
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 September 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01285
 Chengsheng Meng†
Chengsheng Meng† Yuanyuan Yan†
Yuanyuan Yan† Zhengwen Liu
Zhengwen Liu Liting Chen
Liting Chen Yan Zhang
Yan Zhang Xiuxin Li
Xiuxin Li Liqiang Wu
Liqiang Wu Guiyin Zhang
Guiyin Zhang Xingfen Wang*
Xingfen Wang* Zhiying Ma*
Zhiying Ma*Non-specific lipid transfer proteins (nsLTPs) had been previously isolated from cotton fiber but their functions were unclear so far. Bioinformatic analysis of the tetraploid cotton genome database identified 138 nsLTP genes, falling into the 11 groups as reported previously. Different from Arabidopsis, cacao, and other crops, cotton type XI genes were considerably expanded and diverged earlier on chromosome At11, Dt11, and Dt08. Corresponding to the type XI genes, the type XI proteins (GhLtpXIs) all contained an extra N-terminal cap resulting in larger molecular weight. The research revealed that the expression of type XI genes was dramatically increased in fibers of tetraploid cotton compared with the two diploid progenitors. High-level of GhLtpXIs expression was observed in long-fibered cotton cultivars during fiber elongation. Ectopic expression of GhLtpXIs in Arabidopsis significantly enhanced trichome length, suggesting that GhLtpXIs promoted fiber elongation. Overall, the findings of this research provide insights into phenotypic evolution of Gossypium species and regulatory mechanism of nsLTPs during fiber development.
HIGHLIGHT
A specific group, type XI nsLTPs, was identified with predominant expression in elongating fibers of Gossypium hirsutum based on evolutionary, transcriptional, and functional analyses.
Cotton is one of the important commercial fiber crops worldwide. Cotton fiber initiates from the outer epidermis of the ovules, followed by extensive cell elongation (about 1,000~3,000 times) and cell wall synthesis (Basra and Malik, 1984; Kim and Triplett, 2001). In G. hirsutum, lint fibers start to develop prior to or on the day of anthesis, and fuzz fibers develop a few days later (Joshi et al., 1967; Stewart, 1975). Fiber quantity and length, to a large extent, determine cotton yield and quality of the resulting spun thread. Long-chain fatty acid (LCFA) functions as a regulator of endogenous ethylene biosynthesis in cotton ovules, which can maximize the extensibility of cotton fibers (Qin et al., 2007). However, it is unclear how LCFA is transported and accumulated on the outer epidermis of ovules to regulate fiber development. The nsLTP is one of the well-known protein families with bound lipids (Boutrot et al., 2008) such as LCFA. Detailed studies of gene expression and function of nsLTPs are paramount for addressing the above question.
The nsLTPs are widely distributed in the plant kingdom and are capable to bind to acyl groups, various phospholipids and other fatty acid groups with a broad binding affinity (Ostergaard et al., 1993; Kader, 1996; Kragelund et al., 1997; Sodano et al., 1997; Charvolin et al., 1999; Han et al., 2001). It has been revealed that most nsLTPs have a small molecular weight of about 9 kDa in higher plants (Kader, 1996). All known plant nsLTPs contain an N-terminal signal peptide which is characterized by an eight cysteine motif (ECM) backbone forming four conserved disulfide bonds that stabilize a hydrophobic cavity to enclose the lipid to shield the hydrophobic portions of the lipid (Carvalho and Gomes, 2007; Boutrot et al., 2008; Wong et al., 2017). Plant nsLTPs are deemed to be responsible for the shuttling of lipids across cytoplasm and between membranes and regulate the beta-oxidation of fatty acids in glyoxysomes and the intracellular fatty acid pools (Kader, 1996; Cheng et al., 2004). Multiple physiological and biological functions of nsLTPs have been suggested, including membrane and liposome biogenesis (Pyee et al., 1994), somatic embryogenesis (Lee et al., 2009; Chae et al., 2010; Edstam and Edqvist, 2014), pollen development (Zhang et al., 2010; Chen et al., 2011), stress resistance (Zhang et al., 2016), defense (Molina et al., 1993; Schweiger et al., 2013), and signal transduction (Sarowar et al., 2009).
Cotton nsLTP genes have been isolated from fibers decades ago (Ma et al., 1995). GH3, Ltp3, Ltp6, and GhLTPG1 are proved to be specifically expressed in fiber cells and Ltp3 expression reaches to a maximum at the late fiber elongation stage (Ma et al., 1995; Han et al., 2013; Deng et al., 2016). Expression analysis showed that GhLtp6, GhLtp7, GhLtp8, and GhLtp11 are highly expressed during fiber initiation (Han et al., 2013). Regarded as seed trichomes, fiber initiation, and elongation are regulated by MYB genes as leaf trichomes (Guan et al., 2008; Pu et al., 2008; Machado et al., 2009; Walford et al., 2011; Huang et al., 2013; Wang et al., 2013; Liu et al., 2015). And cotton LTP3 was regulated by a MYB protein (Hsu et al., 2005). Thus it is supported that nsLTPs are involved in fiber development. However, the gene family members and the biological function during fiber development are largely unknown of these a few cotton nsLTPs cloned previously. The availability of genome sequence of G. hirsutum and insights into trichome developmental mechanism enable us to understand these scientific issues.
In Arabidopsis and rice, nsLTPs can be classified into several types (I, II, III, IV, V, VI, VII, VIII, IX, XI, and nsLTPy) based on the sequence similarity (Boutrot et al., 2008). Currently, 51 nsLTP genes have been identified in Arabidopsis (Boutrot et al., 2008; Li et al., 2014), 63 in rape and maize (Li et al., 2014; Wei and Zhong, 2014), 58 in sorghum (Wei and Zhong, 2014), and 52 in rice (Boutrot et al., 2008). In the present study, 138 nsLTP genes were identified in G. hirsutum that considerably expanded in Gossypium species during fiber evolution. Both transcriptional and functional analyses suggested an important role of GhLtpXIs in promoting fiber elongation.
The genome sequences of G. hirsutum TM-1, Gossypium arboreum, Gossypium raimondii, Arabidopsis thaliana, Brassica rapa, Theobroma cacao, Oryza sativa, and Vitis vinifera were downloaded from CottonGen (https://www.cottongen.org), CottonFGD (https://cottonfgd.org), TAIR (http://www.arabidopsis.org), BRAD (http://brassicadb.org), Cacao Genome Database (https://www.cacaogenomedb.org), RAP (http://rapdb.dna.affrc.go.jp/), and Grape Genome Browser (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/) networks, respectively. The local genomes including coding sequences and protein sequences were constructed with the blast-2.2.9 program downloaded from the national center for biotechnology information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/2.2.9/blast-2.2.9-ia32-win32.exe). Candidate nsLTP genes were identified by BLASTP against the local databases using published nsLTP protein sequences of Arabidopsis and Brassica (Li et al., 2014) as queries with a cut-off value of e−5. The deduced protein sequences were analyzed for their signal peptides using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP). Then the mature proteins with more than 120 amino acids were discarded, followed by analysis of the ECM domains with BioEdit. The structural domains were further analyzed with Batch Web CD-Search Tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the members of nsLTPs were finally confirmed with HMMER. ProtParam (http://web.expasy.org/protparam/) was used to calculate the molecular weights and isoelectric points of GhLtps.
Multiple alignments of the nsLTP protein sequences of ECMs were performed using Clustal X version 2.0 program. Phylogenetic trees were constructed with the method of Maximum likelihood or Neighbor-Joining using MEGA 6.0 in pairwise complete deletion and Amino Acid P-distance model. For statistical reliability, bootstrap tests were carried out with 1,000 replicates.
The downloaded CDS and genomic sequences of GhLtps were used to construct gene structures on Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). Mapinspect software was used to generate chromosome location image of each GhLtp according to their starting position on chromosomes. The segmental duplication was defined as the following: (1) the length of aligned sequences cover >80% of the longer gene; (2) the identity of the aligned regions >80%; (3) only one duplication event was counted for tightly linked genes (Kong et al., 2013; Wei et al., 2013; Liu et al., 2014). Tandem duplication was defined on the basis of the criteria that tandem duplicated genes are located within 15 predicted open reading frames or within 30 kb of each other (Shin and Bleecker, 2003; Wang et al., 2011). The non-synonymous to synonymous substitution ratio (Ka/Ks) for each duplicated GhLtp gene pairs was calculated using KaKs_Calculator 2.0 software.
The RPKM (Reads Per Kilobase of gene model per Million mapped reads) values of GhLtps were extracted from our RNA-seq data using ovules [0 and 5 days post anthesis (DPA)] and fibers (10–30 DPA) collected from G. hirsutum cultivars including HY405, CCRI8, and ND601 (Ma et al., 2018). Expression pattern analysis and differential expression analysis were subsequently applied with online tools Trend (http://www.omicshare.com/tools/Home/Soft/trend) and Diffanalysis (http://www.omicshare.com/tools/Home/Soft/diffanalysis), respectively. The volcano plot was generated with Excel (Microsoft, United States) to display differentially expressed GhLtps. Transcriptome data of genome-wide gene expression in different organs and cotton species was obtained from cottonFGD (file name: fpkm.Ghir.NAU.txt.gz) and the FPKM (Fragments Per Kilobase of exon per Million fragments mapped) values of cotton nsLTPs were extracted for comparison between different tissues or organs and different Gossypium species. The heatmaps were generated with HemI version 1.0.
