
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 23 May 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00668
This article is part of the Research TopicAntagonistic Interactions Between Plant Hormones Abscisic Acid (ABA) and Gibberellins (GAs)View all 5 articles
Seed dormancy is an adaptive trait that does not allow the germination of an intact viable seed under favorable environmental conditions. Non-dormant seeds or seeds with low level of dormancy can germinate readily under optimal environmental conditions, and such a trait leads to preharvest sprouting, germination of seeds on the mother plant prior to harvest, which significantly reduces the yield and quality of cereal crops. High level of dormancy, on the other hand, may lead to non-uniform germination and seedling establishment. Therefore, intermediate dormancy is considered to be a desirable trait as it prevents the problems of sprouting and allows uniformity of postharvest germination of seeds. Induction, maintenance, and release of seed dormancy are complex physiological processes that are influenced by a wide range of endogenous and environmental factors. Plant hormones, mainly abscisic acid (ABA) and gibberellin (GA), are the major endogenous factors that act antagonistically in the control of seed dormancy and germination; ABA positively regulates the induction and maintenance of dormancy, while GA enhances germination. Significant progress has been made in recent years in the elucidation of molecular mechanisms regulating ABA/GA balance and thereby dormancy and germination in cereal seeds, and this review summarizes the current state of knowledge on the topic.
Cereals are among the most economically important crops worldwide with annual production of over 2500 million tons (Food and Agriculture Organization of the United Nations, 2017). However, their production is challenged by a wide range of biotic and abiotic stress factors including the occurrence of high humidity and wet conditions prior to harvest that causes germination of the grain on the spike, which is also referred to as preharvest sprouting. Grain yield and end use quality losses due to preharvest sprouting are reported to cause an annual loss of around $1 billion worldwide (Black et al., 2006). Preharvest sprouting of cereal grains is closely associated with the degree of dormancy, an adaptive trait that inhibits the germination of seeds under optimal environmental conditions (Gao and Ayele, 2014). However, domestication and selection of cereal crops from their wild relatives have been focused on ensuring uniform germination and seedling establishment, and this has led to the development of modern cultivars with low level of dormancy (Lenser and Theißen, 2013; Meyer and Purugganan, 2013; Gao and Ayele, 2014), making the seeds susceptible to field sprouting when moist and wet conditions occur before harvest. Since high level of dormancy in cereal seeds can also cause undesirable consequences such as uneven and slow postharvest germination of seeds, optimum level of seed dormancy is always required to enhance the yield and quality of cereal crops (Gao and Ayele, 2014).
Loss of seed dormancy can be induced by different treatments, including after-ripening, cold stratification, and light (Bewley and Black, 1994). Dormancy loss in the seeds of cereals such as wheat has been shown to be associated with changes in the physiological state of the seed, which involves alterations in gene and protein expression, oxidative modification of gene transcripts and proteins, and epigenetic modifications (Gao et al., 2012, 2013; Gao and Ayele, 2014). Previous studies have also provided important insights into the significance of the metabolic and signaling aspects of different plant hormones and their potential interaction in the maintenance and release of dormancy in cereal seeds (Liu et al., 2013; Chitnis et al., 2014; Shu et al., 2016b). Among the different plant hormones, abscisic acid (ABA) and gibberellin (GA) are considered as major players in the regulation of dormancy and germination; ABA regulates dormancy induction and maintenance positively while GA promotes seed germination (Kucera et al., 2005; Finkelstein et al., 2008). Therefore, changes in the balance of seed ABA/GA levels and sensitivity constitute a central regulatory mechanism underlying the maintenance and release of seed dormancy (Shu et al., 2016b; Finch-Savage and Footitt, 2017). Previous studies have indicated that change in the ABA/GA balance is regulated at least partly by the reciprocal regulation of the expression of genes involved in ABA and GA metabolism and signaling (Seo et al., 2006; Piskurewicz et al., 2008; Izydorczyk et al., 2017). Other endogenous signaling factors such as reactive oxygen species (ROS) and environmental factors such as temperature and light can also influence the balance between ABA and GA, and therefore dormancy and germination in cereal seeds (Gubler et al., 2008; Ishibashi et al., 2015; Izydorczyk et al., 2017).
Recent progress in the genomics of cereal crops has led to the identification of genes involved in ABA and GA metabolism and signaling pathways, and those mediating ABA-GA interactions in cereal crops, and this has opened up new opportunities for elucidating the molecular mechanisms underlying the roles of ABA and GA in the regulation of dormancy and germination in cereal seeds. This review highlights the recent advances made in this regard.
Abscisic acid is one of the major plant hormones involved in many aspects of plant growth and developmental processes including seed dormancy and germination. The level of ABA in plant tissues/seeds is regulated by its biosynthesis and catabolism (Nambara and Marion-Poll, 2005; Nambara et al., 2010). Of the several steps involved in the biosynthesis of ABA, oxidative cleavage of 9-cis-neoxanthin and violaxanthin by 9-cis-epoxycarotenoid dioxygenase (NCED) is reported to be the rate-limiting step (Schwartz et al., 2003) while ABA catabolism mainly takes place through hydroxylation of ABA at the 8′ position by the action of ABA 8′-hydroxylase (ABA8′OH) (Cutler and Krochko, 1999), which is encoded by CYP707A genes (Kushiro et al., 2004; Saito et al., 2004) (Figure 1). Therefore, the expressions of NCED and CYP707A genes play significant roles in the control of seed ABA level, and therefore dormancy and germination.
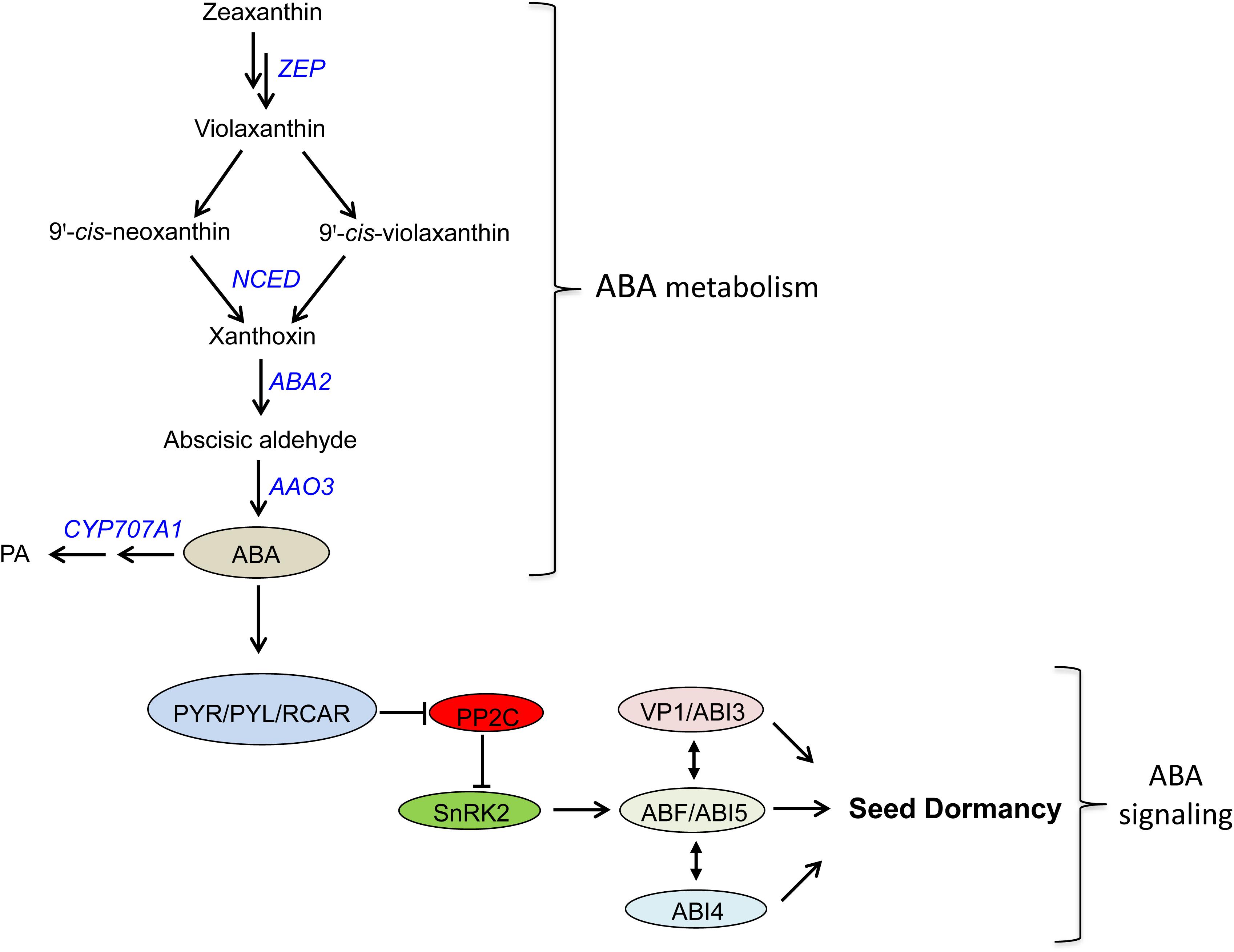
FIGURE 1. Abscisic acid metabolism and signaling pathway in plants. ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; ABA2, ABA deficient 2 (short chain alcohol dehydrogenase); AAO3, abscisic aldehyde oxidase; CYP707A1, a cytochrome P450 monooxygenase gene encoding ABA-8′-hydroxylases; PA, phaseic acid; PYR/PYL/RCAR, pyrabactin resistance/pyrabactin-like/regulatory components of ABA receptors; PP2C, protein phosphatase 2C; SnRK2, SNF1-related protein kinase2; ABI3, abscisic acid insensitive 3; ABI4, abscisic acid insensitive 4; ABI5, abscisic acid insensitive 5; VP1, viviparous 1; ABF, ABRE binding factor.
