
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Plant Sci. , 22 June 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00843
This article is part of the Research Topic From Functional Genomics to Biotechnology in Ornamental Plants View all 14 articles
The enigmatic nature of the specialized developmental programs of orchids has fascinated plant biologists for centuries. The recent releases of orchid genomes indicate that orchids possess new gene families and family expansions and contractions to regulate a diverse suite of developmental processes. However, the extremely long orchid life cycle and lack of molecular toolkit have hampered the advancement of orchid biology research. To overcome the technical difficulties and establish a platform for rapid gene regulation studies, in this study, we developed an efficient protoplast isolation and transient expression system for Phalaenopsis aphrodite. This protocol was successfully applied to protein subcellular localization and protein–protein interaction studies. Moreover, it was confirmed to be useful in delineating the PaE2F/PaDP-dependent cell cycle pathway and studying auxin response. In summary, the established orchid protoplast transient expression system provides a means to functionally characterize orchid genes at the molecular level allowing assessment of transcriptome responses to transgene expression and widening the scope of molecular studies in orchids.
Orchidaceae represent one of the largest angiosperm families comprising more than 25,000 species that are grown in a wide range of habitats including rainforest, grassland, and even mangrove swamp and low arctic tundra. Orchids have distinct morphological and physiological characteristics such as the co-evolution of pollinators and distinct floral structure (Waterman and Bidartondo, 2008), lack of cotyledon development during embryogenesis (Kull and Arditti, 2002), formation of pollen dispersal units (pollinia) (Pacini and Hesse, 2002), and unique growth and development coupled with mycotrophic strategies (Rasmussen, 2002). These unique developmental programs or strategies have drawn the attention of many evolutionary and plant biologists. Additionally, the wide use of certain orchids as medicinal plants indicates that orchids may have a repertoire of secondary metabolites whose functionality still remains to be explored. (Kong et al., 2003; Bulpitt et al., 2007; Bory et al., 2008). Despite the enormous interest in understanding the molecular mechanisms of the specialized developmental or physiological programs in orchids, the lack of a robust molecular toolkit hampers the advancement of orchid biology.
Recent efforts using next generation sequencing have started to unravel the complexity of the orchid genome and transcriptome atlas (Rao et al., 2014; Cai et al., 2015; Fang et al., 2016; Zhang et al., 2016, 2017; Chao et al., 2017). Progress in the development of tools for manipulation and analysis of cellular processes has promoted research in various orchid species (Yu et al., 2001; Liau et al., 2003; Hsu et al., 2011; Lu et al., 2012; Chen and Fang, 2016; Chen et al., 2016; Hsing et al., 2016). However, studies of orchid gene functions and genetic networks are highly challenging because of a lack of mutant collections and the large amounts of time needed to obtain transgenic orchids.
Protoplast transient expression systems have been widely used to study gene regulation, protein localization, protein–protein interactions, and cell signaling pathways in response to hormones, environmental cues, and pathogen-derived elicitors in model systems (Sheen, 2001; Fraiture et al., 2014). Because of their versatility and ability to detect cell-autonomous responses, protoplast transient expression systems have also been developed and applied to many non-model plants whose transformation platforms are not yet available or for which regeneration of transgenic plants is difficult (Nyman and Wallin, 1992; Hirata et al., 2012; Bu et al., 2014; Lin Y.C. et al., 2014; Kidokoro et al., 2015; Muchero et al., 2015; Kanofsky et al., 2016; Lu et al., 2016; Nanjareddy et al., 2016; Thevenin et al., 2016; Shen et al., 2017; Wang et al., 2017). In addition, protoplast-based transient expression systems allow the study of immediate transcriptome responses to expression of the genes of interest and provide an alternative means to characterize and analyze the cellular functions and regulatory networks of such genes.
The protoplast system has also been used to investigate and discover signaling transduction pathways in various plants (Sheen, 2001; Asai et al., 2002; Baena-Gonzalez et al., 2007; Müller and Sheen, 2008; Boudsocq et al., 2010; Fraiture et al., 2014). Even though protoplast isolation has been reported in Phalaenopsis orchids, the requirement of callus induction for protoplast preparation makes the procedure difficult to implement (Kobayashi et al., 1993; Shrestha et al., 2007). A recent study reported a protoplast-based transient gene expression protocol in a Phalaenopsis hybrid cultivar (Li et al., 2018). Inconveniently, this protocol requires young leaves of shoots induced from flower nodal buds that may not be readily available in different Phalaenopsis cultivars. Also, the transfection efficiency is below 50%, a minimum threshold (Yoo et al., 2007) required to obtain reliable and repeatable data for molecular studies. Furthermore, the protoplast transient expression protocol and its broad usage for functional genomics studies have not been rigorously tested. To simplify the protoplast preparation procedure and establish a system for rapid gene regulation studies for orchids, here an optimized petal-based protoplast isolation and transient expression protocol was established. This protoplast transient expression system worked successfully in investigating subcellular localization of proteins and protein–protein interaction. In addition, our results demonstrate its amenability for studies of transcription activity of PaE2F3/PaDP transcription factors and auxin response. Taken together, development of an orchid protoplast transient expression assay provides a versatile experimental platform to enable molecular, cellular, and functional studies of orchids. Because experimental settings and empirical experience are provided, the testing parameters may easily be adjusted for different orchid species.
Tetraploid Phalaenopsis aphrodite subsp. formosana (m1663) plants in 3.5-inch pots were purchased from Chain Port Orchid Nursery (Ping Tung, Taiwan). Plants were grown and maintained as previously described (Chen and Fang, 2016). Under flowering inductive conditions [alternating 12 h light (23°C)/12 h dark (18°C) cycles], the floral stalks (∼0.5 to 1 cm long) became visible approximately 2 months after treatment. The first open flower appeared approximately 3–4 months after treatment.
Fully open flower petals were used for protoplast isolation. Petal protoplasts were successfully isolated from petals collected 1–15 days after full bloom. Orchid petals were cut into 0.5–1.0-mm strips using a fresh sharp razor blade. The petal strips were transferred to a petri dish containing freshly prepared enzyme solution. The enzyme solution was made as follows: 1% (w/v) cellulase R-10 (Yakult Pharmaceutical), 0.25% (w/v) macerozyme R-10 (Yakult Pharmaceutical), 0.7 M (or otherwise described in the Results) mannitol (Sigma), 20 mM KCl (Sigma), and 20 mM MES (pH 5.7, Sigma) were warmed up to 55°C for 10 min to enhance enzyme solubility and to inactivate DNase and protease. The enzyme solution was allowed to cool to room temperature before 10 mM CaCl2 and 0.1% BSA (Sigma cat #A7906) were added. The enzyme mixture was then filtered and sterilized by 0.45 μm Millex-HP filter (Millipore). The petal strips were then completely submerged in the enzyme mixture and allowed to digest without agitation in the dark for approximately 16 h (or as described in the section “Results”). Carbenicillin was added to a final concentration of 50 μg/ml to avoid bacterial contamination. Adding carbenicillin during the protoplast preparation is strongly recommended for petals collected from the greenhouse.
After digestion, the enzyme mixture was gently agitated to release the protoplasts and the protoplast/enzyme suspension was diluted with equal volume of wash and incubation solution (WI-0.7) that contained 0.7 M mannitol (or as described in the section “Results”), 20 mM KCl, and 4 mM MES (pH 5.7). The protoplast/enzyme solution was then filtered through a 100-μm nylon mesh (BD Falcon) to remove tissue debris. (Note: the mesh is normally kept in 95% ethanol and rinsed with WI-0.7 solution before use). The flow-through was then centrifuged at 200 g for 2 min in a desktop centrifuge (Eppendorf 5810R) to pellet the protoplasts. The acceleration ramp was set to 2 and deceleration ramp was set to 0. The supernatant was removed and the pellet was gently resuspended in 3 ml WI-0.7 solution. The protoplast suspension was washed gently one more time with 3 ml WI-0.7 solution. The cell concentration was measured using a hemocytometer. The protoplast suspension was kept on ice for 30 min. The protoplast suspension was briefly centrifuged at 200 g to pellet protoplasts. WI-0.7 solution was carefully removed and the pellet was resuspended in pre-chilled MMG-0.7 solution (0.7 M mannitol, 15 mM MgCl2, and 4 mM MES, pH5.7) to obtain a cell concentration of approximately 1.0 × 106/ml. Based on our protocol, 20 ml of enzyme solution can digest up to 20 orchid petals (10 flowers) and yield 5 ml of ∼1.0 × 106/ml protoplasts before transfection. Approximately 1.0 × 106 cells (from 4 petals) are required for each RNA preparation.
