
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 July 2018
Sec. Computational Genomics
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00963
Jasmonates are critical plant hormones, mediating stress response in plants and regulating plant growth and development. The jasmonate receptor is a multi-component complex, composed of Arabidopsis SKP-LIKE PROTEIN1 (ASK1), CORONATINE INSENSITIVE 1 (COI1), inositol phosphate (InsP), and jasmonate ZIM-domain protein (JAZ). COI1 acts as multi-component signaling hub that binds with each component. InsP is suggested to play important roles in the hormone perception. How InsP binds with COI1 and the structural changes in COI1 upon binding with InsP, JA-Ile, and JAZ are not well understood. In this study, we integrated multiple computational methods, such as molecular docking, molecular dynamics simulations, residue interaction network analysis and binding free energy calculation, to explore the effect of InsP on the dynamic behavior of COI1 and the recognition mechanism of each component of the jasmonate receptor complex. We found that upon binding with InsP, JA-Ile, and JAZ1, the structure of COI1 becomes more compact. The binding of InsP with COI1 stabilizes the conformation of COI1 and promotes the binding between JA-Ile or JAZ1 and COI1. Analysis of the network parameters led to the identification of some hub nodes in this network, including Met88, His118, Arg120, Arg121, Arg346, Tyr382, Arg409, Trp467, and Lys492. The structural and dynamic details will be helpful for understanding the recognition mechanism of each component and the discovery and design of novel jasmonate signaling pathway modulators.
Jasmonates, a family of plant hormones, regulate a variety of plant physiology processes, such as plant development, wound response, as well as defense response to insect herbivory and pathogens (Browse, 2008). It is suggested that CORONATINE INSENSITIVE 1 (COI1) is the receptor of jasmonates (Katsir et al., 2008; Fonseca et al., 2009; Yan et al., 2009). Subsequently, Sheard et al. determined the structure of the Arabidopsis jasmonate receptor (Sheard et al., 2010), a multiple component complex of Arabidopsis SKP-LIKE PROTEIN1 (ASK1), COI1, and Jasmonate ZIM-domain protein 1 (JAZ1). COI1 is the F-box component of a Skp1/Cullin/F-box protein (SCF), which is an E3 ubiquitin ligase complex.
The JA-induced gene expression process can be briefly described as follows: in the resting state, with low concentration of bioactive jasmonic acid (JA) conjugate JA-Ile, JAZ binds with MYC2/3/4 transcription factors to repress the expression of jasmonate-responsive genes (Figure 1). The endogenous JA-Ile concentration increases upon environmental or developmental stimulation. COI1 binds with JA-Ile and recruits JAZ transcriptional repressors, which leads to ubiquitination and proteasomal degradation of the JAZ repressors (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Sheard et al., 2010). MYC or other transcription factors are released after the degradation of JAZ. Subsequently, the expression of jasmonate-responsive genes was initialized (Chini et al., 2007).
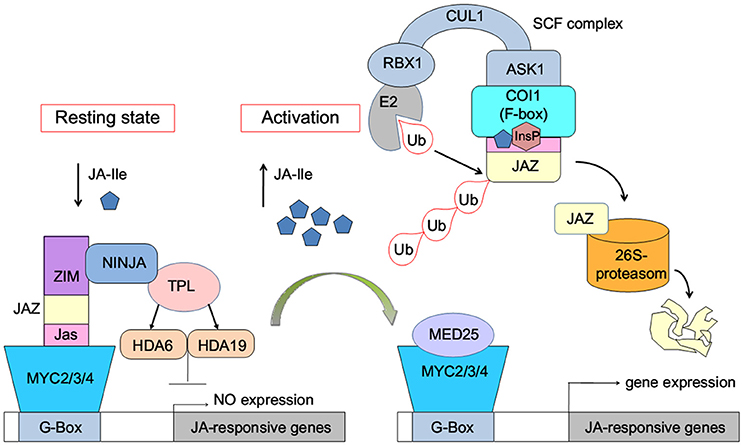
Figure 1. Model of JA-Ile perception via the SCFCOI1-JAZ receptor complex (Wasternack and Hause, 2013; Wasternack and Strnad, 2015; Wasternack and Song, 2016). MYC2/3/4, binding to the G-box of a JA-Ile-responsive gene, is repressed by negative regulators such as jasmonate ZIM-domain proteins (JAZs). Novel Interactor of JAZ1 (NINJA) and TOPLESS (TPL), which act via HISTONE DEACETYLASE6 (HDA6) and HDA19, act as co-repressors. Ub, ubiquitin. InsP, inositol phosphate. E2, RBX1, CULLIN1 (CUL1), ASK1, and the F-box protein COI1 are components of the SCF-complex. MED25, subunit 25 of the Mediator complex.
There exists two domains of COI1, the N-terminal tri-helical F-Box domain, binding with ASK1, and a large C-terminal domain containing leucine-rich repeats (LRR) (Figure 2) (Xie et al., 1998; Sheard et al., 2010). This domain contains 18 tandem LRRs that form a horseshoe-shaped solenoid. Three long loops, the β2-α5 loop, β12-α15 loop, and β14-α17 loop at the top surface of this domain are involved in hormone and JAZ1 binding. JA-Ile binds into the solenoid of COI1. JAZ1 binds at the top surface of the solenoid. Sheard et al. also found that inositol pentakisphosphate (InsP5) binds to the bottom of COI1 solenoid and is involved in plant hormone perception (Sheard et al., 2010).
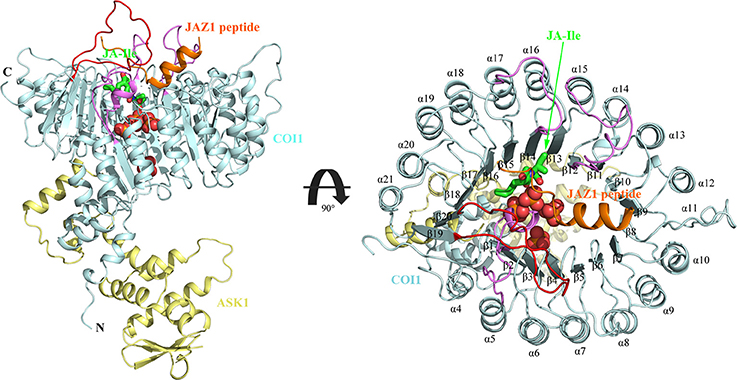
Figure 2. The overall structure of the ASK1-COI1 complex with JA-Ile, inorganic phosphate molecules, and the JAZ1 degron peptide (Sheard et al., 2010). ASK1-COI1 (yellow and cyan ribbons, respectively) with the JAZ1 degron peptide (orange ribbons), inorganic phosphate molecules are depicted as spheres (orange for phosphorus atoms and red for oxygen atoms) and JA-Ile in sticks (green for carbon atoms). Loops 2, 12, and 14 are colored in pink and loop C is colored in red.
InsP5[3-OH] was purified together with the ASK1-COI1 expressed in insect cells by mass spectrometry and NMR analyses (Sheard et al., 2010). It also suggests Ins(1,4,5,6)P4, InsP5, InsP6 can promote jasmonate receptor assembly based on reconstitution assays, while it remains unknown which one is the physiologically relevant form. InsP5[3-OH] was displaced by phosphate molecules in the crystallization process. How it binds with COI1 remains unclear. A subsequent study suggested that InsP5 is involved in the perception of JA-Ile (Mosblech et al., 2011). Recently, Laha et al. found that inositol pyrophosphate InsP7 and InsP8 also promotes the binding between COI1 and JAZ1, which exert important functions in regulation of jasmonate-dependent responses (Laha et al., 2015). In addition, they found that JA treatment leads to InsP8 accumulation in plant, but not InsP5. Subsequently, the binding specificity of different inositol phosphates (InsPs) with ASK1-COI1 was explored by in vitro reconstitution experiments and molecular docking (Laha et al., 2016). The predicted binding mode suggests that InsP8 can form more favorable interaction with COI1 than InsP5, which explains that InsP5 acts as a weaker co-factor than InsP8 in vivo (in yeast). While the dynamics behavior of COI1 responding to InsP8, JA-Ile, and JAZ1 and the recognition mechanism of each component of jasmonate receptor are not well-understood.
