
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 June 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00867
This article is part of the Research Topic Signal Transduction Pathways in Wheat View all 5 articles
Receptor-like kinases form the largest family of receptors in plants and play an important role in recognizing pathogen-associated molecular patterns and modulating the plant immune responses to invasive fungi, including cereal defenses against fungal diseases. But hitherto, none have been shown to modulate the wheat response to the economically important Fusarium head blight (FHB) disease of small-grain cereals. Homologous genes were identified on barley chromosome 6H (HvLRRK-6H) and wheat chromosome 6DL (TaLRRK-6D), which encode the characteristic domains of surface-localized receptor like kinases. Gene expression studies validated that the wheat TaLRRK-6D is highly induced in heads as an early response to both the causal pathogen of FHB disease, Fusarium graminearum, and its’ mycotoxic virulence factor deoxynivalenol. The transcription of other wheat homeologs of this gene, located on chromosomes 6A and 6B, was also up-regulated in response to F. graminearum. Virus-induced gene silencing (VIGS) of the barley HvLRRK-6H compromised leaf defense against F. graminearum. VIGS of TaLRRK-6D in two wheat cultivars, CM82036 (resistant to FHB disease) and cv. Remus (susceptible to FHB), confirmed that TaLRRK-6D contributes to basal resistance to FHB disease in both genotypes. Although the effect of VIGS did not generally reduce grain losses due to FHB, this experiment did reveal that TaLRRK-6D positively contributes to grain development. Further gene expression studies in wheat cv. Remus indicated that VIGS of TaLRRK-6D suppressed the expression of genes involved in salicylic acid signaling, which is a key hormonal pathway involved in defense. Thus, this study provides the first evidence of receptor like kinases as an important component of cereal defense against Fusarium and highlights this gene as a target for enhancing cereal resistance to FHB disease.
Plants have evolved specialized mechanisms to defend themselves against invading microbial pathogens. Defense signaling cascades are induced upon the recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) (Cohn et al., 2001; Dangl and Jones, 2001; Jones and Dangl, 2006). PRRs are either surface-localized receptor-like kinases (RLKs) or receptor-like proteins (RLPs). RLKs comprise a ligand-binding ectodomain, transmembrane domain, and an intracellular kinase domain, while RLPs contains both an ecto-ligand binding and transmembrane domain with only a short cytoplasmic domain that lacks an obvious signaling intracellular kinase domain (Tör et al., 2009; Sanabria et al., 2010; Antolín-Llovera et al., 2012; Macho and Zipfel, 2014). Surface-localized PRRs lead to PRR-triggered immunity. This is complemented by a second layer of intracellular resistance driven by NOD-like receptor (NLR) PRRs, which recognize virulence effectors secreted within host cells by pathogens, thereby inducing effector-triggered immunity (ETI) (Boller and Felix, 2009; Cui et al., 2015; Delga et al., 2015).
RLKs with leucine-rich repeat (LRR) ectodomains form the largest family of receptors in plants (Torii, 2004; Matsushima and Miyashita, 2012; Fischer et al., 2016). The classic LRR-RLK example is the Arabidopsis flagellin sensing 2 (FLS2), which binds bacterial flagellin (flg22) (Gómez-Gómez and Boller, 2000; Chinchilla et al., 2006). LRR-RLKs can act together in mediating the trade-off between growth and immunity (Belkhadir et al., 2014; Lozano-Durán and Zipfel, 2015). For example, the LRR-RLK brassinosteroid-insensitive 1 (BRI1) -associated kinase (BAK1) forms a complex with FLS2 in order to bind the plant growth regulating brassinosteroid hormones, but BAK1 also complexes with FLS2 to initiate PAMP-triggered immunity (Gómez-Gómez and Boller, 2000; Vert et al., 2005; Chinchilla et al., 2007; Boller and Felix, 2009; Chinchilla et al., 2009; Sun et al., 2013; Wu and Zhou, 2013). The LRR-RLK BAK1-interacting receptor-like kinase (BIR2) was reported to interact with BAK1 in the absence of PAMPs to inhibit autoimmune cell-death responses, thus keeping BAK1 under control. Only upon ligand binding to FLS2 is BAK1 released from BIR2 and recruited to the FLS2 complex to induce PAMP-triggered immune signaling (Halter et al., 2014). Overexpression of Arabidopsis BAK1 led to increased accumulation of salicylic acid (SA) and the deregulation of cell death control genes (Kim et al., 2017).
Fungal chitin is an important ligand for LRR-RLKs. The rice lysine-motif (LysM) receptor kinase has been reported to recognize the fungal elicitor chitin (Kaku et al., 2006) and the Arabidopsis receptor kinase CERK1 binds fungal chitin (Miya et al., 2007). Upon binding chitin, there is a rapid phosphorylation of CERK1 and the activation of early defense responses (Petutschnig et al., 2010). Similarly, CEBiP (chitin elicitor binding protein), which functions in cooperation with CERK1, also activates defense signaling against the rice blast fungus Magnaporthe oryzae and the pathogen overcomes this first line of defense by secreting an effector protein, Secreted LysM Protein1 (Slp1), to cause the disease (Chen et al., 2014). In cereal crops, there are several recent examples of RLKs playing a role in the defense against fungal pathogens. CEBiP and CERK1 play a role in the defense against Septoria tritici blotch (STB) disease; a mutant of the causal pathogen disrupted in the effector Mg3LysM [chitin-binding lysin motif (LysM) containing fungal effector homologue of Ecp6] is normally non-pathogenic, but virus-induced gene silencing (VIGS) of the CEBiP and CERK1 genes enabled the mutant to colonize leaf tissue (Lee et al., 2014, 2015). A recent study demonstrated that the silencing of barley HvCERK1 compromised resistance to Fusarium head blight (FHB) disease and complementary gene expression and metabolomics studies elucidated its impact on downstream resistance-related metabolite accumulation (Karre et al., 2017). Semidwarf “uzu” barley lines encode a spontaneous mutation in the kinase domain of BRI1, which renders them more resistant to the economically important FHB disease and stem base disease caused by Fusarium fungi (Ali et al., 2014). Gene expression studies were used to determine that the uzu derivatives are attenuated in downstream brassinosteroid signaling. The reduction of BRI1 RNA levels via VIGS compromised uzu disease resistance, suggesting that mutated BRI1 in uzu is still in some way functional and that the altered function confers uzu lines with the enhanced disease resistance. The authors concluded that the pathogen resistance of uzu derivatives might be due to pleiotropic effects of BRI1 or the cascade effects of their repressed BR signaling.
A gene expression study highlighted genes involved in the uzu barley response to Fusarium inoculation at both 24 h (Ali et al., 2014) and 48 h post-pathogen inoculaton (Ali et al., unpubl. data). The expression of an uncharacterised LRR-RLK gene was seven-fold higher in spikelets of “uzu” versus wild type barley in response to inoculation with Fusarium culmorum at 48 h post-fungal treatment. Herein, we characterize this barley LRR-RLK gene and its Fusarium-responsive wheat homeologs and their role in cereal resistance to the FHB causal agent Fusarium graminearum. FHB is a devastating disease of wheat that causes yield loss and contaminates grain with the mycotoxin deoxynivalenol (DON). In addition to being harmful to human and animal health, DON is also a fungal virulence factor, aiding pathogen colonization of the wheat tissue (Proctor et al., 1995). Using gene expression analysis, we studied the responsive of the wheat homeologs to DON and DON-producing F. graminearum. Using VIGS, we determined the contribution of the Fusarium-responsive barley and wheat LRR-RLK genes to host resistance to FHB disease and associated yield loss. SA is key hormonal pathway activated as an early response to FHB disease (Makandar et al., 2012; Sorahinobar et al., 2016) and gene expression studies analyzed the effect of LRR-RLK gene silencing in cv. Remus on the transcription of key genes involved in SA accumulation, perception and signaling. Our results highlight this LRR-RLK as a contributor to basal defense against FHB disease and point to its importance as an upstream component of SA signaling in wheat.
Wheat cultivars (cvs.) CM82036 [resistant to both FHB disease and DON; (Buerstmayr et al., 2003)], Remus [susceptible to FHB; (Buerstmayr et al., 1996)], Chinese Spring and its derivative nullisomic tetrasomic lines (obtained from Germplasm Resources Unit, JIC, Norwich1) (Supplementary Table S1) were grown under contained environment conditions, as previously described (Ansari et al., 2007). The barley cv. Akashinriki is susceptible to FHB, while its uzu derivative is more resistant (Khan and Doohan, 2009; Ali et al., 2014). Fresh asexual conidial inoculum (macroconidia) of F. graminearum wild type GZ3639 (Bai et al., 2002) and its’ trichothecene-minus mutant derivative GZT40 (Proctor et al., 1995) and F. culmorum strain FCF200 (kindly provided by Dr. Paul Nicholson, John Innes Centre, Norwich, United Kingdom) were cultured on potato dextrose agar (PDA) (Difco, United Kingdom) plates, incubated at 25°C for 5 days. For conidial production, fungi were cultured in mung bean broth (Bai and Shaner, 1996), harvested, washed and adjusted to 106 conidia ml-1, as previously described (Brennan et al., 2005).
DNA was extracted from wheat spikelets and barley leaves using the HP plant DNA mini kit (OMEGA) following the manufacturer’s instructions. RNA was extracted from wheat heads and barley leaves as previously described (Ansari et al., 2007) and was DNase-treated using the TURBO DNA-free TM kit (Ambion Inc., United States). The quality, yield and integrity of the RNA was analyzed using both the ND-1000 spectrophotometer (NanoDrop, Thermo Fisher Scientific, United States) and electrophoresis. Reverse transcription of total RNA and the quality check of synthesized cDNA for DNA contamination was conducted as previously described (Walter et al., 2008).
The barley LRR-RLK (MLOC_12033.1) on chromosome 6H (Matsumoto et al., 2011) was used as a model for gene cloning. We identified a wheat homolog on chromosome 6DL of wheat cv. Chinese Spring (TRIAE_CS42_6DL_TGACv1_-527217_AA1700660.1) via BLASTn analysis of the wheat genome sequence.2 This gene is hereafter referred to as TaLRRK-6D. TaLRRK-6D from wheat cvs. CM82036 and Remus was cloned and sequenced from mRNA following several rounds of 5′/3′ RACE, using gene-specific primers designed along the coding sequence of TaLRRK-6D (Supplementary Table S2). PCR reactions (25 μl volume) contained 0.5 μl of cDNA template, 2 μM each of gene-specific reverse/forward primer and either 5′ GeneRacer TM primer or 5′ nested/3′ forward GeneRacer TM primer, 1.25 U of Takara LA Taq TM and 1X LA buffer II (Mg2+ plus) (Takara Bio Inc., Japan), and 0.4 mM of each dNTP. Reaction conditions were as follows: 94°C for 2 min, 30 cycles of 94°C for 30 s and 68°C for 3 min and a final extension step at 72°C for 10 min and were conducted in a ProFlex PCR System (Applied Biosystems by Life Technologies, United States). The amplified PCR products were cloned into the pGEM-T vector (pGEM-XL Easy cloning kit; Promega, United Kingdom) and sequenced using both gene-specific and plasmid-specific primers (Supplementary Table S2). The reads were aligned with the deduced full-length gene sequences, which were confirmed to be on chromosome 6DL based on BLASTn analysis against the wheat genome.3
TaLRRK-6D sequences from cvs. CM82036 and Remus were used to extract homologs via BLASTp against the EnsemblPlants database,3 Triticum aestivum (TGACv1) (Kersey et al., 2016) and Gramene (Monaco et al., 2014). These sequences were MAFFT-aligned (Katoh et al., 2002) using Blosum62 matrix (Eddy, 2004), a gap opening penalty of 1.53 and an offset value of 0.123. Then the mean pairwise identity values were used to generate a phylogenetic tree using Jukes Cantor genetic distant model and the Neighbor joining tree building method within the Geneious Tree builder4 (Geneious R9 v.9.1.3) (Kearse et al., 2012). For domain analysis, the amino-acid sequence of TaLRRK-6D was analysis via BLASTp against the plant.ensembl.org and integrated InterPro protein database of all annotated eukaryotic genes in Geneious R9 v.9.1.3. LRR-RLKs were further scanned in Geneious R9 v.9.1.3 for the presence of signature domains, transmembrane domains and signal peptide using SMART5 (Letunic et al., 2015), PROSITE analysis6 (Sigrist et al., 2012), HAMAP7 (Pedruzzi et al., 2015), PRINTS8 (Attwood et al., 2012), PFAM9 (Finn et al., 2013), SUPERFAMILY10 (Wilson et al., 2009) and InterPro11 (Mitchell et al., 2014). Signalp v4.1 (Petersen et al., 2011), TMHMM v2.0 (Krogh et al., 2001) and Phobius (Käll et al., 2007).
