
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 April 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00559
Numerous studies have been focusing on breeding tomato plants with enhanced lycopene accumulation, considering its positive effects of fruits on the visual and functional properties. In this study, we used a bidirectional strategy: promoting the biosynthesis of lycopene, while inhibiting the conversion from lycopene to β- and α-carotene. The accumulation of lycopene was promoted by knocking down some genes associated with the carotenoid metabolic pathway. Finally, five genes were selected to be edited in genome by CRISPR/Cas9 system using Agrobacterium tumefaciens-mediated transformation. Our findings indicated that CRISPR/Cas9 is a site-specific genome editing technology that allows highly efficient target mutagenesis in multiple genes of interest. Surprisingly, the lycopene content in tomato fruit subjected to genome editing was successfully increased to about 5.1-fold. The homozygous mutations were stably transmitted to subsequent generations. Taken together, our results suggest that CRISPR/Cas9 system can be used for significantly improving lycopene content in tomato fruit with advantages such as high efficiency, rare off-target mutations, and stable heredity.
Precise genome editing provides remarkable advantages in plant agricultural trait improvement by generating tailored modifications at a target sequence. Clustered regularly interspaced palindromic repeats/associated 9 (CRISPR/Cas9) has been developed from studies on the prokaryote-specific adaptive immune system, where endonuclease Cas9 is coupled with a synthetic single guide RNA (sgRNA), generating an RNA-guide nuclease. The specificity of Cas9-directed DNA double-strand cleavage, causing double-strand breaks (DSBs) in nuclear DNA, is defined by Watson–Crick base-pairing of a 20-bp guide sequence on the guide RNA (gRNA) and a PAM, (“NGG” motif) immediately downstream of the target region. DSBs induced by CRISPR/Cas9 can trigger two independent endogenous DNA repair pathways: NHEJ and HR. NHEJ is error-prone and frequently causes indels around the DSBs, whereas HR accurately repairs DSBs by using the homologous flanking sequence or an exogenous donor DNA as a template, which can frequently result in small or large chromosomal changes (Sonoda et al., 2006; Wyman and Kanaar, 2006; Gaj et al., 2013; Voytas, 2013). CRISPR/Cas9 genome site-specific editing technology, a new genome modification tool, has been successfully applied to various plants, including rice (Miao et al., 2013), wheat (Shan et al., 2013; Wang et al., 2014), maize (Liang et al., 2014), and tomato (Brooks et al., 2014; Cermak et al., 2015; Ma et al., 2015; Ito et al., 2015; Jacobs and Martin, 2016; Lin et al., 2016; Xu et al., 2016). Unlike previous genome editing tools such as ZFN and TALEN, the CRISPR/Cas9 is easier to use, design flexible, and very cost-effective. Moreover, CRISPR/Cas9 genome editing tool is characterized by high mutation efficiency as well as rare off-target mutations (Xu et al., 2015), and homozygous mutations at the desired sites can be transferred to the subsequent generation owing to stable inheritance (Zhang et al., 2014; Fan et al., 2015). Furthermore, transgene-negative plants were observed in nearly all low-copy T1 generation of rice after CRISPR/Cas9-mediated targeted mutagenesis (Xu et al., 2015), which promoted the application of CRISPR/Cas9. CRISPR/Cas9 is also widely used in tomatoes. The mutations mediated by CRISPR/Cas9 are diverse; Rodriguez-Leal et al. (2017) reported that CRISPR/Cas9 can carry multiple gRNAs to rapidly and efficiently generate dozens of alleles for genes in order to interpret their function better. In addition, functional knockout mediated by CRISPR/Cas9 is significant. Ito et al. (2015) reported the CRISPR/Cas9 system can efficiently induce mutations in the tomato RIN gene and affected the accumulation or structure of the RIN protein. Yang et al. (2017) showed that maturation was significantly inhibited in mutants after RNA editing factor SlORRM4 was knocked out by CRISPR/Cas9 in tomato. However, CRISPR/Cas9 genome editing also has certain limitations, the genotypes cannot be artificially controlled, and production of large numbers of homozygotes often requires propagation over several generations.
Lycopene is considered as a bioactive component for treating chronic diseases and lowering the risk of cancer and cardiovascular diseases (Li and Xu, 2014; Pouchieu et al., 2014; Tang et al., 2014); numerous studies have attempted to elucidate the pathways associated with lycopene metabolism. It is a C40 carotenoid and is synthesized via carotenoid metabolism during fleshy fruit ripening (Figure 1A). Carotenoid biosynthesis depends on isopentenyl diphosphate (IPP) and its isomer DMAPP (Chappell et al., 1995). In plastids, four molecules of IPP were condensed to a molecule of GGPP. Then, two molecules of GGPP can be catalyzed by phytoene synthase 1 (PSY1) to form a molecule of colorless 15-cis-phytoene, which are head-to-head condensed, leading to the generation of ζ-carotene and pink prolycopene. Prolycopene is then converted to all-trans-lycopene by carotenoid isomerase (CRTISO). Next, LCY-B and LCY-E catalyze the cyclisation of lycopene to produce β-carotene or α-carotene, respectively. Eventually, these substances can be degraded into lutein, zeaxanthin, and other carotenoids. Lycopene, a major component of carotenoids in ripe tomato, confers the attractive color and economical quality to the fruit. Thus, enhanced lycopene accumulation in fruit is essential to improve the visual and functional properties of tomato.
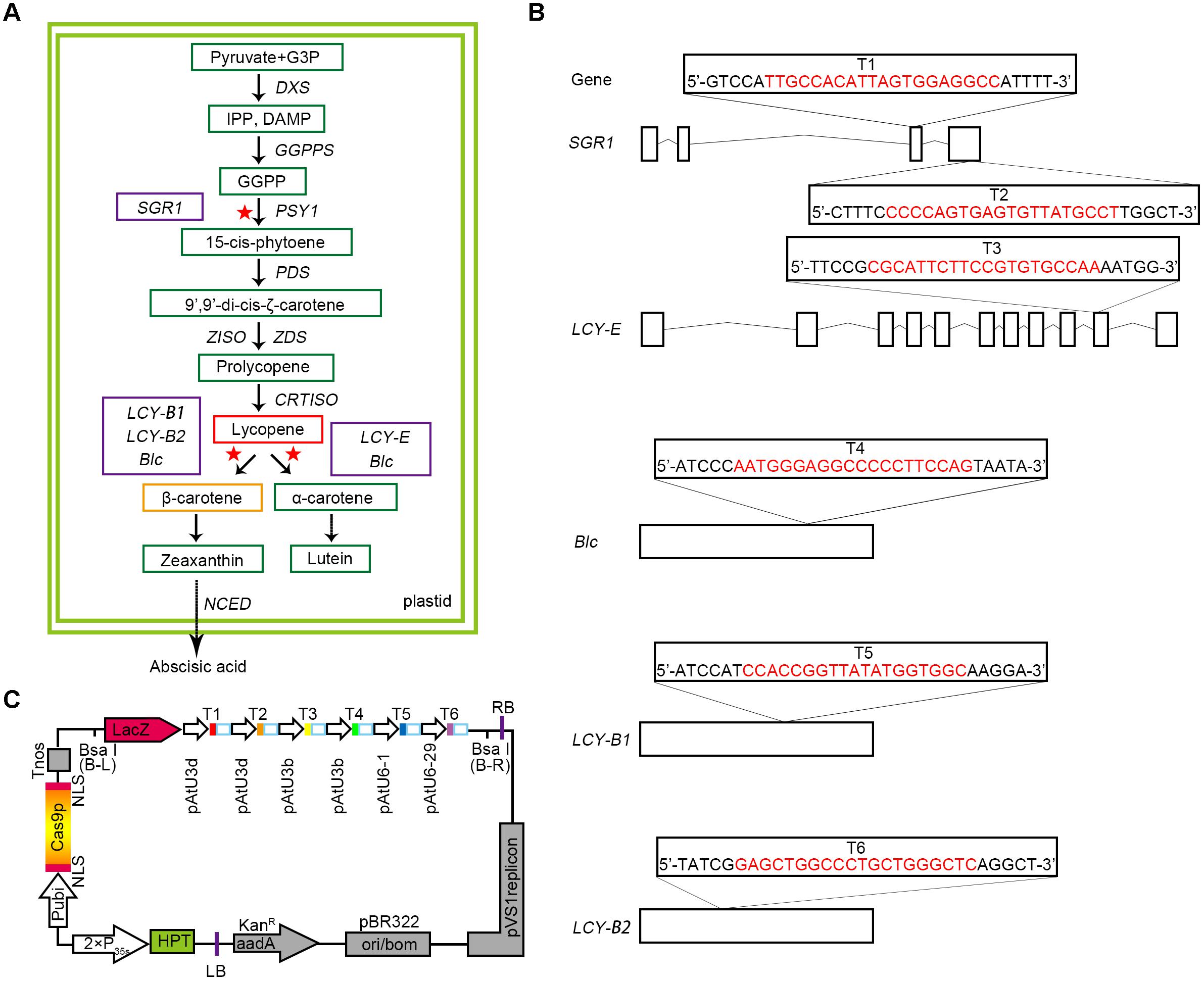
