- 1MOE Key Laboratory of Bioinformatics, Center for Plant Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China
- 2Texas A&M AgriLife Research and Extension Center at Dallas, Texas A&M University, Dallas, TX, United States
The tomato resistance gene Tm-22 encodes a coiled coil-nucleotide binding site-leucine rich repeat type resistance protein and confers effective immune response against tobamoviruses by detecting the presence of viral movement proteins (MPs). In this study, we show that the Nicotiana benthamiana Heat shock protein 90-kD (Hsp90) interacts with Tm-22. Silencing of Hsp90 reduced Tm-22-mediated resistance to Tobacco mosaic virus (TMV) and the steady-state levels of Tm-22 protein. Further, Hsp90 associates with SGT1 in yeast and in plant cells. These results suggest that Hsp90-SGT1 complex takes part in Tm-22-mediated TMV resistance by functioning as chaperone to regulate Tm-22 stability.
Introduction
In the natural environment, pathogen microbes, such as viruses, bacteria, fungi, oomycetes, and nematodes, can cause disease in host plants. To counteract the pathogen attack, plants have evolved multilevel and sophisticated mechanisms to protect them from potential pathogen invasion. One of them is resistance (R) gene-mediated immunity (Dangl and Jones, 2001). R gene or its product recognizes the cognate pathogen avirulence protein directly or indirectly and activates powerful, specific resistance response. The activation of resistance pathways often culminate in a rapid hypersensitive response (HR) cell death at the pathogen infection sites. However, some R genes can induce resistance response without visible HR (Heath, 2000; Jones and Dangl, 2006). A well-known example is the Rx gene from potato, which mediates extreme resistance against Potato virus X (PVX) without any visible cell death at the initial infection sites (Bendahmane et al., 1999). Besides, tomato resistance gene Tm-22 can mediate extreme resistance against tobamoviruses (Zhang et al., 2013). Nevertheless, they still have the potential to induce local cell death response in some conditions (Hall, 1980; Bendahmane et al., 1999; Du et al., 2013; Zhang et al., 2013).
Three tomato genes Tm-1, Tm-2, and Tm-22 mediate resistance against tobamoviruses including Tomato mosaic virus (ToMV) and Tobacco mosaic virus (TMV). In contrast to Tm-1 and Tm-2, Tm-22 mediates much more durable resistance and has been applied in crop cultivation for several decades (Lanfermeijer et al., 2003). Tm-22 contains a coiled-coil (CC) domain, a nucleotide binding site (NBS) domain and a leu-rich repeat (LRR) domain. Tm-22 detects the presence of tobamovirus MPs (Weber and Pfitzner, 1998; Lanfermeijer et al., 2004) and functions on the plasma membrane (Chen et al., 2017). The Tyr-767 in Tm-22 LRR domain is essential for the recognition of the MP of ToMV strain B7 (Kobayashi et al., 2011), suggesting that Tm-22 recognizes viral MP through the LRR domain. In addition, Tm-22 requires all domains for its activity and PM localization (Chen et al., 2017). The N-terminus of ToMV MP is important for Tm-22 recognition (Weber et al., 2004; Chen et al., 2017). The expression of N-terminus (1-187aa) of viral MP is able to trigger Tm-22-dependent cell death (Weber et al., 2004; Chen et al., 2017), although the two amino acid substitutions (S238R and K244E) in the C-terminus of ToMV MP lead to the overcoming of Tm-22-mediated resistance (Weber et al., 1993). RuBisCO small subunit positively involves in Tm-22-mediated extreme resistance (Zhao et al., 2013). Type I J-domain Hsp40 proteins (called NbMIP1s) and co-chaperone SGT1 are also indispensable for Tm-22-mediated extreme resistance (Du et al., 2013; Zhao et al., 2013). Nevertheless, the molecular mechanism of Tm-22-mediated virus resistance is largely unknown.
Heat shock protein 90-kD (Hsp90) is a molecular chaperone required for the stability and activity of many proteins during a variety of cellular processes, such as protein maturation, complex assembly, signal transduction and genetic buffering. For examples, plant Hsp90 can facilitate the folding of mammalian glucocorticoid receptors in vitro (Stancato et al., 1996). Hsp90 associates with the 26S proteasome and is critical for ATP-dependent assembly and maintenance of the 26S proteasome (Imai et al., 2003). Hsp90 is also essential for plant disease resistance. Hsp90 modulates RPS2- and RPM1-mediated resistance in Arabidopsis (Hubert et al., 2003; Takahashi et al., 2003). Silencing Hsp90 using viral-induced gene silencing (VIGS) suppressed the plant resistance conferred by several R genes including N, Rx and Pto in N. benthamiana (Kanzaki et al., 2003; Lu et al., 2003; Liu et al., 2004). Suppression of TaHsp90.2 or TaHsp90.3 compromised the resistance against stripe rust fungus in common wheat (Wang et al., 2011). Knock down of Hsp90 compromised I-2 mediated cell death completely, suggesting that Hsp90 is essential for the tomato I-2-mediated resistance (de la Fuente van Bentem et al., 2005). In addition, Hsp90 is also involved in Mi-1-mediated pest immune response (Bhattarai et al., 2007). SGT1 interacts with Hsp90, and functions as a co-chaperone of Hsp90 to modulate the immune response by regulating R protein stability (Lu et al., 2003; Liu et al., 2004; Zhang et al., 2004).
In this study, we report that N. benthamiana Hsp90 associates with Tm-22 in vitro and in vivo, and plays an essential role in Tm-22-mediated TMV resistance by regulating its protein stability.
Materials and Methods
Plant Materials, Plasmids and Pathogens
Wild type N. benthamiana and transgenic Tm-22 N. benthamiana plants were described (Zhang et al., 2013). All N. benthamiana plants were grown in greenhouse at 23–25°C under a 16 h light/8 h dark cycle with 40–60% relative humidity and 40 umol m-2 s-1 white light illumination.
