
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 April 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00422
This article is part of the Research TopicCrop Breeding for Drought ResistanceView all 16 articles
Drought is one of the major abiotic stresses affecting world agriculture. Breeding drought-resistant crops is one of the most important challenges for plant biologists. PYR1/PYL/RCARs, which encode the abscisic acid (ABA) receptors, play pivotal roles in ABA signaling, but how these genes function in crop drought response remains largely unknown. Here we identified 13 PYL family members in maize (ZmPYL1-13). Changes in expression of these genes under different stresses indicated that ZmPYLs played important roles in responding to multiple abiotic stresses. Transgenic analyses of ZmPYL genes in Arabidopsis showed that overexpression of ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13 significantly enhanced the sensitivity of transgenic plants to ABA. Additionally, transgenic lines overexpressing ZmPYL8, ZmPYL9, and ZmPYL12 were more resistant to drought. Accumulation of proline and enhanced expression of drought-related marker genes in transgenic lines further confirmed the positive roles of ZmPYL genes in plant drought resistance. Association analyses with a panel of 368 maize inbred lines identified natural variants in ZmPYL8 and ZmPYL12 that were significantly associated with maize drought resistance. Our results deepen the knowledge of the function of maize PYL genes in responses to abiotic stresses, and the natural variants identified in ZmPYL genes may serve as potential molecular markers for breeding drought-resistant maize cultivars.
Due to their sessile lifestyle, plants cannot escape from environmental stresses which include biotic and abiotic stresses. Drought is one of the major abiotic stresses that negatively affect plant growth and development. Plants have evolved sophisticated mechanisms to respond to and survive drought. Generally, plants have two major mechanisms for drought resistance, drought avoidance and drought tolerance (Hu and Xiong, 2014). Drought avoidance includes increasing cuticular wax and abscisic acid (ABA) content, controlling relative water content and water potential, along with leaf rolling and stomatal aperture control. General criteria for drought tolerance are content of osmolytes such as proline and sugar, and membrane system stability (Hu and Xiong, 2014).
Abscisic acid is a phytohormone that plays critical roles in plant stress response. Under drought stress, plants increase the production of ABA, which initiates stomatal closure and thereby reduces transpirational water loss, thus helping the plant avoid drought stress (Kim et al., 2010). Plants can perceive ABA via ABA receptors, the regulatory components of the ABA receptor (RCAR) or PYRABACTIN RESISTANCE 1 (PYR1)/PYR1-like protein (PYL) family of START proteins (Ma et al., 2009; Park et al., 2009). The clade A PP2C phosphatases and SnRK2 kinases are also important in mediating ABA signaling (Miyazono et al., 2009; Fujita et al., 2011; Joshi-Saha et al., 2011; Antoni et al., 2012; Nakashima et al., 2012, 2014; Rushton et al., 2012; Roychoudhury et al., 2013). A number of studies revealed the important roles of SnRK2 kinases and clade A PP2C phosphatases in regulation of plant drought resistance. Mutation of SnRK2.6/OST1 caused constitutive stomatal opening and water loss (Mustilli et al., 2002). Triple mutants of snrk2.2/3/6 lost the ability to respond to ABA and were hypersensitive to drought (Fujii and Zhu, 2009). Recent study has revealed that SnRK2.6 regulates the ubiquitin E3 ligase activity of RZFP34/CHYR1, a positive regulator of plant drought resistance (Ding et al., 2015). In Arabidopsis, the HAI PP2Cs function in drought responses by regulating the accumulation of osmoregulatory solutes such as proline (Bhaskara et al., 2012). ZmPP2C-A10, encoding a clade A PP2C phosphatase, negatively regulates maize drought response (Xiang et al., 2017).
There are 14 Arabidopsis genes encoding PYR/PYL/RCAR ABA receptors (Li et al., 2013). Recently, these Arabidopsis PYR/PYL genes have been extensively studied and reported to be important for plant drought responses. By using an ABA inducible promoter, researchers have revealed that overexpressing PYL9 upon stress induction greatly enhanced drought resistance and drought-induced leaf senescence in transgenic Arabidopsis and rice plants (Zhao et al., 2016). Overexpression of PYL5 also enhanced plant drought tolerance (Santiago et al., 2009). The A194T site mutant of PYL4 formed stable complex with PP2CA in the absence of ABA, and 35S:PYL4 (A194T) plants showed dramatically enhanced tolerance to drought and dehydration (Pizzio et al., 2013). In guard cells, PYL/RCAR ABA receptors interact with PP2C phosphatases, thus releasing active SnRK2 kinase to activate the SLAC1 channel which leads to stomatal closure by reducing guard cell turgor (Lee et al., 2013). This shows the involvement of PYL/RCAR ABA receptors in plant drought avoidance. In maize, a previous study reported the expression profiles of ZmPYL genes in response to ABA and dehydration stress (Fan et al., 2016). However, the function of ZmPYL genes in regulation of maize drought tolerance remains elusive.
Association analysis based on linkage disequilibrium (LD) is widely used for dissecting complex traits of crops (Wang and Qin, 2017). By using genome wide association analysis (GWAS), a number of loci involved in regulation of complex plant traits, such as flowering time, oil synthesis, salt tolerance, were indentified recently (Huang et al., 2011; Meyer et al., 2016; Fang et al., 2017). Maize is a model plant for genetic studies due to its huge genetic diversity among various accessions (Strable and Scanlon, 2009). Due to the fast LD decay, researchers can detect trait-locus associations in maize at single gene resolution. For example, recent studies used GWAS to detect two drought-tolerant genes, ZmNAC111 and ZmVPP1, from a maize population consisting of 368 accessions (Mao et al., 2015; Wang et al., 2016). Based on the same maize population, we and another group identified two drought-resistance genes, ZmPP2C-A10 and ZmDREB2.7, and their natural variations through candidate gene association analyses (Liu et al., 2013; Xiang et al., 2017). Although the function of PYL/RCAR ABA receptors in regulation of drought resistance has been revealed in several plant species, how the natural variations in PYL/RCAR genes are associated with plant drought responses remains essentially unknown.
In this study, we identified and cloned the maize PYL family genes. Expression analyses showed that ZmPYL genes responded to multiple abiotic stresses including ABA, drought and salt. Transgenic analyses revealed the roles of several ZmPYLs in regulation of plant ABA and drought responses. Association analysis with a large maize association population identified several favorable alleles of two ZmPYL genes, ZmPYL8 and ZmPYL12, for maize drought resistance.
Maize B73 seeds were used for gene cloning and expression analysis. The Arabidopsis ecotype Col-0 was used as the wild-type. The pRCS2(Bar)-ZmPYLs plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Arabidopsis Col-0 ecotype using the floral dip method (Clough and Bent, 1998). Seeds of transformed Arabidopsis were selected on MS plates containing the appropriate antibiotics. Homozygous lines of T4 generations with one copy of inserted transgene were used for further analysis.
All maize seedlings were grown under 16 h light: 8 h dark photoperiod at 28°C. For drought treatment, maize seedlings were germinated and grown in soil under normal watering conditions until the three-leaf stage. Plants were then transferred onto filter paper and dried at 28°C. Shoot and root samples were harvested after 0, 1, 3, 6, 12, 24 h, respectively. For ABA and salt treatments, maize seeds were sown and grown in Hoagland’s solution until the three-leaf stage. Then seedling roots were soaked in Hoagland’s solution containing various concentrations of ABA (0, 1, 10, 50, 100, and 150 μM) for 3 h. For salt treatments, plant roots were immersed in Hoagland’s solution containing 150 mM NaCl and sampled after 0, 1, 2, 3, and 4 days. Upon harvesting the samples were immediately frozen in liquid nitrogen for RNA extraction.
