
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 June 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00786
 Donald James1
Donald James1 Bhabesh Borphukan1
Bhabesh Borphukan1 Dhirendra Fartyal1,2
Dhirendra Fartyal1,2 Babu Ram1,2
Babu Ram1,2 Jitender Singh1,3
Jitender Singh1,3 Mrinalini Manna1
Mrinalini Manna1 Vijay Sheri1
Vijay Sheri1 Varakumar Panditi1
Varakumar Panditi1 Renu Yadav1
Renu Yadav1 V. Mohan M. Achary1
V. Mohan M. Achary1 Mallireddy K. Reddy1*
Mallireddy K. Reddy1*Glutamine synthetase (GS) is a key enzyme involved in the nitrogen metabolism of higher plants. Abiotic stresses have adverse effects on crop production and pose a serious threat to global food security. GS activity and expression is known to be significantly modulated by various abiotic stresses. However, very few transgenic overexpression studies of GS have studied its impact on abiotic stress tolerance. GS is also the target enzyme of the broad spectrum herbicide Glufosinate (active ingredient: phosphinothricin). In this study, we investigated the effect of concurrent overexpression of the rice cytosolic GS1 (OsGS1;1) and chloroplastic GS2 (OsGS2) genes in transgenic rice on its tolerance to abiotic stresses and the herbicide Glufosinate. Our results demonstrate that the co-overexpression of OsGS1;1 and OsGS2 isoforms in transgenic rice plants enhanced its tolerance to osmotic and salinity stress at the seedling stage. The transgenic lines maintained significantly higher fresh weight, chlorophyll content, and relative water content than wild type (wt) and null segregant (ns) controls, under both osmotic and salinity stress. The OsGS1;1/OsGS2 co-overexpressing transgenic plants accumulated higher levels of proline but showed lower electrolyte leakage and had lower malondialdehyde (MDA) content under the stress treatments. The transgenic lines showed considerably enhanced photosynthetic and agronomic performance under drought and salinity stress imposed during the reproductive stage, as compared to wt and ns control plants. The grain filling rates of the transgenic rice plants under reproductive stage drought stress (64.6 ± 4.7%) and salinity stress (58.2 ± 4.5%) were significantly higher than control plants, thereby leading to higher yields under these abiotic stress conditions. Preliminary analysis also revealed that the transgenic lines had improved tolerance to methyl viologen induced photo-oxidative stress. Taken together, our results demonstrate that the concurrent overexpression of OsGS1;1 and OsGS2 isoforms in rice enhanced physiological tolerance and agronomic performance under adverse abiotic stress conditions, apparently acting through multiple mechanistic routes. The transgenic rice plants also showed limited tolerance to the herbicide Glufosinate. The advantages and limitations of glutamine synthetase overexpression in crop plants, along with future strategies to overcome these limitations for utilization in crop improvement have also been discussed briefly.
Glutamine synthetase (GS; L-glutamate-ammonia ligase; EC 6.3.1.2) is a key enzyme involved in nitrogen (N) metabolism of plants, as it catalyzes the critical incorporation of inorganic ammonium into glutamine in an ATP-dependent manner (Miflin and Habash, 2002; Bernard and Habash, 2009). GS assimilates ammonia (NH), a cytotoxic and reactive metabolite, produced from the fixation of atmospheric N, and or during direct nitrate or ammonia uptake from soil (Hirel and Lea, 2001). GS is also responsible for the re-assimilation of NH, produced during various cellular metabolic processes including, photorespiration and protein degradation, which are further enhanced particularly during stress or senescence (Bernard and Habash, 2009). Along with glutamate synthase (GOGAT; EC 1.4.7.1 and EC 1.4.1.14), GS takes part in the GS/GOGAT cycle which is the focal point of N metabolism in higher plants. The amino acids glutamine and glutamate thus produced, are used to synthesize all other organo-nitrogen compounds including nucleotides, chlorophyll, and also other amino acids like proline etc. (Forde and Lea, 2007; Bernard and Habash, 2009). The efficient functioning of GS is crucial, as the buildup of NH can cause cell death and severe damage to plant tissues (Wild and Manderscheid, 1984; Tachibana et al., 1986). Two isoforms of GS, the cytosolic GS1, and the chloroplastic GS2 are generally present in higher plants. The smaller cytosolic isoform GS1 is responsible for the primary assimilation of inorganic N availed from the soil in the form of nitrate or ammonia, and the re-assimilation of NH released by protein degradation in senescing leaves (Bernard and Habash, 2009). Whereas, the larger chloroplast localized isoform, GS2, is responsible for re-assimilation of NH released during photorespiration and nitrate (NO) reduction in plastids (Wallsgrove et al., 1987; Leegood et al., 1995; Lam et al., 1996). In most plants, a multigene family encodes the cytosolic GS1, while only a single gene encodes the chloroplastic GS2. In rice, one gene encodes the plastidic GS2 (OsGS2) and three encode cytosolic GS1 (OsGS1;1, OsGS1;2, and OsGS1;3). OsGS1;1 is ubiquitous but expressed more in the shoot and stem, OsGS1;2 is expressed mostly in the root, OsGS1;3 is limited to the spikelets, whereas the OsGS2 isoform is abundant in the leaf (Tabuchi et al., 2005). Due to its central role in N metabolism, GS is considered a prime target for transgenic approaches to increase nitrogen use efficiency (NUE) and yield, which is paramount in achieving sustainability in agriculture and ensuring food security for our burgeoning population (Brauer and Shelp, 2010; Swarbreck et al., 2011). Hence most studies have largely focused attention on the impact of transgenic overexpression of GS on the parameters such as yield and NUE in various plants such as tobacco (Migge et al., 2000; Fuentes et al., 2001; Oliveira et al., 2002; Wang et al., 2013), Arabidopsis (Zhu et al., 2014, 2015), maize (Martin et al., 2006); poplar (Gallardo et al., 1999; Fu et al., 2003; Jing et al., 2004; Man et al., 2005), sorghum (Urriola and Rathore, 2015); wheat (Habash et al., 2001), and rice (Cai et al., 2009; Brauer et al., 2011; Bao et al., 2014).
However, genes involved in N metabolism, including GS, are also known to be significantly modulated during various stress responses in different plants (Wang et al., 2012; Goel and Singh, 2015). The expression and activity of GS isoforms have been reported to be modulated in various plants in response to abiotic stresses like drought (Bauer et al., 1997; Nagy et al., 2013; Singh and Ghosh, 2013; Yousfi et al., 2015; Cheng et al., 2016), cold (Lu et al., 2005; Kwon et al., 2007), salinity (Silveira et al., 2001, 2003; Yan et al., 2005; Ouyang et al., 2007; Teixeira and Pereira, 2007; Wang et al., 2007, 2012), and metal toxicity (Chaffei et al., 2004; Rana et al., 2008). Several studies on the GS enzyme have postulated its role in improving tolerance to various abiotic stresses. For example, a comparative study on the expression and activity of various GS isoforms in rice under drought stress inferred that a relatively maintained OsGS2 level and the over-expression of OsGS1;1 might contribute to the enhanced drought tolerance characteristics of the drought tolerant rice cultivar Khitish (Singh and Ghosh, 2013). In addition, gene expression analysis under salinity and drought stress between contrasting durum wheat genotypes showed that the most tolerant genotype exhibited the highest GS activity and had enhanced expression of both GS1 and GS2 isoforms under stress conditions as compared to the control plants (Yousfi et al., 2015). Nagy et al. (2013) observed that drought tolerant wheat genotypes maintained higher GS activity in the flag leaf under drought stress than sensitive cultivars. Furthermore, a comprehensive QTL analysis in potato revealed that the cytosolic GS is essential for improving photosynthetic efficiency and water use efficiency (WUE). It was observed that the GS activity was more enhanced in the high WUE bulk population than in the low WUE bulk population (Kaminski et al., 2015). On similar lines, a recent proteomic study of wheat cultivars under drought stress found that the chloroplastic GS2 was significantly up-regulated in a drought tolerant cultivar as compared to the sensitive cultivar (Cheng et al., 2016).
Hence, several lines of evidence have propounded the ability of GS to confer enhanced tolerance and higher yields of crops under various abiotic stresses. However, only a few transgenic overexpression studies of GS isoforms have studied its impact on tolerance to abiotic stresses. For instance, overexpression of a cytoplasmic GS1 gene from conifer in transgenic poplar trees was observed to confer enhanced tolerance to drought stress (El-Khatib et al., 2004), while transgenic rice lines overexpressing the chloroplastic OsGS2 gene displayed enhanced salinity tolerance (Hoshida et al., 2000). In addition, the constitutive overexpression of the cytosolic OsGS1;1 gene in rice improved tolerance to oxidative stress induced by the heavy metal cadmium (Lee et al., 2013).
GS is also the target enzyme of the widely used post emergent broad spectrum herbicide Glufosinate (active ingredient: L-Phosphinothricin/PPT; common trade name: Basta™). Phosphinothricin (PPT) is a structural analog of the substrate of GS viz. glutamate, and occupies the substrate pocket of the enzyme and blocks glutamate binding to GS (Gill and Eisenberg, 2001). Inhibition of the GS enzyme by PPT causes the buildup of ammonia in plant cells, inhibition of amino acid synthesis and inhibition of photosynthesis, which ultimately leads to the death of the plants (Donn and Köcher, 2002). Several studies have observed that transgenic overexpression of GS confers tolerance to PPT in plants like poplar (Pascual et al., 2008) and rice (Cai et al., 2009). Also, the co-overexpression of a cytosolic GS1 and chloroplastic GS2 from pea was reported to confer tolerance to PPT in wheat and rice transgenics (Huang et al., 2005; Sun et al., 2005).
Since GS is a potential target in multiple avenues of transgenic crop improvement including yield, NUE, abiotic stress tolerance and herbicide resistance, the co-overexpression of multiple GS isoforms through gene stacking was considered expedient to study its effects on these multiple areas. The effect of OsGS1;1/OsGS2 co-overexpression on yield and NUE of rice under varying N regimens is currently being investigated, and will be reported separately. In this study, through an in vitro gene pyramiding approach utilizing a Multi-Round Gateway cloning technology, we were able to concurrently overexpress the cytosolic GSl;1 (OsGS1;1) and chloroplastic GS2 (OsGS2) in transgenic rice. Its impact on abiotic stresses tolerance and resistance to the herbicide Glufosinate (phosphinothricin) is discussed here within.
Gene specific primers were designed based on the sequence of rice cytosolic GS1;1 (OsGS1;1; LOC_Os02g50240) and rice chloroplastic GS2 (OsGS2; LOC_Os04g56400) available in the Rice Genome Annotation database (Supplementary Table 1). The full-length coding sequences of OsGS1;1 (1113 bp) and OsGS2 (1287 bp) were amplified by PCR from rice (Oryza sativa L. ssp japonica cv Nipponbare) cDNA using a high fidelity DNA polymerase (KOD plus, Toyobo, Japan) and cloned into pCR-4-TOPO vector (Invitrogen, USA). Following verification by sequencing, the OsGS1;1 gene was sub-cloned into a Gateway compatible entry vector EV-1 (pL12R34-Ap) in between the rice Actin 2 (OsAct2) promoter and rice Actin 2 (OsAct2) terminator, whereas the OsGS2 gene was cloned into the Gateway compatible EV-2 (pL34R12-Cm-ccdB) vector under the rice Actin 1 (OsAct1) promoter and rice Actin 1 (OsAct1) terminator. The plant expression cassettes of rice cytosolic GS (OsAct2 promoter: OsGS1;1: OsAct2 terminator) and plastidic GS (OsAct1 promoter: OsGS2: OsAct1 terminator) from the entry vectors were sequentially cloned into a Gateway compatible destination vector (pMDC99) for plant transformation using a Multi-Round LR recombinase mediated Gateway™ (Invitrogen, USA) cloning process as described previously (Chen et al., 2006) (Figure 1). The pMDC99 vector contains the hygromycin resistance gene hygromycin phosphotransferase (hpt) as plant selection marker (Curtis and Grossniklaus, 2003). The in vitro gene pyramided construct containing both OsGS1;1 and OsGS2 under constitutive rice actin promoters (pMDC99- OsAct2 P: OsGS1;1: OsAct2 T:: OsAct1 P: OsGS2: OsAct1 T) was transformed into rice (O.sativa ssp Japonica) cultivar Nipponbare through Agrobacterium mediated transformation using a protocol described earlier (Ravikumar et al., 2014). Briefly, twenty-one-day-old scutellum-derived embryogenic calli were infected with Agrobacterium EHA105 strain harboring the above construct and co-cultivated for 2 days on Chu-N6 medium (Duchefa, Germany) supplemented with acetosyringone (200 μM). The transformed calli were selected on Chu-N6 medium supplemented with 50 mg/L Hygromycin (Invitrogen, USA) and 1 mg/L PPT (Duchefa, Germany). Secondary calli that developed after four rounds of selection were transferred to regeneration media for the development of shoots. The regenerated shoots were transferred to rooting media. The transgenic OsGS1;1/OsGS2 co-overexpressing primary transformants (T0) were transferred to soilrite for hardening and then transferred to soil pots in a greenhouse maintained at 28 ± 2°C temperature and relative humidity of 70 ± 5%.
