- 1College of Life Science, Luoyang Normal University, Luoyang, China
- 2Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Chinese Academy of Sciences, Xining, China
- 3Qinghai Provincial Key Laboratory of Crop Molecular Breeding, Xining, China
The chloroplast (cp) genome is useful in the study of phylogenomics, molecular dating, and molecular evolution. Gentiana sect. Kudoa is a predominantly alpine flowering plant that is valued for its contributions to medicine, ecology, and horticulture. Previous evolutionary studies showed that the plastid gene loss pattern and intra-sectional phylogenetics in sect. Kudoa are still unclear. In this study, we compared 11 Gentiana plastomes, including 7 newly sequenced plastomes from sect. Kudoa, to represent its three serious: ser. Ornatae, ser. Verticillatae, and ser. Monanthae. The cp genome sizes of the seven species ranged from 137,278 to 147,156 bp. The plastome size variation mainly occurred in the small single-copy and long single-copy regions rather than the inverted repeat regions. Compared with sect. Cruciata, the plastomes in ser. Ornatae and ser. Verticillatae had lost approximately 11 kb of sequences containing 11 ndh genes. Conversely, far fewer losses were observed in ser. Monanthae. The phylogenetic tree revealed that sect. Kudoa was not monophyletic and that ser. Monanthae was more closely related to other sections rather than sect. Kudoa. The molecular dating analysis indicated that ser. Monanthae and sect. Kudoa diverged around 8.23 Ma. In ser. Ornatae and ser. Verticillatae, the divergence occurred at around 0.07–1.78 Ma. The nucleotide diversity analysis indicated that the intergenic regions trnH-psbA, trnK-trnQ, ycf3-trnS and rpl32-trnL constituted divergence hotspots in both sect. Kudoa and Gentiana, and would be useful for future phylogenetic and population genetic studies.
Introduction
Encompassing 15 sections and 362 species (Ho and Liu, 2001), Gentiana is the largest genus in the family Gentianaceae (He, 1988). Gentiana is predominantly alpine and occurs in numerous mountain systems of the world. Hosting c. 250 species (Ho and Pringle, 1995), the mountain ranges surrounding the Qinghai-Tibetan Plateau (QTP) are the main diversity center of Gentiana. Gentiana has been widely used in traditional Chinese and Tibetan medicines and acts as an edificator in the QTP alpine meadows (Ho and Liu, 2001). In light of their chemical and horticultural value, several species have already been cultured (Rybczyński et al., 2015). Section Kudoa (Masamune) Satake & Toyokuni ex Toyokuni is characterized by roots arising from a collar and stems branching monopodially. This section contains three series and 28 species (Ho and Liu, 2001). Series Ornatae Marquand is the biggest series in sect. Kudoa and contains 16 species. The species in ser. Ornatae have showy flowers, and several of them have been domesticated for horticultural gardening (Rybczyński et al., 2015). ser. Monanthae (H. Smith) T. H. Ho and Series Verticillatae Marquand contain four and eight species, respectively (Ho and Liu, 2001).
A comparative analysis of the chloroplast genomes between sect. Kudoa and sect. Cruciata Gaudin indicated a loss of an approximately 10 kb sequence that mainly comprised 11 ndh genes in Gentiana lawrencei var. farreri, which belongs to ser. Ornatae (Fu et al., 2016). Variable ndh gene loss has been reported in other plant groups such as orchid (Barrett and Davis, 2012; Yang et al., 2013; Lin et al., 2015; Niu et al., 2017). To further assess whether the ndh gene has been lost in sect. Kudoa, additional taxa should be sequenced and included in the comparative analysis. In addition, the phylogenetic relationships in sect. Kudoa are currently controversial. In the latest classification system proposed by Ho and Liu (2001), sect. Kudoa contained three series. A phylogenetic study of the subtribe Gentianinae based on the intergenic spacer (ITS) region and a plastid fragment included only four taxa from sect. Kudoa, which clustered into a monophyletic group (Favre et al., 2010). In another phylogenetic study on Gentiana based on ITS and two plastid fragments, 18 taxa from sect. Kudoa were included and grouped into three clades (Favre et al., 2016), indicating that sect. Kudoa was not monophyletic, and that ser. Verticillatae was embedded in ser. Ornatae. The molecular phylogenetic relationship is not consistent with the classification system proposed by Ho and Liu (2001). Whether the plastid ndh gene loss pattern in sect. Kudoa is helpful to the intra-sectional phylogenetics is worth exploring.
The cp genomes are circular DNA molecules in angiosperms that range in size from 120 to 160 kb and contain 110–130 genes (Palmer, 1985). In land plants, the cp genome typically contains a pair of inverted repeats (IRs) that separate the remaining regions into one large single-copy region (LSC) and one small single-copy region (SSC) (Palmer, 1985; Jansen et al., 2005). The cp genome is recognized as the “workhorse” in plant systematics research due to its uniparental inheritance, haploid nature, highly conserved structures, and slower evolutionary rate of change compared to nuclear genomes (Wolfe et al., 1987; Shaw et al., 2014). Plastid phylogenomics has been widely applied to reassess classifications, for example, the reassessment of Alismatales (Ross et al., 2015), Rosaceae (Zhang S.D. et al., 2017), Gaultheria series Trichophyllae (Zhang M.Y. et al., 2017), and Leptaspis and Streptochaeta in Poaceae (Burke et al., 2016). In addition to phylogenetic classification, the cp genome is widely used in studies of molecular identification, divergence dating, and molecular evolution (Nikiforova et al., 2013; Carbonell-Caballero et al., 2015). Comparative cp genome analysis can reveal insights into the evolution of the cp genome, including sequence inversion (Cho et al., 2015) and gene loss (Wakasugi et al., 1994; Millen et al., 2001), and has been used in the identification of mutational hotspots for the screening of the most informative regions (e.g., Ahmed et al., 2013; Niu et al., 2017).
Presently, only four complete cp genomes have been sequenced in Gentiana, in which three belong to sect. Cruciata (Ni et al., 2016a,b) and one belongs to sect. Kudoa (Fu et al., 2016). The development of more genomic resources for Gentiana should inform our understanding of the phylogenetic relationships and evolutionary history of this large genus. In this study, we focused on sect. Kudoa and sequenced the complete cp genomes of seven species in this section. Based on a comparative analysis of these seven species, as well as four species in sect. Cruciata and sect. Kudoa with available genomes, the genome structure, gene loss, phylogenetic relationships, divergence times and mutational hotspots of sect. Kudoa were analyzed to discover (1) gene loss pattern, particularly ndh, in sect. Kudoa, and (2) plastome phylogenetic implication in sect. Kudoa. This study also makes available sequence information for phylogenetic and evolutionary studies of Gentiana.
