- 1Centro de Biotecnología y Genómica de Plantas, Universidad Politécnica de Madrid, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Madrid, Spain
- 2Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid, Madrid, Spain
Tetranychus urticae (two-spotted spider mite) is a striking example of polyphagy among herbivores with an extreme record of pesticide resistance and one of the most significant pests in agriculture. The T. urticae genome contains a large number of cysteine- and serine-proteases indicating their importance in the spider mite physiology. This work is focused on the potential role of the Kunitz trypsin inhibitor (KTI) family on plant defense responses against spider mites. The molecular characterization of two of these genes, AtKTI4 and AtKTI5, combined with feeding bioassays using T-DNA insertion lines for both genes was carried out. Spider mite performance assays showed that independent KTI silencing Arabidopsis lines conferred higher susceptibility to T. urticae than WT plants. Additionally, transient overexpression of these inhibitors in Nicotiana benthamiana demonstrated their ability to inhibit not only serine- but also cysteine-proteases, indicating the bifunctional inhibitory role against both types of enzymes. These inhibitory properties could be involved in the modulation of the proteases that participate in the hydrolysis of dietary proteins in the spider mite gut, as well as in other proteolytic processes.
Introduction
The fact that higher plants are sessile organisms has favored the acquisition of sophisticated resources to prevent or hamper pest feeding (Walling, 2000; Wu and Baldwin, 2010). Such defenses can be constitutive and/or induced upon attack by herbivore pests. Induced defenses include morphological and metabolic changes with a negative impact on phytophagous arthropod behavior (Walling, 2000; Howe and Jander, 2008; Alba et al., 2011) or the attraction of natural enemies of the herbivore (Sabelis et al., 1999; Dicke and Baldwin, 2010). Plant receptors recognize Herbivore-Associated Molecular Patterns (HAMPs), Microbe-Associated Molecular Patterns (MAMPs) and Damage-Associated Molecular Patterns (DAMPs) and trigger the induction of defenses (Mithöfer et al., 2005; Frost et al., 2007; Staudacher et al., 2017; Santamaria et al., 2018a). Plant responses are specific to the phytophagous pest species (Stout et al., 1998; de Vos et al., 2005; Rodriguez-Saona et al., 2010) and dependent on the duration of the infestation (Kant et al., 2004). However, the perception of herbivory is not well understood and few plant receptors have been identified (Bonaventure, 2012; Santamaria et al., 2018a).
Among plant defenses, protease inhibitors (PIs) exert direct effects on herbivores by interfering with their physiology (Diaz and Santamaria, 2012; Martinez et al., 2016). When an arthropod ingests PIs in its diet, the inhibition of proteolytic activities takes place in its gut avoiding the degradation of proteins. In this context, PI classification may be based on the type of protease they inhibit. There are three main subclasses of proteases involved in arthropod digestion, serine-, cysteine-, and aspartic-proteases, grouped according to the reactive amino acid of their active site group (Terra and Ferreira, 2012). The proteolytic activity is dependent on the pH of the gut (Ortego, 2012; Martinez et al., 2016). Most lepidopteran, orthopteran and hymenopteran and some coleopteran possess alkaline midguts and their digestive systems are largely based on serine-proteases and exopeptidases (Wolfson and Murdock, 1990; Ortego et al., 1996; Johnson and Rabosky, 2000). The majority of coleopteran, hemipteran and some phytophagous acari have slightly acidic midguts providing cysteine- and aspartic-proteases and exopeptidases their major proteolytic activity (Murdock et al., 1987; Cristofoletti et al., 2003; Carrillo et al., 2011). Since Green and Ryan (1972) reported that wound-inducible PIs inhibited digestive herbivore gut proteases, numerous plant PIs have been characterized for their potential to control herbivorous insects (Hilder et al., 1987; Alfonso-Rubi et al., 2003; Tamhane et al., 2005; Telang et al., 2009; Chen et al., 2014). According to MEROPS database, there are currently 85 families of PIs (Rawlings et al., 2018), being the Kazal, Kunitz, Bowman-Birk, Potato I and II, Cystatin, Cereal trypsin/α-amylase, and Serpin families the most represented in plants (Santamaria et al., 2014). Most of them specifically inhibit a mechanistic class of proteases but some may act as multifunctional inhibitors (Grosse-Holz and van der Hoorn, 2016). The first successful PI gene used to improve resistance against larvae of Heliothis virescens when expressed in transgenic tobacco was the cowpea trypsin inhibitor gene (CPTI) from the Bowman–Birk family (Hilder et al., 1987). Then, the CPTI gene was inserted in the genome of other plants like cotton, rice, cabbage, strawberry, sweet potato, potato or pigeon pea enhancing the resistance to different lepidopteran species (reviewed in Diaz and Santamaria, 2012). The I3 Kunitz Trypsin Inhibitor (KTI) gene family is a complex family composed by versatile protease inhibitors. Most of them inhibit serine proteases (families S1 and S8), but some of them are able to inhibit cysteine proteases (families C1 and C13) as well as other hydrolases (Renko et al., 2012). This family has been studied in different plants and contexts but most works have been focused on their potential role in defense against insect attack since their gene expression is up-regulated in response to wounding, jasmonates and insect feeding (Major and Constabel, 2008; Philippe et al., 2009; Botelho-Junior et al., 2014). In vitro assays with KTIs from poplar and soybean expressed as recombinant proteins differentially inhibited midgut proteases from Mamestra configurata and Malacosoma disstria, lepidopteran pests from Populus and crucifers, respectively (Major and Constabel, 2008). KTIs from the passion fruit displayed activity against midgut serine and cysteine proteases from the sugarcane borer Diatraea saccharalis and the coleopteran Callosobruchus maculatus on artificial diets (Botelho-Junior et al., 2014). In addition, the heterologous expression of a good number of KTIs in poplar, sweet corn, potato, rice, tobacco, and tomato conferred resistance to lepidopteran (Confalonieri et al., 1998; Cipriani et al., 1999; Gatehouse et al., 1999; Lee et al., 1999; McManus et al., 1999; Nandi et al., 1999; Marchetti et al., 2000; Rufino et al., 2013; Guimarães et al., 2015), coleopteran (Major and Constabel, 2008) and acari (Castagnoli et al., 2003). Likewise, serine-PIs from other different families overexpressed in several plant species have conferred resistance to lepidopteran, coleopteran, homopteran (reviewed in Diaz and Santamaria, 2012) and acari (Santamaria et al., 2012).
Tetranychus urticae is an extreme polyphagous pest with more than 1,100 documented host plants and an extraordinary ability to develop pesticide resistance (Van Leeuwen and Dermauw, 2016). These features, along with the predicted expansion of spider mites under climate change conditions, make T. urticae one of the most significant pests in the agriculture (Luedeling et al., 2011). Phytophagous mites pierce parenchymatic plant cells using stylets to suck their nutrients, and cause severe chlorosis leading to a reduction in crop yield (Park and Lee, 2002; Farouk and Osman, 2011; Bensoussan et al., 2016). T. urticae is a model within chelicerate herbivores with its genome sequenced and a broad range of tools and protocols developed (Grbic et al., 2011; Cazaux et al., 2014; Suzuki et al., 2017). Besides, mite ability to feed on Arabidopsis thaliana and the wide available toolkits for this plant species have provided an outstanding opportunity for functional studies of plant-mite interaction (Santamaria et al., 2012, 2015a, 2017; Zhurov et al., 2014). Among plant PIs, cystatins and serine-protease inhibitors have been reported to be involved in Arabidopsis defense against spider mite. According to Santamaria et al. (2012), the over-expression of barley cystatin (Icy6 gene) and/or trypsin inhibitor (Itr1 gene) conferred Arabidopsis resistance by producing an increase in mite mortality. In addition, members from I3 and I13 Potato Inhibitor I families are induced upon T. urticae infestation in tomato and Arabidopsis (Martel et al., 2015). In the case of plant cystatins, it is well known that their targets in mites are digestive cysteine-proteases (Carrillo et al., 2011; Santamaria et al., 2012, 2015b, 2018b). In contrast, mite targets for serine-protease inhibitors remain unknown. The fact that an Arabidopsis KTI is able to inhibit papain-like cysteine proteases and participates in the defense against herbivorous arthropods (Boex-Fontvieille et al., 2016; Rustgi et al., 2017) prompted us to examine the role of the Arabidopsis KTI protease inhibitor family in plant defense against mites. Among the seven KTIs identified in Arabidopsis, AtKTI4 and AtKTI5 were selected because of the induction of their corresponding genes after spider mite feeding, and the differences in their amino acid sequences suggesting tridimensional structure dissimilarities. Knock down lines for these two Arabidopsis KTIs genes were used to analyze plant phenotypes after spider mite infestation. Behavior of mites fed on knock down lines was also evaluated to verify KTI effect on mite performance. Furthermore, transient overexpression of these inhibitors in Nicotiana benthamiana was performed to test their ability to inhibit both serine- and cysteine-proteases.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col-0), Kondara (Kon) and Bla-2 (Bla-2) accessions (Nottingham Arabidopsis Seed Collection) were used as wild-types (WT). A. thaliana T-DNA mutants (SALK_131716C, SALK_067224, SALK_115805C and SALK_009101C, referred as kti4.1, kti4.2, kti5.1, and kti5.2, respectively) were obtained from the Arabidopsis Biological Resource Centre, through the European Arabidopsis Stock Centre. For soil growth, a mixture of peat moss and vermiculite (2:1 v/v) was used. Sterilized seeds were stratified in the dark at 4°C for 5 days. Plants were then grown in growth chambers (Sanyo MLR-350-H) under control conditions (23°C ± 1°C, >70% relative humidity and a 16 h/8 h day/night photoperiod).
Spider Mite Maintenance
A colony of T. urticae, London strain (Acari: Tetranychidae), provided by Dr. Miodrag Grbic (UWO, Canada), was reared on beans (Phaseolus vulgaris) and maintained on growth chambers (Sanyo MLR-350-H, Sanyo, Japan) at 25°C ± 1°C, >70% relative humidity and a 16 h/8 h day/night photoperiod.
Sequence Analysis and Molecular Modeling
Multiple protein alignment was performed by MUSCLE program (Edgar, 2004). Arabidopsis Kunitz proteins were named following Santamaria et al. (2014) changing the Kun family term for the more commonly used KTI term. The KTI of Delonix regia (PDB ID 1R8N) was included in the alignment to infer secondary structure locations. Displayed multiple sequence alignment was made by the ESPript 3.0 web server (Robert and Gouet, 2014). 3D modeling was performed using SWISS-MODEL online protein structure prediction tool (Biasini et al., 2014). The known structures of two KTIs (PDB IDs: 3I2A and 3IIR) were used to construct the models for AtKTI4 (At1g73260) and AtKTI5 (At1g17860), respectively. Predictions on papain–AtKTI4 and papain–AtKTI5 interactions were made by using the obtained 3D models and the 3D structure of papain (PDB ID: 1PPN) in the ClusPro 2.0 server (Kozakov et al., 2017). Molecular models were visualized and analyzed by Chimera 1.12 program (Pettersen et al., 2004).
Nucleic Acid Analysis
Genomic DNA was isolated from Arabidopsis T-DNA insertion and control lines essentially as described by Sambrook and Russell (2001). The presence and homozygous status of the T-DNA insertion lines were validated by conventional PCR (Bio-Rad) (Supplementary Figure S1). Specific primers were designed through the Salk Institute website1. Primer sequences are indicated in Supplementary Table S1.
For quantitative real time PCR (RT-qPCR) studies, Arabidopsis rosettes from T-DNA insertion and control lines were collected, frozen into liquid N2 and stored at –80°C until used for RNA isolation. Total RNA was extracted by the phenol/chloroform method, followed by precipitation with 8 M LiCl (Oñate-Sánchez and Vicente-Carbajosa, 2008). Regarding N. benthamiana assays, total RNA was extracted from plants agroinfiltrated with 35S::GFP, 35S::KTI4-GFP and 35S::KTI5-GFP by the TRIZOL reagent following manufacturer instructions (Ambion, Austin, TX, United States). Complementary DNAs (cDNAs) were synthesized from 2 μg of RNA using the Revert AidTM H Minus First Strand cDNA Synthesis Kit (Fermentas) following manufacturer’s instructions. The RT-qPCR conditions used were 40 cycles with 15 s at 95°C, 1 min at 60°C and 5 s at 65°C using FastStart Universal SYBR Green Master (Rox) (Roche). RT-qPCR was performed for three samples coming from three independent experiments as previously described (Santamaria et al., 2017) using a SYBR Green Detection System (Roche) and the CFX Manager Software 2.0 (Bio-Rad). mRNA quantification was expressed as relative expression levels (2-dCt) or fold change (2-ddct) normalized to ubiquitin or actin for Arabidopsis and Nicotiana samples, respectively (Livak and Schmittgen, 2001). Specific primers were designed through PRIMER32. Primer sequences are indicated in Supplementary Table S1.
