- Key Laboratory of Crop Ecophysiology and Farming System in Southwest China, Institute of Ecological Agriculture, Sichuan Agricultural University, Chengdu, China
Phytohormones regulate numerous important biological processes in plant development and biotic/abiotic stress response cascades. More than 50 and 100 years have passed since the initial discoveries of the phytohormones abscisic acid (ABA) and gibberellins (GA), respectively. Over the past several decades, numerous elegant studies have demonstrated that ABA and GA antagonistically regulate many plant developmental processes, including seed maturation, seed dormancy and germination, root initiation, hypocotyl and stem elongation, and floral transition. Furthermore, as a well-established stress hormone, ABA plays a key role in plant responses to abiotic stresses, such as drought, flooding, salinity and low temperature. Interestingly, recent evidence revealed that GA are also involved in plant response to adverse environmental conditions. Consequently, the complex crosstalk networks between ABA and GA, mediated by diverse key regulators, have been extensively investigated and documented. In this updated mini-review, we summarize the most recent advances in our understanding of the antagonistically regulatory roles of ABA and GA in different stages of plant development and in various plant–environment interactions, focusing on the crosstalk between ABA and GA at the levels of phytohormone metabolism and signal transduction.
Introduction
Various plant hormones play key and distinct roles in the plant life cycle, from seed maturation, seed germination to the floral transition and abiotic/biotic stress responses (Shu et al., 2016c; Yang and Li, 2017). Which phytohormones regulate plant developmental processes and stress adaptation, and what are the mechanisms? In the past decades, tremendous progress has been achieved in the field of plant hormone biology to answer these questions, in particular in the model plant Arabidopsis thaliana. The anabolism, catabolism, transport and signal transduction pathways of the phytohormones have been documented (Novak et al., 2017). Numerous elegant studies have demonstrated that different phytohormones interact antagonistically and/or synergistically with one other, forming complicated crosstalk networks (Shan et al., 2012). As a result of such crosstalk networks, different phytohormones regulate distinct biological processes precisely throughout a plant’s life cycle.
Abscisic acid (ABA) and gibberellins (GA) are one pair of classic phytohormones, which antagonistically mediate several plant developmental processes, including seed maturation, seed dormancy and germination, primary root growth, and flowering time control (Wang et al., 2013; Luo et al., 2014; Yang et al., 2014; Shu et al., 2016a,c). Thus, the crosstalk between ABA and GA is a research hotspot in the fields of plant molecular biology and plant genetics, with numerous key regulators, including DELLAs and AP2-domain-containing transcription factors, having been extensively investigated and reviewed (Liu X. et al., 2016; Shu et al., 2018b). These factors carry out functions, central to ABA and GA antagonism, affecting phytohormone biosynthesis and/or signal transduction pathways (Lin et al., 2015; Liu X. et al., 2016; Shu et al., 2018b).
Furthermore, as a near-universal abiotic stress hormone, ABA is involved in diverse abiotic stress response cascades, including those related to drought, flooding, salinity and low temperatures (Zhu et al., 2017). Although GA are primarily regarded as plant growth regulators involved in a number of developmental processes, including stem elongation (Jan et al., 2006; Iwamoto et al., 2011; Li et al., 2011), and flowering time control (Ding et al., 2013; Hyun et al., 2016), recently published data demonstrate that GA also control certain biological processes in response to stress (Qin et al., 2011; Hamayun et al., 2017; Urano et al., 2017; Wang et al., 2017). Consequently, the detailed mechanisms by which ABA and GA precisely mediate plant development and stress responses are ripe for further exploration.
Given the significant progress which has taken place in research into ABA and GA antagonism field, this mini-review will highlight the most recent advances (primarily, over the past 4 years) in the regulatory roles of ABA and GA in plant development and stress responses, focusing on the crosstalk between ABA and GA, as mediated by the key regulators. Finally, future research directions and challenges in this field will be discussed.
ABA and GA: Focusing on Seed Dormancy and Germination
Seed science is one of the most important research fields in plant molecular biology, including seed maturation, seed dormancy and germination, and seed longevity (Waterworth et al., 2015; Sano et al., 2016; Shu et al., 2016c; Nee et al., 2017). Crop seed is the direct product of the agricultural system, and thus optimal germination and seedling emergence in the field are important to achieve the final yield. ABA and GA play different key roles in the regulation of seed dormancy and germination, and the metabolism of and signaling by both phytohormones also changes during seed development, namely seed maturation, seed dormancy and germination, and seedling establishment (Shu et al., 2015, 2016c).
