
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 July 2018
Sec. Plant Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01048
This article is part of the Research Topic New Insights into Mechanisms of Epigenetic Modifiers in Plant Growth and Development View all 15 articles
Epigenetic regulation is referred to as changes in gene function that do not involve changes in the DNA sequence, it is usually accomplished by DNA methylation, histone modifications (repressive marks such as H3K9me, H3K27me, H2Aub, or active marks such as H3K4me, H3K36me, H3Ac), and chromatin remodeling (nucleosome composition, occupancy, and location). In plants, the shoot apex produces different lateral organs during development to give rise to distinguishable phases of a juvenile, an adult and a reproductive phase after embryogenesis. The juvenile-to-adult transition is a key developmental event in plant life cycle, and it is regulated by a decrease in the expression of a conserved microRNA-miR156/157, and a corresponding increase in the expression of its target genes encoding a set of plant specific SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) proteins. Recent work has revealed that the miR156/157-SPL pathway is the master regulator of juvenile-to-adult transition in plants, and genes in this pathway are subjected to epigenetic regulation, such as DNA methylation, histone modifications, and chromatin remodeling. In this review, we summarized the recent progress in understanding the epigenetic regulation of the miR156/157-SPL pathway during juvenile-to-adult transition and bring forward some perspectives of future research in this field.
Unlike mammals, in which organ formation is completed during embryonic development, plants produce new organs from self-sustaining stem cell populations known as meristems in different developmental processes. In plants, post-embryonic development can be divided into a juvenile vegetative phase, an adult vegetative phase and a reproductive phase, and each developmental phase is marked by changes in a series of distinct phase-specific traits (Poethig, 1990; Kerstetter and Poethig, 1998). The transition from the juvenile vegetative phase to the adult vegetative phase was referred to as the juvenile-to-adult transition or vegetative phase change.
In Arabidopsis, the juvenile-to-adult transition is characterized by the formation of leaf abaxial trichomes, an increase in leaf length/width ratio and serration, and a decrease in cell size (Telfer et al., 1997; Tsukaya et al., 2000; Usami et al., 2009). Genetic and molecular analyses demonstrated that the conserved miRNA-miR156/157 and its target genes-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes act sequentially with miR172, another miRNA that targets a class of AP2-like transcription factors (TFs), to regulate juvenile-to-adult transition in plants (Wu and Poethig, 2006; Wu et al., 2009; He et al., 2018). miR156/157 is highly expressed in juvenile phase and its abundance declines gradually, while its target SPL genes increases during shoot development. miR156/157 negatively regulates SPL gene expression through transcript cleavage or translational inhibition. SPLs were also responsive to photoperiodic induction and exhibited an miR156/157-independent expression pattern (Schmid et al., 2003; Jung et al., 2012). Therefore, the outcome of SPL levels fine-tuned by both miR156/157 and exogeneous cues orchestrates the timing of juvenile-to-adult transition (Huijser and Schmid, 2011; Poethig, 2013).
The Arabidopsis genome encodes eight miR156 genes (MIR156A∼H) and four miR157 genes (MIR156A∼D), and those genes function redundantly. The mir156a mir156c double mutant exhibited a similar phenotype to the 35S::MIMICRY156 transgenic plants with significantly reduced levels of miR156, which indicates that MIR156A and MIR156C are the two main loci contributing to the level of miR156 and have dominant roles in vegetative phase change within the miR156 family in Arabidopsis (Yang L. et al., 2013; Yu et al., 2013). miR157 functions redundantly with miR156, but has a much smaller effect on shoot morphology and SPL gene expression than miR156 (He et al., 2018). miR156/157 targets 10 out of 16 different SPL genes in Arabidopsis. Based on the amino acid sequence of the SBP domain, the miR156/157-targeted SPL genes can be classified into five clades, SPL3/SPL4/SPL5, SPL9/SPL15, SPL2/SPL10/SPL11, SPL6, and SPL13A/B (Xie et al., 2006; Riese et al., 2007; Preston and Hileman, 2013). Genetic and functional analysis of the role of SPL genes in vegetative phase change indicated that SPL2/SPL9/SPL10/SPL11/SPL13/SPL15, but not SPL3/4/5/6, contribute to the juvenile-to-adult transition with SPL9/SPL13/SPL15 being more important for juvenile-to-adult transition than SPL2/SPL10/SPL11 (Xu et al., 2016a).
As the master regulator of the juvenile-to-adult transition, miR156/157-SPL pathway has been shown to be subjected to transcriptional and post-transcriptional regulation. Those include the transcriptional regulation of pri-MIR156/157 and SPLs genes, the regulation of miR156/157 biogenesis, and post-transcriptional regulation of SPL genes (Figure 1). Here, we review our current understanding of epigenetic regulation of the miR156/157-SPL pathway and the roles of corresponding players in juvenile-to-adult transition in plants.
