
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 July 2018
Sec. Plant Metabolism and Chemodiversity
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00941
 Emanuela Blanco1*
Emanuela Blanco1* Wilma Sabetta1†
Wilma Sabetta1† Donatella Danzi1
Donatella Danzi1 Donatella Negro1
Donatella Negro1 Valentina Passeri2†
Valentina Passeri2† Antonino De Lisi1
Antonino De Lisi1 Francesco Paolocci2
Francesco Paolocci2 Gabriella Sonnante1
Gabriella Sonnante1Flavonoids are a well-studied group of secondary metabolites, belonging to the phenylpropanoid pathway. Flavonoids are known to exhibit health promoting effects such as antioxidant capacities, anti-cancer and anti-inflammatory activity. Globe artichoke is an important source of bioactive phenolic compounds, including flavonoids. To study the regulation of their biosynthesis, a R2R3-MYB transcription factor, CcMYB12, was isolated from artichoke leaves. Phylogenetic analysis showed that this protein belongs to the MYB subgroup 7 (flavonol-specific MYB), which includes Arabidopsis AtMYB12, grapevine VvMYBF1, and tomato SlMYB12. CcMYB12 transcripts were detected specifically in artichoke immature inflorescence and young leaves and overlapped with the profiles of flavonol biosynthetic genes. Electrophoretic mobility shift assays (EMSAs) revealed that recombinant CcMYB12 protein is able to bind to ACII element, a DNA binding site ubiquitously present in the promoters of genes encoding flavonol biosynthetic enzymes. In transgenic Arabidopsis plants, the overexpression of CcMYB12 activated the expression of endogenous flavonol biosynthesis genes, leading to an increase of flavonol accumulation and a decrease of anthocyanins in leaves. Likewise, in transgenic tobacco petals and leaves, the overexpression of CcMYB12 decreased anthocyanin levels and increased flavonols.
The artichoke CcMYB12 is expressed in flavonol-rich organs. Its overexpression in Arabidopsis and tobacco causes an activation of early flavonoid biosynthetic genes and a metabolic flux diversion from anthocyanins to flavonols.
In the last decades, an increasing attention has been paid to nutrition and its effects on the development of chronic degenerative diseases. The correct nutritional habits in a population can act as important factors in primary and secondary levels of prevention, thus reducing health risks (Sofi et al., 2013). The Mediterranean diet, recognized to be protective against the occurrence of numerous diseases, is characterized by a high consumption of plant-based food, such as legumes, cereals, olive oil, fruits and vegetables, and many of these plants contain functional components with positive effects on health (Ortega, 2006).
Several fruits and vegetables are especially rich in polyphenols, bioactive secondary metabolites, which constitute a highly diverse group with an extremely large structural diversity (flavonoids, phenolic acids, hydroxycinnamic acid derivatives, lignans). In particular, flavonoids are considered as health-protective components as they are able to protect against cardiovascular diseases, some types of cancers (Ravishankar et al., 2013), degenerative disorders (Korkina, 2007), platelet aggregation (Graf et al., 2005), and osteoporosis (Trivedi et al., 2009).
The globe artichoke [Cynara cardunculus var. scolymus (L.) Fiori] represents a relevant constituent of the Mediterranean diet (Gatto et al., 2013), as it is a natural functional food rich in bioactive polyphenolic compounds, inulin, fibers and minerals. Nutritional and pharmaceutical properties of the edible part of artichoke, its immature inflorescences, also called capitula or heads (receptacles with inner and intermediate bracts), can be ascribed to high levels of health-promoting polyphenolic compounds, such as caffeoylquinic derivatives and flavonoids (Sonnante et al., 2010), known to have a marked antioxidant activity (Rice-Evans et al., 1997; Azzini et al., 2007). Moreover, artichoke leaf extracts are known to possess antioxidative, anticarcinogenic, anti immunodeficiency virus, cholesterol-lowering, bile-expelling, and hepatoprotective activities, along with antifungal and antibacterial properties (Agarwal and Mukhtar, 1996; Gebhardt, 1997; Kraft, 1997; Brown and Rice-Evans, 1998; McDougall et al., 1998; Shimoda et al., 2003; Zhua et al., 2005).
The flavonoid biosynthetic pathway is well-established, the structural genes of the enzymes involved in flavonoid formation being mainly regulated at the transcription level (Ververidis et al., 2007; Liu et al., 2015). Transcription factors (TFs) usually constitute gene families and regulate target genes in a tissue- and species-related manner, and/or in response to various biotic and abiotic stress factors (Bovy et al., 2002; Koes et al., 2005; Xu et al., 2015). TFs regulating metabolic networks have been considered as promising tools for engineering the levels of metabolites (Grotewold, 2008). Moreover, the association analyses between the expression of TFs and metabolic profiles could lead to the selection and promotion of specific genotypes with a healthier metabolite accumulation and better stress tolerance.
Flavonoid transcriptional regulators mostly belong to protein families containing R2R3-MYB domains, basic helix-loop-helix (bHLH) domains (MYC proteins), and conserved WD amino acids repeats (Ververidis et al., 2007). ZmP1 from maize was the first MYB factor, involved in flavonoids accumulation, displaying activity without binding a bHLH protein (Grotewold et al., 1994). The Arabidopsis ortholog, AtMYB12, was isolated and fully characterized as flavonol-specific regulator (Mehrtens et al., 2005; Stracke et al., 2007). It was heterologously expressed in plants, like tobacco and tomato, producing high-level accumulation of polyphenolic compounds, particularly flavonols and chlorogenic acid (CGA; Luo et al., 2008).
Flavonoid composition among plant species can be highly different and TF activity varies depending on the investigated plant, highlighting the importance of determining the specificity of TF for their target genes in various species (Luo et al., 2008; Czemmel et al., 2017). The role of MYB12 homologues in flavonoid regulation has been established in many plant species, e.g., grapevine, tomato and Japanese gentian (Czemmel et al., 2009; Ballester et al., 2010; Nakatsuka et al., 2012; Czemmel et al., 2017).
Despite the known beneficial properties of phenylpropanoids from artichoke, their biosynthetic routes in this crop are not well defined yet. The biosynthesis of chlorogenic acid and its derivatives was recently studied (Comino et al., 2007, 2009; De Paolis et al., 2008; Moglia et al., 2009; Sonnante et al., 2010), whereas more recently, a flavonoid 3′-hydroxylase, CcF3′H, implicated in dihydroxylated flavone and flavonol biosynthesis, and in anthocyanin accumulation, was identified (De Palma et al., 2014). To shed light on the accumulation of beneficial compounds in artichoke, regulatory mechanisms of flavonoid synthesis need to be examined.
To this end, here we report on the isolation and functional characterization of CcMYB12, the artichoke putative homologue of R2R3-MYBs controlling the biosynthesis of flavonols in a number of species. We show that CcMYB12 recombinant protein can bind canonical AC element present on the promoters of artichoke early flavonoid genes and that its transcript expression profile overlaps with those of several early flavonoid genes in flavonol- rich artichoke bracts. Finally, by targeted metabolic and molecular analyses we show that the ectopic expression of CcMYB12 in Arabidopsis and tobacco plants promotes flavonol biosynthesis at expenses of anthocyanins.
Plants of globe artichoke (cv. Mola) were grown in the experimental field of the Institute of Biosciences and Bioresources (IBBR-CNR), Bari, Italy. Material from three independent plants was collected from stems, young and adult leaves; flower heads were harvested at the commercial maturity stage and separated into external, intermediate, internal bracts and receptacle (Sonnante et al., 2010).
Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) seeds were germinated on Murashige and Skoog plates (Murashige and Skoog, 1962) containing 1% sucrose in a growth chamber (250 μmol photons m-2 s-1; 22°C) under a long-day regime (16 h of light/8 h of dark). To study the gene expression and metabolite profiles in WT and CcMYB12 transgenic Arabidopsis leaves, seedlings were transferred to soil in a climate-controlled chamber under the same photoperiod and leaves collected 4 weeks after germination. To analyze the effects of sucrose on anthocyanins accumulation in the seedlings, WT and CcMYB12 transgenic seeds were germinated in vitro either on 1% or 6% (w/v) sucrose concentration and 2-week old seedlings collected for molecular and metabolic analyses. Nicotiana tabacum cv. ‘SR-1’ plants were grown in vitro on Murashige and Skoog solid medium supplemented with 3% (w/v) Suc at 23°C and 16/8-h light/dark photoperiod for rooting and then transferred to soil with the same environmental conditions.
Total RNA was isolated from 0.2 g of frozen young leaf of artichoke using RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). cDNA was synthesized from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (QIAGEN).
The NCBI1 globe artichoke EST database was screened to find AtMYB12 homologues (Accession No. CAB09172; Stracke et al., 2001). On the retrieved sequence the primer pair MYB12F1-MYB12R1 (Supplementary Table S1) was designed and employed to amplify a partial cDNA sequence from leaves. Amplification reactions contained 1 μL of cDNA, 1X PCR buffer, 0.4 mM deoxyribonucleotide triphosphates (dNTPs), 400 nM each primer, and 1 unit of JumpStart Taq DNA Polymerase (Sigma–Aldrich, Saint Louis, MO, United States), in a final volume of 25 μl. The thermal cycling program included a step at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. PCR fragments were cloned using the GENEJet PCR Cloning kit (Thermo Fisher Scientific, Waltham, MA United States) and subsequently sequenced. New primers pair designed on the resulting amplicon were then employed to clone the 5′ and 3′ CcMYB12 ends (Supplementary Table S1) with the 5′ and 3′ RACE kits, respectively (Invitrogen, Thermo Fisher Scientific, Waltham, MA United States). Then, the complete CcMYB12 open reading frame (GenBank Accession No. MG517449) was PCR amplified (CcMyb12-FWatg - CcMyb12-RVstop, Supplementary Table S1) and confirmed by sequencing.
The GenomeWalker Universal kit (Clontech Laboratories, Inc. United States) was used to isolate the CcMYB12 promoter (GenBank Accession No. MG517450), following manufacturer’s instructions. After artichoke genomic DNA digestion with DraI, EcoRV, PvuII, and StuI, and ligation to adaptors, nested PCRs were performed for each digested-ligated DNA library, using the supplied AP1 primer from the kit and CcMYB12 specific primers (GWCcMYB12_GSP1 and GWCcMYB12_GSP2; Supplementary Table S1). Amplicons were resolved on a low-melting point agarose gel, and fragments longer than 1 kb were excised from the gel, purified, cloned, and sequenced.
The CcMYB12 full-length cDNA was analyzed by NCBI BLAST2. For the multiple sequence alignment analysis, the amino acid sequence of CcMYB12 and other MYB homologs from different plant species retrieved from NCBI were aligned using Clustal Omega3, Jalview v. 2.10.1 (Waterhouse et al., 2009) and Bioedit v. 7.2.5 softwares4. A phylogenetic tree was constructed using the neighbor-joining method with the minimum evolution test and p-distance model with 10,000 bootstrap replicates, using the MEGA package v. 6.5 The theoretical molecular weight and isoelectronic point (pI) of CcMYB12 protein were calculated using ProtParam tool.6 The conserved domain of CcMYB12 protein was scanned by the InterProScan program7. PlantPAN Interface8 and PlantCARE9 were used to analyze promoter sequence.
The full length coding sequence of CcMYB12 was amplified using specific primers (GW-MYB12-F and CcMYB12-RVstop, Supplementary Table S1). CcMYB12 was firstly cloned into the entry vector pENTR/D-TOPO (Invitrogen) using the Gateway recombination system, and then recombined into the destination vectors pK2GW7 (Karimi et al., 2002) between the cauliflower mosaic virus 35S promoter and the t-nos terminator construct, and pMDC32 (Curtis and Grossniklaus, 2003), between a double mosaic virus 35S promoter and the t-nos terminator. In both cassettes the cDNA insertion was proven to be in the sense orientation by sequencing. pK2GWF7-CcMYB12 and pMDC32-CcMYB12 plasmids were introduced into Agrobacterium tumefaciens strains GV3101, and LBA4404, respectively, by the freeze-thaw method (Hofgen and Willmitzer, 1988).
The recombinant A. tumefaciens GV3101 strain harboring either the pK2GW7-CcMYB12 plasmid or the empty vector were used to stably transform Arabidopsis plants according to the floral dip method (Clough and Bent, 1998), and the A. tumefaciens LBA4404 strain harboring the pMDC32-CcMYB12 plasmid was used to stably transform leaf disks of N. tabacum cv SR1 according to Horsch et al. (1985).