The cotton cultivars (HY405 and ND601) were grown in the growing season in the field in Baoding, Hebei (Latitude 38°48′N, 115°25′E). Cotton ovules (0 DPA) and fibers (5, 10, 15, and 20 DPA) were collected at 14:00–16:00 and fast frozen in liquid nitrogen for qPCR. The cotton cultivar TM-1 were grown at 28°C day/25°C night with 10 h light/14 h dark cycles in greenhouse of Hebei Agricultural University and ovules and fibers were collected at 10 DPA followed by fast frozen in liquid nitrogen for gene amplification.
Arabidopsis including Col wild-type (WT) plants and transgenic plants were grown at 22°C with 16 h light/8 h dark cycles in growth chamber at the North China Key Laboratory for Germplasm Resources of Education Ministry. Mature rosette leaves and cauline leaves were collected from flowering plants and frozen in liquid nitrogen before RNA extraction.
Frozen tissues were ground to a fine powder in liquid nitrogen and the total RNA was extracted with RN09-EASYspin Plant RNA purification kit (Aidlab). After quality and quantity analysis with gel electrophoresis and NanoDrop 2000, the first cDNA strand was synthesized using the PrimerScriptTM RT Master Mix (TaKaRa). Then quantitative real-time PCR was performed with Fast Super EvaGreen qPCR Master Mix (US EVERBRIGHT®INC) on an ABI 7500 Real-Time PCR machine. Three biological replicates of each sample and three technical replicates of each biological replicate were used. The relative expression level of the target gene was calculated with the difference between the cycle threshold (Ct) of the target gene and reference gene (ΔCt = Cttargetgene − Ctreferencegene) and corresponded to 2−ΔCt. Primers used for qPCR were designed against gene specific sequences of CDS and listed in Supplemental Table S1.
The full-length of GhLtpXIs coding sequences were PCR-amplified from cotton cDNA (RNA was extracted from ovules and fibers of TM-1) with primers listed in Supplemental Table S1, and the resulting products were cloned into pGreen 0229 harboring 35S promoter (Yu et al., 2004). After verification by nucleotide sequencing, the resulting vectors were transformed into Agrobacterium followed by transformation with floral dip into Arabidopsis WT (Col) plants. The transgenic plants were selected by BASTA and the first three mature rosette leaves were cut from the petioles after bolting for discoloration with gradient ethanol followed by observation of trichomes under microscope with 20 × objective lens. The trichome length was measured using cellSens Standard (Olympus, Japan). Statistical analyses were performed using Excel (Microsoft, United States) with T-test (tail = 1, type = 3).
To identify the entire collection of putative nsLTP genes in the G. hirsutum genome, local BlastP searches were conducted using 53 and 63 nsLTP protein sequences of Arabidopsis and B. rapa as queries on the whole genome of G. hirsutum. A total of 219 GhLtp genes were obtained. Each of the deduced protein sequences was analyzed with SingalP, after which 18 proteins lacking N-terminal signal sequences (NSS) were omitted. Since low molecular weight is a common character of nsLTP proteins, 46 mature proteins whose peptide length was more than 120 amino acids were not taken into consideration. Then 16 proteins lacking the Cys residues were excluded following manually scanning for the presence of the eight essential Cys residues and 1 protein without the M residue (starting residue) was removed. The remaining protein sequences were detected for the LTPAAI_LTSS structure using the Batch Web CD-Search tool. Subsequently, the GhLtp candidates were further verified according to the HMMER method. Finally, 138 genes of G. hirsutum coding the nsLTP were confirmed (Table 1).
The sequence similarity method (Boutrot et al., 2008) was employed to classify the identified GhLtps and our results showed that the 138 GhLtps could be divided into 10 groups after multiple sequence alignments (Table 1 and Supplemental Table S2). The results showed that the ECMs were highly conserved in all of the 138 GhLtps, which could form four disulfide bonds to stabilize the tertiary structure of hydrophobic cavity (Figure 1). Different from rice, wheat, rape, and Arabidopsis, in which the majority of nsLTPs belongs to type I or II (Boutrot et al., 2008; Li et al., 2014), type XI is the largest group in G. hirsutum (Figure 1 and Table 1). The flanking amino acid residues of each CXC motif were conserved within the members of the same group, except for type I. A more conserved consensus pentapeptide Thr/Ser-X1-X2-Asp-Arg/Lys and a more variable one Pro-Tyr-X-Ile-Ser were found in type I GhLtps as reported previously (Douliez et al., 2000; Li et al., 2014). As small molecules, nsLTPs lack supernumerary sequences at C/N-terminal or contain a short C-terminal tail. However, GhLtpXIs contained a relatively long N-terminal cap.
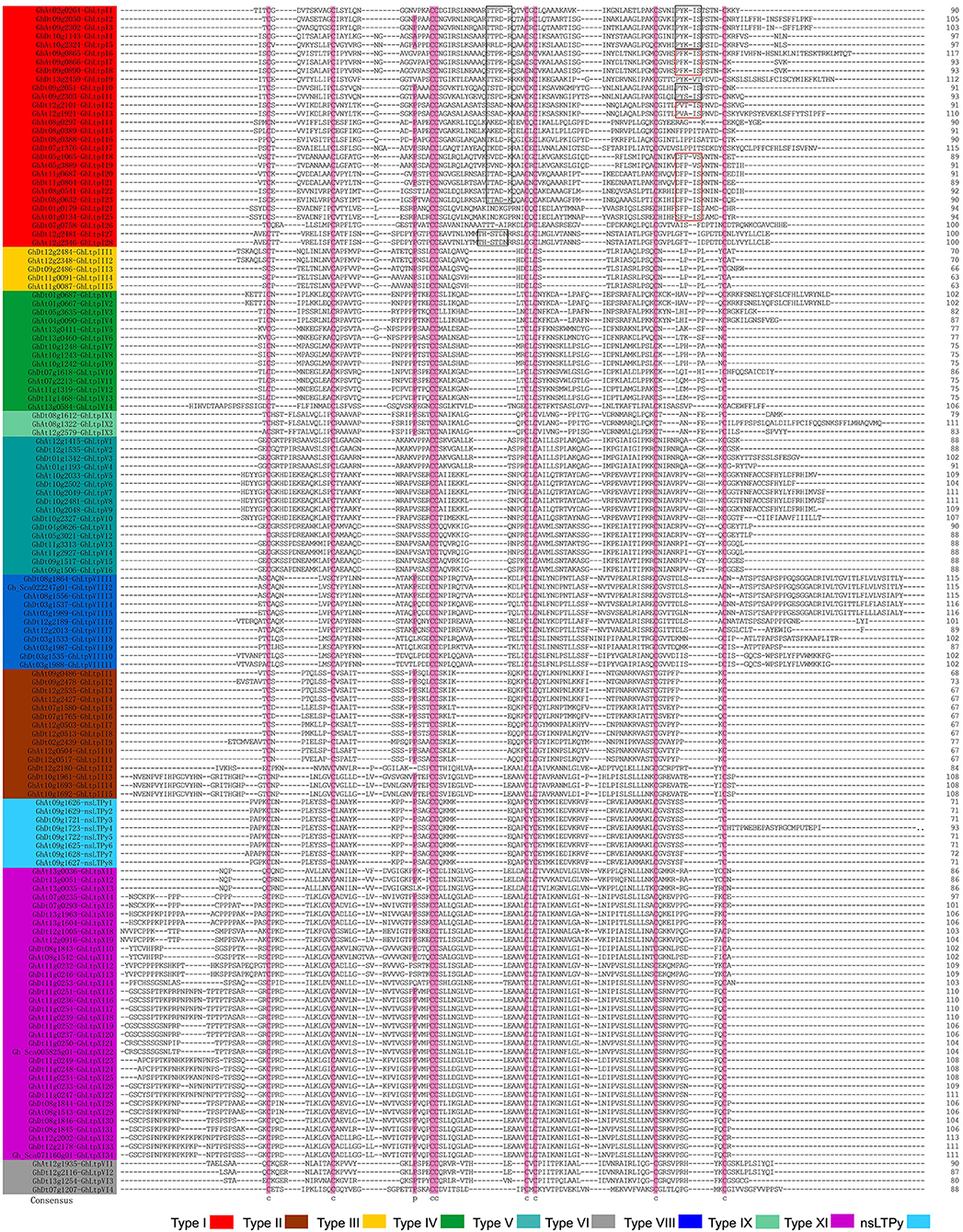
Figure 1. Multiple sequence alignment of GhLtp proteins. The gene ID and names of different types of GhLtps are presented with different colors. The conserved cysteine residues are marked against pink backgrounds. Consensus residues are marked by rectangles.
The molecular weight (MW) and theoretical pI (isoelectric point) of each GhLtp were calculated and summarized in Table 1. Low MW is a common characteristic of plant nsLTPs and very few nsLTPs were found with a MW above 11 kDa in rice, rape and Arabidopsis. Interestingly, all GhLtpXIs displayed high MW (about 11–13 kDa). The high MWs of all the type XI members were due to the presence of supernumerary amino acid residues located at the N-terminal.