Arabidopsis seeds of wild-type and independent transgenic ZmPYL-overexpression lines were sterilized with 10% bleach and then kept at 4°C in the dark for 3 days. For the germination rate assay, sterilized seeds of wild-type and lines overexpressing ZmPYLs were sown on MS plates or MS plates supplemented with 1 μM ABA and grown under 16-h light: 8-h dark photoperiod at 22°C. Germination was first scored on the second day and counted continuously for 5 days. For the root growth assay, the seeds of wild-type and ZmPYLs-overexpression lines were sown perpendicularly on aaa MS agar medium and grown under 16-h light: 8-h dark photoperiod at 22°C for 5 days, then transferred to aaa MS agar medium containing 0 or 10 μM ABA. They were then grown vertically for 7 days, after which root lengths and leaf weights were measured. For the drought treatment, sterilized seeds of wild-type and ZmPYL-overexpression lines were sown on MS growth medium for 9 days, then transferred to small pots containing potting soil. Arabidopsis plants grew under normal watering conditions with 12 h light: 12 h dark photoperiod at 22°C for about 3 weeks. Watering was then halted. Plants were sampled for marker gene assays after 10 days when they began to exhibit lethal effects of dehydration. Samples were taken for proline and malondialdehyde (MDA) assays after 12 days. Watering was then resumed and the fraction surviving was determined.
All PYL protein sequences in Arabidopsis were obtained from the Ensemble Plants database footnotehttp://plants.ensembl.org/. To identify ZmPYL genes in maize, each Arabidopsis PYL sequence was analyzed by BLAST at maizeGDB hidetext1 against the B73 working gene set translations 5a.59 for RefGen_v2. After removing redundant results, all sequences were submitted to SMART2 to test whether the sequence had the Polyketide_cyc domain. Thirteen ZmPYL genes were retained for analysis in this study.
For Arabidopsis transformation, the 13 ZmPYL genes were individually cloned from cDNA of maize line B73. The cDNAs were sequenced after insertion into pJET1.2 vector. The ZmPYL cDNAs were then released from the pJET vectors by digestion with EcoR I and Xho I, and inserted into the pSAT6 vector to produce pSAT6-ZmPYLs. The expression cassettes using 2X35S promoters to drive expression of the ZmPYL cDNAs were released from the pSAT6 vectors by digestion with PI-Psp I and inserted into the pRCS2-Bar-OCS binary vector (Dai et al., 2012).
Samples were collected after stress treatments. Total RNA was isolated using Trizol reagent (TransGen) from more than three seedlings for each treatment. RNA was treated with RNase-free DNase I (Thermo Scientific), and single-stranded cDNA was synthesized using recombinant M-MLV reverse transcriptase (Promega). The maize and Arabidopsis Actin genes were used as internal control to normalize the data.
Arabidopsis seedlings that were dehydrated for 12 days were used for MDA and proline assays. To assay proline, 50 mg fresh weight (FW) of leaves were shredded into 10 ml centrifuge tubes using scissors. Five milliliters of 3% sulfosalicylic acid solution were added to each sample and then the mixture was placed in boiling water for 10–30 min to obtain the extract solution. After cooling to room temperature, 2 ml of supernatant were pipetted into a new 10 ml centrifuge tube and mixed with 2 ml acetic acid and 2 ml ninhydrin. The mixture was placed in a boiling water bath for 30 min. Four milliliters of methylbenzene were added to the extract after cooling to room temperature, followed by centrifugation for 1 min and stewing for 10 min. The upper red solution was transferred to a 1.5 ml centrifuge tube. After centrifugation at 12000 rpm for 15 min, the upper proline solution was pipetted into a cuvette to measure absorbance at 520 nm by UV-vis spectrophotometry with methylbenzene as blank.
To assay MDA content, 50 mg FW of leaves were shredded using scissors and placed in 5 ml 5% TCA. After centrifugation at 3000 rpm and 4°C for 10 min, 2 ml of the supernatants were transferred into new 10 ml centrifuge tubes then 2 ml 0.67% thiobarbituric acid (TBA) were added and the extract was placed in a boiling water bath for 30 min. After cooling to room temperature, 1.5 ml of the extracts were transferred into 1.5 ml tubes and centrifuged at 12000 rpm and room temperature. Absorbance of the supernatants were measured at 440, 532, and 600 nm by UV-vis spectrophotometry. The MDA content was calculated as nmol/g FW tissue. C/umol/L = 6.45 (A532-A600) -0.56A450
Association analyses for the ZmPYL genes were performed using a maize association mapping population that contains 368 inbred lines and corresponding drought tolerance phenotypic data obtained in a previous study. Among 525105 high-quality SNPs data with Minor Allele Frequency (MAF) ≥ 0.05, 112 SNPs were found in the regions of ZmPYL3, ZmPYL8, ZmPYL9, ZmPYL10, and ZmPYL12 genes. The mixed linear model (MLM) was used to detect SNPs significantly associated with drought tolerance using the program TASSEL5.0 (Bradbury et al., 2007).
Statistical analyses were performed using Excel (Microsoft, United States). Figures were plotted by using Photoshop software (Adobe Systems, United States).
To determine how many genes encoding ZmPYL proteins are in the maize genome, each Arabidopsis PYL sequence was used as a BlastP query against the maize genome database (version B73 working gene set translations 5a.59 for RefGen_V2). Thirteen genes encoding ZmPYLs (ZmPYL1-13) were identified (Supplementary Table S1). These genes are located on most of the chromosomes with 1 or 2 genes per chromosome, except that no ZmPYL gene was detected on chromosome 7 (Supplementary Figure S1). A previous study reported 11 ZmPYL genes (Fan et al., 2016). The involvement of ZmPYL3 in ABA signaling has been reported (Wang et al., 2014). To be consistent, the names of ZmPYL1-11 in this study are same as those in the previous report (Fan et al., 2016). In our study, two more ZmPYL genes, GRMZM2G169695 (ZmPYL13, named in our study) and GRMZM2G405064 (ZmPYL12), were identified based on the conserved motifs and important amino acids of PYL family proteins (Li et al., 2013). Based on the protein sequences, a phylogenetic tree was generated using the NJ algorithm (Figure 1A). The ZmPYL proteins clustered into two subgroups, which is very similar to the Arabidopsis PYL proteins (Figure 1A; Li et al., 2013), suggesting the conservation of plant PYL proteins. Sequence analyses revealed that subgroup I genes had fewer introns than subgroup II ZmPYL genes (Figure 1B). In the promoter regions, a number of ABA responsive elements (ABREs) and MYB binding sites (MBS), which are involved in ABA and drought response (Seo et al., 2011; Singh and Laxmi, 2015), were detected in most ZmPYL genes except ZmPYL8 and ZmPYL10 (Figure 1B). The full-length coding sequences of the ZmPYL genes were cloned and confirmed by sequencing for further analyses.