To select Arabidopsis transgenic plants, T1 seeds were allowed to germinate on selected Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1% Sucrose and 50 mg/l kanamycin at 250 μmol photons m-2 s-1 and 22°C. Stable insertion of the exogenous expression cassette was checked by genomic DNA amplification with primers for the neomycin phosphotransferase (nptII) gene (Supplementary Table S1) and for the CcMYB12 cDNA sequence (MYB1_F and CcMyb12-RVstop; Supplementary Table S1).
Transgenic tobacco lines were in vitro selected on MS medium supplemented with 20 mg/L hygromycin. Hygromycin-resistant tobacco plantlets were recovered and screened by PCR for the presence of CcMYB12 and hptII genes (Supplementary Table S1) and then moved to glasshouse under outdoor environmental conditions. As control, CcMYB12 primary transgenic tobacco plants (T1) were grown side by side in the same glasshouse with SR1 untransformed plants. Seeds from self-pollinated tobacco transgenic T1, and control lines were collected, surface sterilized, plated onto MS medium and then grown in the glasshouse as reported above.
Leaves from tobacco and Arabidopsis, and tobacco flowers from T2 and T3 plants were used for molecular and metabolic analyses.
Total RNA was isolated from artichoke as described above. Total RNA of wild type (WT), transgenic Arabidopsis leaves and seedlings and tobacco leaves and petals of fully opened flowers, at stage 12 as reported in Koltunow et al. (1990), was isolated using RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. Primers for quantitative Real-Time PCR (qRT-PCR) were designed on CcMYB12, or on known gene sequences retrieved from NCBI database or taken from the literature (Supplementary Table S2). As for putative artichoke flavonol synthase (FLS), the NCBI Cynara EST database was screened using the putative Helianthus annuus FLS coding sequence (XM_022126949.1) as query. The artichoke GE604603.1 sequence, showing the highest similarity, was retrieved and primers therein designed (Supplementary Table S2).
Reactions were performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, United States), using iTaq Universal SYBR Green Supermix (Bio-Rad). Reaction mixtures (10 μl) included the following components: 1Xmaster mix, 0.2 μM of each primer and 1 μl cDNA. Cycle conditions were 95°C for 20 s and then 40 cycles of 95°C for 15 s and 60°C for 30 s. Normalization against different reference genes was tested prior to qRT-PCR analyses. Actin and Elongation Factor1α (EF1α) genes expression were analyzed for both tobacco and Arabidopsis experiments, in addition tobacco tubulin expression was tested. The expression levels of each gene were normalized by the expression of actin genes both in Arabidopsis and tobacco (At3g18780 and AF154640, respectively). The artichoke EF1α gene (EU442190) was used as reference gene in artichoke analyses, as previously tested (Sonnante et al., 2010). Separation of real-time PCR products on 2% (w/v) agarose gels revealed single bands of the expected size whose identities were confirmed by direct sequencing. Relative quantification was performed according to the comparative Ct (threshold cycle) method (2-ΔΔCt; Bustin, 2000; Palmieri et al., 2008). In all experiments, appropriate negative controls containing no template were used. Analysis was performed on three independent biological replicates per plant sample, and three technical replicates for each sample, and reactions were repeated twice to verify reproducibility.
CcMYB12 recombinant protein in frame with a C-terminal (6XHis)-tag was expressed in E. coli BL-21 Rosetta cells. The coding sequence without the terminal codon was PCR amplified from cDNA of artichoke young leaf (GW-MYB12-F and pE-MYB12-R; Supplementary Table S1) and cloned into the pENTRTM/D-TOPOR Vector (cat. No. K2400-20, Invitrogen, United States) using the Gateway recombination system. The insert was sequenced to confirm identity. Subsequently, the CcMYB12 entry clone was used to generate the expression clone by a recombination reaction with the pET-DEST42 GatewayTM destination vector, encoding a C-terminal (6XHis)-tag, under the control of the Cauliflower Mosaic Virus 35S promoter (Cat. No. 12276-010, Invitrogen, United States). The insert was sequenced prior to protein expression. Growth of E. coli expression cultures in selective liquid LB (100 μg/ml ampicillin) and protein induction were performed as described in the QIAexpressionist manual (protocol 7; QIAGEN, Germany).
Recombinant protein was batch-purified by affinity chromatography using a Ni-NTA resin under denaturing conditions according to instructions (QIAexpressionist, Protocol 10; QIAGEN, Germany) with the exception that lysis buffer B added with imidazole (100 mM NaH2PO4; 10 mM Tris⋅Cl; 8 M urea; 10 mM Imidazole, pH to 8.0 using NaOH) was used to prepare cleared E. coli lysates. The eluted fractions were collected, concentrated with 3 volumes of 100% acetone and re-suspended in Phosphate Buffered Saline pH 7.4. Total protein content from crude extracts and eluted fractions was quantified by Bradford Protein Assay (Bio-Rad, United States) (Bradford, 1976), while purification efficiency was verified by SDS-PAGE (Laemmli, 1970) and Coomassie Blue staining. E. coli extracts harboring no vector/plasmid were used as negative control.
To perform electrophoretic mobility shift assays (EMSA), the AC-rich sequences ACI and ACII containing the MYB-binding cis-motifs ACCAACC and ACCTACC, respectively, were used as MYB-binding sites in two different probes (Patzlaff et al., 2003; Xu et al., 2013). Related mutated probes (mACI and mACII) were used as negative controls. The oligonucleotide probes ACI (gatcctttatacccACCTACCagacacg) mACI (gatcctttatacccCAAGCAAagacacg), ACII (gatccttctccACCAACCcccttcacttcccg) and mACII (gatccttctccGAAGGAAcccttcacttcccg) were labeled with Biotin-11-UTP onto the 3′ end using the Pierce Biotin 3′-End DNA Labeling Kit (Life Technologies), following manufactory’s instructions. About 0.6 μg purified recombinant CcMYB12 protein was allowed to re-nature prior to the binding experiment. Each binding assay was carried out using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, United States), with few modifications to the general instructions: the incubation time for binding reaction was extended up to 60 min, while the incubation time of membrane with the substrate working solution was reduced to 2 min. The bound complexes were subjected to electrophoresis on native 7% polyacrylamide gels in 0.5% Tris–borate EDTA (TBE) buffer (pH 8.0) at 90 V for 60–80 min at 4°C, and then transferred to a positively charged nylon membrane (Biodyne B Nylon Membrane for Chemiluminescent EMSA, Thermo Scientific) using the Trans-Blot® SD Semi-Dry Transfer Cell (Bio-Rad). The biotin-labeled signals were detected using the Streptavidin-Horseradish Peroxidase Conjugate, the chemiluminescent substrate and the chemilumino-image analyzer Chemi Doc (Bio-Rad).
Flavonoids were extracted from freeze-dried samples with 40 μL of 70% MeOH per mg of dry weight (DW) of tissue. Extracts were centrifuged at 16,000 × g and supernatants were filter spun using 0.2 μm filters analyses. HPLC analyses were carried out with a Beckman-Coulter (Fullerton, CA, United States) HPLC System Gold chromatograph equipped with a DAD programmable detector (System Gold, series 166) operated by a 32 Karat software package (Beckman-Coulter).
Arabidopsis flavonols were analyzed by the HPLC method proposed by Mehrtens et al. (2005), modified by Negro et al. (2012) using the reversed phase column Luna C18 (250 × 4.6 mm i.d., particle size 5 μm; Phenomenex, Torrance, CA, United States). Diode array detection was between 200 and 600 nm, and absorbance was recorded at 280, 310, and 350 nm. Peaks were classified as corresponding to kaempferol or quercetin derivatives by UV spectral analysis.
HPLC analyses of tobacco flavonoids and monocaffeoylquinic acids and Arabidopsis anthocyanins were carried out as described in Luo et al. (2007), using a Synergi 4 μm Hydro-RP 80A column (250 × 4,60 mm internal diameter; Phenomenex, Torrance, CA, United States). Anthocyanin contents were calculated from their peak areas at 520 nm in the chromatograms and compared with the external standard cyanidin chloride; flavonoid absorbance was recorded at 325 and 520 nm.
Flavonoid and monocaffeoylquinic acid identification was reached by combining the following information: positions of absorption maxima (λ max) and retention times (min) were compared with those from pure standards and/or interpreted with the help of structural models already available in the literature, the latter identities considered as probable rather than proven. Quantification was made by using six points (10 – 1000 ppm) calibration curves standards of 5-O-caffeoylquinic acid (CGA), quercetin, kaempferol, apigenin 7-O-glucoside and cianidin chloride, with R2 values ranging between 0.9997 and 0.9999. Monocaffeoylquinic acids were expressed as CGA, kaempferol and quercetin derivatives as kaempferol and quercetin equivalents, anthocyanins as cyanidin equivalents. Pure flavonoids, CGA, quercetin and apigenin 7-O-glucoside were obtained from Extrasynthèse (Lyon, France)10, kaempferol and cyanidin chloride were obtained from Sigma–Aldrich (Saint Louis, MO, United States). Formic acid, acetonitrile and HPLC grade water were purchased from J. T. Baker (Deventer, Holland). All analyses were performed on Arabidopsis and tobacco young leaves and on tobacco petals of fully opened flowers (stage 12, Koltunow et al., 1990). Spectrophotometric analysis of anthocyanins in Arabidopsis seedlings was carried out using the acidified methanol method, as described in Mahmood et al. (2016), except for starting material: 100 mg of frozen seedlings from each genotype (2 weeks after germination) were pooled in a single replicate and homogenized in 500 μl of methanol-HCl (1% v/v). Samples were analyzed spectrophotometrically at 530 and 657 nm wavelengths. Quantification of anthocyanin was calculated by the following equation: A530-0.25∗ A657.
Statistical significance was tested with Student’s t-tests using Sigmaplot 10.1 software or with one-way analysis of variance (ANOVA) with Tukey’s comparison tests, where differences were considered statistically significant at p < 0.05.
The artichoke EST database in NCBI was used to retrieve the homologs of AtMYB12. The EST with the highest nucleotide sequence similarity, namely GE612454.1 (76% of identity), was used to design a specific primer pair to amplify a 556 bp long cDNA fragment from artichoke leaves. To clone the full length cDNA for CcMYB12, 5′ and 3′ RACE analyses were performed and then a forward primer starting from the first codon and a reverse primer containing the stop codon were employed in an end-to-end RT-PCR.
The complete CcMYB12 coding sequence (GenBank Accession No. MG517449) was 1176 bp in length and predicted to encode a protein of 391 amino-acids (molecular weight: 43.99 kDa theoretical pI: 4.83). A multi-alignment comparison of the deduced protein with MYB TFs annotated in other plant species revealed a high level of sequence identity at the N-terminus, where the characteristic modular structure of the DNA-binding R2R3-MYB domain was identified. In contrast, the rest of the sequence was extremely variable, as for all plant R2R3-MYB transcription factors. The newly isolated artichoke sequence was named CcMYB12 (Cynara cardunculus MYB12).
The artichoke CcMYB12 R2 and R3 repeats consist of 53 amino acids (from G11to D64 residues) and 49 amino-acids (from K66 to K115), respectively (Figure 1A), with the typical helix-turn-helix structure and the highly conserved tryptophan residues implicated in DNA binding [-W-(X19)-W-(X19)-W-(X12)-I-(X18)-W-(X18)-W-] (Stracke et al., 2001). We scanned the CcMYB12 R2R3 amino acid sequence to search key residues probably involved in target promoter specificity. Recently, Heppel et al. (2013) identified key amino acid residues in the MYB R2R3-domain responsible for the specificity of MYB involvement into anthocyanin and proanthocyanidin (PA) biosynthesis. Briefly, the known anthocyanin MYB regulators reported in Figure 1A (AtMYB75, AtMYB90, VvMYBA1, and LeANT1) contain an Arg (R) residue at position 52 of the alignment, a Val (V) residue at position 77 and a four amino-acid motif represented by Asp-Asn-Glu-Ile (ANDV) at the position 103–106. In PA and other R2R3 type MYB factors including regulators of flavonol synthesis and CcMYB12, the above mentioned key amino-acid residues are substituted by a Gly (G) at position 52, a Glu (E)/ASP (D) at position 77, and an Asp-Asn-Gln-Val motif (DNEI) at position 103-106, respectively.
