
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 June 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00882
 Shou-Hong Zhu1
Shou-Hong Zhu1 Fei Xue1
Fei Xue1 Yan-Jun Li1
Yan-Jun Li1 Feng Liu1
Feng Liu1 Xin-Yu Zhang1
Xin-Yu Zhang1 Lan-Jie Zhao2
Lan-Jie Zhao2 Yu-Qiang Sun3
Yu-Qiang Sun3 Qian-Hao Zhu4*
Qian-Hao Zhu4* Jie Sun1*
Jie Sun1*Cytoplasmic linker-associated proteins (CLASPs) are microtubule-associated proteins (MAPs) involved in regulation of dynamics of microtubules (MTs) that play an important role in plant growth and development. In this study, we identified cotton CLASP genes and investigated the function of GhCLASP2. GhCLASP2 was mainly expressed in stem and developing fibers, especially in fibers of the secondary cell wall deposition stage. Ectopic expression of GhCLASP2 in Arabidopsis increased the branching number of leaf trichomes and rescued the defective phenotypes of clasp-1. In cotton, overexpression of GhCLASP2 increased fiber strength, probably related to enhanced expression levels of tubulin, cellulose synthase, and expansin genes. Suppression of GhCLASP2 caused shorter internodes and semi-dwarfism, abnormal flower stigma, aborted anthers without pollen grains, and sterility. These changed phenotypes were similar to those observed in the Arabidopsis clasp-1 mutant. GhCLASP2 was co-localized with MTs according to transient experiment. These results suggest that GhCLASP2 functions similarly as AtCLASP, acting as a MAP and controlling cotton growth and development by regulating MTs.
As a component of the cytoskeleton in eukaryotic cells, microtubules (MTs) play a vital role in cell life, such as cell morphogenesis and maintenance, intracellular transport, cell migration, cell division, and signal transduction. Their functions are accomplished with a variety of MT-associated proteins (MAPs). Interaction between MTs and MAPs regulates the aggregation and depolymerization of MTs, the orientation changes of MT arrays, and the dynamic activities of MTs through spatial organization of MT arrays. MAPs control cell division and expansion, two fundamental biological processes crucial for growth and development of all eukaryotes (Gardiner, 2013; Hamada, 2014).
Cytoplasmic linker-associated proteins (CLASPs) are members of the ORBIT/MAST/CLASP family of MAPs, which bind to MTs and associate with the cytoplasmic linker proteins (CLIPs), such as CLIP-170 and EB1 (Akhmanova et al., 2001). CLASP was observed anywhere along the entire length of MTs, but mostly concentrated at the plus end of MTs (Kirik et al., 2007; Muller et al., 2009), they thus belong to a diverse group of molecules named MT plus-end-tracking proteins (+TIPs) that specifically accumulate at the plus ends of growing MTs (Akhmanova and Hoogenraad, 2005; Ruan and Wasteneys, 2014). CLASPs have been shown to play various functions in animals and yeast, including stabilizing the MT plus ends (Mimori-Kiyosue et al., 2005), promoting MT rescue by recruiting tubulin dimers (Al-Bassam et al., 2010), attaching MT plus ends to the cell cortex (Lansbergen et al., 2006), regulating persistent motility of MT bundling (Stramer et al., 2010), and nucleating noncentrosomal MTs from trans-Golgi bodies (Efimov et al., 2007). It has been proposed that CLASPs mediate interaction between the plus ends of cytoplasmic MTs radiating from the centrosome to specific cortical sites at the plasma membrane, where they act as cortical anchors for the plus ends of MTs (Mimori-Kiyosue et al., 2005; Lansbergen et al., 2006). CLASPs also stabilize cortical MT at the cell edge through inhibition of MT depolymerization (Ambrose and Wasteneys, 2008; Ambrose et al., 2011). Kinesin-13s, members of the kinesin superfamily of motor proteins, make MTs unstable and increase the chance of catastrophe as a result of MT depolymerization. In contrast, CLASPs promote MT rescue, i.e., preventing MTs from shrinkage or catastrophe and regulate MT’s polymerization status and the stability of MT dynamics to maintain the activities of cell life (Al-Bassam et al., 2010; Al-Bassam and Chang, 2011; De Forges et al., 2016).
The Arabidopsis genome contains only a single copy of CLASP and no CLIP (Ambrose et al., 2007). As in animals and yeast (Kirik et al., 2007; Ambrose and Wasteneys, 2008; Ambrose et al., 2011), AtCLASP is localized at the plus ends of growing MTs and is essential for the proper organization of spindle and phragmoplast (Hamada, 2014). The Arabidopsis clasp-1 mutant exhibited dwarfism, no obvious apical dominance, lower seed setting percentage, shorter pods, reduced trichome branching, slower growth of hypocotyls and roots, reduced size of division and elongation zone in roots, and slower cell elongation rate (Ambrose et al., 2007, 2013). When treated with MT depolymerization drugs, the roots of clasp-1 showed abnormal swelling at a lower concentration of drugs compared to that of wild-type, suggesting the mutant is more sensitive to MT depolymerization drugs than wild-type (Ambrose et al., 2007). These results indicated that CLASP is essential for proper cell division and expansion, and involved in establishment of plant cell morphology. The clasp-1 mutant also showed excessive root branching, a phenomenon related to defective auxin pathways (Ambrose et al., 2007; Kirik et al., 2007). Further investigations demonstrated that clasp-1 displayed auxin-related phenotypes and AtCLASP interacts with the retromer component sorting nexin 1 (SNX1) to regulate PIN2 auxin transporter trafficking and stability (Ambrose et al., 2013; Brandizzi and Wasteneys, 2013; Kakar et al., 2013; Zhang et al., 2013). The relationship between the transport of the plasma membrane substances and MTs found in these studies revealed an auxin signaling mechanism regulated by MT cytoskeleton (Brandizzi and Wasteneys, 2013; Kakar et al., 2013; Zhang et al., 2013; Ruan and Wasteneys, 2014; Brumbarova and Ivanov, 2016). Furthermore, AtCLASP interacts with AtSABRE to stabilize MTs and guides the orientation of plant cell division and cell polarity (Pietra et al., 2013; Zhang et al., 2013; Ruan and Wasteneys, 2014; Van Dop et al., 2015; Brumbarova and Ivanov, 2016; Qi and Greb, 2017).
Cotton is a globally important economic crop. Its fiber is a valuable natural material for the textile industry. Each cotton fiber is an extremely elongated single cell initiated and developed from the epidermal layer of ovule. Cotton fiber serves as an excellent biological model for investigation of mechanisms underlying the process of cell development, particularly cell elongation. Both elongation and synthesis of secondary cell walls of fiber cells are closely related to the arrangement of the periplasmic MTs (Dixit and Cyr, 2004a,b). The number of MTs increased significantly during thickening of secondary cell walls, and the orientation of MT arrays was consistent with the direction of the newly formed fibrous layer (Yatsu and Jacks, 1981). Cortical MTs provide spatial information necessary for the alignment of cellulose microfibrils that confine and regulate cell elongation. During elongation of cotton fiber cells, reorientation of microfibrils caused by orientation change of MT arrays increases accumulation of microfibrils in the fiber cell walls (Seagull, 1986, 1992). MTs are cytoskeletal structures for orbital transport of substances within cells, when MTs are destroyed; substance transportation within cells is inhibited. MTs thus are expected to play an important role in the development of cotton fibers, so are CLASPs because they are a type of MAPs involved in the formation and development of MTs. However, little is known about cotton CLASP genes, including their numbers in the cotton genomes and functions in cotton development, particularly in cotton fiber development.
In this study, we identified cotton CLASP genes and investigated the function of GhCLASP2 using transgenic cottons with an elevated or knocked down level of GhCLASP2. Our results demonstrated a conserved function of GhCLASP2 and AtCLASP, and revealed a potential role of GhCLASP2 in enhancing fiber strength.
Cotton (Gossypium hirsutum L.) cultivars of “Xinluzao33” and “YZ-1” were used in this study. Cotton plants were grown in the experimental field of Shihezi University (Shihezi, Xinjiang, China). Flower buds on the anthesis day were tagged and marked as 0 day post anthesis (DPA). Fibers of 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30 DPA bolls were peeled off from the ovules and used in gene expression analysis. Seeds of Xinluzao33 were decoated and surface sterilized with 0.1% (w/v) HgCl2 solution for 10 min, and then washed three to four times with sterile water and grown on the 1/2 Murashige and Skoog medium (MS medium) under a 16 h light/8 h dark cycle at 28°C. Roots, stems, and leaves were collected from 2-week-old seedlings. The collected materials were immediately frozen in liquid nitrogen for RNA extraction.
The ecotype Columbia (Col-0) of Arabidopsis thaliana and the mutant clasp-1 (Salk_120061) were grown in the growth chambers with a 16 h light/8 h dark cycle at 22°C for observation and transformation by floral dip (Clough and Bent, 1998). Phenotypic observation was performed at the same growth period of different transgenic lines. Thirty leaves collected from the same position (the eighth rosette leave) of each plant were used in counting the number of epidermal trichomes and data analysis. Nicotiana benthamiana plants were grown in the growth chambers with a 16 h light/8 h dark cycle at 22°C and used in observation of subcellular localization of GhCLASP2.