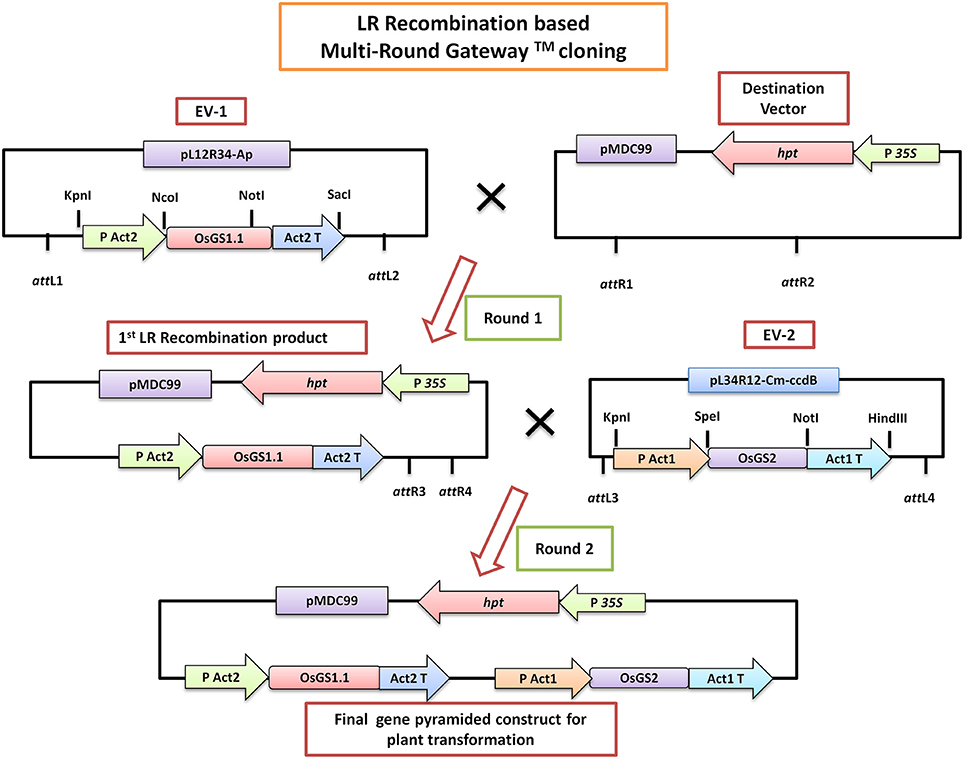
Figure 1. A simplified schematic diagram of the LR-Recombination based Multi-Round Gateway™ technology used for in vitro pyramiding of the OsGS1;1 and OsGS2 genes. The OsGS1;1 gene was cloned into the Entry vector 1 (EV-1) by restriction enzyme based cloning under the rice Actin 2 promoter (PAct2) and Act2 terminator (Act2T) whereas the OsGS2 gene was cloned into the entry vector 2 (EV-2) under the rice Actin 1 promoter (PAct1) and Act1 terminator (Act1T). In the first round of LR cloning, EV-1 was recombined with the destination vector (pMDC99) to obtain the first LR recombined product. Subsequently, EV-2 was recombined with the first LR product in a second round of LR cloning to obtain the final gene pyramided pMDC99 construct containing both OsGS1;1 and OsGS2 genes to be used for plant transformation (For a detailed scheme see Chen et al., 2006).
Putative T0 transgenics seeds were screened by germinating them on Murashige and Skoog media containing 50 mg/L hygromycin, followed by PCR confirmation using hpt, OsGs1;1 and OsGS2 screening primers (Supplementary Table 1). Transgenic plants were progressed to the T2 generation in the greenhouse following standard agronomic practices. T2 generation plants were again verified by PCR using hpt, OsGS1;1: OsAct2T and OsGS2: OsAct1T screening primers (Supplementary Table 1). Null segregants (ns) (azygous lines from the T1 generation of transgenics) identified using PCR were used as controls along with wild type (wt) rice plants to account for probable in vitro regeneration and transformation effects. Transgene insertion and copy number assessment was done by Southern blot analysis in T2 generation plants using DIG non-radioactive nucleic acid labeling and detection system (Roche, QC, Canada) as per the protocol followed by Manna et al. (2016). Briefly, 20 μg of genomic DNA of control and transgenic rice lines was digested with HindIII and genomic DNA fragments were separated on a 0.8% agarose gel, blotted onto Hybond™ N+ nylon membrane (GE Healthcare Limited, UK), and subsequently hybridized with a 900 bp hpt gene specific probe. The probe was synthesized through PCR using hpt gene specific primers (Supplementary Table 1) labeled with DIG (PCR DIG Probe Synthesis Kit, Roche, Germany). The blot was washed and detected according to the manufacturer's instructions (DIG High Prime DNA Labeling and Detection Starter Kit II, Roche, Germany).
Gene expression analysis by semi-quantitative RT-PCR was carried out using OsSG1;1 and OsGS2 RT-PCR specific primers (Supplementary Table 1) using standard protocols. The eEF-1α gene from rice was used as endogenous reference gene for normalizing the relative expression (Jain et al., 2006). The RT-PCR amplified products were resolved on a 1.2% agarose gel. Densitometric analysis of the bands was performed using the Image J software, to quantify relative transcript expression levels (Schneider et al., 2012).
For total GS enzyme activity assay, 2-week-old seedlings of T2 transgenic plants and wild type Nipponbare (wt) controls were homogenized in a GS extraction buffer (15 mL/g fresh weight) containing 50 mM Tris-HCl (pH 8), 5 mM MgC12, 5% (w/v) insoluble PVP, and 15% glycerol. The crude extract was filtered through five layers of gauze and the filtrate was centrifuged at 12,000 rpm for 30 min at 4°C (Singh and Ghosh, 2013). The total protein in the crude supernatant was quantified by Bradford assay, and used for both assay of GS activity and immunoblotting analyses of GS1 and GS2.
Total GS activity was measured using the semi-biosynthetic assay protocol modified from Singh and Ghosh (2013) by quantifying the formation of γ-glutamylhydroxamate. Briefly, the reaction mixture of total 1 mL volume consisted of 50 mM Tris–HCl (pH 7.5), 250 mM glutamate, 20 mM ATP, 10 mM hydroxylamine hydrochloride, 20 mM MgCl2, and 200 μL of crude extract. The reaction mixture was kept at 37°C for 20 min and terminated by adding 2 mL of FeCl3 reagent (0.67 M FeCl3, 0.37 M HCl and 20% (w/v) tri-chloroacetic acid). The reaction mixture was incubated at room temp for 10 min for color development. The reaction mixture was then centrifuged at 4,000 g at room temperature for 10 min, and 1 mL of the supernatant was transferred into a quartz cuvette and the absorbance measured via a spectrophotometer (Ultrospec 2100 pro, Amersham Biosciences, UK) at 540 nm. Values of GS activity were extrapolated from a standard calibration curve made from different known concentrations of γ-glutamylhydroxamate (Sigma-Aldrich, USA). One unit of GS activity represents 1 μmol of γ-glutamylhydroxamate produced in 20 min.
Immunoblotting was done using a protocol modified from Kamachi et al. (1992) using recombinant GS specific antibodies that detect both GS1 and GS2 isoforms (Ishiyama et al., 2004). Briefly, 5 μg total protein from wt and transgenic plants extracted as described above, were separated on 12.5% (w/v) SDS PAGE and blotted onto a PVDF membrane (Millipore, USA) using as Mini transblot electrophoretic cell (Biorad, USA). The blot was incubated in blocking solution (5% non-fat dry milk in PBS) for 1 h. The blot was then washed with PBS containing 0.1% Tween-20 (PBST) for three times at 10 min intervals. Following incubation with recombinant antibody at 1:1,000 dilution in blocking solution, the blot was washed with PBST three times at 10 min intervals. Subsequently, the blot was incubated with alkaline phosphatase conjugated anti-rabbit IgG secondary antibody (Sigma, USA) (1:5,000 dilution in PBS) for 1 h, and then was washed three times with PBS containing 0.3% Tween-20 and again three times with PBST at 10 min intervals. Bands were detected using a ready to use BCIP/NBT solution (Sigma, USA) in a dark room until color development and thereafter the reaction was stopped by washing in PBS. The bands were relatively quantified using densitometric analysis of their intensities using the Image J software (Schneider et al., 2012).
The surface sterilized OsGS1;1/OsGS2 co-overexpressing transgenic rice seeds along with seeds of wild type (wt) and null segregant (ns) plants (both hereafter referred to as “control plants”) were sown on Murashige and Skoog medium or sterile germination paper rolls, and then transferred to a growth chamber at 28°C under a 16-h-light/8-h dark photoperiod and 70% relative humidity. For various analyses, both OsGS1;1/OsGS2-overexpressing transgenic and control rice seeds were germinated on moist paper rolls and then transferred to YS hydroponic culture solution (Yoshida nutrient solution; Yoshida et al., 1976) at the two-leaf stage. The culture solution was replaced once every 2 days and pH set to 5.5. For stress treatments, 2-week-old seedlings were transferred to YS supplemented with 150 mM NaCl (EC ~ 14 dS/m) for imposing salinity stress and 15% PEG (MW 6000) for inducing osmotic or physiological drought stress. The plants were grown for 7 days to assess phenotypic variation in physiological tolerance. In a separate experiment, 2-week-old seedlings of transgenic and control plants were grown in YS supplemented with 20% PEG (MW 6000) or 200 mM NaCl (EC ~ 19 dS/m) for 4 days followed by recovery in normal YS for 4 days to determine tolerance to high drought and salinity stresses. Various biochemical and physiological parameters were assessed after 2 days of stress treatments. The electrical conductivity (EC) of the hydroponic solutions was measured using an electrical conductivity meter (WTW Cond 315i, Germany) according to manufacturer's instructions.
For assessing tolerance to drought at reproductive stage, transgenic plants along with control plants were grown in 500 mL pots filled with a 1:1 mix of topsoil and manure compost (quartered and mixed to homogeneity), and placed in water-filled trays to simulate paddy conditions. The rice plants were grown to panicle initiation stage under optimum agronomic conditions in a green house at (16-h-light/8-h-dark cycles) at 28 to 30°C. Drought stress was imposed by withdrawing water using the gravimetric method for a period of 12 days post panicle initiation, followed by recovery by regular irrigation until maturity to assess yield parameters (protocol modified from Ambavaram et al., 2014). For assessing the impact of salinity stress on growth and yield parameters, ~2-month-old OsGS1;1/OsGS2 co-overexpressing transgenic rice plants along with controls were subjected to moderate salinity stress by irrigating the pots every fortnight with deionized water supplemented with 50 mM NaCl (EC ~ 6 dS/m) (protocol modified from Tripathi et al., 2016). The treatment was continued till initiation of the booting stage after which the plants were recovered and grown until maturity, and thereafter their agronomic performance assessed. Untreated control pots were irrigated with normal deionized water.
For oxidative stress treatment assays, leaf strips of 2 cm length and uniform width from flag leaf of mature T2 generation transgenic lines along with the control plants were incubated in 10 μM methyl viologen (MV; paraquat) for 6 h under dark at 28°C to allow diffusion of MV into the tissue and then exposed to 4 h of sunlight (Mahanty et al., 2012). The total chlorophyll content after MV treatment was determined spectrophotometrically following the protocol of Hiscox and Israelstam (1979). Oxidative stress induced generation of H2O2 after treatment with MV was detected by incubating the leaf strips with 3,3-diaminobenzidine (DAB) (pH 3.8), until a reddish-brown color developed (Thordal-Christensen et al., 1997). In-vivo generation of O in leaves after MV treatment was detected by staining with 1% nitro blue tetrazolium (NBT) in 10 mM sodium phosphate buffer, until a purple-blue color was observed (Lin et al., 2009). After NBT and DAB staining, the chlorophyll of the treated samples was removed by boiling in 95% ethanol, and the images of leaf strips were photographed.
To assess tolerance to PPT, mature leaves of both wt and T2 transgenic rice plants were painted with solutions of 0.5, 1, and 2% Basta (v/v) (Bayer, 13.5% ai) supplemented with 0.01% Tween-20. The leaves were scored for tolerance after 5 days of treatment based on the degree of leaf burning, bleaching and necrosis (Tsai et al., 2006). In addition, T2 transgenic and wt plants at four-six leaf stage were sprayed with a 0.5% (v/v) solution of Basta. After 1 week, the growth of the plants was assessed and survival rates were calculated.