DNA fragments of Tm-22-nLUC, cLUC-NbHsp90, Tm-22-4 × myc and 3 × HA-NbHsp90 were generated by overlapping PCR, and then cloned into T-DNA vector pJG045, a pCAMBIA1300-based T-DNA vector (Zhao et al., 2013). PVX-based vector PVX-LIC was described (Zhao et al., 2016). The coding sequences of NbHsp90 (AY368904: nt1859-2103) was RT-PCR amplified and cloned into PVX-LIC vector for VIGS. All constructs were verified by DNA sequencing.
GFP-tagged TMV (TMV-GFP) was described (Liu et al., 2002a).
Yeast Two-Hybrid Assays
The full-length Tm-22, Tm-22-LRR were PCR amplified and cloned into the LexA DNA binding domain (BD) containing yeast vector pYL302 (Liu et al., 2002b) to generate the bait vectors BD-Tm-22 and BD-Tm-22-LRR. The full-length NbHsp90 cDNA was amplified by RT-PCR and cloned into the B42 activation domain (AD)-containing yeast vector pJG4-5 NbHsp90 to generate AD-NbHsp90. The yeast two-hybrid prey library containing tomato cDNAs (Liu et al., 2002b) was used to screen Tm-22-LRR-binding proteins. The yeast two-hybrid screening and interaction assay were performed as described (Liu et al., 2002b).
Luciferase Complementation Imaging (LCI) Assays
Luciferase complementation imaging assay was conducted as described (Chen et al., 2008; Du et al., 2013). All tested combinations were agroinfiltrated into leaves of 4 weeks old N. benthamiana. The leaves were collected 48 h post infiltration (hpi) and sprayed with luciferin (1 mM) followed by capturing the LUC image using a cooled CCD imaging apparatus (iXon, Andor Technology).
Protein Analysis and Co-Immunoprecipitation (Co-IP)
We used Agrobacterium-mediated transient expression approach for protein expression. The GV2260 strains containing the relevant expression vector were infiltrated into leaves of N. benthamiana. The leaves were collected at 48 hpi for protein extraction. Protein samples were extracted with Laemmli buffer (Laemmli, 1970) and subjected to electrophoresis on SDS–PAGE gel followed by western blot assays using anti-myc (Abmart) or anti-Hsp90 (Santa Cruz Biotechnology) primary antibodies and were detected using Pierce ECL western blotting substrate (Pierce).
For Co-IP assays, HA-NbHsp90 was co-expressed with Tm22-myc or cLUC-myc control in N. benthamiana. The infiltrated leaf tissues were collected 48 hpi and total protein extracts were subjected to IP procedure using anti-HA beads under agitation at 4°C for 2 h, then the beads were washed four times with ice-cold extraction buffer at 4°C (Du et al., 2013). The immunoprecipitates and input were extracted with Laemmli buffer and subjected to electrophoresis on SDS–PAGE gel followed by western blot assays using anti-myc or anti-HA antibody (Cell Signaling Technology) and detected using Pierce ECL western blotting substrate (Pierce).
Gene Expression Assays
RT-PCR and quantitative RT-PCR were conducted, respectively, as described (Liu et al., 2002a; Wang et al., 2013). NbActin mRNA was served as an internal control for normalization. Primers were designed with Primer3web1.
VIGS, Virus Inoculation and GFP Imaging
For VIGS assays, PVX: NbHsp90 or control plasmid was transformed into Agrobacterium tumefaciens strains GV2260 and then infiltrated into the leaves of 4 weeks old N. benthamiana plants. For TMV infection, TMV-GFP was agroinfiltrated into the plant leaves (Liu et al., 2002a). Each silencing experiment was repeated using at least five independent plants at least four times Pictures were photographed under white and UV light using a Canon 650D camera.
Results
Identification of NbHsp90 as Tm-22-Interacting Partner
Tm-22 LRR domain is reported to be involved in virus recognition (Kobayashi et al., 2011). To understand Tm-22 action, we conducted a yeast two-hybrid screen of a tomato cDNA library using Tm-22-LRR (aa: 444-961) as bait, and identified several host proteins interacted with Tm-22 (Liu et al., 2004; Du et al., 2013). In this screen, we identified SGT1 and NbMIP1s as partners interacting with Tm-22 (Liu et al., 2002b; Du et al., 2013). In addition, Hsp90 (AY368906) (Liu et al., 2004) was also identified to interact with Tm-22. Further, two N. benthamiana Hsp90 homologs (AY368904, AY368905) (Wang et al., 2011) were identified to share high identity with tomato Hsp90 (AY368906). It should be noted that two NbHsp90 homologs are almost identical to one another. Because N. benthamiana is an allotetraploid, we believe that these two NbHsp90 homologs are two alleles of same gene.
NbHsp90 Interacts With Tm-22 in Yeast
Further, we verified the interaction of NbHsp90 with Tm-22 using LexA based yeast two-hybrid system (Du et al., 2013). Both BD- and AD- fusion genes were driven by a galactose-inducible promoter. Yeasts transformed AD-NbHsp90 and BD-Tm-22 or BD-Tm-22-LRR grew on galactose medium lacking leucine, and became blue on medium containing X-gal and galactose/raffinose but not glucose (Figure 1). In contrast, control yeasts containing AD or BD alone did not grow on the medium lacking leucine or turn blue on X-gal medium (Figure 1). Therefore, both Tm-22 and Tm-22-LRR interact with NbHsp90 in yeast.