Type XI nsLPTs are rare in plants. None of them was identified in rice and wheat, and only two and three nsLTPs belong to this group in Arabidopsis and B. rapa, respectively (Boutrot et al., 2008; Li et al., 2014). On the contrary, type XI contained most members in G. hirsutum. To study the evolution of cotton nsLTP genes, this gene family was additionally identified and classified from G. arboreum, G. raimondii, Th. cacao and V. vinifera using the same method described above (Supplemental Table S3). An unrooted phylogenetic tree was subsequently built with nsLTP proteins from 8 species including G. hirsutum, G. arboreum, G. raimondii, A. thaliana, B. rapa, Th. cacao, O. sativa, and V. vinifera with neighbor-joining method (Figure 2). None of these proteins formed distinct monophyletic clusters. Type I was the largest group in plants and most sequences of this group formed a separated cluster in the tree. Cotton species have evolved a large number of type XI genes though this group is also identified in grape, cacao and rape. Two type I genes (Bra024938 and Tc11g016320) and five cotton type II genes (Ga3164g39880 and GhLtpII12-15) were close to type XI genes, indicating that cotton type XI genes maybe evolved from type I or II genes. Most importantly, only type XI sequences formed a specific cluster in the tree, indicating that these genes shared a common ancestor.
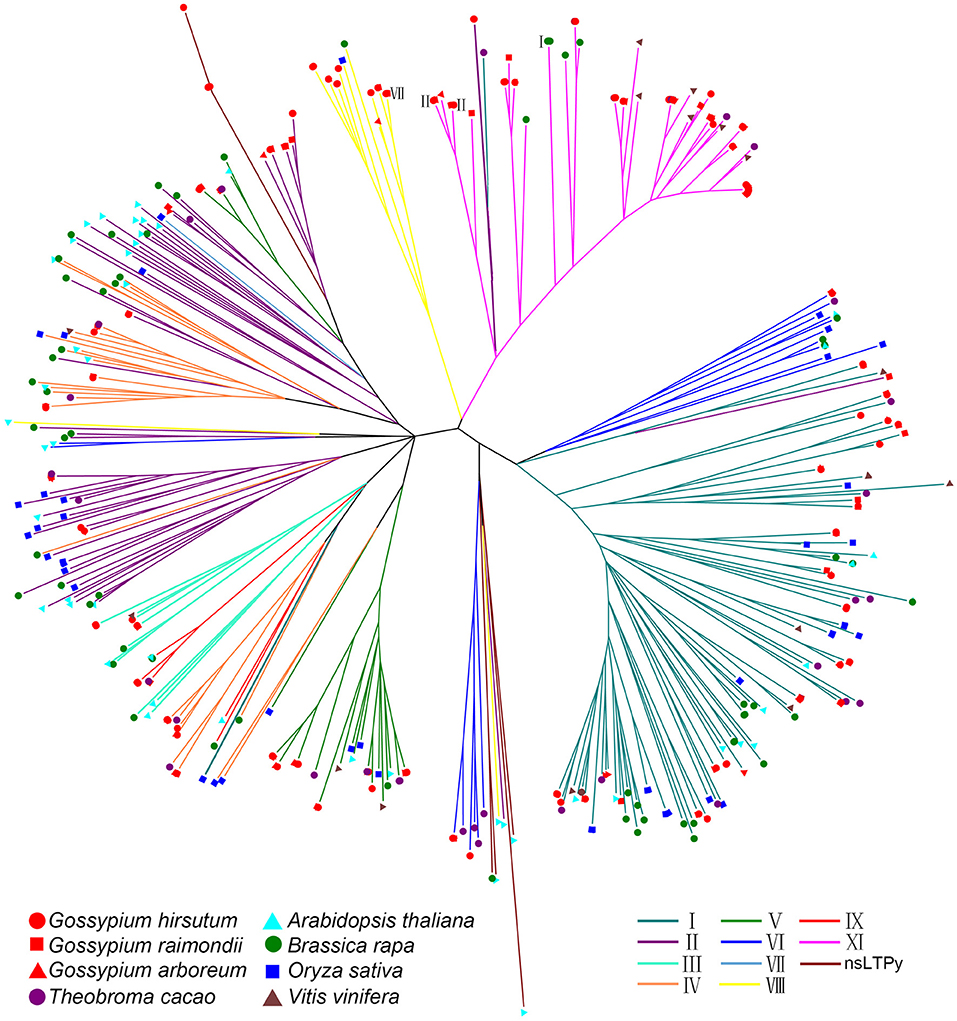
Figure 2. Phylogenetic tree of nsLtps in eight species. The full length of mature protein sequences of nsLTPs from G. hirsutum, G. arboreum, G. raimondii, Arabidopsis, B. rapa, Th. cacao, O. sativa, and V. vinifera were used to construct the phylogenetic tree using a Neighbor-Joining method. Lines of different colors represent classification of nsLTPs. Different species were marked at the end of the lines. Greek numerals present the corresponding type of genes that cannot be displayed by lines.
To further study the phylogenetic organization of the GhLtpXIs, a phylogenetic tree was constructed using Maximum-likehood inference from the alignment of respective 138 and 53 protein sequences of G. hirsutum and Arabidopsis. The results showed that all the nsLTPs can be divided into four clusters from A-D (Supplemental Figure S1). All the type I and III GhLtps belonged to cluster C. Type V, VIII, and IX GhLtps fell into cluster D, and cluster A contained all members of type VI and XI. Phylogenetic analysis had shown that the same type AtLtps constituted monophyletic groups except for the type II (Boutrot et al., 2008; Li et al., 2014). Consistently, GhLtpII12-15 were more distantly related to other type II GhLtps. GhLtpII13, GhLtpII14, and GhLtpII15 were more close to GhLtpXIs, while GhLtpII12 was more related to GhLtpVIs and GhLtpXI1/2/3.
Gene structures of the GhLtps were obtained by comparing the predicted CDS with their corresponding genomic DNA sequences (Supplemental Figure S2). The results showed that only 16 GhLtps had introns. Among these GhLtps, six GhLtps were interrupted by two introns, and the others were interrupted by a single intron. According to the previous studies in some other plants, 35 out of 52 OsLtps, 25 out of 51 AtLtps, and 17 out of 63 BrnsLtps contain introns (Boutrot et al., 2008; Li et al., 2014). These results revealed that the percentage of GhLtps lacking introns was much higher among rice, Arabidopsis, B. rapa, and G. hirsutum, and the diverse distribution of intronic regions was quite low.
To determine the chromosomal distribution of GhLtps, the approximate position of each GhLtp was marked on the physical map of the 26 G. hirsutum chromosomes (Figure 3). It was found that the majority of the GhLtp genes located at the ends of the chromosomes. Chromosome Dt11 harbored the most GhLtps with 13 genes including 9 GhLtpXIs and none of the GhLtps located on chromosome At06 and Dt06. Since G. hirsutum is tetraploid with subgenome At and Dt, comparison was performed between each pair of homologous chromosomes. Ignoring three genes that are unable to locate, the GhLtps almost equally positioned on each genome and the number of genes on chromosomes At and its relative homologous chromosome Dt was uniform or close. Additionally, the distribution patterns of GhLtps were quite similar within each pair of homologous chromosomes, which would probably result from complex evolutional process including recombination, DNA exchanges and gene duplications after merge of genome A and D. These results showed that GhLtps distributed congruently within subgenome At and Dt.
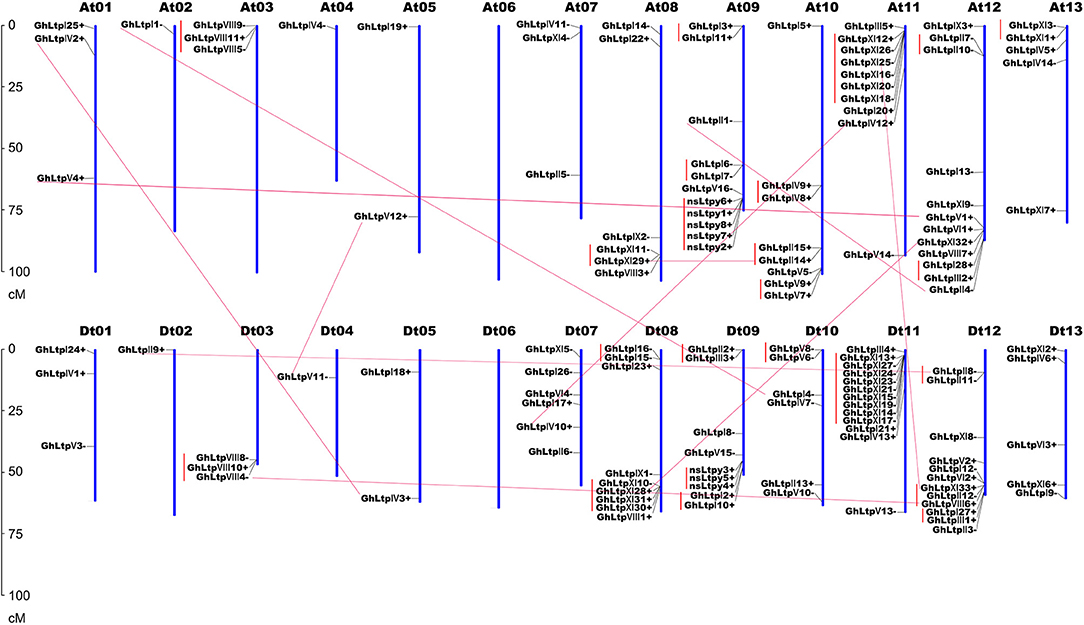
Figure 3. Distribution of GhLtps in 26 chromosomes. Chromosome numbers are indicated above each vertical bar. The scale represents centimorgan (cM). The segmental duplicated gene pairs are connected with red lines. The tandem duplicated gene clusters are marked in red perpendicular lines.
