017). The dominant allele of E genes usually confers later flowering and later maturity, except for E6, E9, and J (Ray et al., 1995; Bonato and Vello, 1999; Kong et al., 2014), while photoperiod sensitivity decreases with numbers of recessive alleles. Owen (1927) described the effect of E1 gene in soybean in his inheritance studies, while Bernard (1971) described two major genes (E1 and E2) which control flowering time and maturity. Buzzell (1971) and Kilen and Hartwig (1971) investigated soybean flowering response to fluorescent daylength conditions and concluded that E3 gene determines late flowering, while e3 causes insensitivity to fluorescent light. Buzzell and Voldeng (1980) studied inheritance of insensitivity to extended daylength and found that E4 allele confers late flowering and sensitivity to extended daylength, while e4 allele influences early flowering and insensitivity to extended daylength. McBlain and Bernard (1987) described E5 gene and its genetic effect similar to E2. However, in the study conducted in order to map E5 locus, the results obtained from different F2 mapping populations expected to segregate for E5 were not consistent, and no candidate QTL was found, indicating that a unique E5 gene may not exist (Dissanayaka et al., 2016). Bonato and Vello (1999) described the dominant allele of E6 gene which confers early flowering and maturity in soybeans. Cober and Voldeng (2001) described E7 - soybean maturity and photoperiod-sensitivity locus - and its link to E1 and T. Cober et al. (2010) mapped E8 gene, with the dominant allele resulting in later maturity and recessive allele conferring early maturity. The E9 locus was subjected to detailed molecular analysis and gene FT2a was identified, with the recessive allele delaying flowering (Kong et al., 2014; Zhao et al., 2016). Samanfar et al. (2017) identified E10, new maturity locus in soybean, with FT4 being the predicted functional gene underlying this locus.
Maturity genes E1 (Xia et al., 2012), E2 (Watanabe et al., 2011), E3 (Watanabe et al., 2009), E4 (Liu et al., 2008), E9 (Zhao et al., 2016), and J (Yue et al., 2017) have recently been characterized at the molecular level. Tsubokura et al. (2014) estimated that maturity genes E1–E4 contribute to 62–66% of the flowering time variation. Association analysis and the study of genetic architecture and networks underlying agronomical traits revealed that E1 and E2 loci have pleiotropic effects across the traits related to yield and seed quality, as the key nodes in the regulation of different traits (Fang et al., 2017). E1 gene was located on chromosome 6 and identified as legume-specific transcription factor which functions as the flowering repressor with putative nuclear localization signal and B3-related domain (Xia et al., 2012). E1 allele is functional, e1-as allele with missense mutation is not fully functional, while e1-fs with a frameshift mutation and e1-nl with deletion of entire E1 gene are both non-functional (Xia et al., 2012). Mutant e1-nl allele was identified in MG000 European (Swedish) cultivar Fiskeby III, along with the major mutant alleles for other maturity genes (E2 and E3) (Valliyodan et al., 2016). Out of the known E loci in soybean, E1 gene is considered to have the largest effect on determination of flowering time under field conditions (Xia et al., 2012). For improvement of varietal adaptability, the function of E1 gene should be reduced due to its strong impact on delaying maturity. Besides E1, allelic variability of other E genes is also important in adaptation and can provide significant genetic plasticity that could enable cultivation of soybeans in wider geographic areas (Cober et al., 1996). Located on chromosome 10, E2 gene is an ortholog of Arabidopsis flowering gene GIGANTEA involved in the circadian rhythm and flowering time pathway (Watanabe et al., 2011). The dominant E2 allele is functional, while e2 with nonsense mutation presents the non-functional allele with an early-flowering phenotype (Watanabe et al., 2011). E3 (GmPhyA3) and E4 (GmPhyA2) are located on chromosomes 19 and 20, respectively, and they represent phytochrome A genes. E3-Ha and E3-Mi are functional alleles, while e3-tr, e3-ns, and e3-fs are non-functional with mutations causing truncated proteins (Watanabe et al., 2009). E3 delays flowering under long-day conditions which ultimately affects maturity. E4 gene has a functional allele E4 and five non-functional alleles (e4-SORE-1, e4-oto, e4-tsu, e4-kam, and e4-kes) (Liu et al., 2008; Tsubokura et al., 2013). Being photoperiodically insensitive, varieties with non-functional alleles are mostly adapted to high latitudes, which indicates the importance of e4 allele for high latitude adaptation. E1 and E2 genes have significant impact on vegetative development, while loci E3 and E4 might affect post-flowering reproductive development by increasing pod filling duration, and the number of nodes and pods by up-regulating the expression of growth habit gene Dt1 (Xu et al., 2013).