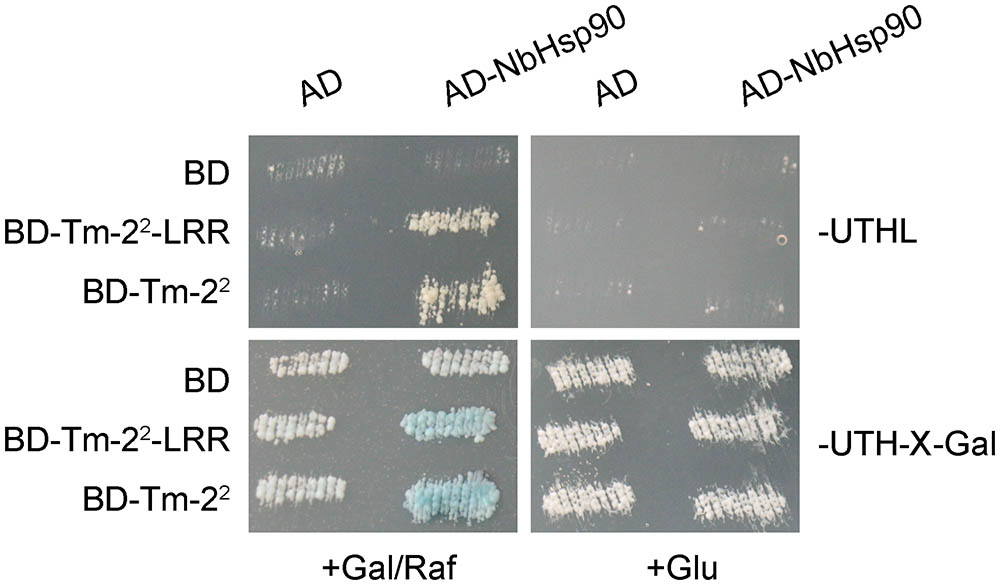
FIGURE 1. NbHsp90 Interacts with Tm-22 in Yeast. Yeast cells containing NLS-LexA BD-Tm-22 or BD-Tm-22-LRR baits transformed with AD-NbHsp90 grew on Leucine deficient medium (Leu-) and turned blue on X-gal medium containing galactose (Gal) and raffinose (Raf) but not on medium containing glucose (Glu) at 28°C for 4 days. Yeast cells transformed with either AD or BD empty vector alone were used as negative control.
NbHsp90 Interacts With Tm-22 in Plant Cells
To examine whether NbHsp90 interacts with Tm-22 in plant cells, we conducted Co-IP assay. The HA-tagged NbHsp90 (HA-NbHsp90) was co-expressed with myc-tagged Tm-22 (Tm-22-myc) or cLUC-myc (as a negative control) in N. benthamiana leaves. Leaf tissues were detached 48 hpi. Total protein was extracted and immunoprecipitated using anti-HA agarose, followed by western blot assays with anti-HA and anti-myc antibodies. We found that NbHsp90 co-immunoprecipitated with Tm-22, but not with cLUC-myc (Figure 2A).
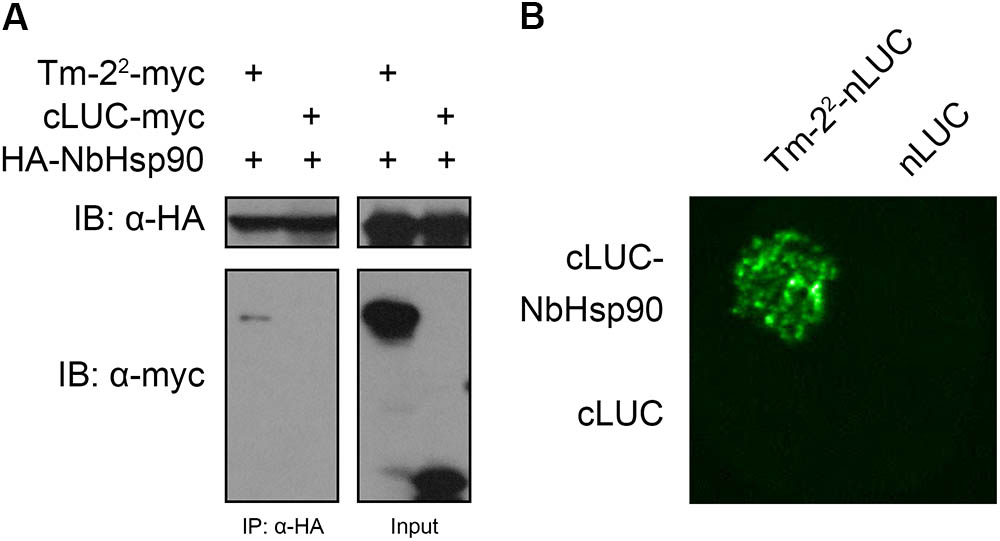
FIGURE 2. NbHsp90 Interacts with Tm-22 in Plant Cells. (A) NbHsp90 co-immunoprecipitated (Co-IP) with Tm-22. HA-NbHsp90 was transiently expressed with Tm-22-myc in Nicotiana benthamiana leaves by agroinfiltration. HA-NbHsp90 co-expressed with cLUC-myc was used as negative control. At 48 h post infiltration (hpi), total protein extracts were immunoprecipitated (IP) with anti-HA beads and the resulting precipitates were assessed by western blotting using anti-HA antibodies (upper panel, left) and anti-myc antibodies (lower panel, left). The presence of HA-NbHsp90, Tm-22-myc and cLUC-myc in the immunoprecipitates and cell lysates were also analyzed (right). IB, Immunoblot. (B) Firefly luciferase complementation imaging (LCI) assays for the in vivo interaction of NbHsp90 with Tm-22. cLUC-NbHsp90 was transiently co-expressed with Tm-22-nLUC or nLUC in N. benthamiana leaves followed by LCI assay.
We further validated the in vivo interaction of NbHsp90 with Tm-22 via LCI assay (Chen et al., 2008). N-terminus (nLUC) and C-terminus (cLUC) of the firefly luciferase can reconstitute active enzyme when they are fused, respectively, with two interacting proteins. To this end, we generated Tm-22-nLUC and cLUC-NbHsp90 and co-expressed them in N. benthamiana leaves. Positive signals were observed for the combination of cLUC-NbHsp90 with Tm-22-nLUC (Figure 2B). However, no signals were observed for the control combinations (cLUC-NbHsp90 plus nLUC, cLUC plus Tm-22-nLUC) (Figure 2B). These results, along with our Co-IP data, suggest that NbHsp90 interacts with Tm-22 in plant cells.
