
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 July 2018
Sec. Plant Systems and Synthetic Biology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00954
This article is part of the Research Topic Systems Biology and Synthetic Biology in Relation to Drought Tolerance or Avoidance in Plants View all 9 articles
A correction has been applied to this article in:
Corrigendum: Populus trichocarpa PtNF-YA9, a Multifunctional Transcription Factor, Regulates Seed Germination, Abiotic Stress, Plant Growth and Development in Arabidopsis
 Conglong Lian1,2,3,4
Conglong Lian1,2,3,4 Qing Li1,2,3,4
Qing Li1,2,3,4 Kun Yao1,2,3,4
Kun Yao1,2,3,4 Ying Zhang1,2,3,4
Ying Zhang1,2,3,4 Sen Meng1,2,3,4
Sen Meng1,2,3,4 Weilun Yin1,2,3,4*
Weilun Yin1,2,3,4* Xinli Xia1,2,3,4*
Xinli Xia1,2,3,4*NF-YAs play important roles in abiotic stress. However, their characteristics and functions in abiotic stress of poplar, a model woody plant, have not been fully investigated. Here, the biological functions of PtNF-YA9 (Potri.011G101000), an NF-YA gene from Populus trichocarpa, were first fully investigated. PtNF-YA9 is located in the nucleus. The expression of PtNF-YA9 was reduced by mannitol, NaCl, and abscisic acid (ABA). The GUS staining of ProNF-YA9::GUS transgenic lines was also reduced by mannitol treatments. In the PtNF-YA9-overexpressed Arabidopsis (OxPtNA9), OxPtNA9 lines exhibited sensitivity to simulated drought, ABA, and salinity stress during germination stage, and growth arrest emerged at post-germination stage. These phenomena might involve the ABA signaling pathway via the regulation of ABI3, ABI4, and ABI5. At vegetative stages, OxPtNA9 lines decreased in water loss via promoting stomatal closure and displayed high instantaneous water-use efficiency (WUE) of the leaf to exhibit enhanced drought tolerance. Furthermore, OxPtNA9 lines exhibited long primary root in the half-strength Murashige–Skoog agar medium supplemented with NaCl and conferred strong tolerance in the soil under salt stress. Additionally, PtNF-YA9 exhibited dwarf phenotype, short hypocotyl, small leaf area and biomass, delayed flowering, and increased chlorophyll content. Above all, our research proposes a model in which PtNF-YA9 not only plays a key role in reducing plant growth but also can play a primary role in the mechanism of an acclimatization strategy in response to adverse environmental conditions.
Plants are usually subjected to various abiotic challenges from the environment, especially in extreme temperature, high salinity, and long-term drought. Drought is a major disadvantageous environmental factor limiting plant development and growth; it is widespread in many regions and is expected to progressively increase (Burke et al., 2006). With the development of biotechnology, the identification and application of genetic transformation technology to enhance stress tolerance of plants are essential for screening and breeding new resistant plants (Valliyodan and Nguyen, 2006). In recent decades, the complicated signaling network response to different abiotic stresses has been relatively and thoroughly defined and characterized. In terms of gene regulation, transcription factor families (Zhu, 2002), including NAC (Lu et al., 2017), MYB (Wei et al., 2017), AP2/ERF (Ahn et al., 2017), bHLH (Dong et al., 2014), and WRKY (Jiang et al., 2014), all play significant roles in regulating the expression level of functional genes to enhance drought tolerance. Thus, many studies have investigated the identification and application of transcription factors in plant genetic engineering to increase plant resistance.
NUCLEAR FACTOR Y (NF-Y) transcription factor, also called the Heme Activator Protein (HAP) or CCAAT-binding factor (CBF), is pervasive in high eukaryotes. The NF-Y member is deemed as a heterotrimeric transcription factor and includes three subunits, namely, NF-YA (HAP2 or CBF-B), NF-YB (HAP3 or CBF-A), and NF-YC (HAP5 or CBF-C) (Forsburg and Guarente, 1989). The heterodimers of NF-YB/NF-YC were supposed to translocate into the nucleus from the cytoplasm and then interact with NF-YA in the nucleus to form heterotrimers (Frontini et al., 2004; Steidl et al., 2004). NF-Ys participate in a series of biological processes, such as flower development (Cao et al., 2014), primary and secondary metabolism (Li et al., 2015), embryo and seed development (Lee et al., 2003), root development (Sorin et al., 2014), and nutrition balance (Pant et al., 2009), and they are also involved in multiple abiotic stresses (Leyva-González et al., 2012; Han et al., 2013). Notably, NF-YA transcription factors, have been shown to be crucial for responses to plant abiotic stress. For instance, AtNFYA5 confers drought resistance via transcriptional and posttranscriptional regulation (Li et al., 2008). AtNF-YA2, AtNF-YA7, and AtNF-YA10 overexpressing lines all result in dwarf phenotypes and confer several types of abiotic stress tolerance (Leyva-González et al., 2012). Furthermore, overexpression of OsHAP2E gene increases multifunction resistance, such as pathogen tolerance and resistance to high drought and salinity in rice (Alam et al., 2015). Moreover, some NF-YA genes negatively regulate several types of stress tolerance. For instance, wheat TaNF-YA10 increases sensitivity to salinity in Arabidopsis thaliana (Ma et al., 2015). In addition, NF-YAs also play a distinct role under different stresses. For instance, overexpression of GmNFYA3 confers enhanced drought resistance but exhibits sensitivity to high salt stress (Ni et al., 2013). Thus, it is extremely rewarding to screen and study the functions of members in the NF-Y transcription factor family.
Populus trichocarpa has a modest genome size and acts as a model for the study of woody plants (Tuskan et al., 2006). Only a few NF-Y genes from woody plants have been functionally characterized, especially in poplar. PtrHAP2 is involved in vegetative bud dormancy in Populus (Potkar et al., 2013), whereas PagHAP2-6 is involved in poplar cambium dormancy and the regulation of ABA (Ding et al., 2016). Our previous studies on NF-YB7 showed that PdNF-YB7 overexpression increases Arabidopsis water-use efficiency (WUE) and drought resistance (Han et al., 2013). Meanwhile, the function of poplar NF-YA in abiotic stresses has not yet been studied. The expression of PtNF-YA9 is reduced under drought condition based on RNAseq data from Popgenie1, but its Arabidopsis homolog AtNF-YA7 confers different abiotic stress tolerance (Leyva-González et al., 2012). These findings pose an interesting question on how this gene acts under drought condition in Populus. To answer this question, quantitative reverse transcription polymerase chain reaction (RT-qPCR) was used, and overexpression of PtNF-YA9 transgenic Arabidopsis lines was generated for function analysis. Here, multifunctional phenotypes of PtNF-YA9 were observed, and the drought tolerance and possible function mechanisms of PtNF-YA9 were further investigated.
In this study, wild-type (WT) Col-0 of A. thaliana and mutant line nfya7 (SALK_121158.47.00.x) on the Col-0 background were used. Seeds of different Arabidopsis lines were sterilized with ethanol (75%) for 1 min followed by NaClO (1%) for 10 min. The seeds were subsequently washed with distilled water for four times before sowing. After sowing, Arabidopsis seeds were kept at 4°C for 48 h for vernalization. The seeds, except those used in germination tests, were grown on half-strength Murashige–Skoog (1/2 MS) medium containing 2% sucrose (0.6% agar, pH 5.8) in a plant growth chamber (22°C) under 16 h light photoperiods of white light (120 μmol m-2 s-1). After germination, 12-day-old Arabidopsis seedlings were transplanted to a pot with a mixture of turfy soil, perlite, and vermiculite (2:1:1) and grown in an illumination incubator with relative humidity of 70% under a 16 h/8 h light/dark photoperiod (120 μmol m-2 s-1) at 23°C.
Populus trichocarpa plantlets were preserved in the Beijing Forestry University by tissue culture. P. trichocarpa mature plants were obtained through 3 months of natural growth after transplanting from in vitro plantlet. The plantlets were transplanted in 4 L pots with a mixture of loam. Hoagland nutrient solutions were used to water every 2 weeks in the greenhouse for 3 months until the experiments. For dehydration stress treatment, uniformly grown P. trichocarpa plants were washed from pots, and natural dehydration was perfomed at similar conditions of room temperature. Moreover, between six and eight nodes of leaves were harvested at four time points of 0, 0.5, 1, and 4 h. For salt stress treatment, uniformly developed P. trichocarpa plants were fully watered using 200 mM NaCl solution. For ABA treatment, 300 μmol ABA was evenly sprayed on the leaves of poplar. The leaves between six and eight nodes of NaCl and ABA treatments were harvested at four time points at 0, 1, 4, and 8 h. All harvested samples were frozen in liquid nitrogen immediately and then preserved at -80°C for later use.
To obtain the cDNA sequence of PtNF-YA9, total RNA was extracted from P. trichocarpa by using the EASYspin Plus Plant RNA Kit (AidLab, Beijing, China) following the manufacturer’s instructions. Then, cDNA was synthesized using the TIANGEN FastQuant RT Kit (Qiagen, Hilden, Germany). The specific primers NA9-F and NA9-R (Supplementary Table S1) were designed according to the full-length cDNA reference sequence obtained from PopGenIE, and the open reading frame (ORF) sequence of PtNF-YA9 (Potri.011G101000) was amplified from the cDNA of P. trichocarpa via PCR and then cloned into the pMD18-T vector. The new vector was named as PtNA9-T. The functional region of PtNF-YA9 was analyzed by InterPro2. Physical and chemical parameters were analyzed by ExPASy3. Multiple sequence alignment of amino acid sequences was analyzed using ClustalW. Phylogenetic trees between P. trichocarpa NF-YA homology proteins were constructed using MEGA software based on neighbor-joining method with 1000 bootstrap replications.
The full-length coding sequence (CDS) of PtNF-YA9 without the stop codon was amplified from PtNA9-T by PCR using gene-specific primers NA9-GFP-Fz and NA9-GFP-Rz (Supplementary Table S1). For the expression of the 35S::PtNF-YA9-GFP fusion protein, the PCR product was cloned between the Cauliflower mosaic virus (CaMV) 35S promoter and GFP gene in the pCambia1304 vector to form NF-YA9 and GFP fusion protein. The construct fusion vector was confirmed by sequencing and then transformed into Agrobacterium strain LBA4404 and subsequently infiltrated into the Nicotiana benthamiana leaves for transient expression via Agrobacterium-mediated gene transformation (Li, 2011). The 3 days N. benthamiana-transformed leaves after infiltration were observed under a confocal microscope (Nikon).