Enzymatic Assays
Total protein extracts from the T-DNA insertion lines and control Arabidopsis rosettes were resuspended in 50 mM sodium phosphate pH 6.0, 0.15 M NaCl 2 mM EDTA, for 1 h at 4°C and treated as described in Santamaria et al. (2012). Total protein content was determined according to the method of Bradford (1976). Cathepsin B- and L-like activities were assayed using N-carbobenzoxy-Arg-Arg-7-amido-4-methylcoumarin (Z-RR-AMC) and N-carbobenzoxy-Phe-Arg-AMC (Z-FR-AMC) commercial substrates, respectively. Trypsin- and chymotrypsin-like activities were analyzed using Z-L-Arg-AMC (ZLA-AMC) and Suc-Ala-Ala-Pro-Phe-AMC (Suc-A-A-P-F-AMC) commercial substrates, respectively.
Inhibitory activity of plant protein extracts was tested in vitro against commercial trypsin (EC 3.4.21.4), chymotrypsin (EC 3.4.21.1), papain (EC 3.4.22.2), and bovine cathepsin B (EC 3.4.22.1) from Sigma. Basically, 20 μg of protein extracts were preincubated for 10 min with 100 ng of cathepsin L- and B-like in a buffer containing 100 mM sodium phosphate pH 6.0, L-cysteine, 10 mM EDTA, and 0.01% (v/v) Brij35, or with 100 ng of trypsin/chymotrypsin in the buffer 0.1 M Tris-HCl pH 7.5. Subsequently, substrates were added at a final concentration of 25 μM and incubated for 1 h at 28°C or 37°C for cysteine and serine proteases, respectively. Fluorescence was measured using an excitation filter of 365 nm and an emission filter of 465 nm (Tecan GeniusPro). The system was calibrated with known amounts of 7-amido-4-methylcoumarin (AMC) hydrolysis product in a standard reaction mixture. Specific enzymatic activity was represented as nmoles of substrate hydrolyzed/min/mg of protease. Inhibitory activity was expressed as percentage of protease activity relative to that in the absence of the inhibitor. All assays were carried out in triplicates and blanks were used to account for spontaneous breakdown of substrates.
Subcellular Location
To create the translational fusions of AtKTI4 and AtKTI5 genes to the Green Fluorescent Protein (GFP) reporter gene, the corresponding cDNAs were amplified by conventional PCR using specific primers (Supplementary Table S1). The amplicons were independently cloned in-frame with the GFP gene into the Gateway binary vector pGWB5 (Invitrogen), under the CaMV35S promoter. 35S-Red Fluorescent Protein (RFP)-HDEL plasmid was used as a control of ER location (Shockey et al., 2006). Transient transformation of onion (Allium cepa) epidermal cells was performed by particle bombardment with a biolistic helium gun device (DuPont PDS-1000; Bio-Rad) as described by Diaz et al. (2005). Fluorescent images were acquired after 24 h of incubation at 22°C in the dark, using a Leica TCS-SP8 confocal microscope. GFP and RFP signals were acquired sequentially using the following settings: GFP, excitation 488 nm and emission 492–552 nm; RFP, excitation 561 nm, emission 581–665 nm.
For N. benthamiana agroinfiltration, the Agrobacterium tumefaciens strain C58CI RifR (GV3101) carrying the constructs 35S::KTI4-GFP (pGWB5), 35S::KTI5-GFP (pGWB5) or 35S::GFP (pEAQ-HT-GFP) was co-incubated with the construct 35S::P19 (pBIN61) that carries the helper P19 (Voinnet et al., 2015) to a final optical density of 0.3 nm. Bacterial suspensions were infiltrated into the abaxial side of the third-youngest fully expanded N. benthamiana leaf using a syringe. Fluorescent images were acquired 3 days post-infiltration (dpi), using a Leica TCS-SP8 confocal microscope. GFP and chlorophyll autofluorescence signals were acquired sequentially using the following settings: GFP, excitation 488 nm and emission 500–600 nm; chlorophyll, excitation 633 nm, emission 639–727 nm.
Plant Damage Determination
Damage quantification analyses were done on A. thaliana plants from T-DNA insertion lines and WT control. Three week-old plants were infested with 20 T. urticae adults per plant. After 4 days of infestation, leaf damage was assessed by scanning the entire rosette using a hp scanjet (HP Scanjet 5590 Digital Flatbed Scanner series), according to Cazaux et al. (2014). Leaf damage was calculated in mm2 using Adobe Photoshop CS software. Six replicates were used for each genotype. Plant damage was also evaluated by analyzing accumulation of hydrogen peroxide (H2O2) and the cell death in response to spider mite attack. Leaf disks (9 mm diameter) from 3 week-old plants from the five studied genotypes, were infested with 10 mites during 24 h. The H2O2 accumulation was analyzed using 3,3-diaminobenzidine tetrachloride hydrate (DAB) substrate which produces a brown precipitate after oxidation in the presence of H2O2 (Martinez de Ilarduya et al., 2003). The staining procedure was performed according to Rodríguez-Herva et al. (2012), and observed under a Zeiss Axiophot microscope. Damage was quantified using Image J software.
Cell death was quantified by trypan blue staining as described by Sanchez-Vallet et al. (2010). Leaves were boiled in trypan blue solution [10 ml lactic acid (Sigma)], 10 ml phenol (Sigma), 10 ml glycerol (Duchefa), 10 ml water and 10 mg trypan blue (Sigma) diluted with 96% (1:2 v/v) ethanol in a 15 ml tube for 1 min. Tissues were incubated in the staining solution overnight at room temperature. Subsequently, leaves were cleared four times with 2.5 g/ml of chloral hydrate solution (PRS). Cleared leaves were prepared for imaging in 50% (v/v) glycerol, 0.1 M sodium phosphate buffer at pH 7.0, and observed under a Zeiss Axiophot microscope. Damage was quantified using Adobe Photoshop CS6 and values relativized to control plants. Six replicates were used for each genotype.
Spider Mite Performance
Mite bioassays were conducted on entire detached Arabidopsis leaves from the T-DNA insertion and control plants. Entire leaves were fit into a closed system with 100 eggs. Samples were maintained under controlled conditions at 25 ± 1°C, >70% relative humidity and a 16 h/8 h day/night photoperiod. To study mortality, the percentage of neonate larvae (<24 h of hatching) that became adults was recorded after 10 days of feeding. Eight replicates were used for each genotype. For fecundity assays detached entire leaves were infested with 12 synchronized females each, and the number of eggs laid was counted after 36 h. Female mite synchronization was conducted on bean leaves. Entire detached leaves were placed onto wet cotton, surrounded by wet filter paper to avoid mite escape in confined special dishes (11.5 cm diameter with ventilation). 50 female mites were placed on each leaf and removed after 36 h. After 11 days, same age females were used to infest Arabidopsis leaves for the fecundity assay.
Inhibitory Ability of KTIs Overexpressed in Nicotiana
To test the ability of AtKTI4 and AtKTI5 to inhibit cysteine/serine proteases, we used extracts from N. benthamiana plants co-agroinfiltrated with 35S::KTI4-GFP (pGWB5), 35S::KTI5-GFP (pGWB5) or 35S::GFP (pGWB5), and 35S::P19 (pBIN61). Entire agroinfiltrated leaves were collected at 3 dpi (days post infiltration). Fluorescent images were acquired using a Leica Fluorescence Stereoscope MZ10F. Leaves were homogenized in a buffer containing 50 mM Tris-HCl pH 7.25, 150 mM NaCl, 2 mM EDTA and 0.1% (v/v) Triton X-100, centrifuged at 16,630 g at 4°C for 10 min and the supernatants were pooled to obtain soluble protein extracts for inhibition assays. The ability to inhibit commercial papain, cathepsin-B, trypsin and chymotrypsin activities was in vitro tested as indicated above but using 15 μg of plant protein extracts for the assays. In addition, E-64 [trans-Epoxysuccinyl-L-leucylamido (4-guanidino)butane], TLCK (Nα-Tosyl-L-lysine chloromethyl ketone hydrochloride) and chymostatin [N-(Nα-Carbonyl-Cpd-X-Phe-al)-Phe] inhibitors (Sigma) were added as positive inhibitory controls for papain and cathepsin-B, trypsin and chymotrypsin inhibition, at a final concentrations of 0.02, 100, and 0.1 μM, respectively, and incubated for 10 min before the addition of substrates.
Statistical Analysis
Differences in gene expression, leaf damage, mortality and fecundity assays were compared by One-Way ANOVA, followed by Student Newman–Keuls multiple comparison test (p < 0.05). Compensation effects and enzymatic assays were compared by One-Way ANOVA followed by Dunnett’s multiple comparison test (p < 0.05).
Results
Gene Expression of Arabidopsis I3 Kunitz Inhibitors in Response to T. urticae Infestation
As a first estimation of the importance of the I3 Kunitz inhibitors in Arabidopsis defense against spider mite, the gene expression of the whole Kunitz family members in infested and non-infested Col-0 plants was studied. This family included seven genes previously identified (Ma et al., 2011; Santamaria et al., 2014). One of these genes, AtKTI4 was previously reported as a differential expressed gene between the resistant Bla-2 and the susceptible Kondara Arabidopsis accessions upon spider mite feeding (Zhurov et al., 2014). RT-qPCR results showed that AtKTI5 and AtKTI3 genes were induced at 12 hpi, whereas AtKTI4 and AtKTI1 genes were up-regulated at 24 hpi, and presented their highest gene expression peak at 48 hpi (Figure 1). In contrast, AtKTI2, AtKTI6, and AtKTI7 gene expression was not detected in Arabidopsis leaves neither in non-infested plants nor upon mite infestation.
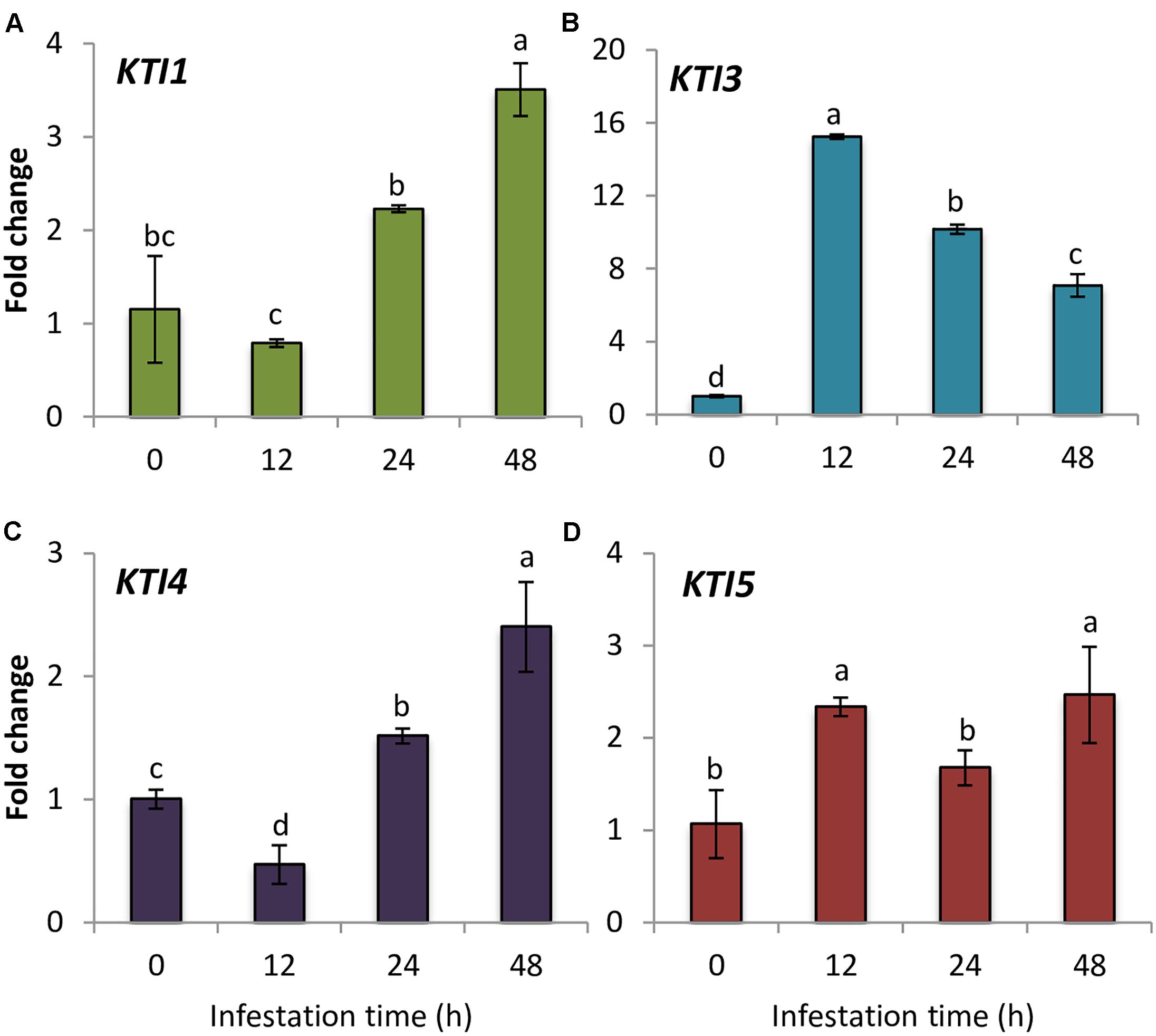
FIGURE 1. Gene expression of Arabidopsis Kunitz-type inhibitors upon spider mite infestation. Fold change in Col-0 plants at 0, 12, 24, and 48 hpi using time 0 as a calibrator sample. (A) AtKTI1 (B) AtKTI3 (C) AtKTI4, and (D) AtKTI5. Data are means of three replicates ± SE. Different letters indicate significant differences (P < 0.05, One-way ANOVA followed by Student–Newman–Keuls test).
