- MOE Key Laboratory of Crop Heterosis and Utilization, National Maize Improvement Center of China, China Agricultural University, Beijing, China
CtIP/Ctp1/Sae2/Com1, a highly conserved protein from yeast to higher eukaryotes, is required for DNA double-strand break repair through homologous recombination (HR). In this study, we identified and characterized the COM1 homolog in maize. The ZmCom1 gene is abundantly expressed in reproductive tissues at meiosis stages. In ZmCom1-deficient plants, meiotic chromosomes are constantly entangled as a formation of multivalents and accompanied with chromosome fragmentation at anaphase I. In addition, the formation of telomere bouquet, homologous pairing and synapsis were disturbed. The immunostaining assay showed that the localization of ASY1 and DSY2 was normal, while ZYP1 signals were severely disrupted in Zmcom1 meiocytes, indicating that ZmCom1 is critically required for the proper SC assembly. Moreover, RAD51 signals were almost completely absent in Zmcom1 meiocytes, implying that COM1 is required for RAD51 loading. Surprisingly, in contrast to the Atcom1 and Oscom1 mutants, Zmcom1 mutant plants exhibited a number of vegetative phenotypes under normal growth condition, which may be partly attributed to mitotic aberrations including chromosomal fragmentation and anaphase bridges. Taken together, our results suggest that although the roles of COM1 in HR process seem to be primarily conserved, the COM1 dysfunction can result in the marked dissimilarity in mitotic and meiotic outcomes in maize compared to Arabidopsis and rice. We suggest that this character may be related to the discrete genome context.
Introduction
Meiosis is a highly conserved process producing haploid germ cells from diploid progenitors and is essential for all sexually reproductive organisms. It includes one round of DNA replication followed by two sequential rounds of cell division containing meiosis I and meiosis II (Zickler and Kleckner, 1999). During meiosis I, crossovers (COs) are formed to ensure the accurate segregation of homologous chromosomes (Mercier et al., 2015; Gray and Cohen, 2016). Homologous recombination (HR) is a prerequisite to the generation of COs. In plants, meiotic recombination is initiated by the programmed introduction of double-strand breaks (DSBs) mediated by SPO11, a conserved type II topoisomerase, and several accessory proteins (Keeney et al., 1997). The resulting DSB ends are resected by a protein complex, MRX/N (Mre11-Rad50-Xrs2/Nbs1) and Sae2/Com1/CtIP/Ctp1, to generate extended single-stranded DNA (ssDNA) overhangs, which are subsequently stabilized by replication protein A (RPA) (Borde, 2007). Next, RPA is displaced by RAD51 and DMC1 to form nucleoprotein filaments that can facilitate homologous pairing and single-end invasion of a non-sister chromatid resulting in the formation of joint molecule (JM) intermediates (Hunter and Kleckner, 2001). Ultimately, these events give rise to either COs or non-crossovers (NCOs) (Allers and Lichten, 2001).
The evolutionarily conserved MRX/N complex functions as one of the critical guardians of genome integrity in eukaryotes and is required for DNA damage repair, cell-cycle checkpoint and telomere maintenance during both mitosis and meiosis (Daoudal-Cotterell et al., 2002; Borde, 2007; Amiard et al., 2010). The three proteins (Mre11, Rad50, and Nbs1/Xrs2) in MRX/N complex play distinct roles. Mre11 specifies 3′ to 5′ exonuclease activity on the double-stranded DNA and endonuclease activity on the single-stranded DNA as well as limited helicase activity (Puizina et al., 2004; Altun, 2008). Rad50 has two long coiled-coil domains that interact with one another to form a head-to-tail dimer to enable the binding of Mre11 and DNA (Carney et al., 1998; Hopfner et al., 2002). NBS1 is phosphorylated by ATM to link the detection of DSBs to signaling events (Waterworth et al., 2007). Null mutations in genes encoding any component of MRX/N complex result in lethality in mammals (Paull and Gellert, 1998), whereas Arabidopsis mre11 and rad50 mutants are viable but fully sterile (Daoudal-Cotterell et al., 2002; Puizina et al., 2004; Samanic et al., 2013). In contrast, the loss-of-function of Arabidopsis NBS1 displays normal growth under standard conditions and shows no defects in fertility (Waterworth et al., 2007). In addition, Arabidopsis mutants defective MRX/N complex in exhibit distinct hypersensitivity to various genotoxic stresses, reflecting both common and unique features of each component of MRX/N complex acting in the different spectrum of DNA lesions and mechanism of their repair (Vannier et al., 2006; Cassani et al., 2018).
As a cofactor for MRX/N, the mammalian CtIP and its fission yeast (Ctp1), budding yeast (Sae2), and plant (Com1) orthologs play the multifunctional roles in directing DSB repair pathway choice and modulate repair activities (McKee and Kleckner, 1997; Prinz et al., 1997; Baroni et al., 2004; Chen et al., 2005; Lengsfeld et al., 2007; Limbo et al., 2007; Penkner et al., 2007; Sartori et al., 2007; Uanschou et al., 2007; Lee-Theilen et al., 2010; Ji et al., 2012). The plant homolog of CtIP/Ctp1/Sae2/Com1 was first identified in Arabidopsis (Uanschou et al., 2007) and later in rice (Ji et al., 2012). Atcom1 and Oscom1 mutant plants exhibit normal vegetative growth but complete male and female sterility (Uanschou et al., 2007; Ji et al., 2012). Cytological investigations revealed that meiosis is severely inhibited, due to the defective homologous pairing and massive chromosome fragmentation (Uanschou et al., 2007; Ji et al., 2012). These studies demonstrate that the function of Com1 homolog in controlling DSB resection is conserved in plants as in other organisms.
In contrast to Arabidopsis and rice, maize has a large genome (ca. 2.3 Gb) and fairly complex genome organization. Here, we characterize the Com1 in maize using a reverse genetic approach. Our results demonstrate that ZmCom1 is essential for DSB repair and HR, establishing the telomere bouquet and SC assembly in maize meiosis. We also show that ZmCom1 is required for mitosis to occur normally in vegetative cells. These results imply that although the roles of Com1 in DSB repair seem to be fundamentally conserved in diverse plant species, the precise behavior of Com1 may vary in the different plant organisms.
Materials and Methods
Plant Materials and Genotyping
UniformMu mutant lines, UFMu-01240 (Zmcom1-1) and UFMu-09026 (Zmcom1-2) induced by Robertsons Mutator transposons in the uniform W22 inbred line were obtained from Maize Stock Center and backcrossed with the W22 inbred line four times before the further analysis. All plants were grown in field or greenhouse in 2014–2017 under the normal growth condition. Genomic DNA extraction and genotyping were conducted as described previously (Li et al., 2013). To confirm a presence of the Mutator insertion, genomic DNA of both mutant lines was amplified with the primer pair of MuTIR and COM1-L2 (Supplementary Table S1) and then PCR product was subject to Sanger sequencing.
Observation of Pollen Viability
Pollen viability was assessed by Alexander staining using previously described methods (Alexander, 1969; Johnson-Brousseau and McCormick, 2004). Anthers were collected from the wild type and Zmcom1 mutants during anthesis stage. Pollen grains were dissected out of anthers in Alexander solution and analyzed under Leica EZ4 HD. The pictures of strained pollen grains were taken using the microscope (Leica DM2000 LED).
cDNA Cloning, RT-PCR and RT-qPCR Analysis
Total RNA was extracted from roots, stems, leaves, developing embryos (16 days after pollination), endosperm (16 days after pollination), meiotic ears as well as anthers of B73 plants and young ear of Zmcom1 plants, and was then reverse-transcribed into cDNA by TaKaRa kits following manufacturer’s instructions. The full-length cDNA was generated using the TransStart FastPfu Fly DNA Polymerase kit (TransGen). PCR primers used for RT-PCR and RT-qPCR are listed in Supplementary Table S1. The maize UBIQUITIN gene was used as a control standard for all tissues. RT-qPCR analysis was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems).
Subcellular Localization
The coding sequence of ZmCOM1 was amplified with the primer pair PCUN-COM1 (Supplementary Table S1) and sub-cloned into of the pCUN+GFP vector using the BamHI and SpeI sites to create an ORF encoding an EGFP fusion protein driven by the 35S promoter. Mesophyll protoplasts were isolated from the second leaves of 2-week-old etiolated B73 seedlings according to the method described previously (Yoo et al., 2007) and transformed with the prepared plasmids using the polyethylene glycol (PEG) mediated transformation method as previously described (Yoo et al., 2007). The protoplasts were cultured at 25°C in the dark for 18 h and observed under a confocal laser scanning microscope (Leica sp5).
Preparation of Mitotic Chromosome Spreads
Chromosome spreads were prepared as described previously (Kato et al., 2004). Kernels were soaked for a night in sterile water before germinating at 30°C for 2–3 days. Root tips of 1–2 cm length were dissected and fixed in a 3:1 mixture of 95% ethanol: glacial acetic acid for 30 min in a vacuum environment and finally stored in 70% ethanol at -20°C until use. After washing in water on ice, the root apical meristem containing dividing cells was dissected and digested in 50 μl enzyme mix containing 1% pectolyase Y23 (ICN) and 2% cellulase Onozuka R-10 (Yakult Pharmaceutical, Tokyo) for 65 min at 37°C. After digestion, the root sections were washed in ice-cold distilled water and then briefly washed in 70% ethanol for three times. The root sections were carefully broken using a needle and vortexed at maximum speed in 75% ethanol for 30 s at room temperature to separate cells from each other. Cells were collected at the bottom of the tube by centrifugation and resuspended in 100% glacial acetic acid solution. Ten microliter of the cell suspension was dropped onto glass slides in a box lined with wet paper towels and dried slowly.
