
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 July 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01066
This article is part of the Research TopicActivation and Suppression of Plant ImmunityView all 27 articles
Although production of tomato (Solanum lycopersicum) is threatened by a number of major diseases worldwide, it has been difficult to identify effective and durable management measures against these diseases. In this study, we attempted to improve tomato disease resistance by transgenic overexpression of genes encoding the Arabidopsis thaliana Elongator (AtELP) complex subunits AtELP3 and AtELP4. We show that overexpression of AtELP3 and AtELP4 significantly enhanced resistance to tomato bacterial speck caused by the Pseudomonas syringae pv. tomato strain J4 (Pst J4) without clear detrimental effects on plant growth and development. Interestingly, the transgenic plants exhibited resistance to Pst J4 only when inoculated through foliar sprays but not through infiltration into the leaf apoplast. Although this result suggested possible involvement of stomatal immunity, we found that Pst J4 inoculation did not induce stomatal closure and there were no differences in stomatal apertures and conductance between the transgenic and control plants. Further RNA sequencing and real-time quantitative PCR analyses revealed a group of defense-related genes to be induced to higher levels after infection in the AtELP4 transgenic tomato plants than in the control, suggesting that the enhanced disease resistance of the transgenic plants may be attributed to elevated induction of defense responses. Additionally, we show that the tomato genome contains single-copy genes encoding all six Elongator subunits (SlELPs), which share high identities with the AtELP proteins, and that SlELP3 and SlELP4 complemented the Arabidopsis Atelp3 and Atelp4 mutants, respectively, indicating that the function of tomato Elongator is probably conserved. Taken together, our results not only shed new light on the tomato Elongator complex, but also revealed potential candidate genes for engineering disease resistance in tomato.
Tomato (Solanum lycopersicum) fruit was once thought to be poisonous, but since being integrated as part of the human diet, its popularity and consumption have increased over the years. Tomato production has an economic impact worldwide, but it is also a costly crop to produce. It is a labor-intensive crop that requires significant amount of chemical inputs to be protected from a wide variety of pests and diseases. There are a number of diseases that affect tomatoes, including bacterial speck, which is caused by the bacterial pathogen Pseudomonas syringae pv. tomato. Bacterial speck can cause up to 75% losses in yield, if present early in the production cycle (Yunis et al., 1980). Control of the disease is primarily based on application of bactericides and sanitary measures. Pathogen-free seeds and resistant varieties carrying the resistance (R) gene Pto have been implemented to control the disease (Monroe and Sasser, 1980; Pedley and Martin, 2003). However, P. syringae pv. tomato strains have evolved to overcome the R gene-mediated resistance in tomato (Thapa and Coaker, 2016).
Since tomatoes are susceptible to many diseases, studies involving identification of disease resistance-related genes in model plants have increased dramatically (Piquerez et al., 2014). Currently, one strategy that is being pursued is to utilize resistance-related genes identified in Arabidopsis and their orthologs in other plant species (Jones et al., 2014). Arabidopsis is a well-established model system, with the complete genome sequenced.1 Furthermore, multiple Arabidopsis genes have been cloned, characterized, and reported to confer resistance to diseases when overexpressed in diverse crop species (Lin et al., 2004; Chan et al., 2005; Lacombe et al., 2010; Schwessinger et al., 2015; Silva et al., 2017), making Arabidopsis a suitable source of defense-related genes for engineering resistance in tomato.
The Elongator protein (ELP) complex is a highly conserved multitasking protein complex in eukaryotes (Otero et al., 1999; Wittschieben et al., 1999; Hawkes et al., 2002; Nelissen et al., 2010; Woloszynska et al., 2016). It consists of six subunits, including ELP1 and ELP2 (scaffolds for complex assembly), ELP3 (catalytic subunit), and an accessory complex formed by ELP4–ELP6 (Svejstrup, 2007). Elongator has been shown to be involved in several distinct cellular processes, such as exocytosis, histone modification, tRNA modification, α-tubulin acetylation, zygotic paternal DNA demethylation, and miRNA biogenesis (Hawkes et al., 2002; Huang et al., 2005; Rahl et al., 2005; Creppe et al., 2009; Okada et al., 2010; Ding and Mou, 2015; Fang et al., 2015). It has been clearly demonstrated that Elongator functions in both the nucleus and the cytoplasm (Versées et al., 2010). In the nucleus, Elongator regulates histone acetylation and DNA methylation/demethylation (Winkler et al., 2002; Lin et al., 2012; Wang et al., 2013), thus being involved in gene transcription. In the cytoplasm, it is responsible for tRNA modification, which consequently regulates protein translation (Huang et al., 2005; Esberg et al., 2006; Glatt et al., 2012).
It has been well documented that the A. thaliana Elongator protein (AtELP) complex plays an important role in plant immunity, likely by regulating the transcription of defense genes (Ding and Mou, 2015; Wang et al., 2015). However, whether Elongator has a similar role in plant species other than Arabidopsis remains to be determined. Although it has been reported that silencing of a tomato AtELP2-like gene, SlELP2L, resulted in pleiotropic phenotypes similar to those of the Atelp mutants, defense phenotypes of the SlELP2L-RNAi lines were not tested (Zhu et al., 2015). Furthermore, some of the phenotypes displayed by the SlELP2L-RNAi lines appear to be different from those of the Atelp mutants. For instance, while ethylene signaling and auxin levels are elevated in the Atelp mutants, both are reduced in the SlELP2L-RNAi lines (Nelissen et al., 2010; Zhu et al., 2015). These differences suggest that the function of Elongator in tomato might not be exactly the same as that in Arabidopsis. Further characterization of genes encoding the Elongator subunits in tomato will not only help in understanding the function of Elongator in plants, but may also identify new strategies for improving disease resistance in tomato.
In this study, we characterized transgenic tomato plants expressing the Arabidopsis AtELP3 and AtELP4 genes. We show that overexpression of AtELP3 and AtELP4 significantly enhanced resistance to tomato bacterial speck caused by the P. syringae pv. tomato strain J4 (Pst J4) without clear detrimental effects on plant growth and development. Interestingly, the enhanced resistance was detected only when plants were inoculated via foliar sprays of bacterial suspensions but not infiltration into the apoplast, suggesting possible involvement of stomatal immunity. However, Pst J4 inoculation did not induce stomatal closure and there were no differences in stomatal apertures and conductance between the transgenic and control plants, indicating that a defense mechanism other than stomatal immunity was activated in the transgenic plants. Indeed, further RNA sequencing (RNA-seq) revealed a group of defense-related genes that were confirmed by real-time quantitative PCR (qPCR) analysis to be induced to higher levels after infection in the AtELP4 transgenic tomato plants than in the control, suggesting that the enhanced disease resistance of the transgenic plants may be attributed to elevated induction of defense responses. Additionally, we show that the tomato genome encodes all six Elongator subunits (SlELPs) and that the tomato SlELP3 and SLELP4 genes complemented the Arabidopsis Atelp3 and Atelp4 mutants, respectively. Thus, the tomato Elongator is most likely functional and AtELP3, AtELP4 as well as their tomato orthologs SlELP3 and SlELP4 could potentially be employed in the improvement of disease resistance in tomato plants.
The T-DNA plasmids (pK7WG2D, 1-AtELP3 and pK7WG2D, 1-AtELP4) reported previously (Silva et al., 2017) were used to transform the tomato cultivar “Moneymaker” following an Agrobacterium tumefaciens-mediated genetic transformation protocol (Lin et al., 2004). The tomato genetic transformation experiment was conducted by the UNL Plant Transformation Facility2. For complementation of the Arabidopsis Atelp3 and Atelp4 mutants, the coding regions of the tomato orthologs (SlELP3 and SlELP4) were amplified from “Moneymaker” cDNAs by PCR using gene specific primers (Supplementary Table S1) and cloned into the binary vector pBI1.4T (Mindrinos et al., 1994). The resulting plasmids were introduced into the A. tumefaciens strain GV3101(pMP90) by electroporation (Shen and Forde, 1989). The Arabidopsis Atelp3-10 and Atelp4/elo1-1 mutant alleles (Nelissen et al., 2005; Defraia et al., 2010), which are in the Columbia (Col-0) and Landsberg erecta ecotype backgrounds, respectively, were used for A. tumefaciens-mediated genetic transformation following the floral dip method (Clough and Bent, 1998).
The T1 transgenic tomato plants obtained from the UNL Plant Transformation Facility were allowed to set seeds. The T2 plants were subjected to PCR analysis using gene-specific primers (Supplementary Table S1) to analyze T-DNA insertion copy numbers based on the expected ratio of 3:1 for a single T-DNA insertion. The transgenic lines that showed the expected ratio for a single T-DNA insertion were kept and seeds from the individual T2 plants were collected separately. The T3 progeny plants from each individual T2 plants were subjected to PCR analysis to identify homozygous plants for each transgenic line. Seeds from the homozygous T3 plants were pooled for further analysis. For Arabidopsis transgenic lines, T2 seeds from individual T1 plants were plated on Murashige and Skoog (MS) medium with 50 μg/mL kanamycin to identify single T-DNA insertion lines based on the segregation ratio of the neomycin phosphotransferase II (nptII) gene. Homozygous plants were similarly identified in the T3 generation.