FIGURE 1. Selection of target genes and designing of CRISPR/Cas9 binary expression cassette. (A) A map of the target genes in the carotenoid metabolic pathway. The green boxes represent the key substances in the metabolic pathway. The red and orange boxes show the two substances, lycopene and β-carotene, respectively. A solid arrow indicates a direct effect, and a dashed arrow indicates an indirect effect. The selected target genes are represented by purple boxes, and the red asterisks represent the sites at which the target genes act on the pathway. G3P, glyceraldehyde 3-phosphate; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; GGPPS, geranylgeranyl pyrophosphate synthase; PDS, phytoene desaturase; ZISO, z-carotene isomerase. (B) Five target genes were selected according to the synthesis and metabolism pathways of lycopene, and six target sites were designed. The target sequences are marked in red, and small rectangle frames indicate the PAM. Straight lines and boxes are the introns and exons of the target genes, respectively. (C) Structures of the pYLCRISPR/Cas9-Lycopene binary vectors. HPT(-H) encodes hygromycin B phosphotransferase. The six targets designed are represented by solid boxes in different colors, and the promoters used for each target are shown.
Modulation of the expression of key genes in the lycopene metabolism pathway is an effective way to increase lycopene content. Recently, significant progress has been achieved in genome modification in plants (Giuliano et al., 1993; Fraser et al., 1994, 1999; Rosati et al., 2000; Galpaz et al., 2008; Chen et al., 2015). For example, null mutation of the gene lycopene β-cyclase 2 (LCY-B2) in transgenic plants increased the lycopene content by about 5% on average and led to the formation of deep-red colored tomato fruits (Ronen et al., 2000). Further, the overexpression of phytoene synthase 1 gene (PSY1) significantly increased the level of lycopene in tomato fruit (Fraser et al., 2007). In addition, fruit-specific RNAi-mediated suppression of 9-cis-epoxycarotenoid dioxygenase 1 (NCED1) produced deep-red colored fruits with high accumulation of lycopene and β-carotene (Sun et al., 2012). Moreover, silencing of stay-green 1 (SGR1) promoted the activity of PSY1 via direct physiological interaction during fruit maturation, which significantly elevated the accumulation of lycopene and β-carotene (four and ninefold, respectively) in fruit (Luo et al., 2013). However, recent studies on enhancing lycopene accumulation in tomato fruit by regulating the carotenoid metabolic pathway are mainly focused on the modification of a single related gene. Modulation of multiple genes in metabolic networks to achieve the accumulation of lycopene has been rarely reported, owing to the complexity and integrity of the metabolic mechanism. Thus, in this study, CRISPR/Cas9 was applied to regulate multiple genes associated with the carotenoid metabolic pathway of tomato, in order to increase the lycopene content in fruits and explore the roles of specific key genes in carotenoid metabolism. On the one hand, we lay the foundation of research for the application with CRISPR/Cas9 multiplex genome editing in metabolic pathway. On the other hand, we provide a new research idea for engineering the content of some target substance in the metabolic pathway.
The study was conducted using Solanum lycopersicum cv. AC. All plants were grown in the same greenhouse (greenhouse conditions: 16 h light/8 h dark, 25°C). Tissue samples at 7 days after breaker stage of ripening (Br+7) of fruits were collected and immediately frozen in liquid nitrogen and stored at -80°C. Mature seeds were collected from T0 plants, dried, and shaken in a 25°C shaker. The germinated seeds were then planted in the soil, and seedlings were grown under the above-mentioned culture conditions.
CRISPR-P1 was used to select specific sgRNAs targeting SGR1 (GenBank accession no. DQ100158), lycopene ε-cyclase (LCY-E; GenBank accession no., EU533951), beta-lycopene cyclase (Blc; GenBank accession no., XM_010313794), lycopene β-cyclase 1 (LCY-B1; GenBank accession no., EF650013), and LCY-B2 (GenBank accession no., AF254793) (Supplementary Table S1). Improper GC content has been shown to lead to inefficient editing (Wang et al., 2014); therefore, the content of GC was between 55% and 75%. In addition, 4 or more consecutive T nucleotides in the target sequence were avoided since they would be recognized as transcriptional termination signal by RNA polymerase III. Furthermore, RNA Folding program2 was used to ensure that no more than five base pairings occurred between the target sequence and sgRNA sequence since the secondary structure of sgRNA was remarkably affected by the editing efficiency (Makarova et al., 2011). In order to obtain a more obvious effect on gene function, we selected target sites in the exon of the target genes. Finally, targets with fewer putative off-target loci were preferred.
The pYLCRISPR/Cas9-Lycopene vector was constructed as described previously (Ma et al., 2015). First, each target sequence was ligated to its corresponding sgRNA expression cassette during the first PCR. This was followed by a second PCR to amplify the fragments as well as induce BsaI restriction sites in the target for generating of sgRNA expression cassettes with target sequences. The standard PCR condition was as follows: 94°C for 2 min; 94°C for 10 s; 55°C for 30 s; 68°C for 20 s for 28 cycles and 72°C for 7 min. Finally, the sgRNA expression cassettes were assembled to pYLCRISPR/Cas9 binary plasmid in one round of cloning by using Golden Gate ligation for six sgRNAs expression cassettes in pYLCRISPR/Cas9-Lycopene vector. The reactions were incubated for three cycles (37°C, 10 min; 10°C, 5 min; 20°C, 5 min) and a subsequent 10 cycles (37°C, 3 min; 10°C, 5 min; 20°C, 5 min) was run. Then the reaction was incubated at 37°C, 5 min. The oligonucleotide primers used are listed in Supplementary Table S2.
The Agrobacterium-mediated transformation method (Van Eck et al., 2006) was used, and pYLCRISPR/Cas9-Lycopene plasmid was transformed into AC. In brief, tomato seeds were germinated on MS medium after sterilization with 4% NaClO. After 7–10 days culture, the apical segments of hypocotyls were punctured with OD600 = 0.5–0.6 of Agrobacterium suspension. Then, the explants were inoculated on selective plates with hygromycin (10 μg/mL) until transgenic plants were regenerated from the calluses. After in vitro regeneration, plants were transplanted into soil in light growth chamber.