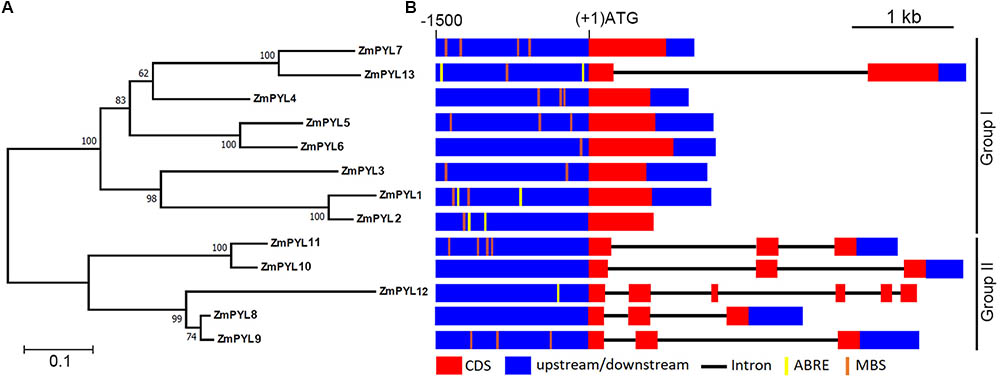
FIGURE 1. Evolutionary relationships of taxa. (A) Phylogenetic tree of ZmPYL proteins. The phylogenetic tree was generated based on the alignments of the full-length ZmPYL protein sequences. Bootstrap values from 1,000 replicates are indicated at each node. The scale bar represents branch lengths. (B) ZmPYL gene structures. The upstream regions (0∼–1500 bp) of these genes were used to search the public data base (PlantCARE). Most of the ZmPYLs except ZmPYL8 and ZmPYL10 contain one or more drought response elements (MBS or ABRE) in the upstream regions. MBS, MYB binding site; ABRE, ABA responsive element.
In order to determine the functions of ZmPYL genes in plant development, we studied their tissue-specific expression patterns. An expression heatmap of ZmPYL genes was created using the publically available transcription data for fifteen maize tissues (Figure 2A). According to the heatmap, the expression levels of ZmPYL genes were divided into two patterns. Subgroup II genes showed higher expression levels than those of subgroup I in most tissues (Figure 2A), indicating that subgroup II ZmPYL genes may have more important roles in regulating plant development. There are no expression data for ZmPYL2 and ZmPYL3 in the public database, therefore these genes were not included in the heatmap. Next we measured the expression levels of these genes in maize seedlings subjected to various levels of drought stress. ZmPYL4 was not detectable in our experiments, may be due to the extremely low expression of this gene in the tissues tested. Based on the expression patterns, the genes can be roughly divided into three types (Figure 2B). Type I genes (ZmPYL1, ZmPYL3, and ZmPYL11) were slightly down-regulated initially but up-regulated at later stages of drought stress in shoots, while in roots they were down-regulated. Type II genes (ZmPYL5, ZmPYL6, ZmPYL7, and ZmPYL10), were down-regulated in both shoots and roots after drought stress. Type III genes (ZmPYL2, ZmPYL8, ZmPYL9, and ZmPYL12) were up-regulated by drought in shoots and slightly up-regulated or unchanged in roots. These results suggested different roles of ZmPYL genes in regulation of drought responses. We next tested the responses of ZmPYL genes to other abiotic stresses, including ABA and salt stress. The results showed that ABA inhibited the expression of most ZmPYL genes in shoots and roots except for ZmPYL7 and ZmPYL8, whose expression levels were enhanced in either roots (ZmPYL8) or both shoots and roots (ZmPYL7) (Supplementary Figure S2). After salt stress, the expression levels of ZmPYL1, ZmPYL2, ZmPYL3, ZmPYL5, ZmPYL6, ZmPYL7, ZmPYL10, and ZmPYL13 were down-regulated, while the expression levels of ZmPYL8, ZmPYL9, ZmPYL11, and ZmPYL12 were slightly up-regulated or not changed (Supplementary Figure S3). Taken together, these results suggested diverse roles of ZmPYL genes in regulation of maize abiotic stress responses.
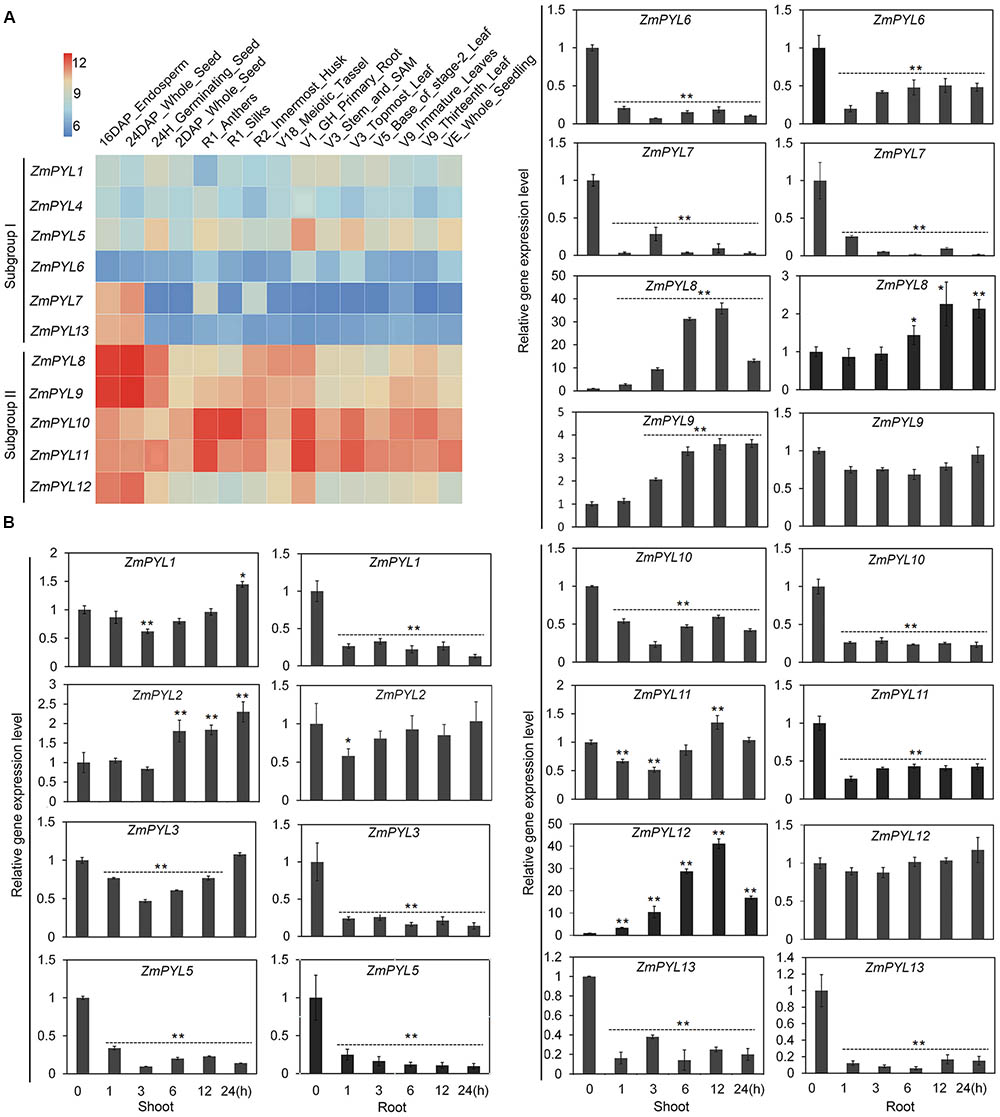
FIGURE 2. Expression Patterns of Maize ZmPYL Genes. (A) Gene expression heat map of 11 ZmPYL genes in 15 different tissues from various developmental stages. Two ZmPYL genes have no available data. The gene expression levels are shown in different colors indicated by the scale bar. (B) Expression patterns of ZmPYL genes in maize B73 leaves and roots under normal and drought treatments. ZmActin5 gene was used as an internal control. Both leaf and root tissues were collected after treatment at time points 0, 1, 3, 6, 12, and 24 h. Data represent the mean ± SD of three biological replicates. Asterisks indicate the significance of T-test, ∗p < 0.05, ∗∗p < 0.01.