Total RNA was extracted from cotton roots, stems, leaves, flowers, and fibers of different developmental stages. cDNA was synthesized by using an EASYspin Plus Plant RNA Kit (Aidlab) with gDNA Eraser (Takara). The expression levels of GhCLASPs were analyzed by semi-quantitative PCR (sqPCR) and quantitative real-time RT-PCR (qRT-PCR). The cotton UBQ7 gene was used as a standard control in both sqPCR and qRT-PCR. The gene-specific primers used in the study were shown in Supplementary Table 1. sqPCR was performed using cDNA template and ExTaq DNA Polymerase (Takara) by 28 cycles at an annealing temperature of 58°C. The qRT-PCR reactions were conducted using a SYBR Green I Master mixture (Roche, Basel, Switzerland) according to the manufacturer’s protocol on a Light Cycler 480II system (Roche, Switzerland) under conditions of an initial denaturation at 95°C for 2 min followed by 40 cycles of denaturing at 95°C for 15 s, annealing at 58°C for 20 s, and extending at 72°C for 15 s. For each tissue, three biological replicates each with three technical replicates were analyzed. The relative expression levels of genes were performed using the 2-ΔΔct method (Livak and Schmittgen, 2001).
The protein sequence of AtCLASP (At2g20190) was used as a query to search against the annotated proteins of G. arboreum (A2, BGI, version 1.0), G. raimondii (D5, JGI, version 2.0), and G. hirsutum (AD1, NAU, version 1.1) to identify CLASP genes in these three cotton species. The blastp search was done in COTTONGEN1 (Yu et al., 2014). The full-length cDNA of GhCLASP2 was amplified using primers (Supplementary Table 2) designed based on the sequence of its G. raimondii homolog Gorai.001G235400.1.
Multiple alignment of the deduced amino acid sequences of the identified CLASP genes was performed using the ClustalW2 at EMBL-EBI2. The phylogenetic relationship tree was constructed by the neighbor-joining (NJ) method implemented in the MEGA (version 6.0) software. Prediction of the CLASP domain, theoretical isoelectric point (pI), and molecular weight was done using the web-based protein database, Expert Protein Analysis System (ExPASy3).
The GhCLASP2-overexpression vector was constructed using the PCR product amplified using primers of GhCLASP2-OE-F (with the KpnI restriction site) and GhCLASP2-OE-R (with the BamHI restriction site) (Supplementary Table 2). The amplified KpnI-GhCLASP2-BamHI product was recombined into the KpnI and BamHI restriction sites of the pCAMBIA2300-ubiquitin-ocs vector. The successful construction of the GhCLASP2-overexpression vector was confirmed by digestion with KpnI and BamHI and sequencing. Agrobacterium (strain GV3101) containing the GhCLASP2-overexpression vector was used to transform Arabidopsis wild-type Col-0 and its mutant clasp-1 plants (Clough and Bent, 1998) to generate Arabidopsis lines overexpressing GhCLASP2 and complementing the clasp-1 mutation, respectively. Homozygous T3 transgenic Arabidopsis lines were identified by PCR and kanamycin (50 mg/l) selection.
Quantitative real-time RT-PCR was performed to evaluate the expression level of GhCLASP2 in the transgenic Arabidopsis lines. Each 10 μl PCR reaction contained 5 μl 2× SYBR Green I Master Mix (Roche, Basel, Switzerland), 50 ng cDNA, and 0.4 μM of each primer specific for GhCLASP2 (Supplementary Table 1). The Arabidopsis Actin gene was used as an internal control for expression normalization. qRT-PCR was performed in a Light Cycler 480II system (Roche, Switzerland) and the relative expression levels of genes were performed using the 2-ΔΔct method (Livak and Schmittgen, 2001).
To investigate the localization of the GhCLASP2 protein, the coding region of GhCLASP2 was amplified using the primers of GFP-GhCLASP2-F (with the adaptor CACC) and GFP-GhCLASP2-R (Supplementary Table 2). The pGWB6-GFP-GhCLASP2 vector was generated by inserting the coding region of GhCLASP2 into the pGWB6 vector using the Gateway technology. The BP reaction was catalyzed by the BP Clonase enzyme mix and the LR reaction was catalyzed by the LR Clonase enzyme (Invitrogen, United States) (Miki and Shimamoto, 2004; Nakagawa et al., 2007). The pGWB6-GFP-GhCLASP2 plasmid was then transformed into Agrobacterium (strain GV3101). Three-week-old tobacco (N. benthamiana) leaves were infiltrated with Agrobacteria (Kong et al., 2015). Subcellular localization was examined with a confocal microscope (Leica TCS SP5) at 2–4 d (days) after infiltration.
To further verify GhCLASP2 acting as a MAP and co-localized with MTs, the mCherry-TUB6 with a mCherry tag and the pGWB6-GFP-GhCLASP2 with a GFP tag were separately transformed into Agrobacterium (strain GV3101) and used in infiltration. Tobacco leaves were infiltrated as described above (the volume ratio of pGWB6-GFP-GhCLASP2 and mCherry-TUB6 Agrobacteria was 1:1, OD = 0.5) and the infiltrated leaves were investigated under a confocal microscope at 2–4 d after infiltration (Kong et al., 2015).
The surface sterilized seeds of “YZ-1” were cultured on the ½ MS medium at 28°C for 5–6 d in the dark. Hypocotyls of seedlings were cut into sections of 6–8 mm and used as explants for Agrobacterium-mediated transformation. The GhCLASP2-RNAi vector was constructed using a PCR fragment amplified by GhCLASP2 specific primers GhCLASP2-RNAi-F (with the adaptor CACC) and GhCLASP2-RNAi-R (Supplementary Table 2). The amplified GhCLASP2 product was recombined into the PANDA35HK vector using the Gateway technology as mentioned above (Miki and Shimamoto, 2004; Nakagawa et al., 2007). Agrobacterium (strain LB4404) containing the GhCLASP2-overexpression vector or the GhCLASP2-RNAi vector was used to infect cotton hypocotyls. Calli induction and plant regeneration were performed based on a published protocol (Jin et al., 2006). Transgenic cotton plants were screened by PCR and antibiotic kanamycin (50 mg/l). The expression level of GhCLASP2 in the transgenic plants was analyzed using sqRT-PCR and qRT-PCR. GhUBQ7 was used as a standard control in both sqRT-PCR and qRT-PCR.
Fiber samples (10 g each) were chosen randomly from each T3 transgenic line overexpressing GhCLASP2 and wild-type plants after ginning. Fiber quality analysis was done using the High Volume Instrument in the Center of Cotton Fiber Quality Inspection and Testing, Chinese Ministry of Agriculture (Anyang, Henan, China).
We started cotton CLASP gene-related work when only the G. raimondii genome sequence was available. We searched the annotated proteins of G. raimondii (D5) (Paterson et al., 2012) using the protein sequence of AtCLASP (At2g20190), and identified four putative cotton CLASP genes (Gorai.001G183300, Gorai.001G235400, Gorai.004G223500, and Gorai.006G054600). Two CLASP genes, GhCLASP1 (GenBank ID: KP742966) (Zhu et al., 2015) and GhCLASP2 (GenBank ID: MF991943), were then cloned in G. hirsutum cv. Xinluzao33 using primers based on the G. raimondii CLASP genes (Supplementary Table 2). GhCLASP1 and GhCLASP2 were similar to Gorai.006G054600 and Gorai.001G235400, respectively. We further searched the annotated proteins of G. arboreum (A2) and G. hirsutum (AD1) when their genome sequences were published (Li et al., 2014; Zhang et al., 2015), and found four and seven homologous CLASPs in G. arboreum and G. hirsutum, respectively. Based on phylogenetic analysis, these cotton CLASP genes formed two distinct groups: group I and group II (Figure 1A). Each G. arboreum and G. raimondii CLASP, except Gorai.006G054600 of G. raimondii from group II, had a homolog identified in G. hirsutum cv. TM-1 (Zhang et al., 2015; Figure 1A). Because GhCLASP1 we cloned from another G. hirsutum cv. Xinluzao33 is a homolog of Gorai.006G054600 (Zhu et al., 2015), we reasoned that TM-1 should also contain GhCLASP1. To confirm this, we searched the annotated proteins of the US version of the TM-1 genome (unpublished data4) using GhCLASP1 as a query and found its corresponding gene, Gohir.1Z037600. The chromosome location of Gohir.1Z037600 has not been determined (located on scaffold_255), but it likely belongs to chromosome D09 based on the location (chromosome A09) of its homoeolog Gh_A09G0520. We also searched another version of the TM-1 genome annotation (Li et al., 2015) and found only six CLASP genes with CotAD_68468 and CotAD_04861 corresponding to GhCLASP1 and GhCLASP2, respectively (Zhu et al., 2017). Gohir.1Z037600 and GhCLASP1 had identical length and 11 single-nucleotide polymorphisms (SNPs; three synonymous and eight non-synonymous ones). Compared to GhCLASP1, CotAD_68468 was 123-bp longer probably due to sequence misassembly and/or inclusion of introns with non-canonical splicing signals (Supplementary Figure 1). These results indicate that the assembly and/or annotation of different version of the TM-1 genome sequences were inconsistent, but G. hirsutum has retained all CLASPs derived from its ancestral diploid species and the CLASP genes are well conserved during cotton evolution.