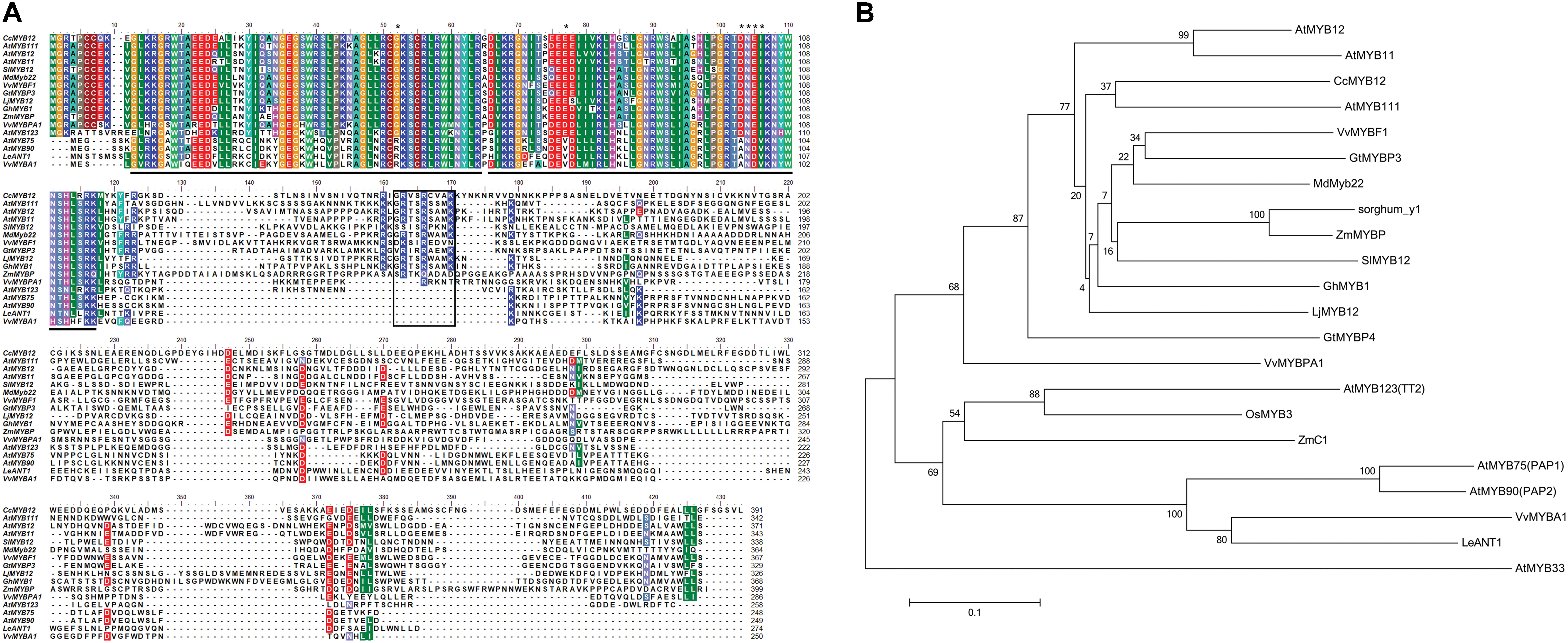
FIGURE 1. Protein sequence alignment of CcMYB12 deduced amino acid sequence with plant R2R3-MYB transcription factors and phylogenetic analysis. (A) Protein sequence alignment of CcMYB12 with representative members of R2R3-MYB-type transcriptional regulators of flavonols, proanthocyanidins and anthocyanins from other plant species. The position of the R2 and R3-type MYB domains is indicated below the alignment by gray bars, the SG7 domain is highlighted by a black box. Key amino acid residues are indicated by asterisks. (B) Phylogenetic analysis of deduced amino acid sequences of R2R3-MYB transcription factors present in higher plants. The Neighbor-Joining phylogenetic tree was generated using MEGA software (Tamura et al., 2013). Numerals next to branch nodes indicate bootstrap values from 10000 replications. The bar indicates an evolutionary distance of 0.1%. Protein accession numbers are as follows: apple MdMYB22 (AAZ20438 [Malus domestica]); Arabidopsis: MYB11 (NP_191820), MYB12 (CAB09172), MYB111 (AAK97396), AtMYB123/TT2, (CAC40021), AtMYB75/PAP1 (AAG42001), AtMYB90/PAP2 (AAG42002), AtMYB33 (NP_850779); grape: MYBF1 (ACT88298), MYBPA1 (CAJ90831.1) and MYBA1 (BAD18977); gentian: GtMYBP3 (BAM71801) and GtMYBP4 (BAF96934); gerbera: GhMYB1 (CAD87007); lotus LjMYB12 (BAF74782 MYB-related protein [Lotus japonicus]); maize: ZmMYBP (P27898) and ZmC1 (AAA33482); rice OsMYB3 (EAY89678 (hypothetical protein [Oryza sativa])); sorghum: Y1 (AAX44239); tomato: SlMYB12 (ACB46530) and LeANT1 (AAQ55181). AtMYB33, a GAMYB-like protein, was used as outgroup.
Phylogenetic analysis revealed the similarity of CcMYB12 to the flavonol regulators from Arabidopsis, AtMYB111, AtMYB12 and AtMYB11, and other plant MYB proteins such as VvMYBF1, GtMYBP3, MdMyb22, SlMYB12 and ZmMYBP (Figure 1B). Similarity was also found to other putative plant flavonol-specific TFs, previously identified (Czemmel et al., 2009), such as Sorghum bicolor sorghum_y1, Lotus japonicus LjMYB12 and Gerbera hybrida GhMYB1 (Figure 1B).
The R2R3 domain of CcMYB12 showed the greatest levels of identity (86%) to Arabidopsis AtMYB111 and grape VvMYBF1, to Arabidopsis AtMYB12 and AtMYB11 (84 and 81%, respectively), and to putative gerbera and Lotus MYB12 homologs (83 and 79%, respectively). The complete CcMYB12 protein sequence exhibited lower percentages of identities to other proteins, being the C-terminal end very different among MYB TFs. It showed 38% identity to AtMYB111 and 34, 33, 31, 32, and 31% identity to VvMYBF1, LjMYB12, GhMYB1, AtMYB12 and AtMYB11, respectively.
Moreover, CcMYB12 R3 domain did not contain the conserved amino acid signature [DE]Lx2[RK]x3Lx6Lx3R for the interaction with BHLH proteins (Zimmermann et al., 2004), while the SG7 motif (GRTxRSxMK; Stracke et al., 2001) characteristic of flavonol regulators of Arabidopsis was found at the C-terminus of CcMYB12, although partially conserved, given the presence of three amino acid substitutions (GRVSRCVAK).
By a genome walking approach a 3,002-bp-long sequence containing the CcMYB12 putative promoter region was cloned (GenBank Accession No. MG517450). The search for conserved cis elements in the 1,468 bp upstream the ATG codon using PlantPan and PlantCARE databases revealed the presence of cis-regulatory elements putatively involved in the response to hormones (ABA and GA responsive elements), water stress (Myb binding sites MBS, MYBcore), light (ACE elements, GT1-motifs, I-box, HY5 ACGT-containing elements, G-box), biotic stresses (Box-W1) and low temperatures (LTR elements) which are also present in genes of the phenylpropanoid pathway such as phenylalanine ammonia lyase (PAL), chalcone synthase (CHS), chalcone isomerase (CHI) dihydroflavonol 4-reductase (DFR), coumaryl-CoA ligase (CL) in many plant species (Supplementary Table S2; Patzlaff et al., 2003; Hartmann et al., 2005; Prouse and Campbell, 2012).
To reveal the expression of CcMYB12 in artichoke organs, qRT-PCR analysis was performed using total RNA isolated from artichoke heads, leaves and stems. CcMYB12 transcripts were detected at a higher level in intermediate bracts followed by internal bracts and young leaves; lower levels were detected in external bracts, receptacle, adult leaves and stem (Figure 2). To investigate whether CcMYB12 gene is involved in the regulation of flavonoid production in vivo, the expression levels of known artichoke biosynthetic genes, namely, phenylalanine ammonia lyase (PAL1, PAL3), p-coumaroyl ester 3′ hydroxylase (C3′H), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase (HQT, HQT1, HQT2), flavone 3′-hydroxylase (F3′H) and of a putative flavonol synthase homolog (FLS), were analyzed in all organs (Supplementary Figure S1). In intermediate bracts, where the expression of endogenous CcMYB12 peaked, the highest levels of HQT2 and C3′H were found, and PAL1 and HCT were highly expressed; in internal bracts HQT1, HQT2 and C3′H were among the more expressed genes, while in young leaves HQT, F3′H and FLS showed the highest levels.
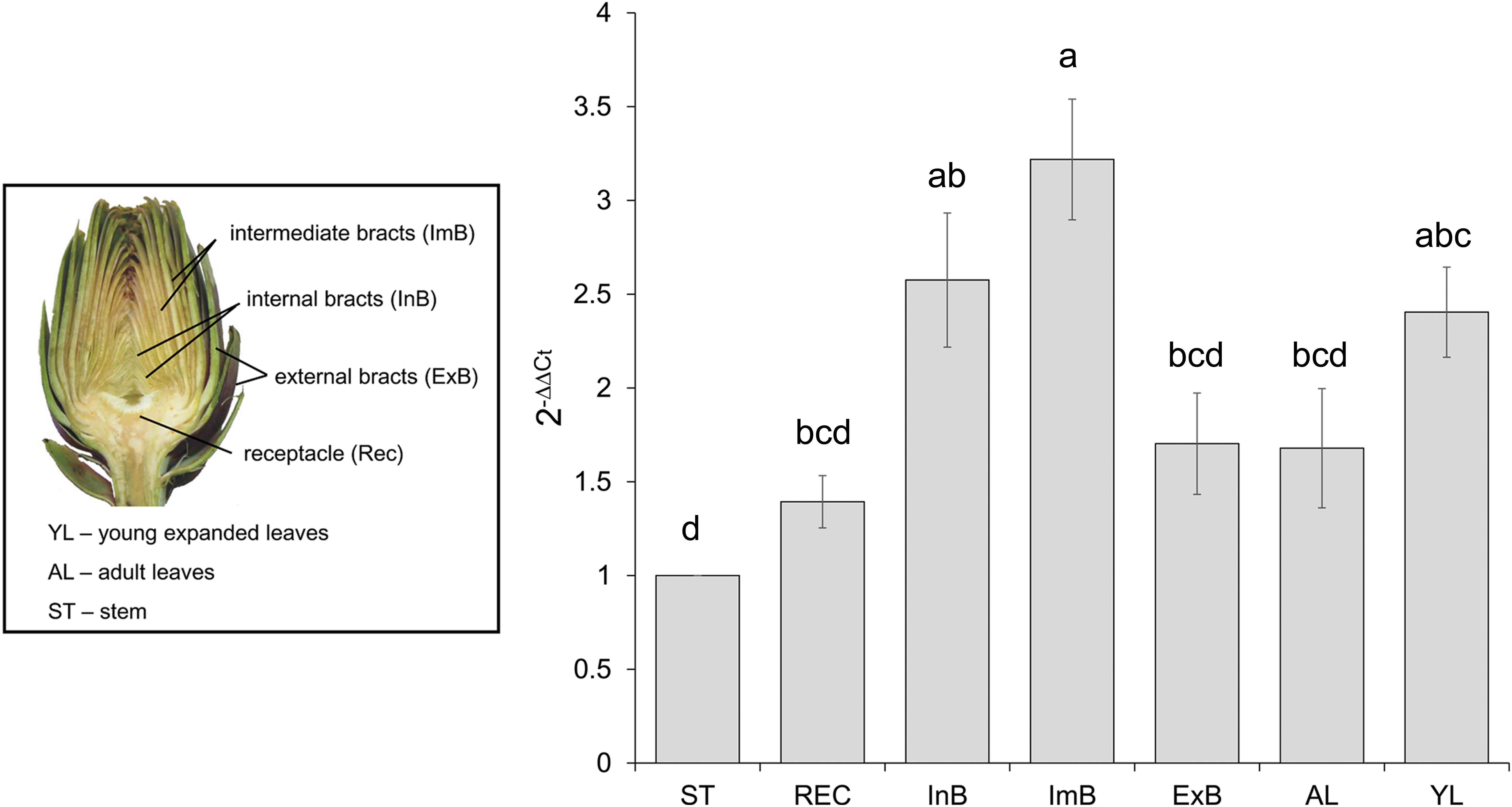
FIGURE 2. CcMYB12 gene expression in different organs of artichoke. Quantitative Real Time PCR analysis was performed in the organs depicted in the photograph: stem (St), receptacle (Rec), internal bracts (InB), intermediate bracts (ImB), external bracts (ExB), adult leaves (AL), young leaves (YL), St is the calibrator. Elongation Factor EF1α was employed as reference gene. Values are means ± SD of three biological replicates. Bars with different letters are statistically different to each other according to one-way ANOVA Tukey test (p < 0.05).