In order to decipher the biological functions of ZmPYL genes, we generated transgenic Arabidopsis plants overexpressing the ZmPYL genes. A total of 12 ZmPYL genes were introduced into and overexpressed in Arabidopsis plants except ZmPYL4. Our failure to clone ZmPYL4 may be due to its extremely low expression in most of maize tissues. In order to determine how these ZmPYL genes function in regulating ABA responses, we germinated ZmPYL transgenic seeds on MS plates with or without ABA, and then measured the germination rates at various time points. To our surprise, we only observed that transgenic seeds overexpressing ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13 responded to ABA treatment (Figure 3 and Supplementary Figure S4). These transgenic seeds were hypersensitive to ABA, suggesting negative roles of ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13 in regulation of ABA-mediated seed germination. It is well known that ABA inhibits plant growth (Luo et al., 2014). To determine how ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13 regulate plant growth in response to ABA, we grew the control and transgenic seedlings on MS plates with or without ABA, and then measured the root lengths and FW. The results showed that the transgenic seedlings overexpressing ZmPYL3 or ZmPYL9 had more severely inhibited plant growth than the controls, suggesting that they were more sensitive to ABA than the controls (Figure 4). These results indicated that the activities of ZmPYL3 and ZmPYL9 were critical for both seed germination and post-germination growth. Intriguingly, we did not observe any phenotypic changes in ZmPYL13 and ZmPYL10 transgenic plants as compared to controls after ABA treatment (Data not shown), suggesting a specific role of ZmPYL10 and ZmPYL13 in regulating Arabidopsis seed germination rather than seedling growth.
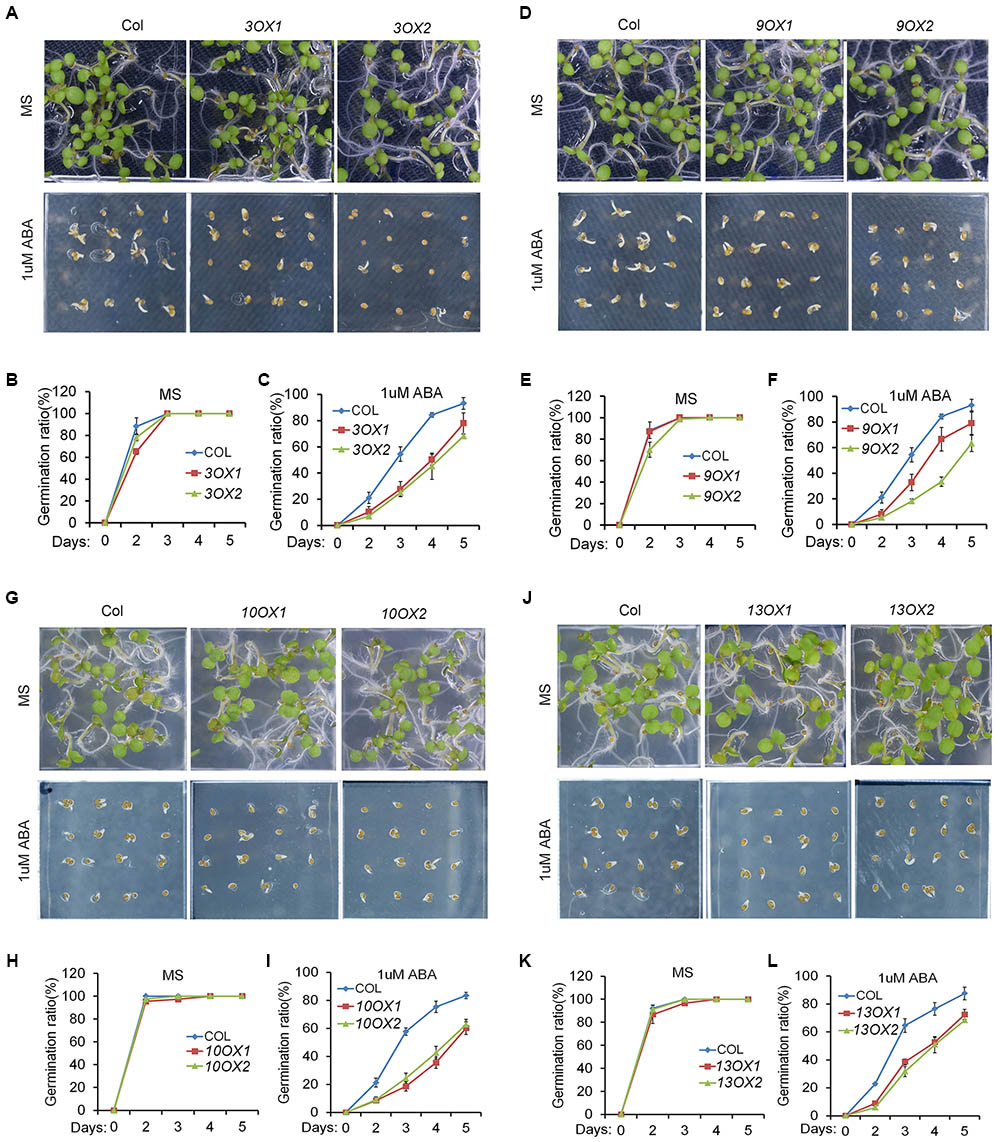
FIGURE 3. Overexpression of ZmPYL3, ZmPYL9, ZmPYL10, ZmPYL13 in Arabidopsis increased the sensitivity of seed germination to abscisic acid (ABA). (A–C) Germination phenotypes (A) and statistical analyses (B,C) of Col and two transgenic seeds overexpressing ZmPYL3 sown on MS medium or MS medium supplemented with 1 μM ABA. (D–F) Germination phenotypes (D) and statistical analyses (E,F) of Col and two transgenic seeds overexpressing ZmPYL9 sown on MS medium or MS medium supplemented with 1 μM ABA. (G–I) Germination phenotypes (G) and statistical analyses (H,I) of Col and two transgenic seeds overexpressing ZmPYL10 sown on MS medium or MS medium supplemented with 1 μM ABA. (J–L) Germination phenotypes (J) and statistical analyses (K,L) of Col and two transgenic seeds overexpressing ZmPYL13 sown on MS medium or MS medium supplemented with 1 μM ABA. Data represent the mean ± SD of three replicates in (B), (C), (E), (F), (H), (I), (K), and (L).