To examine the expression levels of PtNF-YA9 under different stresses and tissues, real-time qPCR was performed using primers NA7-qF and NA7-qR (Supplementary Table S1) by SuperReal PreMix Plus (SYBR Green) (TIANGEN, Beijing, China). The procedure followed the manufacturer’s instructions, the relative quantification value was calculated using the 2-ΔΔCt method, and the kinetics of PCR product was monitored using SYBR Green (Steibel et al., 2005). The transcript levels of GAPDH (AT1G16300) or Actin2 (AT3G18780) were used to quantify the expression of detected genes in samples. RT-PCR was also carried out to investigate the expression of PtNF-YA9 in transgenic Arabidopsis lines. PCR amplification (94°C for 30 s, 56 °C for 30 s, and 72°C for 1 min) was performed for 40 cycles, and each PCR assay was replicated for three biological replicates. All the primers used for RT-PCR and RT-qPCR are shown in Supplementary Table S1.
To isolate the promoter sequence of PtNF-YA9, DNA was extracted from P. trichocarpa by CTAB method, and the reference genomic DNA sequence of PtNF-YA9 was searched in Phytozome. Based on the sequence identified from Populus trichocarpa v3.0 database, gene-specific primers NA9Pro-F and NA9Pro-R (Supplementary Table S1) were designed, and the sequence was amplified by a PCR cover at approximately 2000 bp upstream of the start codon. PlantCARE online database was used to predict cis-acting regulatory elements (Lescot et al., 2002). The PtNF-YA9 promoter adapter sequence was obtained by gene-specific primers NA9Pro-Fz and NA9Pro-Rz (Supplementary Table S1) and was then cloned into pCambia1301 vector instead of CaMV 35S promoter with the Seamless Assembly Cloning Kit (CloneSmarter). The constructed vector was sequenced and then transferred into Agrobacterium strain GV3101. The transgenic Arabidopsis plants were obtained by the floral dip method (Clough and Bent, 1998). To detect the promoter of GUS staining, 16-day-old transgenic Arabidopsis seedlings grown on 1/2 MS agar medium plates were transplanted into 1/2 MS agar medium plates with or without 200 and 250 mM mannitol for 24 h.
The ORF sequence of PtNF-YA9 was obtained from PtNA9-T vector using the specific primers NA7-Fz and NA7-Rz by PCR (Supplementary Table S1). The PCR product was cloned into the pCambia1301 vector containing the CaMV 35S promoter instead of GUS gene with the Seamless Assembly Cloning Kit (CloneSmarter). The constructed vector was sequenced and then transferred into Agrobacterium strain GV3101. The Arabidopsis plants were also transformed by the floral dip method (Clough and Bent, 1998).
For the germination assay, three replicates of 50 seeds from different lines of Arabidopsis were surface-sterilized. The surface-sterilized seeds were sown on 1/2 MS agar medium supplemented with mannitol, NaCl, or ABA. The germinated seeds were counted based on the radicles protruding from the seed coat and statistically analyzed every day until 11 days. Moreover, cotyledon greening rate was analyzed when the germinated seeds established seedlings and cotyledon turned green.
For the experiment of stress treatment at seedling stage, 4-day-old seedlings on 1/2 MS agar medium were transplanted to 1/2 MS agar medium plates supplemented with 200 mM mannitol and 100 mM NaCl. The growth phenotypes were compared, and the length of the primary roots was calculated. For long-term salt stress, 2-week-old nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 lines were subjected to salt stress by pouring saltwater every 5 days until significant difference phenotypes were achieved, and the different phenotypes were photographed and recorded. For long-term drought treatment, the seedlings were watered for 2 weeks after transplanting into the pot. Then, the water was withheld until the difference phenotypes were achieved. Moreover, the differences in plant phenotypes were analyzed and photographed. Subsequently, the pots were re-watered and recovered for 3 days. With water deficit for 7 days, the photosynthetic indexes, which contain net CO2 assimilation, transpiration, and stomatal conductance, were measured by using the LI-6400 photosynthesis system (LI-COR 6400, Lincoln, NE, United States). Instantaneous leaf WUE was calculated as the ratio of net CO2 assimilation/transpiration. The leaf relative water content (RWC) was calculated as (FW - DW)/(TW - DW) × 100, which was described by Sade et al. (2014). The leaf chlorophyll content was extracted by 80% acetone, and the absorbance of extracting solution was measured at 663 and 645 nm. The chlorophyll a (Ca) content was calculated by Ca = 12.7 × A663 - 2.69 × A645, the chlorophyll b (Cb) content was calculated by Cb = 22.9 × A645 - 4.64 × A663, and the total chlorophyll content was defined as the sum of Ca + Cb.
Detached rosette leaves of different lines grown for 3 weeks after transplanting into soil were weighed immediately and incubated on a white paper at room temperature. Losses of fresh weight in leaves were monitored at different time points of 0, 0.5, 1, 1.5, 2, 3, 5, and 8 h. Water loss is expressed as the percentage of initial fresh weight. To analyze the stomatal apertures, the leaves were incubated in a solution containing 50 mM KCl, 50 mM CaCl2, and 10 mM MES/KOH (pH 6.1) for 2 h under light condition. Final concentration of 30 μM ABA was then added. Stomatal apertures were observed with a microscope and measured using Photoshop software after 1 h of ABA treatment. For each experimental repeat, at least 50 stomata were calculated and measured. The values of stomatal width to length ratios acted as the indicator of stomatal phase and were divided into three phases. The stomata was defined as open when the ratio was greater than 0.5 μm, partially closed between 0.5 and 0.2 μm, and closed with less than 0.2 μm.
All data were subjected to SPSS Statistics and Excel for analysis. Student’s t-test was used to detect the significant differences between individual means. Differences at the 1% level were considered significant and denoted by lowercase letters or asterisk among different groups.
A significant portion of the members were found to be involved in various abiotic stresses based on the systematic analysis of the NF-Y family in P. trichocarpa. Thirteen members of the NF-YA subfamily were identified in P. trichocarpa from Popgenie4 and PlantTFDB (Jin et al., 2017). Moreover, 11 members of HAP2/NF-YA genes have been already named in P. trichocarpa (Potkar et al., 2013). We added the remaining two members and used the same gene names in the study to avoid nomenclature confusion. One of them, designated as PtNF-YA9, the closest of Arabidopsis homolog AtNF-YA7 with acclimatization strategy for abiotic stress tolerance, was selected for further functional characterization in this study.
The cDNA sequence of PtNF-YA9 (Potri.011G101000) was identified from the genome of P. trichocarpa. A 648-bp sequence containing a 627-bp CDS in length was obtained by specific primers of NA9-F and NA9-R. PtNF-YA9 was predicted to encode 208 amino acids with a pI of 9.17, molecular mass of 22.76 kDa, and an instability index of 66.41 and is deemed as an unstable protein in nature. Moreover, the atomic composition forms a formula of C986H1541N301O308S7. The genomic sequence of PtNF-YA9 was 4801 bp, including four introns and five exons (Figure 1A).

FIGURE 1. Gene structure, phylogenetic tree, and multiple sequence alignment of the PtNF-YA9 homology proteins across different plant species. (A) Gene structure diagram of PtNF-YA9. (B) Phylogenetic analysis of NF-YA proteins across different plant species. The amino acid sequences were aligned using MAFFT, and the phylogenetic tree was constructed using MEGA 6.0 software by neighbor-joining method with bootstrap analysis of 1000 replicates. The “#” label indicates target protein. (C) Multiple sequence alignment of PtNF-YA4, PtNF-YA9, PtNF-YA12, AtNF-YA4, and AtNF-YA7 proteins.
Phylogenetic tree of the PtNF-YA9 homology proteins across different plant species showed that these proteins were divided into two branches. The PtNF-YA9 of P. trichocarpa showed recent homology to P. euphratica XP_011012079 and Eucalyptus grandis XP_010036606 (Figure 1B). Multiple protein sequence alignment (Figure 1C) revealed that PtNF-YA9 and PtNF-YA12 proteins have the same amino acid sequence in the same chromosome, which is a phenomenon of gene duplication. Furthermore, they share the highest amino acid sequence homology to PtNF-YA4 and heterologous to AtNF-YA7 and AtNF-YA4 of Arabidopsis (Figure 1C). Phylogenetic tree also showed that PtNF-YA1, PtNF-YA3, and PtNF-YA10 are in the same small branch with PtNF-YA9 (Supplementary Figure S1). These proteins clustered in the same branch, indicating functional similarities and functional redundancy. These results generally indicated that PtNF-YA9 belongs to the NF-YA transcription factor family, which has a highly conserved CBFB_NFYA domain.
Most transcription factors are localized in the nucleus. To determine the subcellular localization of PtNF-YA9 protein, the 35S:PtNFYA9-GFP and 35S:GFP vectors were constructed and introduced into epidermal cells of tobacco by using injection method. The fluorescence signals in the inner epidermal cells were observed using confocal laser scanning microscopy. PtNFYA9-GFP fusion protein was visualized in the nucleus, whereas the 35S:GFP (control) was observed throughout the cells (Figure 2). The results indicated that PtNF-YA9 is located in the nucleus.