About 1–5 mg fresh frozen leaves were used for DNA extraction by using hi-DNA secure plant kit (Tiangen, Beijing, China). The extracted genomic DNA was then used as a template to amplify the relevant fragments from each of the target genes by using primers (Supplementary Table S3) flanking the target sites. The standard PCR condition was as follows: 94°C for 3 min; 94°C for 30 s; 55°C for 30 s; 72°C for 30 s for 35 cycles and 72°C for 7 min. The PCR products were sequenced directly by using internal sequencing primers (Supplementary Table S3) or cloned into the pEasy-T1 (TransGen Biotech, China) vector and then sequenced using the Sanger method to identify mutations. Superimposed sequence chromatograms produced by biallelic and heterozygous mutations were decoded using DSDecode3 and manual analysis. The mutation rate of each target is calculated by the ratio, which is the number of transgene tomato plants edited at each target to 24 (the number of total transgenic tomato plants obtained).
In all, 10 mutants were selected randomly from lycopene mutants used for the detection of Cas9 gene by PCR. The standard PCR condition was as follows: 94°C for 3 min; 94°C for 30 s; 55°C for 30 s; 72°C for 30 s for 35 cycles, and 72°C for 7 min. Primers used for amplification of Cas9 gene are listed in Supplementary Table S6.
The lines were selected from the plants in which the targets were edited, based on the first two candidate off-target sites predicted by the online tool (CRISPR-P4), to sequence the changes in the fragment amplified by PCR from its corresponding gene locus. The standard PCR condition was the same as Cas9 analysis. Primers used for off-target site mutation analysis are listed in Supplementary Table S4.
The carotenoid was determined by grounding the samples into powder after freezing in liquid nitrogen. About 0.2 to 0.5 g tissue powder was dissolved in 1 mL of acetone and centrifuged. The tissue debris was dissolved again in 1 mL of dichloromethane and centrifuged. The supernatant was pooled using an acetone filtrate. The dissolving and collection of solvents was repeated until the tissue lost its color. Pigments were extracted by partitioning the solvent mixture against an equal volume of diethyl ether and 0.2 volume of 12% w/v NaCl/H2O. The colored organic fraction (upper phase) was collected and dried under a stream of N2 and the dry lipid extract was re-dissolved in 1 mL acetone for further analysis.
Carotenoid were analyzed using reverse-phase high-performance liquid chromatography (RT-HPLC) as described in a previous study with some modifications (Neuman et al., 2014). Chromatography was performed using an Agilent liquid chromatography system (Agilent Technologies, Santa Clara, CA, United States) equipped with a model G1322A degasser, model G1311A infusion pump, model G1313A auto sampler, model G1316A column thermoformer, and model G1314A detector.
The static phase consisted of a Diamonsil C18(2) reversed-phase column (4.6 mm × 250 mm, 5 μm; Dikma). The mobile phase consisted of eluent A (acetonitrile: H2O; v/v; 9:1) and eluent B (ethyl acetate) at a constant flow of 1 mL⋅min-1. The linear gradient program was performed as follows: initial condition was 55% A to 100% B within 30 min and back to the initial condition for re-equilibration. Analysis was conducted under subdued light to avoid carotenoid degradation.
HPLC-grade β-carotene and lycopene standards were obtained from Sigma (St. Louis, MO, United States).
Carotenoid extractions were performed as described previously (Fantini et al., 2013). Briefly, lyophilised tomato fruit powder was extracted with chloroform and methanol (2:1 by volume). Subsequently, 1 volume of 50 mM Tris buffer (pH 7.5, containing 1 M NaCl) was added, and the samples were kept for 20 min on ice. After centrifugation (15 000 g for 10 min at 4°C), the organic phase was collected and re-extracted. The combined organic phases for each sample were then dried by nitrogen blowing and re-suspended in 100 μL of ethyl acetate. For each group, at least three independent extractions were performed. The carotenoids were identified and quantified using an Accurate-Mass HPLC1200/MS-QTOF 6520A (Agilent Technologies, United States) system packed with a reversed-phase column (4.6∗150 mm, 3 μm; YMC, Japan), and the carotenoid was washed out with the mobile phase: A = 81% MeOH + 15% MTBE + 4% H2O; B = 8% MeOH + 90% MTBE + 2% H2O at a flow rate 0.4 mL/min. The settings were as follows: DAD, 260–550 nm; mass range, 200–800; APCI ion source, drying gas of N2 at the pressure of 40 psi, 350°C, 8 L/min; VCAP, 3500 V; Fragmentor, 160 V; Skimmer, 65 V; and OCT RF Vpp, 750 V, with the negative MS scan mode 2GHzExt Dyn (3200). HPLC peak areas at 260–550 nm was integrated and calibrated using external standards (e.g., α-carotene purchased from Sigma-Aldrich). The calibrated samples were mixed to generate multiple-diluted external calibration curves for the quantification of the pigments. Carotenoids were identified based on typical retention times and specific published absorption spectra (Mialoundama et al., 2010; Regnier et al., 2015).
Transmission electron microscopy (TEM) results were analyzed as described in a previous study (Yang et al., 2017); the pericarps of fruits from WT and mutants at Br+7 were fixed with 2.5% glutaraldehyde for over 2 h and washed three times with 0.1 mL phosphate buffer. Next, the sample was fixed with 1% osmic acid for 2 h and washed three times with 0.1 mL phosphate buffer. The sample was dehydrated using different concentrations of acetone (30, 50, 70, 90, and 100%). Next, it was embedded and aggregated with ethoxy line resin and sliced up using LEICA UC6 ultra microtome. After the sample was dyed with both uranyl acetate and lead citrate, the prepared sample was observed under a JEM-1230 transmission electron microscope.
Based on the advantages of CRISPR/Cas9 multiplex genome editing system, we carefully selected a bidirectional strategy (Figure 1A). Some sites were designed to target gene SGR1 for promoting the synthesis of lycopene, whereas others were selected from genes that catalyze the cyclisation of lycopene, such as LCY-E, LCY-B1, and LCY-B2, as well as Blc. LCY-E was used to prevent the cyclisation from lycopene to α-carotene and LCY-B1 and LCY-B2 were used to prevent the cyclisation from lycopene to β-carotene (Pecker et al., 1996; Ronen et al., 1999, 2000; Moreno et al., 2013; Ralley et al., 2016), whereas Blc is a gene that has both β-cyclase and ε-cyclase activities predicted by InterPro Database5.
Since SGR1 gene plays a relatively important role in the accumulation of lycopene, two target sites (targets T1 and T2) were designed to ameliorate the targeting efficiency of this gene (Figures 1A,B). In addition, the other corresponding target sites were selected (targets T3, T4, T5, and T6) at the exons of genes LCY-E, Blc, LCY-B1, and LCY-B2, respectively (Figures 1A,B). Finally, 6-sgRNA of the CRISPR/Cas9 binary expression system (Figure 1C and Supplementary Table S1) was constructed to target distinct tomato genomic sites of genes associated with the synthesis and metabolic pathways of lycopene, including both multiple genes in a pathway and multiple sites in a single gene.
The editing efficiency of CRISPR/Cas9 for the selected target genes has been shown to be remarkable (Brooks et al., 2014; Zhang et al., 2014; Fan et al., 2015). In all, 24 lines of T0 transgenic plants (CR-lycopene-1 to 24) were obtained; specific editing types of each target were identified and analyzed. Fortunately, the 24 transgenic lines showed different types of genome editing at the target sites (Supplementary Figure S1), which indicated that multiplex CRISPR/Cas9 is extremely efficient in tomato fruit to generate tailor-made modifications at target sequences. The mutation rates varied widely among different target sites, from 0 to 95.83% (Figure 2A). The editing efficiencies of targets T2 and T4, especially target T2 (95.83%), were considerably higher than those of the others. However, target T5 did not show gene editing in any of the obtained transgenic lines (Figure 2A).