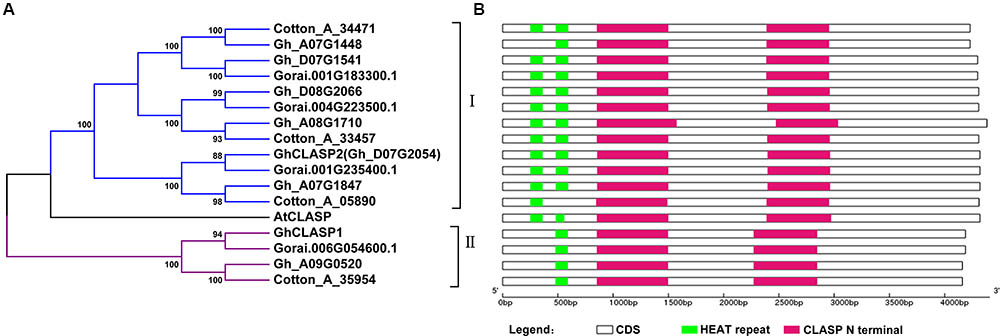
FIGURE 1. Phylogenetic relationship and protein domain organization of CLASP family member from three cotton species and Arabidopsis. (A) Phylogenetic relationship of CLASP proteins from three cotton species and Arabidopsis. The tree was generated using the NJ approach based on the amino acid sequences of CLASPs. The numbers next to each node indicate bootstrap values from 1000 replicates. Cotton_A_35954, Cotton_A_34471, Cotton_A_33457, and Cotton_A_05890 are from G. arboreum; Gorai.001G183300.1, Gorai.001G235400.1, Gorai.006G054600.1, and Gorai.004G223500.1 are from G. raimondii; Gh_A07G1448, Gh_D07G1541, Gh_A09G0520, Gh_D08G2066, Gh_A08G1710, and Gh_A07G1847 are from G. hirsutum acc.TM-1; GhCLASP1 and GhCLASP2 (Gh_D07G2054) are from G. hirsutum acc.Xinluzao33; and AtCLASP is from Arabidopsis. (B) Protein domain organization of the CLASP family members in cotton. The CLASP N terminal domain and HEAT repeats are represented by red and green bars, respectively.
Based on our previous results, the expression levels of the three G. hirsutum CLASP genes, including GhCLASP2 or CotAD_04861, were gradually increased in 6–24 DPA fibers (Zhu et al., 2017). Inspired by this observation, in this study, we aimed to investigate the functionality of GhCLASP2 and to know whether GhCLASP2 plays a role in fiber development. GhCLASP2 has a 4320-bp long coding sequence, encodes 1439 amino acids with a molecular mass of 158.95 kD, and a pI of 6.79 (Supplementary Table 3 and Figure 1B). GhCLASP2 and AtCLASP have a protein sequence identity of 75.16%.
The corresponding genes of GhCLASP2 cloned from Xinluzao33 in the three versions of the TM-1 genome are CotAD_04861 (Li et al., 2015), Gh_D07G2054 (Zhang et al., 2015), and Gohir.D07G208500 (unpublished data), which were annotated quite differently with a length of 3795, 4395, and 4320 bp (Supplementary Table 3), respectively. Gohir.D07G208500 and GhCLASP2 had identical length and four SNPs (two synonymous and two non-synonymous ones). Compared to GhCLASP2, Gh_D07G2054 was 75-bp longer, and CotAD_04861 was 525-bp shorter. Based on sequence alignment, it seems that the longer Gh_D07G2054 was due to inclusion of a 75-bp intron although the exact reason for the shorter CotAD_04861 was unclear. In addition, there were more “SNPs” between GhCLASP2 and Gh_D07G2054 and CotAD_04861 (Supplementary Figures 2, 3). Based on these results, we believe that the sequence of Gohir.D07G208500 was properly annotated while those of Gh_D07G2054 and CotAD_04861 were mis-annotated.
Cytoplasmic linker-associated proteins have two conserved domains: the HEAT repeat domain and the CLASP N terminal domain. The HEAT repeat domains have a helical structure, regulate interaction between proteins, and are involved in intracellular transport (Andrade and Bork, 1995; Groves et al., 1999). The CLASP N terminal domain is highly conserved, and contains MT-binding sites regulating the stability of MT dynamic (Grallert et al., 2006; Pereira et al., 2006). GhCLASPs showed high sequence identity among each other, particularly in the CLASP N terminal domain (Supplementary Figure 4). Like AtCLASP, most GhCLASPs contained two HEAT repeats and two CLASP N terminal domains (Figure 1B). However, all group II homoeologs contained only one HEAT repeat (Zhu et al., 2015) (Figure 1B). Two group I GhCLASPs, Gh_A07G1448 and Cotton_A_05890, also contained a single HEAT domain, and have lost the first and second HEAT domain due to non-synonymous nucleotide change and short sequence deletion, respectively (Figure 1B).
Each cotton fiber is a single celled seed trichome or hair. Cotton fibers have been used as a model for studying cell differentiation and development. The development of cotton fiber generally consists of four distinct but overlapping stages: fiber initiation, fiber cell elongation, biosynthesis of secondary cell wall cellulose, and maturation (Lee et al., 2007). Here, we first analyzed the expression patterns of GhCLASP2 in vegetative, reproductive tissues, and more fiber samples to confirm our previous observation (Zhu et al., 2017). The expression level of GhCLASP2 was much higher in stem and flower than in root and leaf. In developing fibers, the expression level of GhCLASP2 increased gradually from 3 to 30 DPA, and a significantly increased expression was observed from 24 to 27 DPA, coincident with the transition from fiber elongation to secondary cell wall thickening (Figure 2). The expression patterns suggested a role of GhCLASP2 in fiber development, especially in fiber secondary cell wall biosynthesis.
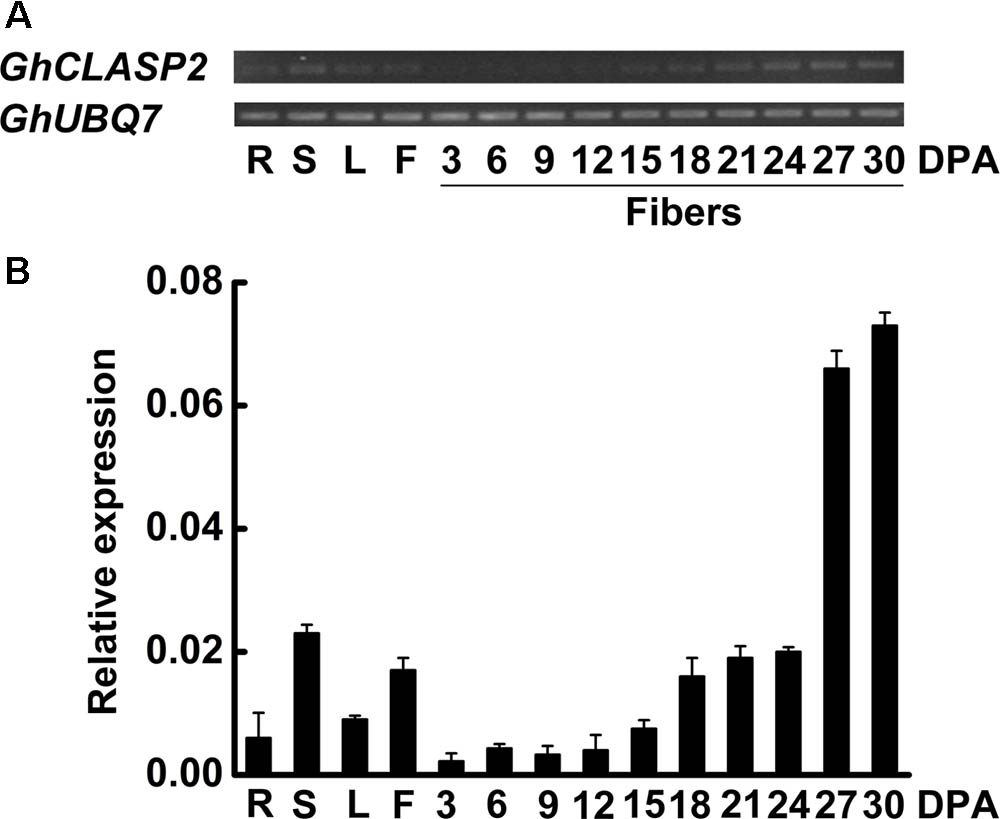
FIGURE 2. Expression patterns of GhCLASP2. (A) Expression level of GhCLASP2 in various cotton tissues measured by sqPCR. (B) Expression level of GhCLASP2 in various cotton tissues measured by qRT-PCR. R, root; S, stem; L, leaf; F, flower (0 DPA), and fibers of 3, 6, 9, 12, 15, 18, 21, 24, 27, and 30 DPA. Data are means ± SD. Error bars represent the standard deviation of triplicate experiments, and GhUBQ7 was used as an internal control.
We also analyzed the expression patterns of other seven GhCLASP2 homologs in different cotton organs (Supplementary Figure 5). The expression levels of the four pairs of homoeologs were quite different. For three homoeologous pairs (Gh_A07G1448/Gh_D07G1541, Gh_A08G171/Gh_D08G2066, and Gh_A07G1847/GhCLASP2), their At and Dt sub-genome homoeologs had a similar expression pattern in the tissues analyzed. The highest expression levels of Gh_A07G1448 and Gh_D07G1541 were observed in stem. Both pairs of Gh_A08G1710/Gh_D08G2066 and Gh_A07G1847/GhCLASP2 were predominantly expressed in fibers. But the expression patterns of Gh_A09G0520 and its homoeolog GhCLASP1 were different in the tissues analyzed (Supplementary Figure 5). Gh_A09G0520 was highly expressed in flowers, whereas GhCLASP1 was preferentially expressed in stem and 27 DPA fibers (Supplementary Figure 5). Different expression patterns suggest that these CLASPs might have different functions in cotton.