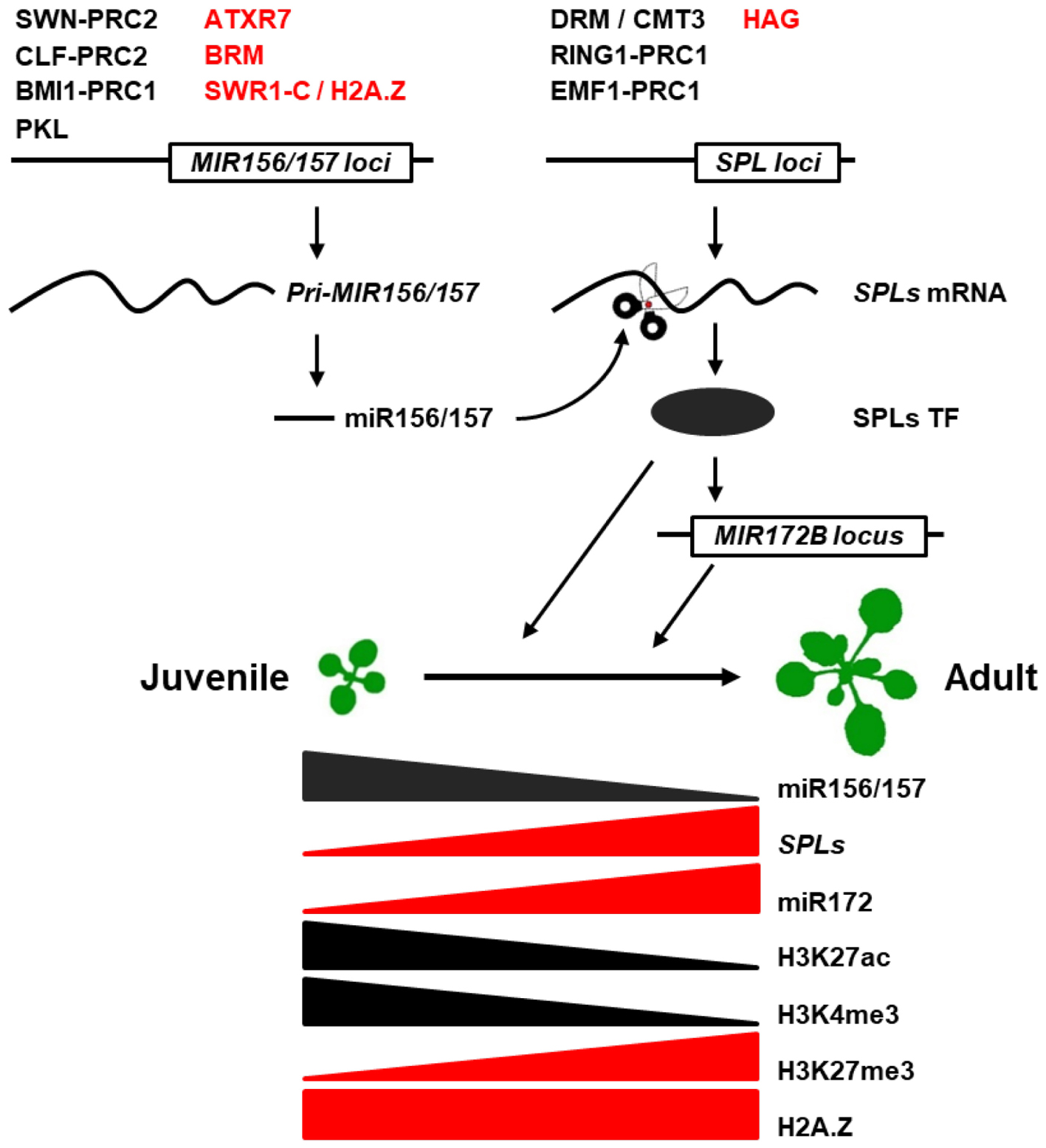
FIGURE 1. Epigenetic regulation of juvenile-to-adult transition in plants. Active and repressive epigenetic regulators were marked in red and black at MIR156/157 and SPL loci, respectively. Triangle indicates gradual increase or decrease in the epigenetic modification levels of MIR156 loci.
DNA methylation [5-Methylcytosine (5mC)] is a hallmark of epigenetic gene silencing in both plants and mammals (Feng et al., 2010; Law and Jacobsen, 2010). DNA methylation is found at CG or non-CG sites including CHH and CHG (H represents A, T, or C) in plants in contrast to CG sites only in mammals (Henderson and Jacobsen, 2007; Cokus et al., 2008). In plants, CG methylation is carried out by DNA METHYLTRANSFERASE 1 (MET1), whereas DOMAINS-REARRANGED METHYLTRANSFERASEs (DRM) and CHROMOMETHYLASE 3 (CMT3) are responsible for the non-CG methylation (Law and Jacobsen, 2010).
The first indication of DNA methylation plays a role in phases of shoot development comes from the work done by Brink. In the 1950s, Brink noticed the similarity between phase change in plants and changes in cell states in non-plant organisms, he proposed that phases of shoot development might be regulated by reversible changes in chromatin based on his research on paramutation in maize (Brink, 1962). Subsequent work on Spm transposable elements (Banks and Fedoroff, 1989) and the Robertson’s Mutator (Mu) element (Martienssen et al., 1990) suggest that DNA methylation may be the underlying mechanism for maintaining phases of shoot development in plants. Recent work in peach also demonstrated that levels of nuclear DNA methylation was higher in adult meristems than that in juvenile and juvenile-like meristems (Bitonti et al., 2002), and an increase in DNA methylation during development seems widespread in plants (Fraga et al., 2002; Ruiz-García et al., 2005). In Arabidopsis, the triple DNA methyltransferase mutant drm1 drm2 cmt3 exhibited a developmental retardation phenotype (Cao and Jacobsen, 2002), indicating that DNA methylation is important for normal growth and development in plants. However, genome-wide DNA methylation analysis of 5-week-old Columbia wild type, met1 and drm1 drm2 cmt3 triple mutant (Zhang et al., 2006), and 25-day-old Columbia wild type (Zilberman et al., 2007) indicated that only the coding sequence of the SPL10 gene contains non-CG methylation. These results suggest that genes upstream or downstream of the miR156/157-SPL pathway, instead of miR156/157 or SPL genes, might be regulated by DNA methylation. Therefore, phenotypic characterization of vegetative phase change phenotype of mutants of DNA methyltransferases (MET1, DRM, and DNMT2) or demethylation enzymes (ROS1, DME, DML2, and DML3), as well as bisulfite sequencing of MIR156/157 and SPLs loci, will facilitate to uncover the role of DNA methylation in regulation of miR156/157-SPL pathway and juvenile-to-adult transition in plants.