A modified PEG-mediated protoplast transfection protocol (Yoo et al., 2007) was used. Orchid protoplasts were adjusted to a final concentration of ∼0.5–2 × 105 cells/ml with MMG-0.7 solution. Twenty microliters of 10 to 20 μg plasmid DNA was mixed with 200 μl protoplasts (∼1–4 × 104 cells) in MMG-0.7 solution and an equal volume (220 μl) of freshly prepared PEG-calcium transfection solution was added. PEG-calcium transfection solution (40% w/v PEG4000, 0.6 M mannitol, and 0.1 M CaCl2) was prepared as follows: PEG4000 (Fluka, cat. no. 81240) and mannitol were first dissolved in water by heating up to 60°C for approximately 10–20 min. After the solution was cooled down to room temperature, CaCl2 was added. The DNA-PEG-calcium-protoplast solution was mixed gently and incubated at room temperature for 6–10 min. After transfection, the transfected protoplast mixture was immediately diluted with 2–3 ml of WI-0.7 solution and centrifuged at 200 g for 2 min. Then the supernatant was carefully removed. The transfected protoplasts were washed one more time with 2–3 ml of WI-0.7 solution followed by centrifugation. The protoplasts were gently resuspended in 1 ml WI-0.7 solution. The protoplast mixture was carefully removed and the transfected protoplasts were incubated in WI-0.7 solution in 12-well tissue culture plates or Eppendorf tubes pre-rinsed with 1% BSA solution for the desired amount of time before further analysis. It is recommended that ∼1 × 106 cells/ml are used and the amount of plasmid DNA is scaled up if transfected protoplasts need to be harvested for RNA or protein extraction.
For hormone treatment, transfected protoplast cells were treated with 1 μM 1-naphthaleneacetic acid (NAA), 100 nM trans-zeatin, 50 μM gibberellic acid 3 (GA3), or 100 μM abscisic acid (ABA) (Müller and Sheen, 2008) for 2 h before microscopic observation. Fluorescence images were photographed on a LSM 710 Confocal Microscope (Zeiss) or Zeiss Axio Scope A1 microscope equipped with an AxioCam HRc camera.
Propidium iodide (PI) was dissolved in 0.65 M mannitol to make 0.5 mg/ml stock solution. Fluorescein diacetate (FDA) was dissolved in acetone to make 5 mg /ml stock solution. A total of 20 μl of PI and 20 μl FDA stock solutions were added in 1 ml 0.65 M mannitol as the staining solution (this has to be made fresh). For staining, 10 μl of staining solution was added into 20 μl of isolated protoplast cells and incubated at room temperature for 1–2 min. The living protoplast cells (green, stained with FDA) and dead cells (red, stained with PI) were visualized and photographed by LSM 710 confocal microscope (Zeiss). Five to twelve snapshots were taken for each sample. For viability measurement, at least 150 protoplast cells were examined from each sample. Viability was measured as green cells/green + red cells × 100%. These experiments were repeated at least three times.
The transfected protoplasts were flash frozen in liquid nitrogen and stored in a freezer at –80°C. RNA was isolated using RNA extraction reagent (3-Zol, MDBio, Inc.) according to the manufacturer’s instructions. To remove DNA, total RNA was treated with RNase-free DNase (Qiagen) followed by RNeasy column purification (Qiagen) according to the manufacturer’s instruction.
RNA (0.4 to 1 micrograms) was reverse transcribed in the presence of a mixture of oligo dT and random primers (9:1 ratio) using the GoScript Reverse Transcription System (Promega) as described previously (Lin H.Y. et al., 2014). Ten microliters of quantitative RT-PCR reaction contained 2.5 μl of 1/20 diluted cDNA, 0.2 μM of primers, and 5 μl of 2X KAPA SYBR FAST master mix (KAPA Biosystems). Real-time RT-PCR was carried out using a Bio-Rad CFX96 (Bio-Rad). The following program was used for amplification: 95°C for 1 min, 40 cycles of 95°C for 5 s, and 58°C for 20 s. PCR was performed in triplicate, and the experiments were repeated with RNA isolated from three independent samples. Fold change in expression was calculated as 2-ΔΔCT. A melting curve of each PCR was examined to ensure no spurious products were present. Primer pairs used for quantitative PCR are listed in Supplementary Table S1. Because expression level of ubiquitin (PaUBI1) remained relatively constant across the tissues examined (Lin H.Y. et al., 2014), it was used as an internal control.
Petal protoplast isolation has been reported in Dendrobium orchid (Hu et al., 1998). We therefore chose petals of Phalaenopsis aphrodite as our starting materials. The release and integrity of petal protoplasts was visually inspected in cellulose- and macerozyme-containing enzyme solution adjusted to different osmotic conditions (in 0.4 M, 0.6 M, 0.7 M, or 0.8 M mannitol). Protoplasts were successfully released from petal tissues and remained intact after overnight (∼16 h) enzyme digestion (Figure 1A) regardless of the concentrations of mannitol tested (Figure 1B). To survey the viability of the petal protoplasts, PI and FDA, which mark dead and live cells, respectively (Huang et al., 1986), were used to stain the isolated protoplasts. Petal tissue digested with enzymes for 16 h gave a better yield than that digested for only 8 h (Table 1). Moreover, the protoplast viability was not compromised after 16 h of digestion (Table 1). Approximately 90–94% of protoplasts were viable after resuspension in WI buffer supplemented with various concentrations of mannitol (Table 2 and Figure 1C). More than 80% of protoplasts remained viable after resuspension in MMG-0.6 and MMG-0.7 solution (Table 2 and Figure 1D). Approximately 75.3% and 77.5% of protoplast cells prepared in 0.6 M and 0.7 M mannitol-based solution, respectively, remained viable after the transfection procedure (Table 2). Even though there was no drastic difference in cell viability with preparation in 0.6 M or 0.7 M mannitol-based buffer, 0.7 M mannitol seemed to work slightly better in protecting cells during the transient expression procedure. Therefore, 0.7 M mannitol was chosen for the rest of the experiments. Under this condition, the yield of viable protoplasts from one petal after transfection was estimated to be approximately 1.9 × 105 cells [2.5 × 105 cells (number of protoplast cells per petal) × 77.5% (viability of transfected protoplast prepared in 0.7 M mannitol condition)]. The size of petal protoplasts was calculated and it ranged from 20 to 50 μm in diameter with an average of approximately 34 ± 7.1 μm in diameter (Supplementary Datasheet S1). These results demonstrated the feasibility of using orchid protoplasts for further molecular biology analyses.
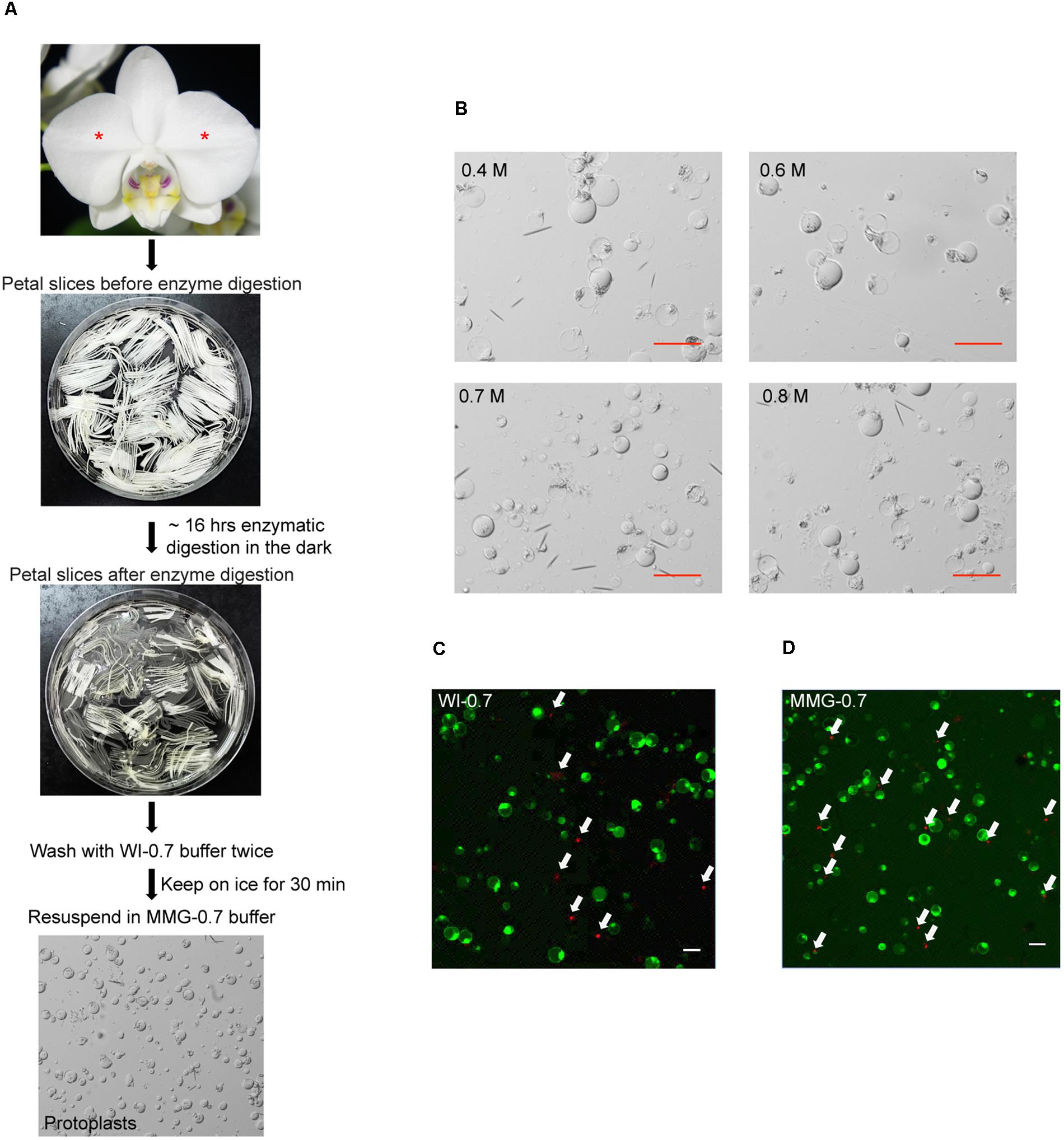
FIGURE 1. Isolation of protoplasts from petals of P. aphrodite. (A) A flowchart showing the procedure of protoplast isolation. Asterisks mark the petals used for protoplast isolation. (B) Protoplast morphology remained intact in enzyme solution supplemented with 0.4 M, 0.6 M, 0.7 M, or 0.8 M mannitol. (C) A snapshot of cell viability test after resuspending cells in WI-0.7 solution. (D) A snapshot of cell viability test after resuspending cells in MGG-0.7 solution. White arrows indicate the dead cells stained by PI. Red and white scale bars, 50 μm.
