- 1Department of Life Sciences, National Cheng Kung University, Tainan, Taiwan
- 2Division of Strategic Research and Development, Graduate School of Science and Engineering, Saitama University, Saitama, Japan
- 3Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan
- 4Institute of Tropical Plant Sciences, National Cheng Kung University, Tainan, Taiwan
- 5Orchid Research and Development Center, National Cheng Kung University, Tainan, Taiwan
Phalaenopsis bellina is a scented orchid emitting large amount of monoterpenes. GERANYL DIPHOSPHATE SYNTHASE (PbGDPS) is the key enzyme for monoterpene biosynthesis, and shows concomitant expression with the emission of monoterpenes during flower development in P. bellina. Here, we identified a dual repeat cis-element in the GDPS promoter that is critical for monoterpene biosynthesis in Phalaenopsis orchids. A strong correlation between the dual repeat and the monoterpene production was revealed by examination of the GDPS promoter fragments over 12 Phalaenopsis species. Serial-deletion of the 2-kb GDPS promoter fragments demonstrated that the integrity of the dual repeat was crucial for its promoter activities. By screening the Arabidopsis transcription factors (TFs) cDNA library using yeast one-hybrid assay, AtbZIP18, a member of group I of bZIP TFs, was identified to be able to bind the dual repeat. We then identified PbbZIP4 in the transcriptome of P. bellina, showing 83% identity in the DNA binding region with that of AtbZIP18, and the expression level of PbbZIP4 was higher in the scented orchids. In addition, PbbZIP4 transactivated the GDPS promoter fragment containing the dual repeat in dual luciferase assay. Furthermore, transient ectopic expression of PbbZIP4 induced a 10-fold production of monoterpenoids in the scentless orchid. In conclusion, these results indicate that the dual repeat is a real TF-bound cis-element significant for GDPS gene expression, and thus subsequent monoterpene biosynthesis in the scented Phalaenopsis orchids.
Introduction
Phalaenopsis species are widespread in the tropical Asia regions and includes approximately 56 native species (Christenson, 2001). Numerous Phalaenopsis cultivars with diverse floral appearance are obtained via breeding and have become popular orchids due to their outstanding floral display and longevity (Hsiao et al., 2011a). In addition, some of the Phalaenopsis cultivars with pleasant fragrance improve their ornamental value in the floriculture market. However, breeding scented orchid cultivars under traditional breeding is difficult compared to other favorable traits (Yeh et al., 2014). The bottlenecks include long generation time (Hsiao et al., 2011a), cross-incompatibility due to the differences in genome size and chromosome size among species (Hsiao et al., 2011a,b; Yeh et al., 2014), and negative correlation between floral scent and other favorable traits (Hsiao et al., 2011b), which is also occurred in other modern floriculture varieties (Vainstein et al., 2001; Dudareva and Negre, 2005). In such circumstances, alternative approaches to facilitate scented orchid breeding are needed.
The majorities of Phalaenopsis orchids are scentless but some do emit scent volatile organic compounds (VOCs) (Kaiser, 1993). These scented species have been extensively used as breeding parents for production of scent cultivars, such as P. amboinensis, P. bellina, P. javanica, P. lueddemanniana, P. schilleriana, P. stuartiana, P. venosa, and P. violace (Hsiao et al., 2011b; Yeh et al., 2014). Both P. bellina and P. violacea are two very close species popular in breeding scented phenotype and emits similar but distinct floral VOCs. P. bellina emits mainly monoterpenoids, including citronellol, geraniol, linalool, myrcene, nerol, and ocimene (Hsiao et al., 2006, 2011b), while P. violacea emits monoterpenoids accompanied with a phenylpropanoid, cinnamyl alcohol (Kaiser, 1993). The VOCs of P. schilleriana contain monoterpenoids as well, including citronellol, nerol and neryl acetate (Awano et al., 1997).
Monoterpenoids, the most abundant constituent in volatile terpenoids (Knudsen and Gershenzon, 2006; Nagegowda et al., 2010), are involved in specialized interactions with other organisms and surrounding environment (Tholl, 2015), for example, to function as attractants for pollinators (Byers et al., 2014), as antibacterial and antifungal compounds (Hammer et al., 2003; Yamasaki et al., 2007; Marei et al., 2012; Mirzaei-Najafgholi et al., 2017), and for defense against herbivores (Paré and Tumlinson, 1999; Mumm et al., 2008; Huang et al., 2018). Apart from their roles in nature, monoterpenoids are widely used in flavor, cosmetics, and perfumery industries due to their unique and pleasant fragrance characteristics (Schwab et al., 2008). In addition, they are exploited as health-promoting compounds and have potential to be applied in cancer therapeutics because of their anti-cancer activities (Gould, 1997; Crowell, 1999; Carnesecchi et al., 2001; Jun et al., 2006; Murthy et al., 2012; Cho et al., 2016; Nakayama et al., 2017).
The precursors of monoterpenoids, IDP and its isomer, DMADP, are produced from the methylerythritol phosphate (MEP) pathway in the plastid. The short-chain prenyltransferases, GDPS, is responsible for the head-to-tail condensation of IDP and DMADP to generate the direct substrate GDP for monoterpene synthases (Dudareva et al., 2004). In Phalaenopsis orchids, PbGDPS is characterized as the key enzyme to provide precursors for monoterpene biosynthesis in P. bellina (Hsiao et al., 2008). Interestingly, recombinant PbGDPS possesses dual prenyltransferase activities for the production of both GDP and farnesyl diphosphate (FDP), the precursor for monoterpeneoids, and sesquiterpenoids, respectively (Hsiao et al., 2008). Expression of PbGDPS is concomitant with the emission of monoterpenoids during flower developments, peaked on day 5 post anthesis (D + 5) (Hsiao et al., 2008).
To date, promoters of the genes in the terpenoid biosynthesis pathway have been investigated and functionally assayed in several species. The vascular-specific expression of a hydroxymethylbutenyl 4-diphosphate synthase gene (HDS) promoter is identified from the MEP pathway in Catharanthus roseus (Ginis et al., 2012). The leaf-specific expression of a geranylgeranyl diphosphate synthase gene (SmGGPPs) promoter is analyzed in Salvia miltiorrhiza (Hua et al., 2012). The vegetative organ-specific expression of a mevalonate diphosphate decarboxylase (GbMVD) from MVA pathway is examined in Ginkgo biloba (Liao et al., 2016). For defense responses, the promoters of four sesquiterpene synthase genes for β-caryophyllene (CPS), epi-cedrol (ECS), β-farnesene (FS), and amorpha-4,11-diene synthase (ADS), and one monoterpene synthase gene (LIS) are studied in Artemisia annua (Wang et al., 2013, 2014). In addition, a number of cis-acting elements on the promoters of terpenoid biosynthesis pathway genes have been identified. A W-box palindrome is identified in the promoter of a cotton sesquiterpene synthase gene (CAD1-A) for GaWRKY1 binding (Xu et al., 2004). The E-box on the promoters of two Arabidopsis sesquiterpene synthase genes TPS21 and TPS11 is recognized by MYC2 involved in gibberellin and jasmonate induction (Hong et al., 2012). The NAC binding sites are found in the promoter of a monoterpene synthase gene (AaTPS1) in the scented kiwifruit Actinidia arguta for AaNAC2, 3 and 4 binding, which are mutated in the scentless A. chinensis (Nieuwenhuizen et al., 2015). The GCC-box is identified in the promoter of the terpene synthase gene (TPS10) for ZmEREB58 binding in Zea mays (Li et al., 2015), and in the promoter of CitTPS16 for CitERF71 in Citrus fruit (Li et al., 2017).
In this study, we reported that a dual repeat in the upstream promoter fragments of GDPS is essential for its transcriptional activation in Phalaenopsis orchids. The full dual repeat was present only in the Phalaenopsis orchids emitting monoterpenes, and its integrity showed strong association with the transactivation of a bZIP TF, bZIP4. As this dual repeat was closely related to the monoterpene production in Phalaenopsis orchids, it could be developed as a promising molecular marker for early detection of monoterpene phenotype in the offspring and thus facilitate scented orchid breeding in future.
Materials and Methods
Plant Materials and Growth Condition
Ten native and two cultivar hybrids were used in the study (Supplementary Figure 1), including P. amboinensis var. yellow (abbreviated as P. amboinensis), P. aphrodite subsp. formosana (abbreviated as P. aphrodite), P. bellina, P. cornu-cervi var. red (abbreviated as P. cornu-cervi), P. equestris ‘RO-5’, P. equestris ‘WY-7’, P. javanica, P. lueddemanniana, P. mannii, P. schilleriana, P. I-Hsin Venus ‘KHM2212’ (abbreviated as P. I-Hsin Venus), and P. Meidarland Bellina Age ‘LM128’ (abbreviated as P. Meidarland Bellina Age). These individual plants were collected from various orchid nurseries across Taiwan (details in Supplementary Table 1). Both P. I-Hsin Venus and P. Meidarland Bellina Age are commercial scented cultivars with monoterpenes as their major scent compounds (Chuang et al., 2017), whose ancestries contain multiple scent species as the following: P. I-Hsin Venus - P. amboinensis (25%), P. equestris (25%), P. venosa (18.75%), P. violacea (15.63%), P. lueddemanniana (3.13%); P. Meidarland Bellina Age - P. bellina (50%), P. violacea (24.22%), P. venosa (15.63%), P. amboinensis (3.91%), P. lueddemanniana (3.13%) via ORCHIDEYA.CA1. All the plant materials were grown in the greenhouse at National Cheng Kung University (NCKU) under natural light and surrounding temperature from 27 to 30°C in spring and summer with 75–85% humidity.
Gas Chromatographic Analysis of Floral Volatiles
Analysis of the floral VOCs of 12 Phalaenopsis orchids was carried out according to the previous studies (Hsiao et al., 2006; Chuang et al., 2017). The VOCs were collected during the most emitted scent period (from 10:00 to 16:00) by using solid phase extraction system (DSC-Si and DCS-18, Supelco, United States) as described (Chuang et al., 2017), and the compounds were then identified by using gas chromatography/high-resolution mass spectrometry (GC/HRMS) at the NCKU Instrument Center (Hsiao et al., 2006). To assess the amounts of each compound, 1 μg of ethyl myristate was recruited as the internal standard (Fluka, Honeywell, United States).
Detection of GDPS Gene Sequence, Upstream Regulatory Fragment and the Dual Repeat Region in 12 Orchid Genomes
To detect the GDPS gene and its upstream regulatory fragment, the genomic DNA of 12 Phalaenopsis orchids were extracted by using Plant Genomic DNA Purification Kit (Bio-GPD50, Biokit, Taiwan). Standard PCRs were applied to amplify the N-terminal region of GDPS (∼400-bp) with the primer designed based on PbGDPS genomic sequence (all the primers used here and thereafter were listed in Supplementary Table 2) since PbGDPS is an intronless gene (Hsiao et al., 2008). The 1-kb upstream promoter fragments of GDPS were also isolated from the 12 Phalaenopsis orchids using the designed primers based on the genomic DNA of P. bellina (Chuang et al., 2017). The dual repeat region was then amplified and cloned with ZeroBack Fast Ligation Kit (TIANGEN, China). Six to eight colonies were selected randomly for sequencing. The presence of the cis-elements in the dual repeats was predicted using PlantPAN (Chow et al., 2015), with 100% similar score accepted as the predicted results.
Quantitative Real-Time RT-PCR
Total RNA was extracted from the flowers on D + 5 stage of the 12 Phalaenopsis orchids following the protocol of Plant Total RNA Miniprep Purification Kit (TR02, GeneMark, Taiwan). Reverse transcription to cDNA involved use of SuperScript III (Thermo Fisher Scientific, United States). Primers were designed by using Primer Express 3.0 (Thermo Fisher Scientific, United States). Quantitative RT-PCR (qRT-PCR) was performed by using the ABI StepOne Plus Quantitative real-time PCR instrument and SYBR Green kit (Applied Biosystems, United States) as described (Hsu et al., 2015). All expression results were normalized to the reference gene, PbActin1 (Chuang et al., unpublished). Mean and standard error were calculated from triplicate repeats.