Materials and Methods
Sample Collection, Genome Sequencing, and Assembly
A total of seven species were sampled in the QTP (Supplementary Table S1) to represent all three series of sect. Kudoa. Five species (G. veitchiorum Hemsley, G. ornata Grisebach, G. caelestis H. Smith, G. obconica T. N. Ho, and G. oreodoxa H. Smith) belong to ser. Ornatae, one (G. stipitata Edgeworth) belongs to ser. Monanthae, and one (G. hexaphylla Maximowicz ex Kusnezow) belongs to ser. Verticillatae. The species were identified by Dr. Peng-Cheng Fu and Dr. Shi-Long Chen. Voucher specimens were deposited in the herbarium of the College of Life Science, Luoyang Normal University. The samples were collected from a single plant of each species. Total genomic DNA isolation, DNA fragmentation, and sequencing library construction followed the process described in Fu et al. (2016). Based on the genome size of some Gentiana taxa (Mishiba et al., 2009) and reported examples of sequenced cp genomes (Fu et al., 2016), we expected to obtain approximately 5 Gb raw data for each species. The fragmented genomic DNA of the seven Gentiana species was sequenced using the Illumina HiSeq 4000 platform (Novogene, Tianjing, China), yielding 150-bp paired-end reads from a library of approximately 300-bp DNA fragments.
Reads corresponding to plastid DNA were identified using a BLASTN (E-value: 10-6) search against the plastome sequences of G. lawrencei var. farreri (GenBank accession no. KX096882). The recovered reads were assembled using Velvet 1.2.10 (Zerbino and Birney, 2008). Detailed information regarding the raw reads for each taxon is presented in Supplementary Table S2. All the genomic regions located at the junction between the two assembled contigs were verified by Sanger sequencing. The primers used were designed by PRIMER V. 5.0 software and are listed in Supplementary Table S3. The plastome sequences of the seven species were deposited in GenBank (MG192304–MG192310).
Genome Annotation
For each species, the protein coding genes (PCGs), rRNAs, and tRNAs in the cp genome were predicted and annotated using Dual Organellar GenoMe Annotator (DOGMA) using the default parameters (Wyman et al., 2004). The positions of the start and stop codons, or intron/exon junctions of the PCGs, were manually corrected using a BLAST search against reported cp genomes of other closely related species. The cp gene maps of the seven species were drawn using OGDraw V. 1.2 (Lohse et al., 2007).
Comparative Analysis
In addition to the seven newly sequenced species, the cp genome sequences of G. lawrencei var. farreri (KX096882), which belongs to ser. Ornata, and G. straminea (KJ657732), G. robusta (KT159969), and G. crassicaulis (KJ676538), which belong to sect. Cruciata, were obtained for comparative analysis from the National Center for Biotechnology Information. Genome comparisons were performed to identify the differences among the 11 taxa using mVISTA (Frazer et al., 2004) and Geneious Basic 5.6.4 (Kearse et al., 2012). To identify divergence hotspots, nucleotide diversity (Pi) was determined using DnaSP V. 5.10 (Librado and Rozas, 2009).
Phylogenetic Analysis
To investigate the phylogenetic relationships of the genome sequences of sect. Kudoa, an additional 14 available complete cp genomes in the order Gentianales were retrieved from GenBank (Supplementary Table S4). Forty-six PCGs (atpA, atpB, atpE, atpH, atpI, cemA, matK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaI, psaJ, psbA, psbC, psbD, psbE, psbF, psbH, psbI, psbI, psbK, psbL, psbM, psbN, psbT, rbcL, rpl14, rpl16, rpl20, rpl22, rpl33, rpl36, rpoA, rps2, rps3, rps4, rps8, rps11, rps14, rps15, and rps18) present in all of the species were extracted from the selected cp genomes. Phylogenetic analyses were performed using the concatenated nucleotide sequences and PhyML3.1 software (Guindon and Gascuel, 2003) using the maximum likelihood (ML) method. Based upon the Akaike information criterion in the software jModelTest 2.1.7 (Guindon and Gascuel, 2003; Posada, 2008), the selected best-fitting model of sequence evolution was the GTR+I+G model with a p-inv of 0.404 and gamma shape of 0.808. A bootstrap analysis was performed with 100 replications.
Molecular Dating
The PCG dataset was used to estimate divergence times using the Bayesian method implemented in the program BEAST 1.7.5 (Drummond et al., 2012) under the GTR+I+G substitution model, the Yule model, and an uncorrelated lognormal clock model (Drummond et al., 2006). Due to the limited fossils available for Gentiana, we constrained only one of the nodes with a seed fossil of sect. Cruciata. For the seed fossil, we used lognormal priors with an offset at 5.0 Ma, a mean of 0.7, and a standard deviation of 1.0, as applied by Pirie et al. (2015) and Favre et al. (2016). We did not use uniform priors for sect. Cruciata, as they were rejected following comparison with the lognormal priors provided by Favre et al. (2016). We ran three independent Markov chain Monte Carlo analyses for 50 million generations, sampling every 5,000th generation. We assessed the convergence of the estimated parameters in Tracer 1.5 (Rambaut and Drummond, 2010), ensuring that the effective sample size values exceeded 200. Trees were summarized in TreeAnnotator 1.7.5 (Drummond et al., 2012) after setting the burn-in to 10%, and then visualized in FigTree 1.4.01.
Results
Features of the Seven Newly Sequenced Plastomes
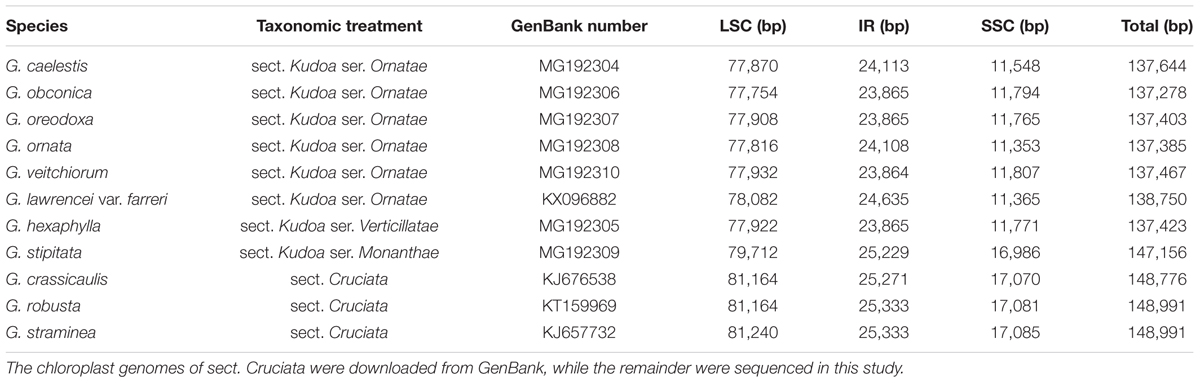
Complete plastome sequences of seven Gentiana species were newly sequenced in this study and deposited in GenBank. The seven plastid genomes constituted closed circular molecules whose sizes ranged from 137,278 to 147,156 bp with an average of 138,822 bp (Table 1). Each cp genome comprised a pair of IR regions (IRa and IRb), one LSC region, and one SSC region. They all possessed the overall typical quadripartite structure that resembles the majority of land plant cp genomes (Shinozaki et al., 1986). The IR regions of the 7 species ranged from 23,864 to 25,229 bp; the LSC regions ranged from 77,754 to 79,712 bp; and the SSC regions ranged from 11,353 to 16,986 bp (Table 1). Gentiana stipitata possessed the longest LSC, SSC, and IR regions of the seven species. The average GC contents of the LSC, SSC, and IR regions and the whole cp genome in the seven species were 35.7, 30.6, 43.8, and 36.8%, respectively, which corroborates other reported Gentiana cp genomes (Fu et al., 2016; Ni et al., 2016a,b). Furthermore, these plastid genomes were similar in structure and gene arrangement to previously published Gentiana plastomes (Fu et al., 2016; Ni et al., 2016a,b). All the plastome maps are presented in Supplementary Figures S1–S7.