To understand the function of GhCLASP2, we tried to complement the defective phenotypes of the Arabidopsis mutant clasp-1 by transforming a copy of GhCLASP2, which was driven by the Ubiquitin promoter. For comparison, transgenic Arabidopsis plants with overexpressed GhCLASP2 were also generated (Figure 3). Arabidopsis leaf trichomes have variable number of branches. In the wild-type, most trichomes (88.56%) had three branches. In contrast, most trichomes of clasp-1 had two branches (71.26%). clasp-1 also had more one-branch trichomes than the wild-type (Figures 3D,E and Supplementary Table 4). These observations were consistent with previous findings (Ambrose et al., 2007; Kirik et al., 2007). Compared to clasp-1, the complementary lines (CL3, CL6, and CL7) restored the phenotype of dominant three-branch trichomes (82.52–84.37%), although the percentage of three-branch trichomes was still not as high as that of the wild-type. These results suggested that GhCLASP2 could rescue the defective leaf trichome phenotype of clasp-1. In the GhCLASP2-overexpressed lines, as in the wild-type, the majority leaf trichomes had three branches (89.80–91.41%), but the percentage was higher than that of the wild-type (88.56%). Furthermore, trichomes with four branches were significantly higher in the GhCLASP2-overexpressed lines than in the wild-type (Figures 3D,E and Supplementary Table 4). Trichomes with five branches were also observed in some GhCLASP2-overexpressed lines (Supplementary Table 4). These results suggest that the ectopic expression of GhCLASP2 promoted trichome branching in Arabidopsis leaves and that trichome branching is positively correlated with the expression level of GhCLASP2 (Figures 3B,C).
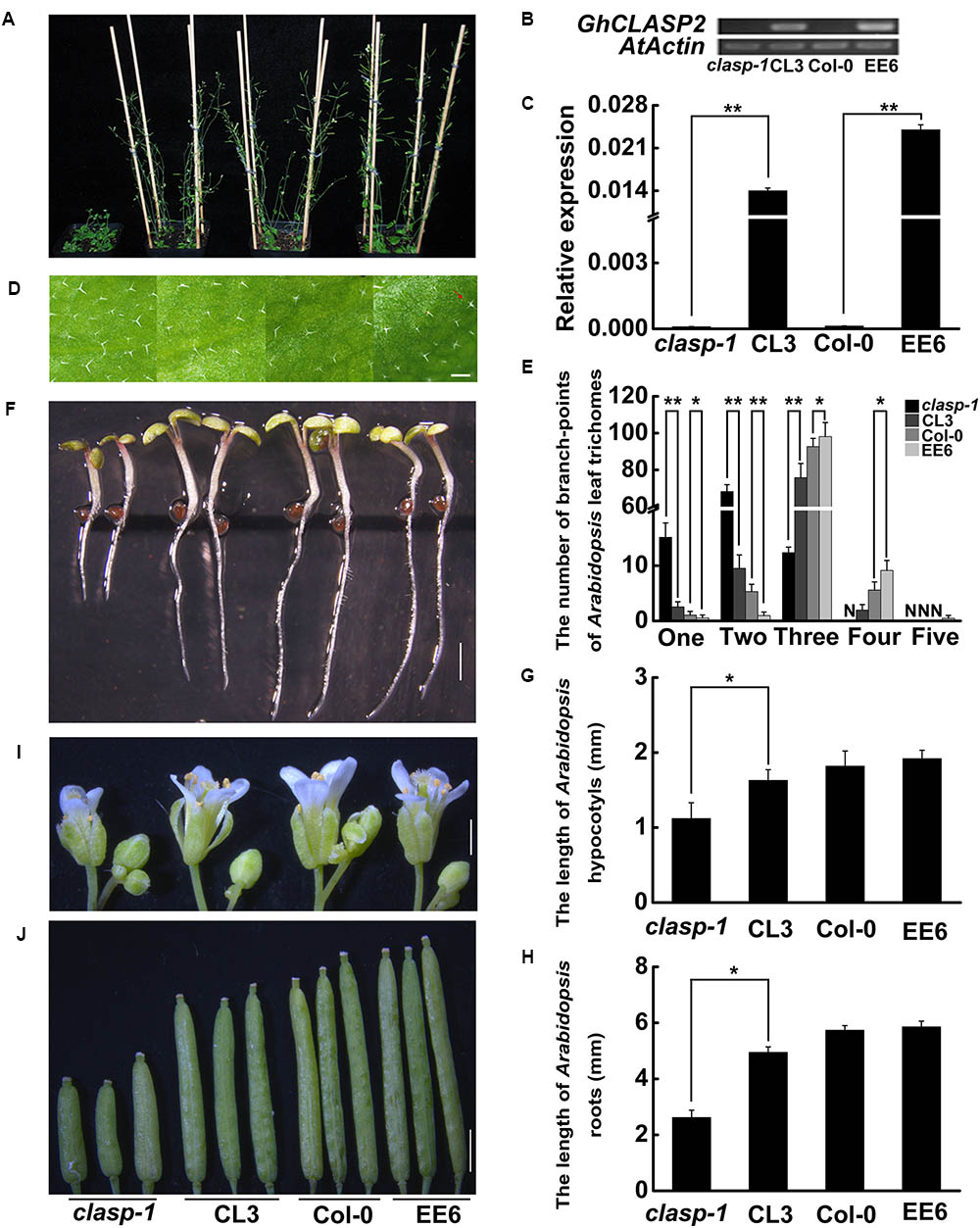
FIGURE 3. The phenotypes of clasp-1, complementing line (CL3), wild-type (Col-0), and the transgenic line with ectopic expression of GhCLASP2 (EE6) in Arabidopsis. (A) Thirty-two-day-old plants. (B) sqPCR analysis of GhCLASP2 expression in the plants shown in (A). (C) qRT-PCR analysis of GhCLASP2 expression in the plants shown in (A). (D) Leaf trichomes of the four types of Arabidopsis plants. The red arrow indicates a leaf trichome with five branches (Bar = 200 μm). (E) The graph depicts the number of trichomes with different numbers of branch-points per leaf shown in (D) (n = 30). N, none; one, one branch; two, two branches; three, three branches; four, four branches; five, five branches in per leaf. (F) Seven-day-old Arabidopsis seedlings. Bar = 1 mm. (G) Hypocotyls length of the 7-day-old Arabidopsis seedlings shown in (F) (n = 20). (H) Root length of the 7-day-old Arabidopsis seedlings shown in (F) (n = 20). (I) Thirty-two-day-old Arabidopsis flowers (Bar = 1 mm). (J) Thirty-two-day-old Arabidopsis pods (Bar = 1 mm). For both sqPCR and qRT-PCR, AtActin was used as a reference gene. Data are means ± SD. P-values were determined based on Student’s t-tests. ∗ and ∗∗ indicate significant differences at p < 0.05 and p < 0.01, respectively.
In addition, we investigated hypocotyl elongation and root growth in the same materials (Figures 3F–H). Compared to clasp-1, the three complementary lines had significantly elongated hypocotyls (1.50–1.63 mm versus 1.12 mm) and roots (4.95–5.01 mm versus 2.62 mm). However, transgenic lines with overexpressed-GhCLASP2 and wild-type had a similar length of hypocotyls and roots (Figures 3F–H and Supplementary Table 5). Furthermore, GhCLASP2 was able to rescue the defective phenotypes of flower and silique of clasp-1, although Arabidopsis lines overexpressing GhCLASP2 and wild-type had similar flowers and siliques (Figures 3I,J). Together, these results suggest that GhCLASP2 functions similarly as AtCLASP in plant growth and development.
To investigate the subcellular localization of GhCLASP2, we fused the full-length GhCLASP2 coding sequence to the C terminus of the GFP gene driven by the 35S promoter (pGWB6-GFP-GhCLASP2) and infiltrated the construct into tobacco (N. benthamiana) leaf. Enriched GFP signal was observed along with the entire length of the filamentous structures in the leaf epidermal pavement cells (Figure 4A).