Plasmid Construction
The 2-kb promoter fragment upstream from the translation start site of PbGDPS (denoted as PbGp-2010) and its nine serial deletion fragments (PbGp-1076, PbGp-859, PbGp-784, PbGp-710, PbGp-584, PbGp-410, PbGp-354, PbGp-297, PbGp-216) (Figure 4A) were amplified from the genomic DNA of P. bellina. Further serial-deletion fragments (PbGp-836, PbGp-822, PbGp-760, PbGp-747, PbGp-729) (Figure 4B) were amplified for detail analysis of the region between -836 bp to -710 bp. Specific primers with the restriction endonuclease sites of BamH I and Nco I were designed to amplify these truncated fragments. The amplified fragments were double-digested with the restriction enzyme BamH I and Nco I, and cloned into the corresponding enzyme digestion sites of pJD301(f) to drive the firefly (Photinus pyralis) luciferase gene (Hsu et al., 2014). All constructs were verified by DNA sequencing. The promoter-LUC constructs were schematically presented in Figure 4.
Transactivation Assay of PbGDPS Promoter Fragments in Planta
The promoter-LUC constructs were bombarded into the floral tissues of P. I-Hsin Venus with an internal control plasmid, pJD301(R), containing the Renilla luciferase gene driven by cauliflower mosaic virus (CaMV) 35S promoter. For normalization, the luciferase activity of the reporter construct was divided by that of the internal control. The involvement of internal control reduced experimental variability resulted from differential bombardment efficiency and transformation efficiency among various experimental groups. The amount of the reporter plasmid and the internal control was 10 and 0.1 μg, respectively. At least six individual flowers of P. I-Hsin Venus were employed for replicates. Luciferase activity of each sample was measured (Hsu et al., 2014). For statistics analysis between two groups, pairwise comparisons were performed by using Tukey’s honestly significant difference test at α = 0.05.
Yeast One-Hybrid (Y1H) Library Screening
Systematic screening of Y1H TF library composed of approximately 1,350 Arabidopsis TFs (Mitsuda et al., 2010) was performed in yeast strain YM4271 to identify the TFs that are able to bind the dual repeat. For construction of bait plasmid, the dual repeat was amplified from the genomic DNA of P. bellina by using appropriate primers with the restriction endonuclease site of Xma I and Xba I, cloned into pHisi2, and integrated it into the yeast genome. The Y1H assay was performed as previously described (Mitsuda et al., 2010). In the screenings of Arabidopsis TF library, the degree of positive interaction between a prey TF and a bait sequences is scored between 0 and 3 in each screening according to the yeast growth status under selective media so that each TF has its own “sticky score” as the sum of this score. The sticky score of AtbZIP29 and AtbZIP30 was 50 and 24, respectively, among 247 TFs isolated so far in 105 screenings including this study and therefore were considered as sticky TFs in this Y1H system.
Identification of bZIP Group I TFs in Phalaenopsis Transcriptome
The bZIP group I TFs was identified in the P. bellina transcriptomic data of four floral development stages at anthesis day (Dd), D + 3, D + 5, and D + 7, which corresponds to the four periods of the floral monoterpene emission pattern, including onset, increase, peak and decline (Chuang et al., unpublished), by using those in Arabidopsis as queries with E-value cutoff of 1e-5 (Jakoby et al., 2002; Pyo et al., 2006). The expression level of each individual TF gene was presented by fragments per kilobase of transcript per million mapped reads (FPKM). FPKM values of the P. bellina transcriptome were transformed by log 3.22 to achieve equivalent expression levels of reference genes as those in the transcriptomic data of P. aphrodite (Su et al., 2013b), including Actin4 (Chen et al., 2005; Hsieh et al., 2013; Pan et al., 2014; Hsu et al., 2015), Actin9 (Hsiao et al., 2008; Pan et al., 2011, 2014; Hsu et al., 2015), and Ubiquitin10 (Lu et al., 2007; Hsiao et al., 2008) (Chuang et al., unpublished). A multiple sequence alignment of bZIP domains was generated by using Clustal Omega2 and displayed by using BOXSHADE3. The phylogenetic tree was built with the neighbor-joining method with 1000 bootstrap trials by using MEGA6 (Tamura et al., 2013).
Examination of the Transactivation of PbbZIP4 and PbbZIP26
Promoter fragments of PbGDPS and PaGDPSpA/PaGDPSpB were isolated from P. bellina and P. aphrodite genomic DNA, respectively (Chuang et al., unpublished). After sequence confirmation, these promoter fragments were cloned into pJD301(f) to drive the firefly luciferase gene. Coding sequences for both PbbZIP4 and PbbZIP26 were amplified with gene-specific primers from full-bloom flowers of P. bellina and cloned into pBI221 and under the control of CaMV 35S promoter. Three separate plasmids, including pBI221 with PbbZIP4/26, pJD301(f) with promoter fragments, and internal control pJD301(R), were co-bombarded into P. aphrodite floral tissues at a ratio of 1.5: 1.5: 0.15 (total 3.15 μg) as described previously (Hsu et al., 2014). The luciferase activity of each sample was measured after 20 h post bombardment. The relative fold change in activity was calculated by the comparison to the control assay with GUS in pBI221 for at least triplicate biological repeats. For statistics analysis between two groups, pairwise comparisons were performed by using Tukey’s honestly significant difference test at α = 0.05.
Transient Ectopic Expression of PbbZIP4 in the Scentless Orchid
The coding sequence of PbbZIP4 was isolated from the pBI221 plasmid containing PbbZIP4 described above, and cloned into the p1304NhXb vector under a duplicated CaMV 35S promoter. The Agrobacterium tumefaciens EHA105 cells taking the resulting plasmids were infiltrated into the perianths of P. aphrodite at the Dd stages (Hsu et al., 2015). The empty plasmid vector containing GUS served as a negative control. Three individual flowers of P. aphrodite were employed for replicates. The volatiles of infiltrated flowers were collected on day 4 post infiltration for 6 hr (from 10:00 to 16:00) and the compounds were identified by GC/HRMS as described above. Total RNA was isolated from the infiltrated tissues following the protocol of RNeasy Plant Mini Kit (QIAGEN, Germany). Reverse transcription to cDNA and quantitative RT-PCR was performed as described above.
Results
Isolation of a Dual Repeat in the GDPS Upstream Promoter
Previously, two individual 1-kb fragments of GDPS promoters were isolated from the scentless P. aphrodite, namely PaGDPSpA and PaGDPSpB (Chuang et al., unpublished). Compared to the GDPS promoter from the scented P. bellina (PbGDPSp), two GDPS promoters identified from P. aphrodite, PaGDPSpA and PaGDPSpB contained an 11-bp deletion and a 75-bp deletion, respectively (Figure 1). PaGDPSpB also had two 14-bp insertions in addition to numerous nucleotide substitutions. By performing the luciferase promoter assays in planta, PaGDPSpA showed the similar promoter activity as PbGDPSp either in scented or scentless flowers, while PaGDPSpB revealed very low promoter activity even in the scented P. bellina floral tissues. These results indicated that the lack of the 75-bp region in the PaGDPSpB is detrimental for its activity (Chuang et al., unpublished). Further sequence analysis of the PbGDPS promoter showed that a second 75-bp repeat is present downstream from the original 75-bp repeat and formed a dual repeat consisted of the two 75-bp units. The first and second 75-bp units were then denoted as ‘R1’, and ‘R2’, respectively, located from -859 to -710 nt upstream from the ATG (Figure 1).
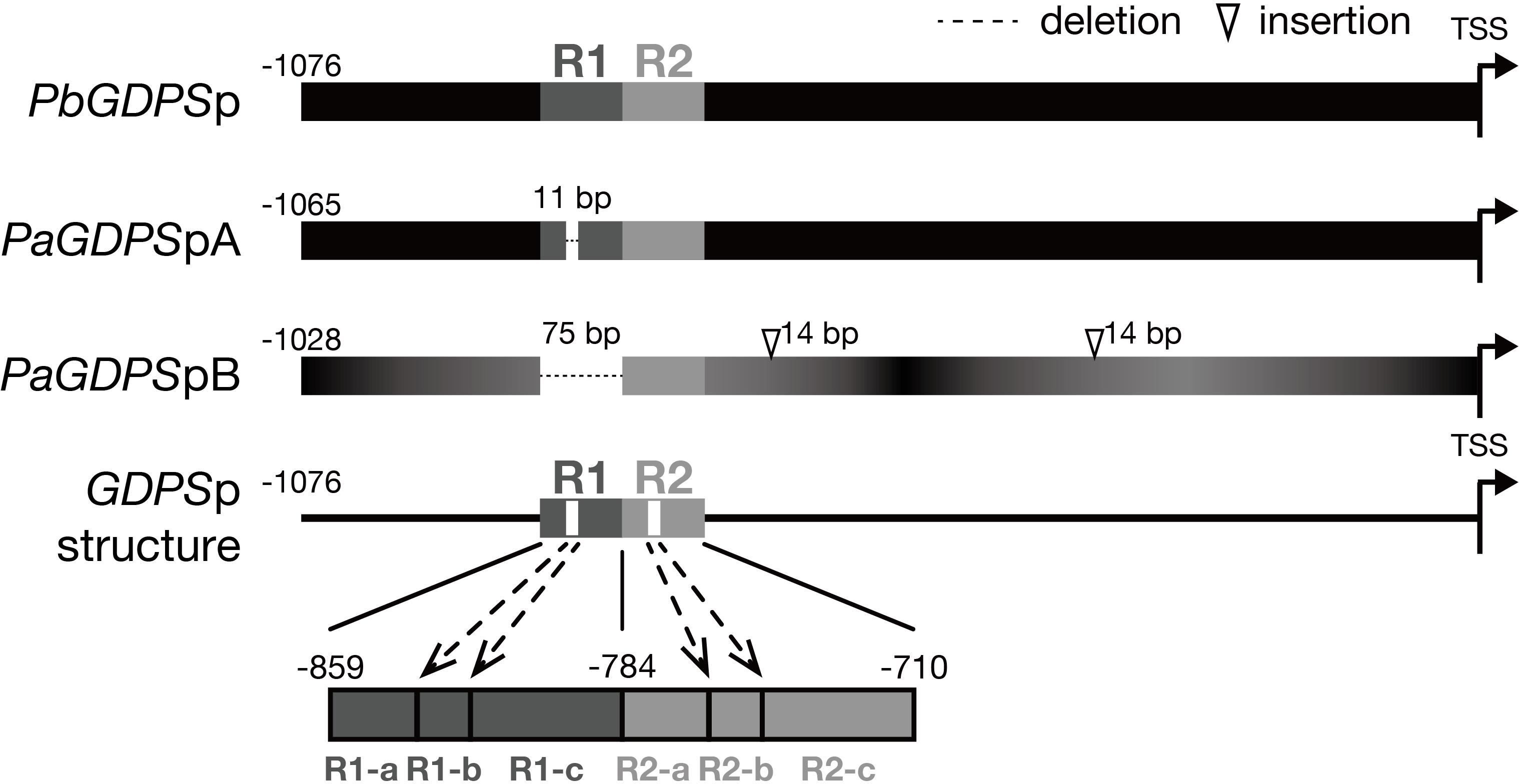
FIGURE 1. Promoter structure of GDPS. Promoter structure of GDPS was revealed by the comparison of three sequences of PbGDPSp, PaGDPSpA, and PaGDPSpB. Two repeats located from –859 to –710 of PbGDPSp were named as R1 and R2. The repeat was further dissected into three subunits based on 11-bp deletion located in the center of R1. This deletion was referred as R1-b, and the sequences prior to and behind R1-b was R1-a and R1-c, respectively. The corresponding dissection in R2 was R2-a, R2-b, and R2-c. TSS indicates the translation start site (ATG). Black color gradient in PaGDPSpB indicated its numerous substitutions compared to PbGDPSp.