Gene duplication events including segmental and tandem duplications were thought to be essential for the expansion of gene family in the genome (Maere et al., 2005) and thus gene duplication events of nsLTP family were investigated in G. hirsutum. A total of 32 duplication events were found, including 9 segmental duplication pairs and 23 tandem duplication pairs (Figure 3 and Supplemental Table S4). The duplication events were mainly concentrated in the same subfamily except for three pairs of tandem duplication (GhLtpII2/III3, GhLtpI28/III2, and GhLtpXI33/II12/VIII6). It was worthy to note that tandem duplication pairs distributed in the proximate locations of each paralogous blocks and the expansion of type XI group was largely contributed by tandem duplication. We subsequently calculated the non-synonymous to synonymous substitution ratio (Ka/Ks) for each duplicated GhLtp gene pairs. Several duplication pairs of GhLtpXIs displayed larger Ks values and these genes constituted four gene clusters located in chromosomes 08 and 11 in subgenome At and Dt, implying an early divergence time of these GhLtpXIs. Additionally, most Ka/Ks ratios were below 1, except for five duplication pairs (Supplemental Table S4), suggesting that GhLtps had mainly experienced strong purifying selection pressure with limited functional divergence. These results revealed that GhLtpXIs expanded early on chromosome At11 and Dt11 and the functions of the duplicated GhLtps did not diverge much during evolution.
In this study, expression of GhLtps were analyzed in various organs/tissues (Supplemental Figure S3). It was worthy to point out that all the type nsLTPy genes showed specific expression in stamen, while other group members did not show any expression preference. Thirty four genes showed abundant transcription profile during fiber development and nearly 30% of them pertained to type XI.
Due to the irreplaceable economical value of cotton fibers, breeders have been making persistent efforts to develop various types of cotton (Gossypium spp.) with desirable fiber characteristics, which provides materials for investigation of the regulation of fiber development (Han et al., 2016). To further elucidate the function of GhLtps during fiber development, the expression of GhLtps in cultivars HY405, CCRI8, and ND601 was analyzed using our RNA-seq data. It was noted that 110 genes out of 138 GhLtps were expressed during fiber development (Supplemental Figure S4). The other 28 GhLtps with scarcely any transcripts covered each group. GhLtpI1/10/11 and GhLtpXI32 were highly transcribed during fiber development. And seven GhLtpXIs out of 18 GhLtps demonstrated high transcripts only in fiber initiation and early elongation stage. Further, the 138 GhLtps could be clustered into four groups based on the expression trend during fiber development (Supplemental Figure S5). The majority of GhLtps were expressed in the fiber initiation stage, and kept a relatively high expression level during the early stage of elongation. It was noticeable that the expression of nearly half of the GhLtps decreased gradually, suggesting important roles in cotton fiber initiation and elongation which are essential for lint fiber formation. Additionally, over 60% of GhLtpXIs displayed higher expression during fiber initiation and early elongation. It seemed that type XI GhLtps were important for fiber initiation and elongation.
Further comparison of the transcription level was made between longer (HY405) and shorter (CCRI8 and ND601) fibers. The results between comparison of HY405 vs. CCRI8 and HY405 vs. ND601 were consistent (Figure 4A). More GhLtps were significantly activated in cultivar HY405 with longer fibers, especially at 5, 15, and 25 DPA (Figure 4A). Among these genes, GhLtpIs and GhLtpXIs occupied 54% proportion (Figure 4B). The transcripts of GhLtpI26, V14, VI4, XI12, XI13, XI30, XI32, and XI33 dramatically accumulated in long-fibered cultivars when fiber started to elongate (Figures 4A,C), suggesting a possible contribution of GhLtps (especially GhLtpXIs) to fiber length improvement.
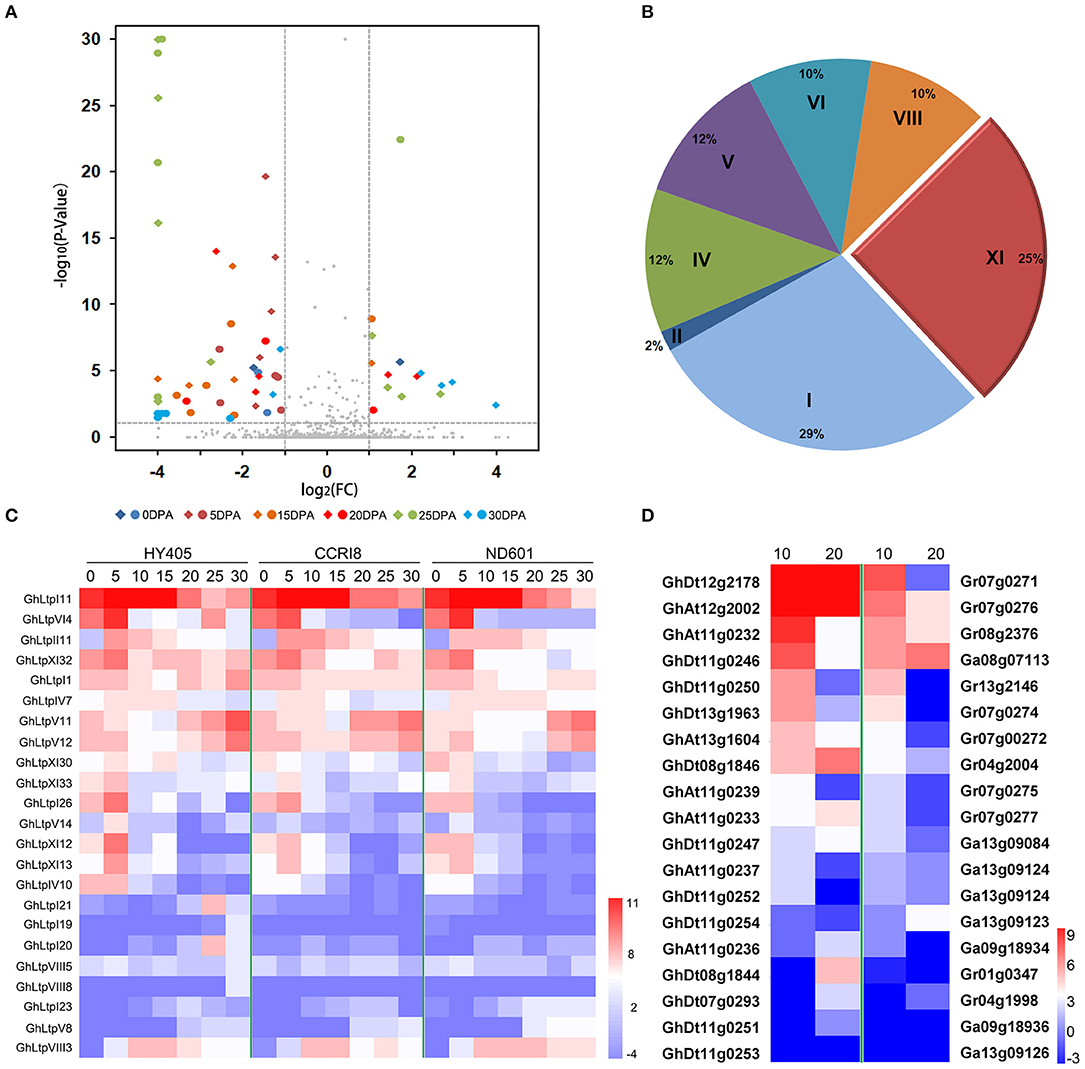
Figure 4. Differential expression analysis of GhLtps. (A) Volcano plot shows expression comparison between HY405 vs. CCRI8 and HY405 vs. ND601 represented by round and square points, respectively. FC stands for fold change. (B) Pie chart shows the proportion of gene types in differentially expressed GhLtps. (C) Heatmap shows expression of differentially expressed genes between cultivars with longer and shorter fibers. Data shown were log2-transformed RPKM. Developmental stages of fiber are indicated in days post anthesis (DPA) above. The color bar represents the relative expression level. (D) Heatmap shows expression of differentially expressed GhLtps compared with their orthologs in G. raimondii or G. arboreum. Data shown were log2-transformed FPKM of each gene which was quantified using RNA-seq data downloaded from CottonFGD. Developmental stages of fiber are indicated in days post anthesis (DPA) above. The color bar represents the relative expression level.