Seed development in dicot species such as Arabidopsis is characterized by two peaks of ABA accumulation that occur during the mid- and late-phases of seed maturation; ABA in the latter peak, which is mainly synthesized in the zygotic tissues (Kanno et al., 2010), plays a key role in the induction and maintenance of seed dormancy (Kermode, 2005; Nambara et al., 2010). Similarly, two peaks of embryonic ABA level have been detected during wheat seed development (King, 1976; Suzuki et al., 2000). The first peak of ABA occurs at about 25 days after pollination (DAP) while the second peak occurs at about 35 DAP. However, the second ABA peak appears to occur for an extended duration, that is, up to 40 DAP in the seeds of dormant wheat line as compared to the non-dormant line (Suzuki et al., 2000), suggesting the involvement of ABA synthesized during the late phase of seed maturation for the establishment of seed dormancy. Furthermore, maintenance of elevated embryonic ABA level for prolonged duration during the late-maturation phase of wheat seed development has been reported to be associated with the higher level of dormancy (Walker-Simmons and Sesing, 1990) while maturing seeds (at about 30 DAP) of a non-dormant mutant line of wheat have been shown to exhibit a significantly lower level of embryonic ABA than that observed in the dormant line from which the mutant is generated (Kawakami et al., 1997). In agreement with this, mutations in the two homolog of TaABA8′OH1 (TaABA8′OH1A and TaABA8′OH1D) that cause an increase in embryonic ABA content during the middle and later stages of seed development (40–60 DAA) have been shown to result in an enhanced degree of seed dormancy (Chono et al., 2013), reinforcing the significance of elevated embryonic ABA level during the maturation phase of seed development in the induction of dormancy in wheat seeds. Although these studies provided important insights into the role of ABA in dormancy induction in wheat seeds, detailed understanding of the molecular mechanisms underlying the regulation of ABA metabolism during seed maturation and therefore dormancy induction awaits further study.
A study in rice seeds, however, implicated that the accumulation of ABA involved in the induction of dormancy occurs at the earlier stage of seed development; rice cultivars with deep dormancy exhibit higher seed ABA level during the early and middle stage of seed development (10–20 DAP) than that observed in cultivars with medium and low levels of dormancy (Liu et al., 2014). In addition, ABA accumulation during seed development in weedy red rice has been shown to peak at 10 DAP; however, seeds of dormant lines contain over twofold more ABA than that observed in non-dormant lines and this variation in seed ABA content appeared to be associated at least partly with the expression patterns of NCED1 paralogs (Gu et al., 2011). In barley, developing seeds of both dormant and non-dormant cultivars have been shown to exhibit a high amount of embryonic ABA through the physiological maturity phase after which embryonic ABA level declined substantially in the non-dormant but not in the dormant cultivar (Benech-Arnold et al., 1999). Analysis of the expression patterns of ABA metabolic genes in developing barley seeds revealed that HvNCED2 regulates ABA level during the early- to mid-phases of seed maturation while HvABA8′OH1 (a barley homolog of CYP707A) is responsible for controlling seed ABA level thereafter (Chono et al., 2006). These results suggest the importance of ABA catabolism in controlling seed ABA level and dormancy induction in maturing barley seeds. Furthermore, seed development in a triticale cultivar that is less susceptible to preharvest sprouting has been shown to be characterized by higher TsNCED1 expression and ABA level than that observed in a cultivar that is more sensitive to preharvest sprouting, suggesting the role of ABA biosynthesis in the regulation of dormancy establishment in cereal seeds (Fidler et al., 2016).
Seed dormancy is partly determined by the level of ABA contained in mature seeds (Finkelstein et al., 2008; Nambara et al., 2010). Previous studies have shown that dormancy loss and germination of cereal seeds are associated with a decrease in seed ABA level during imbibition, and the change in seed ABA level appears to be related with the expression patterns of NCED and CYP707A genes. For example, after-ripening induces loss of seed dormancy in barley, and this is mediated by imbibition-induced reduction in seed ABA content to a level lower than that observed in the corresponding dormant seeds. This difference in ABA content and therefore level of dormancy is reported to be associated mainly with the expression pattern of HvABA8′OH1 (Millar et al., 2006; Gubler et al., 2008). Consistently, RNAi-mediated knock-down of HvABA8′OH1 leads to enhanced seed ABA level and dormancy (Gubler et al., 2008). Studies in rice also indicated that the reduction of ABA level during imbibition of non-dormant seeds of rice is related mainly with increased expression of OsABA8ox (rice homologs of CYP707A) genes (Zhu et al., 2009; Du et al., 2015). These results suggest the significance of ABA catabolism in the regulation of ABA level and seed dormancy maintenance and release in the seeds of cereals such as rice and barley.
Other studies in barley and Brachypodium have also demonstrated the importance of ABA biosynthesis and/or catabolism in the regulation of seed dormancy. For example, after-ripening of dormant Brachypodium seeds leads to a decrease in the expression of BdNCED1 along with a transient induction in the expression of BdABA8′OH1 (a Brachypodium homolog of CYP707A) during imbibition, resulting in a reduced ABA level and enhanced seed germination (Barrero et al., 2012). Furthermore, imbibition of dormant seeds of barley under white light is reported to enhance the expression of HvNCED1 with no marked effect on that of HvABA8′OH1, leading to increased embryonic ABA content and inhibition of seed germination (Gubler et al., 2008). White light has also been reported to promote dormancy in both dormant and non-dormant seeds of Brachypodium, although this effect does not appear to be closely associated with changes in ABA metabolism/seed ABA level (Barrero et al., 2012). Exposure of partially after-ripened barley seeds to blue light has also been shown to induce the expression of HvNCED1 and repress that of HvABA8′OH1 in the embryo, resulting in an increase in embryonic ABA level and suppression of germination (Barrero et al., 2014). Blue light also induces secondary dormancy in barley seeds, and this appears to be associated with an increase in embryonic ABA content, which is mediated by enhanced expression of HvNCED1 and HvNCED2, and decreased expression of HvABA8′OH1 (Hoang et al., 2014). Recent progress in the molecular aspects of ABA metabolism and dormancy regulation in wheat seeds is highlighted in the following section.
Similar to that observed in the seeds of other cereal crops, after-ripening of dormant wheat seeds leads to imbibition-mediated reduction of ABA level, while maintenance of elevated ABA level was evident in the corresponding dormant seeds, and these differences in ABA level are mediated by changes in the expression patterns of TaNCED1 and TaABA8′OH1 (a wheat homolog of CYP707A) genes (Jacobsen et al., 2013), implying the contribution of both ABA biosynthesis and catabolism in the regulation of seed ABA level and dormancy in wheat. Furthermore, seeds from a dormant wheat genotype have been shown to exhibit higher and lower expression of TaNCED2 and TaABA8′OH1, respectively, in both dry and imbibed states than those derived from a non-dormant genotype (Son et al., 2016). Consistently, ectopic expression of TaNCED2A (a homeolog of TaNCED2 from the A genome of wheat) and TaABA8′OH1B (a homeolog of TaABA8′OH1 from the B genome of wheat) in Arabidopsis caused alteration of seed ABA level and dormancy (Son et al., 2016), and a double mutation in the A and D genome copies of TaABA8′OH1 resulted in an increase in embryonic ABA level and a decrease in seed germination (Chono et al., 2013). Other treatments that lead to dormancy loss in wheat seeds also affect the expression patterns of ABA metabolic genes and ABA level. For example, dormancy loss due to imbibition of wheat seeds at lower temperature (15°C) decreases ABA level in the embryos via increased expression of TaABA8′OHs (Kashiwakura et al., 2016). Furthermore, after-ripening and cold stratification mediated dormancy loss in wheat seeds is reported to be associated with decreased ABA levels in the embryos and aleurone (Tuttle et al., 2015). Moreover, the effect of different seed imbibition temperature regimes on wheat seed germination is reported to be mediated by changes in the expressions of TaNCED and TaABA8′OH genes, and seed ABA level (Izydorczyk et al., 2017). For example, delay in the germination of wheat seeds due to imbibition under supraoptimal temperature (35°C) is associated with elevated embryonic ABA level, which is mediated via enhanced expression of TaNCEDs; whereas inhibition of germination by suboptimal temperature (4°C) appears to be caused by increased ABA level in both embryo and endosperm tissues, which is regulated by increased and decreased expression of TaNCEDs and TaABA8′OHs, respectively, in both tissues (Izydorczyk et al., 2017).
ABA signaling involves three major core components; PYRABACTIN RESISTANCE1/PYRABACTIN-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR), PROTEIN PHOSPHATASE 2Cs (PP2Cs; negative regulators), and SNF1-RELATED PROTEIN KINASE 2s (SnRK2s; positive regulators) (Figure 1). In the absence of ABA, PP2Cs inhibits the activities of SnRK2s through dephosphorylation of their kinase activation loop; however, when ABA is present, the ABA receptors PYR/PYL/RCAR form a complex with PP2C, and this inhibits the phosphatase activity of PP2C and thereby activate SnRK2 (Finkelstein, 2013; Ng et al., 2014). The activated form of SnRK2 subsequently turns on ABRE-binding protein/ABRE-binding factor (AREB/ABF) transcription factors, which in turn activates the transcription of ABA responsive genes (Ng et al., 2014). Among AREB/ABF transcription factors, ABA insensitive 5 (ABI5), a member of the basic leucine zipper transcription factor family, plays a central role in regulating ABA-responsive genes in seeds (Nambara et al., 2010; Finkelstein, 2013). In addition, ABI4 and ABI3, AP2-type and B3-type transcription factors, respectively, have been reported to function along with ABI5 to induce the expression of ABA responsive genes, and thereby regulate seed dormancy and germination (Figure 1).
Dormancy maintenance in cereal seeds is also determined by seed sensitivity to ABA (Walker-Simmons, 1987; Steinbach et al., 1995; Tuttle et al., 2015), which is regulated by the expression of genes involved in the ABA signaling pathway including those encoding the three central components (PYR/PYL/RCAR5, PP2C, and SnRK2) and the downstream transcription factors including ABI3, ABI4, and ABI5 (Nambara et al., 2010). Previous studies have demonstrated that mutants of ABI3, ABI4, and ABI5 exhibit ABA resistant germination phenotype, and these transcription factors appear to interact extensively (Söderman et al., 2000) and control the expression of ABA responsive genes that are involved in the regulation of seed dormancy/germination, including those involved in ABA catabolism such as CYP707A1 and CYP707A2 (Shu et al., 2013), GA catabolism such as GA2ox3 (Cantoro et al., 2013), and starch degradation such as α-amylase (Hoecker et al., 1995) genes. One of the first key ABA signaling components identified and functionally characterized with respect to seed dormancy in cereals is the maize Viviparous1 (Vp1) gene, an ortholog of the ABI3 of Arabidopsis (McCarty et al., 1991). Embryos of maize seeds that exhibit varying degree of resistance to preharvest sprouting have been reported to exhibit temporal difference in the expression of Vp1 during embryogenesis (Carrari et al., 2001), suggesting its role in the control of dormancy. Consistently, mutations in Vp1 lead to disruption of embryo maturation and induction of its germination while still on the cob (McCarty et al., 1991; Hoecker et al., 1995). On the other hand, ectopic expression of the maize Vp1 gene in wheat results in increased level of seed dormancy and tolerance to preharvest sprouting (Huang et al., 2012). It has been shown previously in pea that alternative splicing of the ABI3 homolog results in the formation of non-functional/truncated protein products from mis-spliced transcripts (Gagete et al., 2009), and such an event has been observed in rice and wheat genotypes exhibiting early dormancy release (McKibbin et al., 2002; Fan et al., 2007). Furthermore, the level of dormancy in wheat seeds has been shown to be associated with the expression pattern of Vp1 (Nakamura and Toyama, 2001; Laethauwer et al., 2012).