The PaGDPSpB lacked the entire R1 unit, and PaGDPSpA harbored a 11-bp deletion in the center of R1, which was defined as R1-b subunit (Figure 1). The region (25-bp) prior to the R1-b was denoted as R1-a, and those (39-bp) behind was R1-c, and the corresponding divisions in R2 were denoted as R2-a, R2-b and R2-c (Figure 1). The dual repeat structure was schematically represented in Figure 1, and the sequence of the dual repeat is in Supplementary Figure 2. The difference between the GDPS promoters of the scent P. bellina and the scentless P. aphrodite resided in the dual repeat, and this is well correlated with the monoterpene phenotype.
Concomitance of the Integrity of the Dual Repeat With the Monoterpene Production
According to the promoter analysis results of PaGDPS and PbGDPS from scented and scentless Phalaenopsis orchids, we hypothesized that the dual repeat is associated with the monoterpene production. To confirm this, another 10 frequently used breeding parents of Phalaenopsis orchids (Supplementary Figure 1) were recruited and assessed for the correlation analysis between the dual repeat and the monoterpene production.
We first examined the floral scent profile (Supplementary Table 3) and found that four orchids emitted monoterpenoids, including P. Meidarland Bellina Age, P. bellina, P. I-Hsin Venus, and P. lueddemanniana. In contrast, the major VOCs of P. javanica and P. amboinensis were sesquiterpenoids and benzenoids, and that of P. mannii was phenylpropanoids and fatty acid derivatives (Supplementary Table 3). P. schilleriana emitted trace amounts of benzenoids. P. aphrodite, P. cornu-cervi, P. equestris ‘RO-5’, and P. equestris ‘WY-7’ were considered as “scentless” since no scent compounds were detected (Supplementary Table 3). For brief, the relative amounts of monoterpenoids emitted from these Phalaenopsis orchids were symbolized in Figure 2A.
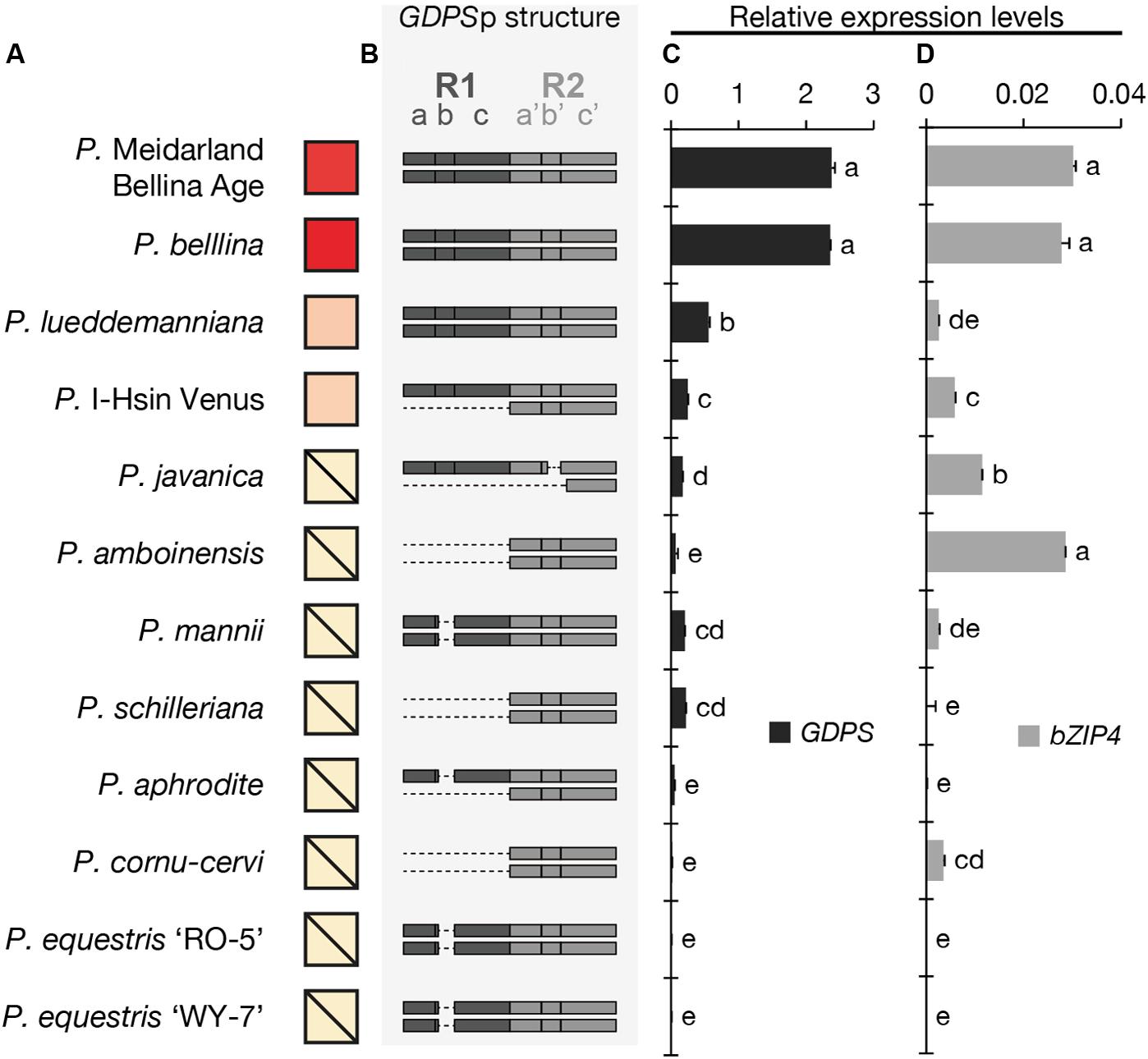
FIGURE 2. The analysis of the floral volatiles, dual-repeat and gene expression in 12 Phalaenopsis orchids. (A) The relative amounts of floral volatiles were represented by a red color gradient. The block with a diagonal line indicates that no monoterpenoid is identified in this orchid. (B) The dual repeat structure of GDPS promoter. (C,D) The expression levels of GDPS (C), and bZIP4 (D). Expression level was normalized to that of PbActin1 by real-time RT-PCR. Data are mean ± SE from triplicate measurements. Statistic tests were performed by using Tukey’s honestly significant difference test at α = 0.05.
The presence of the GDPS gene and its promoter sequence in the 12 Phalaenopsis orchids were then analyzed (Figure 3). Intriguingly, the GDPS gene was present in all of these orchids regardless of being scent or scentless phenotype (Figure 3). It is plausible that the defects are resided in the promoter region (GDPSp, Figure 3). We then amplified the dual repeat on GDPSp and a polymorphism of the dual repeat fragment length was detected among the 12 Phalaenopsis orchids (Figure 3). The four scented orchids with monoterpene production contain the complete dual repeat (Figure 3, the black arrowheads). In contrast, the amplified dual repeat fragments of the other orchids were reduced to various extents with various deletions in the dual repeat region. These 12 fragments were cloned and sequenced. Deletions in the dual repeats were detected between nucleotides 11 and 110, which appear to cause defects in GDPS promoter activities in the orchids without monoterpene production (Figures 2A,B). Strikingly, most defects occurred in the R1 region (Figure 2B).
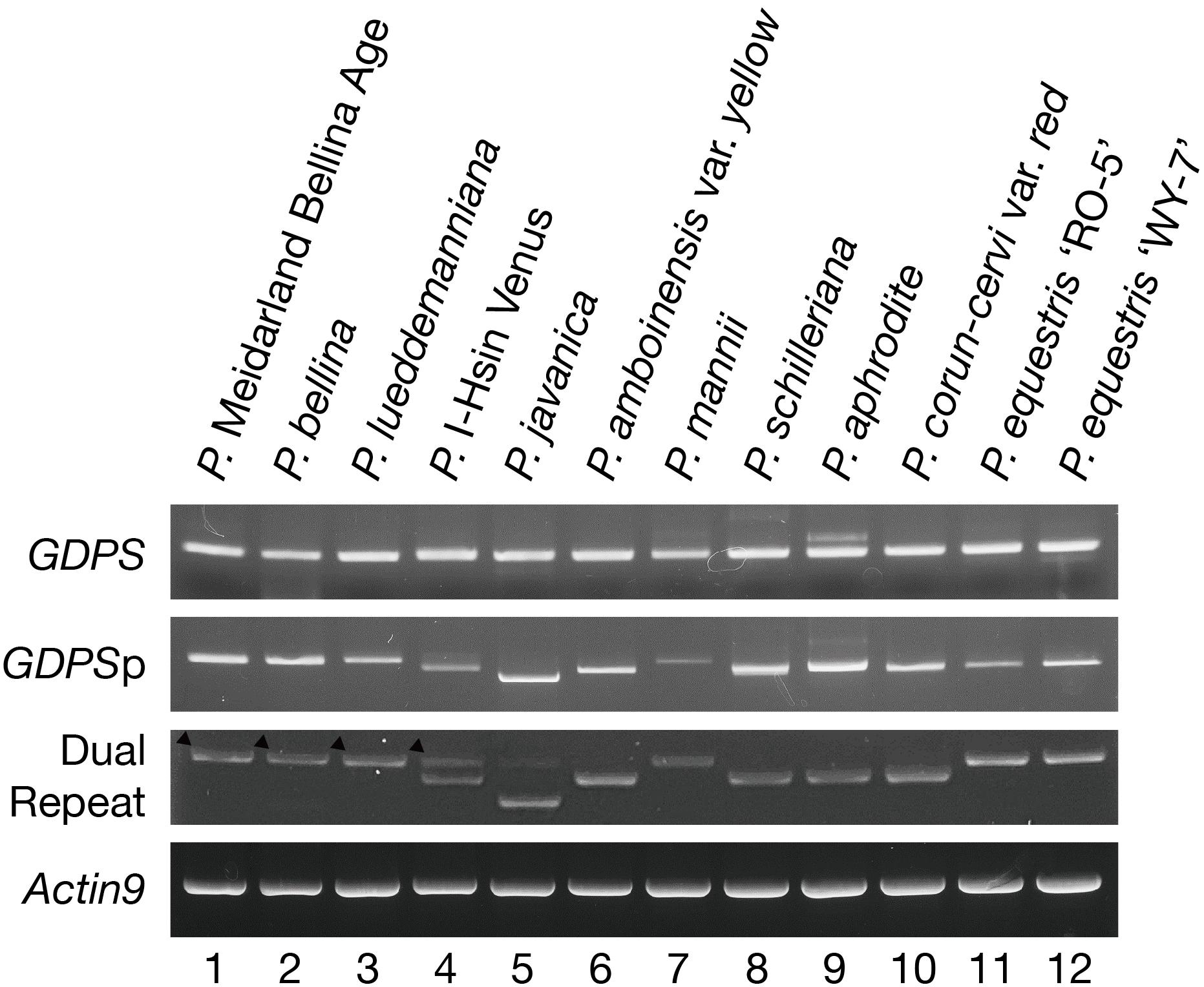
FIGURE 3. PCR Amplification of the GDPS gene, its 1-kb promoter fragment and the dual repeat in the genomic DNA of 12 Phalaenopsis orchids. Actin was used as a control. Amplification of 1-kb promoter and dual repeat showed polymorphisms.
We then examined the expression levels of GDPS for 12 Phalaenopsis orchids (Figure 2C). The four orchids emitting monoterpenoids especially both P. Meidarland Bellina Age and P. bellina showed higher GDPS expression levels than the others (Figure 2C). Taken together, we concluded that the integrity of the dual repeat in the GDPS promoter is strongly correlated with its elevated expression and thus the monoterpene production.
The Dual Repeat Is Crucial for GDPS Promoter Activity
To investigate the role of the dual repeat in the promoter activity of GDPS, the ∼2-kb promoter fragment (denoted PbGp-2010) upstream from the start site of PbGDPS was isolated and subjected to serial deletions. The activity of PbGp-2010 and the nine truncated promoter fragments were evaluated in P. I-Hsin Venus flowers via particle bombardment for dual luciferase assays. It was legitimate that we should examine PbGDPS promoter activity in the original species P. bellina. However, the supply of P. bellina flowers fell short of demand for experiments as P. bellina commonly produces only one flower per 20 days. Instead, P. I-Hsin Venus, the offspring of P. bellina emitting similar scents, was micropropagated to large quantities with the identical genetic background and would help to reduce variation.