Sequence and Structural Features of Arabidopsis KTIs
To obtain some clues on the inhibitory capacities of the seven Arabidopsis KTIs, their amino acid sequences were aligned. The seven inhibitors presented a signal peptide in the N-terminal region and their sequences were more similar in the regions aligned with the amino acids involved in secondary structures of a Kunitz inhibitor from D. regia (Figure 2A). The AtKTI2 did not present the positively charged residue (Lys or Arg) in the loop between strands β4 and β5, essential to inhibit trypsin. This residue was at the right position in AtKTI3, 4, 5, and 6. The amino acid pair Trp-Pro, located in the loop between strands β5 and β6, and putatively involved in cysteine-protease inhibitory capability of AtKTI2 was only partially conserved in AtKTI1 and AtKTI3. Whereas AtKTI2, AtKTI6, and AtKTI7 lacked the two cysteines involved in the first disulphide bridge, AtKTI5 is deficient in one of them but had two additional cysteines between strands β9 and β10 that could form a novel disulphide bridge. To determine the possible effect of this variation on the protein structure, tridimensional structures for AtKTI4 and 5 were made by homology modeling. Predictions showed structural differences. This predicted structural dissimilarity was mainly observed in the loops connecting secondary structures and leads to a distinct spatial orientation of the Lys reactive residue in the β4–β5 loop (Figure 2B). The predicted sequence-structure plasticity of these inhibitors could lead to different inhibitory properties. Thus, AtKTI4 and 5 were selected to further characterization.
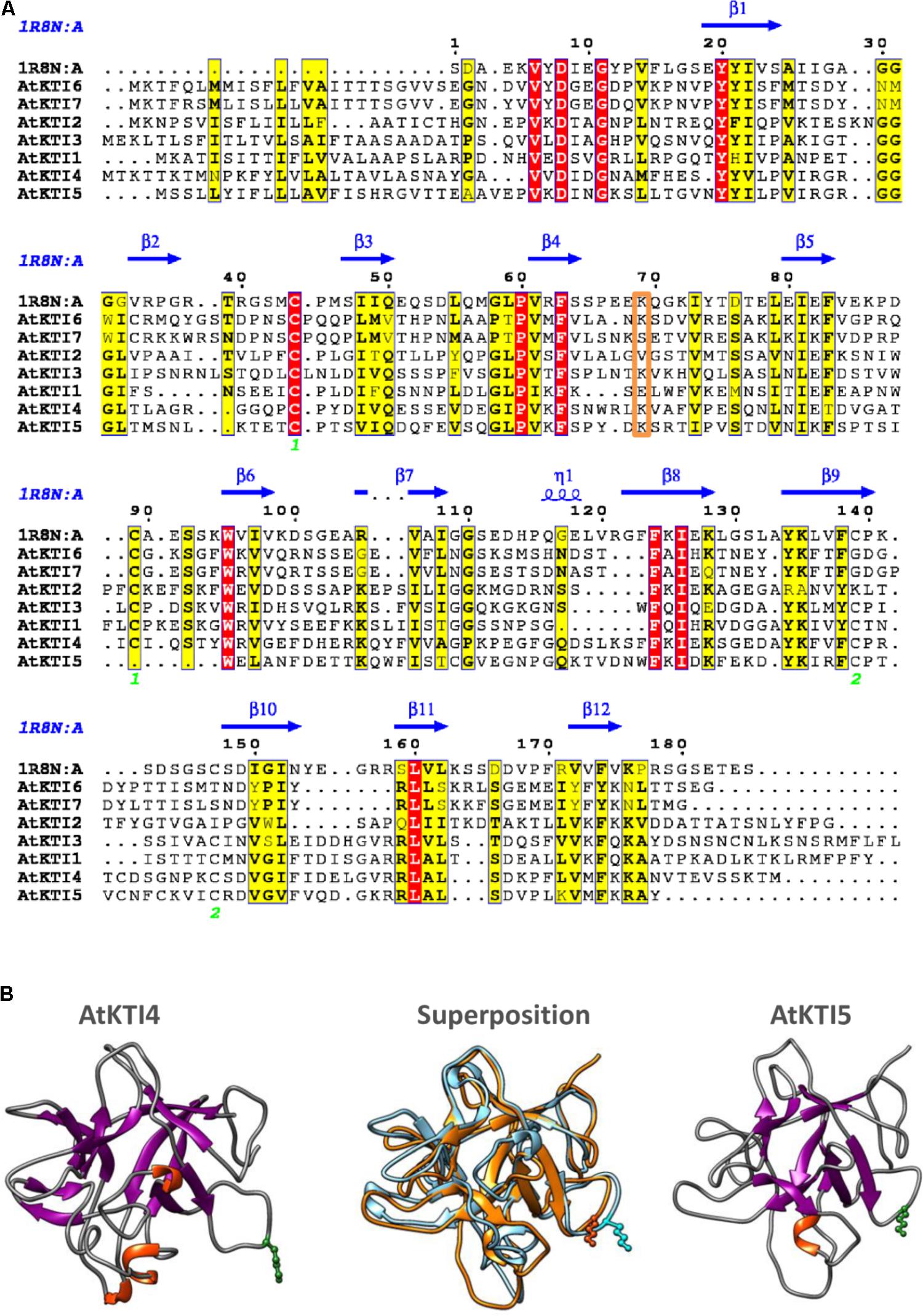
FIGURE 2. Sequence-structure analysis of Kunitz-type inhibitors. (A) Multiple sequence alignment of Arabidopsis KTIs. The Kunitz type inhibitor of Delonix regia (PDB ID 1R8N) was included in the alignment to infer secondary structure locations. Conserved residues are in red boxes. Similar residues are indicated in black bold characters and boxed in yellow. Green numbers at the bottom indicate disulphide bridge topology. Putative reactive site is boxed in orange. The figure was made with the ESPript 3.0 web server. (B) Ribbon diagrams showing structural models for AtKTI4 and AtKTI5, and their superposition (blue, AtKTI4; orange, AtKTI5). Conserved reactive Lys residue is colored in green. Modelization and visualization were made by SWISS-Model and Chimera tools.
AtKTI4 and AtKTI5 Protein Subcellular Location
Signal peptides found in every AtKTI indicate a targeted transport to the endoplasmatic reticulum. To determine the final AtKTI4 and AtKTI5 subcellular localization, transient expression assays were performed in onion epidermal layers by microparticle bombardment. AtKTI4 and AtKTI5 were fused to GFP. To determine the subcellular localization of the GFP signal, the cells were co-transfected with the 35S::RFP-HDEL plasmid to reveal the ER. Both green and red signals were found in the nuclear area, tracking along threads of cytoskeleton elements and the cell periphery. These locations are consistent with the subcellular distribution of the ER (Figures 3A–C,E–G). The 35S::GFP control showed an intense fluorescence in the cell nucleus, entering due to its small size, whereas the ER marker is seen in the nuclear periphery (Figures 3I,J). Agroinfiltration of N. benthamiana plants with the same constructs confirmed the location of both AtKTI4 and AtKTI5 inhibitors in the endomembrane system. In this case, the red signal corresponds to the autofluorescence of chlorophyll (Figures 3M–V,X,Y).
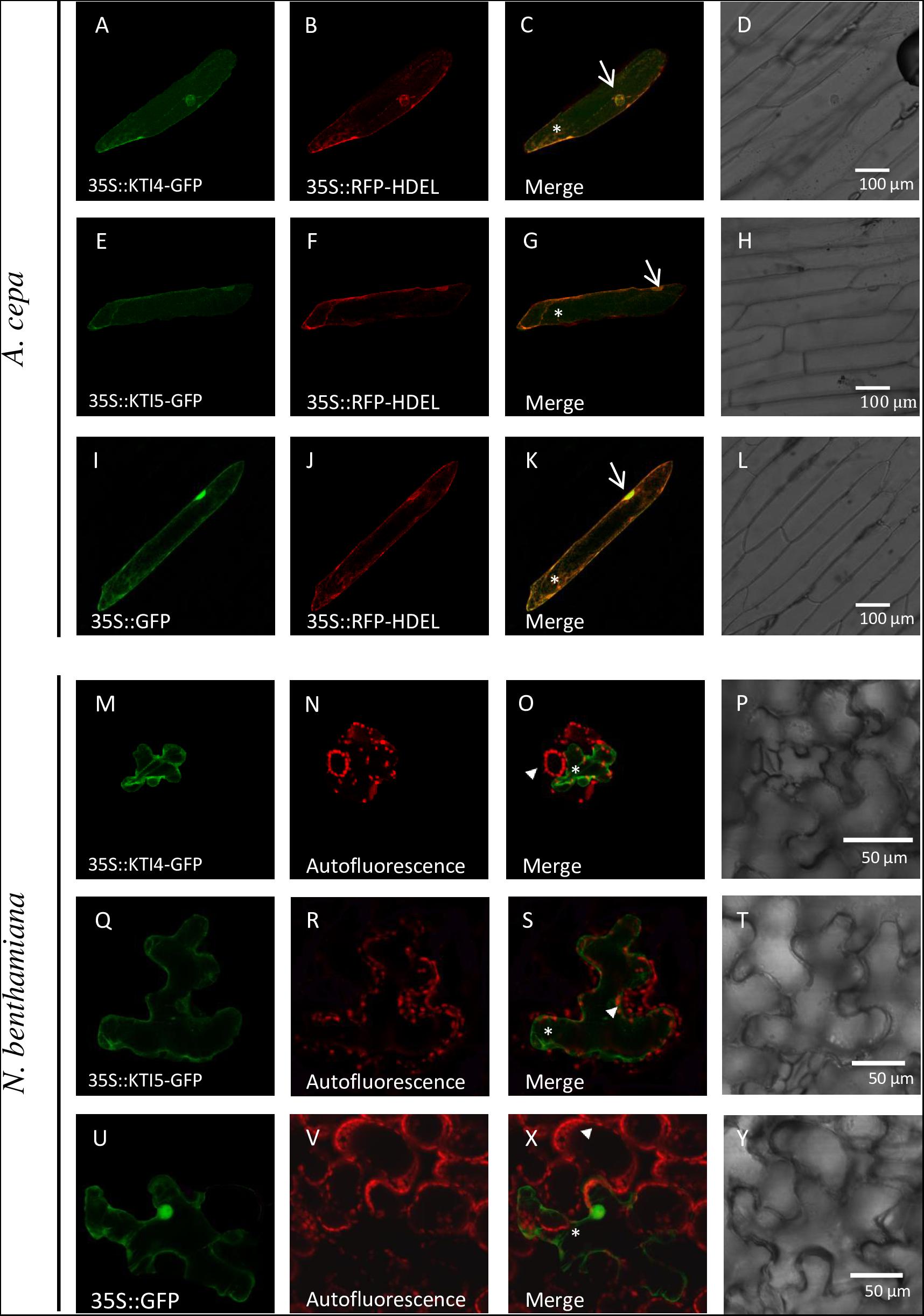
FIGURE 3. AtKTI4 and AtKTI5 subcellular localization. Confocal stacks spanning epidermal onion cells co-transformed with 35S::KTI4-GFP, 35S::KTI5-GFP and 35S::GFP and 35S::RFP-HDELcontrols. Confocal images and projections of the KTI4 (A) and KTI5 (E) are shown. Projections from GFP (A,E,I), RFP (B,F,J), merged (C,G,K) and the corresponding Nomarski snapshots (D,H,L). Confocal stacks spanning N. benthamiana cells agroinfiltrated with 35S::KTI4-GFP (M), 35S::KTI5-GFP (Q) and 35S::GFP control (U). Projections from GFP (M,Q,U), chlorophyll auto fluorescence (N,R,V), merged (O,S,X) and the corresponding Nomarski snapshots (P,T,Y). Bars are indicated in images. Arrows indicate nuclei, asterisks signal ER and arrowheads the chlorophyll autofluorescence.