Comparison of cp Genomes
The comparative analysis indicated that the six species in Gentiana ser. Ornatae possessed very similar plastomes, with genome sizes ranging from 137,278 to 138,750 bp. The only obvious difference was located at the boundary between IRb and SSC (Figure 1A), in which four sequence patterns were detected. The first pattern, which appeared in G. lawrencei var. farreri, possessed almost all the sequences of ycf1 and most of the forward sequences of ndhF (Figure 1A). The second pattern, appearing in G. caelestis and G. ornata, possessed the forward 416 bp of ycf1, but had lost the forward sequence of ndhF (Figure 1A). In comparison with the second pattern, the third pattern only possessed the forward 181 bp of ycf1 (Figure 1A). The third pattern appeared in G. obconica, G. veitchiorum, and G. oreodoxa. In sect. Kudoa, G. hexaphylla from ser. Verticillatae also possessed the third pattern. However, G. stipitata from ser. Monanthae possessed all of the whole sequences of ycf1 and ndhF, constituting the fourth pattern. This species differed from all the other taxa in sect. Kudoa, but did not differ from the three taxa in sect. Cruciata (Figure 1A).

FIGURE 1. The sequence length variation of four regions across 11 Gentiana chloroplast genomes. (A) The region between ycf1 and rpl32; (B) the region between ccsA and rps15; (C) the region between trnF-GAA and trnV-UAC; (D) the region between trnL-CAA and rps7.
The comparative analysis in Gentiana revealed that the variation in plastid genome size in the 11 plastomes could mainly be attributed to sequence loss in four locations. One of these locations was the IRb-SSC boundary mentioned above. The second was the region between ccsA to rps15, in which about 5 kb had been lost in some taxa. The lost sequences mainly contained a small section of ndhD, the majority of ndhE and ndhH, and all of ndhG, ndhI, and ndhA (Figure 1B). These ndh genes are shown in yellow in the Supplementary Figures S1–S7. The third was the region between trnF-GAA and trnV-UAC, in which about 2.2 kb had been lost in some taxa. The lost sequences mainly contained all of ndhJ, ndhK, and ndhC (Figure 1C). The fourth was the region between trnL-CAA and rps7, in which about 1 kb had been lost in some taxa. The lost sequences mainly contained the whole exon 1 and part of the intron of ndhB (Figure 1D). All the sequences that have been lost in the three regions were missing in the species of ser. Ornatae and ser. Verticillatae, but present in ser. Monanthae and sect. Cruciata.
Phylogenetic and Molecular Dating Analyses
The ML phylogenetic tree constructed using the 46 PCGs clearly identified the three families (Gentianaceae, Apocynaceae, and Rubiaceae) as being monophyletic with high bootstrap support (Figure 2). Two monophyletic groups were identified within the Gentianaceae clade in these analyses. Taxa from ser. Ornatae and ser. Verticillatae clustered into one monophyletic group whose monophyletic sister group contained taxa from sect. Cruciata and ser. Monanthae (Figure 2). The ML tree showed that G. stipitata was more closely related to other sections rather than sect. Kudoa.
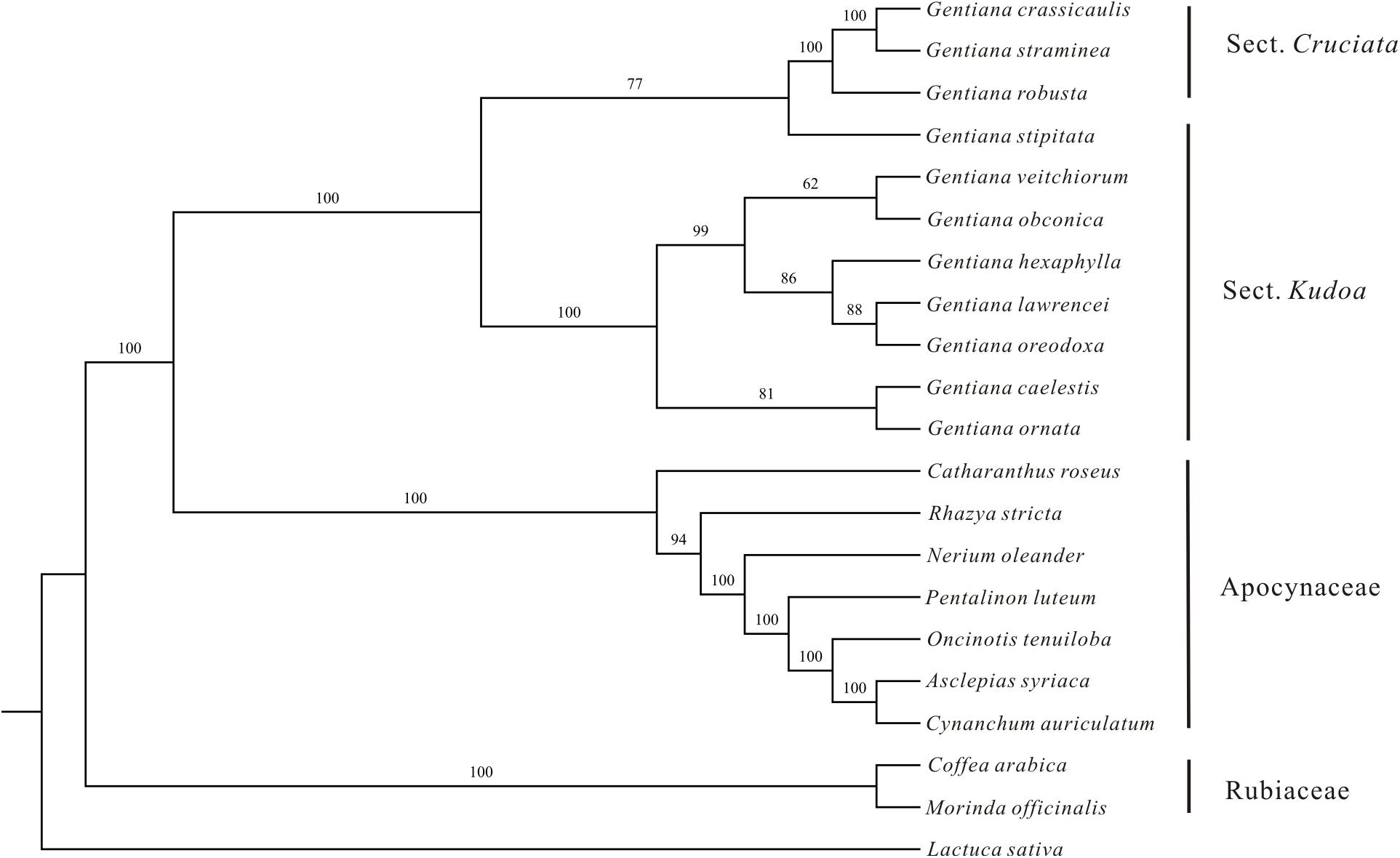
FIGURE 2. Maximum likelihood tree of Gentianales based on 46 PCGs of the chloroplast genomes. Numbers above the nodes indicate bootstrap support values.