An adult plant experiment was conducted to analyze the temporal response of TaLRRK-6D and its homeologs to both DON and FHB disease in the wheat cv. CM82036. The wheat plants were grown under contained glasshouse environment conditions, two plants per pot, as previously described (Ansari et al., 2007), with minor modifications. At mid anthesis (growth stage 65) (Zadoks et al., 1974), the two central spikelets of wheat heads were treated with 20 μl (40 μl per head) of either DON (Santa Cruz, Texas, United States) (5 mg ml-1 in 0.02% Tween-20,), or 106 conidia ml-1 of either wild type F. graminearum strain GZ3639 (WT) or its DON-minus derivative GZT40 which is mutated in the key mycotoxin biosynthesis gene Tri5 (Proctor et al., 1995) or 0.02% Tween-20 (mock treatment). One head was treated per plant. After treatment, the heads were covered with a plastic bag for 2 days to maintain high humidity. Treated spikelets were harvested at either 0, 1, 2, 3, or 5 days post-treatment, flash-frozen in liquid N2 and stored at -70°C prior to RNA extraction. The experiment comprised a total of eight heads per treatment combination (two trials, each containing four heads per treatment combination). For gene expression studies, RNA was extracted from the two treated spikelets per head and, for each treatment combination, RNA samples were bulked to give a total of four RNA samples (two per trial).
The barley stripe mosaic virus (BSMV)-derived VIGS vectors used in this study consisted of the wild type BSMV ND18 α, β, and γ tripartite genome (Holzberg et al., 2002; Scofield et al., 2005). Two constructs were independently used for gene silencing. The two constructs, BSMV:LRR1 and BSMV:LRR2, were designed to preferentially target the kinase domains of barley LRR-RLK on chromosome 6H, its wheat homolog TaLRRK-6D, but not the wheat homologs on chromosome 6A, 6B, 2A, 2B, and 2D (Supplementary Table S3). Construct specificity was determined via a combination of (i) homology of at least 25 nt long with VIGS fragment to the LRR-RLK variant gene sequence in wheat of (cv. Chinese Spring, CS) or barley (cv. Morex), (ii) siRNA finder si-fi tool12 and (iii) quantitative reverse transcriptase PCR analysis (qRT-PCR) with variant-specific primers in wheat (cvs. CM82036 and Remus) or barley (cv. Akashinriki) (Table S3). Fragments were amplified from wheat cv. CM82036 cDNA of TaLRRK-6D using the VIGS primers (Supplementary Table S2; see qRT-PCR section below) and were ligated in the antisense orientation into NotI/Pac1-digested BSMV γ vector pSL038-1 (Scofield et al., 2005). The construct authenticity was verified by sequencing. A BSMV γ vector construct containing a 185 bp-fragment of the barley phytoene desaturase gene (BSMV:PDS) was used as positive control for VIGS, as previously described (Scofield et al., 2005). Prior to RNA synthesis, the vectors were linearized (the vectors containing the BSMV α and γ genomes and the γ genome vectors containing either BSMV:LRR1, BSMV:LRR2, or BSMV:PDS were linearized with MluI, while the BSMV β genome was linearized with SpeI). Capped in vitro transcripts were prepared from the linearized plasmids using the mMessage mMachine T7 in vitro transcription kit (AM1344, Ambion) following the manufacturer’s protocol. RNA quantity and quality were evaluated using ND-1000 spectrophotometer (NanoDrop, Thermo Fisher Scientific, United States) measurement and agarose (Sigma-Aldrich, United States, 1.5%) gel electrophoresis.
Ali et al. (2014) reported the use of a detached leaf assay to analyze the response of barley to F. culmorum. This assay was used to assess the effect of VIGS of the barley LRR-RLK (MLOC_12033.1) on the response of leaves to F. culmorum. Perochon et al. (2015) reported the use of VIGS to silence genes in wheat heads and this assay was used to assess the effect of VIGS of wheat TaLRRK6D on the development of FHB disease caused by F. graminearum on wheat heads. In both the barley and wheat experiments, the VIGS treatment applied were either the VIGS buffer FES or this buffer containing a 1:1:1 mixture of the in vitro transcripts of BSMV α, β, and γ RNA (BSMV:00), or of BSMV α and β plus the γ RNA that contained the appropriate gene fragment (BSMV:PDS, BSMV:LRR1, or BSMV:LRR2) (Scofield et al., 2005).
For the detached leaf barley experiment, the second leaf of 10-day-old barley cv. Akashinriki or its uzu derivative plants were rub-inoculated with the VIGS treatment and, after 7 days, the third leaf of each treated plant was harvested and cut into three sections of 2, 3, and 3 cm in length. The 2 cm section was flash frozen for subsequent gene expression analysis (to determine the efficacy of gene silencing). The two 3 cm long leaf sections were used in the detached leaf phenotyping assay. The center of each leaf section was punctured with a glass Pasteur pipette and treated with a 5 μl droplet of either 0.02% Tween-20 (mock treatment) or conidia of F. culmorum strain FCF200 (106 spores ml-1) as described earlier (Ali et al., 2014). The experiment included three trials, each of which included 10 plates per treatment with two leaf sections per plate. The plates were incubated at 22°C under a 16 h light/8 h dark cycle and all leaf sections were analyzed 4 days post-inoculation for both disease severity and macroconidial production (60 leaves analyzed per treatment combination). Diseased leaf area was estimated using IMAGE-J software analysis of the photographed leaf sections (Perochon and Doohan, 2016). Leaf segments were suspended in 2 ml of distilled water and vortexed and the macroconidial concentration in the water was determined using a haemocytometer (Hycor Biomedical, United States). For gene expression analysis for the barley VIGS leaf experiment, RNA was extracted from individual leaves and then equivalent amounts were bulked from the two leaves within the same plate, resulting in 30 bulk RNA samples per treatment combination (10 per trial).
For the wheat VIGS experiment, just before the emergence of heads of cvs. CM82036 and Remus, the flag leaf was rub-inoculated with the VIGS treatment. At mid-anthesis two central spikelets of heads on VIGS-treated tillers were treated with either 106 conidia ml-1 of F. graminearum strain GZ3639 or of 0.02% Tween-20 (mock treatment) as described above in the time course experiment. After 24 h, the third spikelet above the treated spikelet was harvested, flash frozen in liquid N2 and stored at -70°C for subsequent gene expression analysis (to determine the efficacy of silencing; note that preliminary optimization experiments indicated that in cv. Remus the gene was significantly activated in this distal tissue at 1 dpi). Thereafter, the number of diseased (discolored) spikelets (including treated spikelets) was assessed at 7 and 14 dpi. At harvest, the number of seeds per head and average seed weight (mg) was determined. For disease and yield assessment, the experiment comprised three trials, each including 20 heads/biological replicates per treatment combination (five plants, two per pot and two heads per plant). For gene expression analysis, RNA was extracted from individual spikelets (each representing an independent head) from two of the trials and equivalent amounts of RNA representing the four treated heads per pot were bulked to give a total of ten bulk RNA samples per treatment combination (five per trial).
Real time qRT-PCR was conducted using the Mx3000p Real-Time PCR (Stratagene, Germany). Each PCR reaction contained 1.25 μl of 1:5 (vv-1) dilution of cDNA and 0.2 μM each of the forward and reverse primers (Supplementary Table S2), 1X SYBR® Premix Ex TaqTM (Tli RNase H plus, RR420A, Takara) in a total reaction volume of 12.5 μl. PCR reaction conditions were: 1 cycle of 1 min at 95°C; 40 cycles of 5 s at 95°C and 20 s at 58°C; a final cycle of 1 min at 95°C, 30 s at 58°C, and 30 s at 95°C for the dissociation curve. To analyze the temporal expression and VIGS of TaLRRK-6D and its variants via VIGS, primers were designed for each homeologue that were both variant-specific and targeted a region distinct from the VIGS targets; primers specific to HvLRRK-6H were used to analyze gene silencing via VIGS (Supplementary Table S2). The specificity of the primers was validated via sequence analysis of PCR product clones and by confirming the lack of PCR amplification in DNA from the relevant wheat cv. Chinese Spring nullisomic-tetrasomic lines. The wheat α-tubulin (GenBank no. U76558.1) (Xiang et al., 2011) and TaGAPDH2 (Perochon et al., 2015) were used as housekeeping genes for all wheat qRT-PCR analysis (verified to be unaffected by either VIGS or Fusarium treatments). The barley actin (HvActin, Accession number: AY145451.1) (Ferdous et al., 2015) and Hvα-tubulin (Affymetrix Contig127_s_at) (Ali et al., 2014) were used as housekeeping genes for barley VIGS qRT-PCR analysis. To analyze the effect of silencing of TaLRRK-6D on SA signaling in wheat defense against FHB, primers were designed to amplify variants/homeologs of the SA biosynthesis genes ICS1 and PAL1, the SA regulator NPR1 and the SA receptors NPR3-like and NPR4 (see Supplementary Table S4). Genes from Arabidopsis and Rice were used to identify wheat cDNA homologs via BLASTn in Ensembl Plants [Triticum aestivum (TGACv1)13. The Fusarium responsiveness and expression profile of target ICS1, NPR1, NPR3-like, and NPR4 genes was analyzed using the Wheat Expression Browser,14 PLEXdb (Plant Expression Database), Expression Atlas15 and the SRA database.16 The bulked RNA sample numbers analyzed per treatment combination were: 4 (wheat time course experiment, representing 8 replicates), 30 (VIGS barley leafs, representing 60 replicates), and 10 (VIGS wheat heads for both validation of gene silencing and SA genes, representing 40 replicates). All qRT-PCR analyses were conducted in duplicate for each sample For plant gene expression studies, the threshold cycle (CT) values obtained by qRT-PCR were used to calculate the relative gene expression using the formula = [2-(Ct target gene-Ct of the housekeeping gene1) + (2-(Ct target gene-Ct of the housekeeping gene2)]/2. The relative expression of the ICS1, NPR1, NPR3-like, and NPR4 in samples was expressed as the fold change relative to FES (VIGS buffer) mock treatment and was calculated using the formula [(Etarget)ΔCt target (control-sample)/(Ehousekeeping) ΔCt housekeeping (control-sample)] (Souaze et al., 1996).