Leaves are readily accessible and mesophyll protoplast cells have been successfully isolated from leaf tissues of various plant species (Sheen, 2001; Chen et al., 2006; Yoo et al., 2007; Mazarei et al., 2008; Lung et al., 2011; Masani et al., 2014; Nanjareddy et al., 2016; Shen et al., 2017). Therefore, we also tested the conditions for mesophyll protoplast preparation. The youngest fully expanded leaves were used for this test. Similarly, different osmotic conditions of enzyme solution (in 0.4 M, 0.6 M, 0.7 M, or 0.8 M mannitol) were tested and the integrity of leaf mesophyll protoplasts during enzyme digestion was inspected under a microscope over time. Unlike petal protoplasts, the integrity of mesophyll protoplasts declined quickly over time. For the cells that retained relative integrity, the interior content of protoplasts including chloroplasts was gradually concentrated and pushed to one side of the cell (Supplementary Figure S1A). Mesophyll protoplast cells started to rupture after 2 h of incubation in enzyme solution (Supplementary Figure S1B). The cell integrity was completely disrupted after transfection (Supplementary Figure S1C).
To test whether petal protoplasts were suitable for subcellular localization studies, nuclear and plasma membrane markers were transformed into protoplasts by PEG-mediated transformation (see Methods). As expected, nuclear marker mCherry-VirD2NLS (Lee et al., 2008), which carries the nuclear localization signal, started to appear in the nucleus 4 h after transfection (Figure 2A) and accumulated in almost all the inspected nuclei 20 h after transfection (Figure 2B). Unlike biolistic transient assay where cells with fluorescence signals were sparsely distributed on the petal due to uneven spraying of the gold particles (Supplementary Figure S2), the majority of protoplasts (>80%, Supplementary Datasheet S2) had fluorescence signals, indicating protoplast-based transfection is relatively homogenous and enables broader molecular and biochemical analyses of the transgene-encoded protein product (Figure 2B). This transfection efficiency is almost equivalent or better than protoplast transfection efficiency reported in the model systems (Yoo et al., 2007; Wu et al., 2009; Faraco et al., 2011). In addition, the plasma membrane marker aquaporin AtPIP2a-YFP (Nelson et al., 2007) was correctly targeted to the plasma membrane (Figure 2C). These experiments demonstrated that Phalaenopsis orchid protoplasts are suitable for protein localization studies.
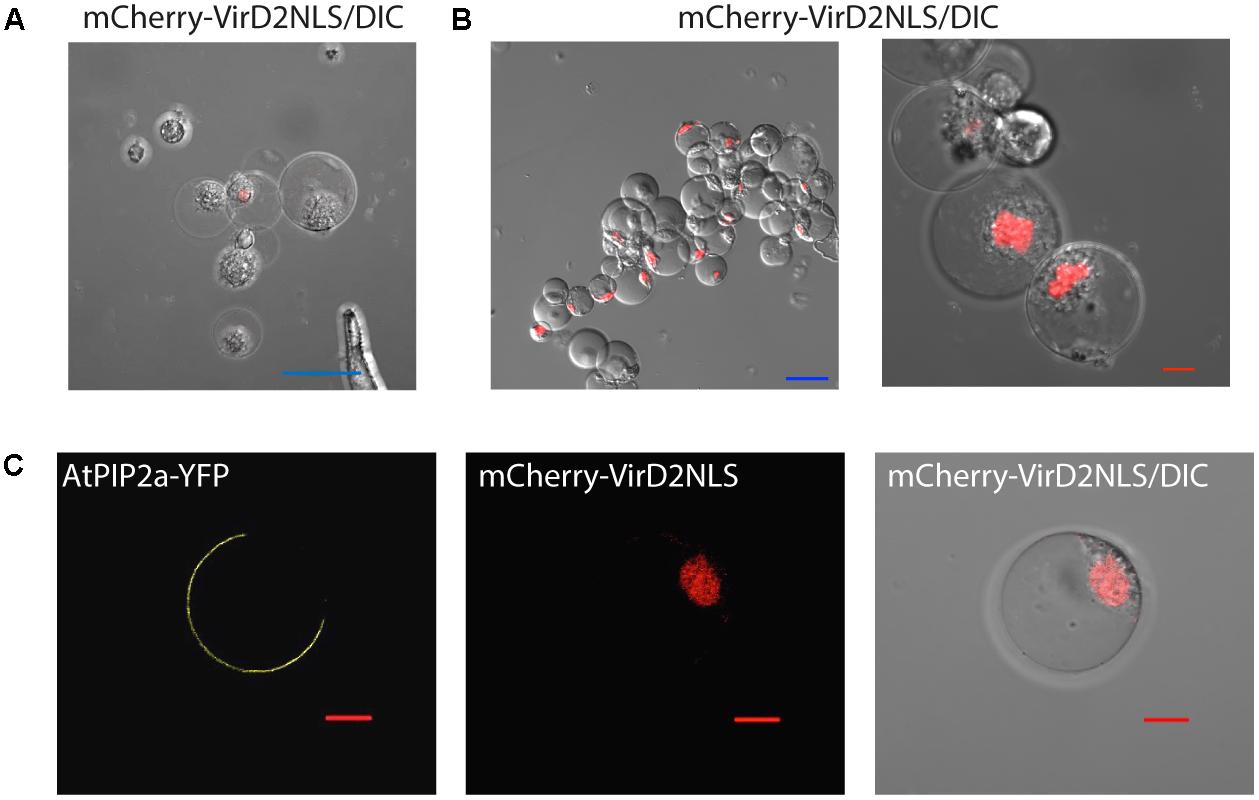
FIGURE 2. Subcellular localization of protein markers. (A) Nuclear localization of mCherry-VirD2NLS marker 4 h after transfection. (B) The vast majority of cells expressed the nuclear marker 20 h after transfection. (C) Plasma membrane localization of AtPIP2a-YFP aquaporin marker. Note that the protoplasts were co-transfected with AtPIP2a-YFP and mCherry-VirD2NLS markers. Blue scale bar, 50 μm; red scale bar, 10 μm. DIC, differential interference contrast image of cells superimposed with the fluorescence marker.
The application of petal protoplasts to bimolecular fluorescence complementation (BIFC) analysis for protein–protein interaction study was also tested. Cyclin-dependent kinases (CDKs) are central cell-cycle regulators whose activities are regulated by physical interaction with the cell-cycle phase specific cyclins (CYCs) (Morgan, 2007). To test the feasibility of using orchid protoplasts for protein–protein interaction studies, the construct containing the PaCDKA gene fused N-terminal half of the EYFP (N-(nEYFP)-CDKA1) was co-transfected with a construct containing PaCYCD3;1 fused to a C-terminal half of the EYFP (N-cEYFP-CYCA3;1). The direct interaction of PaCDKA and PaCYCD3;1 scored by the presence of yellow fluorescence was verified by confocal microscopy (Figure 3A and Supplementary Figure S3A). As a negative control, the construct containing N-(nEYFP)-CDKA1 was co-transfected with only the C-terminal half of the EYFP (N-cEYFP) construct. No fluorescence was detected when the N-cEYFP construct was co-transfected with the N-(nEYFP)-CDKA1 construct (Figure 3B and Supplementary Figure S3B). This is consistent with biolistic-based BIFC assay in petal cells (Lin H.Y. et al., 2014).