The molecular dating analysis of the PCG dataset (Figure 3) estimated that the two lineages in the Gentiana tree diverged approximately 8.23 Ma (95% highest posterior density [HPD]: 5.40–15.60 Ma). G. stipitata and sect. Cruciata diverged about 6.11 Ma (95% HPD: 5.06–9.25 Ma). The divergence in ser. Ornatae and ser. Verticillatae occurred at around 0.07–1.78 Ma.
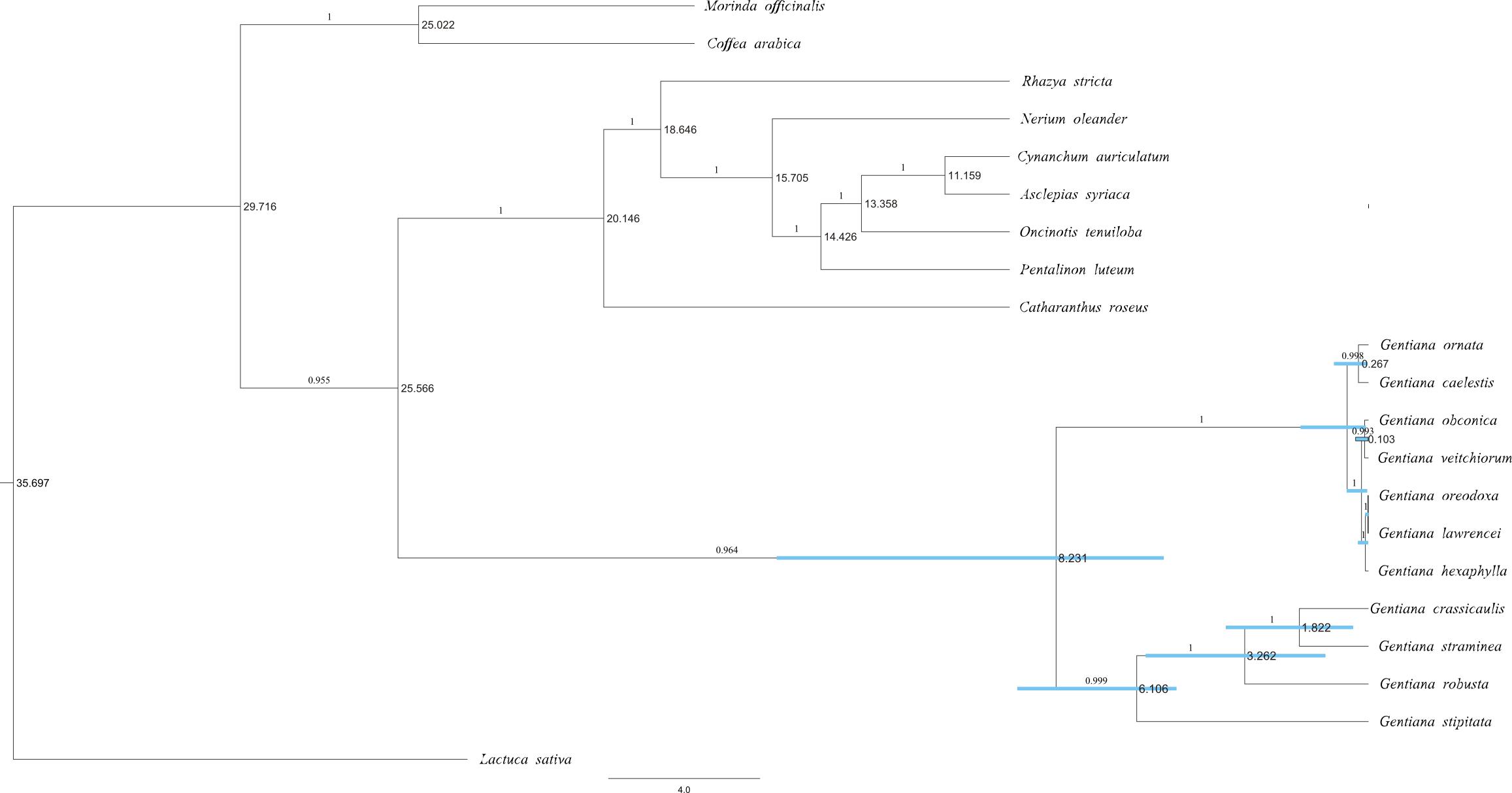
FIGURE 3. Majority-rule consensus phylogenetic tree of Gentianales based on 46 PCGs of the chloroplast genomes based on Bayesian inference. Numbers on the branches indicate Bayesian posterior probabilities. Node age estimates are marked dextrally. Blue bars represent 95% HPDs.
Divergence Hotspots in Gentiana
The coding genes, introns, and non-coding regions were compared to detect divergence hotspots. We compared all 11 species mentioned above as well as the 8 species in sect. Kudoa. A total of 114 regions (49 coding genes, 9 intron regions, and 55 intergenic regions) greater than 200 bp were generated in both comparisons.
Among the 11 species in Gentiana, Pi ranged from 0.00163 (rpl2 gene) to 0.13984 (trnH-psbA region). The average Pi in LSC, SSC, and IR was 0.02696, 0.02624, and 0.011, respectively. Five regions (trnH-psbA, trnK-trnQ, accD-psaI, ycf3-trnS, and psbK-psbI), all located in the LSC region, showed high levels of variation (Figure 4A and Table 2).
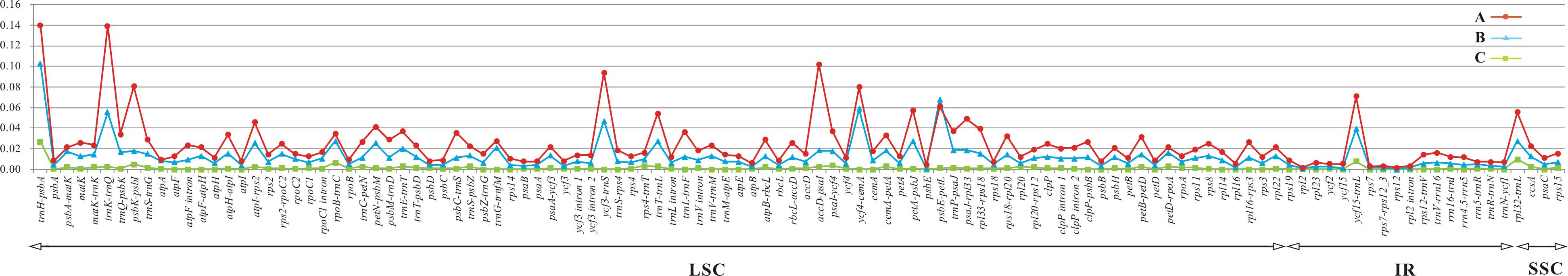
FIGURE 4. Comparison of the nucleotide variability (Pi) values in (A) Gentiana, (B) sect. Kudoa, and (C) sect. Kudoa except ser. Monanthae. A total of 114 regions (49 coding genes, 9 intron regions, and 55 intergenic regions) greater than 200 bp in size were generated in both comparisons.