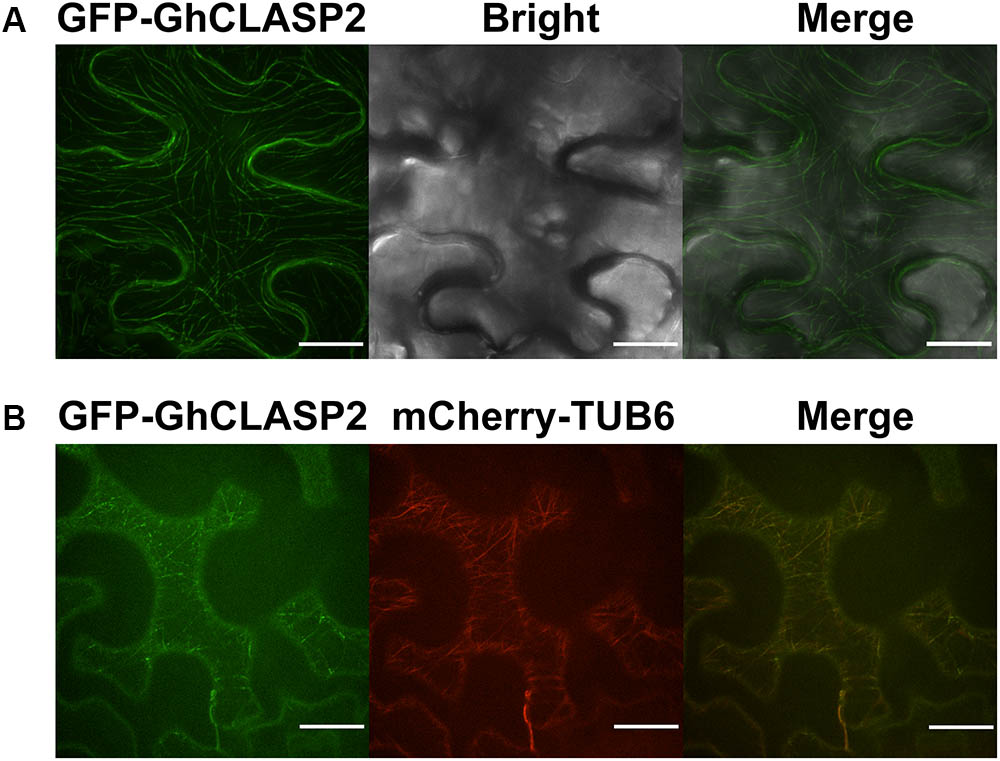
FIGURE 4. GFP-GhCLASP2 co-localized with MTs in the epidermal pavement cell of tobacco leaf. (A) Transiently expressed GFP-GhCLASP2 was localized in all MT arrays and induced stable MT bundles (Bar = 20 μm). (B) GFP-GhCLASP2 was co-expressed with a tubulin marker, mCherry-TUB6, in tobacco leaf epidermal cells (Bar = 25 μm).
To investigate whether GhCLASP2 and MTs are co-localized, the pGWB6-GFP-GhCLASP2 construct and mCherry-TUB6, which labels β-tubulin, were transiently co-transformed into tobacco (N. benthamiana) leaf. GFP signal was found to be co-localized with MTs in the filamentous structures of pavement cells (Figure 4B). In the leaf epidermal cells, GFP-GhCLASP2 distributed in a nonuniform manner along the full length of MTs and seemed to be induced by extensive MT bundling (Figure 4). Just like AtCLASP, GhCLASP2 was prominently observed as small and immobile spots with variable intensity at MTs, indicating that GhCLASP2 interacts with MTs potentially through regulating MTs as a +TIP (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005).
To know the function of GhCLASP2 in cotton development, we generated transgenic cotton plants with the GhCLASP2-overexpressed construct driven by the Ubiquitin promoter or the RNAi-GhCLASP2 construct driven by the 35S promoter (Supplementary Figures 6A, 7A). Six GhCLASP2-overexpressed T0 transgenic lines were obtained. T3 homozygous transgenic lines were identified and used in further investigation. The up-regulated level of GhCLASP2 in these plants was confirmed by sqPCR and qRT-PCR (Figures 5B,C and Supplementary Figure 6). Compared to the wild-type YZ-1, none of the GhCLASP2-overexpressed transgenic lines showed significant differences in both vegetative and reproductive growth (Figure 5). However, GhCLASP2-overexpressed lines had significantly increased fiber strength (3.85–4.67%) compared to the wild-type YZ-1 (Table 1), although they had a similar fiber length and micronaire value (Figure 6).
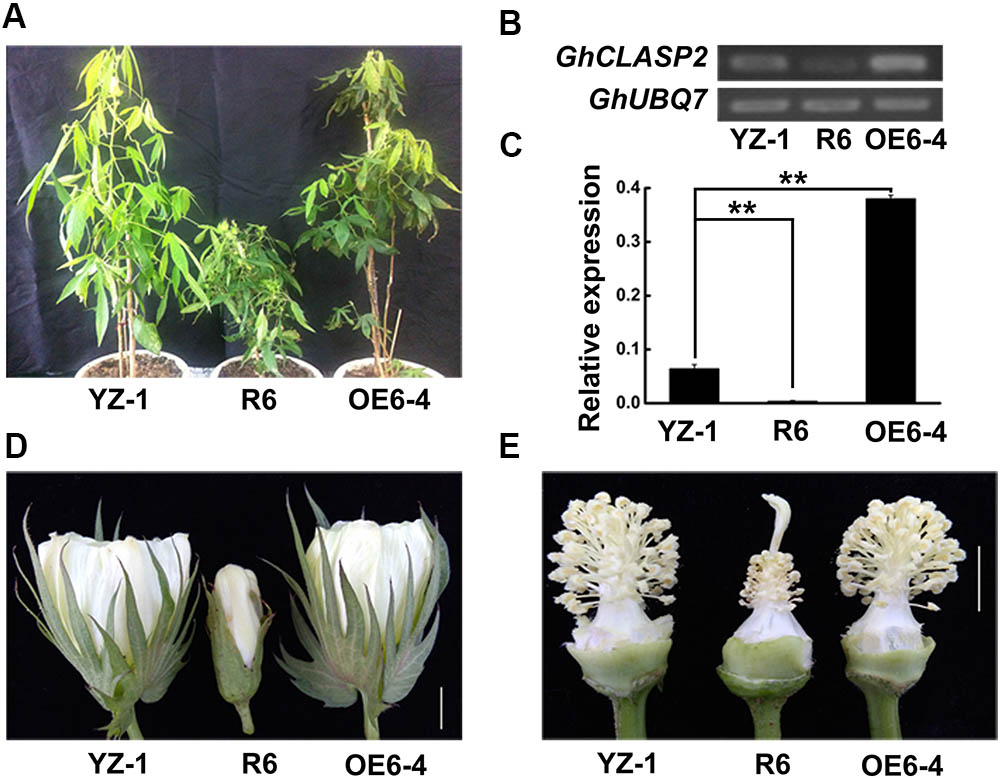
FIGURE 5. GhCLASP2 affected cotton plant growth and development. (A) From left to right, the control plant YZ-1, the GhCLASP2-RNAi transgenic plant (R6), and the GhCLASP2-overexpressed transgenic plant (OE6-4). (B) sqPCR analysis of GhCLASP2 expression in the plants shown in (A). (C) qRT-PCR analysis of GhCLASP2 expression in the plants shown in (A). (D) Flower of YZ-1, R6, and OE6-4 on the anthesis day (0 DPA) (Bar = 1 cm). (E) Anther, pistil, and stamen from 0 DPA flower of YZ-1, R6, and OE6-4 (Bar = 1 cm). For both sqPCR and qRT-PCR, GhUBQ7 was used as a reference gene. Data are means ± SD. P-values were determined based on Student’s t-tests. ∗ and ∗∗ indicate significant differences at p < 0.05 and p < 0.01, respectively.
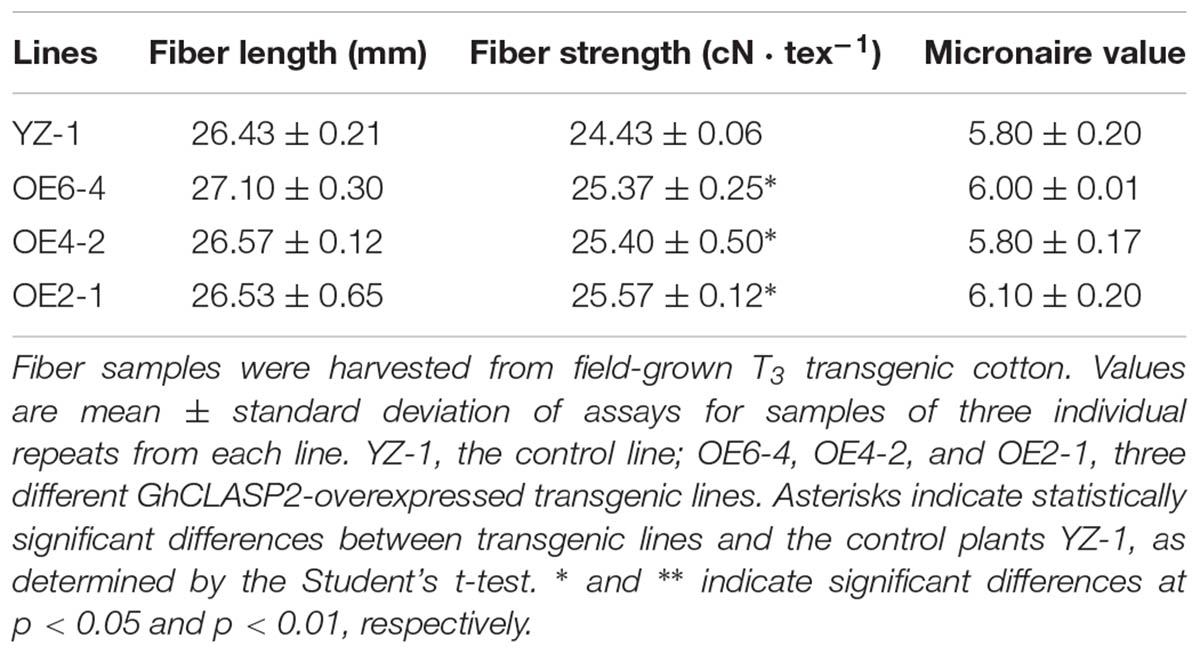
TABLE 1. Analysis of fiber quality between field-grown GhCLASP2-overexpressed transgenic lines and controls.