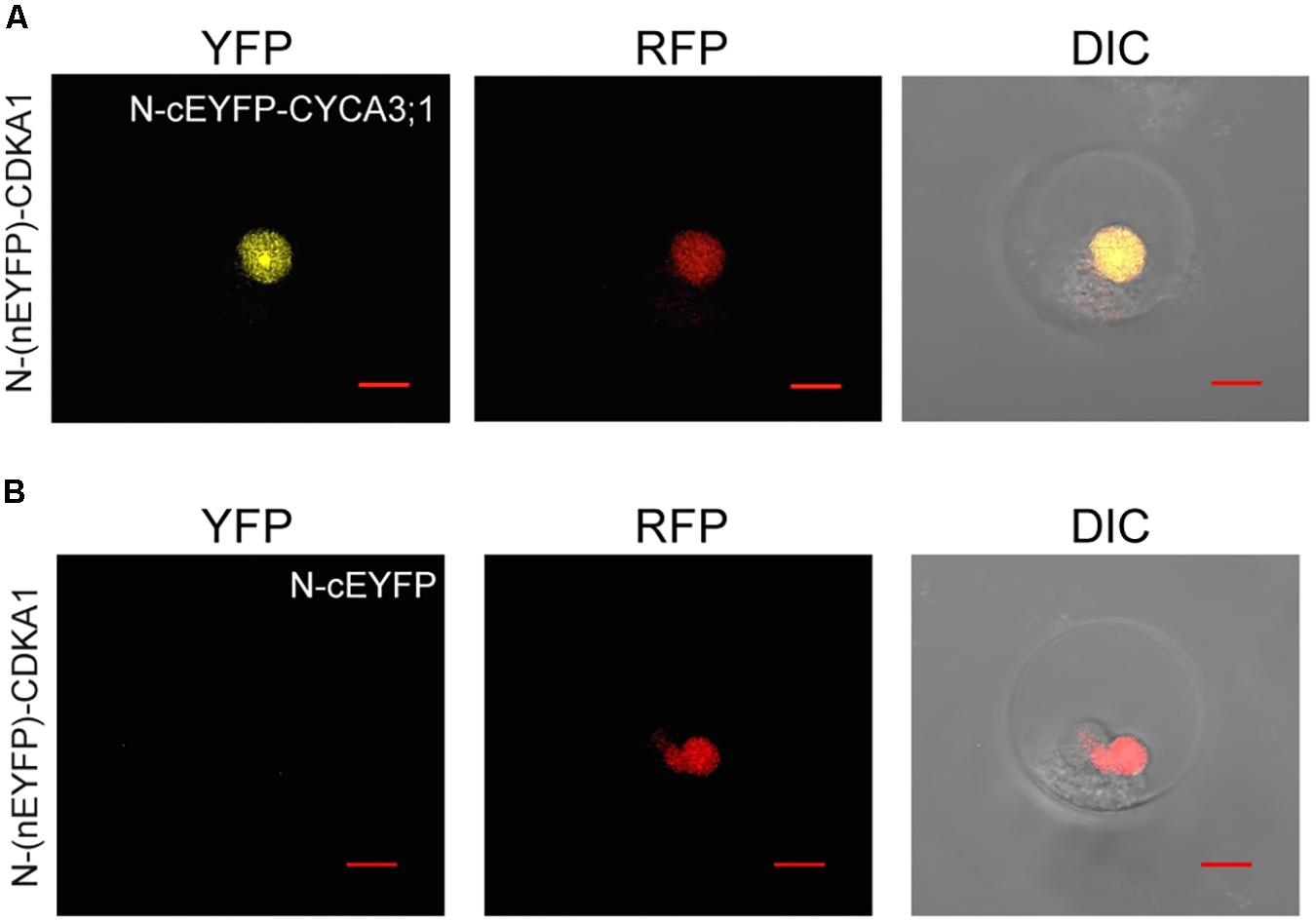
FIGURE 3. Protein–protein interactions of CDKA1/CYCA3;1 proteins visualized using BiFC. (A) The BiFC signal was detected in protoplast cells co-transfected with N-(nEYFP)-CDKA1 and N-cEYFP-CYCA3;1 constructs. (B) No signal was detected in protoplast cells co-transfected with N-(nEYFP)-CDKA1 and N-cEYFP constructs. mCherry-VirD2NLS was used as a nuclear marker. DIC, differential interference contrast images of cells superimposed with YFP and RFP channels. Scale bar, 10 μm.
The cell division cycle is fundamental for the growth of organisms (Hall et al., 2004). In plants, cell cycle genes are duplicated and diverged to accommodate complex developmental programs (Gutierrez, 2016). Our previous study showed that expression of the core cell cycle genes is coordinately regulated from ovule development to embryogenesis during sexual reproduction in P. aphrodite (Lin H.Y. et al., 2014). Moreover, transcripts associated with cell cycle-associated biological processes, such as DNA replication initiation, cell division, and regulation of the cell cycle are strongly enriched in interior ovary tissues 30–40 days after pollination (DAP) when ovules are developing (Fang et al., 2016). Among the enriched cell cycle regulators identified, PaE2F3 encodes an evolutionarily conserved transcription factor that heterodimerizes with the DP protein to control G1/S transition as cells enter the division cycle. Being the cell cycle activator, E2F/DP transcription factor is reported to activate genes involved in DNA replication and mitotic functions (Ishida et al., 2001; Ramirez-Parra et al., 2003; Fung and Poon, 2005; Vandepoele et al., 2005; Dante et al., 2014). Furthermore, overexpression of Arabidopsis E2F (AtE2Fb) and its dimerization partner AtDPa is sufficient to activate downstream cell cycle genes and drive cell proliferation in differentiated tissues (De Veylder et al., 2002; Rossignol et al., 2002; Magyar et al., 2005; Sozzani et al., 2006). To investigate the potential targets of PaE2F3, the expression levels of selected cell cycle regulators enriched simultaneously in ovary tissues at 30–40 DAP (Table 3) were analyzed in protoplast cells co-transfected with PaE2F3 and its interaction partner PaDP1 or PaDP2 (Lin H.Y. et al., 2014).
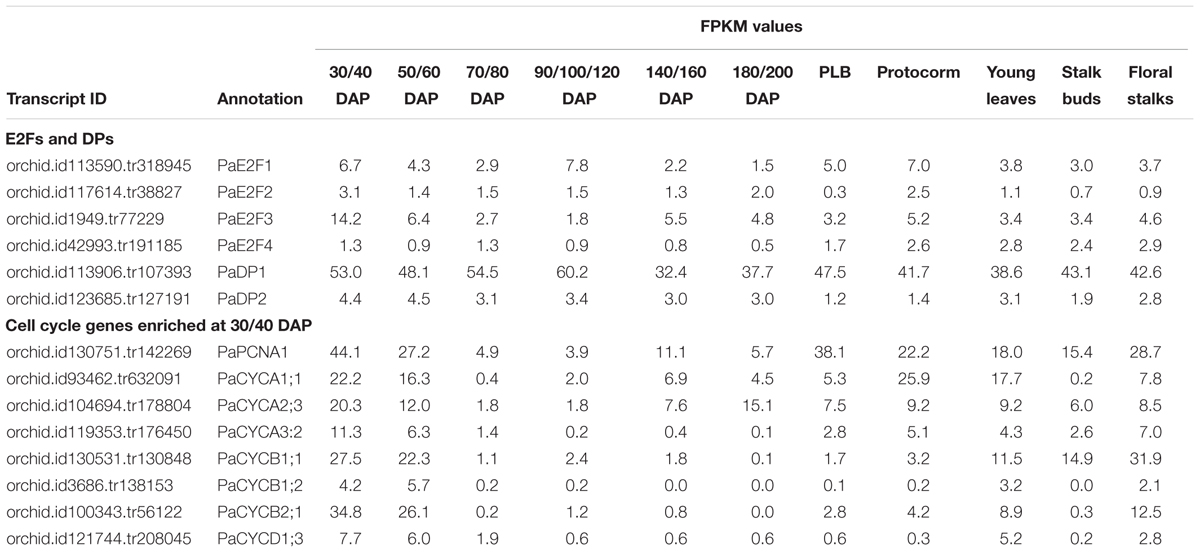
TABLE 3. Comparative transcript abundances of cell cycle genes in reproductive tissues of P. aphrodite by RNA-sequencing analysis.