Previous reports have also implicated other ABA signaling components in the regulation of dormancy and germination in cereal seeds. For example, seeds of rice plants overexpressing PYL/RCAR5 exhibit hypersensitivity to ABA and delayed germination phenotype (Kim et al., 2012). Moreover, immature seeds (30 DAP, before physiological maturity) from a dormant line of sorghum exhibit transcriptional induction of PKABA1 (SnRK2 ortholog), ABI3, ABI4, and ABI5, and enhanced expression of ABI5 protein during imbibition as compared to those derived from the less-dormant line (Rodríguez et al., 2009). A recent transcriptomic analysis of maturing seeds from dormant and non-dormant genotypes of wheat revealed the enrichment of several ABRE motifs, which act as binding sites for ABI5, in the embryo and endosperm gene co-expression clusters of both genotypes, of which G-box-like motif is specifically enriched in both tissues of seeds derived from the dormant genotype, suggesting importance of ABI5 in the control dormancy establishment is mediated by specific ABRE motifs (Yamasaki et al., 2017). Increased expression of TaABF1, a wheat ortholog of ABI5, has also been reported during imbibition of dormant wheat grains (Johnson et al., 2002). On the other hand, dormancy release due to after-ripening in wheat and barley seeds is associated with transcriptional repression of SnRK2, ABI5, and ABI3-interacting protein2 (AIP2) during imbibition (Barrero et al., 2009; Liu et al., 2013), suggesting a decrease in seed ABA sensitivity. A recent study has also elucidated the role of ABI4 in regulating ABA sensitivity and thereby dormancy and germination in Arabidopsis seed (Shu et al., 2013); however, the physiological role of this ABA signaling components has yet to be determined in cereal seeds.
Environmental factors such as temperature also modulate the expression of ABA signaling genes and thereby ABA sensitivity of wheat seeds during imbibition. It has been shown recently that imbibition of non-dormant wheat seeds under supraoptimal temperature leads to delay in germination via inducing the expression of PYL5, SnRK2, ABI3, and ABI5 genes in the embryo tissue and thereby enhancing its sensitivity to ABA. Similarly, the inhibitory effect of suboptimal imbibition temperature on germination appears to be mediated by increased ABA sensitivity of both embryo and endosperm tissues through transcriptional activation of the ABA signaling genes; PYL5, SnRK2, ABI3, and ABI5 in the embryo, and SnRK2 and ABI5 in the endosperm (Izydorczyk et al., 2017).
Gibberellin is the other major phytohormone with an important role in the regulation of seed dormancy and germination (Finch-Savage and Leubner-Metzger, 2006). The level of biologically active GAs in plant tissues is determined by the balance between its biosynthesis and inactivation (Yamaguchi, 2008). The biosynthesis of GA is regulated mainly by reactions catalyzed by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) while its inactivation is controlled primarily by GA 2-oxidase (GA2ox) (Figure 2). Genes encoding these enzymes have been identified from several crop species including cereals such as rice, barley, and wheat (Spielmeyer et al., 2004; Yamaguchi, 2008; Pearce et al., 2015), and their expression play important roles in regulating seed GA level, and therefore dormancy and germination.
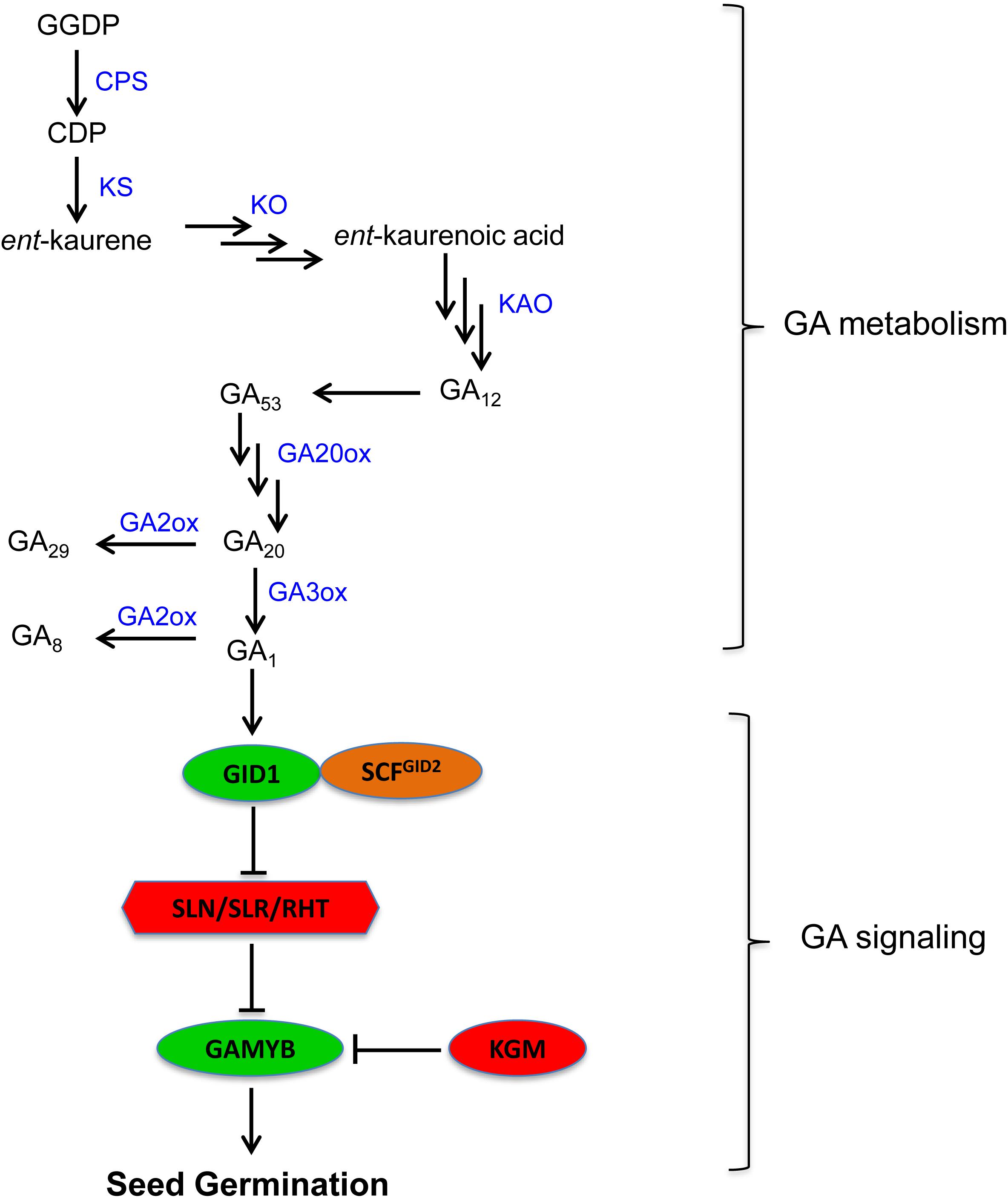
FIGURE 2. Gibberellin metabolism and signaling pathway in plants (GA1 is the major bioactive GA in seeds of cereals such as wheat). GGDP, geranylgeranyl diphosphate; CDP, ent-copalyl diphosphate; CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase; GA20ox, GA20 oxidase; GA3ox, GA3 oxidase; GA2ox, GA2 oxidase; GID1, gibberellin insensitive dwarf 1; GID2, gibberellin insensitive dwarf 2; SLN, slender1 in barley; SLR1, slender rice1; RHT, reduced height; GAMYB, GA regulated MYB transcriptional regulator; KGM, kinase associated with GAMYB.
Several genetic and physiological analyses of the seeds of dicot species such as Arabidopsis and tomato have demonstrated the requirement of GA for germination; GA promotes germination through enhancing the growth potential of the embryo and overcoming the mechanical barriers imposed by covering layers surrounding embryo (Debeaujon and Koornneef, 2000; Kucera et al., 2005). With respect to cereals, reports that implicate GA in the control of seed dormancy and germination are mainly based on comparative analysis of dormant and non-dormant seeds. For example, dormancy loss in wheat and barley seeds due to after-ripening has been shown to be associated with enhanced expression of the TaGA20ox and TaGA3ox genes and increased level of bioactive GA1 during imbibition (Gubler et al., 2008; Liu et al., 2013; Kashiwakura et al., 2016). Consistently, comparative genomic analysis among barley, wheat, and rice identified HvGA20ox as a candidate gene regulating a seed dormancy QTL in barley (Li et al., 2004). Likewise, the expression of the GA biosynthetic genes (GA20ox and GA3ox2) is induced while that of GA inactivating genes (GA2ox) is repressed during imbibition of non-dormant rice and sorghum seeds, and this has been shown to be associated with increased level of bioactive GA (Kaneko et al., 2002; Rodríguez et al., 2012; Du et al., 2015). Recent genetic studies in rice have identified OsGA20ox2 and OsGA2ox3 as candidate genes for controlling seed germination (Ye et al., 2015; Magwa et al., 2016), and loss of function mutation in OsGA20ox2 leads to reduced seed GA level and enhanced dormancy (Ye et al., 2015).
Exogenous factors that are implicated in the regulation of dormancy in cereal seeds such as temperature can also induce changes in the expression patterns of GA metabolic genes. For example, dormancy decay in wheat seeds due to imbibition at lower than optimal temperature (15°C) is related with enhanced expression of TaGA3ox2 and thereby increased level of bioactive GA in the embryo (Kashiwakura et al., 2016) while inhibition of germination due to imbibition of non-dormant wheat seeds at suboptimal temperature (4°C) is related with decreased expression of TaGA20oxs and TaGA3ox2, and reduced bioactive GA level in both embryo and endosperm tissues (Izydorczyk et al., 2017). However, further genetic analyses are required for detailed understanding of the physiological roles of GA metabolic genes in the regulation of seed dormancy in cereals.
Gibberellin signaling in plants is triggered when bioactive GA is perceived by its receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al., 2005) (Figure 2). The prevalence of GA responses such as seed germination requires GA induced degradation of DELLA protein, which acts as a negative regulator of GA signaling (Sun, 2011). The binding of GA to GID1 promotes the formation of GA-GID1-DELLA complex (Murase et al., 2008), which in turn associates with F-box protein, the central component of SCFSLY 1/GID2 E3 ubiquitin ligase, leading to DELLA degradation via the ubiquitin-26S proteasome pathway (McGinnis et al., 2003; Sasaki et al., 2003). The degradation of DELLA by GA activates GAMYB, a downstream GA signaling component that mediates GA signaling effects (Gubler et al., 1995, 1999). The function of GAMYB in cereal aleurone cells has been reported to be repressed by another downstream GA signaling component designated as KINASE ASSOCIATED WITH GAMYB1 (KGM1) (Woodger et al., 2003).