Total chlorophyll contents of untreated and stressed seedlings were measured spectrophotometrically following the protocol of Hiscox and Israelstam (1979). Relative chlorophyll content of plants under reproductive stage stress experiments was measured on the third fully opened leaf from the top using a SPAD 502 portable chlorophyll meter (Minolta, Japan). RWC (Relative Water Content) was measured according to the protocol of Barrs and Weatherley (1962). Electrolyte leakage measurements after stress treatments were performed as previously described by Sairam et al. (2005). Measurement of proline was performed according to the protocol of Bates et al. (1973). Estimation of lipid peroxidation was done by determining the amount of MDA (malondialdehyde) using the TBARS (thiobarbituric acid-reactive-substances) assay following the protocol of Hodges et al. (1999). Mean NH liberation with and without PPT application was determined by a modified Berthelot reaction assay given by (Rasco-Gaunt et al., 1999). The plant traits and yield components of rice namely plant height (cm), number of panicles, grain filling rate (%), and grain yield per plant (g), were measured as described by Yoshida et al. (1976).
Photosynthetic gas exchange parameters were measured in the morning (9:00–11:00 a.m.), on flag leaves, using an Infrared Gas Analyzer (IRGA; LI-6400XT portable photosynthesis system equipped with a LI-6400-40 Leaf Chamber Fluorometer, LICOR, Lincoln NB). The measurements were made at a CO2 concentration of 400 μmol mol−1, PPFD of 1200 μmol m−2 s−1 and a chamber temperature of 30°C. The chlorophyll fluorescence (Fv/Fm) during various stress conditions were measured after dark adaptation for 30 min following the protocol outlined by Murchie and Lawson (2013). The measurements of photosynthetic parameters for drought stress treated plants were made after 4 days of recovery following 12 days of drought stress treatment. Measurements for salinity stress treatments were taken after 7 days of recovery following final salinity stress treatment. All the measurements were repeated five times on fully expanded flag leaves of control and transgenic plants and the means ± SD are represented.
All data represented are means ± SD (n = 3). Data were analyzed using ANOVA and post-hoc Tukey-Kramer multiple comparisons tests to assess significance among means using the statistical tools available in GraphPad Prism 6 TM software.
The rice GS isoforms OsGS1;1 and OsGS2 were successfully PCR amplified from rice cDNA. The rice GS1;1 encoding sequence was successfully cloned between the OsAct2 promoter and OsAct2 terminator as a plant expression cassette in EV-1 while the rice GS2 encoding sequence was cloned between the rice OsAct1 promoter and OsAct1 terminator in EV-2 to constitutively overexpress the rice cytosolic and plastidic GS isoforms. Both the expression cassettes were in vitro pyramided on to a single plant transformation vector (pMDC99) using the Multi-Round Gateway cloning technology (Figure 1). The recombinant construct was then transformed into EHA 105 strain of Agrobacterium by electroporation and used for rice transformation. A total of around 150 putative T0 transgenic rice lines were generated. Screening of the putative transgenics to check the integration of hpt, OsGS1;1 and OsGS2 genes into rice genome showed that more than 80% of the putative transformants were PCR positive and showed integration of all the transgenes. The PCR analysis of five selected positive T2 transgenics (L1, L2, L3, L4, and L5) showed the expected ~1 Kb DNA fragment with hpt primer set, ~1.1 kb DNA fragment with OsGS1;1: OsAct2 terminator primer set, and ~1.3 kb fragment with OsGS2: OsAct1 terminator primer set respectively from the transgenic rice genomic DNA, whereas no such DNA fragments were amplified from untransformed wild type (wt) rice genomic DNA (Figure 2A). Southern analysis of selected T2 transgenic lines probed with hpt gene sequence showed a single hybridization band with a distinct pattern for each transgenic line suggesting independent single copy integration of the transgene in each transgenic event (Figure 2B). The untransformed wt plants did not show any hybridization signal. Three independent T2 transgenic lines (L1, L4, and L5), which showed PCR positive amplification for hpt, OsGS1;1 and OsGS2 transgene cassettes and single copy positive southern hybridization were selected for semi quantitative RT-PCR analysis. All the selected transgenic lines (L1, L4, and L5) showed higher expression of OsGS1;1 and OsGS2 transcripts compared to wt controls as quantified by densitometric analysis of semi-quantitative RT-PCR bands (Figure 2C). Transgenic line L1 had 2.5-fold increased OsGS1;1 and 2.7-fold increased OsGS2 expression levels while L4 had 3.1-fold increased OsGS1;1 expression and 2.9-fold increased OsGS2 expression. Whereas, transgenic line L5 showed only 2-fold increase in expression of OsGS1;1 and OsGS2 compared to wt controls (Supplementary Figure S1). The immunoblotting analysis also showed the increased accumulation of OsGS1;1 and OsGS2 protein in all the selected transgenic lines (Figure 2D). An immunopositive 39 kDa polypeptide signal which corresponded to the OsGS1;1 isoform and a 42 kDa polypeptide signal which corresponded to the chloroplastic OsGS2 isoform were observed (Figure 2D). Densitometric analysis of the immunopositive signals using the Image J software showed that transgenic lines L1 and L4 had around 3-fold higher OsGS2 protein content and 2-fold increase in OsGS1;1 protein content in total soluble protein extracts from the transgenic rice seedlings (Supplementary Figure S2). Total GS enzyme activity varied between the selected transgenic lines with L1 and L4 having around 4-folds and L5 having close to 2-fold higher activities as compared to wt controls (Figure 2E). Our results demonstrate that the increase in expression of the GS isoforms in the OsGS1;1/OsGS2 co-overexpressing transgenic Nipponbare rice, led to a corresponding increase in GS protein as well as total GS enzymatic activity (Figures 2C–E). However, it has been reported that higher content of GS protein did not always correspond to higher enzymatic activities of GS in indica rice varieties, and this was attributed to post translational regulation (Obara et al., 2000).
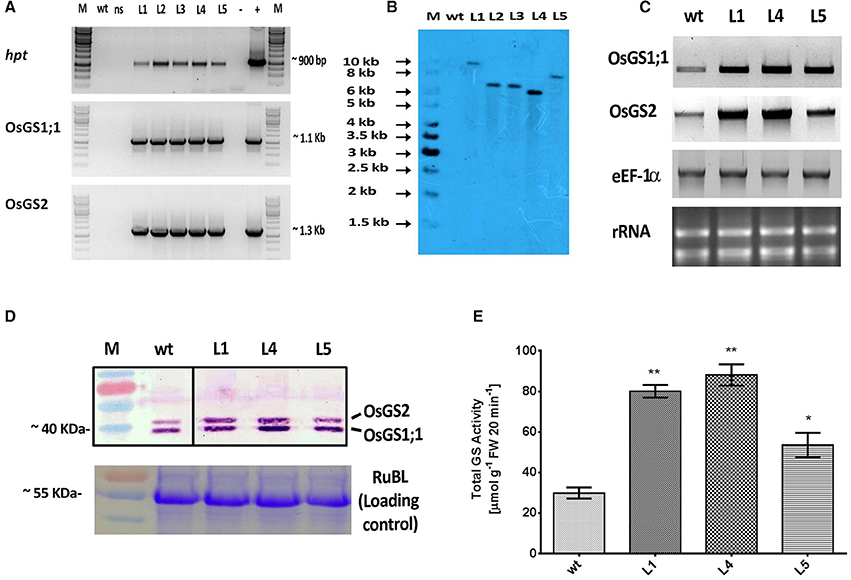
Figure 2. Molecular and biochemical analysis of transgenic rice lines co-overexpressing OsGS1;1 and OsGS2.(A) PCR amplification of hygromycin phosphotransferase (hpt), OsGS1;1 and OSGS2 genes using specific primers in wild type (wt), null segregant (ns), and five positive T2 transgenic lines (L1-L5). M: 1Kb DNA ladder (+): positive PCR control (pMDC99) and (–) water blank. (B) Southern blot analysis of wt and five T2 transgenic lines (L1, L2, L3, L4, and L5), probed with hpt gene probe showing single copy insertion. (C) Semi quantitative RT-PCR showing overexpression of OsGS1;1 and OsGS2 in transgenic lines (L1, L4, and L5) as compared to wt. The rice eEF1α gene was used as a reference gene and rRNA was used as loading control. (D; top panel) Immunoblot analysis of three transgenic rice lines (L1, L4, and L5) and wt using a recombinant antibody which detects both OsGS1;1 and OsGS2 isoforms (black lines separate spliced regions from same blot) (D; bottom panel) Coomassie blue stained Rubisco large subunit (RubL) was used as loading control. (E) Total GS activity of three transgenic rice lines (L1, L4, and L5) in comparison to wt as assayed by a modified semi-biosynthetic assay (Singh and Ghosh, 2013). One unit of GS activity represents 1.0 μmol of γ-glutamylhydroxamate produced in 20 min. Asterisks above bars indicate significant differences from wt (* at p ≤ 0.05 and ** at p ≤ 0.01).
Two-week-old transgenic T2 lines, wt and ns seedlings were grown hydroponically either in untreated YS solution (untreated control) or grown in YS supplemented with 150 mM NaCl (EC~12 dS/m) (salinity stress) or in YS supplemented with 15% PEG (osmotic/physiological drought stress) to study the physiological tolerance of OsGS1;1/OsGS2 overexpressing transgenic rice lines to abiotic stresses. The transgenic, wt and ns seedlings grew well and produced new leaves in the untreated control set (Figure 3A). However, when grown in YS supplemented with either 15% PEG or 150 mM NaCl, there were apparent differences in visible symptoms between OsGS1;1/OsGS2 co-overexpressing transgenic rice seedlings and control seedlings after 7 days of treatments (Figures 3B,C). In the presence of 15% PEG, the wt and ns control plants showed wilting and leaf rolling within 7 days, whereas the transgenic seedlings were able to maintain turgidity and had no symptoms of leaf rolling or wilting (Figure 3B). The OsGS1;1/OsGS2 co-overexpressing transgenic plants were able to survive 15% PEG induced osmotic stress up to 12 days while the control plants underwent severe wilting or died (Supplementary Figure S3). Under salinity stress (150 mM NaCl), both transgenic and control plants showed retarded seedling growth and reduced leaf size compared to the corresponding seedlings grown in untreated YS. However, visible symptoms like leaf tip burning, necrosis, yellowing, wilting and leaf rolling were observed in wt and ns seedlings, whereas such symptoms were not very significant in the transgenic seedlings (Figure 3C). Overall, the OsGS1;1/OsGS2 co-overexpressing transgenic seedlings displayed better growth and increased tolerance to salinity stress in comparison to both wt and ns control plants.

Figure 3. Phenotype of OsGS1;1/OsGS2 co-overexpressing transgenic rice under moderate osmotic and salinity stress at seedling stage. Phenotype of 2-week-old seedlings of wt, ns and three transgenic rice lines (L1, L4, and L5), grown hydroponically in (A) normal Yoshida solution (untreated control) or (B) Yoshida solution supplemented with 15% PEG (osmotic stress) or (C) 150 mM NaCl (EC ~ 12 dS/m) (salinity stress) before and after 7 days of treatment. Visual phenotypic variation amongst seedlings of wt, ns and three transgenic rice lines (L1, L4, and L5), following 4 days of recovery after being grown hydroponically for 4 days in (D) Yoshida solution supplemented with 20% PEG (osmotic stress) or (E) 200 mM NaCl (EC ~ 19 dS/m) (salt stress). Scale bar = 1 cm.
To further study the physiological impact of OsGS1;1/OsGS2 co-overexpression on drought and salinity tolerance in rice, 2-week-old transgenic seedlings and control seedlings were grown hydroponically and subjected to high osmotic stress (20% PEG for 4 days) or high salinity stress [200 mM NaCl (EC ~19 dS/m) for 4 days] followed by 4 days of recovery in untreated YS. Various physiological and biochemical parameters were assessed after 2 days of high osmotic and salinity stress treatments. The OsGS1;1/OsGS2 co-overexpressing transgenic plants showed tolerant phenotypes and were able to recover better after high osmotic and salinity treatments, whereas the wt and ns control seedlings wilted and died (Figures 3D,E). Furthermore, the total fresh weight and chlorophyll content was significantly higher in the transgenic plants under both high salinity and osmotic stresses in comparison to the control. Transgenic lines had 61–67% higher fresh weight and 25–34% higher chlorophyll content compared to the corresponding control seedlings in the presence of 20% PEG induced osmotic stress (Figures 4A,B). Similarly, the transgenic seedlings maintained 29–44% higher fresh weight and 35–48% higher chlorophyll compared to the control seedlings under 200 mM NaCl induced salinity stress (Figures 4A,B). Interestingly, even under control conditions, the transgenic lines had around 12–17% higher fresh weight and 4–15% increased in chlorophyll content as compared to the corresponding non-transgenic control seedlings (Figures 4A,B).