Histone modification at specific lysine sites functions as transcription repressive marks such as H3K9me, H3K27me, H2Aub, etc., or active marks such as H3K4me, H3K36me, H3Ac, etc., this modification is catalyzed by Polycomb group (PcG) protein complexes and Trithorax group (TrxG) protein complexes, respectively (Pien and Grossniklaus, 2007; Köhler and Hennig, 2010; Grossniklaus and Paro, 2014; Kingston and Tamkun, 2014). PcG complexes are repressors of gene transcription, and function in multi-subunit complexes, such as Polycomb Repressor Complex 1 (PRC1) or Polycomb Repressor Complex 2 (PRC2) (Grossniklaus and Paro, 2014).
PRC2 is a highly conserved and well-characterized PcG complex, and it represses target gene expression by trimethylating histone H3 at lysine 27 (H3K27me3) through the E(z) SET domain (Köhler and Hennig, 2010; Grossniklaus and Paro, 2014). In the Arabidopsis genome, three paralogous genes MEDEA (MEA), SWINGER (SWN), and CURLY LEAF (CLF) are orthologs of the Drosophila E(z) gene, which function as a histone methyltransferase subunit in the PRC2 complex. MEA appears to function in embryogenesis specifically, and CLF and SWN are broadly expressed and partially redundant in vegetative and reproductive development (Zheng and Chen, 2011; Bemer and Grossniklaus, 2012; Xu et al., 2016b).
Whole genome analysis in Arabidopsis uncovered 1000s of gene loci carrying the H3K27me3 mark catalyzed by the PRC2 complex, indicating that H3K27me3 is a major epigenetic silencing mechanism in plants (Zhang et al., 2007; Lafos et al., 2011). Among them, most MIR156/157 loci, especially the dominant loci (MIR156A, MIR156C, and MIR157A), also carry H3K27me3 mark. However, except for SPL4 and SPL6 which play no obvious roles in juvenile-to-adult transition, miR156/157-targeted SPL genes are largely devoid of the H3K27me3 mark. These results imply that the PRC2 complex promotes SPL gene transcription indirectly by repressing the transcription of MIR156/157 loci (Lafos et al., 2011).
During juvenile-to-adult transition in Arabidopsis, the decrease in the transcription of MIR156A and MIR156C loci is associated with an increase in the binding of the PRC2 complex to these two loci, causing an increase in the H3K27me3 mark in their promoter and transcribed regions as well as a decrease in the H3K27ac mark in the region immediately after transcription start sites (TSS) (Xu et al., 2016b,c). Loss-of-function mutant of SWN, but not the loss-of-function mutant of CLF, exhibited an obvious delayed juvenile-to-adult transition phenotype (Xu et al., 2016b,c). H3K27me3 was completely lost in clf swn double mutant and it eventually dedifferentiated into a callus-like tissue, making it impossible to determine the phenotype of juvenile-to-adult transition (Xu et al., 2016b). Therefore, the question of if SWN and CLF functions redundantly in vegetative phase change remains unknown. However, the H3K27me3 mark at MIR156A/MIR156C loci was significantly reduced in clf mutants, but that in swn mutant remains controversial, which indicates that SWN and CLF may function redundantly to repress MIR156A/MIR156C by catalyzing H3K27me3 (Xu et al., 2016b,c).
PRC1 is thought to recognize the H3K27me3 mark to confer stable transcriptional repression (Lund and van Lohuizen, 2004). PRC1 is more dissimilar between Arabidopsis and animals, but it has related functions. In Arabidopsis, the function of PRC1 can be histone 2A mono-ubiquitination (H2Aub) dependent or independent. H2Aub dependent group requires the E3 ubiquitin ligase activity of Arabidopsis B lymphoma Moloney murine leukemia virus insertion region1 homolog 1A (AtBMI1A)/B/C or AtRING1A/B, while H2Aub independent group requires the activity of the EMBRYONIC FLOWER 1 (EMF1) (Yang C. et al., 2013; Calonje, 2014). BMI1-PRC1 and RING1-PRC1 are required for the repression of seed maturation program after germination, whereas EMF1-PRC1 is required for floral repression (Moon et al., 2003; Calonje et al., 2008; Chen et al., 2010).
PRC1 has been shown to be involved in juvenile-to-adult transition in Arabidopsis. BMI1-PRC1 maintains the repression of miR156 and accelerates juvenile-to-adult transition (Picó et al., 2015). The levels of MIR156A and MIR156C were upregulated in atbmi1a/b mutant and the juvenile phase was prolonged with the H2Aub and H3K27me3 marks being decreased in the TSS region of MIR156A and MIR156C (Picó et al., 2015).
