- Department of Biology, University of Rome Tor Vergata, Rome, Italy
In this review we highlight the advances achieved in the investigation of the role of 14-3-3 proteins in hormone signaling, biosynthesis, and transport. 14-3-3 proteins are a family of conserved molecules that target a number of protein clients through their ability to recognize well-defined phosphorylated motifs. As a result, they regulate several cellular processes, ranging from metabolism to transport, growth, development, and stress response. High-throughput proteomic data and two-hybrid screen demonstrate that 14-3-3 proteins physically interact with many protein clients involved in the biosynthesis or signaling pathways of the main plant hormones, while increasing functional evidence indicates that 14-3-3-target interactions play pivotal regulatory roles. These advances provide a framework of our understanding of plant hormone action, suggesting that 14-3-3 proteins act as hubs of a cellular web encompassing different signaling pathways, transducing and integrating diverse hormone signals in the regulation of physiological processes.
Introduction
14-3-3 proteins are highly conserved dimeric proteins with a subunit mass of 30 kDa, widespread in eukaryotic organisms (Aitken et al., 1992; Fu et al., 2000; Huber et al., 2002). They exist in multiple isoforms that form homo- and hetero-dimers (Jones et al., 1995). Among eukaryotes, plants have the largest number of 14-3-3 genes, such as 15 in Arabidopsis, 5 in barley and 8 in rice. In Arabidopsis, the 13 expressed isoforms are designated by Greek letters (χ,ω,ψ,ϕ,υ,λ,ν,κ,μ,ε,o,ι,π) and classified, according to their amino acid sequence similarities, into two distinct groups: the 𝜀 and the non-𝜀 group (Chevalier et al., 2009; Denison et al., 2011).
Although the high degree of sequence conservation among isoforms suggests a corresponding functional redundancy, increasing evidence demonstrates that 14-3-3 isoforms bind to individual targets with different affinities, thereby opening the possibility that regulation of specific processes could be accomplished by single 14-3-3 isoforms (Paul et al., 2012; Pallucca et al., 2014).
Moreover, the large number of isoforms suggests a very high combinatorial complexity in dimer arrangement, which in turn could underlie a fine tuning of their cellular functions.
14-3-3 proteins are, together with the FHA domain-containing proteins, the only phospho-binding regulators identified so far in plants (Chevalier et al., 2009). The common trait of 14-3-3 proteins is their ability to bind target proteins through the recognition of phosphorylated consensus motifs. So far, three 14-3-3 consensus motifs have been proposed: mode I (R/K)XX(pS/pT)XP, mode II (R/K)XXX(pS/pT)XP (Muslin et al., 1996; Yaffe et al., 1997) and the C-terminal mode III (pS/pT)X1-2-COOH (Coblitz et al., 2006; Paiardini et al., 2014), where X is any amino acid and pS/pT represents a phosphoserine or phosphothreonine.