Higher temperatures at reproductive stages were found to affect plant vigor and overall yield (Egli et al., 2005; Ren et al., 2009). It has also been reported that N and P concentrations in mature seeds increase with the increment of day/night temperatures (Thomas, 2001). This observation implicates that there might be some loci which show optimum activity of N uptake and transport at elevated temperatures (Patil et al., 2017). Protein content was also positively correlated with higher temperature (Wolf et al., 1982). Higher protein content was observed in late maturity group lines (MGV-MGX) compared to early maturity group lines (MG000-MGII). Since genetic variation in seed composition is more affected by geographic regions (∼5%) than by MG (∼2%) (Bandillo et al., 2015), it can be concluded that the effect of lower temperatures on seed composition at higher latitudes might be overcome by the selection of more diverse parental lines in early MGs (Piper and Boote, 1999). Hence, achieving higher protein content without affecting yield or oil content could be obtained by exploring genetic variants of E genes.
Soybean and E Genes Allelic Variation
Study of allelic variation and diversity of soybean flowering-time and maturity genes may enhance soybean breeding for particular environments, since achieving appropriate maturity in a target environment maximizes crop yield potential (Langewisch et al., 2017). However, there are not many studies focused on soybean maturity and genes affecting it. This especially stands for European soybean varieties, as most of the studies published so far deal with North-American and Chinese genotypes (Tsubokura et al., 2013, 2014; Jiang et al., 2014; Langewisch et al., 2014, 2017; Zhai et al., 2014; Abugalieva et al., 2016; Valliyodan et al., 2016; Kurasch et al., 2017; Li et al., 2017; Wolfgang and An, 2017)
Soybean collection maintained at Institute of Field and Vegetable Crops Novi Sad, Serbia (IFVCNS), consists of more than 1200 cultivars and lines originating from America (50.1%), Asia (15.6%), and Europe (34.4%). Most of the genotypes belong to MG0 and MGI, but the collection also includes genotypes from MG000 to MGV (Hrustić and Miladinović, 2011). In order to characterize allelic variation and distribution of the major maturity genes (E1–E4) we conducted the study on 445 genotypes from soybean collections of North-American (NA) ancestral lines (27), Chinese germplasm (14), European varieties (12), genotypes developed and released at IFVCNS - NS varieties (56), NS breeding lines (229), 57 high-protein and 50 high-oil genotypes from NS collection. Our research was mainly focused on identification of the most prominent E allelic combinations in NS varieties and breeding lines, with the aim of applying the obtained results for the improvement and targeted breeding of soybean varieties adapted to the environments of Central-Eastern Europe. MGs of the analyzed soybean genotypes ranged from MG000 to MGIII, including several genotypes from NA ancestral population which belong to later MGs MGV-MGVII. All 445 soybean genotypes were characterized using genotyping-by-sequencing approach (Elshire et al., 2011). Sequenced libraries produced a total of 145x107 raw 100-bp DNA reads that were aligned against soybean reference genome Williams 82. Reads aligned in the same genomic region were used for SNP calling, producing initially more than 85,000 SNP markers, where mean sequencing depth per SNP locus was 5. After filtering and application of a combination of criteria, such as minor allele frequency (MAF < 0.05) and the percentage of missing values (PMV < 20%), the obtained marker dataset was used for prediction of the allelic variation of four maturity genes (E1/e1-as, E2/e2, E3/e3-tr, and E4/e4). Genes E1 and E2 possess SNP that gives rise to alleles e1-as (G/C; Glyma.W82.a2: 20,207,322 bp) and e2 (A/T; Glyma.W82.a2: 45,310,781 bp), while E3 and E4 genes do not have a functional SNP, causing corresponding occurrence of variant alleles e3-tr and e4 correspondingly. The determinant SNP makers for genes E1 and E2 were not present in our dataset, as marker analysis was performed with reduced representation genotyping approach. Thus we examined groups of markers positioned in specific genomic regions which include entire E genes (positions given in Supplementary Table S1), by characterizing haplotypes aiming to identify the causative haplotype for different alleles, functional, and variant (Supplementary Table S1). SNPViz tool was used for haplotype visualization and comparison, in order to provide information on genetic neighborhood in which variable alleles appeared, as described by Langewisch et al. (2014). By comparing the obtained haplotypes with previously published profiles of E genes for NA ancestral lines and European varieties (Kurasch et al., 2017; Langewisch et al., 2017), we managed to identify the groups of haplotypes for each allele. Within these regions, lines with known different alleles were distinctively separated to specific haplotypes, which allows assignment of alleles of each gene to specific haplotypes. Moreover, it was made possible to further specify the identified haplotypes and select groups of diagnostic SNP markers able to determine different alleles of E genes (Supplementary Table S1 and Supplementary Figures S1–S4). Four SNP markers were identified for E1 gene which could be distinguished among alleles E1, e1-as, and e1-nl Supplementary Figure S1. Two SNP markers were defined for genes E2 and E3, causing the difference between E2 and e2 alleles, as well as between E3 and e3-tr Supplementary Figures S2, S3. According to the five identified SNP markers, lines could be classified as E4 or e4 allele (Supplementary Figure S4). The obtained results were applied for further allele scoring of E genes of previously non-genotyped lines, such as NS-varieties and NS-breeding lines. Predictions did not include the non-functional el-fs allele, and no distinction could be made between E3-Ha and E3-Mi alleles, or e4-oto, e4-tsu, e4-kam, and e4-kes.