ABI4 (Abscisic acid-insensitive 4), an enhancer of the ABA signal transduction pathway, deepened seed dormancy in Arabidopsis by increasing ABA biosynthesis while decreasing GA biosynthesis (Shu et al., 2013). Detailed biochemical analysis showed that ABI4 directly interacts with promoter regions of NCED6, an ABA biosynthesis gene, and of GA2ox7, a GA-inactivator gene (Shu et al., 2016b). Consequently, ABI4 is the central factor which mediates the antagonism between ABA and GA by regulating the biosynthesis of both phytohormones, resulting in the precise control of the degree of seed dormancy and post-germination seedling growth (Shu et al., 2016c, 2018b). Interestingly, ABI4 also inhibits seed germination and cotyledon greening through the mediation of cytokinin signaling (Huang et al., 2017). Investigations revealed that, in Sorghum bicolor, transcription factors SbABI4 and SbABI5 enhanced the transcription of SbGA2ox3, a GA-inactivator gene, through directly binding to its promoter, and consequently extended seed dormancy (Cantoro et al., 2013). During the post-germination seedling growth stage, ABI4 also enhanced ANAC060 transcription by directly interacting with its promoter, with ANAC060 reducing ABA sensitivity and glucose-mediated ABA accumulation (Li et al., 2014). Another gene, CK2 (Casein Kinase 2), positively mediated ABA signaling and stress responses during seed germination and early seedling establishment, the partial mechanism being that CK2 indirectly enhanced ABI4 expression (Wang et al., 2014). Furthermore, diverse factors, including miRNA 165/166, E3 ubiquitin ligase CER9 (ECERIFERUM 9), transcription factors RAV1, OsAP2-39 and MYB96, nuclear C2H2 zinc-finger protein ZFP3, and AtGLR3.5 (glutamate receptor homolog 3.5), regulated ABA signaling during seed germination and post-germination seedling growth through the ABI4-mediated cascades (Yaish et al., 2010; Feng et al., 2014; Joseph et al., 2014; Zhao et al., 2014; Kong et al., 2015; Lee et al., 2015; Yan et al., 2016). Consequently, ABI4 is a key factor with regard to ABA-mediated regulation of seed germination and early seedling establishment.
Recently, several key components, which regulate seed germination, were dissected by analyzing their effect on the balance between ABA and GA. GIM 2 (Germination Insensitive to ABA mutant 2) promoted GA biosynthesis while reducing ABA biosynthesis, and subsequently the gim2 mutant seeds showed the ABA-insensitive phenotype during seed germination and post-germination seedling growth (Xiong et al., 2017). In addition, exogenous auxin or NaCl treatment delayed soybean seed germination through decreasing the GA/ABA ratio (Shu et al., 2017; Shuai et al., 2017). A similar effect of NaCl on seed germination of the halophyte Suaeda salsa has been reported, which is also mediated through ABA and GA pathways (Li et al., 2015). Aluminum (Al) in contaminated soil inhibited rice seed germination, while exogenous H2 (hydrogen) alleviated the Al toxicity through increasing the GA/ABA ratio (Xu et al., 2017). Further detailed analysis showed that H2 promoted the expression of GA20ox1 and GA20ox2, two GA biosynthesis genes, and of ABA8ox1 and ABA8ox2, two ABA catabolism genes (Xu et al., 2017).
In maize, under chilling stress, the combination of the phytohormone salicylic acid (SA) and the reactive oxygen species hydrogen peroxide (H2O2) up-regulated transcription of both the GA biosynthesis gene ZmGA20ox1 and the ABA catabolism gene ZmCYP707A2, while down-regulating the expression of the GA catabolism gene ZmGA2ox1 (Li Z. et al., 2017). Exogenous application of SA and H2O2 increased the GA/ABA ratio and accelerated maize seed germination under chilling stress conditions (Li Z. et al., 2017). Karrikins, a group of plant growth regulators present in the smoke of burning plant material, mediated soybean seed germination through regulating GA and ABA biosynthesis and signal balance (Meng et al., 2016a,b). Moreover, the NF-YC-RGL2-ABI5 cascades, which integrate GA and ABA signaling pathways to precisely regulate seed germination (Liu X. et al., 2016), have been dissected. FOA2 (F-box overexpressed/oppressed ABA signaling), and the transcription factors FUSCA3 and DAG1 (DOF AFFECTING GERMINATION 1) also regulate biosynthesis and signal transduction of GA and ABA during seed germination (Boccaccini et al., 2014a,b, 2016; Chiu et al., 2016a,b; He et al., 2016). Overall, diverse key genes regulate seed germination by mediating ABA and GA biosynthesis and/or signal transduction pathways (Table 1).