RING1-PRC1 and EMF1-PRC1 function to repress SPLs to delay juvenile-to-adult transition (Li et al., 2017). In ring 1a ring 1b double mutant, the H2Aub mark was obviously decreased in the promoter and coding region of SPL3, SPL9 and SPL10, causing upregulation of these genes to accelerate the appearance of adult traits (Li et al., 2017). Therefore, PRC1 variants function in vegetative phase change mainly by targeting different MIR156/157 loci or SPL genes in the miR156/157-SPL pathway, and they have opposing roles in this process. However, how PRC1 variants recognize distinct targets still remains unclear, and more work is required to explore the mechanism of how PRC1 works.
The Arabidopsis genome encodes three H3K4 methyltransferase, namely ARABIDOPSIS TRITHORAX1 (ATX1), ATX2, and ATXR7 (Avramova, 2009). ATX1 and ATX7 are members of the Trithorax family, and ATXR7 is the only member of the SET1 subfamily in Arabidopsis (Tamada et al., 2009). atxr7-1, but not atx1-1, atx2-1, or atx1 atx2 double mutant, exhibits a precocious juvenile-to-adult transition phenotype. Chromatin immunoprecipitation (ChIP) analyses indicated that ATXR7 binds to a region adjacent to the TSS of MIR156A and deposits the H3K4me3 mark to activate MIR156A transcription (Xu et al., 2018).
Histone acetylation is generally considered as an active epigenetic mark, which is a balanced process regulated by histone acetyltransferases (HAG1) and histone deacetylases (HDA1, HAD6). Spt–Ada–Gcn5–acetyltransferase-like histone acetyltransferase complex (SAGA-like complex) is conserved in mammals, plants, files and yeast, and General Control Non-repressed 5 (GCN5) functions as the catalytic component for this complex (Turner, 2000).
In Arabidopsis, loss-of-function mutants in HAG1 (the Arabidopsis homolog of GCN5), hag1-6 and hag1-7, exhibited a significantly delayed juvenile-to-adult transition phenotype (Kim et al., 2015). In hag1-6 mutant, transcripts of MIR156 loci and mature miR156 remained stable; however, those of SPL3, SPL4, SPL5, SPL9, SPL11, SPL13, SPL15, and SPL8 were greatly reduced, suggesting that the regulation of SPLs by HAG1 is independent of miR156. ChIP results showed HAG1 was bound to the promoters and transcribed regions of SPL3 and SPL9 directly, leading to histone acetylation at the H3K9, H3K14, and H3K27 sites in these genes (Kim et al., 2015). HAG1-mediated H3 acetylation (H3Ac) of SPL9 is also responsive to light signals, which indicates that HAG1-mediated H3Ac of SPL9 might function as a sensor of environmental conditions to modulate the developmental process in plants (Kim et al., 2015).
Chromatin remodeling includes changes in nucleosome composition, nucleosome occupancy, nucleosome location, and the accessibility of the DNA to other transcriptional regulators.
ATP-dependent SWR1 chromatin remodeling complex (SWR1-C) functions in exchanging the histone H2A-H2B dimer with the H2A.Z-H2B dimer, and then produces nucleosome variant (Mizuguchi et al., 2004; Luk et al., 2010). In Arabidopsis, mutations in the SWR1-C subunit coding genes (ARP6, SEF, and PIE1) and H2A.Z coding genes (HTA8, HTA9, and HTA11) exhibited a similar pleiotropic phenotype, which indicates that the primary function of SWR1-C is to deposit H2A.Z (Mizuguchi et al., 2004; Wu et al., 2005). However, the mechanism of H2A.Z modification by SWR1-C to regulate different target gene expression is distinguishable in that H2A.Z can change the nucleosome occupancy to destabilize nucleosomes or to increase nucleosome stability and/or to function with H3K4me3 mark together (Martin-Trillo et al., 2006; Kumar and Wigge, 2010; Choi et al., 2013).
In arp6 and hat9/hat11 mutants, MIR156A/MIR156C transcripts were reduced and juvenile-to-adult transition was accelerated (Choi et al., 2016; Xu et al., 2018). ChIP with H2A.Z antibody showed that H2A.Z was enriched at the first 500 nucleotides after TSS of MIR156A/MIR156C, and the level of H2A.Z was significantly reduced in arp6 mutant. However, H2A.Z level does not change significantly during juvenile-to-adult transition, suggesting that H2A.Z and SWR1-C contribute to maintaining the expression of MIR156A/MIR156C early in shoot development, but do not regulate the timing of juvenile-to-adult transition (Xu et al., 2018). MIR156A transcript was reduced in arp6 mutant due to higher nucleosome occupancy in its promoter region (Choi et al., 2016); however, it was suggested that H2A.Z increases the expression of MIR156A/MIR156C by promoting the deposition of H3K4me3 rather than by decreasing nucleosome occupancy in the MIR156A promoter region (Xu et al., 2018).
BRAHMA (BRM) is the ATPase subunit of the most widely studied SWI2/SNF2 chromatin remodeling protein complex. It uses the energy derived from ATP hydrolysis to change the histone octamer-DNA interaction (Saha et al., 2006; Clapier and Cairns, 2009). BRM regulates MIR156A transcription by directly binding to the promoter region and maintaining low occupancy of the -2 and +1 nucleosomes proximal to the TSS. brm mutants exhibit an accelerated juvenile-to-adult transition phenotype by reducing the transcription of MIR156A (Xu et al., 2016c). BRM also antagonizes the function of SWN in the PRC2 complex to remove H3K27me3 repressive mark in MIR156A (Xu et al., 2016c).