All examined genotypes were classified into 12 E haplogroups which represent a unique combination of E1, E2, E3, and E4 alleles (Supplementary Table S2). Each collection had one major E haplogroup which included most of the lines (≥50%) (Figure 1). The exception were European varieties with two predominant groups and NA ancestral lines which were distributed almost evenly among several haplogroups. The most abundant NA ancestral haplogroup was e1-as/e2/E3/E4 with 26% of genotypes. Over 20% of genotypes had E1/E2/E3/E4, which coincides with the high number of late-maturing genotypes included in the NA collection. This is concordant with molecular maturity model proposed by Langewisch et al. (2017) where early-maturing lines had non-functional e1/e2/e3-tr and late-maturing lines had E1/E2/E3. None of the lines from the Chinese collection carried functional photoperiod-sensitive E1 allele, while two photoperiod-insensitive alleles (e1-nl and e1-as) were identified at E1 locus. Contrary to previous reports, where most of the tested accessions of breeding collections from China were E1/e2/E3/E4 followed by e1-as/e2/e3/E4 (Jia et al., 2014; Jiang et al., 2014), the most abundant haplogroup in Chinese germplasm in our collection was e1-as/e2/e3-tr/E4 including 57% of lines. The remaining lines mostly contained e1-nl allele with e1-nl/e2/e3-tr/E4 and e1-nl/e2/e3-tr/e4, covering 21 and 14% of genotypes, respectively. Of all examined genotypes, only three lines from the NA ancestral and Chinese germplasm had the non-functional e4 allele, which is in agreement with the fact that e4 alleles were found in small geographical locations with high latitudes and low temperature (Tsubokura et al., 2013; Langewisch et al., 2014). European varieties from our collection differed from the varieties studied by Kurasch et al. (2017) in their E haplotype, as more than 30% of varieties from our collection were e1-as/E2/E3/E4 and the same number of genotypes had e1-as/E2/e3-tr/E4, compared to 1.3 and 12%, respectively, in the study of Kurasch et al. (2017). The haplogroup e1-as/e2/E3/E4 comprised 17% of lines in European varieties. Similar to the results of Kurasch et al. (2017), NS varieties primarily had e1-as/e2/E3/E4 haplotype identified in 63% of genotypes. The same haplotype was found to be predominant in the Kazakh breeding collection (Abugalieva et al., 2016), which was attributed to the frequent exchange of germplasm with partners from former USSR countries. The second largest haplotype in NS varieties is e1-as/e2/e3-tr/E4 with 11% of genotypes. All other allelic combinations were found in less than 10% of genotypes. For the E1 locus, allele el-as was detected in 88% of NS varieties classified into MG000-MGIII. Nearly half of the NS breeding lines were predicted to be e1-as/e2/E3/E4, with the additional 13% of genotypes predicted as e1-as/e2/e3-tr/E4 and 13% of e1-as/E2/e3-tr/E4.
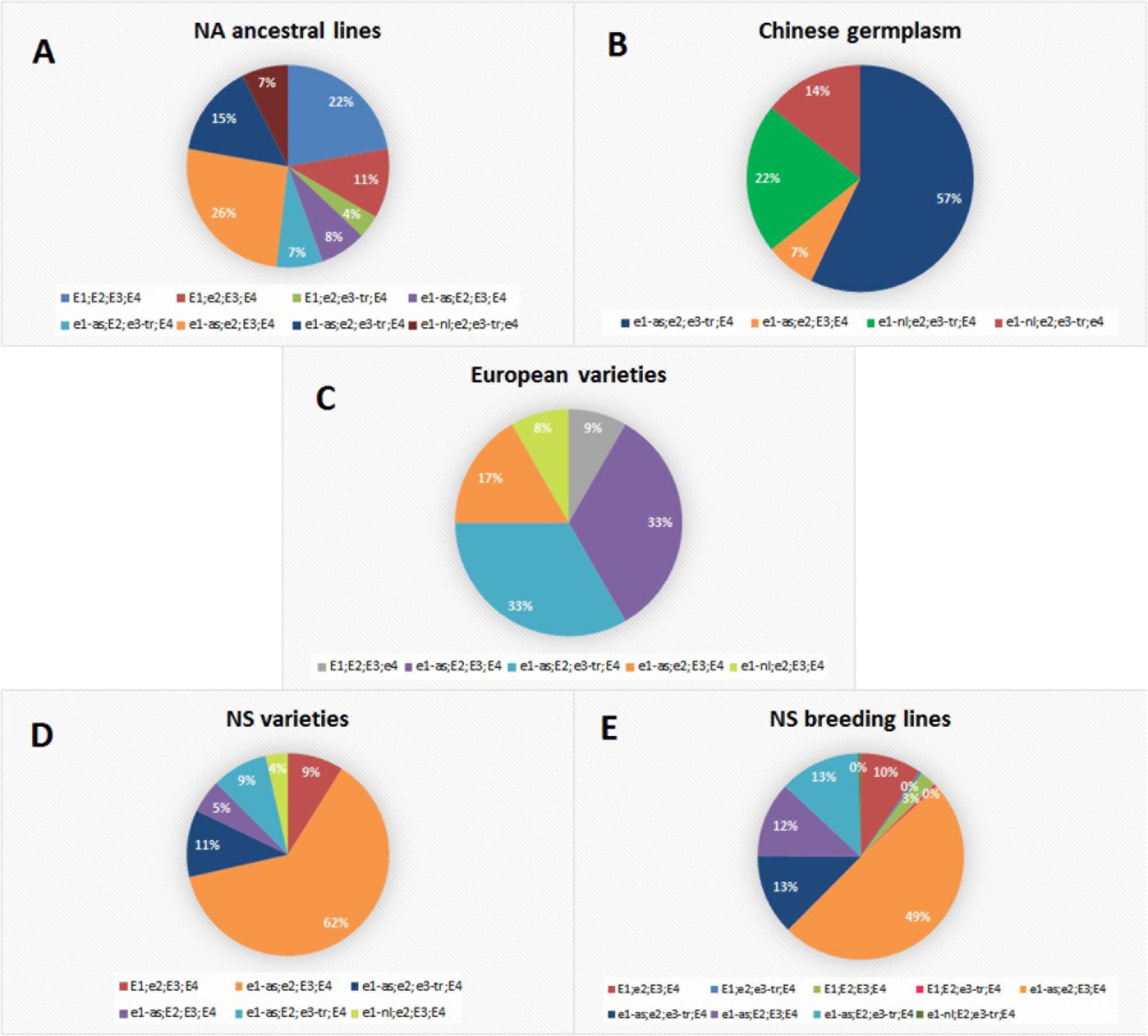