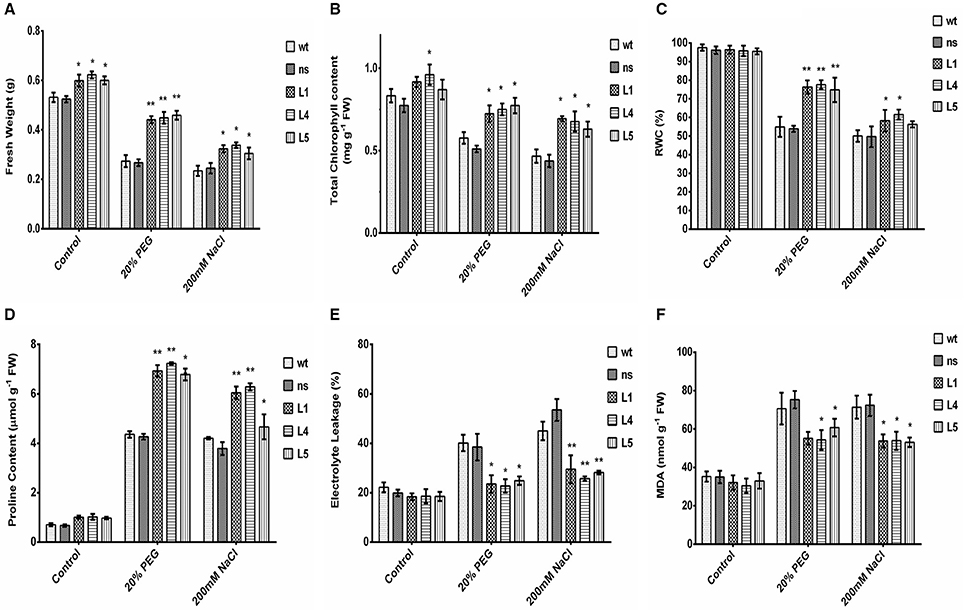
Figure 4. Various biochemical and physiological parameters of 2-week-old seedlings of wt, ns, and three OsGS1;1/OsGS2 co-overexpressing transgenic rice lines (L1, L4, and L5) assessed after 2 days of osmotic (20% PEG) or salinity stress (200 mM NaCl) treatments as compared to untreated control conditions. (A) Fresh weight (FW) (in g). (B) Total chlorophyll content (in mg/g FW). (C) Relative water content (RWC) (in %). (D) Proline content (in μmol/g FW). (E) Electrolyte leakage (in %) (F) Malondialdehyde (MDA) content (in nmol/g FW). All data represented are means ± SD (n = 3). Asterisks above bars indicate significant differences from wt (*p-value ≤ 0.05 and **p-value ≤ 0.01).
Relative water content (RWC) is a quick method to determine the plant water status and gives an estimate of the cellular hydration levels after stress treatments (Barrs and Weatherley, 1962). The OsGS1;1/OsGS2 co-overexpressing transgenic lines were able to maintain 74–78% RWC as compared to the corresponding control seedlings which maintained only 53–55% of RWC under 20% PEG induced high osmotic stress (Figure 4C). Whereas, under high salinity stress (200 mM NaCl), the transgenic seedlings maintained 56–61% of RWC compared to the corresponding control seedlings which had around 49–51% of RWC (Figure 4C). The higher RWC in transgenic seedlings under stressed conditions highlight the enhanced cellular hydration levels and physiological fitness compared to the corresponding non-transgenic control seedlings. Proline accumulation has been correlated with stress tolerant phenotype in many plants and acts as a compatible solute to protect the cellular machinery from stress induced damage (Hayat et al., 2012). In the present study, the relative proline content increased significantly (59–69%) in transgenic as compared to control seedlings in the presence of high osmotic stress (Figure 4D). Similarly, under high salinity stress, the transgenic lines showed increased proline content (59–65%) compared to wt control plants.
Electrolyte leakage provides an estimate of the cellular membrane stability during stress conditions (Bajji et al., 2002). The OsGS1;1/OsGS2 co-overexpressing transgenic rice seedlings showed 34–43% lower rate of electrolyte leakage after treatment with either 20% PEG or 200 mM NaCl, in comparison to corresponding wt control seedlings exposed to the same stress conditions (Figure 4E). Also, the transgenic seedlings had 14–23% reduced MDA contents than wt control seedlings after high osmotic stress treatment and also 24–26% reduced MDA content than wt control seedlings after high salinity stress (Figure 4F). Malondialdehyde (MDA) is the product of decomposition of polyunsaturated fatty acids present in membranes as a result of free radical chain reactions and lipid peroxidation of biomembranes during stress conditions. The amount MDA content gives an estimate of the extent of lipid peroxidation and membrane injury that has occurred during adverse abiotic stresses. Overall, the assessment of various stress responsive physiological and biochemical parameters showed that OsGS1;1/OsGS2 co-overexpressing transgenic rice seedlings had enhanced tolerance to osmotic and salinity stresses.
To assess the impact of photo-oxidative stress on the transgenic plants, uniform leaf strips harvested from the fully expanded leaves from OsGS1;1/OsGS2 co-overexpressing rice seedlings and their corresponding ns and wt control rice seedlings were pre incubated with 10 μM methyl viologen (MV; paraquat) and exposed to sunlight. The leaf strips harvested from wt and ns control seedlings showed faster chlorosis than the transgenic seedlings (Figure 5A). To estimate the amount of in vivo ROS formation after treatment, the MV treated leaf strips were histochemically stained with DAB and NBT to estimate the amount of the ROS generated in response to the photo-oxidative treatment. H2O2 in the presence of peroxidases oxidizes DAB which leads to the production of a reddish brown precipitate. Similarly, the O free radical reacts with NBT to form a dark blue insoluble formazan compound (Kumar et al., 2014). The staining intensity of these tissues provides an estimate of in vivo H2O2 and O2−− formation in response to MV or in response to any other stress. The transgenic lines showed significantly lower ROS production than wt and ns controls after MV treatment as evident by DAB and NBT staining (Figure 5A). The transgenic plants could also retain higher chlorophyll content due to protection from photo-oxidative damage through lesser ROS production after the MV treatment compared to wt and ns controls (Figure 5B). Thus, our preliminary results suggests that OsGS1;1/OsGS2 co-overexpression in transgenic rice plants enhanced tolerance against MV induced photo-oxidative stress by modulating oxidative stress responses that mitigate excessive ROS formation.
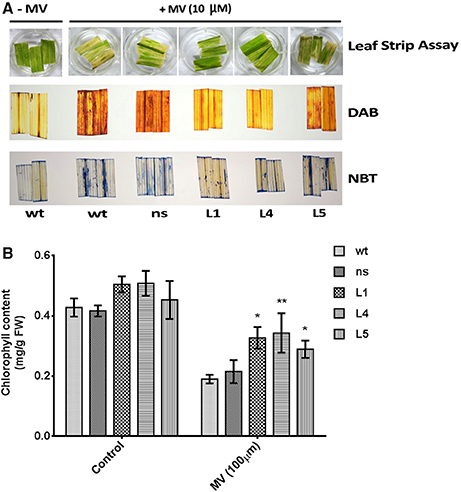
Figure 5. Assessment of tolerance of OsGS1;1/OsGS2 co-overexpressing transgenic rice to methyl viologen (MV) induced photo-oxidative stress. (A; top panel) Leaf strips of wt, ns, and three T2 transgenic lines (L1, L4, and L5) after photo-oxidative stress treatment (incubation in 10 μM MV). Untreated (–MV) wt was used as a control (A; middle panel) Histochemical assessment of in vivo H2O2 formation following MV treatment by DAB staining. (A; bottom panel) In vivo generation of O in leaf strips after MV treatment as detected by NBT staining. (B) Total chlorophyll contents (in mg/g FW) after MV treatment as compared to untreated controls. Data represented are means ± SD (n = 3). Asterisks above bars indicate significant differences from wt (*p-value ≤ 0.05 and **p-value ≤ 0.01).
We imposed terminal drought stress at the post panicle emergence stage by withdrawing irrigation for a period of 12 days to a set of OsGS1;1/OsGS2 co-overexpressing transgenic and corresponding control rice plants until apparent differences in wilting and leaf rolling occurred. Following the drought treatment, the plants were recovered by regular irrigation. Another set of plants, which were regularly irrigated were kept as untreated control (Figure 6A). The transgenic plants were able to withstand the drought stress treatment and recovered vigorously post irrigation, whereas the wt and ns controls which had pronounced leaf rolling and wilting, recovered slowly (Figure 6B). The transgenic lines were able to maintain significantly (19–38%) higher net photosynthetic rates (PN) than the control lines after recovery from the drought stress (Figure 6D). The chlorophyll content as estimated by SPAD meter reading after drought treatment showed that the transgenic lines had 8–20% higher chlorophyll content than the control plants (Figure 6E). Chlorophyll fluorescence (Fv/Fm) gives an estimate of photodamage related effects of abiotic stress on the photosynthetic machinery. The chlorophyll fluorescence (Fv/Fm) ratios for the transgenic rice plants were significantly higher compared to both wt and ns controls under drought stress (Figure 6F). After drought stress recovery, transgenic rice lines showed significantly better agronomic performance with higher panicle number, grain filling rates and yield than wt and ns controls (Figure 6H). The transgenic lines had 28–48% higher panicle numbers than control plants and also showed significantly higher (64.6 ± 4.7%) grain filling rate compared to that controls (35.7 ± 2.8%) after recovery from drought stress (Figures 6H,J). The overall yield gain by the transgenic lines was 62.32 ± 9.65% higher than control plants after drought stress recovery (Figure 6H).
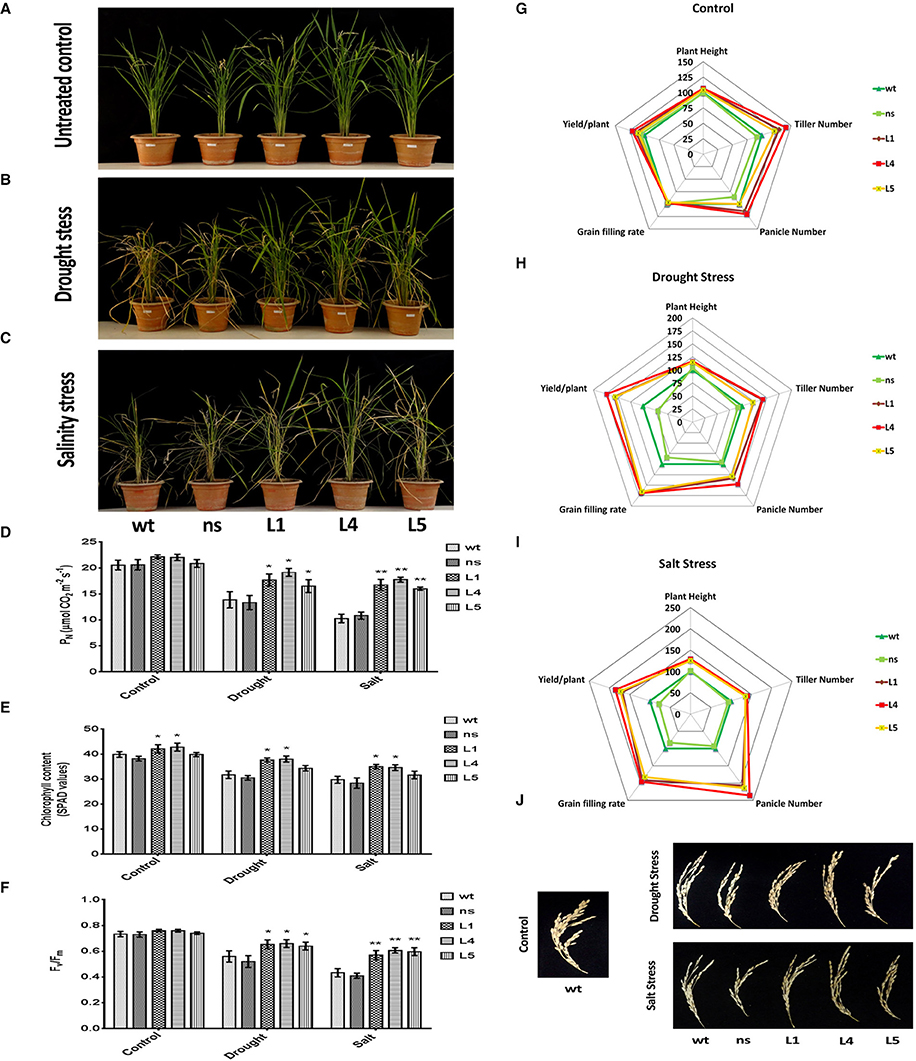
Figure 6. Agronomic and physiological performance of OsGS1;1/OsGS2 co-overexpressing transgenic rice plants under abiotic stresses at reproductive stage. (A) Phenotypes of wild type (wt), null segregant (ns), and three transgenic lines (L1, L4, and L5) at reproductive stage under untreated control conditions. (B) Phenotypes after recovery for 15 days following drought stress treatment imposed by water withdrawal for 12 days post panicle initiation. (C) Phenotypes after recovery following moderate salinity stress imposed on ~2-month-old plants by irrigating pots every fortnight with water supplemented with 50 mM NaCl (EC~6 dS/m) until booting stage. Various physiological parameters such as (D) net photosynthetic rate (PN) (in μmol CO2/m2/s) (E) chlorophyll content (in SPAD values) and (F) chlorophyll fluorescence (Fv/Fm) assessed under control, drought and salinity stress conditions. All data represented are means ± SD (n = 3). Asterisks above bars indicate significant differences from wt (*p-value ≤ 0.05 and **p-value ≤ 0.01). Spider plots of agronomic traits of three independent T2 transgenic lines (L1, L4, and L5) and corresponding ns and wt controls under (G) untreated control (H) drought and, (I) salinity stress conditions respectively. Data plotted are percentages of mean values (n = 5). Mean values from wt plants were set at 100% as reference. (J) Grain filling phenotypes in wt, ns, and three transgenic lines (L1, L4, and L5) after recovery from drought and salinity stress as compared to untreated wt control.