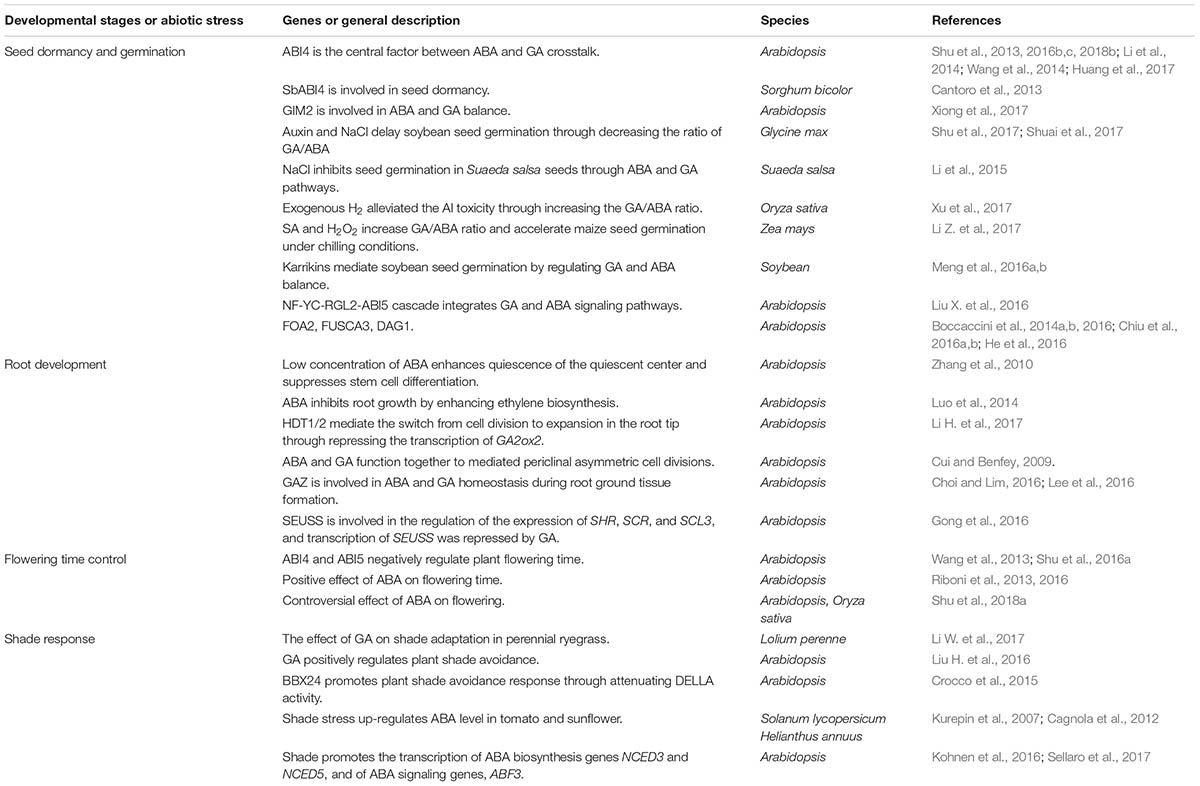
TABLE 1. General descriptions of recent reported factors roles in plant development and abiotic stress responses in regard to ABA and GA crosstalk network.
ABA and GA in the Regulation of Root Development
The root system is of great significance in both plant stress response and nutrient absorption. Numerous studies have demonstrated that auxins play important roles in the regulation of root growth, especially in the maintenance of the root stem cell niche (Liu et al., 2017; Du and Scheres, 2018). However, ABA and GA are also involved in the control of root development, although the detailed molecular mechanisms involved require further investigation.
A previous study had revealed that low concentrations of ABA enhanced quiescence of the quiescent center and suppressed stem cell differentiation in the primary root meristem niche of Arabidopsis (Zhang et al., 2010). Applications of high concentrations of exogenous ABA or the abiotic stress-induced accumulation of ABA inhibited Arabidopsis primary root growth, but the molecular mechanisms involved are not fully understood. An earlier study showed that ABA promoted the transcription of ICK1/KRP1, which encode a negative regulator of the cell cycle, so that ABA delays cell expansion and proliferation (Wang et al., 1998). Recently, the Gong lab showed that ABA inhibited root growth by enhancing ethylene biosynthesis (Luo et al., 2014). The ethylene biosynthesis inhibitor L-alpha-(2-aminoethoxyvinyl)-glycine reduced ABA-mediated inhibition of root growth. Further biochemical analysis revealed that CPK4 and CPK11, two ABA-activated calcium-dependent protein kinases, phosphorylate the C-terminus of ACS6, increasing the stability of this protein, and promote ethylene biosynthesis (Luo et al., 2014). The identification of this ABA-ethylene cascade represents a recent breakthrough in the regulation of root development mediated by ABA.
Previous studies had demonstrated that GA exhibited a positive effect on root growth in Arabidopsis (Ubeda-Tomas et al., 2008, 2009). A recent study revealed that HDT1/2 (histone deacetylases) mediated the switch from cell division to expansion in the root tip through repressing the transcription of GA2ox2, a GA-inactivator gene (Li H. et al., 2017). Further genetic analysis showed that upregulation of GA2ox2 in hdt1 and hdt2 background caused a decrease in GA concentration, which then resulted in an earlier switch from cell division to cell expansion of the transit-amplifying cells developing from the root stem cells (Li H. et al., 2017). Precise control of the timing and extent of asymmetric cell divisions is crucial for correct patterning. Previous studies had demonstrated that ABA and GA function together to mediate periclinal asymmetric cell divisions of the endodermis during ground tissue formation (Cui and Benfey, 2009). Recent studies revealed that the GAZ (GA- AND ABA-RESPONSIVE ZINC FINGER) gene is involved in the regulatory pathways, while transcription of GAZ is regulated by GA and ABA (Lee et al., 2016). Transgenic GAZ-overexpressed plants were sensitive to both ABA and GA during the middle cortex formation stage, whereas RNAi-GAZ lines displayed the opposite phenotype. Further transcriptional analysis showed that GAZ is also involved in ABA and GA homeostasis during root ground tissue formation (Choi and Lim, 2016; Lee et al., 2016).