Effects of Knock-Down AtKTI4 and AtKTI5 Lines on KTI Expression and Protease Inhibitory Properties
To investigate the role of AtKTI4 and AtKTI5 proteins in plant defense, silenced lines for these genes (kti4.1, kti4.2, kti5.1, kti5.2) were ordered. The characterization of the homozygous mutant lines revealed the insertion of the T-DNA at the promotor region for kti4.1 and kti5.2 lines, at the coding region for kti5.1 line and at the 3′UTR for the kti4.2 line (Supplementary Figure S1A).
The expression of AtKTI1, AtKTI3, AtKTI4, and AtKTI5 genes was analyzed in the kti4 and kti5 mutants and in the WT plants. As expected, the expression of AtKTI4 and AtKTI5 genes was reduced in their cognate T-DNA insertion lines (Figures 4C,D). Moreover, statistical differences in mRNA quantification revealed variations in the expression of other AtKTI genes in these lines. Regarding the mutant lines for AtKTI4 gene, kti4.1 plants were knocked down for AtKTI3, AtKTI4, and AtKTI5 genes and up regulated for AtKTI1 gene (Figures 4A–D) while kti4.2 plants were down regulated for AtKTI3 and AtKTI4 genes (Figure 4C). In the case of the T-DNA insertion lines for AtKTI5 gene, kti5.1 plants were knocked down for AtKTI3 and AtKTI5 genes while the expression of AtKTI1 gene was induced (Figures 4A,B,D), and kti5.2 mutant plants were down regulated for AtKTI4 and AtKTI5 genes (Figures 4B–D).
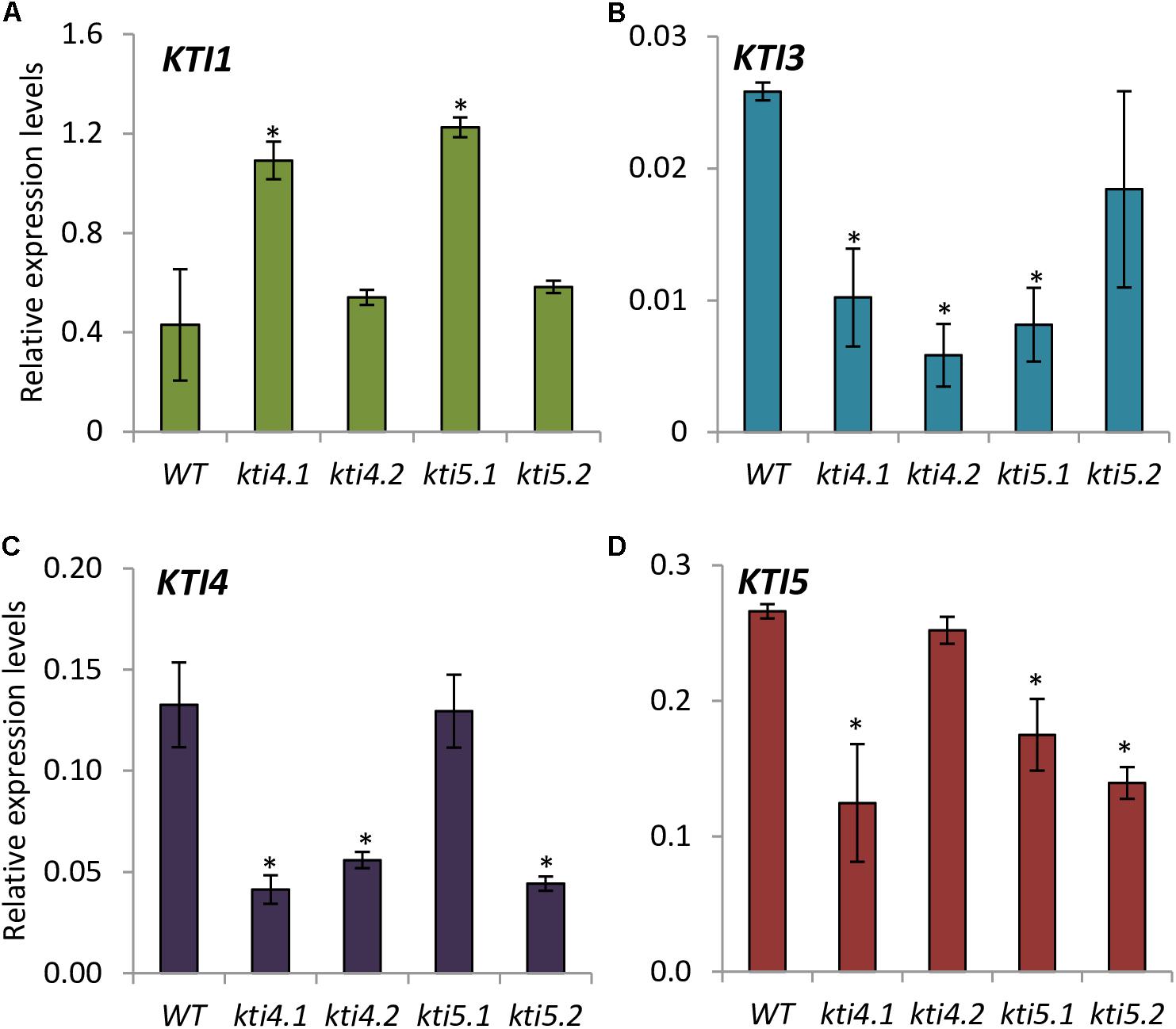
FIGURE 4. Gene expression of Kunitz-type inhibitors in Arabidopsis T-DNA insertion lines and WT plants. Relative gene expression levels of: (A) AtKTI1 (B) AtKTI3 (C) AtKTI4 and (D) AtKTI5. Data are means ± SE of three replicates. Asterisk indicates significant differences with the WT (P < 0.05, One-way ANOVA followed by Dunnett’s test).
Compensatory effects described above were also studied by analyzing the effect of T-DNA insertions on the protease activities of these plants. Results showed higher trypsin activity in all mutants than in the WT plants (Figure 5A). For chymotrypsin activity, kti4.1 and kti4.2 plants presented higher proteolytic levels in comparison to WT plants while kti5.1 and kti5.2 did not (Figure 5A). In contrast, no significant differences on both, cathepsin L- and B-like activities were detected between mutant and WT plants. Only the kti4.1 line showed slightly higher levels of cathepsin B-like activity than the WT plants (Figure 5B). The capability of the different Arabidopsis genotypes to inhibit serine- and cysteine-proteases was also tested by using commercial proteases. Significant differences were not found for trypsin, chymotrypsin and bovine cathepsin B assays (Figures 5C,D). Interestingly, the commercial papain (cathepsin L-like) was less inhibited by protein extracts from mutant lines than from the WT plants (Figure 5D).
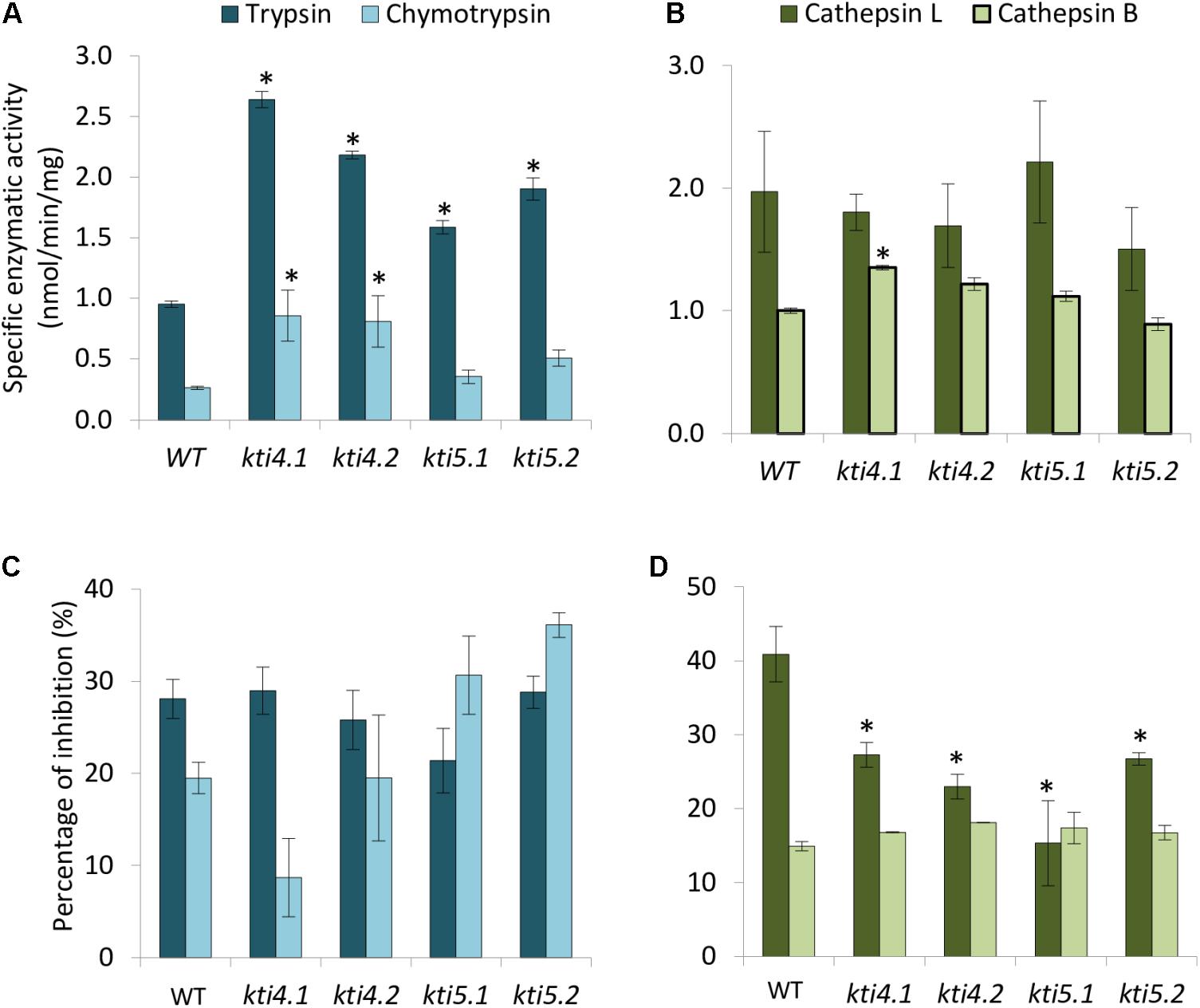
FIGURE 5. Proteolytic patterns of T-DNA insertion lines. Specific proteolytic activities of protein extracts from Arabidopsis T-DNA insertion lines and control WT using specific substrates. (A) Trypsin- and chymotrypsin-like specific activities. (B) Cathepsin L- and B-like specific activities. Data are expressed as nmoles/min/mg. Inhibitory activity of protein extracts from T-DNA insertion lines and control plants against commercial proteases. (C) Inhibitory activity against trypsin and chymotrypsin. (D) Inhibitory activity against commercial papain (cathepsin L-like) and bovine cathepsin B. Data are expressed as a percentage of inhibition. Data are means ± SE of three replicates. Asterisk indicates significant differences with the WT (P < 0.05, One-way ANOVA followed by Dunnett’s test).
AtKTI4 and AtKTI5 Are Able to Inhibit Both Serine and Cysteine Proteases
To further explore the serine- and cysteine-protease inhibitory capacity of AtKTI4 and AtKTI5 inhibitors, Agrobacterium-mediated transient over-expression for AtKTI4 and AtKTI5 genes fused to GFP was performed in N. benthamiana plants. GFP detection demonstrated the expression of the GFP gene and the location of the GFP protein in all agroinfiltrated plants (Figures 6A,B). Protein extracts from plants expressing the AtKTI4 and AtKTI5 proteins fused to GFP presented higher capability to inhibit serine- (trypsin and chymotrypsin) and cysteine- proteases (papain and bovine cathepsin B) compared with the extracts from the plants expressing only the GFP protein (Figure 6C). These results support the consideration of AtKTI4 and AtKTI5 as bifunctional inhibitors of both serine and cysteine proteases.