14-3-3 structure and the mechanism of interaction with target proteins has been elucidated upon the determination of X-ray structures in different eukaryotic organisms (Liu et al., 1995; Xiao et al., 1995). As shown in Figure 1, monomers consist of nine anti-parallel α-helices and associate each other through the N-terminal region to assemble the dimeric protein. The 14-3-3 dimer has a characteristic cup-like shape with a highly conserved internal surface and a variable external surface. A conserved amphipathic groove, where the interaction with the phosphorylated target takes place, is present on the concave surface of each monomer, thus implicating that a 14-3-3 dimer can potentially bind two targets at the same time (Yaffe et al., 1997; Ottmann et al., 2007; Taoka et al., 2011).
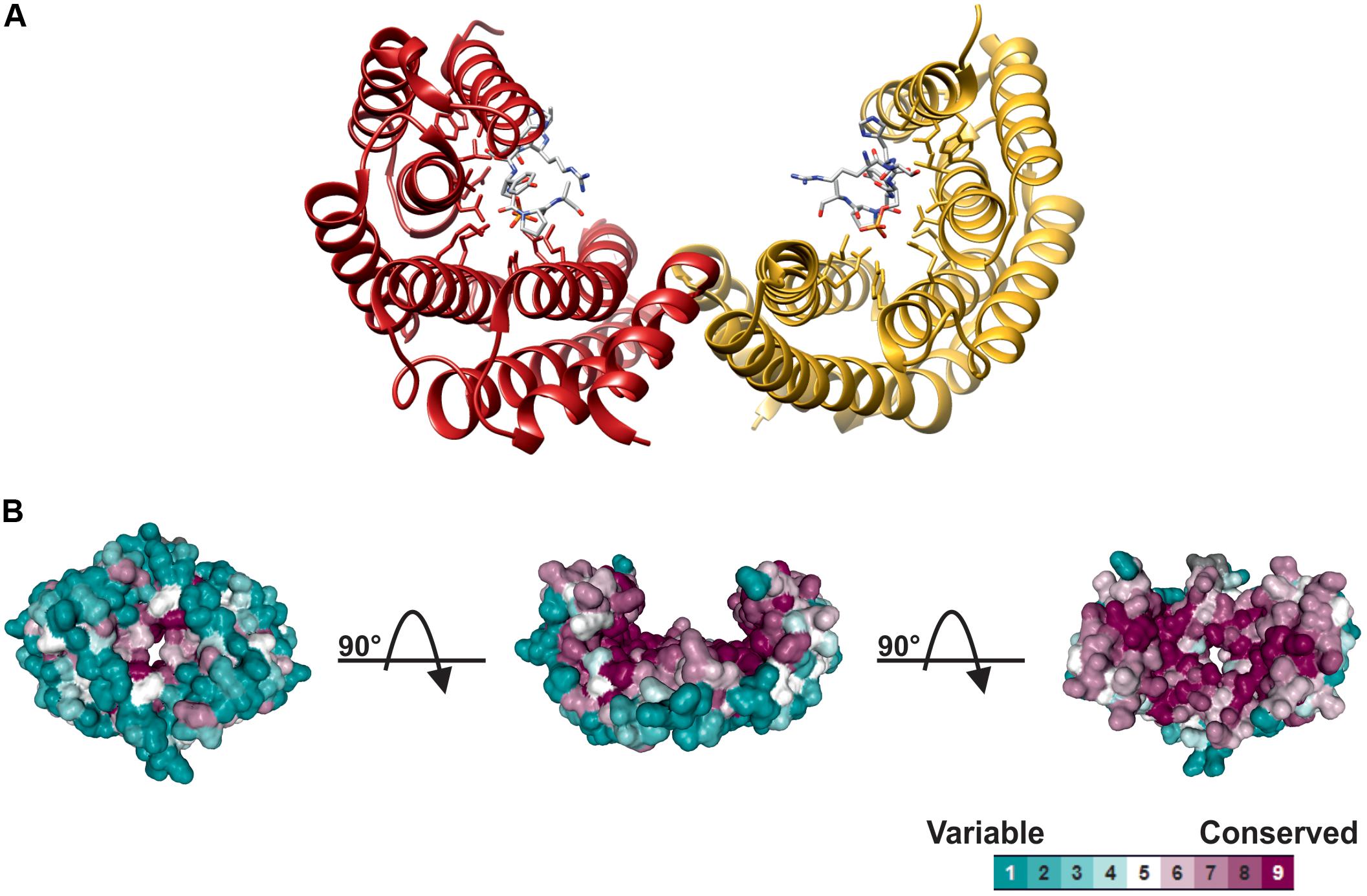
FIGURE 1. Structure of 14-3-3 proteins. (A) Ribbon plot of the human 14-3-3ζ dimer (PDB 1QJB), showing the two monomers (red and yellow ribbon, respectively) complexed with the mode I Raf-1 phosphopeptide (stick models, with carbon atoms colored in gray, oxygen in red, nitrogen in blue and phosphorus in yellow, respectively), which is bound in an extended conformation to the amphipathic groove of each monomer. (B) Spacefill structure of 14-3-3ζ shaded according to residue conservation. Structure was analyzed using Consurf (Ashkenazy et al., 2016), aligning 150 14-3-3 isoforms from different eukaryotic organisms. Multiple Sequence Alignment was built using MAFFT (Katoh and Standley, 2013) with residue conservation determined by a maximum likelihood method within Consurf. 14-3-3 structure is shown in three different orientations by sequential 90° rotations, to highlight the high conservation of internal cavity and the variability of the outer surface.
Depending on the biochemical feature of the phosphorylated target, association of 14-3-3 proteins can have different functional consequences, leading to regulation of its enzymatic activity, subcellular localization, protein stability or alteration of protein-protein interactions (Figure 2; Hermeking, 2003; Wilson et al., 2016). In plants, 14-3-3 proteins have been originally identified as component of DNA-protein complexes (Lu et al., 1992) and as co-receptors of the fungal phytotoxin fusicoccin (Korthout and de Boer, 1994; Marra et al., 1994; Oecking et al., 1994). Thereafter, they were found to regulate the plasma membrane H+-ATPase (Jahn et al., 1997; Baunsgaard et al., 1998; Fullone et al., 1998) and enzymes of carbon and nitrogen metabolism (Bachmann et al., 1996a,b; Douglas et al., 1997; Toroser et al., 1998; Huber et al., 2002).
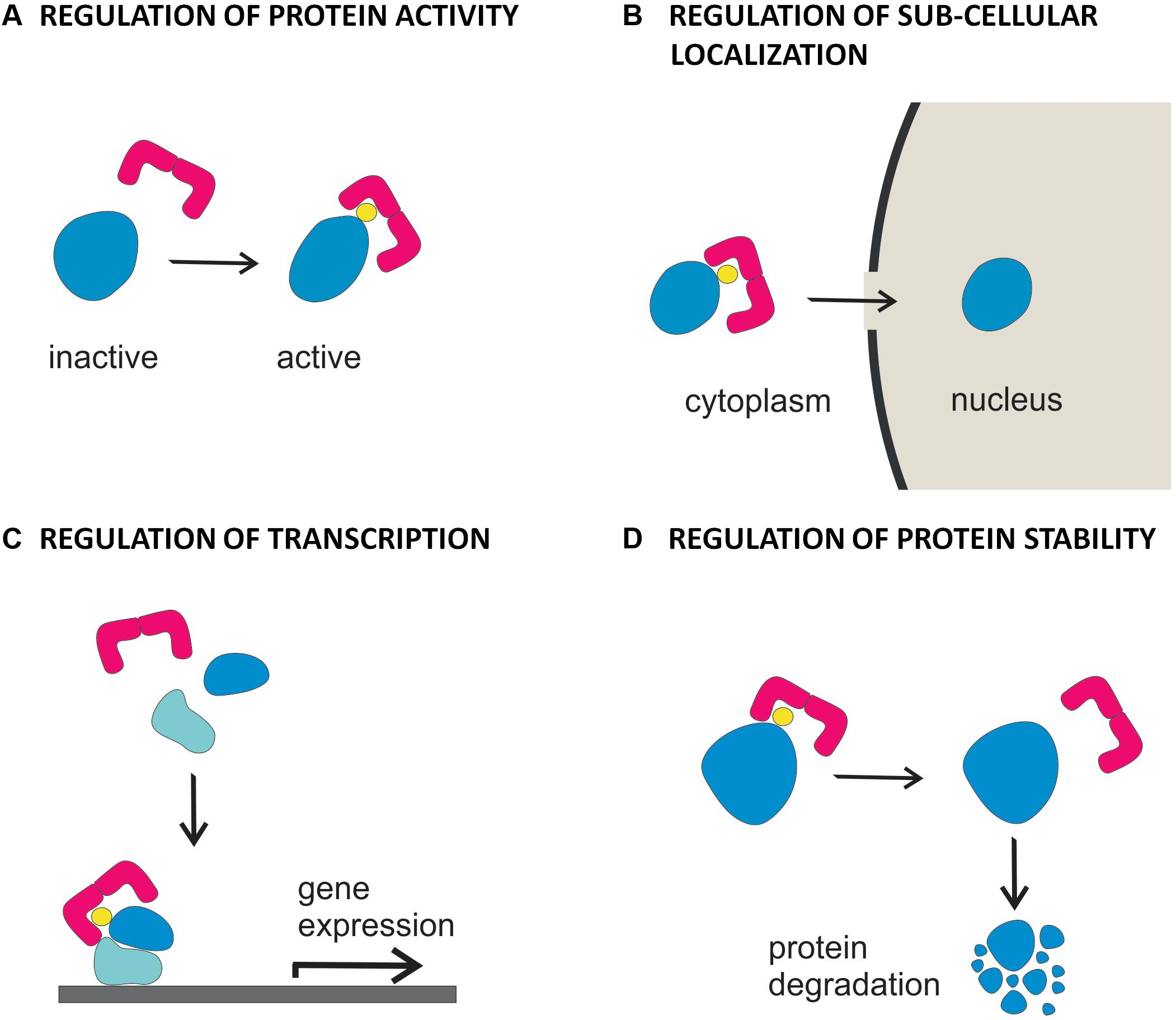
FIGURE 2. Functional diversity of 14-3-3 proteins in hormone signaling. Binding of 14-3-3 proteins to targets involved in hormone action can result in (A) regulation of the activity of the protein, as demonstrated for BKI1 in the BR signaling pathway, for H+-ATPase, in IAA and ABA signaling pathways and KAT1, in the IAA signaling pathway (B) regulation of the cytoplasmic vs nuclear localization of the protein, as demonstrated for BES1 and BZR1 in the BR signaling pathway and KAT1 in the IAA signaling pathway (C) regulation of the assembly and/or the activity of transcriptional elements, as demonstrated for EMBP1/VP1, ABF 1-2 and HvABI5 in the ABA signaling pathway (D) control of the turnover of the protein, as demonstrated for ACC synthase and ETO/ETO1-like proteins in the ethylene biosynthesis and signaling pathways.
