- State Key Laboratory of Wheat and Maize Crop Science, Collaborative Innovation Center of Henan Grain Crops, College of Life Sciences, Henan Agricultural University, Zhengzhou, China
Missing Proteins in Plant Proteomic Analysis
Recently, technical advances, especially in liquid chromatography (LC) and mass spectrometry (MS), have improved the sensitivity, coverage, reliability, and throughput of proteome analysis (Boersema et al., 2015). Novel proteomics methods, such as targeted proteomics (Marx, 2013), degradomics (Stoehr et al., 2013), structural proteomics (Walzthoeni et al., 2013), chemical proteomics (Rudolf et al., 2013), and microproteomics (Kasuga et al., 2017), are becoming essential tools for in-depth analyses of biological systems and phenomena, such as plant growth, development, and responses to stress factors.
The numbers of plant proteins detected using MS-based proteomics remains much lower than expected. For example, the improved maize reference genome contains 39,324 protein-coding genes, with an average of 3.3 transcripts per gene (Jiao et al., 2017), each of which may produce at least several different proteins. Moreover, additional proteins may be synthesized by proteolysis of other existing proteins. To date only 947 reviewed and 169,813 unreviewed maize protein entries have been collected in the UniProtKB (http://www.uniprot.org/uniprot/?query=organism:“maize”) (retrieved on Feb 6, 2018). Similarly, analysis of the UniProtKB entries for maize organelle proteins reveals few reviewed proteins compared with a large number of unreviewed entries (Supplementary Table 1). An important reason for this phenomenon is that maize proteomic data has not be curated and collected as the annotation of unreviewed protein entries. Definitely, numerous “missing (hidden) proteins” that are predicted at the transcript level remain unidentified at the protein level in plants.
While many factors contribute to missing proteins, one major cause is using inefficient protein extraction methods, especially for hydrophobic membrane proteins and low-abundant proteins (LAPs) (Thelen and Peck, 2007; Libault et al., 2017). Sample quality is critical for the coverage, reliability, and throughput of plant proteomic analysis, although advanced detection approaches (especially LC-MS/MS) can greatly enhance the sensitivity and reliability of protein identification. Here, in view of the current approaches and trends in plant proteomics, we highlight the importance of using multiple protein extraction methods to obtain a more complete picture of plant proteome. Moreover, to promote the identification of more “missing proteins,” we discuss the key aspects of protein extraction methods at the tissue, single-cell, and organelle levels.
Extraction of Total Proteins for Comparative Proteomic Analysis
Comparative proteomic analysis is mostly conducted using total proteins extracted from tissues, organs, or whole plants. Such approaches are effective to understand plant activities at the corresponding level, but suffer from a “dilution” effect that masks the unique biological properties of individual cells and cell-types (Libault et al., 2017).
A major challenge in plant proteomics is the effective and comprehensive extraction of proteins from plant tissues, due to the high dynamic range of plant proteins and the high levels of interfering substances (e.g., phenolics, lipids, organic acids, carbohydrates, terpenes, and pigments) (Wang et al., 2008). Therefore, for total proteins extraction from plant tissues, it is important to consider each of the following steps.
First, the extraction scale should be decided at an early stage. Plant tissues can be easily homogenized with quartz sand in the extraction buffer or pulverized with liquid N2 in a mortar. A small amount of plant materials (0.1–1.0 g fresh weight, depending on tissue type) is usually sufficient for proteomic analysis (Wu et al., 2014a).
Second, removal of interfering substances is necessary for preparing high-quality protein samples. To this purpose, two approaches are currently used: based on acetone/TCA precipitation and based on phenol extraction (Wang et al., 2008). Many pioneering works have contributed to the development, evaluation, and optimization of these approaches (Santoni et al., 2000; Giavalisco et al., 2003; Wang et al., 2003; Friso et al., 2004; Rose et al., 2004; Carpentier et al., 2005; Isaacson et al., 2006). Acetone/TCA precipitation method works well for almost plant tissues (Wang et al., 2008). Following acetone/TCA precipitation, organic-soluble substances are rinsed away, leaving proteins and other insoluble substances in the precipitate. Proteins are extracted using a buffer suitable for 2DE, iTRAQ, or LC-based separation. Phenol extraction method works by selectively extracting proteins from aqueous extracts during phase separation (Wu et al., 2014a). The profiles of the extracted proteome are highly dependent on the extraction buffers used (Chatterjee et al., 2012; Petriccione et al., 2013; Wu et al., 2014b). In addition, when using this approach one must also consider temperature (Wu et al., 2014b), pH (Sari et al., 2015), and extraction times (Feiz et al., 2006). Changing any of these parameters will affect the profile of the extracted proteome (e.g., Sari et al., 2014, 2015; Zhang et al., 2014). The success of the acetone/TCA precipitation and the phenol extraction approaches relies on the plant tissue being completely pulverized (Wu et al., 2014a).