The equality of the variance assumption and the normality of data set distribution was assessed using the Levene’s test (P < 0.05) (Stanton and Slinker, 1990). For normally distributed data (time course and VIGS gene expression data), comparative analysis was conducted ANOVA incorporating Tukey’s significant difference test at the 0.05% level of significance. For non-normally distributed data (disease scoring data from the VIGS experiment and spore data from the detached leaf experiment), the significance of differences between treatments was assessed using the Kruskal–Wallis test with Dunn’s post-test (Dunn, 1964) in GraphPad Prism (version 5.03 for Windows; GraphPad Software, San Diego, CA, United States).
A previous barley microarray study highlighted that a LRR gene on chromosome 6H, HvLRRK-6H, was activated in response to F. culmorum in seedling tissue at 48 h post-inoculation (Ali et al., unpubl. data). Bioinformatics analyses showed that the most homologous gene in wheat was on the long arm of chromosome 6D of the cv. Chinese Spring genome and hence it was named TaLRRK-6D. We cloned and sequenced the TaLRRK-6D mRNA sequence from the FHB resistant wheat cv. CM82936 and the FHB susceptible wheat cv. Remus (see Supplementary Table S3 for gene IDs). The ORFs from the three wheat cultivars (cvs. Chinese Spring, Remus and CM82036) shared >97% nucleotide identity. However, at the protein level, the homology was lower, particularly for the cv. Chinese Spring (Supplementary Table S5 and Supplementary Figure S1). The deduced TaLRRK-6D protein from cv. Chinese Spring shared 73% identity with those from cvs. CM82036 and Remus, and 94% homology with a deduced protein encoded by the unpublished stripe rust-responsive gene ID GU84176 (wheat genotype not specified). Deduced protein homeologues on chromosomes 6A and 6B shared 13-53% identity with the chromosome 6D homeologs from cvs. Chinese Spring, CM82036 and Remus, the 6B variant being particularly divergent at the N terminal region (∼13% identity to the cvs. Chinese Spring, CM82036 and Remus chromosome 6D variants) (Supplementary Table S5 and Supplementary Figure S1). Another group of homologous genes were detected on cv. Chinese Spring chromosome 2AL (TaLRRK-2A:CS), 2BL (TaLRRK-2B:CS), and 2DL (TaLRRK-2D:CS), which shared 40.4-58.9% identity with TaLRRK-6D variants from the three wheat genotypes (Supplementary Table S5).
Phylogenetic analysis shows the TaLRRK-6D protein from cvs. CM82036, Remus and Chinese Spring clusters with proteins from other Pooaceae plants (Figure 1). All homologs from other non-Poaceae families clustered with the 6B homeolog from cv. Chinese Spring, which is a shorter protein that is completely devoid of the signature LRR containing ectodomain of LRR-RLKs (Supplementary Figure S2). The deduced TaLRRK-6D, TaLRRK-6A, TaLRRK-2B, and TaLRRK-2D proteins from CS and TaLRRK-6D from cvs. CM82036 and Remus contains leucine rich repeats in its’ ectodomain (that are crucial for protein–protein interactions; (Kobe and Deisenhofer, 1994), a transmembrane domain and a signal peptide, all signatures of membrane-localized RLK proteins (Shiu and Bleecker, 2001; Rameneni et al., 2015) (Supplementary Figure S2). The protein also encoded a putative concanavalin A-like lectin/glucanase domain (IPR013320) sandwiched within the transmembrane domain (Supplementary Figure S2). Phylogenetic analysis, protein domain and motif analysis firmly place TaLRRK-6D in the LRR-RLK receptor protein subfamily XII (Liu et al., 2017), the closest rice and Arabidopsis homologs being LOC Os02g12440 and AT3G47090.1, respectively.
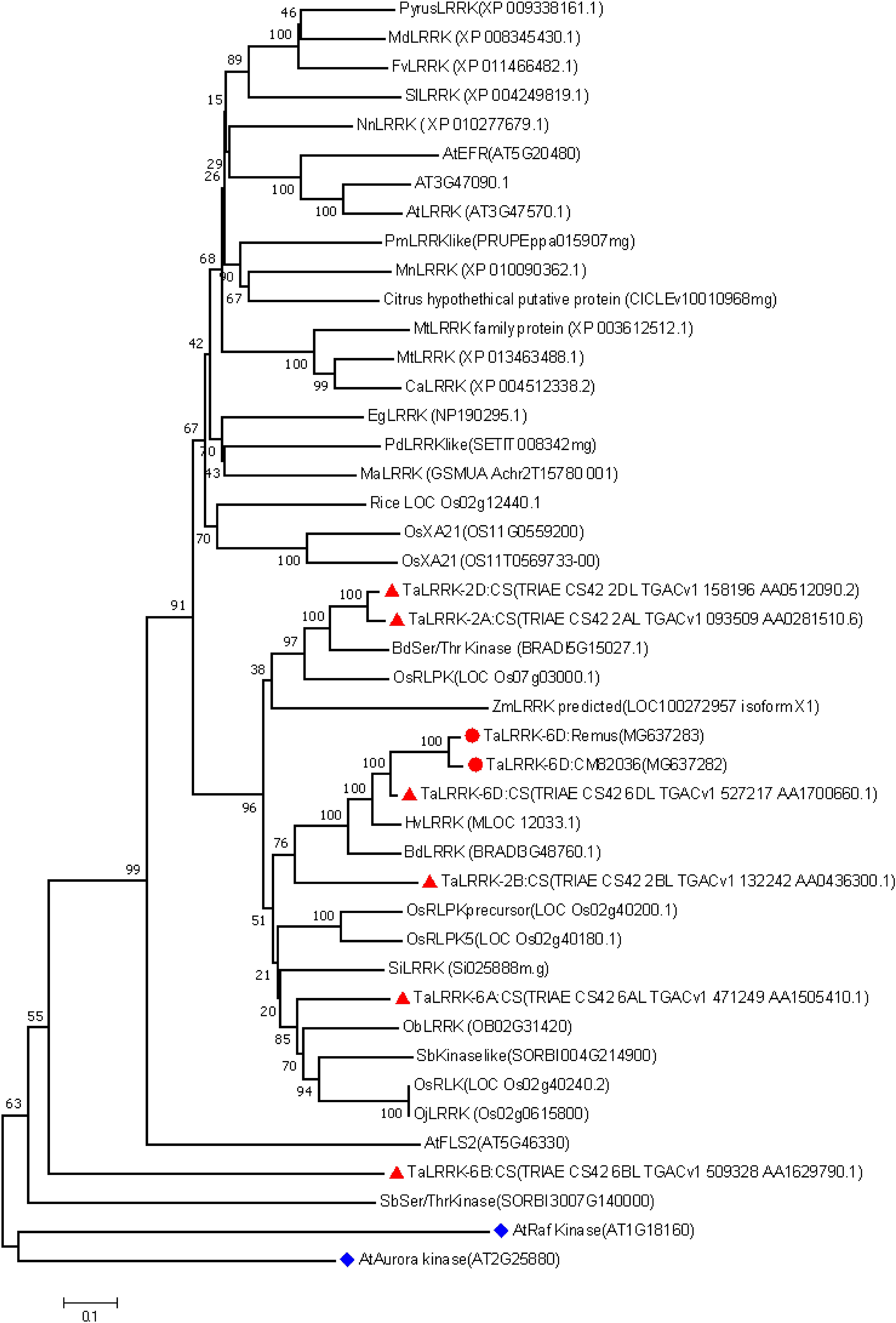
FIGURE 1. Phylogenetic analysis of the relationship between wheat TaLRRK-6D and homologs from across the plant kingdom. Plant homologs of TaLRRK-6D were identified by BLASTp using EnsemblPlants database.3 The cv. Chinese Spring (CS) chromosome 6A, 6B, 6D, 2A, 2B, and 2D TaLRRK variants are highlighted by red triangles; the TaLRRK-6D variants from cvs. CM82036 and Remus are highlighted by red circles; outgroups are highlighted by blue squares. Sequences were MAFFT-aligned (Katoh et al., 2002) using Blosum62 matrix (Eddy, 2004) and mean pairwise identity values were used to generate a phylogenetic tree using Jukes Cantor genetic distant model and the Neighbor joining tree building method within the Geneious Tree builder (Kearse et al., 2012) with bootstrapping (1000 replications). Species abbreviations: Sb, Sorghum bicolor; Os, Oryza sativa; Ob, Oryza brachyantha; Oj, Oryza japonica; Bd, Brachypodium distachyon; At, Arabidopsis thaliana; Hv, Hordeum vulgare; Pc, Pyrus calleryana; Ta, Triticum aestivum; Eg, Elaeis guineensis; Fg, Fragaria vesca; Ma, Musa acuminate; Md, Malus domestica; Mn, Morus notabilis; Ca, Cicer arietinum; Cc, Citrus clementina; Pm, Prunus mume; Pd, Phoenix dactylifera; Mt, Medicago truncatula; Nn, Nalumbo nucifera; Zm, Zea mays, and Si, Setaria italica.
Gene variant-specific qRT-PCR assays were developed and used to determine whether TaLRRK-6D or its homeologs/homologs on chromosome 6A, 6B, 2A, 2B, and 2D were expressed in heads of the FHB resistant cv. CM82036 in response to F. graminearum. TaLRRK-6D expression was induced at 2 dpi, with the effect of the fungus decreasing thereafter (Figure 2A; note that in wheat cv. Remus, it was activated earlier at 1 dpi as determined via subsequent gene silencing experiments illustrated in Figures 3 and 4). The 6A and 6B homeologs were also up-regulated in cv. CM82036 in response to FHB (Figures 2B,C). But for the 6B homeolog, the pathogen induction peaked earlier (1 dpi) as compared to the 6A and 6D homeologs. Although the basal expression of Ta-LRRK-6D was lower as compared to either the 6A or 6B homeologs, the responsiveness to the pathogen was higher than that of 6B (peaks of 3.10 and 1.01-fold induction at 1 and 2 dpi, respectively) and lower than that observed for the 6A homeolog (peaking at 4.3-fold induction at 2 dpi) (Figure 2). The response of the 6A homeolog to FHB was more sustained than that of the chromosome 6B and 6D variants. The FHB-responsiveness of the TaLRRK-2A, TaLRRK-2B, and TaLRRK-2D homologs was also analyzed. None of these homeologs were responsive to the pathogen from 1 to 5 dpi (Figures 2D–F).