After transfection, overexpression of PaE2F3 and PaDP1 was verified by quantitative RT-PCR analysis (Figure 4A). Overexpression of PaE2F3 and PaDP1 resulted in an increase in expression levels of PaPCNA1 in three independent experiments (Figure 4A), but did not have significant effects on expression of PaCYCB2;1, PaCYCA1:1, PaCYCA2:3, PaCYCA3:2, PaCYCB1:1, and PaCYCD1:3 (Supplementary Figure S4A). Because PaCYCB1;2 mRNA could not be reliably detected in transfected protoplast cells, it was omitted from further analysis.
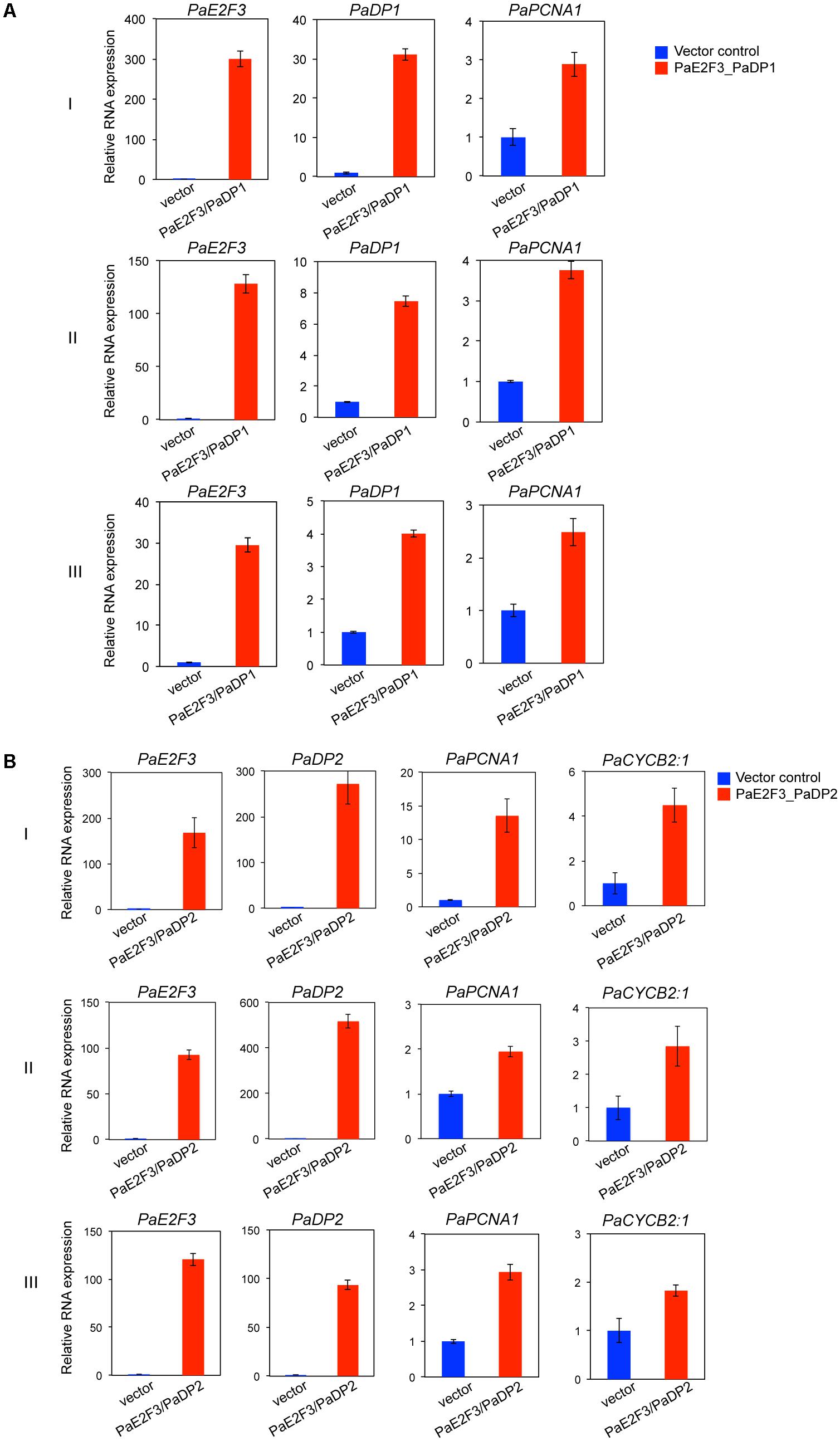
FIGURE 4. Overexpression of PaE2F3 and PaDP2 up-regulates expression of the specific cell cycle genes. (A) Validation of expression of PaE2F3 and PaDP2 mRNAs in protoplast cells co-transfected with the PaE2F3 and PaDP2 constructs. (B) Relative expression levels of PaPCNA1 and PaCYCB2:1 in protoplast cells co-transfected with the PaE2F3 and PaDP2 constructs. Protoplasts transfected with the empty vectors were used as a negative control. Expression levels of the indicated genes from protoplasts transfected with the empty vectors (vector) were arbitrarily set to be one. Three independent experiments I, II, and III are shown.
For protoplast cells transfected with PaE2F3 and PaDP2 constructs, accumulation of PaE2F3 and PaDP2 mRNAs was also verified by quantitative RT-PCR analysis (Figure 4B). Overexpression of PaE2F3 and PaDP2 up-regulated expression of PaPCNA1 and PaCYCB2;1 (Figure 4B) but did not have significant effects on accumulation of PaCYCA1:1, PaCYCA2:3, PaCYCA3:2, PaCYCB1:1, and PaCYCD1:3 (Supplementary Figure S4B). Activation of PaPCNA1 and PaCYCB2;1 by co-overexpression of PaE2F3 and PaDP2 was validated in three independent experiments (Figure 4B). Hence, our data provide evidence that PaPCNA1 and PaCYCB2;1 are the potential targets of PaE2F3.
To assess the potential of using Phalaenopsis protoplasts to investigate the hormone response, the auxin reporter DR5v2 (Liao et al., 2015) was transfected into the petal protoplasts. While treatment with a synthetic auxin, NAA, resulted in accumulation of GFP and ntdTomato florescent proteins in the nuclei, only the background signal was detected in the absence of NAA treatment (Figure 5A and Supplementary Figure S5). Furthermore, treatment with trans-zeatin, GA, or ABA had no effect on accumulation of GFP and ntdTomato florescent proteins in the transfected protoplasts (Figure 5B and Supplementary Figure S5). Taken together, these results demonstrated that DR5v2 reporter responds to the auxin signal in petal protoplasts and suggests the potential of using DR5v2 reporter to map the auxin regulatory pathway in Phalaenopsis orchids.
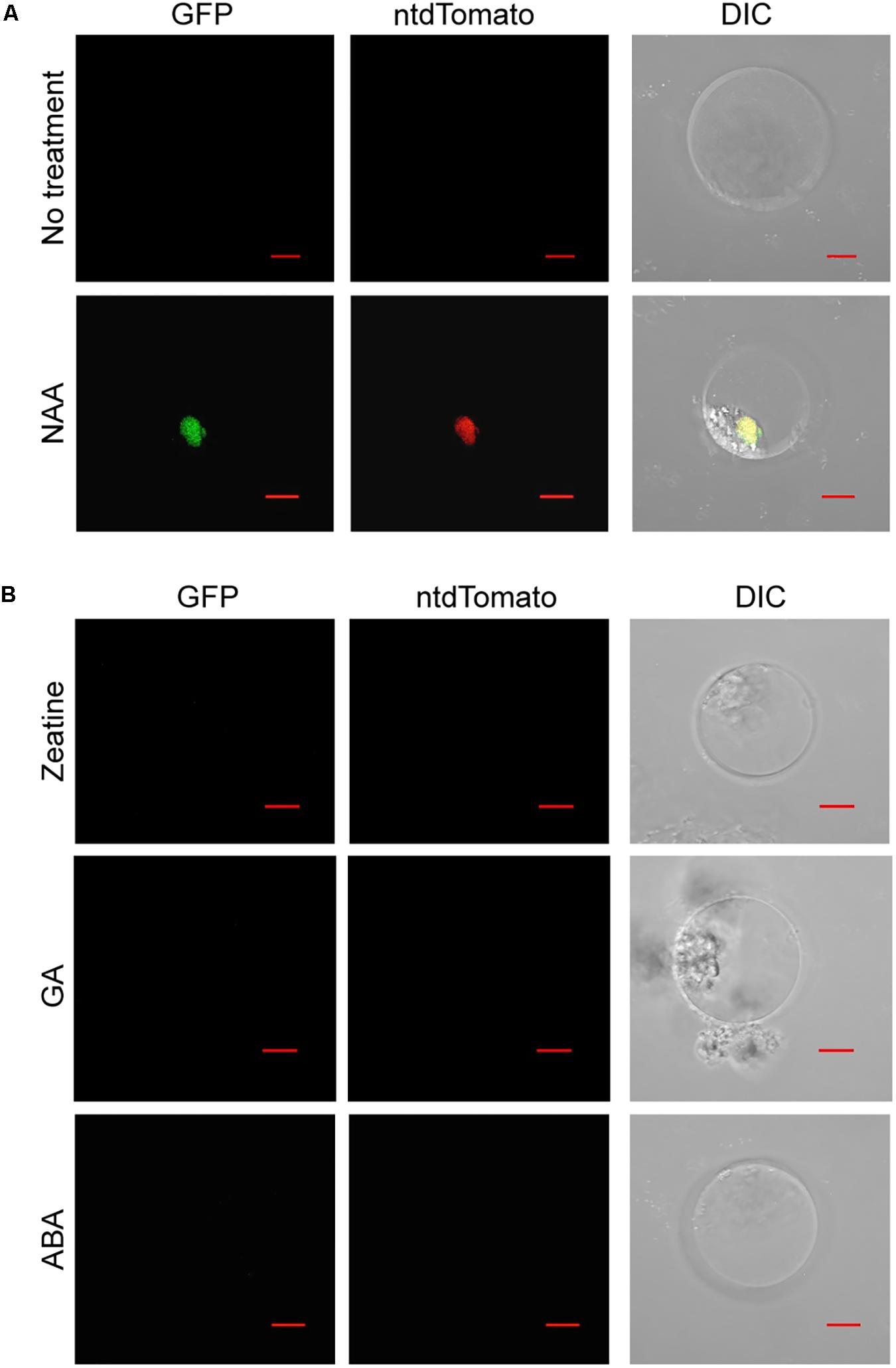
FIGURE 5. Auxin activates the DR5v2 reporter in Phalaenopsis protoplast transient expression assay. (A) NAA activated DR5v2 reporter. (B) Zeatin, GA, and ABA did not activate DR5v2 reporter. DIC, differential interference contrast images of cells superimposed with GFP and ntdTomato signals. Scale bar, 10 μm.