Preparation of Meiotic Chromosome Spreads
Chromosome spreads were prepared from tassels fixed in Carnoy’s solution (3:1 ethanol: acetic acid, v/v). After infiltration for 30 min at room temperature, the tassels were stored in 75% ethanol at 4°C until observation. Squashes were made in a drop of 45% acetic acid. The microscope slides were frozen in liquid nitrogen and the coverslips were removed immediately. The slides were then dehydrated through an ethanol series (70% to 90% to 100%) for 5 min each and air dried. The chromosomes were stained with 4′, 6-diamidino-2-phenylindole (DAPI) in an antifade solution (Vector, H-1200, CA, United States). Images were captured using a Ci-S-FL microscope (Nikon, Tokyo) equipped with a DS-Qi2 Microscope Camera system.
FISH Analysis
The FISH procedure was performed as described previously (Li and Arumuganathan, 2001; Cheng, 2013). Plasmids carrying 5S rDNA repeats (pTa794) or the telomere-specific repeats (pAtT4) were used as FISH probes (Richards and Ausubel, 1988; Ji et al., 2012). The 5S rDNA-specific and telomere-specific probes were individually labeled with digoxigenin by nick translation (Roche, Cat.No.11745808910) and detected with a fluorescein isothiocyanate (FITC) conjugated anti-digoxigenin antibody (Vector Laboratories). The chromosomes were counterstained with DAPI in Vectashield antifade solution (Vector laboratories). Chromosome spreads were observed under a Ci-S-FL fluorescence microscope (Nikon) and captured with a DS-Qi2 Microscope Camera.
Fluorescence Immunolocalization
Young anthers at the meiotic prophase (∼1.5–2.5 mm, Zhang et al., 2014) were fixed with 4% (w/v) paraformaldehyde in 1 × Buffer A for 30 min at room temperature (25°C), washed in 1 × Buffer A at room temperature and stored in 1 × Buffer A at 4°C for several months. The procedure of immunolocalization was performed as described previously (Pawlowski et al., 2003; Cheng, 2013). All primary (ASY1, DSY2, ZYP1, and RAD51) and secondary antibodies were used at a dilution of 1:100. The images were viewed with software NIS-Elements to generate 2D projected images. Surface rendered images were colored by the ImageJ software through the Merge Channels.
Results
Identification of ZmCom1 and Isolation of Its Mutants
A BLASTP search using the rice Com1 amino acid sequence was conducted in the maize genome database (MaizeGDB) and only one candidate gene model (GRMZM2G076617) with significant similarity was identified. The cDNA sequence, which was redefined by rapid amplification of cDNA ends (RACE) PCR, contains 2,134 bp with an open reading frame of 1,668 bp. The gene has two exons and one intron (Figure 1A). The protein sequence consists of 555 amino acids and shows 62% of identity and 72% of similarity to OsCom1 (363/583 residues identical and 421/583 residues positive, Supplementary Figure S1). ZmCom1 protein harbors an N-terminal SMC-N domain and a C-terminal SAE2 superfamily domain, both of which conventionally exist in Com1 homologs of other organisms (Supplementary Figure S1). Phylogeny analyses revealed that Com1 homologs form two distinct clades reflecting the divergence between monocots and dicots (Supplementary Figure S2).
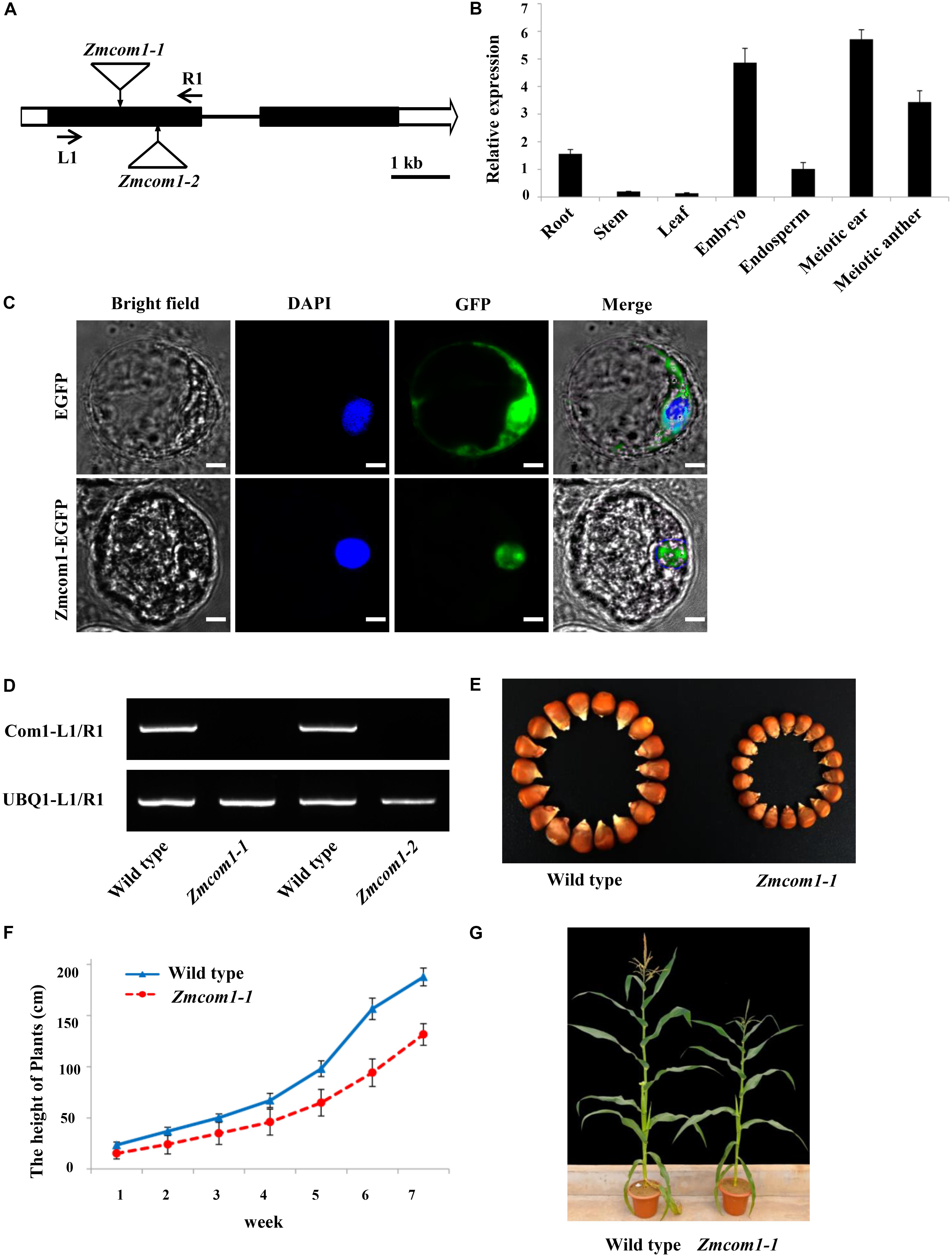
FIGURE 1. Characterization of the ZmCom1 gene. (A) Diagram of genomic structures of the ZmCom1 genes with Mutator transposon insertion sites marked with triangles. Bars indicate exons and lines represent introns. Scale bar = 1 kb. (B) RT-qPCR analysis of ZmCom1 transcription in different tissues. Expression levels relative to the expression level of the UBQ1 gene are presented with standard errors. Values are means of three independent experiments. (C) ZmCom1 protein is localized to nucleus in maize protoplasts. Scale bar = 5 μm. (D) RT-PCR analysis of ZmCom1 transcript in different genotypes. The maize UBQ1 gene was used as an internal control. (E) Morphological comparison of mature seeds between wild type and Zmcom1-1 mutant. (F) Growth-curve of plant height in wild type and zmcom1-1 mutant plants. Values are means of 10 individual plants. (G) Morphological comparison of mature plants between wild type and Zmcom1-1 mutant.
By means of quantitative RT-PCR, we examined the tissue-specific expression pattern of ZmCom1. We found that ZmCom1 is expressed most highly in meiotic ears and anthers, as well as in developing embryo, less in root and endosperm, and extremely low in leaf and stem (Figure 1B). These results support the function of ZmCom1 in meiosis and mitosis. To elucidate the cellular localization of ZmCom1, we induced expression of the ZmCom1 fused to the EGFP under the control of the CaMV35S promoter in maize protoplasts. The GFP signal was revealed in nuclei (Figure 1C).
Two independent Mutator transposon insertion lines, UFMu-01240 (Zmcom1-1) and UFMu-09026 (Zmcom1-2), were identified in the public maize Mutator line database (Harper et al., 2016). By conducting locus-specific PCR amplification followed by Sanger sequencing, we confirmed that both insertion sites are within the first exon of ZmCom1 (Figure 1A). RT-PCR with primers flanking the Mutator insertion sites failed to detect the ZmCom1 transcripts (Figure 1D), indicating that both mutants are null. The heterozygous alleles of both mutants did not exhibit any obvious defects during either the vegetative or reproductive stages in comparison to the wild type. However, we constantly observed a proportion of small kernels in the offspring of self-pollinated heterozygous plants for both mutations (Figure 1E and Supplementary Figure S3A), and the ratio of small to normal seeds was not significantly different from the expected 1:3 ratio (Chi-square test, P > 0.05; Supplementary Table S2). More importantly, PCR analyses confirmed that these small kernels co-segregated with homozygous Zmcom1 genotype (Supplementary Figure S4), indicating that the loss-of-function of ZmCom1 has an effect on maize seed development. The overall statue of Zmcom1 plants seemed to be comparable to wild type, but obvious dwarf phenotype started appearing from first weeks until the maturity (Figures 1F,G and Supplementary Figures S3B,C).