FIGURE 1. Allelic combinations of maturity loci E1, E2, E3, and E4 over different soybean populations: (A) North-American ancestral lines; (B) Chinese germplasm; (C) European varieties; (D) NS varieties; (E) NS breeding lines.
Novi Sad collection exhibited a high diversity of allelic combinations of E genes. The most abundant haplogroup in NS varieties and breeding lines was e1-as/e2/E3/E4. The same haplogroup comprised 26% of genotypes in NA ancestral lines which represent more than 85% of the pedigree of NS varieties, indicating that e1-as/e2/E3/E4 haplotype was favored by artificial selection during soybean breeding under the environmental conditions of Central-Eastern Europe. NS varieties and breeding lines had predominantly e1-as allele, indicating that they might have been purposely selected for similar maturity and high-yield capacity. This finding is compatible with identified genomic regions affected by positive selection during breeding of NS soybean varieties, where two loci (Satt357 and Satt557) on chromosome 6 were identified as strong positive selection candidates (Tomičić et al., 2015). Similar results were obtained during evaluation of the allelic variants at the E genes in 75 European cultivars from five MGs (MG000-MGII), where none of the cultivars carried functional E1 allele (Kurasch et al., 2017). It was concluded that the E1 locus plays a major role in early flowering and maturity, and that photoperiod insensitivity at this locus is a probable requirement for soybeans adapted to Central or Northern Europe.
Based on the used SNP markers, two major clusters can be identified and described as early material (EM) consisting of MG000-MGI and late material (LM) consisting mainly of MGII-MGIV genotypes (Figure 2). The first subgroup from EM included Chinese germplasm, NA ancestral lines and NS varieties. Recessive haplotype e1-as/e2/e3-tr/E4 was the dominant haplotype in this subgroup. The majority of NS varieties belonged to the second subgroup, along with a few NA and European varieties. The most abundant allelic combination in this subcluster was e1-as/e2/E3/E4, which represents the major haplogroup within environmental conditions of Central-Eastern Europe. The second group mostly comprised LM genotypes, including NA ancestral lines, European and NS varieties, from MGI to MGIV, with the exception of NS Princeza which belongs to MG0. Most lines from the LM cluster had three or four dominant alleles. The observed grouping of genotypes into late and early maturing clusters indicates that alleles of E1–E4 genes can be used in the assessment and classification of the assortment according to maturity. However, four genes used in this study are not sufficient to make the precise distinction between different MGs, as there is no unique allelic combination specific for each group. Further work is needed to identify molecular mechanisms behind soybean photoperiod sensitivity, and linking specific allelic combination with a certain MG.