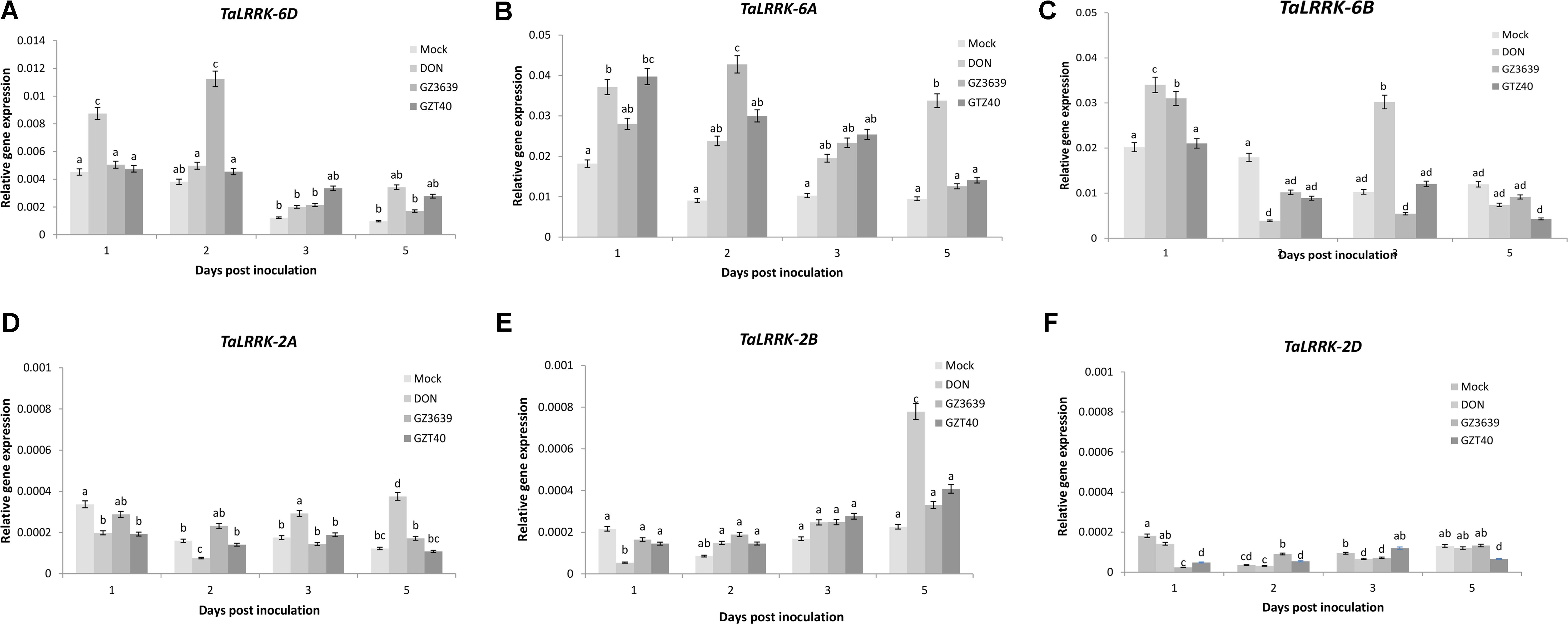
FIGURE 2. Accumulation of transcript from TaLRRK-6D and its homeologs/homologs on chromosomes 2A, 2B, 2D, 6A, and 6B in wheat heads in response to Fusarium graminearum or its toxigenic virulence factor deoxynivalenol (DON). Transcripts: (A) TaLRRK-6D, (B) TaLRRK-6A, (C) TaLRRK-6B, (D) TaLRRK-2A (E) TaLRRK-2B, and (F) TaLRRK-2D. Spikes of wheat heads (Fusarium head blight resistant cv. CM82036) were treated with either mock (Tween20), DON or conidia of wild type DON-producing F. graminearum strain GZ3639 or its DON-minus mutant derivative GZT40. Treated spikelets were harvested at different time points post-treatment as indicated in the figure legend. Expression of TaLRRK-6D and its variants was measured relative to that of the housekeeping genes α-tubulin and GAPDH2. Results represent mean data obtained from 2 trials for FHB and DON (and in each biological replicate, RNA was extracted from a bulk of four heads per treatment per time point and qRT-PCR was conducted twice per bulk RNA sample). Bars indicate SEM. Treatments with the same letter are not significantly different (P > 0.05).
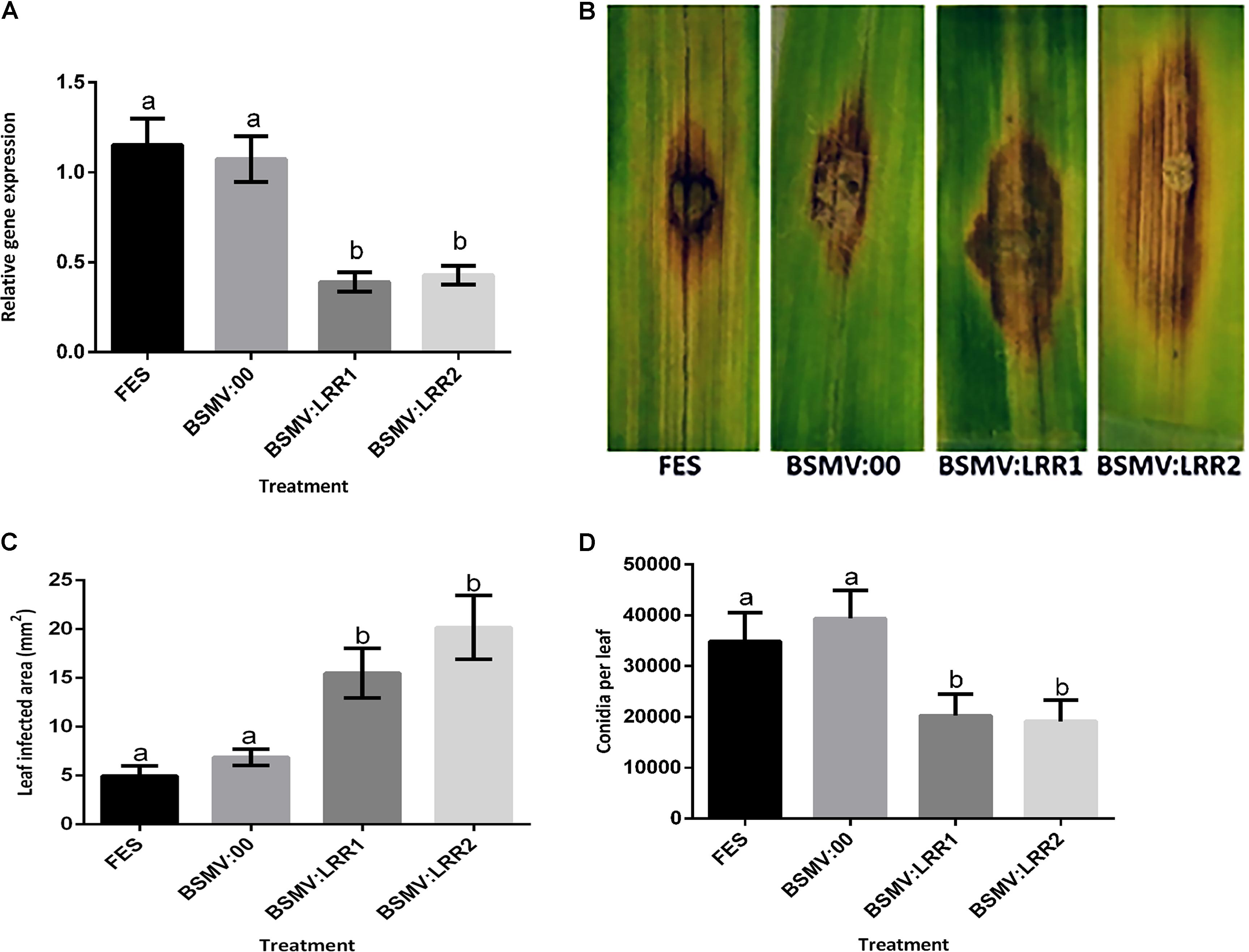
FIGURE 3. Effect of virus induced gene silencing (VIGS) of HvLRRK-6H on the susceptibility of detached barley (cv. Akashinriki) leaves to Fusarium culmorum. Plants were treated with either FES (the VIGS buffer), BSMV:00 (empty vector) or BSMV:LRR1 or BSMV:LRR2 (constructs targeting HvLRRK-6H). VIGS treatments were applied to the second leaf and the third leaf was detached and treated with a droplet of F. culmorum conidia. (A) Gene silencing of HvLRRK-6H in barley leaves was quantified by real-time PCR analysis using reference genes barley actin (HvActin) and α-tubulin (Hvα-tubulin) and the 2ˆ-ΔCt method (Livak and Schmittgen, 2001). (B) Symptoms of leaf necrosis 4 days post-inoculation of spores on HvLRRK-6H silenced Akashinriki lines. (C) Quantification of area of infection using image J software in pixel count and converted to area of (2000 pixel = 0.1 cm2). (D) Macroconidia production by Fusarium on the inoculated leaf segments. Results represent mean data obtained from 3 trials (RNA was extracted from individual leaves and then equivalent amounts were bulked from the two leafs from each plate, resulting in 10 bulked RNA samples per treatment combination per trial). Bars in graphs indicate SEM. Treatments with the same letter are not significantly different (P > 0.05).
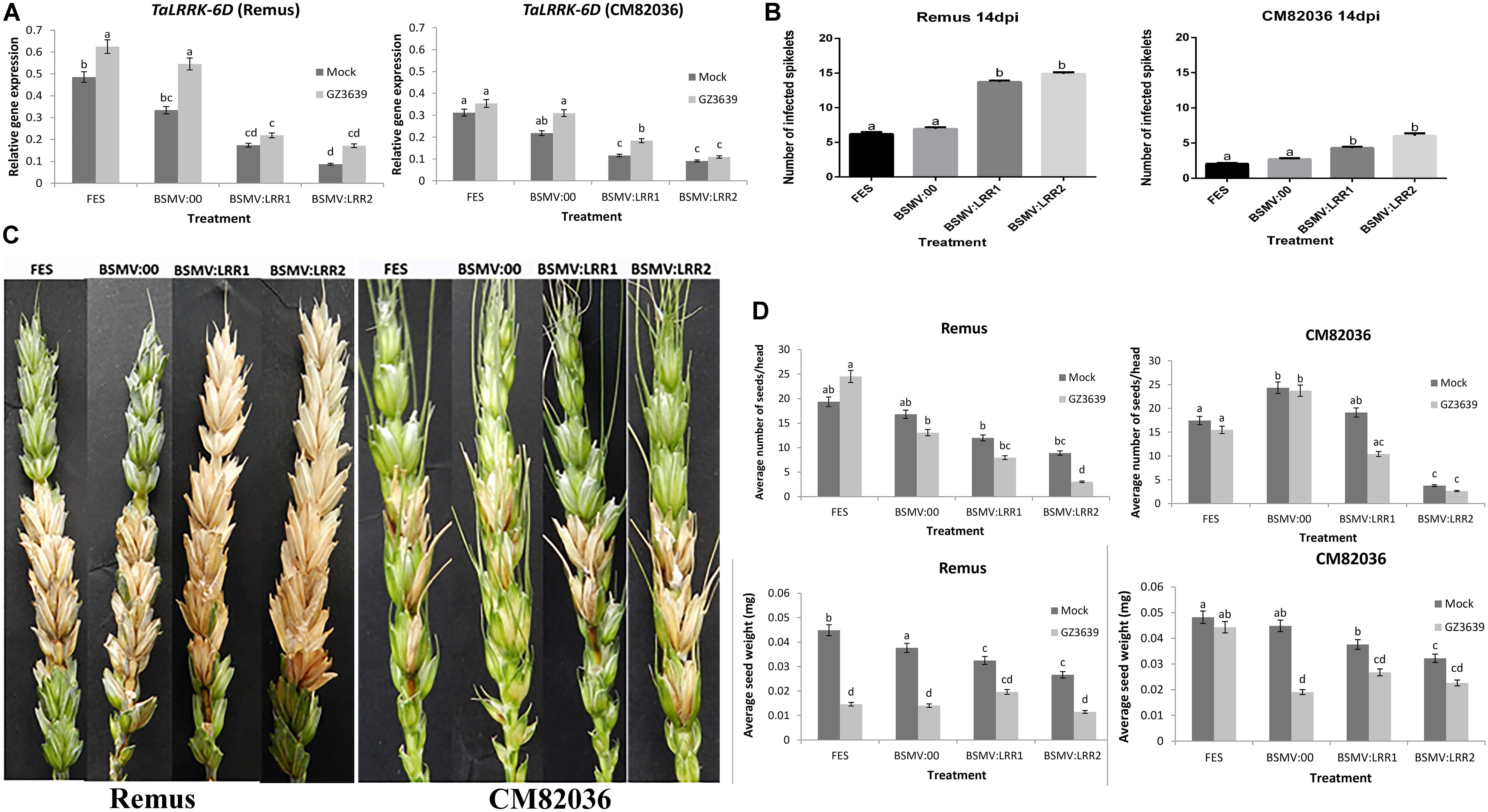
FIGURE 4. Virus-induced gene silencing (VIGS) of TaLRRK-6D in wheat spikelets. Plants of wheat cv. CM82036 and Remus were subjected to VIGS using barley stripe mosaic virus (BSMV) constructs. Plants were treated with either FES (the VIGS buffer), BSMV:00 (empty vector) or BSMV:LRR1 or BSMV:LRR2 (constructs targeting TaLRRK-6D). Flag leaves were treated with virus prior to emergence of the first head, and at mid anthesis (growth stage Zadoks 65) the two central florets of the spikelet were inoculated with either mock (Tween20) or conidia of wild type DON-producing F. graminearum strain GZ3639. The third spikelet above the treated spikelets was collected for gene expression studies. (A) TaLRRK-6D gene silencing in wheat spikelets was quantified by real-time PCR analysis using reference genes α-tubulin and GAPDH2 and the 2ˆ-ΔCt method (Livak and Schmittgen, 2001). Disease symptoms were scored at 14 days post-Fusarium treatment. (B) Images displaying typical disease symptoms. (C) Quantification of the number of diseased spikelet per head to assess the disease progression in both CM82036 and Remus cultivars. (D) Yield analysis expressed as average number of seeds per head and average seed weight. Disease and yield results represent mean data obtained from 60 heads (20 heads per treatment combination in each of three trials) while gene expression represents 40 heads (RNA from five bulks of four heads per treatment combination from two of the three trials). Bars in graphs indicate SEM. Treatments with the same letter are not significantly different (P > 0.05).