The Zmcom1 plants reached the anthesis stage at the same time as wild type but were completely male sterile (Figure 2A and Supplementary Figure S3D). When pollinated with normal pollen grains from the wild type, the Zmcom1 plants could not set any seeds (Figure 2B and Supplementary Figure S3E), suggesting that megagametogenesis was aborted. To investigate the male sterility, Zmcom1 and wild type pollen grains were stained with the Alexander solution (Figures 2C,D and Supplementary Figure S3F,G). Pollen grains from the wild type were round (Figure 2C and Supplementary Figure S3F), while those from Zmcom1 plants were empty and shrunken (Figure 2D and Supplementary Figure S3G), indicating that microspore development was also aborted. Taken together, these results indicate that the disruption of ZmCom1 gene leads to defects in both vegetative and reproductive development.
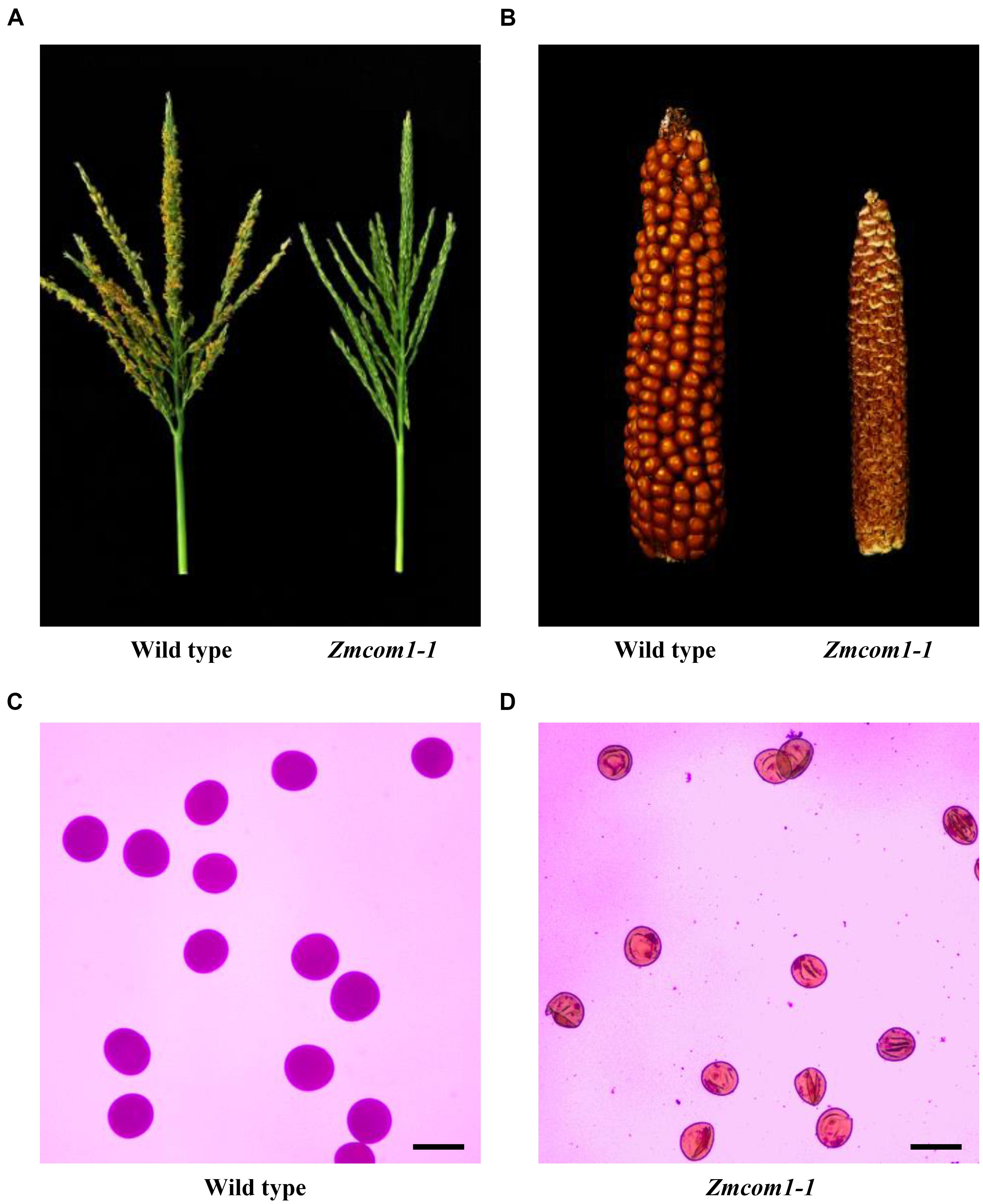
FIGURE 2. Male and female sterility of Zmcom-1 mutant plants. (A) Comparison of a wild type tassel and a Zmcom1-1 tassel at the flowering stage. (B) Comparison of a wild type ear and a Zmcom1-1 ear. (C) Normal pollen grains of the wild type. Scale bar = 100 μm. (D) Complete sterile pollen grains of the Zmcom1-1 plant. Scale bar = 100 μm.
Abnormal Chromosome Behavior in Zmcom1
To establish the cause of sterility in the Zmcom1 mutant, we examined the meiotic chromosome behavior in pollen mother cells (PMCs) of both wild type and Zmcom1-1 plants using 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 3). In the wild type, the chromosomes appeared as thin threads at leptotene (Figure 3A), and homologous chromosomes underwent pairing and synapsis during zygotene (Figure 3B). At pachytene, homologous chromosomes completed synapsis and chromosomes appeared as thick threads (Figure 3C). During diakinesis, chromosomes became condensed, and 10 short bivalents connected by chiasmata were observed in nuclei (Figure 3D). At metaphase I, all 10 bivalents aligned in an orderly manner on the equatorial plate (Figure 3E), and homologous chromosomes separated and migrated to opposite poles at anaphase I (Figure 3F). Finally, all chromosomes reached the two poles at telophase I to form regular dyads (Figure 3G). During meiosis II, sister chromatids separated from each other, ultimately giving rise to tetrad (Figure 3H).
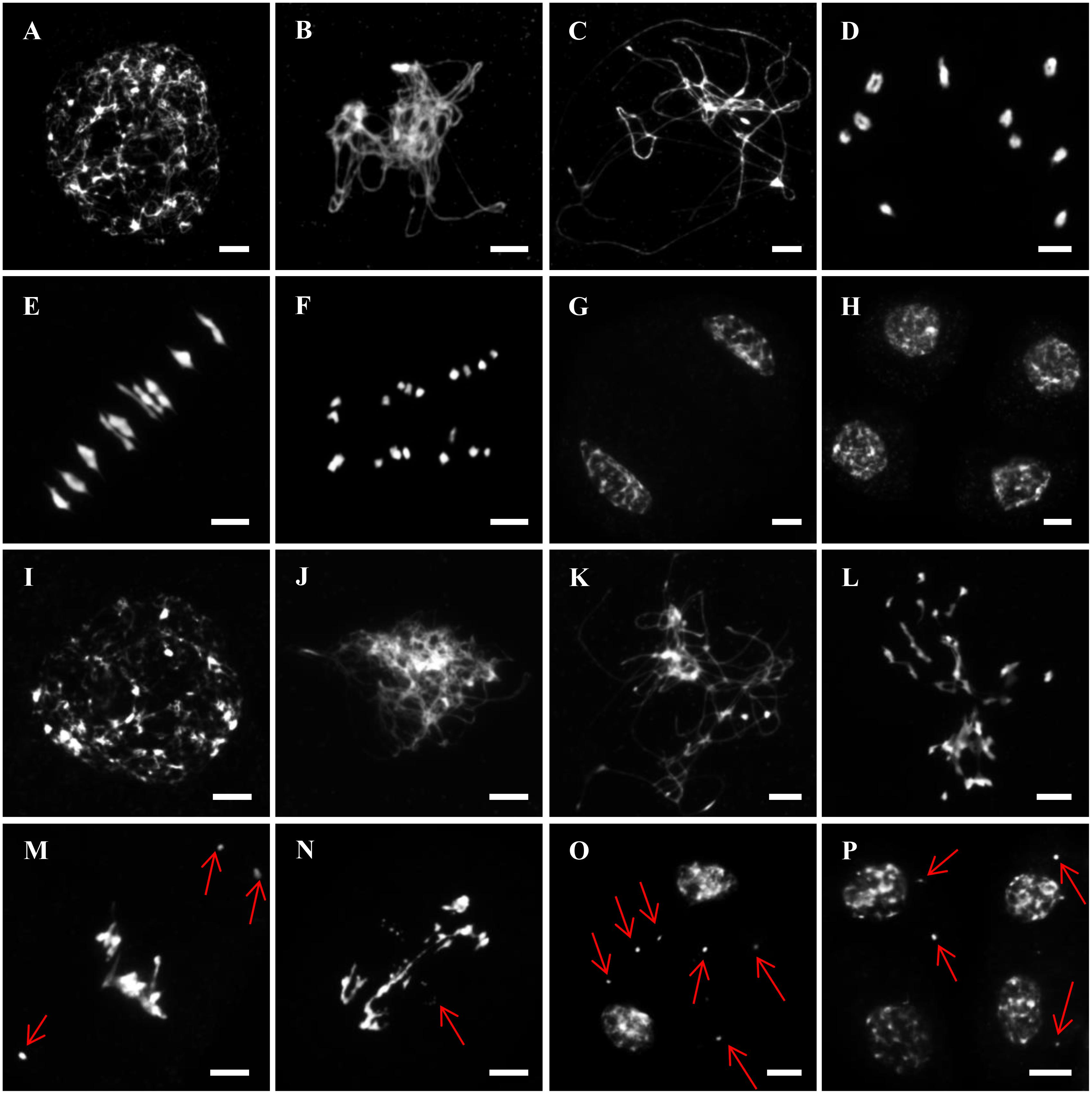
FIGURE 3. Male meiosis in wild type and Zmcom1-1. (A–H) Meiosis in wild type. (A) Leptotene; (B) Zygotene; (C) Pachytene; (D) Diakinesis; (E) Metaphase I; (F) Anaphase I; (G) Dyad; (H) Tetrads. Scale bars = 10 μm. (I–P) Meiosis in Zmcom1-1. (I) Leptotene; (J) Zygotene; (K) Pachytene; (L) Diakinesis; (M) Metaphase I; (N) Anaphase I; (O) Dyad; (P) Tetrads. The red arrows pointed out the chromosomal fragments and abnormal bridges. Scale bars = 10 μm.