Nowadays, a wide range of 14-3-3 clients with a pivotal role in various physiological processes, including growth and development and response to stress has been identified (Moorhead et al., 1999; Aducci et al., 2002; Chevalier et al., 2009; Denison et al., 2011; Jaspert et al., 2011).
In the last years, a growing body of evidence has emerged regarding the involvement of 14-3-3 proteins as key players of different aspects of plant hormone physiology. In this review, we highlight novel insights into the role of 14-3-3 proteins in the regulation of hormonal signaling, biosynthesis and transport.
Brassinosteroids
Brassinosteroids (BRs) are steroid hormones regulating fundamental functions in plant growth and development, including cell division and elongation, vascular differentiation, flowering, photomorphogenesis, senescence, and responses to environmental stresses. (Clouse and Sasse, 1998).
Genetic and molecular studies in Arabidopsis have greatly advanced the understanding of the BR mode of action, and revealed that 14-3-3 proteins play a complex role in BR signaling by interacting with different members of the BR transduction machinery. Proteomics and yeast two-hybrid screen studies identified the BR receptor Brassinosteroid-Insensitive1 (BRI1), the BRI1 Kinase Inhibitor (BKI1), the BRI1 Suppressor phosphatase (BSU1), the transcription factors BRI1-EMS-Suppressor1 (BES1) and Brassinazole-Resistant1 (BZR1) as 14-3-3 client proteins (Milne et al., 2002; Schoonheim et al., 2007b; Chang et al., 2009).
The first evidence that demonstrated a functional role of 14-3-3 proteins in BR signaling concerned their interaction with BES1/BZR1, which regulate the expression of BR-responsive genes. In the absence of BR, binding of 14-3-3 proteins to phosphorylated BZR1 and BES1 results in their cytoplasmic sequestration and in BR-signaling inhibition. Binding of 14-3-3 proteins occurs upon phosphorylation of Ser173 whithin the mode II-type motif RISNpSCP. Accordingly, mutations in Ser173 suppress the dwarf phenotype of the receptor mutant (Gampala et al., 2007; Ryu et al., 2007, 2010).
More recently, the interaction with the negative regulator of BR signaling BKI1 has been unveiled (Wang et al., 2011). In the presence of BR, BKI1 is phosphorylated by the BR receptor BRI1 in its C-terminal domain and released into the cytosol, where it associates and antagonizes 14-3-3 proteins, thus promoting BZR1/BES1 translocation into the nucleus. Intriguingly, the interaction with 14-3-3 proteins occurs via an uncommon 14-3-3 mode II motif (RGELFpS270APApS274), which is also involved in the association with BRI1.
Remarkably, these data suggest that 14-3-3 proteins can act both as negative and positive regulators of the pathway, depending on the BR levels. At low BR levels, they function as negative regulators, whereas at higher BR levels BKI1 and 14-3-3 are released from BRI1 and BES1/BZR1, bind and inhibit each other, thereby allowing the full activation of the BR signaling pathway.
Auxin
Auxin (indole-3-acetic acid, IAA) is a key regulator of nearly every aspect of plant growth and development, including embryogenesis, lateral root development, vascular tissue differentiation, apical dominance, flower development and tropisms (Teale et al., 2006; Weijers and Wagner, 2016). At the cellular level, IAA controls cell expansion by stimulating the H+-ATPase dependent proton extrusion into the cell wall (Rayle and Cleland, 1992). Enzyme activation involves the binding of 14-3-3 proteins to a conserved Thr residue (Thr947 in the Arabidopsis AHA1 isoform) at the extreme C terminal end of the autoinhibitory domain of the enzyme, which brings about its displacement, thereby releasing enzyme activity (Fuglsang et al., 1999; Svennelid et al., 1999; Camoni et al., 2000). It was proposed that IAA activates H+-ATPase gene transcription (Hager et al., 1991). Successively, it was demonstrated that IAA activates the H+-ATPase by a post-translational mechanism. IAA promotes Thr947 phosphorylation and subsequent 14-3-3 binding to the Tyr-pThr-Val mode III motif (Takahashi et al., 2012). Although it is not clear whether IAA can activate protein kinases responsible of Thr947 phosphorylation, it has recently been demonstrated that IAA action involves the inhibition of a PP2C-D subfamily of type 2C protein phosphatases, that negatively regulates H+-ATPase activity (Spartz et al., 2014). In fact, IAA induces the transcription of Small Auxin Up RNA19 (SAUR19), a protein encoded by the SAUR19-24 subfamily of auxin-induced genes. SAUR19 interacts and inhibits PP2C-D phosphatases, thus promoting H+-ATPase phosphorylation and activation. Accordingly, constitutive AtSAUR19 overexpression promotes hypocotyl elongation in tomato plants by a mechanism involving PP2C-D inhibition (Spartz et al., 2014).