NbHsp90 Is Essential for Tm-22-Mediated TMV Resistance
To determine the role of NbHsp90 in N. benthamiana plants, we cloned a partial fragment of NbHsp90 (nt: 1859-2103) into PVX VIGS vector PVX-LIC (Zhao et al., 2016) to generate PVX-NbHsp90, and the PVX vector alone was used as negative control. Silencing of NbHsp90 induced developmental abnormalities including stopping growing and severely stunted (Supplementary Figure S1) (Liu et al., 2004), and quantitative RT-PCR data showed that the NbHsp90 mRNA level was greatly reduced in the PVX-NbHsp90 plants compared to the PVX vector plants (Figure 3D). Further, western blot assays using Hsp90-specific antibody showed that silencing of NbHsp90 greatly reduced Hsp90 protein level (Figures 3E,F).
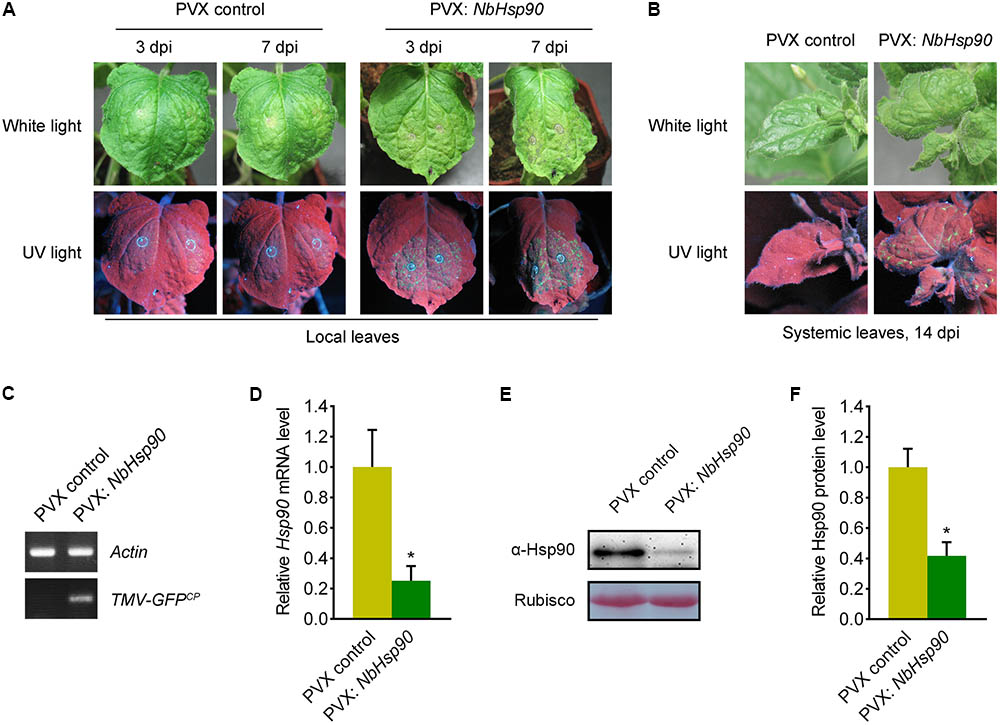
FIGURE 3. Silencing of NbHsp90 Compromised Tm-22-mediated Resistance to TMV. (A) Silencing of NbHsp90 caused the appearance of TMV-GFP infection foci and visible HR lesions (right) in the inoculated leaves of NbHsp90-silenced Tm-22-containing plants. PVX-infected Tm-22-containing plants were used as negative controls (left). (B) TMV-GFP spread into the systemic leaves of NbHsp90-silenced Tm-22-containing plants (right) but not the PVX control plants (left). Photos were taken at 14 days post TMV-GFP infection (dpi). (C) RT-PCR to confirm the presence of TMV-GFP in systemic leaves of NbHsp90-silenced Tm-22-containing plants (right) but not the PVX control plants (left). (D) Quantitative RT-PCR assays to confirm the reduction in NbHsp90 mRNA (means ± SEM, n = 3). ∗P < 0.05, Student’s t-test. NbActin mRNA levels were used as the internal control. (E,F) Western blot assays to confirm the reduction in NbHsp90 protein level (means ± SEM, n = 4). ∗P < 0.05, Student’s t-test. Equal loading of protein samples was validated by Ponceau Red staining of Rubisco subunit.
Then we investigated the role of NbHsp90 in Tm-22-mediated TMV resistance. To this end, we performed this experiment in transgenic Tm-22 N. benthamiana plants (thereafter called Tm-22 plants) that show effective resistance against TMV-GFP (Zhang et al., 2013). We agroinfiltrated the NbHsp90-silenced and PVX control non-silenced Tm-22 plants with Agrobacterium containing TMV-GFP plasmid (Liu et al., 2002a) and observed virus infection foci in inoculated leaves at 3 dpi (Figure 3A, left). Compared to the non-silenced Tm-22 plants, NbHsp90-silenced Tm-22 plants developed more TMV-GFP foci and subsequently developed obvious necrosis lesions at 7 dpi in inoculated leaves (Figure 3A, right). Furthermore, at 14 dpi TMV-GFP spread into the systemic leaves of NbHsp90-silenced Tm-22 plants but not that of non-silenced control Tm-22 plants (Figure 3B). RT-PCR showed that TMV RNA was readily detected in the systemic leaves of NbHsp90-silenced Tm-22 plants but not in the systemic leaves of non-silenced Tm-22 plants (Figure 3C). Taken together, these findings suggest that Tm-22-mediated TMV resistance requires NbHsp90.
NbHsp90 Is Essential for Stability of Tm-22 Protein
NbHsp90 is essential for Rx-mediated PVX resistance by regulating the protein level of Rx-4 × HA in N. benthamiana (Lu et al., 2003). RPM1 level is also reduced in Arabidopsis hsp90.2 mutant (Hubert et al., 2003). To investigate how NbHsp90 regulates Tm-22-mediated TMV resistance, we expressed Tm-22-myc in NbHsp90-silenced and non-silenced N. benthamiana plants to investigate the effect of NbHsp90 silencing on Tm-22 protein accumulation (Du et al., 2013). Western blot assays showed that NbHsp90-silenced plants accumulated less Tm-22 protein compared with non-silenced control plants (Figure 4A). However, quantitative RT-PCR assay indicated that NbHsp90 silencing had no significant effect on Tm-22 mRNA level (Figure 4B). Taken together, these findings indicate that NbHsp90 is indispensable for Tm-22 protein stability.