The highest luciferase activity was observed for PbGp-859, which showed approximately threefold increase as compared to that PbGp-784, and fivefold increase as compared to that of PbGp-710 (Figure 4A). Thus, the cis-element responsible for high promoter activity was between nucleotide (nt) -859 and nt -710 (150-bp), in which the dual repeat located. Further dissection of PbGp-710 to generate PbGp-584, PbGp-410, PbGp-354, PbGp-297, and PbGp-216 showed that no extra enhancers were present at the downstream of the dual repeat (Figure 4A). Compared with PbGp-859, the promoter activity of PbGp-2010 and PbGp-1076 was much decreased (Figure 4A), suggesting the presence of repressor elements at the upstream region of PbGp-859. These results verified that the dual repeat plays a crucial role for PbGDPS promoter activity.
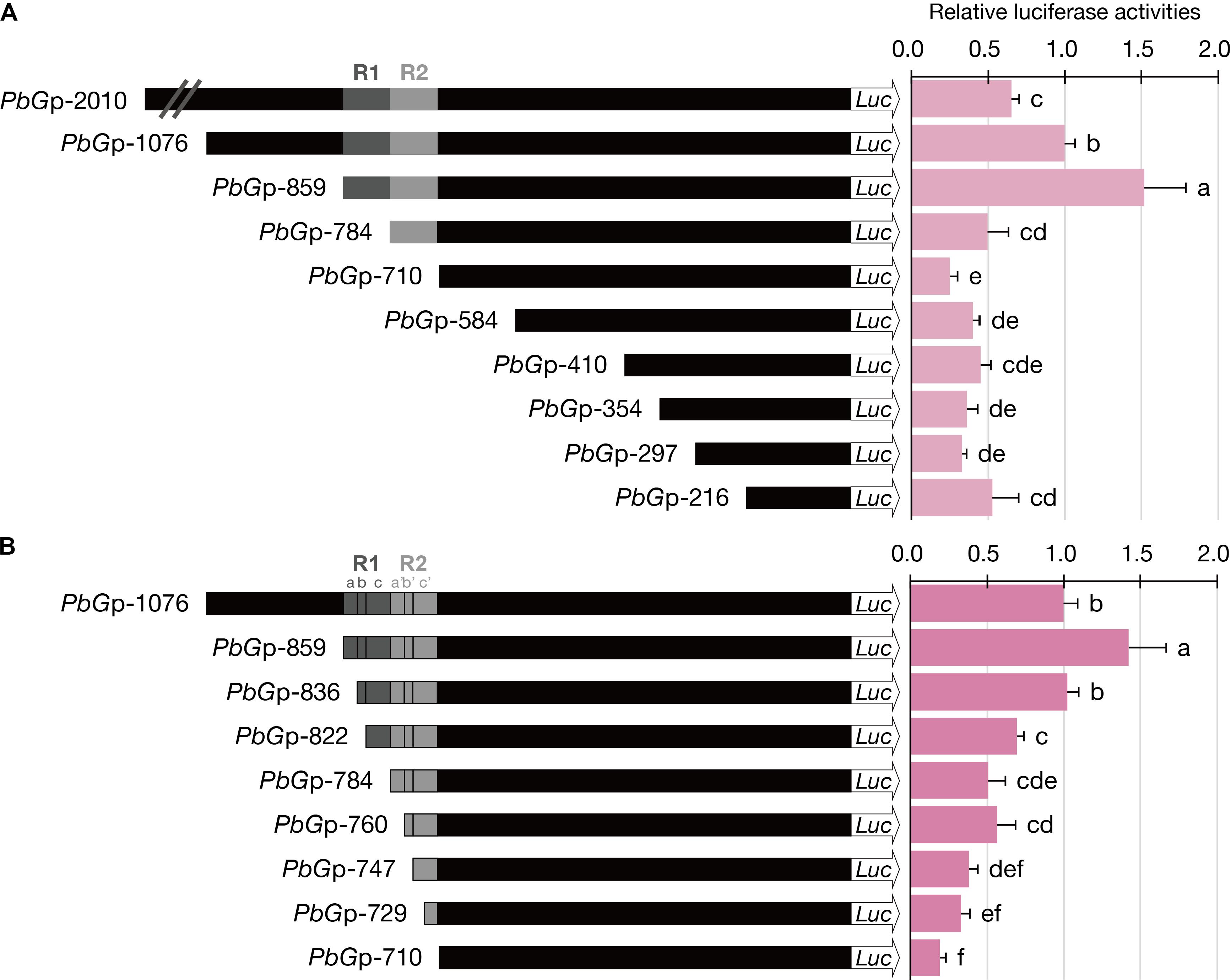
FIGURE 4. Luciferase activities of dissected PbGDPS promoters in P. I-Hsin Venus by particle bombardment. (A) Serial deletion of PbGDPS promoter to analyze the putative cis-element. (B) Identification of the subunit of the dual repeat responsible for promoter driving abilities. Activation level was given by the ratio of Luc/RLuc and the level of PbGp-1076 was set as 1. Experiments are based on at least sextuplicate biological replicate. Statistic tests were performed by using Tukey’s honestly significant difference test at α = 0.05.
To further define the cis-element in the 150-bp dual repeat region for transcriptional regulation, a more detailed analysis was performed based on the subunit division, namely R1a, R1b, R1c, R2a, R2b, and R2c. These subunits were serial deleted to generate a series of truncated promoter constructs. The luciferase activities were reduced further with gradual deletions of the subunit, indicating the essential nature of the complete dual repeat for full promoter activity of PbGDPS (Figure 4B).
Yeast One-Hybrid Screening of the Transcription Factor Bound Onto the Dual Repeat
To identify the candidate upstream trans-activators, Y1H screening was performed using P. bellina floral cDNA as preys. However, the dual repeat bait produced extensive background growth of yeast, and it could not be eliminated even under the addition of inhibitor 3-Amino-1,2,4-triazole (3-AT). Alternatively, Y1H screening against prey library composed of approximately 1,350 TFs of Arabidopsis thaliana was performed (Mitsuda et al., 2010). The dual repeat was amplified from P. bellina genomic DNA and fused to a minimal promoter of HISTIDINE SYNTHASE3 (Supplementary Figure 3A). In this approach, the leaky expression of the reporter gene could be overcome by the addition of 4 mM 3-AT (Supplementary Figure 3B). Total four positive TFs were obtained from this screening (Table 1) and three of them encoded bZIP family proteins including AtbZIP18, AtbZIP29, and AtbZIP30. The other one candidate AtAGL81, a MADS-box TF, failed to be isolated from P. bellina floral transcriptome.
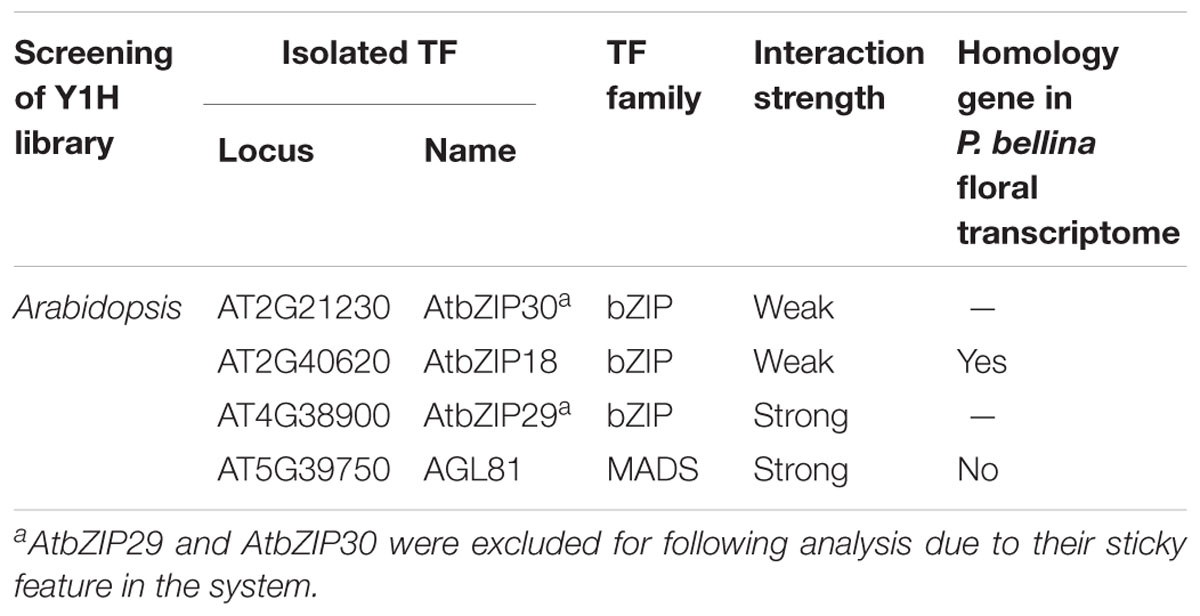
TABLE 1. Transcription factors binding to the dual repeat by screening of yeast one-hybrid libraries.
The three bZIP TFs belong to the group I of bZIP family composed of a conserved bZIP domain for DNA binding and a leucine zipper motif for dimerization (Jakoby et al., 2002). The P. bellina floral transcriptomic data (Chuang et al., unpublished) showed nine proteins belong to group I, and can be divided into three subgroups (i, ii, and iii) according to their phylogenetic relationship with the Arabidopsis ones (Figure 5A). We selected subgroup i including AtbZIP18 for further analysis since AtbZIP29 and AtbZIP30 (subgroup ii) were repeatedly isolated in other unrelated screenings and therefore considered as sticky factors in the Y1H system. No PbbZIPs were classified into subgroup iii (Figure 5A). Multiple alignments showed that five PbbZIPs namely PbbZIP4, PbbZIP10, PbbZIP26, PbbZIP29, and PbbZIP32 in the subgroup i shared 83–86% identity with AtbZIP18 in the basic region (Figure 5B).