In this study, the interaction mechanism between InsPs and the jasmonate receptor complex was investigated by multiple computational methods, such as molecular docking, molecular dynamics simulation, and binding free energy calculation. The recognition of each component of the complex was investigated by residue interaction network analysis. Our results provide information on the molecular recognition of InsP8 by COI1 and describe how InsP8 modulate the conformational dynamics of COI1. The detailed interaction mechanism of each component of the jasmonate receptor was also illustrated. This information will be helpful for the understanding of the recognition mechanism of each component of the jasmonate receptor complex. The results will facilitate the discovery and design of novel jasmonate signaling pathway modulators.
The X-ray crystal structure of the jasmonate receptor (PDB ID: 3OGM) was downloaded as the initial structure. The missing regions in this structure were built by Rosetta (De et al., 2015) and Modeller (Eswar et al., 2016), which comprises residue numbers 68–79 in ASK1 and 550–562 (loop C) in COI1 (68-SKAEAVEGAATS-79, 550-VPEVNQQGEIREM-565). This structure was then refined in vacuo by using Amber 12 (Pearlman et al., 1995). AMBER ff03 force field was assigned to the protein (Wang et al., 2004; Hornak et al., 2006). In the first stage, the side chains were relaxed by restraining the backbone atoms of the protein (5,000 cycles of steepest descent and 2,000 cycles of conjugate gradient minimizations); second, the whole protein were relaxed without any restrains (5,000 cycles of steepest descent and 2,000 cycles of conjugate gradient minimizations).
The structures of inositol phosphates used for docking were extracted from the protein structures in the Protein Data Bank (PDB). The structures of InsP5[3-OH], InsP6, and 1,5-InsP8 were extracted from the protein coded by 1FHW (Ferguson et al., 2012), 2P1P (Tan et al., 2007), and 3T9F (Wang et al., 2011), respectively. The other isoforms of InsP5 and InsP8 were sketched by Accelrys Discovery Studio 2.5 2010. The subsequent energy minimization of InsPs and JA-Ile was performed at the HF/6-31G* level of Gaussian09 program (Frisch et al., 2009).
AutoDock Vina program (Trott and Olson, 2010) was used to dock all the forms of InsPs into the binding pocket of COI1. Autodock tools (ADT) (Morris et al., 2009) was employed to prepare the protein and InsPs. All water molecules were deleted, polar hydrogens were added and Gasteiger partial charges were assigned for the protein. The torsions were determined for InsPs. The protein was kept rigid. The grid center was determined according to the center of four phosphate molecules, with searching space size of 15 Å. The global search exhaustiveness value was set to 50. The maximum energy difference between the optimal binding mode and the worst-case was set to 5 kcal/mol. The structures were visualized using PyMOL (Schrödinger LLC., 2010).
In order to explore the underlying dynamics simulation and interaction mechanism of jasmonate receptor and 1,5-InsP8, JA-Ile, and JAZ1, we set up six systems. The composition, water molecules and total atoms of all the systems are included in Table 1. MD simulations were performed for each system by NAMD2.9 (Phillips et al., 2005) with CHARMM27 force field (Mackerell et al, 2004) for the protein. The force field parameters for the 1,5-InsP8 and JA-Ile were built by the SwissParam service (Zoete et al., 2011). The protonation state of the ionized residue at PH 8 was determined on the basis of the predicted pKa value by the H++ server (Gordon et al., 2005). An explicit water solvent box with TIP3P waters was used to systems, and 0.15 M NaCl were added to neutralize electric charge of the box. For each system, 50,000 time steps of energy minimization were conducted with a harmonic force constraint on the ligand (InsP8 and JA-Ile) and protein, ligand and backbone of protein and ligand and Cα atoms of protein, respectively. After that the temperature was raised from 0 to 300 K gradually within 200 ps under the NVT ensemble. After 2 ns equilibration under the NPT ensemble condition, 100 ns MD production was performed. The SHAKE algorithm was applied to restrain the bond length relating with hydrogen atoms (Coleman et al., 1977) and Particle Mesh Ewald (PME) (Tom et al., 1993) method was used to calculate the long electrostatic interactions. A 12.0 Å cut-off was set to calculate the van der Waals interactions. The integration step was set to 2 fs. The coordinates were preserved every 10 ps for trajectory analysis (Humphrey et al., 1996). The MD simulation data has been submitted to the zendo database (https://zenodo.org/) with accession links: https://zenodo.org/record/1255820.
For each system, the structures extracted from the final 20-ns MD simulation trajectory, were submitted to Residue Interaction Network Generator 2.0 (RING 2.0) server (Martin et al., 2011; Piovesan et al., 2016) for residue interaction network (RIN) analysis. The network was visualized by the plugin RINalyzer (Doncheva et al., 2011) integrated in Cytoscape (Shannon et al., 2003). The nodes denote amino acid residues or ligands and the edges represent residue-residue interactions, which include the interaction between the closest atoms, hydrogen bonding, salt bridge, ionic interaction, π-π stacking, and van der Waals interaction.
In addition, to characterize the residue interaction network of COI1, the parameters including shortest path betweenness and closeness centrality were calculated by the NetworkAnalyzer (Assenov et al., 2008) plugin of Cytoscape. The value of them is between 0 and 1 (Doncheva et al., 2012). It is suggested that residues with high shortest path betweenness values play an important role in stabilizing the protein structures, as well as those with high closeness values are likely significant for the protein function (Vendruscolo et al., 2002; Amitai et al., 2004; Brinda and Vishveshwara, 2005). Residue-residue communication is critical for the function of a protein (Süel et al., 2003). The closeness centrality measures how fast the information flows from a node to other reachable nodes in a network (Freeman, 1978).
The binding free energy of JA-Ile or JAZ1 with COI1 was calculated according to Molecular Mechanics/Generalized Born Surface Area (MM-GBSA) method using Prime/MM-GBSA module (Prime) of Schrödinger (Schrödinger, LLC, New York, NY)(2005). The total of 100 COI1—JA-Ile and COI1—JAZ1 complexes was extracted from the equilibrated MD trajectories. The complexes were minimized and the energies of the complex were calculated using the OPLS_2005 force field and VSGB solvation model (Jianing et al., 2011). The binding free energy is estimated according to the following equation:
Where Gcomplex is the minimized free energy for the complex, Greceptor and Gligand are the minimized free energy for the free COI1 and free ligand (JA-Ile or JAZ1). Each energy term was calculated by a summation of molecular mechanics energy (ΔEMM), GBSA solvation energy (ΔGGB), and surface area energy (ΔGSA). Strain energy was calculated for both COI1 and JA-Ile or JAZ1.
Statistical analysis was performed to affirm the significance of the difference of binding free energy. A one-way ANOVA was conducted here to test for significant differences (P < 0.05) between system 3/4 (COI1+ASK1+1,5-InsP8+JA-Ile/ COI1+ASK1+JAZ1+JA-Ile) and system 6 for the binding free energy of COI1—JA- Ile, and system 4/5 (COI1+ASK1 +JAZ1 +JA-Ile/ COI1+ASK1+ JAZ1+1,5-InsP8) and system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile) for the binding free energy of COI1—JAZ1.
The structure of Loop C from COI1 and missing regions ASK1 were built by Rosetta and Modeler, which is shown in Figure 2. The built model was subsequently minimized by NAMD. NAMD is one of the most efficient open-source program for molecular dynamics simulation, which is widely applied to explore the dynamic behavior of large systems (millions of atoms). To compare the binding affinity between COI1 and different forms of InsPs, InsP5s, InsP6, and InsP8s were all docked to the binding site of phosphate ions in the refined full structure of jasmonate receptor complex. Autodock vina is an efficient and accurate docking program widely used in predicting the binding mode and binding affinity between protein and ligand (Trott and Olson, 2010; Wang et al., 2016). The vina score of each InsP is summarized in Table 2. It shows that InsP8s exhibits lower binding free energy than InsP5s and InsP6.