Seed dormancy or germination in cereals such as wheat is also reported to be associated with changes in seed sensitivity to GA (Tuttle et al., 2015), which is influenced by the expression of genes encoding the GA signaling components. The gene encoding GID1 was first identified from rice and later in other cereals such as barley (Chandler et al., 2008) and wheat (Li et al., 2013). Genetic analysis of rice mutant lines showed that GA responses are mediated by GID1 in the nucleus of aleurone cells and there is no alternative GA receptor in the rice aleurone (Yano et al., 2015). However, mutation in GID1 of rice does not appear to affect seed germination although it causes repression of the activity of α-amylase (Ueguchi-Tanaka et al., 2005). Furthermore, the expression pattern of GID1 is found not to be associated with the dormancy or germination phenotype of wheat and barley seeds (Barrero et al., 2009; Liu et al., 2013; Izydorczyk et al., 2017). Studies in rice and wheat have shown that GID2 is a part of SCF complex and positively regulates the GA response (Hirano et al., 2010; Lou et al., 2016); however, mutation in GID2 of rice leads to repression of the α-amylase activity with no any effect on seed germination (Ueguchi-Tanaka et al., 2005). Similar to that observed for GID1, the expression of GID2 is not associated with the seed germination phenotype (Ueguchi-Tanaka et al., 2008). These results overall suggest that GA signaling in the seeds of cereals such as wheat, barley, and rice can operate independent of GID1 and GID2 levels; however, more studies are required to validate these observations.
Cereals appear to have a single or fewer DELLA proteins that are reported to be highly conserved (Davière and Achard, 2013) including SLENDER RICE1 (SLR1) of rice (Ikeda et al., 2001), SLENDER1 (SLN1) of barley (Chandler et al., 2002), REDUCED HEIGHT (RHT) of wheat (Peng et al., 1999), and D8 and D9 of maize (Lawit et al., 2010). Overexpression of SLN1 leads to repression of GA induced α-amylase expression in barley seeds (Zentella et al., 2002), and in agreement with this, the SLN mutant of barley is characterized by non-dormant seeds with high amylase activity in the aleurone (Chandler, 1988). A recent report in wheat showed that inhibition of the germination of non-dormant wheat seeds due to imbibition at suboptimal and supraoptimal temperatures is associated with increased expression of RHT1, suggesting the role of GA signaling or seed GA sensitivity in the induction of dormancy (Izydorczyk et al., 2017). Another evidence for the role of RHT1 in seed dormancy and germination comes from the identification of RHT1 alleles that are able to produce different levels of seed dormancy (Velde et al., 2017). In contrast, no differential expression of RHT was observed during imbibition of dormant and after-ripened seeds of wheat (Liu et al., 2013); however, this discrepancy could be due to the fact that the whole seed tissue instead seed embryo was analyzed in this particular study.
The degradation of DELLA (SLN/SLR) by GA activates GAMYB, which in turn induces the transcription of α-amylase in the aleurone of barley and rice seeds via binding to the GA-responsive elements (GARE) present in its promoter (Gubler et al., 1995, 1999; Sun and Gubler, 2004). Furthermore, GAMYB has been shown to interact synergistically with other transcription factors such as DNA binding with one finger (DOF) proteins and regulate the expression of α-amylase in barley (Zou et al., 2008). On the other hand, GAMYB regulates other transcription factors such as GATA type transcription factors that act as positive regulators of seed dormancy (Ravindran et al., 2017), and transcription factors such as KGM regulates GAMYB negatively and thereby lead to repression of genes encoding hydrolases in the aleurone (Woodger et al., 2003). Despite their reported functionalities, no differential expression of GAMYB and KGM1 was evident between dormant and after ripened/non-dormant seeds of wheat (Liu et al., 2013). The induction in the expression of hydrolases during imbibition of after-ripened wheat seeds irrespective of the absence of differential expression of GAMYB and KGM between the dormant and after-ripened seed samples might suggest that GA signaling in cereal seeds is not dependent on the level of the two transcriptional regulators. Functional analysis of GAMYB with respect to dormancy, and identification and characterization of other molecular features that regulate or interact with GAMYB are crucial to enhance our understanding of the role of downstream GA signaling elements in the control of seed dormancy and germination in cereals.
Several studies have indicated that the dynamic balance between ABA and GA metabolism and thereby the developmental switch between dormancy and germination can be modulated by their reciprocal regulation (Seo et al., 2006; Piskurewicz et al., 2008; Izydorczyk et al., 2017). It has been shown previously that vivipary phenotype in maize kernels due to ABA deficiency can be reversed through inhibition of GA synthesis, and this demonstrates the role of GA in antagonizing the action of ABA and thereby seed transition from dormancy to germination (White et al., 2000). Changes in the balance between ABA and GA levels, which modulate seed dormancy status, are associated with alterations in the expression patterns of their metabolic genes; for example, imbibition of non-dormant barley seeds induces the expression of ABA catabolic gene HvABA8′OH1 and GA biosynthetic gene HvGA3ox2 (Suzuki et al., 2005; Millar et al., 2006), which leads to reduced ABA and increased GA levels. Furthermore, imbibition of non-dormant wheat seeds has been shown to lead to transcriptional induction of TaGA3ox2 and HvABA8′OH1 genes (Liu et al., 2013; Son et al., 2016). Further genetic studies such as mutational and gain of function studies are required to precisely elucidate the antagonistic relationship between ABA and GA metabolic pathways in regulating the switch of cereal seeds between dormancy and germination.
In Arabidopsis, a PP2C protein, HONSU, which acts as a negative regulator of ABA signaling, modulates the expression of GA metabolism and signaling genes and thereby enhance the transition of seeds from dormant to germinating state (Kim et al., 2013). In cereals such as rice, PP2C protein represses OsbZIP10 (ABI5 homolog of rice) via dephosphorylation and thereby promote seed germination (Bhatnagar et al., 2017). Consistently, overexpression of OsPP2C51 leads to increased expression of α-amylase and activity of the corresponding enzyme. Since SbABI4 and SbABI5 from sorghum have been shown to interact with SbGA2ox promoter in vitro (Cantoro et al., 2013), the transcriptional repression of OsbZIP10 (ABI5) by OsPP2C51 may imply decreased GA inactivation that leads to an increase in bioactive GA level and enhanced dormancy loss and germination. Furthermore, the expression of HvPP2C in the aleurone of barley appears to be differentially regulated by GA and ABA (Chen and An, 2006). All these results underline the significance of PP2C in mediating ABA and GA responses, thereby modulating the equilibrium between germination and dormancy; however, more studies are required to further elucidate this role of PP2C.
A recent study has also shown that Tiller Enhancer (TE) of rice, which acts as an activator of the APC/CTE E3 ubiquitin ligase complex, mediates the balance between ABA and GA signaling and thereby the developmental switch of seeds between dormancy and germination (Lin et al., 2015; Shu et al., 2018). The interaction of TE with the ABA receptor OsPYLs/RCARs enhances proteasome mediated degradation of the receptor. However, ABA mediated activation of SnRK2s, which induces the phosphorylation of TE, ultimately represses the TE-OsPYLs/RCARs interaction and thereby stabilizes OsPYLs/RCARs, leading to increased seed sensitivity to ABA and enhanced dormancy. In contrast, GA enhances TE-OsPYLs/RCARs interaction and thereby the degradation of OsPYLs/RCARs through repression of the activity of SnRK2s, and this leads to decreased seed sensitivity to ABA and enhanced germination. Furthermore, a rice AP2-domain containing transcription factor, designated as OsAP2-39, regulates a switch between seed dormancy and germination through modulating the balance between ABA and GA levels via modulating the expression levels of OsNCED1 and Elongation of Uppermost Internode (EUI) genes (Yaish et al., 2010; Shu et al., 2018). Studies in Arabidopsis have also shown that ABI4 regulates the equilibrium between seed dormancy and germination through modulating the balance between ABA and GA levels (Shu et al., 2013). For example, dry seeds of the abi4 mutant of Arabidopsis contain lower and higher levels of ABA and GA, respectively, as compared to that of the wild-type. Consistently, ABI4 has been shown to enhance the expression of ABA biosynthetic (NCED6) and GA catabolic (GA2ox7) genes, although in a post-germination stage (Shu et al., 2016a), while repressing the expression of ABA catabolic (CYP707A1 and CYP707A2) genes (Shu et al., 2013). However, further studies are needed to determine if a similar mechanism underlies the role of ABI4 in regulating seed dormancy and germination in cereals seeds.
It has been shown that the DELLA proteins of Arabidopsis act as regulators of GA and ABA crosstalk and therefore the balance between dormancy and germination via its interaction with ABI3 and ABI5 (Lim et al., 2013). Furthermore, NUCLEAR FACTOR-YC (NF-YC) modulates GA and ABA signaling via the NF-YC–DELLA (RGL2)–ABI5 cascade independent of ABA (Liu et al., 2016); the NF-YCs binds to RGL2, which in turn regulates ABI5, a downstream component of ABA signaling, via specific CCAAT elements that are also shown to be present in the promoter of GA inducible GAMYB (Washio and Morikawa, 2006). The role of DELLA in mediating the balance between GA and ABA responses and in turn dormancy and germination can take place through its interaction with XERICO, a RING-H2 zinc finger E3 ubiquitin ligase that acts as important regulator of ABA signaling (Zentella et al., 2007; Piskurewicz et al., 2008) (Figure 3). When GA level is low, DELLA activates the expression of XERICO, which in turn enhances ABA accumulation and activates the transcription of ABI5, leading to dormancy maintenance/inhibition of seed germination. Consistently, overexpression of XERICO in rice results in increased ABA level and response via enhancing the expression of OsNCED1 and OsABI5, respectively, causing a delay in germination (Zeng et al., 2014, 2015). Its overexpression in maize also leads to ABA accumulation via modulating the expression of ZmCYP707A rather than that of the ZmNCED genes as observed in rice (Brugière et al., 2017). However, detailed understanding of the role of XERICO in regulating the balance between ABA and GA response, and in turn seed transition between dormancy and germination awaits further study. It is well established that the germination of cereal seeds is characterized by transcriptional induction of hydrolytic enzymes such as α-amylase, which are required for the degradation of starch and its subsequent mobilization to the expanding embryo (Weiss and Ori, 2007). The expression of α-amylase in the aleurone of barley seeds can be activated by the GA inducible GAMYB (Gubler et al., 1995) but repressed by ABA induced PKABA1 (Ser-Thr kinase), which binds to GAMYB and represses its transcription (Gómez-Cadenas et al., 1999, 2001). However, RNAi mediated suppression of PKABA1 in barley does not affect ABA mediated repression of α-amylase in the seeds, suggesting the presence of PKABA1-independent ABA signaling pathway (Zentella et al., 2002). Indeed, a study in rice has shown that ABA mediated inhibition of GA inducible responses such as the expression of α-amylase can take place via an alternative pathway that involves WRKY transcription factors (Xie et al., 2006) (Figure 3).