In addition to GAZ, another key factor, SEUSS, is also involved in middle cortex formation. The seu mutants exhibited clearly reduced expression of SHR (SHORT-ROOT), SCR (SCARECROW), and SCL3 (SCARECROW-LIKE3), suggesting that SEUSS positively regulated the transcription of these genes (Gong et al., 2016). In addition, SEUSS transcription was repressed by GA and enhanced by the GA biosynthesis inhibitor, paclobutrazol, indicating that SUESS regulated middle cortex formation via the GA pathway (Gong et al., 2016) (Table 1). However, the antagonism between ABA and GA in the regulation of root initiation needs further exploration and dissection.
Flowering Time Control: the Other Battlefield Between ABA and GA
During the plant life cycle, the appropriate flowering time is a crucial and important checkpoint for growth and survival, especially under diverse environmental stress conditions. The positive effect of GA on plant floral transition has been extensively and intensively explored and documented (Ding et al., 2013; Hyun et al., 2016; Zhu et al., 2016; Brambilla et al., 2017; Conti, 2017; Gong et al., 2017; Sawettalake et al., 2017).
ABA is also involved in the regulation of flowering time (Wang et al., 2013; Shu et al., 2016a). However, the contribution of ABA to the control of flowering time is still controversial, as both positive and negative effects have been documented (Riboni et al., 2013, 2016; Wang et al., 2013; Shu et al., 2016a). With regard to the effect of ABA on floral transition, we have recently reviewed the literature and proposed a working model (Shu et al., 2018a). The positive and negative effects of ABA on floral transition may be associated with environmental cues, such as drought, salt and other abiotic stresses (Riboni et al., 2016; Shu et al., 2018a) (Table 1). However, the detailed mechanisms through which ABA and GA antagonistically mediate flowering need further investigation.
Shade Response: the New Battlefield Between ABA and GA?
Numerous investigations have demonstrated that ABA is involved in diverse abiotic/biotic stress response cascades, including drought, salt, low temperature and pathogens, with many research articles and reviews covering the effects of ABA on stress responses (Edel and Kudla, 2016; Skubacz et al., 2016; Lievens et al., 2017; Saradadevi et al., 2017). Although GA is primarily regarded as a hormone contributing to the control of plant growth and development, recent data showed that GA also plays roles in plant adaptation to stresses, such as dehydration stress (Qin et al., 2011; Plaza-Wuthrich et al., 2016; Urano et al., 2017). A negative regulator of GA signaling, SPINDLY, plays a negative role in drought stress tolerance by integrating GA and cytokinin crosstalk (Qin et al., 2011). Under early osmotic stress, differential levels of GA biosynthesis gene expression, DELLA-regulated transcription and RGA protein accumulation were reported in proliferating cells (Skirycz et al., 2011; Claeys et al., 2012). GA is also involved in plant shade and flooding stress responses (Bailey-Serres and Voesenek, 2010; Colebrook et al., 2014; Li W. et al., 2017). In this review, we focus mainly on the effects of ABA and GA on shade response.
Shade stress is expressed as reductions in both photosynthetically active radiation and the ratio between red and far-red (R/FR) light, resulting from sunlight passing through the leaves of neighboring plants under dense planting systems (Yang and Li, 2017; Yang et al., 2018). Auxin biosynthesis and signal transduction plays a key role with regard to plant shade response, and the phytochromes PHYA, PHYB, and the phytochrome-interacting factors (PIFs) are involved in these cascades (Casal, 2012, 2013; Yang et al., 2018). A recent study detected an effect of GA on shade adaptation in perennial ryegrass (Lolium perenne) (Li W. et al., 2017). The ryegrass shadow-1 mutant exhibited the dwarf and shade-insensitive phenotype, while transcriptome analysis revealed that the transcription of GA biosynthesis and response genes was down-regulated in shadow-1 plants, compared to the wild type (Li W. et al., 2017). This study highlighted the important roles of GA biosynthesis under shade conditions. In line with this, phenotypic analysis of GA-related mutants also suggested that GA positively regulated plant shade avoidance in Arabidopsis (Liu H. et al., 2016). Another study demonstrated that the transcriptional regulator BBX24 promoted the plant shade avoidance response through attenuating the activity of DELLA proteins, negative regulators of the GA signaling pathway (Crocco et al., 2015). The shade-response defect in bbx24 mutants was fully restored by exogenous GA application, a treatment which promotes DELLA degradation (Crocco et al., 2015). These investigations highlighted the important functions which GA play in the plant shade response (Table 1).
Although ABA is involved in diverse abiotic stress response pathways, the detailed roles of ABA in shade avoidance have not been elucidated to date. Earlier studies had shown that shade stress up-regulated ABA concentration in tomato and sunflower (Kurepin et al., 2007; Cagnola et al., 2012). Recent studies revealed that shade stress promoted the transcription of several ABA biosynthesis genes, including NCED3 and NCED5, and of the ABA signaling gene, abscisic acid-responsive element-binding factor 3 (ABF3) (Kohnen et al., 2016; Sellaro et al., 2017). However, a better understanding of the function of ABA in the regulation of shade response is needed.
Concluding Remarks
The antagonism between ABA and GA in the control of diverse aspects of plant development and abiotic stress response is an attractive target in the research field of plant molecular biology. Significant progress has been made in the model plant Arabidopsis to understanding the underlying mechanisms. However, several key questions remain to be answered.