Sustained H+-ATPase-mediated proton extrusion and growth is dependent on K+ influx mediated by inward-rectifier K+ channels, required to depolarize the negative potential that thermodynamically inhibits the proton pump. Interestingly, 14-3-3 proteins are also involved in K+ channel post-translational regulation. In fact, 14-3-3 proteins bind to and activate the inward-rectifier K+ channel1 in Arabidopsis thaliana 1 (KAT1) by modifying its open probability (Sottocornola et al., 2006) and by increasing the number of channel delivered at the plasma membrane (Sottocornola et al., 2008). Binding occurs at the mode III motif HLYFSpS676N (Saponaro et al., 2017). The overall data suggest that 14-3-3 proteins may function as pivotal regulators of ion transport, integrating different stimuli in the generation and maintenance of the plasma membrane potential.
Localized IAA concentration gradients are essential in different aspects of plant physiology, including tropisms and organ formation. Pin-formed (PIN) proteins, which can be relocated in the cell by endocytic recycling, are a family of IAA transporters essential in the generation of IAA gradients (Naramoto, 2017). It has been recently shown that RNA-interference repression of 𝜀 members of 14-3-3 protein family in Arabidopsis seedlings caused altered polar distribution of IAA and produced related IAA-transport phenotypes (Keicher et al., 2017). These data, despite lack of information concerning molecular 14-3-3 interactors involved in PIN repositioning, clearly point to a fundamental role of the 𝜀 group of 14-3-3 proteins in the regulation of PIN distribution, and IAA transport.
On the whole, accumulated evidence indicates that 14-3-3 proteins are versatile regulators of IAA action, intervening at very different points of the IAA regulatory network: In fact, 14-3-3 proteins can function both downstream, as final transducer of IAA growth-promoting signaling as well as upstream, as wardens of hormone traffic, controlling the formation of IAA gradients.
Abscisic Acid
Abscisic acid (ABA) is involved in the regulation of key processes of plant development, such as embryogenesis, seed maturation, dormancy, and germination. At the same time, it mediates the response to environmental stresses, including salinity, cold, and drought (Zeevaart and Creelman, 1988).
Knowledge about pathways of ABA signaling has for a long time been fragmentary, until recent studies have shed light on the molecular functions of genetically identified components, including receptors, protein kinases/phosphatases, and different ABA-Responsive Element Binding Factors (ABFs), so that a core model of ABA signaling can be envisaged. In Arabidopsis, ABA is perceived by the Pyrabactin Resistance1 (PYR1)/PYR1-Like (PYL) multigenic family of receptors (Miyakawa et al., 2013). Upon hormone binding, they undergo a conformational change that allow them to associate and inhibit members of clade A of type 2C protein phosphatases (PP2Cs), negative regulators of ABA signaling. PP2C inhibition in turn allows Sucrose non-fermenting-Related kinase2 (SnRK2) kinase activation and phosphorylation of different ABFs, thereby inducing the response (Melcher et al., 2010; Miyakawa et al., 2013). In vitro evidence indicates that the SnRK2-type kinase Open Stomata1 (OST1) in Arabidopsis guard cells phosphorylates Thr451 of ABF3 within the 14-3-3 binding motif RXX(S/T)XP, conserved in ABFs, thus promoting 14-3-3 association (Sirichandra et al., 2010). Notably, ABA treatment induces ABF3 phosphorylation in planta and indirect evidence suggests that Thr451 phosphorylation is correlated to enhanced ABF3 stability (Sirichandra et al., 2010).
In Arabidopsis stomata cells and hypocotyls, ABA inhibits the plasma membrane H+-ATPase by inducing its dephosphorylation and 14-3-3 release (Hayashi et al., 2011, 2014), while in barley embryonic roots ABA inhibits 14-3-3-activated inward K+ channels (van den Wijngaard et al., 2005).
14-3-3 proteins also play a key role in ABA regulated transcription. In fact, they were found as part of transcriptional complexes of ABA-regulated genes. In rice embryogenic cultures and maize embryos, 14-3-3 proteins are part of the complex between the basic leucine zipper (bZIP) transcription factor EmBP1 and Viviparus1 (VP1), which binds to the ABA responsive element Em1a (Schultz et al., 1998), while in Arabidopsis embryos they are associated to ABI3 regulated AtEm1 promoter (del Viso et al., 2007). Moreover, in embryonic barley roots 14-3-3 proteins have a function in the ABA regulated transcriptional cascade. In fact, RNAi-mediated silencing of individual 14-3-3 isoforms resulted in reduction of the expression of a reporter gene controlled by the ABA-inducible promoter ABA-Response Complex3 (ABRC3). Yeast two-hybrid screen allowed to identify the seed specific ABI1-3 and ABI5 proteins, belonging to the ABF family of bZip transcription factors, as 14-3-3 interactors (Schoonheim et al., 2007a, 2009). Interestingly, in this system ABA also increases the expression of four out of five 14-3-3 barley genes, thus revealing a reciprocal relationship between ABA and 14-3-3 proteins: they act as signaling effectors and in turn are under transcriptional control by ABA, according to a positive feedback circuit.