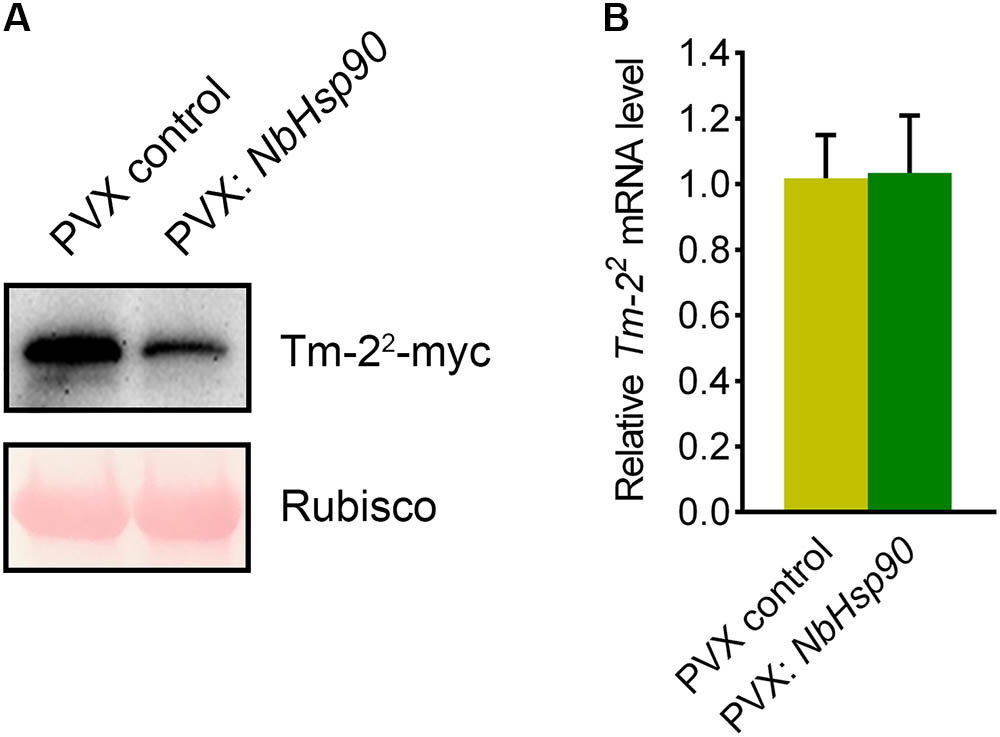
FIGURE 4. NbHsp90 is Required for Tm-22 Protein Stability. (A) Tm-22-myc was transiently agroinfiltrated into leaves of NbHsp90 silenced plants and control plants, respectively, proteins were extracted and analyzed by SDS–PAGE, followed by western blot using anti-myc antibody (upper panels). Equal loading of protein samples was validated by Ponceau Red staining of Rubisco subunit (lower panels). (B) Real-time RT-PCR assays showed that silencing of NbHsp90 did not affect Tm-22 mRNA level (means ± SEM, n = 5). NbActin mRNA levels were used as the internal control.
NbHsp90 Interacts With NbSGT1 in Yeast and in Plant Cells
We had reported that NbSGT1 interacts with Tm-22 and is essential for Tm-22-mediated TMV resistance by regulating Tm-22 protein stability (Du et al., 2013). To investigate whether Hsp90 regulates Tm-22 protein stability through NbHsp90-NbSGT1 chaperone complex, we first tested the interaction between NbHsp90 and NbSGT1 using yeast two-hybrid system. Yeast cells harboring both AD-NbHsp90 and BD-SGT1 grew on medium lacking leucine, and became blue on medium containing X-gal and galactose/raffinose but not glucose (Figure 5A). However, control yeasts containing AD or BD vector alone neither grew on the medium lacking leucine nor turned blue on X-gal medium (Figure 5A). NbHsp90 therefore interacts with NbSGT1 in yeast. Further, we performed LCI assays to investigate whether NbHsp90 interacts with NbSGT1 in plant cells. We found that cLUC-NbHsp90 interacts with NbSGT1-nLUC, but not with empty cLUC control (Figure 5B). These experiments show that NbHsp90 interacts with NbSGT1 in both yeast and plant.