PICKLE (PKL) is a CHD3 ATP-dependent nucleosome remodeling protein, which is physically associated with the nucleosome remodeling and deacetylation complex (Perruc et al., 2007; Zhang et al., 2008; Ho et al., 2013). PKL is bound to the TSS adjacent region of MIR156A/MIR156C to promote the juvenile-to-adult transition by repressing the transcription of MIR156A/MIR156C. In pkl mutants, MIR156A/MIR156C transcripts were elevated due to the reduction in nucleosome occupancy at the +1 position, an increase in the H3K27ac mark, and a corresponding decrease in the H3K27me3 mark in the promoter and transcribed region (Xu et al., 2016b).
Although the miR156/157-SPL pathway has been shown to be the master regulator of juvenile-to-adult transition in plants, yet little is known about the upstream regulator of this pathway, especially for miR157. Recent studies have revealed that DNA methylation, histone modification, chromatin remodeling play important roles in regulating the expression of some components in the miR156/157-SPL pathway. However, there are still some critical questions remain to be solved as illustrated in Figure 2.
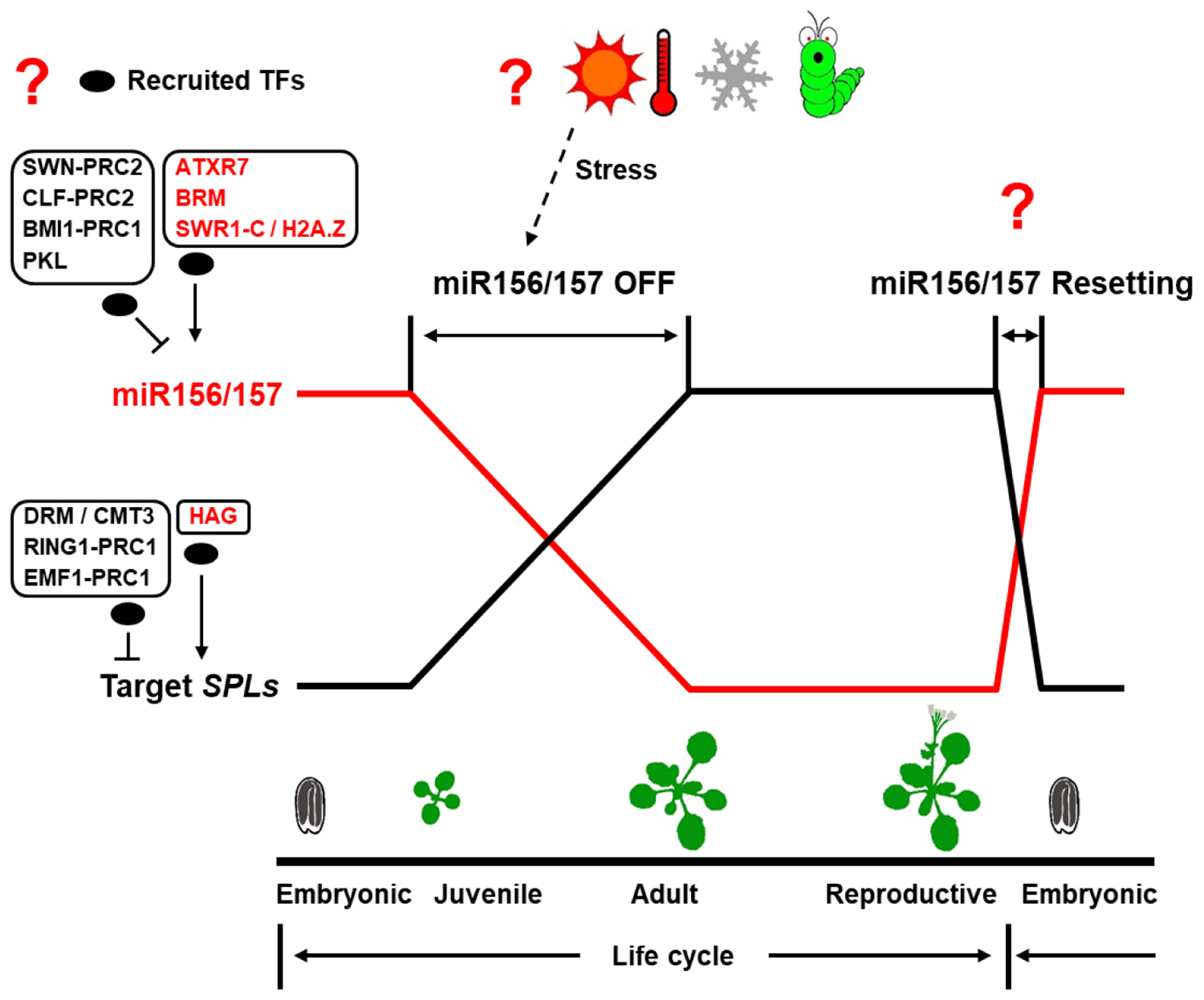
FIGURE 2. Epigenetic regulation of the miR156/157-SPL pathway in plant lifecycle. In plant lifecycle, the transcription of genes in the miR156/157-SPL pathway exhibits a fixed temporal expression pattern. The major unknown parts in epigenetic regulation of the miR156/157-SPL pathway are shown in question mark. Oval represents recruited transcription factors (TFs).