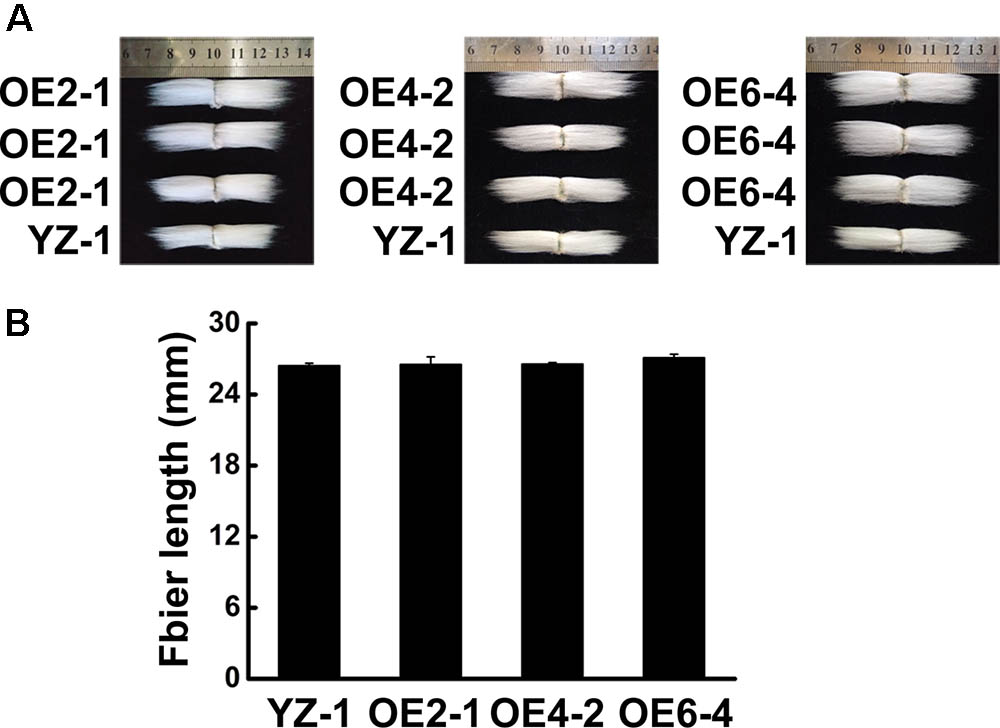
FIGURE 6. Phenotype of fiber in GhCLASP2-overexpressed transgenic cotton lines. (A) Phenotype of final mature fiber in GhCLASP2-overexpressed transgenic cotton lines (OE2-1, OE4-2, and OE6-4), comparing with YZ-1. (B) Average fiber length from YZ-1- and GhCLASP2-overexpressed transgenic cotton lines (OE2-1, OE4-2, and OE6-4). Data are means ± SD. Error bars indicate standard deviation of triplicate experiments.
The GhCLASP2-specific down-regulation in the six GhCLASP2-RNAi T0 transgenic lines was confirmed by sqPCR and qRT-PCR (Figures 5B,C and Supplementary Figure 7). We did not detected down-regulation of other GhCLASP2 homologs in the T0 transgenic lines (Supplementary Figure 8). The plant height of the T0 GhCLASP2-RNAi lines was shorter than that of the wild-type YZ-1 due to shorter internodes (Figure 5). In addition, the GhCLASP2-RNAi lines showed smaller flower, aborted anthers (without pollen grains), slightly elongated stigma, and sterility (Figures 5A,D,E). These phenotypes were similar to those observed in the Arabidopsis mutant clasp-1 (Ambrose et al., 2007; Kirik et al., 2007). CLASP, acting as a MAP in cell, is involved in cell division and expansion (Ambrose et al., 2007). Due to the reduced expression level of GhCLASP2 in the GhCLASP2-RNAi transgenic cotton plants, the stability of MTs was probably decreased, which affected the characteristics of cell wall and formation of cell cytoskeleton to cause the abnormal phenotypes observed in the GhCLASP2-RNAi lines.
Because GhCLASP2-overexpressed transgenic plants showed increased fiber strength, we analyzed the expression levels of eight selected genes related to fiber development in those plants by qRT-PCR (Supplementary Table 1) (Li et al., 2002, 2013; Xu et al., 2013). Compared to the wild-type, all three GhCLASP2-overexpressed lines showed an elevated level of MT-associated genes, i.e., GhTUB1, GhTUB12, GhTUA6, and GhTUA9, although the increased levels were gene and line dependent (Figure 7), suggesting that GhCLASP2 might have the ability to induce transcription of MT-associated genes and to promote development of MTs, and finally development of cotton fibers.
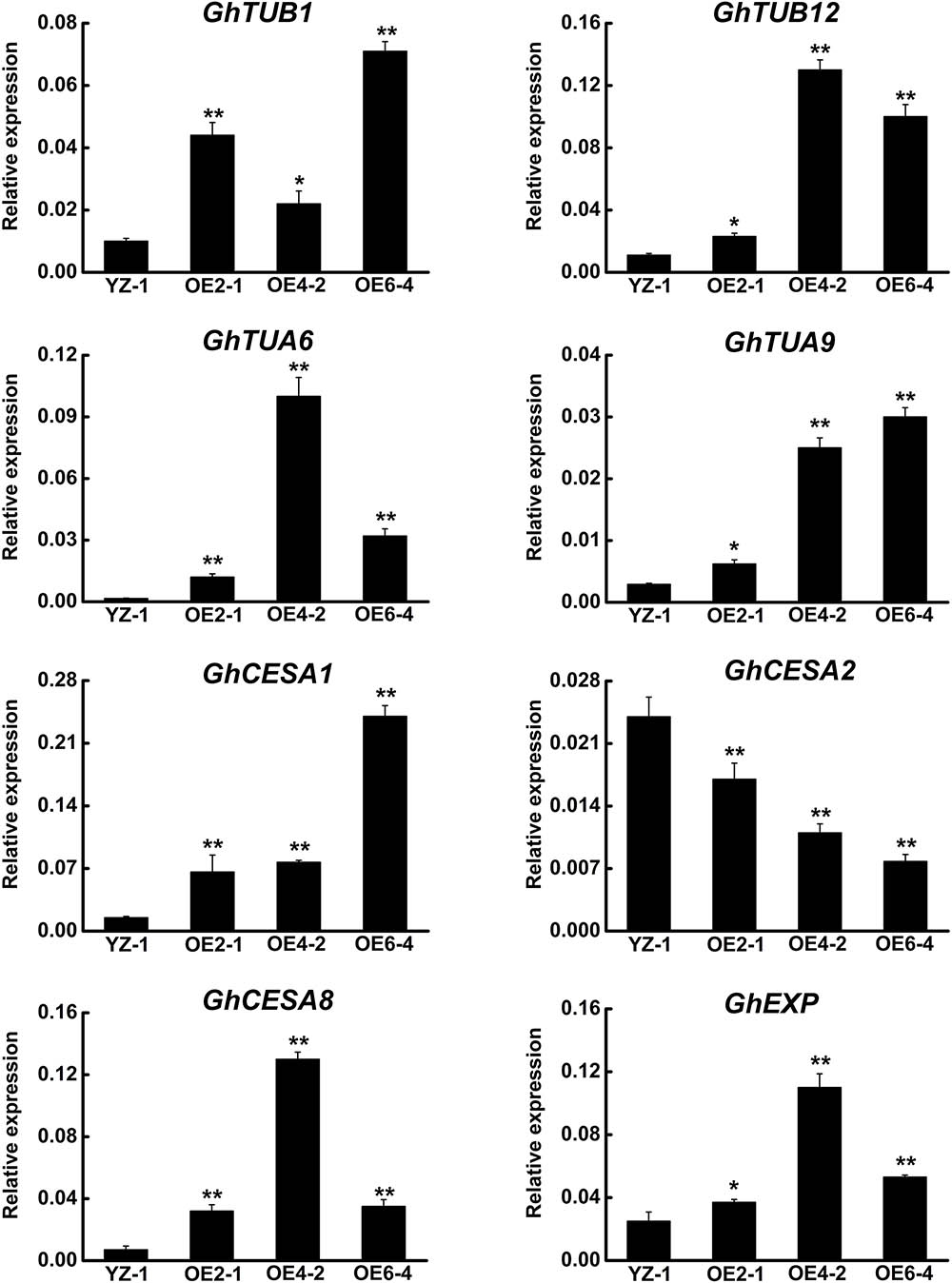
FIGURE 7. qRT-PCR analysis of fiber development-related genes in the GhCLASP2-overexpressed transgenic cotton lines. Fiber of 27DPA from the wild-type YZ-1 and three transgenic lines was used. Data are means ± SD. Error bars represent the standard deviation of triplicate experiments, and GhUBQ7 was used as an internal control. P-values were determined based on Student’s t-tests. ∗ and ∗∗ indicate significant differences at p < 0.05 and p < 0.01, respectively.
