- 1Hybrid Rapeseed Research Center of Shaanxi Province, Yangling, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
Many vegetable and oilseed crops belong to Brassica species. The seed production of these crops is hampered often by abnormal floral organs, especially under the conditions of abiotic conditions. However, the molecular reasons for these abnormal floral organs remains poorly understood. Here, we report a novel pistil-like flower mutant of B. rapa. In the flower of this mutant, the four sepals are modified to one merged carpel that look like a ring in the sepal positions, enveloping some abnormal stamens and a pistil, and resulting in poor seed production. This novel mutant is named sepal-carpel modification (scm). DNA sequencing showed that the BrAP2a gene, the ortholog of Arabidopsis APETALA2 (AP2) that specifies sepal identity, losses the function of in scm mutant due to a 119-bp repeated sequence insertion that resulted in an early transcription termination. BrAP2b, the paralog of BrAP2a featured two single-nucleotide substitutions that cause a single amino acid substitution in the highly conserved acidic serine-rich transcriptional activation domain. Each of the two BrAP2 genes rescues the sepal defective phenotype of the ap2-5 mutant of Arabidopsis. Furthermore, the knockout mutation of the corresponding BnAP2 genes of oilseed rape (B. napus) by CRISPR/Cas9-mediated genome editing system resulted in scm-like phenotype. These results suggest that BrAP2 gene plays a key role in sepal modification. Our finding provides an insight into molecular mechanism underlying morphological modification of floral organs and is useful for genetic manipulation of flower modification and improvement of seed production of Brassica crops.
Introduction
Eudicot flowers are composed of four distinct floral organs, sepals, petals, stamens, and carpels, which are arranged in four concentric whorls from the outer to the inner layer of the flower. Genetic analysis of floral homeotic mutants in Arabidopsis thaliana and Antirrhinum majus led to the development of the classic ABC model of floral organ identity (Bowman et al., 1991; Coen and Meyerowitz, 1991). This model proposes three classes of floral homeotic genes that act alone or cooperate with each other to specify the four floral organs: class A specifies the sepals, A and B specify the petals, B and C specify the stamens, and C specifies the carpels. A mutation in the A-function gene generates a homeotic transition of sepals to carpels and petals to stamens. A B-functional gene mutation gives rise to a homeotic transformation of petals to sepals and stamens to carpels. A C-functional gene mutation leads to a homeotic translation of stamens to petals and carpels to sepals (Meyerowitz et al., 1991; Weigel and Meyerowitz, 1994).
In Arabidopsis, transcription factor APETALA1 (AP1) and APETALA2 (AP2) are A-functional genes, APETALA3 (AP3) and PISTILLATA (PI) are B-functional genes, while AGAMOUS (AG) is a C-functional gene (Yanofsky et al., 1990; Bowman et al., 1993; Jofuku et al., 1994; Krizek and Meyerowitz, 1996). In addition, it has been shown that SEPALLATA orthologs (SEP1–SEP4) are also required to specify floral organ identity (Pelaz et al., 2000). These genes are defined as E-function genes, and their discovery led to the proposal of the ABCE model (Jack, 2001). All of these well-characterized genes, except for AP2, are members of the MADS-box family encoding MIKC-type MADS-box transcription factors (Saedler et al., 2001; Theissen, 2001; Kaufmann et al., 2005). These encoded proteins constitute four different tetrameric complexes that specify the various corresponding floral organs. Accordingly, the AP1, AP1, SEP, and SEP tetramer determines the sepal development; AP1, AP3, PI, and SEP complex determines the petal development; AP3, PI, AG, and SEP determines the stamen development; while the AG, AG, SEP, and SEP tetramer determines the development of carpel (Yanofsky et al., 1990; Honma and Goto, 2001; Pelaz et al., 2001; Melzer and Theißen, 2009). This protein complexes-based model of floral organ specification is referred to as the quartet model, and is an improved successor of the ABCE theory (Theissen and Saedler, 2001). So far, the ABC model and its improved, more detailed versions have proven to be useful frameworks for interpreting floral development in a variety of species. However, there is little evidence to support the fact that the A-function genes specify the development of sepals and petals in non-Arabidopsis plant species.
In Arabidopsis, both A-functional genes AP1 and AP2 are required for sepal and petal development (Kunst et al., 1989; Irish and Sussex, 1990; Drews et al., 1991; Bowman et al., 1993; Gustafson-Brown et al., 1994). Indeed, Arabidopsis is the only species in which floral organ identity defects have been linked to A-functional genes (Davies et al., 1999; Litt, 2007; Causier et al., 2010). Interestingly, the SQUAMOSA (SQUA) gene, an AP1 ortholog in Antirrhinum majus, plays a key role in determining floral meristem identity, but does not affect the specification of sepals and petals in this species (Huijser et al., 1992). Meanwhile, AP2 was originally identified in Arabidopsis as a floral homeotic gene involved both in the A-functional roles of specifying organ identity and in restricted C-functional activities. Loss of AP2 function has previously been shown to cause a homeotic transition of sepals to carpels, and petals to stamens (Drews et al., 1991; Bowman et al., 1993; Causier et al., 2010). Further investigations revealed that the AP2 gene has numerous additional functions in the regulation of flowering time (Aukerman and Sakai, 2003), maintenance of the shoot apical meristem (Wurschum et al., 2006), and the determination of seed and fruit development (Maes et al., 2001; Jofuku et al., 2005; Ohto et al., 2005; Ripoll et al., 2011; Zhou et al., 2012). However, the exact role of the AP2 gene in flower development, especially in sepal and petal specification, has not been determined. Therefore, exploring of novel A-functional homeotic mutants in non-Arabidopsis species is currently critical to verify the functions of the AP1 and AP2 genes in sepal and petal specification, which could contribute new details to complement the classic ABC model of floral organ identity.