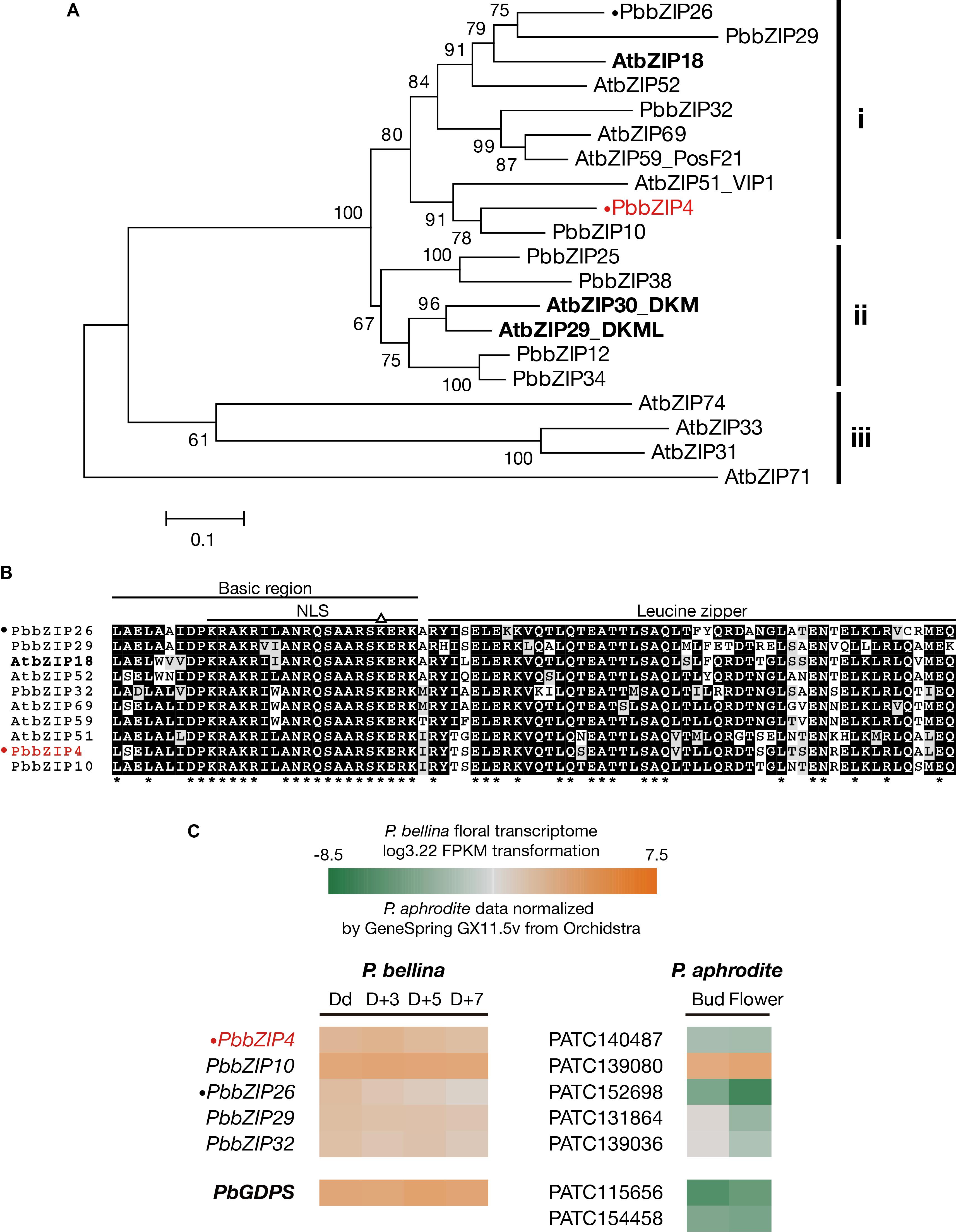
FIGURE 5. Analysis of bZIP group I TFs in P. bellina floral transcriptome. (A) Phylogenetic tree inferred from the amino sequences of the nine bZIP group I TFs with the ones in Arabidopsis. The three subgroups were divided based on their phylogenetic relationship. Three AtbZIPs isolated by Y1H screening were seen in bold. (B) Amino acid sequence alignment of subgroup i proteins in (A). Asterisks (∗) below the alignment indicate the consensus residues in all sequences. The triangle (∆) above the alignment indicates the lysine replacement of the conserved arginine residue in the basic region of group I of bZIP family. (C) Comparison of the expression levels of five bZIP TFs between two Phalaenopsis transcriptomes. Gene expression levels in P. bellina transcriptomes were transformed by log3.22 FPKM in four floral developmental stages, including Dd, D + 3, D + 5, and D + 7 (Chuang et al., unpublished), while the levels of two floral stages of flower bud and flower in P. aphrodite transcriptomes were analyzed via microarray experiments stored in Orchidstra (Su et al., 2013a). Expression levels of putative genes are represented by a color gradient from orange to gray to green. Two PbbZIPs for transient assays, PbbZIP4 and PbbZIP26, are labeled with a dot (•). PbbZIP4 was especially shown in red. The expression levels of PbGDPS (in bold) was also included for comparison.
Previously, we found that the promoter activity of GDPS was much higher in the scented P. bellina than in the scentless P. aphrodite. In addition, PaGDPSpA was a functional promoter since it showed similar promoter activity as PbGDPSp in the scented P. bellina flower tissues (Chuang et al., unpublished). Both results indicated that the down-regulation of the corresponding upstream activators of GDPS was responsible for the extremely low GDPS expression in P. aphrodite (Figure 5C).
The possibility of the five bZIPs regulating GDPS was evaluated by comparing their gene expression between P. bellina and P. aphrodite (Figure 5C). Among them, bZIP4, bZIP26, bZIP29, and bZIP32 showed higher expression in P. bellina than in P. aphrodite. However, both bZIP29 and bZIP32 also expressed in floral bud of P. aphrodite but still GDPS did not express (Figure 5C), suggesting that both bZIP29 and bZIP32 did not transactivate PaGDPSpA promoter. Both PbbZIP29 and PbbZIP32 thus were excluded for further analysis.
PbbZIP4 Was Able to Distinguish the Promoter Containing the Dual Repeat
Previously, we have shown that PbbZIP4 was able to transactivate PbGDPS promoter (Chuang et al., unpublished). Here, to further examine the transactivating ability of PbbZIP4 and PbbZIP26 on various GDPS promoters, dual luciferase assay was performed in the floral tissues of the scentless P. aphrodite. In the presence of PbbZIP4, it enhanced the promoter activities of both PbGDPSp and PaGDPSpA, but revealed no effects on PaGDPSpB (Figure 6). This was consistent with the previous results, in which the activities of both PbGDPSp and PaGDPSpA were higher than PaGDPSpB in the scented P. bellina floral tissues (Chuang et al., unpublished). In contrast, PbbZIP26 did not transactivate PbGDPSp, PaGDPSpA, or PaGDPSpB (Figure 6). These results indicated that PbbZIP4 was able to distinguish the GDPS promoter containing the full dual repeat (PbGDPSp), or near full-dual-repeat (PaGDPSpA), from the solely one repeat unit (PaGDPSpB).
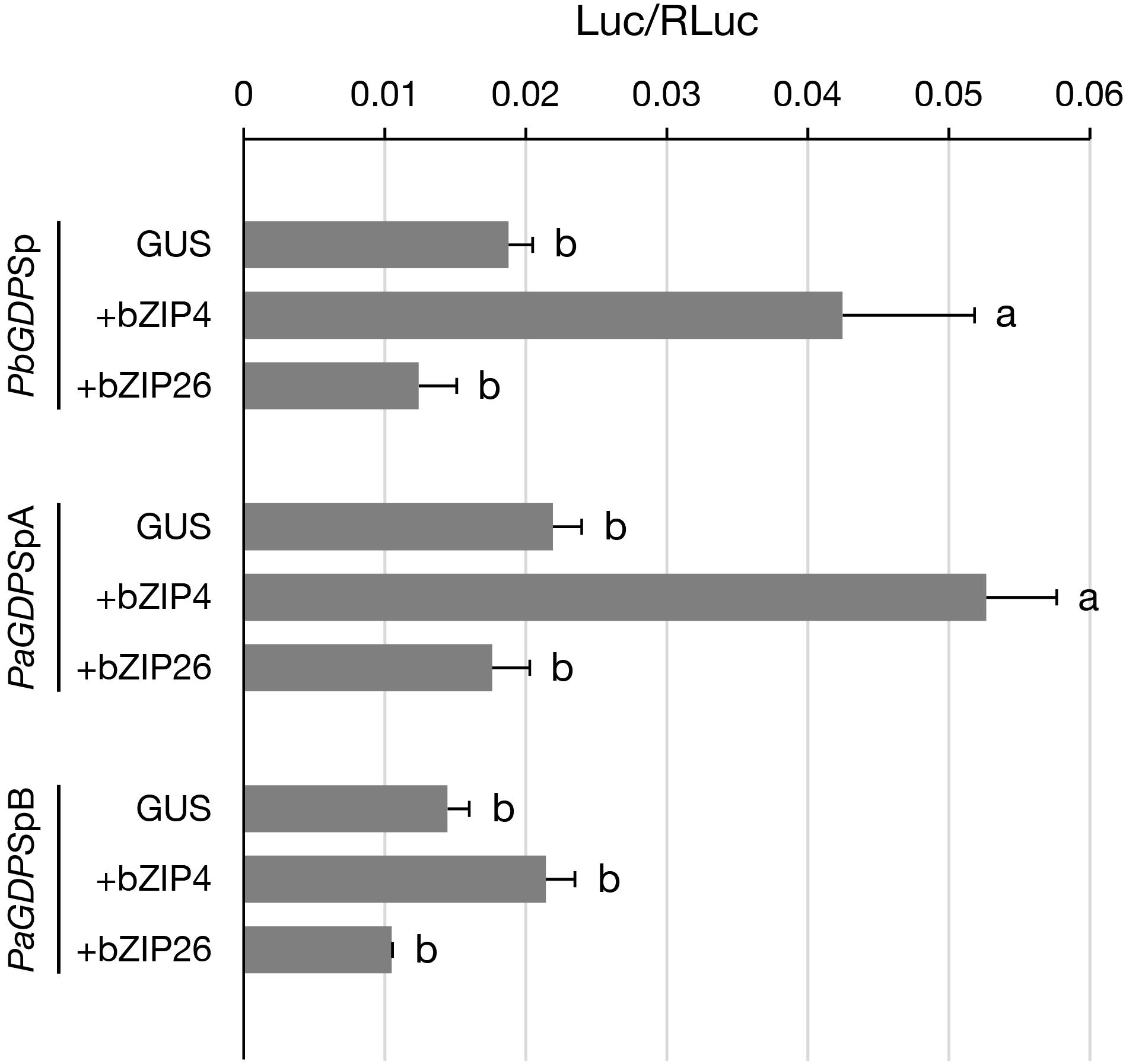
FIGURE 6. Transient assay for analyzing the transactivation ability of PbbZIP4 and PbbZIP26 on PbGDPSp, PaGDPSpA, and PaGDPSpB. The promoter structure of PbGDPSp, PaGDPSpA, and PaGDPSpB was schematically represented in Figure 1. Activation level was given by the ratio of Luc/RLuc. GUS indicated that the assay was performed without the transcription factor. Experiments were based on at least three biological replicates. Statistic tests were performed by using Tukey’s honestly significant difference test at α = 0.05.
The Close Association Between the Trans-Factor and the Cis-Element for Monoterpene Phenotype
The cis-element and trans-factor for GDPS promoter were both identified in the scented P. bellina. To investigate how these two factors affected the monoterpene phenotype among the 12 Phalaenopsis orchids, we examined the expression levels of bZIP4 (Figure 2D) to establish its correlation with the presence of the dual repeat on GDPS promoter (Figure 2B) and GDPS expression levels (Figure 2C) for monoterpene production (Figure 2A). Interestingly, bZIP4 expressed to various extents among these orchids (Figure 2D). Strikingly, only the orchid plants concomitantly harbored bZIP4 expression and the dual repeat on GDPS promoter exhibited high GDPS expression, and thus produced monoterpenes, including P. Meidarland Bellina Age, P. bellina, P. lueddemanniana, and P. I-Hsin Venus (Figure 2).
However, orchid species with similar or even higher bZIP4 expression levels but without the dual repeat showed low GDPS expression, and thus did not emit monoterpenes, including P. javanica, P. amboinensis, P. mannii, and P. cornu-cervi (Figure 2). In contrast, another four species with extremely low expression levels of bZIP4, together with their incomplete dual repeat, contributed to their scentless phenotype, including P. schilleriana, P. aphrodite, P. equestris ‘RO-5’, and P. equestris ‘WY-7’ (Figure 2). Collectively, these results indicated that not only the dual repeat in GDPS promoter but also the TFs are crucial for monoterpene production in Phalaenopsis orchids.
Transient Ectopic Expression of PbbZIP4 in the Scentless P. aphrodite
So far, the stable genetic transformation for Phalaenopsis orchids is with low efficiency. In addition, Phalaenopsis orchids have a long-life cycle with the regeneration time is about 2–3 years, especially if we want to examine the floral phenotype. Instead, the transient ectopic expression system in floral tissues has been successfully established for the study of three MYB TFs regulating the pigmentation patterning in Phalaenopsis orchids (Hsu et al., 2015). Thus, to confirm the role of PbbZIP4 in planta, we performed a transient assay by infiltrating the Agrobacterium into the flower tissues of the scentless P. aphrodite. We analyzed the volatile terpenes emitted from PbbZIP4-expressing P. aphrodite flowers and detected a 10-fold induction of α-terpineol (a monoterpenoid) as compared to the GUS control (Figures 7A,B). Furthermore, the raise in the levels of monoterpenes in the infiltrated tissues was indeed resulted from the large increase of PbbZIP4 transcripts (Figure 7C). Therefore, we concluded that PbbZIP4 was involved in the scent production in Phalaenopsis orchids.