To evaluate the resistance of the transgenic tomato plants to bacterial speck, a bacterial suspension of Pst J4, adjusted to 1 × 108 colony-forming units (cfu)/mL, was sprayed on 4-week-old tomato plants in pots with a diameter of 10 cm. This inoculum is able to induce consistent levels of disease severity on tomato plants (Kozik and Sobiczewski, 2000). The plants were then immediately covered with bags and a rubber band was placed around the base of the pot to seal the bag in order to maintain high humidity for 40 h. A total of six plants per line were tested and non-transformed “Moneymaker” was included as the control. Inoculated plants were incubated in the growth chamber and maintained at 22°C under a regimen of 12 h dark and 12 h light. The disease symptoms were evaluated 6 days post-inoculation. The disease assessment consisted of the following disease scores: 0 indicates no symptom development; 1 indicates few slightly visible lesions; 2 indicates a significant number of discernible lesions; 3 indicates a higher amount of discernible necrotic and chlorotic lesions; and 4 indicates extensive necrotic and chlorotic lesions and extensive dead tissue.
For quantifying bacterial populations of Pst J4 in the inoculated tomato plants, leaf tissues were sampled every 6 days. Three leaf disks with an area of 1 cm2 were obtained from each transgenic line using a cork borer. The leaf disks were placed into glass tubes and ground in 1 mL of sterile water. The resulting suspensions were diluted by making five 10-fold dilutions. The dilutions were plated on nutrient agar medium and then the plates were incubated at 28°C for 2 days. Colonies typical of P. syringae pv. tomato were counted and the bacterial number per cm2 of leaf tissue was calculated.
For testing growth of the bacterial pathogen Psm ES4326 in Arabidopsis, leaves were infiltrated with a suspension of Psm ES4326 (OD600 = 0.0001) using a 1 mL needleless syringe as described previously (Defraia et al., 2010). Leaf disks were collected from eight leaves 3 days post-inoculation using a cork borer. Each leaf disk was placed into a tube containing 500 μL of 10 mM MgCl2 and ground with a sterile pellet pestle. The resulting suspensions were serially diluted 20-fold four times. The dilutions were then plated on Trypticase Soy Agar medium supplemented with 25 μg/mL streptomycin and incubated at 28°C for 2 days. Colonies that grew on the plates were counted and the bacterial number per leaf disk tissue was calculated.
Tomato plants were sprayed with a Pst J4 bacterial suspension (108 cfu/mL). Stomatal conductance was measured before the inoculation (time 0) and every 30 min post-inoculation using a portable photosynthesis system (LI-6800, LI-COR Biosciences3). The principle of the measurement is that the time required to force a certain volume of air through the plant leaf is inversely proportional to leaf stomatal conductance (Rebetzke et al., 2000). Ten fully expanded leaves per plant were used for the measurement at each time point, and the readings from the abaxial side of the leaves were recorded.
Tomato plants were sprayed with a Pst J4 suspension (108 cfu/mL). Three replicates of leaf tissues from six plants per genotype were collected at 0, 8, and 24 h post-inoculation. Total RNA was extracted from the collected leaf tissues using the RNeasy plant mini kit following the manufacturer’s protocol (Qiagen4). RNA concentration and quality were determined using a Qubit 2.0 Fluorometer (ThermoFisher5) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.6), respectively. Total RNA samples with 28S/18S >1 and RNA integrity number ≥7 were used for RNA-seq analysis. The RNA samples from the three biological replicates were pooled and equal amounts of RNA from the pooled samples were used for RNA-seq library preparation. Briefly, 1 μg of total RNA together with 2 μL of 1:200 diluted ERCC (External RNA Controls Consortium) RNA spike-in mix was used for mRNA extraction with 15 μL of NEBNext Magnetic Oligo d(T)25 and fragmented in NEBNext First Strand Synthesis Buffer by heating at 94°C for 8 min, then followed by first strand cDNA synthesis using reverse transcriptase and random primers. Synthesis of double-stranded cDNA was done using the second strand master mix provided in the kit. The resulting double-stranded cDNA was subjected to end-repair and dA-tailing and then ligated with NEBNext adaptors. Finally, the library was enriched by PCR amplification and purified by Agencourt AMPure beads (Beckman Coulter7). Barcoded libraries were sized and quantitated. qPCR was used to validate the library’s functionality, using the KAPA library quantification kit (Kapa Biosystems8). The six individual samples were pooled equimolarly for one lane of HiSeq 3000 2 × 100 cycles run. Sequencing was performed on the Illumina HiSeq 3000 instrument at the University of Florida Interdisciplinary Center for Biotechnology (UF ICBR) NextGen DNA Sequencing core. The reads that passed Illumina quality control filtering were cleaned up with the Cutadapt program (Martin, 2011) to trim off sequencing adaptors and low quality bases with a quality Phred-like score <20. Reads <40 bases were excluded from RNA-seq analysis. The transcripts of tomato from National Center for Biotechnology Information (NCBI) were used as reference sequences for RNA-seq analysis. The cleaned reads of each sample were mapped independently to the reference sequences using the mapper of bowtie2 with a maximum of three mismatches for each read. The mapping results were processed with the samtools and scripts developed in house at the UF ICBR to remove potential PCR duplicates and select uniquely mapping reads for gene expression estimation. The number of mapped reads for each individual gene was counted. Comparison was made between the AtELP4 transgenic line 61-5 and the control samples collected at the same time point.
For qPCR analysis, total RNA was extracted from tomato plants using Trizol reagent (Thermo Fisher Scientific) and treated with RNAse-free DNAse I (Thermo Fisher Scientific). First strand complementary DNA was synthetized using 10 μg of total RNA with oligo (dT) primer and Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific). Gene-specific primers used for qPCR analysis were listed in Supplementary Table S1. qPCR was performed using ABsolute SYBR Green PCR master mix (Thermo Fisher Scientific) using the SYBR Green protocol (Applied Biosystems9). Reactions were run and analyzed on a MX3000P qPCR system (Agilent10). The relative mRNA levels of the target genes were expressed relative to the tomato Actin gene (Zhou et al., 2014a,b), and calculated using the ΔCT method (Wittwer et al., 2001).
Statistical analyses were performed using one-way ANOVA followed by a Tukey’s multiple comparisons test in Prism 7 (GraphPad Software11).
Sequence data from this article can be found in the Arabidopsis Genome Initiative, the Tomato Genome Sequencing Project, or GenBank/EMBL databases under the following accession numbers: AtELP3 (At5g50320); AtELP4 (At3g11220); PR1b1 (Y08804.1); PR-5x (AY093595); DES (AF317515); ER1 (J04099.1); SlELP1 (Solyc05g054630); SlELP2 (Solyc06g008310); SlELP3 (Solyc03g110910); SlELP4 (Solyc11g010950); SlELP5 (Solyc02g086100); SlELP6 (Solyc12g009500); and NCBI Gene Expression Omnibus Series number GSE97697 (RNA-seq data).
Based on PCR amplification of the cDNA of AtELP3 or AtELP4 using gene specific primers (Supplementary Table S1), out of 80 T1 putative transgenic plants produced by the University of Nebraska–Lincoln (UNL) Plant Transformation Facility, 36 carried the AtELP3 transgene and 35 the AtELP4 gene. Note that the PCR reactions did not amplify any products from the non-transformed “Moneymaker” plants, which were used as the negative control in the experiment, demonstrating the specificity of the primers. The transgenic plants with T-DNA insertion were kept for seed production. Single T-DNA insertion lines were identified in the T2 generation based on the 3:1 segregation ratio expected for a single T-DNA insertion, and homozygosity of the single insertion lines was determined in the T3 generation. In total, five and four single insertion homozygous lines were identified for AtELP3 and AtELP4, respectively. Expression levels of AtELP3 and AtELP4 in the single insertion homozygous lines were determined in the T4 generation by qPCR using gene-specific primers (Supplementary Table S1). While AtELP3 and AtELP4 transcripts were barely detectable in the control plants, the transgenes were expressed at varied levels in the different lines (Figures 1A,B). For AtELP3, expression levels of the transgene were significantly higher in lines 56-9, 60-5, and 51-9 than in lines 51-2, 44-2, and the control plants (Figure 1A). For AtELP4, the transgenic lines could clearly be classified into three groups: one high expresser (line 37-3), two medium expressers (lines 23-1 and 61-5), and one low expresser (line 28-6) (Figure 1B).