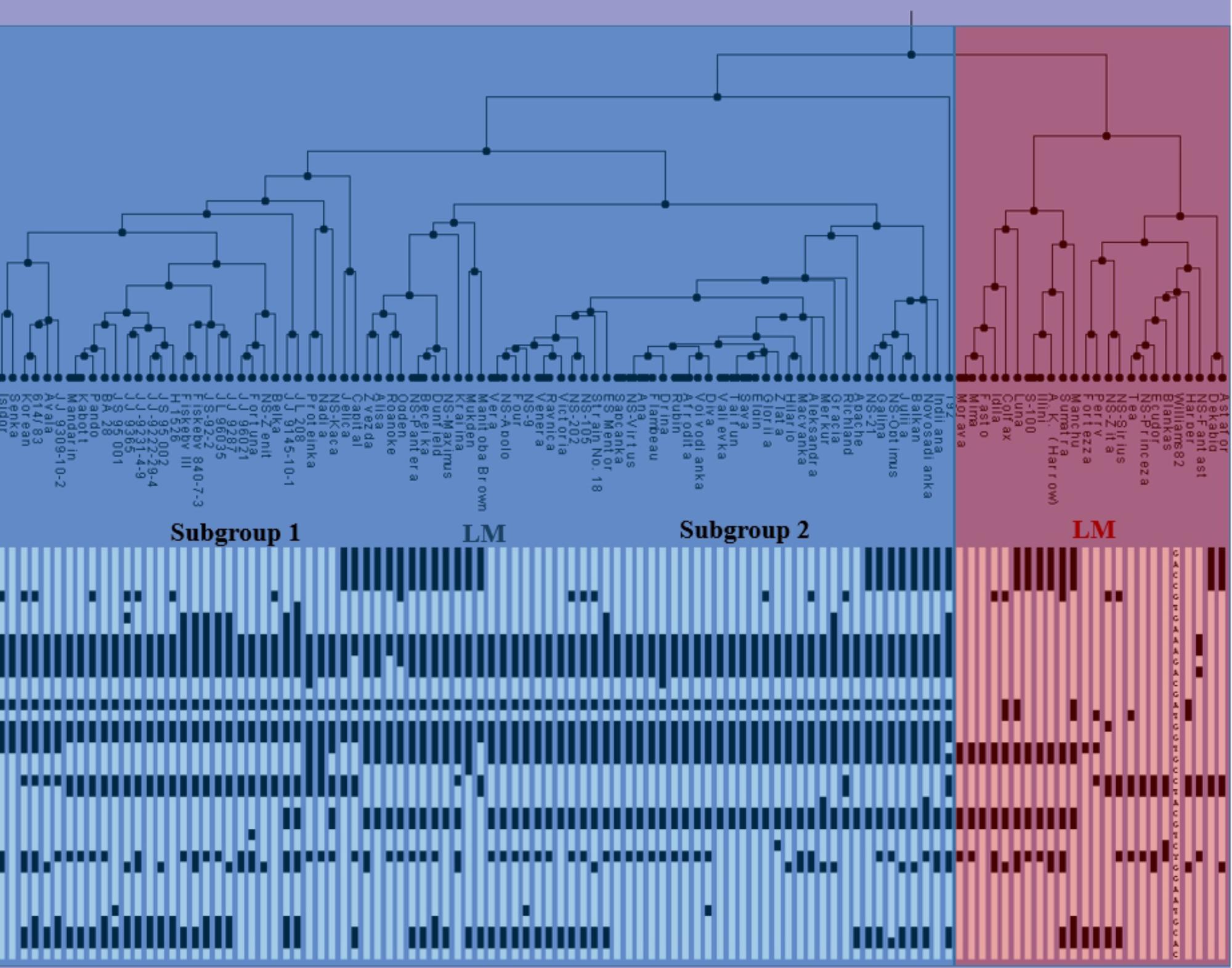
FIGURE 2. Clustering tree and haplotypes of NA ancestral lines, Chinese germplasm, European varieties, and NS varieties, based on the diagnostic SNP markers for four analyzed E genes, (EM, early maturing; LM, late maturing).
Future Prospects
Flowering time and maturity are agronomically important traits, which affect soybean adaptation, quality traits, and yield. Complete understanding of their regulation will therefore allow breeding of varieties with optimal flowering or maturity for particular geographic regions. Considering this, we have studied the distribution of different haplogroups of E genes in soybean collections of NA ancestral lines, Chinese germplasm, European varieties, NS varieties, and NS breeding lines. Overall, the observed allelic combinations of E1–E4 genes significantly determined the adaptation of varieties to different geographic regions, although they have different impacts on maturity.
The study was also the first attempt to assess NS soybean varieties and breeding line adaptation based on E gene variation, as well as comparatively assess E gene variation in NA, Chinese and European genotypes. The study aims to identify the most prominent E allelic combinations in NS genotypes and apply the obtained results in target breeding and introgression of yield and quality traits for environments of Central-Eastern Europe. As e1-as/e2/E3/E4 was the most common haplotype in the NS population comprising high-yielding varieties grown throughout Central and Eastern Europe, the specific allele combination was proposed as the optimal combination for this environment.
The obtained data provide useful information about the selection of parental genotypes most appropriate for use in local breeding programs for improved soybean productivity. This allows plant breeders to transfer traits more effectively into different MGs, enhance germplasm exploitation, and increase overall efficiency of soybean breeding. Furthermore, the obtained knowledge will significantly contribute to the marker-assisted selection and molecular breeding of soybean, and increase breeding efficiency by enhancing genomic prediction models.