Gibberellins
Gibberellins (GAs) are a wide family of tetracyclic diterpenoid molecules that regulates fundamental plant processes, like germination and stem elongation, besides many other aspects of plant growth and development, such as floral initiation, pollen development, leaf expansion, trichome and anther development (Richards et al., 2001). Initial research demonstrated that 14-3-3 proteins are involved in the control of the GA biosynthetic pathway. GAs regulate their own biosynthesis by a negative feedback mechanism involving the bZip transcriptional activator Repression of Shoot Growth (RSG), which binds to the promoter of the biosynthetic enzyme ent kaurene oxidase (GA3) gene (Fukazawa et al., 2000). In transgenic tobacco plants, 14-3-3 proteins were co-precipitated with RSG. Mutation of Ser114 whithin the sequence RSLpSVD impaired 14-3-3 binding, inducing RSG translocation into the nucleus and increased transcription (Igarashi et al., 2001). Moreover, RSG translocation into the nucleus was promoted by a reduction of GA levels (Ishida et al., 2004). These lines of evidence clearly indicate that 14-3-3 proteins participate to GA biosynthesis as negative regulators, sequestering in the cytoplasm the transcriptional regulator RSG. 14-3-3 binding is mediated by RSG phosphorylation promoted by Nicotiana tabacum Calcium-Dependent Protein Kinase (NtCDPK1), which also functions as a scaffold protein, bridging 14-3-3 proteins to RSG (Ito et al., 2014).
More recent work demonstrates that 14-3-3 proteins are also involved in GA signaling. In barley aleurone cells, isoform-specific 14-3-3 RNAi-mediated silencing inhibits GA activation of a reporter gene under the control of the α-amylase promoter. In this system, a possible role for 14-3-3 proteins in the coordination of GA and ABA signaling has emerged. In fact, the overexpression of ABA responsive, 14-3-3-interacting transcription factors ABF1-3 impairs GA action, indicating that they act as negative regulators of GA signaling and that 14-3-3 proteins may function by sequestering ABF1-3 in the cytoplasm. However, the mechanism of 14-3-3 action is still unclear, since abolition of 14-3-3/ABF1-2 interaction affects their ABA-dependent transactivation activity, while the same deletion does not influence their inhibitory activity in GA signaling (Schoonheim et al., 2009).
Ethylene
The gaseous hormone ethylene influences several aspects of plant growth and development, including germination, cell expansion, leaf and flower senescence and abscission, fruit ripening, resistance to pathogen infection and adaptation to stress conditions (Bleecker and Kende, 2000). Ethylene is synthesized from the amino acid methionine. Conversion of S-adenosyl-methionine (SAM) in 1-aminocyclopropane-1-carboxylic acid (ACC), catalyzed by a family of ACC synthase enzymes (ACS), is the rate-limiting step of ethylene synthesis (Wang et al., 2002). In the last years, the role of 14-3-3 proteins in the post-translational regulation of ethylene biosynthesis has emerged. In fact, it has been demonstrated the ability of 14-3-3 proteins to interact in vivo with different ACS isoforms (Chang et al., 2009; Huang et al., 2013; Yoon and Kieber, 2013; Catalá et al., 2014). 14-3-3 proteins likely bind to ACS through non-canonical binding sites as neither the mode I nor mode II binding sites are present in ACS proteins. Interestingly, 14-3-3 proteins interact also with components involved in the regulation of ACS stability, the Ethylene-Over-producer1 (ETO1)/ETO1-Like (EOLs) proteins. They are part of a Cullin-3 E3 ubiquitin ligase complex that targets ACS protein for 26S-proteasome-mediated degradation. Binding of 14-3-3 proteins destabilizes ETO1/EOLs, thereby blocking the Cullin-3 E3 ubiquitin ligase activity and consequently the proteasome-mediated ACS degradation (Yoon and Kieber, 2013).
However, contrasting results have been obtained studying the Arabidopsis mechanism which regulates freezing tolerance and cold acclimation. The 14-3-3 ψ isoform, encoded by the RARE COLD INDUCIBLE 1A (RCA1) gene, interacts with ACS, negatively regulating its stability and consequently lowering ethylene production (Catalá et al., 2014).
Cytokinins and Other Hormones
In the last years, data on the interaction of 14-3-3 proteins with components of signaling pathways of other hormones have been reported. Although the physiological relevance of these interactions is still to be ascertained, these data allow to envisage a regulatory role of 14-3-3 proteins in the action of cytokinin, jasmonate (JA) and salicylic acid (SA).
Affinity chromatography/mass spectrometry and yeast two hybrid screen (Schoonheim et al., 2007b; Chang et al., 2009; Jaspert et al., 2011) in barley and Arabidopsis allowed to identify different enzymes of cytokinin metabolism (CKX3, cytokinin oxidase) and signaling components (Arabidopsis Response Regulators ARR2 and ARR12 and Cytokinin Response Factor CRF6) as 14-3-3 interacting proteins.
Immunoprecipitation and Surface Plasmon Resonance (SPR) experiments demonstrated that 13-lipoxygenase (13-LOX) interacts with 14-3-3 proteins in barley embryos. This enzyme controls lipid metabolism, which is a key process not only in germination, but also in the biosynthesis of the stress responsive hormone JA (Holtman et al., 2000a,b). Furthermore, the 14-3-3 λ isoform was identified by a yeast two-hybrid screen as an interactor of the RPW8.2 gene product, a R receptor that mediates SA-dependent resistance to the biotrophic fungal pathogens Golovinomyces spp. Accordingly, overexpression of GF14λ gene enhanced, whereas downregulation hampered, the SA-dependent resistance (Yang et al., 2009).
Concluding Remarks
This review highlighted the involvement of 14-3-3 proteins in plant hormone regulation, an emerging topic concerning 14-3-3 functions. In fact, whereas a direct regulatory role of 14-3-3 proteins in diverse aspects of plant physiology, from primary metabolism to ion transport, has been well documented, a growing body of evidence indicates that 14-3-3 participate also to a secondary level of regulation, i.e., by affecting hormone signal transduction pathways and biosynthesis. The emerging picture is complex, reflecting the high number of targets and the multiplicity of the 14-3-3 effects. In fact, their distinctive trait to bind to phosphorylated targets ensures that 14-3-3 proteins interact simultaneously with multiple components and/or at different steps of hormone signaling networks, implicating that they can carry out diverse and even opposing functions in different pathways (e.g., GA, ABA), or in the same pathway (e.g., BR). Furthermore, additional complexity may arise from reciprocal regulation by hormones of 14-3-3 concentrations, as well as from specificity/redundancy of functions of the numerous plant 14-3-3 isoforms, for which information is still scarce. However, even though a unique rationale of 14-3-3 mode of action in hormone regulation cannot be envisaged, some common traits can be inferred from so far available data.