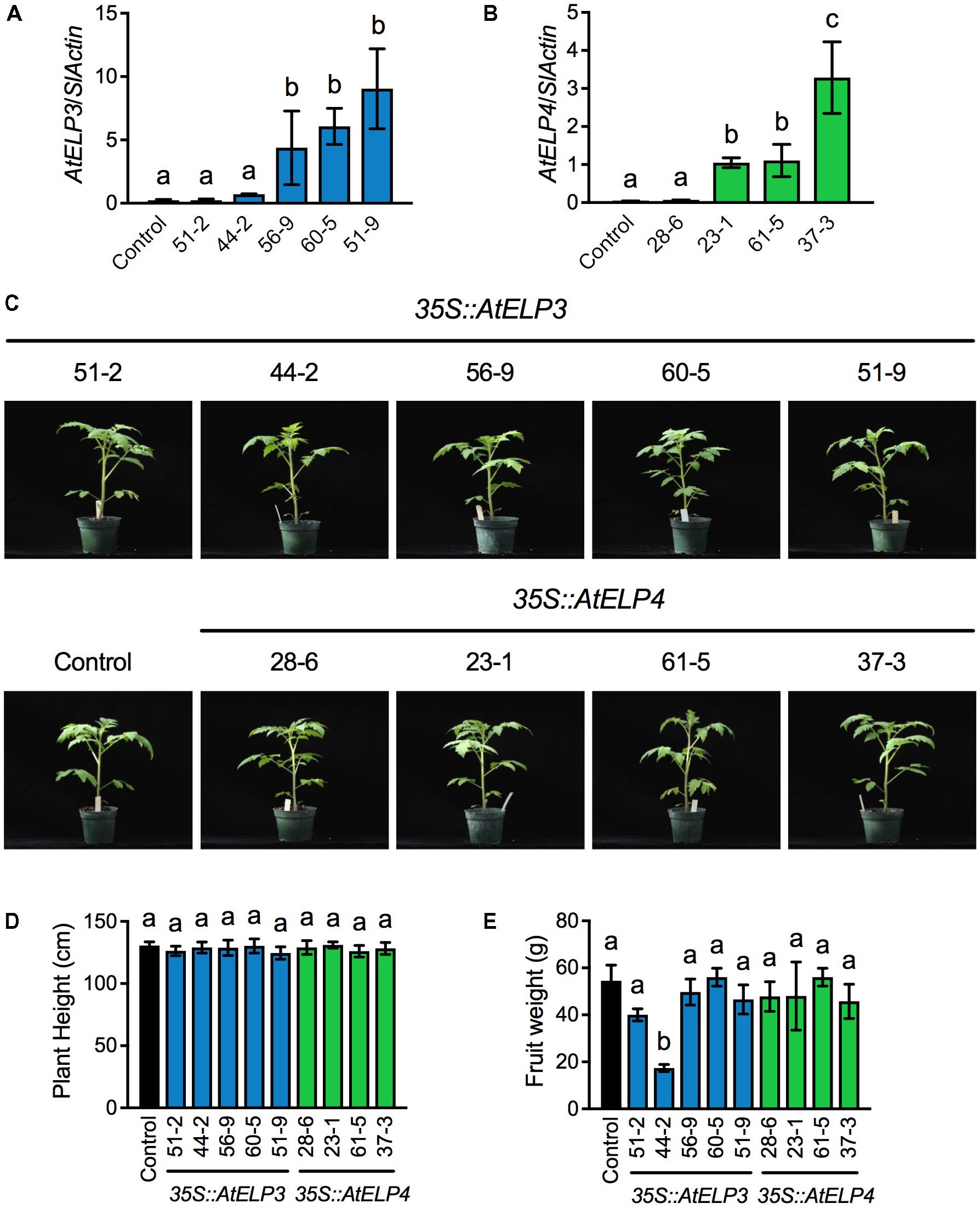
FIGURE 1. Molecular and morphological characterization of transgenic tomato lines expressing AtELP3 and AtELP4. (A,B) Expression levels of AtELP3 (A) and AtELP4 (B) in independent AtELP3 and AtELP4 transgenic lines, respectively. Expression of the transgene was normalized against the constitutively expressed SlActin gene. Almost no AtELP3 and AtELP4 expression was detected in the non-transformed control. Data represent the mean of three biological replicates with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05). (C) Morphological phenotypes of the transgenic tomato lines and the control. Photos were taken 30 days after germination. (D) Plant height of the transgenic lines and the control. Data represent the average of six plants with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05). (E) Fruit weight of the transgenic lines and the control. Data represent the average weight of fruit from six plants with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05).
The overall morphology and development of the transgenic tomato plants were very similar to those of the control plants under standard greenhouse conditions (Figure 1C). There were no significant differences in plant height between the transgenic lines and the control (Figure 1D). Furthermore, all of the transgenic lines formed flowers and fruits. The fruit weight of all the transgenic lines except 44-2 was not significantly different from that of the control (Figure 1E). The fruit from the transgenic line 44-2 was very small and similar to that produced by cherry tomato varieties. The small-fruit phenotype of line 44-2 was unlikely caused by overexpression of AtELP3, since other lines (56-9, 60-5, and 51-9) that expressed higher levels of AtELP3 than line 44-2 did not show such a phenotype. It might be possible that the small-fruit phenotype was caused by a T-DNA insertion mutation. Alternatively, there might be seed contamination during the development of transgenic plants. Nevertheless, these results indicate that transgenic overexpression of AtELP3 and AtELP4 does not affect tomato plant growth and development.
To test whether transgenic overexpression of AtELP3 or AtELP4 in tomato improves disease resistance, we inoculated the single insertion homozygous transgenic lines with the bacterial pathogen Pst J4, which causes bacterial speck on tomato plants. Both leaf infiltration and foliar sprays were employed in the experiment, since the transgenic plants might respond differently to these two commonly used inoculation methods. The bacterial speck disease symptoms, characterized by small, black, or brown necrotic lesions surrounded by a chlorotic halo, appeared 3 days post-inoculation on the transgenic plants and the control for both inoculation methods. When the plants were inoculated using the leaf infiltration method, no significant differences were observed between the bacterial speck disease symptoms developed on any of the transgenic lines and the control, and Pst J4 grew to similar levels in all the tested plants (Supplementary Figure S1). In contrast, when foliar sprays were used, the disease symptoms on different transgenic lines and the control differed drastically. To quantify the disease symptoms, different disease scores were assigned to the transgenic lines based on the disease severity on the leaves (Figures 2A,B). The disease symptoms on the AtELP3 transgenic lines 44-2 and 60-5 were similar to those on the control, the disease symptoms on the AtELP3 transgenic lines 51-2, 56-9, and 51-9 as well as the AtELP4 transgenic lines 28-6 and 37-3 were slightly less severe than those on the control, and the disease symptoms on the AtELP4 transgenic lines 23-1 and 61-5 were much less severe than those on the control (Figure 2B). Interestingly, there was no clear correlation between the disease severity and the expression levels of the transgenes (Figures 1A,B, 2B), which is not without precedent (Luhua et al., 2008). The transgenic line 61-5, which was a medium expresser of AtELP4 (Figure 1B), exhibited the strongest resistance to Pst J4 (Figure 2). The bacterial speck disease progression on the transgenic line 61-5 was markedly slower than that on the control plants (Figure 2C). At 3 days post-inoculation, leaves on the control had already wilted, whereas those on the transgenic line 61-5 still stayed uncurled (Figure 2D). We also determined bacterial titers in the AtELP4 transgenic lines, since, based on disease symptoms, two independent AtELP4 transgenic lines (23-1 and 61-5) displayed clear resistance to Pst J4. Consistent with the observed disease symptoms, the bacterial titers in the transgenic lines 61-5 and 23-1 were the lowest and the second lowest, respectively, among the AtELP4 transgenic lines as well as the control (Figure 2E). These results indicate that overexpression of the AtELP4 gene in tomato is able to significantly improve resistance to Pst J4-caused bacterial speck disease.