Fusarium graminearum is a hemibiotroph (Sutton, 1982; Kazan and Lyons, 2014). In the initial stages of infection, it behaves as a biotroph, feeding off living host tissue, and later (approx. 72 h post-infection), it switches to a necrotrophic lifestyle, feeding off dead host tissue (Oliver and Ipcho, 2004; Król et al., 2015). DON has been shown to facilitate the necrotrophic phase of disease (Bai et al., 2002; Maier et al., 2006; Cuzick et al., 2008) and resistance to the phytotoxic effects of DON is an innate component of FHB resistance (Miller et al., 1985; Mesterházy, 2002; Gunupuru et al., 2017). Thus, in the temporal gene expression experiment, we also analyzed the effect of DON on TaLRRK-6D gene expression and found that the gene was induced as an early response to the toxin at 1 day post-treatment, expression returning to levels comparable to the mock treatment by 2 days (Figure 2A). In order to confirm that endogenous fungal DON levels produced during wheat infection could induce TaLRRK-6D, we included an additional fungal treatment in the time course experiment – i.e., we treated wheat heads with GZT40 which is a non-DON-producing mutant of wild type strain GZ3639 (Proctor et al., 1995). Unlike wild type strain GZ3639, mutant GZT40 did not significantly induce TaLRRK-6D expression at 1 dpi or at any other time point assessed (Figure 2A). Previously, QPCR was used to analyze fungal actin levels as an indicator of fungal biomass in the same RNA extracts and levels for the mutant and wild type was not significantly different up to 5 dpi (Perochon et al., 2015). Thus, the lack of TaLRRK-6D induction by the mutant was not reflective of biomass levels. Hence, we conclude that DON production by F. graminearum facilitates the early induction of TaLRRK-6D as part of the wheat defense response against FHB disease. TaLRRK-6B was similar to the chromosome 6D gene in that both were induced by DON and wild type-F. graminearum, but not by the non-DON producing mutant strain. However, unlike the 6D variant, induction of TaLRRK-6B by DON was biphasic, occurring at both 1 and 3 dpi (Figure 2C). In contrast to TaLRRK-6B and TaLRRK-6D, the TaLRRK-6A homeolog was induced by wild type and DON-minus mutant fungus (and by DON) (Figure 2B). The 6A variant was significantly induced at both day one and day five by DON, but the biphasic nature was not as clear as for the 6B gene (expression levels in response to DON being statistically similar at all 4 days’ P > 0.05). The expression of the 2A, 2B, and 2D homeologs were very low relative to that of 6A, 6B, and 6D variants: none were induced by F. graminearum and induction of the 2A and 2B homeologs by DON did not occur until 5 dpi (Figures 2D–F).
Virus-Induced Gene Silencing was used to determine if silencing the barley HvLRRK-6H altered the hosts’ ability to resist Fusarium infection. We designed two independent non-overlapping, gene-specific VIGS constructs (BSMV:LRR1 and BSMV:LRR2; Supplementary Table S3) that can target both the barley HvLRRK-6H and wheat TaLRRK-6D for silencing. We used the detached leaf assay to assess the effect of HvLRRK-6H silencing on the response of cv. Akashinriki to F. culmorum. Gene-specific qRT-PCR of leaves at 7 days post-pathogen inoculation confirmed that VIGS worked efficiently. Fusarium treatment induced HvLRRK-6H, but in gene-silenced plants (BSMV:LRR1 and BSMV:LRR2) the expression of HvLRRK-6H was significantly reduced by 64%, as compared to the effect of F. culmorum on plants treated with the mock virus (BSMV:00) (P ≤ 0.05) (Figure 3A). At a phenotypic level, treatment with either BSMV:LRR1 or BSMV:LRR2 led to respective 2.6 and 3.2-fold increases in disease lesion size by 4 dpi, relative to BSMV:00-treated plants (Figures 3B,C). These results suggest that a functional HvLRRK-6H is important for barley resistance to Fusarium infection. Thus, we concluded that HvLRRK-6H contributes to barley resistance to F. culmorum. As stated earlier, HvLRRK-6H was originally identified as being up-regulated in the uzu derivative of cv. Akashinriki, as compared to the parental line, in response to Fusarium. A VIGS study in uzu (conducted concurrently with the VIGS study in cv. Akashinriki) validated that HvLRRK-6H also contributed to leaf resistance in this kinase mutant barley derivative (Supplementary Figure S3). Notably, there was no evidence that uzu leaves were more resistant to Fusarium than the wild type parent (comparing Figure 3 and Supplementary Figure S3), suggesting that the Fusarium resistance of uzu is not manifested in the leaves.
Completion of the disease cycle requires the production of conidia that serve as inoculum for the infection of new plants Perochon et al. (2015) recently found that the Pooideae–specific orphan gene TaFROG inhibited lesion development and the number of spores produced by Fusarium on wheat leaves. We also assessed the effect of HvLRRK-6H silencing on the quantity of spores produced by F. culmorum on barley leaves and found that BSMV:LRR1 and BSMV:LRR2–treated sections both contained two-fold less rather than more conidia as compared to BSMV:00 treated plants (P < 0.05) (Figure 3D). Thus we conclude that either the increased lesion size due to gene silencing did not positively affect sporulation at the time point analyzed (i.e., there was no positive association between disease lesion size and conidia as seen for other resistance genes, e.g., Perochon et al., 2015) and/or that the controls contained more ungerminated conidia at this time, relative to the gene-silenced leaves.
We used VIGS to determine if TaLRRK-6D contributes to wheat defense against FHB in heads of both a disease resistant and susceptible genotype (cvs. CM82036 and Remus, respectively). The VIGS constructs BSMV:LRR1 and BSMV:LRR2 used above for barley were also used for wheat as they can also target TaLRRK-6D for silencing (Supplementary Table S3). The constructs specifically targeted the wheat variant on chromosome 6D for silencing (Figure 4A), as compared to either the chromosome 6A, 6B, 2A, 2B, or 2D variants (see Supplementary Figure S4). VIGS did not silence the 6A and 6B genes and the expression of the 2A, 2B, and 2D variants was very low, irrespective of treatment. TaLRRK-6D-specific qRT-PCR of heads at 1-day post-Fusarium treatment validated that, in the absence of gene silencing (FES buffer treatment or empty virus BSMV:00 treatment), the expression of TaLRRK-6D was lower in cv. CM82036 than in cv. Remus, and at this time point the gene was significantly upregulated by Fusarium in cv. Remus but not in cv. CM82036 (P ≤ 0.05). Silencing by either VIGS construct (BSMV:LRR1 or BSMV:LRR2) reduced the transcription of TaLRRK-6D in the two wheat genotypes (Figure 4A). In non-fungal treated heads, treatment with BSMV:LRR1 or BSMV:LRR2 reduced transcript levels by 40–86% in cvs. CM82036 and Remus, and relative to plants treated with the empty virus (BSMV:00). Effects of VIGS on gene expression in fungal treated tissue reflected the effects observed in mock-treated tissue (reductions of 42–69%, relative to BSMV:00; Figure 4A). At the phenotype level, the assessment of heads at 14 days post pathogen treatment showed that, in both wheat genotypes, BSMV:LRR1 and BSMV:LRR2 plants were 2.7-fold more diseased than BSMV:00 plants (Figures 4B,C; Supplementary Figure S5 shows that similar results were obtained at 7 dpi). By 21 dpi, it should be noted that pink fungal growth was visible on diseased spikelets of plants wherein the LRR gene was silenced, often embedded with black sexual spores structures (Supplementary Figure S6).
At harvest, seed numbers and seed dry weight were calculated. The mock virus treatment BSMV itself affected grain development, but to a lesser extent than the gene silencing constructs, particularly BSMV:LRR2 (Figure 4D). The most striking results for grain was that gene silencing, relative to BSMV, retarded grain development in healthy non-diseased heads. In non-fungal treated heads of the two cultivars, silencing of TaLRRK-6D resulted in a 21–85% reduction in the average number of seeds per head and a 28–69% reduction in grain weight, as compared to BSMV:00 treatment (Figure 4D). Thus, we conclude that TaLRRK-6D reduces the severity of the disease symptoms caused by FHB and it also positively contributes to grain development. FHB effects on grain number and weight were usually not significantly exacerbated due to gene silencing (an interesting aside was that the empty virus BSMV negated the FHB resistance in cv. CM82036 in terms of the effect of disease on seed weight).
The involvement of SA signaling in crop defense against the biotrophic phase of FHB disease has been demonstrated via studies on the effect of exogenous SA on key signaling genes (Makandar et al., 2012; Sorahinobar et al., 2016). As shown above, gene expression studies validated that TaLRRK-6D is systemically activated by F. graminearum during this phase, as early as 1 dpi in cv. Remus (Figure 4) and that VIGS of this gene enhanced FHB severity; resulting in very high disease levels in cv. Remus (Figure 4). We thus postulated that TaLRRK-6D might be an upstream and important component of SA defense against FHB. To test this hypothesis, we analyzed the effect of VIGS of TaLRRK-6D in cv. Remus on the expression of genes involved in SA accumulation, signaling and perception (using qRT-PCR studies of the RNA from tissue harvested 1-day post F. graminearum treatment). ICS1 and PAL are key SA accumulation genes as shown in Arabidopsis (Dempsey et al., 2011); PAL1, but not ICS1, was activated during wheat defense against Fusarium (Ding et al., 2011; Makandar et al., 2012; Sorahinobar et al., 2016). The silencing of TaLRRK-6D resulted in the down-regulation of both ICS1 and PAL in both mock and F. graminearum-treated tissue (Figures 5A,B). It is noteworthy that ICS1 was activated by Fusarium in this wheat genotype. The non-induction of PAL1 as compared to ICS1 upon Fusarium infection may be a time factor as the genes targeted by the qRT-PCR primers have been shown to be activated in wheat in response to FHB disease at 30 and 48 or 50 h post-inoculation (Kugler et al., 2013; Xiao et al., 2013) (see Supplemental Results). The key SA regulator gene NPR1 and negative regulators NPR3-like and NPR4 are all induced by either SA (Liu et al., 2005) or by the SA analog 2,6-dichloroisonicotinic acid (INA) (Zhang et al., 2006) (in Arabidopsis) and the silencing of TaLRRK-6D in wheat cv. Remus led to the downregulation of these three genes (Figures 5C–E), indicative of reduced SA signaling. At the time point assessed (1 dpi) these genes were repressed rather than induced by Fusarium (see FES treatment; an interesting aside was that the results for the BSMV:00 mock viral treatment demonstrates that both NPR1 and NPR3-like were down-regulated by the virus used for VIGS but induced by the combination of virus and Fusarium). Like the PAL1 results, findings must be interpreted with caution as the time point (1 dpi) may be too early for Fusarium induction of these genes in this genotype.