To investigate whether CcMYB12 protein is able to bind in vitro to oligonucleotides enclosing MYB-binding cis-motifs, the recombinant CcMYB12 protein was expressed in E. coli and its interaction with the MYBPLANT motifs (CCTACC and CCAACC) was investigated by electrophoretic mobility shift assays (EMSA), making use of the ACI and ACII probes (Patzlaff et al., 2003). MYBPLANT motifs were found in the promoters of artichoke structural genes PAL3a, HQT1, HQT2, and C3′H, involved in hydroxycinnammic acids and flavonoids synthesis, (De Paolis et al., 2008; Moglia et al., 2009; Sonnante et al., 2010), as well as of several other plant flavonoids biosynthetic genes (Prouse and Campbell, 2012) and CcMYB12 gene itself.
EMSA analyses revealed that recombinant CcMYB12 was able to bind to ACII probe with high affinity (Figure 3), while there was no affinity for ACI probe (Supplementary Figure S2). The CcMYB12 protein did not bind to mutated version of both AC-rich elements, differing by single nucleotide changes, and its binding to biotin-labeled probe ACII∗ was outcompeted by cold competitor ACII. Thus, CcMYB12 binds to AC-rich canonical R2R3 MYB-binding site motif MYBPLANT in vitro, although in a specifically different manner, preferring ACII to ACI element.
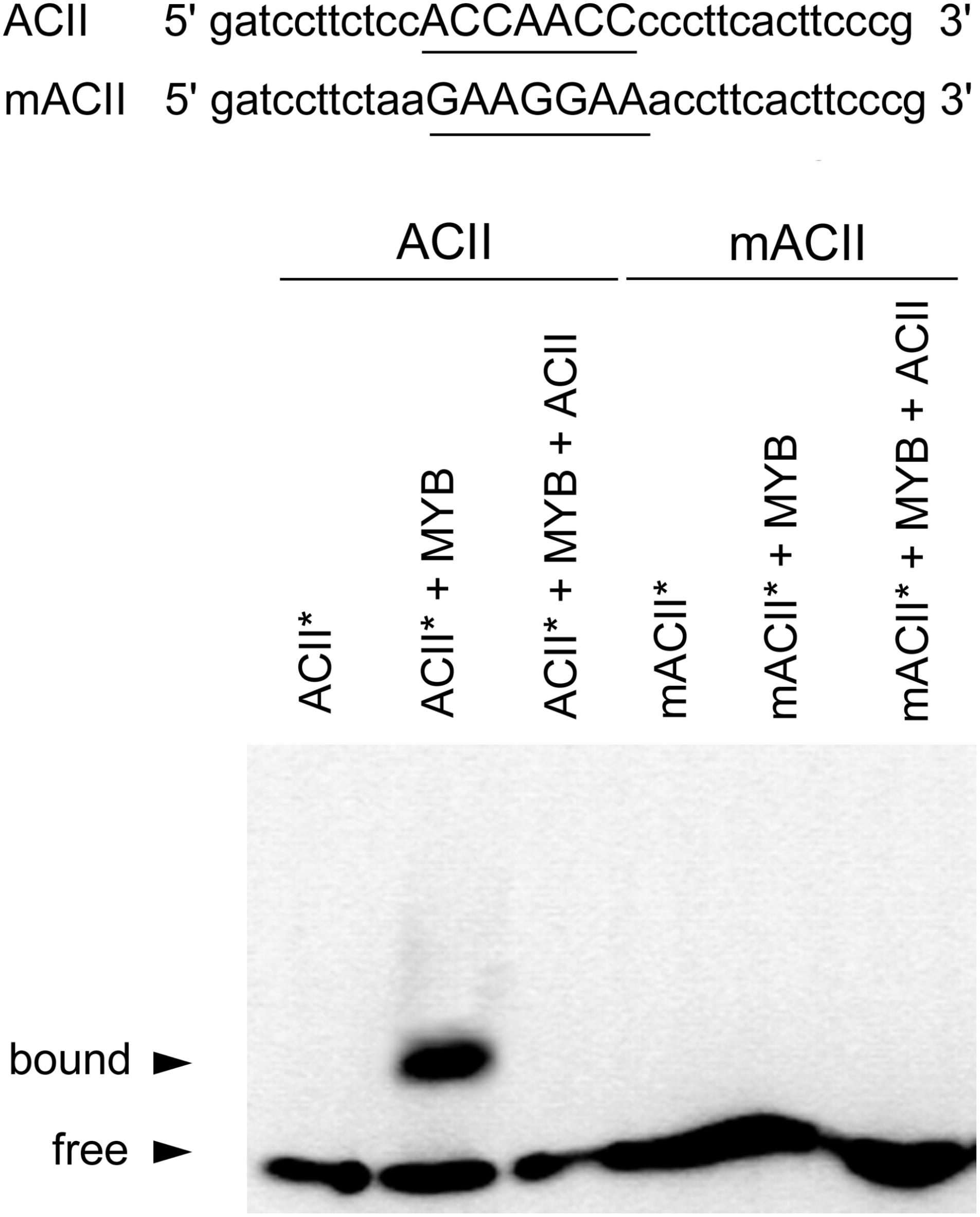
FIGURE 3. Analyses of CcMYB12 protein binding to AC elements. EMSA output of the purified recombinant CcMYB12 binding to the AC element ACII (left) or to its mutated counterpart (mACII). The biotin-labeled free probe (ACII∗) without added protein is designed as control. Binding of recombinant CcMYB12 to biotin-labeled probe ACII∗ can be outcompeted by cold competitor ACII. Nucleotide sequences of probes are shown on the top of the figure. The AC or mAC element is underlined.
In order to gain functional evidence in vivo, CcMYB12 was overexpressed in two different plant systems, Arabidopsis and tobacco. Progenies from twelve independent Arabidopsis primary CcMYB12 transgenic lines were obtained. No morphological changes were observed between WT and empty-vector control plants as well as between transgenic lines, their progenies and WT (data not shown). The expression levels of Arabidopsis genes involved in phenylpropanoid pathway were analyzed by qRT-PCR in leaves of progenies of four CcMYB12 transgenic and WT lines, each sample corresponding to a pool of leaves from four different plants.
Although to a different extent, all transgenic lines showed the overexpression of CcMYB12, which was absent in WT, as expected (Figure 4A). The expression levels of CHS, CHI and FLS were considerably enhanced in the leaves of transgenic plants compared to WT. F3′H and F3H levels showed a lower but still significant increase, followed by DFR which was only subtly upregulated in transgenic vs. WT plants. Conversely, the expression of PAL and ANS was unaffected by CcMYB12. The expression of endogenous AtMYB12 and AtMYB111 were tested in each transgenic line; a modest increase, not exceeding 1.5 fold, was recorded for AtMYB12 in three out of the four investigated lines (Figure 4B), while no effects was recorded for AtMYB111.
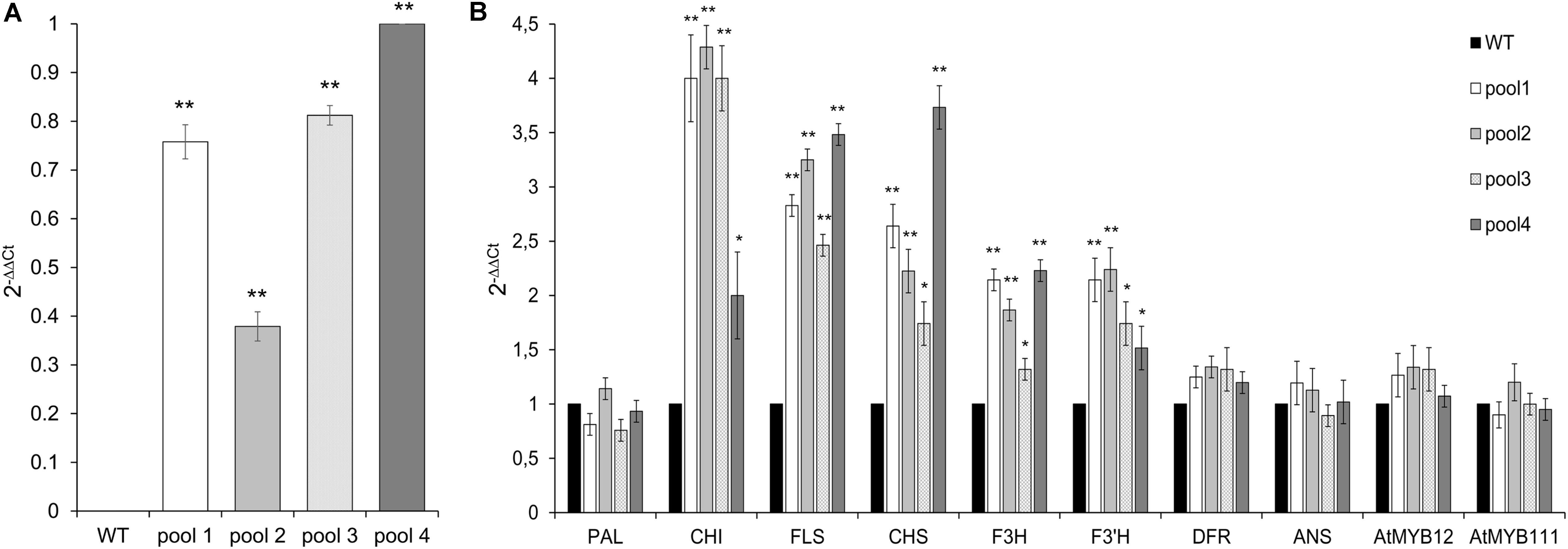
FIGURE 4. Expression of CcMYB12 transgene (A) and of endogenous phenylpropanoid biosynthetic genes (B) in leaves of transgenic Arabidopsis plants. qRT-PCR experiments were conducted on cDNAs from various transgenic Arabidopsis T3 plants grouped in 4 different pools (pool 1, 2, 3, and 4), each constituted by 4 plants of the same transgenic T3 line. Actin was employed as reference gene. (A) qRT-PCR of CcMYB12 in transgenic Arabidopsis plants. Pool 4 as calibrator. (B) Expression levels of PAL, CHI, FLS, CHS, F3H, F3′H, DFR, ANS, AtMYB12 and AtMYB111 in CcMYB12 transgenic Arabidopsis plants. WT as calibrator. Values are means ± SD of three biological replicates. Asterisks indicate a statistical difference (∗p-value < 0.05; ∗∗p-value < 0.01) between the means for WT and tested transgenic samples, according to Student’s t-test.
The upregulation of these early phenylpropanoid genes led to an altered polyphenolic profile in the leaves of transgenic vs. Arabidopsis WT plants. HPLC analyses, performed on methanolic extracts of the same four independent transgenic lines, showed that all plants contained higher amounts of flavonols than WT, in terms of total kaempferols and total quercetins (Figures 5A,B). Apigenin content was also investigated, but barely detectable amounts were found in both WT and transgenic plants (data not shown). To check whether these biochemical alterations extend to anthocyanins, the content of these compounds was also determined by HPLC. All transgenic lines showed a significant reduction in anthocyanin levels (Figure 5C).