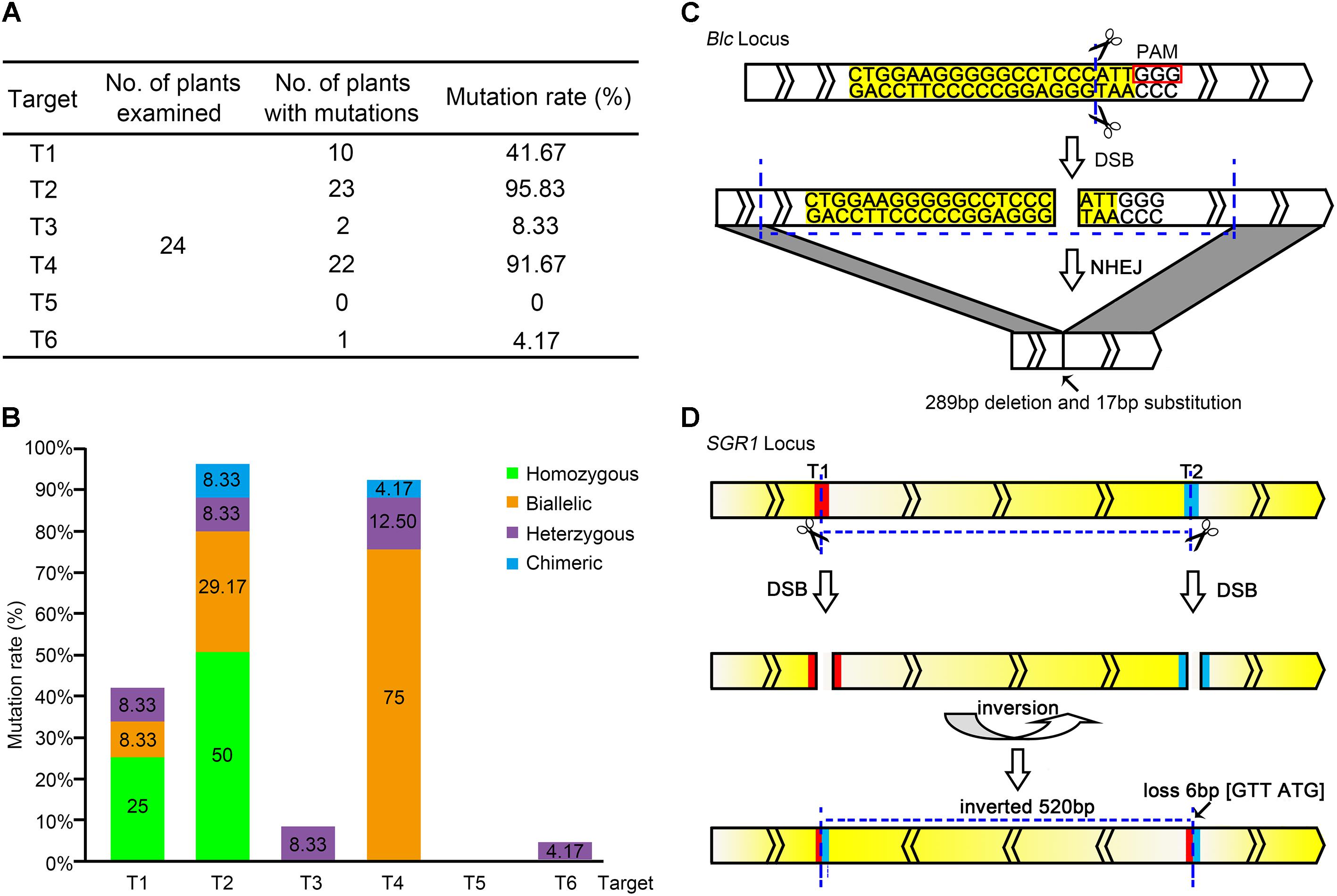
FIGURE 2. Editing of each target site in the pYLCRISPR/Cas9-lycopene expression cassette. (A) Total editing efficiency of the six targets in the pYLCRISPR/Cas9-lycopene expression cassette. The mutation rate is the ratio of the number of mutations detected to that of the total number of plants in which mutations are detected. (B) Specific types of each target among the six target sites in the pYLCRISPR/Cas9-lycopene expression cassette. Green, orange, purple, and blue represent homozygous, biallelic, heterozygous, and chimeric mutations, respectively. (C) Large fragment deletion on Blc. Target sequence is labeled with yellow. Small rectangle frames indicate the protospacer adjacent motifs. The transcriptional direction of the Blc is shown. (D) Special DNA inversion on SGR1. The red and blue parts represent the targets T1 and T2, respectively. Gradient change of color between yellow and white indicates the transcriptional direction of SGR1.
Further analysis of the RNA-guided genome-editing events indicated that the editing type also varied among distinct targets (Figure 2B). Two kinds of mutations, homozygous and biallelic mutations, were the most common among six target sites. Generating homozygous mutations at targets T1 and T2 with higher editing efficiency was possible. However, target T4 was slightly distinct, and the biallelic mutation was dominant. Correspondingly, the mutation type of targets T3 and T6 with low editing efficiency was not ideal; it was dominated by heterozygous mutations. In our study, targets T1 and T2 driven by the AtU3b promoter and target T4 driven by the AtU3d promoter showed high editing efficiencies. In contrast, target T5 regulated by the AtU6-1 promoter and target T6 regulated by the AtU6-29 promoter showed very low editing effects.
Although most gene mutant alleles were small indels (less than 10 bp) at the desired target sites, two interesting mutation types were observed. One was a large DNA fragment deletion. The length of PCR fragments in target T4 was obviously shorter, suggesting the deletion of a large DNA fragment with different sizes on target gene (Figure 2C and Supplementary Figure S2). Another was DNA inversion (Figure 2D), where the direct and complementary strand exchange occurred between targets T1 and T2 of SGR1 gene.
In addition, we found that the percentage of T0 plants carrying mutations at two target sites was similar to the expected double mutation rate (Supplementary Table S5), indicating that mutations at two sites targeted by one construct might occur independently of each other. This finding is similar to those reported previously (Zhang et al., 2014). This result suggests that the levels of Cas9 and sgRNAs were not limited in transgenic tomato plants.
The presence of Cas9 in lycopene mutants was confirmed by PCR for Cas9 gene in the randomly selected lycopene mutation lines (primers listed in Supplementary Table S6). The Cas9 gene was detected in the lycopene mutants (Supplementary Figure S3), indicating that the CRISPR/Cas9 binary expression cassette was indeed delivered to the tomato plant cells by Agrobacterium tumefaciens.
The potential off-target effects of CRISPR/Cas9 in tomato were evaluated by detecting the editing of putative off-target sites (Supplementary Table S7). No clear off-target events were detected, suggesting that CRISPR/Cas9-induced mutagenesis was highly specific in tomato plants. Previous studies have also shown similar results that CRISPR/Cas9 system has obvious advantages of avoiding off-target mutations in tomato (Zhang et al., 2014). The CRISPR/Cas9 multi-target gene editing technology with well-designed specific sgRNAs has distinct characteristics of significant editing and low off-target mutations.
The levels of carotenoids were determined by classifying the 24 transgenic tomato plants into 5 mutant groups according to the different mutant target genes, including single, double, triple, and quadruple mutants, which were named as Lycopene-1 to 5 (Figure 3A). Representative transgenic lines were selected from each group for further analysis. The tomato fruits of different mutant groups at Br+7 of ripening were sampled separately for determining lycopene and β-carotene contents by using HPLC. The levels of two main components of the tomato fruit were determined according to the respective peak of the commercial standard of lycopene and β-carotene (Figure 3B). HPLC analysis showed that the contents of lycopene and β-carotene in all lycopene mutants were higher than those in the WT plants (Figures 3C,D). In particular, the lycopene content of group Lycopene-1, in which the SGR1 gene was targeted alone, was the highest. Correspondingly, the color of Lycopene-1 fruit was also the most vivid (Figure 3B), and the lycopene level (5.1-fold) in Lycopene-1 mutants was slightly higher than that (4-fold) in a previous transgenic tomato fruit subjected to RNAi of SGR1 (Luo et al., 2013). However, mutation of the Blc gene alone (Lycopene-2) did not remarkably improve lycopene accumulation compared with that in the Lycopene-1 group (Figure 3C), indicating that the effect of SGR1 gene on the regulation of lycopene content in tomato fruit was more pronounced than that of Blc gene.