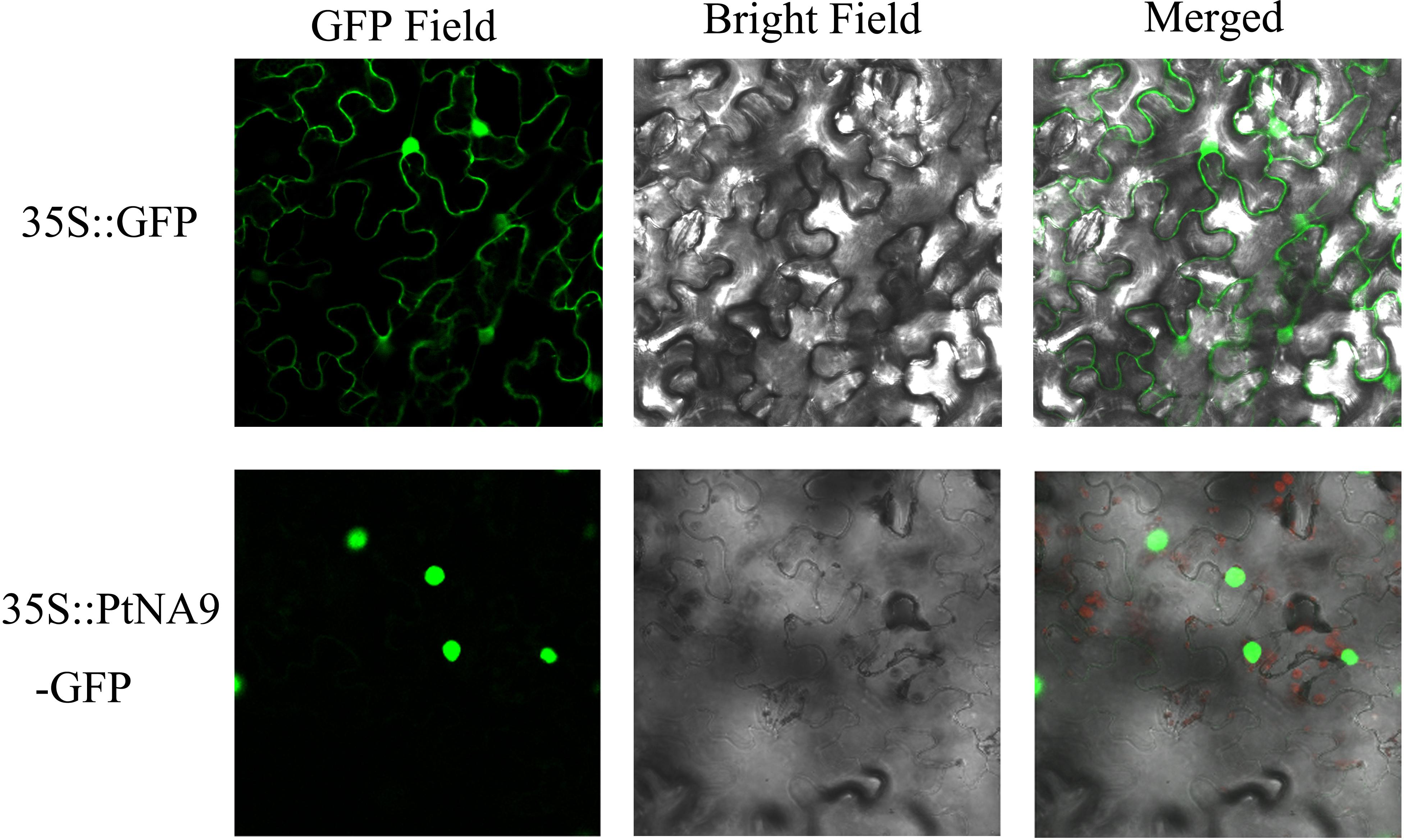
FIGURE 2. Subcellular localization of PtNF-YA9 protein. The 35S::GFP and 35S::PtNFYA9-GFP fusion protein transiently expressed in tobacco. Microscopic images contain green fluorescence, bright field, and merged microscope images.
PtNF-YA9 showed stress-response expression in the Popgenie database. The database showed that PtNF-YA9 transcript was inhibited by drought stress and highly expressed in mature leaves than in the roots (Figure 3H). To further confirm the potential functions of PtNF-YA9 in response to different abiotic stresses, the transcript abundance of PtNF-YA9 in multiple organs and under a variety of abiotic stress treatments was detected by RT-qPCR. The results indicated that PtNF-YA9 was actually inhibited by drought (Figure 3A). PtNF-YA9 was also significantly downregulated by NaCl (Figure 3C) and ABA (Figure 3B), especially NaCl for 1 h and ABA for 8 h. These results indicated that PtNF-YA9 might participate in the response of ABA, salt, and osmotic stresses.
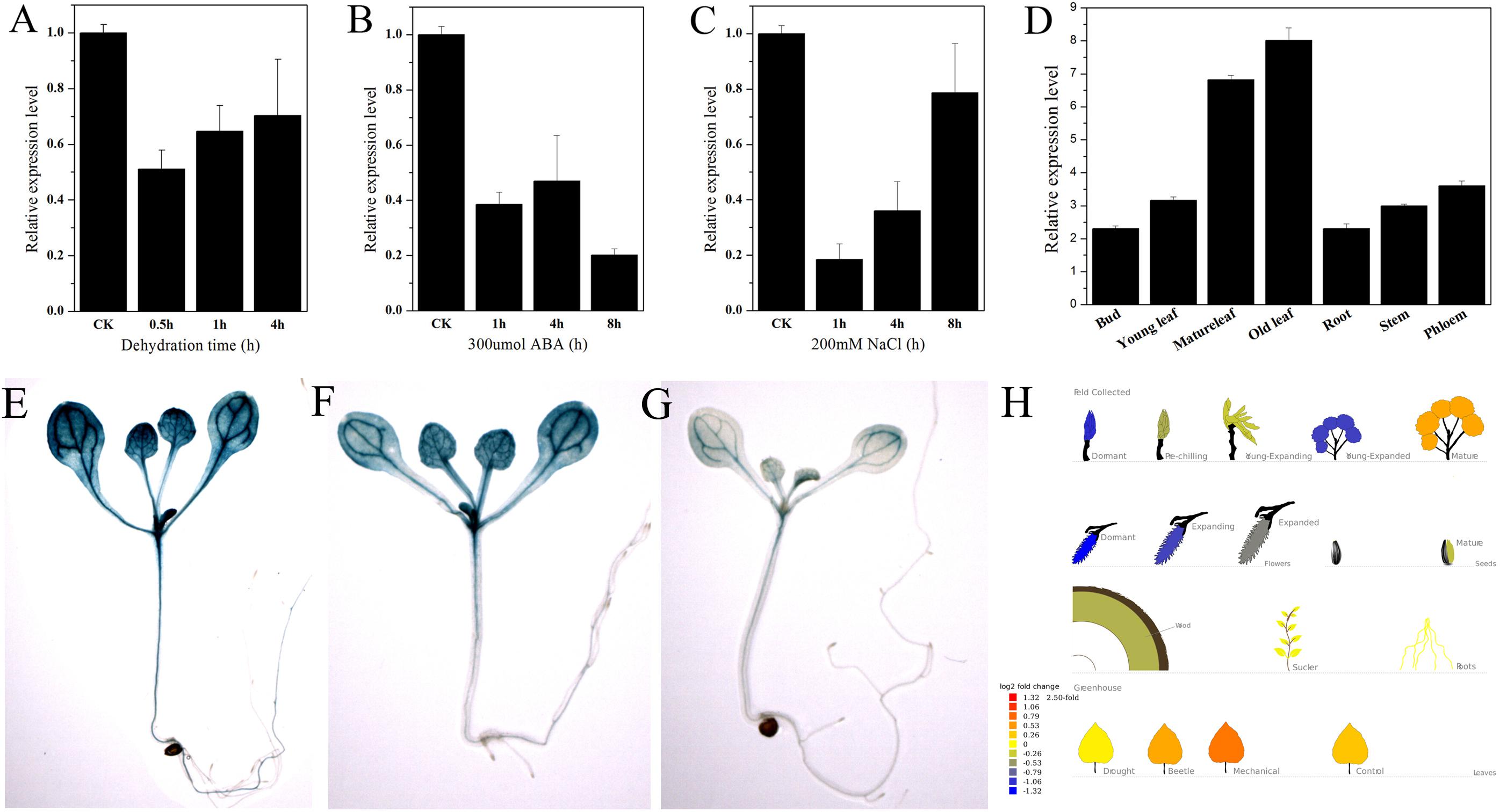
FIGURE 3. Expression profile of P. trichocarpa NF-YA9. (A) Expression pattern of PtNF-YA9 in P. trichocarpa leaves under dehydration stress. (B) Expression pattern of PtNF-YA9 in P. trichocarpa leaves under 300 μM ABA. (C) Expression pattern of PtNF-YA9 in P. trichocarpa leaves under 200 mM NaCl. (D) Tissue expression pattern of PtNF-YA9 in poplar. The expression levels were normalized to that of UBQ, and the level of PtNF-YA9 transcript in the control was set at 1. Three biological repeats were performed, and each RT-qPCR was also performed thrice. (E) Seventeen-day-old ProNF-YA9::GUS transgenic Arabidopsis seedlings grown on 1/2 MS agar medium plates without treatment. (F) Sixteen-day-old ProNF-YA9::GUS transgenic Arabidopsis seedlings grown on 1/2 MS agar medium plates were transplanted into 1/2 MS agar medium with 200 mM mannitol for 24 h. (G) Sixteen-day-old ProNF-YA9::GUS transgenic Arabidopsis seedlings grown on 1/2 MS agar medium plates were transplanted into 1/2 MS agar medium with 250 mM mannitol for 24 h. (H) The expression patterns of PtNF-YA9 in the Popgenie database (http://popgenie.org/gene?id=Potri.011G101000).
We explored the expression pattern of PtNF-YA9 at the transcriptional level regulated by abiotic stresses. The promoter of PtNF-YA9 containing the 1985-bp genomic sequence upstream from their initiation codons was cloned, and cis-elements were analyzed (Supplementary Figure S2). There were some abiotic stress elements, such as HSE, MBS, TC-rich repeats, and WUN motif, and hormone response elements, such as CGTCA motif, TGACG motif, GA motif, and TCA-element. The prediction of possible transcription core promoter region sequence was AAGAACTTCAAA AAAATGCTGGTTTAGCCACATTTTGCTCATGCAAATGA from -808 bp to -658 bp by BDGP (Supplementary Figure S2). Furthermore, plant expression vector ProNF-YA9::GUS expressing the GUS gene under the control of the PtNF-YA9 promoter was constructed, and the ProNF-YA9::GUS transgenic Arabidopsis lines were generated. GUS staining was used to detect the transcript abundance of the GUS reporter gene. As shown in Figure 3, the GUS signal was observed in almost all of the analyzed tissues/organs of seedlings, especially in leaf vasculature and shoot apical meristem (Figure 3E). A significant decrease of GUS staining of 16-day-old seedlings was observed for ProNF-YA9::GUS lines under 200 (Figure 3F) and 250 mM mannitol (Figure 3G) of 1/2 MS agar medium for 24 h. From the above findings, the expression patterns of ProNF-YA9::GUS were consistent with the results of RT-qPCR.