The detailed binding mode of each form of InsPs with the lowest binding free energy is also shown in Figure S1. InsP5 forms hydrogen bond interaction with Lys79, Lys81, Arg85, Arg120, Lys147, Arg409, and Arg440 from COI1 and Arg206 from JAZ1 (Figure S1A). It also interacts with Trp467 of COI1 through hydrophobic interaction. Regarding InsP6, it forms hydrogen bond interaction with Lys79, Lys81, Arg85, Arg120, Arg121, Arg346, Arg409, and Arg440 from COI1 and Arg206 from JAZ1 and hydrophobic interaction with Met88 and Trp467 from COI1. For InsP8s, 1,2-InsP8 shows the lowest binding free energy (−7.1 kcal/mol) with COI1 in all the isoforms. The second-lowest binding free energy is for 1,5-InsP8, with the score of −7.0 kcal/mol. Comparison of the detailed binding mode of these two isoforms, shows that 1,2-InsP8 forms hydrogen bond interaction with Lys79, Lys81, Arg85, Arg120, Arg346, Arg409, and Arg440 from COI1 and Arg206 from JAZ1 and hydrophobic interaction with Trp467 of COI1. 1,5-InsP8 forms hydrogen bond interaction with Lys79, Lys81, Arg85, His118, Arg120, Arg121, Lys144, Lys147, Arg346, Arg409, and Lys492 from COI1 and Arg206 from JAZ1 and hydrophobic interaction with Met88 of COI1. A previous study shows that His118, Arg346, and Lys492 from COI1 are important for the binding of InsP8 with COI1 (Laha et al., 2015); and 1,5-InsP8 can form hydrogen bond interaction with all of these residues. However, 1,2-InsP8 does not interact with His118 and Lys492. Based on the predicted binding free energy and binding mode, 1,5-InsP8 was selected for the further MD simulations.
To investigate the stability and conformational differences of the jasmonate receptor in response to the binding with InsP8, JA-Ile, and JAZ1, we carried out molecular dynamics simulations of the jasmonate receptor structures in six systems. The conformational stability of six systems during MD simulations was evaluated by calculating the root mean square deviation (RMSD) of the backbone atoms. Figure 3 reflected the trend of RMSD in the six systems relative to the crystal structure.
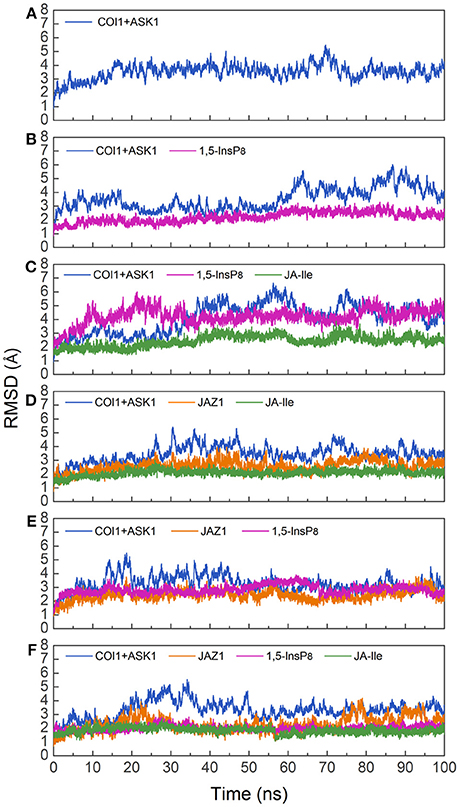
Figure 3. RMSD plots of the complex during molecular dynamics simulation with respect to the crystal structure. (A) system 1, COI1+ASK1; (B) system 2, COI1+ASK1+1,5-InsP8; (C) system 3, COI1+ASK1+1,5-InsP8+JA-Ile; (D) system 4, COI1+ASK1+JAZ1 +JA-Ile; (E) system 5, COI1 +ASK1 +JAZ1+1, 5-InsP8; (F) system 6, COI1+ ASK1+JAZ1 +1,5-InsP8+JA-Ile.
Since there was no InsP molecule in the structure of jasmonate receptor (PDB ID: 3OGM), the complex was constructed based on the docked pose with the lowest vina score of 1,5-InsP8. MD simulations were constructed to examine the stability and the effect of 1,5-InsP8, JA-Ile, and JAZ1 with the jasmonate receptor. The six systems (systems 1–6) achieved equilibrium at 20, 65, 55, 30, 20, and 35 ns, respectively. The RMSD values of the backbone atoms converged at 3.27–4.68 Å in six systems. The average RMSD values of 1,5-InsP8 are 2.52, 4.38, 2.87, and 2.00 Å in system 2 (COI1+ASK1+1,5-InsP8), system 3 (COI1+ASK1+1,5-InsP8+JA-Ile), system 5 (COI1+ASK1+JAZ1+1,5-InsP8), and system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), respectively. For JA-Ile, the average RMSD values are 2.59, 2.13, and 1.79 Å in system 3, system 4 (COI1+ASK1+JAZ1+JA-Ile), and system 6, respectively. In addition, the averaged RMSD of JAZ1 is 2.67, 2.49, and 2.33 in the system 4, system 5, and system 6, respectively. Thus, based on the RMSD results, our MD simulations are reliable for further investigation.
Furthermore, the radius of gyration (Rg) of COI1 in six systems was also calculated to detect the compactness of its structure, as shown in Figure 4. We calculated the average Rg ranging from 40 to 100 ns in each system. The average Rg of COI1 is the highest in system 1 (COI1+ASK1), with the value of 26.03 ± 0.11 Å. The next is that in system 5 (COI1+ASK1+JAZ1+1,5-InsP8), with the value of 25.90 ± 0.09 Å. The COI1 shows average Rg values in system 2 (COI1+ASK1+1,5-InsP8) of 25.75 ± 0.16 Å and system 4 (COI1+ASK1+JAZ1+JA-Ile) of 25.73 ± 0.11 Å. The mean Rg values of COI1 was 25.60 ± 0.09 Å and 25.71 ± 0.09 Å in systems 3 (COI1+ASK1+1,5-InsP8+JA-Ile) and 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), respectively. Comparing with other systems, it indicates that the structure of COI1 in system 1 (COI1+ASK1) without any ligands, also termed as the apo system, was less compact.
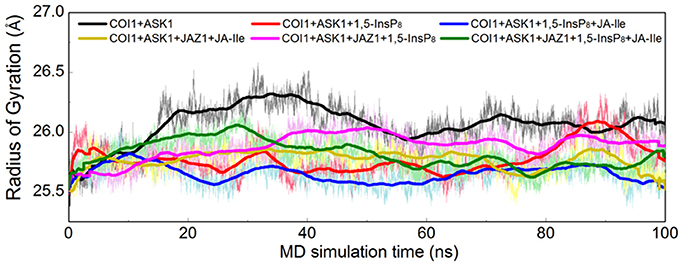
Figure 4. Rg plots of backbone atoms of protein for the six systems. Black is used for system 1 (COI1+ASK1), red for system 2 (COI1+ASK1+1,5-InsP8), blue for system 3 (COI1+ASK1+1,5-InsP8+JA-Ile), yellow for system 4 (COI1+ASK1+JAZ1+JA-Ile), magenta for system 5 (COI1+ASK1+JAZ1+1,5-InsP8), and green for system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile).