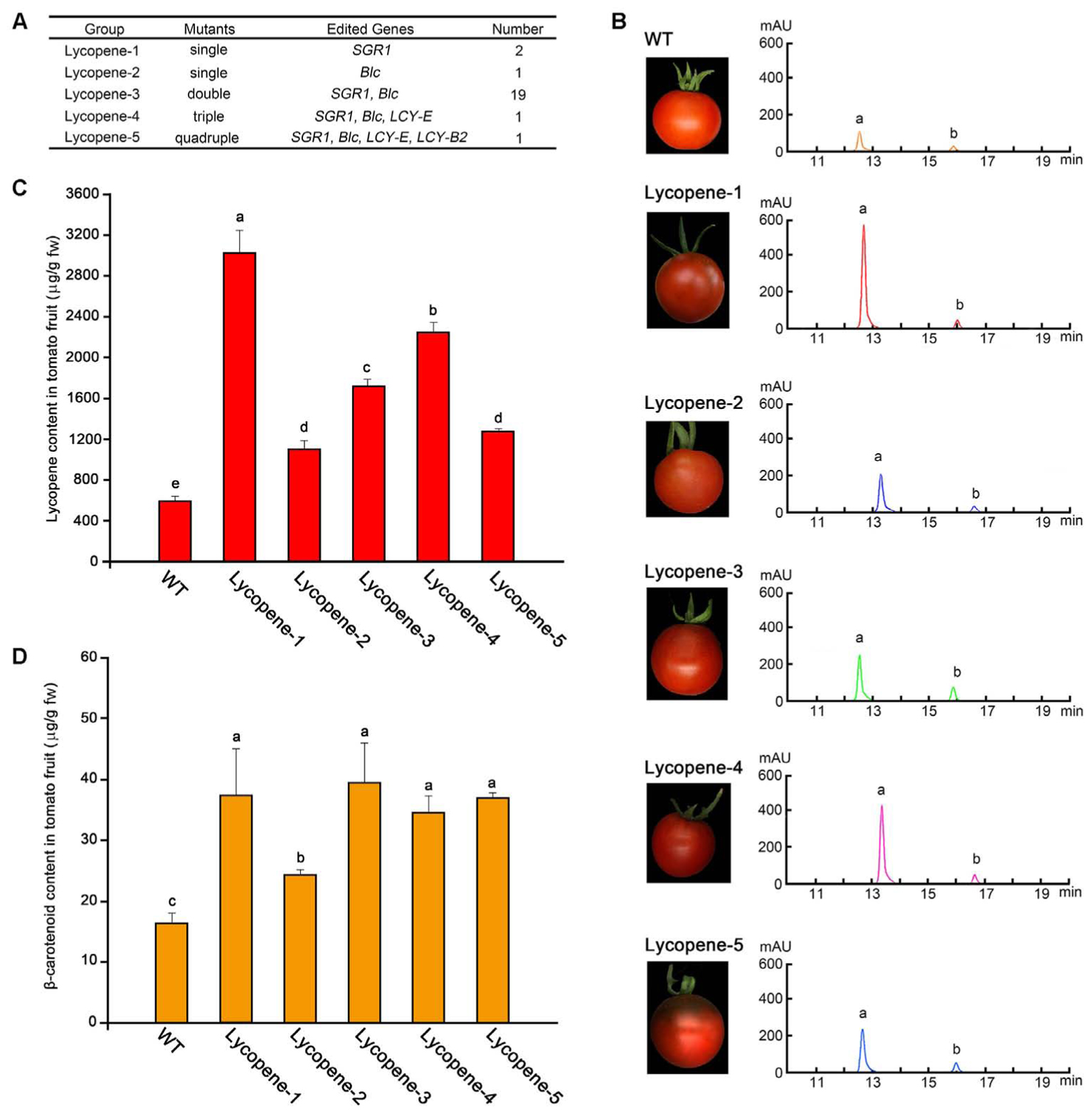
FIGURE 3. Determination of lycopene content in tomato fruit at Br+7 from different mutant groups. (A) List of different groups of lycopene mutants according to the different combinations of mutant genes. (B) HPLC results of crude extracts from tomato fruit samples in different groups. a, b, lycopene and β-carotenoids, respectively. (C,D) Contents of lycopene and β-carotenoids of tomato fruit in the five different groups and WT, respectively. Error bars represent standard deviation. Different lowercase letters show statistically significant difference according to ANOVA followed by Duncan’s test (p < 0.05).
The content of other carotenoids in the metabolic pathway was determined using HPLC-MS (Supplementary Figure S4). The results showed that the content of phytoene, prolycopene, α-carotene, and lutein in most of the lycopene mutants enriched significantly compared with in WT. Even the contents of these carotenoids in the single mutants also increased significantly, such as Lycopene-1 and Lycopene-2. However, the Lycopene-4 mutant showed a significant decrease in α-carotene content compared to that in WT. Moreover, unlike in other mutants, prolycopene in Lycopene-4 mutant showed no significant change from that in WT.
To further confirm the improvement of lycopene accumulation at the cellular level, we used TEM to observe the tomato fruits of WT, Lycopene-1, and Lycopene-5 (Figure 4). Unlike in WT, in Lycopene-1 and Lycopene-5, the plastid numbers in the pericarp cells were significantly higher at Br+7. Further, carotenoid-containing structures (osmiophilic globules) and crystal lines were higher in the plastids of lycopene mutant fruits than in WT fruits (Figure 4), resulting in the higher lycopene content of mutant tomatoes. In addition, the TEM images were different between the mutant and WT fruits at Br+7 (Figure 4). At the ripening stage, the vacuolisation of chloroplast was evident in the WT fruits, but a complete membrane was still observed. However, only plastids, but not chloroplast, were observed in the mutants. Our findings indicated that, with increased lycopene concentration in fruits, the conversion of chloroplasts to chromoplasts might occur earlier in transgenic fruits than in WT. This is sufficient to show that CRISPR/Cas9 multi-target genome editing can be successfully applied to regulating the metabolism of carotenoids to increase the accumulation of lycopene in the T0 generation of tomato.