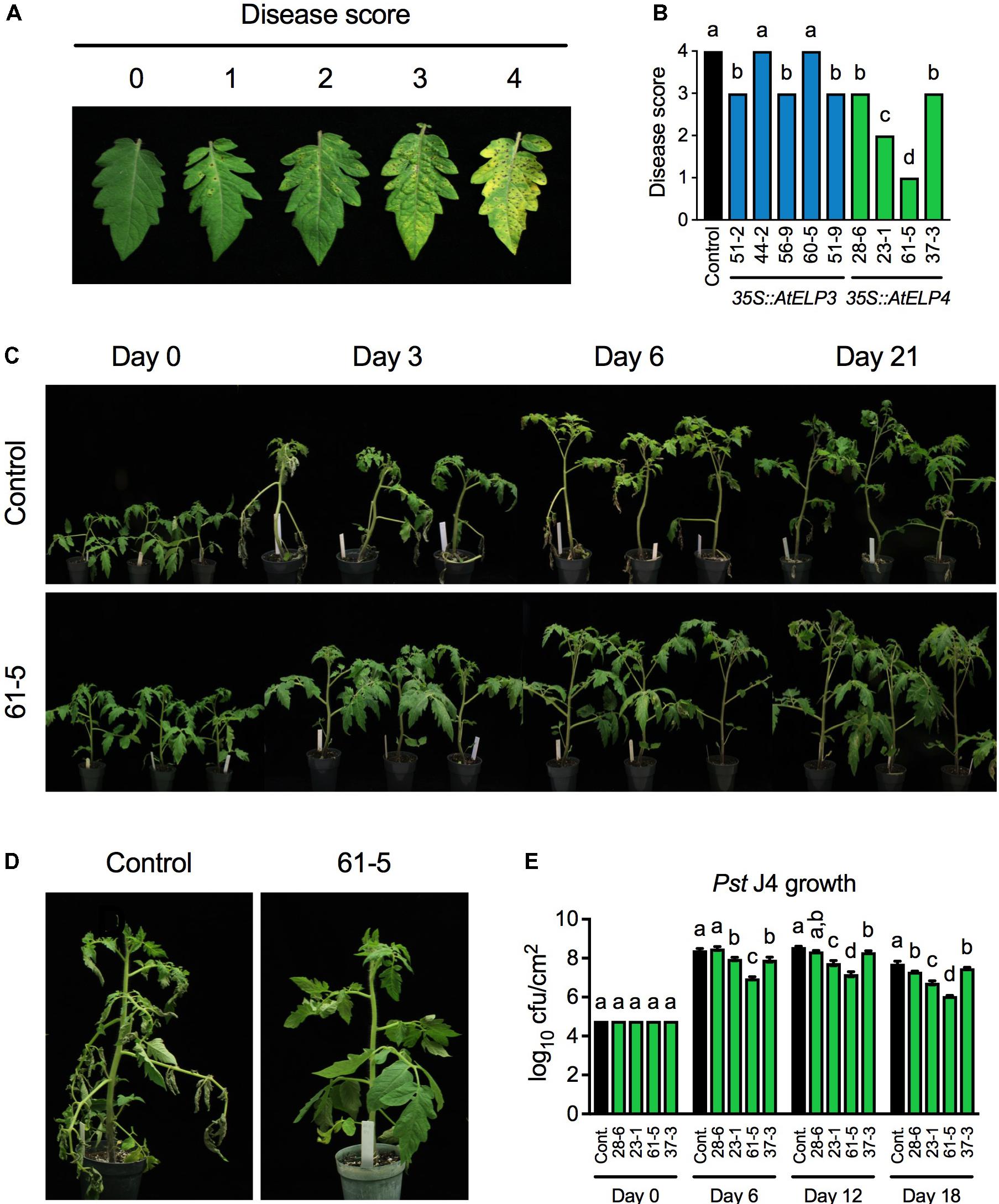
FIGURE 2. Resistance of the transgenic lines to bacterial speck disease caused by Pst J4. (A) Responses on the leaves to which different disease scores were assigned after spray inoculation with Pst J4. Score 0: no symptoms; Score 1: few slightly visible lesions; Score 2: significant number of discernible lesions; Score 3: higher amount of discernible necrotic and chlorotic lesions; and Score 4: extensive necrotic and chlorotic lesions and extensive dead tissue. (B) Disease scores of bacterial speck disease on five AtELP3 transgenic lines, four AtELP4 transgenic lines, and the control. Data represent the mean of six plants with SD. The disease symptoms on each line were highly uniform and the SD values were 0. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05). (C) Bacterial speck disease progression on the most resistant transgenic line 61-5 and the control following spray inoculation with Pst J4. (D) Close-up pictures of the most resistant transgenic line 61-5 and the control at 3 days post-inoculation. (E) Bacterial titers in the AtELP4 transgenic lines and the control. cfu, colony forming unit. Data represent the mean of three biological replicates with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05). Note that the comparison was made separately among the transgenic lines as well the control (cont.) for each time point.
The different responses of the transgenic lines to leaf infiltration and foliar sprays suggested a possible involvement of stomatal immunity (Melotto et al., 2006). To test this possibility, we assessed stomatal morphology and movement after foliar sprays of Pst J4 under optical microscope. Interestingly, inoculation of tomato plants with Pst J4 did not induce stomatal closure and no appreciable differences in stomatal apertures between the most resistant transgenic line 65-1 and the control were observed (Supplementary Figure S2). We further measured stomatal conductance using a portable photosynthesis system. Overall, there were no clear differences in stomatal conductance between the transgenic line 61-5 and the control in a period of 4.5 h following foliar sprays of Pst J4 (Figure 3). Taken together, these results suggest that alteration of stomatal immunity may not be a predominant factor for the enhanced disease resistance observed in the AtELP4 transgenic plants.
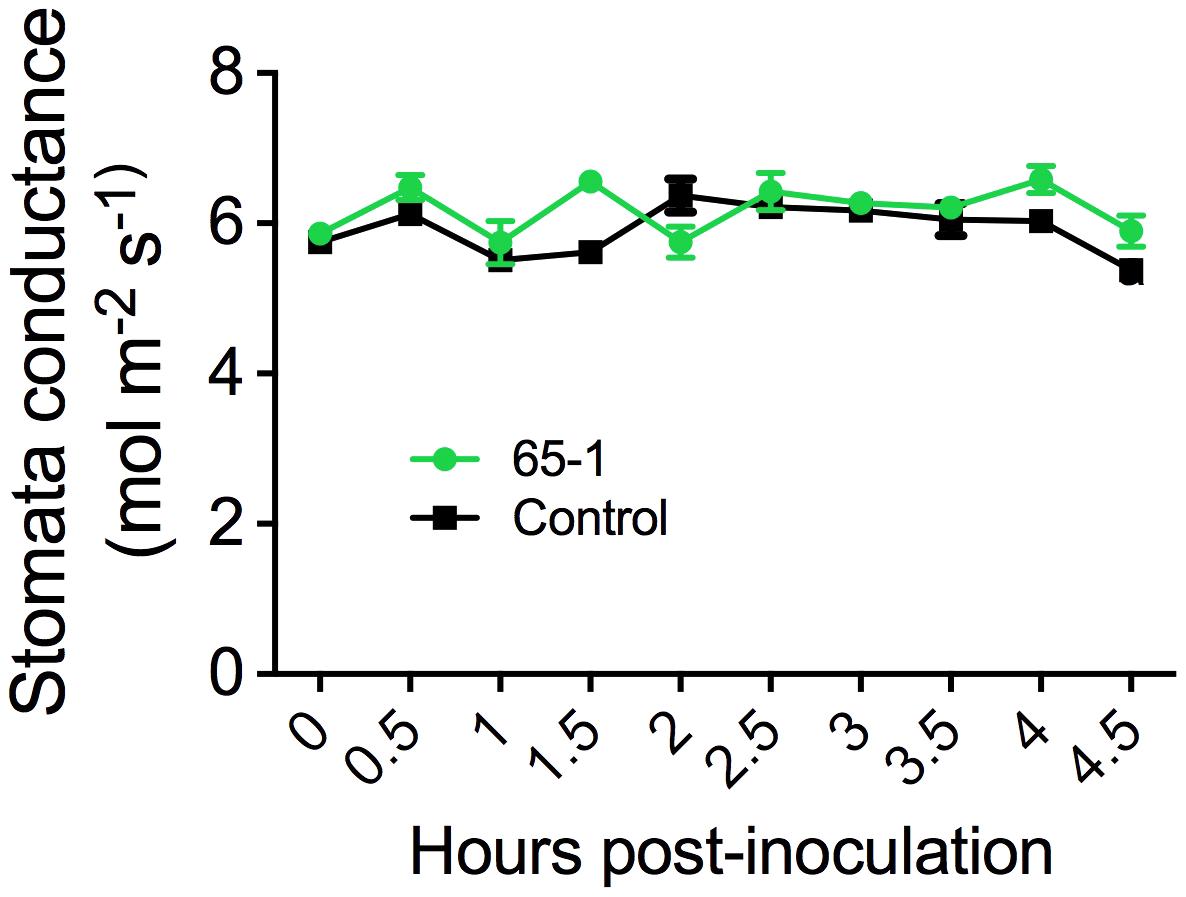
FIGURE 3. Stomatal conductance of the transgenic line 61-5 and the control. The assessment was done every 30 min after spray inoculation with Pst J4. Data represent the mean of five biological replicates with SD.
To uncover potential mechanisms underlying the enhanced disease resistance of the transgenic tomato plants overexpressing AtELP4, we performed an RNA-seq experiment to compare Pst J4-induced transcriptome changes in the transgenic line 61-5 and the control, and then examined genes that were potentially induced to higher levels in the transgenic line 61-5 than in the control (NCBI Gene Expression Omnibus Series number GSE97697). Interestingly, a group of defense-related genes, including PATHOGENESIS-RELATED (PR) gene PR1b1, PR-5 family member PR-5x, DIVINYL ETHER SYNTHASE (DES), and ETHYLENE-RESPONSIVE (ER) PROTEASE INHIBITOR 1 (ER1) (GenBank accession numbers: Y08804.1, AY093595, AF317515, and J04099.1, respectively), which have previously been shown to be associated with disease resistance in tomato (Pautot et al., 1991; Ishihara et al., 2012), were potentially induced to higher levels in the transgenic line 61-5 than in the control. Since the RNA-seq experiment did not include biological replicates, statistical significance could not be evaluated. To confirm the RNA-seq results for the selected genes, we used qPCR to monitor the induction of PR1b1, PR-5x, DES, and ER1 in the transgenic line 61-5 and the control after Pst J4 infection. Consistent with the RNA-seq results, the induction of the four selected genes in the transgenic line 61-5 was significantly higher than that in the control plants (Figure 4). These results suggest that the enhanced disease resistance of the transgenic plants overexpressing AtELP4 may be due to increased induction of defense genes.