Protoplast transient expression systems are a powerful tool for studying the molecular mechanisms underlying signal transduction pathways. Here, we described a streamlined protocol for petal protoplast isolation and polyethylene glycol-calcium transfection for P. aphrodite (Figure 1A) and demonstrated its feasibility for analyses of protein subcellular localization and protein–protein interaction. Moreover, our reported transfection efficiency (> 80%) is significantly improved in comparison to the transfection efficiency of Phalaenopsis protoplasts reported recently (Li et al., 2018). The ability to isolate large numbers of viable protoplasts (Table 1), high transfection efficiency, and high numbers of viable transfected cells (Table 2) enable construction of a hierarchical gene regulatory pathway and studies of hormone responses. Hence, this protocol provides a useful platform for in-depth studies of gene functions and molecular networks that were previously difficult in Phalaenopsis orchids. Recently, protoplast transient expression systems have been extended to studies on dissection of miRNA pathways (Martinho et al., 2015), protein-DNA binding (Lee et al., 2017), microbe-associated molecular patterns-triggered immunity (Fraiture et al., 2014; Kanofsky et al., 2016), ribonuclease-mediated mRNA decay (Hayashi et al., 2016), and auxin-mediated transcriptional regulatory networks (Wehner et al., 2017). It is therefore conceivable that our established protocol may be applied to diverse aspects of orchid biology studies.
PaE2F3 has been shown to interact with PaDP1 and PaDP2 in yeast two-hybrid and petal transient expression assays (Lin H.Y. et al., 2014). As proof-of-principle for gene regulation study, we found that transient overexpression of PaE2F3/PaDP2 transcription factor was capable of up-regulating expression of PaPCNA1 and PaCYCB2;1 that are co-expressed with PaE2F3 in ovary tissues of P. aphrodite. Transient overexpression of PaE2F3/PaDP1 transcription factor, on the other hand, only up-regulated expression of PaPCNA1 and did not seem to have a significant effect on the expression of the other genes tested. It is possible that PaE2F3 interacts with different partners to regulate both overlapping and distinct sets of cell cycle genes in P. aphrodite.
It is not surprising to see that co-expression of PaE2F3 and PaDPs induced accumulation of PaPCNA1 mRNA because PCNA, which is required for DNA replication in both plant and animal systems, is identified as the direct downstream target of E2F/DP protein (Yamaguchi et al., 1995; Egelkrout et al., 2002; Kosugi and Ohashi, 2002; Vandepoele et al., 2005). It is intriguing to discover that PaE2F3/PaDP2 transcription factor induced accumulation of B-type cyclin, PaCYCB2;1, which is normally considered to be an M phase gene (Van Leene et al., 2011). The functional roles of E2F/DP at the G2-M checkpoint and mitotic activity have been reported in animals (Ren et al., 2002; Li et al., 2012). In Arabidopsis, E2Fa-DPa transcription factor has been reported to regulate G2-M phase gene CDKB1;1 to control stomatal development (Boudolf et al., 2004). Our findings illustrate a potential PaE2F3/PaDP2-mediated cross talk between the G1-S and G2-M stages in ovary development of P. aphrodite and support the role of E2F in G2-M phase. It will be interesting to investigate how PaE2F3/PaDP2 regulates expression of PaCYCB2;1 to coordinate the cell cycle progression in Phalaenopsis orchids. Taken together, our data demonstrate the possibility of using the orchid protoplast transient expression system to quickly screen for potential E2F/DP downstream targets in developing ovary tissues.
Auxin is reported to play important roles in orchid development (Arditti et al., 1971; Tsavkelova et al., 2007; Novak and Whitehouse, 2013; Novak et al., 2014). However, it is not clear how auxin responses are regulated to coordinate specialized orchid developmental programs. DR5v2 reporter assay provides evidence that petal protoplasts may be potentially conducive to dissecting the auxin response pathway of Phalaenopsis orchids. In addition, the orchid protoplast transient expression system could be used to assess the transcriptional responses of auxin signaling molecules or regulatory components.
In addition to Phalaenopsis orchids, petal protoplasts have successfully been used for gene functional studies in petunia and rose (Faraco et al., 2011; Hirata et al., 2012) supporting the general applicability of the petal system. However, conceivable limitations of the petal protoplast transient expression system may exist. It is reported that protoplasts retain their differentiation state and gene expression program within the time frame of transient expression experiments (Faraco et al., 2011; Lin Y.C. et al., 2014). Accordingly, protoplasts with the right cellular context providing the ideal gene expression program may be required to address tissue- or context-specific questions. It is possible that petal protoplasts are not suitable to analyze light- and sugar-dependent responses of the photosynthetic genes. In such cases, results should be interpreted with caution. It is presently difficult to isolate active protoplasts from mature leaf tissues, the most used source for protoplast isolation. During the attempts to isolate mesophyll protoplasts, protoplasts collapsed hours after isolation regardless the mannitol concentration tested. It is not clear why leaf mesophyll protoplasts are sensitive to the isolation procedure. Generally, protoplasts rupture in hypotonic solution and collapse in hypertonic solution (Ohshima and Toyama, 1989). It is possible that the cell wall of mesophyll cells releases phytotoxic factors that poison the isolated protoplasts (Hahne and Lorz, 1988). The presence of abundant calcium oxalate crystals may also contribute to the damage to protoplasts. The establishment of orchid protoplast transient expression protocols for different tissue types in the future may be important to address tissue- or context-specific questions.
Protoplast-based high-throughput systems have been established for dissection of signaling pathways, analysis of transcription factor functions, and identification of kinase-associated phosphoisoforms (Wehner et al., 2011; Zhang et al., 2015; Dory et al., 2016; Wehner et al., 2017). Therefore, the orchid protoplast transient expression system provides a molecular tool to characterize functions of proteins identified from open reading frames from recently released transcriptome datasets (Cai et al., 2015; Fang et al., 2016; Chao et al., 2017) and potentially allows discovery and/or validation of hierarchical gene regulatory networks. In summary, the protocol presented here will enable in-depth studies of the molecular networks governing the unique developmental processes and hormone regulating pathways in orchids and provide an alternative method to conduct functional genomic studies in Phalaenopsis orchids.
S-CF wrote the manuscript and designed the experiments. H-YL developed the protoplast isolation and transfection system. J-CC maintained the orchid plants and did the RT-PCR experiments. All authors read and approved the final manuscript.