Microtubules are intimately associated with cellulose synthesis activity, especially during secondary wall deposition (Wasteneys, 2004; Panteris and Galatis, 2005; Paredez et al., 2006). The secondary wall thickening of cotton fiber is mainly a process of cellulose synthesis and precipitation, which is crucial to the quality of cotton fibers, especially fiber strength. Because the expression level of GhCLAP2 was rapidly increased in the developmental stage of secondary wall thickening (after 27 DPA), we examined the expression levels of three cellulose synthase genes (CESAs) in GhCLASP2-overexpressed transgenic lines. Compared to the wild-type, GhCLASP2-overexpressed transgenic lines had a higher expression level of GhCESA1 and GhCESA8, but a lower expression level of GhCESA2 (Figure 7). Since AtCLASP could be involved in cell expansion and division (Ambrose et al., 2007), we also analyzed the expression level of GhEXP in cotton fiber, which showed an increased level in all three transgenic lines, particularly in OE4-2 (Figure 7). These data suggest that overexpression of GhCLASP2 may alter the expression level of genes related to fiber development and consequently promotes cotton fiber development.
Cytoplasmic linker-associated proteins are one of the MAP families that interact with and regulate MTs, an essential component of plant cytoskeleton. Genes encoding CLASPs have been bioinformatically identified in several species (Gardiner, 2013) but only the Arabidopsis CLASP has been functionally characterized so far (Ambrose et al., 2007; Kirik et al., 2007). In this work, we identified cotton CLASP genes and investigated the function of one of the eight CLASPs identified in G. hirsutum, GhCLASP2.
Upland cotton (G. hirsutum) is an allotetraploid and thought to have originated from a relatively recent interspecific hybridization event between an A genome-like ancestral species similar to G. arboreum or G. herbaceum and a D-genome-like species similar to G. raimondii. Each of the two diploid cotton species, G. arboreum and G. raimondii, contains four CLASP genes. All of the eight genes from the two diploids have been retained in G. hirsutum, suggesting conservation of the CLASP genes during cotton evolution. One reason for this conservation could be that these genes are essential for normal cotton development. Functional conservation of CLASPs is further supported by our observation that the GhCLASP2-RNAi cotton lines showed similar phenotypes as those of the Arabidopsis clasp-1 mutant (Figure 5) and that the defective phenotypes of clasp-1 could be rescued by GhCLASP2 (Figure 3).
Microtubules play an important role in plant cell morphology and development (Hamada, 2014). It has been shown that MTs participated in the initiation and localization of epidermal protrusions (Mathur and Chua, 2000). Mutations in the factors regulating MTs caused misassembly of α/β tubulin dimers resulted in disruption of the formation of new MTs and the branching pattern of leaf trichomes (Kirik et al., 2002). Leaf trichomes of Arabidopsis plants overexpressing GhCLASP2 had more branch numbers than the wild-type (Figure 3). GhCLASP2 was also co-localized with MTs (Figure 4). Therefore, like AtCLASP, GhCLASP2 might achieve its role through interacting with and regulating MTs.
Cotton fiber is an ideal model for studying cell development (Guan et al., 2014). In view of the similarity between cotton fiber development and Arabidopsis leaf trichome development (Machado et al., 2009). We reasoned that CLASPs would have a role in cotton fiber formation. Cellulose is the main component of cotton fiber, which accounts for more than 90% of dry weight of mature cotton fiber cells (Meinert and Delmer, 1977). Cellulose is synthesized by cellulose synthase complexes, mainly in the form of microfibrils (Kudlicka and Brown, 1997; Doblin et al., 2002). At the onset of secondary wall thickening, the rate of cellulose synthesis increases significantly. Studies have shown that GhCESA1, GhCESA2, and GhCESA8 are mainly participating in secondary wall formation of cotton fiber. Cortical MTs are associated with cellulose synthase complexes and correlated with accumulation of cellulose microfibrils, especially during the period of secondary wall deposition (Gardiner et al., 2003; Wasteneys, 2004; Panteris and Galatis, 2005; Smith and Oppenheimer, 2005; Paredez et al., 2006). An involvement of cortical MTs in the movement of the cellulose synthase complex is evident in the plasma membrane (Mcfarlane et al., 2014). Secondary wall thickening of cotton fibers is crucial for the formation of cotton fiber quality, such as fiber fineness, maturity, and strength. We found that the expression level of GhCLASP2 in cotton fibers increased dramatically from 24 DPA to 27 DPA and stayed high thereafter (Figure 2), and that the GhCLASP2-overexpressed lines had stronger fibers than the wild-type (Figure 6 and Table 1). These results suggest a potential relationship between the expression level of GhCLASP2 and fiber strength. We found that the expression levels of three cellulose synthase genes (GhCESA1, GhCESA2, and GhCESA8) have altered expression levels in the 27 DPA fibers of GhCLASP2-overexpressed transgenic cotton plants (Figure 7). Previous studies have shown that AtCLASP, as a MT binding protein, is most likely to be involved in cell expansion by affecting the mechanical properties of the cell wall, and thus affecting the organ morphology and axial extension in the plants (Ambrose et al., 2007). A gene encoding expansion (EXP) protein and four MT-associated genes (GhTUA6, GhTUA9, GhTUB1, and GhTUB12) showed up-regulated expression in the 27 DPA fibers of the GhCLASP2-overexpressed transgenic cotton plants (Figure 7). Based on these results, we speculate that increased fiber strength observed in the GhCLASP2-overexpressed plants is a result of coordinated up-regulation and down-regulation of a number of genes (such as TUAs, TUBs, CESAs, and expansin) involved in fiber development. It indicates that GhCLASP2 has direct or indirect relationship with these genes. We thus propose that GhCLASP2 might be involved in regulating fiber development, particularly fiber strength, by interacting with MTs and affecting cellulose biosynthesis and deposition. The network involving GhCLAP2 that participates in fiber development is needed to be further investigated and analyzed in the future. GhCLASP2 was relatively highly expressed in stem (Figure 2). Downregulation of GhCLASP2 caused dwarfism due to shortened internode length, suggesting that cell division and/or elongation of stem were affected by the reduced expression level of GhCLASP2. This function of CLASP has been reported in Arabidopsis, where shorter and wider epidermal cells were observed in the hypocotyls of clasp-1 (Ambrose et al., 2007). Cotton fiber is an extremely elongated single cell. We were unable to analyze the effect of knock-down GhCLASP2 on fiber elongation, as all transgenic plants harboring the GhCLASP2-RNAi construct generated in this study were sterile and failed to produce any cotton boll; however, we did not observe fiber length difference between the plants overexpressing GhCLASP2 and the wild-type (Figure 6 and Table 1). Whether CLASP having negative effects on fiber cell elongation is yet to be investigated using other CLASP genes, such as Gh_A08G1710/Gh_D08G2066, that are preferentially expressed in cotton fibers and very weakly expressed in flowers.
There are eight CLASPs in G. hirsutum. Like GhCLASP2, some of them were also predominantly expressed in fibers, such as the At sub-genome homoeolog (Gh_A07G1847) of GhCLASP2 and Gh_A08G1710/Gh_D08G2066. In this study we were unable to address their redundant roles in fiber development due to sterility of the GhCLASP-RNAi lines, but demonstrated the functional conservativeness between GhCLASP2 and AtCLASP. In addition, our results suggest that it might be possible to improve cotton fiber strength by overexpressing CLASPs.
S-HZ, FX, Q-HZ, and JS conceived and designed the experiments. S-HZ and L-JZ performed the experiments. S-HZ analyzed the data. JS, Y-JL, FL, X-YZ, and Y-QS contributed reagents, materials, and analysis tools. S-HZ and Q-HZ wrote the paper.
This work was supported by the National Natural Science Foundation of China (Grant No. 31760401), the National Key Research and Development Program of China (Grant Nos. 2016YFD0101900 and 2016YFD0100200), and Specific Project for Crops Breeding of Shihezi University (Grant No. gxjs-yz03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HM and the handling Editor declared their shared affiliation.
We thank Professor Zhao Sheng Kong (Chinese Academy of Sciences, China) for his kind assistance in experiments and for providing us with the mCherry-TUB6 plasmid vector. The seeds of the mutant clasp-1 (Salk_120061) of Arabidopsis were kindly provided by Professor Bo Liu (Section of Plant Biology, University of California, United States). We also thank Dr. William J. Gale (College of Agriculture, Shihezi University, China) for critical reading and professional editing of the manuscript. We thank Professor Gui Xian Xia (Chinese Academy of Sciences, China) for insightful comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00882/full#supplementary-material
Akhmanova, A., and Hoogenraad, C. C. (2005). Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 17, 47–54.
Akhmanova, A., Hoogenraad, C. C., Drabek, K., Stepanova, T., Dortland, B., Verkerk, T., et al. (2001). CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923–935.
Al-Bassam, J., and Chang, F. (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604–614. doi: 10.1016/j.tcb.2011.06.007
Al-Bassam, J., Kim, H., Brouhard, G. J., Van Oijen, A. M., Harrison, S. C., and Chang, F. (2010). CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell 19, 245–258. doi: 10.1016/j.devcel.2010.07.016
Ambrose, C., Allard, J. F., Cytrynbaum, E. N., and Wasteneys, G. O. (2011). A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2:430. doi: 10.1038/ncomms1444
Ambrose, C., Ruan, Y., Gardiner, J., Tamblyn, L. M., Catching, A., Kirik, V., et al. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24, 649–659. doi: 10.1016/j.devcel.2013.02.007
Ambrose, J. C., Shoji, T., Kotzer, A. M., Pighin, J. A., and Wasteneys, G. O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19,2763–2775.