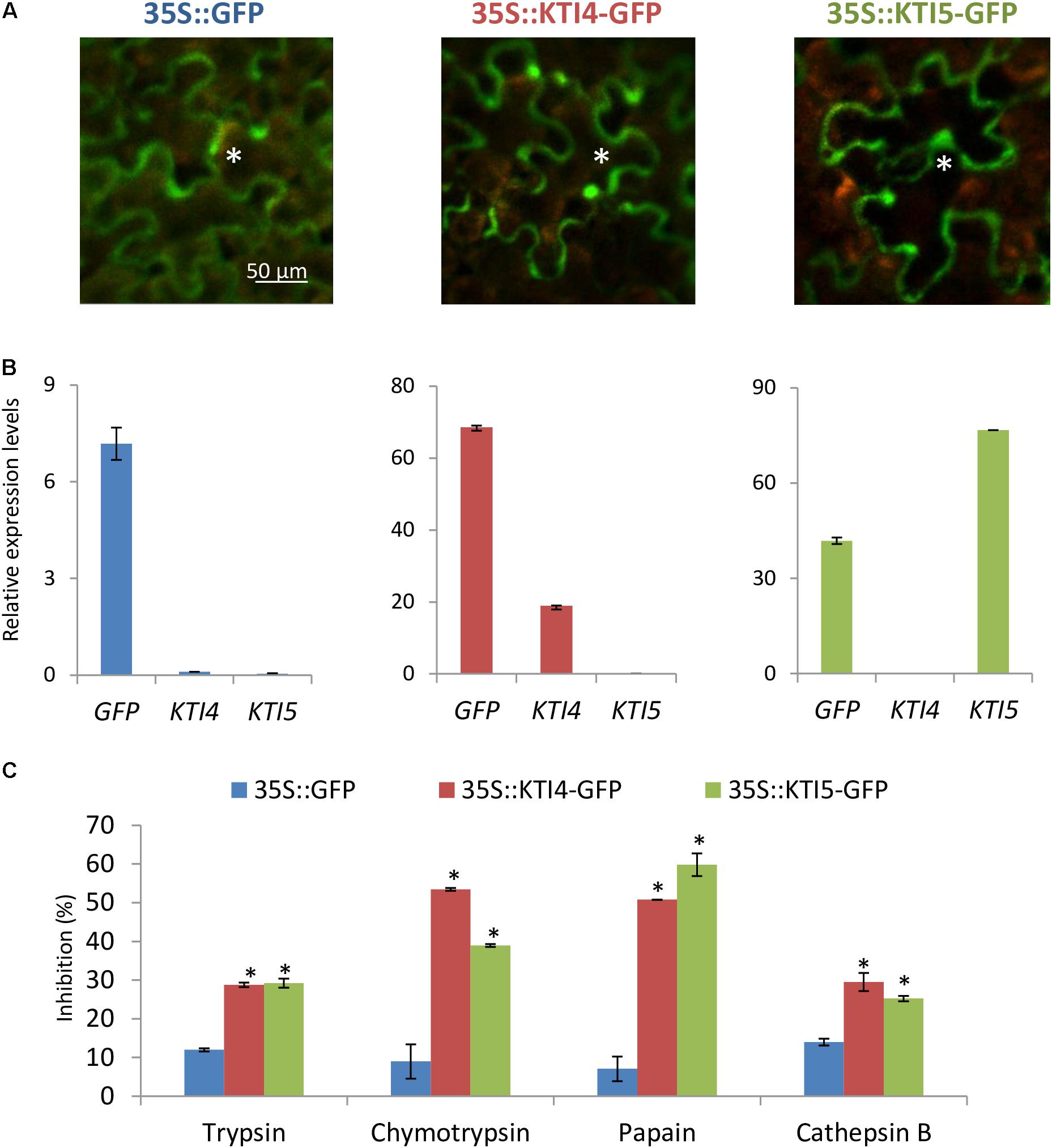
FIGURE 6. Characterization of the expression, location and inhibitory properties of AtKTI4 and AtKTI5 genes by using transient expression assays in N. benthamiana plants. (A) GFP signal after 3 days of agroinfiltration. Asterisks indicate GFP signal of the epidermal cells. (B) Relative expression levels of GFP, KTI4, and KTI5 genes in the N. benthamiana plants after 3 days of infiltration. (C) Inhibitory ability of N. benthamiana protein extracts expressing KTI4 and KTI5 genes against commercial trypsin, chymotrypsin, papain and bovine cathepsin B 3 days post-agroinfiltration. Data are mean ± SE of triplicate measurements of each sample. Asterisk indicates significant differences with the 35S::GFP plants (P < 0.05, One-way ANOVA followed by Dunnett’s test).
Docking analyses using ClusPro program were carried out to figure out the putative interaction between papain and the inhibitors AtKTI4 and AtKTI5. Models suggest an intrusion of different residues into the reactive site of papain stabilized by hydrogen bonds (Supplementary Figure S2). In the AtKTI4-papain interaction, the residues Glu122 and the Lys175 at the β6–β7 and β9–β10 loops, respectively, would form hydrogen bonds with the Cys and His amino acids of the papain reactive site. A similar interaction by the residues Val164 and Lys165 at the β9–β10 loop of AtKTI5 was predicted. Additional hydrogen bonds between residues of protease and inhibitor are predicted, which probably are involved in the preservation of the interaction (data not shown).
Effects of AtKTI4 and AtKTI5 Genes on Plant Resistance and Mite Performance
To further explore into the role of AtKTI4 and AtKTI5 in plant defense against T. urticae, homozygous T-DNA insertion lines and WT plants were infested with spider mites and the plant damage (chlorotic area) was visualized and quantified 4 days upon mite infestation. All knock down lines showed more damage than the WT plants. The injury was between 1.2 and 1.5 times higher in the knock down lines than in the WT plants (Figures 7A,D). Cell death caused by spider mite feeding was evaluated by trypan blue staining of leaves from all Arabidopsis genotypes after 24 h of infestation. All knock down lines showed higher levels of staining than WT plants with kti5.1 and the kti5.2 lines being the most prominent (Figures 7B,D). As the production of H2O2 is used as a plant damage indicator, H2O2 concentrations were determined in the five Col-0 genotypes. The quantification of H2O2 in infested plants, expressed as DAB relative units, demonstrated that kti5.1 and kti5.2 lines accumulated more H2O2 than kti4.1, kti4.2 lines and WT plants (Figures 7C,D).
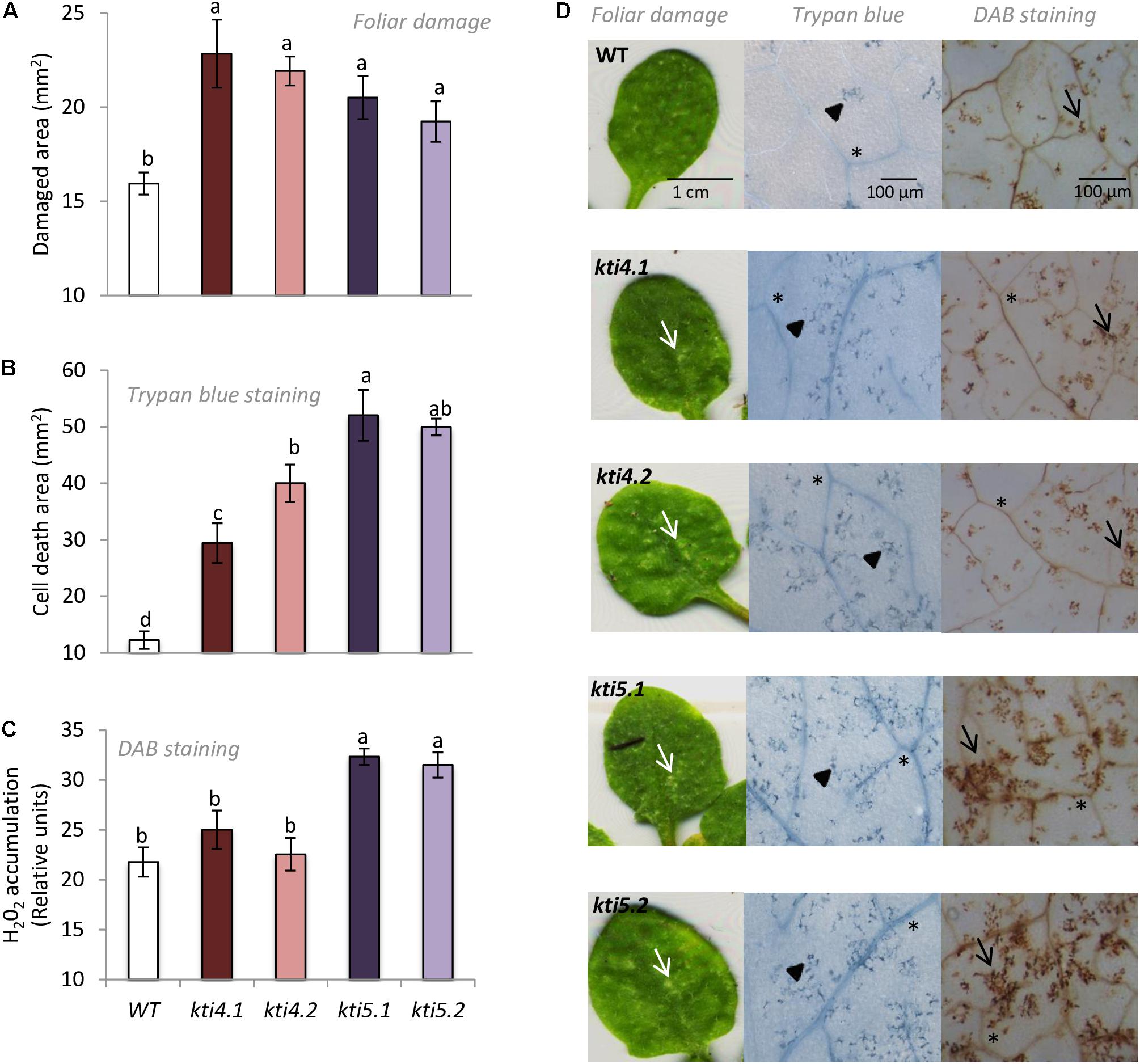
FIGURE 7. Plant damage and hydrogen peroxide production of Arabidopsis genotypes infested with T. urticae. (A) Foliar area damaged on Arabidopsis T-DNA insertion lines (kti4.1, kti4.2, kti5.1, kti5.2) and WT plants after 4 days of spider mite infestation. (B) Trypan blue stained area on leaf disks from Arabidopsis T-DNA insertion lines and WT plants after 1 day of spider mite infestation. (C) Hydrogen peroxide production on leaf disks from Arabidopsis T-DNA insertion lines and WT plants after 1 day of spider mite infestation. (D) Example of leaf phenotypes of Col-0 genotypes after spider mite infestation. Data are means ± SE of six replicates. Different letters indicate significant differences (P < 0.05, One-way ANOVA followed by Student–Newman–Keuls test). Bars are as indicated in images. White arrows indicate plant damage, arrowheads cell death, black arrows H2O2 accumulation and asterisks signal indicate vascular tissues.
To ensure that the chlorotic area correlated with mite feeding, mite performance was analyzed after feeding on mutants for AtKTI4 and AtKTI5 lines. Fecundity assays carried out on leaves from different Col-0 genotypes showed that synchronized mites fed on insertion lines had higher fecundity rates than the ones fed on WT plants (Figure 8A). To evaluate the KTI toxicity for mites, the mortality was recorded after feeding. Mites fed on mutant lines exhibited lower mortality rates than those on the WT plants (Figure 8B).