Third, complex protein samples can be pre-fractionated to deplete high-abundance proteins, to enhance the detection of “missing” low-abundant proteins (LAPs). For example, the depletion of RuBisCO in leaves (Kim et al., 2013; Gupta and Kim, 2015) and of storage proteins in seeds (Xiong et al., 2014) and tubers (Wu et al., 2012; Kim et al., 2015; Lee et al., 2015; Gupta et al., 2016) significantly improved the separation and detection of LAPs.
Finally, each extraction method produces distinct protein complements. Therefore, integrating the application of different extraction methods will improve proteome coverage. Indeed, the importance of using multiple protein extraction methods to obtain comprehensive proteome coverage has been highlighted by several researchers (e.g., Karthikaichamy et al., 2017; Takác et al., 2017).
Protein Extraction for Organelle Proteomics
The low abundance of proteins in specific subcellular locations can result in their missing from tissue, organ, or whole plant protein samples (Libault et al., 2017). Therefore, the isolation of pure organelles allows for the analysis of LAPs that are specifically accumulated within them.
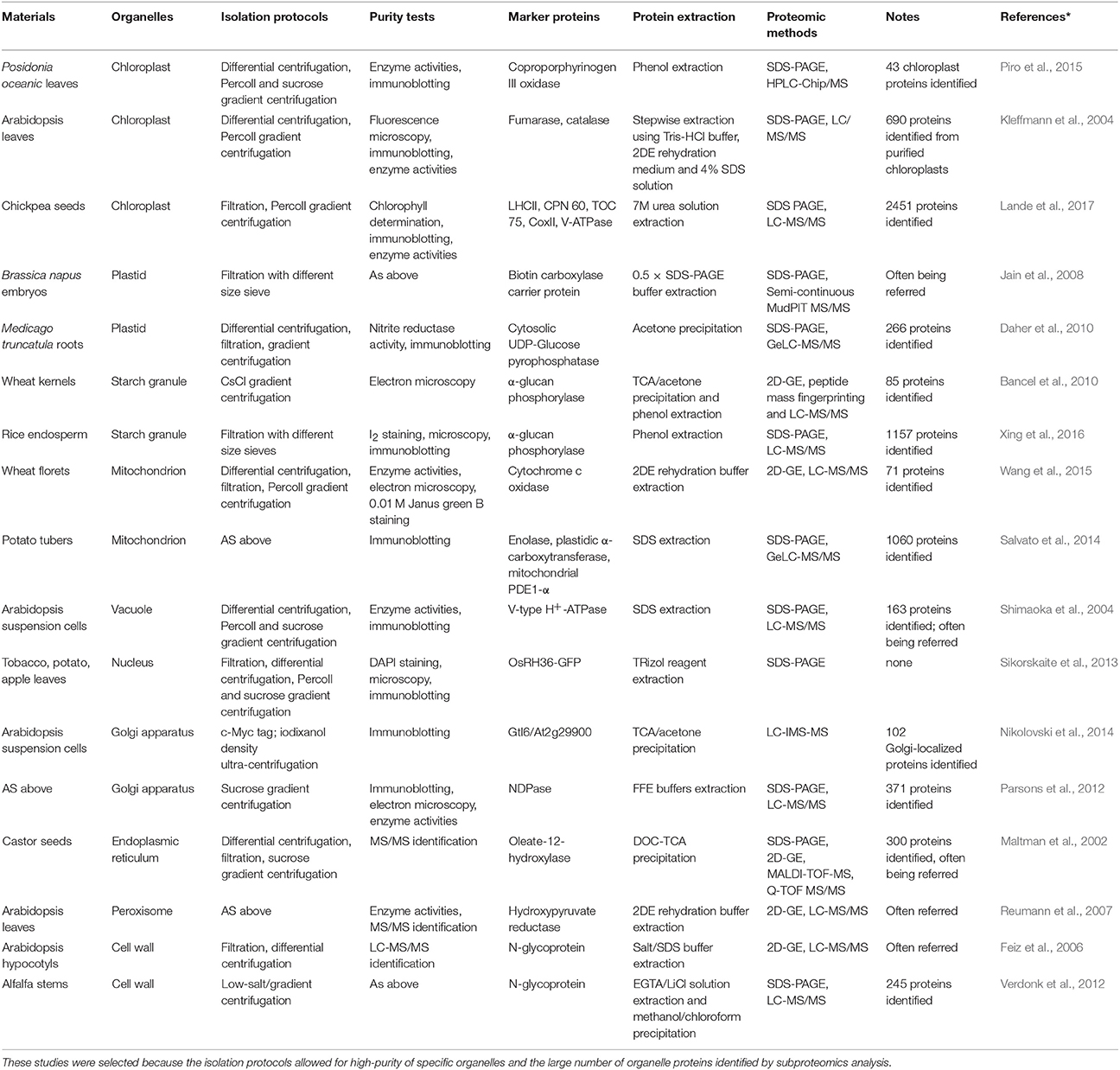
Using isolated organelles for protein extraction significantly reduces the complexity of the extracted proteome. This approach also enriches the LAP fraction in protein extracts, allowing for their improved separation and detection. Extensive proteomic studies of purified organelles, such as chloroplasts (Hall et al., 2011; Piro et al., 2015), nuclei (Sikorskaite et al., 2013), mitochondria (Lang et al., 2011; Salvato et al., 2014), and starch granules (Xing et al., 2016), have characterized a number of organelle proteins, and defined their localization information.
Previous cell biology and biochemistry studies have developed protocols for the isolation and purification of organelles including via homogenate filtration, differential centrifugation, and density gradients centrifugation (Table 1). The purity and integrity of extracted organelles can be tested by enzyme activity assay, light and electron microscopy, immunoblotting, and MS/MS identification. In contrast to the pulverization of plant tissue for total protein extraction, the extraction of organelle proteins requires gentle grinding to obtain pure and/or intact organelles before protein extraction.