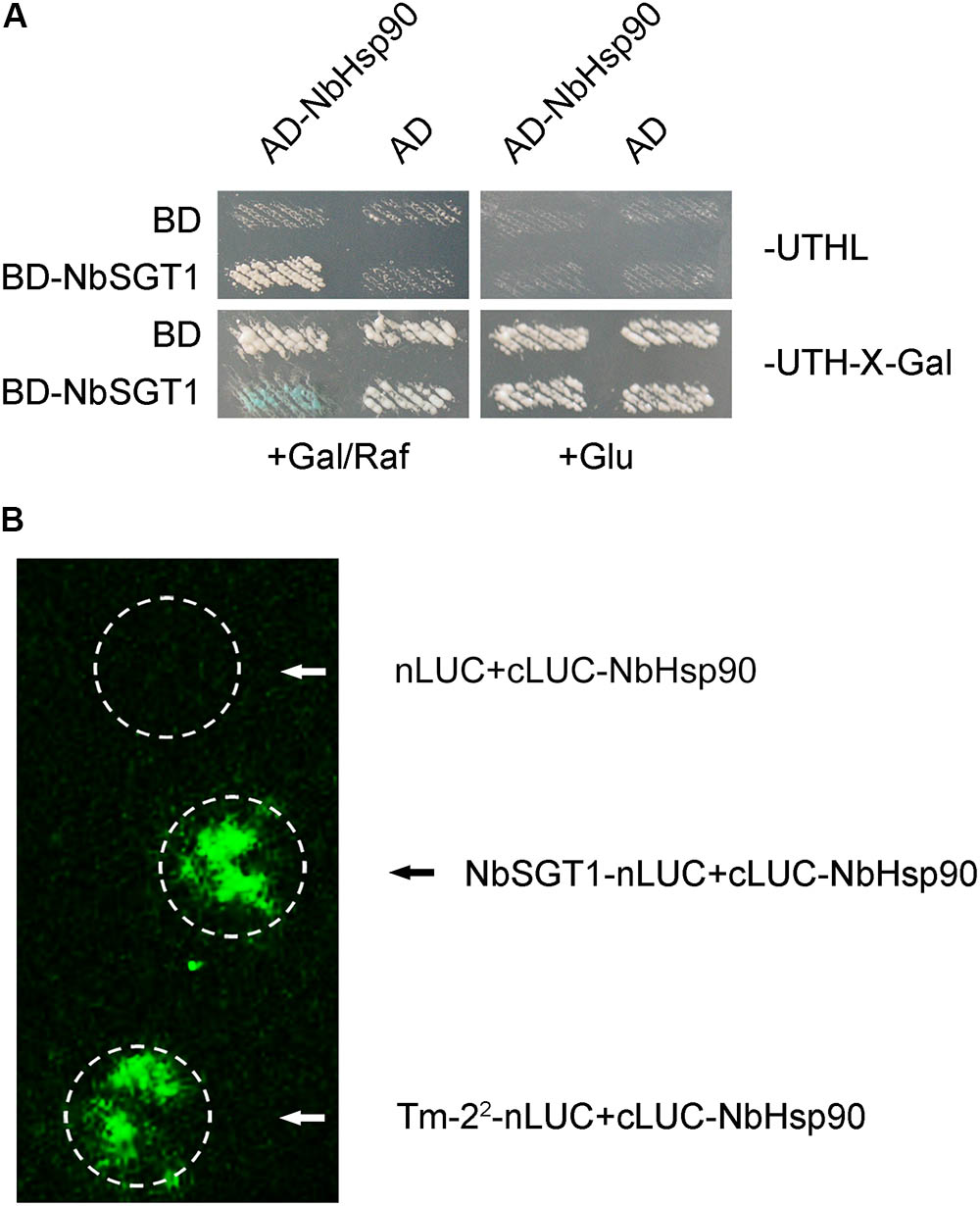
FIGURE 5. NbHsp90 Interacts with NbSGT1 in Yeast and in Plant Cells. (A) Yeast cells harboring NLS-LexA BD-SGT1 baits transformed with AD-NbHsp90 grew on Leu- deficient medium and turned blue on X-gal medium containing galactose (Gal) and raffinose (Raf) but not on medium containing glucose (Glu) at 28°C for 4 days. Yeast cells transformed with either AD or BD empty vector alone were used as negative control. (B) Firefly LCI assays for the in vivo interaction between NbHsp90 with SGT1. cLUC-NbHsp90 was transiently co-expressed with SGT1-nLUC or nLUC in N. benthamiana leaves followed by LCI assay.
Discussion
The Role of Hsp90 in Plant R Gene-Mediated Resistance Against Viruses
Hsp90 is a highly abundant and conserved cellular chaperone known to regulate various biological processes, and is reported to play crucial roles in plant disease resistance (Hubert et al., 2003; Lu et al., 2003; Takahashi et al., 2003; Liu et al., 2004). Using high throughput VIGS assay, Hsp90 was characterized to be a cofactor of Rx protein to stabilize its protein level (Lu et al., 2003). Association of Hsp90 with NBS-LRR proteins has been reported (Hubert et al., 2003; Liu et al., 2004; Zhu et al., 2017). Hsp90 associates with N protein directly to modulate the immune response to TMV (Liu et al., 2004). Hsp90, SGT1 and Rar1 form a complex and act as co-chaperones during virus disease resistance (Picard, 2002). In fact, the structurally conserved Hsp90-SGT1 complex (Seo et al., 2008; Shirasu, 2009) are functionally required for different NBS-LRR proteins’ function as immune modulator against various pathogens including bacteria (Takahashi et al., 2003; Zhang et al., 2004), fungi (Bieri et al., 2004; Thao et al., 2007), oomycetes (Michael Weaver et al., 2006; Bhaskar et al., 2008; Oh et al., 2014), nematodes (Bhattarai et al., 2007; Zhu et al., 2017). Here we reported that Hsp90 directly interacts with Tm-22, a CC-NBS-LRR type of resistance protein, confers robust immune response against tobamoviruses. Besides, we found that Hsp90 interacts with SGT1 in yeast and in plant cells. This finding is consistent with our earlier report that SGT1 participates in Tm-22-mediated resistance against TMV by regulating protein stability of Tm-22 through its interaction with Tm-22 (Du et al., 2013). Hsp90 and its co-chaperone SGT1 may facilitate the folding and maturation of R proteins. The misfolded R proteins can be eliminated by protein quality control machine. In such a scenario, knock down of Hsp90 or SGT1 decreases the amount of R protein and compromises R protein function (Lu et al., 2003; Liu et al., 2004; Zhang et al., 2004). Accordingly, silencing of Hsp90 suppressed Tm-22-mediated TMV resistance and reduced the stability of Tm-22 protein. Taken together, our findings further support that Hsp90-SGT1 chaperone mediates the stabilization and maturation of R proteins.
The Role of Hsp90-Related Co-chaperones in Plant-Virus Interaction
The DnaJ/Hsp40 works as a co-chaperone in Hsp90-Hsp70-Hsp40 complex, and can also form complex with Hsp90 during protein folding process (Verchot, 2012). DnaJ/Hsp40 proteins play dual roles in plant virus infection and host resistance. Via directly interaction with virus effectors, varied DnaJ/Hsp40 type proteins positively or negatively affect the replication and/or movement of several plant viruses including PVX, PVY, TSWV, and TMV (Soellick et al., 2000; Hofius et al., 2007; Cho et al., 2012; Shimizu et al., 2009). In addition, type I DnaJ/Hsp40 protein NbMIP1s also interact with Tm-22 and SGT1 and are required for Tm-22-mediated resistance by sustaining the protein stability (Du et al., 2013). Type-III DnaJ/Hsp40 plays a positive role in plant defense against Soybean mosaic virus in soybean (Liu and Whitham, 2013).