Author Contributions
JM, MĆ, SB-T, and VuĐ planned and designed the experiment. MĆ, KP, VuĐ, and VoĐ performed the research. JM, DM, MĆ, and VuĐ contributed to the interpretation and analysis of results. JM, MĆ, and DM wrote and approved the manuscript.
Funding
This research was part of the project TR31022, which is supported by Ministry of Education, Science and Technological Development of the Republic of Serbia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01286/full#supplementary-material
FIGURE S1 | Diagnostic haplotypes of NA ancestral lines, Chinese germplasm, European varieties and NS varieties, based on selected SNP markers for E1 gene.
FIGURE S2 | Diagnostic haplotypes of NA ancestral lines, Chinese germplasm, European varieties and NS varieties, based on selected SNP markers for E2 gene.
FIGURE S3 | Diagnostic haplotypes of NA ancestral lines, Chinese germplasm, European varieties and NS varieties, based on selected SNP markers for E3 gene.
FIGURE S4 | Diagnostic haplotypes of NA ancestral lines, Chinese germplasm, European varieties and NS varieties, based on selected SNP markers for E4 gene.
TABLE S1 | SNP calls located in the genomic regions of E1, E2, E3, and E4 genes in soybean collections of North-American (NA) ancestral lines, Chinese germplasm, European varieties, and genotypes developed and released at Institute of Field and Vegetable Crops – NS (Novi Sad) varieties, NS breeding lines, high-protein and high-oil genotypes from NS collection.
TABLE S2 | Allelic variation on the E1, E2, E3, and E4 maturity genes in soybean collections of North-American (NA) ancestral lines, Chinese germplasm, European varieties, and genotypes developed and released at Institute of Field and Vegetable Crops – NS (Novi Sad) varieties, NS breeding lines, high-protein and high-oil genotypes from NS collection.
References
Abugalieva, S., Didorenko, S., Anuarbek, S., Volkova, L., Gerasimova, Y., Sidorik, I., et al. (2016). Assessment of soybean flowering and seed maturation time in different latitude regions of Kazakhstan. PLoS One 11:e0166894. doi: 10.1371/journal.pone.0166894
Bandillo, N., Jarquin, D., Song, Q., Nelson, R., Cregan, P., Specht, J., et al. (2015). A population structure and genome-wide association analysis on the USDA soybean germplasm collection. Plant Genome 8, 1–13. doi: 10.3835/plantgenome2015.04.0024
Bernard, R. L. (1971). Two major genes for time of flowering and maturity in soybeans. Crop Sci. 11, 242–244. doi: 10.2135/cropsci1971.0011183X001100020022x
Board, J. E., and Hall, W. (1984). Premature flowering in soybean yield reductions at nonoptimal planting dates as influenced by temperature and photoperiod. Agron. J. 76, 700–704. doi: 10.2134/agronj1984.00021962007600040043x
Bonato, E. R., and Vello, N. A. (1999). E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22, 229–232. doi: 10.1590/S1415-47571999000200016
Buzzell, R. I. (1971). Inheritance of a soybean flowering response to fluorescent daylength conditions. Can. J. Genet. Cytol. 13, 703–707. doi: 10.1139/g71-100
Buzzell, R. I., and Voldeng, H. D. (1980). Inheritance of insensitivity to long daylength. Soyb. Genet. Newsl. 7, 26–29. doi: 10.1093/jhered/esp113
Cober, E. R., Molnar, S. J., Charette, M., and Voldeng, H. D. (2010). A new locus for early maturity in soybean. Crop Sci. 50, 524–527. doi: 10.2135/cropsci2009.04.0174
Cober, E. R., Tanner, J. W., and Voldeng, H. D. (1996). Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci. 36, 606–610. doi: 10.2135/cropsci1996.0011183X003600030014x
Cober, E. R., and Voldeng, H. D. (2001). A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701. doi: 10.2135/cropsci2001.413698x
Colson, J., Bouniols, A., and Jones, J. W. (1995). Soybean reproductive development: adapting a model for european varieties. Agron. J. 87, 1129–1139. doi: 10.2134/agronj1995.00021962008700060015x
Davidson, H. R., and Campbell, C. A. (1983). The effect of temperature, moisture and nitrogen on the rate of development of spring wheat as measured by degree days. Can. J. Plant Sci. 63, 833–846. doi: 10.4141/cjps83-106
Dissanayaka, A., Rodriguez, T. O., Di, S., Yan, F., Githiri, S. M., Rodas, F. R., et al. (2016). Quantitative trait locus mapping of soybean maturity gene E5. Breed. Sci. 66, 407–415. doi: 10.1270/jsbbs.15160
Egli, D., TeKrony, D., Heitholt, J., and Rupe, J. (2005). Air temperature during seed filling and soybean seed germination and vigor. Crop Sci. 45, 1329–1335. doi: 10.2135/cropsci2004.0029
Egli, D. B., and Wardlaw, I. F. (1980). Temperature response of seed growth characteristics of soybeans. Agron. J. 72, 560–564. doi: 10.2134/agronj1980.00021962007200030036x
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Fang, C., Ma, Y., Wu, S., Liu, Z., Wang, Z., Yang, R., et al. (2017). Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18:161. doi: 10.1186/s13059-017-1289-9
Garner, W. W., and Allard, H. A. (1930). Photoperiodic response of soybeans in relation to temperature and other environmental factors. J. Agric. Res. 41, 719–735.