Similarly, we monitored the physiological fitness and agronomic performance of OsGS1;1/OsGS2 co-overexpressing transgenic lines and the corresponding control plants under salinity stress by irrigating with 50 mM NaCl solution until the booting stage, followed by recovery. Another set of plants were irrigated with normal deionized water (untreated controls), grown to maturity and agronomic parameters assessed (Figure 6G). The transgenic lines showed improved growth parameters like significantly higher plant height, tiller number, panicle number, and yield than wt and ns controls under the salinity stress (Figure 6C). Under the salinity stress, the transgenic lines showed 25–29% higher plant height, 35–40% more tiller numbers, and 107–135% higher panicle number compared to wt and ns control plants (Figure 6I). Moreover, grain filling rates were significantly improved in transgenic lines, which had around 58.2 ± 4.5% as compared to 28 ± 10.6% in the controls (Figures 6I,J). The overall grain yield was 75.4 ± 8.7% higher in transgenic lines compared to wt and ns control plants under the salinity stress (Figure 6I). The transgenic lines were also able to maintain significantly (55–73%) higher net photosynthetic rates (PN) than the control lines after recovery from salinity stress (Figure 6D). The chlorophyll content of the transgenic lines was 6–17% higher in comparison to controls (Figure 6E). Also, the chlorophyll fluorescence, i.e., the Fv/Fm ratio was comparatively higher in transgenic plants as compared to both wt and ns control rice plants under salinity stress (Figure 6F). Our results show that the enhanced agronomic yield parameters in the OsGS1;1/OsGS2 co-overexpressing transgenic rice plants correlated with the increased photosynthetic rates of transgenic lines in comparison to control plants, which suggests that the co-overexpression of OsGS1;1/OsSG2 provided efficient photo-oxidative protection to the transgenic rice plants under reproductive stage drought and salinity stress.
The OsGS1;1/OsGS2 co-overexpression in transgenic rice and control seedlings at four-six leaf stage were sprayed with 0.5, 1, and 2% (v/v) solution of Basta (Bayer, 13.5% ai). Transgenic lines had increased survival rates of 79–88% as compared to 10–22% of control plants after spraying with 0.5% (v/v) Basta solution (Figure 7A). The survival rate of line L5 was comparatively lower than the other lines, which could be as a result of the lower total GS enzymatic activity of this transgenic line. However, it was observed that the transgenic lines could not tolerate higher concentrations of 1% and 2% (v/v) Basta spray and all the transgenic lines along with the controls wilted by the 4th day after spraying (not shown). The mature leaves of both control and transgenic plants were also painted with a solution of 0.5% Basta supplemented with 0.01% Tween-20. After 5 days of treatment, the leaves were scored for visible injuries such as chlorosis, curling, bleaching and leaf burning on the painted areas; these symptoms were more prominent in wt and ns control seedlings than transgenic lines (Figure 7B). Thus, our results showed that the OsGs1;1/OsGS2 co-overexpressing transgenic rice plants had limited tolerance to 0.5% (v/v) Basta (Glufosinate/phosphinothricin) herbicide.
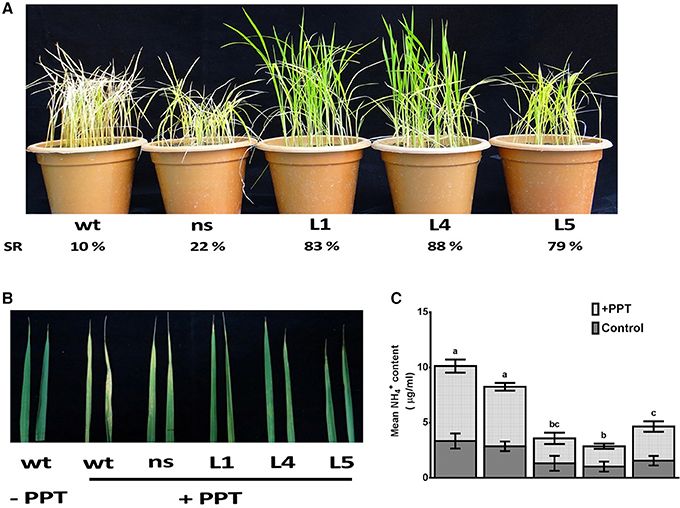
Figure 7. Assessment of tolerance of OsGS1;1/OsGS2 co-overexpressing transgenic rice to phosphinothricin (PPT). (A) Phenotypes of wt, ns, and three T2 transgenic (L1, L4, and L5) rice seedlings after 0.5% (v/v) Basta herbicide (Glufosinate/PPT) spraying. Survival rates (SR) after spraying are indicated as percentages. (B) Mature leaves of wt, ns, and three T2 transgenic lines (L1, L4, and L5) painted with a solution of 0.5% Basta (v/v) (Bayer, 13.5% ai) supplemented with 0.01% Tween-20. (C) Mean NH liberation from leaves before and after PPT treatment. Data represented are the means ± SD (n = 3). Different letters above bars indicate significant difference among means of PPT treated group (Tukey-Kramer tests, p-value < 0.05).
The active ingredient of Basta (PPT/phosphinothricin) inhibits GS activity and prevents the re-assimilation of cytotoxic ammonium produced during various cellular metabolic processes including photorespiration. We monitored the accumulation of ammonium content in transgenic and control rice leaves before and after Basta (PPT) spray. We estimated 3.1 ± 0.3 μg/mL free ammonium in control rice leaf samples, whereas, 1.2 ± 0.2 μg/mL in OsGS1;1/OsGS2 co-overexpressing transgenic rice leaf samples before spraying the 0.5% Basta solution (Figure 7C). However, after Basta spraying, the ammonium content significantly accumulated to 9.1 ± 3.6 μg/mL in control rice leaf samples, whereas, only a marginal increase (3.6 ± 0.8 μg/mL) was observed in OsGS1;1/OsGS2 co-overexpressing transgenic rice (Figure 7C).
Our results demonstrate that concurrently overexpressing both the cytosolic OsGS1;1 and chloroplastic OsGS2 isoforms in transgenic rice, enhanced its tolerance against drought and salinity stresses during seedling and reproductive stages, and against MV induced photo-oxidative stress. The transgenic plants showed higher chlorophyll fluorescence (Fv/Fm) under drought and salinity stress as compared to the corresponding control rice plants (Figure 6F), suggesting that the transgenic lines had enhanced protection of the photosynthetic machinery, which thereby led to improved recovery post stress. These results corroborate several other previous studies, which highlight the role of GS overexpression in conferring photoprotection of the photosynthetic machinery during various abiotic stresses. For instance, transgenic rice overexpressing the chloroplastic OsGS2 gene was shown to have improved tolerance to salinity and chilling stress due to increased photorespiratory capacities (Hoshida et al., 2000), while the ectopic overexpression of a pine cytoplasmic GS1 gene in transgenic poplar conferred improved tolerance to drought stress (El-Khatib et al., 2004). Similarly, Kozaki and Takeba (1996) reported that transgenic tobacco plants over-expressing the chloroplastic GS2 displayed enhanced tolerance to high light intensity. They determined that the overexpression of GS2 led to improved re-assimilation of photorespiratory ammonia, resulting in better protection of photosynthesis by reducing the damage due to photo-oxidation on the photosynthetic apparatus. Furthermore, several lines of evidence show that the rate-limiting step in photorespiration is likely to be the re-assimilation of NH catalyzed by the chloroplastic GS2 (Wallsgrove et al., 1987; Häusler et al., 1994; Kozaki and Takeba, 1996). For instance, mutant barley plants lacking the chloroplastic GS2 isoform were found to have severely reduced photorespiratory capacities (Wallsgrove et al., 1987). Photorespiration is thought to function as a potential route for the dissipation of excess light energy or reducing power (NADPH) generated during various abiotic stresses (Osmond and Grace, 1995; Willekens et al., 1997; Wingler et al., 2000; Voss et al., 2013). During various abiotic stress conditions, such as drought, salinity, or high light, the reducing equivalents (NADPH) generated through photosynthetic light reactions often surpasses the demand of the Calvin Benson Bassham cycle. The excess energy thus generated is dissipated as heat or the electrons are transferred from various complexes in the electron transport chain to other acceptor molecules such as O2, which would thereby result in the production of excessive reactive oxygen species (ROS) (Peterhansel et al., 2010). Thus, under such stress conditions, photorespiration can apparently act as an electron sink by consuming the excess electrons or reducing equivalents through the re-assimilation of photorespiratory NH by chloroplastic GS2 (Wingler et al., 2000; Peterhansel et al., 2010). The overexpression of GS2 would thus enhance photoprotection since excess electrons generated by photo-oxidation during stresses would be diverted from O2 to the photorespiratory cycle which would lead to the reduction in ROS generation, and thereby enhance tolerance to abiotic stresses (Hoshida et al., 2000). Our findings reveal that under photo-oxidative stress induced by methyl viologen, OsGS1;1/OsGS2 co-overexpressing transgenics had significantly enhanced tolerance and reduced ROS production as evidenced by leaf strip assays and DAB, NBT staining for H2O2 and O free radicals. Moreover, GS overexpression may also lead to reduction of ROS levels by modulating the oxidative stress response enzymes involved in antioxidant formation. Lee et al. (2013) previously reported that OsGS1;1 overexpressing transgenic rice had enhanced tolerance to cadmium induced oxidative stress through the modulation of oxidative stress responses. In addition, transgenic poplar plants overexpressing GS were shown to differentially regulate the expression of genes involved in mitigation against ROS, such as copper-dependent super oxide dismutases, thereby significantly improving their tolerance against drought stress (Molina-Rueda et al., 2013; Molina-Rueda and Kirby, 2015).
Also, GS overexpression is likely to increase the flux through the GS-GOGAT cycle, which would thereby increase net glutamate and glutamine amino acid levels. Several GS overexpression studies in plants have reported associated increases in total free amino acid levels especially of glutamate and glutamine (Migge et al., 2000; Fuentes et al., 2001). The amino acid glutamate is required for the synthesis of osmoprotectants such as proline, polyamines, as well as glutathione (GSH) (a tripeptide containing glutamate, cysteine, and glycine), a major intracellular antioxidant. A higher free glutamate amino acid level is essential for enhanced GSH levels which would in turn help in mitigation of excessive ROS production during stress conditions. Moreover, increased photorespiratory activity of GS overexpressing plants would also result in the synthesis of sufficient amounts of glycine, which has enhanced demand during GSH synthesis under abiotic stress conditions (Noctor et al., 1997). In addition, higher glutamine levels have also been shown to induce the transcription of stress responsive transcription factors such as DREB1A, IRO2, and NAC5 involved in various abiotic stresses (Kan et al., 2015), which is another possible mechanism by which GS overexpression leads to enhanced tolerance to abiotic stresses.
Membrane lipid peroxidation due to enhanced ROS activity and subsequent membrane leakage is common during various abiotic stresses and causes severe damage to plant cells leading eventually to cell death. However, the OsGS1;1 and OsGS2 co-overexpressing rice transgenic lines in our study showed significantly lower MDA content and lesser electrolyte leakage under drought and salinity stress compared to control rice plants under the same stress condition (Figures 4E,F). The observed membrane stability and lower lipid peroxidation in transgenic rice lines is likely to be due to lower ROS generation under drought and salinity stress. Our result corroborates reports that transgenic rice lines constitutively overexpressing the cytosolic OsGS1;1 gene had significantly lower MDA levels under cadmium stress (which is considered to be an oxidative stress enhancing factor), thus suggesting that GS overexpression modulates oxidative stress responses (Lee et al., 2013).
The OsGS1;1/OSGS2 co-overexpressing rice transgenics in our study, also accumulated significantly higher proline contents than controls under salinity and drought stresses (Figure 4D). The accumulation of proline and polyamines is a common response to various abiotic stresses (Vinocur and Altman, 2005). They act as osmolytes and help in protecting cellular membranes and proteins under various stresses (Yoshiba et al., 1997; Sengupta et al., 2016). It has been demonstrated that GS plays a key role in regulating proline production in the phloem and that higher GS activity is essential to synthesizing proline under water stress (Larher et al., 1998; Brugiere et al., 1999). Mutants of Lotus plants, deficient in chloroplastic GS2 had significantly lower proline contents than wild type plants during drought stress, which led to their compromised recovery following re-watering (Díaz et al., 2010). Moreover, poplar cells treated with a GS enzyme inhibitor (methionine sulphoximine) caused a reduction of the polyamine content which suggested that polyamine levels in plants are also primarily regulated by GS (Bhatnagar et al., 2001). Thus, increased accumulation of such osmoprotectants may be another possible mechanism of tolerance in GS overexpressing plants to abiotic stress.