This work was supported by the Innovative Translational Agricultural Research Grants (to S-CF); and in part by a grant(to S-CF) from the Biotechnology Center in Southern Taiwan, Academia Sinica.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Dolf Weijers for sharing the DR5v2 construct, Dr. Choun-Sea Lin for sharing the subcellular localization constructs; the AS-BCST Greenhouse and Confocal Microscopy Core Facilities for core services; Mr. Chin-Lin Chung and Ms. Yen-Ling Lin for critical reading of the manuscript; and Ms. Miranda Loney for English editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00843/full#supplementary-material
Arditti, J., Jeffrey, D. C., and Flick, B. H. (1971). Post-pollination phenomena in orchid flowers. 3. Effects and interactions of auxin, kinetin or gibberellin. New Phytol. 70, 1125–1141. doi: 10.1111/j.1469-8137.1971.tb04595.x
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Bory, S., Grisoni, M., Duval, M. F., and Besse, P. (2008). Biodiversity and preservation of vanilla: present state of knowledge. Genet. Resour. Crop Evol. 55, 551–571. doi: 10.1007/s10722-007-9260-3
Boudolf, V., Vlieghe, K., Beemster, G. T., Magyar, Z., Torres Acosta, J. A., Maes, S., et al. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16, 2683–2692. doi: 10.1105/tpc.104.024398
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Bu, Y., Zhao, M., Sun, B., Zhang, X., Takano, T., and Liu, S. (2014). An efficient method for stable protein targeting in grasses (Poaceae): a case study in Puccinellia tenuiflora. BMC Biotechnol. 14:52. doi: 10.1186/1472-6750-14-52
Bulpitt, C. J., Li, Y., Bulpitt, P. F., and Wang, J. (2007). The use of orchids in Chinese medicine. J. R. Soc. Med. 100, 558–563. doi: 10.1258/jrsm.100.12.558
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Chao, Y. T., Yen, S. H., Yeh, J. H., Chen, W. C., and Shih, M. C. (2017). Orchidstra 2.0-a transcriptomics resource for the orchid family. Plant Cell Physiol. 58, 1–13. doi: 10.1093/pcp/pcw220
Chen, J. C., and Fang, S. C. (2016). The long pollen tube journey and in vitro pollen germination of Phalaenopsis orchids. Plant Reprod. 29, 179–188. doi: 10.1007/s00497-016-0280-z
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., and Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Chen, T. K., Yang, H. T., Fang, S. C., Lien, Y. C., Yang, T. T., and Ko, S. S. (2016). Hybrid-Cut: an improved sectioning method for recalcitrant plant tissue samples. J. Vis. Exp. 117:54754. doi: 10.3791/54754
Dante, R. A., Larkins, B. A., and Sabelli, P. A. (2014). Cell cycle control and seed development. Front. Plant Sci. 5:493. doi: 10.3389/fpls.2014.00493
De Veylder, L., Beeckman, T., Beemster, G. T., de Almeida Engler, J., Ormenese, S., Maes, S., et al. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. doi: 10.1093/emboj/21.6.1360
Dory, M., Doleschall, Z., Nagy, S. K., Ambrus, H., Mészáros, T., Barnabás, B., et al. (2016). Kinase-associated phosphoisoform assay: a novel candidate-based method to detect specific kinase-substrate phosphorylation interactions in vivo. BMC Plant Biol. 16:204. doi: 10.1186/s12870-016-0894-1
Egelkrout, E. M., Mariconti, L., Settlage, S. B., Cella, R., Robertson, D., and Hanley-Bowdoin, L. (2002). Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14, 3225–3236. doi: 10.1105/tpc.006403
Fang, S. C., Chen, J. C., and Wei, M. J. (2016). Protocorms and protocorm-like bodies are molecularly distinct from zygotic embryonic tissues in Phalaenopsis aphrodite. Plant Physiol. 171, 2682–2700. doi: 10.1104/pp.16.00841
Faraco, M., Di Sansebastiano, G. P., Spelt, K., Koes, R. E., and Quattrocchio, F. M. (2011). One protoplast is not the other! Plant Physiol. 156, 474–478. doi: 10.1104/pp.111.173708
Fraiture, M., Zheng, X., and Brunner, F. (2014). An Arabidopsis and tomato mesophyll protoplast system for fast identification of early MAMP-triggered immunity-suppressing effectors. Methods Mol. Biol. 1127, 213–230. doi: 10.1007/978-1-62703-986-4_17
Fung, T. K., and Poon, R. Y. (2005). A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 16, 335–342. doi: 10.1016/j.semcdb.2005.02.014
Gutierrez, C. (2016). 25 Years of cell cycle research: What’s ahead? Trends Plant Sci. 21, 823–833. doi: 10.1016/j.tplants.2016.06.007
Hahne, G., and Lorz, H. (1988). Release of phytotoxic factors from plant-cell walls during protoplast isolation. J. Plant Physiol. 132, 345–350. doi: 10.1016/S0176-1617(88)80118-4
Hall, M. N., Raff, M., and Thomas, G. (2004). Cell Growth - Control of Cell Size. Cold Spring Harbor, NY: John Inglis.
Hayashi, S., Wakasa, Y., Ozawa, K., and Takaiwa, F. (2016). Characterization of IRE1 ribonuclease-mediated mRNA decay in plants using transient expression analyses in rice protoplasts. New Phytol. 210, 1259–1268. doi: 10.1111/nph.13845
Hirata, H., Ohnishi, T., Ishida, H., Tomida, K., Sakai, M., Hara, M., et al. (2012). Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J. Plant Physiol. 169, 444–451. doi: 10.1016/j.jplph.2011.12.005
Hsing, H. X., Lin, Y. J., Tong, C. G., Li, M. J., Chen, Y. J., and Ko, S. S. (2016). Efficient and heritable transformation of Phalaenopsis orchids. Bot. Stud. 57:30. doi: 10.1186/s40529-016-0146-6
Hsu, C. T., Liao, D. C., Wu, F. H., Liu, N. T., Shen, S. C., Chou, S. J., et al. (2011). Integration of molecular biology tools for identifying promoters and genes abundantly expressed in flowers of Oncidium Gower Ramsey. BMC Plant Biol. 11:60. doi: 10.1186/1471-2229-11-60
Hu, W. W., Wong, S. M., Loh, C. S., and Goh, C. J. (1998). Synergism in replication of cymbidium mosaic potexvirus (CymMV) and odontoglossum ringspot tobamovirus (ORSV) RNA in orchid protoplasts. Arch. Virol. 143, 1265–1275. doi: 10.1007/s007050050374
Huang, C. N., Cornejo, M. J., Bush, D. S., and Jones, R. L. (1986). Estimating viability of plant protoplasts using double and single staining. Protoplasma 135, 80–87. doi: 10.1007/Bf01277001
Ishida, S., Huang, E., Zuzan, H., Spang, R., Leone, G., West, M., et al. (2001). Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21, 4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001
Kanofsky, K., Lehmeyer, M., Schulze, J., and Hehl, R. (2016). Analysis of microbe-associated molecular pattern-responsive synthetic promoters with the parsley protoplast system. Methods Mol. Biol. 1482, 163–174. doi: 10.1007/978-1-4939-6396-6_11
Kidokoro, S., Watanabe, K., Ohori, T., Moriwaki, T., Maruyama, K., Mizoi, J., et al. (2015). Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81, 505–518. doi: 10.1111/tpj.12746
Kobayashi, T., Kameya, T., and Ichihashi, S. (1993). Plant regeneration from protoplasts derived from callus of Phalaenopsis. Plant Tissue Cult. Lett. 10, 267–270. doi: 10.5511/plantbiotechnology1984.10.267
Kong, J. M., Goh, N. K., Chia, L. S., and Chia, T. F. (2003). Recent advances in traditional plant drugs and orchids. Acta Pharmacol. Sin. 24, 7–21.