To manifest the function of PtNF-YA9 in the regulation of plant responses to abiotic stresses, overexpression of PtNF-YA9 transgenic Arabidopsis (OxPtNA9) under the control of the CaMV 35S promoter was generated (Supplementary Figure S3A). Various expression levels of PtNF-YA9 in several independent lines were obtained. OxPtNA9-7 and OxPtNA9-3 showed the highest mRNA levels by RT-PCR (Supplementary Figure S3B) and RT-qPCR (Supplementary Figure S3C) and were selected for further study. On the 1/2 MS agar medium, the seeds of OxPtNA9 lines exhibited slightly delayed germination than WT, and all of them showed good germination (Figure 4D).
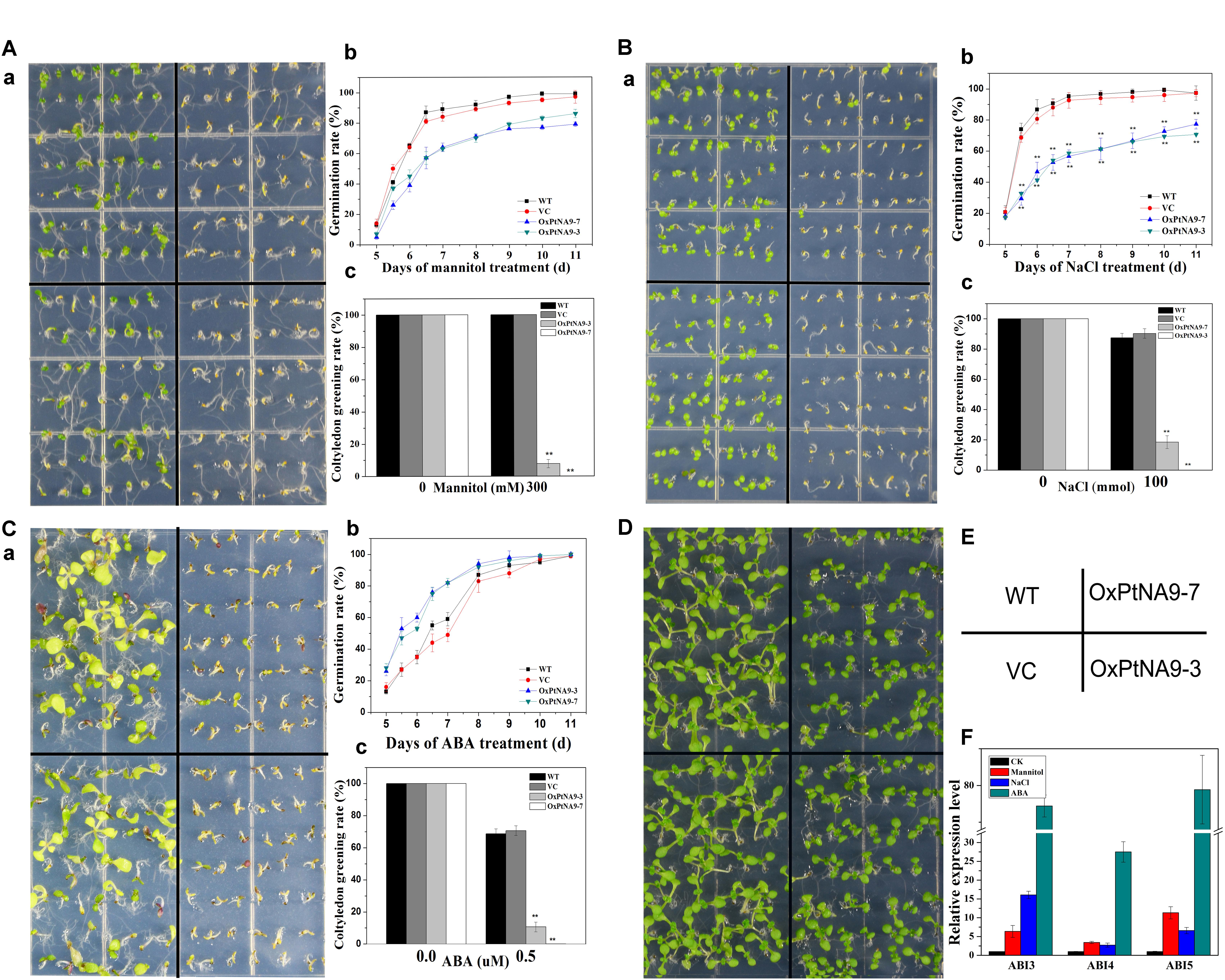
FIGURE 4. 35S::PtNF-YA9 transgenic lines are hypersensitive to mannitol, salt stress, and ABA during germination growth. The 35S::PtNF-YA9 transgenic lines are hypersensitive to 300 mM mannitol (A), 100 mM NaCl (B), and 0.5 μM ABA (C) during germination stage. (a) Phenotypes of 35S::PtNF-YA9 transgenic lines are hypersensitive to mannitol, salt, and ABA. (b) Germination rate of different seeds on the mannitol, salt, and ABA medium counted for 11 days after sowing. (c) Cotyledon greening rate of different seeds on the mannitol, salt stress, and ABA medium after post-germination. (D) Control phenotype of 35S::PtNF-YA9 transgenic lines under 1/2 MS agar medium. (E) Sketch map of diagram a. (F) Accumulation of ABI3 and ABI5 in germinated seeds under mannitol, ABA, and salt treatment. The35S::PtNF-YA9 transgenic seeds were germinated on 1/2 MS liquid medium-moistened filter paper for 24 h after stratification and then were transferred onto filtered paper moistened with water (control), 300 mM mannitol, 100 mM NaCl, and 0.5 μM ABA. After treating for 8 h, the samples were harvested, and RNA was extracted. Here, three biological repeats were performed, and each RT-qPCR was also performed thrice.
The germination of progeny homozygous seeds were analyzed under osmotic stress. Less than 87% of PtNF-YA9 transgenic seeds germinated in 300 mM mannitol-supplemented 1/2 MS agar medium after 11 days, whereas the germination of WT and VC exhibited more than 99 and 97%, respectively (Figure 4A). In addition, PtNF-YA9 transgenic line OxPtNA9-7 failed to establish seedling, and OxPtNA9-3 showed less and smaller seedlings of approximately 7.69% compared with 100% seedlings of WT and VC (Figure 4A).
The seed germination and post-germination growth under salt stress was detected. As shown in Figure 4B, the OxPtNA9 germinated slowly, and the rates of germination were significantly reduced. Similarly, after seed germination, a notable post-germination growth arrest was also observed in OxPtNA9 in the 150 mM NaCl of 1/2 MS agar medium (Figure 4B). Then, OxPtNA9-7 germinated but could not develop into seedlings, and OxPtNA9-3 exhibited 18.52% smaller seedlings compared with 100% seedlings of WT and VC (Figure 4B).
Additionally, when we sowed the same WT and PtNF-YA9 overexpressing lines on 1/2 MS agar medium supplemented with 0.5 μM ABA, the post-germination growth arrest was also exhibited, and a severe phenotype of arrest was found in the OxPtNA9 lines, establishing only 10.67% seedlings. Meanwhile, WT and VC can establish approximately 70% seedlings in the early stages (Figure 4C).
Furthermore, to determine the potential molecular mechanisms responsible for the post-germination growth arrest of PtNF-YA9, RT-qPCR was performed to detect the abundance of ABI3, ABI4, and ABI5 at germination stage under different abiotic stresses. The results showed that the expression levels of ABI3, ABI4, and ABI5 were significantly elevated under 300 mM mannitol, 100 mM NaCl, and 0.5 μM ABA at germination stage (Figure 4F). Overall, PtNF-YA9 overexpression acted as a negative regulator of seed germination and led to post-germination growth arrest under different abiotic stresses possibly through ABIs of ABA signaling pathway.
PtNF-YA9 transgenic seeds displayed a drought-sensitive phenotype (Figure 4A) and an ABA-hypersensitive phenotype (Figure 4C). OxPtNA9 was deemed as the resistance phenotype because of the small and green rosette leaves (Figure 5) (Achard et al., 2008). Hence, we investigated whether PtNF-YA9 transgenic plants displayed an altered phenotype in response to drought stress (Figure 5). At seedling stages, 7-day-old seedlings of nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 lines were transferred to 1/2 MS agar medium with or without 200 mM mannitol. Without 200 mM mannitol, the primary roots were shorter but showed no significant differences, whereas the lateral roots were significantly increased in the OxPtNA9 lines (Supplementary Figure S4). With 200 mM mannitol, the results indicated no significant differences in the length of the primary root, but OxPtNA9 lines showed more lateral roots than nfya7 and WT (Supplementary Figure S5).