In Zmcom1 mutant, meiotic chromosomes behaved normally at leptotene (Figure 3I and Supplementary Figure S5A). However, the chromosomes remained as single threads and did not pair with their homologs during zygotene and pachytene (Figures 3J,K and Supplementary Figures S5B,C). Irregularly shaped univalents were scattered throughout the entire nucleus during diakinesis (Figure 3L and Supplementary Figure S5D). At metaphase I, chromosomes intertwined into a block, and chromosome fragments appeared as small dots (Figure 3M and Supplementary Figure S5E). During anaphase I, these entangled chromosomes separated, resulting in unequal segregation of chromosomes to the opposite poles. Chromosome bridges were constantly observed and chromosome fragments remained at the equatorial plate (Figure 3N and Supplementary Figure S5F). Although most chromosomes had arrived in the two poles at telophase I, many lagging fragments were still randomly scatted within the nucleus (Figure 3O and Supplementary Figure S5G). After the second division, abnormal tetrads with several micronuclei were eventually generated (Figure 3P and Supplementary Figure S5H). Therefore, we concluded that the sterility of the Zmcom1 mutant may be caused by deficiency in homologous chromosome pairing, synapsis, and profound chromosomal fragmentation.
Defective Telomere Bouquet Formation and Homologous Pairing in Zmcom1
Telomere bouquet clustering, a particular event in early prophase I, may facilitate the initiation of homologous pairing (Golubovskaya et al., 2002; Harper et al., 2004; Klutstein et al., 2015). To explore the pairing defects in Zmcom1, we conducted fluorescent in situ hybridization (FISH) analysis using the telomere-specific probe in wild type and Zmcom1 meiocytes (Figures 4A,B and Supplementary Figure S6A). In wild type meiocytes (n = 46), 97.8% of the telomere signals attached to the nuclear envelop and were clustered at early zygotene stage, displaying a typical telomere bouquet formation (Figure 4A). However, in Zmcom1 meiocytes (n = 55 and 41 for Zmcom1-1 and Zmcom1-2, respectively), telomeres did not cluster within a certain region but scattered throughout the nucleus (Figure 4B and Supplementary Figure S6A), indicating that the telomere bouquet formation was defective in the Zmcom1 mutants.
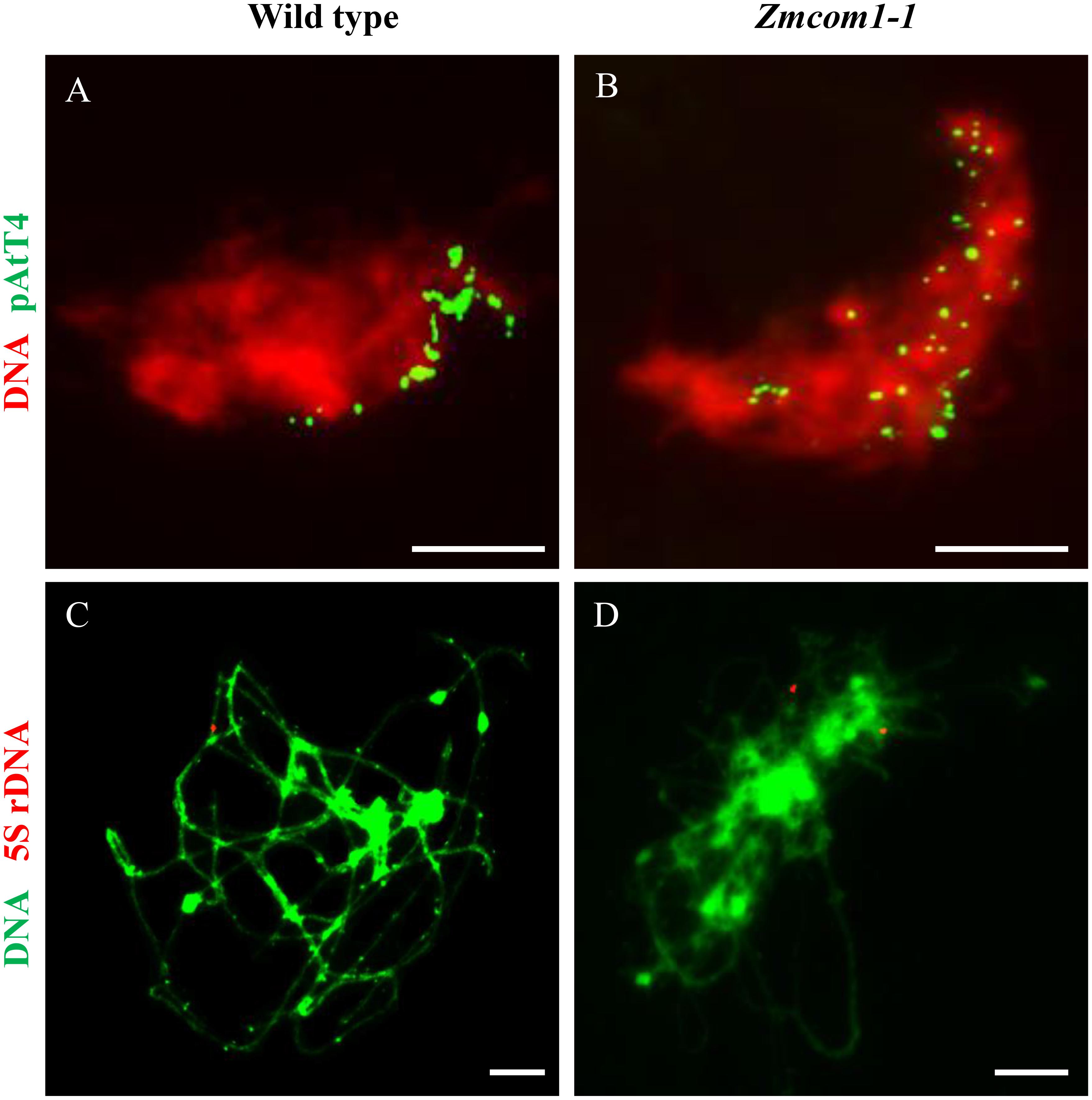
FIGURE 4. ZmCom1 is essential for bouquet formation and homologous pairing. (A,B) Bouquet formation analysis using FISH with the telomere-specific pAtT4 probe in in the wild type (A) and Zmcom1-1 (B). Scale bars = 10 μm. (C,D) Homologous pairing analysis using FISH with 5S rDNA probe in in the wild type (C) and Zmcom1-1 (D). Scale bars = 10 μm.
To further determine chromosome pairing behavior in Zmcom1 mutant, we performed FISH experiments using 5S rDNA as probes in wild type and Zmcom1 meiocytes (Figures 4C,D and Supplementary Figure S6B). 5S rDNA is a tandemly repetitive sequence that only locates on the distal regions of the long arm of chromosome 2 (Li and Arumuganathan, 2001). In wild type meiocytes (n = 32), only one 5S rDNA signal was detected at pachytene stage (Figure 4C), indicating that two homologous chromosomes had been well paired. In contrast, two separate 5S rDNA signals were detected in Zmcom1 meiocytes (n = 38 and 27 for Zmcom1-1 and Zmcom1-2, respectively) (Figure 4D and Supplementary Figure S6B). These results further confirmed that homologous chromosome paring was deficient in Zmcom1.
Normal Axial Element Installation but Deficient Central Element Installation in Zmcom1
The SC consists of two parallel lateral elements (LEs – former axial elements – AEs) and one central element (CE). To investigate whether the SC was properly assembled in Zmcom1-1, we conducted immunostaining analysis using antibodies against the maize SC components ASY1, DSY2, and ZYP1. ASY1, a homolog of rice PAIR2, is the AE protein which plays pivotal roles in bouquet formation, homologous pairing and the SC assembly (Sanchez-Moran et al., 2008; Wang et al., 2011). We found that ASY1 distribution on chromosomes in Zmcom1-1 meiocytes (n = 42) was similar to that in wild type meiocytes (n = 53) (Figures 5A,D). DSY2, a homolog of rice PAIR3 and Arabidopsis ASY3, acts as a structural protein to connect the AE/LEs to the CE for the SC assembly (Wang et al., 2011; Ferdous et al., 2012; Lee et al., 2015). In Zmcom1-1 meiocytes (n = 56), DSY2 also loaded regularly onto chromosomes during zygotene, and did not show any difference from the wild type (n = 47) (Figures 5B,E). We also investigated the installation of the AE components in Zmcom1-2, which was similar to those of Zmcom1-1 (Supplementary Figures S7A,B). Therefore, we conclude that the loss-of-function of ZmCom1 has no significant effect on the installation of the AEs.