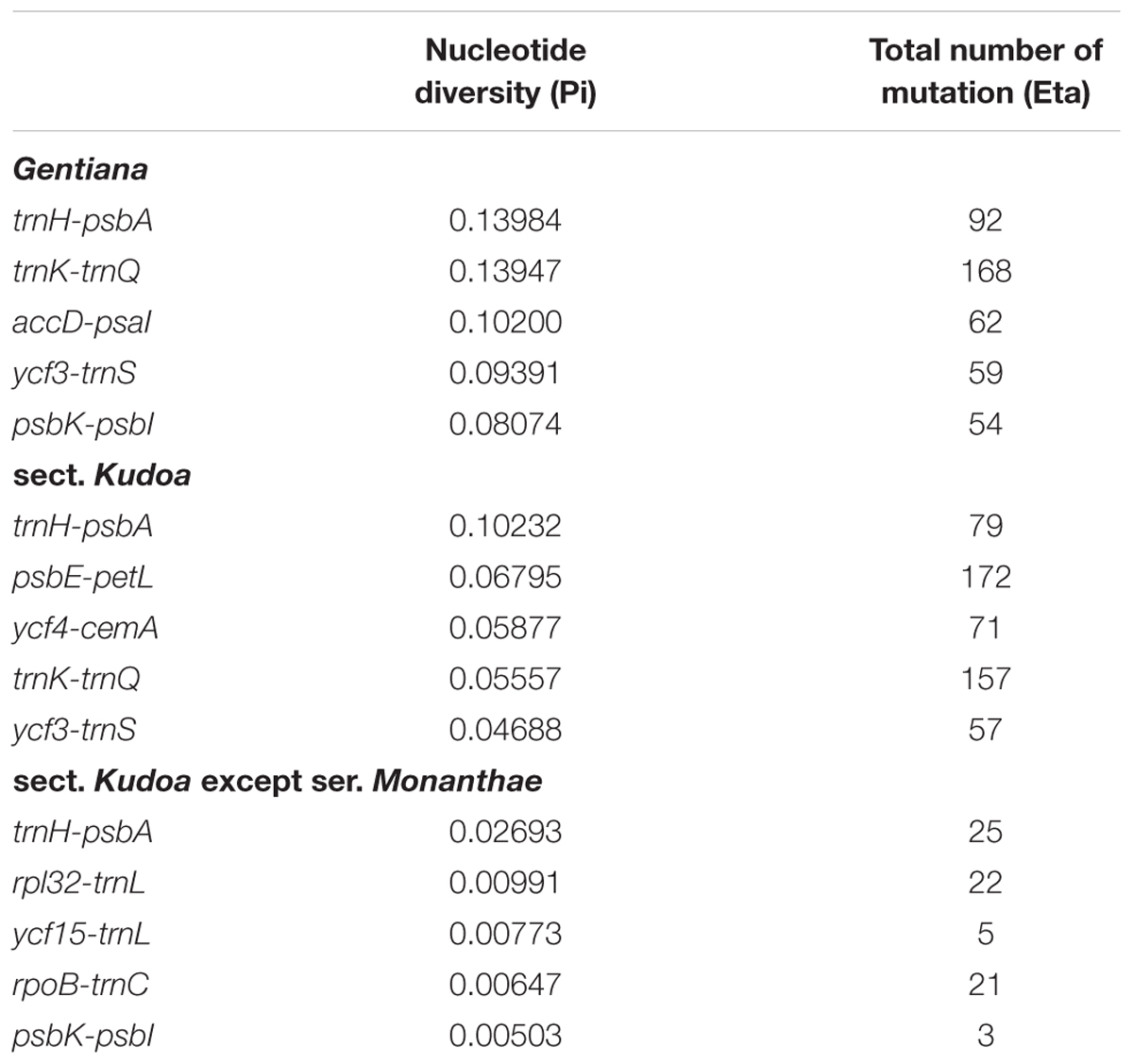
TABLE 2. Five regions of highly variable sequences of Gentiana, sect. Kudoa and sect. Kudoa except ser. Monanthae.
The sequence divergence among the eight species in sect. Kudoa ranged from 0.00100 (psbE gene) to 0.10232 (trnH-psbA region). The average Pi in LSC, SSC, and IR was 0.01406, 0.01320, and 0.00543, respectively. Five of these, trnH-psbA, psbE-petL, ycf4-cemA, trnK-trnQ, and ycf3-trnS, constituted highly divergent hotspots (Table 2). All of the five regions were located in the LSC region (Figure 4B).
The sequence divergence among the seven species in sect. Kudoa except ser. Monanthae is much lower than the sequence divergence among sect. Kudoa and Gentiana (Figure 4C). The average Pi in LSC, SSC, and IR was 0.00145, 0.00362, and 0.00067, respectively. Five of these, trnH-psbA, rpl32-trnL, ycf15-trnL, rpoB-trnC, and psbK-psbI, constituted highly divergent hotspots (Table 2).
Discussion
Loss of Plastid ndh Genes in Gentiana sect. Kudoa
Variations in plastome length were detected in Gentiana. Compared with sect. Cruciata, ser. Ornatae and ser. Verticillatae lost approximately 11 kb of sequences, while ser. Monanthae lost almost none. The plastomes of ser. Ornatae and ser. Verticillatae were highly similar in size and structure. The majority of length variation of the newly sequenced plastomes of ser. Ornatae and ser. Verticillatae in this study mainly occurred in the SSC and LSC regions, rather than the two IR regions. The length variation pattern is similar to that observed in G. lawrencei var. farreri (Fu et al., 2016), whose genome size variation was not caused by deletions in the IR regions, but by deletions in the SSC and LSC regions. An explanation for the size variation pattern is that the junction between the IR and LSC region is located within the rps19 gene, which is a coding gene, and thus contributes to the more constant size of the IRs than the SSC and LSC region in the great majority of angiosperms (Palmer, 1985; Fu et al., 2016). As noted in G. lawrencei var. farreri (Fu et al., 2016), the genome size variation led to the loss of plastid ndh genes.