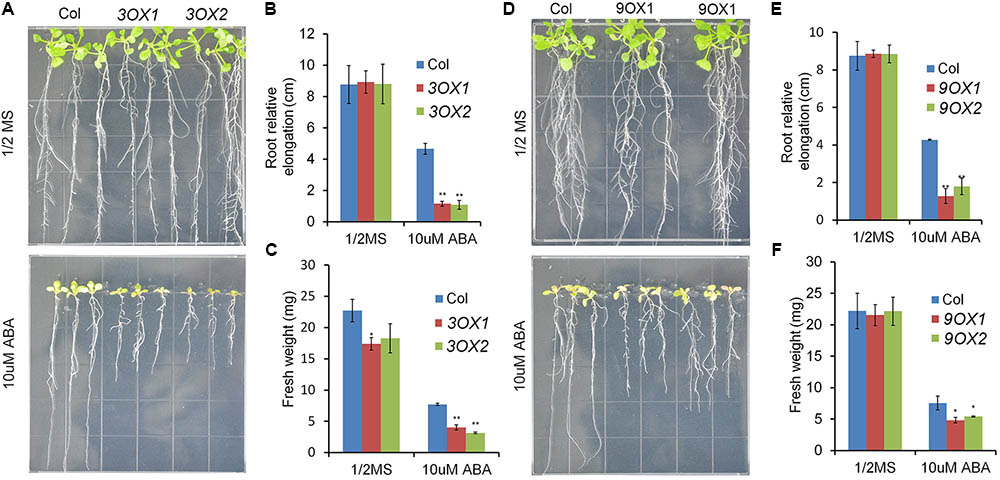
FIGURE 4. Overexpression of ZmPYL3, ZmPYL9 in Arabidopsis increased the sensitivity of plant growth to ABA. (A–C) Root growth phenotypes (A), statistical analyses of root elongation (B), and fresh weigh (C) of Col and ZmPYL3 overexpression transgenic lines grown on aaa MS medium or aaa MS medium containing ABA (10 μM). (D–F) Root growth phenotypes (D), statistical analyses of root elongation (E), and fresh weigh (F) of Col and ZmPYL9 overexpression transgenic lines grown on aaa MS medium or aaa MS medium containing ABA (10 μM). Five-day seedlings were transferred from aaa MS medium to aaa MS medium containing ABA (10 μM), then grown vertically for 7 days before phenotype. Data represent the mean ± SD of three replicates for (B), (C), (E), and (F). Asterisks indicate the significance of T-test, ∗p < 0.05, ∗∗p < 0.01.
Next, we performed experiments to test how ZmPYL genes regulate plant drought responses. All the transgenic lines overexpressing ZmPYL genes were subjected to severe drought stress. After stress, the plants were re-watered and survival rates of the transgenic lines were determined. We observed that most plants overexpressing ZmPYL8 recovered upon rewatering after severe drought stress, whereas most control plants died (Figures 5A,B). The survival rates of plants overexpressing ZmPYL8 were twofold higher than those of controls (Figure 5C). Further analyses showed that more than twofold higher of proline (Figure 5D), and 30% less of MDA accumulated in plants overexpressing ZmPYL8 after drought stress as compared to controls (Figure 5E). These results suggested that ZmPYL8 plays a positive role in regulation of plant drought tolerance. We observed similar phenotypic changes in plants overexpressing ZmPYL9 and ZmPYL12. After drought stress, the transgenic plants overexpressing ZmPYL9 and ZmPYL12 had higher survival rates than the controls (Supplementary Figure S5), suggesting that ZmPYL9 and ZmPYL12 also have positive roles in plant drought tolerance.
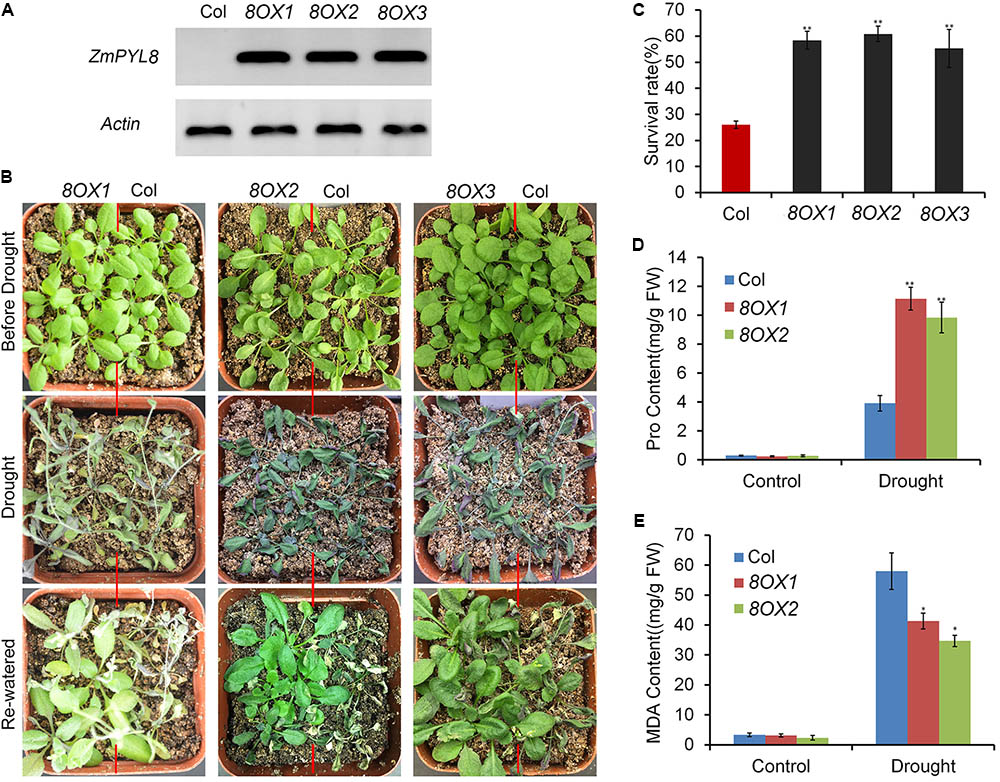
FIGURE 5. Overexpression of ZmPYL8 in Arabidopsis enhanced drought tolerance. (A) RT-PCR shows overexpression of ZmPYL8 (8OX) in three independent transgenic lines. Arabidopsis actin7 gene was used as control. (B) Drought tolerance assay of three transgenic lines overexpressing ZmPYL8. Three-week old plants were drought-stressed for 2 weeks and then rewatered for 1 day. (C) Survival rates of Col and ZmPYL8 transgenic plants after drought stress. Data represent the mean ± SD of three replicates. (D) Proline contents of watered and drought-stressed Col and ZmPYL8 transgenic plants. (E) Malondialdehyde (MDA) contents of watered and drought-stressed Col and ZmPYL8 transgenic plants. Data represent the mean ± SD of three biological replicates. Asterisks indicate the significance of T-test, ∗p < 0.05, ∗∗p < 0.01.