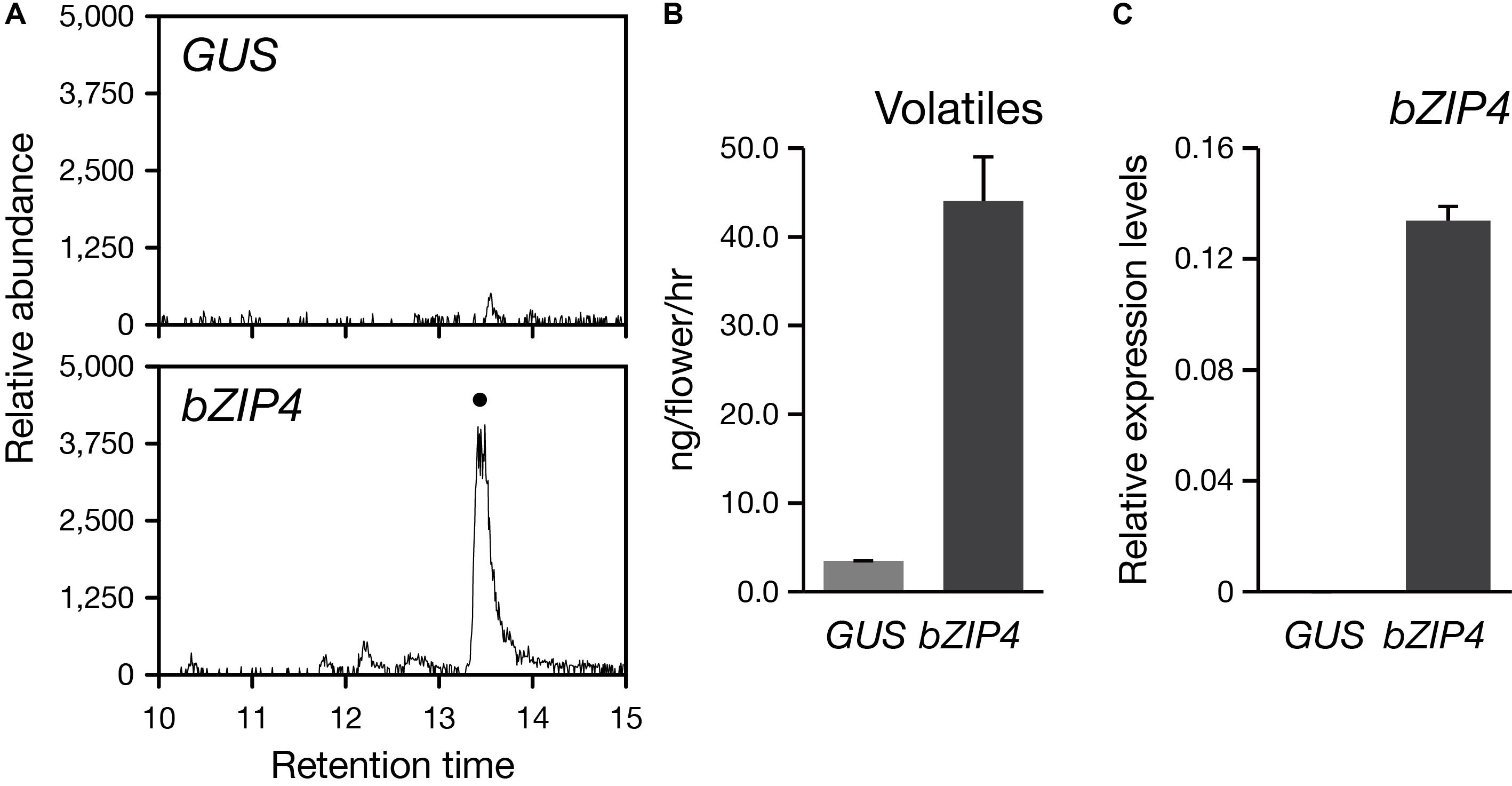
FIGURE 7. Transient expression of PbbZIP4 in the scentless P. aphrodite flowers induced the production of monoterpeneoids. (A) Selected ion (m/z 136) GC-MS profiles of volatiles detected from GUS- and PbZIP4-expression flowers. The peak with a dot indicates α-terpineol. The products were confirmed by comparing the mass spectra of the indicated peak with the one of the library hit. (B) The levels of volatiles emitted from the flowers infiltrated with GUS (gray) and PbbZIP4 (dark-gray). (C) Relative expression levels of PbbZIP4 of the flowers infiltrated with GUS (gray) and PbbZIP4 (dark-gray). Error bars on each column represent ± SE from three biological replicates.
Discussion
The Key Role of the Repeat Unit in the GDPS Promoter Activity
In this study, we studied the molecular mechanism determining the scent phenotype in Phalaenopsis orchids. We found that a dual repeat of cis-element on the GDPS promoter played a vital role for floral monoterpene production. The orchids without monoterpene production harbored defective dual repeat and noticeably, most occurring in the R1 region. Serial deletion analysis showed that the removal of R1 unit decreased the promoter activity by 67%, and the further deletion of R2 unit caused an additional 16% reduction, which implies that the R1 unit is crucial for high levels of GDPS promoter activity and that the R2 unit is required for the 100% transactivation activity.
The Evolutionarily Conserved GDPS Promoter Sequences Among Phalaenopsis Orchids
The GDPS promoter fragments isolated from the 10 native Phalaenopsis species shared extremely high similarities (90–100% identities, data not shown), which indicates the conservation of the GDPS promoter sequences among the Phalaenopsis orchids. The 10 native Phalaenopsis orchids belong to two-pollinia Polychilos and subgenus Phalaenopsis (Supplementary Table 4) (Christenson, 2001). According to the evolutionary trend deduced by pollinia number, molecular evidences and biogeography, the four-pollinia basal subgenus Aphyllae in South China and Indochina is developed into two-pollina groups during the dispersal into Southeast Asia, including subgenus Polychilos in Indonesia and Malaysia, and subgenus Phalaenopsis in the Philippines, respectively (Tsai, 2003, 2011; Tsai et al., 2010). As both two-pollina subgenus shared similar GDPS promoter sequences, it is possible that this conserved GDPS promoter was inherited from their common ancestor of four-pollinia basal group.
Possible Origin of the Dual Repeats
Repetitive DNA is accounting for a substantial proportion in the whole genomic DNA in most eukaryotes. A number of genetic diseases are related to the large copy number of repetitive sequences, such as Huntington’s disease, fragile X syndrome, and myotonic dystrophy (Pelley, 2007). There are two main classes of repetitive DNA, interspersed repeats dispersed throughout the genome, and tandem repeats located in one area of DNA (also known as satellite DNA) (Bhagavan and Ha, 2011). The interspersed repeats include short interspersed nuclear elements (100- to 500-bp in length) and long interspersed nuclear elements (6000- to 7000-bp in length), and both belong to transposons (Pelley, 2007). On the other hand, satellite DNA is divided into three groups based on the length of the repeat unit, including macrosatellites, minisatellites, and microsatellites. Minisatellites are consisted of repeat sequences ranged from 9 bp to 80 bp (Bhagavan and Ha, 2011), and the size normally ranged from 1-kb to 20-kb. In cauliflower, a Harbinger DNA transposon in the promoter of an R2R3-MYB TF, purple, leads to an increase in the gene expression and produces the purple phenotype (Chiu et al., 2010). Two reports have described that a minisatellite-like structure on the promoter of an anthocyanin-regulating TF, MYB10, is required for the red color formation in the fruit flesh and leaves in apple and crabapple, respectively (Espley et al., 2009; Tian et al., 2017).
The 150-bp dual repeat identified here consisted of two 75-bp repeat units adjacent to each other, and thus was not considered as a transposon but defined as a minisatellite-like structure. Indeed, this dual repeat did not respond to any known transposon sequences by BLAST against the repetitive sequence database of RepBase (Jurka, 1998; Bao et al., 2015). We speculate that this dual repeat was generated by a tandem duplication event, and have not undergone additional mutation yet for multiple copies.
Six out of 10 Phalaenopsis orchids had the dual repeat or near-full dual repeat, including P. bellina and P. luddemanian, and P. javanica, P. mannii, P. aphrodite, and P. equestris, respectively. These six orchids either belong to subgenus Polychilos or subgenus Phalaenopsis (Supplementary Table 4), suggesting that the dual repeat was retained from their common predecessors, subjected to further mutation, and became defective in the orchids without monoterpene production. As two species of the basal four-pollina subgenus Aphyllae (Tsai et al., 2010), P. hainanensis and P. wilsonii, also emits fragrance (information from http://www.orchid.url.tw/), it will be interesting to study whether they have the dual repeat as well, just similar to the cases in Capsella (Brandvain et al., 2013; Sas et al., 2016) and Petunia genus (Amrad et al., 2016) describing the loss of the floral scent during the shift of pollinator types.
Tandem Repeats Correlated to Transcriptional Regulation
In Phalaenopsis orchids, the dual repeat on PbGDPS promoter is required for its 100% promoter activity. Several studies have also shown that the increasing number of tandem repeats lead to a stepwise increase in promoter activity, and subsequently in gene expression levels. In citrus, three copies of a 20-bp enhancer element on the promoter of lycopene β-cyclases (CsLCYb1), an enzyme representing a branch point for carotenoid biosynthesis, are important for its promoter activity (Lu et al., 2016). In C. roseus, the copy number of a simple sequence repeats (CT) on the promoter of Tryptophan decarboxylase, the first step in the indole alkaloids biosynthesis pathway, is strongly associated its expression levels (Kumar and Bhatia, 2016). Furthermore, the copy number of the cis-elements even determines a specific phenotype. For instance, in apple, five direct tandem repeats on the promoter of MYB10, an anthocyanin-regulating TF, is only present in the red-fleshed apple varieties, and cause itself autoregulation (Espley et al., 2009). In cotton, two 228-bp tandem repeats on the promoter of an anthocyanin-regulating TF, Red Leaf Cotton 1, is critical for its promoter activity, and only present in the red leaf variety (Gao et al., 2013). Similarly, in our study, the dual repeat is essential for the monoterpene production in Phalaenopsis orchids.
The Dual Repeat Could Be a Novel Cis-Acting Sequence
We performed Y1H screening to identify the TFs binding to the dual repeat, and member of bZIP, and MADS-box TFs were isolated. Here, we excluded the possibility of AtAGL81 (MADS-box TF) to interact with PbGDPS promoter since it was failed to be isolated from P. bellina floral transcriptome in E-value cutoff of 1e-5. Moreover, we were not able to isolate the similar sequences to AtAGL81 from P. aphrodite transcriptome, whose transcript profiles were analyzed in leaf, root, flower bud, and fully open flower (Orchidstra 2.0) (Chao et al., 2017) and P. equestris genome (Cai et al., 2015).
The presence of cis-elements on the dual repeat was examined by PlantPan analysis (Chow et al., 2015). Several types of TFs were predicted to interact with the dual repeat, including MYB-related, Dof zinc finger protein, GATA, zinc-finger homeodomain protein, squamosa promoter binding protein, and nuclear TF Y subunit beta. Interestingly, we did not find any bZIP-binding cis-elements, suggesting that the dual repeat could be a novel cis-acting sequence. As the length of the dual repeat is 150-bp, it is possible that other types of TFs may be involved in the scent regulating phenotype, and it awaits further analysis.
Application for Molecular Marker-Assisted Breeding
The molecular markers to distinguish scent traits have been well developed in rice (Garland et al., 2000; Cordeiro et al., 2002; Jin et al., 2003; Bradbury et al., 2005; Shi et al., 2008; Sakthivel et al., 2009; Myint et al., 2012), and also in other crops, such as soybean (Arikit et al., 2011; Juwattanasomran et al., 2011, 2012), sorghum (Yundaeng et al., 2013), cucumber (Pramnoi et al., 2013; Yundaeng et al., 2015), coconut (Vongvanrungruang et al., 2016), and winter melon (Ruangnam et al., 2017). Just like other modern floriculture cultivars, negative correlation between floral scent and other favorable traits resulted in the difficulty in the scented orchids breeding. The introduction of the scent trait to a well-commercialized cultivar is an alternative approach (Hsiao et al., 2011b). However, the long duration from seedling to first blooming for confirming the floral traits are time consuming and cost inefficient under conventional breeding.