The OsGS1;1/OsGS2 co-overexpressing rice lines were also able to withstand terminal drought stress and salinity stress and recovered vigorously with significant increases in tiller number, panicle number, better grain filling, and showed overall yield improvement in comparison to corresponding control plants (Figures 6A–I). As we observed better grain filling rates in transgenic plants over controls under abiotic stresses, we postulate that sufficient GS1;1 activity in the shoots combined with enhanced GS2 activity in the chloroplast in OsGS1;1/OsGS2 co-overexpressing transgenic rice plants, may lead to improved N re-assimilation into sink tissues and better photoprotection of photosynthetic machinery thereby leading to better recovery post abiotic stresses and consequently to an increase in grain filling and yield under stresses. Furthermore, under abiotic stress conditions, increased cellular processes like proteolysis can result in high intracellular ammonia, causing toxicity if not removed efficiently (Lutts et al., 1999). Enhanced GS activity would potentially alleviate this toxicity and at the same time improve N recycling efficiency, maintain photosynthetic enzymes and thereby lead to better growth and yield (Fuentes et al., 2001). The maintenance of photosynthesis at higher rates post recovery from abiotic stresses as observed in the OsGS1;1/OsGS2 co-overexpressing transgenic rice plants in comparison to stressed control plants also adds credence to this view (Figure 6D). Several GS overexpression studies have reported similar results in various transgenic plants (Migge et al., 2000; Fuentes et al., 2001; Habash et al., 2001; Oliveira et al., 2002; Martin et al., 2006; Cañas et al., 2010; Brauer et al., 2011). In addition, several quantitative trait loci (QTL) previously mapped and implicated in crop yield and growth components were shown to co-localize to the GS loci in maize (Hirel et al., 2001, 2007; Gallais and Hirel, 2004), rice (Obara et al., 2001, 2004; Yamaya et al., 2002), wheat (Habash et al., 2007; Bernard et al., 2008), and barley (See et al., 2002). Besides, the overexpression of the OsGS1;2 isoform in rice was shown to increase spikelet number and percentage grain filling in comparison to azygous controls (Brauer et al., 2011). Thus, the enhanced GS activity in transgenic rice plants presumptively leads to improved photoprotection of the photosynthetic machinery, thereby leading to better recovery post abiotic stresses and consequently to an increase in grain filling and yield. Efficient partitioning and remobilization of N resources under stress conditions is likely to be enhanced by OsGS1;1/OsGS2 co-overexpression which could also be the reason behind the enhanced agronomic performance of the transgenics in comparison to control plants.
In contrast, Cai et al. (2009) reported the transgenic overexpression of cytosolic OsGS1;1 or OsGS1;2 in rice showed very poor plant growth and reduced yield and had no significant tolerance to any abiotic stresses. This phenotype was seen to be due to imbalances in the carbon-nitrogen (C-N) metabolic status (Bao et al., 2014). It is important to note that the C-N metabolisms in the plant system are tightly interlinked and work in a concerted and regulated manner, evident from the fact that GS metabolism needs energy in the form of ATP and reduced ferredoxin or NADH, and also requires C-skeletons in the form of 2-oxoglutarate (2-OG), which are provided by C metabolism (Hodges, 2002). The GS-GOGAT cycle thus is believed to act as a linker between C and N metabolic cycles. It is very likely that GS overexpression leading to improvement of growth, yield and or abiotic stress tolerance would only be feasible in plants with higher GOGAT activity and/or enhanced supply of energy (ATP and NADH) and 2-oxoglutarate from C metabolism. The involvement of a highly coordinated carbon-nitrogen (C-N) metabolic system along with the intricate regulation of GS at various levels is often associated with the inconsistent GS overexpression studies under various plant genetic backgrounds (for a complete review see Thomsen et al., 2014).
We observed that OsGS1;1/OsGS2 co-overexpressing rice plants could tolerate Glufosinate herbicide (PPT) application to a limited extent (0.5% v/v Basta spray and painting) (Figure 7A). The lower accumulation of ammonia after PPT treatment in the transgenics as compared to wt showed that the overexpression of GS isoforms was able to partially detoxify the excess ammonia produced by PPT application to a limited extent. Several overexpression studies of GS in crops have been reported to show tolerance to the herbicide Glufosinate (PPT) which is a potent inhibitor of the GS enzyme. In rice, transgenic plants overexpressing the OsGS1;2 gene under a CaMV 35S promoter showed resistance to 10 mg/L of Basta in vitro and 0.5% (v/v) solution of Basta applied as a foliar spray. However, it was seen that the overexpression of the OsGS1;1 isoform alone did not result in Basta tolerance (Cai et al., 2009). Sun et al. (2005) reported that simultaneous overexpression of pea GS1;1 and GS2 in rice plants conferred resistance to 0.3% Basta solution painted on leaves. Similar results were seen in wheat plants simultaneously overexpressing both cytosolic and chloroplastic pea GS isoforms which tolerated upto 0.3% (v/v) Basta when painted on leaves (Huang et al., 2005). Also, transgenic poplar overexpressing a cytosolic pine GS showed considerable tolerance to a foliar application of PPT with enhanced growth in transgenic over controls (Pascual et al., 2008). Commercially available transgenic herbicide resistant crops have till date utilized the bar (bialaphos resistance) or pat (phosphinothricin acetyl transferase) derived from Streptomyces species for conferring resistance to the Glufosinate herbicide. For instance, Bayer's Liberty link Glufosinate resistant transgenic crops are one of the most successfully commercialized transgenics currently in use in modern agriculture. However, given the increasing bio-safety concerns dissuading the use of bacterial genes in food crops and the general public resentment against them, an alternative strategy for Glufosinate resistant herbicide crops would be the overexpression of the target gene (GS) or introducing mutations in GS for conferring tolerance against Glufosinate. But, when compared to resistance levels in plants obtained using the PPT detoxifying bar or pat genes the overexpression of GS as a strategy for developing Glufosinate resistant crops has shown unsuitably low resistance levels for commercial viability. One of the limitations facing this is the presence of multiple GS isozymes in different crop species (Donn and Köcher, 2002). As seen in our studies, gene stacking of multiple GS isoforms using the Multi-Round Gateway technology may help in overcoming this problem. Besides, several mutations in the GS enzyme have been seen to confer improved resistance to Glufosinate (Supplementary Table 2). For instance, recently a DNA shuffling of the OsGS1;1 gene of rice under selective pressure of high concentrations of PPT, identified an arginine at 295 to lysine mutation (R295K) as responsible for conferring PPT resistance. Further complementation studies of this mutation in a GS mutant Saccharomyces cerevisiae and transgenic overexpression of the OsGS1;1 R295K mutant gene in Arabidopsis confirmed its ability to confer high levels of tolerance to PPT (Tian et al., 2015). Therefore, multiple-gene stacking of such mutant GS isoforms may help in further developing the commercial viability of this strategy. However, due to the presence of multiple gene loci encoding for GS isozyme in plant species, with non-overlapping and non-redundant physiological roles in plant development, the introduction of targeted mutations to confer Glufosinate resistance in all the isoforms is a daunting task.
Taken together, our results suggest that OsGS1;1/OsGS2 co-overexpression in rice conferred enhanced physiological tolerance and increased agronomic performance under abiotic stresses, apparently acting through multiple mechanistic routes (Figure 8). In addition, the co-overexpression of OsGS1;1 and OsGS2 in rice also conferred limited tolerance to the herbicide Glufosinate. However, a more comprehensive understanding of the regulation of GS and the functions of various isoforms in abiotic stresses would be needed before consistent results can be obtained across species and varieties. Moreover, since chloroplastic GS2 is itself prone to oxidative degradation by ROS under higher levels of stress (Palatnik et al., 1999; Ishida et al., 2002); the overexpression of a mutant GS resistant to inactivation under oxidative conditions might help in increasing abiotic stress tolerance of crop plants. Future overexpression strategies using GS for crop improvement will also have to take into consideration the complex regulation of GS and its intimate interaction with the C-N metabolic pathway to overcome potential metabolic bottlenecks. The use of more refined strategies, such as gene stacking in combination with developmental-stage and/or tissue specific expression, is expected to provide improved and consistent results (Thomsen et al., 2014). However, the prospect of transgenically manipulating GS to enhance yield and NUE under abiotic stress conditions, and provide field level herbicide resistance is lucrative enough to continue such directed efforts to tap the full potential of this unique enzyme in crop improvement for sustainable agriculture.
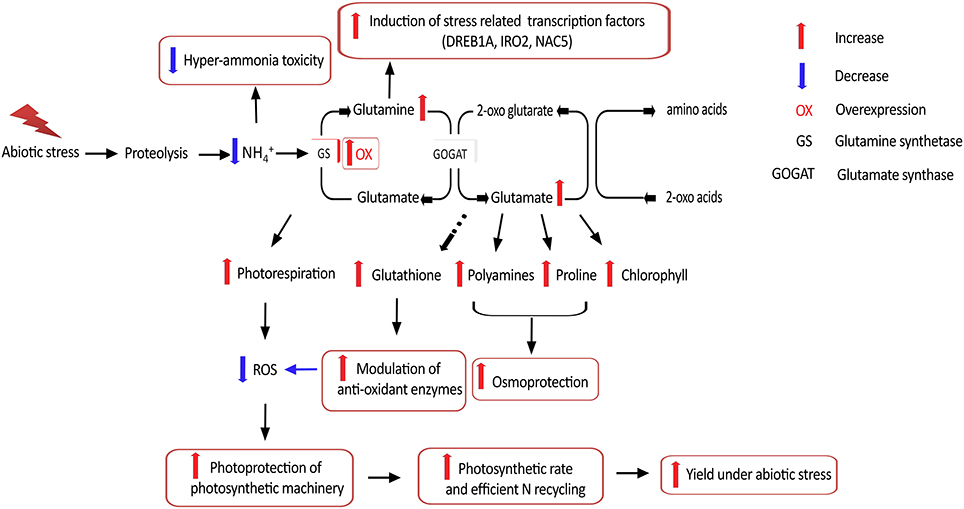
Figure 8. Putative mechanistic roles of GS in abiotic stress tolerance. Overexpression of GS alleviates hyper-ammonia toxicity caused due to proteolysis associated with various abiotic stresses. Since GS is considered the rate limiting step for photorespiration, GS overexpression can increase photorespiratory capacities and thereby ensuring photoprotection of the photosynthetic machinery via reduced reactive oxygen species (ROS) generation. GS overexpression has been reported to enhance production of the amino acids glutamine and glutamate, which is necessary for production of proline and polyamines which provide osmoprotection, as well as the anti-oxidant glutathione which can modulate anti-oxidant enzyme responses and thereby alleviate oxidative stress. Higher glutamine content is reported to induce expression of stress related transcription factors. Overexpression of GS is also reported to increase photosynthetic rates and is likely to improve N recycling efficiency, thereby leading to better yield under abiotic stress.
MR and DJ conceived the project and all co-authors were involved in planning the experiments. DJ, DF, BB, BR, RY, and MM conducted the experiments and collected the data. DJ, JS, VP, VA, and VS performed data analysis and drafting of the manuscript. DJ, VA, and MR revised the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The entry vectors (EV-1 and EV-2) were courtesy of Dr. Xue Chen Wang, State Key Lab. of Plant Physiology and Biochemistry, China. The plant transformation destination vector (pMDC99) was a kind gift provided by Dr. Mark Curtis, Department of Plant and Microbial Biology, UZH, Switzerland. Antibodies against recombinant GS detecting both OsGS1;1; and OsGS2 isoforms were a kind gift from Dr. Toshihiko Hayakawa, Lab. of Plant Cell Biochemistry, Tohoku University, Japan. DJ acknowledges DBT, India for providing research fellowship.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00786/full#supplementary-material
Ai, Active ingredient; BCIP, 5-Bromo-4-chloro-3-indolyl phosphate; DAB, 3, 3-Diaminobenzidine; DIG, Digoxigenin; EC, Electrical conductivity; GS, Glutamine synthetase; MDA, Malondialdehyde; MW, Molecular Weight; NBT, Nitroblue tetrazolium; NUE, Nitrogen use efficiency; PAGE, Polyacrylamide gel electrophoresis; PBS, Phosphate buffered saline; PEG, Poly Ethylene Glycol; PPT, Phosphinothricin; QTL, Quantitative trait loci; ROS, Reactive oxygen species; RWC, Relative water content; SDS, Sodium dodecyl sulfate; WUE, Water use efficiency; YS, Yoshida hydroponics nutrient solution.