Some organelles are relatively easy to isolate from others, especially those with storage functions (e.g., lipid-bodies and starch granules) and large organelles with membranous structures (e.g., chloroplasts and mitochondria). Novel methods are constantly being developed to isolate difficult organelles for subproteomic analysis, e.g., a combination of density centrifugation and surface charge separation techniques to isolate pure Golgi membranes (Parsons et al., 2012), Percoll gradient centrifugation followed by sucrose gradient centrifugation to isolate peroxisomes (Reumann and Singhal, 2014), and a simple density gradient (ultra-)centrifugation protocol to isolate intact vacuoles (Ohnishi et al., 2018) from Arabidopsis suspension cultured cells.
Once organelles are isolated, standard protein extraction approaches can be used. The composition of protein extraction buffers can be altered to suit the properties of target proteins (e.g., solubility, hydrophobicity or hydrophilicity, pI, and the degree associated with membranes). Importantly, for organelles with membrane structures, the membranes need to be broken by grinding, sonication, enzyme digestion, or detergent lysis to release soluble proteins (Lang et al., 2011; Piro et al., 2015).
For the organelles with complex structures, the separate extraction of proteins from each suborganelle fraction enables producing more detailed subproteome profiles. For example, subproteomic analysis involving the isolation of Arabidopsis chloroplasts as stroma, thylakoid membrane, and lumen fractions (Hall et al., 2011) and the separate isolation of inner and outer mitochondrial membrane fractions (Duncan et al., 2011; Schikowsky et al., 2018) have also provided information about the specific localization of proteins within the organelles.
Finally, compared with proteins in intracellular compartments, a major technical challenge in extracting cell wall proteins (CWPs) is the preparation of a pure cell wall sample. This is particularly challenging because substantial amounts of intracellular proteins inevitably associate with the cell wall during the process of tissue or cell homogenization (Rose and Lee, 2010). Cell wall isolation methods have been optimized (Feiz et al., 2006; Zhang et al., 2011; Printz et al., 2015) and, in general, the cell wall proteome consists of sensu stricto CWPs, apoplast proteins, secreted proteins, and xylem sap proteins (Wu et al., 2018). Most loosely bound cell wall proteins can be dissolved using a low ionic strength solution, while strongly bound cell wall proteins are resistant to salt-extraction (Jamet et al., 2008). Besides, the extraction and proteomic analysis of apoplast proteins, secreted proteins and xylem sap proteins (Soares et al., 2007; Kim et al., 2014) have made important achievements.