To determine whether binding with InsP8, JA-Ile, and JAZ1 affected the dynamic behavior of COI1 residues, the mobility of the protein residues was examined by plotting the root-mean-square fluctuation (RMSF) of the backbone atoms of COI1 versus the residue number (Figure 5). We can observe that most of high flexible regions are the loops linking the regular secondary elements or those on the surface of COI1, such as β2-α5 (termed as loop 2), β12-α15 (loop 12), β14-α17 (loop 14), β19-β20 loop (loop C), α11, and α21. The large fluctuations may be caused by the intrinsic flexibility of this protein, in the light of these regions exhibit high flexibility in all the systems. The main flexible regions were identified for all systems. The locations of these regions in the protein structure are shown in Figure 6. The β16-α19 loop and β17-α20 loop located at the top surface of the COI1 solenoid, exhibit high flexibility in system 1 (COI1+ASK1) and system 2 (COI1+ASK1+1,5-InsP8). The α16-β14 loop located close to 1,5-InsP8 also shows high flexibility in system 2 and system 3 (COI1+ASK1+1,5-InsP8+JA-Ile). The β13-α16 and β16-α19 loops, at the top of the COI1, exhibit high flexibility in system 4 (COI1+ASK1+JAZ1+JA-Ile). In system 5 (COI1+ASK1+JAZ1+1,5-InsP8), loop β17-α20 at the top of the COI1 shows high flexibility. In addition, the α18-β16 loop is close to 1,5-InsP8, exhibiting high flexibility. In system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), the β6-α8 loop is located close to JAZ1, the α14-β12 loop and α17-β15 loop are close to the bottom of 1,5-InsP8, exhibiting high flexibility.
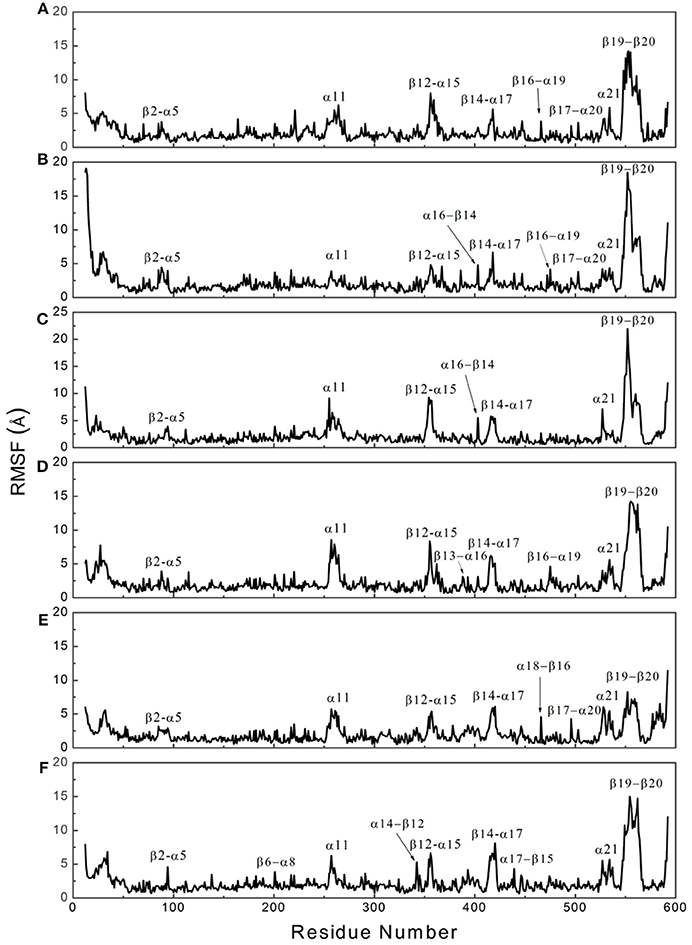
Figure 5. RMSF plots of Cα atoms of COI1 in the six systems. (A) system 1, COI1+ASK1; (B) system 2, COI1+ASK1 +1,5-InsP8; (C) system 3, COI1+ASK1+1,5-InsP8+JA-Ile; (D) system 4, COI1+ASK1+JAZ1 +JA-Ile; (E) system 5, COI1+ASK1+JAZ1+1,5-InsP8; (F) system 6, COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile.
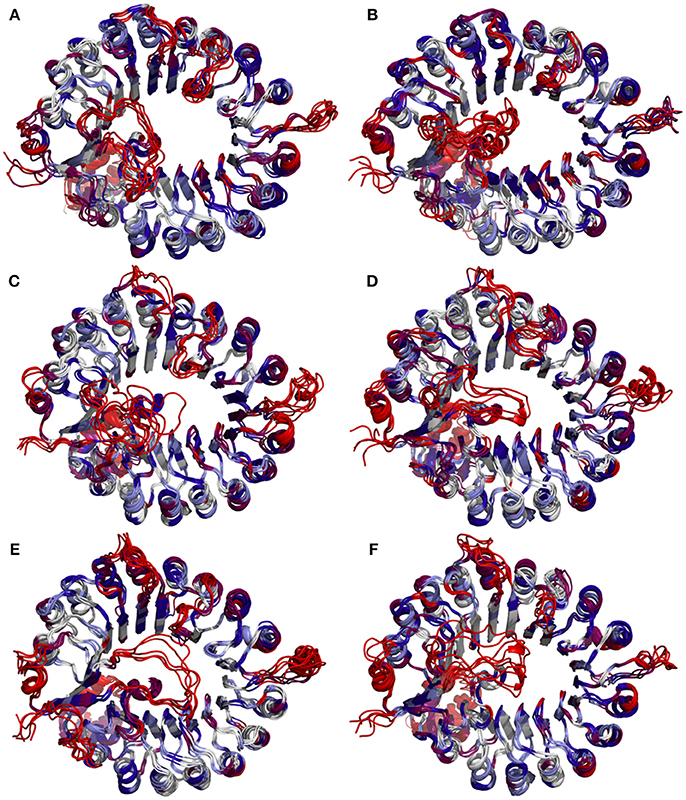
Figure 6. Snapshots of COI1 structures from MD trajectories. The color is coded by the RMSF value, with blue being the lowest fluctuations to light blue to white to magenta to red being the highest fluctuations. (A) system 1, COI1+ASK1; (B) system 2, COI1+ASK1 +1,5-InsP8; (C) system 3, COI1+ASK1+1,5-InsP8+JA-Ile; (D) system 4, COI1+ASK1+JAZ1 +JA-Ile; (E) system 5, COI1+ASK1+JAZ1+1,5-InsP8; (F) system 6, COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile.
In order to analyze the interaction mode between 1,5-InsP8 and COI1 after MD simulations, we extracted the representative structures from the final 20-ns MD trajectory (Figure 7). The detailed binding mode of 1,5-InsP8 with COI1 reveals that the components, JA-Ile and JAZ1, are also important for the whole interaction network in the solenoid of COI1. In system 2 (COI1+ASK1+1,5-InsP8), 1,5-InsP8 maintains the hydrogen bond interaction with Lys79, Lys81, Arg120, Lys144, Lys147, and Lys492 from COI1 (Figure 7A). Additionally, 1,5-InsP8 forms novel hydrogen bond interaction with Arg348, as well as hydrophobic interaction with Try382 and Trp467 of COI1. The hydrogen bond interaction between 1,5-InsP8 and His118 or Arg346 from COI1 disappeared, which were shown to be important for the binding of InsP8 with COI1. In system 3 (COI1+ASK1+1,5-InsP8+JA-Ile), its interaction with Lys81, Arg85, Arg120, Arg121, Arg409, Arg440, and Lys492 from COI1 was maintained (Figure 7B), but the hydrogen bond interaction with His118 and Arg346 from COI1 was lost. Additionally, 1,5-InsP8 forms hydrogen bond with Asp407 and hydrophobic interaction with Tyr382 and Trp467 of COI1. In system 5 (COI1+ASK1+JAZ1+1,5-InsP8), the interaction with Lys81, Arg85, Arg120, Arg121, Lys147, Arg346, Arg440, and Lys492 from COI1 was maintained, as well as with Arg206 from JAZ1 and the hydrophobic interaction with Met88 (Figure 7C). The hydrogen interaction with His118 and Arg409 from COI1 was lost. It forms hydrogen bond with Arg348 and hydrophobic interaction with Tyr382. In system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), the hydrogen interaction with Lys79, Lys81, Arg85, Arg120, Arg121, Lys147, Arg346, Arg440, Arg409, and Lys492 from COI1 was maintained, as well as that with Arg206 from JAZ1 (Figure 7D). The hydrogen interaction with His118 from COI1 was lost and hydrogen bond interaction with Arg348 and hydrophobic interaction with Tyr382 and Trp467 of COI1 was formed.