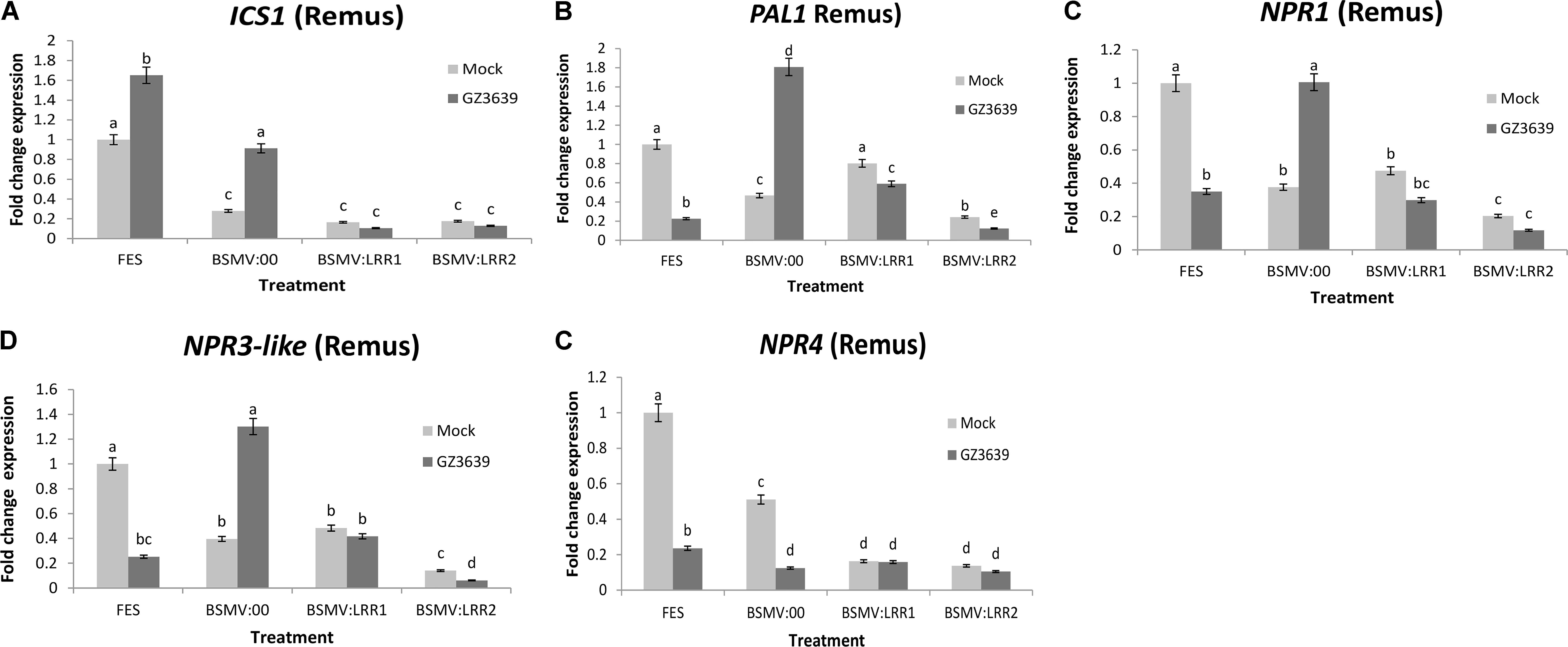
FIGURE 5. Effect of TaLRRK-6D silencing on the expression pattern of SA signaling genes in wheat heads challenged with Fusarium graminearum. The relative expression of SA signaling pathway genes were analyzed in VIGS TaLRRK-6D silenced spikelets samples of wheat cv. Remus 1 dpi of Fusarium. SA signaling transcripts: (A) ICS1, (B) PAL1, (C) NPR1, (D) NPR3-like, and (E) NPR4 showing independent expression with Fusarium treatment and effects of TaLRRK-6D silencing on its expression. The relative expression was calculated using an efficiency corrected model using the formula [(Etarget)ΔCt target (control-sample)/ (Ehousekeeping)ΔCt housekeeping (control-sample)] (Souaze et al., 1996). Thereafter, the values are expressed relative to the fold change for the FES mock treatment. Results represent mean data obtained from 40 heads (two trials and in each RNA was obtained from five bulks of four heads per treatment combination). Bars in graphs indicate SEM. Treatments with the same letter are not significantly different (P > 0.05).
Herein we have identified and characterized a wheat LRR gene and deduced that it and its’ barley homolog both contribute to cereal disease resistance. The results for TaLRRK-6D concur with previous studies, which suggest that LRR genes are induced as part of the early cereal response to FHB disease. A LRR-RLK gene and NBS-LRR genes were induced as part of the early response to FHB disease in wheat (Guo et al., 2006; Subramaniam et al., 2009; Ravensdale et al., 2014; Kosaka et al., 2015) and barley (Huang et al., 2016). TaLRRK-6D induction by F. graminearum was in direct response to the fungal virulence factor DON. We thus deduce that TaLRRK-6D is a signaling molecule involved in the wheat response to DON. It is possible that the induction might be due to the immediate downstream defense responses that are activated in response to DON production by the fungus. It is known that DON has the ability to trigger the production of reactive oxygen species (ROS) and defense gene induction in wheat (Desmond et al., 2006). Two LRR-RLKs (TaRLK1 and TaRLK2) from wheat have been linked to altered ROS homeostasis in the defense response to Blumeria graminis f.sp. tritici (Bgt) infection. The expression of these TaRLK genes was induced upon hydrogen peroxide application and in wheat overexpressing TaRLK there was increased hydrogen peroxide accumulation at the Bgt penetration sites (Chen et al., 2016). The enhanced ROS production during F. culmorum infection of uzu as compared to parent barley lines (Ali et al., 2014) suggest that the kinase activity of the LRR-RLK BRI1 might not be essential for defense responses, but this requires further validation.
ROS and defense gene induction are downstream components of PTI which is activated as a result of PAMPs interacting with RLKs such as TaLRRK-6D (Zipfel, 2014; Silva Couto and Zipfel, 2016). The Fusarium induction of TaLRRK-6D (and the 6A and 6B homeologs) peaks during the biotrophic phase of FHB disease, i.e., within the first 72 h post-inoculation. ROS accumulation would be a valuable counter-attack against biotrophism. It remains to be determined whether TaLRRK-6D plays a role in ROS accumulation or cell death signaling. Similarly, SA is a downstream component of PTI (Newman et al., 2013) and the reduced transcription of SA biosynthesis, regulator and receptor genes as a result of TALRRK-6D silencing, together with the effects of exogenous SA in the early wheat defense against FHB (Makandar et al., 2012), further validate the importance of the RLK as an important component of early cereal defense against F. graminearum. VIGS of TALRRK-6D reduced the amount of ICS1, PAL1, NPR1 NPR3-like, and NPR4 trancript in wheat heads at 1 dpi. VIGS of ICS1 also negated the Fusarium induction of the ICS1 gene at 1 dpi. Reduced ICS1 transcript is indicative of reduced SA acummulation in TALRRK-6D-silenced plants; defective ICS1 in Arabidopsis led to a 90% reduction in SA accumulation in wild-type plants upon pathogen challenge (Dewdney et al., 2000). Reduced SA levels leading to reduced signaling upon TALRRK-6D silencing is supported by reduced basal PAL1, NPR1, NPR3-like, and NPR4 transcript levels, but the time assessed (1 dpi) was likely too early to analyze any Fusarium induction of these genes; indeed, it is interesting to note that they were repressed by the pathogen at this time. The negative effect of VIGS on the basal expression of all SA pathway genes analyzed (and on the early Fusarium induction of ICS1) leads us to hypothesize that TaLRRK-6D is upstream of SA signaling and that silencing of this gene downregulates or attenuates SA signaling in wheat.
The sequence diversity between TaLRRK-6D from different species, genotypes and between this protein and its homeologs is not unexpected. A high level of intrachromosomal segmental (SD) and tandem (TD) duplication among wheat TaLRRKs from chromosomes 6 and 2 has been reported (Shumayla et al., 2016). Similar LRR-RLKs sequence diversity has been reported for homologs of a maize wall-associated receptor-like kinase (ZmWAK-RLK1); the extracellular domain of ZmWAK-RLK1 is highly diverse between different maize genotypes (Hurni et al., 2015). Such diversity in domain composition has also been reported for LRR-RLKs from rice (Sun and Wang, 2011) and brassica (Rameneni et al., 2015). The variation in domain composition found in LRR-RLKs may support the deviation in signal perception and diverse biological roles. There was a clear distinction between the three chromosome 6 TaLRRK homeologs with respect to their temporal and DON-dependent responsiveness to F. graminearum. Unlike 6D, the chromosome 6A homeolog also induced by the DON-minus mutant of the fungus (therefore it was not specific to DON), and the 6B homeolog was clearly induced by DON in a biphasic manner (with a less defined biphasic pattern for the 6A homeolog). The biphasic induction of the 6B homeolog is reminiscent of the biphasic oxidative burst that occurs in many incompatible plant-pathogen interactions, whereby an initial localized burst of ROS is linked to a secondary systemic phase of ROS production (Ren et al., 2006; Torres et al., 2006; Liu et al., 2007; Zurbriggen et al., 2010). Sub-functionalisation of homeologous wheat genes is a relatively new area of study: Powell et al. (2017) recently showed that homeolog expression bias underpins a large proportion of the wheat transcriptome. The temporal and stimulus-specific differences in the FHB induction of these homeologous LRRK genes suggests that the study of their role in disease resistance provides an interesting model to improve our understanding the subtleties of how polyploidy has contributed to the sophistication of wheat defense responses.
When comparing the TaLRRK-6D expression in Remus (mock and Fusarium) versus CM82036, we found that the expression of TaLRRK-6D was always higher. The VIGS analysis indicated that wheat TaLRRK-6D and its barley homolog HvLRRK-6H positively contributes to resistance to F. graminearum. The VIGS study in wheat also confirmed that TaLRRK-6D contributed to defense in both FHB-resistant and susceptible wheat genotypes, with gene silencing enhancing visual disease symptom development. Whether this is true for the Fusarium-responsive wheat A and B homeologs remains to be determined. HvLRRK-6H is the second LRR shown to contribute to barley resistance to FHB disease. HvLRRK-6H was originally highlighted as being overexpressed in uzu barley lines in which the gene encoding the LRR receptor kinase brassinosteroid-insentitive 1 (BRI1) is mutated (Ali et al., unpubl. data). Ali et al. (2014) demonstrated that BRI1 contributes to barley resistance to Fusarium in both seedling and flowering tissue. VIGS of another barley LRR receptor kinase responsive to both powdery mildew (Blumeria graminis f. sp. hordei) and stem rust (Puccinia graminis f. sp. tritici) resulted in reduced defense genes expression, suggesting a tentative PRR role against these fungal pathogens (Parrott et al., 2016). TaLRRK-6D and HvLRRK-6H add a cereal gene to the list of LRR-RLK sub-family LRR XII genes with proven roles in defense against pathogens; this sub-family also includes Arabidopsis FLS2, Arabidopsis EF-Tu receptor and rice Xa21 (Shiu et al., 2004; Schoonbeek et al., 2015; Schwessinger et al., 2015; Thomas et al., 2017).