Protein Extraction for Single Cell-Level Proteomics
Another reason that proteins can be missing from plant proteomic analysis is that some LAPs (e.g., transcription and regulatory factors) accumulate in specialized cell or tissue types and at specific development stages (Dubos et al., 2010). In entire organ, or whole plant analyses, the presence of these proteins is often masked by that of high-abundance proteins. Therefore, single cell level proteomics or microproteomics will minimize the cellular complexity of the analyzed sample (Libault et al., 2017). However, sample preparation and protein extraction techniques for microproteomic analysis of plant tissues remain challenging.
Microproteomic techniques rely on accurate and precise sample collection, preparation, excision, and protein extraction (Feist and Hummon, 2015). Laser capture microdissection (LCM) is a promising method for cell level sampling. LCM allows cell types of interest to be isolated of from a fixed sample under direct microscopic visualization with the assistance of a laser beam. LCM has been successfully used in the proteomic analysis of Arabidopsis (Schad et al., 2005), maize (Dembinsky et al., 2007), barley (Kaspar et al., 2010), and tomato (Zhu et al., 2016). The best example of the application of LCM, combined with pressure catapulting, was to isolate the nucellar projection and endosperm transfer cells of an developing barley grain at 8 days post-flowering. The protein extracts were analyzed by nanoUPLC separation combined with ESI-Q-TOF MS, which successfully identified 137 and 44 proteins in nucellar projection and endosperm transfer cells, respectively (Kaspar et al., 2010). In addition, a method of mechanical separation of leaf epidermal, vascular, and mesophyll tissues has been developed in Arabidopsis (Falter et al., 2015), tomato, and cassava (Svozil et al., 2016), and the separated tissue samples can be used for quantitative LCM-assisted microproteomic analysis.
It takes a lot of time and effort to obtain sufficient numbers of cells from limited samples using LCM. Therefore, it is necessary to develop micro-scale protein extraction methods, compatible with decreased sample size (100 μg and less), to use in parallel with this approach to generate high-quality MS data for “missing” LAPs.
Concluding Remarks
Many “missing” proteins have not been proven at the protein level. Therefore, we have emphasized the importance of optimization of protein extraction methods to enhance the detection of the missing proteins in plant proteomics. Surely, MS-based proteomics alone is not sufficient to explore and identify all missing proteins. Integrated multi-omics approaches will facilitate the identification of many of the missing proteins (Chang et al., 2014).
It is necessary to note that the aim of the Opinion article is not to review previous studies, but to highlight the importance of developing novel approaches to establish plant proteomes. Special attention should always be paid to developing quantitative, reproducible, and comparable methodologies for plant proteomics. Particularly, suitable protein extraction methods integrating with isolation techniques for organelles, specific cells and tissues will greatly enhance plant proteomic analysis and allow to identify more “missing proteins.” Good protein extraction makes for a good proteome.