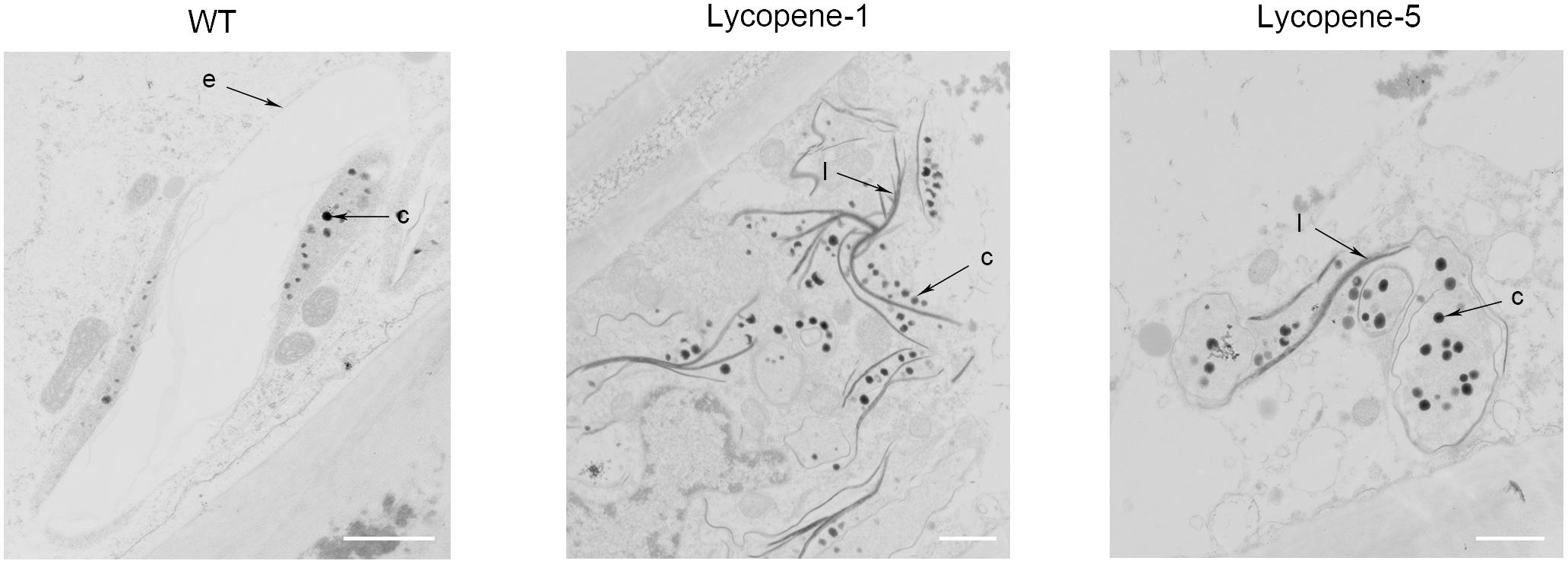
FIGURE 4. TEM images of epidermal cells in tomato fruit of WT and lycopene mutants at Br+7. c, carotenoid containing structures; e, plastid envelope; l, crystal line. Bar, 1 μm.
During the growth of lycopene mutants, most mutants in which the SGR1 gene was targeted (alone or along with other genes) showed the desired rust color in fruit after the breaker stage of ripening (Figure 5). The SGR1 gene silencing in tomato fruit has been shown to lead to the development of rust color containing both red and green (Luo et al., 2013). In Lycopene-1 and Lycopene-5, the differences in the cross-section of lycopene mutant fruits between Br+7 and Br+14 could be clearly observed (Supplementary Figure S5). The red portion of the entire surface of the fruit of Lycopene-1 and Lycopene-5 at Br+14 was significantly increased compared with that at Br+7, when the green portion was correspondingly reduced. Therefore, the green color in the endocarp of the fruit at Br+7 refers to the immature stage. However, the flesh color of Lycopene-5 was still green rather than red at Br+14. This residual green was distinct from that noted in the previous RNAi-SGR1 lines, but was similar to the apparent trait of the SGR1 natural mutant (Luo et al., 2013). Thus, CRISPR/Cas9 specific-site DNA editing is more complete and comprehensive than the traditional RNAi technology of RNA silencing.

FIGURE 5. Phenotype of tomato fruit at different ripening stages. Three groups of transgenic tomato fruits were photographed at different times after the breaker stage of ripening and compared with that in WT. Sections were obtained at Br+7. Each group photograph was obtained from the same tomato fruit. e, exocarp; i, endocarp and f, flesh of the tomato fruit.
Interestingly, we also found a clear trend: higher was the number of genome sites that were modified in plants including SGR1 gene targeted, the greater was the green portion in the cross-section of the fruit. Lycopene-1, Lycopene-3, and Lycopene-5 fruit cross-sections are shown in Figure 5. We found that the surface color of lycopene mutant fruits was significantly different from that of the WT fruits after the breaker stage of ripening; the green portion was clearly visible in the former. Further, the green colouration was more obviously closer to the stalk. In the same growth stage, the proportion of green part of fruit of Lycopene-1, Lycopene-3, and Lycopene-5 increased in turn. These results indicate that the combination of different target editing types affects the surface color of the tomato fruit.
In order to obtain tomato varieties with high lycopene content and stable genetic traits, we selected the progeny of two lines to investigate the transmission pattern of CRISPR/Cas9-mediated mutations. As expected, Lycopene-1 T1 generation of targets T1-SGR1 and T2-SGR1 homozygotes were homozygous for the same mutations (Table 1), indicating that the mutations in the homozygotes were stably transferred to the next generation in a Mendelian fashion, which was consistent with the findings of previous studies (Feng et al., 2014; Zhang et al., 2014; Pan et al., 2016). However, an unexpected segregation ratio of 1:4:1 was observed in the T1 generation of the target T3 (LCY-E) lycopene-5 lines. These results indicated that two alleles in one biallelic mutant might not be inherited with equal frequencies (Xu et al., 2015). However, the segregation patterns of most heterozygotes and bialleles were less predictable, and many new mutants were found in the T1 lines, which might be attributed to somatic mutations. Studies in Arabidopsis have shown that most mutations in early generations are somatic mutations, leading to the difficulty in predetermining the targeted genotype in the subsequent generation (Jiang et al., 2013; Jacobs et al., 2015; Xu et al., 2015).
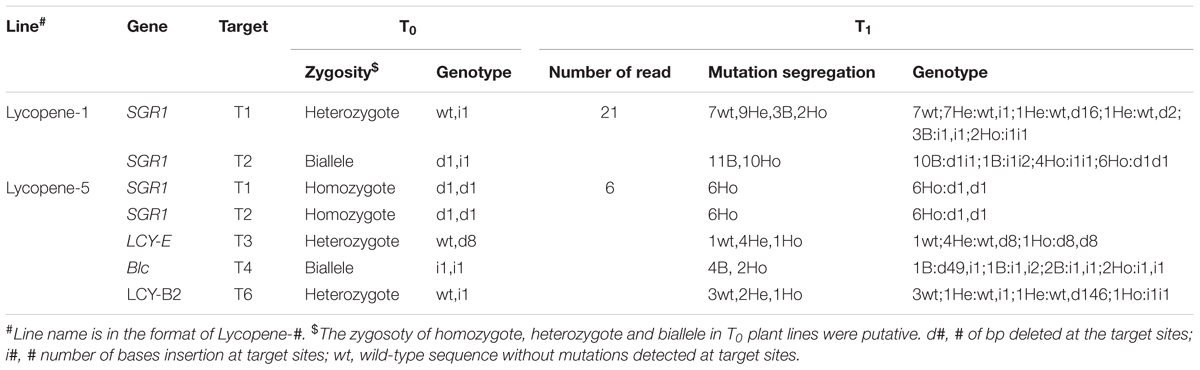
TABLE 1. Segregation patterns of CRISPRCas9-medicated targeted mutagenesis during the T0 to T1 generation.