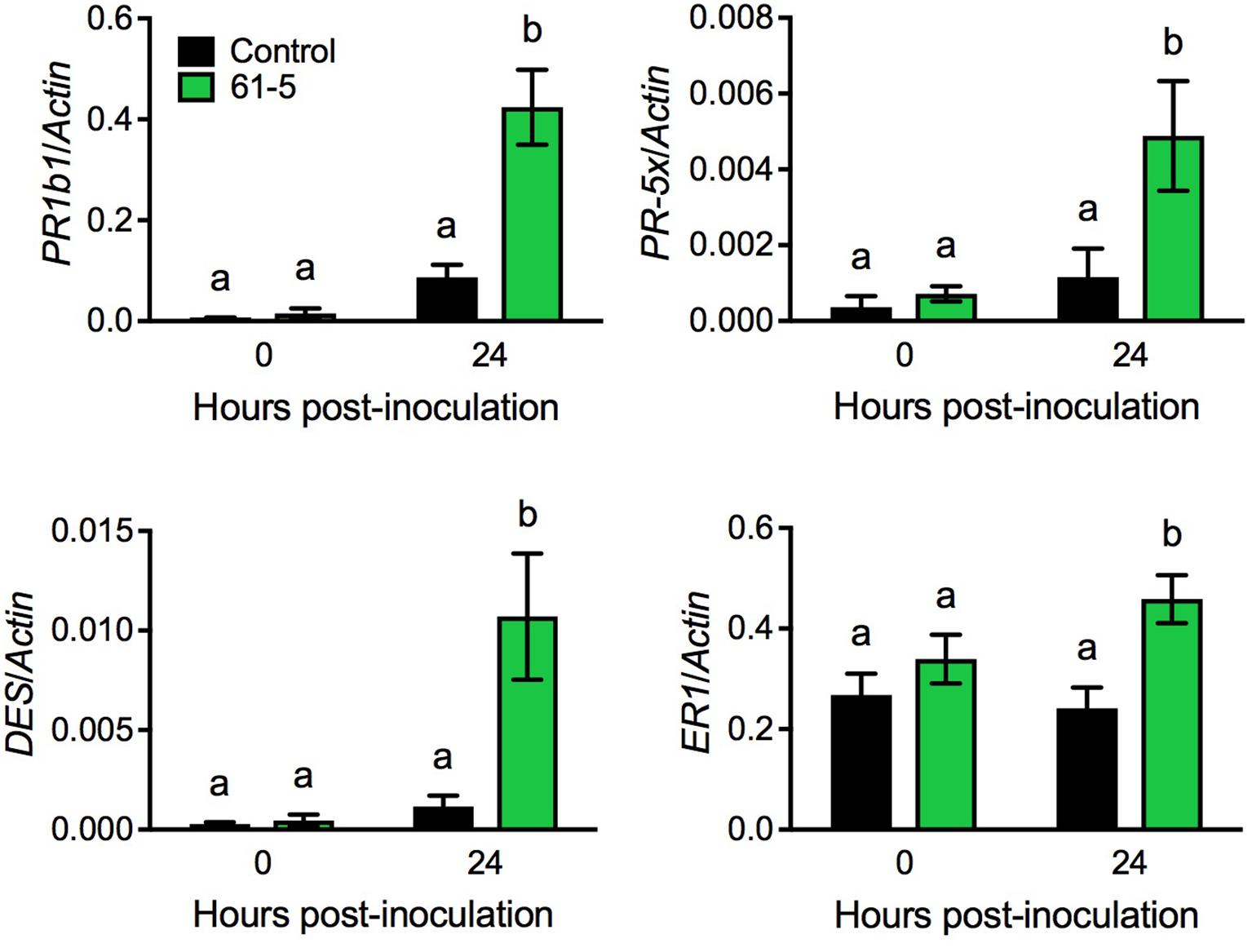
FIGURE 4. Pst J4-induced expression of PR1b1, PR-5x, DES, and ER1 in the transgenic line 61-5. Plants were spray inoculated with the bacterial pathogen Pst J4 (108 cfu/mL). Leaf tissues were collected at the indicated time points. Total RNA was extracted from the inoculated leaves and analyzed for the expression of the indicated genes using qPCR. Expression was normalized against the constitutively expressed Actin gene. Data represent the mean of three biological replicates with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05).
It is generally believed that the Elongator complex is highly conserved in eukaryotes. In agreement with this belief, Zhu et al. (2015) have shown that silencing of a tomato AtELP2-like gene, SlELP2L, resulted in pleiotropic phenotypes similar to those of Atelp mutants. Thus, the tomato genome should encode all SlELPs. To test this, BLAST searches were conducted on the tomato (S. lycopersicum) genome12 using AtELP protein sequences as the queries. The results showed that each subunit of the Elongator complex is encoded by a single-copy gene in the tomato genome. Amino acid sequence alignments indicated that the SlELP proteins all share high identities (>53%) and similarities (>70%) with the AtELP proteins (Supplementary Figure S3). Particularly, SlELP3 has very high amino acid sequence identities (93%) and similarities (96%) with AtELP3, indicating that the Elongator complex catalytic subunits are highly conserved in tomato and Arabidopsis. Therefore, tomato probably has a functional Elongator complex.
To assess the functionality of the SlELP3 and SlELP4 proteins, the SlELP3 and SlELP4 open reading frames driven by the 35S promoter were introduced into the Atelp3 and Atelp4 mutants, respectively, via A. tumefaciens-mediated genetic transformation. Multiple single insertion homozygous lines were obtained for both SlELP3 and SlELP4. Morphologically, the Atelp3 and Atelp4 mutant plants have narrow leaves, long petioles, and shortened siliques (Nelissen et al., 2005). These morphological phenotypes all were completely restored to wild type in the transgenic Atelp3 and Atelp4 plants expressing SlELP3 and SlELP4, respectively (Figures 5, 6). Thus, the functions of SlELP3 and SlELP4 are conserved.
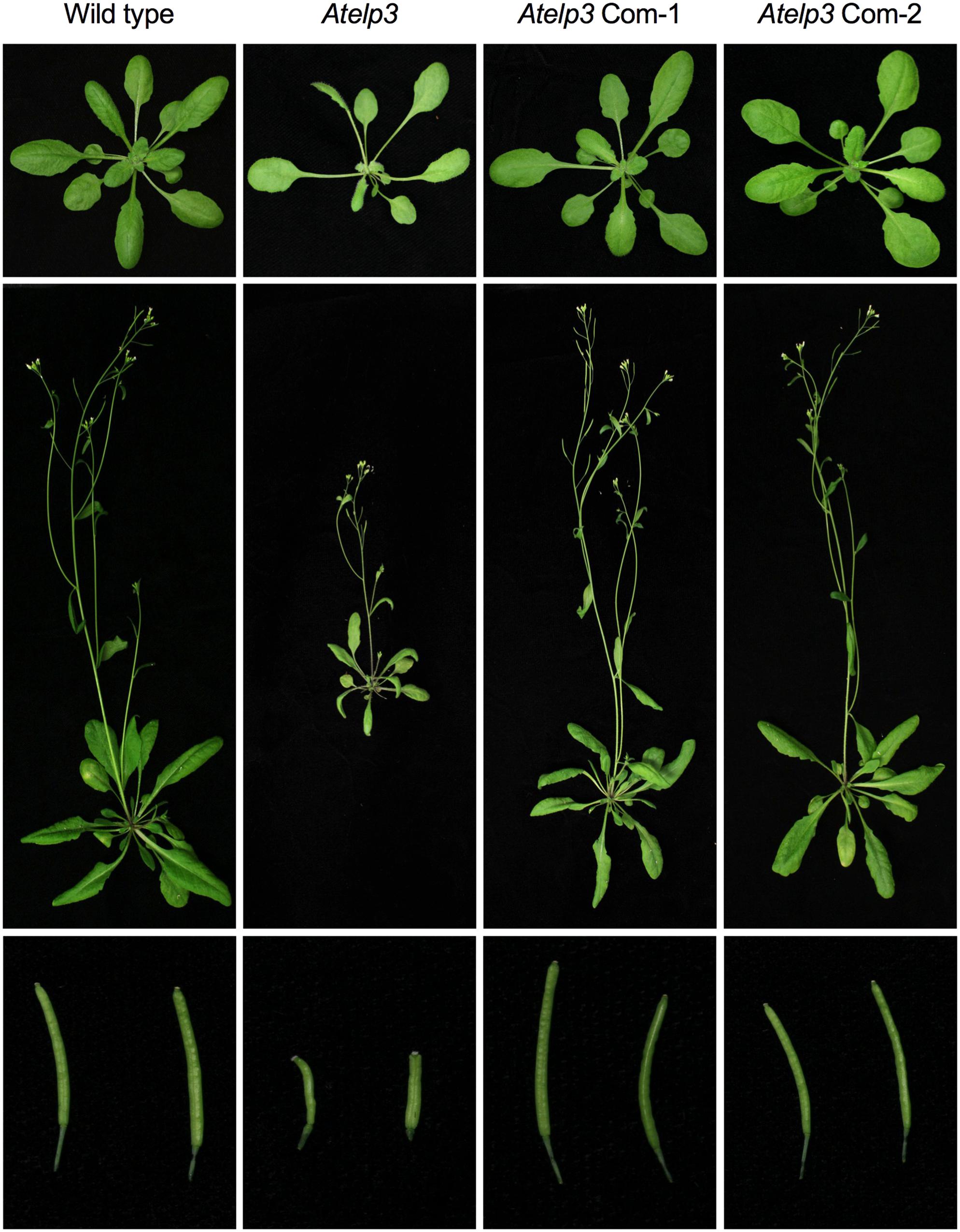
FIGURE 5. Complementation of the Arabidopsis Atelp3 mutant by SlELP3. Morphological phenotypes of Arabidopsis wild type, Atelp3, and two independent complementation lines (35S::SlELP3 Atelp3): Atelp3 Com-1 and Atelp3 Com-2. (Top) 3-weak-old plants; (Middle) 6-week-old plants; (Bottom) siliques.