After its divergence from an ancestor shared with Th. cacao (Carvalho et al., 2011), the cotton lineage evolved into D-genome and A-genome. However, spinnable fiber only evolved in the A-genome and was further elongated after the merger of A and D genome (Paterson et al., 2012). In order to further clarify the function of GhLtpXIs in fiber development, the expression of cotton type XI genes was analyzed. Firstly, orthologs of GhLtpXIs were identified from D-genome and A-genome according to the sequence similarity, which was further confirmed by the syntenic relationships between diploid and tetraploid cotton species identified previously (Supplemental Figure S6) (Zhang et al., 2015). Ten type XI genes in A and D genome lost during evolution and 4 pairs of non-reciprocal DNA exchanges were identified including Gr13g0054/GhAt13g0035, GhAt11g0237/GhDt11g0252, GhAt11g0234/GhDt12g1005, and GhDt12g1005/GhAt12g0916. Then the expression of orthologous gene pairs in ovules was compared at 10 and 20 DPA, respectively, using the downloaded transcriptome data. The results demonstrated that the transcriptional level of most GhLtpXIs, discarding 12 GhLtpXIs with scarce transcripts, significantly varied from their diploid progenitors (Figure 4D), among which the expression of 15 GhLtpXIs varied at 10 DPA and 12 varied at 20 DPA. It was notable that the expression of most GhLtpXIs was higher than their diploid progenitors except for GhLtpXI14-17. Especially, expression of GhLtpXI32 and GhLtpXI33 was extremely higher in tetraploid cotton than their orthologs in diploid cotton. Additionally, transcripts of GhLtpXI6 and GhLtpXI12 were dramatically abundant during fiber elongation in tetraploid cotton and GhLtpXI27, GhLtpXI28, and GhLtpXI30 increased sharply at 20 DPA.
The previous studies showed that polar lipids increased and reached the maximum level and were incorporated into the cell wall from 3 to 20 DPA (Wan et al., 2005; Edstam et al., 2013; Kumar et al., 2013). Our results suggested that GhLtps, especially GhLtpXIs play important roles during fiber elongation. Those differentially expressed GhLtps are likely to delivering lipids to the outer integument of cotton ovules (Edstam et al., 2013) and thus contribute to the variance of fiber length.
Based on the analysis of the RNA-seq data, it is possible that the considerably expanded GhLtpXIs play important roles in different fiber developmental stages. On account of this, we selected nine genes to verify their expression by qPCR (Figure 5). The expression trend of the selected genes were consistent with RNA-seq data except for GhLtpXI12 and GhLtpXI33, which would be due to that samples at 5 DPA used for RNA-seq and qRT-PCR were different. All of these GhLtpXIs showed higher transcription levels in cultivar HY405 that produces longer fibers. The expression profile of each gene varied in different stages of fiber development, suggesting different roles of GhLtpXIs. GhLtpXI27, GhLtpXI28, and GhLtpXI30 exhibited highest transcripts at 0 DPA and decreased dramatically at 5 DPA, revealing a role in fiber initiation. The expression of GhLtpXI6/7 decreased gradually during fiber elongation. Noticeably, GhLtpXI6/7 maintained a high level in HY405 until 10 DPA when its transcription in ND601 has decreased to a relatively low level, and GhLtpXI14 and GhLtpXI32 demonstrated a constantly high level during fiber elongation in HY405, which would probably contribute to long fiber quality of the cultivar. GhLtpXI1, GhLtpXI12, and GhLtpXI33 showed a transcriptional peak during fiber elongation. The expression trend of GhLtpXI1 was consistent in both cultivars with a higher expression in HY405. Though the expression of GhLtpXI12 was comparable in two cultivars, its expression peaked earlier in HY405 at 10 DPA that is essential to fiber elongation. There is also an advanced expression peak of GhLtpXI33 in HY405 with a significantly abundant transcripts compared with that in ND601. It might be another important effecter on fiber length.
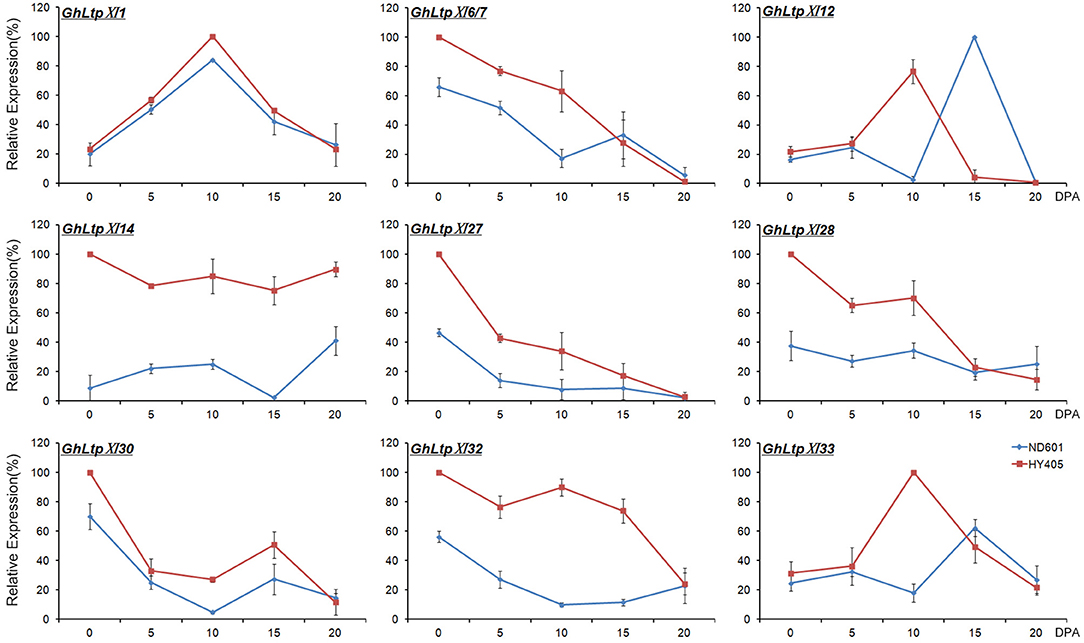
Figure 5. Temporal expression of nine GhLtpXIs in developing fibers. Ovules were collected on the day of anthesis and fibers were harvested on 5, 10, 15, and 20 DPA. Gene expression levels determined by qPCR were normalized to UBQ14 expression and shown as relative values to the maximal gene expression levels set at 100%. Error bars indicate SD of three biological replicates.
It is believed that the regulatory mechanism is shared by fiber and leaf trichome development (Kim and Triplett, 2001; Qin and Zhu, 2011; Lei et al., 2014), and Arabidopsis continues to serve as a useful experimental system for dissecting the mechanisms of cotton fiber development (Guan et al., 2014; Shangguan et al., 2016; Ma et al., 2018). Since GhLtpXIs considerably expanded in Gossypium species, several members with early divergent time were cloned and ectopically expressed in Arabidopsis to further verify the function of GhLtpXIs in fiber development. Expression analysis of T1 transgenic plants revealed that these cotton genes were successfully expressed in Arabidopsis (Figure 6A). The trichomes of the mature rosette leaves were observed under microscope. The results showed that the trichome morphology was not affected by overexpressing of different GhLtpXIs, whereas the trichome length of all the transgenic plants was significantly longer than that of the WT plants (Figure 6B), and the improvement of trichome length was positively correlated to the expression of GhLtpXIs. These results suggested that GhLtpXIs function to promote trichome elongation. Cotton fiber which is derived from the epidermal cells is a single-celled trichome of seed and thus these GhLtpXIs might probably regulate fiber elongation.
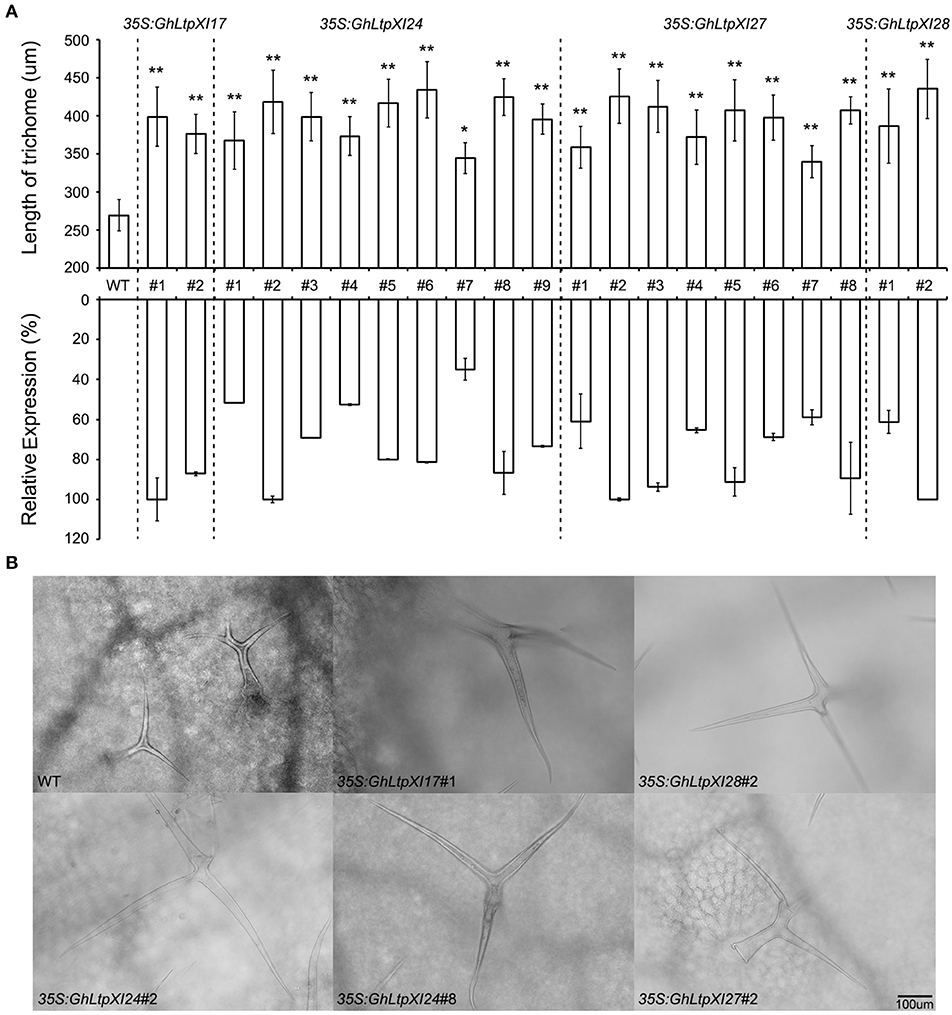
Figure 6. GhLtpXIs regulated trichome development in Arabidopsis. (A) Upregulation of GhLtpXIs in independent overexpression transgenic plants correlated to the length of trichomes on the surface of mature rosette leaves. Trichome length was measured under microscope and calculated for average of 30~50 trichomes. Asterisks indicate the significant difference in relation to WT (T-test, *p < 0.05, **p < 0.01). Expression of GhLtpXIs was determined by qPCR in leaves of each independent transgenic plants. Transcripts of GhLtpXIs were undetectable in WT. Results were normalized against the expression of TUB2. The maximum expression of each GhLtpXI is set as 100%. Error bars indicate SD. (B) Overexpressing of GhLtpXIs in transgenic plants promoted trichome length. Trichomes were observed under microscope after decolorization of leaves. The bar represents 100 μm.