Multiplex CRISPR/Cas9 genome editing has been used for efficiently editing targets in various species (Miao et al., 2013; Brooks et al., 2014; Feng et al., 2014; Zhang et al., 2014; Fan et al., 2015; Ma et al., 2015; Xu et al., 2015). In this study, all the T0 lycopene mutants were genetically modified (Supplementary Figure S1), which confirms that the CRISPR/cas9 system has a high editing efficiency for well-designed sites at genes of interest in tomato. Ma et al. (2015) suggested that the GC contents and sgRNA secondary structure are important for improving target editing efficiency. Since, the GC content and base pair number (between sgRNA sequence and target sequence) were not significantly different among six target sites (Supplementary Table S1) in this study based on strict optimisation. Therefore, the distinct mutant efficiencies among tomato plants could likely be attributed to different promoters. AtU3b, AtU3d, AtU6-1, AtU6-29 are four different snRNA promoters cloned from Arabidopsis thaliana to avoid HR between sgRNA expression cassettes in Agrobacterium or plant genome (Ma et al., 2015). Specifically, there is a difference in length between them. AtU3b promoter is the longest, up to 344 bp; AtU3d promoter is the shortest, only 102 bp. The results in this study suggest that there may be a distinction in the efficiency of target editing between different promoter, but the exact mechanism is not clear. Unfortunately, none of the indel mutations were detected at the T5 target in all transgenic plants obtained. We speculated two reasons for this. One is the failure of gene targeting because of a series of complex and unclear mechanisms. The other is probably that the gene Blc selected by the T5 target has a very important role in the conversion of lycopene to α- and β-carotenoids. Mutation of this gene might produce lethal phenotypes, but the specific mechanism is not yet clear. In addition, target-sgRNAs with extended nucleotides at the 5′ end (derived from the vector ligation site) have been shown to guide genome editing in plants (Xie and Yang, 2013). Studies in rice suggest that this target-sgRNAs do not affect the editing efficiency (Ma et al., 2015). However, in this study, target T5 that did not show target editing and target T6 with significantly lower editing efficiency were so-called regular targets (using the U6 promoter, while the target 5′ end for G); whether this kind of target-sgRNAs affects the editing efficiency in tomato is not yet known.
To improve yield and stress resistance, researchers are paying considerable attention to the enhancement fruit quality of tomato. The content of lycopene, which is a kind of carotenoid, is one of the most important quality traits of tomato fruit, with industrial, health, and nutritional attributes. Since the amounts of fruit volatiles are correlated to carotenoid levels, the higher the lycopene content in a fruit, the more essential flavor volatiles are generated, rendering the fruit to be more nutritious (Klee and Giovannoni, 2011). The high demand for natural lycopene to serve as healthy antioxidant requires that tomato plants are converted to green factories for the economical production of high-value lycopene; this has triggered increasing interest in the up-regulation of lycopene content in fruit. In this study, we successfully enhanced lycopene in tomato fruits by using multiplex CRISPR/Cas9 genome editing. However, with an increase in mutant genes, no concomitant increase in lycopene content was noted in the lycopene mutants (Figure 3C); this could be speculated by two reasons: bottom-up of the carotenoid accumulation model associated with carotenoid metabolism in tomato (Luo et al., 2013) and the mechanism of the feedback regulation of the carotenoid metabolic pathway. However, it is obvious that whether such a feedback mechanism really works or not remains to be verified. The transformation pathway of lycopene to α-carotene and β-carotene was inhibited in groups Lycopene-3–5, unlike in groups in which only the SGR1 gene was targeted (Lycopene-1). Therefore, the accumulation of the two above mentioned carotenoids and thus that of lycopene was affected in the tomato fruit. Further, we concluded that the promotion of lycopene synthesis may be better than the inhibition of its cyclisation to enhance the accumulation of lycopene by regulating the carotenoid metabolism networks in tomato.
The green coloration in the fruits of some lycopene mutants was mainly caused by target editing of the gene SGR1 (Figure 5). SGR is a positively regulated target of RIN, and its proteins have been shown to play a critical role in the initiation of chlorophyll degradation and senescence (Jiang et al., 2007; Ren et al., 2007; Barry et al., 2008; Hortensteiner, 2009; Fujisawa et al., 2013). We found that the green color in the fruit surface of lycopene mutants would gradually weaken with time; however, some green residues remained within the fruit compared to that in WT. The reason for this phenomenon is not yet clear. Furthermore, previous studies suggested that the use of RNAi to silence the SGR1 gene did not affect the maturation of tomato fruit. The time required for transgenic fruit diameter to reach 1 cm was reported to be not significantly different from that in WT (Luo et al., 2013). Thus, the combination of a gene or many genes such as the four genes (SGR1, LCY-E, Blc, and LCY-B2) might have directly or indirectly affected the maintenance of the tomato fruit transitions in color.
More recently, studies have shown that the CRISPR/cas9 system can be used for genome editing of transcription factors (Ito et al., 2015) and PDS as the reporter gene (Pan et al., 2016) in the model plant tomato. In addition, the CRISPR/cas9 system has also recently been used to enhance tomato fruit quality, resistance to disease, and to extend the shelf life of tomato plants. The content of γ-aminobutyric acid in tomato fruit has been reported to be effectively increased by CRISPR/cas9 (Li et al., 2017; Nonaka et al., 2017). Nekrasov et al. (2017) obtained tomato plants with stronger resistance to the powdery mildew fungal pathogen by using the CRISPR/Cas9 technology. Recent studies have also shown that CRISPR/Cas9-induced gene replacement via homology-directed repair provided a valuable method for generating tomato lines having a long shelf life (Yu et al., 2017). In this study, the pYLCRISPR/Cas9 multi-target editing system was successfully applied to create mutations of many related genes in carotenoid synthesis and metabolic pathways in tomato plants by artificially introducing a transformation vector containing multiple sgRNA expression cassettes to increase the content of lycopene in fruit. This study provides the basis for acquiring new tomato varieties with improved agricultural traits.
XL and HZ designed the research. YW and SC provided help for carotenoid analysis using HPLC. XL performed the experiments and drafted the manuscripts. HT, DF, BZ, and YL provided materials and intellectual input for the work.
This work was supported by National Key R&D Program of China (2016YFD0400901), grants from Chinese Universities Scientific Fund (2018QC100), and National Natural Science Foundation of China (31622050, 91540118, 31471921, and 31672208) to HZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Yaoguang Liu (South China Agricultural University, Guangzhou, China) for kindly providing the binary vector pYLCRISPR/Cas9 and the sgRNA plasmids.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00559/full#supplementary-material
AC, Ailsa Craig; Br+7, 7 days after breaker stage of ripening; CRISPR/Cas9, clustered regularly interspaced palindromic repeat associated protein 9; CRTISO, carotenoid isomerase; DMAPP, dimethylallyl diphosphate; DPA, days post anthesis; DSB, double-strand breaks; DXS, 1-deoxy-d-xylulose 5-phosphate synthase; G3P, glyceraldehyde 3-phosphatephytoene desaturase; GGPP, geranylgeranyl pyrophosphate; GGPPS, geranylgeranyl pyrophosphate synthase; HR, homologous recombination; indels, insertions or deletions; IPP, isopentenyl diphosphate; LCY-B, lycopene β-cyclase; LCY-E, lycopene δ-cyclase; MCR, mutagenic chain reaction; NHEJ, non-homologous end joining; PAM, protospacer-adjacent motif; PDS, phytoene desaturase; PSY1, phytoene synthase 1; RP-HPLC, reverse phase high-performance liquid chromatography; (s)gRNA, (single) guide RNA; TALEN, transcription activator-like effector nuclease; TEM, transmission electron microscopy; WT, wild type; ZFN, zinc finger nuclease; ZISO, z-carotene isomerase.