Plastid ndh genes have been retained in the majority of higher plants (Martín and Sabater, 2010), and appear to have been frequently lost in parasitic and epiphytic plants (e.g., Stefanović and Olmstead, 2005). Along with the publication of numerous plastomes, the independent loss of ndh genes has been detected in increasing numbers of higher plants including orchids (Chang et al., 2006; Yang et al., 2013; Lin et al., 2015; Niu et al., 2017), gnetophytes (Braukmann et al., 2009; Wu et al., 2011), slender naiads (Peredo et al., 2013), and saguaros (Sanderson et al., 2015). The independent ndh gene loss in various groups could be an example of convergent evolution in plants. In Gentiana, the loss of ndh genes was previously detected in G. lawrencei var. farreri, which belongs to sect. Kudoa, but was not detected in the other three previously sequenced plastomes (Fu et al., 2016; Ni et al., 2016a). Upon analysis of the plastomes of sect. Kudoa, we discovered that the loss of ndh genes was common in ser. Ornatae and ser. Verticillatae. The 11 ndh genes in the plastome encode a protein complex that catalyzes the transfer of electrons from NADH to plastoquinone at photosystem I (Sazanov et al., 1998; Martín and Sabater, 2010). However, the PGR5-dependent cyclic electron transport pathway already exists in cells. Transgenic plants defective in ndh genes showed impaired photosynthesis rates, demonstrating that the NDH complex is required for the optimization of photophosphorylation rates and might play an important role in regulating CO2 assimilation under stress conditions (Wang et al., 2006; Martín and Sabater, 2010). However, no deleterious effects have been observed in ndh-deficient mutants under favorable growing conditions (Ruhlman et al., 2015). This suggests that the plastid ndh genes might be dispensable in contemporary plants. A plastome study in orchids proposed that the expansion/contraction of IR boundaries might be associated with the loss of ndh genes, especially ndhF (Kim et al., 2015; Niu et al., 2017). Previous studies have shown that the expansion of IRs is common to ndh-absent plastomes (Niu et al., 2017). In Gentiana, it is likely that the expansion/contraction of IR boundaries is correlated with the deletion of the ndh gene, particularly ndhF. However, the contraction, rather than expansion, of IRs was observed in ndh-deleted Gentiana plastomes. This suggests that the evolution of ndh genes in plastomes may vary between different taxa, and thus requires further exploration.
The loss of plastid ndh genes was common in sect. Kudoa, with the exception of ser. Monanthae. Compared with sect. Cruciata, ser. Monanthae did not exhibit significant size variation. As observed in sect. Cruciata, G. stipitata in ser. Monanthae maintained all 11 plastid ndh genes. This suggests that ser. Monanthae might be evolutionarily more closely related to other sections rather than sect. Kudoa (although cp genome sequences from additional sections are necessary to confirm this). Considering that gene loss in plastomes is an ongoing process in evolution (Martin et al., 1998), ser. Ornatae and ser. Verticillatae may have shorter evolutionary histories than ser. Monanthae.
Phylogenetic Relationships and Divergence Times
Plastid phylogenomics has been successfully applied in the phylogenetic reassessment of several groups (Burke et al., 2016; Zhang M.Y. et al., 2017; Zhang S.D. et al., 2017). In this study, the phylogenetic relationships constructed using the 46 PCGs were consistent with previous studies whereby the three families (namely Gentianaceae, Apocynaceae, and Rubiaceae) were classified as three monophyletic clades, and also identified Rubiaceae as the base group in Gentianales (Backlund et al., 2000).
The delimitation of Gentiana sect. Kudoa has been controversial. Ho (1985) and Ho and Pringle (1995) erected four serious in sect. Kudoa: ser. Monanthae, ser. Ornatae, ser. Verticillatae, and ser. Apteroideae (H. Smith) T. N. Ho. Based on 61 informative characters from morphology, palynology, and cytology, Ho et al. (1996) supported three series in sect. Kudoa and treated ser. Apteroideae as sect. Monopodiae (H. Smith) T. N. Ho. The treatment of three series in sect. Kudoa was accepted by Ho and Liu (2001), and was also corroborated by the phylogenetic analysis of Favre et al. (2016) based on atpB-rbcL, trnL–trnF, and ITS. The taxa from sect. Monopodiae, namely Kudoa I in Favre et al. (2016), constituted a monophyletic group and were paraphyletic with the other sections (Favre et al., 2016). We therefore adopted the treatment of three series in sect. Kudoa as the starting point of this study.
Contrary to the classical classification, molecular phylogeny shows that sect. Kudoa is paraphyletic. Although the phylogeny of sect. Kudoa was not mainly discussed in Favre et al. (2016), the phylogenetic trees in their study also indicated that the three series are not a monophyletic group. Our plastid phylogenomic relationships is in accord with this. The cp-based phylogenomic tree suggested that ser. Monanthae was more closely related to other sections rather than sect. Kudoa. The previous phylogenetic trees reconstructed based on atpB-rbcL, trnL–trnF and ITS data indicated that ser. Monanthae, sect. Isomeria and sect. Microsperma clustered into one clade (Favre et al., 2016). The instability of ser. Monanthae in the phylogenetic tree may suggest a potential hybrid origin of this group, since they have a partly sympatric distribution and nearly contemporaneous flowering times to other groups like sect. Kudoa and sect. Cruciata (Ho and Liu, 2001), and furthermore, hybridization is an important means of speciation in the QTP (Li et al., 2007; Liu et al., 2014; Wen et al., 2014). In terms of morphology, ser. Verticillatae is characterized by whorled phyllotaxy. However, our phylogenomic tree showed that ser. Verticillatae did not constitute a monophyletic group, but was rather inlaid in ser. Ornatae. This is also supported by the phylogenetic tree based on atpB-rbcL, trnL–trnF, and ITS (Favre et al., 2016). The results suggested that the present classification in sect. Kudoa based on morphology is not consistent with the molecular phylogenetic reconstruction. Therefore, the phylogenetic relationships in sect. Kudoa require further evaluation.
Molecular dating analysis in this study showed that ser. Monanthae and sect. Cruciata diverged around 6.11 Ma (95% HPD: 5.06–9.25 Ma) when sect. Cruciata diverged from its sister groups (Favre et al., 2016). The divergence of the ser. Verticillatae and ser. Ornatae clade occurred at around 0.07–1.78 Ma, which is much younger than 1.34–9.55 Ma estimated in Favre et al. (2016). Since the ndh gene loss indicated that ser. Verticillatae and ser. Ornatae were younger than ser. Monanthae, we believe that the time of divergence between ser. Verticillatae and ser. Ornatae found in this study may be true. We recommend that the plastomes and ndh gene remain/loss patterns of more taxa that are closely related to sect. Kudoa are included in future studies.
Mutational Hotspot
The comparative analysis indicated that the plastomes of ser. Ornatae and ser. Verticillatae exhibited little variation. The popular barcoding plastid markers such as matK and rbcL (Li et al., 2015; Coissac et al., 2016) exhibited very poor sequence variation in the two series. Some popular genetic markers used in intra-species population genetics such as trnS-trnG, rps15-ycf1, and rpl20-rps12 (e.g., Burnier et al., 2009; Takayama et al., 2013; Khan et al., 2016) show very limited variation in the populations of some species such as G. veitchiorum and G. lawrencei var. farreri (Fu, Unpublished Data). Additionally, trnQ-rps16 (Shepherd et al., 2017) was not detected as rps16 is a pseudogene in Gentiana (Ni et al., 2016a). Since the plastome-wide comparisons could facilitate the screening of mutational hotspots used for inter-species phylogenetics (Shaw et al., 2014) and intra-species discrimination (Ahmed et al., 2013), suitable molecular markers for phylogenetic and population genetic studies could be identified in the mutational hotspots in Gentiana, particularly in sect. Kudoa.