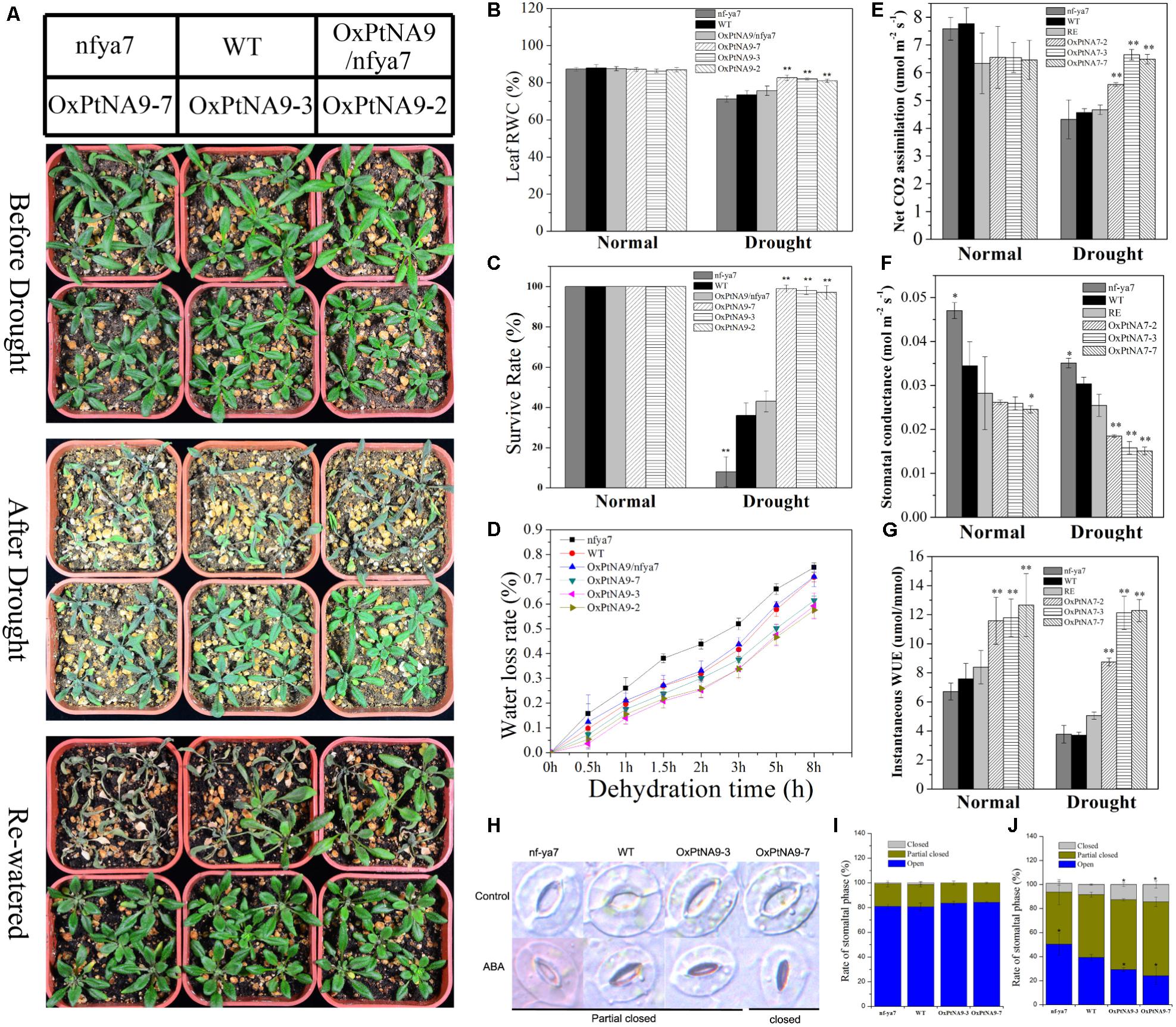
FIGURE 5. PtNF-YA9 overexpression confers drought tolerance in Arabidopsis. (A) Morphological differences in drought experiments. (B) RWC of the leaves under normal and drought conditions. (C) Survival rate of seedlings under normal and drought conditions. (D) Water loss from detached leaves; water loss is expressed as the percentage of initial fresh weight of detached leaves. (E) Net photosynthetic rate. (F) Stomatal conductance. (G) Instantaneous WUE of different lines under normal and drought conditions. Under normal conditions, the data were collected on day 21 after being transferred to soil. Under drought conditions, the data were collected after 7 days of water withholding condition. All data are means from the six leaves for each of three independent experiments. (H) Typical phenotype of stomatal opening phase in transgenic lines, nfya7, and WT with or without ABA (30 μM) treatment for 2 h. (I) Percentages of the three types of stomata in transgenic lines, nfya7 mutant, and WT plants are calculated in stomatal-induced liquid without ABA (30 μM) treatment for 1 h. (J) Percentages of the three types of stomata in transgenic lines, nfya7 mutant, and WT plants are calculated with ABA (30 μM) treatment for 2 h.
To examine the effects of nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 lines on long-term drought condition, we subjected 2-week-old nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 plants to water withholding conditions until significant phenotype appeared, and they were then re-watered. Under normal growth condition, smaller rosette phenotypes of OxPtNA9 transgenic lines were observed compared with WT plants (Figure 5A). However, with intensifying and lasting drought stress, nfya7 mutant, WT, and OxPtNA9/nfya7 plants displayed a more withered phenotype than PtNF-YA9 transgenic plants (Figure 5A). Photosynthesis analysis showed that nfya7 mutant and WT were significantly reduced and maintained a significantly lower photosynthetic rate than OxPtNA9 transgenic lines under drought treatment (Figure 5E). Under normal growth conditions, the stomatal conductance of nfya7 mutant was the highest, followed by the WT and then OxPtNA9 transgenic lines. Meanwhile, under drought condition, the stomatal conductance of OxPtNA9 lines was significantly lower than that of WT and nfya7 mutant (Figure 5F). The transpiration rates of OxPtNA9 lines were also significantly lower than those of WT and nfya7 mutant under both well-watered and drought conditions. These photosynthesis results showed a significant increase in the instantaneous leaf WUE of OxPtNA9 lines (Figure 5G). Furthermore, the OxPtNA9 lines had higher leaf RWC compared with WT and nfya7 mutant under drought condition (Figure 5B). Thus, PtNF-YA9 overexpression was demonstrated to improve water deficiency tolerance in Arabidopsis at the vegetative growth stage. In addition, OxPtNA9 lines recovered more quickly than WT plants after re-watering for 3 days (Figure 5A). The survival rate of OxPtNA9 lines was 97–100%. Meanwhile, the survival rates of WT and OxPtNA9/nfya7 were 36% ± 6.3% and 43% ± 5.2%, respectively. The nfya7 mutant plants almost could not recover (Figure 5C).
To explore the physiological mechanism of the drought-resistance phenotype displayed by PtNF-YA9, a water loss assay with the fresh weight of detached rosette leaves by times as an indirect indication of the transpiration rate was performed. We discovered that the rate of water loss was lower in PtNF-YA9 overexpressing lines than in WT. After dehydration treatment for 8 h, the fresh weight of WT decreased by 70.83% ± 1.85%, but that of the three OxPtNA9 lines only lost 59.28 ± 5.22%, 57.54 ± 3.15%, and 61.40 ± 1.81% (Figure 5D). This result indicated that the drought-tolerant phenotype of OxPtNA9 lines was attributed to enhance the water retention capacity.
In plants, water loss is regulated by guard cells, which cause the stomata to open and close. ABA as an exogenous hormone can induce stomatal closure. Studies have showed that PtNF-YA9 is involved in ABA signaling. To further demonstrate whether PtNF-YA9 participates in the regulation of stomatal aperture, the phenotypes of the stomata of nfya7, WT, and OxPtNA9 lines were compared. According to the stomatal aperture, we divided the stomatal types into three categories as open, partially closed, and closed (Figure 5H). After inducing full stomatal opening, the stomas in plants were primarily completely open and partially closed, and the rates of these three stomatal phases were similar among WT, nfya7, and OxPtNA9 lines (Figure 5I). However, after 2 h of ABA treatment, the rates of closed and partially closed stomata increased remarkably, especially in the OxPtNA9 lines. By contrast, the nfya7 mutant and WT plants still maintained several opened stomata (Figure 5J). Overall, these results indicated that PtNF-YA9 overexpression in Arabidopsis confers drought resistance via an increased ABA sensitive to induced stomatal closure.
To detect the enhanced drought resistance by altered gene expression level of PtNF-YA9, the transcript abundance of some stress-related genes, including ABA-activated signaling pathway genes ABF1 and ABI5 (Figures 6A,B), dehydration-responsive element binding protein genes DREB2A and DREB2B (Figures 6C,D), and desiccation-responsive genes RD29A and RD29B (Figures 6E,F), in the leaves of nfya7 mutant, WT, OxPtNA9-7, and OxPtNA9-3 under well-watered and drought conditions were analyzed by RT-qPCR. The results showed that the stress-related genes were differentially expressed in the OxPtNA9 lines compared with the WT and nfya7 mutant lines. Under well-watered condition, the expression levels of DREB2A and RD29A in OxPtNA9 lines were similar to WT, and the expression levels of ABF1, ABI5, DREB2B, and RD29B were highly expressed. Under water deficit condition for 7 days, these stress-responsive genes were strongly induced. Moreover, the expression levels of these genes were induced higher in OxPtNA9 lines than in WT and nfya7 plants.
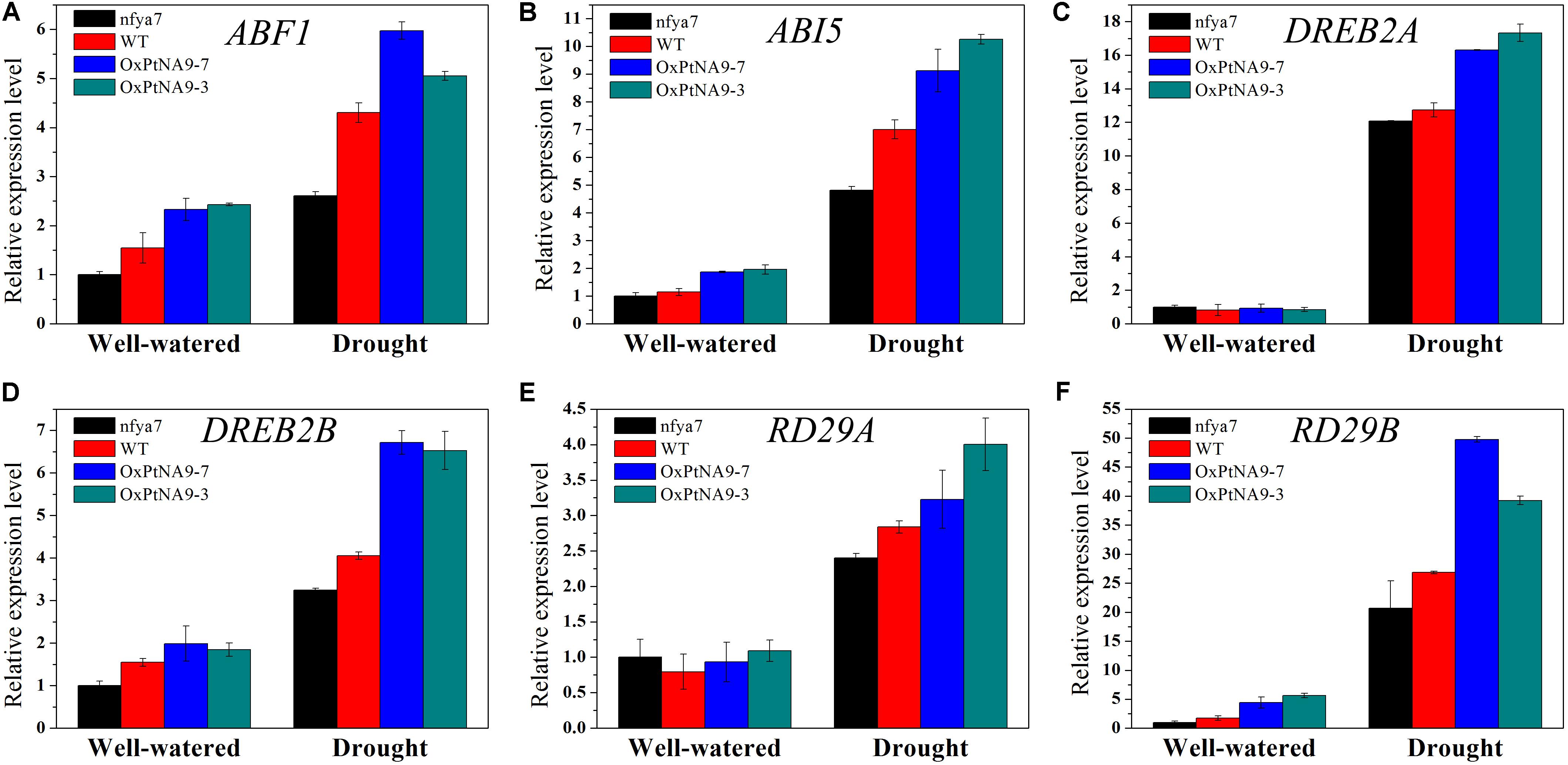
FIGURE 6. Transcript level analysis of drought-related genes in the leaves of nfya7 mutant, WT, and three different OxPtNA9 lines under well-watered and drought conditions by RT-qPCR. Under well-watered conditions, the seedlings and leaves were sampled on day 21 after being transferred to soil. Under drought conditions, the seedlings and leaves were sampled after 7 days of water withholding condition. The drought-related genes including ABF1 (A), ABI5 (B), DREB2A (C), DREB2B (D), RD29A (E), and RD29B (F).