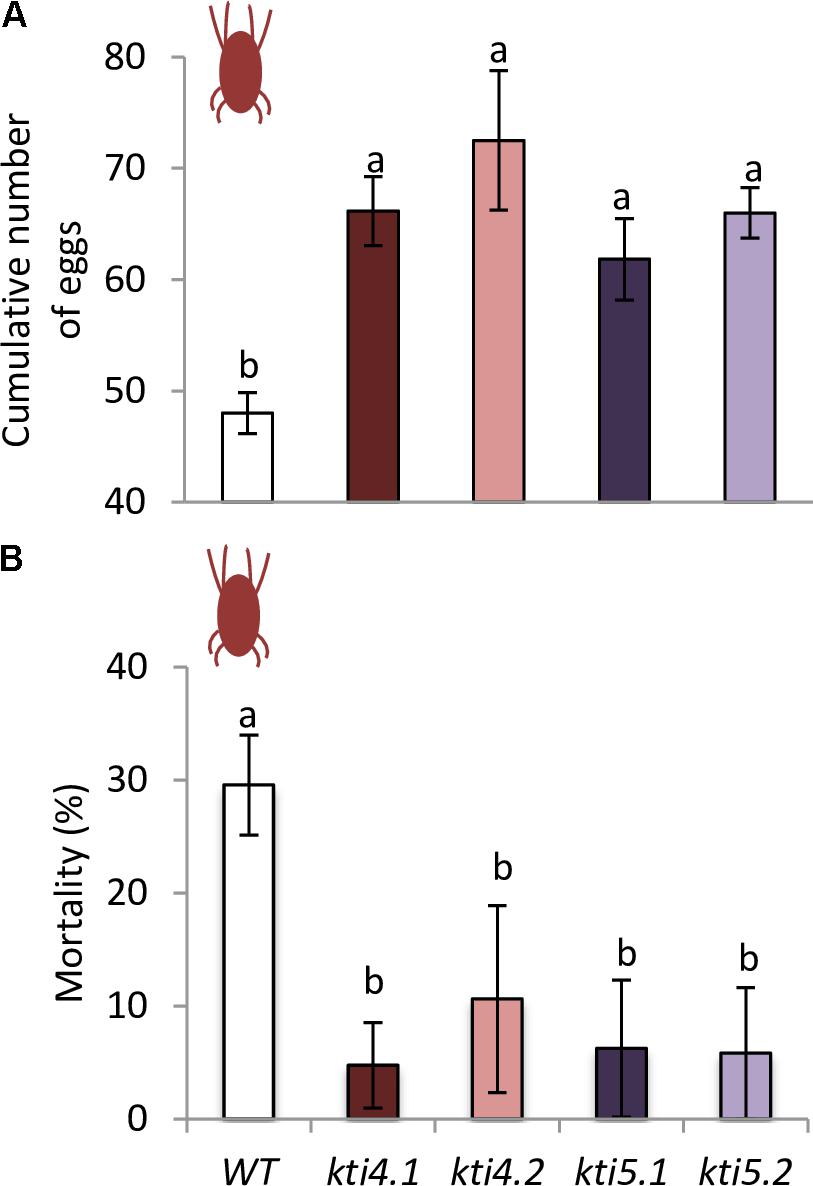
FIGURE 8. Analysis of spider mite performance. (A) Effects of T-DNA insertion lines and control plants on T. urticae fecundity, 36 h after infestation with synchronized females. (B) Effects of T-DNA insertion lines and control plants on T. urticae mortality, 10 days after infestation with neonate larvae. Data are means ± SE of eight replicates. Different letters indicate significant differences (P < 0.05, One-way ANOVA followed by Student–Newman–Keuls test).
Discussion
PIs from plants are proteins of particular interest because of their putative involvement in the natural defense system to phytophagous pests and pathogens. Particularly, the defense role of the plant KTI family against insects has been proven, although most studies involved individual Kunitz-type members. Determining the functional diversity of all members of the AtKTI gene family against an important pest such as the polyphagous mite T. urticae may provide alternatives to spider mite control. From the 7 AtKTI genes identified in Arabidopsis, four of them, AtKTI1, AtKTI3, AtKTI4, and AtKTI 5 responded to spider mite feeding (Figure 1). AtKTI2, AtKTI6, and AtKTI7 genes were not expressed in Arabidopsis rosettes either under control or infested conditions. In the case of the AtKTI2, this result was in agreement with a recent report showing a restricted expression to flowers and etiolated seedlings (Boex-Fontvieille et al., 2016).
Generally, KTIs are small proteins of about 20 kDa folded in β-trefoil manner with two disulphide bonds, but small differences in sequence or structure may make them different in their role as inhibitors of serine- and/or cysteine-proteases (Bendre et al., 2018). The low similarity observed among the amino acids residues of the 7 AtKTI members when their sequences were aligned may justify specific inhibitory roles against mite proteases (Figure 2). Trypsin inhibition is expected as the S1-binding site is negatively charged and accepts the conserved Lys at the P1 position. Likewise, chymotrypsin inhibition could be due to the presence of Tyr, Leu, and Phe residues in the β4–β5 reactive loop. The ability to inhibit papain could not be a priori predicted from the analysis of the amino acid sequences due to the multiple ways that Kunitz and Kunitz-structurally related inhibitors follow to inhibit C1 cysteine-proteases (Renko et al., 2012). These inhibitors may use different loops to occlude the reactive site of the cysteine protease. For instance, residues Trp88 and Pro89 of the AtKTI2 β5–β6 loop have been proposed to intrude into the catalytic triad of the Arabidopsis cysteine-protease RD21, blocking its protease activity (Boex-Fontvieille et al., 2015). This interaction is stabilized by hydrogen bonds between RD21 amino acids and residues in the AtKTI2 β2–β3 and β5–β6 loops. The comparative alignment of amino acid sequences of the AtKTI members revealed that AtKTI2, AtKTI6, and AtKTI7 were the most different among the whole family. These inhibitors presented the cysteine residues essential to form only one disulphide bridge instead of the two bridges predicted in the rest of AtKTIs. Besides, the AtKTI2 lacked the positive residue in the loop between strands β4 and β5 essential for trypsin inhibition, which correlated with its inability to inhibit serine-proteases. In contrast, although several dissimilarities were found in the tridimensional structures of AtKTI4 and AtKTI5, both proteins conserved the Lys residue in the β4–β5 loop and are putatively able to interact with the catalytic site of papain. This plasticity of the loops coming out of the stable β-trefoil scaffold maybe the reason of the versatility of these inhibitors, which display several different mechanisms of inhibition involving different positions of the loops and their combinations (Renko et al., 2012). Thus, the wide sequence-structure variability of KTIs supports the bifunctional action shown for AtKTI4 and AtKTI5 against trypsin and chymotrypsin serine-proteases as well as against cathepsin B- and L-like cysteine-proteases. Many protease inhibitors have to be targeted to the endomembrane system to reach their functional location (Martinez et al., 2009). As expected for proteins with signal peptide that would end up as vesicle-secreted proteins, AtKTI4 and AtKTI5 were subcellular located in the endomembrane system (Figure 3). Previous reports have detected AtKTI2 in the cell wall, apoplast spaces and tonoplast (Boex-Fontvieille et al., 2015).
To elucidate the response of AtKTI4 and AtKTI5 genes after mite feeding, we studied their effect on the spider mite performance using T-DNA insertion lines for both genes. First, we confirmed that the reduction of AtKTI4 and AtKTI5 transcripts detected in the mutant lines was associated to an increase of commercial trypsin activity, detected by in vitro assays using plant extracts, a promising result to perform bioassays (Figure 5). Besides, we found compensation effects in T-DNA insertion lines through the alteration of the expression of other AtKTI genes, suggesting that AtKTI1, AtKTI3, AtKTI4, and AtKTI5 proteins might participate in the defense process against spider mites in a concerted manner to modulate protease target activities (Figure 4).
Feeding assays conducted with the spider mite resulted in a significant increase of leaf damage either quantified as total chlorotic area or detected by trypan blue staining in comparison to control plants (Figure 7). However, only the knock down lines for the AtKTI5 gene accumulated higher levels of H2O2 than the WT plants. These findings indicate that these mutant lines either produced more H2O2 when infested, or alternatively, were not able to detoxify H2O2 efficiently, and triggered the cell death. Santamaria et al. (2017) demonstrated that an increase in H2O2 during T. urticae feeding was associated to sharp reduction in the accumulation of thiol groups and a parallel promotion of cell death. It is generally accepted that moderate levels of reactive oxygen and/or nitrogen species may differentially sense defense signaling while an excess of oxidative stress results to programmed cell death (Foyer and Noctor, 2005; Baxter et al., 2014). Therefore, these changes in the redox status have a potential impact on mite behavior. Accordingly, mite performance improved when fed on knock down lines (Figure 8). Our results confirm a significant reduction in mite mortality and higher fecundity rates upon feeding on mutant lines compared to control plants. Previous literature indicated that the role of AtKTI4 and AtKTI5 in Arabidopsis defense was not specifically associated to mites. Both genes were induced by Botrytis cinerea treatment (Coolen et al., 2016). AtKTI4 was also triggered by pathogen-derived elicitors and antagonized pathogen-associated cell death in Arabidopsis (Li et al., 2008), which may explain the higher cell death observed in our T-DNA insertion lines after spider mite infestations. Additionally, the expression of AtKTI4 and AtKTI5 was modulated by Pieris rapae infestation (Coolen et al., 2016). Interestingly, the expression pattern of AtKTI4 gene showed a highly localized response to P. brassicae eggs (Little et al., 2007) and was also induced upon nematode infestations in Arabidopsis (Jammes et al., 2005).
A second, but probably the most important mechanism of defense mediated by the AtKTIs is based on their capability to inhibit mite protease activities. The sequence and annotation of T. urticae genome revealed a large proliferation of serine- and cysteine-protease gene families in comparison to other sequenced arthropod species (Grbic et al., 2011). The 70 gene members identified in the serine-protease family pointed out an essential role in the spider mite physiology (Grbic et al., 2011). In many phytophagous insects, particularly in lepidopteran, the participation of serine proteases in the gut digestion has been demonstrated (Lara et al., 2000; Patankar et al., 2001; Fan and Wu, 2005; Srinivasan et al., 2006; Chougule et al., 2008; Tabatabaei et al., 2011). However, the lack of detection of trypsin- and chymotrypsin activities in mite extracts (Carrillo et al., 2011) together with the fact that serine-protease genes did not show a clear developmental pattern of expression correlated with feeding stages (Santamaria et al., 2012, 2015b), and the absence of this activity in mite feces suggested the association of this protease type with physiological processes other than the hydrolysis of dietary proteins (Santamaria et al., 2015b). Jonckheere et al. (2016) found some genes that presumably code for serine-proteases expressed in the salivary glands of T. urticae suggesting a pre-digestive function in the saliva. Based on these data, an alternative target for KTIs could be the serine-proteases present in the mite saliva. However, other putative roles involved in the regulation of mite growth and development have been suggested for these enzymes (Santamaria et al., 2012). In any case, plant inhibitors might get access to the endogenous proteases through the mite gut, as has been described for some insects (Down et al., 1999; Azzouz et al., 2005). The induction of members of the PIN-I and PIN-II serine PI families after spider mite attack (Li et al., 2002; Kant et al., 2004, 2008; Martel et al., 2015) and the enhanced resistance to spider mites showed by Arabidopsis when overexpress a barley trypsin inhibitor (Carrillo et al., 2011; Santamaria et al., 2012) strongly support the potential defense role of KTIs. Interestingly, the transient expression of AtKTI4 and AtKTI5 genes in Nicotiana plants showed their bifunctional features to inhibit cysteine- and serine-protease activities (Figure 6), which it should substantiate the impact of AtKTIs on cysteine-proteases from the mite gut involved in the hydrolysis of dietary proteins.
Conclusion
Our results confirm a wide role of the Arabidopsis KTI proteins in defense against spider mite based on: (i) four out of the seven KTIs identified in A. thaliana are induced upon spider mite infestation; (ii) transcriptional KTI compensation effects take place among the silencing AtKTI4 and AtKTI5 lines; (iii) T. urticae inflicts more leaf damage in kti4 and kti5 mutant lines than in WT plants; (iv) mites feeding on AtKTI silencing lines improve their performance; (v) AtKTI4 and AtKTI5 show a bifunctional inhibitory activity against both serine and cysteine proteases. In consequence, the inhibition of proteolytic process mediated by AtKTIs may decrease the mite access to essential amino acids and consequently to impair protein functions and to disrupt crucial physiological events needed by T. urticae performance. These effects finally increase mite mortality and reduce mite reproduction. Further research is needed to elucidate if the ability to inhibit serine protease activity contributes to the defense role of these inhibitors against spider mite.
Author Contributions
ID and MM conceived the research. AA, LT-M, and MS performed most of the experimental research. ID, MM, PG-M, and MS participated in the design, the acquisition, analysis, or interpretation of data for the work. MS, MM, and ID wrote the manuscript. All authors contributed to final version of the manuscript.