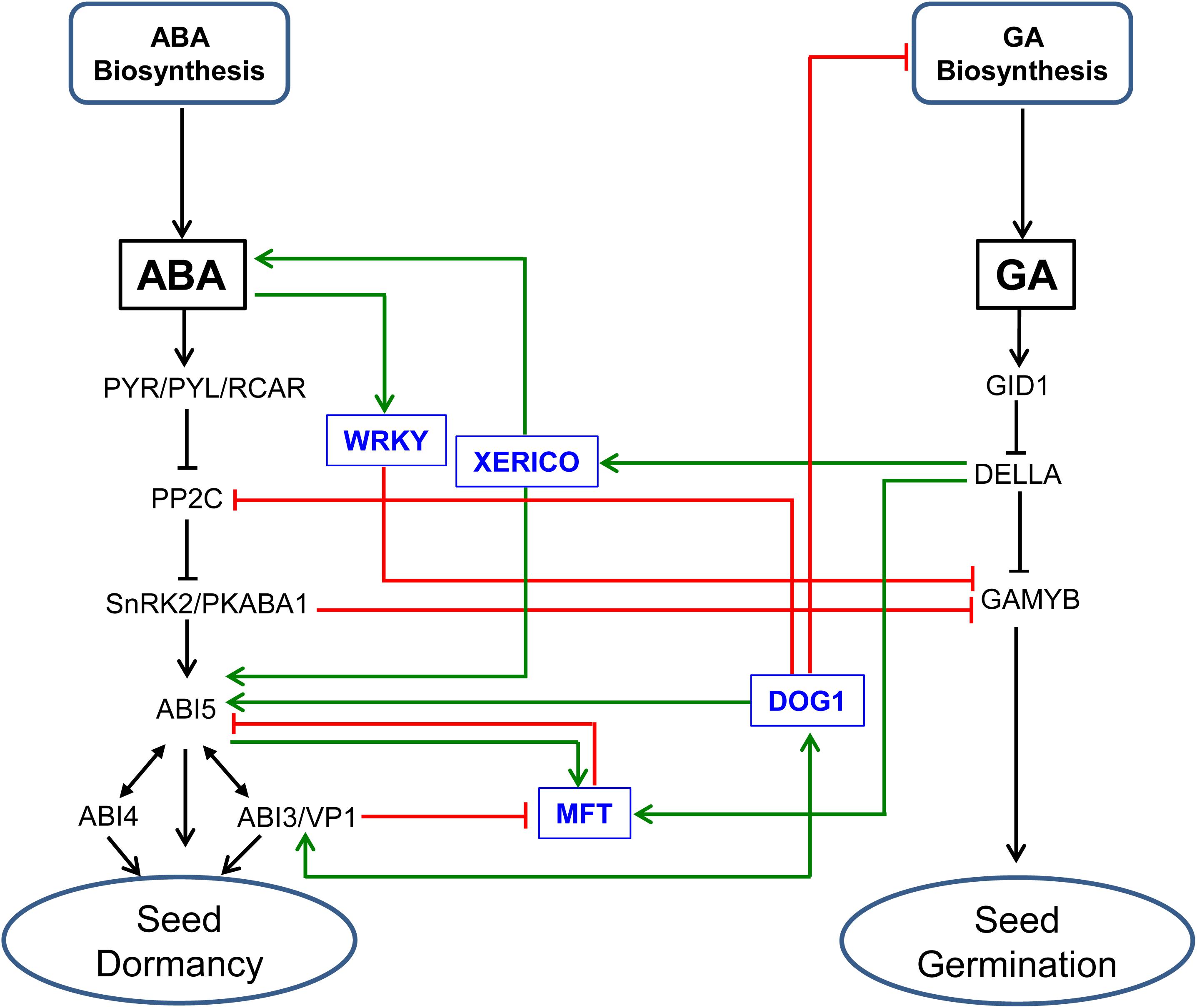
FIGURE 3. Genetic/molecular elements implicated in the regulation of ABA/GA balance and seed transition between dormancy and germination. ABA inhibits GAMYB-mediated GA responses through modulating the expression of WRKY transcription factors (Xie et al., 2006). DELLA protein regulates the balance between GA and ABA responses, and thereby seed dormancy and germination through its interaction with XERICO, a RING-H2 zinc finger E3 ubiquitin ligase. In the absence of GA, DELLA induces the expression of XERICO, which in turn enhances ABA accumulation, and ABI5 activity (Piskurewicz et al., 2008), leading to dormancy maintenance/inhibition of seed germination. MOTHER OF FT AND TF1 (MFT) represses ABI5 and therefore ABA signaling through a negative feedback mechanism (Xi et al., 2010). The role of MFT in regulating the balance between ABA and GA responses and thereby seed dormancy and germination involves ABI3 and ABI5 that act as repressor and activator of MFT expression, respectively, and DELLA that acts as activator of MFT expression (Xi et al., 2010). DELAY OF GERMINATION1 (DOG1) regulates ABA signaling and therefore seed dormancy through its interaction with PP2C (Née et al., 2017), and likely through modulating the expression of ABI5 and interacting with ABI3 (Dekkers et al., 2016). DOG1 also regulates GA metabolism through the expressions of GA biosynthetic and inactivation genes in temperature dependent manner (Kendall et al., 2011; Graeber et al., 2014). SnRK2/PKABA1 binds to GAMYB and repress its transcription (Gómez-Cadenas et al., 2001). See the legends of Figure 1 and Figure 2 for descriptions of ABA and GA signaling components.
Previous studies have also identified other genetic/molecular elements that are implicated in the regulation of seed dormancy and germination in cereals such as MOTHER OF FT AND TFL1 (MFT) and DELAY OF GERMINATION 1 (DOG1) (Ashikawa et al., 2010, 2014; Nakamura et al., 2011). Studies with the model plant Arabidopsis have demonstrated that MFT enhances seed transition from the state of dormancy to germination via feedback transcriptional repression of ABI5 and ABA signaling (Xi et al., 2010). MFT also mediates ABA and GA responses, and this appears to involve the ABA signaling components ABI3 and ABI5 that act as repressor and activator of MFT expression, respectively, and the GA signaling component DELLA, which acts as transcriptional activator of MFT (Xi et al., 2010) (Figure 3). Recent reports have shown that DOG1 enhances ABA signaling and thereby seed dormancy through its interaction with PP2C (Née et al., 2017) and presumably by modulating the expression of ABI5 and genetically interacting with ABI3 (Dekkers et al., 2016), implying that the action of DOG1 in regulating seed dormancy is coordinated with ABA (Nakabayashi et al., 2012). Mutational analysis of DOG1, furthermore, revealed that its role in dormancy maintenance is mediated at least partly by modulation of GA metabolism via altering the expressions of GA biosynthetic and inactivation genes in a temperature dependent manner (Kendall et al., 2011; Graeber et al., 2014) (Figure 3). However, it is yet unclear if the role of MFT and DOG1 in the regulation of the developmental switch of cereal seeds between dormancy and germination is mediated by their interference with the balance between ABA and GA metabolism and signaling.
Seed imbibition environmental conditions such as temperature and light can also modulate the balance between seed dormancy and germination through altering the metabolic and signaling equilibrium between ABA and GA. Recent reports in wheat have demonstrated the role of imbibition temperature in regulating the balance of ABA/GA ratio and thereby seed transition between dormancy and germination (Kashiwakura et al., 2016; Izydorczyk et al., 2017). Imbibition of dormant seeds at lower temperature (15°C) induces germination through enhancing the expression of ABA catabolic (TaABA8′OH1 and TaABA8′OH2) and GA biosynthesis (TaGA3ox2) genes that lead to a decrease in embryo ABA level while increasing that of GA (Kashiwakura et al., 2016). However, delay in germination as a result of seed imbibition at supraoptimal temperature (35°C) is mediated mainly by enhanced expression of ABA biosynthesis genes (TaNCED1 and TaNCED2) that leads to increased ABA content in the embryo, and this ABA is also suggested to interfere with GA effects (Izydorczyk et al., 2017). Similarly, induction of dormancy/inhibition of germination as a result of imbibition at suboptimal temperature (4°C) appeared to be associated with increased expression of the NCED genes in both embryo and endosperm tissues and decreased expression of CYP707A genes in the endosperm, which ultimately led to increased seed ABA level. This effect of suboptimal temperature has been shown to be accompanied by repression of GA biosynthetic genes and GA levels in both embryo and endosperm tissues (Izydorczyk et al., 2017). The dynamic balance between ABA and GA signaling in regulating the switch of seeds between dormancy and germination can also be influenced by temperature. For example, the transition of wheat seeds from non-dormant/germinating to dormant state under supraoptimal imbibition temperature has been shown to be mediated by enhanced expression of embryonic genes that positively and negatively regulate ABA and GA signaling (Izydorczyk et al., 2017), respectively, leading to alteration of the balance between seed sensitivity to ABA and GA. In addition, seed imbibition at suboptimal temperature induces the expression of genes that regulate ABA signaling positively in both embryo and endosperm tissues, and this effect was accompanied by transcriptional activation of genes that negatively regulate GA signaling in the embryo.