To detect whether PtNF-YA9 is also resistant to salt stress, 7-day-old seedlings of nfya7 mutant, WT, OxPtNA9/nfya7, and OxPtNA9 lines were transferred to 100 mM NaCl of 1/2 MS agar medium. The results indicated a significant difference in the length of the primary root, and OxPtNA9 lines showed much longer primary root than nfya7, WT, and OxPtNA9/nfya7 plants (Supplementary Figures S6A,B). Furthermore, to examine the different effects of nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 lines on long-term salt treatment, we subjected 2-week-old nfya7, WT, OxPtNA9/nfya7, and OxPtNA9 plants to salt stress by pouring saltwater every 5 days until significant difference phenotype was achieved. With the salt stress lasting, nfya7 mutant, WT, and OxPtNA9/nfya7 plants displayed much more serious phenotypes than OxPtNA9 lines. As plants withered, the leaves turned yellow and a portion of them became white. The nfya7 mutant, WT, and OxPtNA9/nfya7 plants stopped bolting, and almost all lodged, whereas OxPtNA9 lines showed slow growth with erect bolting (Supplementary Figure S6C). Taking together, salt treatment studies showed that PtNF-YA9 confers high-salt tolerance after seedling-established stages.
PtNF-YA9 altered the lateral root growth (Supplementary Figure S4) and exhibited a dwarf phenotype (Figures 4, 5) both at the seedling from post-germination growth and adult stages (Figures 8). A very obvious phenotype is the shortened hypocotyl in the seedling stage of PtNF-YA9 overexpressing lines (Figure 7A). The nfya7 mutant lines had the longest length of hypocotyl (4.37 ± 0.17 mm), WT showed the middle phenotype (3.65 ± 0.25 mm), whereas OxPtNA9 lines had the significantly shortest length of hypocotyl (1.87–2.60 mm) (Figure 7B).
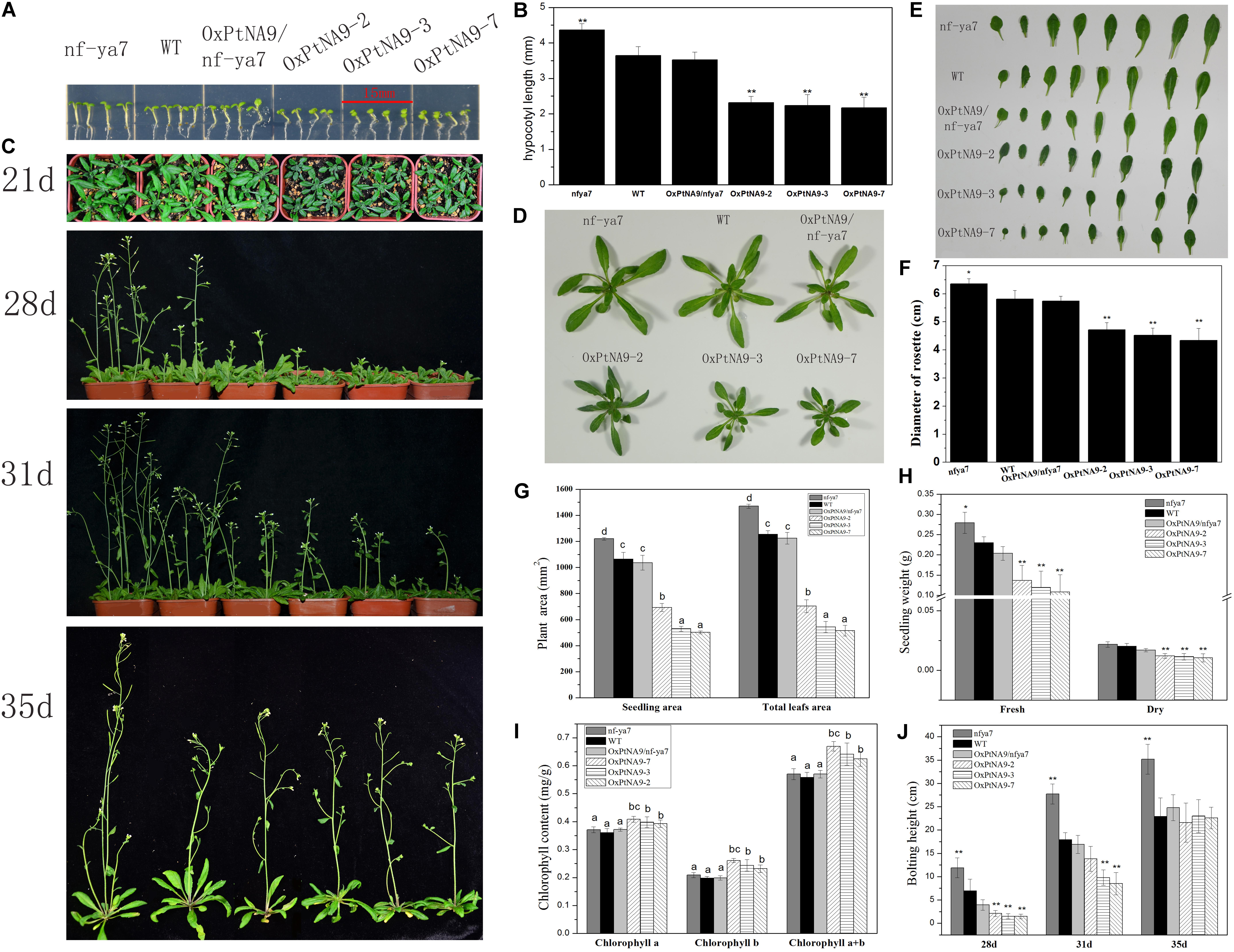
FIGURE 7. Growth difference of OxPtNA9 lines at different stages. (A) Morphology difference of hypocotyl of 10-day-old seedlings. (B) Hypocotyl length statistics. (C) Phenotypes of differences in Arabidopsis growth process. (D) Morphology of rosettes of 3-week-old seedlings. (E) Morphology of detached leaves of 3-week-old seedlings. (F) Diameter of rosettes of 3-week-old seedlings. (G) Plant area of 3-week-old seedlings. (H) Fresh and dry weights of 3-week-old seedlings. (I) Chlorophyll a and b and total chlorophyll contents of 3-week-old seedlings. (J) Height of inflorescence at different days.
Another dwarf phenotype of OxPtNA9 lines was reflected in the diameter and the area of OxPtNA9 rosettes, which were markedly reduced (Figures 7C–G). Three lines of OxPtNA9 seedling area were 693.08 ± 30.97, 529.65 ± 20.02, and 502.71 ± 11.04 mm2, showing almost half the area of nfya7 mutant seedlings, 1220.42 ± 10.22 mm2. WT and OxPtNA9/nfya7 complementary lines showed the middle phenotypes of approximately 1063.05 ± 52.42 and 1036.44 ± 57.31 mm2, respectively. Calculating the sum of all detached leaf areas also showed similar results (Figure 7G). The dwarf phenotype also resulted in biomass reduction. Both the fresh and dry weights of nfya7 mutant lines were heavier than WT, whereas the weight of OxPtNA9 lines was significantly low (Figure 7H).
In addition, the leaves of OxPtNA9 lines exhibited darker green color than WT. This phenomenon correlated with a higher chlorophyll content of OxPtNA9 lines compared with WT (Figure 7I). It indicated significantly higher chlorophyll a (Ca: 0.39–0.41 mg/g) and chlorophyll b (Cb: 0.23–0.26 mg/g) in the OxPtNA9 lines than in WT (Ca: 0.36 mg/g, Cb: 0.19 mg/g) and nfya7 mutant plants (Ca: 0.37 mg/g, Cb: 0.20 mg/g), resulting in a significantly increased total chlorophyll content of OxPtNA9 lines.