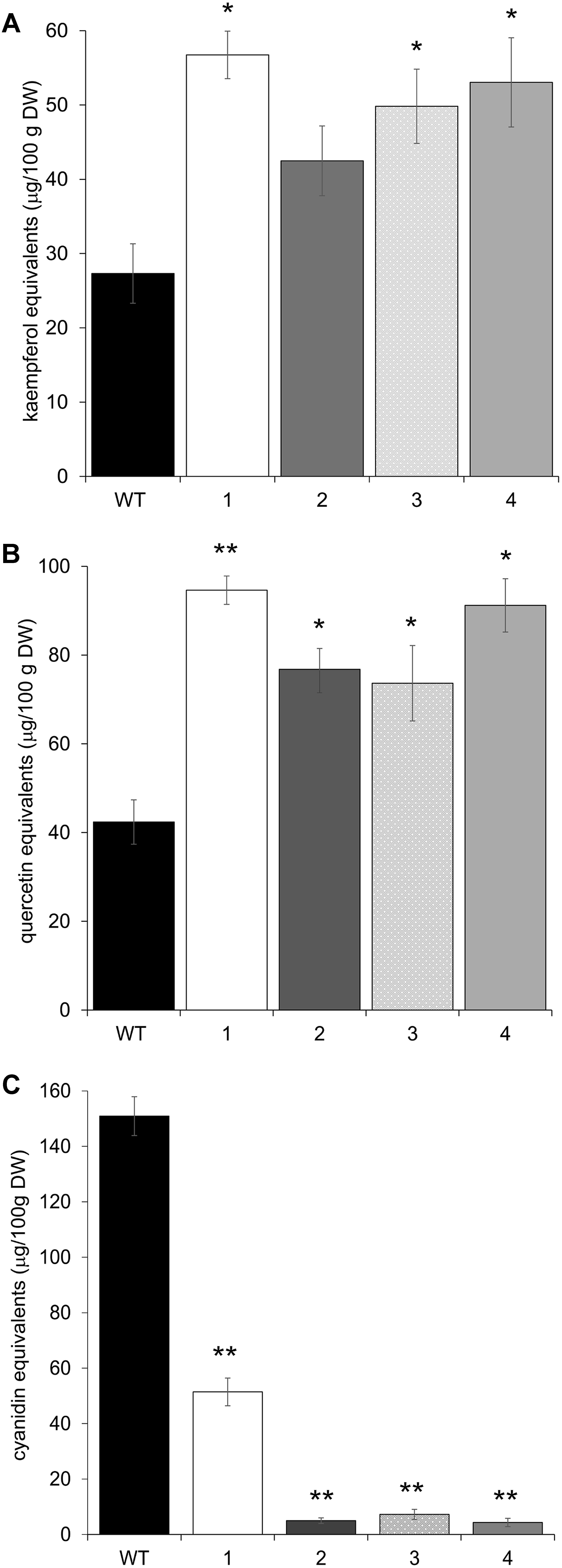
FIGURE 5. HPLC flavonol and anthocyanin quantification in Arabidopsis plants overexpressing CcMYB12 protein. Leaves were analyzed from wild type (WT) plants and from four different Arabidopsis pools (1–4), each constituted by four plants of the same transgenic T3 line. (A) total kaempferols, (B) total quercetins, (C) total anthocyanins. Error bars indicate the SD of the average of quercetin, kaempferol or cyanidin equivalents determined as triplicates in three independent biological replicates. Asterisks ∗ and ∗∗ indicate statistically significant differences (P < 0.05 and P < 0.01 respectively) between the means for WT and tested transgenic samples according to Student’s t-test. DW, dry weight.
To further investigate whether and to what extent CcMYB12 is a negative regulator of anthocyanin biosynthesis in Arabidopsis, WT and CcMYB12 overexpressing lines were grown either on control (1%) or on anthocyanin inductive medium (6% sucrose) (Teng et al., 2005; Solfanelli et al., 2006; Rubin et al., 2009). Under inductive condition anthocyanin production increased, in both WT and transgenic seedlings with respect to the control, but the levels of these metabolites remained significantly lower in CcMYB12 than in WT seedlings (Figure 6A). On anthocyanin inductive medium all the flavonoid early (EBGs) and late (LBGs) biosynthetic genes tested were up-regulated in WT and CcMYB12 seedlings. Nevertheless, while in control condition, CHI, FLS, CHS, F3H, F3′H transcript levels were significantly higher in CcMYB12 than in WT, on 6% sucrose the transcript levels of these genes were not significantly different between WT and CcMYB12 lines (Figure 6B). Interestingly, LBGs showed comparable transcript levels between WT and CcMYB12 transgenic seedlings on 1% sucrose (panel in Figure 6C), whereas they were significant downregulated in the latter samples on high sucrose condition (Figure 6C). The anthocyanin regulatory gene PAP1 showed an expression pattern similar to that of LBGs, whereas the endogenous flavonol regulator AtMYB12 was unaffected by the transgene (Figure 6D).
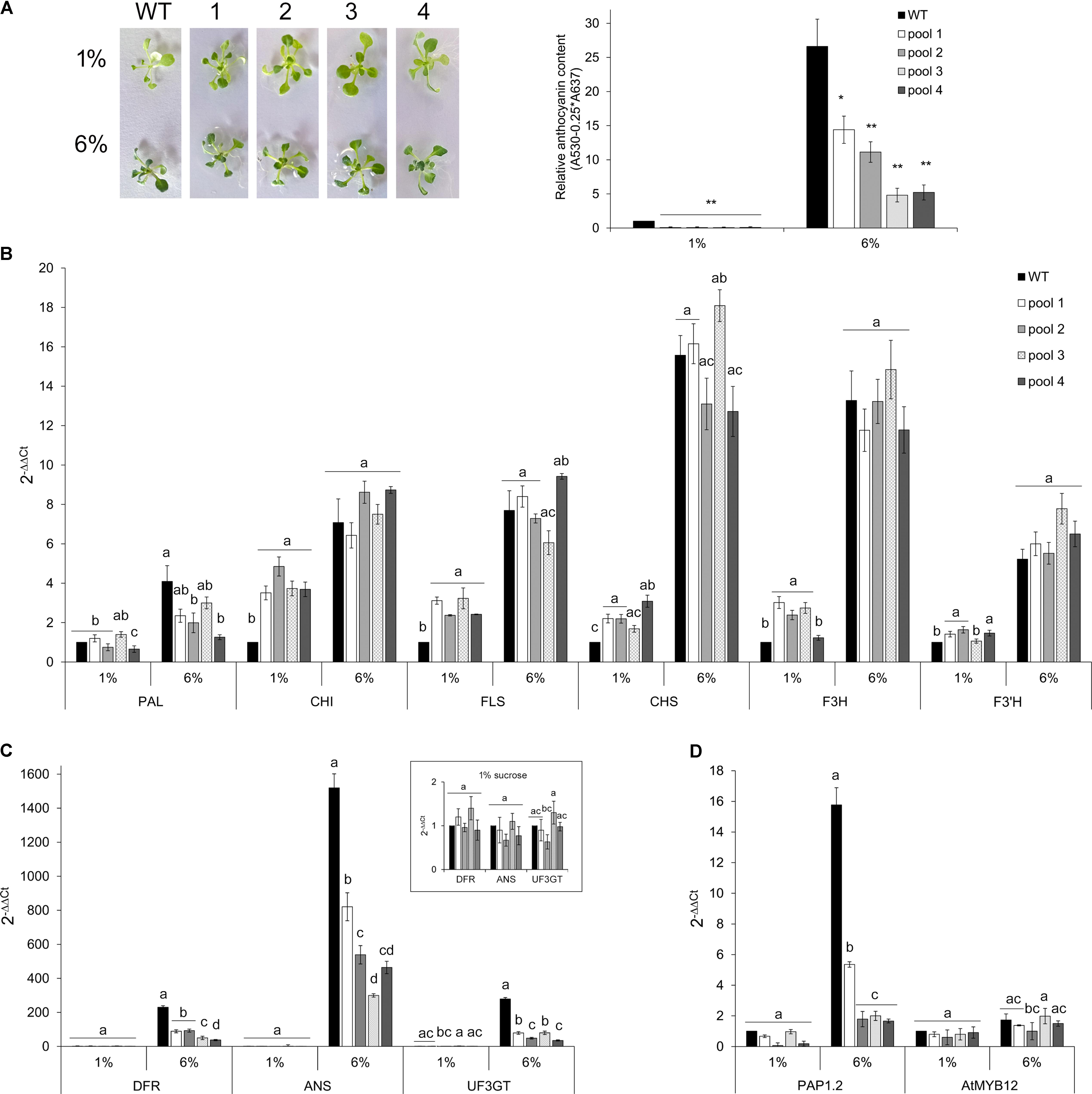
FIGURE 6. Anthocyanin content and qRT-PCR analysis in Arabidopsis seedlings grown on control and anthocyanin inductive medium. (A) Left panel: 20-days-old Arabidopsis wild type (WT) and CcMYB12 seedlings (1, 2, 3, and 4) grown in MS medium supplemented with 1 or 6% sucrose under long day condition. Right panel: Anthocyanin content in 2-weeks-old WT and CcMYB12 seedlings grown in 1 and 6% sucrose. Values are reported as relative to WT 1% sucrose, set as 1. Data represent mean values (+SD, n = 3). Within each growth condition, asterisks ∗ and ∗∗ indicate statistically significant differences (P < 0.05 and P < 0.01 respectively), between the means for WT and tested transgenic samples according to Student’s t-test. (B–D) qRT-PCR analysis of early (B) and late (C) flavonoid biosynthetic genes, and AtPAP1 and AtMYB12 TFs (D) in seedlings grown in 1 and 6% sucrose. Upper panel in (C) represents late biosynthetic genes with a low expression level in 1% sucrose condition, with the appropriate axis scale. Actin was employed as reference gene. WT grown in 1% sucrose is the calibrator. Values are means ± SD of three biological replicates. Bars with different letters are statistically different to each other according to one-way ANOVA Tukey test (p < 0.05).
Tobacco primary transformants and their progenies showed clear pigmentation alterations in fully opened flowers: the floral limbs of CcMYB12 plants were markedly less colored compared to WT plants or tobacco plants carrying the GUS reporter gene, indicating reduced levels of anthocyanins (Figure 7A).
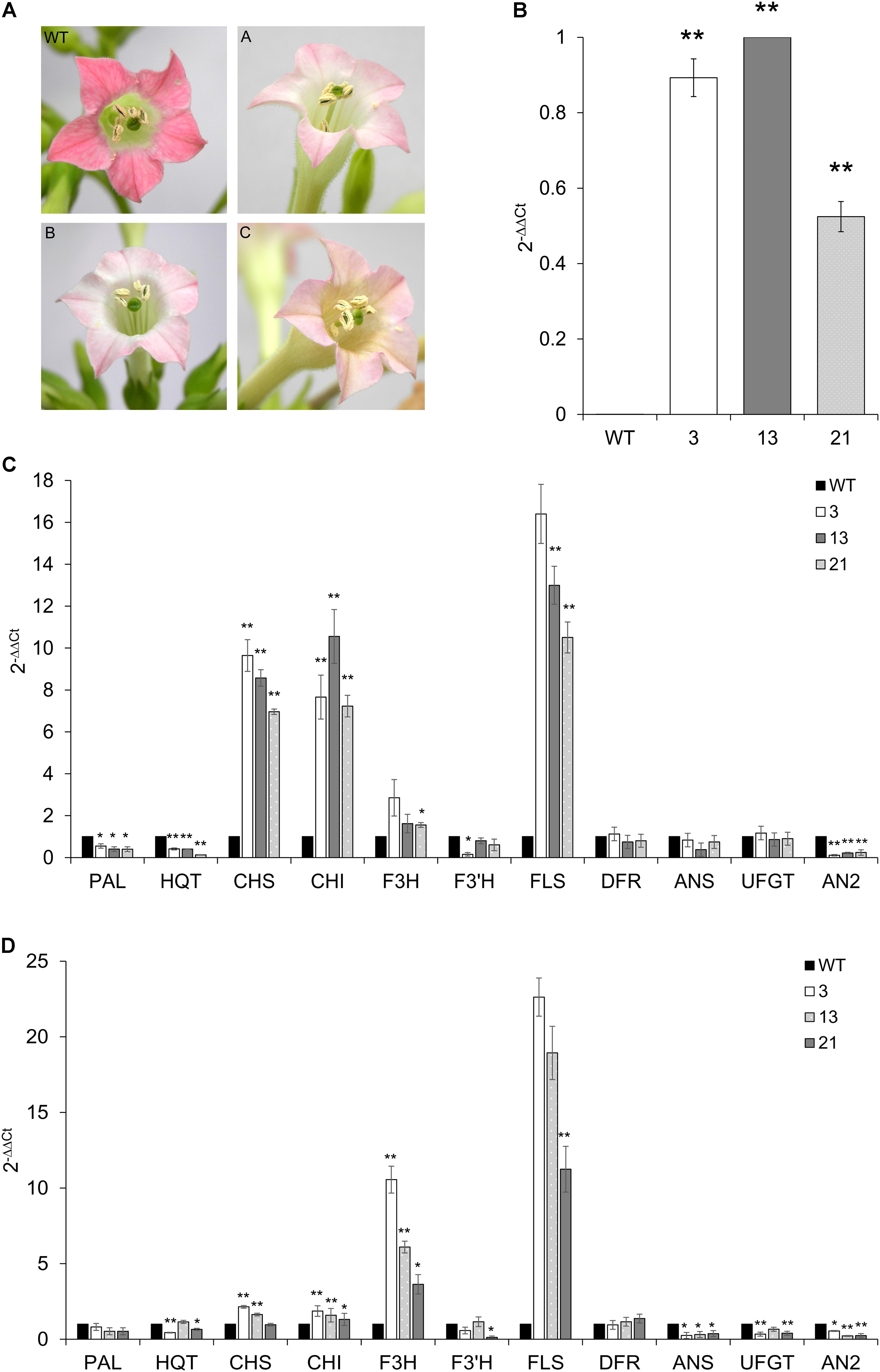
FIGURE 7. Expression of CcMYB12 transgene and of endogenous phenylpropanoid biosynthetic genes in CcMYB12-expressing tobacco plants. (A) Color of tobacco flowers in transgenic CcMYB12 lines. The pigmentation of corolla limbs of three different transgenic lines expressing CcMYB12 was compared with corolla from WT. (A) line 3, (B) line 21, and (C) line 13. (B) qRT-PCR of exogenous CcMYB12 in transgenic tobacco plants. Line 13 as calibrator. (C,D), Expression levels of the early biosynthetic genes, namely PAL, HQT, CHS, CHI, F3H, F3′H, FLS, and ANS, DFR, UFGT and AN2in transgenic tobacco leaves (C) and flowers (D). WT as calibrator. qRT-PCR experiments were conducted on cDNAs from various transgenic tobacco T3 plants. Three independent preparations of total RNA from each line were assayed in triplicate. Actin was employed as reference gene. Values are means ± SD of three biological replicates. Asterisks indicate a statistical difference (∗p-value < 0.05; ∗∗p-value < 0.01) between the means for WT and tested transgenic samples, according to Student’s t-test.