In this study, we found that Hsp90, like NbMIP1, is required for Tm-22-mediated resistance against TMV. However, no interaction between TMV MP and Hsp90 is detected (data not shown). In addition, NbHsp90 expression (at mRNA and protein levels) was not induced by TMV infection and Tm-22-mediate resistance (Supplementary Figure S2). Considered that NbMIP1 interacts with TMV MP, and NbMIP1s is induced by TMV infection and Tm-22-mediate resistance (Du et al., 2013), Hsp90 and NbMIP1s may exist in different cellular protein complexes during plant-virus interaction (Cintron and Toft, 2006). Indeed, Hsp90 and DnaJ/HsP40 proteins are not necessarily linked in their role as chaperones to facilitate the folding of diverse client proteins during different biological processes such as virus infection and plant resistance (Li et al., 2012; Verchot, 2012), and the chaperone machinery Hsp90-Sgt1 and Hsp90-Hsp40 is of different partnership for client recruitment and folding (Park and Seo, 2015).
Author Contributions
YL, JZ, LQ, and YD designed the experiments, analyzed the data, and prepared the manuscript. JZ, LQ, YD, XZ, and MH carried out the experiments. All authors contributed the revision of manuscript through the discussion.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31530059, 31421001, and 31470254).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Jianmin Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing LCI vectors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00411/full#supplementary-material
Footnote
References
Bendahmane, A., Kanyuka, K., and Baulcombe, D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–791. doi: 10.1105/tpc.11.5.781
Bhaskar, P. B., Raasch, J. A., Kramer, L. C., Neumann, P., Wielgus, S. M., Austin-Phillips, S., et al. (2008). Sgt1, but not Rar1, is essential for the RB-mediated broad-spectrum resistance to potato late blight. BMC Plant Biol. 8:8. doi: 10.1186/1471-2229-8-8
Bhattarai, K. K., Li, Q., Liu, Y., Dinesh-Kumar, S. P., and Kaloshian, I. (2007). The Mi-1-mediated pest resistance requires Hsp90 and Sgt1. Plant Physiol. 144, 312–323. doi: 10.1104/pp.107.097246
Bieri, S., Mauch, S., Shen, Q. H., Peart, J., Devoto, A., Casais, C., et al. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16, 3480–3495. doi: 10.1105/tpc.104.026682
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Chen, T., Liu, D., Niu, X., Wang, J., Qian, L., Han, L., et al. (2017). Antiviral resistance protein Tm-22 functions on the plasma membrane. Plant Physiol. 173, 2399–2410. doi: 10.1104/pp.16.01512
Cho, S. Y., Cho, W. K., Sohn, S. H., and Kim, K. H. (2012). Interaction of the host protein NbDnaJ with Potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 417, 451–456. doi: 10.1016/j.bbrc.2011.11.137
Cintron, N. S., and Toft, D. (2006). Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem. 281, 26235–26244. doi: 10.1074/jbc.M605417200
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
de la Fuente van Bentem, S., Vossen, J. H., Vries, K. J., Wees, S., Tameling, W. I., Dekker, H. L., et al. (2005). Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 43, 284–298. doi: 10.1111/j.1365-313X.2005.02450.x
Du, Y., Zhao, J., Chen, T., Liu, Q., Zhang, H., Wang, Y., et al. (2013). Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Pathog. 9:e1003659. doi: 10.1371/journal.ppat.1003659
Hall, T. J. (1980). Resistance at the TM-2 locus in the tomato to tomato mosaic virus. Euphytica 29, 189–197. doi: 10.1007/BF00037266
Heath, M. C. (2000). “Hypersensitive response-related death,” in Programmed Cell Death in Higher Plants, eds E. Lam, H. Fukuda, and J. Greenberg (Dordrecht: Springer), 70–90.
Hofius, D., Maier, A. T., Dietrich, C., Jungkunz, I., Börnke, F., Maiss, E., et al. (2007). Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 81, 11870–11880. doi: 10.1128/JVI.01525-07
Hubert, D. A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., et al. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22, 5679–5689. doi: 10.1093/emboj/cdg547
Imai, J., Maruya, M., Yashiroda, H., Yahara, I., and Tanaka, K. (2003). The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 22, 3557–3567. doi: 10.1093/emboj/cdg349
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kanzaki, H., Saitoh, H., Ito, A., Fujisawa, S., Kamoun, S., Katou, S., et al. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4, 383–391. doi: 10.1046/j.1364-3703.2003.00186.x
Kobayashi, M., Yamamoto-Katou, A., Katou, S., Hirai, K., Meshi, T., Ohashi, Y., et al. (2011). Identification of an amino acid residue required for differential recognition of a viral movement protein by the Tomato mosaic virus resistance gene Tm-22. J. Plant Physiol. 168, 1142–1145. doi: 10.1016/j.jplph.2011.01.002
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lanfermeijer, F. C., Dijkhuis, J., Sturre, M. J., de Haan, P., and Hille, J. (2003). Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 52, 1039–1051. doi: 10.1023/A:1025434519282
Lanfermeijer, F. C., Jiang, G., Ferwerda, M. A., Dijkhuis, J., de Haan, P., Yang, R., et al. (2004). The durable resistance gene Tm-22 from tomato confers resistance against ToMV in tobacco and preserves its viral specificity. Plant Sci. 167, 687–692. doi: 10.1016/j.plantsci.2004.04.027
Li, J., Soroka, J., and Buchner, J. (2012). The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823, 624–635. doi: 10.1016/j.bbamcr.2011.09.003