Many vegetable and oilseed crops belong to Brassica species. The seed production of these crops is hampered often by abnormal floral organs, especially under conditions of abiotic stress (Zhou et al., 2008; Huang et al., 2010). Similar to Arabidopsis, both B. rapa and B. napus are members of the Cruciferae family. B. napus is an allodiploid species derived from spontaneous hybridization between B. rapa and B. oleracea, and possesses the complete diploid chromosome sets of the parental genomes (Parkin et al., 1995; Snowdon et al., 2002). In our previous study, we recovered a novel pistil-like flower mutant with sepal-to-carpel modification (scm) in an F3 selfing line of an interspecific cross between B. napus and B. rapa, which prompted us to further explore the molecular mechanism underlying its phenotype. Here we report that the scm mutant was apetalous resulting in poor seed production. Further studies indicated that the mutations in the two AP2 orthologs in B. rapa (BrAP2 genes) genes of B. rapa (Song et al., 2013) were the fundamental causes of the defects.
Materials and Methods
Plant Materials
The scm and scm2 from the same F3 selfing line Nc116 were derived by an interspecific cross between B. napus and B. rapa. Both wild-type B. rapa (WTr) and B. napus (WTn) were used as controls. The AP2 weak mutant ap2-5 of Arabidopsis thaliana (stock name CS6239, purchased from the website1) with a mild sepal-to-carpel and petal-to-stamen phenotype was used as transgenic receptor material.
RNA Extraction and Gene Expression Assays
Total RNA was extracted from young inflorescence at the budding stage using RNAiso Reagent (TaKaRa, Dalian, China). The first strand cDNA was synthesized using the Prime Script RT-PCR kit (TaKaRa, Dalian, China). Primers specific for the AP1, AP2, AP3, PI, AG, and SEP3 genes were designed based on sequences retrieved from the Brassica database2 and the NCBI nucleotide and EST databases (details of primers are given in Supplementary Table 1). The expression of these genes was determined using semi-quantitative RT-PCR. The expression levels of the target genes were normalized to the internal control, the cytosolic 18S rRNA gene. The PCR products were separated on a 1% agarose gel, EtBr stained, purified using the Omega DNA Gel Extraction Kit (Omega Bio-Tek), and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, United States) for sequencing (Beijing Sunbiotech Co., Ltd.).
Generation of Transgenic Arabidopsis Lines of the Two B. rapa AP2 Genes
To produce GFP-tagged AP2 genes from B. rapa (Br) and Arabidopsis (At), the coding sequences (CDSs) without the stop codon of the BrAP2a, BrAP2b, and AtAP2 (control) were PCR-amplified using the following primer sets: BAP2-F+Sac I/BAP2-R2a+Xba I, BAP2-F+Sac I/BAP2-R2b+Xba I, and AtAP2-F+Kpn I/AtAP2-R+Xba I, respectively (Supplementary Table 1). The amplicons were checked by sequencing, and the full-length CDSs were then cloned into the Sac I and Xba I sites of Cam-35S-GFP-containing vectors, resulting in the C-terminal fusion to GFP.
The resultant Cam-35S-BrAP2a-GFP, Cam-35S-BrAP2b-GFP, and Cam-35S-AtAP2-GFP fusion constructs were transformed into the Agrobacterium tumefaciens strain GV3101 using electroporation and subsequently infiltrated into Arabidopsis ap2-5 and Col-0 plants are using the floral dip method (Clough and Bent, 1998). T1 seeds from the infiltrated plants were selected and plated on 1/2 MS medium containing 33 mg/L hygromycin B.
Western Blot Assay
To examine the expression of the AtAP2-GFP, BrAP2a-GFP, and BrAP2b-GFP genes in independent transgenic lines, 50 mg of young leaves were ground in liquid N2 and treated with 200 μL 1×SDS loading buffer without Bromophenol Blue (50 mM pH 6.8 Tris–HCl, 5% β-mercaptoethanol, 10% Glycerol, and 2% SDS), followed by further grinding. The homogenate was boiled for 5 min and centrifuged for 5 min at 10,000 g. The supernatant was collected, and its protein concentration was determined using a Bradford assay. Samples containing 50 μg of protein were subjected to 10% SDS–PAGE and transferred to a PVDF membrane (Roche, Mannheim, Germany). Immunoblot analysis was performed using an anti-GFP (1:2000) monoclonal antibody (Beijing TransGen Biotech Co., Ltd.). The chemiluminescent HRP substrate (Millipore) was then added, incubated for 5 min, and the blots were imaged using Image Lab Software (Bio-Rad, Hercules, CA, United States).
Generation of ap2 Quadruple Mutants of B. napus by CRISPR/Cas9-Mediated Genome Editing System
To construct the desired recombinant plasmids using CRISPR/Cas9-based system, the same target sequence of AP2 for the four cDNAs of B. napus was designed using the web tool CRISPR-P3. The target dsDNA molecules with sticky ends were generated via direct annealing of two oligonucleotides primers in which the sticky ends sequences match those of the Bsa I ends in the pHSN401 vector (Xing et al., 2014). The resultant pHSN401-AP2 constructs were transformed into the Agrobacterium tumefaciens strain GV3101:pSoup using electroporation, and subsequently transformed into B. napus by the previously described method (Bhalla and Singh, 2008). The genomic DNA extracted from T1 plants was used for PCR amplification with primer sets AP2-F250/AP2-R507, and the PCR products were cloned into pMD19-T Vector (Takara) for sequencing using M13-47 primer (Beijing Sunbiotech Co., Ltd.).