The reduced grain weight due to the silencing of TaLRRK-6D suggests that it may have role to play in wheat yield components, as previously reported for a rice LRR-RLK. The overexpression of rice LRR-RLK OsLRK1 gene led to a 27% increase in total grain yield per plant (Zha et al., 2009). Recently, overexpression of the wheat LRR-RLK TaBRI1 in Arabidopsis was found to induce early flowering, increased silique size and increased seed yield (Singh et al., 2016). TaLRRK-6D is not the first FHB resistance gene associated with grain development; a wheat ABC transporter (Walter et al., 2015) and cytochrome P450 (Gunupuru et al., unpubl. data) were found to contribute to grain formation. And a genome-wide gene expression profiling of the wheat (in a FHB susceptible cultivar) found that the FHB responsive transcriptome was enriched in genes involved in grain development (Chetouhi et al., 2016). A global expression analysis of rice RLK indicated that they are important players during embryo and endosperm development (Gao and Xue, 2012).
To conclude, this study highlighted the contribution of specific wheat and barley leucine rich repeat receptor like kinase homologs to Fusarium resistance. These should be further investigated as potential candidate genes for both GM and breeding programs that aim to enhance Fusarium resistance in cereals. Based on the current wheat genome sequence, these genes do not collocate with known FHB resistance genetic loci on chromosome 6D. Ongoing studies will determine if variation in the TaLRRK-6D gene/gene promoter or its homeologs contribute to quantitative resistance to FHB. The gene expression studies suggest that the 6A and 6B variants are also important components of the FHB response in wheat and these also merit further study to determine if they contribute to disease resistance. Ultimately, the determination of the interacting partners and potential ligand(s) of TaLRRK proteins will help us better understand the cellular mechanisms underlying FHB resistance and the durability of such defense strategies.
GT and FD planned and designed the research. GT, LG, JH, and AK performed the experiments. GT, EM, and FD analyzed the data. GT and FD wrote the manuscript.
This work was supported by Department of Agriculture, Food and the Marine Research Stimulus Project Wheatenhance (11/S/103) and by Science Foundation Ireland research projects 10/IN.1/B3028 and 14IA2508.
The authors have a patent pending related to this material.
We thank (i) Dr. K. Sato, Barley Germplasm Centre, Okayama University, Japan for providing us with the barley cv. Akashinriki and its’ uzu derivative seed, (ii) Prof. Buerstmayr (BOKU, Austria) for providing us with the wheat cvs. CM82036 and Remus seed, (iii) Dr. Proctor (USDA, United States) for F. graminearum fungi, and (iv) Dr. Scofield (USDA, United States) for providing the VIGS vectors.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00867/full#supplementary-material
Ali, S. S., Gunupuru, L. R., Kumar, G. S., Khan, M., Scofield, S., Nicholson, P., et al. (2014). Plant disease resistance is augmented in uzu barley lines modified in the brassinosteroid receptor BRI1. BMC Plant Biol. 14:1. doi: 10.1186/s12870-014-0227-1
Ansari, K. I., Walter, S., Brennan, J. M., Lemmens, M., Kessans, S., Mcgahern, A., et al. (2007). Retrotransposon and gene activation in wheat in response to mycotoxigenic and non-mycotoxigenic-associated Fusarium stress. Theor. Appl. Genet. 114, 927–937. doi: 10.1007/s00122-006-0490-0
Antolín-Llovera, M., Ried, M. K., Binder, A., and Parniske, M. (2012). Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol. 50, 451–473. doi: 10.1146/annurev-phyto-081211-173002
Attwood, T. K., Coletta, A., Muirhead, G., Pavlopoulou, A., Philippou, P. B., Popov, I., et al. (2012). The PRINTS database: a fine-grained protein sequence annotation and analysis resource—its status in 2012. Database 2012:bas019. doi: 10.1093/database/bas019
Bai, G.-H., Desjardins, A., and Plattner, R. (2002). Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause DiseaseSpread in wheat spikes. Mycopathologia 153, 91–98. doi: 10.1023/A:1014419323550
Bai, G.-H., and Shaner, G. (1996). Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80, 975–979. doi: 10.1094/PD-80-0975
Belkhadir, Y., Yang, L., Hetzel, J., Dangl, J. L., and Chory, J. (2014). The growth–defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem. Sci. 39, 447–456. doi: 10.1016/j.tibs.2014.06.006
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brennan, J., Egan, D., Cooke, B., and Doohan, F. (2005). Effect of temperature on head blight of wheat caused by Fusarium culmorum and F. graminearum. Plant Pathol. 54, 156–160. doi: 10.1111/j.1365-3059.2005.01157.x
Buerstmayr, H., Lemmens, M., Grausgruber, H., and Ruckenbauer, P. (1996). Scab resistance of international wheat germplasm. Cereal Res. Commun. 24, 195–202.
Buerstmayr, H., Steiner, B., Hartl, L., Griesser, M., Angerer, N., Lengauer, D., et al. (2003). Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 107, 503–508. doi: 10.1007/s00122-003-1272-6
Chen, F., Dong, Z., Zhang, N., Zhang, X., and Cui, D. (2014). Different profile of transcriptome between wheat Yunong 201 and its high-yield mutant Yunong 3114. bioRxiv [Preprint]. doi: 10.1101/005496
Chen, T., Xiao, J., Xu, J., Wan, W., Qin, B., Cao, A., et al. (2016). Two members of TaRLK family confer powdery mildew resistance in common wheat. BMC Plant Biol. 16:27. doi: 10.1186/s12870-016-0713-8
Chetouhi, C., Bonhomme, L., Lasserre-Zuber, P., Cambon, F., Pelletier, S., Renou, J.-P., et al. (2016). Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct. Integr. Genomics 16, 183–201. doi: 10.1007/s10142-016-0476-1
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Chinchilla, D., Shan, L., He, P., De Vries, S., and Kemmerling, B. (2009). One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535–541. doi: 10.1016/j.tplants.2009.08.002
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Cohn, J., Sessa, G., and Martin, G. B. (2001). Innate immunity in plants. Curr. Opin. Immunol. 13, 55–62. doi: 10.1016/S0952-7915(00)00182-5
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Cuzick, A., Urban, M., and Hammond-Kosack, K. (2008). Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. New Phytol. 177, 990–1000. doi: 10.1111/j.1469-8137.2007.02333.x
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Delga, A., Le Roux, C., and Deslandes, L. (2015). Plant immune receptor decoy: pathogens in their own trap. Oncotarget 6, 15748–15749. doi: 10.18632/oncotarget.4717
Dempsey, D. M. A., Vlot, A. C., Wildermuth, M. C., and Klessig, D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. doi: 10.1199/tab.0156
Desmond, O. J., Edgar, C. I., Manners, J. M., Maclean, D. J., Schenk, P. M., and Kazan, K. (2006). Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 67, 171–179. doi: 10.1016/j.pmpp.2005.12.007
Dewdney, J., Reuber, T. L., Wildermuth, M. C., Devoto, A., Cui, J., Stutius, L. M., et al. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. doi: 10.1046/j.1365-313x.2000.00870.x
Ding, L., Xu, H., Yi, H., Yang, L., Kong, Z., Zhang, L., et al. (2011). Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One 6:e19008. doi: 10.1371/journal.pone.0019008
Dunn, O. J. (1964). Multiple comparisons using rank sums. Technometrics 6, 241–252. doi: 10.1080/00401706.1964.10490181
Eddy, S. R. (2004). Where did the BLOSUM62 alignment score matrix come from? Nat. Biotechnol. 22, 1035–1036.
Ferdous, J., Li, Y., Reid, N., Langridge, P., Shi, B.-J., and Tricker, P. J. (2015). Identification of reference genes for quantitative expression analysis of MicroRNAs and mRNAs in barley under various stress conditions. PLoS One 10:e0118503. doi: 10.1371/journal.pone.0118503
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2013). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Fischer, I., Diévart, A., Droc, G., Dufayard, J.-F., and Chantret, N. (2016). Evolutionary dynamics of the leucine-rich repeats receptor-like kinase (LRR-RLK) subfamily in angiosperms. Plant Physiol. 170, 1595–1610. doi: 10.1104/pp.15.01470
Gao, L.-L., and Xue, H.-W. (2012). Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 5, 143–153. doi: 10.1093/mp/ssr062
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gunupuru, L., Perochon, A., and Doohan, F. (2017). Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 42, 175–183. doi: 10.1007/s40858-017-0147-3
Guo, P.-G., Bai, G.-H., Li, R.-H., Shaner, G., and Baum, M. (2006). Resistance gene analogs associated with Fusarium head blight resistance in wheat. Euphytica 151, 251–261. doi: 10.1007/s10681-006-9153-0
Halter, T., Imkampe, J., Mazzotta, S., Wierzba, M., Postel, S., Bücherl, C., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. doi: 10.1016/j.cub.2013.11.047
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Huang, Y., Li, L., Smith, K. P., and Muehlbauer, G. J. (2016). Differential transcriptomic responses to Fusarium graminearum infection in two barley quantitative trait loci associated with Fusarium head blight resistance. BMC Genomics 17:1. doi: 10.1186/s12864-016-2716-0
Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785. doi: 10.1073/pnas.1502522112
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Käll, L., Krogh, A., and Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35, W429–W432. doi: 10.1093/nar/gkm256
Karre, S., Kumar, A., Dhokane, D., and Kushalappa, A. C. (2017). Metabolo-transcriptome profiling of barley reveals induction of chitin elicitor receptor kinase gene (HvCERK1) conferring resistance against Fusarium graminearum. Plant Mol. Biol. 93, 247–267. doi: 10.1007/s11103-016-0559-3
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kazan, K., and Lyons, R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26, 2285–2309. doi: 10.1105/tpc.114.125419
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kersey, P. J., Allen, J. E., Armean, I., Boddu, S., Bolt, B. J., Carvalho-Silva, D., et al. (2016). Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44, D574–D580. doi: 10.1093/nar/gkv1209
Khan, M. R., and Doohan, F. M. (2009). Bacterium-mediated control of Fusarium head blight disease of wheat and barley and associated mycotoxin contamination of grain. Biol. Control 48, 42–47. doi: 10.1016/j.biocontrol.2008.08.015
Kim, S. Y., Shang, Y., Joo, S.-H., Kim, S.-K., and Nam, K. H. (2017). Overexpression of brassinosteroid insensitive1-associated receptor kinase 1 causes salicylic acid accumulation and deregulation of cell death control genes. Biochem. Biophys. Res. Commun. 484:4. doi: 10.1016/j.bbrc.2017.01.166
Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19, 415–421. doi: 10.1016/0968-0004(94)90090-6
Kosaka, A., Ban, T., and Manickavelu, A. (2015). Genome-wide transcriptional profiling of wheat infected with Fusarium graminearum. Genomics Data 5, 260–262. doi: 10.1016/j.gdata.2015.06.020
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Król, P., Igielski, R., Pollmann, S., and Kȩpczyńska, E. (2015). Priming of seeds with methyl jasmonate induced resistance to hemi-biotroph Fusarium oxysporum f. sp. lycopersici in tomato via 12-oxo-phytodienoic acid, salicylic acid, and flavonol accumulation. J. Plant Physiol. 179, 122–132. doi: 10.1016/j.jplph.2015.01.018