The identified sequence divergence hotspot regions in Gentiana and sect. Kudoa in this study were trnH-psbA, trnK-trnQ, accD-psaI, ycf3-trnS, psbK-psbI, psbE-petL, and ycf4-cemA. Focusing on ser. Ornatae and ser. Verticillatae, the divergence hotspot regions were trnH-psbA, rpl32-trnL, ycf15-trnL, rpoB-trnC, and psbK-psbI. These findings should inform future studies on the inter-species phylogenetics and intra-species population genetics in Gentiana and sect. Kudoa.
Conclusion
The complete cp genome sequences of seven species from Gentiana sect. Kudoa were reported in this study, and the evolutionary characteristics of 11 Gentiana plastomes from two sections were described. We discovered that the loss of plastid ndh genes is common in ser. Ornatae and ser. Verticillatae, but not in ser. Monanthae. The phylogenetic tree and deletion patterns in the plastid ndh genes indicated that ser. Monanthae is more closely related to other sections rather than sect. Kudoa, which is not monophyletic. The sequence and divergence hotspot information presented here could be useful in future studies on the population genetics, phylogenetics, and evolution of Gentiana.
Author Contributions
S-SS designed the experiments, performed the experiments, analyzed the data, prepared the figures and/or tables, and revised the manuscript. P-CF conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, and prepared the figures and/or tables. X-JZ and Y-WC performed the experiments and analyzed the data. F-QZ and S-LC collected the samples and reviewed the drafts of the paper. Q-BG analyzed the data, prepared figures and/or tables, reviewed the drafts of the paper, and revised the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 31600296 and 31400322), Science and Technology Project of Henan Province, China (No. 162102110097), Educational Commission of Henan Province (No. 16A210033), and The Open Project of Qinghai Provincial Key Laboratory of Crop Molecular Breeding (No. 2017-ZJ-Y14).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhi-Zhong Li of the Wuhan Botanical Garden, Chinese Academy of Sciences, for providing laboratory support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00493/full#supplementary-material
Footnotes
References
Ahmed, I., Matthews, P. J., Biggs, P. J., Naeem, M., Mclenachan, P. A., and Lockhart, P. J. (2013). Identification of chloroplast genome loci suitable for high-resolution phylogeographic studies of Colocasia esculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour. 13, 929–937. doi: 10.1111/1755-0998.12128
Backlund, M., Oxelman, B., and Bremer, B. (2000). Phylogenetic relationships within the Gentianales based on ndhF and rbcL sequences, with particular reference to the Loganiaceae. Am. J. Bot. 87, 1029–1043. doi: 10.2307/2657003
Barrett, C. F., and Davis, J. I. (2012). The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 99, 1513–1523. doi: 10.3732/ajb.1200256
Braukmann, T. W., Kuzmina, M., and Stefanović, S. (2009). Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 55, 323–337. doi: 10.1007/s00294-009-0249-7
Burke, S. V., Lin, C. S., Wysocki, W. P., Clark, L. G., and Duvall, M. R. (2016). Phylogenomics and plastome evolution of tropical forest grasses (Leptaspis, Streptochaeta: Poaceae). Front. Plant. Sci. 7:1993. doi: 10.3389/fpls.2016.01993
Burnier, J., Buerki, S., Arrigo, N., Küpfer, P., and Alvarez, N. (2009). Genetic structure and evolution of Alpine polyploid complexes: Ranunculus kuepferi (Ranunculaceae) as a case study. Mol. Ecol. 18, 3730–3744. doi: 10.1111/j.1365-294X.2009.04281.x
Carbonell-Caballero, J., Alonso, R., Ibañez, V., Terol, J., Talon, M., and Dopazo, J. (2015). A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol. Biol. Evol. 32, 2015–2035. doi: 10.1093/molbev/msv082
Chang, C. C., Lin, H. C., Lin, I. P., Chow, T. Y., Chen, H. H., Chen, W. H., et al. (2006). The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 23, 279–291. doi: 10.1093/molbev/msj029
Cho, K. S., Yun, B. K., Yoon, Y. H., Hong, S. Y., Mekapogu, M., Kim, K. H., et al. (2015). Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One 10:e0125332. doi: 10.1371/journal.pone.0125332
Coissac, E., Hollingsworth, P. M., Lavergne, S., and Taberlet, P. (2016). From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 25, 1423–1428. doi: 10.1111/mec.13549
Drummond, A. J., Ho, S. Y., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Favre, A., Michalak, I., Chen, C. H., Wang, J. C., Pringle, J. S., Matuszak, H., et al. (2016). Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J. Biogeogr. 43, 1967–1978. doi: 10.1111/jbi.12840
Favre, A., Yuan, Y. M., Küpfer, P., and Alvarez, N. (2010). Phylogeny of subtribe Gentianinae (Gentianaceae): biogeographic inferences despite limitations in temporal calibration points. Taxon 59, 1701–1711. doi: 10.2307/41059867
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Fu, P. C., Zhang, Y. Z., Geng, H. M., and Chen, S. L. (2016). The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ 4:e2540. doi: 10.7717/peerj.2540
Guindon, S., and Gascuel, O. (2003). A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
He, T. N. (1988). “Sect. Cruciata,” in Flora Reipublicae Popularis Sinicae 62. Gentianaceae, ed. T. N. He (Beijing: Science Press), 1–75.
Ho, T. N., Liu, S. W., and Lu, X. F. (1996). A phylogenetic analysis of Gentiana (Gentianaceae). Acta Phytotaxonomica Sin. 34, 505–506.
Ho, T. N., and Pringle, J. S. (1995). “Gentianaceae,” in Flora of China, Vol. 16, eds Z. Y. Wu and P. H. Raven (Beijing: Science Press), 1–140.