Results
Origin and Floral Identities of scm, a Novel Flower Mutant of B. rapa
During our breeding experiments, we recovered a novel pistil-like flower mutant in the F3 selfing line Nc116, which was generated from an interspecific cross between B. napus and B. rapa. Unlike the wild-type Brassica (Figures 1A,D,G,I,L1,L2, WT), the flowers of the mutant consisted of a very large pistil and lacked sepals, petals, or stamens (Figures 1B,E1). After 2 days of growth, the mutants showed a small pistil stretching out of the large, initial pistil (Figure 1E2). When the large pistil was torn, it appeared to be separated into three parts: the outer layer was a large pistil consisting of four fused carpels with green ovules inside; the intermediate layer only consisted of only 1–4 abnormal stamens with chimeric carpel-like homeotic identity; and the inner layer was a normal pistil (Figures 1H,J,M1,M2). Surprisingly, it appeared that the four whorls and four types floral organs (sepals, petals, stamens, and carpels) found in normal flowers had transformed into only three whorls containing just two types of organs (carpels, stamens, and carpels, respectively), with the petals being completely abolished. Clearly, the mutant had severe defects in both the sepal and petal whorls; the ring of four merged carpels developed at the four sepal positions representing a typical homeotic transition of sepals to carpels, and the complete loss of the four petals indicating a typical apetalous trait. Evidently, the mutant had typical sepal carpeloid and apetalous characteristics, which we therefore designated as scm. Due to the reduced number of stamens in the scm flower compared to wild-type Brassica flowers, we could not conclude whether scm had undergone a homeotic transition of petals to stamens.
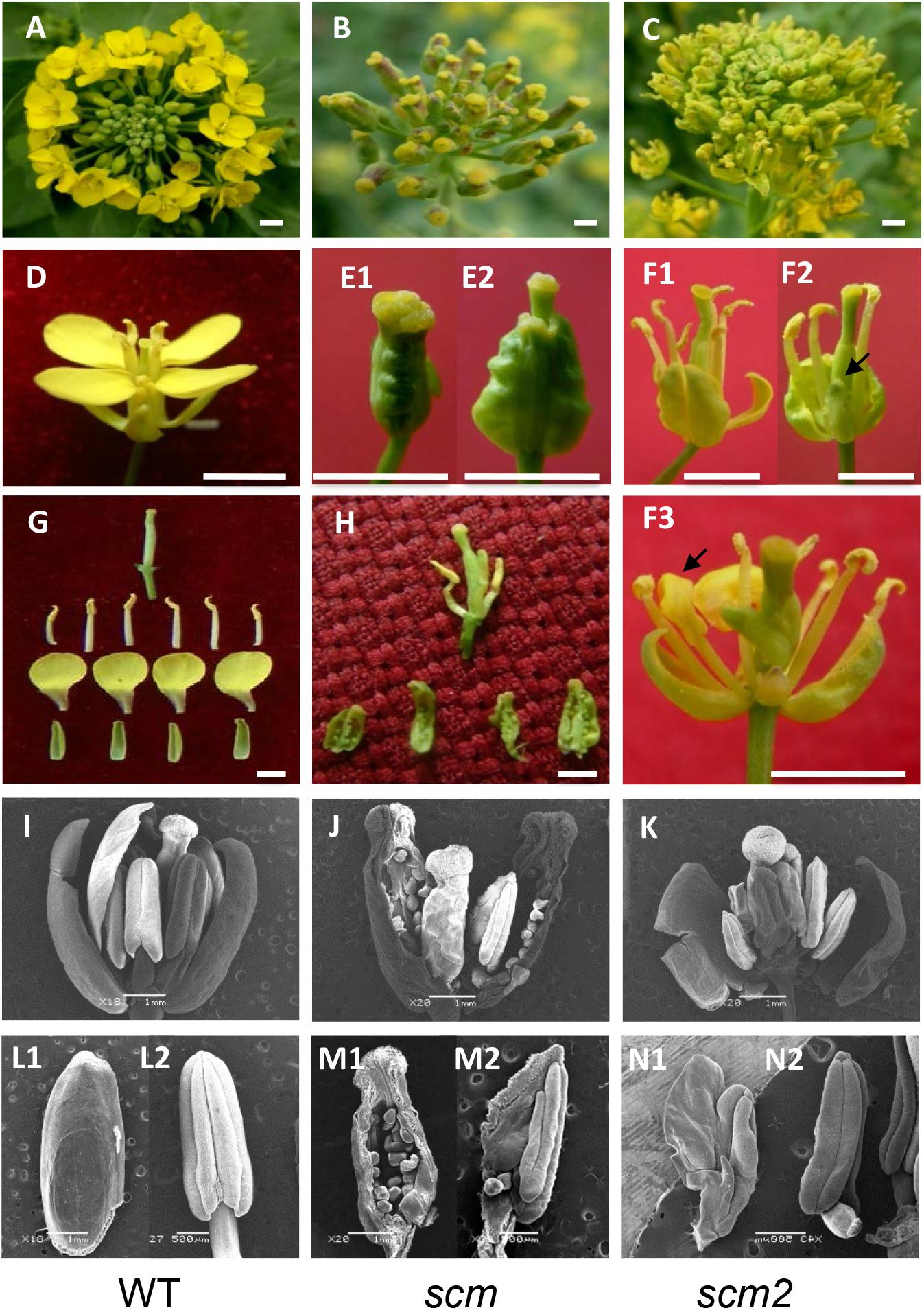
FIGURE 1. Characteristics the sepal carpeloid mutants of Brassica. (A–C): inflorescence; (D,E1,E2,F1–F3): flower; (G,H): floral organs; (I–K): SEM photos of flowers; (L1,L2,M1,M2,N1,N2): SEM photos of floral organs; (A,D,G,I,L1,L2): wild type Brassica (WT). (L1,L2) Are sepal (left) and stamen (right); (B,E1,E2,H,J,M1,M2): the typical sepal carpeloid mutant (scm). M1 is a sepal carpeloid organ, and M2 is an incomplete stamen to carpel organ with one ovule; (C,F1–F3,K,N1,N2): the incomplete sepal carpeloid mutant (scm2). Arrows indicate sepal carpel-like transition in F2 and petal stamen-like transition in F3. N1 is an incomplete petal to stamen organ, and N2 is a stamen of scm2. Bars in E1 and E2 are 5 mm; bars in I–K,L1,M1 are 1 mm; bars in L2,M2,N1,N2 are 500 μm; and bars in the others are 10 mm.