To assay the bolted and flowering time, the nfya7 mutant, WT, and OxPtNA9 lines were planted and grown under similar conditions. The result showed that the WT bolted at 24–25 days, and nfya7 mutant plants bolted early at 22–23 days. Meanwhile, the OxPtNA9 lines bolted at 26–27 days after sowing the seeds (Figure 7C). Additionally, the nfya7 mutant plants showed higher stem elongation at the early shooting stage compared with WT, and OxPtNA9 lines showed the lowest stem elongation. At the 31st day, the inflorescence length of OxPtNA9 lines varied from 6.2 cm to 16.8 cm, WT varied from 16.2 cm to 20.2 cm, and the nfya7 mutant varied from 24.3 cm to 30.2 cm. However, at the 35th day, the inflorescence length of OxPtNA9 lines was 17.0–27.0 cm and exhibited no significant difference compared with WT, 18.2–27.9 cm. Meanwhile, the nfya7 mutant lines also had high inflorescence of 30.6–39.4 cm (Figure 7J). These results indicated that overexpression of PtNF-YA9 lines exhibits slow stem elongation at the early shooting stage and delayed flowering.
Populus trichocarpa NF-YA is an unknown function member of the NF-Y family. In this study, the roles of PtNF-YA9 at different stages were reported comprehensively. At the germination stage, the PtNF-YA9 overexpressing lines are hypersensitive to mannitol, salt, and ABA treatments and exhibited growth arrest during the post-germination stage (Figure 4). At seedling stages, OxPtNA9 lines showed dwarf phenotype of short hypocotyl, small leaf, less biomass, and delayed flowering but promoted the lateral root growth and increased chlorophyll content. These different observations and multiple functions demonstrated that PtNF-YA9 participates in multiple signaling pathways at different growth stages (Figure 8).
At the seed germination and post-germination growth stages, OxPtNA9 lines showed hypersensitive phenotype to salt, ABA, and mannitol stresses. The germination rate of OxPtNA9 lines was decreased and delayed, and dramatic arrest development of the post-germination growth before the cotyledons turned green and developed into seedlings under the treatment of ABA, mannitol, and NaCl treatment (Figure 4). Lopez-Molina et al. found that post-germination growth arrest is one of the adverse strategies to protect the germinated seeds from water deficit, and the application of ABA can also result in growth arrest after germination (Lopez-Molina et al., 2001). Both mannitol and NaCl treatments could cause water deficit and induce ABA production. Thus, PtNF-YA9 played a positive role to protect the germinated seeds from water deficit under abiotic stress by arresting growth after germination. Genes of cpr5 (Gao et al., 2011), wrky2 (Jiang and Yu, 2009), and hyl1 (Lu et al., 2002) were also involved in ABA signaling pathways and participated in the regulation of post-germination growth arrest. In Arabidopsis, AtNF-YA1 also regulates post-germination growth arrest and is involved in ABA signaling pathways under salt stress (Li et al., 2013).
Abscisic acid, an important stress phytohormone, not only acted as a pivotal signaling molecule in stomatal movement and abiotic stress but was also involved in seed germination and post-germination growth. In this study, to reveal whether the post-germination growth arrest of PtNF-YA9 was also involved in ABA signaling, the ABA treatment of OxPtNA9 seeds was further studied, and the result showed that ABA acted positively in post-germination growth arrest under exogenous ABA treatment (Figure 4C). ABI3 and ABI5, as important genes in the ABA signaling pathway, play an important regulatory role in the growth arrest during seed germination and post-germination growth (Lopez-Molina et al., 2001). ABI5 played a positive role in keeping germinated seeds in a quiescent state to protect them from drought stress (Lopez-Molina et al., 2002). ABI4 directly promotes ABI5 transcription (Bossi et al., 2010), positively regulates primary seed dormancy, while negatively regulating cotyledon greening, by mediating the biogenesis of ABA and GA (Shu et al., 2013). Furthermore, to acquire further insight into the mechanism of PtNF-YA9 during germination stage, the transcript abundances of the ABI3, ABI4, and ABI5 in OxPtNA9 lines under abiotic stresses were analyzed. Interestingly, the transcript abundances of ABI3, ABI4, and ABI5 were significantly increased in OxPtNA9 lines under abiotic stresses (Figure 4E). The high expression of ABI3, ABI4, and ABI5 in OxPtNA9 lines under abiotic stress might stimulate post-germination growth arrest. In conclusion, our study revealed that during the post-germination growth arrest, PtNF-YA9 might participate in the regulation of the ABA signaling pathway via the regulation of ABIs.
Plants have complicated signaling regulatory mechanisms to acclimatize in adverse environments. Many transcription factors had been reported to bind specifically the motif in the promoters of the stress-responsive genes and are involved in drought tolerance. Some NF-YA genes had been identified as regulators of drought stress in different plants. AtNF-YA5 (Li et al., 2008), GmNF-YA3 (Ni et al., 2013), and OsHAP2E (Alam et al., 2015) were demonstrated to enhance drought tolerance in Arabidopsis, soybeans, and rice, respectively. In our study, PtNF-YA9 overexpression in Arabidopsis exhibited typical drought-resistant phenotypes, such as dwarf phenotype, small seedling areas, increased chlorophyll content, high photosynthetic rate, and improved WUE under drought condition. Substantial evidence demonstrated that growth reduction is a part of an acclimatization strategy in plants to adapt to adversity. CBF1 (Achard et al., 2008), AtNF-YA5 (Li et al., 2008), AtNF-YA2, AtNF-YA7, and AtNF-YA10 genes (Leyva-González et al., 2012) all exhibited dwarfism phenotypes and confers drought stress. Based on the phylogenetic analysis, PtNF-YA9 clustered with AtNF-YA7, and they shared high similarity. Consistent with this observation, PtNF-YA9 overexpression in Arabidopsis also caused dwarf phenotype and enhanced drought tolerance at the vegetative stage (Figure 5A).
Osmotic stress induces a large number of stress-response genes and is involved in an ABA-dependent or ABA-independent manner (Yoshida et al., 2014). In an ABA-dependent manner, endogenous ABA was induced to modulate stomatal aperture under drought stress, and the expression levels of many drought-response genes were also altered (Zhu, 2002; Seki et al., 2007). RT-qPCR analysis showed that PtNF-YA9 expression was downregulated under ABA treatment (Figure 3). Moreover, seed germination assay provided further evidence to support that PtNF-YA9 is involved in ABA regulation in stress tolerance (Figure 4C). Thus, whether PtNF-YA9-enhanced drought tolerance in Arabidopsis was regulated by ABA to induce stomatal closure remains to be elucidated. To clarify this issue, first, water loss assay was performed and revealed that OxPtNA9 lines enhance water retention (Figure 5D). Second, OxPtNA9 lines promoted stomatal closure in Arabidopsis under ABA treatment (Figure 5J). Third, the expression of ABA-related genes ABI5 and ABF was significantly changed in the OxPtNA9 lines compared with WT both at normal and drought conditions (Figure 6). In conclusion, our results indicated that PtNF-YA9 transgenic plants confer drought tolerance by reducing water loss and promoting stomatal closure via the ABA signaling pathway.
In Arabidopsis, most NF-YA overexpressing lines exhibited dwarf phenotype with smaller rosette diameters than WT plants except NF-YA4 and NF-YA6 (Siriwardana et al., 2014). In this study, we found that OxPtNA9 lines also had dwarf phenotype (Figure 7), short hypocotyl (Figure 7A), small leaf area and biomass (Figures 7C–G), and delayed flowering (Figure 7C). Plant growth predominantly depends on cellular elongation, and EXPs and XTHs play an important role in cell wall loosening with realignment of cell wall components (Cosgrove, 2005). In addition, XTH and EXP genes were repressed in several PXVE:NF-YA lines (Leyva-González et al., 2012). Similar to PXVE:NF-YA lines in Arabidopsis (Leyva-González et al., 2012), PtNF-YA9 or the PtNF-YA9 heterotrimeric complex might participate in XTHs and EXPs to regulate cell elongation.
AtNF-YA2 and AtNF-YA10 promoting leaf development and cell expansion were regulated by the auxin signaling pathway. Auxin homeostasis, polar transport, and signaling affect the entire progress of leaf development (Scarpella et al., 2010). Mutations of ARF19 and NPH4/ARF7 interact with each other and cause a reduced leaf cell expansion (Wilmoth et al., 2005). Furthermore, during leaf–primordium development, auxin production shifts and plays an important role in leaf morphogenesis and vascular differentiation (Mattsson et al., 1999; Aloni et al., 2003). Previous findings suggested that MP, IAA12/BDL, and AXR6 genes participate in auxin signal transduction during vascular development (Hamann et al., 2002). In our studies, PtNF-YA9 altered the leaf size, and GUS staining of ProNF-YA9::GUS showed especially high expression level patterns in leaf vasculature. Thus, PtNF-YA9 might mediate the expression of auxin-related genes to alter the leaf size. However, several additional experiments are required to directly demonstrate the interaction of PtNF-YA9 with auxin-related genes.
In conclusion, PtNF-YA9 plays pleiotropic functions, such as seed germination, post-germination growth, abiotic stress, plant growth, and development (Figure 8). The CCAAT-box element, which presents approximately 30% of eukaryotic promoter sequence, can be recognized by NF-Y activating transcription (Bucher, 1990). NF-Ys are encoded by three multigene subfamilies, which contain 10 NF-YAs, 13 NF-YBs, and 13 NF-YCs genes in Arabidopsis (Siefers et al., 2009). With PtNF-YA9 alone or heterotrimeric complex formed in Arabidopsis, possibilities of at least 1 × 13 × 13 combinations would exist. Thus, the pleiotropic functions of PtNF-YA9 can be explained by the various combinations of PtNF-YA9 with other NF-YB/NF-YC factors at different stages in Arabidopsis. In a word, it is meaningful to gain deep insight into the molecular mechanism of PtNF-YA9 at different stages, and further investigation on the interactions of protein–DNA or protein–protein is also needed to establish PtNF-YA9 regulation network at each individual function.
We have demonstrated that PtNF-YA9 plays an important role in the regulation of drought stress in Arabidopsis via the ABA-mediated signaling pathway. In our gain-of-function genetic studies, PtNF-YA9 overexpressing lines were hypersensitive to simulated drought, ABA, and salt stresses during the early post-germination growth stages and showed a severe post-germination growth arrest involved in ABA signaling pathway by elevating the expression levels of ABI3 and ABI5. Meanwhile, PtNF-YA9 overexpressing lines displayed high instantaneous leaf WUE and decreased in water loss via ABA-induced stomatal closure to exhibit enhanced drought tolerance at seedling stages. In addition, OxPtNA9 lines also confer strong tolerance to salt stress. Furthermore, PtNF-YA9 was also involved in the regulation of plant growth and development in exhibiting dwarf phenotype, as reduced hypocotyl, leaf area, and biomass and delayed flowering, while promoting lateral root growth and increasing chlorophyll content. Our findings propose a model in which PtNF-YA9 plays an important role in reducing plant growth and provide a valuable and complex insight into the plant adaption to abiotic stress.