Kugler, K. G., Siegwart, G., Nussbaumer, T., Ametz, C., Spannagl, M., Steiner, B., et al. (2013). Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genomics 14:728. doi: 10.1186/1471-2164-14-728
Lee, W.-S., Rudd, J. J., Hammond-Kosack, K. E., and Kanyuka, K. (2014). Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 27, 236–243. doi: 10.1094/MPMI-07-13-0201-R
Lee, W.-S., Rudd, J. J., and Kanyuka, K. (2015). Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 79, 84–88. doi: 10.1016/j.fgb.2015.04.006
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Liu, G., Holub, E. B., Alonso, J. M., Ecker, J. R., and Fobert, P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. doi: 10.1111/j.1365-313X.2004.02296.x
Liu, P.-L., Du, L., Huang, Y., Gao, S.-M., and Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17:47. doi: 10.1186/s12862-017-0891-5
Liu, Y., Ren, D., Pike, S., Pallardy, S., Gassmann, W., and Zhang, S. (2007). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51, 941–954. doi: 10.1111/j.1365-313X.2007.03191.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lozano-Durán, R., and Zipfel, C. (2015). Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. doi: 10.1016/j.tplants.2014.09.003
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Maier, F. J., Miedaner, T., Hadeler, B., Felk, A., Salomon, S., Lemmens, M., et al. (2006). Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. doi: 10.1111/j.1364-3703.2006.00351.x
Makandar, R., Nalam, V. J., Lee, H., Trick, H. N., Dong, Y., and Shah, J. (2012). Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant Microbe Interact. 25, 431–439. doi: 10.1094/MPMI-09-11-0232
Matsumoto, T., Tanaka, T., Sakai, H., Amano, N., Kanamori, H., Kurita, K., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 156, 20–28. doi: 10.1104/pp.110.171579
Matsushima, N., and Miyashita, H. (2012). Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules 2, 288–311. doi: 10.3390/biom2020288
Mesterházy, Á. (2002). Role of deoxynivalenol in aggressiveness of Fusarium graminearum and F. culmorum and in resistance to Fusarium head blight. Eur. J. Plant Pathol. 108, 675–684. doi: 10.1023/A:1020631114063
Miller, J., Young, J., and Sampson, D. (1985). Deoxynivalenol and Fusarium head blight resistance in spring cereals. J. Phytopathol. 113, 359–367. doi: 10.1111/j.1439-0434.1985.tb04837.x
Mitchell, A., Chang, H.-Y., Daugherty, L., Fraser, M., Hunter, S., Lopez, R., et al. (2014). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221. doi: 10.1093/nar/gku1243
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Monaco, M. K., Stein, J., Naithani, S., Wei, S., Dharmawardhana, P., Kumari, S., et al. (2014). Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 42, D1193–D1199. doi: 10.1093/nar/gkt1110
Newman, M.-A., Sundelin, T., Nielsen, J. T., and Erbs, G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant. Sci. 4:139. doi: 10.3389/fpls.2013.00139
Oliver, R. P., and Ipcho, S. V. (2004). Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens. Mol. Plant Pathol. 5, 347–352. doi: 10.1111/j.1364-3703.2004.00228.x
Parrott, D. L., Huang, L., and Fischer, A. M. (2016). Downregulation of a barley (Hordeum vulgare) leucine-rich repeat, non-arginine-aspartate receptor-like protein kinase reduces expression of numerous genes involved in plant pathogen defense. Plant Physiol. Biochem. 100, 130–140. doi: 10.1016/j.plaphy.2016.01.005
Pedruzzi, I., Rivoire, C., Auchincloss, A. H., Coudert, E., Keller, G., De Castro, E., et al. (2015). HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res. 43, D1064–D1070. doi: 10.1093/nar/gku1002
Perochon, A., and Doohan, F. M. (2016). Assessment of wheat resistance to Fusarium graminearum by automated image analysis of detached leaves assay. Bioprotocol 6:e2065. doi: 10.21769/BioProtoc.2065
Perochon, A., Jianguang, J., Kahla, A., Arunachalam, C., Scofield, S. R., Bowden, S., et al. (2015). TaFROG encodes a Pooideae orphan protein that interacts with SnRK1 and enhances resistance to the mycotoxigenic fungus Fusarium graminearum. Plant Physiol. 169, 2895–2906. doi: 10.1104/pp.15.01056
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Petutschnig, E. K., Jones, A. M., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Powell, J. J., Fitzgerald, T. L., Stiller, J., Berkman, P. J., Gardiner, D. M., Manners, J. M., et al. (2017). The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 15, 533–543. doi: 10.1111/pbi.12651
Proctor, R., Hohn, T., and Mccormick, S. (1995). Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene: MPMI. Mol. Plant Microbe Interact. 8, 593–601. doi: 10.1094/MPMI-8-0593
Rameneni, J. J., Lee, Y., Dhandapani, V., Yu, X., Choi, S. R., Oh, M.-H., et al. (2015). Genomic and Post-translational modification analysis of leucine-rich-repeat receptor-like kinases in Brassica rapa. PLoS One 10:e0142255. doi: 10.1371/journal.pone.0142255
Ravensdale, M., Rocheleau, H., Wang, L., Nasmith, C., Ouellet, T., and Subramaniam, R. (2014). Components of priming-induced resistance to Fusarium head blight in wheat revealed by two distinct mutants of Fusarium graminearum. Mol. Plant Pathol. 15, 948–956. doi: 10.1111/mpp.12145
Ren, D., Yang, K.-Y., Li, G.-J., Liu, Y., and Zhang, S. (2006). Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 141, 1482–1493. doi: 10.1104/pp.106.080697
Sanabria, N. M., Huang, J.-C., and Dubery, I. A. (2010). Self/non-self perception in plants in innate immunity and defense. Self Nonself 1, 40–54. doi: 10.4161/self.1.1.10442
Schoonbeek, H. J., Wang, H. H., Stefanato, F. L., Craze, M., Bowden, S., Wallington, E., et al. (2015). Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. doi: 10.1111/nph.13356
Schwessinger, B., Bahar, O., Thomas, N., Holton, N., Nekrasov, V., Ruan, D., et al. (2015). Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 11:e1004809. doi: 10.1371/journal.ppat.1004809
Scofield, S. R., Huang, L., Brandt, A. S., and Gill, B. S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. doi: 10.1104/pp.105.061861
Shiu, S.-H., and Bleecker, A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 113, 1–13. doi: 10.1126/stke.2001.113.re22
Shiu, S.-H., Karlowski, W. M., Pan, R., Tzeng, Y.-H., Mayer, K. F., and Li, W.-H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Shumayla, S. S., Kumar, R., Mendu, V., Singh, K., and Upadhyay, S. K. (2016). Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front. Plant Sci. 7:1374. doi: 10.3389/fpls.2016.01374
Sigrist, C. J., De Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2012). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Silva Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Singh, A., Breja, P., Khurana, J. P., and Khurana, P. (2016). Wheat brassinosteroid-insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in Arabidopsis. PLoS One 11:e0153273. doi: 10.1371/journal.pone.0153273
Sorahinobar, M., Niknam, V., Ebrahimzadeh, H., Soltanloo, H., Behmanesh, M., and Enferadi, S. T. (2016). Central role of salicylic acid in resistance of wheat against Fusarium graminearum. J. Plant Growth Regul. 35, 477–491. doi: 10.1007/s00344-015-9554-1
Souaze, F., Ntodou-Thome, A., Tran, C., Rostene, W., and Forgez, P. (1996). Quantitative RT-PCR: limits and accuracy. Biotechniques 21, 280–285.
Stanton, A., and Slinker, B. (1990). Primer of Applied Regression and Analysis of Variance. New York, NY: McGraw-Hill.
Subramaniam, R., Nasmith, C. G., Harris, J., Ouellet, T., Bouarab, K., Brisson, N., et al. (2009). Insight into Fusarium-cereal Pathogenesis. Molecular Plant-Microbe Interactions. Wallingford: CAB International, 319–336.
Sun, X., and Wang, G.-L. (2011). Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PLoS One 6:e16079. doi: 10.1371/journal.pone.0016079
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. doi: 10.1126/science.1243825
Sutton, J. (1982). Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can. J. Plant Pathol. 4, 195–209. doi: 10.1080/07060668209501326
Thomas, N. C., Oksenberg, N., Liu, F., Caddell, D., Nalyvayko, A., Nguyen, Y., et al. (2017). The Rice XA21 ectodomain fused to the Arabidopsis EFR cytoplasmic domain confers resistance to Xanthomonas oryzae pv. oryzae. Peer J. 6:e4456. doi: 10.7717/peerj.4456
Tör, M., Lotze, M. T., and Holton, N. (2009). Receptor-mediated signalling in plants: molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654. doi: 10.1093/jxb/erp233
Torii, K. U. (2004). Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234, 1–46. doi: 10.1016/S0074-7696(04)34001-5
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Vert, G., Nemhauser, J. L., Geldner, N., Hong, F., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201. doi: 10.1146/annurev.cellbio.21.090704.151241
Walter, S., Brennan, J. M., Arunachalam, C., Ansari, K. I., Hu, X., Khan, M. R., et al. (2008). Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct. Integr. Genomics 8, 421–427. doi: 10.1007/s10142-008-0089-4
Walter, S., Kahla, A., Arunachalam, C., Perochon, A., Khan, M. R., Scofield, S. R., et al. (2015). A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 66, 2583–2593. doi: 10.1093/jxb/erv048
Wilson, D., Pethica, R., Zhou, Y., Talbot, C., Vogel, C., Madera, M., et al. (2009). SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37, D380–D386. doi: 10.1093/nar/gkn762
Wu, Y., and Zhou, J. M. (2013). Receptor-like kinases in plant innate immunity. J. Integr. Plant Biol. 55, 1271–1286. doi: 10.1111/jipb.12123
Xiang, Y., Song, M., Wei, Z., Tong, J., Zhang, L., Xiao, L., et al. (2011). A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 62, 5471–5483. doi: 10.1093/jxb/err226
Xiao, J., Jin, X., Jia, X., Wang, H., Cao, A., Zhao, W., et al. (2013). Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:1. doi: 10.1186/1471-2164-14-197
Zadoks, J. C., Chang, T. T., and Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zha, X., Luo, X., Qian, X., He, G., Yang, M., Li, Y., et al. (2009). Over-expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 7, 611–620. doi: 10.1111/j.1467-7652.2009.00428.x
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., and Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi: 10.1111/j.1365-313X.2006.02903.x
Zipfel, C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35, 345–351. doi: 10.1016/j.it.2014.05.004
Keywords: Leucine rich repeat receptor like kinase (LRR-RLK), Triticum aestivum, Fusarium, Pathogen-associated molecular pattern (PAMP), Virus-induced gene silencing (VIGS)
Citation: Thapa G, Gunupuru LR, Hehir JG, Kahla A, Mullins E and Doohan FM (2018) A Pathogen-Responsive Leucine Rich Receptor Like Kinase Contributes to Fusarium Resistance in Cereals. Front. Plant Sci. 9:867. doi: 10.3389/fpls.2018.00867
Received: 03 November 2017; Accepted: 04 June 2018;
Published: 26 June 2018.
Edited by:
Nora A. Foroud, Agriculture and Agri-Food Canada, CanadaReviewed by:
Shawn Clark, National Research Council Canada (NRC-CNRC), CanadaCopyright © 2018 Thapa, Gunupuru, Hehir, Kahla, Mullins and Doohan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fiona M. Doohan, ZmlvbmEuZG9vaGFuQHVjZC5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.