We also obtained another incomplete sepal carpeloid mutant, named scm2, in the same population. Compared to scm, this mutant lost the large pistil shape (Figures 1C,K), and only a few sepals developed into carpels (Figures 1F1,F2). Moreover, some flowers of scm2 were also apetalous, some of which containing few small abnormal petals, and some showing stamen-like petals (Figures 1F3,N1,N2). These results indicated that scm2 was an incomplete mutant with the homeotic transitions of sepals to carpels and petals to stamens.
Expression of ABCE Class of Genes in the scm Mutant
According to the classical ABC model, floral organ identity genes, especially those of class A and B, specify sepal and petal development. Mutations in the A class gene AP2 in Arabidopsis lead to homeotic transitions of sepals to carpels and petals to stamens (Kunst et al., 1989; Coen and Meyerowitz, 1991). Consistent with the previous findings, our mutants displayed a similar homeotic transition. This prompted us to examine the AP2 gene expression pattern, as well as the expression of the other ABCE classes of genes, in scm and in wild-type B. rapa (WTr) and B. napus (WTn).
Thus, the expression of AP1, AP2, AP3, PI, AG, and SEP3 genes in these samples was studied by semi-quantitative RT-PCR. When AP1-specific primers were applied, all the three samples formed two identical PCR amplicons (Figure 2, top left). By contrast, when BrAP2-specific primers were used, both WTr and WTn each produced one clear band for amplicons of identical size, whereas scm formed two bands (Figure 2, top right). The length of one of the bands was similar to both controls, while the other band was slightly larger than its counterpart. This larger fragment was named SCM-a, and the smaller fragment was defined as SCM-b. Using AP3 primers, we detected fragments of the same size in the WTr and scm samples, but an additional larger band appeared in the gel of the WTn sample (Figure 2, second from the top left). Meanwhile, the amplification with PI, AG, and SEP3 primers resulted in the production of identical amplicons in all the three samples, with no significant differences (Figure 2). These data indicate that a Brassica BrAP2 mutation, like the sepal carpeloid AP2-gene mutant in Arabidopsis, is probably relevant to the scm mutant. Moreover, the results from the AP3 gene amplification indicated that the genetic background of scm was very similar to that of its B. rapa parent.
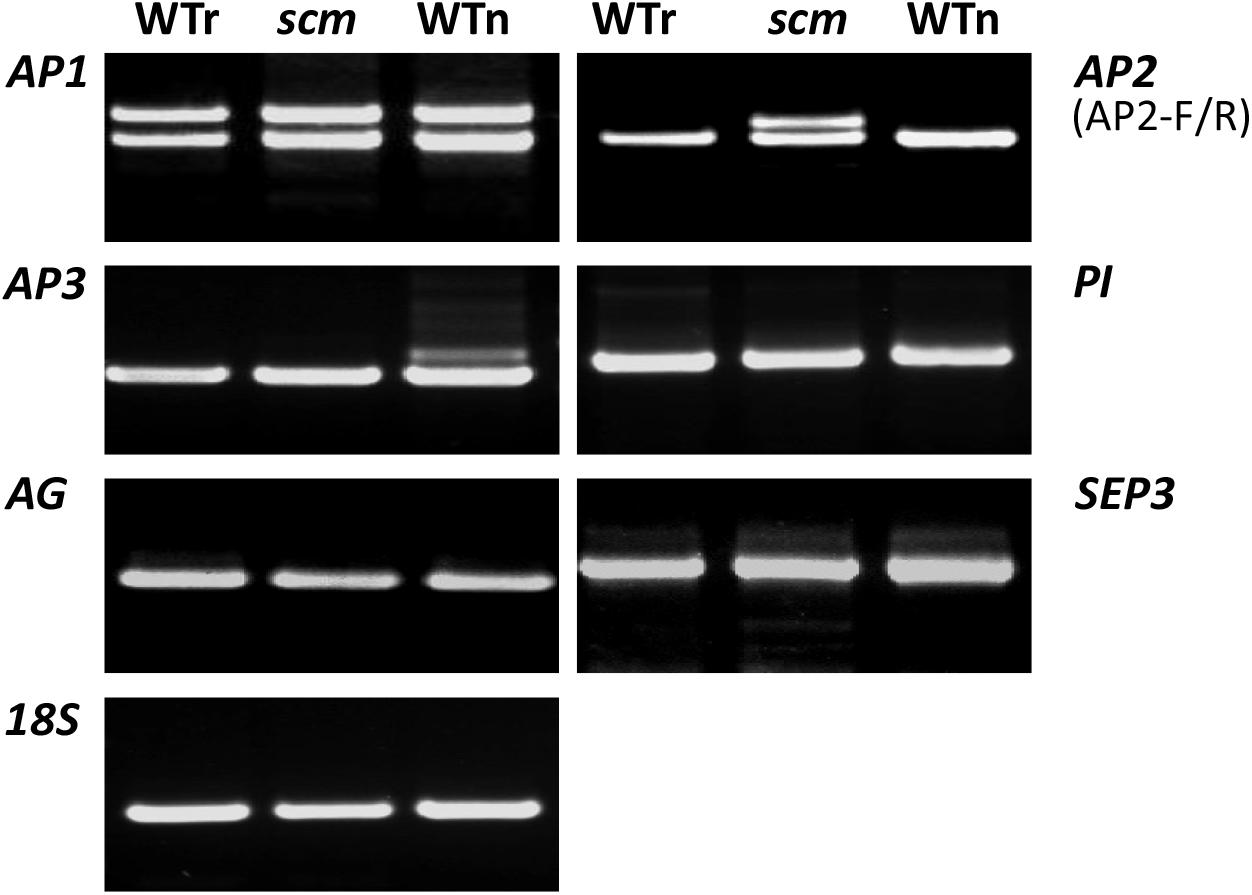
FIGURE 2. Expression of ABCE classic genes in the scm mutant and wild-type Brassica. WTr, wild-type B. rapa; WTn, wild-type B. napus; scm, sepal carpeloid mutant of Brassica.