Ambavaram, M. M. R., Basu, S., Krishnan, A., Ramegowda, V., Batlang, U., Rahman, L., et al. (2014). Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 5:5302. doi: 10.1038/ncomms6302
Bajji, M., Kinet, J. M., and Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. doi: 10.1023/A:1014732714549
Bao, A., Zhao, Z., Ding, G., Shi, L., Xu, F., and Cai, H. (2014). Accumulated expression level of cytosolic glutamine synthetase1 gene (OsGS1;1 or OsGS1;2) alter plant development and the carbon-nitrogen metabolic status in rice. PLoS ONE 9:e95581. doi: 10.1371/journal.pone.0095581
Barrs, H., and Weatherley, P. (1962). A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 15:413. doi: 10.1071/BI9620413
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bauer, D., Biehler, K., Fock, H., Carrayol, E., Hirel, B., Migge, A., et al. (1997). A role for cytosolic glutamine synthetase in the remobilization of leaf nitrogen during water stress in tomato. Physiol. Plant. 99, 241–248. doi: 10.1111/j.1399-3054.1997.tb05408.x
Bernard, S. M., and Habash, D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620. doi: 10.1111/j.1469-8137.2009.02823.x
Bernard, S. M., Møller, A. L. B., Dionisio, G., Kichey, T., Jahn, T. P., Dubois, F., et al. (2008). Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.). Plant Mol. Biol. 67, 89–105. doi: 10.1007/s11103-008-9303-y
Bhatnagar, P., Glasheen, B. M., Bains, S. K., Long, S. L., Minocha, R., Walter, C., et al. (2001). Transgenic manipulation of the metabolism of polyamines in poplar cells. Plant Physiol. 125, 2139–2153. doi: 10.1104/pp.125.4.2139
Brauer, E. K., and Shelp, B. J. (2010). Nitrogen use efficiency: re-consideration of the bioengineering approach. Botany 88, 103–109. doi: 10.1139/B09-111
Brauer, E. K., Rochon, A., Bi, Y. M., Bozzo, G. G., Rothstein, S. J., and Shelp, B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant. 141, 361–372. doi: 10.1111/j.1399-3054.2011.01443.x
Brugiere, N., Dubois, F., Limami, A., Lelandais, M., Roux, Y., Sangwan, R., et al. (1999). Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11, 1995–2012. doi: 10.1105/tpc.11.10.1995
Cai, H., Zhou, Y., Xiao, J., Li, X., Zhang, Q., and Lian, X. (2009). Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep. 28, 527–537. doi: 10.1007/s00299-008-0665-z
Cañas, R. A., Quilleré, I., Lea, P. J., and Hirel, B. (2010). Analysis of amino acid metabolism in the ear of maize mutants deficient in two cytosolic glutamine synthetase isoenzymes highlights the importance of asparagine for nitrogen translocation within sink organs. Plant Biotechnol. J. 8, 966–978. doi: 10.1111/j.1467-7652.2010.00524.x
Chaffei, C., Pageau, K., Suzuki, A., Gouia, H., Ghorbel, M. H., and Masclaux-Daubresse, C. (2004). Cadmium toxicity induced changes in Nitrogen management in Lycopersicon esculentum leading to a metabolic safeguard through an amino acid storage strategy. Plant Cell Physiol. 45, 1681–1693. doi: 10.1093/pcp/pch192
Chen, Q. J., Zhou, H. M., Chen, J., and Wang, X. C. (2006). A Gateway-based platform for multigene plant transformation. Plant Mol. Biol. 62, 927–936. doi: 10.1007/s11103-006-9065-3
Cheng, L., Wang, Y., He, Q., Li, H., Zhang, X., and Zhang, F. (2016). Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 16:188. doi: 10.1186/s12870-016-0871-8
Curtis, M. D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in-planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Díaz, P., Betti, M., Sánchez, D. H., Udvardi, M. K., Monza, J., and Márquez, A. J. (2010). Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188, 1001–1013. doi: 10.1111/j.1469-8137.2010.03440.x
Donn, G., and Köcher, H. (2002). “Inhibitors of glutamine synthetase,” in Herbicide Classes in Development, eds P. Böger, K. Wakabayashi, and K. Hirai (Berlin; Heidelberg: Springer), 87–101.
El-Khatib, R. T., Hamerlynck, E. P., Gallardo, F., and Kirby, E. G. (2004). Transgenic poplar characterized by ectopic expression of a pine cytosolic glutamine synthetase gene exhibits enhanced tolerance to water stress. Tree Physiol. 24, 729–736. doi: 10.1093/treephys/24.7.729
Forde, B. G., and Lea, P. J. (2007). Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58, 2339–2358. doi: 10.1093/jxb/erm121
Fu, J., Sampalo, R., Gallardo, F., Cánovas, F. M., and Kirby, E. G. (2003). Assembly of a cytosolic pine glutamine synthetase holoenzyme in leaves of transgenic poplar leads to enhanced vegetative growth in young plants. Plant Cell Environ. 26, 411–418. doi: 10.1046/j.1365-3040.2003.00972.x
Fuentes, S. I., Allen, D. J., Ortiz-Lopez, A., and Hernández, G. (2001). Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 52, 1071–1081. doi: 10.1093/jexbot/52.358.1071
Gallais, A., and Hirel, B. (2004). An approach to the genetics of nitrogen use efficiency in maize. J. Exp. Bot. 55, 295–306. doi: 10.1093/jxb/erh006
Gallardo, F., Fu, J., Cantón, F. R., García-Gutiérrez, A., Cánovas, F. M., and Kirby, E. G. (1999). Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta 210, 19–26. doi: 10.1007/s004250050649
Gill, H. S., and Eisenberg, D. (2001). The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 40, 1903–1912. doi: 10.1021/bi002438h
Goel, P., and Singh, A. K. (2015). Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS ONE 10:e0143645. doi: 10.1371/journal.pone.0143645
Habash, D. Z., Bernard, S., Schondelmaier, J., Weyen, J., and Quarrie, S. A. (2007). The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theor. Appl. Genet. 114, 403–419. doi: 10.1007/s00122-006-0429-5
Habash, D. Z., Massiah, A. J., Rong, H. L., Wallsgrove, R. M., and Leigh, R. A. (2001). The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Biol. 138, 83–89. doi: 10.1111/j.1744-7348.2001.tb00087.x
Häusler, R. E., Lea, P. J., and Leegood, R. C. (1994). Control of photosynthesis in barley leaves with reduced activities of glutamine synthetase or glutamate synthase - II. Control of electron transport and CO2 assimilation. Planta 194, 418–435. doi: 10.1007/BF00197543
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Hirel, B., and Lea, P. J. (2001). “Ammonia assimilation,” in Plant Nitrogen, eds P. J. Lea and J. F. Morot-Gaudry (Berlin; Heidelberg: Springer), 79–99.
Hirel, B., Bertin, P., Quilleré, I., Bourdoncle, W., Attagnant, C., Dellay, C., et al. (2001). Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 125, 1258–1270. doi: 10.1104/PP.125.3.1258
Hirel, B., Le Gouis, J., Ney, B., and Gallais, A. (2007). The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 58, 2369–2387. doi: 10.1093/jxb/erm097
Hiscox, J. D., and Israelstam, G. F. (1979). A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334. doi: 10.1139/b79-163
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Hodges, M. (2002). Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot. 53, 905–916. doi: 10.1104/pp.123.3.817
Hoshida, H., Tanaka, Y., Hibino, T., Hayashi, Y., Tanaka, A., Takabe, T., et al. (2000). Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 43, 103–111. doi: 10.1023/A:1006408712416
Huang, Q. M., Liu, W. H., Sun, H., Deng, X., and Su, J. (2005). Agrobacterium tumefaciens mediated transgenic wheat plants with glutamine synthetases confer tolerance to herbicide (in Chinese). J. Plant Ecol. 29, 338–344. doi: 10.17521/cjpe.2005.0044
Ishida, H., Anzawa, D., Kokubun, N., Makino, A., and Mae, T. (2002). Direct evidence for non-enzymatic fragmentation of chloroplastic glutamine synthetase by a reactive oxygen species. Plant Cell Environ. 25, 625–631. doi: 10.1046/j.1365-3040.2002.00851.x
Ishiyama, K., Inoue, E., Tabuchi, M., Yamaya, T., and Takahashi, H. (2004). Biochemical background and compartmentalized functions of cytosolic glutamine synthetase for active ammonium assimilation in rice roots. Plant Cell Physiol. 45, 1640–1647. doi: 10.1093/pcp/pch190
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Jing, Z. P., Gallardo, F., Pascual, M. B., Sampalo, R., Romero, J., De Navarra, A. T., et al. (2004). Improved growth in a field trial of transgenic hybrid poplar overexpressing glutamine synthetase. New Phytol. 164, 137–145. doi: 10.1111/j.1469-8137.2004.01173.x
Kamachi, K., Yamaya, T., Hayakawa, T., Mae, T., and Ojima, K. (1992). Changes in cytosolic glutamine synthetase polypeptide and its mRNA in a leaf blade of rice plants during natural senescence. Plant Physiol. 98, 1323–1329. doi: 10.1104/pp.98.4.1323
Kaminski, K. P., Kørup, K., Andersen, M. N., Sønderkær, M., Andersen, M. S., Kirk, H. G., et al. (2015). Cytosolic glutamine synthetase is important for photosynthetic efficiency and water use efficiency in potato as revealed by high-throughput sequencing QTL analysis. Theor. Appl. Genet. 128, 2143–2153. doi: 10.1007/s00122-015-2573-2
Kan, C. C., Chung, T. Y., Juo, Y. A., Hsieh, M. H., Xu, G., Fan, X., et al. (2015). Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genomics 16:731. doi: 10.1186/S12864-015-1892-7
Kozaki, A., and Takeba, G. (1996). Photorespiration protects C 3 plants from photooxidation. Nature 384, 557–560. doi: 10.1038/384557a0
Kumar, D., Yusuf, M. A., Singh, P., Sardar, M., and Sarin, N. B. (2014). Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bioprotocol 4, 1–4. doi: 10.21769/BioProtoc.1108
Kwon, S. J., Kwon, S., Il Bae, M. S., Cho, E. J., and Park, O. K. (2007). Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol. 48, 1713–1723. doi: 10.1093/pcp/pcm143
Lam, H.-M., Coschigano, K. T., Oliveira, I. C., Melo-Oliveira, R., and Coruzzi, G. M. (1996). The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Larher, F., Aziz, A., Deleu, C., Lemesle, P., Ghaffar, A., Bouchard, F., et al. (1998). Suppression of the osmoinduced proline response of rapeseed leaf discs by polyamines. Physiol. Plant. 102, 139–147. doi: 10.1034/j.1399-3054.1998.1020118.x
Lee, H. J., Abdula, S. E., Jang, D. W., Park, S.-H., Yoon, U.-H., Jung, Y. J., et al. (2013). Overexpression of the glutamine synthetase gene modulates oxidative stress response in rice after exposure to cadmium stress. Plant Cell Rep. 32, 1521–1529. doi: 10.1007/s00299-013-1464-8
Leegood, R. C., Lea, P. J., Adcock, M. D., and Häusler, R. E. (1995). The regulation and control of photorespiration. J. Exp. Bot. 46, 1397–1414. doi: 10.1093/jxb/46.special_issue.1397
Lin, Z. F., Liu, N., Lin, G. Z., and Peng, C. L. (2009). In situ localisation of superoxide generated in leaves of Alocasia macrorrhiza (L.) shott under various stresses. J. Plant Biol. 52, 340–347. doi: 10.1007/s12374-009-9044-8
Lu, B., Yuan, Y., Zhang, C., Ou, J., Zhou, W., and Lin, Q. (2005). Modulation of key enzymes involved in ammonium assimilation and carbon metabolism by low temperature in rice (Oryza sativa L.) roots. Plant Sci. 169, 295–302. doi: 10.1016/j.plantsci.2004.09.031
Lutts, S., Majerus, V., and Kinet, J. M. (1999). NaCl effects on proline metabolism in rice (Oryza sativa) seedlings. Physiol. Plant. 105, 450–458. doi: 10.1034/j.1399-3054.1999.105309.x
Mahanty, S., Kaul, T., Pandey, P., Reddy, R. A., Mallikarjuna, G., Reddy, C. S., et al. (2012). Biochemical and molecular analyses of copper–zinc superoxide dismutase from a C4 plant Pennisetum glaucum reveals an adaptive role in response to oxidative stress. Gene 505, 309–317. doi: 10.1016/j.gene.2012.06.001
Man, H. M., Boriel, R., El-Khatib, R., and Kirby, E. G. (2005). Characterization of transgenic poplar with ectopic expression of pine cytosolic glutamine synthetase under conditions of varying nitrogen availability. New Phytol. 167, 31–39. doi: 10.1111/j.1469-8137.2005.01461.x