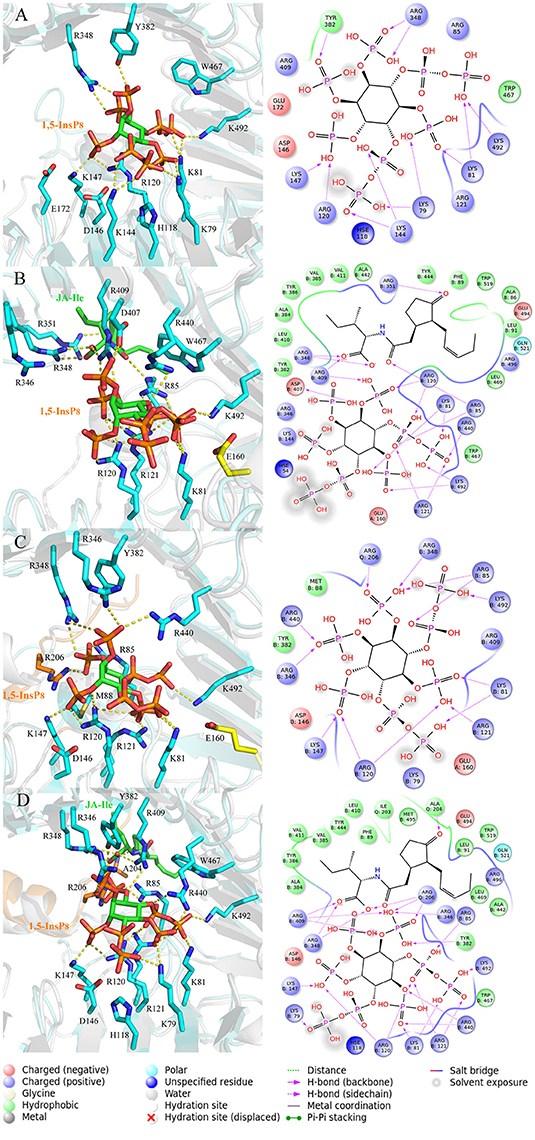
Figure 7. The representative snapshots from the last 20 ns of the MD trajectories. (A) system 2, COI1+ASK1 +1,5-InsP8; (B) system 3, COI1+ASK1+1,5-InsP8+JA-Ile; (C) system 5, COI1+ASK1+JAZ1+1,5-InsP8; (D) system 6, COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile. For panels (A–D), the proteins are in white and cyan for the docked and MD structures, respectively. Hydrogens are omitted for clarity.
Investigation and resolution of residue interaction network (RIN) is imperative for the understanding protein structure-function relationships (Amitai et al., 2004; Del et al., 2006; Vishveshwara et al., 2009). Recently, RIN analysis has been successfully applied to investigate mutation effects, protein folding, domain-domain communication, and catalytic activity (Dokholyan et al., 2002; Swintkruse, 2004; Del et al., 2006; Soundararajan et al., 2010; Boehr et al., 2013; Scaini et al., 2014; Biswas et al., 2017). Based on the interaction mode analysis, we found that 1,5-InsP8 lost some interactions and formed novel interactions with the jasmonate receptor complex in different systems. To comprehensively understand the differences between RINs in the solenoid of COI1 binding with different ligands, we explored the relationship between crucial residues of COI1 solenoid and the ligands, including InsP8, JA-Ile, and JAZ1. The residue interaction networks were generated based on the representative structures extracted from the final 20-ns of MD trajectory.
RIN plots shown in Figure 8 depicts the presence of interaction network among His118, Arg120, Arg121, Lys144, Asp146, and Met88 or Arg346, Arg348, Tyr382, Arg409, Trp467, and Lys492 in system 1 (COI1+ASK1, Figure 8A). The competent form of the apo structure exhibits the weakest network (system 1). Upon binding with 1,5-InsP8 (Figure 8B), 17 interactions are observed between 1,5-InsP8 and COI1, including 8 hydrogen bond interactions and 9 interactions between closest atoms. Upon addition of a new component, JA-Ile, the network became stronger in system 3 (COI1+ASK1+1,5-InsP8+JA-Ile); there are 24 interactions between 1,5-InsP8 and COI1, including 8 hydrogen bond interactions and 16 interactions between closest atoms (Figure 8C). In system 5 (COI1+ASK1+JAZ1+1,5-InsP8), 1,5-InsP8 forms 22 interactions with COI1, ASK1, and JAZ1, including 10 hydrogen bond interactions and 12 interactions between closest atoms (Figure 8E). In system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), 1,5-InsP8 forms 22 interactions with COI1, ASK1, JA-Ile, and JAZ1, including 12 hydrogen bond interactions and 10 interactions between closest atoms (Figure 8F).
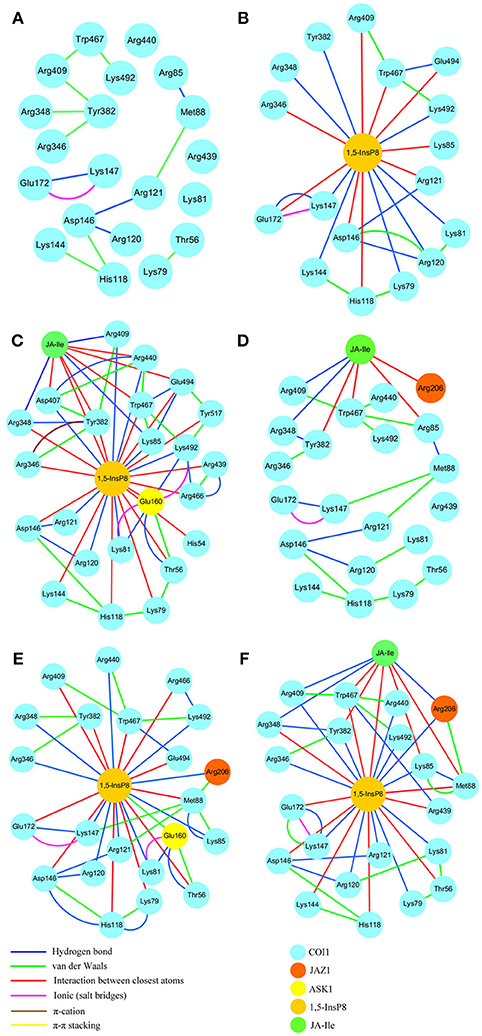
Figure 8. The residue interaction network of amino acids within 5Å around 1,5-InsP8. (A) system 1, COI1+ASK1, without 1,5-InsP8 and JA-Ile, the residue interaction network in the cavity of COI1; (B) system 2, COI1+ASK1+1,5-InsP8, the residue interaction network between 1,5-InsP8 and COI1; (C) system 3, COI1+ASK1+1,5-InsP8+JA-Ile, the residue interaction network among 1,5-InsP8, JA-Ile and COI1; (D) system 4, COI1+ASK1+JAZ1+JA-Ile, the residue interaction network in the cavity of COI1 and between JA-Ile and COI1; (E) system 5, COI1 +ASK1 +JAZ1 +1,5-InsP8, the residue interaction network between 1,5-InsP8 and COI1; (F) system 6, COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile, the residue interaction network among 1,5-InsP8, JA-Ile, and COI1.
Regarding systems 1 (COI1+ASK1) and 4 (COI1+ASK1+JAZ1+JA-Ile) that lack of 1,5-InsP8, the interaction network is obviously less stronger than that of other systems (Figures 8A,D). Compared with system 4 (Figure 8D), the van der Waals interaction between Met88 and Arg121 or Lys147 disappeared, which resulted in conformational variation of Met88 in system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile, Figure 8F). Met88 forms interaction of closest atom with JA-Ile and van der Waals interaction with JAZ1. Regarding system 5 (COI1+ASK1+JAZ1+1,5-InsP8), which lack JA-Ile, the conformation of 1,5-InsP8 changed greatly. Met88 forms hydrogen bond interaction with Ala207 of JAZ1 and van der Waals interaction with Arg121 or Lys147 (Figure 9). Hub nodes can clearly be found in the network inside COI1, including 1,5-InsP8, JA-Ile, Met88, His118, Arg120, Arg121, Arg346, Tyr382, Arg409, Trp467, and Lys492. The binding with 1,5-InsP8, JA-Ile, and JAZ1 significantly changes the network in COI1 solenoid. Most importantly, 1,5-InsP8 promotes and stabilizes the interaction of COI1—JA-Ile or COI1—JAZ1. The system lacking 1,5-InsP8 or JAZ1 also affects the hydrogen bond interaction of COI1—JA-Ile (Figure 10). The distance between Arg85 of COI1 and JA-Ile decreases in the presence of InsP8 or JAZ1.