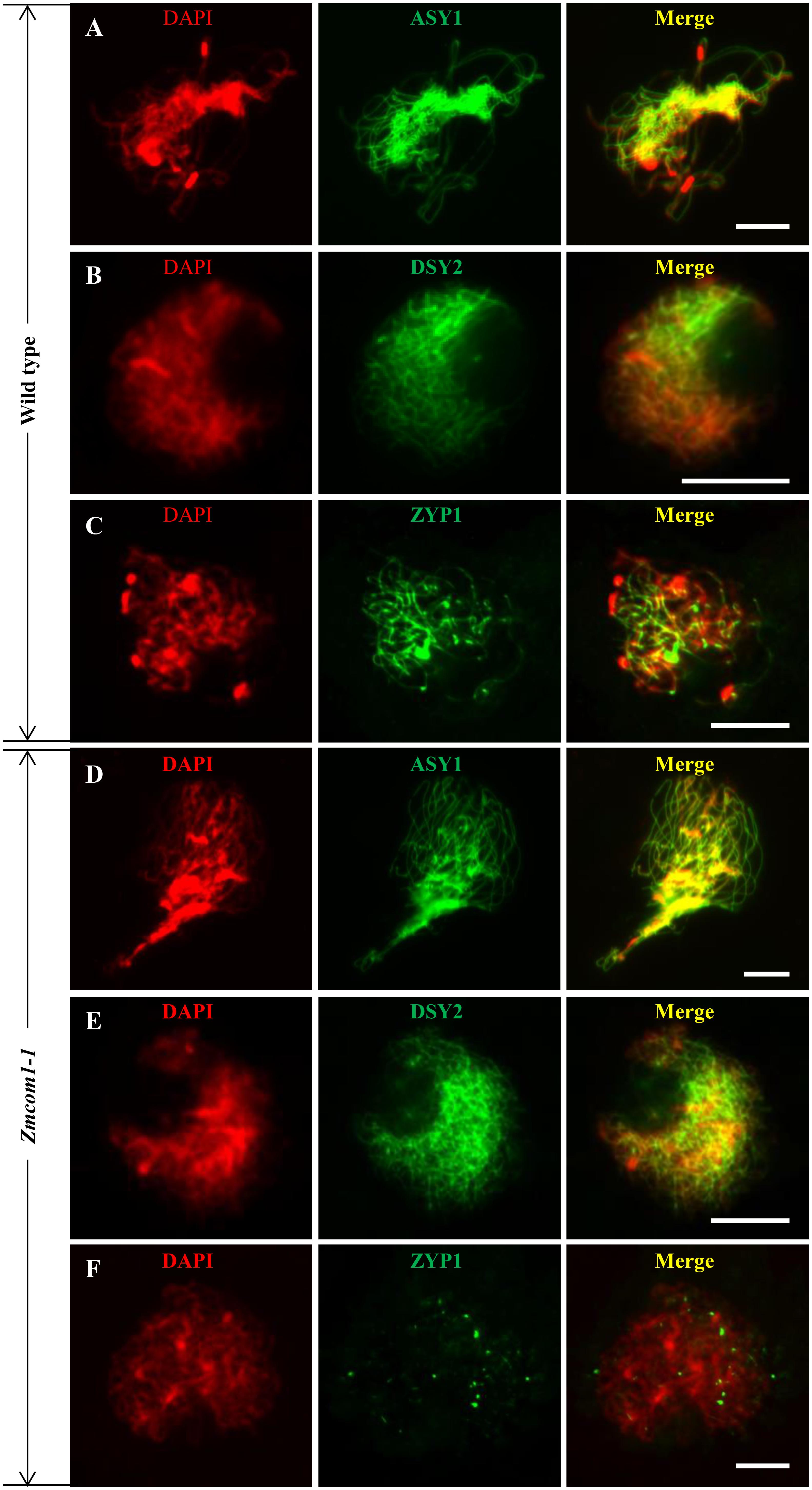
FIGURE 5. Immunolocalization of ASY1, DSY2, and ZYP1 antibodies in the wild type and Zmcom1-1. (A–C) ASY1 (A), DSY2 (B), and ZYP1 (C) on prophase I chromosomes in the wild type. DAPI staining is used to indicate the chromosomes. Scale bars = 10 μm. (D–F) ASY1 (D), DSY2 (E), and ZYP1 (F) on prophase I chromosomes in Zmcom1-1. Scale bars = 10 μm.
ZYP1, a transverse filament protein, constitutes the CE of the SC in maize (Higgins et al., 2005; Wang et al., 2010; Barakate et al., 2014). In the wild type, ZYP1 was first detected as discontinuous foci at the leptotene stage during zygotene, and it gradually formed discontinuous linear signals. At pachytene, ZYP1 signals were aligned perfectly along the entire chromosome length (n = 35; Figure 5C). In the Zmcom1-1 meiocytes, ZYP1 signals could not elongate to form linear signals and only present as punctate signals (n = 63; Figure 5F). We also investigated the installation of the CE component in Zmcom1-2, which was similar to those of Zmcom1-1 (Supplementary Figure S7C). Taken together, we conclude that the SC assembly is deficient in Zmcom1.
ZmCom1 Is Critical for DSB Repair
The chromosome fragmentation observed in Zmcom1-1 meiocytes suggests that DSBs could still maintain due to loss-of-function of ZmCom1, as it was observed in Oscom1 (Ji et al., 2012) and Atcom1 (Uanschou et al., 2007). To ascertain whether defective homologous pairing and synapsis in Zmcom1 were correlated with the improper DSB repair, the RAD51 immunostaining experiment was performed in wild type and Zmcom1-1 meiocytes. Loading of RAD51 onto the ssDNA serves as a cytological marker for DSB repair via HR in different organisms (Pawlowski et al., 2003). In the wild type zygotene meiocytes, a substantial number of RAD51 foci was observed (n = 33, Figure 6A). In contrast, a parallel experiment did not detect any RAD51 foci in Zmcom1-1 (n = 49, Figure 6B) or Zmcom1-2 meiocytes (n = 36, Supplementary Figure S8). These results indicate that ZmCom1 is required for the proper recruitment/loading of RAD51 onto the chromosomes and further demonstrate a serious defect in DSB repair in Zmcom1.
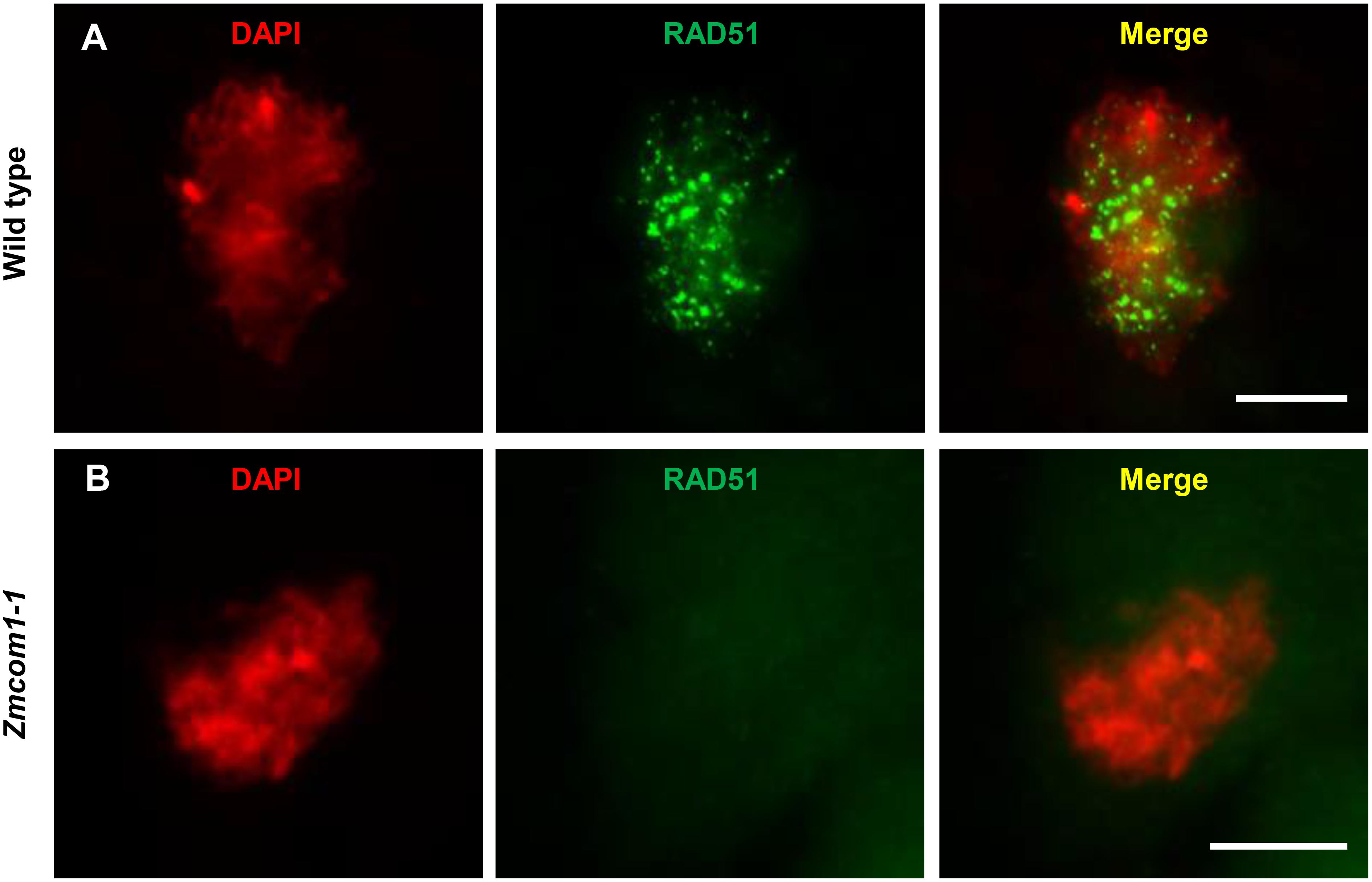
FIGURE 6. Immunolocalization of RAD51 antibodies in the wild type and Zmcom1-1 meioctyes at zygotene. (A) Wild type, (B) Zmcom1-1 mutant. DAPI staining is used to indicate the chromosomes. Scale bars = 10 μm.