CL, WY, and XX conceived the research and designed the experiments. CL performed the experiments with help from KY, YZ, and QL. CL analyzed the experimental results with the assistance of SM. All authors discussed the results. CL and XX wrote the manuscript. And all the authors approved the final manuscript.
This research was supported by grants from the National Key Program on Transgenic Research (2018ZX08020002) and the National Natural Science Foundation of China (31570308, 31600484, 31770649).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00954/full#supplementary-material
FIGURE S1 | Phylogenetic tree analysis of the NF-YA subfamily proteins in P. trichocarpa and Arabidopsis. The amino acid sequences were aligned using MAFFT, and the phylogenetic tree was constructed using MEGA 6.0 software by neighbor-joining method with bootstrap analysis of 1000 replicates.
FIGURE S2 | Promoter sequence and cis-element analysis of PtNF-YA9. Arrow lines are cis-elements, and the direction represents sense and antisense strand. ATG is the translation initiation site, and the predicted core promoter region is marked by the black box.
FIGURE S3 | Molecular analysis of PtNF-YA9 transgenic Arabidopsis plants via RT-PCR and RT-qPCR. (A) Simple diagram of 35S:PtNF-YA9 vector. (B,C) mRNA level identification via RT-PCR (B) and RT-qPCR (C) of PtNF-YA9 overexpression lines in Arabidopsis.
FIGURE S4 | Overexpression of PtNF-YA9 grown under 1/2 MS agar medium at seedling stage in Arabidopsis. (A) Morphological differences of root phenotypes in 1/2 MS agar medium plates. (B) Primary root length calculated at different lines. (C) Lateral root numbers calculated at different lines.
FIGURE S5 | Overexpression of PtNF-YA9 confers drought tolerance at seedling stage in Arabidopsis. (A) Morphological differences in 1/2 MS agar medium plates with 200 mM mannitol. (B) Primary root length of different plants calculated under treatment of 200 mM mannitol.
FIGURE S6 |PtNF-YA9 overexpression confers salt tolerance in Arabidopsis. (A) Morphological differences in 1/2 MS agar medium plates with 100 mM NaCl. (B) Primary root length of different plants calculated under treatment of 100 mM NaCl. (C) Morphological differences in long-term salt treatment. The seedlings were grown in soil for 2 weeks under well-watered conditions and thereafter subjected to salt stress by pouring 100 mM NaCl–water every 5 days until 4 weeks.
TABLE S1 | Primers used in this research.
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Ahn, H., Jung, I., Shin, S. J., Park, J., Rhee, S., Kim, J. K., et al. (2017). Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front. Plant Sci. 8:1044. doi: 10.3389/fpls.2017.01044
Alam, M. M., Tanaka, T., Nakamura, H., Ichikawa, H., Kobayashi, K., Yaeno, T., et al. (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 13, 85–96. doi: 10.1111/pbi.12239
Aloni, R., Schwalm, K., Langhans, M., and Ullrich, C. I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216, 841–853.
Bossi, F., Cordoba, E., Dupré, P., Mendoza, M. S., Román, C. S., and León, P. (2010). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59, 359–374. doi: 10.1111/j.1365-313X.2009.03877.x
Bucher, P. (1990). Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212, 563–578. doi: 10.1016/0022-2836(90)90223-9
Burke, E. J., Brown, S. J., and Christidis, N. (2006). Modeling the recent evolution of global drought and projections for the twenty-first century with the hadley centre climate model. J. Hydrometeorol. 7, 1113–1125. doi: 10.1175/Jhm544.1
Cao, S., Kumimoto, R. W., Gnesutta, N., Calogero, A. M., Mantovani, R., and Holt, B. F. III (2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26, 1009–1017. doi: 10.1105/tpc.113.120352
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Ding, Q., Zeng, J., and He, X. Q. (2016). MiR169 and its target PagHAP2-6 regulated by ABA are involved in poplar cambium dormancy. J. Plant Physiol. 198, 1–9. doi: 10.1016/j.jplph.2016.03.017
Dong, Y., Wang, C., Han, X., Tang, S., Liu, S., Xia, X., et al. (2014). A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Biophys. Res. Commun. 450, 453–458. doi: 10.1016/j.bbrc.2014.05.139
Forsburg, S. L., and Guarente, L. (1989). Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3, 1166–1178. doi: 10.1101/gad.3.8.1166
Frontini, M., Imbriano, C., Manni, I., and Mantovani, R. (2004). Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 3, 217–222. doi: 10.4161/cc.3.2.654
Gao, G., Zhang, S., Wang, C., Yang, X., Wang, Y., Su, X., et al. (2011). Arabidopsis CPR5 independently regulates seed germination and postgermination arrest of development through LOX pathway and ABA signaling. PLoS One 6:e19406. doi: 10.1371/journal.pone.0019406
Hamann, T., Benkova, E., Baurle, I., Kientz, M., and Jurgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. doi: 10.1101/gad.229402
Han, X., Tang, S., An, Y., Zheng, D.-C., Xia, X.-L., and Yin, W.-L. (2013). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. doi: 10.1093/jxb/ert262
Jiang, W., and Yu, D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9:96. doi: 10.1186/1471-2229-9-96
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65, 6629–6644. doi: 10.1093/jxb/eru381
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Lee, H., Fischer, R. L., Goldberg, R. B., and Harada, J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100, 2152–2156. doi: 10.1073/pnas.0437909100
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramírez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7:e48138. doi: 10.1371/journal.pone.0048138
Li, L., Zheng, W., Zhu, Y., Ye, H., Tang, B., Arendsee, Z. W., et al. (2015). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. U.S.A. 112, 14734–14739. doi: 10.1073/pnas.1514670112
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Li, X. (2011). Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio Protoc. 1:e95. doi: 10.21769/BioProtoc.95
Li, Y.-J., Fang, Y., Fu, Y.-R., Huang, J.-G., Wu, C.-A., and Zheng, C.-C. (2013). NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS One 8:e61289. doi: 10.1371/journal.pone.0061289
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Lu, C., Han, M. H., Guevara-Garcia, A., and Fedoroff, N. V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 99, 15812–15817. doi: 10.1073/pnas.242607499
Lu, X., Zhang, X., Duan, H., Lian, C., Yin, W., and Xia, X. (2017). Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 162, 73–97. doi: 10.1111/ppl.12613
Ma, X., Zhu, X., Li, C., Song, Y., Zhang, W., Xia, G., et al. (2015). Overexpression of wheat NF-YA10 gene regulates the salinity stress response in Arabidopsis thaliana. Plant Physiol. Biochem. 86, 34–43. doi: 10.1016/j.plaphy.2014.11.011
Mattsson, J., Sung, Z. R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991.
Ni, Z. Y., Hu, Z., Jiang, Q. Y., and Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Pant, B. D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J., et al. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555. doi: 10.1104/pp.109.139139
Potkar, R., Recla, J., and Busov, V. (2013). ptr-MIR169 is a posttranscriptional repressor of PtrHAP2 during vegetative bud dormancy period of aspen (Populus tremuloides) trees. Biochem. Biophys. Res. Commun. 431, 512–518. doi: 10.1016/j.bbrc.2013.01.027
Sade, D., Sade, N., Shriki, O., Lerner, S., Gebremedhin, A., Karavani, A., et al. (2014). Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to tomato yellow leaf curl virus. Plant Physiol. 165, 1684–1697. doi: 10.1104/pp.114.243402
Scarpella, E., Barkoulas, M., and Tsiantis, M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2:a001511. doi: 10.1101/cshperspect.a001511
Seki, M., Umezawa, T., Urano, K., and Shinozaki, K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10, 296–302. doi: 10.1016/j.pbi.2007.04.014
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Siefers, N., Dang, K. K., Kumimoto, R. W., Bynum, W. E. IV, Tayrose, G., and Holt, B. F. III (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. doi: 10.1104/pp.108.130591
Siriwardana, C. L., Kumimoto, R. W., Jones, D. S., and Holt, B. F. III (2014). Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol. Biol. Rep. 32, 971–986. doi: 10.1007/s11105-014-0704-6
Sorin, C., Declerck, M., Christ, A., Blein, T., Ma, L., Lelandais-Briere, C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 202, 1197–1211. doi: 10.1111/nph.12735
Steibel, J., Poletto, R., and Rosa, G. (2005). Statistical analysis of relative quantification of gene expression using real time RT-PCR data. J. Anim. Sci. 83, 104–104.
Steidl, S., Tuncher, A., Goda, H., Guder, C., Papadopoulou, N., Kobayashi, T., et al. (2004). A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 342, 515–524. doi: 10.1016/j.jmb.2004.07.011
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Valliyodan, B., and Nguyen, H. T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 9, 189–195. doi: 10.1016/j.pbi.2006.01.019
Wei, Q., Luo, Q., Wang, R., Zhang, F., He, Y., Zhang, Y., et al. (2017). A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 8:1374. doi: 10.3389/fpls.2017.01374
Wilmoth, J. C., Wang, S. C., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130. doi: 10.1111/j.1365-313X.2005.02432.x
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Keywords: Populus trichocarpa, NF-YA, post-germination growth arrest, drought tolerance, plant development
Citation: Lian C, Li Q, Yao K, Zhang Y, Meng S, Yin W and Xia X (2018) Populus trichocarpa PtNF-YA9, A Multifunctional Transcription Factor, Regulates Seed Germination, Abiotic Stress, Plant Growth and Development in Arabidopsis. Front. Plant Sci. 9:954. doi: 10.3389/fpls.2018.00954
Received: 30 January 2018; Accepted: 13 June 2018;
Published: 09 July 2018.
Edited by:
Xiaohan Yang, Oak Ridge National Laboratory (DOE), United StatesReviewed by:
Kai Shu, Sichuan Agricultural University, ChinaCopyright © 2018 Lian, Li, Yao, Zhang, Meng, Yin and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weilun Yin, eWlud2xAYmpmdS5lZHUuY24= Xinli Xia, eGlheGxAYmpmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.