FIGURE 6. Complementation of the Arabidopsis Atelp4 mutant by SlELP4. Morphological phenotypes of Arabidopsis wild type, Atelp4, and two independent complementation lines (35S::SlELP4 Atelp4): Atelp4 Com-1 and Atelp4 Com-2. (Top) 3-weak-old plants; (Middle) 6-week-old plants; (Bottom) siliques.
To further confirm the morphological complementation results, we inoculated the wild-type Col-0, Atelp3, and two independent Atelp3 complementation lines, Com-1 and Com-2, with the bacterial pathogen P. syringae pv. maculicola (Psm) ES4326 to test whether the enhanced disease susceptibility phenotype of Atelp3 was also complemented by SlELP3 (Defraia et al., 2013). As shown in Figure 7, while the Atelp3 mutant was significantly more susceptible than the wild type to Psm ES4326, the growth of Psm ES4326 in the complementation lines was comparable to that in the wild type. This result indicates that the SlELP3 gene can also complement the defense phenotype of the Atelp3 mutant.
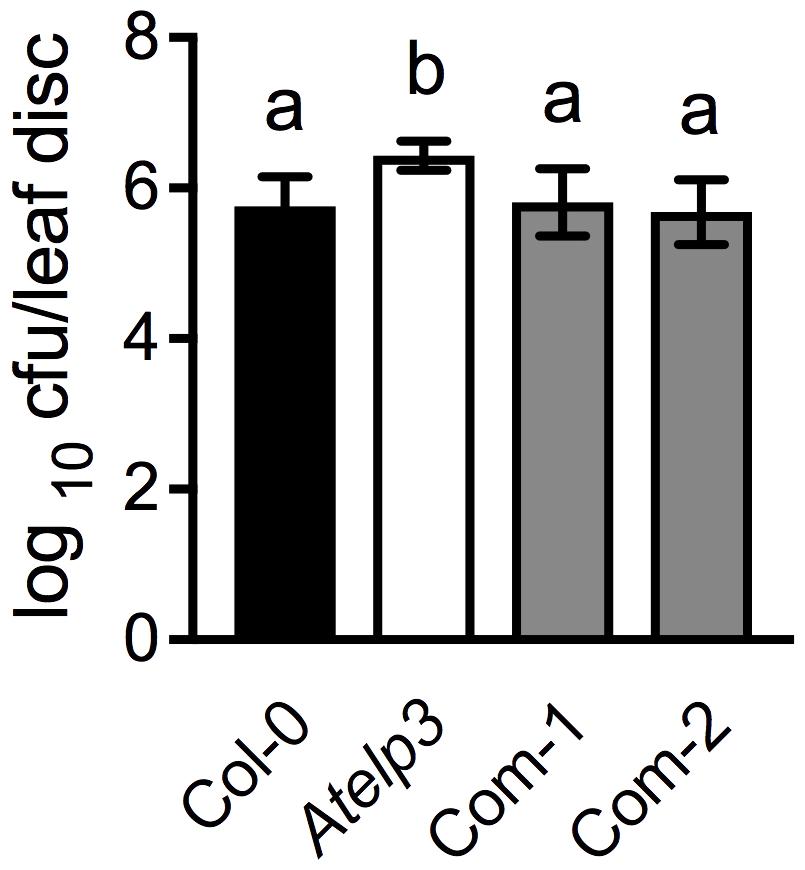
FIGURE 7. Complementation of the disease susceptibility phenotype of the Atelp3 mutant by SlELP3. Four-week-old wild-type Col-0, Atelp3, and two independent complementation lines (Com-1 and Com-2) were inoculated with the bacterial pathogen Psm ES4326 (OD600 = 0.0001). The in planta bacterial titers were determined 3 days post-inoculation. Data represent the mean of eight independent samples with SD. Different letters above the bars indicate significant differences (Tukey’s test, P < 0.05).
Bacterial speck, caused by Pst, is an important disease that concerns tomato growers worldwide (Bashan et al., 1978; Devash et al., 1980; Smitley and McCarter, 1982). Because of the lack of an effective control for the disease (Smitley and McCarter, 1982; Ramos et al., 1989), we investigated the potential of Arabidopsis defense-related genes for improvement of disease resistance against Pst J4 in tomato. The Arabidopsis genes encoding the Elongator subunits were chosen as the candidates for tomato transformation, as their effectiveness in mediating resistance to bacterial diseases has previously been demonstrated in Arabidopsis (Defraia et al., 2010, 2013; Wang et al., 2015; An et al., 2017) and strawberry (Silva et al., 2017). Additionally, given that Elongator functions at the chromatin level and is not directly involved in specific recognition of pathogens (Wang et al., 2013), the possibility of the pathogen’s ability to overcome the resistance is remote, which is critical for generating durable resistance in crop plants.
The aim of this work was to investigate the disease resistance of single insertion homozygous AtELP3 and AtELP4 transgenic tomato plants under growth chamber conditions. In total, we identified nine single insertion homozygous transgenic lines (Figures 1A,B). Although Arabidopsis Elongator mutants display striking morphological phenotypes (Nelissen et al., 2005), overexpression of AtELP3 and AtELP4 in tomato did not cause any abnormality. All transgenic lines except line 44-2 displayed morphological and developmental traits similar to those of the control (Figures 1C–E). The observed small-fruit phenotype of line 44-2 is not associated with the expression of the transgene AtELP3 (Figure 1A), and may thus be caused by a T-DNA insertion mutation or seed contamination during transgenic plant development.
Overexpression of AtELP3 and AtELP4 both improved tomato resistance to bacterial speck, which is in agreement with the result reported in strawberry (Silva et al., 2017). However, the improvement conferred by AtELP3 was marginal (Figure 2B). This result suggests that AtELP3 may not be effective in improving disease resistance in tomato or that the AtELP3 protein levels accumulated in the five AtELP3 transgenic lines used in this study may not be sufficient for activation of strong disease resistance. On the other hand, two AtELP4 transgenic lines (23-1 and 61-5) displayed considerable enhanced resistance to bacterial speck under growth chamber conditions. The bacterial speck disease symptoms on both lines were dramatically alleviated (Figures 2B–D). Consistently, the bacterial populations in these transgenic lines were significantly lower over an 18-day period than those in the control (Figure 2E). We noticed that the enhanced disease resistance was not tightly correlated with the expression levels of the transgene, which is a common phenomenon in transgenic studies (Luhua et al., 2008). Unfortunately, no anti-AtELP4 antibody is available to determine the protein levels in the transgenic lines. Nevertheless, our results indicate that overexpression of AtELP4 in tomato is able to significantly enhance resistance to Pst J4.
Interestingly, increased disease resistance was not observed when the bacterial suspension was infiltrated into the apoplast of the transgenic tomato plants (Supplementary Figure S1). This suggested that AtELP3 and AtELP4 might improve stomatal immunity in tomato. We therefore investigated if stomatal morphology and conductance were affected in the most resistant transgenic line. Surprisingly, inoculation of tomato plants with Pst J4 did not cause stomatal closure (Supplementary Figure S2), suggesting that Pst J4 has some mechanisms to keep stomata open (Cai et al., 2011). Moreover, no clear differences in stomatal morphology and conductance were detected between the transgenic line and the control (Figure 3; Supplementary Figure. S2). Further investigation is thus needed to understand the mechanisms underlying the observed resistance in the AtELP3 and AtELP4 transgenic lines, which is effective only when a spray inoculation method is used.