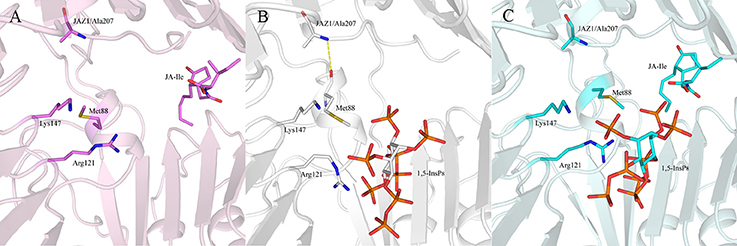
Figure 9. The interaction between 1,5-InsP8, JA-Ile, JAZ1 and COI1 (Met88, Arg121, Lys147) from the last 20 ns of the MD trajectories. (A) system 4 (COI1+ASK1+JAZ1+JA-Ile); (B) system 5 (COI1+ASK1+JAZ1+1,5-InsP8); (C) system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile). The carbon atoms of residues or ligands are colored by pink, white, and cyan in systems 4, 5, and 6, respectively. The COI1 and JAZ1 are colored by light pink, white, and palecyan in system 4, system 5, and system 6, respectively.
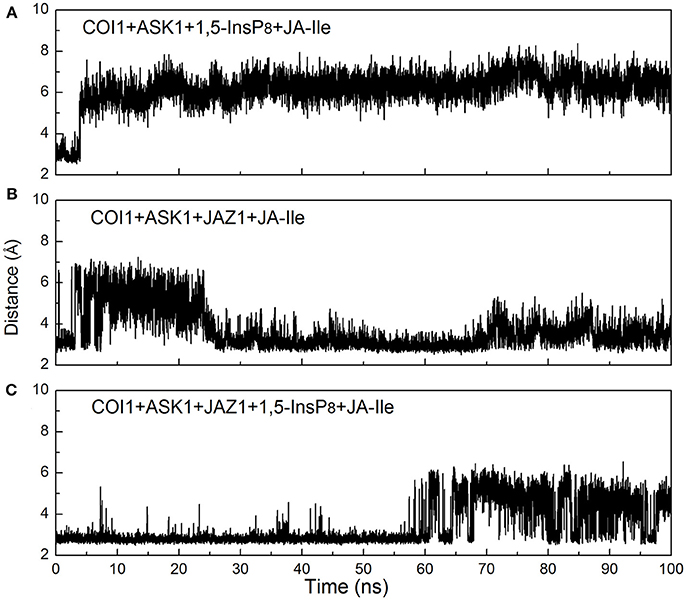
Figure 10. Monitoring of the intermolecular hydrogen bonds between JA-Ile and COI1 during the MD simulation, (A) system 3 (COI1+ASK1+1,5-InsP8+JA-Ile); (B) system 4 (COI1+ASK1+JAZ1+JA-Ile); (C) system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile).
To further compare the residue network changes in different systems, we calculated the two parameters of each node in the network: the shortest path betweenness and closeness centrality. Table 3 summarizes the value of critical residues shortest path betweenness and closeness centrality in the network in each system. It is found that 1,5-InsP8 possesses the highest value of betweenness in all the systems, suggesting that it is crucial in the COI1 network. The next highest value is found for JA-Ile. Both of them are crucial for stabilizing the conformation of COI1, which is important for the binding with JAZ1. In addition, the closeness centrality of the key residues increases with upon addition of each component of the jasmonate receptor complex, by comparing systems 1 (COI1+ASK1), 2 (COI1+ASK1+1,5-InsP8), 3 (COI1+ASK1+1,5-InsP8+JA-Ile), and 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile). While the closeness centrality of the key residues is smaller in system 4 (COI1+ASK1+JAZ1+JA-Ile) and 5 (COI1+ASK1+JAZ1+1,5-InsP8) than that in system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile). This implies that the information flows more rapidly with the help of 1,5-InsP8, JA-Ile, and JAZ1 in the COI1 solenoid.
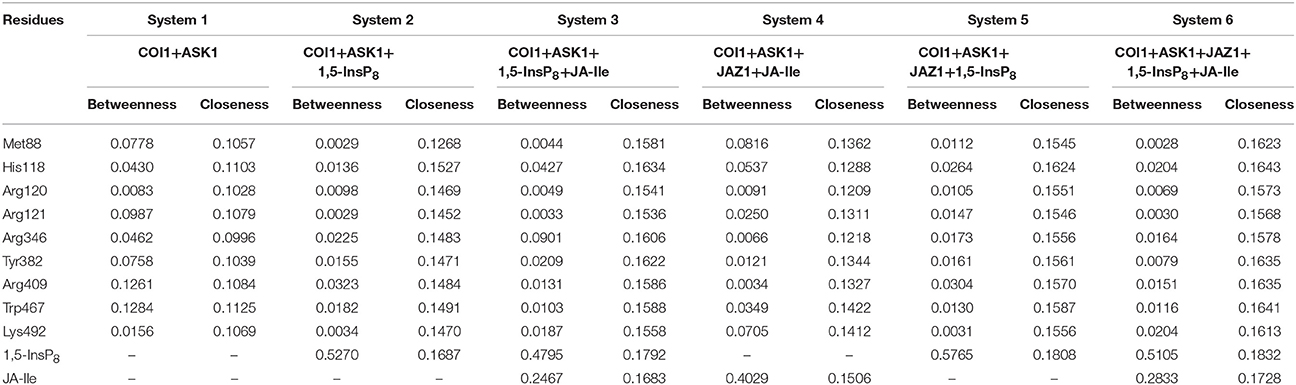
Table 3. Summary of the shortest path betweenness and closeness centrality of selected residues in the six simulation systems.
To obtain the quantitativeestimation of the binding affinity of JA-Ile or JAZ1 with COI1, we performed binding free energy calculation by extracting 100 snapshots from the last 50 ns of the MD trajectories. The results were summarized in Table 4. The output energy terms include: the coulomb energy (ΔGCoulomb), the covalent binding energy (ΔGCovalent), the hydrogen-bonding energy (ΔGHbond), the lipophilic energy (ΔGLipo), the Van der Waals energy (ΔGvdw), the generalized Born electrostatic solvation energy ΔGGB, the Pi-pi packing energy (ΔGPacking), and the self-contact correction (ΔGSelfCont).
The free energy components showed in Table 4 suggest that ΔGcoulomb, ΔGLipo, and ΔGvdw are the majority of the favorable contributions for the binding of JA-Ile and JAZ1, whereas the polar solvation energies (ΔGGB) and ΔGCovalent generate the unfavorable contributions. The predicted binding free energy for COI1—JA-Ile is −109.83 ± 3.98, −88.37 ± 3.35, and −110.77 ± 4.46 kcal/mol in system COI1+ASK1+1,5-InsP8+JA-Ile, COI1+ASK1+JAZ1+JA-Ile, and COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile, respectively. The predicted binding free energy for COI1—JAZ1 is −188.66 ± 5.72, −195.53 ± 3.98, and −207.04 ± 4.94 kcal/mol in system COI1+ASK1+JAZ1+1,5-InsP8, COI1+ASK1+JAZ1+JA-Ile, and COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile, respectively.
Based on the calculated binding free energy, the binding between COI1 and JA-Ile or JAZ1 is stronger in system with 1,5-InsP8 than those systems without 1,5-InsP8. The statistical analysis affirmed that the differences are significant (P < 0.05). The results indicate that the binding of InsP8 with COI1 promotes the binding between COI1 and JA-Ile or JAZ1.