Funding
This work was supported by projects from Ministerio de Economía y Competitividad of Spain (projects BIO2014-53508-R, 618105-FACCE-Era Net Plus; BIO2017-83472-R).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00986/full#supplementary-material
Footnotes
References
Alba, J. M., Glas, J. J., Schimmel, B. C. J., and Kant, M. R. (2011). Avoidance and suppression of plant defenses by herbivores and pathogens. J. Plant Interact. 6, 221–227. doi:10.1080/17429145.2010.551670
Alfonso-Rubi, J., Ortego, F., Castanera, P., Carbonero, P., and Diaz, I. (2003). Transgenic expression of trypsin inhibitor CMe from barley in indica and japonica rice, confers resistance to the rice weevil Sitophilus oryzae. Transgenic Res. 12, 23–31. doi: 10.1023/A:1022176207180
PubMed Abstract | CrossRef Full Text | Google Scholar
Azzouz, H., Cherqui, A., Campan, E. D., Rahbé, Y., Duport, G., Jouanin, L., et al. (2005). Effects of plant protease inhibitors, oryzacystatin I and soybean Bowman-Birk inhibitor, on the aphid Macrosiphum euphorbiae (Homoptera, Aphididae) and its parasitoid Aphelinus abdominalis (Hymenoptera, Aphelinidae). J. Insect Physiol. 51, 75–86. doi: 10.1016/j.jinsphys.2004.11.010
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bendre, A. D., Ramasamya, S., and Suresh, C. G. (2018). Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int. J. Macromol. 113, 933–943. doi: 10.1016/j.ijbiomac.2018.02.148
Bensoussan, N., Santamaria, M. E., Zhurov, V., Diaz, I., Grbic, M., and Grbic, V. (2016). Plant-herbivore interaction: dissection of the cellular pattern of Tetranychus urticae: toward understanding cell biology of plant-pest interaction. Front. Plant Sci. 7:1105. doi: 10.3389/fpls.2016.01105
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Boex-Fontvieille, E., Rustgi, S., von Wettstein, D., Pollmann, S., Reinbothe, S., and Reinbothe, C. (2016). An ethylene-protected achilles’ heel of etiolated seedlings for arthropod deterrence. Front. Plant Sci. 7:1246. doi: 10.3389/fpls.2016.01246
Boex-Fontvieille, E., Rustgi, S., von Wettstein, D., Reinbothe, S., and Reinbothe, C. (2015). Water-soluble chlorophyll protein is involved in herbivore resistance activation during greening of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 7303–7308. doi: 10.1073/pnas.1507714112
Bonaventure, G. (2012). Perception of insect feeding by plants. Plant Biol. 14, 872–880. doi: 10.1111/j.1438-8677.2012.00650.x
Botelho-Junior, S., Machado, O. L., Fernandes, K. V., Lemos, F. J., Perdizio, V. A., Oliveira, A. E., et al. (2014). Defense response in non-genomic model species: methyl jasmonate exposure reveals the passion fruit leaves ability to assemble a cocktail of functionally diversified Kunitz-type trypsin inhibitors and recruit two of them against papain. Planta 240, 345–356. doi: 10.1007/s00425-014-2085-3
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Carrillo, L., Martinez, M., Ramessar, K., Cambra, I., Castañera, P., Ortego, F., et al. (2011). Expression of a barley cystatin gene in maize enhances resistance against phytophagous mites by altering their cysteine-proteases. Plant Cell Rep. 30, 101–112. doi: 10.1007/s00299-010-0948-z
Castagnoli, M., Caccia, R., Liguori, M., Simoni, S., Marinari, S., and Soressi, G. P. (2003). Tomato transgenic lines and Tetranychus urticae: changes in plant suitability and susceptibility. Exp. Appl. Acarol. 31, 177–189. doi: 10.1023/B:APPA.0000010387.48323
Cazaux, M., Navarro, M., Bruinsma, K. A., Zhurov, V., Negrave, T., Van Leeuwen, T., et al. (2014). Application of two-spotted spider mite Tetranychus urticae for plant-pest interaction studies. J. Vis. Exp. 89:e51738. doi: 10.1104/pp.113.231555
Chen, P., Senthilkumar, R., Jane, W., He, Y., Tian, Z., and Yeh, K. (2014). Transplastomic Nicotiana benthamiana plants expressing multiple defence genes encoding protease inhibitors and chitinase display broad-spectrum resistance against insects, pathogens and abiotic stresses. Plant Biotechnol. J. 12, 503–515. doi: 10.1111/pbi.12157
Chougule, N. P., Doyle, E., Fitches, E., and Gatehouse, J. A. (2008). Biochemical characterization of midgut digestive proteases from Mamestra brassicae (cabbage moth; Lepidoptera: Noctuidae) and effect of soybean Kunitz inhibitor (SKTI) in feeding assays. J. Insect Physiol. 54, 563–572. doi: 10.1016/j.jinsphys.2007.12.005
Cipriani, G., Michaud, D., Brunelle, F., Golmirzaie, A., and Zhang, D. P. (1999). Expression of Soybean Proteinase Inhibitor in Sweet Potato. Impact on a Changing World. CIP Program Report 1997-1998. Lima: International Potato Center, 271–277.
Confalonieri, M., Allegro, G., Balestrazzi, A., Fogher, C., and Delledonne, M. (1998). Regeneration of Populus nigra transgenic plants expressing a Kunitz proteinase inhibitor (KTi3) gene. Mol. Breed. 4, 137–145. doi: 10.1023/A:1009640204314
Coolen, S., Proietti, S., Hickman, R., Davila Olivas, N. H., Huang, P. P., Van Verk, M. C., et al. (2016). Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 86, 249–267. doi: 10.1111/tpj.13167
Cristofoletti, P. T., Ribeiro, A. F., Deraison, C., Rahbé, Y., and Terra, W. R. (2003). Midgut adaptation and digestive enzyme distribution in a phloem feeding insect, the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 49, 11–24. doi: 10.1016/S0022-1910(02)00222-6
de Vos, M., van Oosten, V. R., van Poecke, R. M. P., van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18, 923–937. doi: 10.1094/MPMI-18-0923
Diaz, I., Martinez, M., Isabel-LaMoneda, I., Rubio-Somoza, I., and Carbonero, P. (2005). The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 42, 652–662. doi: 10.1111/j.1365-313X.2005.02402.x
Diaz, I., and Santamaria, M. E. (2012). “Biotechnological approaches to combat phytophagous arthropods,” in Arthropod-Plant Interactions: Novel Insights and Approaches for IPM, eds G. Smagghe and I. Diaz (Dordrecht: Springer), 159–176. doi: 10.1007/978-94-007-3873-7
Dicke, M., and Baldwin, I. T. (2010). The evolutionary context for herbivore-induced plant volatiles: beyond the “cry for help”. Trends Plant Sci. 15, 167–175. doi: 10.1016/j.tplants.2009.12.002
Down, R. E., Ford, L., Mosson, H. J., Fitches, E., Gatehouse, J. A., and Gatehouse, A. M. R. (1999). Protease activity in the larval stage of the parasitoid wasp, Eulophus pennicornis (Nees) (Hymenoptera: Eulophidae); effects of protease inhibitors. Parasitology 119, 157–166. doi: 10.1016/j.jinsphys.2004.11.010
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fan, S. G., and Wu, G. J. (2005). Characteristics of plant proteinase inhibitors and their applications in combating phytophagous insects. Bot. Bull. Acad. Sin. 46, 273–292.
Farouk, S., and Osman, M. A. (2011). The effect of plant defence elicitors on common bean (Phaseolus vulgaris L.) growth and yield in absence or presence of spider mite (Tetranychus urticae Koch) infestation. J. Stress Physiol. Biochem. 7, 5–22.
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Frost, C. J., Appel, H. M., Carlson, J. E., DeMoraes, C. M., Mescher, M. C., and Schultz, J. C. (2007). Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol. Lett. 10, 490–498. doi: 10.1111/j.1461-0248.2007.01043.x
Gatehouse, A. M. R., Norton, E., Davison, G. M., Babbé, S. M., Newell, C. A., and Gatehouse, J. A. (1999). Digestive proteolytic activity in larvae of tomato moth, Lacanobia oleracea; effects of plant protease inhibitors in vitro and in vivo. J. Insect Physiol. 45, 545–558. doi: 10.1016/S0022-1910(98)00161-9
Grbic, M., Van Leeuwen, T., Clark, R. M., Rombauts, S., Rouzé, P., Grbic, V., et al. (2011). The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492. doi: 10.1038/nature10640
Green, T. R., and Ryan, C. A. (1972). Wound induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175, 776–777. doi: 10.1126/science.175.4023.776
Grosse-Holz, F. M., and van der Hoorn, R. A. L. (2016). Juggling jobs: roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 210, 794–807. doi: 10.1111/nph.13839
Guimarães, L. C., Oliveira, C. F. R., Marangoni, S., Oliveira, D. G. L., and Macedo, M. L. R. (2015). Purification and characterization of a Kunitz inhibitor from Poincianella pyramidalis with insecticide activity against the Mediterranean flour moth. Pest. Biochem. Physiol. 118, 1–9. doi: 10.1016/j.pestbp.2014.12.001
Hilder, V. A., Gatehouse, A. M. R., Sheerman, S. E., Barker, R. F., and Boulter, D. (1987). A novel mechanism of insect resistance engineered into tobacco. Nature 330, 160–163. doi: 10.1038/330160a0
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Ann. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Jammes, F., Lecomte, P., de Almeida-Engler, J., Bitton, F., Martin-Magniette, M. L., Renou, J. P., et al. (2005). Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 44, 447–458. doi: 10.1111/j.1365-313X.2005.02532.x
Johnson, K. S., and Rabosky, D. (2000). Phylogenetic distribution of cysteine proteinases in beetles: evidence for an evolutionary shift to an alkaline digestive strategy in Cerambycidae. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126, 609–619. doi: 10.1016/S0305-0491(00)00232-7
Jonckheere, W., Dermauw, W., Zhurov, V., Wybouw, N., Van den Bulcke, J., Villarroel, C. A., et al. (2016). The salivary protein repertoire of the polyphagous spider mite Tetranychus urticae: a quest for effectors. Mol. Cell Proteom. 15, 3594–3613. doi: 10.1074/mcp.M116.058081
Kant, M. R., Ament, K., Sabelis, M. W., Haring, M. A., and Schuurink, R. C. (2004). Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 135, 483–495. doi: 10.1104/pp.103.038315
Kant, M. R., Sabelis, M. W., Haring, M. A., and Schuurink, R. C. (2008). Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proc. Biol. Sci. 275, 443–452. doi: 10.1098/rspb.2007.1277
Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., et al. (2017). The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278. doi: 10.1038/nprot.2016.169
Lara, P., Ortego, F., Gonzalez-Hidalgo, E., Castañera, P., Carbonero, P., and Diaz, I. (2000). Adaptation of Spodoptera exigua (Lepidoptera: Noctuidae) to barley trypsin inhibitor BTI-CMe expressed in transgenic tobacco. Transgenic Res. 9, 169–178. doi: 10.3390/ijms17101747
Lee, S. I., Lee, S. H., Koo, J. C., Chun, H. J., Lim, C. O., Mun, J. H., et al. (1999). Soybean Kunitz trypsin inhibitor (SKTI) confers resistance to the brown planthopper (Nilaparvata lugens Stal) in transgenic rice. Mol. Breed. 5, 1–9. doi: 10.1023/A:1009660712382
Li, C., Williams, M. M., Loh, Y. T., Lee, G. I., and Howe, G. A. (2002). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130, 494–503. doi: 10.1104/pp.005314
Li, J., Bradera, G., and Palvaa, E. T. (2008). Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin B1 in Arabidopsis. Mol. Plant 1, 482–495. doi: 10.1093/mp/ssn013
Little, D., Gouhier-Darimont, C., Bruessow, F., and Reymond, P. (2007). Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiol. 143, 784–800. doi: 10.1104/pp.106.090837
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luedeling, E., Steinmann, K. P., Zhang, M., Brown, P. H., Grant, J., Girvetz, E. H., et al. (2011). Climate change effects on walnut pests in California. Glob. Chang. Biol. 17, 228–238. doi: 10.1111/j.1365-2486.2010.02227.x
Ma, Y., Zhao, Q., Lu, M., and Wang, J. (2011). Kunitz-type trypsin inhibitor gene family in Arabidopsis and Populus trichocarpa and its expression response to wounding and herbivore in Populus nigra. Tree Genet. Genomes 7, 431–441. doi: 10.1007/s11295-010-0345-3