Previous studies with the seeds of dicot species such as Arabidopsis have shown that red and far-red lights regulate dormancy and germination by altering the ABA/GA balance (Seo et al., 2006, 2009). The roles of these two lights in modulating the balance between ABA and GA metabolisms and therefore the transition of seeds between dormant and germinating states have also been investigated in monocot species. For example, the transition from dormant to germinating state of Brachypodium and photoblastic weedy rice seeds appeared to be promoted by red light but repressed by far-red light (Kim et al., 2009; Barrero et al., 2012), and these effects of the two lights in weedy rice seeds have been shown to be associated with changes in ABA and GA levels (Kim et al., 2009). Contrary to these observations, red and far-red lights are reported not to have an effect on the expression of ABA biosynthetic genes, the level of ABA and germination in barley seeds (Gubler et al., 2008). In addition to the two light qualities, previous studies investigated the roles of white and blue lights in regulation of seed dormancy and germination in cereals seeds. White and blue light are reported to inhibit the germination of barley and wheat seeds through increasing ABA content, and this appears to be mediated by transcriptional activation of the ABA biosynthetic NCED genes and repression of the ABA catabolic ABA8′OH1 gene in the embryo (Gubler et al., 2008; Jacobsen et al., 2013; Hoang et al., 2014). Furthermore, the inhibitory effect of blue light but not that of white light on the germination of barley and rice seeds is associated with transcriptional repressions of GA biosynthetic gene and activation of GA inactivation genes (Gubler et al., 2008; Hirose et al., 2012; Hoang et al., 2014). Consistently, knocking-down of a rice gene encoding cryptochrome 1 (CRY1), which is a blue/ultraviolet-A photoreceptor, induces the expression of the GA inactivation OsGA2ox genes (Hirose et al., 2012). However, further mutational and genetic studies are required for detailed elucidation of the molecular mechanisms underlying the role of temperature and light in modulating ABA/GA balance and thereby developmental switch of seeds between dormancy and germination.
Antagonistic interaction between ABA and GA and thereby the transition of seeds between dormancy and germination can also be mediated by ROS including hydrogen peroxide (H2O2), superoxide anion (O2-), and hydroxyl radical (OH-). The role of ROS in alleviating seed dormancy in cereals such as barley has been reported to be mediated mainly by the modulation of GA metabolism with almost no effect on ABA metabolism (Bahin et al., 2011). Consistently, ABA induced reduction of ROS production in rice seeds appeared to repress GA accumulation via repression of specific OsGA20ox and OsGA3ox genes, and thereby seed germination (Ye et al., 2012). Comparative analysis of dormant and non-dormant seeds revealed that the germination of non-dormant wheat and barley seeds is associated with the production of more ROS during imbibition (Caliskan and Cuming, 1998; Ishibashi et al., 2017), which in turn enhance the expression of ABA catabolic gene (HvABA8′OH1) and thereby reduce ABA level (Ishibashi et al., 2017). In agreement with these results, inhibition of the germination of non-dormant barley seeds through repression of ROS production has been shown to decrease embryonic GA level via repression of the expression of TaGA20ox1 and TaGA3ox1 genes, and increase ABA level via decreased expression of HvABA8′OH1 (Ishibashi et al., 2015). Furthermore, GA induced promotion of H2O2 production in barley aleurone enhances the expression of α-amylase via activating the transcription of GAMYB and repressing the expression and activity of ABA inducible PKABA (Ishibashi et al., 2012). Despite these reports, the molecular bases underlying ROS mediated alteration of ABA/GA balance and the equilibrium between dormancy and germination in cereal seeds are still elusive.
To date, considerable progress has been made in the dissection of molecular mechanisms underlying the control of ABA/GA balance and thereby seed transition between dormancy and germination in dicot species such as the model plant Arabidopsis. However, this phenomenon is still poorly understood in cereal seeds, and this emphasizes the need for further studies to determine if the molecular mechanisms identified in the seeds of dicot species are conserved in cereal seeds and identify novel mechanisms that are specific to cereal seeds. Although a limited number of molecular elements have been reported as regulators of ABA/GA interplay in cereal seeds, these elements are identified mainly based on comparative analysis of seeds from dormant and non-dormant lines or in vitro assays. Therefore, further genetic studies such as mutational and gain-of-function analyses are necessary to precisely pinpoint their physiological functions. Several studies have also provided important insights into the roles of other plant hormones, and genetic and epigenetic mechanisms in the control of dormancy and germination in cereal seeds. It is therefore interesting to explore if these factors influence dormancy status in cereal seeds through modulation of the ABA/GA balance. In recent years, there have been significant advances in the availability of genomic resources of cereal crops, and these resources are providing important platforms for the identification of molecular elements regulating ABA/GA balance in cereal seeds and elucidation of their roles in controlling seed transition between dormancy and germination. It is well established that preharvest sprouting, which causes substantial yield and quality losses in cereal crops, is closely associated with the level of dormancy manifested in the seeds. Therefore, a detailed understanding of molecular mechanisms underlying the regulation of ABA/GA balance and thereby dormancy and germination in cereal seeds will have a significant contribution in developing molecular/genomic tools that can enhance the breeding of cultivars with improved preharvest sprouting tolerance, and in turn lead to increased yield and quality of cereal crops.
BA conceived the topic and organized the manuscript. PAT, RK, and BA wrote and prepared the manuscript. PR and PKT were involved in the collection of literature and writing of Sections “ABA Signaling” and “Gibberellin Metabolism” of the manuscript.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada, the Western Grains Research Foundation, and the Manitoba Wheat and Barley Growers Association to BA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ashikawa, I., Abe, F., and Nakamura, S. (2010). Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 179, 536–542. doi: 10.1016/j.plantsci.2010.08.002
Ashikawa, I., Mori, M., Nakamura, S., and Abe, F. (2014). A transgenic approach to controlling wheat seed dormancy level by using Triticeae DOG1-like genes. Transgenic Res. 23, 621–629. doi: 10.1007/s11248-014-9800-5
Bahin, E., Bailly, C., Sotta, B., Kranner, I., Corbineau, F., and Leymarie, J. (2011). Crosstalk between reactive oxygen species and hormonal signaling pathways regulates grain dormancy in barley. Plant Cell Environ. 34, 980–993. doi: 10.1111/j.1365-3040.2011.02298.x
Barrero, J. M., Downie, A. B., Xu, Q., and Gubler, F. (2014). A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 26, 1094–1104. doi: 10.1105/tpc.113.121830
Barrero, J. M., Jacobsen, J. V., Talbot, M. J., White, R. G., Swain, S. M., Garvin, D. F., et al. (2012). Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 193, 376–386. doi: 10.1111/j.1469-8137.2011.03938.x
Barrero, J. M., Talbot, M. J., White, R. G., Jacobsen, J. V., and Gubler, F. (2009). Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150, 1006–1021. doi: 10.1104/pp.109.137901
Benech-Arnold, R. L., Cristina Giallorenzi, M., Frank, J., and Rodriguez, V. (1999). Termination of hull-imposed dormancy in developing barley grains is correlated with changes in embryonic ABA levels and sensitivity. Seed Sci. Res. 9, 39–47. doi: 10.1017/s0960258599000045
Bewley, J. D., and Black, M. (1994). Seeds: Physiology of Development and Germination. Berlin: Springer. doi: 10.1007/978-1-4899-1002-8
Bhatnagar, N., Min, M. K., Choi, E. H., Kim, N., Moon, S. J., Yoon, I., et al. (2017). The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 93, 389–401. doi: 10.1007/s11103-016-0568-2
Black, M., Bewley, J. D., and Halmer, P. (2006). The Encyclopedia of Seeds: Science, Technology and Uses. Wallingford: CABI Publishing. doi: 10.1079/9780851997230.0000
Brugière, N., Zhang, W., Xu, Q., Scolaro, E. J., Lu, C., Kahsay, R. Y., et al. (2017). Overexpression of RING domain E3 Ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis. Plant Physiol. 175, 1350–1369. doi: 10.1104/pp.17.01072
Caliskan, M., and Cuming, A. C. (1998). Spatial specificity of H2O2-generating oxalate oxidase gene expression during wheat embryo germination. Plant J. 15, 165–171. doi: 10.1046/j.1365-313X.1998.00191.x
Cantoro, R., Crocco, C. D., Benech-Arnold, R. L., and Rodríguez, M. V. (2013). In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 64, 5721–5735. doi: 10.1093/jxb/ert347
Carrari, F., Perez-Flores, L., Lijavetzky, D., Enciso, S., Sanchez, R., Benech-Arnold, R., et al. (2001). Cloning and expression of a sorghum gene with homology to maize vp1. Its potential involvement in pre-harvest sprouting resistance. Plant Mol. Biol. 45, 631–640. doi: 10.1023/a:1010648420669
Chandler, P. M. (1988). Hormonal regulation of gene expression in the “slender” mutant of barley (Hordeum vulgare L.). Planta 175, 115–120. doi: 10.1007/bf00402888
Chandler, P. M., Harding, C. A., Ashton, A. R., Mulcair, M. D., Dixon, N. E., and Mander, L. N. (2008). Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol. Plant 1, 285–294. doi: 10.1093/mp/ssn002
Chandler, P. M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. molecular and physiological characterization. Plant Physiol. 129, 181–190. doi: 10.1104/pp.010917
Chen, K., and An, Y.-Q. C. (2006). Transcriptional responses to gibberellin and abscisic acid in barley aleurone. J. Int. Plant Biol. 48, 591–612. doi: 10.1111/j.1744-7909.2006.00270.x
Chitnis, V. R., Gao, F., Yao, Z., Jordan, M. C., Park, S., and Ayele, B. T. (2014). After-ripening induced transcriptional changes of hormonal genes in wheat seeds: the cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS One 9:e87543. doi: 10.1371/journal.pone.0087543
Chono, M., Honda, I., Shinoda, S., Kushiro, T., Kamiya, Y., Nambara, E., et al. (2006). Field studies on the regulation of abscisic acid content and germinability during grain development of barley: molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 57, 2421–2434. doi: 10.1093/jxb/erj215