The jasmonate receptor, a multi-component complex, consists of ASK1, COI1, inositol phosphate, and JAZ1. COI1, as a multi-component signaling hub, shares a conserved 3D structure with the auxin receptor TIR1. InsP plays important roles in hormone perception. How InsP binds with COI1 and the structural variations in COI1 that occur upon binding with InsP, JA-Ile, and JAZ are not well understood. Thus, a computational study was performed to elucidate the binding mechanism between the jasmonate receptor and InsPs.
Since there is no published crystal structure available for the InsP bound form of COI1, the different forms of InsPs were docked into the solenoid of COI1 in this study. The results suggest that 1,5-InsP8 binds to COI1 with the second lowest binding free energy and reasonable binding mode. 1,5-InsP8 forms hydrogen bond interaction with Lys79, Lys81, Arg85, His118, Arg120, Arg121, Arg346, Arg409, Lys492, from COI1 and Arg206 from JAZ1, which is also observed in Laha's work (Laha et al., 2015, 2016). Besides, it forms hydrogen bond interaction with Lys144 and Lys147 of COI1, which is not observed in Laha's work. It is not observed that 1,5-InsP8 forms hydrogen bond interaction with Arg440 of COI1 in our model, which is different from Laha's work. Residues His118, Arg346, and Lys492 of COI1 are observed to coordinate several phosphorus atoms of 1,5-InsP8, which is also reported in Laha's work (Laha et al., 2015). While Tyr382 of COI1 doesn't interact with 1,5-InsP8 in our model, which is different with Laha's model. Analysis of the detailed binding mode revealed that 1,5-InsP8 forms hydrogen bond interaction with His118, Arg346, Arg492 of COI1 and Arg206 of JAZ1, which is previously suggested to be important for the binding with InsP8 in the experimental work (Laha et al., 2015). Therefore, 1,5-InsP8 was chosen for further MD simulations.
MD simulations were conducted to elucidate the structural variations of COI1 that occur in response to binding with 1,5-InsP8, JA-Ile, and JAZ1. We found that the systems lacking JAZ1 [system 2 (COI1+ASK1+1,5-InsP8) and system 3 (COI1+ASK1+1,5-InsP8+JA-Ile)] exhibit higher variation than that in the other systems [system 4 (COI1+ASK1+JAZ1+JA-Ile), system 5 (COI1+ASK1+JAZ1+1,5-InsP8), and system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile)]. The JA-Ile also exhibits higher variation in system 3 than those in system 5 and system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile).
To further identify the flexible region of COI1, the RMSF of COI1 was plotted in each system. Through the RMSF plots (Figure 6), the flexible region of COI1 in response to binding with InsP, JA-Ile, and JAZ1 can be observed. Compared with the apo system and system 4 in the absence of InsP8, the α16-β14 loop, the α18-β16 loop, and the α14-β12 and the α17-β15 loop of COI1 in systems 2 and 3, system5 and system 6, respectively, exhibited high flexibility upon binding with InsP8. The β13-α16 and β16-α19 loops, the β17-α20 loop, and the β6-α8 loop in system 4, system 5, and system 6, respectively, showed higher flexibility upon binding with JAZ1. The flexible regions indicated their involvement in InsP8 or JAZ1 binding or entering the binding pocket in COI1. Due to JAZ1 is an intrinsically disordered protein (Chini et al., 2016), the flexible regions of COI1 found here would be helpful to suppose the binding mode of COI1 with other part of JAZ1, besides its conserved jas domain. Loop C exhibits high flexibility in all systems, which is based on the initial conformation by Rosetta and Modeler. Loop C forms tight interaction with JAZ1 in systems 3 to system 6, which supports the results of Sheard's study (Sheard et al., 2010), that loop C covers loop 2 and is involved in the binding with JAZ1. This conformation differs from that in the auxin receptor (Tan et al., 2007).
The interaction network in COI1 solenoid changes greatly upon the introduction of 1,5-InsP8, JA-Ile, and JAZ1. The detailed binding mode was analyzed for each system after MD simulation. We can find that some interactions disappear and some novel interactions form between 1,5-InsP8 and COI1. The conformation of 1,5-InsP8 also changes in those systems with or without JA-Ile and JAZ1 compared with system 6 (COI1+ASK1+JAZ1+1,5-InsP8+JA-Ile), implying that those components all affect the residue network in the COI1 solenoid. To further understand the residue interaction network, we performed the RIN analysis based on the structures extracted from the MD trajectories. The results indicated that 1,5-InsP8 and JA-Ile, with the highest betweenness values, act as the hub nodes and play crucial roles in the stabilizing the conformation of COI1 solenoid. The introduction of 1,5-InsP8 alters the residue interaction network in the COI1 solenoid, especially the interaction between Met88 and Arg121 or Lys147. Met88 then forms interaction of closest atom with JA-Ile and van der Waals interaction with JAZ1 (Figure 8F). Additionally, 1,5-InsP8 forms hydrogen bond interaction with Arg121, which explains why COI1 with a mutation of Met88 or Arg121 to alanine can't form a complex with JAZ1 even under the induction of coronatine (Sheard et al., 2010).
In view the importance of InsP8 in the network of COI1, it acts a necessary constituent and promotes the COI1-JAZ interaction when the levels of JA-Ile rise in response to a stimulus. In addition, based on the interaction between InsP8 and JAZ1, the doses of InsP8 would also modulate the conformation of COI1, which lead to its binding with different JAZ repressors in different conditions or responses. It was suggested that the detection of COI1—JAZ1, COI1—JAZ3, and COI1—JAZ9 required 60, 15, and 1.5 μM JA-Ile, respectively (Melotto et al., 2008; Chini et al., 2016). It can be conceived that InsP8 concentration would be another important factor in the specificity of COI1 among binding with different JAZ repressors.
In the present study, the binding mechanism between InsP and COI1 was explored by molecular docking, molecular dynamics simulations, residue interaction network analysis and binding free energy calculation. The binding with 1,5-InsP8, JA-Ile, and JAZ1 makes the structure of COI1 more compact. In addition, the binding of 1,5-InsP8 with COI1 not only stabilizes the conformation of COI1 solenoid but also promotes the binding between JA-Ile or JAZ1 and COI1. Analysis the network parameters led to the identification of some hub nodes in this network, including Met88, His118, Arg120, Arg121, Arg346, Tyr382, Arg409, Trp467, and Lys492 of COI1. This study provides molecular basis on the recognition mechanism of each component of the jasmonate receptor complex. These results will facilitate the discovery and design of novel jasmonate signaling pathway modulators.
JD conceived and supervised the experiments. MC performed MD simulations. MC and JD analyzed the data. MC, JD, and XY wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the Natural Science Foundation of Shandong Province, China (Grant No. ZR2018QB004), the Talents of High Level Scientific Research Foundation (Grant No. 6631113318) and the Graduate Research Plan (QYC201702) of Qingdao Agricultural University. This work was carried out on TianHe-1 (A) of the National Super Computer Center in Tianjin. We acknowledge the National Super Computer Center in Tianjin for the computational resource support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00963/full#supplementary-material
Figure S1. Binding mode between different forms of inositol phosphates and the COI1-ASK1-JAZ1 complex predicted by molecular docking. The carbon atom and oxygen atoms of inositol phosphates and JA-Ile are colored in green and red, phosphorus atoms of inositol phosphates colored by orange, respectively. Hydrogens are omitted for clarity. (A) COI1+ASK1+JAZ1+JA-Ile+InsP5[3-OH]. (B) COI1+ASK1+JAZ1+JA-Ile+InsP6. (C) COI1+ASK1+JAZ1+JA-Ile+1,2-InsP8. (D) COI1+ASK1+JAZ1+JA-Ile+1,5-InsP8.