Somatic Aberrations in Zmcom1
Unlike Atcom1 and Oscom1, Zmcom1 exhibited vegetative aberrations under standard growth conditions. To explore whether and how the loss-of-function of ZmCom1 influences the mitotic process, we assessed the frequency of chromosomal instability in root apical meristem for wild type and Zmcom1 plants. At prophase, there was no obvious deviation between the wild type and Zmcom1-1 (Figures 7A,D). However, we consistently observed an increased occurrence of acentric fragments at mitotic metaphase in Zmcom1-1 (10.5%, n = 238; Figure 7E and Table 1) compared to that in the wild type (0.3%, n = 323; Figure 7B and Table 1). Later, ∼12.8% of mitotic cells had bridges or chromosome fragments in Zmcom1-1 anaphase (n = 258; Figure 7F and Table 1), significantly higher than ∼0.3% of that in the wild type (n = 351; Figure 7C and Table 1). We also investigated the mitotic process in Zmcom1-2, which was similar to that of Zmcom1-1 (Supplementary Figure S9). These results suggest that Zmcom1 mutant suffers somatic chromosomal destabilization even under the normal growth condition.
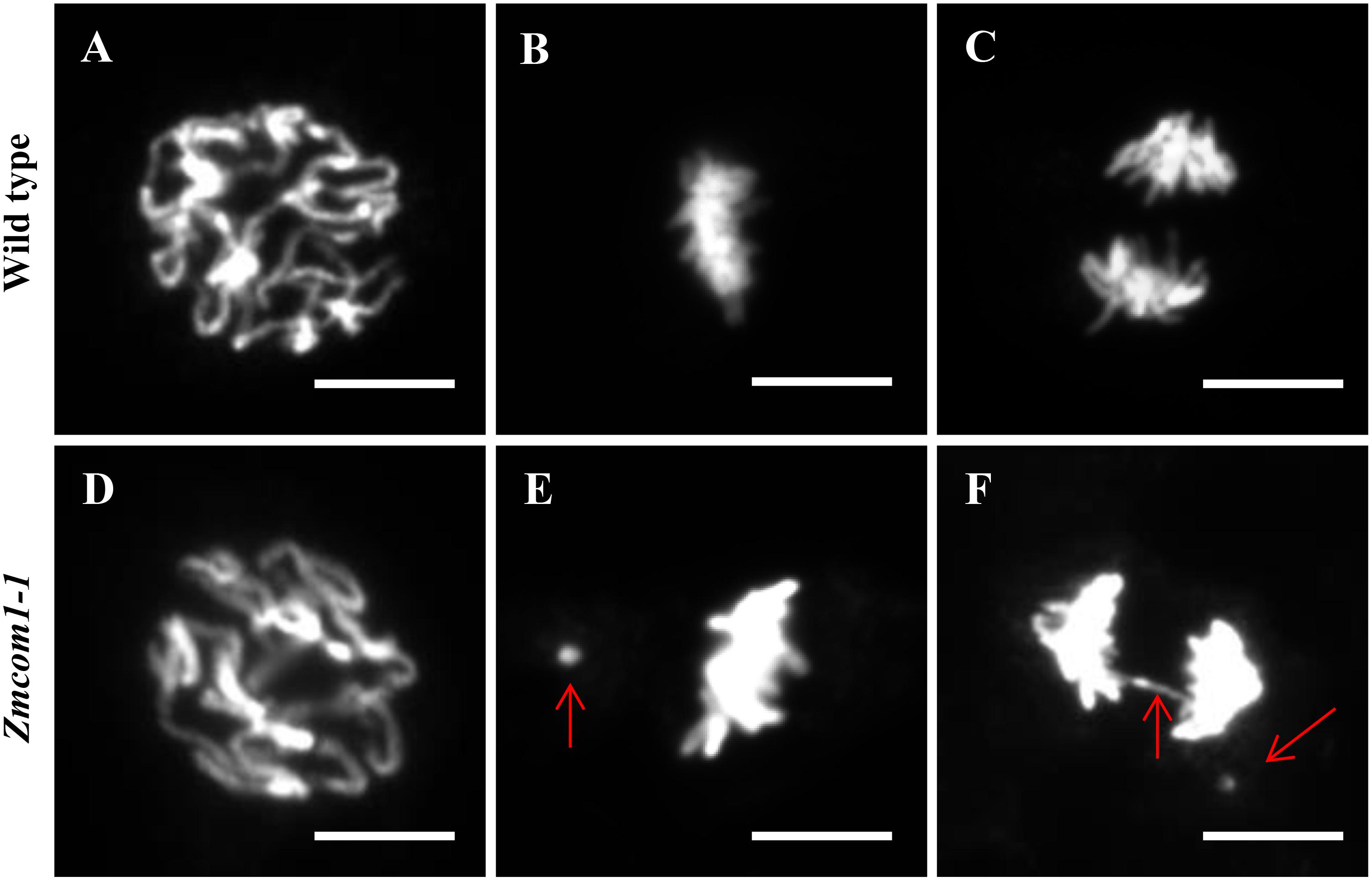
FIGURE 7. Genome instability in mitotic cells from Zmcom1-1 plants. (A–C) Wild type. (A) Prophase; (B) Metaphase; (C) Anaphase. Scale bars = 10 μm. (D–F) Zmcom1-1. (D) Prophase; (E) Metaphase; (F) Anaphase. Lagging.
Discussion
Role of ZmCom1 in Maize Meiosis
The conserved roles of CtIP/Ctp1/Sae2/Com1 in meiosis have been identified in several organisms. Consistent with this, in the present study we show that the loss-of-function of the ZmCom1 gene leads to chromosome fragmentation and defects in homologous pairing and synapsis during meiosis, indicating that also in maize, Com1 is an essential element in DSB repair. However, the precise effect of Com1 homolog on chromosome behavior differs from other plant organisms.
Telomere bouquet formation, a specialized arrangement of chromosomes during early prophase of meiosis in which telomeres are clustered on the nuclear envelope, has been observed in some plant species, animals and fungi (Niwa et al., 2000; Harper et al., 2004; Ding et al., 2007). Numerous maize mutants exhibit the defective bouquet including pam1 (Golubovskaya et al., 2002), dy (Murphy and Bass, 2012), dsy1 (Bass et al., 2003), afd1 (Golubovskaya et al., 2006), and phs1 (Pawlowski et al., 2004), as well as the rice pair3 (Wang et al., 2011) and zygo1 (Zhang et al., 2017). Meanwhile, all these mutants also show the concurrent abnormality in homologous pairing, suggesting that the proper bouquet formation is a key event to facilitate homologous chromosome pairing (Zhang et al., 2017).
It is interesting that, the telomere bouquet formation is unaffected in the rice com1 mutant and the telomere clustering is indistinguishable from that in the wild type (Ji et al., 2012), whereas in the maize com1 mutants, a typical telomere clustering was never observed indicating that the ZmCom1 gene is critically required for bouquet formation. Therefore, the remarkable difference between ZmCom1 and OsCom1 in mediating bouquet formation highlights the questions of why and how such character is conferred in different plant species. Also, the other intriguing question raised is whether the participation of ZmCom1 in bouquet formation is restricted to its own character, or other members of MRN complex are also involved. Those questions would be of great interest in future studies.
Chromosome fragmentation and entanglements is a typical phenomenon observed in mutants deficient in DSB repair machinery. Our data showed that the Zmcom1 mutant phenotype is similar to that of the Oscom1 and Atcom1 mutants, as well as other related mutants such as Atmre11 (Samanic et al., 2013), Atrad50 (Vannier et al., 2006), Osxrcc3 (Zhang et al., 2015), and Osrad51c (Tang et al., 2014). However, chromosome segregation and the integrity of the tetrads seems to be less severe in Zmcom1 compared to the Oscom1 or Atcom1 mutants. A simple explanation for this dissimilarity could be that the alternative DSB repair pathway, such as non-homologous end-joining (NHEJ) or microhomology-mediated end-joining (MMEJ) (McVey and Lee, 2008; Shrivastav et al., 2008), may be more actively stimulated in the absence of HR pathway in maize. In this scenario, ZmCom1 act as a regulator to balance the different DSB repair pathways, a mechanism suggested in the previous studies (Ji et al., 2012). Therefore, it would be worth to investigate the meiotic consequences after combining mutation in ZmCom11 with mutations in the genes involved in NHEJ and MMEJ pathway, which are largely unexplored yet in maize.