Major, I. T., and Constabel, C. P. (2008). Functional analysis of the Kunitz trypsin inhibitor family in poplar reveals biochemical diversity and multiplicity in defense against herbivores. Plant Physiol. 146, 888–903. doi: 10.1104/pp.107.106229
Marchetti, S., Delledonne, M., Fogher, C., Chiaba, C., Chiesa, F., Savazzini, F., et al. (2000). Soybean Kunitz, C-II and PI-IV inhibitor genes confer different levels of insect resistance to tobacco and potato transgenic plants. Theor. Appl. Genet. 101, 519–526. doi: 10.1007/s001220051511
Martel, C., Zhurov, V., Navarro, M., Martinez, M., Cazaux, M., Auger, P., et al. (2015). Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol. Plant Microbe Interact. 28, 343–361. doi: 10.1094/MPMI-09-14-0291-FI
Martinez, M., Cambra, I., Carrillo, L., Diaz-Mendoza, M., and Diaz, I. (2009). Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 151, 1531–1545. doi: 10.1104/pp.109.146019
Martinez, M., Santamaria, M. E., Diaz-Mendoza, M., Arnaiz, A., Carrillo, L., Ortego, F., et al. (2016). Phytocystatins: defense proteins against phytophagous insects and Acari. Int. J. Mol. Sci. 17:E1747. doi: 10.3390/ijms17101747
Martinez de Ilarduya, O., Xie, Q., and Kaloshian, I. (2003). Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant Microbe Interact. 16, 699–708. doi: 10.1094/MPMI.2003.16.8.699
McManus, M. T., Burgess, E. P. J., Philip, B., Watson, L. M., Laing, W. A., Voisey, C. R., et al. (1999). Expression of the soybean (Kunitz) trypsin inhibitor in transgenic tobacco: effects on larval development of Spodoptera litura. Transgenic Res. 8, 383–395. doi: 10.1023/A:1008957610872
Mithöfer, A., Wanner, G., and Boland, W. (2005). Effects of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiol. 137, 1160–1168. doi: 10.1104/pp.104.054460
Murdock, L. L., Brookhart, G., Dunn, P. E., Foard, D. E., Kelley, S., Kitch, L., et al. (1987). Cysteine digestive proteinases in Coleoptera. Comp. Biochem. Physiol. B Comp. Biochem. 87, 783–787. doi: 10.1016/0305-0491(87)90388-9
Nandi, A. K., Basu, D., Das, S., and Sen, S. K. (1999). High level expression of soybean trypsin inhibitor gene in transgenic tobacco plants failed to confer resistance against damage caused by Helicoverpa armigera. J. Biosci. 24, 445–452. doi: 10.1007/BF02942655
Oñate-Sánchez, L., and Vicente-Carbajosa, J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1:93. doi: 10.1186/1756-0500-1-93
Ortego, F. (2012). “Physiological adaptations of the insect gut to herbivory,” in Arthropod-Plant Interactions: Novel Insights and Approaches for IPM, eds G. Smagghe and I. Diaz (Dordrecht: Springer), 75–88. doi: 10.1007/978-94-007-3873-7
Ortego, F., Novillo, C., and Castañera, P. (1996). Characterization and distribution of digestive proteases of the stalk corn borer, Sesamia nonagrioides Lef. (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 33, 163–180. doi: 10.1002/(SICI)1520-6327 (1996)33:2<163::AID-ARCH6>3.0.CO;2-Z
Park, Y. L., and Lee, J. H. (2002). Leaf cell and tissue damage of cucumber caused by two spotted spider mite (Acari: Tetranychidae). J. Econ. Entomol. 95, 952–957. doi: 10.1093/jee/95.5.952
Patankar, A. G., Giri, A. P., Harsulkar, A. M., Sainani, M. N., Deshpande, V. V., Ranjekar, P. K., et al. (2001). Complexity in specificities and expression of Helicoverpa armigera gut proteinases explains polyphagous nature of the insect pest. Insect Biochem. Mol. 31, 453–464. doi: 10.1016/S0965-1748(00)00150-8
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera- a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Philippe, R. N., Ralph, S. G., Kulheim, C., Jancsik, S. I., and Bohlmann, J. (2009). Poplar defense against insects: genome analysis, full-length cDNA cloning, and transcriptome and protein analysis of the poplar Kunitz-type protease inhibitor family. New Phytol. 184, 865–884. doi: 10.1111/j.1469-8137.2009.03028
Rawlings, N. D., Barrett, A. J., Thomas, P. D., Huang, X., Bateman, A., and Finn, R. D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632. doi: 10.1093/nar/gkx1134
Renko, M., Sabotiè, J., and Turk, D. (2012). β-trefoil inhibitors – from the work of Kunitz onward. Biol. Chem. 393, 1043–1054. doi: 10.1515/hsz-2012-0159
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rodríguez-Herva, J. J., Gonzalez-Melendi, P., Cuartas-Lanza, R., Antunez-Lamas, M., Rio-Alvarez, I., Li, Z., et al. (2012). A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 14, 669–681. doi: 10.1111/j.1462-5822.2012.01749.x
Rodriguez-Saona, C. R., Musser, R. O., Vogel, H., Hum-Musser, S. M., and Thaler, J. S. (2010). Molecular, biochemical, and organismal analyses of tomato plants simultaneously attacked by herbivores from two feeding guilds. J. Chem. Ecol. 36, 1043–1057. doi: 10.1007/s10886-010-9854-7
Rufino, F. P., Pedroso, V. M., Araujo, J. N., França, A. F., Rabêlo, L. M., Migliolo, L., et al. (2013). Inhibitory effects of a Kunitz-type inhibitor from Pithecellobium dumosum (Benth) seeds against insect-pests’ digestive proteinases. Plant Physiol. Biochem. 63, 70–76. doi: 10.1016/j.plaphy.2012.11.013
Rustgi, S., Boex-Fontvieille, E., Reinbothe, C., von Wettstein, D., and Reinbothe, S. (2017). Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, 2212–2217. doi: 10.1073/pnas.1621496114
Sabelis, M. W., Janssen, A., Bruin, J., Bakker, F. M., Drukker, B., Scutareanu, P., et al. (1999). “Interactions between arthropod predators and plants: a conspiracy against herbivorous arthropods?,” in Ecology and Evolution of the Acari, eds J. Bruin, L. P. S. van der Geest, and M. W. Sabelis (Dordrecht: Kluwer), 207–229.
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sanchez-Vallet, A., Ramos, B., Bednarek, P., López, G., Pislewska-Bednarek, M., Schulze-Lefert, P., et al. (2010). Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 63, 115–127. doi: 10.1111/j.1365-313X.2010.04224.x
Santamaria, M. E., Arnaiz, A., Diaz-Mendoza, M., Martinez, M., and Diaz, I. (2015a). Inhibitory properties of cysteine protease pro-peptides from barley confer resistance to spider mite feeding. PLoS One 10:e0128323. doi: 10.1371/journal.pone.0128323
Santamaria, M. E., Arnaiz, A., Gonzalez-Melendi, P., Martinez, M., and Diaz, I. (2018a). Plant perception and short-term responses to phytophagous insects and mites. Int. J. Mol. Sci. 19:1356. doi: 10.3390/ijms19051356
Santamaria, M. E., Cambra, I., Martinez, M., Pozancos, C., Gonzalez-Melendi, P., Grbic, V., et al. (2012). Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PLoS One 7:e43011. doi: 10.1371/journal.pone.0043011
Santamaria, M. E., Diaz-Mendoza, M., Diaz, I., and Martinez, M. (2014). Plant protein peptidase inhibitors: an evolutionary overview based on comparative genomics. BMC Genomics 15:812. doi: 10.1186/1471-2164-15-812
Santamaria, M. E., Diaz-Mendoza, M., Perez-Herguedas, D., Hensel, G., Kumlehn, J., Diaz, I., et al. (2018b). Overexpression of HvIcy6 in barley enhances resistance against Tetranychus urticae and entails partial transcriptomic reprogramming. Int. J. Mol. Sci. 19:E697. doi: 10.3390/ijms19030697
Santamaria, M. E., Gonzalez-Cabrera, J., Martinez, M., Grbic, V., Castañera, P., Diaz, I., et al. (2015b). Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae. J. Insect Physiol. 78, 69–77. doi: 10.1016/j.jinsphys.2015.05.002
Santamaria, M. E., Martinez, M., Arnaiz, A., Ortego, F., Grbic, V., and Diaz, I. (2017). MATI, a novel protein involved in the regulation of herbivore-associated signaling pathways. Front. Plant Sci. 8:975. doi: 10.3389/fpls.2017.00975
Shockey, J. M., Gidda, S. K., Chapital, D. C., Kuan, J. C., Dhanoa, P. K., Bland, J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. doi: 10.1105/tpc.106.043695
Srinivasan, A., Giri, A. P., and Gupta, V. S. (2006). Structural and functional diversities in lepidopteran serine proteases. Cell Mol. Biol. Lett. 11, 132–154. doi: 10.2478/s11658-006-0012-8
Staudacher, H., Bernardus, C. J., Schimmel, B. C., Lamers, M. M., Wybouw, N., Groot, A. T., et al. (2017). Independent effects of a herbivore’s bacterial symbionts on its performance and induced plant defences. Int. J. Mol. Sci. 18:182. doi: 10.3390/ijms18010182
Stout, M. J., Workman, K. V., Bostock, R. M., and Duffey, S. S. (1998). Specificity of induced resistance in the tomato, Lycopersicon esculentum. Oecologia 113, 74–81. doi: 10.1007/s004420050355
Suzuki, T., España, M. U., Nunes, M. A., Zhurov, V., Dermauw, W., Osakabe, M., et al. (2017). Protocols for the delivery of small molecules to the two-spotted spider mite, Tetranychus urticae. PLoS One 12:e0180658. doi: 10.1371/journal.pone.0180658
Tabatabaei, P. R., Hosseininaveh, V., Goldansaz, S. H., and Talebi, K. H. (2011). Biochemical characterization of digestive proteases and carbohydrases of the carob moth, Ectomyelois ceratoniae (Zeller) (Lepidoptera: Pyralidae). J. Asia Pac. Entomol. 14, 187–194. doi: 10.1016/j.aspen.2010.12.010
Tamhane, V. A., Chougule, N. P., Giri, A. P., Dixit, A. R., Sainani, M. N., and Gupta, V. S. (2005). In vivo and in vitro effect of Capsicum annum proteinase inhibitors on Helicoverpa armigera gut proteinases. Biochem. Biophys. Acta 1722, 156–167. doi: 10.1016/j.bbagen.2004.12.017
Telang, M. A., Giri, A. P., Pyati, P. S., Gupta, V. S., Tegeder, M., and Franceschi, V. R. (2009). Winged bean chymotrypsin inhibitors retard growth of Helicoverpa armigera. Gene 431, 80–85. doi: 10.1016/j.gene.2008.10.026
Terra, W. R., and Ferreira, C. (2012). “Biochemistry of digestion,” in Comprehensive Molecular Insect Science, eds L. I. Gilbert, K. Iatrou, and S. S. Gill (Oxford: Elsevier), 171–224. doi: 10.1016/B0-44-451924-6/00053-3
Van Leeuwen, T., and Dermauw, W. (2016). The molecular evolution of xenobiotic metabolism and resistance in Chelicerate mites. Annu. Rev. Entomol. 61, 475–498. doi: 10.1146/annurev-ento-010715-023907
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2015). Retraction: “an enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus”. Plant J. 84:846. doi: 10.1111/tpj.13066
Walling, L. L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216. doi: 10.1007/s003440000026
Wolfson, J. L., and Murdock, L. L. (1990). Diversity in digestive proteinase activity among insects. J. Chem. Ecol. 16, 1089–1102. doi: 10.1007/BF01021013
Wu, J., and Baldwin, I. T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. doi: 10.1146/annurev-genet-102209-163500
Keywords: plant-herbivore interphase, Tetranychus urticae, Arabidopsis thaliana, serine protease inhibitors, cysteine protease inhibitors, spider mite digestion
Citation: Arnaiz A, Talavera-Mateo L, Gonzalez -Melendi P, Martinez M, Diaz I and Santamaria ME (2018) Arabidopsis Kunitz Trypsin Inhibitors in Defense Against Spider Mites. Front. Plant Sci. 9:986. doi: 10.3389/fpls.2018.00986
Received: 27 April 2018; Accepted: 18 June 2018;
Published: 10 July 2018.
Edited by:
Andrea Chini, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Flávio Henrique-Silva, Universidade Federal de São Carlos, BrazilTakeshi Suzuki, Tokyo University of Agriculture and Technology, Japan
Copyright © 2018 Arnaiz, Talavera-Mateo, Gonzalez-Melendi, Martinez, Diaz and Santamaria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. E. Santamaria, bWUuc2FudGFtYXJpYUB1cG0uZXM=
 Ana Arnaiz
Ana Arnaiz Lucia Talavera-Mateo
Lucia Talavera-Mateo Pablo Gonzalez-Melendi
Pablo Gonzalez-Melendi Manuel Martinez
Manuel Martinez Isabel Diaz
Isabel Diaz M. E. Santamaria
M. E. Santamaria