Quantifications of transcripts of phenylpropanoid genes and of flavonoids were performed in WT and T3 tobacco leaves and petals from flowers at stage 12. Three independent T3 transgenic lines were assayed, and all of them showed the expression of CcMYB12, with plants 3 and 13 displaying the highest levels (Figure 7B). As in transgenic Arabidopsis, the expression of CHS, CHI, FLS and, although less markedly, of F3H, increased in tobacco transgenic leaves compared to WT, whereas DFR and ANS (anthocyanin synthase) genes were not affected. PAL, HQT and AN2 (a R2R3-MYB TF that regulates anthocyanin accumulation in tobacco) were significantly downregulated, and F3′H was significantly downregulated in plant 3 only (Figure 7C). In tobacco transgenic flowers, high expression of F3H and FLS were found, CHS and CHI transcripts slightly, but still significantly, increased, while no differences were found in the expression levels of PAL and DFR, with respect to WT (Figure 7D). Furthermore, the expression of CcMYB12 caused a significant downregulation of ANS, UFGT (glucose: flavonoid 3-O-glucosyltransferase) and AN2 in the flowers of all the transgenic plants assayed, whereas HQT and F3′H did not show a trend fully matching with the expression of the transgene (Figure 7D).
HPLC analyses of flavonoids in tobacco leaves showed a significant increase of kaempferol in the transgenic lines, and of quercetin in two of the three tested lines (Figures 8A,B and Supplementary Figure S4). The apigenin 7-O-glucoside content was also increased in all transgenic lines, although significantly only in plant 3 (Supplementary Figure S3), whereas the CQA content, in particular CGA, was similar to WT (Figure 8C and Supplementary Figure S4).
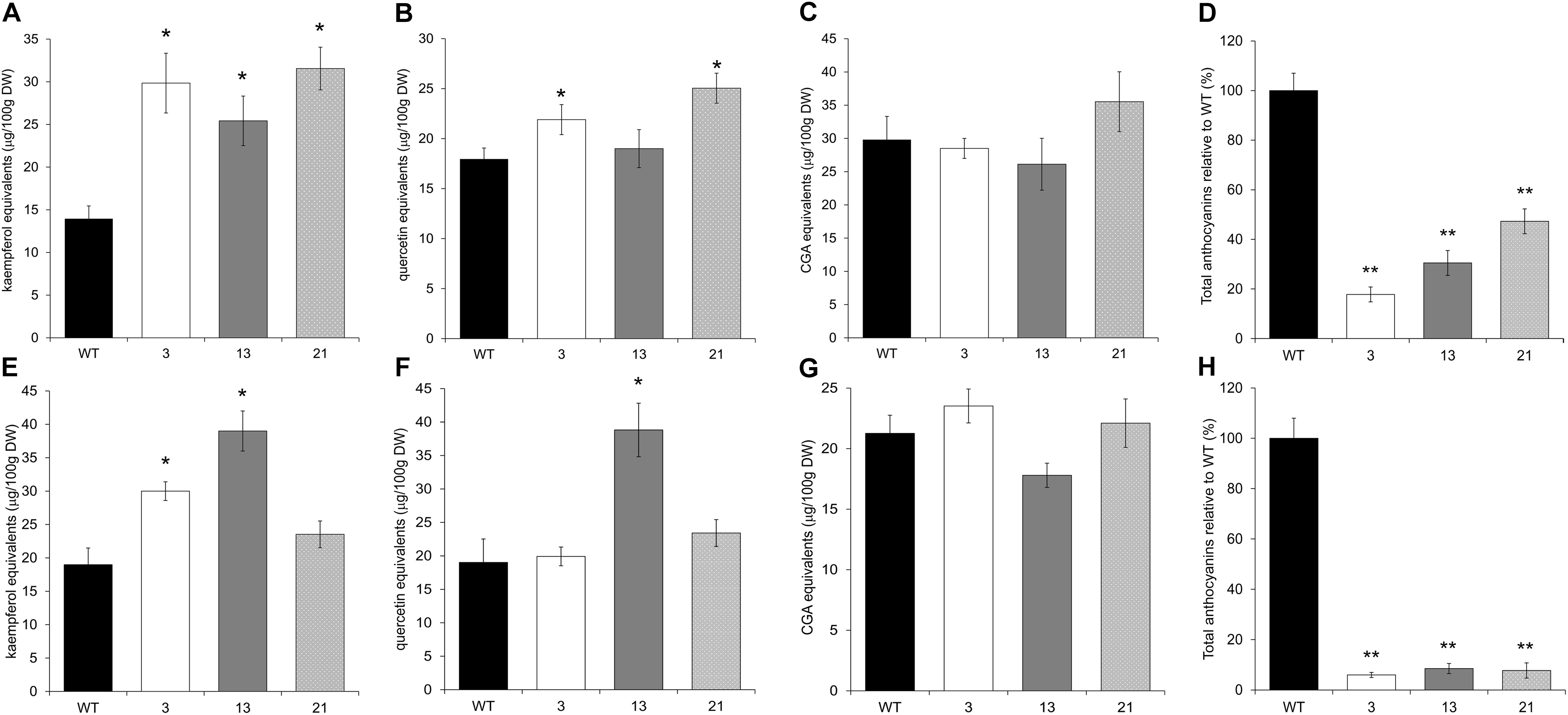
FIGURE 8. HPLC analyses of flavonoids and caffeoyl-quinic acids in leaves and flowers of tobacco plants overexpressing CcMYB12 protein. Leaves (A–D) and flowers (E–H) from WT and T3 transgenic tobacco lines 3, 13, and 21 were tested. Total kaempferols (A,E), total quercetins (B,F), chlorogenic acid (CGA; C), total caffeoyl-quinic acids (CQAs; G), total anthocyanins (D,H). Total anthocyanin content is relative (in percentage) to the mean levels of the WT line. Error bars indicate the SD of the average of kaempferol, quercetin, CGA or cyanidin equivalents, and CGA determined as triplicates in three independent biological replicates. Asterisks indicate a statistical difference (∗p-value < 0.05; ∗∗p-value < 0.01) between the means for WT and for tested transgenic samples, according to Student’s t-test. DW, dry weight.
Fully developed flowers of CcMYB12 transgenic tobacco lines showed a high content of flavonols as kaempferol derivatives when compared to WT flowers, whereas a clear trend did not emerge for quercetin derivatives (Figures 8E,F). Total CQAs were not affected (Figure 8G), while apigenin 7-O-glucoside was not detected in both WT and transgenic flowers.
HPLC analysis revealed that the depigmentation exhibited by CcMYB12 transgenic flowers was due to a severe reduction, up to 90%, of anthocyanin levels if compared to WT flowers (Figure 8H and Supplementary Figure S4). A strong reduction of anthocyanins, spanning from 50 to 80%, was also observed in the leaves of transgenic plants (Figure 8D).
Therefore, the results obtained from qRT-PCR and HPLC analyses suggest that, while promoting the accumulation of flavonols, mainly kaempeferol derivatives, CcMYB12 interferes with anthocyanin accumulation in both Arabidopsis and tobacco heterologous models.
Direct consumption of artichoke heads represents an important source of polyphenols, inulin and flavonoids, healthy compounds with antioxidant and therapeutic activity. Artichoke leaves are also rich in these metabolites. The profiling of nutraceuticals, their qualitative - quantitative variation according to environmental conditions on artichoke landraces, genotypes and plant organs are issues which have been approached only recently (Lombardo et al., 2010; Negro et al., 2012; de Falco et al., 2016). A fine metabolic characterization is the prerequisite for programs of germplasm preservation, selection and propagation of this healthy Mediterranean crop.
Regulation of phenylpropanoid accumulation is achieved at the transcription level, and R2R3-MYB proteins act as flavonoid transcriptional regulators (Liu et al., 2015). The first R2R3-MYB controlling flavonol biosynthesis, MYB12, has been characterized in Arabidopsis, where AtMYB12 mainly controls flavonol production, acting as a regulator on CHS, CHI, F3H and FLS (Mehrtens et al., 2005; Stracke et al., 2007). AtMYB12 shows a high degree of functional similarity to AtMYB11 and AtMYB111, with differential spatial activities (Stracke et al., 2007). Interestingly, AtMYB12 overexpression in other plant species emphasized the complex regulation of plant secondary metabolism. If AtMYB12 overexpression in tomato leads to flavonol and CGA accumulation (Luo et al., 2008), in kale it triggers over accumulation of total phenolics and flavonols (Lännenpää, 2014), while in tobacco plants and cell cultures its influence is restricted to flavonol and rutin accumulation (Luo et al., 2008; Misra et al., 2010). Moreover, characterization of endogenous MYB12 TF in plants as grape (VvMYBF1, Czemmel et al., 2009; Czemmel et al., 2017), tomato (SlMYB12, Ballester et al., 2010) and gentian (GtMYBP3 and GtMYBP4, Nakatsuka et al., 2012) showed how flavonoid accumulation, their compositional diversity and targeted biosynthetic genes could be differentially regulated across plant species. Hence, in order to improve flavonoid content and composition, there is an effective need to examine regulatory mechanisms of flavonoid biosynthesis, species by species. In face of its relevance for the Mediterranean diet, only genes related to chlorogenic acid (CGA) biosynthesis and early phenylpropanoid biosynthetic pathways have been investigated in artichoke thus far (Comino et al., 2007, 2009; De Paolis et al., 2008; Moglia et al., 2009; Sonnante et al., 2010; De Palma et al., 2014). Hence, in this study we aimed at identifying and characterizing MYB12 from artichoke, as one of the main regulators of flavonol biosynthesis.
Plant MYB factors can be categorized into subgroups, based on Arabidopsis conserved amino acid motifs previously detected in the C-terminus, outside the MYB domain (Stracke et al., 2001). These subgroups may cluster factors that share the same function. Our phylogenetic analysis reveals that CcMYB12 joins the clade of putative flavonol MYB factors from different plant species. It shows high level of similarity to the Arabidopsis flavonol regulator AtMYB111, to the functionally redundant AtMYB12 and AtMYB11, and to grape VvMYBF1, gerbera GhMYB1, gentian GtMYBP3, tomato SlMYB12 and maize ZmP1.
A detailed analysis of protein conserved domains shows that CcMYB12 owns key amino acid residues, within the canonical R2R3 repeat domains, probably involved in target promoter specificity for flavonol biosynthesis (i.e., Gly52, E77, and DNEI103-106), also conserved in AtMYB12 and its homologs. CcMYB12 lacks the bHLH binding site for interaction with bHLH proteins (Zimmermann et al., 2004), as other plant flavonol specific MYB TF, thus showing cofactor independency. Although the sequence similarity between R2R3-MYB proteins is generally confined to the N-terminus, the CcMYB12 presents a motif (GRVSRCVAK) outside the R2R3 domains that shares 67% identity with the SG7 motif (GRTxRSxMK). This motif typifies R2R3-MYB proteins that regulate flavonol biosynthesis (Stracke et al., 2001). These evidences are in support of the hypothesis that CcMYB12 is indeed a MYB TF involved in regulation of flavonol biosynthesis. This hypothesis is also corroborated by transcriptional data, as they reveal that CcMYB12 is highly expressed in artichoke intermediate and internal bracts, and in young leaves. The intermediate bracts, where CcMYB12 is more expressed, show high levels of the known biosynthetic flavonoid genes HQT1, HQT2, C3′H, HCT and to a lesser extent of PAL3a. Although the correlation of CcMYB12 expression with biosynthetic flavonoid gene levels and flavonoid accumulation in artichoke tissues cannot be established directly, as not all structural genes have been isolated and characterized yet, transcriptional and metabolic data seem to overlap quite nicely. We note in fact that artichoke heads are very rich in caffeoylquinic acids and in some apigenin derivatives (apigenin, luteolin, naringenin and their related glycosides), mainly in receptacles and inner bracts (Fratianni et al., 2007; Negro et al., 2012). Anthocyanins (specifically cyanidin 3-O-β-glucoside) are present in the water-soluble fractions of artichoke head extracts (de Falco et al., 2016), whereas flavonoids are more concentrated in leaves (Lattanzio et al., 2009; Negro et al., 2012; de Falco et al., 2016). Accordingly, CcMYB12 is expressed both in flavonoid rich leaves and in capitula tissues, the latter characterized by high levels of caffeoylquinic acids and apigenin derivatives.