The Two AP2 Orthologs in B. rapa Are Mutated
To explore the AP2 orthologs in B. rapa, a BLAST search was performed in the Brassica database, and two genes, Bra011741 and Bra017809, were identified. Bra011741 is located on chromosome A01 between base pairs 00793728 and 00795868, while Bra017809 is found on the chromosome A03 between base pairs 30612490 and 30614651. We also detected two AP2 orthologs in B. oleracea, Bol028934 and Bol018627, which are located on chromosomes C01 and C06, respectively (Supplementary Figure 1). Our results showed that B. rapa has two BrAP2 genes, whereas B. napus might have four BnAP2 genes, two of which from B. rapa and two from B. oleracea (Parkin et al., 1995; Snowdon et al., 2002).
According to the BrAP2 sequences from B. rapa and B. oleracea, primers spanning the full length cDNA of the two BrAP2 genes were designed: BrAP2-F and BrAP2-R2a target to the Bra011741 and Bol028394 genes; and BrAP2-F and BrAP2-R2b specific for the Bra017809 and Bol018627 genes. PCR products obtained from WTr samples using AP2-F/R2a primers were 1302-bp in length, and were named BrAP2a, sharing 99.4% nucleotide identity and 99.1% amino acid identity with Bra011741. Using the same AP2-F/R2a primer pair, the amplified genes from scm were 100% identical to BrAP2a except for a 119-bp insertion, which was a repeat of the sequence from nucleotides 485–603 (Figure 3A and Supplementary Figure 2).
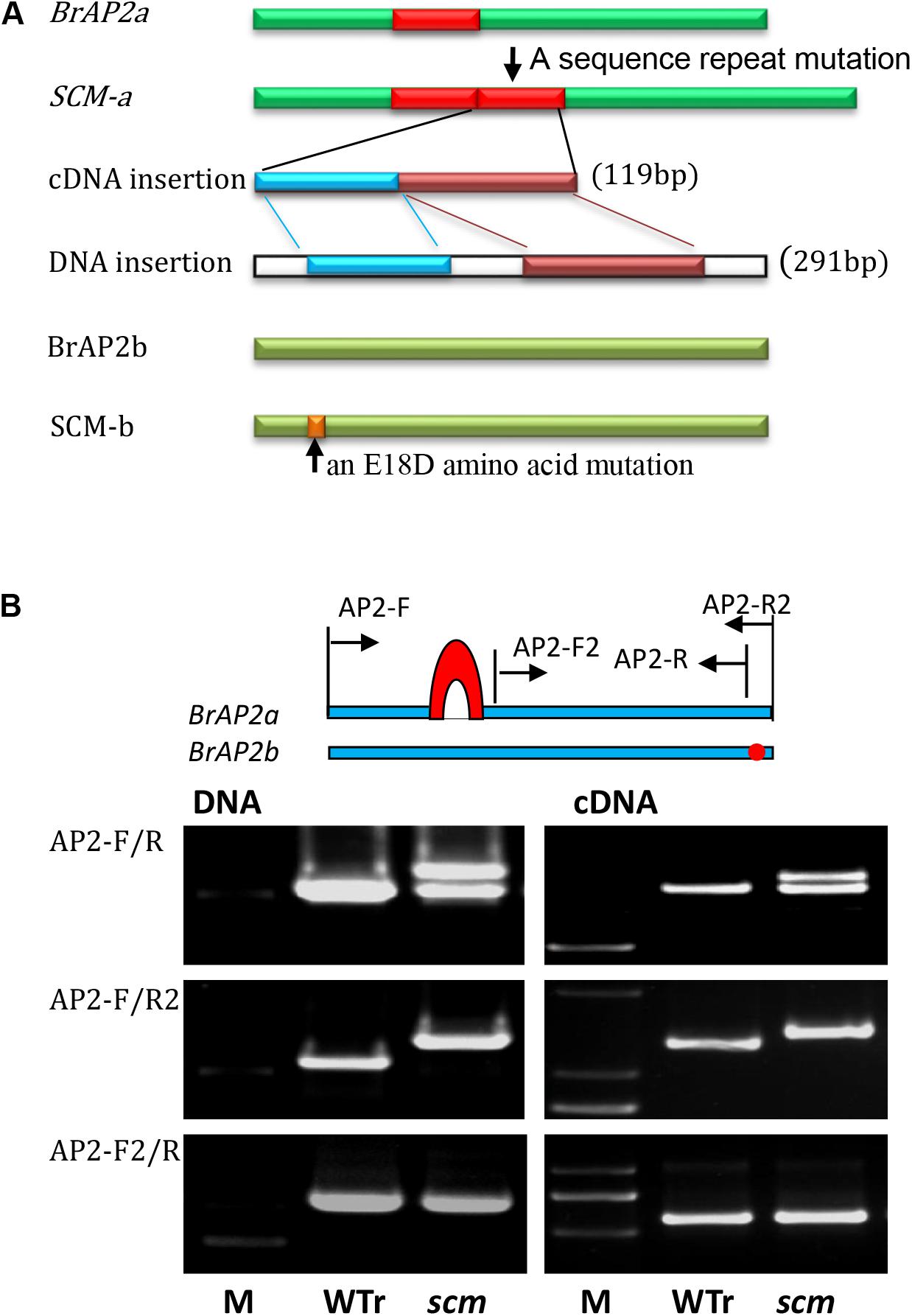
FIGURE 3. Mutations of the two AP2 orthologs in the scm of B. rapa. (A) Schematic of the two AP2 gene mutations in scm. The BrAP2a cDNA from scm (SCM-a) had a 119-bp sequence repeat mutation, which was derived from the transcription of a 291-bp repeat in genomic DNA that consisted of nucleotides 648–938 repeated after nucleotide 938. BrAP2b cDNA of scm (SCM-b) had an A to C single nucleotide mutation that transformed the 18th glutamic acid into aspartic acid (E18D). (B) A schematic of the location of the designed primers. The relative locations of the AP2 primers in the scm and wild-type B. rapa (WTr) sequences are shown (upper part). PCR amplification of genome DNA and cDNA of WTr and the scm using primer pairs of AP2-F/R, AP2-F/R2, and AP2-F2/R (lower part).