By using RNA-seq and qPCR, we identified a group of genes (PR1b1, PR-5x, DES, and ER1) that were induced to higher levels after Pst J4 infection in the most resistant AtELP4 transgenic line than in the control (Figure 4). Note that these genes were not constitutively expressed in the transgenic tomato plants, which is different from the constitutive defense gene expression seen in the transgenic strawberry plants (Silva et al., 2017). These tomato genes have been reported to be involved in defense responses to pathogen infections. For instance, PR1b1, PR-5x, and DES have been shown to be associated with resistance to Ralstonia solanacearum in tomato (Ishihara et al., 2012). PR1b1 and PR-5x proteins were also found to accumulate in tomato xylem upon infection by Fusarium oxysporum (Rep et al., 2002), and divinyl ethers, the products of DES, have been reported to accumulate more rapidly in potato cultivars with increased resistance to late blight, a disease caused by Phytophthora infestans (Weber et al., 1999). Furthermore, The ER1 gene has been shown to be associated with bacterial speck disease in tomato (Pautot et al., 1991). Therefore, the enhanced disease resistance in the AtELP4 transgenic tomato plants is likely attributed to elevated induction of defense-related genes. Although it has been well demonstrated that Elongator regulates gene transcription by modifying chromatin structure (histone acetylation and/or DNA methylation) (Wang et al., 2013, 2015), whether overexpression of AtELP4 in tomato alters the chromatin structure of the identified defense-related genes requires further investigation.
It is interesting that overexpression of a single Elongator subunit can dramatically improve tomato disease resistance. This phenomenon has also been seen in strawberry where overexpression of AtELP3 or AtELP4 significantly increased resistance to several bacterial and fungal pathogens (Silva et al., 2017). Such results appear to be in conflict with the notion that the Elongator complex functions as a whole and mutations in any of the subunits compromise the activity of the complex (Versées et al., 2010; An et al., 2016). However, it has been shown that overexpression of ELP3, but not ELP4, in human 293 T cells suppressed cell growth and enhanced transcription, and that overexpression of ELP4 and ELP3 together exhibited a synergistic effect on transcription activation (Gu et al., 2009). Moreover, elevating ELP3 expression in yeast suppressed the anaphase-promoting complex 5 mutant defects (Turner et al., 2010). These results together strongly suggest that individual Elongator subunits may either be able to increase the Elongator complex activity or have some Elongator complex-independent functions. Further investigation is clearly warranted to pinpoint the underlying molecular mechanisms.
Tomato appears to have a functionally conserved Elongator complex. The tomato genome contains single-copy genes encoding all SlELPs and the SlELP proteins share high identities and similarities with their corresponding AtELP proteins (Supplementary Figure S3). Furthermore, tomato orthologs of the Arabidopsis AtELP3 and AtELP4 genes, when transformed into the Arabidopsis Atelp3 and Atelp4 mutants, were able to restore wild-type morphology to the mutants (Figures 5, 6). Resistance to Psm ES4326 was also completely restored in the Atelp3 mutant plants expressing the SlELP3 gene (Figure 7). These results indicate that the functions of SlELP3 and SlELP4 are conserved and suggest that the function of the Elongator complex may be conserved in tomato. Indeed, silencing of SlELP2 in tomato plants resulted in pleiotropic phenotypes similar to those of the Atelp mutants (Zhu et al., 2015). These results taken together indicate that the tomato Elongator complex not only is essential for plant fitness, but may also play an important roles in immunity. Although it has been shown that Elongator modulates the transcription of thousands of genes in Arabidopsis and Saccharomyces cerevisiae (Krogan and Greenblatt, 2001; Wang et al., 2013), how Elongator functions in tomato remains to be elucidated.
This study, together with our previous study (Silva et al., 2017), revealed several dramatic differences between the transgenic tomato and transgenic strawberry AtELP3 and AtELP4 plants. First, overexpression of AtELP3 and AtELP4 did not clearly impact tomato plant growth and development, which is in sharp contrast to the collateral effects observed in strawberry. Second, overexpression of AtELP3 and AtELP4 conferred resistance in tomato to bacterial speck caused by Pst J4 only when inoculated through foliar sprays but not through infiltration into the leaf apoplast, but in strawberry it provided resistance to the angular leaf spot-causing bacterial pathogen Xanthomonas fragariae when the pathogen was infiltrated into the apoplast. And finally, the elevated resistance in tomato is likely attributed to a stronger induction of defense responses in the transgenic plants than in the control, whereas in the transgenic strawberry plants resistance was associated with constitutive expression of defense genes. These results suggest that different plant species may respond differently to overexpression of genes encoding the subunits of the evolutionarily conserved Elongator complex. Further investigations are required to fully understand this interesting phenomenon.
JP, JJ, and ZM designed the research and wrote the manuscript. JP characterized transgenic plants and conducted qPCR assay. YZ performed RNA-seq analysis. FY analyzed RNA-seq data.
This work was supported by a doctoral fellowship to JP from CNPq (Brazilian National Council for Scientific and Technological Development – Grant Procs. 245345/2012-4) and grants from the Florida Tomato Committee to JJ and ZM.
ZM is a co-inventor on a patent application titled “Use of Elongator genes to improve plant disease resistance.”The remaining authors declare that the research was conducted in the absence of any other commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the University of Nebraska–Lincoln Plant Transformation Facility for generation of transgenic tomato plants, Mr. Gerald Minsavage for assistance with greenhouse work and bacterial population assays, Dr. Chenggang Wang for assistance with qPCR analysis, Ms. Xudong Zhang for assistance with T-DNA vector construction, and Dr. Sixue Chen (University of Florida) for access to the LI-6800 Portable Photosynthesis System.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01066/full#supplementary-material
An, C., Ding, Y., Zhang, X., Wang, C., and Mou, Z. (2016). Elongator plays a positive role in exogenous NAD-induced defense responses in Arabidopsis. Mol. Plant Microbe Interact. 29, 396–404. doi: 10.1094/MPMI-01-16-0005-R
An, C., Wang, C., and Mou, Z. (2017). The Arabidopsis Elongator complex is required for nonhost resistance against the bacterial pathohen Xanthomonas citri subsp. citri and Pseudomonas syringae pv. Phaseolicola NPS3121. New Phytol. 214, 1245–1259. doi: 10.1111/nph.14442
Bashan, Y., Okon, Y., and Henis, Y. (1978). Infection studies of Pseudomonas tomato causal agent of bacterial speck of tomato. Phytoparasitica 6, 135–143. doi: 10.1007/BF02981213
Cai, R., Lewis, J., Yan, S., Liu, H., Clarke, C. R., Campanile, F., et al. (2011). The plant pathogen Pseudomonsa syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 7:e1002130. doi: 10.1371/journal.ppat.1002130
Chan, Y. L., Prasad, V., Sanjaya, Chen, K. H., Liu, P. C., Chan, M. T., et al. (2005). Transgenic tomato plants expressing an Arabidopsis thionin (Thi2.1) driven by fruit-inactive promoter battle against phytopathogenic attack. Planta 221, 386–393. doi: 10.1007/s00425-004-1459-3
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Creppe, C., Malinouskaya, L., Volvert, M. L., Gillard, M., Close, P., Malaise, O., et al. (2009). Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136, 551–564. doi: 10.1016/j.cell.2008.11.043
Defraia, C. T., Wang, Y., Yao, J., and Mou, Z. (2013). Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 13:102. doi: 10.1186/1471-2229-13-102
Defraia, C. T., Zhang, X., and Mou, Z. (2010). Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 64, 511–523. doi: 10.1111/j.1365-313X.2010.04345.x
Devash, Y., Okon, Y., and Henis, Y. (1980). Survival of Pseudomonas tomato in soil and seeds. Phytopathology 99, 175–185. doi: 10.1111/j.1439-0434.1980.tb03777.x
Ding, Y., and Mou, Z. (2015). Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front. Plant Sci. 6:296. doi: 10.3389/fpls.2015.00296
Esberg, A., Huang, B., Johansson, M. J., and Bystrom, A. S. (2006). Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148. doi: 10.1016/j.molcel.2006.07.031