Manna, M., Achary, V. M. M., Islam, T., Agrawal, P. K., and Reddy, M. K. (2016). The development of a phosphite-mediated fertilization and weed control system for rice. Sci. Rep. 6:24941. doi: 10.1038/srep24941
Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. doi: 10.1105/tpc.106.042689
Miflin, B. J., and Habash, D. Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987. doi: 10.1093/jexbot/53.370.979
Migge, A., Carrayol, E., Hirel, B., and Becker, T. W. (2000). Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210, 252–260. doi: 10.1007/PL00008132
Molina-Rueda, J. J., and Kirby, E. G. (2015). Transgenic poplar expressing the pine GS1a show alterations in nitrogen homeostasis during drought. Plant Physiol. Biochem. 94, 181–190. doi: 10.1016/j.plaphy.2015.06.009
Molina-Rueda, J. J., Tsai, C. J., and Kirby, E. G. (2013). The Populus superoxide dismutase gene family and its responses to drought stress in transgenic Poplar overexpressing a Pine cytosolic glutamine Synthetase (GS1a). PLoS ONE 8:e56421. doi: 10.1371/journal.pone.0056421
Murchie, E. H., and Lawson, T. (2013). Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 64, 3983–3998. doi: 10.1093/jxb/ert208
Nagy, Z., Németh, E., Guóth, A., Bona, L., Wodala, B., and Pécsváradi, A. (2013). Metabolic indicators of drought stress tolerance in wheat: Glutamine synthetase isoenzymes and Rubisco. Plant Physiol. Biochem. 67, 48–54. doi: 10.1016/j.plaphy.2013.03.001
Noctor, G., Arisi, A.-C. M., Jouanin, L., Valadier, M.-H., Roux, Y., and Foyer, C. H. (1997). The role of glycine in determining the rate of glutathione synthesis in poplar. Possible implications for glutathione production during stress. Physiol. Plant. 100, 255–263. doi: 10.1034/j.1399-3054.1997.1000206.x
Obara, M., Kajiura, M., Fukuta, Y., Yano, M., Hayashi, M., Yamaya, T., et al. (2001). Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (Oryza sativa L.). J. Exp. Bot. 52, 1209–1217. doi: 10.1093/jexbot/52.359.1209
Obara, M., Sato, T., and Yamaya, T. (2000). High content of cytosolic glutamine synthetase does not accompany a high activity of the enzyme in rice (Oryza sativa) leaves of indica cultivars. Physiol. Plant. 108, 11–18. doi: 10.1034/j.1399-3054.2000.108001011x
Obara, M., Sato, T., Sasaki, S., Kashiba, K., Nagano, A., Nakamura, I., et al. (2004). Identification and characterization of a QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice. Theor. Appl. Genet. 110, 1–11. doi: 10.1007/s00122-004-1828-0
Oliveira, I. C., Brears, T., Knight, T. J., Clark, A., and Coruzzi, G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1170–1180. doi: 10.1104/pp.020013
Osmond, C. B., and Grace, S. C. (1995). Perspectives on photoinhibition and photorespiration in the field: quintessential inefficiencies of the light and dark reactions of photosynthesis? J. Exp. Bot. 46, 1351–1362. doi: 10.1093/jxb/46.special_issue.1351
Ouyang, B., Yang, T., Li, H., Zhang, L., Zhang, Y., Zhang, J., et al. (2007). Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J. Exp. Bot. 58, 507–520. doi: 10.1093/jxb/erl258
Palatnik, J., Carrillo, N., and Valle, E. (1999). The role of photosynthetic electron transport in the oxidative degradation of chloroplastic glutamine synthetase. Plant Physiol. 121, 471–478. doi: 10.1104/pp.121.2.471
Pascual, M. B., Jing, Z. P., Kirby, E. G., Cánovas, F. M., and Gallardo, F. (2008). Response of transgenic poplar overexpressing cytosolic glutamine synthetase to phosphinothricin. Phytochemistry 69, 382–389. doi: 10.1016/j.phytochem.2007.07.031
Peterhansel, C., Horst, I., Niessen, M., Blume, C., Kebeish, R., Kürkcüoglu, S., et al. (2010). Photorespiration. Arab B 8:e0130. doi: 10.1199/tab.0130
Rana, N. K., Mohanpuria, P., and Yadav, S. K. (2008). Cloning and characterization of a cytosolic glutamine synthetase from Camellia sinensis (L.) O. Kuntze that is upregulated by ABA, SA, and H2O2. Mol. Biotechnol. 39, 49–56. doi: 10.1007/s12033-007-9027-2
Rasco-Gaunt, S., Riley, A., Lazzeri, P., and Barcelo, P. (1999). A facile method for screening for phosphinothricin (PPT)-resistant transgenic wheats. Mol. Breed. 5, 255–262.
Ravikumar, G., Manimaran, P., Voleti, S. R., Subrahmanyam, D., Sundaram, R. M., Bansal, K. C., et al. (2014). Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res. 23, 421–439. doi: 10.1007/s11248-013-9776-6
Sairam, R. K., Srivastava, G. C., Agarwal, S., and Meena, R. C. (2005). Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol. Plant 49, 85–91. doi: 10.1007/s10535-005-5091-2
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to Image J: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
See, D., Kanazin, V., Kephart, K., and Blake, T. (2002). Mapping genes controlling variation in barley grain protein concentration. Crop Sci. 42, 680–685. doi: 10.2135/cropsci2002.6800
Sengupta, A., Chakraborty, M., Saha, J., Gupta, B., and Gupta, K. (2016). “Polyamines: osmoprotectants in plant abiotic stress adaptation,” in Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies, eds N. Iqbal, R. Nazar, and A. N. Khan (New Delhi: Springer), 97–127.
Silveira, J. A. G., Melo, A. R. B., Viégas, R. A., and Oliveira, J. T. A. (2001). Salinity-induced effects on nitrogen assimilation related to growth in cowpea plants. Environ. Exp. Bot. 46, 171–179. doi: 10.1016/S0098-8472(01)00095-8
Silveira, J. A. G., de Viégas, R. A., da Rocha, I. M. A., Moreira, A. C. D. O. M., de Moreira, R. A., and Oliveira, J. T. A. (2003). Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J. Plant Physiol. 160, 115–123. doi: 10.1078/0176-1617-00890
Singh, K. K., and Ghosh, S. (2013). Regulation of glutamine synthetase isoforms in two differentially drought-tolerant rice (Oryza sativa L.) cultivars under water deficit conditions. Plant Cell Rep. 32, 183–193. doi: 10.1007/s00299-012-1353-6
Sun, H., Huang, Q., and Su, J. (2005). High Technology Letters. English China Online Journals, ECOJ. Available online at: http://europepmc.org/abstract/cba/498301 (Accessed June 10, 2017).
Swarbreck, S. M., Defoin-Platel, M., Hindle, M., Saqi, M., and Habash, D. Z. (2011). New perspectives on glutamine synthetase in grasses. J. Exp. Bot. 62, 1511–1522. doi: 10.1093/jxb/erq356
Tabuchi, M., Sugiyama, K., Ishiyama, K., Inoue, E., Sato, T., Takahashi, H., et al. (2005). Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J. 42, 641–651. doi: 10.1111/j.1365-313X.2005.02406.x
Tachibana, K., Watanabe, T., Sekizawa, Y., and Takematsu, T. (1986). Accumulation of ammonia in plants treated with bialaphos. J. Pestic. Sci. 11, 33–37. doi: 10.1584/jpestics.11.33
Teixeira, J., and Pereira, S. (2007). High salinity and drought act on an organ-dependent manner on potato glutamine synthetase expression and accumulation. Environ. Exp. Bot. 60, 121–126. doi: 10.1016/j.envexpbot.2006.09.003
Thomsen, H. C., Eriksson, D., Møller, I. S., and Schjoerring, J. K. (2014). Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 19, 656–663. doi: 10.1016/j.tplants.2014.06.002
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tian, Y.-S., Xu, J., Zhao, W., Xing, X.-J., Fu, X.-Y., Peng, R.-H., et al. (2015). Identification of a phosphinothricin-resistant mutant of rice glutamine synthetase using DNA shuffling. Sci. Rep. 5:15495. doi: 10.1038/srep15495
Tripathi, A. K., Pareek, A., and Singla-Pareek, S. L. (2016). Evidence for the role of an H3/H4-histone chaperone OsNAPL6 in abiotic stress response and adaptation in rice. Plant Physiol. 171, 2854–2868. doi: 10.1104/pp.16.00408
Tsai, C. J., Wang, C. S., and Wang, C. Y. (2006). Physiological characteristics of glufosinate resistance in rice. Weed Sci. 54, 634–640. doi: 10.1614/WS-06-017R.1
Urriola, J., and Rathore, K. S. (2015). Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res. 24, 397–407. doi: 10.1007/s11248-014-9852-6
Vinocur, B., and Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Voss, I., Sunil, B., Scheibe, R., and Raghavendra, A. S. (2013). Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 15, 713–722. doi: 10.1111/j.1438-8677.2012.00710.x
Wallsgrove, R. M., Turner, J. C., Hall, N. P., Kendall, A. C., and Bright, S. W. (1987). Barley mutants lacking chloroplast glutamine synthetase-biochemical and genetic analysis. Plant Physiol. 83, 155–158. doi: 10.1104/pp.83.1.155
Wang, H., Wu, Z., Zhou, Y., Han, J., and Shi, D. (2012). Effects of salt stress on ion balance and nitrogen metabolism in rice. Plant Soil Environ. 58, 62–67. doi: 10.1186/1471-2229-12-194
Wang, Y., Fu, B., Pan, L., Chen, L., Fu, X., and Li, K. (2013). Overexpression of Arabidopsis Dof1, GS1 and GS2 enhanced nitrogen assimilation in transgenic tobacco grown under low-nitrogen conditions. Plant Mol. Biol. Rep. 31, 886–900. doi: 10.1007/s11105-013-0561-8
Wang, Z. Q., Yuan, Y. Z., Ou, J. Q., Lin, Q. H., and Zhang, C. F. (2007). Glutamine synthetase and glutamate dehydrogenase contribute differentially to proline accumulation in leaves of wheat (Triticum aestivum) seedlings exposed to different salinity. J. Plant Physiol. 164, 695–701. doi: 10.1016/j.jplph.2006.05.001
Wild, A., and Manderscheid, R. (1984). The effect of phosphinothricin on the assimilation of ammonia in plants. Z. Naturforsch. Sect. C Biosci. 39, 500–504. doi: 10.1515/znc-1984-0539
Willekens, H., Chamnongpol, S., Davey, M., Schraudner, M., Langebartels, C., Van Montagu, M., et al. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16, 4806–4816. doi: 10.1093/emboj/16.16.4806
Wingler, A., Lea, P. J., Quick, W. P., and Leegood, R. C. (2000). Photorespiration: metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1517–1529. doi: 10.1098/rstb.2000.0712
Yamaya, T., Obara, M., Nakajima, H., Sasaki, S., Hayakawa, T., and Sato, T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53, 917–925. doi: 10.1093/jexbot/53.370.917
Yan, S., Tang, Z., Su, W., and Sun, W. (2005). Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5, 235–244. doi: 10.1002/pmic.200400853
Yoshiba, Y., Kiyosue, T., Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 38, 1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). Laboratory Manual for Plant Physiological Studies of Rice, 3rd Edn. Manila: IRRI Press.
Yousfi, S., Márquez, A. J., Betti, M., Araus, J. L., and Serret, M. D. (2015). Gene expression and physiological responses to salinity and water stress of contrasting durum wheat genotypes. J. Integr. Plant Biol. 58, 48–66. doi: 10.1111/jipb.12359
Zhu, C., Fan, Q., Wang, W., Shen, C., Meng, X., Tang, Y., et al. (2014). Characterization of a glutamine synthetase gene DvGS2 from Dunaliella viridis and biochemical identification of DvGS2-transgenic Arabidopsis thaliana. Gene 536, 407–415. doi: 10.1016/j.gene.2013.11.009
Keywords: glutamine synthetase, abiotic stress, herbicide tolerance, Glufosinate, in vitro gene pyramiding, Multi-Round Gateway technology
Citation: James D, Borphukan B, Fartyal D, Ram B, Singh J, Manna M, Sheri V, Panditi V, Yadav R, Achary VMM and Reddy MK (2018) Concurrent Overexpression of OsGS1;1 and OsGS2 Genes in Transgenic Rice (Oryza sativa L.): Impact on Tolerance to Abiotic Stresses. Front. Plant Sci. 9:786. doi: 10.3389/fpls.2018.00786
Received: 31 August 2017; Accepted: 23 May 2018;
Published: 21 June 2018.
Edited by:
Sagadevan G. Mundree, Queensland University of Technology, AustraliaReviewed by:
Anirudha R. Dixit, Kennedy Space Center, United StatesCopyright © 2018 James, Borphukan, Fartyal, Ram, Singh, Manna, Sheri, Panditi, Yadav, Achary and Reddy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mallireddy K. Reddy, cmVkZHlAaWNnZWIucmVzLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.