The amplicons obtained from WTr samples using the BrAP2-F/R2b primer set were identified as a 1299-bp fragment named BrAP2b. This gene was 100% identical to Bra017809. The corresponding sequences from scm were 100% identical to BrAP2b except for two single-nucleotide substitutions, among which only one A to C mutation, translating into the alteration of the 18th glutamic acid of the corresponding protein into aspartic acid, was observed (Figure 3A and Supplementary Figure 3 E18D). Among all the tested AP2 sequences obtained from scm, we could not detect any AP2 genes derived from B. oleracea, thus confirming that the genetic background of scm was very similar to its B. rapa parent.
The amplification of genomic DNA using the AP2-F/R2a primer set revealed that the BrAP2a gene of scm had an additional 291-bp fragment, which was a repeat of BrAP2a nucleotides 648–938, inserted after nucleotide 938. The above-mentioned 119-bp cDNA insertion was derived from transcription of this 291-bp DNA repeat sequence (Figure 3A and Supplementary Figure 2). Further sequence analysis indicated that this insertion led to an early translation termination of the BrAP2a mRNA.
We then designed additional specific primers to further confirm the above results. Using the AP2-F/R primer set, both DNA and cDNA of scm displayed an additional band compared to WTr (Figure 3B). When the BrAP2a specific primer set AP2-F/R2a was applied, both DNA and cDNA of scm produced one band that was larger than that of WTr. Using a mutual primer set (AP2-F2/R) for both of the two AP2 genes, both DNA and cDNA of scm formed one amplicon, the band for which was the same as that of the WTr-derived product. These results showed that the BrAP2a gene of scm underwent an insertion mutation that led to the loss of function of this gene. Altogether, these results suggested that mutations in BrAP2a and BrAP2b are most likely associated with the sepal carpeloid mutant scm. However, it was still unclear whether the loss of function of the two AP2 genes was the key cause of the observed scm phenotype.
The Two AP2 Genes of B. rapa Can Both Rescue the ap2-5 Mutant of Arabidopsis
The loss of function of AP2 in Arabidopsis has been previously demonstrated to cause a homeotic transition of sepals to carpels and petals to stamens (Kunst et al., 1989). Moreover, a weak AP2-defective Arabidopsis mutant ap2-5 shows a few sepal-to-carpel and petal-to-stamen homeotic transitions (Kunst et al., 1989). Both AP2 genes in B. rapa, BrAP2a and BrAP2b, shared high similarity with the Arabidopsis AP2 gene in their protein coding sequences (Supplementary Figure 3). To clarify whether BrAP2a and/or BrAP2b have biologically significant roles in floral organ specification, we transformed the two B. rapa AP2 genes and Arabidopsis AP2 (AtAP2) gene (control) separately into the wild-type and ap2-5 mutant Arabidopsis.
All the three AP2 genes were fused to green fluorescent protein (GFP) under the control of constitutive 35S promoter and transferred into Arabidopsis Col-0 to generate p35S::AtAP2/Col, p35S::BrAP2a/Col, and p35S::BrAP2b/Col over-expressing transgenic lines. These three transgenic lines displayed normal flower phenotypes (Figure 4A), suggesting that over-expression of the AP2 gene does not affect floral organ development.

FIGURE 4. Wild type and an ap2 weak mutant of Arabidopsis transformed with AP2 genes from B. rapa. (A) Wild-type Arabidopsis (Col-0) transformed with AtAP2 (control) and two AP2 genes of B. rapa. Their over-expressed lines p35S:AtAP2/Col, p35S::BrAP2a/Col, and p35S::BrAP2b/Col had normal flowers similar to the wild-type flower. (B) The AP2 mutant of Arabidopsis (ap2-5), which has a mild sepal-to-carpel and petal-to-stamen phenotype, transformed with AtAP2 gene and two B. rapa AP2 genes. All three transgenic plants, p35S:AtAP2/ap2-5, p35S::BrAP2a/ap2-5, and p35S::BrAP2a/ap2-5, rescued the ap2-5 mutant defect and developed normal sepals and petals. (C) The knockdown lines, p35S::AtAP2/ap2-5 KD, p35S::BrAP2a/ap2-5 KD, and p35S::BrAP2b/ap2-5 KD, from three transgenic group of p35S:AtAP2/ap2-5, p35S::BrAP2a/ap2-5, and p35S::BrAP2a/ap2-5. (D) semi-quantitative RT-PCR and Western blot assays to validate the transgenic lines. Bars, 500 μm.
When the three AP2 fusion constructs were transformed into the ap2-5 mutant of Arabidopsis, the transgenic lines p35S::AtAP2/ap2-5, p35S::BrAP2a/ap2-5, and p35S::BrAP2b/ap2-5 displayed normal development of the sepals and petals (Figure 4B), and thus the AP2 defect was rescued. Moreover, we recovered some strong sepal carpeloid mutants not only in the three knockdown transgenic groups: p35S::AtAP2/ap2-5 KD, p35S::BrAP2a/ap2-5 KD, and p35S::BrAP2b/ap2-5 KD (Figure 4C), but also in the three over-expressing groups caused by a cosuppression effect (data not shown). These mutants had some common characteristics: the sepals were transformed into carpels, the petals were absent, and the number of stamens was reduced to 1–4, but the pistil showed normal development. These lines displayed a strong AP2-knockdown phenotype, which is most likely also observed in scm. We further carried out RT-PCR and Western blot assays on these transgenic lines. Our RT-PCR results confirmed that the three AP2 rescued lines exhibited high expression whereas the three knockdown phenotypes showed low expression level of the AP2 gene. Interestingly, strong protein bands were present in the Western blot analyses of all the three AP2 rescued lines but could not be detected for the three knock-down lines (Figure 4D). Therefore, we inferred that the exogenous AP2 gene could interfere with the expression of endogenous AtAP2 gene through RNAi effect. These results confirmed that the AP2 gene is involved in the regulation of sepal and petal development, and the knockdown of these genes could affect sepal and petal development to generate sepal carpeloid and apetalous mutants.