Role of ZmCom1 in Maize Mitosis
Unrepaired DSBs are one of the most lethal types of DNA damage and highly threaten on chromosome stability and cell survival (Edlinger and Schlögelhofer, 2011). Beside the programmed induction during meiosis, DSBs can be triggered by both endogenous (e.g., transposition events of transposable elements (TEs), errors of oxidative metabolism, stalled, or collapsed replication forks) and exogenous sources (e.g., ionizing radiation or genotoxic stresses) in the vegetative growth period. Organisms have evolved two major pathways, HR and NHEJ, for repairing DSBs and maintaining genome integrity. Coordinated with MRX/N complex, CtIP/Ctp1/Sae2/Com1 plays a critical role in HR. Therefore it is not surprised to find that the mutation in those genes will result in the increased sensitivity toward various genotoxic stresses. Indeed, Atcom1 mutants showed the retarded development of true leaves after treatment with mitomycin C (Uanschou et al., 2007). Meanwhile, without the special treatment, Atcom1 mutant plants grew well, and did not show any vegetative phenotypes compared to the wild type. This is also the case for Oscom1 mutant plants. Those results suggest that under the normal condition, the endogenous DSBs can be efficiently repaired in spite of lack of intact Com1–dependent HR in both Arabidopsis and rice. However, we observed some mitotic and vegetative abnormalities in Zmcom1 mutants when plants were grown under standard environmental conditions. As the appearance of those vegetative phenotypes seems to be unique for Zmcom1 mutants, we speculate that it can be attributed to the special feature of maize chromosomes. In contrast to Arabidopsis and rice, maize has a large genome with over 85% of TEs (Schnable et al., 2009; Andres and Williams, 2017). Although the mobility for the majority of TEs would be principally silenced by DNA methylation, a fraction of TEs still remains the activity for jumping around genome and driving genetic evolution (Mirouze and Vitte, 2014). In this context, maize genome may suffer from a greater frequency of transposition-derived DSBs compared to rice and Arabidopsis. Alternatively, alike mammalian cells(Feng et al., 2016), the pervasive distribution of repetitive element on maize chromosomes may have a tendency to cause replication fork stalling and subsequent collapse of stalk fork, and can induce replication-associated DSBs (Nikolov and Taddei, 2016). In both scenarios, the Com1 activity is hypothetically required to maintain the genome integrity. Meanwhile, as both TE-transposition and the collapse of stalk fork frequently occur during the S-phase of the cell cycle, it would be also conceivable to explain how the disruption of ZmCom1 led to the abnormality in the seed development, a period when cells fast divide and proliferate.
Author Contributions
YH conceived and supervised the project. YW, LJ, TZ, and JJ conducted the experiments. YW and YH prepared the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31471172).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of our laboratories for helpful discussion and assistance during this project. We are thankful to Chung-Ju Rachel Wang (Academia Sinica, Taiwan) for kindly providing us with ASY1, DSY2, and ZYP1 antibodies. In addition, we greatly thank Wojciech Pawlowski (Cornell University, United States) for gifting us RAD51 antibody. We thank Ljudmilla Timofejeva (Tallinn University of Technology, Estonia) for embellishing and critical reading of the article. Finally, we also greatly thank Weiwei Jin (China Agricultural University, China) for offering us the plasmids with 5S rDNA repeat and pAtT4 with telomeric repeats.
Supplementary Materials
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01005/full#supplementary-material
FIGURE S1 | Protein sequence alignment of ZmCom1 and OsCom1. The proteins were aligned with CLUSTALW and image was made by MultAlign (http://multalin.toulouse.inra.fr/multalin/). Conserved (>90% conservation) amino acid residues are red, variable (<50% conservation) are blue. Species abbreviation: Zm, Zea mays; Os, Oryza sativa. The red and green underlines indicate the conserved SMC-N and SAE2 domain, respectively.
FIGURE S2 | Neighbor-joining phylogeny reconstruction of Com1 homologs from different plant species. Numbers next to branches indicate posterior probability values. The scale indicates number of substitutions per site. Protein sequences were aligned using ClustalX (Jeanmougin et al., 1998) and phylogeny reconstruction was conducted using the online software (http://www.phylogeny.fr/, Dereeper et al., 2010). Species abbreviation: Zm, Zea mays; Os, Oryza sativa; Sb, Sorghum bicolor; At, Arabidopsis thaliana; Bn, Brassica napa; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Ta, Triticum aestivum; Cs, Camelina sativa; Rs, Raphanus sativus; Es, Eutrema salsugineum.
FIGURE S3 | Morphological comparison between wild type and Zmcom1-2 mutant. (A) Morphological comparison of mature seeds between wild type and Zmcom1-2 mutant. (B) Growth-curve of plant height in wild type and Zmcom1-2 mutant plants. Values are means of 10 individual plants. (C) Comparison of Morphological comparison of mature plants between wild type and Zmcom1-2 mutant. (D) Comparison of a wild type tassel and a Zmcom1-2 tassel at the flowering stage. (E) Comparison of a wild type ear and a Zmcom1-2 ear. (F) Normal pollen grains of the wild type. Scale bar = 100 μm. (G) Complete sterile pollen grains of the Zmcom1-2 plant. Scale bar = 100μm.
FIGURE S4 | PCR-based genotyping of seeds from self-propagated heterozygous Zmcom1 plants. (A) F2 progeny of Zmcom1-1. 1–6: Seeds with normal size; 7–24: Seeds with small size. (B) F2 progeny of Zmcom1-2. 25–30: Seeds with normal size; 31–48: Seeds with small size.
FIGURE S5 | Male meiosis in Zmcom1-2. (A) Leptotene; (B) Zygotene; (C) Pachytene; (D) Diakinesis; (E) Metaphase I; (G) Anaphase I; (F) Dyad; (H) Tetrads. The red arrows pointed out the chromosomal fragments and abnormal bridges. Scale bars = 10 μm.
FIGURE S6 | The defective bouquet formation and homologous pairing in Zmcom1-2. (A) Bouquet formation analysis using FISH with the telomere-specific pAtT4 probe in Zmcom1-2 (n = 41). Scale bars = 10 μm. (B) Homologous pairing analysis using FISH with 5S rDNA probe in in Zmcom1-2 (n = 27). Scale bars = 10 μm.
FIGURE S7 | Immunolocalization of ASY1, DSY2, and ZYP1 antibodies in Zmcom1-2. ASY1 (A, n = 23), DSY2 (B, n = 33), and ZYP1 (C, n = 37) on prophase I chromosomes in Zmcom1-2. Scale bars = 10 μm.
FIGURE S8 | Immunolocalization of RAD51 antibodies in Zmcom1-2 meioctyes (n = 36) at zygotene. DAPI staining is used to indicate the chromosomes. Scale bars = 10 μm.
FIGURE S9 | Genome instability in mitotic cells from Zmcom1-2 plants. (A) Prophase; (B) Metaphase; (C) Anaphase. Lagging chromosome fragments and anaphase bridges were highlighted by red arrows. Scale bars = 10 μm.
TABLE S1 | Primers used in this study.
TABLE S2 | Segregation ratio of small seeds versus normal seeds.
References
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. doi: 10.1016/S0092-8674(01)00416-0
Altun, C. (2008). Maize Mre11 DNA Repair and Recombination Complex. West Lafayette, IN: Purdue University.
Amiard, S., Charbonnel, C., Allain, E., Depeiges, A., White, C. I., and Gallego, M. E. (2010). Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22, 3020–3033. doi: 10.1105/tpc.110.078527
Andres, S. N., and Williams, R. S. (2017). CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair 56, 109–117. doi: 10.1016/j.dnarep.2017.06.013
Barakate, A., Higgins, J. D., Vivera, S., Stephens, J., Perry, R. M., Ramsay, L., et al. (2014). The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740. doi: 10.1105/tpc.113.121269
Baroni, E., Viscardi, V., Cartagena-Lirola, H., Lucchini, G., and Longhese, M. P. (2004). The functions of budding yeast SAE2 in the dna damage response require Mec1- and tel1-dependent phosphorylation. Mol. Cell. Biol. 24,4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004
Bass, H. W., Bordoli, S. J., and Foss, E. M. (2003). The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L) cause distinct telomere-misplacement phenotypes during meiotic prophase. J. Exp. Bot. 54, 39–46. doi: 10.1093/jxb/erg032
Borde, V. (2007). The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 15, 551–563. doi: 10.1007/s10577-007-1147-9
Carney, J. P., Maser, R. S., Olivares, H., Davis, E. M., Le Beau, M., Yates, J. R., et al. (1998). The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–486. doi: 10.1016/S0092-8674(00)81175-7
Cassani, C., Gobbini, E., Vertemara, J., Wang, W., Marsella, A., Sung, P., et al. (2018). Structurally distinct Mre11 domains mediate MRX functions in resection, end-tethering and DNA damage resistance. Nucleic Acids Res. 46, 2990–3008. doi: 10.1093/nar/gky086
Chen, P. L., Liu, F., Cai, S., Lin, X., Li, A., Chen, Y., et al. (2005). Inactivation of CtIP Leads to early embryonic lethality mediated by g1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25, 3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005
Cheng, Z. (2013). Analyzing meiotic chromosomes in rice. Methods Mol. Biol. 990, 125–134. doi: 10.1007/978-1-62703-333-6_13
Daoudal-Cotterell, S., Gallego, M. E., and White, C. I. (2002). The plant Rad50-Mre11 protein complex. FEBS Lett. 516, 164–166. doi: 10.1016/S0014-5793(02)02536-X
Dereeper, A., Audic, S., Claverie, J. M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Ding, X., Xu, R., Yu, J., Xu, T., Zhuang, Y., and Han, M. (2007). SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872. doi: 10.1016/j.devcel.2007.03.018
Edlinger, B., and Schlögelhofer, P. (2011). Have a break: determinants of meiotic DNA double strand break (DSB) formation and processing in plants. J. Exp. Bot. 62, 1545–1563. doi: 10.1093/jxb/erq421
Feng, Y. L., Xiang, J. F., Kong, N., Cai, X. J., and Xie, A. Y. (2016). Buried territories: heterochromatic response to DNA double-strand breaks. Acta Biochim. Biophys. Sin. 48, 594–602. doi: 10.1093/abbs/gmw033
Ferdous, M., Higgins, J. D., Osman, K., Lambing, C., Roitinger, E., Mechtler, K., et al. (2012). Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8:e1002507. doi: 10.1371/journal.pgen.1002507
Golubovskaya, I. N., Hamant, O., Timofejeva, L., Wang, C. J., Braun, D., Meeley, R., et al. (2006). Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119, 3306–3315. doi: 10.1242/jcs.03054
Golubovskaya, I. N., Harper, L. C., Pawlowski, W. P., Schichnes, D., and Cande, W. Z. (2002). The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162, 1979–1993.