Plant nsLTPs are characterized by the ECM backbone forming a stabilized hydrophobic cavity and could be classified into 11 groups including I, II, III, IV, V, VI, VII, VIII, IX, XI, and nsLTPy according to the number of flanking amino acids within the conserved ECM domain (Kader, 1996; Carvalho and Gomes, 2007; Boutrot et al., 2008; Li et al., 2014). The nsLTP family has been identified in Arabidopsis, rice, rape, maize, and sorghum in previous studies (Boutrot et al., 2008; Li et al., 2014; Wei and Zhong, 2014) and in G. hirsutum, G. arboreum, G. raimondii, Th. Cacao, and V. vinifera in this study (Supplemental Table S3). As an ancient species, V. vinifera genome contained 3 type XI genes, and this group is likely to disappear evolutionally because of the lack of type XI genes in rice and Arabidopsis. Although several type XI genes were identified in cacao, grape and rape, type I or II consistently possess most members in grape, cacao, rape, rice and Arabidopsis. It is interesting that cotton type XI nsLTPs harbor an extra N-terminal cap and are larger in molecular weight, which are different from other nsLTPs. These outcomes reveal that type XI nsLTPs specifically expanded in the Gossypium species (Figure 2).
Cotton type XI genes were close to type II genes in the phylogenetic tree and duplication pairs were found between GhLtpXIs and GhLtpIIs (Figures 2, 3 and Supplemental Table S4). The special protein structure of GhLtpXIs was only similar to GhLtpII13-15 (Figure 1). These findings imply that cotton type XI genes might diverge from type II genes. A large amount of tandem duplication events was found within GhLtpXIs (Figure 3), suggesting an essential contribution to the tremendous expansion of type XI genes.
Considered as an ideal model for cell elongation, cotton fiber is the single-celled seed trichome developed from the outer integument of ovule and cotton fiber quality is quantitative traits controlled by multiple genes. Due to their economic importance and biological feature, cotton fiber development has been the subject of much scientific interest. Two decades ago, scientists have cloned nsLTPs from fibers and believed that they should play roles during fiber development (Ma et al., 1995, 1997; Liu et al., 2000; Orford and Timmis, 2000). Later, 12 nsLTPs were found highly expressed during fiber elongation in upland cotton fibers compared with fuzzless mutant ovules (Ji et al., 2003). However, little progress was achieved on how these small proteins affect fiber development. The complement of whole genome sequence of G. hirsutum (Li et al., 2015; Zhang et al., 2015) facilitates the study of gene families contributing to fiber development. In the present study, 138, 65, and 70 nsLTPs were strictly identified from G. hirsutum, G. arboretum, and G. raimondii, respectively. RNA-seq data suggested that most GhLtps were transcribed during fiber development and some GhLtpXIs displayed much abundant transcripts during fiber initiation and elongation (Figure 5 and Supplemental Figure S4). Significant transcriptional differences of GhLtps were found between cultivated upland cotton varieties with longer and shorter fibers. And many GhLtpXIs displayed preponderance of expression in longer fibers, which was likely to contribute to the variation of fiber quality (Figures 4, 5). The transcription of several GhLtpXIs was verified by qPCR with a higher level or advanced peak in long-fibered cultivar (Figure 5). Furthermore, significant elongation of Arabidopsis leaf trichomes induced by overexpression of GhLtpXIs suggested functional roles of GhLtpXIs in promoting cell elongation (Figure 6).
Since the first genetic map was constructed in cotton, many QTLs for fiber quality traits have been identified (Shen et al., 2005; Wang et al., 2006; Qin et al., 2008; Zhang et al., 2012; Ning et al., 2014; Shao et al., 2014; Shang et al., 2015; Jamshed et al., 2016). After blast with the QTLs for fiber quality traits, GhLtpI24, GhLtpIV6 and a cluster of GhLtpXIs located in chromosome Dt11 (GhLtpXI13/14/15/17/19/21/23/24/27) fell in QTL qFL15.1, qFS-D13-1, and qFM21.1, respectively (Zhang et al., 2012; Ning et al., 2014; Shao et al., 2014). These findings further proved that GhLtps especially GhLtpXIs are important regulators of fiber development and thus contribute to fiber quality.
LCFA regulates endogenous ethylene biosynthesis in cotton ovules to promote the extensibility of fibers (Qin et al., 2007). The basic function of nsLTPs is to transfer lipids. GhLtps were speculated to bind with various lipids to be responsible for the intracellular and intercellular movement of lipids including fatty acids and thus affect ethylene production and fiber elongation. A GPI-anchored lipid transport protein (GhLTPG1) has been identified to regulate cotton fiber elongation through mediating the transport of phosphatidylinositol monophosphates (Ostergaard et al., 1993; Deng et al., 2016), which supports the speculation. As a consequence of the transportation of lipids, the intercellular fatty acid pools will be regulated by GhLtps, which would cause a feedback regulation on reactions enrolling fatty acids, including LCFA biosynthesis and accumulation. In addition to transferring lipids, nsLTPs can also act as lipid sensors and lipid chaperones (Wong et al., 2017). Plant nsLTPs are supposed to bind a lipid molecular competing with elicitin and mediate signaling (Blein et al., 2002; Maldonado et al., 2002). When nsLTPs present part of a lipid (typically the hydrophilic head group) to another protein, they act as lipid chaperones to mediate signaling (Wang et al., 2005). Thus a hypothesis that GhLtps participant in the signaling of phytohormones involved in fiber development and other signaling pathways mediating fiber development could be proved when functional lipids were isolated as signaling molecules from fibers or the outer integument of ovules.
Shortly after the divergence from the same ancestor as Th. cacao at least 60 Myr ago (Carvalho et al., 2011), the genus Gossypium experienced an abrupt polyploidization with a maternal A-genome propagule resembling G. herbaceum and a pollen parent D-genome species resembling G. raimondii which diverged ~5–10 Myr ago (Senchina et al., 2003). The nascent AtDt allopolyploid diverged into at least five species, of which two major cultivated species G. hirsutum and G. barbadense were domesticated independently to spawn textile industry and became a major oilseed. Although G. hirsutum has been well domesticated and bred for many cultivated varieties that vary in fiber quality, the evolution of fiber and regulatory mechanism of fiber development are unclarified. Our results suggested that the expanded type XI genes in G. hirsutum genome are involved in fiber development. Although GhLtpXIs were phylogenetically close to GhLtpIIs, they evolved to be different in protein structure and MWs. Members of this group contain an extra N-terminal cap (Figure 1). Protein sequences of GhLtpXIs were highly conserved within the ECMs and only varied at the N-terminal (Figure 1). Additionally, the divergence time of GhLtpXIs was earlier than other GhLtps. It is clear that GhLtpXIs form a unique group distinctive from other GhLtps.
The sequence of a G. hirsutum cultivar reveals many non-reciprocal DNA exchanges between subgenomes that may have contributed to phenotypic innovation that spinnable fiber evolved from the merger of A genome and D genome determining a fibered and a fibreless phenotype, respectively (Paterson et al., 2012). Noticeably, non-reciprocal DNA exchanges were found in cotton type XI genes and most transcribed GhLtpXIs in ovules displayed significant expression difference during fiber elongation compared with their orthologs in A or D genome, indicating a correlation between cotton type XI genes and fiber evolution. Therefore the cotton XI genes are speculated to comprise a possible “fiber clade” (Paterson et al., 2012) evolving from polyploidy, non-reciprocal DNA exchanges and duplication events, which would be supported by further sequence comparison of nsLTPs in different cotton species and their evolutional close species and molecular investigation on how these candidates regulate fiber development.