When compared to control plants, we did not observe any phenotypic changes in transgenic plants overexpressing other ZmPYL genes in our drought stress experiments (Data not shown), suggesting these genes do not play major roles in regulating drought resistance of the transgenic plants. ZmPYL8, ZmPYL9, and ZmPYL12 clustered together in the phylogenetic tree (Figure 1A), suggesting conserved roles of these genes at least in drought resistance. We therefore chose plants overexpressing ZmPYL8 as representatives to measure the expression of drought-responsive marker genes. The results showed that the expression of all of these marker genes was enhanced in transgenic plants after drought stress (Figure 6). The increased expression of ABA3, COR47, RD26, RD29A, RD29B, ABI1, ABI2, DREB2A, which are known to respond to ABA, suggested enhanced ABA signaling in transgenic plants. P5CS1 is an enzyme responsible for proline biosynthesis (Kesari et al., 2012). The enhanced expression of P5CS1 is consistent with the elevated accumulation of proline in ZmPYL8 transgenic plants (Figure 5D). Taken together, these results suggested overexpression of ZmPYL8 promoted ABA signaling, which, in turn, enhanced drought resistance of the transgenic plants.
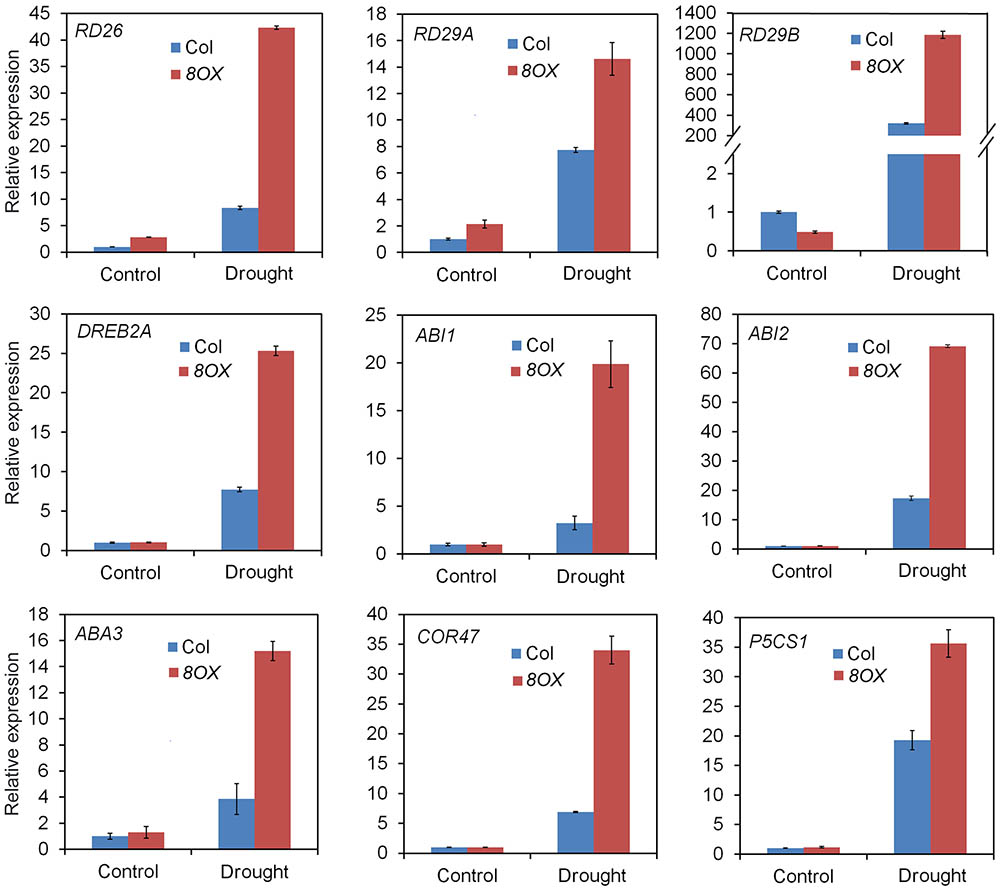
FIGURE 6. Overexpression of ZmPYL8 raised the expression of drought responsible marker genes. The expression of all marker genes under normal and drought conditions was examined by qRT-PCR. Three-week old plants were either watered or drought-stressed for 10 days.
Previous studies have identified one million SNPs in a maize panel consisting of 368 accessions (Fu et al., 2013). Using survival rates after severe drought stress as indices, the drought-resistance phenotypes of these maize accessions were reported in several studies (Liu et al., 2013; Wang et al., 2016). To determine whether natural variation in ZmPYL genes was associated with maize drought resistance, we performed candidate gene association analyses with the MLM on ZmPYL3, ZmPYL8, ZmPYL9, ZmPYL10, ZmPYL12, which we identified as playing roles in regulation of ABA responses and/or drought resistance (Figures 3–5 and Supplementary Figure S5). By using the genotypic and phenotypic data of the 368 accessions, we observed from association analyses that two genes, ZmPYL8 and ZmPYL12, had natural variants that were significantly associated with drought resistance (p ≤ 0.01, Table 1). The most significant p-value (lead p-value) was 0.00277 for ZmPYL8 and 0.0018 for ZmPYL12 (Table 1). Further analyses showed that the lead SNP (SNP1543, with lead p-value) in ZmPYL8 was located in the third exon of this gene, and showed strong LD with another significant SNP (SNP1640) located downstream of the lead SNP (Supplementary Figure S6). The lead SNP of ZmPYL12 (SNP415) was located in the second exon and showed strong LD with another significant SNP (SNP-52) located upstream of the lead SNP (Figures 7A,B). Further analyses identified the drought-resistant alleles of SNP415 and SNP-52 in ZmPYL12 (Figures 7C,D). In the association populations, there were four major haplotypes (Hap), among which Hap4 was most drought-resistant (Figure 7E). In ZmPYL8, we did not detect drought-resistant alleles from the associated SNPs (data not shown).
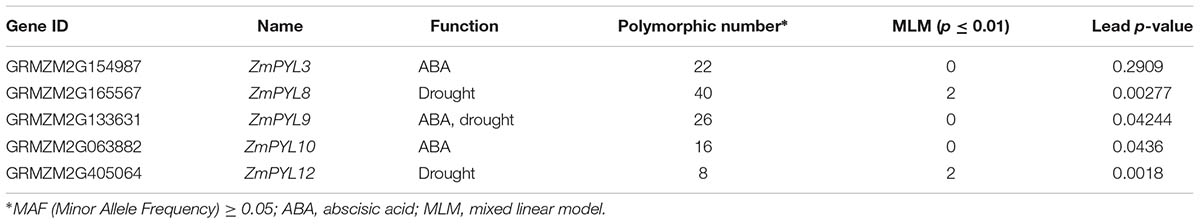
TABLE 1. Natural variations in ZmPYL genes associated with drought resistance in 368 maize inbred lines.