Liu, J. Z., and Whitham, S. A. (2013). Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J. 74, 110–121. doi: 10.1111/tpj.12108
Liu, Y., Burch-Smith, T., Schiff, M., Feng, S., and Dinesh-Kumar, S. P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. doi: 10.1074/jbc.M310029200
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002a). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Liu, Y., Schiff, M., Serino, G., Deng, X. W., and Dinesh-Kumar, S. P. (2002b). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14, 1483–1496. doi: 10.1105/tpc.002493
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546s
Michael Weaver, L., Swiderski, M. R., Li, Y., and Jones, J. D. (2006). The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant Cell 47, 829–840. doi: 10.1111/j.1365-313X.2006.02834.x
Oh, S. K., Kim, H., and Choi, D. (2014). Rpi-blb2-mediated late blight resistance in Nicotiana benthamiana requires SGT1 and salicylic acid-mediated signaling but not RAR1 or HSP90. FEBS Lett. 588, 1109–1115. doi: 10.1016/j.febslet.2014.02.028
Park, C. J., and Seo, Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323–333. doi: 10.5423/PPJ.RW.08.2015.0150
Picard, D. (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. 59, 1640–1648. doi: 10.1007/s00018-012-1177-0
Seo, Y. S., Lee, S. K., Song, M. Y., Suh, J. P., Hahn, T. R., Ronald, P., et al. (2008). The HSP90-SGT1-RAR1 molecular chaperone complex: a core modulator in plant immunity. J. Plant Biol. 51, 1–10. doi: 10.1007/BF03030734
Shimizu, T., Yoshii, A., Sakurai, K., Hamada, K., Yamaji, Y., Suzuki, M., et al. (2009). Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of tobacco mosaic virus. Arch. Virol. 154, 959–967. doi: 10.1007/s00705-009-0397-6
Shirasu, K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. doi: 10.1146/annurev.arplant.59.032607.092906
Soellick, T. R., Uhrig, J. F., Bucher, G. L., Kellmann, J. W., and Schreier, P. H. (2000). The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 2373–2378. doi: 10.1073/pnas.030548397
Stancato, L. F., Hutchison, K. A., Krishna, P., and Pratt, W. B. (1996). Animal and plant cell lysates share a conserved chaperone system that assembles the glucocorticoid receptor into a functional heterocomplex with hsp90. Biochemistry 35, 554–561. doi: 10.1021/bi9511649
Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 11777–11782. doi: 10.1073/pnas.2033934100
Thao, N. P., Chen, L., Nakashima, A., Hara, S. I., Umemura, K., Takahashi, A., et al. (2007). RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 19, 4035–4045. doi: 10.1105/tpc.107.055517
Verchot, J. (2012). Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 3:275. doi: 10.3389/fpls.2012.00275
Wang, G. F., Wei, X., Fan, R., Zhou, H., Wang, X., Yu, C., et al. (2011). Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 191, 418–431. doi: 10.1111/j.1469-8137.2011.03715.x
Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S., et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383–1399. doi: 10.1105/tpc.112.108993
Weber, H., Ohnesorge, S., Silber, M. V., and Pfitzner, A. J. P. (2004). The Tomato mosaic virus 30 kDa movement protein interacts differentially with the resistance genes Tm-2 and Tm-22. Arch. Virol. 149, 1499–1514. doi: 10.1007/s00705-004-0312-0
Weber, H., and Pfitzner, A. J. (1998). Tm-22 resistance in tomato requires recognition of the carboxy terminus of the movement protein of tomato mosaic virus. Mol. Plant Microbe Interact. 11, 498–503. doi: 10.1094/MPMI.1998.11.6.498
Weber, H., Schultze, S., and Pfitzner, A. J. (1993). Two amino acid substitutions in the tomato mosaic virus 30-kilodalton movement protein confer the ability to overcome the Tm-22 resistance gene in the tomato. J. Virol. 67, 6432–6438.
Zhang, H., Zhao, J., Liu, S., Zhang, D. P., and Liu, Y. (2013). Tm-22 confers different resistance responses against Tobacco mosaic virus dependent on its expression level. Mol. Plant 6, 971–974. doi: 10.1093/mp/sss153
Zhang, Y., Dorey, S., Swiderski, M., and Jones, J. D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224. doi: 10.1111/j.1365-313X.2004.02201.x
Zhao, J., Liu, Q., Hu, P., Jia, Q., Liu, N., Yin, K., et al. (2016). An efficient Potato virus X -based microRNA silencing in Nicotiana benthamiana. Sci. Rep. 6:20573. doi: 10.1038/srep20573
Zhao, J., Liu, Q., Zhang, H., Jia, Q., Hong, Y., and Liu, Y. (2013). The rubisco small subunit is involved in tobamovirus movement and Tm-22-mediated extreme resistance. Plant Physiol. 161, 374–383. doi: 10.1104/pp.112.209213
Keywords: Tobacco mosaic virus, plant-virus interaction, Hsp90, Tm-22, SGT1, NBS-LRRs, Nicotiana benthamiana, immunity
Citation: Qian L, Zhao J, Du Y, Zhao X, Han M and Liu Y (2018) Hsp90 Interacts With Tm-22 and Is Essential for Tm-22-Mediated Resistance to Tobacco mosaic virus. Front. Plant Sci. 9:411. doi: 10.3389/fpls.2018.00411
Received: 01 February 2018; Accepted: 14 March 2018;
Published: 10 April 2018.
Edited by:
Yi Li, Peking University, ChinaReviewed by:
Aiming Wang, Agriculture and Agri-Food Canada (AAFC), CanadaTao Zhou, China Agricultural University, China
Copyright © 2018 Qian, Zhao, Du, Zhao, Han and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Zhao, amlucGluZ3poYW9Ac2luYS5jbg== Yule Liu, eXVsZWxpdUBtYWlsLnRzaW5naHVhLmVkdS5jbg==
†These authors have contributed equally to this work.
 Lichao Qian
Lichao Qian Jinping Zhao
Jinping Zhao Yumei Du1
Yumei Du1 Yule Liu
Yule Liu