The ap2 Quadruple Mutants of B. napus Produced by CRISPR/Cas9 System Resulting in Typical Sepal Carpeloid Phenotype
To further confirm that the AP2 genes are involved in regulating sepal and petal development, a CRISPR/Cas9-mediated genome editing experiment was performed to test whether the loss of AP2 function in B. napus could lead to a similar sepal carpeloid phenotype as in scm. We designed and constructed CRISPR/Cas9 nuclease system to cut at four identical target sites in the conserved sequences of AP2 (Figure 5A and Supplementary Figure 4A).

FIGURE 5. ap2 quadruple mutant in B. napus generated by CRISPR/Cas9 genome editing system. (A) The designed target sequence used for CRISPR/Cas9. The PAM sequence is indicated in red. (B) a1–a3: wild-type Brassica (WT); b1–b3: a strong ap2 quadruple mutant; c1–c4: a weak ap2 quadruple mutant. a1, b1, c1: inflorescence; a2, b2, c2: flower; a3, b3, c3: floral organs; c4: enlargement of the floral organ shown in the red box in c3. The ap2 quadruple mutant of B. napus, which has a similar sepal-to-carpel and petal-to-stamen phenotype as the scm mutant of Brassica. (C) Sequencing results of the target sites in the strong ap2 mutant. Bars are 1 mm in c4 and 10 mm in all the others panels.
After that, we performed Agrobacterium-mediated transformation to introduce CRISPR/Cas9 construct into B. napus to generate 33 transgenic plants (Yan et al., 2012). Among these, some plants showed typical sepal carpeloid phenotypes and some exhibited weak carpeloid phenotypes (Figure 5B). Furthermore, the genetic composition of one typical mutant was identified by sequencing, in which five or four nucleotides were deleted in each single chromosome of BnaC-AP2a gene; a “G” nucleotide was inserted into BnaA-AP2a; a “T” nucleotide was inserted or deleted in each single chromosome of BnaA-AP2b gene; and a “A” nucleotide was inserted into BnaC-AP2b (Figure 5C and Supplementary Figure 4B). In this way, all of the four AP2 genes in B. napus acquired reading frameshift mutations and early translation termination, reflecting the fact that the four AP2 genes completely lost their functions. The resulting ap2 quadruple mutant exhibited carpels sepals, missing petals, and a reduced number of stamens, but showed a normal pistil, which are identical phenotypes to those displayed by scm (Figure 1B). These results suggest that knocking out the AP2 genes could severely affect floral organ development and lead to the generation of a sepal carpeloid and apetalous mutant in Brassica.
Discussion
The Novel Pistil-Like Flower Mutant Is a Sepal Carpeloid and Apetalous Mutant
The scm displayed some distinct characteristics: the sepals and petals were absent; the four merged carpels that looked like a very large pistil developed in the sepal positions; stamens reduced in numbers to 1–4; and only the pistil developed normally. Clearly, the mutant underwent two substantial changes with respect to its floral organs: one was a typical homeotic transformation of sepals into carpels, and the other was an apetalous phenotype. Thus, the pistil-like flower mutant was not only a typical sepal carpeloid mutant but also an apetalous mutant.
Moreover, we found some petals undergoing transformation into stamens in scm2 indicating that the scm2 underwent an incomplete homeotic transition of sepals to carpels and petals to stamens. It has been well-documented in Arabidopsis that the defective A-function AP2 gene affects sepal and petal development and leads to the homeotic transformation of sepals to carpels and petals to stamens (Kunst et al., 1989). However, our weak and strong mutants presented different phenotypes in which the weak mutant exhibited an incomplete transformation of sepals to carpels and petals to stamens, whereas the strong mutant exhibited a complete sepal carpeloid transition and an apetalous trait. These results suggest that the AP2 strong mutant generates only the sepal-carpeloid transformation and the missing petals trait, but could not produce the petal-to-stamen transformation. Although the phenotypes of the two mutants were not identical, both had severe sepal and petal developmental defects.
Defective AP2 Genes Are the Main Cause of the scm Phenotype
Our data revealed that the scm mutant, derived from the F3 generation of an interspecific cross between B. napus and B. rapa, contains only two, B. rapa-similar AP2 genes, suggesting that its genetic background is close to that of the parent B. rapa. Both the AP2 genes in scm were mutated: one contained a partial repeat sequence that led to an early translation termination signal, and the other included an A to C single-nucleotide substitution that transformed the 18th glutamic acid of the corresponding protein into an aspartic acid. This mutation occurred in a highly conserved acidic serine-rich region (amino acids 14–50) that may serve as a transcriptional activation domain (Jofuku et al., 1994). Overall, the mutation of both AP2 genes in scm most likely resulted in the loss of function of these genes. Correspondingly, Yan et al. (2012) showed that knocking out the BnAP2 gene in B. napus generated a sepal carpeloid and apetalous mutant as well as some incomplete mutants. Similarly, we also generated several AP2-knockdown plants in the AP2-overexpressing transgenic Arabidopsis lines, as well as a number of AP2-knockout B. napus transgenic plants with a sepal carpeloid and apetalous phenotype. These results further confirmed that loss-of-function mutations of the AP2 genes resulted in the sepal carpeloid and apetalous phenotypes.
Our transgenic results demonstrated that the two normal AP2 genes from B. rapa can both rescue the sepal and petal developmental defects of the ap2-5 mutant of Arabidopsis, indicating that both normal AP2 genes have the ability to regulate sepal and carpel development, and reflecting that mutation of neither BrAP2a nor BrAP2b alone can lead to the scm phenotype.
Taken together, our results confirm that the AP2 gene plays a key role in regulating sepal and petal development and that its function is highly conserved in Cruciferae family, many of which are important crop plants. The complete loss of AP2 function severely affects sepal and petal development and generates sepal carpeloid and apetalous mutants. The two defective AP2 genes of B. rapa were the core cause for the sepal carpeloid mutant scm phenotype. However, the molecular mechanism of floral development regulated by AP2 gene is still unclear, and needs to be investigated further.