Ambrose, J. C., and Wasteneys, G. O. (2008). CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell 19, 4730–4737. doi: 10.1091/mbc.E08-06-0665
Andrade, M. A., and Bork, P. (1995). HEAT repeats in the Huntington’s disease protein. Nat. Genet. 11, 115–116.
Brandizzi, F., and Wasteneys, G. O. (2013). Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J. 75, 339–349. doi: 10.1111/tpj.12227
Brumbarova, T., and Ivanov, R. (2016). Differential gene expression and protein phosphorylation as factors regulating the state of the Arabidopsis SNX1 protein complexes in response to environmental stimuli. Front. Plant Sci. 7:1456. doi: 10.3389/fpls.2016.01456
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
De Forges, H., Pilon, A., Cantaloube, I., Pallandre, A., Haghirigosnet, A., Perez, F., et al. (2016). Localized mechanical stress promotes microtubule rescue. Curr. Biol. 26, 3399–3406. doi: 10.1016/j.cub.2016.10.048
Dixit, R., and Cyr, R. J. (2004a). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16, 3274–3284.
Dixit, R., and Cyr, R. J. (2004b). The cortical microtubule array: from dynamics to organization. Plant Cell 16, 2546–2552.
Doblin, M. S., Kurek, I., Jacobwilk, D., and Delmer, D. (2002). Cellulose biosynthesis in plants : from genes to rosettes. Plant Cell Physiol. 43, 1407–1420.
Efimov, A., Kharitonov, A., Efimova, N., Loncarek, J., Miller, P. M., Andreyeva, N., et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12, 917–930.
Gardiner, J. (2013). The evolution and diversification of plant microtubule-associated proteins. Plant J. 75, 219–229.
Gardiner, J. C., Taylor, N. G., and Turner, S. R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15, 1740–1748.
Grallert, A., Beuter, C., Craven, R. A., Bagley, S., Wilks, D. P., Fleig, U., et al. (2006). S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 20, 2421–2436.
Groves, M. R., Hanlon, N., Turowski, P., Hemmings, B. A., and Barford, D. (1999). The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110.
Guan, X., Pang, M., Nah, G., Shi, X., Ye, W., Stelly, D. M., et al. (2014). miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5:3050. doi: 10.1038/ncomms4050
Hamada, T. (2014). Microtubule organization and microtubule-associated proteins in plant cells. Int. Rev. Cell Mol. Biol. 312, 1–52.
Jin, S., Zhang, X., Nie, Y., Guo, X., Liang, S., and Zhu, H. (2006). Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 50, 519–524.
Kakar, K., Zhang, H., Scheres, B., and Dhonukshe, P. (2013). CLASP-mediated cortical microtubule organization guides PIN polarization axis. Nature 495, 529–533. doi: 10.1038/nature11980
Kirik, V., Grini, P. E., Mathur, J., Klinkhammer, I., Adler, K., Bechtold, N., et al. (2002). The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. Plant Cell 14, 2265–2276.
Kirik, V., Herrmann, U., Parupalli, C., Sedbrook, J. C., Ehrhardt, D. W., and Hulskamp, M. (2007). CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. J. Cell Sci. 120, 4416–4425.
Kong, Z., Ioki, M., Braybrook, S. A., Li, S., Ye, Z., Lee, Y. J., et al. (2015). Kinesin-4 functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant 8, 1011–1023. doi: 10.1016/j.molp.2015.01.004
Kudlicka, K., and Brown, R. M. (1997). Cellulose and callose biosynthesis in higher plants (I. solubilization and separation of (1- > 3)- and (1- > 4)-[beta]-glucan synthase activities from mung bean). Plant Physiol. 115, 643–656.
Lansbergen, G., Grigoriev, I., Mimorikiyosue, Y., Ohtsuka, T., Higa, S., Kitajima, I., et al. (2006). CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5β. Dev. Cell 11, 21–32.
Lee, J. J., Woodward, A. W., and Chen, Z. J. (2007). Gene expression changes and early events in cotton fibre development. Ann. Bot. 100, 1391–1401.
Li, A., Xia, T., Xu, W., Chen, T., Li, X., Fan, J., et al. (2013). An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense. Planta 237, 1585–1597. doi: 10.1007/s00425-013-1868-2
Li, C., Zhu, Y., Meng, Y., Wang, J., Xu, K., Zhang, T., et al. (2002). Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci. 163, 1113–1120.
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔct Method. Methods 25, 402–408.
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., and Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59, 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
Mathur, J., and Chua, N. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477.
Mcfarlane, H. E., Doring, A., and Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Meinert, M. C., and Delmer, D. P. (1977). Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 59, 1088–1097.
Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495.
Mimori-Kiyosue, Y., Grigoriev, I., Lansbergen, G., Sasaki, H., Matsui, C., Severin, F., et al. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153.
Muller, S., Wright, A. J., and Smith, L. G. (2009). Division plane control in plants: new players in the band. Trends Cell Biol. 19, 180–188. doi: 10.1016/j.tcb.2009.02.002
Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., et al. (2007). Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100.
Panteris, E., and Galatis, B. (2005). The morphogenesis of lobed plant cells in the mesophyll and epidermis: organization and distinct roles of cortical microtubules and actin filaments. New Phytol. 167, 721–732.
Paredez, A. R., Somerville, C., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495.
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Pereira, A. L., Pereira, A. J., Maia, A. R., Drabek, K., Sayas, C. L., Hergert, P., et al. (2006). Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell 17, 4526–4542.
Pietra, S., Gustavsson, A., Kiefer, C. S., Kalmbach, L., Horstedt, P., Ikeda, Y., et al. (2013). Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 4:2779. doi: 10.1038/ncomms3779
Qi, J., and Greb, T. (2017). Cell polarity in plants: the Yin and Yang of cellular functions. Curr. Opin. Plant Biol. 105–110. doi: 10.1016/j.pbi.2016.11.015
Ruan, Y., and Wasteneys, G. O. (2014). CLASP: a microtubule-based integrator of the hormone-mediated transitions from cell division to elongation. Curr. Opin. Plant Biol. 149–158. doi: 10.1016/j.pbi.2014.11.003
Seagull, R. W. (1986). Changes in microtubule organization and wall microfibril orientation during in vitro cotton fiber development: an immunofluorescent study. Botany 64, 1373–1381.
Seagull, R. W. (1992). A quantitative electron microscopic study of changes in microtubule arrays and wall microfibril orientation during in vitro cotton fiber development. J. Cell Sci. 101, 561–577.
Smith, L. G., and Oppenheimer, D. G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21, 271–295.
Stramer, B., Moreira, S., Millard, T. H., Evans, I., Huang, C., Sabet, O., et al. (2010). Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. J. Cell Biol. 189, 681–689. doi: 10.1083/jcb.200912134
Van Dop, M., Liao, C., and Weijers, D. (2015). Control of oriented cell division in the Arabidopsis embryo. Curr. Opin. Plant Biol. 25–30.
Wasteneys, G. O. (2004). Progress in understanding the role of microtubules in plant cells. Curr. Opin. Plant Biol. 7, 651–660.
Wittmann, T., and Waterman-Storer, C. M. (2005). Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells. J. Cell Biol. 169, 929–939. doi: 10.1083/jcb.200412114
Xu, W., Zhang, D., Wu, Y., Qin, L., Huang, G., Li, J., et al. (2013). Cotton PRP5 gene encoding a proline-rich protein is involved in fiber development. Plant Mol. Biol. 353–365. doi: 10.1007/s11103-013-0066-8
Yatsu, L. Y., and Jacks, T. J. (1981). An ultrastructural study of the relationship between microtubules and microfibrils in cotton (Gossypium hirsutum L.) cell wall reversals. Am. J. Bot. 68, 771–777.
Yu, J., Jung, S., Cheng, C., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 1229–1236. doi: 10.1093/nar/gkt1064
Zhang, C., Raikhel, N. V., and Hicks, G. R. (2013). CLASPing microtubules and auxin transport. Dev. Cell 24, 569–571. doi: 10.1016/j.devcel.2013.03.008
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhu, S., Xue, F., Zhao, L., Liu, Y., Li, Y., and Sun, J. (2015). Cloning and expression analysis of GhCLASP1 gene in Gossypium hirsutum L. Acta Bot. Boreali Occidentalia Sin. 35, 1941–1948.
Keywords: GhCLASP2, microtubule, trichome, cotton fiber, fiber strength, G. hirsutum
Citation: Zhu S-H, Xue F, Li Y-J, Liu F, Zhang X-Y, Zhao L-J, Sun Y-Q, Zhu Q-H and Sun J (2018) Identification and Functional Characterization of a Microtubule-Associated Protein, GhCLASP2, From Upland Cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:882. doi: 10.3389/fpls.2018.00882
Received: 10 April 2018; Accepted: 06 June 2018;
Published: 27 June 2018.
Edited by:
Taku Takahashi, Okayama University, JapanReviewed by:
Hiroyasu Motose, Okayama University, JapanCopyright © 2018 Zhu, Xue, Li, Liu, Zhang, Zhao, Sun, Zhu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian-Hao Zhu, cWlhbmhhby56aHVAY3Npcm8uYXU= Jie Sun, c3VuamllQHNoenUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.