Author Contributions
All authors contributed to the writing of the manuscript. LN and WW revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge financial support from the National Natural Science Foundation of China (Grant No. 31230055) the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (Grant no. 15IRTSTHN015).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00802/full#supplementary-material
References
Bancel, E., Rogniaux, H., Debiton, C., Chambon, C., and Branlard, G. (2010). Extraction and proteome analysis of starch granule-associated proteins in mature wheat kernel (Triticum aestivum L.). J. Proteome Res. 9, 3299–3310. doi: 10.1021/pr9010525
Boersema, P. J., Kahraman, A., and Picotti, P. (2015). Proteomics beyond large-scale protein expression analysis. Curr. Opin. Biotech. 34, 162–170. doi: 10.1016/j.copbio.2015.01.005
Carpentier, S. C., Witters, E., Laukens, K., Deckers, P., Swennen, R., and Panis, B. (2005). Preparation of protein extracts from recalcitrant plant tissues: an evaluation of different methods for two dimensional gel electrophoresis analysis. Proteomics 5, 2497–2507. doi: 10.1002/pmic.200401222
Chang, C., Li, L., Zhang, C., Wu, S., Guo, K., Zi, J., et al. (2014). Systematic analyses of the transcriptome, translatome, and proteome provide a global view and potential strategy for the C-HPP. J. Proteome Res. 13, 38–49. doi: 10.1021/pr4009018
Chatterjee, M., Gupta, S., Bhar, A., and Das, S. (2012). Optimization of an efficient protein extraction protocol compatible with two-dimensional electrophoresis and mass spectrometry from recalcitrant phenolic rich roots of chickpea (Cicer arietinum L.). Int. J. Proteomics 2012:536963. doi: 10.1155/2012/536963
Daher, Z., Recorbet, G., Valot, B., Robert, F., Balliau, T., Potin, S., et al. (2010). Proteomic analysis of Medicago truncatula root plastids. Proteomics 10, 2123–2137. doi: 10.1002/pmic.200900345
Dembinsky, D., Woll, K., Saleem, M., Liu, Y., Fu, Y., Borsuk, L. A., et al. (2007). Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 145, 575–588. doi: 10.1104/pp.107.106203
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trend. Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Duncan, O., Taylor, N. L., Carrie, C., Eubel, H., Kubiszewski-Jakubiak, S., Zhang, B., et al. (2011). Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol. 157, 1093–1113. doi: 10.1104/pp.111.183160
Falter, C., Ellinger, D., von Hülsen, B., Heim, R., and Voigt, C. A. (2015). Simple preparation of plant epidermal tissue for laser microdissection and downstream quantitative proteome and carbohydrate analysis. Front. Plant Sci. 6:194. doi: 10.3389/fpls.2015.00194
Feist, P., and Hummon, A. B. (2015). Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int. J. Mol. Sci. 16, 3537–3563. doi: 10.3390/ijms16023537
Feiz, L., Irshad, M., Pont-Lezica, R. F., Canut, H., and Jamet, E. (2006). Evaluation of cell wall preparations for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2:10. doi: 10.1186/1746-4811-2-10
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Giavalisco, P., Nordhoff, E., Lehrach, H., Gobom, J., and Klose, J. (2003). Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophor. 24, 207–216. doi: 10.1002/elps.200390016
Gupta, R., and Kim, S. T. (2015). Depletion of RuBisCO protein using the protamine sulfate precipitation method. Methods Mol. Biol. 1295, 225–233. doi: 10.1007/978-1-4939-2550-6_17
Gupta, R., Min, C. W., Wang, Y., Kim, Y. C., Agrawal, G. K., Rakwal, R., et al. (2016). Expect the unexpected enrichment of “hidden proteome” of seeds and tubers by depletion of storage proteins. Front. Plant Sci. 7:761. doi: 10.3389/fpls.2016.00761
Hall, M., Mishra, Y., and Schröder, W. P. (2011). Preparation of stroma, thylakoid membrane, and lumen fractions from Arabidopsis thaliana chloroplasts for proteomic analysis. Methods Mol. Biol. 775, 207–222. doi: 10.1007/978-1-61779-237-3_11
Isaacson, T., Damasceno, C. M., Saravanan, R. S., He, Y., Catal,á, C., Saladi,é, M., et al. (2006). Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 1, 769–774. doi: 10.1038/nprot.2006.102