Firstly, it is proposed that several key factors regulate the balance between ABA and GA, and subsequently achieve precise mediation of plant development and stress responses. Several transcription factors, including ABI4 and OsAP2-39, belong to this large family, which directly or indirectly controls the transcription pattern of ABA and GA biosynthesis genes (Yaish et al., 2010; Shu et al., 2013, 2016b). Identification and dissection of the modes-of-action of other, currently unknown transcription factors, which mediate ABA and GA antagonism, would be a major step forward in GA/ABA antagonism research, while identification of the target genes of these transcription factors in ABA and GA biosynthesis and/or signaling pathways would also be a most worthwhile project.
Secondly, in addition to transcriptional control, regulation at the post-transcription level also needs an increased focus, whereby the currently unknown transcription factors regulate ABA and GA antagonism through mediating the activity of some of the enzymes involved in ABA- and GA- related biosynthesis and signaling pathways. For instance, different types of protein modification, including protein ubiquitination, acetylation, methylation and phosphorylation, affect ABA/GA biosynthesis and signal transduction, contributing to the control of plant development and stress responses.
Thirdly, although GA has been shown to positively regulate plant shade response (Liu H. et al., 2016; Li W. et al., 2017), the detailed mechanisms involved, especially the relationship between GA and auxin, need further investigation. In addition, the roles of ABA in plant response to shade conditions are also not fully understood. Overall, these remaining scientific questions with regard to ABA and GA antagonism are worthy of further exploration.
Author Contributions
KS conceived and designed this paper and wrote the manuscript. KS, WZ, FC, and XL sorted and discussed those published papers. KS and WY analyzed the database.
Funding
The work in our lab was supported by the funding from the National Natural Science Foundation of China (31701064), National Key Research and Development Program of China (2017YFD0201306), Science and Technology Department of Sichuan Province (18SYXHZ0083), and China Postdoctoral Science Foundation funded project (2014M552377 and 2016T90868).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very thankful to the members in our lab for the critical comments on the manuscript. We sincerely apologize to some colleagues who have contributed to this field but were not cited because of space limitations.
References
Bailey-Serres, J., and Voesenek, L. A. (2010). Life in the balance: a signaling network controlling survival of flooding. Curr. Opin. Plant Biol. 13, 489–494. doi: 10.1016/j.pbi.2010.08.002
Boccaccini, A., Lorrai, R., Ruta, V., Frey, A., Mercey-Boutet, S., Marion-Poll, A., et al. (2016). The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 16:198. doi: 10.1186/s12870-016-0890-5
Boccaccini, A., Santopolo, S., Capauto, D., Lorrai, R., Minutello, E., Belcram, K., et al. (2014a). Independent and interactive effects of DOF affecting germination 1 (DAG1) and the Della proteins GA insensitive (GAI) and Repressor of ga1-3 (RGA) in embryo development and seed germination. BMC Plant Biol. 14:200. doi: 10.1186/s12870-014-0200-z
Boccaccini, A., Santopolo, S., Capauto, D., Lorrai, R., Minutello, E., Serino, G., et al. (2014b). The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 7, 1486–1489. doi: 10.1093/mp/ssu046
Brambilla, V., Gomez-Ariza, J., Cerise, M., and Fornara, F. (2017). The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals. Front. Plant Sci. 8:665. doi: 10.3389/fpls.2017.00665
Cagnola, J. I., Ploschuk, E., Benech-Arnold, T., Finlayson, S. A., and Casal, J. J. (2012). Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol. 160, 1110–1119. doi: 10.1104/pp.112.201921
Cantoro, R., Crocco, C. D., Benech-Arnold, R. L., and Rodriguez, M. V. (2013). In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 64, 5721–5735. doi: 10.1093/jxb/ert347
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Chiu, R. S., Pan, S., Zhao, R., and Gazzarrini, S. (2016a). ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant J. 88, 749–761. doi: 10.1111/tpj.13293
Chiu, R. S., Saleh, Y., and Gazzarrini, S. (2016b). Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signal. Behav. 11:e1247137. doi: 10.1080/15592324.2016.1247137
Choi, J. W., and Lim, J. (2016). Control of asymmetric cell divisions during root ground tissue maturation. Mol. Cells 39, 524–529. doi: 10.14348/molcells.2016.0105
Claeys, H., Skirycz, A., Maleux, K., and Inze, D. (2012). DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol. 159, 739–747. doi: 10.1104/pp.112.195032
