
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 March 2018
Sec. Plant Nutrition
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00292
Induction and secretion of acid phosphatases (APases) is considered to be an important strategy for improving plant growth under conditions of low inorganic phosphate (Pi). Purple acid phosphatases (PAPs), are an important class of plant APases that could be secreted into the rhizosphere to utilize organic phosphorus (Po) for plant growth and development. To date, only a few members of the PAP family have been identified in soybean. In this paper, we identified a secreted PAP in soybean, GmPAP14, and investigated its role in utilizing external phytate, the main form of organic phosphorus in the soil. An analysis of its expression and promoter showed that GmPAP14 was mainly expressed in the root and was strongly induced following Po treatment, during which its expression expanded from meristematic to maturation zones and root hairs. In vitro enzyme assays indicated that GmPAP14 had a relatively high phytase activity. Furthermore, GmPAP14 overexpression increased secreted APase activities and phytase activities, leading to the improved use of external plant phytate, higher phosphorus content, and increased shoot weight. Thus, these results confirmed that GmPAP14 is an important gene induced in response to Po, and that it predominantly participates in utilizing external Po to enhance plant growth and development.
Phosphorus (P) is an essential macronutrient that plays an important role in plant growth and development (Marschner and Rimmington, 1988; Vance, 2001). However, in most soil, the content of inorganic phosphate (Pi), is much lower than that needed for plant growth. The majority of P exists in an organic form (Po) or is fixed with calcium, iron and aluminum, which cannot be directly utilized by plants (George and Richardson, 2008). Thereby, efficiency of P-utilization is a major target for improving crop productivity. In fact, to maintain or obtain high crop yields, phosphate fertilizers are commonly applied in farming; however, most phosphate fertilizers are not completely absorbed and become unavailable to crops. Additionally, large applications of fertilizers cause soil degradation and water eutrophication (Gilbert, 2009). Therefore, improving crop P acquisition and utilization efficiency is essential for sustainable gains in agricultural production (Song et al., 2014).
To survive in low Pi environments, plants have developed a wide range of adaptive responses to improve efficiency of P-utilization (Chiou and Lin, 2011; Wu et al., 2013), such as modification of root architecture (Devaiah et al., 2007), increased transport capabilities of high-affinity Pi transporters (Qin et al., 2012; Fiorilli et al., 2013; Liu et al., 2013), and increased acid phosphatase (APase) (Xiao et al., 2006; Hur et al., 2010; Liang et al., 2010). These evolutionary strategies are precisely controlled by different mediators in a complex P regulatory network. Of these various strategies, improvement of APase activity is considered that enables plants to utilize extracellular and intracellular Po (Kaida et al., 2010; Sun et al., 2013; Wang et al., 2014).
Purple acid phosphatase (PAP) is a type of APase and contains a binuclear metal ion center, which hydrolyses a wide range of phosphate esters and anhydrides under optimal acidic conditions. PAPs have five conserved motifs (DXG/GDXXY/GNH(D/E)/VXXH/GHXH, bold letters represent invariant residues), which can coordinate the binuclear metal center to hydrolyse Po (Schenk et al., 1999; Olczak et al., 2003; Bozzo et al., 2004). Some of the biological functions of PAPs have been described including P acquisition and utilization (Ravichandran et al., 2013, 2015), peroxidation (Zhang et al., 2008), and response to salt tolerance (Liao et al., 2003; Li et al., 2008). Among these functions, P acquisition and utilization is widely studied and considered to be the most main research focus (Hurley et al., 2010; Kaida et al., 2010; Holme et al., 2016).
Multigene families of plant PAPs have been reported in Arabidopsis (Li et al., 2002), rice (Zhang et al., 2011), soybean (Li et al., 2012) and maize (Gonzalez-Munoz et al., 2015). Additionally, several of them have been studied and demonstrated to participate in Po utilization and mobilization. In Arabidopsis, AtPAP26 was induced by Pi starvation, and its ability to utilize Po has been well characterized. Atpap26-mutant plants grew much smaller than the wild-type (WT) plants under a P starvation condition (Veljanovski et al., 2006; Hurley et al., 2010). AtPAP10 is another APase gene that is induced during Pi starvation and plays an important role in plant tolerance to Pi limitation (Wang et al., 2011). In rice, OsPAP10a and OsPAP10c are induced by Pi starvation, and their over-expression increase APase activities and improve the utilization of external Po, suggesting that they are important APase genes in rice (Tian et al., 2012; Lu et al., 2016). In maize, 33 members of the PAP family have been identified, and an analysis of RNA-seq transcriptome data revealed that PAP genes were expressed in different plant tissues at multiple stages of development, suggesting they are generally important throughout the plant life cycle (Gonzalez-Munoz et al., 2015).