Jain, R., Katavic, V., Agrawal, G. K., Guzov, V. M., and Thelen, J. J. (2008). Purification and proteomic characterization of plastids from Brassica napus developing embryos. Proteomics 8, 3397–3405. doi: 10.1002/pmic.200700810
Jamet, E., Albenne, C., Boudart, G., Irshad, M., Canut, H., and Pont-Lezica, R. (2008). Recent advances in plant cell wall proteomics. Proteomics 8, 893–908. doi: 10.1002/pmic.200700938
Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M. C., Wang, B., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546, 524–527. doi: 10.1038/nature22971
Karthikaichamy, A., Deore, P., Rai, V., Bulach, D., Beardall, J., Noronha, S., et al. (2017). Time for multiple extraction methods in proteomics? a comparison of three protein extraction methods in the Eustigmatophyte alga Microchloropsis gaditana CCMP526. OMICS 21, 678–683. doi: 10.1089/omi.2017.0128
Kaspar, S., Weier, D., Weschke, W., Mock, H. P., and Matros, A. (2010). Protein analysis of laser capture micro-dissected tissues revealed cell-type specific biological functions in developing barley grains. Anal. Bioanal. Chem. 398, 2883–2893. doi: 10.1007/s00216-010-4120-y
Kasuga, K., Katoh, Y., Nagase, K., and Igarashi, K. (2017). Microproteomics with microfluidic-based cell sorting: application to 1000 and 100 immune cells. Proteomics 17:1600420. doi: 10.1002/pmic.201600420
Kim, J. Y., Wu, J., Kwon, S. J., Oh, H., Lee, S. E., Kim, S. G., et al. (2014). Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics 14, 2307–2318. doi: 10.1002/pmic.201400066
Kim, Y. J., Lee, H. M., Wang, Y., Wu, J., Kim, S. G., et al. (2013). Depletion of abundant plant RuBisCO protein using the protamine sulfate precipitation method. Proteomics 13, 2176–2179. doi: 10.1002/pmic.201200555
Kim, Y. J., Wang, Y., Gupta, R., Kim, S. W., Min, C. W., Kim, Y. C., et al. (2015). Protamine sulfate precipitation method depletes abundant plant seed-storage proteins: a case study on legume plants. Proteomics 15, 1760–1764. doi: 10.1002/pmic.201400488
Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjölander, K., Gruissem, W., et al. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 9, 354–362. doi: 10.1016/j.cub.2004.02.039
Lande, N. V., Subba, P., Barua, P., Gayen, D., Keshava Prasad, T. S., Chakraborty, S., et al. (2017). Dissecting the chloroplast proteome of chickpea (Cicer arietinum L.) provides new insights into classical and non-classical functions. J. Proteomics 165, 11–20. doi: 10.1016/j.jprot.2017.06.005
Lang, E. G., Mueller, S. J., Hoernstein, S. N., Porankiewicz-Asplund, J., Vervliet-Scheebaum, M., and Reski, R. (2011). Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 30, 205–215. doi: 10.1007/s00299-010-0935-4
Lee, H. M., Gupta, R., Kim, S. H., Wang, Y., Rakwal, R., Agrawal, G. K., et al. (2015). Abundant storage protein depletion from tuber proteins using ethanol precipitation method: suitability to proteomics study. Proteomics 15, 1765–1769. doi: 10.1002/pmic.201400526
Libault, M., Pingault, L., Zogli, P., and Schiefelbein, J. (2017). Plant systems biology at the single-cell level. Trends Plant Sci. 22, 949–960. doi: 10.1016/j.tplants.2017.08.006
Maltman, D. J., Simon, W. J., Wheeler, C. H., Dunn, M. J., Wait, R., and Slabas, A. R. (2002). Proteomic analysis of the endoplasmic reticulum from developing and germinating seed of castor (Ricinus communis). Electrophoresis 23, 626–639. doi: 10.1002/1522-2683(200202)23:4<626::AID-ELPS626>3.0.CO;2-#
Nikolovski, N., Shliaha, P. V., Gatto, L., Dupree, P., and Lilley, K. S. (2014). Label-free protein quantification for plant Golgi protein localization and abundance. Plant Physiol. 166, 1033–1043. doi: 10.1104/pp.114.245589
Ohnishi, M., Yoshida, K., and Mimura, T. (2018). “Analyzing the vacuolar membrane (tonoplast) proteome,” in Plant Membrane Proteomics Methods in Molecular Biology, Vol 1696, eds H. P. Mock, A. Matros, and K. Witzel (New York, NY: Humana Press), 107–116.