Kosugi, S., and Ohashi, Y. (2002). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. doi: 10.1046/j.1365-313x.2002.01196.x
Kull, T., and Arditti, J. (2002). Orchid Biology Reviews and Perspectives. London: Kluwer Academic Publishers. doi: 10.1007/978-94-017-2500-2
Lee, J. H., Jin, S., Kim, S. Y., Kim, W., and Ahn, J. H. (2017). A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods 13:42. doi: 10.1186/s13007-017-0192-4
Lee, L. Y., Fang, M. J., Kuang, L. Y., and Gelvin, S. B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:24. doi: 10.1186/1746-4811-4-24
Li, J. L., Liao, X. Z., Zhou, S. S., Liu, S., Jiang, L., and Wang, G. D. (2018). Efficient protoplast isolation and transient gene expression system for Phalaenopsis hybrid cultivar ’Ruili Beauty’. In Vitro Cell. Dev. Biol. Plant 54, 87–93. doi: 10.1007/s11627-017-9872-z
Li, Y., Zhang, D. Y., Ren, Q., Ye, F., Zhao, X., Daniels, G., et al. (2012). Regulation of a novel androgen receptor target gene, the cyclin B1 gene, through androgen-dependent E2F family member switching. Mol. Cell. Biol. 32, 2454–2466. doi: 10.1128/MCB.06663-11
Liao, C. Y., Smet, W., Brunoud, G., Yoshida, S., Vernoux, T., and Weijers, D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210. doi: 10.1038/nmeth.3279
Liau, C. H., You, S. J., Prasad, V., Hsiao, H. H., Lu, J. C., Yang, N. S., et al. (2003). Agrobacterium tumefaciens-mediated transformation of an Oncidium orchid. Plant Cell Rep. 21, 993–998. doi: 10.1007/s00299-003-0614-9
Lin, H. Y., Chen, J. C., Wei, M. J., Lien, Y. C., Li, H. H., Ko, S. S., et al. (2014). Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite. Plant Mol. Biol. 84, 203–226. doi: 10.1007/s11103-013-0128-y
Lin, Y. C., Li, W., Chen, H., Li, Q., Sun, Y. H., Shi, R., et al. (2014). A simple improved-throughput xylem protoplast system for studying wood formation. Nat. Protoc. 9, 2194–2205. doi: 10.1038/nprot.2014.147
Lu, H. C., Hsieh, M. H., Chen, C. E., Chen, H. H., Wang, H. I., and Yeh, H. H. (2012). A high-throughput virus-induced gene-silencing vector for screening transcription factors in virus-induced plant defense response in orchid. Mol. Plant Microbe Interact. 25, 738–746. doi: 10.1094/MPMI-10-11-0266
Lu, X., Kracher, B., Saur, I. M., Bauer, S., Ellwood, S. R., Wise, R., et al. (2016). Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E6486–E6495. doi: 10.1073/pnas.1612947113
Lung, S. C., Yanagisawa, M., and Chuong, S. D. (2011). Protoplast isolation and transient gene expression in the single-cell C4 species, Bienertia sinuspersici. Plant Cell Rep. 30, 473–484. doi: 10.1007/s00299-010-0953-2
Magyar, Z., De Veylder, L., Atanassova, A., Bakó, L., Inzé, D., and Bögre, L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17, 2527–2541. doi: 10.1105/tpc.105.033761
Martinho, C., Confraria, A., Elias, C. A., Crozet, P., Rubio-Somoza, I., Weigel, D., et al. (2015). Dissection of miRNA pathways using Arabidopsis mesophyll protoplasts. Mol. Plant 8, 261–275. doi: 10.1016/j.molp.2014.10.003
Masani, M. Y., Noll, G. A., Parveez, G. K., Sambanthamurthi, R., and Prufer, D. (2014). Efficient transformation of oil palm protoplasts by PEG-mediated transfection and DNA microinjection. PLoS One 9:e96831. doi: 10.1371/journal.pone.0096831
Mazarei, M., Al-Ahmad, H., Rudis, M. R., and Stewart, C. N. Jr. (2008). Protoplast isolation and transient gene expression in switchgrass, Panicum virgatum L. Biotechnol. J. 3, 354–359. doi: 10.1002/biot.200700189
Muchero, W., Guo, J., DiFazio, S. P., Chen, J. G., Ranjan, P., Slavov, G. T., et al. (2015). High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genomics 16:24. doi: 10.1186/s12864-015-1215-z
Müller, B., and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. doi: 10.1038/nature06943
Nanjareddy, K., Arthikala, M. K., Blanco, L., Arellano, E. S., and Lara, M. (2016). Protoplast isolation, transient transformation of leaf mesophyll protoplasts and improved Agrobacterium-mediated leaf disc infiltration of Phaseolus vulgaris: tools for rapid gene expression analysis. BMC Biotechnol. 16:53. doi: 10.1186/s12896-016-0283-8
Nelson, B. K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Novak, D. S., Luna, L. J., and Gamage, R. N. (2014). Role of auxin in orchid development. Plant Signal. Behav. 9:e972277. doi: 10.4161/psb.32169
Novak, S. D., and Whitehouse, G. A. (2013). Auxin regulates first leaf development and promotes the formation of protocorm trichomes and rhizome-like structures in developing seedlings of Spathoglottis plicata (Orchidaceae). AoB Plants 5:pls053. doi: 10.1093/aobpla/pls053
Nyman, M., and Wallin, A. (1992). Transient gene expression in strawberry (Fragaria x ananassa Duch.) protoplasts and the recovery of transgenic plants. Plant Cell Rep. 11, 105–108. doi: 10.1007/BF00235264
Ohshima, M., and Toyama, S. (1989). Studies on culture of cells and tissues of crop plants: 1. Survey on enzymatic isolation and culture of rice leaf sheath protoplasts. Jpn. J. Crop Sci. 58, 103–110. doi: 10.1626/jcs.58.103
Pacini, E., and Hesse, M. (2002). Types of pollen dispersal units in orchids, and their consequences for germination and fertilization. Ann. Bot. 89, 653–664. doi: 10.1039/aob/mcf138
Ramirez-Parra, E., Frundt, C., and Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33, 801–811. doi: 10.1046/j.1365-313X.2003.01662.x
Rao, X., Krom, N., Tang, Y., Widiez, T., Havkin-Frenkel, D., Belanger, F. C., et al. (2014). A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia). BMC Genomics 15:964. doi: 10.1186/1471-2164-15-964
Rasmussen, H. N. (2002). Recent developments in the study of orchid mycorrhiza. Plant Soil 244, 149–163. doi: 10.1023/A:1020246715436
Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A., et al. (2002). E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256. doi: 10.1101/gad.949802
Rossignol, P., Stevens, R., Perennes, C., Jasinski, S., Cella, R., Tremousaygue, D., et al. (2002). AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol. Genet. Genomics 266, 995–1003. doi: 10.1007/s00438-001-0624-7
Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Shen, Y., Meng, D., McGrouther, K., Zhang, J., and Cheng, L. (2017). Efficient isolation of Magnolia protoplasts and the application to subcellular localization of MdeHSF1. Plant Methods 13:44. doi: 10.1186/s13007-017-0193-3
Shrestha, B. R., Tokuhara, K., and Mii, M. (2007). Plant regeneration from cell suspension-derived protoplasts of Phalaenopsis. Plant Cell Rep. 26, 719–725. doi: 10.1007/s00299-006-0286-3
Sozzani, R., Maggio, C., Varotto, S., Canova, S., Bergounioux, C., Albani, D., et al. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140, 1355–1366. doi: 10.1104/pp.106.077990
Thevenin, J., Xu, W., Vaisman, L., Lepiniec, L., Dubreucq, B., and Dubos, C. (2016). The Physcomitrella patens system for transient gene expression assays. Methods Mol. Biol. 1482, 151–161. doi: 10.1007/978-1-4939-6396-6_10
Tsavkelova, E. A., Cherdyntseva, T. A., Botina, S. G., and Netrusov, A. I. (2007). Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 162, 69–76. doi: 10.1016/j.micres.2006.07.014
Van Leene, J., Boruc, J., De Jaeger, G., Russinova, E., and De Veylder, L. (2011). A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci. 16, 141–150. doi: 10.1016/j.tplants.2010.12.004
Vandepoele, K., Vlieghe, K., Florquin, K., Hennig, L., Beemster, G. T., Gruissem, W., et al. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328. doi: 10.1104/pp.105.066290
Wang, H., Nicolay, B. N., Chick, J. M., Gao, X., Geng, Y., Ren, H., et al. (2017). The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430. doi: 10.1038/nature22797
Waterman, R. J., and Bidartondo, M. I. (2008). Deception above, deception below: linking pollination and mycorrhizal biology of orchids. J. Exp. Bot. 59, 1085–1096. doi: 10.1093/jxb/erm366
Wehner, N., Hartmann, L., Ehlert, A., Bottner, S., Onate-Sanchez, L., and Droge-Laser, W. (2011). High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 68, 560–569. doi: 10.1111/j.1365-313X.2011.04704.x
Wehner, N., Herfert, J., Droge-Laser, W., and Weiste, C. (2017). High-throughput protoplast trans-activation (PTA) screening to define transcription factors in auxin-mediated gene regulation. Methods Mol. Biol. 1569, 187–202. doi: 10.1007/978-1-4939-6831-2_16
Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T., and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. doi: 10.1186/1746-4811-5-16
Yamaguchi, M., Hayashi, Y., and Matsukage, A. (1995). Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270, 25159–25165. doi: 10.1074/jbc.270.42.25159
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, H., Yang, S. H., and Goh, C. J. (2001). Agrobacterium-mediated transformation of a Dendrobium orchid with the class 1 knox gene DOH1. Plant Cell Rep. 20, 301–305. doi: 10.1007/s002990100334
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6:19029. doi: 10.1038/srep19029
Keywords: protoplast, Phalaenopsis aphrodite, orchid, transient expression, gene regulation
Citation: Lin H-Y, Chen J-C and Fang S-C (2018) A Protoplast Transient Expression System to Enable Molecular, Cellular, and Functional Studies in Phalaenopsis orchids. Front. Plant Sci. 9:843. doi: 10.3389/fpls.2018.00843
Received: 27 February 2018; Accepted: 30 May 2018;
Published: 22 June 2018.
Edited by:
Mariana Mondragón-Palomino, University of Regensburg, GermanyReviewed by:
Guangdong Wang, Nanjing Agricultural University, ChinaCopyright © 2018 Lin, Chen and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Su-Chiung Fang, c2NmYW5nQGdhdGUuc2luaWNhLmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.