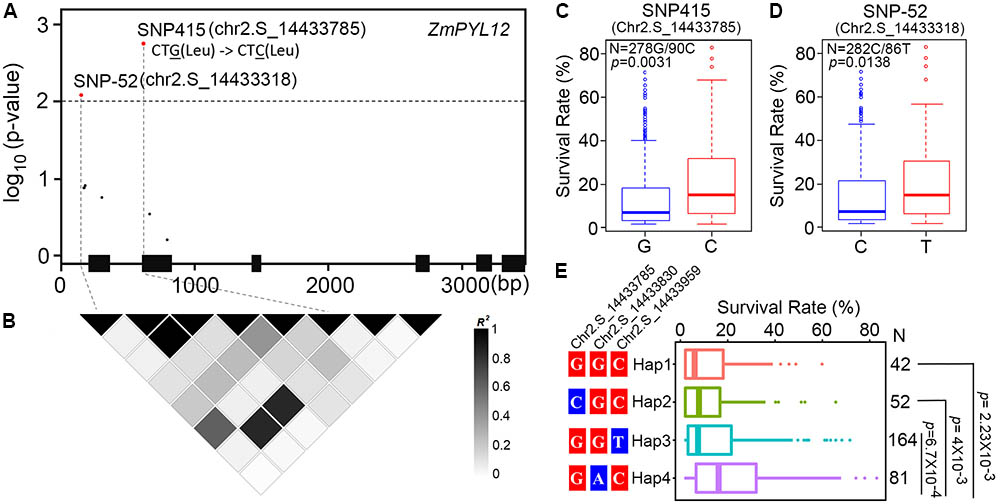
FIGURE 7. Association analysis of genetic variations in ZmPYL12 with maize drought tolerance. (A) Associations of ZmPYL12 SNPs with drought tolerance. The gene structure is shown at the bottom. The significant associations are shown in red (p < 0.01). Exons and introns are shown as filled boxes and dark lines, respectively. SNP415 has G/C natural variations, but does not change the coding information of Leu. (B) LD map for ZmPYL12 SNPs. Color in each block represents the R-squared value between two SNPs. (C) Association of two alleles in SNP415 with drought tolerance. (D) Association of two alleles in SNP-52 with drought tolerance. (E) Association of hyplotypes in ZmPYL12 with drought tolerance.
Abscisic acid is well-known to be an important phytohormone regulating plant stress responses. A recent breakthrough in our understanding of ABA signaling came when two different groups identified the ABA receptors, the RCAR/PYL family proteins (Ma et al., 2009; Park et al., 2009). Subsequently, the functions of PYL receptors in regulation of ABA and stress responses have been widely studied (Klingler et al., 2010; Antoni et al., 2013; Liang et al., 2017; Pri-Tal et al., 2017). Maize is an important food and industrial crop worldwide and every year suffers yield losses due to drought. How ZmPYL ABA receptors are involved in maize drought resistance remains largely unknown. In this study, we comprehensively investigated the expression of ZmPYL genes in response to ABA and drought stresses, the roles of ZmPYLs in regulating ABA and drought responses and natural variants of ZmPYL genes associated with maize drought resistance.
Thirteen ZmPYLs genes were identified in this study. Analyses of the promoter sequences of these genes identified multiple cis elements, including ABRE and MBS, which have been reported to be involved in responses to ABA and drought stress (Seo et al., 2011; Singh and Laxmi, 2015). The changes in ZmPYL gene expression after ABA and drought stresses indicated the roles of these cis elements in regulation of gene expression. Intriguingly, most ZmPYL genes were down-regulated after drought treatment, whereas ZmPYL8, ZmPYL9, ZmPYL2, and ZmPYL12 were up-regulated. After ABA treatment, most ZmPYL genes were down-regulated, while ZmPYL7 gene was up-regulated. Upon salt treatment, about 50% ZmPYL genes were down-regulated, while the rest were slightly affected or not changed. The different response patterns of ZmPYL genes after drought, ABA and salt treatments suggested diverse roles of ZmPYLs in regulation of plant abiotic stress responses. A previous study reported the expression of 11 ZmPYLs in response to ABA and PEG treatments (Fan et al., 2016). Both previous and this studies revealed very similar expression patterns of most ZmPYL genes in response to ABA, except ZmPYL1-3 (Supplementary Figure S2; Fan et al., 2016). The various expression patterns of few ZmPYLs may be caused by different growth stages or environments for the experiments. Interestingly, ZmPYLs showed very different expression patterns in response to PEG or drought treatments. For instance, the expression of half of ZmPYLs was down-regulated under drought in this study, but the expression of most of ZmPYLs was up-regulated in response to PEG (Figure 2; Fan et al., 2016). These results suggested different effects of PEG and drought on ZmPYL expression.
To investigate the biological functions of ZmPYLs, we cloned all of the ZmPYL genes and overexpressed them in Arabidopsis plants, except ZmPYL4 due to failure of detecting its expression in maize leaves. We observed that overexpression of four genes, ZmPYL3, ZmPYL9, ZmPYL10, and ZmPYL13, resulted in hypersensitivity to ABA treatment, suggesting negative roles of these genes in ABA responses in transgenic plants. This is very similar to what was observed in Arabidopsis, where PYL genes also played negative roles in ABA responses (Ma et al., 2009; Park et al., 2009), suggesting that the negative roles of these PYL genes in ABA responses are conserved across various plant species. There are 13 ZmPYL genes in maize, but surprisingly only four of them had roles in regulating ABA responses when ectopically expressed in Arabidopsis. One possibility is that the other ZmPYL genes have other roles rather than mediating ABA signaling. Considering that the expression levels of ZmPYL1, ZmPYL4, ZmPYL5, and ZmPYL6 were also dramatically inhibited by ABA in maize but played no roles in regulation of ABA responses when ectopically expressed in Arabidopsis, another possible reason is that these ZmPYL genes may not function in Arabidopsis. Transgenic studies of these ZmPYL genes in maize may provide more precise evaluation of their roles in plant development and ABA responses.
In drought stress experiments, we observed that overexpressing ZmPYL8, ZmPYL9, and ZmPYL12 in Arabidopsis, resulted in resistance of the transgenic plants to drought treatment, suggesting positive roles of these genes in drought responses. ZmPYL8, ZmPYL9, and ZmPYL12 were clustered together in the phylogenetic tree (Figure 1). After drought treatment, ZmPYL8, ZmPYL9, and ZmPYL12 showed very similar expression patterns in both shoots and roots (Figure 2). These observations indicated conserved roles of these genes in regulation of plant drought resistance. But the fact that only ZmPYL9 was involved in regulation of both drought and ABA responses indicated functional diversity of these genes. A previous study reported the roles of Arabidopsis PYLs in plant drought stress responses by ectopically expressing these genes with various promoters, including the stress-inducible RD29A promoter, the GC1 and ROP11 promoters (guard cell-specific) and the RBCS promoter (green tissue-specific), as well as the constitutive 35S promoter (Zhao et al., 2016). Based on their experiments, different promoters resulted in different levels of drought resistance when these promoters were used to drive the expression of various PYL genes (Zhao et al., 2016), indicating that the roles of plant PYLs in drought resistance depends on their temporal and spatial patterns of expression. In future, it will be worthwhile to use maize stress-inducible or tissue-specific promoters to study the functions of ZmPYL genes in regulation of drought resistance.
Developing new molecular markers is one of the most important topics in plant breeding. Thanks to the advances in high-throughput technologies, millions of SNP markers distributed throughout the whole genome at high density have been developed for various crops, including maize, rice, and other crops (Huang et al., 2011; Jiao et al., 2012; Fu et al., 2013; Zhou et al., 2015; Yano et al., 2016). Candidate gene association analyses, which are based on genetic variants and plant phenotypes, are widely used in dissecting the natural variation involved in plant drought resistance (Liu et al., 2013; Xiang et al., 2017). By using this association method, we detected natural variants of both ZmPYL8 and ZmPYL12 from a maize panel consisting of 368 natural accessions. Considering that these associations were most significant as compared to those detected in other ZmPYL genes which regulated ABA responses, and that ZmPYL8 and ZmPYL12 were proved to play positive roles in plant drought resistance, these associations are not false positive but real associations. Further analyses failed to detect resistant alleles in ZmPYL8, perhaps because they are not causal alleles of drought resistance. However, we detected resistant alleles in ZmPYL12 (Figure 7), indicating that these alleles are causal alleles or have strong LD with the causal alleles.