Here, PCR amplification of the dual repeat showed fragment length polymorphism. The two native Phalaenopsis orchids and two cultivars emitting monoterpenes showed the 150-bp fragment length of dual repeat, which indicates its potential to be developed as a molecular marker for scent trait. After the link between the dual repeat and the monoterpene emission further verified in more Phalaenopsis species and cultivars, the dual repeat could be applied as a molecular marker for early characterization of the scent phenotype in the seedlings and thus benefiting the breeding of scented Phalaenopsis orchids.
Author Contributions
Y-CC, W-CT, W-HC, and H-HC conceived the research plans. Y-CC performed most of the experiment, analyzed the data, and wrote the article with contributions of all the authors. Y-CH and C-YH assisted in the identification of the dual repeat and the analysis in the gene expression and scent compounds in Phalaenopsis orchids. C-MY, NM, and MO-T performed the yeast one hybrid screening analysis and provided valuable comments on the manuscript. H-HC supervised and complemented the writing. All authors read and approved the submitted version.
Funding
This work was supported by grant no. MOST-102-2313-B-006-001-MY3 from the Ministry of Science and Technology, Taiwan, and partly supported by KAKENHI (grant no. 25113001 to NM) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Tuan-Hua David Ho and Dr. Shu-Hsing Wu (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan), and Dr. Yu-Yun Hsiao (Orchid Research and Development Center, NCKU, Taiwan) for helpful discussions. We thank Ya-Lan Chang (Department of Life Sciences, NCKU, Taiwan) for the assistance in the statistics analysis, and Dr. Chia-Chi Hsu (Department of Life Sciences, NCKU, Taiwan) for the transposon analysis. We also thank Pei-Han Lai (Department of Life Sciences, NCKU, Taiwan) and Ms. Fumie Tobe (AIST, Japan) for their technical help.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00765/full#supplementary-material
Abbreviation
bZIP, basic leucine zipper; DMADP, dimethylallyl diphosphate; GDPS, geranyl diphosphate synthase; IDP, isopentenyl diphosphate; P., Phalaenopsis; TFs, transcription factors; Y1H, yeast one-hybrid.
Footnotes
- ^ http://www.orchideya.ca/
- ^ http://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://embnet.vital-it.ch/software/BOX_form.html
References
Amrad, A., Moser, M., Mandel, T., de Vries, M., Schuurink, R. C., Freitas, L., et al. (2016). Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr. Biol. 26, 3303–3312. doi: 10.1016/j.cub.2016.10.023
Arikit, S., Yoshihashi, T., Wanchana, S., Tanya, P., Juwattanasomran, R., Srinives, P., et al. (2011). A PCR-based marker for a locus conferring aroma in vegetable soybean (Glycine max L.). Theor. Appl. Genet. 122, 311–316. doi: 10.1007/s00122-010-1446-y
Awano, K., Honda, T., Ogawa, T., Suzuki, S., and Matsunaga, Y. (1997). Volatile components of Phalaenopsis schilleriana Rehb. f. Flav. Frag. J. 12, 341–344. doi: 10.1002/(SICI)1099-1026(199709/10)12:5<341::AID-FFJ657>3.0.CO;2-L
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6:11. doi: 10.1186/s13100-015-0041-9
Bhagavan, N., and Ha, C.-E. (2011). “Structure and properties of DNA,” in Essentials of Medical Biochemistry: With Clinical Cases, eds C.-E. Ha and N. Bhagavan (Cambridge, MA: Academic Press).
Bradbury, L. M., Henry, R. J., Jin, Q., Reinke, R. F., and Waters, D. L. (2005). A perfect marker for fragrance genotyping in rice. Mol. Breed. 16, 279–283. doi: 10.1007/s11032-005-0776-y
Brandvain, Y., Slotte, T., Hazzouri, K. M., Wright, S. I., and Coop, G. (2013). Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9:e1003754. doi: 10.1371/journal.pgen.1003754
Byers, K. J., Bradshaw, H., and Riffell, J. A. (2014). Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). J. Exp. Biol. 217, 614–623. doi: 10.1242/jeb.092213
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Carnesecchi, S., Schneider, Y., Ceraline, J., Duranton, B., Gosse, F., Seiler, N., et al. (2001). Geraniol, a component of plant essential oils, inhibits growth and polyamine biosynthesis in human colon cancer cells. J. Pharmacol. Exp. Ther. 298, 197–200.
Chao, Y.-T., Yen, S.-H., Yeh, J.-H., Chen, W.-C., and Shih, M.-C. (2017). Orchidstra 2.0-A transcriptomics resource for the orchid family. Plant Cell Physiol. 58:e9. doi: 10.1093/pcp/pcw220
Chen, Y. H., Tsai, Y. J., Huang, J. Z., and Chen, F. C. (2005). Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Res. 15, 639–657. doi: 10.1038/sj.cr.7290334
Chiu, L.-W., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154, 1470–1480. doi: 10.1104/pp.110.164160
Cho, M., So, I., Chun, J. N., and Jeon, J.-H. (2016). The antitumor effects of geraniol: modulation of cancer hallmark pathways. Int. J. Oncol. 48, 1772–1782. doi: 10.3892/ijo.2016.3427
Chow, C. N., Zheng, H. Q., Wu, N. Y., Chien, C. H., Huang, H. D., Lee, T. Y., et al. (2015). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1160. doi: 10.1093/nar/gkv1035
Chuang, Y.-C., Lee, M.-C., Chang, Y.-L., Chen, W.-H., and Chen, H.-H. (2017). Diurnal regulation of the floral scent emission by light and circadian rhythm in the Phalaenopsis orchids. Bot. Stud. 58:50. doi: 10.1186/s40529-017-0204-8
Cordeiro, G. M., Christopher, M. J., Henry, R. J., and Reinke, R. F. (2002). Identification of microsatellite markers for fragrance in rice by analysis of the rice genome sequence. Mol. Breed. 9, 245–250. doi: 10.1023/A:1020350725667
Crowell, P. L. (1999). Prevention and therapy of cancer by dietary monoterpenes. J. Nutr. 129, 775S–778S. doi: 10.1093/jn/129.3.775S
Dudareva, N., and Negre, F. (2005). Practical applications of research into the regulation of plant volatile emission. Curr. Opin. Plant Biol. 8, 113–118. doi: 10.1016/j.pbi.2004.11.007
Dudareva, N., Pichersky, E., and Gershenzon, J. (2004). Biochemistry of plant volatiles. Plant Physiol. 135, 1893–1902. doi: 10.1104/pp.104.049981
Espley, R. V., Brendolise, C., Chagné, D., Kutty-Amma, S., Green, S., Volz, R., et al. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21, 168–183. doi: 10.1105/tpc.108.059329
Gao, Z., Liu, C., Zhang, Y., Li, Y., Yi, K., Zhao, X., et al. (2013). The promoter structure differentiation of a MYB transcription factor RLC1 causes red leaf coloration in empire red leaf cotton under light. PLoS One 8:e77891. doi: 10.1371/journal.pone.0077891
Garland, S., Lewin, L., Blakeney, A., Reinke, R., and Henry, R. (2000). PCR-based molecular markers for the fragrance gene in rice (Oryza sativa. L.). Theor. Appl. Genet. 101, 364–371. doi: 10.1007/s001220051492
Ginis, O., Courdavault, V., Melin, C., Lanoue, A., Giglioli-Guivarc’h, N., St-Pierre, B., et al. (2012). Molecular cloning and functional characterization of Catharanthus roseus hydroxymethylbutenyl 4-diphosphate synthase gene promoter from the methyl erythritol phosphate pathway. Mol. Biol. Rep. 39, 5433–5447. doi: 10.1007/s11033-011-1343-8
Gould, M. N. (1997). Cancer chemoprevention and therapy by monoterpenes. Environ. Health Perspect. 105(Suppl. 4), 977. doi: 10.1289/ehp.97105s4977
Hammer, K., Carson, C., and Riley, T. (2003). Antifungal activity of the components of Melaleuca alternifolia (tea tree) oil. J. Appl. Microbiol. 95, 853–860. doi: 10.1046/j.1365-2672.2003.02059.x
Hong, G.-J., Xue, X.-Y., Mao, Y.-B., Wang, L.-J., and Chen, X.-Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648. doi: 10.1105/tpc.112.098749
Hsiao, Y. Y., Jeng, M. F., Tsai, W. C., Chuang, Y. C., Li, C. Y., Wu, T. S., et al. (2008). A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD (X) 2-4D motif. Plant J. 55, 719–733. doi: 10.1111/j.1365-313X.2008.03547.x
Hsiao, Y.-Y., Pan, Z.-J., Hsu, C.-C., Yang, Y.-P., Hsu, Y.-C., Chuang, Y.-C., et al. (2011a). Research on orchid biology and biotechnology. Plant Cell Physiol. 52, 1467–1486. doi: 10.1093/pcp/pcr100
Hsiao, Y. Y., Tsai, W. C., Chen, W. H., and Chen, H. H. (2011b). “Biosynthetic regulation of floral scent in Phalaenopsis,” in Orchid Biotechnology II, eds W. H. Chen and H. H. Chen (Singapore: World Scientific), 145–180.
Hsiao, Y. Y., Tsai, W. C., Kuoh, C. S., Huang, T. H., Wang, H. C., Wu, T. S., et al. (2006). Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biol. 6:14. doi: 10.1186/1471-2229-6-14
Hsieh, M.-H., Pan, Z.-J., Lai, P.-H., Lu, H.-C., Yeh, H.-H., Hsu, C.-C., et al. (2013). Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J. Exp. Bot. 64, 3869–3884. doi: 10.1093/jxb/ert218
Hsu, C.-C., Chen, Y.-Y., Tsai, W.-C., Chen, W.-H., and Chen, H.-H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Hsu, C. C., Wu, P. S., Chen, T. C., Yu, C. W., Tsai, W. C., Wu, K., et al. (2014). Histone acetylation accompanied with promoter sequences displaying differential expression profiles of B-class MADS-box genes for Phalaenopsis floral morphogenesis. PLoS One 9:e106033. doi: 10.1371/journal.pone.0106033
Hua, W., Song, J., Li, C., and Wang, Z. (2012). Molecular cloning and characterization of the promoter of SmGGPPs and its expression pattern in Salvia miltiorrhiza. Mol. Biol. Rep. 39, 5775–5783. doi: 10.1007/s11033-011-1388-8
Huang, X. Z., Xiao, Y. T., Köllner, T. G., Jing, W. X., Kou, J. F., Chen, J. Y., et al. (2018). The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defense responses against herbivores. Plant Cell Environ. 41, 261–274. doi: 10.1111/pce.13088
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jin, Q., Waters, D., Cordeiro, G. M., Henry, R. J., and Reinke, R. F. (2003). A single nucleotide polymorphism (SNP) marker linked to the fragrance gene in rice (Oryza sativa L.). Plant Sci. 165, 359–364. doi: 10.1016/S0168-9452(03)00195-X
Jun, M., Jeong, W., and Ho, C. (2006). Health promoting properties of natural flavor substances. Food Sci. Biotechnol. 15, 329–338.
Jurka, J. (1998). Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 8, 333–337. doi: 10.1016/S0959-440X(98)80067-5
Juwattanasomran, R., Somta, P., Chankaew, S., Shimizu, T., Wongpornchai, S., Kaga, A., et al. (2011). A SNP in GmBADH2 gene associates with fragrance in vegetable soybean variety “Kaori” and SNAP marker development for the fragrance. Theor. Appl. Genet. 122, 533–541. doi: 10.1007/s00122-010-1467-6
Juwattanasomran, R., Somta, P., Kaga, A., Chankaew, S., Shimizu, T., Sorajjapinun, W., et al. (2012). Identification of a new fragrance allele in soybean and development of its functional marker. Mol. Breed. 29, 13–21. doi: 10.1007/s11032-010-9523-0
Kaiser, R. (1993). The Scent of Orchids: Olfactory and Chemical Investigations. Amsterdam: Elsevier. doi: 10.1021/bk-1993-0525.ch018
Knudsen, J. T., and Gershenzon, J. (2006). “The chemical diversity of floral scent,” in Biology of Floral Scent, eds N. Dudareva, and E. Pichersky (Boca Raton, FL: CRC Press), 27–52.