ZM, CM, and YY designed the experiments. CM and YY conducted the experiments and analyzed the data. ZL, LC, and YZ performed part of the experiments. XL helped in the bioinformatics analyses. LW and GZ helped in growing experimental materials. CM and YY wrote the manuscript. ZM and XW carefully edited the manuscript and analyzed the data. All authors revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Xiyin Wang and Jinpeng Wang for their assistance in bioinformatics analysis. Our thanks also go to Hongping Wu for her help in data analysis. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0101006), the Science and Technology Support Program of Hebei Province (16226307D), the Top Talent Fund of Hebei Province, the High School Science and Technology Research Program of Hebei Province (ZD2018057) and financial support for research from College of Agronomy of Hebei Agricultural University.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2018.01285/full#supplementary-material
Supplemental Figure S1. Phylogenetic tree of GhLtps. The amino acids of the mature proteins of cotton and Arabidopsis nsLtps were used to build the phylogenetic tree using Maximum likelihood method. The 10 types of nsLTPs and another five AtLtps are showed with different color background. Four clusters are marked against different colors.
Supplemental Figure S2. Gene structure of the GhLtps. Exons are represented by green boxes and introns are showed with black lines.
Supplemental Figure S3. Expression profiles of GhLtpXIs in different organs and tissues of G. hirsutum. Data shown were log2-transformed FPKM of each gene which was quantified using RNA-seq data downloaded from CottonFGD. Clustering of expression level is showed on the right. The color bar represents the relative expression level.
Supplemental Figure S4. Expression of GhLtps during fiber development. Data shown were log2-transformed RPKM. Developmental stages of fiber are indicated in days post anthesis (DPA) above. The color bar represents the relative expression level.
Supplemental Figure S5. Expression trend analysis of GhLtps. Number of genes is indicated below the relevant expression pattern.
Supplemental Figure S6. Phylogenetic tree of nsLTPs in cotton species. The full lengths of mature protein sequences were used to construct the phylogenetic tree using a Neighbor-Joining method. The same color was used to present orthologous gene pairs and non-reciprocal DNA exchanges were labeled with different dots.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. Diversity of the eight cysteine motif.
Supplemental Table S3. Classification of nsLTP genes in cotton, Arabidopsis, rape, rice, cacao and grape.
Supplemental Table S4. Ka/Ks analysis for the duplicated gene pairs.
Basra, A. S., and Malik, C. P. (1984). Development of the cotton fiber. Int. Rev. Cytol. 89, 65–113.
Blein, J. P., Coutos-Thévenot, P., Marion, D., and Ponchet, M. (2002). From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends in Plant Sci. 7, 293–296. doi: 10.1016/S1360-1385(02)02284-7
Boutrot, F., Chantret, N., and Gautier, M. F. (2008). Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLTP) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9:86. doi: 10.1186/1471-2164-9-86
Carvalho, A. O., and Gomes, V. M. (2007). Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28, 1144–1153. doi: 10.1016/j.peptides.2007.03.004
Carvalho, M. R., Herrera, F. A., Jaramillo, C. A., Wing, S. L., and Callejas, R. (2011). Paleocene malvaceae from northern south America and their biogeographical implications. Am. J. Bot. 98, 1337–1355. doi: 10.3732/ajb.1000539
Chae, K., Gonong, B. J., Kim, S. C., Kieslich, C. A., Morikis, D., Balasubramanian, S., et al. (2010). A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 61, 4277–4290. doi: 10.1093/jxb/erq228
Charvolin, D., Douliez, J. P., Marion, D., Cohenaddad, C., and Pebaypeyroula, E. (1999). The crystal structure of a wheat non-specific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. Febs J. 264, 562–568.
Chen, C., Chen, G., Hao, X., Cao, B., Chen, Q., Liu, S., et al. (2011). CaMF2, an anther-specific lipid transfer protein (LTP) gene, affects pollen development in Capsicum annuum L. Plant Sci. 181, 439–448. doi: 10.1016/j.plantsci.2011.07.003
Cheng, H. C., Cheng, P. T., Peng, P., Lyu, P. C., and Sun, Y. J. (2004). Lipid binding in rice non-specific lipid transfer protein-1 complexes from Oryza sativa. Protein. Sci. 13:2304. doi: 10.1110/ps.04799704
Deng, T., Yao, H. Y., Wang, J., Wang, J., Xue, H. W., and Zuo, K. J. (2016). GhLTPG1, a cotton GPI-anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Sci. Rep. 6:26829. doi: 10.1038/srep26829
Douliez, J. P., Michon, T., Elmorjani, K., and Marion, D. (2000). Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cer. Sci. 32, 1–20. doi: 10.1006/jcrs.2000.0315
Edstam, M. M., Blomqvist, K., Eklof, A., Wennergren, U., and Edqvist, J. (2013). Co-expression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol. Biol. 83, 625–649. doi: 10.1007/s11103-013-0113-5
Edstam, M. M., and Edqvist, J. (2014). Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 152, 32–42. doi: 10.1111/ppl.12156
Guan, X. Y., Li, Q. J., Shan, C. M., Wang, S., Mao, Y. B., Wang, L. J., et al. (2008). The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant. 134, 174–182. doi: 10.1111/j.1399-3054.2008.01115.x
Guan, X. Y., Pang, M. X., Nah, G., Shi, X. L., Ye, W. X., David, M. S., et al. (2014). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5:3050. doi: 10.1038/ncomms4050
Han, G. W., Lee, J. Y., Song, H. K., Chang, C., Min, K., Moon, J., et al. (2001). Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308, 263–278. doi: 10.1006/jmbi.2001.4559
Han, H. C., Huang, Y. Q., Wang, J., and Zuo, K. J. (2013). Cloning and expression pattern analysis of Gossypium hirsutum lipid transfer protein gene family. J. Agr. Sci. Tech. 15, 84–90.
Han, J., Pan, Y. X., Wang, X. F., Zhang, Y., and Ma, Z. Y. (2016). Antisense expression of Gossypium barbadense UGD6 in Arabidopsis thaliana significantly alters cell wall composition. Sci. China Life Sci. 59, 213–218. doi: 10.1007/s11427-016-5004-y
Hsu, C.-Y., Jenkins, J. N., Saha, S., and Ma, D.-P. (2005). Transcriptional regulation of the lipid transfer protein gene LTP3 in cotton fibers by a novel MYB protein. Plant Sci. 168, 167–181. doi: 10.1016/j.plantsci.2004.07.033
Huang, Y. Q., Liu, X., Tang, K. X., and Zuo, K. J. (2013). Functional analysis of the seed coat-specific gene GbMYB2 from cotton. Plant Physiol. Bioch. 73, 16–22. doi: 10.1016/j.plaphy.2013.08.004
Jamshed, M., Jia, F., Gong, J. W., Palanga, K. K., Shi, Y. Z., Li, J. W., et al. (2016). Identification of stable quantitative trait loci (QTLs) for fiber quality traits across multiple environments in Gossypium hirsutum recombinant inbred line population. BMC Genomics 17:197. doi: 10.1186/s12864-016-2560-2
Ji, S. J., Lu, Y. C., Feng, J. X., Wei, G., Li, J., Shi, Y. H., et al. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucl. Acids Res. 31, 2534–2543. doi: 10.1093/nar/gkg358
Joshi, P. C., Wadhwani, A. M., and Johri, B. M. (1967). Morphological and embryological studies of Gossypium, L. Indian J. Agr. Res. 33, 37–93.
Kader, J. C. (1996). Lipid-transfer proteins in plants. Ann. Plant Physiol. Plant Mol. Biol. 47, 627–654.
Kim, H. J., and Triplett, B. A. (2001). Cotton fiber growth in planta and in vitro models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127, 1361–1366. doi: 10.1104/pp.010724
Kong, X. P., Lv, W., Jiang, S. S., Zhang, D., Cai, G. H., Pan, J. W., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14:433. doi: 10.1186/1471-2164-14-433
Kragelund, B. B., Bech, L. M., and Poulsen, F. M. (1997). Barley lipid-transfer protein complexed with palmitoyl CoA: the structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure 5, 291–306.