Fang, X., Cui, Y., Li, Y., and Qi, Y. (2015). Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1:15075. doi: 10.1038/nplants.2015.75
Glatt, S., Letoquart, J., Faux, C., Taylor, N. M., Seraphin, B., and Muller, C. W. (2012). The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 19, 314–320. doi: 10.1038/nsmb.2234
Gu, J., Sun, D., Zheng, Q., Wang, X., Yang, H., Miao, J., et al. (2009). Human Elongator complex is involved in cell cycle and suppresses cell growth in 293T human embryonic kidney cells. Acta Biochim. Biophys. Sin. 41, 831–838. doi: 10.1093/abbs/gmp072
Hawkes, N. A., Otero, G., Winkler, G. S., Marshall, N., Dahmus, M. E., Krappmann, D., et al. (2002). Purification and characterization of the human elongator complex. J. Biol. Chem. 277, 3047–3052. doi: 10.1074/jbc.M110445200
Huang, B., Johansson, M. J., and Bystrom, A. S. (2005). An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11, 424–436. doi: 10.1261/rna.7247705
Ishihara, T., Mitsuhara, I., Takahashi, H., and Nakaho, K. (2012). Transcriptome analysis of quantitative resistance-specific response upon Ralstonia solanacearum infection in tomato. PLoS One 7:e46763. doi: 10.1371/journal.pone.0046763
Jones, J. D., Witek, K., Verweij, W., Jupe, F., Cooke, D., Dorling, S., et al. (2014). Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130087. doi: 10.1098/rstb.2013.0087
Kozik, E. U., and Sobiczewski, P. (2000). Response of tomato genotypes to bacterial speck (Pesudomonas syringae pv. tomato). Acta Physiol. Plant 22, 243–246. doi: 10.1007/s11738-000-0021-6
Krogan, N. J., and Greenblatt, J. F. (2001). Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., van Esse, H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Lin, W. C., Lu, C. F., Wu, J. W., Cheng, M. L., Lin, Y. M., Yang, N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13, 567–581. doi: 10.1007/s11248-004-2375-9
Lin, Z., Zhao, W., Diao, W., Xie, X., Wang, Z., Zhang, J., et al. (2012). Crystal structure of elongator subcomplex Elp4-6. J. Biol. Chem. 287, 21501–21508. doi: 10.1074/jbc.M112.341560
Luhua, S., Ciftci-Yilmaz, S., Harper, J., Cushman, J., and Mittler, R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 148, 280–292. doi: 10.1104/pp.108.124875
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F. M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rice repeats. Cell 78, 1089–1099. doi: 10.1016/0092-8674(94)90282-8
Monroe, J. G., and Sasser, M. (1980). Prevention-the key to controlling bacterial spot and bacterial speck of tomato. Plant Dis. 64, 831–834. doi: 10.1094/PD-64-831
Nelissen, H., De Groeve, S., Fleury, D., Neyt, P., Bruno, L., Bitonti, M. B., et al. (2010). Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. U.S.A. 107, 1678–1683. doi: 10.1073/pnas.0913559107
Nelissen, H., Fleury, D., Bruno, L., Robles, P., De Veylder, L., Traas, J., et al. (2005). The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. U.S.A. 102, 7754–7759. doi: 10.1073/pnas.0502600102
Okada, Y., Yamagata, K., Hong, K., Wakayama, T., and Zhang, Y. (2010). A role for the elongator complex in zygotic paternal genome demethylation. Nature 463, 554–558. doi: 10.1038/nature08732
Otero, G., Fellows, J., Li, Y., de Bizemont, T., Dirac, A. M., Gustafsson, C. M., et al. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118. doi: 10.1016/S1097-2765(00)80179-3
Pautot, V., Holzer, F. M., and Walling, L. L. (1991). Differential expression of tomato proteinase inhibitor I and II genes during bacterial pathogen invasion and wounding. Mol. Plant Microbe Interact. 4, 284–292. doi: 10.1094/MPMI-4-284
Pedley, K. F., and Martin, G. B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. doi: 10.1146/annurev.phyto.41.121602.143032
Piquerez, S. J., Harvey, S. E., Beynon, J. L., and Ntoukakis, V. (2014). Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front. Plant Sci. 5:671. doi: 10.3389/fpls.2014.00671
Rahl, P. B., Chen, C. Z., and Collins, R. N. (2005). Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17, 841–853. doi: 10.1016/j.molcel.2005.02.018
Ramos, R. S., Sinigaglia, C., and Chiba, S. (1989). Chemical control of bacterial speck (Pseudomonas syringae pv. tomato) of tomato. Biologico 55, 1–3.
Rebetzke, G. J., Read, J. J., Barbour, M. M., Condon, A. G., and Rawson, H. M. (2000). A hand-held porometer for rapid assessment of leaf conductance in wheat. Crop Sci. 40, 277–280. doi: 10.2135/cropsci2000.401277x
Rep, M., Dekker, H. L., Vossen, J. H., de Boer, A. D., Houterman, P. M., Speijer, D., et al. (2002). Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 130, 904–917. doi: 10.1104/pp.007427
Schwessinger, B., Bahar, O., Thomas, N., Holton, N., Nekrasov, V., Ruan, D., et al. (2015). Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 11:e1004809. doi: 10.1371/journal.ppat.1004809
Shen, W. J., and Forde, B. G. (1989). Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res. 17:8385. doi: 10.1093/nar/17.20.8385
Silva, K. J. P., Brunings, A. M., Pereira, J. A., Peres, N. A., Folta, K. M., and Mou, Z. (2017). The Arabidopsis ELP3/ELO3 and ELP4/ELO1 genes enhance disease resistance in Fragaria vesca L. BMC Plant Biol. 17:230. doi: 10.1186/s12870-017-1173-5
Smitley, D. R., and McCarter, S. M. (1982). Spread of Pseudomonas syringae pv. tomato and role of epiphytic populations and environmental conditions in disease development. Plant Dis. 66, 713–717. doi: 10.1094/PD-66-713
Svejstrup, J. Q. (2007). Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19, 331–336. doi: 10.1016/j.ceb.2007.04.005
Thapa, S. P., and Coaker, G. (2016). Genome sequences of two Pseudomonas syringae pv. tomato race 1 strains, Isolated from tomato fields in California. Genome Announc. 4:e1671-15. doi: 10.1128/genomeA.01671-15
Turner, E. L., Malo, M. E., Pisclevich, M. G., Dash, M. D., Davies, G. F., Arnason, T. G., et al. (2010). The Saccharomyces cerevisiae anaphase-promoting complex interacts with multiple histone-modifying enzymes to regulate cell cycle progression. Eukaryot. Cell 9, 1418–1431. doi: 10.1128/EC.00097-10
Versées, W., De Groeve, S., and Van Lijsebettens, M. (2010). Elongator, a conserved multitasking complex? Mol. Microbiol. 76, 1065–1069. doi: 10.1111/j.1365-2958.2010.07162.x
Wang, C., Ding, Y., Yao, J., Zhang, Y., Sun, Y., Colee, J., et al. (2015). Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 83, 1019–1033. doi: 10.1111/tpj.12946
Wang, Y., An, C., Zhang, X., Yao, J., Zhang, Y., Sun, Y., et al. (2013). The Arabidopsis Elongator complex subunit2 epigenetically regulates plant immune respones. Plant Cell 25, 762–776. doi: 10.1105/tpc.113.109116
Weber, H., Chetelat, A., Caldelari, D., and Farmer, E. E. (1999). Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 11, 485–494. doi: 10.2307/3870875
Winkler, G. S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J. Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 3517–3522. doi: 10.1073/pnas.022042899
Wittschieben, B. O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., et al. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128. doi: 10.1016/S1097-2765(00)80194-X
Wittwer, C. T., Herrmann, M. G., Gundry, C. N., and Elenitoba-Johnson, K. S. (2001). Real-time multiplex PCR assays. Methods 25, 430–442. doi: 10.1006/meth.2001.1265
Woloszynska, M., Le Gall, S., and Van Lijsebettens, M. (2016). Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim. Biophys. Acta 1859, 1025–1033. doi: 10.1016/j.bbagrm.2016.06.008
Yunis, H., Bashan, Y., Okon, Y., and Henis, Y. (1980). Weather dependence, yield losses, and control of bacterial speck of tomato caused by Pseudomonas tomato. Plant Dis. 64, 937–939. doi: 10.1094/PD-64-937
Zhou, J., Wang, J., Yu, J. Q., and Chen, Z. (2014a). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5:174. doi: 10.3389/fpls.2014.00174
Zhou, J., Xia, X. J., Zhou, Y. H., Shi, K., Chen, Z., and Yu, J. Q. (2014b). RBOH1-dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J. Exp. Bot. 65, 595–607. doi: 10.1093/jxb/ert404
Keywords: tomato, the Elongator complex, AtELP4, transgenic overexpression, disease resistance, Pseudomonas syringae
Citation: Pereira JA, Yu F, Zhang Y, Jones JB and Mou Z (2018) The Arabidopsis Elongator Subunit ELP3 and ELP4 Confer Resistance to Bacterial Speck in Tomato. Front. Plant Sci. 9:1066. doi: 10.3389/fpls.2018.01066
Received: 09 May 2018; Accepted: 29 June 2018;
Published: 24 July 2018.
Edited by:
Zhengqing Fu, University of South Carolina, United StatesReviewed by:
Yuxin Hu, Institute of Botany (CAS), ChinaCopyright © 2018 Pereira, Yu, Zhang, Jones and Mou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey B. Jones, amJqb25lc0B1ZmwuZWR1 Zhonglin Mou, emhsbW91QHVmbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.