Soybean is an economically important nitrogen-fixing crop that is grown worldwide. Insufficient P is an important limiting factor for its growth and production (Song et al., 2014). Although PAPs are important enzymes for Po utilization that are generally induced in response to low Pi, the functions of the majority of PAPs remain unknown (Li et al., 2012). GmPAP3, the first documented PAP gene in soybean, is induced by NaCl stress, not by Pi starvation (Liao et al., 2003). GmPAP4, the second documented PAP gene, is induced by Po, and its overexpression enhanced Po utilization (Kong et al., 2014). GmPAP21, the third and most recently documented PAP gene, was induced in nodules, roots and old leaves, suggesting a potential role in P recycling in leaves and P metabolism in nodules (Li et al., 2017). In a previous study, we cloned a PAP gene in soybean, GmPAP14. It contained a 1395 bp encoding a polypeptide with 464 amino acid residues. A bioinformatics analysis predicted that GmPAP14 had the characteristics of an APase (Kong et al., 2012). In this paper, we have investigated GmPAP14 more thoroughly and demonstrated its roles in Po utilization.
The soybean cultivar zhonghuang15 (ZH15, P-high efficiency, Figure 1A) and Arabidopsis (Columbia ecotype) were used in this study. The Pi and Po treatments in this study were performed using a modified Hoagland solution containing 22.3 μg/L MnSO4⋅4H2O, 0.83 μg/L KI, 0.025 μg/L CoCl2⋅6H2O, 8.6 μg/L ZnSO4⋅7H2O, 0.025 μg/L CuSO4⋅5H2O, 6.2 μg/L H3BO3, 0.25 μg/L NaMoO4⋅2H2O, 0.278 mg/L FeSO4⋅7H2O, 0.373 mg/L Na2EDTA, 0.505 g/L KNO3, 0.49 g/L MgSO4⋅7H2O, 1.18 g/L Ca(NO3)2⋅7H2O, supplemented with either 0.137 g/L KH2PO4 (1 mmol/L, Pi) or 0.11 g/L phytate (1 mmol/L, Po), and adjusted to a pH of 6.0.
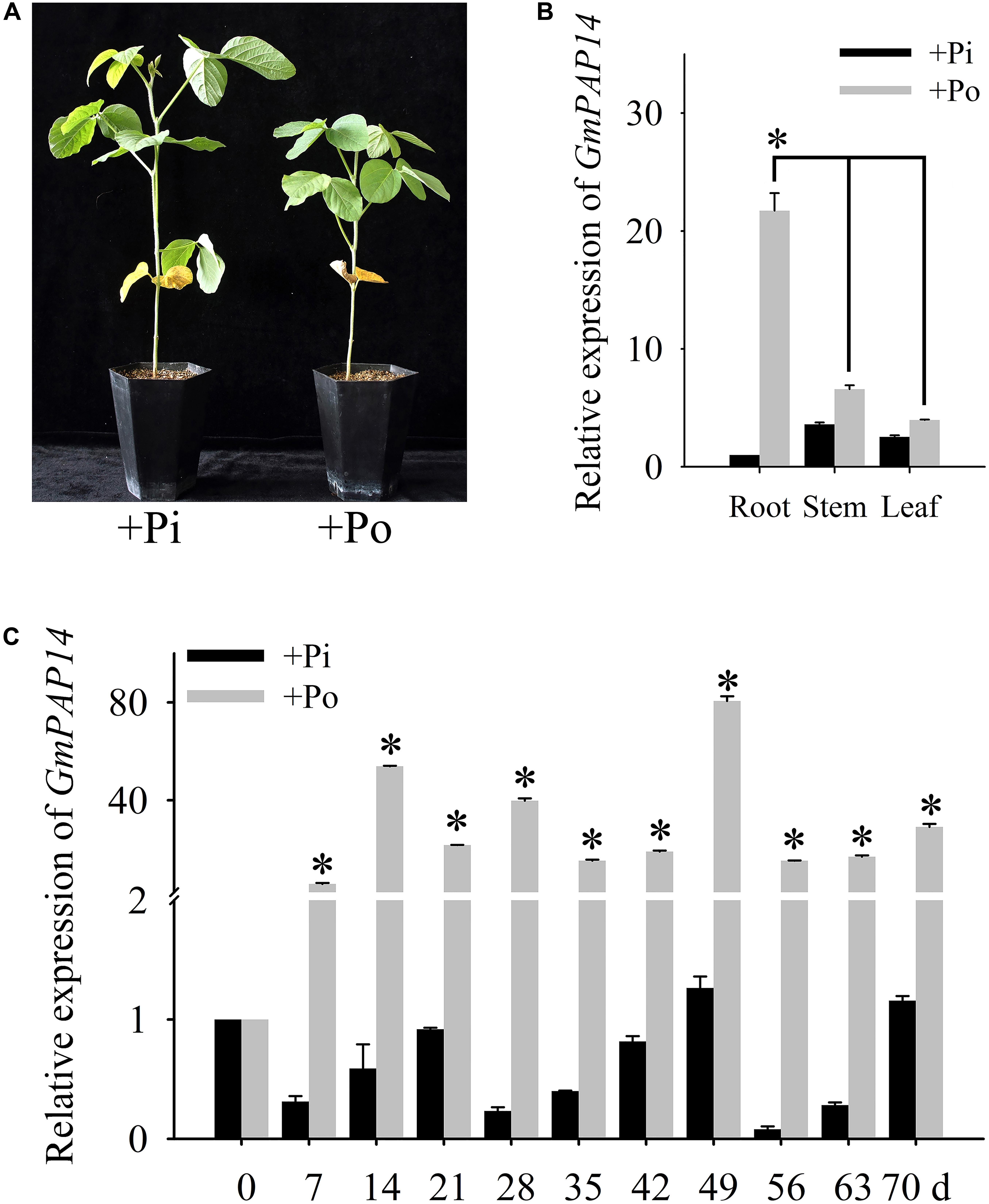
FIGURE 1. Analysis of GmPAP14 expression patterns in soybean. (A) Phenotype of zhonghuang15 (ZH15) under KH2PO4 (+Pi) and Phytate (+Po). (B) Expression analysis of GmPAP14 in different tissues of ZH15 under +Pi and +Po conditions. Seven-day-old seedlings were treated with +Pi and +Po. Seven days after treatment, roots, stems, and leaves were harvested for gene expression in different tissues. Asterisk represents significant difference of GmPAP14 expression in root compared to that in leaf and stem under +Po conditions (P < 0.05, t-test). (C) Expression of GmPAP14 in roots of ZH15. Seven-day-old seedlings were treated with +Pi and +Po (0 day was used as a control). Roots were sampled after 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70 days and were used for temporal expression analysis. Asterisks represent significant differences of GmPAP14 expression at the same growth period under +Pi and +Po conditions (P < 0.05, t-test). The relative expression value was calculated by the ratio of the expression value of GmPAP14 to that of soybean housekeeping gene GmActin11 using the 2-ΔΔCt method. Each bar is the mean of three replicates with the standard error.
The seeds of ZH15 were surface-sterilized and placed in pots with vermiculite in a greenhouse under 16 h light (28°C) /8 h dark (24°C) cycles. After 7 days (d) of growth (0 d was used as a control), the seedlings were separately treated with Pi and Po. After another 7 days, roots, stems, and leaves were harvested to profile the expression of GmPAP14 in different tissues. Roots were sampled after 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70 days and were used for temporal gene expression profiling.
Total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen, China). Then, the first-strand cDNA was synthesized with a PrimeScriptTM Reagent kit and the gDNA Eraser (Takara, Japan). Quantitative RT-PCR (qPCR) was performed with SYBR Green Mix (Takara, Japan) on a CFX96 (Bio-Rad, United States). GmPAP14 primers (5′-TCAAGCAGCCCCTTCATTAG-3′ and 5′-AGTTTTCCTTCGGCAATCTTC-3′) and primers for the housekeeping gene GmActin11 (5′-ATCTTGACTGAGCGTGG TTATTCC-3′ and 5′-GCTGGTCCTGGCTGTCTCC-3′) were used in qPCR (Wang et al., 2012). Relative expression was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). The data are presented as the means of three biological replicates with SE.
The cDNA of GmPAP14 was cloned into the EcoR I and Hind III sites of the pET32a vector and transformed in Transetta (DE3) chemically competent cells (TransGen Biotech, China). The primers used were 5′-GAATTCAAAGTTGAGAAGACAG-3′ and 5′-AAGCTTTTAATGTGAAACATGAG-3′. The transformed cells were induced with 0.6 mmol/L IPTG at 24°C for 12 h to produce the GmPAP14-His fusion protein. Recombinant GmPAP14 was purified using a His-Tagged Protein Purification Kit (CWBIO, China). APase and phytase activity measurements, thermal stability and the effect of pH on GmPAP14 activity were analyzed using the methods described by Kuang et al. (2009). All data are presented as the means of three replicates with SE.