Chono, M., Matsunaka, H., Seki, M., Fujita, M., Kiribuchi-Otobe, C., Oda, S., et al. (2013). Isolation of a wheat (Triticum aestivum L.) mutant in ABA 8′-hydroxylase gene: effect of reduced ABA catabolism on germination inhibition under field condition. Breed. Sci. 63, 104–115. doi: 10.1270/jsbbs.63.104
Cutler, A. J., and Krochko, J. E. (1999). Formation and breakdown of ABA. Trends Plant Sci. 4, 472–478. doi: 10.1016/s1360-1385(99)01497-1
Davière, J.-M., and Achard, P. (2013). Gibberellin signaling in plants. Development 140, 1147–1151. doi: 10.1242/dev.087650
Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. doi: 10.1104/pp.122.2.415
Dekkers, B. J. W., He, H., Hanson, J., Willems, L. A. J., Jamar, D. C. L., Cueff, G., et al. (2016). The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 85, 451–465. doi: 10.1111/tpj.13118
Du, W., Cheng, J., Cheng, Y., Wang, L., He, Y., Wang, Z., et al. (2015). Physiological characteristics and related gene expression of after-ripening on seed dormancy release in rice. Plant Biol. 17, 1156–1164. doi: 10.1111/plb.12371
Fan, J., Niu, X., Wang, Y., Ren, G., Zhuo, T., Yang, Y., et al. (2007). Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa). J. Exp. Bot. 58, 3811–3817. doi: 10.1093/jxb/erm231
Fidler, J., Zdunek-Zastocka, E., Prabucka, B., and Bielawski, W. (2016). Abscisic acid content and the expression of genes related to its metabolism during maturation of triticale grains of cultivars differing in pre-harvest sprouting susceptibility. J. Plant Physiol. 207, 1–9. doi: 10.1016/j.jplph.2016.09.009
Finch-Savage, W. E., and Footitt, S. (2017). Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 68, 843–856. doi: 10.1093/jxb/erw477
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Food and Agriculture Organization of the United Nations (2017). World Food Situation. Available at: http://www.fao.org/worldfoodsituation/csdb/en/
Gagete, A. P., Riera, M., Franco, L., and Rodrigo, M. I. (2009). Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing. J. Exp. Bot. 60, 1703–1714. doi: 10.1093/jxb/erp038
Gao, F., and Ayele, B. T. (2014). Functional genomics of seed dormancy in wheat: advances and prospects. Front. Plant Sci. 5:458. doi: 10.3389/fpls.2014.00458
Gao, F., Jordan, M. C., and Ayele, B. T. (2012). Transcriptional programs regulating seed dormancy and its release by after-ripening in common wheat (Triticum aestivum L.). Plant Biotechnol. J. 10, 465–476. doi: 10.1111/j.1467-7652.2012.00682.x
Gao, F., Rampitsch, C., Chitnis, V. R., Humphreys, G. D., Jordan, M. C., and Ayele, B. T. (2013). Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnol. J. 11, 921–932. doi: 10.1111/pbi.12083
Gómez-Cadenas, A., Verhey, S. D., Holappa, L. D., Shen, Q., Ho, T.-H. D., and Walker-Simmons, M. K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. U.S.A. 96, 1767–1772. doi: 10.1073/pnas.96.4.1767
Gómez-Cadenas, A., Zentella, R., Walker-Simmons, M. K., and Ho, T.-H. D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–680. doi: 10.1105/tpc.13.3.667
Graeber, K., Linkies, A., Steinbrecher, T., Mummenhoff, K., Tarkowská, D., Turečková, V., et al. (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. U.S.A. 111, E3571–E3580. doi: 10.1073/pnas.1403851111
Gu, X.-Y., Foley, M. E., Horvath, D. P., Anderson, J. V., Feng, J., Zhang, L., et al. (2011). Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189, 1515–1524. doi: 10.1534/genetics.111.131169
Gubler, F., Hughes, T., Waterhouse, P., and Jacobsen, J. (2008). Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147, 886–896. doi: 10.1104/pp.107.115469
Gubler, F., Kalla, R., Roberts, J. K., and Jacobsen, J. V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7, 1879–1891. doi: 10.1105/tpc.7.11.1879
Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J. V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17, 1–9. doi: 10.1046/j.1365-313X.1999.00346.x
Hirano, K., Asano, K., Tsuji, H., Kawamura, M., Mori, H., Kitano, H., et al. (2010). Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22, 2680–2696. doi: 10.1105/tpc.110.075549
Hirose, F., Inagaki, N., Hanada, A., Yamaguchi, S., Kamiya, Y., Miyao, A., et al. (2012). Cryptochrome and phytochrome cooperatively but independently reduce active gibberellin content in rice seedlings under light irradiation. Plant Cell Physiol. 53, 1570–1582. doi: 10.1093/pcp/pcs097
Hoang, H. H., Sechet, J., Bailly, C., Leymarie, J., and Corbineau, F. (2014). Inhibition of germination of dormant barley (Hordeum vulgare L.) grains by blue light as related to oxygen and hormonal regulation. Plant Cell Environ. 37, 1393–1403. doi: 10.1111/pce.12239
Hoecker, U., Vasil, I. K., and McCarty, D. R. (1995). Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 9, 2459–2469. doi: 10.1101/gad.9.20.2459
Huang, T., Qu, B., Li, H.-P., Zuo, D.-Y., Zhao, Z.-X., and Liao, Y.-C. (2012). A maize viviparous 1 gene increases seed dormancy and preharvest sprouting tolerance in transgenic wheat. J. Cereal Sci. 55, 166–173. doi: 10.1016/j.jcs.2011.11.003
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., et al. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. doi: 10.1105/tpc.13.5.999
Ishibashi, Y., Aoki, N., Kasa, S., Sakamoto, M., Kai, K., Tomokiyo, R., et al. (2017). The interrelationship between abscisic acid and reactive oxygen species plays a key role in barley seed dormancy and germination. Front. Plant Sci. 8:275. doi: 10.3389/fpls.2017.00275
Ishibashi, Y., Kasa, S., Sakamoto, M., Aoki, N., Kai, K., Yuasa, T., et al. (2015). A role for reactive oxygen species produced by NADPH oxidases in the embryo and aleurone cells in barley seed germination. PLoS One 10:e0143173. doi: 10.1371/journal.pone.0143173
Ishibashi, Y., Tawaratsumida, T., Kondo, K., Kasa, S., Sakamoto, M., Aoki, N., et al. (2012). Reactive oxygen species are involved in gibberellin/abscisic acid signaling in barley aleurone cells. Plant Physiol. 158, 1705–1714. doi: 10.1104/pp.111.192740
Izydorczyk, C., Nguyen, T.-N., Jo, S., Son, S., Tuan, P. A., and Ayele, B. T. (2017). Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signaling mediates the effects of suboptimal and supraoptimal temperatures on seed germination in wheat (Triticum aestivum L.). Plant Cell Environ. 41, 1022–1037. doi: 10.1111/pce.12949
Jacobsen, J. V., Barrero, J. M., Hughes, T., Julkowska, M., Taylor, J. M., Xu, Q., et al. (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 238, 121–138. doi: 10.1007/s00425-013-1878-0
Johnson, R. R., Wagner, R. L., Verhey, S. D., and Walker-Simmons, M. K. (2002). The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130, 837–846. doi: 10.1104/pp.001354
Kaneko, M., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., and Matsuoka, M. (2002). The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 128, 1264–1270. doi: 10.1104/pp.010785
Kanno, Y., Jikumaru, Y., Hanada, A., Nambara, E., Abrams, S. R., Kamiya, Y., et al. (2010). Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51, 1988–2001. doi: 10.1093/pcp/pcq158
Kashiwakura, Y.-I., Kobayashi, D., Jikumaru, Y., Takebayashi, Y., Nambara, E., Seo, M., et al. (2016). Highly sprouting-tolerant wheat grain exhibits extreme dormancy and cold imbibition-resistant accumulation of abscisic acid. Plant Cell Physiol. 57, 715–732. doi: 10.1093/pcp/pcw051
Kawakami, N., Miyake, Y., and Noda, K. (1997). ABA insensitivity and low ABA levels during seed development of non-dormant wheat mutants. J. Exp. Bot. 48, 1415–1421. doi: 10.1093/jxb/48.7.1415
Kendall, S. L., Hellwege, A., Marriot, P., Whalley, C., Graham, I. A., and Penfield, S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23, 2568–2580. doi: 10.1105/tpc.111.087643
Kermode, R. A. (2005). Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24, 319–344. doi: 10.1007/s00344-005-0110-2
Kim, H., Hwang, H., Hong, J.-W., Lee, Y.-N., Ahn, I. P., Yoon, I. S., et al. (2012). A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 63, 1013–1024. doi: 10.1093/jxb/err338
Kim, S.-Y., Hwang, S.-J., Lee, I.-J., Shin, D.-H., Park, S.-T., Yeo, U.-S., et al. (2009). Effect of light on endogenous levels of gibberellin and abscisic acid in seed germination of photoblastic weedy rice (Oryza sativa L.). J. Crop Sci. Biotechnol. 12, 149–152. doi: 10.1007/s12892-009-0110-z
Kim, W., Lee, Y., Park, J., Lee, N., and Choi, G. (2013). HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 54, 555–572. doi: 10.1093/pcp/pct017
King, R. W. (1976). Abscisic acid in developing wheat grains and its relationship to grain growth and maturation. Planta 132, 43–51. doi: 10.1007/bf00390329
Kucera, B., Cohn, M. A., and Leubner-Metzger, G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15, 281–307. doi: 10.1079/SSR2005218
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Laethauwer, S., Reheul, D., De Riek, J., and Haesaert, G. (2012). Vp1 expression profiles during kernel development in six genotypes of wheat, triticale and rye. Euphytica 188, 61–70. doi: 10.1007/s10681-011-0613-9
Lawit, S. J., Wych, H. M., Xu, D., Kundu, S., and Tomes, D. T. (2010). Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 51, 1854–1868. doi: 10.1093/pcp/pcq153
Lenser, T., and Theißen, G. (2013). Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 18, 704–714. doi: 10.1016/j.tplants.2013.08.007
Li, A., Yang, W., Li, S., Liu, D., Guo, X., Sun, J., et al. (2013). Molecular characterization of three GIBBERELLIN-INSENSITIVE DWARF1 homologous genes in hexaploid wheat. J. Plant Physiol. 170, 432–443. doi: 10.1016/j.jplph.2012.11.010
Li, C., Ni, P., Francki, M., Hunter, A., Zhang, Y., Schibeci, D., et al. (2004). Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genomics 4, 84–93. doi: 10.1007/s10142-004-0104-3
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi: 10.1105/tpc.113.118604
Lin, Q., Wu, F., Sheng, P., Zhang, Z., Zhang, X., Guo, X., et al. (2015). The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6:7981. doi: 10.1038/ncomms8981
Liu, A., Gao, F., Kanno, Y., Jordan, M. C., Kamiya, Y., Seo, M., et al. (2013). Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS One 8:e56570. doi: 10.1371/journal.pone.0056570
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC–RGL2 module integrates GA and ABA signaling to regulate seed germination in Arabidopsis. Nat. Commun. 7:12768. doi: 10.1038/ncomms12768
Liu, Y., Fang, J., Xu, F., Chu, J., Yan, C., Schläppi, M. R., et al. (2014). Expression patterns of ABA and GA metabolism genes and hormone levels during rice seed development and imbibition: a comparison of dormant and non-dormant rice cultivars. J. Genet. Genomics 41, 327–338. doi: 10.1016/j.jgg.2014.04.004