MIR156/157 and/or SPL loci are subjected to epigenetic regulation to modulate juvenile-to-adult transition in plants. However, these epigenetic regulators, by their own, have no DNA binding specificity. Therefore, a central question is how these epigenetic regulators are recruited to their target genes.
PRC2-mediated H3K27me3 is a conserved epigenetic modification between plants and the animal kingdom (Mozgova and Hennig, 2015; Xiao and Wagner, 2015). Recent genomic study in Arabidopsis showed that PRC2 components bind to specific DNA motifs called Polycomb response elements (PREs) by interacting with specific TFs (Xiao et al., 2017). Interestingly, six top enriched motifs (CTCC, CCG, G-box, GA repeat, AC-rich, and Telobox motifs) out of 170 computationally defined PREs were present at the MIR156A locus (Xiao et al., 2017). The GA repeat and Telobox motifs were present adjacent to the TSS region of MIR156A and MIR156C loci together, these motifs are the potential binding sites for class I BPC and C1-2iD TFs, respectively. This information will be helpful to identity TFs through which the PRC2 complex interacts to be recruited to the MIR156A/MIR156C loci during juvenile-to-adult transition.
Plants are sessile organisms and they are forced to adapt to the changing environment. The miR156/157-SPL pathway functions as the master regulator of juvenile-to-adult transition and flowering (Wang et al., 2009; Wu et al., 2009). Therefore, plants evolved a precise mechanism to adapt to the environment by shortening or prolonging the juvenile phase or changing the flowering time. Under salt or drought stress conditions, miR156 was induced to maintain plants in the juvenile phase for a relatively longer time; when they were returned to favorable conditions, miR156 was suppressed to accelerate the developmental transition (Cui et al., 2014). Under UV-B radiation conditions, the PRC2-mediated H3K27me3 modification in the MIR156A/MIR156C loci was decreased, and the corresponding up-regulation of miR156 delayed juvenile-to-adult transition (Dotto et al., 2018). Other studies also indicate that the expression of miR156 is responsive to ambient temperature (Stief et al., 2014), phosphate starvation (Hsieh et al., 2009), CO2 treatment (May et al., 2013), suggesting a tight interaction between juvenile-to-adult transition and environment through the miR156/157-SPL pathway.
Epigenetic modification is a reversible mark, which can be removed or deposited to target genes to affect their expression in response to changing environment. It will be of great interest to learn how epigenetic modification patterns of MIR156/157 and/or SPL loci change in response to external cues, especially to environment stresses, as well as how this changing environment affects the juvenile-to-adult transition.
During juvenile-to-adult transition, miR156/157 transcription was reduced or silenced gradually to ensure the plant to enter the adult phase and flower. This is achieved by disposing of active epigenetic marks such as H3K4me3, H3K27ac and depositing some repressive epigenetic marks such as H3K27me3 to miR156 loci. Interestingly, this silencing process needs to be reset to an active state in each generation as miR156/157 is de-repressed again to be highly expressed in the pro-embryo stage (Nodine and Bartel, 2010) after flowering.
A similar example of Off-Resetting pattern in plant lifecycle is the regulation of FLOWERING LOCUS C (FLC). FLC is silenced by depositing H3K27me3 mark under winter cold treatment, and the silenced state was maintained in the mature pollen grains and the egg cells (De Lucia et al., 2008; Sheldon et al., 2008). In pro-embryo stage, FLC is activated by depositing active epigenetic marks such as H3K4me3, H3K36me3, and disposing of repressive marks such as H3K27me3. LEAFY COTYLEDON1 (LEC1), a seed-specific pioneer TF (Tao et al., 2017), and EARLY FLOWERING 6 (ELF6), a H3K27me3 demethylase (Crevillén et al., 2014), were shown to play critical roles in FLC re-activation.
As for MIR156/157, it is still unknown where and when the de novo re-activation occurs. Moreover, whether the resetting of miR156/157 depends on a reversible epigenetic regulation still remains elusive. Further study of when, where and how miR156/157 Off-Reset pattern is initiated during plant life cycle will be an important future task.