Kumar, S., and Bhatia, S. (2016). A polymorphic (GA/CT) n-SSR influences promoter activity of Tryptophan decarboxylase gene in Catharanthus roseus L. Don. Sci. Rep. 6:33280. doi: 10.1038/srep33280
Li, S., Wang, H., Li, F., Chen, Z., Li, X., Zhu, L., et al. (2015). The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 84, 296–308. doi: 10.1111/tpj.12994
Li, X., Xu, Y., Shen, S., Yin, X., Klee, H., Zhang, B., et al. (2017). Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 68, 4929–4938. doi: 10.1093/jxb/erx316
Liao, Y., Xu, F., Huang, X., Zhang, W., Cheng, H., Wang, X., et al. (2016). Characterization and transcriptional profiling of Ginkgo biloba mevalonate diphosphate decarboxylase gene (GbMVD) promoter towards light and exogenous hormone treatments. Plant Mol. Biol. Rep. 34, 566–581. doi: 10.1007/s11105-015-0947-x
Lu, H.-C., Chen, H.-H., Tsai, W.-C., Chen, W.-H., Su, H.-J., Chang, D. C.-N., et al. (2007). Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 143, 558–569. doi: 10.1104/pp.106.092742
Lu, S., Zhang, Y., Zheng, X., Zhu, K., Xu, Q., and Deng, X. (2016). Isolation and functional characterization of a lycopene β-cyclase gene promoter from citrus. Front. Plant Sci. 7:1367. doi: 10.3389/fpls.2016.01367
Marei, G. I. K., Rasoul, M. A. A., and Abdelgaleil, S. A. (2012). Comparative antifungal activities and biochemical effects of monoterpenes on plant pathogenic fungi. Pestic. Biochem. Physiol. 103, 56–61. doi: 10.1016/j.pestbp.2012.03.004
Mirzaei-Najafgholi, H., Tarighi, S., Golmohammadi, M., and Taheri, P. (2017). The effect of citrus essential oils and their constituents on growth of Xanthomonas citri subsp. citri. Molecules 22:E591. doi: 10.3390/molecules22040591
Mitsuda, N., Ikeda, M., Takada, S., Takiguchi, Y., Kondou, Y., Yoshizumi, T., et al. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51, 2145–2151. doi: 10.1093/pcp/pcq161
Mumm, R., Posthumus, M. A., and Dicke, M. (2008). Significance of terpenoids in induced indirect plant defence against herbivorous arthropods. Plant Cell Environ. 31, 575–585. doi: 10.1111/j.1365-3040.2008.01783.x
Murthy, K. N. C., Jayaprakasha, G. K., and Patil, B. S. (2012). D-limonene rich volatile oil from blood oranges inhibits angiogenesis, metastasis and cell death in human colon cancer cells. Life Sci. 91, 429–439. doi: 10.1016/j.lfs.2012.08.016
Myint, K. M., Arikit, S., Wanchana, S., Yoshihashi, T., Choowongkomon, K., and Vanavichit, A. (2012). A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor. Appl. Genet. 125, 887–896. doi: 10.1007/s00122-012-1880-0
Nagegowda, D. A., Rhodes, D., and Dudareva, N. (2010). “The role of the methyl-erythritol-phosphate (MEP) pathway in rhythmic emission of volatiles,” in The Chloroplast: Basics and Application, eds C. A. Rebeiz, C. Benning, H. J. Bohnert, H. Daniell, J. K. Hoober, H. K. Lichtenthaler et al. (Dordrecht: Springer), 139–154.
Nakayama, K., Murata, S., Ito, H., Iwasaki, K., Villareal, M. O., Zheng, Y. W., et al. (2017). Terpinen-4-ol inhibits colorectal cancer growth via reactive oxygen species. Oncol. Lett. 14, 2015–2024. doi: 10.3892/ol.2017.6370
Nieuwenhuizen, N. J., Chen, X., Wang, M. Y., Matich, A. J., Perez, R. L., Allan, A. C., et al. (2015). Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 167, 1243–1258. doi: 10.1104/pp.114.254367
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C., et al. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202, 1024–1042. doi: 10.1111/nph.12723
Pan, Z. J., Cheng, C. C., Tsai, W. C., Chung, M. C., Chen, W. H., Hu, J. M., et al. (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 52, 1515–1531. doi: 10.1093/pcp/pcr092
Paré, P. W., and Tumlinson, J. H. (1999). Plant volatiles as a defense against insect herbivores. Plant Physiol. 121, 325–332. doi: 10.1104/pp.121.2.325
Pelley, J. W. (2007). “Organization, synthesis, and repair of DNA,” in Elsevier’s Integrated Biochemistry, ed. J. W. Pelley (Philadelphia, PA: Mosby).
Pramnoi, P., Somta, P., Chankaew, S., Juwattanasomran, R., and Srinives, P. (2013). A single recessive gene controls fragrance in cucumber (Cucumis sativus L.). J. Genet. 92, 147–149. doi: 10.1007/s12041-013-0228-0
Pyo, H., Demura, T., and Fukuda, H. (2006). Vascular cell expression patterns of Arabidopsis bZIP group I genes. Plant Biotechnol. 23, 497–501. doi: 10.5511/plantbiotechnology.23.497
Ruangnam, S., Wanchana, S., Phoka, N., Saeansuk, C., Mahatheeranont, S., de Hoop, S. J., et al. (2017). A deletion of the gene encoding amino aldehyde dehydrogenase enhances the “pandan-like” aroma of winter melon (Benincasa hispida) and is a functional marker for the development of the aroma. Theor. Appl. Genet. 130, 2557–2565. doi: 10.1007/s00122-017-2976-3
Sakthivel, K., Rani, N. S., Pandey, M. K., Sivaranjani, A., Neeraja, C., Balachandran, S., et al. (2009). Development of a simple functional marker for fragrance in rice and its validation in Indian Basmati and non-Basmati fragrant rice varieties. Mol. Breed. 24, 185–190. doi: 10.1007/s11032-009-9283-x
Sas, C., Müller, F., Kappel, C., Kent, T. V., Wright, S. I., Hilker, M., et al. (2016). Repeated inactivation of the first committed enzyme underlies the loss of benzaldehyde emission after the selfing transition in Capsella. Curr. Biol. 26, 3313–3319. doi: 10.1016/j.cub.2016.10.026
Schwab, W., Davidovich-Rikanati, R., and Lewinsohn, E. (2008). Biosynthesis of plant-derived flavor compounds. Plant J. 54, 712–732. doi: 10.1111/j.1365-313X.2008.03446.x
Shi, W., Yang, Y., Chen, S., and Xu, M. (2008). Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 22, 185–192. doi: 10.1007/s11032-008-9165-7
Su, C.-L., Chao, Y.-T., Yen, S.-H., Chen, C.-Y., Chen, W.-C., Chang, Y.-C. A., et al. (2013a). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54:e11. doi: 10.1093/pcp/pct004
Su, C.-L., Chen, W.-C., Lee, A.-Y., Chen, C.-Y., Chang, Y.-C. A., Chao, Y.-T., et al. (2013b). A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS One 8:e80462. doi: 10.1371/journal.pone.0080462
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30,2725–2729. doi: 10.1093/molbev/mst197
Tholl, D. (2015). “Biosynthesis and biological functions of terpenoids in plants,” in Biotechnology of Isoprenoids, eds J. Schrader, and J. Bohlmann (Cham: Springer), 63–106.
Tian, J., Li, K.-T., Zhang, S.-Y., Zhang, J., Song, T.-T., Zhu, Y.-J., et al. (2017). The structure and methylation level of the McMYB10 promoter determine the leaf color of Malus Crabapple. HortScience 52, 520–526. doi: 10.21273/HORTSCI11563-16
Tsai, C., Chiang, Y., Huang, S., Chen, C., and Chou, C. (2010). Molecular phylogeny of Phalaenopsis Blume (Orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst. Evol. 288, 77–98. doi: 10.1007/s00606-010-0314-1
Tsai, C. C. (2003). Molecular phylogeny, biogeography, and evolutionary trends of the genus Phalaenopsis (Orchidaceae). Ph.D. thesis, National Sun Yat-sen University, Kaohsiung.
Tsai, C.-C. (2011). “Molecular phylogeny and biogeography of Phalaenopsis species,” in Orchid Biotechnology II, eds W.-H. Chen, and H. H. Chen (Singapore: World Scientific), 1–24.
Vainstein, A., Lewinsohn, E., Pichersky, E., and Weiss, D. (2001). Floral fragrance. New inroads into an old commodity. Plant Physiol. 127, 1383–1389. doi: 10.1104/pp.010706
Vongvanrungruang, A., Mongkolsiriwatana, C., Boonkaew, T., Sawatdichaikul, O., Srikulnath, K., and Peyachoknagul, S. (2016). Single base substitution causing the fragrant phenotype and development of a type-specific marker in aromatic coconut (Cocos nucifera). Genet. Mol. Res. 15. doi: 10.4238/gmr.15038748
Wang, H., Han, J., Kanagarajan, S., Lundgren, A., and Brodelius, P. E. (2013). Studies on the expression of sesquiterpene synthases using promoter-β-Glucuronidase fusions in transgenic Artemisia annua L. PLoS One 8:e80643. doi: 10.1371/journal.pone.0080643
Wang, H., Kanagarajan, S., Han, J., Hao, M., Yang, Y., Lundgren, A., et al. (2014). Studies on the expression of linalool synthase using a promoter-β-glucuronidase fusion in transgenic Artemisia annua. J. Plant Physiol. 171, 85–96. doi: 10.1016/j.jplph.2013.09.019
Xu, Y.-H., Wang, J.-W., Wang, S., Wang, J.-Y., and Chen, X.-Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yamasaki, Y., Kunoh, H., Yamamoto, H., and Akimitsu, K. (2007). Biological roles of monoterpene volatiles derived from rough lemon (Citrus jambhiri Lush) in citrus defense. J. Gen. Plant Pathol. 73, 168–179. doi: 10.1007/s10327-007-0013-0
Yeh, Y.-C., Tsay, Y.-S., Shih, C.-T., and Huang, P. (2014). Selection and breeding of scented Phalaenopsis varieties. Agric. Policy Agric. Situation267, 97.
Yundaeng, C., Somta, P., Tangphatsornruang, S., Chankaew, S., and Srinives, P. (2015). A single base substitution in BADH/AMADH is responsible for fragrance in cucumber (Cucumis sativus L.), and development of SNAP markers for the fragrance. Theor. Appl. Genet. 128, 1881–1892. doi: 10.1007/s00122-015-2554-5
Keywords: cis-element, floral scent, monoterpene, orchid, Phalaenopsis, promoter, transcription factor, yeast-one hybrid screening
Citation: Chuang Y-C, Hung Y-C, Hsu C-Y, Yeh C-M, Mitsuda N, Ohme-Takagi M, Tsai W-C, Chen W-H and Chen H-H (2018) A Dual Repeat Cis-Element Determines Expression of GERANYL DIPHOSPHATE SYNTHASE for Monoterpene Production in Phalaenopsis Orchids. Front. Plant Sci. 9:765. doi: 10.3389/fpls.2018.00765
Received: 09 March 2018; Accepted: 17 May 2018;
Published: 05 June 2018.
Edited by:
Swee-Suak Ko, Academia Sinica, TaiwanReviewed by:
Dinesh A. Nagegowda, Central Institute of Medicinal and Aromatic Plants (CIMAP), IndiaTariq Akhtar, University of Guelph, Canada
Copyright © 2018 Chuang, Hung, Hsu, Yeh, Mitsuda, Ohme-Takagi, Tsai, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Hwa Chen, aGhjaGVuQG1haWwubmNrdS5lZHUudHc=
 Yu-Chen Chuang
Yu-Chen Chuang Yi-Chu Hung1
Yi-Chu Hung1 Chuan-Ming Yeh
Chuan-Ming Yeh Nobutaka Mitsuda
Nobutaka Mitsuda Masaru Ohme-Takagi
Masaru Ohme-Takagi Wen-Chieh Tsai
Wen-Chieh Tsai Wen-Huei Chen
Wen-Huei Chen Hong-Hwa Chen
Hong-Hwa Chen