Together, our results revealed the fundamental roles ZmPYLs in mediating ABA signaling and drought resistance in maize. Breeding of drought-resistant maize cultivars is an important goal for maize breeders. The drought-resistant ZmPYLs may be used as genetic resources in drought-tolerant breeding via transgenic approaches. Moreover, the resistant alleles or haplotypes detected in ZmPYL12 have the potential to be used as molecular markers in genetic improvement of maize drought resistance.
MD and ZH designed the experiments. ZH and JZ conducted the experiments and analyzed the data. XS helped in the association and bioinformatic analyses. BW helped in the transgenic experiments. WT carefully edited the manuscript and analyzed the data. MD, ZH, and JZ wrote the manuscript. All authors revised the manuscript.
This study was supported by the Ministry of Science and Technology of China (2015BAD02B01), the Natural Science Foundation of China (31671256), the Fundamental Research Funds for the Central Universities of China (2662015PY170).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00422/full#supplementary-material
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Peirats-Llobet, M., Pizzio, G. A., Fernandez, M. A., et al. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161, 931–941. doi: 10.1104/pp.112.208678
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A., and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158, 970–980. doi: 10.1104/pp.111.188623
Bhaskara, G. B., Nguyen, T. T., and Verslues, P. E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160, 379–395. doi: 10.1104/pp.112.202408
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dai, M., Zhang, C., Kania, U., Chen, F., Xue, Q., McCray, T., et al. (2012). PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24, 2497–2514. doi: 10.1105/tpc.112.098905
Ding, S., Zhang, B., and Qin, F. (2015). Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27, 3228–3244. doi: 10.1105/tpc.15.00321
Fan, W., Zhao, M., Li, S., Bai, X., Li, J., Meng, H., et al. (2016). Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 16:99. doi: 10.1186/s12870-016-0764-x
Fang, C., Ma, Y., Wu, S., Liu, Z., Wang, Z., Yang, R., et al. (2017). Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18:161. doi: 10.1186/s13059-017-1289-9
Fu, J., Cheng, Y., Linghu, J., Yang, X., Kang, L., Zhang, Z., et al. (2013). RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 4:2832. doi: 10.1038/ncomms3832
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Hu, H., and Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Huang, X., Zhao, Y., Wei, X., Li, C., Wang, A., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Jiao, Y., Zhao, H., Ren, L., Song, W., Zeng, B., Guo, J., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44, 812–815. doi: 10.1038/ng.2312
Joshi-Saha, A., Valon, C., and Leung, J. (2011). Abscisic acid signal off the STARting block. Mol. Plant 4, 562–580. doi: 10.1093/mp/ssr055
Kesari, R., Lasky, J. R., Villamor, J. G., Des Marais, D. L., Chen, Y. J., Liu, T. W., et al. (2012). Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc. Natl. Acad. Sci. U.S.A. 109, 9197–9202.
Kim, T. H., Böhmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Klingler, J. P., Batelli, G., and Zhu, J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signaling. J. Exp. Bot. 61, 3199–3210. doi: 10.1093/jxb/erq151
Lee, S. C., Lim, C. W., Lan, W., He, K., and Luan, S. (2013). ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 6, 528–538. doi: 10.1093/mp/sss078
Li, W., Wang, L., Sheng, X., Yan, C., Zhou, R., Hang, J., et al. (2013). Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 23, 1369–1379. doi: 10.1093/mp/sss078
Liang, C., Liu, Y., Li, Y., Meng, Z., Yan, R., Zhu, T., et al. (2017). Activation of ABA receptors gene GhPYL9-11A is positively correlated with cotton drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 8:1453. doi: 10.3389/fpls.2017.01453
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9:e1003790. doi: 10.1371/journal.pgen.1003790
Luo, X., Chen, Z., Gao, J., and Gong, Z. (2014). Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 79, 44–55. doi: 10.1111/tpj.12534
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mao, H., Wang, H., Liu, S., Li, Z., Yang, X., Yan, J., et al. (2015). A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6:8326. doi: 10.1038/ncomms9326
Meyer, R. S., Choi, J. Y., Sanches, M., Plessis, A., Flowers, J. M., Amas, J., et al. (2016). Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat. Genet. 48, 1083–1088. doi: 10.1038/ng.3633
Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H. J., Asano, A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462, 609–614. doi: 10.1038/nature08583
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071.
Pizzio, G. A., Rodriguez, L., Antoni, R., Gonzalez-Guzman, M., Yunta, C., Merilo, E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163, 441–455. doi: 10.1104/pp.113.224162
Pri-Tal, O., Shaar-Moshe, L., Wiseglass, G., Peleg, Z., and Mosquna, A. (2017). Non-redundant functions of the dimeric ABA receptor BdPYL1 in the grass Brachypodium. Plant J. 92, 774–786. doi: 10.1111/tpj.13714
Roychoudhury, A., Paul, S., and Basu, S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32, 985–1006. doi: 10.1007/s00299-013-1414-5
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., et al. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588. doi: 10.1111/j.1365-313X.2009.03981.x
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Singh, D., and Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895. doi: 10.3389/fpls.2015.00895
Strable, J., and Scanlon, M. J. (2009). Maize (Zea mays): a model organism for basic and applied research in plant biology. Cold Spring Harb. Protoc. 2009:pdb.emo132. doi: 10.1101/pdb.emo132
Wang, H., and Qin, F. (2017). Genome-wide association study reveals natural variations contributing to drought resistance in crops. Front. Plant Sci. 8:1110. doi: 10.3389/fpls.2017.01110
Wang, X., Wang, H., Liu, S., Ferjani, A., Li, J., Yan, J., et al. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241. doi: 10.1038/ng.3636
Wang, Y. G., Yu, H. Q., Zhang, Y. Y., Lai, C. X., She, Y. H., Li, W. C., et al. (2014). Interaction between abscisic acid receptor PYL3 and protein phosphatase type 2C in response to ABA signaling in maize. Gene 549, 179–185. doi: 10.1016/j.gene.2014.08.001
Xiang, Y., Sun, X., Gao, S., Qin, F., and Dai, M. (2017). Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol. Plant 10, 456–469. doi: 10.1016/j.molp.2016.10.003
Yano, K., Yamamoto, E., Aya, K., Takeuchi, H., Lo, P. C., Hu, L., et al. (2016). Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 48, 927–934. doi: 10.1038/ng.3596
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Keywords: ABA receptors, drought resistance, natural variation, Zea mays, ABA response
Citation: He Z, Zhong J, Sun X, Wang B, Terzaghi W and Dai M (2018) The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 9:422. doi: 10.3389/fpls.2018.00422
Received: 26 November 2017; Accepted: 16 March 2018;
Published: 04 April 2018.
Edited by:
Lijun Luo, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Yang Zhao, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2018 He, Zhong, Sun, Wang, Terzaghi and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingqiu Dai, bWluZ3FpdWRhaUBtYWlsLmh6YXUuZWR1LmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.