YX and LZ wrote the article, and GW revised it.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31770209 and 31700249), a start-up fund from the Zhejiang Agricultural & Forestry University (Grant No. 2013FR084), and a fund from the Education Department of Zhejiang Province (Grant No. Y201635908).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Avramova, Z. (2009). Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int. J. Dev. Biol. 53, 371–381. doi: 10.1387/ijdb.082664za
Banks, J. A., and Fedoroff, N. (1989). Patterns of developmental and heritable change in methylation of the suppressor-mutator transposable element. Dev. Genet. 10, 425–437. doi: 10.1002/dvg.1020100604
Bemer, M., and Grossniklaus, U. (2012). Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 15, 523–529. doi: 10.1016/j.pbi.2012.09.006
Bitonti, M. B., Cozza, R., Chiappetta, A., Giannino, D., Ruffini, C. M., Dewitte, W., et al. (2002). Distinct nuclear organization, DNA methylation pattern and cytokinin distribution mark juvenile, juvenile-like and adult vegetative apical meristems in peach (Prunus persica (L.) Batsch). J. Exp. Bot. 53, 1047–1054. doi: 10.1093/jexbot/53.371.1047
Brink, R. A. (1962). Phase change in higher plants and somatic cell heredity. Q. Rev. Biol. 37, 1–22. doi: 10.1086/403567
Calonje, M. (2014). PRC1 marks the difference in plant PcG repression. Mol. Plant 7, 459–471. doi: 10.1093/mp/sst150
Calonje, M., Sanchez, R., Chen, L., and Sung, Z. R. (2008). EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20, 277–291. doi: 10.1105/tpc.106.049957
Cao, X., and Jacobsen, S. E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl Acad. Sci. U.S.A. 99, 16491–16498. doi: 10.1073/pnas.162371599
Chen, D., Molitor, A., Liu, C., and Shen, W. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Choi, K., Kim, J., Müller, S. Y., Oh, M., Underwood, C., Henderson, I., et al. (2016). Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex in Arabidopsis thaliana. Plant Physiol. 171, 1128–1143. doi: 10.1104/pp.16.00332
Choi, K., Zhao, X., Kelly, K. A., Venn, O., Higgins, J. D., Yelina, N. E., et al. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336. doi: 10.1038/ng.2766
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Cokus, S. J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C. D., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. doi: 10.1038/nature06745
Crevillén, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Cui, L., Shan, J., Shi, M., Gao, J., and Lin, H. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
De Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl Acad. Sci. U.S.A. 44, 16831–16836. doi: 10.1073/pnas.0808687105
Dotto, M., Gómez, M. S., Soto, M. S., and Casati, P. (2018). UV-B radiation delays flowering time through changes in the PRC2 complex activity and miR156 levels in Arabidopsis thaliana. Plant Cell Environ. 41, 1394–1406. doi: 10.1111/pce.13166
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627. doi: 10.1126/science.1190614
Fraga, M., Canal, M., and Rodriguez, R. (2002). Phase-change related epigenetic and physiological changes in Pinus radiata D. Don. Planta 215, 672–678. doi: 10.1007/s00425-002-0795-4
Grossniklaus, U., and Paro, R. (2014). Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 6:a019331. doi: 10.1101/cshperspect.a019331
He, J., Xu, M., Willmann, M. R., McCormick, K., Hu, T., Yang, L., et al. (2018). Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Gent. 14:e1007337. doi: 10.1371/journal.pgen.1007337
Henderson, I. R., and Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature 447, 418–424. doi: 10.1038/nature05917
Ho, K. K., Zhang, H., Golden, B. L., and Ogas, J. (2013). PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta 1829, 199–210. doi: 10.1016/j.bbagrm.2012.10.011
Hsieh, L.-C., Lin, S.-I., Shih, A. C.-C., Chen, J.-W., Lin, W.-Y., Tseng, C.-Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151, 2120–2132. doi: 10.1104/pp.109.147280
Huijser, P., and Schmid, M. (2011). The control of developmental phase transitions in plants. Development 138, 4117–4129. doi: 10.1242/dev.063511
Jung, J., Ju, Y., Seo, P. J., Lee, J., and Park, C. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69, 577–588. doi: 10.1111/j.1365-313X.2011.04813.x
Kerstetter, R. A., and Poethig, R. S. (1998). The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14, 373–398. doi: 10.1146/annurev.cellbio.14.1.373
Kim, J., Oh, J. E., Noh, Y., and Noh, B. (2015). Epigenetic control of juvenile-to-adult phase transition by the Arabidopsis SAGA-like complex. Plant J. 83, 537–545. doi: 10.1111/tpj.12908
Kingston, R. E., and Tamkun, J. W. (2014). Transcriptional regulation by Trithorax-group proteins. Cold Spring Harb Perspect Biol. 6:a019349. doi: 10.1101/cshperspect.a019349
Köhler, C., and Hennig, L. (2010). Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 20, 541–547. doi: 10.1016/j.gde.2010.04.015
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147.
Lafos, M., Kroll, P., Hohenstatt, M. L., Thorpe, F. L., Clarenz, O., and Schubert, D. (2011). Dynamic regulation of H3K27 Trimethylation during Arabidopsis differentiation. PLoS Genet. 7:e1002040. doi: 10.1371/journal.pgen.1002040
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Li, J., Wang, Z., Hu, Y., Cao, Y., and Ma, L. (2017). Polycomb group proteins RING1A and RING1B regulate the vegetative phase transition in Arabidopsis. Front. Plant Sci. 8:867. doi: 10.3389/fpls.2017.00867
Luk, E., Ranjan, A., FitzGerald, P. C., Mizuguchi, G., Huang, Y., Wei, D., et al. (2010). Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736. doi: 10.1016/j.cell.2010.10.019
Lund, A. H., and van Lohuizen, M. (2004). Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16, 239–246. doi: 10.1016/j.ceb.2004.03.010
Martienssen, R., Barkan, A., Taylor, W. C., and Freeling, M. (1990). Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Gene Dev. 4, 331–343. doi: 10.1101/gad.4.3.331
Martin-Trillo, M., Lázaro, A., Poethig, R. S., Gómez-Mena, C., Piñeiro, M. A., Martinez-Zapater, J. M., et al. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodeling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133, 1241–1252. doi: 10.1242/dev.02301