Amitai, G., Shemesh, A., Sitbon, E., Shklar, M., Netanely, D., Venger, I., et al. (2004). Network analysis of protein structures identifies functional residues. J. Mol. Biol. 344, 1135–1146. doi: 10.1016/j.jmb.2004.10.055
Assenov, Y., Ramírez, F., Schelhorn, S. E., Lengauer, T., and Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics 24, 282–284. doi: 10.1093/bioinformatics/btm554
Biswas, A., Shukla, A., Chaudhary, S. K., Santhosh, R., Jeyakanthan, J., and Sekar, K. (2017). Structural studies of a hyperthermophilic Thymidylate Kinase enzyme reveal conformational sub-states along the reaction coordinate. FEBS J. 284, 2527–2544. doi: 10.1111/febs.14140
Boehr, D. D., Schnell, J. R., Mcelheny, D., Bae, S. H., Duggan, B. M., Benkovic, S. J., et al. (2013). A distal mutation perturbs dynamic amino acid networks in dihydrofolate reductase. Biochemistry 52, 4605–4619. doi: 10.1021/bi400563c
Brinda, K. V., and Vishveshwara, S. (2005). A network representation of protein structures: implications for protein stability. Biophys. J. 89, 4159–4170. doi: 10.1529/biophysj.105.064485
Browse, J. (2008). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Coleman, T. G., Mesick, H. C., and Darby, R. L. (1977). Numerical integration. Ann. Biomed. Eng. 5, 322–328. doi: 10.1007/BF02367312
De, S. O., Shi, J., and Deane, C. M. (2015). Building a better fragment library for de novo protein structure prediction. PLoS ONE 10:e0123998. doi: 10.1371/journal.pone.0123998
Del, S. A., Fujihashi, H., Amoros, D., and Nussinov, R. (2006). Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol. 2:2006.0019. doi: 10.1038/msb4100063
Dokholyan, N. V., Li, L., Feng, D., and Shakhnovich, E. I. (2002). Topological determinants of protein folding. Proc. Natl. Acad. Sci. U.S.A. 99, 8637–8641. doi: 10.1073/pnas.122076099
Doncheva, N. T., Assenov, Y., Domingues, F. S., and Albrecht, M. (2012). Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670–685. doi: 10.1038/nprot.2012.004
Doncheva, N. T., Klein, K., Domingues, F. S., and Albrecht, M. (2011). Analyzing and visualizing residue networks of protein structures. Trends Biochem. Sci. 36, 179–182. doi: 10.1016/j.tibs.2011.01.002
Eswar, N., Webb, B., Martirenom, M. A., Madhusudhan, M. S., Eramian, D., Shen, M., et al. (2016). Comparative protein structure modeling using modeller. Curr. Prot. Prot. Sci. 86, 2.9.1–2.9.37. doi: 10.1002/0471250953.bi0506s15
Ferguson, K. M., Kavran, J. M., Sankaran, V. G., Fournier, E., Isakoff, S. J., Skolnik, E. Y., et al. (2012). Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384. doi: 10.1016/S1097-2765(00)00037-X
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Freeman, L. C. (1978). Centrality in social networks conceptual clarification. Soc. Netw. 1, 215–239. doi: 10.1016/0378-8733(78)90021-7
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2009). Wallingford, CT: Gaussian, Inc.
Gordon, J. C., Myers, J. B., Folta, T., Shoja, V., Heath, L. S., and Onufriev, A. (2005). H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, 368–371. doi: 10.1093/nar/gki464
Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., and Simmerling, C. (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725. doi: 10.1002/prot.21123
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Jianing, L., Robert, A., Kai, Z., Yixiang, C., Suwen, Z., and Friesner, R. A. (2011). The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins 79, 2794–2812. doi: 10.1002/prot.23106
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105. doi: 10.1073/pnas.0802332105
Laha, D., Johnen, P., Azevedo, C., Dynowski, M., Weiß, M., Capolicchio, S., et al. (2015). VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in arabidopsis. Plant Cell 27, 1082–1097. doi: 10.1105/tpc.114.135160
Laha, D., Parvin, N., Dynowski, M., Johnen, P., Mao, H., Bitters, S. T., et al. (2016). Inositol polyphosphate binding specificity of the jasmonate receptor complex. Plant Physiol. 171, 2364–2370. doi: 10.1104/pp.16.00694
Mackerell, A. D. Jr., Feig, M., and Brooks, C. L. III. (2004). Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415. doi: 10.1002/jcc.20065
Martin, A. J., Vidotto, M., Boscariol, F., Di, D. T., Walsh, I., and Tosatto, S. C. (2011). RING: networking interacting residues, evolutionary information and energetics in protein structures. Bioinformatics 27, 2003–2005. doi: 10.1093/bioinformatics/btr191
Melotto, M., Mecey, C., Niu, Y., Chung, H. S., Katsir, L., Yao, J., et al. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988. doi: 10.1111/j.1365-313X.2008.03566.x
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi: 10.1002/jcc.21256
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957. doi: 10.1111/j.1365-313X.2011.04480.x
Pearlman, D. A., Case, D. A., Caldwell, J. W., Ross, W. S., Thomas, .E. C, DeBolt, S., et al. (1995). AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 91, 1–41. doi: 10.1016/0010-4655(95)00041-D
Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., et al. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802. doi: 10.1002/jcc.20289
Piovesan, D., Minervini, G., and Tosatto, S. C. E. (2016). The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 44, W367–W374. doi: 10.1093/nar/gkw315
Scaini, M. C., Minervini, G., Elefanti, L., Ghiorzo, P., Pastorino, L., Tognazzo, S., et al. (2014). CDKN2A unclassified variants in familial malignant melanoma: combining functional and computational approaches for their assessment. Hum. Mutat. 35, 828–840. doi: 10.1002/humu.22550
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Routledge 13, 2498–2504. doi: 10.1101/gr.1239303
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Soundararajan, V., Raman, R., Raguram, S., Sasisekharan, V., and Sasisekharan, R. (2010). Atomic interaction networks in the core of protein domains and their native folds. PLoS ONE 5:e9391. doi: 10.1371/journal.pone.0009391
Süel, G. M., Lockless, S. W., Wall, M. A., and Ranganathan, R. (2003). Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59–69. doi: 10.1038/nsb881
Swintkruse, L. (2004). Using networks to identify fine structural differences between functionally distinct protein states. Biochemistry 43, 10886–10895. doi: 10.1021/bi049450k
Tan, X., Calderon-Villalobos, L. I., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tom, D., Darrin, Y., and Lee, P. (1993). Particle mesh Ewald: an N [center-dot] log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. doi: 10.1063/1.464397
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Vendruscolo, M., Dokholyan, N. V., Paci, E., and Karplus, M. (2002). Small-world view of the amino acids that play a key role in protein folding. Phys. Rev. E Stat Nonlin Soft Matter Phys. 65:061910. doi: 10.1103/PhysRevE.65.061910
Vishveshwara, S., Ghosh, A., and Hansia, P. (2009). Intra and inter-molecular communications through protein structure network. Curr. Protein Pept. Sci. 10, 146–160. doi: 10.2174/138920309787847590
Wang, H., Falck, J. R., Hall, T. M. T., and Shears, S. B. (2011). Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8, 111–116. doi: 10.1038/nchembio.733
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004). Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174. doi: 10.1002/jcc.20035
Wang, Z., Sun, H., Yao, X., Li, D., Xu, L., Li, Y., et al. (2016). Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 18, 64–75. doi: 10.1039/C6CP01555G
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wasternack, C., and Song, S. (2016). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Wasternack, C., and Strnad, M. (2015). Jasmonate signaling in plant stress responses and development – active and inactive compounds. New Biotechnol. 33, 604–613. doi: 10.1016/j.nbt.2015.11.001
Xie, D.-X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: An arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Keywords: jasmonate receptor, Inositol phosphate (InsP), binding mechanism, molecular docking, molecular dynamics simulation
Citation: Cui M, Du J and Yao X (2018) The Binding Mechanism Between Inositol Phosphate (InsP) and the Jasmonate Receptor Complex: A Computational Study. Front. Plant Sci. 9:963. doi: 10.3389/fpls.2018.00963
Received: 26 January 2018; Accepted: 14 June 2018;
Published: 18 July 2018.
Edited by:
Juan Caballero, Universidad Autónoma de Querétaro, MexicoReviewed by:
Simone Brogi, University of Siena, ItalyCopyright © 2018 Cui, Du and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Du, ZHVqdWFubnhAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.