Hartwig, E. E. (1973). “Varietal development,” in Soybeans: Improvement, Production and Uses, ed. B. E. Caldwell (Madison WI: ASA),187–207.
Hodges, T., and French, V. (1985). Soyphen: soybean growth stages modeled from temperature, daylength, and water availability. Agron. J. 77, 500–505. doi: 10.2134/agronj1985.00021962007700030031x
Hrustić, M., and Miladinović, J. (2011). “Importance, origin and expansion of soybean,” in Soybean, eds J. Miladinović, M. Hrustić, and M. Vidić (Bečej: Sojaprotein), 11–40.
Jia, H., Jiang, B., Wu, C., Lu, W., Hou, W., Sun, S., et al. (2014). Maturity group classification and maturity locus genotyping of early-maturing soybean varieties from high-latitude cold regions. PLoS One 9:e94139. doi: 10.1371/journal.pone.0094139
Jiang, B., Nan, H., Gao, Y., Tang, L., Yue, Y., Lu, S., et al. (2014). Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9:e106042. doi: 10.1371/journal.pone.0106042
Jones, P. G., and Laing, D. R. (1978). Simulation of the phenology of soybeans. Agric. Syst. 3, 295–311. doi: 10.1371/journal.pone.0124682
Kilen, T. C., and Hartwig, E. E. (1971). Inheritance of a light-quality sensitive character in soybeans. Crop Sci. 11, 559–561. doi: 10.2135/cropsci1971.0011183X001100040030x
Kong, F., Nan, H., Cao, D., Li, Y., Wu, F., Wang, J., et al. (2014). A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535. doi: 10.2135/cropsci2014.03.0228
Kurasch, A. K., Hahn, V., Leiser, W. L., Vollmann, J., Schori, A., Bétrix, C.-A., et al. (2017). Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant Cell Environ. 40, 765–778. doi: 10.1111/pce.12896
Langewisch, T., Lenis, J., Jiang, G.-L., Wang, D., Pantalone, V., and Bilyeu, K. (2017). The development and use of a molecular model for soybean maturity groups. BMC Plant Biol. 17:91. doi: 10.1186/s12870-017-1040-4
Langewisch, T., Zhang, H., Vincent, R., Joshi, T., Xu, D., and Bilyeu, K. (2014). Major soybean maturity gene haplotypes revealed by SNPViz analysis of 72 sequenced soybean genomes. PLoS One 9:e94150. doi: 10.1371/journal.pone.0094150
Li, J., Wang, X., Song, W., Huang, X., Zhou, J., Zeng, H., et al. (2017). Genetic variation of maturity groups and four E genes in the Chinese soybean mini core collection. PLoS One 12:e0172106. doi: 10.1371/journal.pone.0172106
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., and Jun, A. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180, 995–1007. doi: 10.1534/genetics.108.092742
McBlain, B. A., and Bernard, R. L. (1987). A new gene affecting the time of maturity in soybeans. J. Hered. 78, 160–162. doi: 10.1371/journal.pone.0172106
Meckel, L., Egli, D. B., Phillips, R. E., Radcliffe, D., and Leggett, J. E. (1984). Effect of moisture stress on seed growth in soybean. Agron. J. 76, 647–650. doi: 10.1371/journal.pone.0172106
Miladinović, J., and Đorđević, V. (2011). “Soybean morphology and stages of development,” in Soybean, eds J. Miladinović, M. Hrustić, and M. Vidić (Bečej: Sojaprotein), 45–68. doi: 10.2134/agronj1984.00021962007600040033x
Miladinović, J., Kurosaki, H., Burton, J. W., Hrustić, M., and Miladinović, D. (2006). Adaptability of shortseasons soybeans to varying longitudinal regions. Eur. J. Agron. 25, 243–249. doi: 10.1016/j.eja.2006.05.007
Owen, F. V. (1927). Inheritance studies in soybeans. II. Glabrousness, color of pubescence, time of maturity, and linkage relations. Genetics 12, 519–529.
Patil, G., Mian, R., Vuong, T., Pantalone, V., Song, Q., Chen, P., et al. (2017). Molecular mapping and genomics of soybean seed protein: a review and perspective for the future Theor. Appl. Genet. 130, 1975–1991. doi: 10.1007/s00122-017-2955-8
Piper, E. L., and Boote, K. I. (1999). Temperature and cultivar effects on soybean seed oil and protein concentrations. J. Am. Oil Chem. Soc. 76, 1233–1241. doi: 10.1007/s11746-999-0099-y
Putterill, J., Stockum, C., and Warman, G. (2009). “Photoperiodic flowering in long-day plant Arabidopsis thaliana,” in Photoperiodism: The Biological Calendar, eds R. J. Nelson, D. L. Denlinger, and D. E. Somers (Oxford: Oxford University Press), 9–37.