May, P., Liao, W., Wu, Y., Shuai, B., McCombie, R. W., Zhang, M. Q., et al. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4:2145. doi: 10.1038/ncomms3145
Mizuguchi, G., Shen, X., Landry, J., Wu, W., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. doi: 10.1126/science.1090701
Moon, Y. H., Chen, L., Pan, R. L., Chang, H. S., Zhu, T., Maffeo, D. M., et al. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15, 681–693. doi: 10.1105/tpc.007831
Mozgova, I., and Hennig, L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Nodine, M. D., and Bartel, D. P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Gene Dev. 24, 2678–2692. doi: 10.1101/gad.1986710
Perruc, E., Kinoshita, N., and Lopez-Molina, L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52, 927–936. doi: 10.1111/j.1365-313X.2007.03288.x
Picó, S., Ortiz-Marchena, M. I., Merini, W., and Calonje, M. (2015). Deciphering the role of POLYCOMB REPRESSIVE COMPLEX1 variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol. 168, 1286–1297. doi: 10.1104/pp.15.00073
Pien, S., and Grossniklaus, U. (2007). Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophy. Acta 1769, 375–382. doi: 10.1016/j.bbaexp.2007.01.010
Poethig, R. S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930. doi: 10.1126/science.250.4983.923
Poethig, R. S. (2013). Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 105, 125–152. doi: 10.1016/B978-0-12-396968-2.00005-1
Preston, J. C., and Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4:80. doi: 10.3389/fpls.2013.00080
Riese, M., Höhmann, S., Saedler, H., Münster, T., and Huijser, P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401, 28–37. doi: 10.1016/j.gene.2007.06.018
Ruiz-García, L., Cervera, M. T., and Martínez-Zapater, J. M. (2005). DNA methylation increases throughout Arabidopsis development. Planta 222, 301–306. doi: 10.1007/s00425-005-1524-6
Saha, A., Wittmeyer, J., and Cairns, B. R. (2006). Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7, 437–447. doi: 10.1038/nrm1945
Schmid, M., Uhlenhaut, N. H., Godard, F., Demar, M., Bressan, R., Weigel, D., et al. (2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012. doi: 10.1242/dev.00842
Sheldon, C. C., Hills, M. J., Lister, C., Dean, C., Dennis, E. S., and Peacock, W. J. (2008). Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl Acad. Sci. U.S.A. 105, 2214–2219. doi: 10.1073/pnas.0711453105
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Baurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Tamada, Y., Yun, J. Y., Woo, S. C., and Amasino, R. M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone h3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21, 3257–3269. doi: 10.1105/tpc.109.070060
Tao, Z., Shen, L., Gu, X., Wang, Y., Yu, H., and He, Y. (2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128. doi: 10.1038/nature24300
Telfer, A., Bollman, K. M., and Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654.
Tsukaya, H., Shoda, K., Kim, G., and Uchimiya, H. (2000). Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta 210, 536–542. doi: 10.1007/s004250050042
Turner, B. M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X
Usami, T., Horiguchi, G., Yano, S., and Tsukaya, H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136, 955–964. doi: 10.1242/dev.028613
Wang, J., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wu, G., Park, M. Y., Conway, S. R., Wang, J., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by Mir156 and its target SPL3. Development 18, 3539–3547. doi: 10.1242/dev.02521
Wu, W., Alami, S., Luk, E., Wu, C., Sen, S., Mizuguchi, G., et al. (2005). Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12, 1064–1071. doi: 10.1038/nsmb1023
Xiao, J., Jin, R., Yu, X., Shen, M., Wagner, J. D., Pai, A., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552. doi: 10.1038/ng.3937
Xiao, J., and Wagner, D. (2015). Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23, 15–24. doi: 10.1016/j.pbi.2014.10.003
Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xu, M., Hu, T., Smith, M. R., and Poethig, R. S. (2016a). Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28, 28–41. doi: 10.1105/tpc.15.00854
Xu, M., Hu, T., Zhao, J., Park, M., Earley, K. W., Wu, G., et al. (2016b). Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. doi: 10.1371/journal.pgen.1006263
Xu, M., Leichty, A. R., Hu, T., and Poethig, R. S. (2018). H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 145:dev152868. doi: 10.1242/dev.152868
Xu, Y., Guo, C., Zhou, B., Li, C., Wang, H., Zheng, B., et al. (2016c). Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 172, 2416–2428. doi: 10.1104/pp.16.01588
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., and Calonje, M. (2013). VAL- and AtBMI1-Mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yang, L., Xu, M., Koo, Y., He, J., and Poethig, R. S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2:e00260. doi: 10.7554/eLife.00260
Yu, S., Cao, L., Zhou, C., Zhang, T., Lian, H., Sun, Y., et al. (2013). Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2:e00269. doi: 10.7554/eLife.00269
Zhang, H., Rider, S. D., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V., et al. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283, 22637–22648. doi: 10.1074/jbc.M802129200
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S. W. L., Chen, H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201. doi: 10.1016/j.cell.2006.08.003
Zheng, B., and Chen, X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14, 123–129. doi: 10.1016/j.pbi.2011.01.001
Keywords: epigenetic regulation, miR156, SPL, juvenile-to-adult transition, plants
Citation: Xu Y, Zhang L and Wu G (2018) Epigenetic Regulation of Juvenile-to-Adult Transition in Plants. Front. Plant Sci. 9:1048. doi: 10.3389/fpls.2018.01048
Received: 27 April 2018; Accepted: 28 June 2018;
Published: 17 July 2018.
Edited by:
Ming Luo, South China Botanical Garden (CAS), ChinaReviewed by:
Abdelali Hannoufa, Agriculture and Agri-Food Canada (AAFC), CanadaCopyright © 2018 Xu, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Wu, d3VnYW5nQHphZnUuZWR1LmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.