A recurring regulatory mechanism of 14-3-3 action is exerted at the transcriptional level, that 14-3-3 proteins influence by functioning as adaptor proteins (e.g., ABA) or altering the sub-cellular localization (e.g., GA, BR) or stability (e.g., ABA) of diverse families of transcriptional regulators. Intriguingly, recent work suggests that the control of stability or localization of transcription factors, shared by different pathways, may represent a mechanism which allows 14-3-3 proteins to integrate multiple hormone pathways, thus controlling a specific physiological process (e.g., ABFs factors in GA and ABA signaling). Alternatively, 14-3-3 can regulate hormone action at the post-translational level, by modulating the activity of client proteins in signaling cascades (e.g., IAA, ABA) or in biosynthetic pathways (e.g., ethylene, GA). Simultaneous control of multiple 14-3-3 clients (e.g., H+-ATPase, KAT1) within the same pathway (e.g., IAA) or regulation of targets (e.g., H+-ATPase) participating to different pathways (e.g., IAA, ABA) provides a way for 14-3-3 proteins to coordinate the action of diverse hormones in the control of a specific physiological process.
Data reported in this review depict a complex scenario, where a network of interactions among 14-3-3 proteins and their targets finely regulate hormone signaling and homeostasis. It is conceivable that in the next future the identification of novel clients will increase the complexity of the 14-3-3 signaling web. Hence, in order to get a deeper insight, future work should be addressed toward a detailed biochemical characterization of interactions, including the identification of the binding sequence as well as the functional results, i.e., whether it involves modification of protein activity, sub-cellular localization or stability (Figure 2). Moreover, to solve the 14-3-3 specificity vs. redundancy dilemma, it will be crucial to get information about the relative affinities of different isoforms toward each single 14-3-3 client.
Author Contributions
LC and MM wrote the manuscript. SV and PA contributed to the writing of manuscript and critically revised it for essential intellectual content. LC prepared the figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We have endeavored to cover the more relevant aspects of 14-3-3-hormone cross-talk, but recognize that not all literature has been cited. We apologize in advance with the 14-3-3 community for possible lackings.
References
Aducci, P., Camoni, L., Marra, M., and Visconti, S. (2002). From cytosol to organelles: 14-3-3 proteins as multifunctional regulators of plant cell. IUBMB Life 53, 49–55. doi: 10.1080/15216540210813
Aitken, A., Collinge, D. B., van Heusden, B. P., Isobe, T., Roseboom, P. H., Rosenfeld, G., et al. (1992). 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17, 498–501. doi: 10.1016/0968-0004(92)90339-B
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W450. doi: 10.1093/nar/gkw408
Bachmann, M., Huber, J. L., Athwal, G. S., Wu, K., Ferl, R. J., and Huber, S. C. (1996a). 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398, 26–30.
Bachmann, M., Huber, J. L., Liao, P. C., Gage, D. A., and Huber, S. C. (1996b). The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 387, 127–131.
Baunsgaard, L., Fuglsang, A. T., Jahn, T., Korthout, H. A., de Boer, A. H., and Palmgren, M. G. (1998). The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 13, 661–671. doi: 10.1046/j.1365-313X.1998.00083.x
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell. Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Camoni, L., Iori, V., Marra, M., and Aducci, P. (2000). Phosphorylation-dependent interaction between plant plasma membrane H(+)-ATPase and 14-3-3 proteins. J. Biol. Chem. 275, 9919–9923. doi: 10.1074/jbc.275.14.9919
Catalá, R., López-Cobollo, R., Mar Castellano, M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. doi: 10.1105/tpc.114.127605
Chang, I. F., Curran, A., Woolsey, R., Quilici, D., Cushman, J. C., Mittler, R., et al. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985. doi: 10.1002/pmic.200800445
Chevalier, D., Morris, E. R., and Walker, J. C. (2009). 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60, 67–91. doi: 10.1146/annurev.arplant.59.032607.092844
Clouse, S. D., and Sasse, J. M. (1998). Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
Coblitz, B., Wu, M., Shikano, S., and Li, M. (2006). C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 580, 1531–1535. doi: 10.1016/j.febslet.2006.02.014
del Viso, F., Casaretto, J. A., and Quatrano, R. S. (2007). 14-3-3 Proteins are components of the transcription complex of the ATEM1 promoter in Arabidopsis. Planta 227, 167–175. doi: 10.1007/s00425-007-0604-1
Denison, F. C., Paul, A. L., Zupanska, A. K., and Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. doi: 10.1016/j.semcdb.2011.08.006
Douglas, P., Pigaglio, E., Ferrer, A., Halfords, N. G., and MacKintosh, C. (1997). Three spinach leaf nitrate reductase-3-hydroxy-3-methylglutaryl-CoA reductase kinases that are regulated by reversible phosphorylation and/or Ca2+ ions. Biochem. J. 325, 101–109. doi: 10.1042/bj3250101
Fu, H., Subramanian, R. R., and Masters, S. C. (2000). 14-3-3s: structure function and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647. doi: 10.1146/annurev.pharmtox.40.1.617
Fuglsang, A. T., Visconti, S., Drumm, K., Jahn, T., Stensballe, A., Mattei, B., et al. (1999). Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780. doi: 10.1074/jbc.274.51.36774
Fukazawa, J., Sakai, T., Ishida, S., Yamaguchi, I., Kamiya, Y., and Takahashi, Y. (2000). Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12, 901–915. doi: 10.1105/tpc.12.6.901
Fullone, M. R., Visconti, S., Marra, M., Fogliano, V., and Aducci, P. (1998). Fusicoccin effect on the in vitro interaction between plant 14-3-3 proteins and plasma membrane H+-ATPase. J. Biol. Chem. 273, 7698–7702. doi: 10.1074/jbc.273.13.7698
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189. doi: 10.1016/j.devcel.2007.06.009
Hager, A., Debus, G., Edel, H. G., Stransky, H., and Serrano, R. (1991). Auxin induces exocytosis and the rapid synthesis of a high-turnover pool of plasma-membrane H(+)-ATPase. Planta 185, 527–537. doi: 10.1007/BF00202963
Hayashi, M., Inoue, S., Takahashi, K., and Kinoshita, T. (2011). Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 52, 1238–1248. doi: 10.1093/pcp/pcr072
Hayashi, Y., Takahashi, K., Inoue, S., and Kinoshita, T. (2014). Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H(+)-ATPase in Arabidopsis thaliana. Plant Cell Physiol. 55, 845–853. doi: 10.1093/pcp/pcu028