Ray, J. D., Hinson, K., Mankono, E. B., and Malo, F. M. (1995). Genetic control of a long-juvenile trait in soybean. Crop Sci. 35, 1001–1006. doi: 10.1038/ng.3819
Ren, C., Bilyeu, K. D., and Beuselinck, P. (2009). Composition, vigor, and proteome of mature soybean seeds developed under high temperature. Crop Sci. 49, 1010–1022. doi: 10.2135/cropsci2008.05.0247
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. The. Appl. Genet. 130, 377–390. doi: 10.1007/s00122-016-2819-7
Scott, W. O., and Aldrich, S. R. (1983). Modern soybean production. Champaign: S & A Publications Inc.
Seddigh, M., Jolliff, G. D., and Orf, J. H. (1989). Night temperature effects on soybean phenology. Crop Sci. 29, 400–406. doi: 10.1371/journal.pone.0124682
Thomas, J. M. G. (2001). Impact of Elevated Temperature and Carbon Dioxide on Development and Composition of Soybean Seed. Doctoral dissertation, University of Florida, Gainesville, FL.
Tomičić, M., Đorđević, V., Obreht, D., Miladinović, J., Brbaklić, L., Mikić, A., et al. (2015). Tracking footprints of selection associated with soybean adaptation to Central-East Europe environments. Euphytica 203, 701–713. doi: 10.1007/s10681-014-1313-z
Tsubokura, Y., Matsumura, H., Xu, M., Liu, B., Nakajima, H., Anai, T., et al. (2013). Genetic variation in soybean at maturity locus E4 is involved in adaptation to long days at high latitudes. Agronomy 3, 117–134. doi: 10.3390/agronomy3010117
Tsubokura, Y., Watanabe, S., Xia, Z., Kanamori, H., Yamagata, H., Kaga, A., et al. (2014). Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113, 429–441. doi: 10.1093/aob/mct269
Valliyodan, B., Qiu, D., Patil, G., Zeng, P., Huang, J., Dai, L., et al. (2016). Landscape of genomic diversity and trait discovery in soybean. Sci. Rep. 6:23598. doi: 10.1038/srep23598
Wang, J., McBlain, B. A., Wooley, J. T., Hesketh, J. D., and Bernard, R. L. (1987). A soybean phenology model and associated data base. Biotronics 16, 25–38.
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Watanabe, S., Xia, Z., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Wolf, R., Cavins, J., Kleiman, R., and Black, L. (1982). Effect of temperature on soybean seed constituents: oil, protein, moisture, fatty acids, amino acids and sugars. J. Am. Oil Chem. Soc. 59, 230–232. doi: 10.1007/BF02582182
Wolfgang, G., and An, Y.-Q. C. (2017). Genetic separation of southern and northern soybean breeding programs in North America and their associated allelic variation at four maturity loci. Mol. Breed. 37:8. doi: 10.1007/s11032-016-0611-7
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109, E2155–E2164. doi: 10.1073/pnas.1117982109
Xu, M., Xu, Z., Liu, B., Kong, F., Tsubokura, Y., Watanabe, S., et al. (2013). Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 13:91. doi: 10.1186/1471-2229-13-91
Yue, Y., Liu, N., Jiang, B., Li, M., Wang, H., Jiang, Z., et al. (2017). A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant. 10, 656–658. doi: 10.1016/j.molp.2016.12.004
Zhai, H., Lü, S., Wang, Y., Chen, X., Ren, H., Yang, J., et al. (2014). Allelic variations at four major maturity E genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. PLoS One 9:e97636. doi: 10.1371/journal.pone.0097636
Keywords: photoperiodism, E genes, soybean, maturity, allelic variation, SNP
Citation: Miladinović J, Ćeran M, Đorđević V, Balešević-Tubić S, Petrović K, Đukić V and Miladinović D (2018) Allelic Variation and Distribution of the Major Maturity Genes in Different Soybean Collections. Front. Plant Sci. 9:1286. doi: 10.3389/fpls.2018.01286
Received: 09 October 2017; Accepted: 16 August 2018;
Published: 04 September 2018.
Edited by:
Rafael Lozano, University of Almería, SpainReviewed by:
Gunvant Baliram Patil, University of Minnesota, United StatesMatthew Nicholas Nelson, Royal Botanic Gardens, Kew, United Kingdom
Copyright © 2018 Miladinović, Ćeran, Đorđević, Balešević-Tubić, Petrović, Đukić and Miladinović. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jegor Miladinović, amVnb3IubWlsYWRpbm92aWNAaWZ2Y25zLm5zLmFjLnJz
 Jegor Miladinović
Jegor Miladinović Marina Ćeran
Marina Ćeran Vuk Đorđević
Vuk Đorđević Svetlana Balešević-Tubić
Svetlana Balešević-Tubić Kristina Petrović
Kristina Petrović Vojin Đukić
Vojin Đukić Dragana Miladinović
Dragana Miladinović