Kumar, S., Kumar, K., Pandey, P., Rajamani, V., Padmalatha, K. V., Dhandapani, G., et al. (2013). Glycoproteome of elongating cotton fiber cells. Mol. Cell. Pro. 12, 3677–3689. doi: 10.1074/mcp.M113.030726
Lee, S. B., Go, Y. S., Bae, H. J., Park, J. H., Cho, S. H., Cho, H. J., et al. (2009). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen alternaria brassicicola. Plant Physiol. 150, 42–54. doi: 10.1104/pp.109.137745
Lei, L., Chen, L., Shi, X. F., Li, Y. X., Wang, J. Y., Chen, D. S., et al. (2014). A nodule-specific lipid transfer protein asE246 participates in transport of plant-synthesized lipids to symbiosome membrane and is essential for nodule organogenesis in chinese milk vetch. Plant Physiol. 164, 1045–1058. doi: 10.1104/pp.113.232637
Li, F. G., Fan, G. Y., Lu, C. R., Xiao, G. H., Zou, C. S., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotech. 33, 524–530. doi: 10.1038/nbt.3208
Li, J., Gao, G. Z., Xu, K., Chen, B. Y., Yan, G. X., Li, F., et al. (2014). Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS ONE 9:e84556. doi: 10.1371/journal.pone.0084556
Liu, B. L., Zhu, Y. C., and Zhang, T. T. (2015). The R3-MYB gene GhCPC negatively regulates cotton fiber elongation. PLoS ONE 10:e0116272. doi: 10.1145/2818302
Liu, H. C., Creech, R. G., Jenkins, J. N., and Ma, D. P. (2000). Cloning and promoter analysis of the cotton lipid transfer protein gene Ltp3. Bioch. Bioph. Acta 1487, 106–111. doi: 10.1016/S1388-1981(00)00072-X
Liu, W., Li, W., He, Q., Daud, M. K., Chen, J., and Zhu, S. J. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS ONE 9:e98189. doi: 10.1371/journal.pone.0098189
Ma, D. P., Liu, H. C., Tan, H. P., Creech, R. G., Jenkins, J. N., and Chang, Y. F. (1997). Cloning and characterization of a cotton lipid transfer protein gene specifically expressed in fiber cells. Bioch. Bioph. Acta 1344, 111–114. doi: 10.1016/S0005-2760(96)00166-X
Ma, D. P., Tan, H., Si, Y., Creech, R. G., and Jenkins, J. N. (1995). Differential expression of a lipid transfer protein gene in cotton fiber. Bioch. Bioph. Acta 1257, 81–84.
Ma, Z. Y., He, S. P., Wang, X. F., Sun, J. L., Zhang, Y., Zhang, G. Y., et al. (2018). Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813. doi: 10.1038/s41588-018-0119-7
Machado, A., Wu, Y. R., Yang, Y. M., Llewellyn, D. J., and Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Montagu, M. V., and Kuiper, M. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Nat. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., and Cameron, R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. doi: 10.1038/nature00962
Molina, A., Segura, A., and Garcia-Olmedo, F. (1993). Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. Febs Letters 316, 119–122.
Ning, Z. Y., Chen, H., Mei, H. X., and Zhang, T. Z. (2014). Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema. Euphytica 195, 143–156. doi: 10.1007/s10681-013-0990-3
Orford, S. J., and Timmis, J. N. (2000). Expression of a lipid transfer protein gene family during cotton fibre development. Bioch. Bioph. Acta 1483, 275–284.
Ostergaard, J., Vergnolle, C., Schoentgen, F., and Kader, J. C. (1993). Acyl-binding/lipid-transfer proteins from rape seedlings, a novel category of proteins interacting with lipids. Bioch. Bioph. Acta 1170, 109–117.
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D. C., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Pu, L., Li, Q., Fan, X. P., Yang, W. C., and Xue, Y. B. (2008). The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180, 811–820. doi: 10.1534/genetics.108.093070
Pyee, J., Yu, H. S., and Kolattukudy, P. E. (1994). Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biol. Biol. 311, 460–468.
Qin, H. D., Guo, W. Z., Zhang, Y. M., and Zhang, T. Z. (2008). QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor. Appl. Genet. 117:883. doi: 10.1007/s00122-008-0828-x
Qin, Y. M., Hu, C. Y., Pang, Y., Kastaniotis, A. J., Hiltunen, J. K., and Zhu, Y. X. (2007). Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19, 3692–3704. doi: 10.1105/tpc.107.054437
Qin, Y. M., and Zhu, Y. X. (2011). How cotton fibers elongate: a tale of linear cell-growth mode. Curr. Opin. Plant Biol. 14, 106–111. doi: 10.1016/j.pbi.2010.09.010
Sarowar, S. J., Kim, Y. J., Kim, K. D., Hwang, B. K., Ok, S. H., and Shin, J. S. (2009). Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 28, 419–427. doi: 10.1007/s00299-008-0653-3
Schweiger, W., Steiner, B., Ametz, C., Siegwart, G., Wiesenberger, G., Berthiller, F., et al. (2013). Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.ifa-5A, identifies novel candidate genes. Mol. Plant Pathol. 14, 772–785. doi: 10.1111/mpp.12048
Senchina, D. S., Alvarez, I., Cronn, R. C., Liu, B., Rong, J. K., Noves, R. D., et al. (2003). Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20, 633–643. doi: 10.1093/molbev/msg065
Shang, L. G., Liang, Q. Z., Wang, Y. M., Wang, X. C., Wang, K. B., Abduweli, A., et al. (2015). Identification of stable QTLs controlling fiber traits properties in multi-environment using recombinant inbred lines in Upland cotton (Gossypium hirsutum L.). Euphytica 205, 877–888. doi: 10.1007/s10681-015-1434-z
Shangguan, X. X., Yang, C. Q., Zhang, X. F., and Wang, L. J. (2016). Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol. Plant. 158, 200–212. doi: 10.1111/ppl.12450
Shao, Q. S., Zhang, F. J., Tang, S. Y., Liu, Y., Fang, X. M., Liu, D. X., et al. (2014). Identifying QTL for fiber quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica 198, 43–58. doi: 10.1007/s10681-014-1082-8
Shen, X. L., Guo, W. Z., Zhu, X. F., Yuan, Y. L., Yu, J. Z., Kohel, R. J., et al. (2005). Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breed. 15, 169–181. doi: 10.1007/s11032-004-4731-0
Shin, H. S., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Sodano, P., Caille, A., Sy, D., De, P. G., Marion, D., and Ptak, M. (1997). 1H NMR and fluorescence studies of the complexation of DMPG by wheat non-specific lipid transfer protein. Global fold of the complex. Febs Lett. 416, 130–134.
Stewart, J. M. D. (1975). Fiber initiation on the cotton ovule (Gossypium Hirsutum). Am. J. Bot. 62, 723–730.
Walford, S. A., Wu, Y. R., Llewellyn, D. J., and Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313X.2010.04464.x
Wan, S. W., Welti, R., Moreau, R. A., and Chapman, K. D. (2005). Identification and quantification of glycerolipids in cotton fibers: reconciliation with metabolic pathway predictions from DNA databases. Lipids 40, 773–785. doi: 10.1007/s11745-005-1439-4
Wang, B. H., Guo, W. Z., Zhu, X. F., Wu, Y. T., Huang, N. T., and Zhang, T. (2006). QTL mapping of fiber quality in an elite hybrid derived-RIL population of upland cotton. Euphytica 152, 367–378. doi: 10.1007/s10681-006-9224-2
Wang, G., Zhao, G. H., Jia, Y. H., and Du, X. M. (2013). Identification and characterization of cotton genes involved in fuzz-fiber development. J. Integr. Plant Biol. 55, 619–630. doi: 10.1111/jipb.12072
Wang, P. Y., Weng, J., and Anderson, R. G. (2005). OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science 307, 1472–1476. doi: 10.1126/science.1107710
Wang, Y. P., Wang, X. Y., Tang, H. B., Tan, X., Ficklin, S. P., Feltus, F. A., et al. (2011). Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE 6:e28150. doi: 10.1371/journal.pone.0028150
Wei, H. L., Li, W., Sun, X. W., Zhu, S. J., and Zhu, J. (2013). Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS ONE 8:e68435. doi: 10.1371/journal.pone.0068435
Wei, K., and Zhong, X. (2014). Non-specific lipid transfer proteins in maize. BMC Plant Biol. 14:281. doi: 10.1186/s12870-014-0281-8
Wong, L. H., Čopič, A. K., and Levine, T. P. (2017). Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 42, 516–530. doi: 10.1016/j.tibs.2017.05.001
Yu, H., Ito, T., Wellmer, F., and Meyerowitz, E. M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36, 157–161. doi: 10.1038/ng1286
Zhang, D. S., Liang, W. Q., Yin, C. S., Zong, J., Gu, F. W., and Zhang, D. B. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162. doi: 10.1104/pp.110.158865
Zhang, K., Zhang, J., Ma, J., Tang, S. Y., Liu, D. J., Teng, Z. H., et al. (2012). Genetic mapping and quantitative trait locus analysis of fiber quality traits using a three-parent composite population in upland cotton (Gossypium hirsutum L.). Mol. Breed. 29, 335–348. doi: 10.1007/s11032-011-9549-y
Zhang, T. Z., Hu, Y., Jiang, W. K., Guan, X. Y., Chen, J. D., Zhang, J. Z., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotech. 33, 531–537. doi: 10.1038/nbt.3207
Keywords: cotton, nsLTP, type XI, expansion, fiber development, fiber evolution
Citation: Meng C, Yan Y, Liu Z, Chen L, Zhang Y, Li X, Wu L, Zhang G, Wang X and Ma Z (2018) Systematic Analysis of Cotton Non-specific Lipid Transfer Protein Family Revealed a Special Group That Is Involved in Fiber Elongation. Front. Plant Sci. 9:1285. doi: 10.3389/fpls.2018.01285
Received: 25 April 2018; Accepted: 16 August 2018;
Published: 19 September 2018.
Edited by:
Guijun Yan, University of Western Australia, AustraliaReviewed by:
Shahidul Islam, Murdoch University, AustraliaCopyright © 2018 Meng, Yan, Liu, Chen, Zhang, Li, Wu, Zhang, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiying Ma, bXpoeUBoZWJhdS5lZHUuY24=
Xingfen Wang, Y290dG9uQGhlYmF1LmVkdS5jbg==
† These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.