The maize P protein (Grotewold et al., 1994) and more recently AtMYB11, AtMYB12 and AtMYB111 (Stracke et al., 2007) were shown to bind to ACC(A/T)ACC(A/C/T) motifs, commonly referred to as MYBPLANT, P-box, MRE or AC-elements (Patzlaff et al., 2003; Prouse and Campbell, 2012). Such elements are present in the promoter regions of artichoke phenylpropanoid genes such as HQT1, HQT2, PAL3a and C3′H (De Paolis et al., 2008; Sonnante et al., 2010) as well as in the promoter of CcMYB12 itself, as reported here. We show that the recombinant protein CcMYB12 specifically recognizes the ACII probe containing the MYBPLANT motif “ACCAACC”, whereas it does not bind to the other probe tested (ACI: ACCTACC). Although, data on the interaction between MYBs and their target DNAs could differ moving from in vitro to in vivo analyses (Prouse and Campbell, 2012), we interpret the results of our in vitro mobility shift assay to mean that CcMYB12 is indeed a TF and that MYB12 TFs from different plant species retain a different specificity toward target DNA motifs. Thus, the transcriptional profiles of early biosynthetic genes and in vitro EMSA convey to suggest a role of CcMYB12 in controlling flavonol biosynthesis in artichoke.
Being artichoke recalcitrant to genetic transformation, direct evidence on the role of CcMYB12 cannot be gained. To overcome this problem, we ectopically expressed CcMYB12 into the model systems Arabidopsis and tobacco, and performed targeted transcriptional and metabolic analyses on Arabidopsis and tobacco leaves and tobacco flowers. Differently from Arabidopsis, tobacco produces CGA (Tamagnone et al., 1998; Niggeweg et al., 2004) and its flowers accumulate the flavonols quercetin and kaempferol in the form of various glycosides (Luo et al., 2007; Passeri et al., 2017) as well as anthocyanin derivatives, being cyanidin 3-O-rutinoside the dominant anthocyanin (Aharoni et al., 2001; Luo et al., 2007; Nakatsuka et al., 2007; Passeri et al., 2017).
The main effects shared by Arabidopsis leaves, tobacco leaves and flowers as a result of CcMYB12 ectopic expression consist in the metabolic flux diversion from anthocyanins to flavonols, which is conceivably mainly due to the upregulation of FLS. With a few exceptions (see below), these effects resemble those reported in the same model systems as a consequence of the ectopic expression of AtMYB12, to suggest that CcMYB12 is a putative homologue of AtMYB12.
The expression of CcMYB12 induces the upregulation of the early biosynthetic genes CHS, CHI, F3H and FLS both in Arabidopsis and tobacco leaves. The co-regulation of multiple genes of the flavonoid pathway was previously reported as a result of the overexpression of AtMYB12 in different species such as Arabidopsis, tobacco and tomato (Hartmann et al., 2005; Stracke et al., 2007; Luo et al., 2008; Misra et al., 2010; Pandey et al., 2015). CHS, CHI, F3H and FLS genes were fully characterized in Arabidopsis; they carry identical cis-elements in their promoters which are required for flavonol biosynthesis, including the MYBPLANT motif, also recognized by the newly identified artichoke MYB TF. Differently from what observed in Arabidopsis and tobacco leaves, CcMYB12 overexpression does not induce CHS expression in the flowers of all transgenic plants.
As stated above, few differences characterize CcMYB12 with respect to AtMYB12 function, both in target gene regulation and metabolite profiles of transgenic plants. Firstly, a previous study showed that both AtMYB12 Arabidopsis gain- and loss-of-function mutants do not display seedlings with altered content of anthocyanins with respect to WT in culture medium with no sucrose (Mehrtens et al., 2005), whereas here we show that the levels of these metabolites decrease in CcMYB12 transgenic Arabidopsis leaves as well as in transgenic seedlings, regardless of the sucrose concentration. Moreover, AtMYB12 does not appear to induce F3′H gene expression strongly (Mehrtens et al., 2005; Luo et al., 2008), while CcMYB12 does on leaves and, although not in all transgenic lines, on seedlings too. In tobacco, the inverse correlation between anthocyanins and flavonols in AtMYB12 transgenic flowers was previously interpreted as the result of the competition between DFR and FLS for dihydroflavonol precursors (Luo et al., 2008). Likewise, we argue that the flux diversion between flavonols and anthocyanins in CcMYB12 transgenic tobacco leaves and flowers is also accountable to the competition between FLS and DFR, being FLS gene upregulated and DFR unaffected in these organs by the transgene. Such a competition occurs particularly in floral limbs where CcMYB12 expression leads to a severe reduction in ANS steady state levels. Despite this parallelism, AtMYB12 and CcMYB12 functions also diverge in tobacco: while AtMYB12 increases the levels of CQA, the levels of these metabolites are unaffected both in CcMYB12 transgenic leaves and flowers. In the absence of a clear effect of AtMYB12 on genes encoding HQTs and C3′H, the CQA increment in AtMYB12 transgenic tobacco flowers was previously accounted to the induction of PAL (Luo et al., 2008). Thus, embracing the assumption that PAL activity limits flux through phenylpropanoid metabolism in tobacco (Howles et al., 1996), in accordance with Luo and colleagues we reason that the lack of CQA increment in CcMYB12 transgenic tobacco is due to the inability of this TF in promoting the upregulation of PAL as well as of HQT. Better still, we note that the levels of CGA in transgenic tobacco leaves remain unaltered with respect to the WT, despite the fact that both PAL and HQT are downregulated by CcMYB12.
As stated above, in Arabidopsis leaves CcMYB12 not only clearly increases the steady state levels of CHS, CHI and FLS, but also that of F3′H, which is necessary for the production of quercetin. In turn, both kaempferol and quercetin derivatives are concomitantly increased in transgenic Arabidopsis plants. Conversely, in transgenic tobacco plants we observe a more variegated patter concerning these flavonols, since both in leaves and flowers kaempferol derivatives are increased, whereas the increment of quercetin derivatives is less marked and it does not occur in the leaves and flowers of all CcMYB12 transgenic lines. This is consistent with the qRT-PCR data showing that in tobacco F3′H is not among the CcMYB12 primary target genes. Incidentally, we note that a different and organ-specific regulatory mechanism between the kaempferol and quercetin branches has been highlighted in tobacco flowers as a consequence of the ectopic expression of the proanthocyanidin regulator MYBPA1 from grape (Passeri et al., 2017). Also the different effect exerted in tomato on the production of kaempferol and quercetin type flavonol by the MYBs genes C1 or Lc vs. AtMYB12, was attributed to the different ability of these TFs in inducing the endogenous F3′H gene (Bovy et al., 2002; Luo et al., 2008).
In tobacco, in addition to the flux diversion from anthocyanins to flavonols, CcMYB12 triggers changes in the levels of the flavone apigenin. This is the major flavone present in tobacco leaves and in developing tobacco flowers (Nakatsuka et al., 2006; De Palma et al., 2014). In keeping with this observation, we did not detect apigenin in fully developed flowers (stage 12), however its content increased in the CcMYB12 transgenic leaves if compared to WT, likely as a result of the marked upregulation of CHS and CHI in these organs. Moreover, it negatively regulates UFGT and AN2.
Finally, we note that in Arabidopsis seedlings the decrease in anthocyanins in CcMYB12 overexpressing lines might be accountable to two different, although not mutually exclusive, mechanisms depending on growing conditions: under control condition CcMYB12 promotes FLS upregulation and consequently the flux diversion from anthocyanins to flavonols. Conversely, under anthocyanin inductive condition, CcMYB12 seems to directly interfere with the steady state levels of PAP1, one of the main regulators of the biosynthesis of these pigments and, in turn, it negatively regulates key LBGs such as DFR, ANS and UFGT. Further analyses will be needed to prove this hypothesis and disentangle the mechanism by which CcMYB12 represses PAP1.
In sum, through the isolation of CcMYB12 and its expression in heterologous models, here we shed light on a regulator of flavonol biosynthesis from the globe artichoke, producing an accumulation of flavonols and a reduction of anthocyanins. The differences between host plants (Arabidopsis and tobacco), plant organs (tobacco leaves vs. flowers) and plant growing conditions (anthocyanin inductive vs. control condition) in the sets of the genes targeted by CcMYB12 suggest a different, species- and organ- specific and environmental dependent – capacity of this TF to interact with endogenous genes of the flavonoid pathway. Likewise, in artichoke the interaction of CcMYB12 with other endogenous genes as well as its expression profiles might vary according to the organs and in response to different stimuli and/or developmental stages.
Further analyses on CcMYB12 expression profiles and its interaction with flavonoid regulatory and biosynthetic genes in different organs and under different plant growing conditions will advance our understanding on genetic and environmental determinants underlying flavonol biosynthesis in artichoke. This is the prerequisite to develop agronomic and biotechnological strategies aimed at selecting and breeding artichoke germplasm with enhanced levels of healthy compounds.
EB and GS conceived and designed the research. EB, WS, DD, DN, and VP performed the experiments. EB, WS, DD, DN, ADL, FP, and GS analyzed and interpreted the data. EB, FP, and GS contributed to reagents, materials, and analysis tools. EB, FP, and GS wrote and revised the paper.
This research was funded by the project BiodiverSO (Biodiversità delle Specie Orticole della Puglia) – PSR Puglia 2007–2013 Mis. 214/4 subaz. a), BiodiverSO – PSR Puglia 2014-2020 Mis.10.2, and by a dedicated grant from the Italian Ministry of Economy and Finance to the National Research Council for the project “Innovazione e Sviluppo del Mezzogiorno – Conoscenze Integrate per Sostenibilità ed Innovazione del Made in Italy Agroalimentare (C.I.S.I.A.)” – Legge n. 191/2009.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Anita Morgese for laboratory assistance in qRT-PCR expression analyses.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00941/full#supplementary-material
CGA, chlorogenic acid; CQAs, caffeoyl-quinic acids; TF, transcription factor.
Agarwal, R., and Mukhtar, H. (1996) Cancer chemoprevention by polyphenols in green tea and artichoke. Adv. Exp. Med. Biol. 401, 35–50. doi: 10.1007/978-1-4613-0399-2_4
Aharoni, A., De Vos, C. H., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
Azzini, E., Bugianesi, R., Romano, F., Di Venere, D., Miccadei, S., Durazzo, A., et al. (2007). Absorption and metabolism of bioactive molecules after oral consumption of cooked edible heads of Cynara scolymus L. (cultivar violetto di provenza) in human subjects: a pilot study. Br. J. Nutr. 97, 963–969. doi: 10.1017/S0007114507617218
Ballester, AR., Molthoff, J., de Vos, R., Hekkert, Bt., Orzaez, D., Fernández-Moreno, JP., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. doi: 10.1104/pp.109.147322
Bovy, A., de Vos, R., Kemper, M., Schijlen, E., Almenar Pertejo, M., Muir, S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes Lc and C1. Plant Cell 14, 2509–2526. doi: 10.1105/tpc.004218
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brown, J. E., and Rice-Evans, C. A. (1998). Luteolin-rich artichoke extract protects low density lipoprotein from oxidation in vitro. Free Radic. Res. 29, 247–255. doi: 10.1080/10715769800300281
Bustin, S. A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193. doi: 10.1677/jme.0.0250169
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Comino, C., Hehn, A., Moglia, A., Menin, B., Bourgaud, F., Lanteri, S., et al. (2009). The isolation and mapping of a novel hydroxycinnamoyltransferase in the globe artichoke chlorogenic acid pathway. BMC Plant Biol. 9:30. doi: 10.1186/1471-2229-9-30
Comino, C., Lanteri, S., Portis, E., Acquadro, A., Romani, A., Hehn, A., et al. (2007). Isolation and functional characterization of a cDNA coding a hydroxycinnamoyltransferase involved in phenylpropanoid biosynthesis in Cynara cardunculus L. BMC Plant Biol. 7:14. doi: 10.1186/1471-2229-7-14
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
Czemmel, S., Höll, J., Loyola, R., Arce-Johnson, P., Alcalde, J. A., Matus J. T., et al. (2017). Transcriptome-wide identification of novel uv-b- and light modulated flavonol pathway genes controlled by VviMYBF1. Front. Plant Sci. 8:1084. doi: 10.3389/fpls.2017.01084
de Falco, B., Incerti, G., Pepe, R., Amato, M., and Lanzotti, V. (2016). Metabolomic fingerprinting of romaneschi globe artichokes by nmr spectroscopy and multivariate data analysis. Phytochem. Anal. 27, 304–314. doi: 10.1002/pca.2632
De Palma, M., Fratianni, F., Nazzaro, F., and Tucci, M. (2014). Isolation and functional characterization of a novel gene coding for flavonoid 3′-hydroxylase from globe artichoke. Biol. Plant. 58, 445–455. doi: 10.1007/s10535-014-0424-7
De Paolis, A., Pignone, D., Morgese, A., and Sonnante, G. (2008). Characterization and differential expression analysis of artichoke phenylalanine ammonia-lyase coding sequences. Physiol. Plant. 132, 33–43.