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Bot. 217, 67–75. doi: 10.1242/jeb.089938
Conti, L. (2017). Hormonal control of the floral transition: can one catch them all? Dev. Biol. 430, 288–301. doi: 10.1016/j.ydbio.2017.03.024
Crocco, C. D., Locascio, A., Escudero, C. M., Alabadi, D., Blazquez, M. A., and Botto, J. F. (2015). The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 6:6202. doi: 10.1038/ncomms7202
Cui, H., and Benfey, P. N. (2009). Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 58, 1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x
Ding, L., Wang, Y., and Yu, H. (2013). Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 54, 595–608. doi: 10.1093/pcp/pct026
Du, Y., and Scheres, B. (2018). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. doi: 10.1093/jxb/erx223
Edel, K. H., and Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., and Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. doi: 10.1111/tpj.12670
Gong, X., Flores-Vergara, M. A., Hong, J. H., Chu, H. W., Lim, J., Franks, R. G., et al. (2016). SEUSS integrates gibberellin signaling with transcriptional inputs from the SHR-SCR-SCL3 module to regulate middle cortex formation in the Arabidopsis root. Plant Physiol. 170, 1675–1683. doi: 10.1104/pp.15.01501
Gong, X., Shen, L., Peng, Y. Z., Gan, Y., and Yu, H. (2017). DNA topoisomerase Ialpha affects the floral transition. Plant Physiol. 173, 642–654. doi: 10.1104/pp.16.01603
Hamayun, M., Hussain, A., Khan, S. A., Kim, H. Y., Khan, A. L., Waqas, M., et al. (2017). Gibberellins producing endophytic fungus Porostereum spadiceum AGH786 rescues growth of salt affected soybean. Front. Microbiol. 8:686. doi: 10.3389/fmicb.2017.00686
He, R., Yu, D., Li, X., Duan, G., Zhang, Y., Tang, D., et al. (2016). F-box gene FOA2 regulates GA- and ABA- mediated seed germination in Arabidopsis. Sci. China Life Sci. 59, 1192–1194. doi: 10.1007/s11427-016-0098-3
Huang, X., Zhang, X., Gong, Z., Yang, S., and Shi, Y. (2017). ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 89, 354–365. doi: 10.1111/tpj.13389
Hyun, Y., Richter, R., Vincent, C., Martinez-Gallegos, R., Porri, A., and Coupland, G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37, 254–266. doi: 10.1016/j.devcel.2016.04.001
Iwamoto, M., Kiyota, S., Hanada, A., Yamaguchi, S., and Takano, M. (2011). The multiple contributions of phytochromes to the control of internode elongation in rice. Plant Physiol. 157, 1187–1195. doi: 10.1104/pp.111.184861
Jan, A., Kitano, H., Matsumoto, H., and Komatsu, S. (2006). The rice OsGAE1 is a novel gibberellin-regulated gene and involved in rice growth. Plant Mol. Biol. 62, 439–452. doi: 10.1007/s11103-006-9030-1
Joseph, M. P., Papdi, C., Kozma-Bognar, L., Nagy, I., Lopez-Carbonell, M., Rigo, G., et al. (2014). The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 165, 1203–1220. doi: 10.1104/pp.113.234294
Kohnen, M. V., Schmid-Siegert, E., Trevisan, M., Petrolati, L. A., Senechal, F., Muller-Moule, P., et al. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28, 2889–2904. doi: 10.1105/tpc.16.00463
Kong, D. D., Ju, C. L., Parihar, A., Kim, S., Cho, D., and Kwak, J. M. (2015). Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 167, 1630–1642. doi: 10.1104/pp.114.251298
Kurepin, L. V., Emery, R. J., Pharis, R. P., and Reid, D. M. (2007). Uncoupling light quality from light irradiance effects in Helianthus annuus shoots: putative roles for plant hormones in leaf and internode growth. J. Exp. Bot. 58, 2145–2157. doi: 10.1093/jxb/erm068
Lee, K., Lee, H. G., Yoon, S., Kim, H. U., and Seo, P. J. (2015). The Arabidopsis MYB96 transcription factor is a positive regulator of ABSCISIC ACID-INSENSITIVE4 in the control of seed germination. Plant Physiol. 168, 677–689. doi: 10.1104/pp.15.00162
Lee, S. A., Jang, S., Yoon, E. K., Heo, J. O., Chang, K. S., Choi, J. W., et al. (2016). Interplay between ABA and GA modulates the timing of asymmetric cell divisions in the Arabidopsis root ground tissue. Mol. Plant 9, 870–884. doi: 10.1016/j.molp.2016.02.009
Li, H., Torres-Garcia, J., Latrasse, D., Benhamed, M., Schilderink, S., Zhou, W., et al. (2017). Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number. Plant Cell 29, 2183–2196. doi: 10.1105/tpc.17.00366
Li, J., Jiang, J., Qian, Q., Xu, Y., Zhang, C., Xiao, J., et al. (2011). Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23, 628–640. doi: 10.1105/tpc.110.081901
Li, P., Zhou, H., Shi, X. L., Yu, B., Zhou, Y., Chen, S. L., et al. (2014). The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 10:e1004213. doi: 10.1371/journal.pgen.1004213
Li, W., Katin-Grazzini, L., Gu, X. B., Wang, X. J., El-Tanbouly, R., Yer, H., et al. (2017). Transcriptome analysis reveals differential gene expression and a possible role of gibberellins in a shade-tolerant mutant of perennial ryegrass. Front. Plant Sci. 8:868. doi: 10.3389/Fpls.2017.00868