Jansen, R. K., Raubeson, L. A., Boore, J. L., dePamphilis, C. W., Chumley, T. W., Haberle, R. C., et al. (2005). Methods for obtaining and analyzing chloroplast genome sequences. Methods Enzymol 395, 348–384. doi: 10.1016/S0076-6879(05)95020-9
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khan, G., Zhang, F. Q., Gao, Q. B., Fu, P. C., Xing, R., Wang, J. L., et al. (2016). Phylogenetic analyses of Spiraea, (Rosaceae) distributed in the Qinghai-Tibetan Plateau and adjacent regions: insights from molecular data. Plant Syst. Evol. 302, 11–21. doi: 10.1007/s00606-015-1238-6
Kim, H. T., Kim, J. S., Moore, M. J., Neubig, K. M., Williams, N. H., Whitten, W. M., et al. (2015). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One 10:e0142215. doi: 10.1371/journal.pone.0142215
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Li, X. J., Wang, L. Y., Yang, H. L., and Liu, J. Q. (2007). Confirmation of natural hybrids between Gentiana straminea and G. siphonantha (Gentianaceae) based on molecular evidence. Front. Biol. China 3:470–476. doi: 10.1007/s11515-008-0076-0
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, C. S., Chen, J. J., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5:9040. doi: 10.1038/srep09040
Liu, J. Q., Duan, Y. W., Hao, G., Ge, X. J., and Sun, H. (2014). Evolutionary history and underlying adaptation of alpine plants on the Qinghai-Tibet Plateau. J. Syst. Evol. 52, 241–249. doi: 10.1111/jse.12094
Lohse, M., Drechsel, O., and Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274. doi: 10.1007/s00294-007-0161-y
Martín, M., and Sabater, B. (2010). Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645. doi: 10.1016/j.plaphy.2010.04.009
Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K. V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. doi: 10.1038/30234
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. doi: 10.1105/tpc.13.3.645
Mishiba, K. I., Yamane, K., Nakatsuka, T., Nakano, Y., Yamamura, S., Abe, J., et al. (2009). Genetic relationships in the genus Gentiana based on chloroplast DNA sequence data and nuclear DNA content. Breed. Sci. 59, 119–127. doi: 10.1270/jsbbs.59.119
Ni, L., Zhao, Z., Xu, H., Chen, S., and Dorje, G. (2016a). Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr. Genet. 63, 241–252. doi: 10.1007/s00294-016-0631-1
Ni, L., Zhao, Z., Xu, H., Chen, S., and Dorje, G. (2016b). The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 577, 281–288. doi: 10.1016/j.gene.2015.12.005
Nikiforova, S. V., Cavalieri, D., Velasco, R., and Goremykin, V. (2013). Phylogenetic analysis of 47 chloroplast genomes clarifies the contribution of wild species to the domesticated apple maternal line. Mol. Biol. Evol. 30, 1751–1760. doi: 10.1093/molbev/mst092
Niu, Z., Xue, Q., Zhu, S., Sun, J., Liu, W., and Ding, X. (2017). The complete plastome sequences of four Orchid species: insights into evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 8:715. doi: 10.3389/fpls.2017.00715
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Peredo, E. L., King, U. M., and Les, D. H. (2013). The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS One 8:e68591. doi: 10.1371/journal.pone.0068591
Pirie, M. D., Litsios, G., Bellstedt, D. U., Salamin, N., and Kissling, J. (2015). Back to Gondwanaland: can ancient vicariance explain (some) Indian Ocean disjunct plant distributions? Biol. Lett. 11:20150086. doi: 10.1098/rsbl.2015.0086
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Rambaut, A., and Drummond, A. J. (2010). Tracer, Version 1.5. Available at: http://tree.bio.ed.ac.uk/software/tracer/ [accessed August 25, 2013].
Ross, T. G., Barrett, C. F., Soto Gomez, M., Lam, V. K., Henriquez, C. L., Les, D. H., et al. (2015). Plastid phylogenomics and molecular evolution of Alismatales. Cladistics 32, 160–178. doi: 10.1111/cla.12133
Ruhlman, T. A., Chang, W. J., Chen, J. J., Huang, Y. T., Chan, M. T., Jin, Z., et al. (2015). NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol 15:100. doi: 10.1186/s12870-015-0484-7
Rybczyński, J. J., Davey, M. R., and Mikuła, A. (eds) (2015). The Gentianaceae-Volume 2: Biotechnology and Applications. Berlin: Springer.
Sanderson, M. J., Copetti, D., Búrquez, A., Bustamante, E., Charboneau, J. L., Eguiarte, L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102, 1115–1127. doi: 10.3732/ajb.1500184
Sazanov, L. A., Burrows, P. A., and Nixon, P. J. (1998). The plastid ndh genes code for an NADH-specific dehydrogenase: isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 1319–1324. doi: 10.1073/pnas.95.3.1319
Shaw, J., Shafer, H. L., Leonard, O. R., Kovach, M. J., Schorr, M., and Morris, A. B. (2014). Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. Bot. 101, 1987–2004. doi: 10.3732/ajb.1400398
Shepherd, L. D., Lange, P. J. D., Perrie, L. R., and Heenan, P. B. (2017). Chloroplast phylogeography of new zealand sophora trees (Fabaceae): extensive hybridization and widespread last glacial maximum survival. J. Biogeogr. 44, 1640–1651. doi: 10.1111/jbi.12963
Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5, 2043–2049. doi: 10.1007/BF02669253
Stefanović, S., and Olmstead, R. G. (2005). Down the slippery slope: plastid genome evolution in convolvulaceae. J. Mol. Evol. 61, 292–305. doi: 10.1007/s00239-004-0267-5
Takayama, K., Tamura, M., Tateishi, Y., Webb, E. L., and Kajita, T. (2013). Strong genetic structure over the American continents and transoceanic dispersal in the mangrove genus Rhizophora (Rhizophoraceae) revealed by broad-scale nuclear and chloroplast DNA analysis. Am. J. Bot. 100, 1191–1201. doi: 10.3732/ajb.1200567
Wakasugi, T., Tsudzuki, J., Ito, S., Nakashima, K., Tsudzuki, T., and Sugiura, M. (1994). Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. U.S.A. 91, 9794–9798. doi: 10.1073/pnas.91.21.9794
Wang, P., Duan, W., Takabayashi, A., Endo, T., Shikanai, T., Ye, J. Y., et al. (2006). Chloroplastic NAD (P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141, 465–474. doi: 10.1104/pp.105.070490
Wen, J., Zhang, J.-Q., Nie, Z.-L., Zhong, Y., and Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5:4. doi: 10.3389/fgene.2014.00004
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, C. S., Wang, Y. N., Hsu, C. Y., Lin, C. P., and Chaw, S. M. (2011). Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 3, 1284–1295. doi: 10.1093/gbe/evr095
Wyman, S. K., Janse, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Yang, J. B., Tang, M., Li, H. T., Zhang, Z. R., and Li, D. Z. (2013). Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 13:84. doi: 10.1186/1471-2148-13-84
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Zhang, M. Y., Fritsch, P. W., Ma, P. F., Wang, H., Lu, L., and Li, D. Z. (2017). Plastid phylogenomics and adaptive evolution of Gaultheria series Trichophyllae (Ericaceae), a clade from sky islands of the Himalaya-Hengduan Mountains. Mol. Phylogenet. Evol. 110, 7–18. doi: 10.1016/j.ympev.2017.01.015
Keywords: evolution, gene loss, Gentiana, sect. Kudoa, plastome
Citation: Sun S-S, Fu P-C, Zhou X-J, Cheng Y-W, Zhang F-Q, Chen S-L and Gao Q-B (2018) The Complete Plastome Sequences of Seven Species in Gentiana sect. Kudoa (Gentianaceae): Insights Into Plastid Gene Loss and Molecular Evolution. Front. Plant Sci. 9:493. doi: 10.3389/fpls.2018.00493
Received: 29 October 2017; Accepted: 03 April 2018;
Published: 01 May 2018.
Edited by:
Genlou Sun, Saint Mary’s University, CanadaReviewed by:
Jacob A. Tennessen, Oregon State University, United StatesDavid Richard Nash, University of Copenhagen, Denmark
Copyright © 2018 Sun, Fu, Zhou, Cheng, Zhang, Chen and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Bo Gao, cWJnYW9AbndpcGIuY2FzLmNu
†These authors have contributed equally to this work.
 Shan-Shan Sun
Shan-Shan Sun Peng-Cheng Fu
Peng-Cheng Fu Xiao-Jun Zhou1
Xiao-Jun Zhou1 Yan-Wei Cheng
Yan-Wei Cheng Fa-Qi Zhang
Fa-Qi Zhang Qing-Bo Gao
Qing-Bo Gao