Hermeking, H. (2003). The 14-3-3 cancer connection. Nat. Rev. Cancer 3, 931–943. doi: 10.1038/nrc1230
Holtman, W. L., Roberts, M. R., Oppedijk, B. J., Testerink, C., van Zeijl, M. J., and Wang, M. (2000a). 14-3-3 proteins interact with a 13-lipoxygenase, but not with a 9-lipoxygenase. FEBS Lett. 474, 48–52. doi: 10.1016/S0014-5793(00)01575-1
Holtman, W. L., Roberts, M. R., and Wang, M. (2000b). 14-3-3 proteins and a 13-lipoxygenase form associations in a phosphorylation-dependent manner. Biochem. Soc. Trans. 28, 834–836. doi: 10.1042/bst0280834
Huang, S. J., Chang, C. L., Wang, P. H., Tsai, M. C., Hsu, P. H., and Chang, I. F. (2013). A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 64, 4343–4360. doi: 10.1093/jxb/ert241
Huber, S. C., MacKintosh, C., and Kaiser, W. M. (2002). Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 50, 1053–1063. doi: 10.1023/A:1021284002779
Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13, 2483–2497. doi: 10.1105/tpc.13.11.2483
Ishida, S., Fukazawa, J., Yuasa, T., and Takahashi, Y. (2004). Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator repression of shoot growth by gibberellins. Plant Cell 16, 2641–2651. doi: 10.1105/tpc.104.024604
Ito, T., Nakata, M., Fukazawa, J., Ishida, S., and Takahashi, Y. (2014). Scaffold function of Ca2+-dependent protein kinase: tobacco Ca2+-dependent protein kinase1 transfers 14-3-3 to the substrate repression of shoot growth after phosphorylation. Plant Physiol. 165, 1737–1750. doi: 10.1104/pp.114.236448
Jahn, T., Fuglsang, A. T., Olsson, A., Brüntrup, I. M., Collinge, D. B., Volkmann, D., et al. (1997). The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9, 1805–1814. doi: 10.1105/tpc.9.10.1805
Jaspert, N., Throm, C., and Oecking, C. (2011). Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front. Plant Sci. 2:96. doi: 10.3389/fpls.2011.00096
Jones, D. H. A., Ley, S., and Aitken, A. (1995). Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 368, 55–58. doi: 10.1016/0014-5793(95)00598-4
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keicher, J., Jaspert, N., Weckermann, K., Möller, C., Throm, C., Kintzi, A., et al. (2017). Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. Elife 6:e24336. doi: 10.7554/eLife.24336
Korthout, H. A., and de Boer, A. H. (1994). A fusicoccin binding protein belongs to the family of 14-3-3 brain protein homologs. Plant Cell 6, 1681–1692. doi: 10.1105/tpc.6.11.1681
Liu, D., Bienkowska, J., Petosa, C., Collier, R. J., Fu, H., and Liddington, R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376, 191–194. doi: 10.1038/376191a0
Lu, G., DeLisle, A. J., de Vetten, N. C., and Ferl, R. J. (1992). Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc. Natl. Acad. Sci. U.S.A. 89, 11490–11494. doi: 10.1073/pnas.89.23.11490
Marra, M., Fullone, M. R., Fogliano, V., Pen, J., Mattei, M., Masi, S., et al. (1994). The 30-kilodalton protein present in purified fusicoccin receptor preparations is a 14-3-3-like protein. Plant Physiol. 106, 1497–1501. doi: 10.1104/pp.106.4.1497
Melcher, K., Zhou, X. E., and Xu, H. E. (2010). Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr. Opin. Struct. Biol. 20, 722–729. doi: 10.1016/j.sbi.2010.09.007
Milne, F. C., Moorhead, G., Pozuelo Rubio, M., Wong, B., Kulma, A., Harthill, J. E., et al. (2002). Affinity purification of diverse plant and human 14-3-3-binding partners. Biochem. Soc. Trans. 30, 379–381. doi: 10.1042/bst0300379
Miyakawa, T., Fujita, Y., Yamaguchi-Shinozaki, K., and Tanokura, M. (2013). Structure and function of abscisic acid receptors. Trends Plant Sci. 18, 259–266. doi: 10.1016/j.tplants.2012.11.002
Moorhead, G., Douglas, P., Cotelle, V., Harthill, J., Morrice, N., Meek, S., et al. (1999). Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 18, 1–12. doi: 10.1046/j.1365-313X.1999.00417.x
Muslin, A. J., Tanner, J. W., Allen, P. M., and Shaw, A. S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897. doi: 10.1016/S0092-8674(00)81067-3
Naramoto, S. (2017). Polar transport in plants mediated by membrane transporters: focus on mechanisms of polar auxin transport. Curr. Opin. Plant Biol. 40, 8–14. doi: 10.1016/j.pbi.2017.06.012
Oecking, C., Eckerskorn, C., and Weiler, E. W. (1994). The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 352, 163–166. doi: 10.1016/0014-5793(94)00949-X
Ottmann, C., Marco, S., Jaspert, N., Marcon, C., Schauer, N., Weyand, M., et al. (2007). Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell. 25, 427–440. doi: 10.1016/j.molcel.2006.12.017
Paiardini, A., Aducci, P., Cervoni, L., Cutruzzolà, F., Di Lucente, C., Janson, G., et al. (2014). The phytotoxin fusicoccin differently regulates 14-3-3 proteins association to mode III targets. IUBMB Life 66, 52–62. doi: 10.1002/iub.1239
Pallucca, R., Visconti, S., Camoni, L., Cesareni, G., Melino, S., Panni, S., et al. (2014). Specificity of 𝜀 and non-𝜀 isoforms of Arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets. PLoS One 9:e90764. doi: 10.1371/journal.pone.0090764
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., and Ferl, R. J. (2012). 14-3-3 Phosphoprotein interaction networks - does isoform diversity present functional interaction specification? Front. Plant Sci. 3:190. doi: 10.3389/fpls.2012.00190
Rayle, D. L., and Cleland, R. E. (1992). The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99, 1271–1274. doi: 10.1104/pp.99.4.1271
Richards, D. E., King, K. E., Ait-Ali, T., and Harberd, N. P. (2001). How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. doi: 10.1146/annurev.arplant.52.1.67
Ryu, H., Cho, H., Kim, K., and Hwang, I. (2010). Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 29, 283–290. doi: 10.1007/s10059-010-0035-x
Ryu, H., Kim, K., Cho, H., Park, J., Choe, S., and Hwang, I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762. doi: 10.1105/tpc.107.053728
Saponaro, A., Porro, A., Chaves-Sanjuan, A., Nardini, M., Rauh, O., Thiel, G., et al. (2017). Fusicoccin activates KAT1 channels by stabilizing their interaction with 14-3-3 proteins. Plant Cell 29, 2570–2580. doi: 10.1105/tpc.17.00375
Schoonheim, P. J., Costa Pereira, D. D., and de Boer, A. H. (2009). Dual role for 14-3-3 proteins and ABF transcription factors in gibberellic acid and abscisic acid signalling in barley (Hordeum vulgare) aleurone cells. Plant Cell Environ. 32, 439–447. doi: 10.1111/j.1365-3040.2009.01932.x
Schoonheim, P. J., Sinnige, M. P., Casaretto, J. A., Veiga, H., Bunney, T. D., Quatrano, R. S., et al. (2007a). 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 49, 289–301.