Author Contributions
XiW and YZ designed the experiments. YZ, SH, JL, JM, and XG performed the experiments. YZ and SH wrote and revised the article. XuW and JT bred the scm mutant. All the authors gave the final approval for submission of the manuscript.
Funding
This work was supported by a grant from National Key R&D Program of China (2016YFD0101900), National Science Foundation of Shaanxi Province (2014JQ3087), and the Ph.D. Programs Foundation of China (No. 2011M500441).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Weirong Xu at Ningxia University for comments on the manuscript and Dr. Qijun Chen at China Agricultural University for the kind gift of CRISPR/Cas9 vector pHSN401.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00367/full#supplementary-material
Footnotes
References
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
Bhalla, P. L., and Singh, M. B. (2008). Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat. Protoc. 3, 181–189. doi: 10.1038/nprot.2007.527
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M., and Smyth, D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743.
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20.
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Davies, B., Motte, P., Keck, E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1999). PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18, 4023–4034. doi: 10.1093/emboj/18.14.4023
Drews, G. N., Bowman, J. L., and Meyerowitz, E. M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002. doi: 10.1016/0092-8674(91)90551-9
Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1994). Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 76, 131–143. doi: 10.1016/0092-8674(94)90178-3
Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. doi: 10.1038/35054083
Huang, L., Zhao, X., Liu, T., Dong, H., and Cao, J. (2010). Developmental characteristics of floral organs and pollen of Chinese cabbage (Brassica campestris L. ssp. chinensis). Plant Syst. Evol. 286, 1–2. doi: 10.1007/s00606-010-0283-4
Huijser, P., Klein, J., Lonnig, W. E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum-majus. EMBO J. 11, 1239–1249. doi: 10.1002/j.1460-2075.1992.tb05168.x
Irish, V. F., and Sussex, I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. doi: 10.1105/tpc.2.8.741
Jack, T. (2001). Relearning our ABCs: new twists on an old model. Trends Plant Sci. 6, 310–316. doi: 10.1016/S1360-1385(01)01987-2
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3117–3122. doi: 10.1073/pnas.0409893102
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Krizek, B. A., and Meyerowitz, E. M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22.
Kunst, L., Klenz, J. E., Martinez-Zapater, J., and Haughn, G. W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1, 1195–1208. doi: 10.1105/tpc.1.12.1195
Litt, A. (2007). An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Mol. Sci. 168, 73–91. doi: 10.1086/509662
Maes, T., Van de Steene, N., Zethof, J., Karimi, M., D’Hauw, M., Mares, G., et al. (2001). Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13, 229–244. doi: 10.1105/tpc.13.2.229
Melzer, R., and Theißen, G. (2009). Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 37, 2723–2736. doi: 10.1093/nar/gkp129
Meyerowitz, E. M., Bowman, J. L., Brockman, L. L., Drews, G. N., Jack, T., Sieburth, L. E., et al. (1991). A genetic and molecular model for flower development in Arabidopsis thaliana. Dev. Suppl. 1, 157–167.
Ohto, M., Fischer, R. L., Goldberg, R. B., Nakamura, K., and Harada, J. J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3123–3128. doi: 10.1073/pnas.0409858102
Parkin, I. A. P., Sharpe, A. G., Keith, D. J., and Lydiate, D. J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38, 1122–1131. doi: 10.1139/g95-149
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E. R., and Yanofsky, M. F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182–184. doi: 10.1016/S0960-9822(01)00024-0
Ripoll, J. J., Roeder, A. H. K., Ditta, G. S., and Yanofsky, M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138, 5167–5176. doi: 10.1242/dev.073031
Saedler, H., Becker, A., Winter, K. U., Kirchner, C., and Theissen, G. (2001). MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 48, 351–358.
Snowdon, R. J., Friedrich, T., Friedt, W., and Kohler, W. (2002). Identifying the chromosomes of the A- and C-genome diploid Brassica species B. rapa (syn. campestris) and B. oleracea in their amphidiploid B. napus. Theor. Appl. Genet. 104, 533–538. doi: 10.1007/s00122-001-0787-y
Song, X., Li, Y., and Hou, X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14:573. doi: 10.1186/1471-2164-14-573
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Theissen, G., and Saedler, H. (2001). Plant biology-Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Wurschum, T., Gross-Hardt, R., and Laux, T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18, 295–307. doi: 10.1105/tpc.105.038398
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Yan, X. H., Zhang, L., Chen, B., Xiong, Z. Y., Chen, C. L., Wang, L. J., et al. (2012). Functional identification and characterization of the Brassica napus transcription factor gene BnAP2, the Ortholog of Arabidopsis thaliana APETALA2. PLoS One 7:e33890. doi: 10.1371/journal.pone.0033890
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zhou, Y., Lu, D. F., Li, C. Y., Luo, J. H., Zhu, B. F., Zhu, J. J., et al. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24, 1034–1048. doi: 10.1105/tpc.111.094383
Keywords: APETALA2 (AP2), carpeloid sepal, Brassica, flower development, oilseed rape, organ modification
Citation: Zhang Y, Huang S, Wang X, Liu J, Guo X, Mu J, Tian J and Wang X (2018) Defective APETALA2 Genes Lead to Sepal Modification in Brassica Crops. Front. Plant Sci. 9:367. doi: 10.3389/fpls.2018.00367
Received: 17 January 2018; Accepted: 06 March 2018;
Published: 20 March 2018.
Edited by:
Yuke He, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Xianzhong Feng, Northeast Institute of Geography and Agroecology (CAS), ChinaHui Feng, Shenyang Agricultural University, China
Copyright © 2018 Zhang, Huang, Wang, Liu, Guo, Mu, Tian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Wang, d2FuZ3hmZjk5QG53c3VhZi5lZHUuY24= Jianxin Mu, anhtc3h5Y0AxNjMuY29t
†These authors have contributed equally to this work.
 Yanfeng Zhang1,2†
Yanfeng Zhang1,2† Shuhua Huang
Shuhua Huang Xiaofeng Wang
Xiaofeng Wang