Parsons, H. T., Christiansen, K., Knierim, B., Carroll, A., Ito, J., Batth, T. S., et al. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159, 12–26. doi: 10.1104/pp.111.193151
Petriccione, M., Di Cecco, I., Arena, S., Scaloni, A., and Scortichini, M. (2013). Proteomic changes in Actinidia chinensis shoot during systemic infection with a pandemic Pseudomonas syringae pv. actinidiae strain. J. Proteomics 78, 461–476. doi: 10.1016/j.jprot.2012.10.014
Piro, A., Serra, I. A., Spadafora, A., Cardilio, M., Bianco, L., Perrotta, G., et al. (2015). Purification of intact chloroplasts from marine plant Posidonia oceanica suitable for organelle proteomics. Proteomics 15, 4159–4174. doi: 10.1002/pmic.201500246
Printz, B., Dos Santos Morais, R., Wienkoop, S., Sergeant, K., Lutts, S., Hausman, J. F., et al. (2015). An improved protocol to study the plant cell wall proteome. Front. Plant Sci. 6:237. doi: 10.3389/fpls.2015.00237
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E., et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. doi: 10.1105/tpc.107.050989
Reumann, S., and Singhal, R. (2014). Isolation of leaf peroxisomes from Arabidopsis for organelle proteome analyses. Methods Mol. Biol. 1072, 541–552. doi: 10.1007/978-1-62703-631-3_36
Rose, J. K., Bashir, S., Giovannoni, J. J., Jahn, M. M., and Saravanan, R. S. (2004). Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J. 39, 715–733. doi: 10.1111/j.1365-313X.2004.02182.x
Rose, J. K., and Lee, S. J. (2010). Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 153, 433–436. doi: 10.1104/pp.110.154872
Rudolf, G. C., Heydenreuter, W., and Sieber, S. A. (2013). Chemical proteomics: ligation and cleavage of protein modifications. Curr. Opin. Chem. Biol. 17, 110–117. doi: 10.1016/j.cbpa.2012.11.007
Salvato, F., Havelund, J. F., Chen, M., Rao, R. S., Rogowska-Wrzesinska, A., Jensen, O. N., et al. (2014). The potato tuber mitochondrial proteome. Plant Physiol. 164, 637–653. doi: 10.1104/pp.113.229054
Santoni, V., Kieffer, S., Desclaux, D., Masson, F., and Rabilloud, T. (2000). Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophor 21, 3329–3344. doi: 10.1002/1522-2683(20001001)21:16<3329::AID-ELPS3329>3.0.CO;2-F
Sari, Y. W., Alting, A. C., Floris, R., Sanders, J. P. M., and Bruins, M. E. (2014). Glutamic acid production from wheat by-products using enzymatic and acid hydrolysis. Biomass Bioenerg. 67, 451–459. doi: 10.1016/j.biombioe.2014.05.018
Sari, Y. W., Syafitri, U., Bruins, M. E., and Sanders, J. P. M. (2015). How biomass composition determines protein extractability. Ind. Crop. Prod. 70, 125–133. doi: 10.1016/j.indcrop.2015.03.020
Schad, M., Lipton, M. S., Giavalisco, P., Smith, R. D., and Kehr, J. (2005). Evaluation of two-dimensional electrophoresis and liquid chromatography–tandem mass spectrometry for tissue-specific protein profiling of laser-microdissected plant samples. Electrophoresis 26, 2729–2738. doi: 10.1002/elps.200410399
Schikowsky, C., Thal, B., Braun, H. P., and Eubel, H. (2018). Sample preparation for analysis of the plant mitochondrial membrane proteome. Methods Mol. Biol. 1696, 163–183. doi: 10.1007/978-1-4939-7411-5_11
Shimaoka, T., Ohnishi, M., Sazuka, T., Mitsuhashi, N., Hara-Nishimura, I., Shimazaki, K., et al. (2004). Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol. 45, 672–683. doi: 10.1093/pcp/pch099
Sikorskaite, S., Rajamäki, M. L., Baniulis, D., Stanys, V., and Valkonen, J. P. (2013). Protocol: optimised methodology for isolation of nuclei from leaves of species in the Solanaceae and Rosaceae families. Plant Methods 9, 31–40. doi: 10.1186/1746-4811-9-31
Soares, N. C., Francisco, R., Ricardo, C. P., and Jackson, P. A. (2007). Proteomics of ionically bound and soluble extracellular proteins in Medicago truncatula leaves. Proteomics 7, 2070–2082. doi: 10.1002/pmic.200600953
Stoehr, G., Schaab, C., Graumann, J., and Mann, M. (2013). A SILAC-based approach identifies substrates of caspase-dependent cleavage upon TRAIL-induced apoptosis. Mol. Cell Proteomics 12, 1436–1450. doi: 10.1074/mcp.M112.024679