Schoonheim, P. J., Veiga, H., Pereira Dda, C., Friso, G., van Wijk, K. J., and de Boer, A. H. (2007b). A comprehensive analysis of the 14-3-3 interactome in barley leaves using a complementary proteomics and two-hybrid approach. Plant Physiol. 143, 670–683.
Schultz, T. F., Medina, J., Hill, A., and Quatrano, R. S. (1998). 14-3-3 proteins are part of an abscisic acid-VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10, 837–847. doi: 10.2307/3870669
Sirichandra, C., Davanture, M., Turk, B. E., Zivy, M., Valot, B., Leung, J., et al. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One 5:e13935. doi: 10.1371/journal.pone.0013935
Sottocornola, B., Gazzarrini, S., Olivari, C., Romani, G., Valbuzzi, P., Thiel, G., et al. (2008). 14-3-3 proteins regulate the potassium channel KAT1 by dual modes. Plant Biol. 10, 231–236. doi: 10.1111/j.1438-8677.2007.00028.x
Sottocornola, B., Visconti, S., Orsi, S., Gazzarrini, S., Giacometti, S., Olivari, C., et al. (2006). The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J. Biol. Chem. 281, 35735–35741. doi: 10.1074/jbc.M603361200
Spartz, A. K., Ren, H., Park, M. Y., Grandt, K. N., Lee, S. H., Murphy, A. S., et al. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26, 2129–2142. doi: 10.1105/tpc.114.126037
Svennelid, F., Olsson, A., Piotrowski, M., Rosenquist, M., Ottman, C., Larsson, C., et al. (1999). Phosphorylation of Thr-948 at the C terminus of the plasma membrane H(+)-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11, 2379–2391.
Takahashi, K., Hayashi, K., and Kinoshita, T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159, 632–641. doi: 10.1104/pp.112.196428
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Teale, W. D., Paponov, I. A., and Palme, K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859. doi: 10.1038/nrm2020
Toroser, D., Athwal, G. S., and Huber, S. C. (1998). Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 435, 110–114. doi: 10.1016/S0014-5793(98)01048-5
van den Wijngaard, P. W., Sinnige, M. P., Roobeek, I., Reumer, A., Schoonheim, P. J., Mol, J. N., et al. (2005). Abscisic acid and 14-3-3 proteins control K channel activity in barley embryonic root. Plant J. 41, 43–55. doi: 10.1111/j.1365-313X.2004.02273.x
Wang, H., Yang, C., Zhang, C., Wang, N., Lu, D., Wang, J., et al. (2011). Dual role of BKI1 and 14-3-3s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834. doi: 10.1016/j.devcel.2011.08.018
Wang, K. L., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151. doi: 10.1105/tpc.001768
Weijers, D., and Wagner, D. (2016). Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574. doi: 10.1146/annurev-arplant-043015-112122
Wilson, R. S., Swatek, K. N., and Thelen, J. J. (2016). Regulation of the regulators: post-translational modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front. Plant Sci. 7:611. doi: 10.3389/fpls.2016.00611
Xiao, B., Smerdon, S. J., Jones, D. H., Dodson, G. G., Soneji, Y., Aitken, A., et al. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376, 188–191. doi: 10.1038/376188a0
Yaffe, M. B., Rittinger, K., Volinia, S., Caron, P. R., Aitken, A., Leffers, H., et al. (1997). The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91, 961–971. doi: 10.1016/S0092-8674(00)80487-0
Yang, X., Wang, W., Coleman, M., Orgil, U., Feng, J., Ma, X., et al. (2009). Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 60, 539–550. doi: 10.1111/j.1365-313X.2009.03978.x
Yoon, G. M., and Kieber, J. J. (2013). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028. doi: 10.1105/tpc.113.110106
Keywords: 14-3-3 proteins, hormone signaling, brassinosteroids, auxin, abscisic acid, gibberellins, ethylene
Citation: Camoni L, Visconti S, Aducci P and Marra M (2018) 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 9:297. doi: 10.3389/fpls.2018.00297
Received: 19 December 2017; Accepted: 21 February 2018;
Published: 13 March 2018.
Edited by:
Ján A. Miernyk, Agricultural Research Service (USDA), United StatesCopyright © 2018 Camoni, Visconti, Aducci and Marra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Camoni, Y2Ftb25pQHVuaXJvbWEyLml0
 Lorenzo Camoni
Lorenzo Camoni Sabina Visconti
Sabina Visconti Patrizia Aducci
Patrizia Aducci Mauro Marra
Mauro Marra