Lou, X., Li, X., Li, A., Pu, M., Shoaib, M., Liu, D., et al. (2016). Molecular characterization of three GIBBERELLIN-INSENSITIVE DWARF2 homologous genes in common wheat. PLoS One 11:e0157642. doi: 10.1371/journal.pone.0157642
Magwa, R. A., Zhao, H., and Xing, Y. (2016). Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet. 17:28. doi: 10.1186/s12863-016-0340-2
McCarty, D. R., Hattori, T., Carson, C. B., Vasil, V., Lazar, M., and Vasil, I. K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. doi: 10.1016/0092-8674(91)90436-3
McGinnis, K. M., Thomas, S. G., Soule, J. D., Strader, L. C., Zale, J. M., Sun, T.-P., et al. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15, 1120–1130. doi: 10.1105/tpc.010827
McKibbin, R. S., Wilkinson, M. D., Bailey, P. C., Flintham, J. E., Andrew, L. M., Lazzeri, P. A., et al. (2002). Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. U.S.A. 99, 10203–10208. doi: 10.1073/pnas.152318599
Meyer, R. S., and Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Millar, A. A., Jacobsen, J. V., Ross, J. J., Helliwell, C. A., Poole, A. T., Scofield, G., et al. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 45, 942–954. doi: 10.1111/j.1365-313X.2006.02659.x
Murase, K., Hirano, Y., Sun, T.-P., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Nakabayashi, K., Bartsch, M., Xiang, Y., Miatton, E., Pellengahr, S., Yano, R., et al. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. doi: 10.1105/tpc.112.100214
Nakamura, S., Abe, F., Kawahigashi, H., Nakazono, K., Tagiri, A., Matsumoto, T., et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215–3229. doi: 10.1105/tpc.111.088492
Nakamura, S., and Toyama, T. (2001). Isolation of a VP1 homologue from wheat and analysis of its expression in embryos of dormant and non-dormant cultivars. J. Exp. Bot. 52, 875–876. doi: 10.1093/jexbot/52.362.1952
Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Nambara, E., Okamoto, M., Tatematsu, K., Yano, R., Seo, M., and Kamiya, Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20, 55–67. doi: 10.1017/s0960258510000012
Née, G., Kramer, K., Nakabayashi, K., Yuan, B., Xiang, Y., Miatton, E., et al. (2017). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy. Nat. Commun. 8:72. doi: 10.1038/s41467-017-00113-6
Ng, L. M., Melcher, K., Teh, B. T., and Xu, H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35, 567–584. doi: 10.1038/aps.2014.5
Pearce, S., Huttly, A. K., Prosser, I. M., Li, Y.-D., Vaughan, S. P., Gallova, B., et al. (2015). Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol. 15:130. doi: 10.1186/s12870-015-0520-7
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Ravindran, P., Verma, V., Stamm, P., and Kumar, P. P. (2017). A novel RGL2–DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10, 1307–1320. doi: 10.1016/j.molp.2017.09.004
Rodríguez, M. V., Mendiondo, G. M., Cantoro, R., Auge, G. A., Luna, V., Masciarelli, O., et al. (2012). Expression of seed dormancy in grain sorghum lines with contrasting pre-harvest sprouting behavior involves differential regulation of gibberellin metabolism genes. Plant Cell Physiol. 53, 64–80. doi: 10.1093/pcp/pcr154
Rodríguez, M. V., Mendiondo, G. M., Maskin, L., Gudesblat, G. E., Iusem, N. D., and Benech-Arnold, R. L. (2009). Expression of ABA signaling genes and ABI5 protein levels in imbibed Sorghum bicolor caryopses with contrasting dormancy and at different developmental stages. Ann. Bot. 104, 975–985. doi: 10.1093/aob/mcp184
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707As encode (1)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. doi: 10.1126/science.1081077
Schwartz, S. H., Qin, X., and Zeevaart, J. A. D. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Seo, M., Hanada, A., Kuwahara, A., Endo, A., Okamoto, M., Yamauchi, Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366. doi: 10.1111/j.1365-313X.2006.02881.x
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69, 463–472. doi: 10.1007/s11103-008-9429-y
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016a). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Shu, K., Liu, X.-D., Xie, Q., and He, Z.-H. (2016b). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed Dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Shu, K., Zhou, W., and Yang, W. (2018). APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217, 977–983. doi: 10.1111/nph.14880
Söderman, E. M., Brocard, I. M., Lynch, T. J., and Finkelstein, R. R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. doi: 10.1104/pp.124.4.1752
Son, S., Chitnis, V. R., Liu, A., Gao, F., Nguyen, T.-N., and Ayele, B. T. (2016). Abscisic acid metabolic genes of wheat (Triticum aestivum L.): identification and insights into their functionality in seed dormancy and dehydration tolerance. Planta 244, 429–447. doi: 10.1007/s00425-016-2518-2
Spielmeyer, W., Ellis, M., Robertson, M., Ali, S., Lenton, J. R., and Chandler, P. M. (2004). Isolation of gibberellin metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor. Appl. Genet. 109, 847–855. doi: 10.1007/s00122-004-1689-6
Steinbach, H. S., Benech-Arnold, R. L., Kristof, G., Sanchez, R. A., and Marcucci-Poltri, S. (1995). Physiological basis of pre-harvest sprouting resistance in Sorghum bicolor (L.) Moench. ABA levels and sensitivity in developing embryos of sprouting-resistant and -susceptible varieties. J. Exp. Bot. 46, 701–709. doi: 10.1093/jxb/46.6.701
Sun, T.-P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, 338–345. doi: 10.1016/j.cub.2011.02.036
Sun, T.-P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55, 197–223. doi: 10.1146/annurev.arplant.55.031903.141753
Suzuki, H., Ishiyama, K., Kobayashi, M., and Ogawa, T. (2005). Specific expression of the gibberellin 3β-hydroxylase gene, HvGA3ox2, in the epithelium is important for Amy1 expression in germinating barley seeds. Plant Biotechnol. 22, 195–200. doi: 10.5511/plantbiotechnology.22.195
Suzuki, T., Matsuura, T., Kawakami, N., and Noda, K. (2000). Accumulation and leakage of abscisic acid during embryo development and seed dormancy in wheat. Plant Growth Regul. 30, 253–260. doi: 10.1023/a:1006320614530
Tuttle, K. M., Martinez, S. A., Schramm, E. C., Takebayashi, Y., Seo, M., and Steber, C. M. (2015). Grain dormancy loss is associated with changes in ABA and GA sensitivity and hormone accumulation in bread wheat, Triticum aestivum (L.). Seed Sci. Res. 25, 179–193. doi: 10.1017/s0960258515000057
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not Require SLR1 degradation in the gid2 mutant. Plant Cell 20, 2437–2446. doi: 10.1105/tpc.108.061648
Velde, K. V. D., Chandler, P. M., Van Der Straeten, D., and Rohde, A. (2017). Differential coupling of gibberellin responses by Rht-B1c suppressor alleles and Rht-B1b in wheat highlights a unique role for the DELLA N-terminus in dormancy. J. Exp. Bot. 68, 443–455. doi: 10.1093/jxb/erw471
Walker-Simmons, M. (1987). ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–66. doi: 10.1104/pp.84.1.61
Walker-Simmons, M., and Sesing, J. (1990). Temperature effects on embryonic abscisic acid levels during development of wheat grain dormancy. J. Plant Growth Regul. 9, 51–56. doi: 10.1007/bf02041941
Washio, K., and Morikawa, M. (2006). Common mechanisms regulating expression of rice aleurone genes that contribute to the primary response for gibberellin. Biochim. Biophys. Acta 1759, 478–490. doi: 10.1016/j.bbaexp.2006.09.001
Weiss, D., and Ori, N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246. doi: 10.1104/pp.107.100370
White, C. N., Proebsting, W. M., Hedden, P., and Rivin, C. J. (2000). Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 122, 1081–1088. doi: 10.1104/pp.122.4.1081
Woodger, F. J., Gubler, F., Pogson, B. J., and Jacobsen, J. V. (2003). A Mak-like kinase is a repressor of GAMYB in barley aleurone. Plant J. 33, 707–717. doi: 10.1046/j.1365-313X.2003.01663.x
Xi, W., Liu, C., Hou, X., and Yu, H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733–1748. doi: 10.1105/tpc.109.073072
Xie, Z., Zhang, Z.-L., Zou, X., Yang, G., Komatsu, S., and Shen, Q. J. (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46, 231–242. doi: 10.1111/j.1365-313X.2006.02694.x
Yaish, M. W., El-kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y.-M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. doi: 10.1371/journal.pgen.1001098
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamasaki, Y., Gao, F., Jordan, M. C., and Ayele, B. T. (2017). Seed maturation associated transcriptional programs and regulatory networks underlying genotypic difference in seed dormancy and size/weight in wheat (Triticum aestivum L.). BMC Plant Biol. 17:154. doi: 10.1186/s12870-017-1104-5
Yano, K., Aya, K., Hirano, K., Ordonio, R. L., Ueguchi-Tanaka, M., and Matsuoka, M. (2015). Comprehensive gene expression analysis of rice aleurone cells: probing the existence of an alternative gibberellin receptor. Plant Physiol. 167, 531–544. doi: 10.1104/pp.114.247940
Ye, H., Feng, J., Zhang, L., Zhang, J., Mispan, M. S., Cao, Z., et al. (2015). Map-based cloning of Seed Dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169, 2152–2165. doi: 10.1104/pp.15.01202
Ye, N., Zhu, G., Liu, Y., Zhang, A., Li, Y., Liu, R., et al. (2012). Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 63, 1809–1822. doi: 10.1093/jxb/err336
Zeng, D.-E., Hou, P., Xiao, F., and Liu, Y. (2014). Overexpressing a novel RING-H2 finger protein gene, OsRHP1, enhances drought and salt tolerance in rice (Oryza sativa L.). J. Plant Biol. 57, 357–365. doi: 10.1007/s12374-013-0481-z
Zeng, D.-E., Hou, P., Xiao, F., and Liu, Y. (2015). Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza Sativa L.). J. Plant Biochem. Biotechnol. 24, 56–64. doi: 10.1007/s13562-013-0236-4
Zentella, R., Yamauchi, D., and Ho, T.-H. D. (2002). Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14, 2289–2301. doi: 10.1105/tpc.003376
Zentella, R., Zhang, Z.-L., Park, M., Thomas, S. G., Endo, A., Murase, K., et al. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. doi: 10.1105/tpc.107.054999
Zhu, G., Ye, N., and Zhang, J. (2009). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 50, 644–651. doi: 10.1093/pcp/pcp022
Keywords: abscisic acid, dormancy, germination, gibberellin, plant hormones, preharvest sprouting, seed, cereals
Citation: Tuan PA, Kumar R, Rehal PK, Toora PK and Ayele BT (2018) Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front. Plant Sci. 9:668. doi: 10.3389/fpls.2018.00668
Received: 11 February 2018; Accepted: 30 April 2018;
Published: 23 May 2018.
Edited by:
Paola Vittorioso, Sapienza Università di Roma, ItalyReviewed by:
Alessandra Boccaccini, Université de Lausanne, SwitzerlandCopyright © 2018 Tuan, Kumar, Rehal, Toora and Ayele. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Belay T. Ayele, YmVsYXkuYXllbGVAdW1hbml0b2JhLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.