Svozil, J., Gruissem, W., and Baerenfaller, K. (2016). Meselect – a rapid and effective method for the separation of the main leaf tissue types. Front. Plant Sci. 7:1701. doi: 10.3389/fpls.2016.01701
Takác, T., Šamajová, O., and Šamaj, J. (2017). Integrating cell biology and proteomic approaches in plants. J. Proteomics 169, 165–175. doi: 10.1016/j.jprot.2017.04.020
Thelen, J. J., and Peck, S. C. (2007). Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346. doi: 10.1105/tpc.107.053991
Verdonk, J. C., Hatfield, R. D., and Sullivan, M. L. (2012). Proteomic analysis of cell walls of two developmental stages of alfalfa stems. Front. Plant Sci. 13:279. doi: 10.3389/fpls.2012.00279
Walzthoeni, T., Leitner, A., Stengel, F., and Aebersold, R. (2013). Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struc. Biol. 23, 252–260. doi: 10.1016/j.sbi.2013.02.008
Wang, S., Zhang, G., Zhang, Y., Song, Q., Chen, Z., Wang, J., et al. (2015). Comparative studies of mitochondrial proteomics reveal an intimate protein network of male sterility in wheat (Triticum aestivum L.). J. Exp. Bot. 66, 6191–6203. doi: 10.1093/jxb/erv322
Wang, W., Scali, M., Vignani, R., Spadafora, A., Sensi, E., Mazzuca, S., et al. (2003). Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24, 2369–2375. doi: 10.1002/elps.200305500
Wang, W., Tai, F., and Chen, S. (2008). Optimizing protein extraction from plant tissues for enhanced proteomic analysis. J. Sep. Sci. 31, 2032–2039. doi: 10.1002/jssc.200800087
Wu, X., Gong, F., and Wang, W. (2014a). Protein extraction from plant tissues for 2DE and its application in proteomic analysis. Proteomics 14, 645–658. doi: 10.1002/pmic.201300239
Wu, X., Xiong, E., An, S., Gong, F., and Wang, W. (2012). Sequential extraction results in improved proteome profiling of medicinal plant Pinellia ternata tubers, which contain large amounts of high-abundance proteins. PLoS ONE 7:e50497. doi: 10.1371/Journal.Pone.0050497
Wu, X., Xiong, E., Wang, W., Scali, M., and Cresti, M. (2014b). Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 9, 362–374. doi: 10.1038/nprot.2014.022
Wu, X., Zhang, Q., Wu, Z., Tai, F., and Wang, W. (2018). Subcellular locations of potential cell wall proteins in plants: predictors, databases and cross-referencing. Brief. Bioinform. doi: 10.1093/bib/bbx050
Xing, S., Meng, X., Zhou, L., Mujahid, H., Zhao, C., Zhang, Y., et al. (2016). Proteome profile of starch granules purified from rice (Oryza sativa) endosperm. PLoS ONE 11:e0168467. doi: 10.1371/journal.pone.0168467
Xiong, E., Wu, X., Yang, L., Gong, F., Tai, F., and Wang, W. (2014). Chloroform-assisted phenol extraction improving proteome profiling of maize embryos through selective depletion of high-abundance storage proteins. PLoS ONE 9:e112724. doi: 10.1371/journal.pone.0112724
Zhang, C., Sanders, J. P. M., and Bruins, M. E. (2014). Critical parameters in cost-effective alkaline extraction for high protein yield from leaves. Biomass Bioenerg. 67, 466–472. doi: 10.1016/j.biombioe.2014.05.020
Zhang, Y., Giboulot, A., Zivy, M., Valot, B., Jamet, E., and Albenne, C. (2011). Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72, 1109–1123. doi: 10.1016/j.phytochem.2010.10.019
Keywords: plant proteome, protein extraction, missing proteins, comparative proteomics, organelle subproteomics, single-cell level proteomics
Citation: Niu L, Yuan H, Gong F, Wu X and Wang W (2018) Protein Extraction Methods Shape Much of the Extracted Proteomes. Front. Plant Sci. 9:802. doi: 10.3389/fpls.2018.00802
Received: 12 February 2018; Accepted: 25 May 2018;
Published: 12 June 2018.
Edited by:
Dominique Job, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Maria Cristina Baracat-Pereira, Universidade Federal de Viçosa, BrazilSun Tae Kim, Pusan National University, South Korea
Copyright © 2018 Niu, Yuan, Gong, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, d2FuZ3dlaUBoZW5hdS5lZHUuY24=
 Liangjie Niu
Liangjie Niu Xiaolin Wu
Xiaolin Wu Wei Wang
Wei Wang