Gray, S., and Cohen, P. E. (2016). Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210. doi: 10.1146/annurev-genet-120215-035111
Harper, L., Gardiner, J., Andorf, C., and Lawrence, C. J. (2016). MaizeGDB: the maize genetics and genomics database. Methods Mol. Biol. 1374, 187–202. doi: 10.1007/978-1-4939-3167-5_9
Harper, L., Golubovskaya, I., and Cande, W. Z. (2004). A bouquet of chromosomes. J. Cell Sci. 117, 4025–4032. doi: 10.1242/jcs.01363
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H., and Franklin, F. C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500. doi: 10.1101/gad.354705
Hopfner, K. P., Putnam, C. D., and Tainer, J. A. (2002). DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12, 115–122. doi: 10.1016/S0959-440X(02)00297-X
Hunter, N., and Kleckner, N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70. doi: 10.1016/S0092-8674(01)00430-5
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal x. Trends Biochem. Sci. 23, 403–405. doi: 10.1016/S0968-0004(98)01285-7
Ji, J. H., Tang, D., Wang, K. J., Wang, M., Che, L. X., Li, M., et al. (2012). The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis. Plant J. 72, 18–30. doi: 10.1111/j.1365-313X.2012.05025.x
Johnson-Brousseau, S. A., and McCormick, S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39, 761–775. doi: 10.1111/j.1365-313X.2004.02147.x
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U.S.A. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Keeney, S., Giroux, C. N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. doi: 10.1016/S0092-8674(00)81876-0
Klutstein, M., Fennell, A., Fernandez-Alvarez, A., and Cooper, J. P. (2015). The telomere bouquet regulates meiotic centromere assembly. Nat. Cell Biol. 17, 458–469. doi: 10.1038/ncb3132
Lee-Theilen, M., Matthews, A. J., Kelly, D., Zheng, S., and Chaudhuri, J. (2010). CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18, 75–79. doi: 10.1038/nsmb.1942
Lee, D. H., Kao, Y. H., Ku, J. C., Lin, C. Y., Meeley, R., Jan, Y. S., et al. (2015). The axial element protein desynaptic2 mediates meiotic double-strand break formation and synaptonemal complex assembly in maize. Plant Cell 27,2516–2529. doi: 10.1105/tpc.15.00434
Lengsfeld, B. M., Rattray, A. J., Bhaskara, V., Ghirlando, R., and Paull, T. T. (2007). Sae2 Is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28, 638–651. doi: 10.1016/j.molcel.2007.11.001
Li, H., Li, J., Cong, X. H., Duan, Y. B., Li, L., Wei, P. C., et al. (2013). A high-throughput, high-quality plant genomic DNA extraction protocol. Genet. Mol. Res. 12, 4526–4539. doi: 10.4238/2013.October.15.1
Li, L., and Arumuganathan, K. (2001). Physical mapping of 45S and 5S rDNA on maize metaphase and sorted chromosomes by FISH. Hereditas 134, 141–145. doi: 10.1111/j.1601-5223.2001.00141.x
Limbo, O., Chahwan, C., Yamada, Y., De Bruin, R. A., Wittenberg, C., and Russell, P. (2007). Ctp1 Is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 28, 134–146. doi: 10.1016/j.molcel.2007.09.009
McKee, A. H. Z., and Kleckner, N. (1997). A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146, 797–816.
McVey, M., and Lee, S. E. (2008). MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24, 529–538. doi: 10.1016/j.tig.2008.08.007
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N., and Grelon, M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327. doi: 10.1146/annurev-arplant-050213-035923
Mirouze, M., and Vitte, C. (2014). Transposable elements, a treasure trove to decipher epigenetic variation: insights from Arabidopsis and crop epigenomes. J. Exp. Bot. 65, 2801–2812. doi: 10.1093/jxb/eru120
Murphy, S. P., and Bass, H. W. (2012). The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. J. Cell Sci. 125,3681-3690. doi: 10.1242/jcs.108290
Nikolov, I., and Taddei, A. (2016). Linking replication stress with heterochromatin formation. Chromosoma 125, 523–533. doi: 10.1007/s00412-015-0545-6
Niwa, O., Shimanuki, M., and Miki, F. (2000). Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19, 3831–3840. doi: 10.1093/emboj/19.14.3831
Paull, T. T., and Gellert, M. (1998). The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1, 969–979. doi: 10.1016/S1097-2765(00)80097-0
Pawlowski, W. P., Golubovskaya, I. N., and Cande, W. Z. (2003). Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15,1807–1816. doi: 10.1105/tpc.012898
Pawlowski, W. P., Golubovskaya, I. N., Timofejeva, L., Meeley, R. B., Sheridan, W. F., and Cande, W. Z. (2004). Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92. doi: 10.1126/science.1091110
Penkner, A., Portik-Dobos, Z., Tang, L., Schnabel, R., Novatchkova, M., Jantsch, V., et al. (2007). A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 26, 5071–5082. doi: 10.1038/sj.emboj.7601916
Prinz, S., Amon, A., and Klein, F. (1997). Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146, 781–795.
Puizina, J., Siroky, J., Mokros, P., Schweizer, D., and Riha, K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978. doi: 10.1105/tpc.104.022749
Richards, E. J., and Ausubel, F. M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53, 127–136. doi: 10.1016/0092-8674(88)90494-1
Samanic, I., Simunic, J., Riha, K., and Puizina, J. (2013). Evidence for distinct functions of MRE11 in Arabidopsis meiosis. PLoS One 8:e78760. doi: 10.1371/journal.pone.0078760
Sanchez-Moran, E., Osman, K., Higgins, J. D., Pradillo, M., Cunado, N., Jones, G. H., et al. (2008). ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenet. Genome Res. 120, 302–312. doi: 10.1159/000121079
Sartori, A. A., Lukas, C., Coates, J., Mistrik, J., Fu, S., Bartek, J. (2007). Human CtIP promotes DNA end resection. Nature 450, 509–514. doi: 10.1038/nature06337
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134–147. doi: 10.1038/cr.2007.111
Tang, D., Miao, C. B., Li, Y. F., Wang, H. J., Liu, X. F., Yu, H. X., et al. (2014). OsRAD51C is essential for double-strand break repair in rice meiosis. Front. Plant Sci. 5:167. doi: 10.3389/fpls.2014.00167
Uanschou, C., Siwiec, T., Pedrosa-Harand, A., Kerzendorfer, C., Sanchez-Moran, E., Novatchkova, M., et al. (2007). A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26, 5061–5070. doi: 10.1038/sj.emboj.7601913
Vannier, J. B., Depeiges, A., White, C., and Gallego, M. E. (2006). Two roles for Rad50 in telomere maintenance. EMBO J. 25, 4577–4585. doi: 10.1038/sj.emboj.7601345
Wang, K., Wang, M., Tang, D., Shen, Y., Qin, B., Li, M., et al. (2011). PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 22, 12–19. doi: 10.1091/mbc.e10-08-0667
Wang, M., Wang, K., Tang, D., Wei, C., Li, M., Shen, Y., et al. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430. doi: 10.1105/tpc.109.070789
Waterworth, W. M., Altun, C., Armstrong, S. J., Roberts, N., Dean, P. J., Young, K., et al. (2007). NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 52, 41–52. doi: 10.1111/j.1365-313X.2007.03220.x
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, B. W., Wang, M., Tang, D., Li, Y. F., Xu, M., Gu, M. H., et al. (2015). XRCC3 is essential for proper double-strand break repair and homologous recombination in rice meiosis. J. Exp. Bot. 66, 5713–5725. doi: 10.1093/jxb/erv253
Zhang, F., Tang, D., Shen, Y., Xue, Z., Shi, W., Ren, L., et al. (2017). The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell 29, 2597–2609. doi: 10.1105/tpc.17.00287
Zhang, H., Egger, R. L., Kelliher, T., Morrow, D., Fernandes, J., Nan, G. L., et al. (2014). Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 4, 993–1010. doi: 10.1534/g3.113.009738
Keywords: maize, meiosis, HR, DSB, COM1
Citation: Wang Y, Jiang L, Zhang T, Jing J and He Y (2018) ZmCom1 Is Required for Both Mitotic and Meiotic Recombination in Maize. Front. Plant Sci. 9:1005. doi: 10.3389/fpls.2018.01005
Received: 28 March 2018; Accepted: 20 June 2018;
Published: 16 July 2018.
Edited by:
Mónica Pradillo, Complutense University of Madrid, SpainReviewed by:
Chung-Ju Rachel Wang, Academia Sinica, TaiwanIsabelle Colas, The James Hutton Institute, United Kingdom
Copyright © 2018 Wang, Jiang, Zhang, Jing and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan He, eWgzNTJAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Yazhong Wang
Yazhong Wang Luguang Jiang
Luguang Jiang Ting Zhang
Ting Zhang Juli Jing
Juli Jing Yan He
Yan He