Fratianni, F., Tucci, M., De Palma, M., Pepe, R., and Nazzaro, F. (2007). Polyphenolic composition in different parts of some cultivars of globe artichoke (Cynara cardunculus L. var. scolymus (L.) Fiori). Food Chem. 104, 1282–1286. doi: 10.1016/j.foodchem.2007.01.044
Gatto, A., De Paola, D., Bagnoli, F., Vendramin, G. G., and Sonnante, G. (2013). Population structure of Cynara cardunculus L. complex and the origin of the conspecific crops artichoke and cardoon. Ann. Bot. 112, 855–865. doi: 10.1093/aob/mct150
Gebhardt, R. (1997). Antioxidative and protective properties of extracts from leaves of artichoke (Cynara scolymus L.) against hydroperoxide-induced oxidative stress in cultured rat hepatocytes. Toxicol. Appl. Pharmacol. 144, 279–286. doi: 10.1006/taap.1997.8130
Graf, B. A., Milbury, P. E., and Blumberg, J. B. (2005) Flavonols, flavones, flavanones, and human health: epidemiological evidence. J. Med. Food 8, 281–290. doi: 10.1089/jmf.2005.8.281
Grotewold, E., Drummond, B. J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. doi: 10.1016/0092-8674(94)90117-1
Grotewold, E. (2008). Transcription factors for predictive plant metabolic engineering: are we there yet? Curr. Opin. Biotechnol. 19, 138–144. doi: 10.1016/j.copbio.2008.02.002
Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., and Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171. doi: 10.1007/s11103-004-6910-0
Heppel, S. C., Jaffè, F. W., Takos, A. M., Schellmann, S., Rausch, T., Walker, A. R., et al. (2013). Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 82, 457–471. doi: 10.1007/s11103-013-0074-8
Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for agrobacterium transformation. Nucleic Acids Res. 16, 9877–9877. doi: 10.1093/nar/16.20.9877
Horsch, R. B., Rogers, S. G., and Fraley, R. T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. doi: 10.1101/SQB.1985.050.01.054
Howles, P. A., Sewalt, V., Paiva, N. L., Elkind, Y., Bate, N. J., Lamb, C., et al. (1996). Overexpression of l-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol. 112, 1617–1624. doi: 10.1104/pp.112.4.1617
Karimi, M., Inze, D., and Depicker, A. (2002). Gateway vectors for Agrobacterium mediated plant transformation. Trends Plant Sci. 5, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Koltunow, A. M., Truettner, J., Cox, K. H., Wallroth, M., and Goldberg, R. B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2, 1201–1224. doi: 10.1105/tpc.2.12.1201
Korkina, L. G. (2007). Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell. Mol. Biol. 53, 15–25.
Kraft, K. (1997). Artichoke leaf extract: recent findings reflecting effects on lipid metabolism, liver and gastrointestinal tracts. Phytomedicine 4, 369–378. doi: 10.1016/S0944-7113(97)80049-9
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lännenpää, M. (2014). Heterologous expression of AtMYB12 in kale (Brassica oleracea var. acephala) leads to high flavonol accumulation. Plant Cell Rep. 33, 1377–1388. doi: 10.1007/s00299-014-1623-6
Lattanzio, V., Kroon, P. A., Linsalata, V., and Cardinali, A. (2009). Globe artichoke: a functional food and source of nutraceutical ingredients. J. Funct. Foods 1, 131–144. doi: 10.1016/j.jff.2009.01.002
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Lombardo, S., Pandino, G., Mauromicale, G., Knödler, M., Carle, R., and Schieber, A. (2010). Influence of genotype, harvest time and plant part on polyphenolic composition of globe artichoke (Cynara cardunculus L. var. scolymus (L.) Fiori). Food Chem. 119, 1175–1181. doi: 10.1016/j.foodchem.2009.08.033
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. doi: 10.1111/j.1365-313X.2008.03597.x
Luo, J., Nishiyama, Y., Fuell, C., Taguchi, G., Elliott, K., Hill, L., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50, 678–695. doi: 10.1111/j.1365-313X.2007.03079.x
Mahmood, K., Xu, Z., El-Kereamy, A., Casaretto, J. A., and Rothstein, S. J. (2016). The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 7:1548. doi: 10.3389/fpls.2016.01548
McDougall, B., King, P. J., Wu, B. W., Hostomsky, Z., Manfred, G., and Robinson, W. E. Jr. (1998). Dicaffeoylquinic acid and dicaffeoyltartaric acid are selective inhibitors of human immunodeficiency virus type 1 integrase. Antimicrob. Agents Chemother. 42, 140–146.
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Misra, P., Pandey, A., Tiwari, M., Chandrashekar, K., Sidhu, O. P., Asif, M. H., et al. (2010). Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 152, 2258–2268. doi: 10.1104/pp.109.150979
Moglia, A., Comino, C., Portis, E., Acquadro, A., De Vos, R. C., Beekwilder, J., et al. (2009). Isolation and mapping of a C3′H gene (CYP98A49) from globe artichoke, and its expression upon UV-C stress. Plant Cell Rep. 28, 963–974. doi: 10.1007/s00299-009-0695-1
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nakatsuka, T., Nishihara, M., Mishiba, K., and Yamamura, S. (2006). Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol. Breed. 17, 91–99. doi: 10.1007/s11032-005-2520-z
Nakatsuka, T., Abe, Y., Kakizaki, Y., Yamamura, S., and Nishihara, M. (2007). Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. Plant Cell Rep. 26, 1951–1959. doi: 10.1007/s00299-007-0401-0
Nakatsuka, T., Saito, M., Yamada, E., Fujita, K., Kakizaki, Y., and Nishihara, M. (2012). Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J. Exp. Bot. 63, 6505–6517. doi: 10.1093/jxb/ers306
Negro, D., Montesano, V., Grieco, S., Crupi, P., Sarli, G., De Lisi, A., et al. (2012). Polyphenol compounds in artichoke plant tissues and varieties. J. Food Sci. 77, C244–C252. doi: 10.1111/j.1750-3841.2011.02531.x
Niggeweg, R., Michael, A. J., and Martin, C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746–754. doi: 10.1038/nbt966
Ortega, R. (2006). Importance of functional foods in the Mediterranean diet. Public Health Nutr. 9, 1136–1140. doi: 10.1017/S1368980007668530
Palmieri, L., Santoro, A., Carrari, F., Blanco, E., Nunes-Nesi, A., Arrigoni, R., et al. (2008). Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis. Plant Physiol. 148, 1797–1808. doi: 10.1104/pp.108.130310
Pandey, A., Misra, P., and Trivedi, P. K. (2015). Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L. J., Smith, C., et al. (2003). Characterization of a pine MYB that regulates lignification. Plant J. 36, 743–754. doi: 10.1046/j.1365-313X.2003.01916.x
Passeri, V., Martens, S., Carvalho, E., Bianchet, C., Damiani, F., and Paolocci, F. (2017). The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathwy in tobacco leaves. Planta 246, 185–199. doi: 10.1007/s00425-017-2667-y
Prouse, M. B., and Campbell, M. M. (2012). The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 1819, 67–77. doi: 10.1016/j.bbagrm.2011.10.010
Ravishankar, D., Rajora, A. K., Greco, F., and Osborn, H. M. (2013). Flavonoids as prospective compounds for anti-cancer therapy. Int. J. Biochem. Cell Biol. 45, 2821–2831. doi: 10.1016/j.biocel.2013.10.004
Rice-Evans, C. A., Miller, N. J., and Paganga, G. (1997). Antioxidant properties of phenolic compounds. Trends Plant Sci. 2, 152–159. doi: 10.1016/S1360-1385(97)01018-2
Rubin, G., Tohge, T., Matsuda, F., Saito, K., and Scheible, W.-R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. doi: 10.1105/tpc.109.067041
Shimoda, H., Ninomiya, K., Nishida, N., Yoshino, T., Morikawa, T., Matsuda, H., et al. (2003) Anti-hyperlipidemic sesquiterpenes and new sesquiterpene glycosides from the leaves of artichoke (Cynara scolymus L.): structure requirement and mode of action. Bioorg. Med. Chem. Lett. 13, 223–228. doi: 10.1016/S0960-894X(02)00889-2
Sofi, F., Macchi, C., Abbate, R., Gensini, G. F., and Casini, A. (2013). Mediterranean diet and health. Biofactors 39, 335–342. doi: 10.1002/biof.1096
Solfanelli, C., Poggi, A., Loreti, E., Alpi, A., and Perata, P. (2006). Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140, 637–646. doi: 10.1104/pp.105.072579
Sonnante, G., D’Amore, R., Blanco, E., Pierri, C. L., De Palma, M., Luo, J., et al. (2010). Novel hydroxycinnamoyl- coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol. 153, 1224–1238. doi: 10.1104/pp.109.150144
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F. A., Roberts, K., et al. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10, 135–154. doi: 10.1105/tpc.10.2.135
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). Mega6: molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Trivedi, R., Kumar, A., Gupta, V., Kumar, S., Nagar, G. K., Romero, J. R., et al. (2009). Effects of Egb 761 on bone mineral density, bone microstructure, and osteoblast function: possible roles of quercetin and kaempferol. Mol. Cell Endocrinol. 302, 86–91. doi: 10.1016/j.mce.2009.01.011
Ververidis, F., Trantas, E., Douglas, C., Vollmer, G., Kretzschmar, G., and Panopoulos, N. (2007). Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part II: reconstruction of multienzyme pathways in plants and microbes. Biotechnol. J. 2, 1235–1249. doi: 10.1002/biot.200700184
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, W., Grain, D., Le Gourrierec, J., Harscoët, E., Berger, A., Jauvion, V., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198, 59–70. doi: 10.1111/nph.12142
Zhua, X. F., Zhanga, H. X., and Lob, R. (2005). Antifungal activity of Cynara scolymus L. extracts. Fitoterapia 76, 108–111. doi: 10.1016/j.fitote.2004.10.016
Keywords: artichoke, flavonoid biosynthesis, flavonol, flower color, healthy compounds, qRT-PCR, R2R3-MYB, transcription factor
Citation: Blanco E, Sabetta W, Danzi D, Negro D, Passeri V, De Lisi A, Paolocci F and Sonnante G (2018) Isolation and Characterization of the Flavonol Regulator CcMYB12 From the Globe Artichoke [Cynara cardunculus var. scolymus (L.) Fiori]. Front. Plant Sci. 9:941. doi: 10.3389/fpls.2018.00941
Received: 15 January 2018; Accepted: 12 June 2018;
Published: 04 July 2018.
Edited by:
Kevin Davies, The New Zealand Institute for Plant & Food Research Limited, New ZealandReviewed by:
José Tomás Matus, Centre for Research in Agricultural Genomics (CRAG), SpainCopyright © 2018 Blanco, Sabetta, Danzi, Negro, Passeri, De Lisi, Paolocci and Sonnante. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emanuela Blanco, ZW1hbnVlbGEuYmxhbmNvQGliYnIuY25yLml0
† Present address: Wilma Sabetta, Spin Off Sinagri, Università degli Studi di Bari “Aldo Moro”, Bari, Italy Valentina Passeri, Plant Development and (Epi)Genetics, Swammerdam Institute for Life Science, University of Amsterdam, Amsterdam, Netherlands
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.