Li, W., Yamaguchi, S., Khan, M. A., An, P., Liu, X., and Tran, L. S. (2015). Roles of gibberellins and abscisic acid in regulating germination of Suaeda salsa dimorphic seeds under salt stress. Front. Plant Sci. 6:1235. doi: 10.3389/fpls.2015.01235
Li, Z., Xu, J., Gao, Y., Wang, C., Guo, G., Luo, Y., et al. (2017). The synergistic priming effect of exogenous salicylic acid and H2O2 on chilling tolerance enhancement during maize (Zea mays L.) seed germination. Front. Plant Sci. 8:1153. doi: 10.3389/fpls.2017.01153
Lievens, L., Pollier, J., Goossens, A., Beyaert, R., and Staal, J. (2017). Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 8:587. doi: 10.3389/Fpls.2017.00587
Lin, Q., Wu, F., Sheng, P., Zhang, Z., Zhang, X., Guo, X., et al. (2015). The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6:7981. doi: 10.1038/ncomms8981
Liu, H., Yang, C., and Li, L. (2016). Shade-induced stem elongation in rice seedlings: implication of tissue-specific phytohormone regulation. J. Integr. Plant Biol. 58, 614–617. doi: 10.1111/jipb.12468
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7:12768. doi: 10.1038/ncomms12768
Liu, J. L., Moore, S., Chen, C. L., and Lindsey, K. (2017). Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Mol. Plant 10, 1480–1496. doi: 10.1016/j.molp.2017.11.002
Luo, X., Chen, Z., Gao, J., and Gong, Z. (2014). Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 79, 44–55. doi: 10.1111/tpj.12534
Meng, Y., Chen, F., Shuai, H., Luo, X., Ding, J., Tang, S., et al. (2016a). Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Sci. Rep. 6:22073. doi: 10.1038/srep22073
Meng, Y., Shuai, H., Luo, X., Chen, F., Zhou, W., Yang, W., et al. (2016b). Karrikins: regulators involved in phytohormone signaling networks during seed germination and seedling development. Front. Plant Sci. 7:2021. doi: 10.3389/fpls.2016.02021
Nee, G., Xiang, Y., and Soppe, W. J. (2017). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. doi: 10.1016/j.pb1.2016.09.002
Novak, O., Napier, R., and Ljung, K. (2017). Zooming in on plant hormone analysis: tissue- and cell-specific approaches. Annu. Rev. Plant Biol. 68, 323–348. doi: 10.1146/annurev-arplant-042916-040812
Plaza-Wuthrich, S., Blosch, R., Rindisbacher, A., Cannarozzi, G., and Tadele, Z. (2016). Gibberellin deficiency confers both lodging and drought tolerance in small cereals. Front. Plant Sci. 7:643. doi: 10.3389/fpls.2016.00643
Qin, F., Kodaira, K. S., Maruyama, K., Mizoi, J., Tran, L. S., Fujita, Y., et al. (2011). SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. doi: 10.1104/pp.111.187302
Riboni, M., Galbiati, M., Tonelli, C., and Conti, L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiol. 162, 1706–1719. doi: 10.1104/pp.113.217729
Riboni, M., Test, A. R., Galbiati, M., Tonelli, C., and Conti, L. (2016). ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 67, 6309–6322. doi: 10.1093/jxb/erw384
Sano, N., Rajjou, L., North, H. M., Debeaujon, I., Marion-Poll, A., and Seo, M. (2016). Staying alive: molecular aspects of seed longevity. Plant Cell Physiol. 57, 660–674. doi: 10.1093/pcp/pcv186
Saradadevi, R., Palta, J. A., and Siddique, K. H. M. (2017). ABA-mediated stomatal response in regulating water use during the development of terminal drought in wheat. Front. Plant Sci. 8:1251. doi: 10.3389/Fpls.2017.01251
Sawettalake, N., Bunnag, S., Wang, Y., Shen, L., and Yu, H. (2017). DOAP1 promotes flowering in the orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 8:400. doi: 10.3389/fpls.2017.00400
Sellaro, R., Pacin, M., and Casal, J. J. (2017). Meta-analysis of the transcriptome reveals a core set of shade-avoidance genes in Arabidopsis. Photochem. Photobiol. 93, 692–702. doi: 10.1111/php.12729
Shan, X., Yan, J., and Xie, D. (2012). Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 15, 84–91. doi: 10.1016/j.pbi.2011.09.006
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, S., et al. (2016a). ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 67, 195–205. doi: 10.1093/jxb/erv459
Shu, K., Chen, Q., Wu, Y. R., Liu, R. J., Zhang, H. W., Wang, P. F., et al. (2016b). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016c). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Shu, K., Luo, X., Meng, Y., and Yang, W. (2018a). Toward a molecular understanding of abscisic acid actions in floral transition. Plant Cell Physiol. 59, 215–221. doi: 10.1093/pcp/pcy007
Shu, K., Meng, Y. J., Shuai, H. W., Liu, W. G., Du, J. B., Liu, J., et al. (2015). Dormancy and germination: How does the crop seed decide? Plant Biol. 17, 1104–1112. doi: 10.1111/plb.12356
Shu, K., Qi, Y., Chen, F., Meng, Y., Luo, X., Shuai, H., et al. (2017). Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front. Plant Sci. 8:1372. doi: 10.3389/fpls.2017.01372