Barry, C. S., McQuinn, R. P., Chung, M. Y., Besuden, A., and Giovannoni, J. J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147, 179–187. doi: 10.1104/pp.108.118430
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16:232. doi: 10.1186/s13059-015-0796-9
Chappell, J., Wolf, F., Proulx, J., Cuellar, R., and Saunders, C. (1995). Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme a reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol. 109, 1337–1343. doi: 10.1104/pp.109.4.1337
Chen, W., He, S., Liu, D., Patil, G. B., Zhai, H., Wang, F., et al. (2015). A sweetpotato geranylgeranyl pyrophosphate synthase gene, IbGGPS, increases carotenoid content and enhances osmotic stress tolerance in Arabidopsis thaliana. PLoS One 10:e0137623. doi: 10.1371/journal.pone.0137623
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Fantini, E., Falcone, G., Frusciante, S., Giliberto, L., and Giuliano, G. (2013). Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 163, 986–998. doi: 10.1104/pp.113.224733
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Fraser, P. D., Enfissi, E. M., Halket, J. M., Truesdale, M. R., Yu, D., Gerrish, C., et al. (2007). Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19, 3194–3211. doi: 10.1105/tpc.106.049817
Fraser, P. D., Kiano, J. W., Truesdale, M. R., Schuch, W., and Bramley, P. M. (1999). Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol. Biol. 40, 687–698. doi: 10.1023/A:1006256302570
Fraser, P. D., Truesdale, M. R., Bird, C. R., Schuch, W., and Bramley, P. M. (1994). Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol. 105, 405–413. doi: 10.1104/pp.105.1.405
Fujisawa, M., Nakano, T., Shima, Y., and Ito, Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386. doi: 10.1105/tpc.112.108118
Gaj, T., Gersbach, C. A., and Barbas, C. R. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Galpaz, N., Wang, Q., Menda, N., Zamir, D., and Hirschberg, J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730. doi: 10.1111/j.1365-313X.2007.03362.x
Giuliano, G., Bartley, G. E., and Scolnik, P. A. (1993). Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5, 379–387. doi: 10.1105/tpc.5.4.379
Hortensteiner, S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14, 155–162. doi: 10.1016/j.tplants.2009.01.002
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi: 10.1016/j.bbrc.2015.09.117
Jacobs, T. B., LaFayette, P. R., Schmitz, R. J., and Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. doi: 10.1186/s12896-015-0131-2
Jacobs, T. B., and Martin, G. B. (2016). High-throughput CRISPR vector construction and characterization of DNA modifications by generation of tomato hairy roots. J. Vis. Exp. 110:e53843. doi: 10.3791/53843
Jiang, H., Li, M., Liang, N., Yan, H., Wei, Y., Xu, X., et al. (2007). Molecular cloning and function analysis of the stay green gene in rice. Plant J. 52, 197–209. doi: 10.1111/j.1365-313X.2007.03221.x
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Klee, H. J., and Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59. doi: 10.1146/annurev-genet-110410-132507
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., et al. (2017). Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. doi: 10.1111/pbi.12781
Li, X., and Xu, J. (2014). Meta-analysis of the association between dietary lycopene intake and ovarian cancer risk in postmenopausal women. Sci. Rep. 4:4885. doi: 10.1038/srep04885
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lin, D., Xiang, Y., Xian, Z., and Li, Z. (2016). Ectopic expression of SlAGO7 alters leaf pattern and inflorescence architecture and increases fruit yield in tomato. Physiol. Plant. 157, 490–506. doi: 10.1111/ppl.12425
Luo, Z., Zhang, J., Li, J., Yang, C., Wang, T., Ouyang, B., et al. (2013). A STAY-GREEN protein SlSGR1 regulates lycopene and beta-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 198, 442–452. doi: 10.1111/nph.12175
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Mialoundama, A. S., Heintz, D., Jadid, N., Nkeng, P., Rahier, A., Deli, J., et al. (2010). Characterization of plant carotenoid cyclases as members of the flavoprotein family functioning with no net redox change. Plant Physiol. 153, 970–979. doi: 10.1104/pp.110.155440
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Moreno, J. C., Pizarro, L., Fuentes, P., Handford, M., Cifuentes, V., and Stange, C. (2013). Levels of lycopene beta-cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS One 8:e58144. doi: 10.1371/journal.pone.0058144
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482. doi: 10.1038/s41598-017-00578-x
Neuman, H., Galpaz, N., Cunningham, F. J., Zamir, D., and Hirschberg, J. (2014). The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 78, 80–93. doi: 10.1111/tpj.12451
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient increase of - aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7:7057. doi: 10.1038/s41598-017-06400-y
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6:24765. doi: 10.1038/srep24765
Pecker, I., Gabbay, R., Cunningham, F. J., and Hirschberg, J. (1996). Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol. Biol. 30, 807–819. doi: 10.1007/BF00019013
Pouchieu, C., Galan, P., Ducros, V., Latino-Martel, P., Hercberg, S., and Touvier, M. (2014). Plasma carotenoids and retinol and overall and breast cancer risk: a nested case-control study. Nutr. Cancer 66, 980–988. doi: 10.1080/01635581.2014.936952
Ralley, L., Schuch, W., Fraser, P. D., and Bramley, P. M. (2016). Genetic modification of tomato with the tobacco lycopene beta-cyclase gene produces high beta-carotene and lycopene fruit. Z. Naturforsch. C 71, 295–301. doi: 10.1515/znc-2016-0102
Regnier, P., Bastias, J., Rodriguez-Ruiz, V., Caballero-Casero, N., Caballo, C., Sicilia, D., et al. (2015). Astaxanthin from Haematococcus pluvialis Prevents oxidative stress on human endothelial cells without toxicity. Mar. Drugs 13, 2857–2874. doi: 10.3390/md13052857
Ren, G., An, K., Liao, Y., Zhou, X., Cao, Y., Zhao, H., et al. (2007). Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144, 1429–1441. doi: 10.1104/pp.107.100172
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470.e8–480.e8. doi: 10.1016/j.cell.2017.08.030
Ronen, G., Carmel-Goren, L., Zamir, D., and Hirschberg, J. (2000). An alternative pathway to beta -carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. U.S.A. 97, 11102–11107. doi: 10.1073/pnas.190177497
Ronen, G., Cohen, M., Zamir, D., and Hirschberg, J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17, 341–351. doi: 10.1046/j.1365-313X.1999.00381.x
Rosati, C., Aquilani, R., Dharmapuri, S., Pallara, P., Marusic, C., Tavazza, R., et al. (2000). Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J. 24, 413–419. doi: 10.1046/j.1365-313x.2000.00880.x
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y., and Takeda, S. (2006). Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029. doi: 10.1016/j.dnarep.2006.05.022
Sun, L., Yuan, B., Zhang, M., Wang, L., Cui, M., Wang, Q., et al. (2012). Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and beta-carotene contents in tomato fruit. J. Exp. Bot. 63, 3097–3108. doi: 10.1093/jxb/ers026
Tang, L., Lee, A. H., Su, D., and Binns, C. W. (2014). Fruit and vegetable consumption associated with reduced risk of epithelial ovarian cancer in southern Chinese women. Gynecol. Oncol. 132, 241–247. doi: 10.1016/j.ygyno.2013.10.020
Van Eck, J., Kirk, D. D., and Walmsley, A. M. (2006). Tomato (Lycopersicum esculentum). Methods Mol. Biol. 343, 459–473. doi: 10.1385/1-59745-130-4:459
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi: 10.1146/annurev-arplant-042811-105552
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wyman, C., and Kanaar, R. (2006). DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40, 363–383. doi: 10.1146/annurev.genet.40.110405.090451
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Xu, C., Park, S. J., Van Eck, J., and Lippman, Z. B. (2016). Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 30, 2048–2061. doi: 10.1101/gad.288415.116
Xu, R. F., Li, H., Qin, R. Y., Li, J., Qiu, C. H., Yang, Y. C., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5:11491. doi: 10.1038/srep11491
Yang, Y., Zhu, G., Li, R., Yan, S., Fu, D., Zhu, B., et al. (2017). The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 175, 1690–1702. doi: 10.1104/pp.17.01265
Yu, Q. H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7:11874. doi: 10.1038/s41598-017-12262-1
Keywords: lycopene, CRISPR/Cas9 system, genome editing, tomato fruits, carotenoid metabolic pathway
Citation: Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y and Zhu H (2018) Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Received: 21 November 2017; Accepted: 10 April 2018;
Published: 26 April 2018.
Edited by:
Jose M. Seguí-Simarro, Universitat Politècnica de València, SpainReviewed by:
Ghulam Kadir Ahmad Parveez, Malaysian Palm Oil Board, MalaysiaCopyright © 2018 Li, Wang, Chen, Tian, Fu, Zhu, Luo and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongliang Zhu, aGx6aHVAY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.