Shu, K., Zhang, H. W., Wang, S. F., Chen, M. L., Wu, Y. R., Tang, S. Y., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Shu, K., Zhou, W., and Yang, W. (2018b). APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217, 977–983. doi: 10.1111/nph.14880
Shuai, H., Meng, Y., Luo, X., Chen, F., Zhou, W., Dai, Y., et al. (2017). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 7:12620. doi: 10.1038/s41598-017-13093-w
Skirycz, A., Claeys, H., De Bodt, S., Oikawa, A., Shinoda, S., Andriankaja, M., et al. (2011). Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23, 1876–1888. doi: 10.1105/tpc.111.084160
Skubacz, A., Daszkowska-Golec, A., and Szarejko, L. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/Fpls.2016.01884
Ubeda-Tomas, S., Federici, F., Casimiro, I., Beemster, G. T., Bhalerao, R., Swarup, R., et al. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194–1199. doi: 10.1016/j.cub.2009.06.023
Ubeda-Tomas, S., Swarup, R., Coates, J., Swarup, K., Laplaze, L., Beemster, G. T., et al. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10, 625–628. doi: 10.1038/ncb1726
Urano, K., Maruyama, K., Jikumaru, Y., Kamiya, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2017). Analysis of plant hormone profiles in response to moderate dehydration stress. Plant J. 90, 17–36. doi: 10.1111/tpj.13460
Wang, B., Wei, H., Xue, Z., and Zhang, W. H. (2017). Gibberellins regulate iron deficiency-response by influencing iron transport and translocation in rice seedlings (Oryza sativa). Ann. Bot. 119, 945–956. doi: 10.1093/aob/mcw250
Wang, H., Qi, Q., Schorr, P., Cutler, A. J., Crosby, W. L., and Fowke, L. C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. doi: 10.1046/j.1365-313X.1998.00231.x
Wang, Y., Chang, H. P., Hu, S., Lu, X. T., Yuan, C. Y., Zhang, C., et al. (2014). Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J. Exp. Bot. 65, 4159–4175. doi: 10.1093/jxb/eru190
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Waterworth, W. M., Bray, C. M., and West, C. E. (2015). The importance of safeguarding genome integrity in germination and seed longevity. J. Exp. Bot. 66, 3549–3558. doi: 10.1093/jxb/erv080
Xiong, W., Ye, T., Yao, X., Liu, X., Ma, S., Chen, X., et al. (2017). The dioxygenase GIM2 functions in seed germination by altering gibberellin production in Arabidopsis. J. Integr. Plant Biol. doi: 10.1111/jipb.12619 [Epub ahead of print].
Xu, D., Cao, H., Fang, W., Pan, J., Chen, J., Zhang, J., et al. (2017). Linking hydrogen-enhanced rice aluminum tolerance with the reestablishment of GA/ABA balance and miRNA-modulated gene expression: a case study on germination. Ecotoxicol. Environ. Saf. 145, 303–312. doi: 10.1016/j.ecoenv.2017.07.055
Yaish, M. W., El-kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y. M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. doi: 10.1371/journal.pgen.1001098
Yan, J., Zhao, C. Z., Zhou, J. P., Yang, Y., Wang, P. C., Zhu, X. H., et al. (2016). The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 12:e1006416. doi: 10.1371/journal.pgen.1006416
Yang, C., Xie, F., Jiang, Y., Li, Z., Huang, X., and Li, L. (2018). Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44, 29–41. doi: 10.1016/j.devcel.2017.11.017
Yang, C. W., and Li, L. (2017). Hormonal regulation in shade avoidance. Front. Plant Sci. 8:1527. doi: 10.3389/Fpls.2017.01527
Yang, L., Zhang, J., He, J., Qin, Y., Hua, D., Duan, Y., et al. (2014). ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 10:e1004791. doi: 10.1371/journal.pgen.1004791
Zhang, H., Han, W., De Smet, I., Talboys, P., Loya, R., Hassan, A., et al. (2010). ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64, 764–774. doi: 10.1111/j.1365-313X.2010.04367.x
Zhao, H. Y., Zhang, H. M., Cui, P., Ding, F., Wang, G. C., Li, R. J., et al. (2014). The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiol. 165, 1255–1268. doi: 10.1104/pp.114.239699
Zhu, Y., Liu, L., Shen, L., and Yu, H. (2016). NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2:16075. doi: 10.1038/nplants.2016.75
Keywords: seed dormancy, germination, ABA, GA, antagonism, abiotic stress
Citation: Shu K, Zhou W, Chen F, Luo X and Yang W (2018) Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 9:416. doi: 10.3389/fpls.2018.00416
Received: 25 January 2018; Accepted: 15 March 2018;
Published: 27 March 2018.
Edited by:
Paola Vittorioso, Sapienza Università di Roma, ItalyReviewed by:
Fabrizio Araniti, Università degli Studi Mediterranea di Reggio Calabria, ItalyKeisuke Nagai, Nagoya University, Japan
Copyright © 2018 Shu, Zhou, Chen, Luo and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Shu, a3NodUBzaWNhdS5lZHUuY24= Wenyu Yang, bXNzaXlhbmd3eUBzaWNhdS5lZHUuY24=
 Kai Shu
Kai Shu Wenguan Zhou
Wenguan Zhou Feng Chen
Feng Chen Xiaofeng Luo
Xiaofeng Luo