To construct the overexpression vector, the full-length cDNA of GmPAP14 was cloned and inserted into a pCHAC vector (containing an HA tag) that had been digested with BamH I and Kpn I. The primers used for GmPAP14 cDNA amplification were 5′-GGATCCATGGGTGTTGTGGAGGGTCTCT-3′ and 5′-GGTACCATGTGAAACATGAGCCGTGGAATCAT-3′. To construct the promoter vector, a 2568-bp GmPAP14 promoter fragment was amplified by PCR using the primers 5′-AAGCTTAGTCGAAGATGGGGATGT-3′ and 5′-GGATCCTTTGCTCAAAACCCGTG-3′. The amplified fragment was cloned into the Hind III and BamH I sites of the pBI121 vector in place of the CaMV 35S promoter. The reconstructed plasmids were transferred into Agrobacterium tumefaciens GV3101 using a freeze-thaw procedure and transgenic plants were produced via Agrobacterium-mediated floral dip (Clough and Bent, 1998).
The T3 GmPAP14 promoter-GUS (PGmPAP14-GUS) transgenic plants were grown on agar under Pi and Po conditions. The roots were harvested for GUS staining after 15, 20, and 30 days, and the leaves were harvested for GUS staining after 15 days. The samples were immersed in a reaction buffer containing 50 mmol/L phosphate buffer (NaH2PO4 and Na2HPO4), pH 7.0, 10 mmol/L Na2EDTA, pH 8.0, 1 mmol/L K4Fe(CN)6⋅3H2O, 1 mmol/L K3[Fe(CN)6], 2 mmol/L X-Gluc, 0.1% Triton X-100, and incubated at 37°C overnight. After staining, the tissues were cleared with 70% ethanol. All the samples were observed and imaged using a BX51 microscope (Olympus, Japan).
Western blotting was performed as described by Wang et al. (2011). The primary antibody [HA Tag monoclonal antibody (1:5000, Thermo Fisher, United States)], and the secondary antibody [goat anti-mouse IgG(H+L)-HRP (1:5000, Beijing Protein Innovation, China)] was used for the Western blot. The blotted membrane was detected using an Odyssey FC imaging system (LI-COR, United States).
To measure secreted APase activities, 15-day-old seedlings grown in the Pi condition were transferred to 2-mL Eppendorf tubes containing 1.5 mL of a liquid medium supplemented with 1 mmol/L ρ-NPP. After being held for 1 day at 24°C, 0.5 mL of 0.5 mmol/L NaOH was added to terminate the reaction. Absorbance was measured at 410 nm (Wang et al., 2009). APase activity was expressed as ρ-NP released per hour per plant. All experiments were repeated three times, with five plants per replicate.
To measure the secreted phytase activities, we transferred 15-day-old seedlings from the Pi condition to 2-mL Eppendorf tubes containing 1.5 mL of a liquid medium with 1 mmol/L phytate. After 1 day at 24°C. The liquid medium was incubated with 0.5 mL of malachite green reagent, then measured at 650 nm (Lu et al., 2016). Phytase activity was defined as Pi released per hour per plant. All experiments were repeated three times, with five plants per replicate.
For the quantitative analysis of APase and phytase activities in culture medium, transgenic and WT plants were sown on vermiculite under Pi and Po conditions. After 30 days of growth, the proteins were concentrated from the culture medium and measured using the previously described methods.
Total P measurements were analyzed as previously described (Lu et al., 2016). First the samples were dried under 80°C to a constant weight. Then, the samples were digested in glass tubes with H2SO4 and H2O2 at 180°C. The digested solutions were cooled to room temperature, reacted with malachite green reagent, and measured at 650 nm.
The transgenic and WT plants were grown under Pi and Po conditions. After 30 days of growth the carbon assimilation rates were measured using a CIRAS-3 portable photosynthesis system (PP Systems, United States).
All data were analyzed using SPSS 17.0 software (IBM, United States). Student’s t-test was used to identify differences between the observations.
Pi deficiency significantly inhibited soybean growth (Figure 1A), in the present study, the spatial and temporal expression of GmPAP14 in response to Po was first investigated by qPCR. The results showed that GmPAP14 expression in the roots was much higher than that in the leaves and stems, suggesting that GmPAP14 was mainly expressed in the roots grown under the Po condition (Figure 1B). Subsequently, temporal expression of GmPAP14 in the roots showed that GmPAP14 was rapidly induced at 7 days post Po stress (DPP), and maintained high expression from 14 to 70 DPP, with the peak occurring at 49 DPP. In contrast, GmPAP14 expression was very low and unaffected under the Pi condition (Figure 1C).
To further verify the expression of GmPAP14, a 2568-bp GmPAP14 promoter sequence was cloned, fused to a GUS reporter gene (PGmPAP14-GUS) and transformed into WT Arabidopsis plants. First, we observed intense GUS staining in germinating seeds and cotyledons, and in the meristematic zones of the root during the early stages of seedling development (Figures 2A1,A2). In mature plants, GUS staining was predominantly detected in the root tips (Figure 2A3).
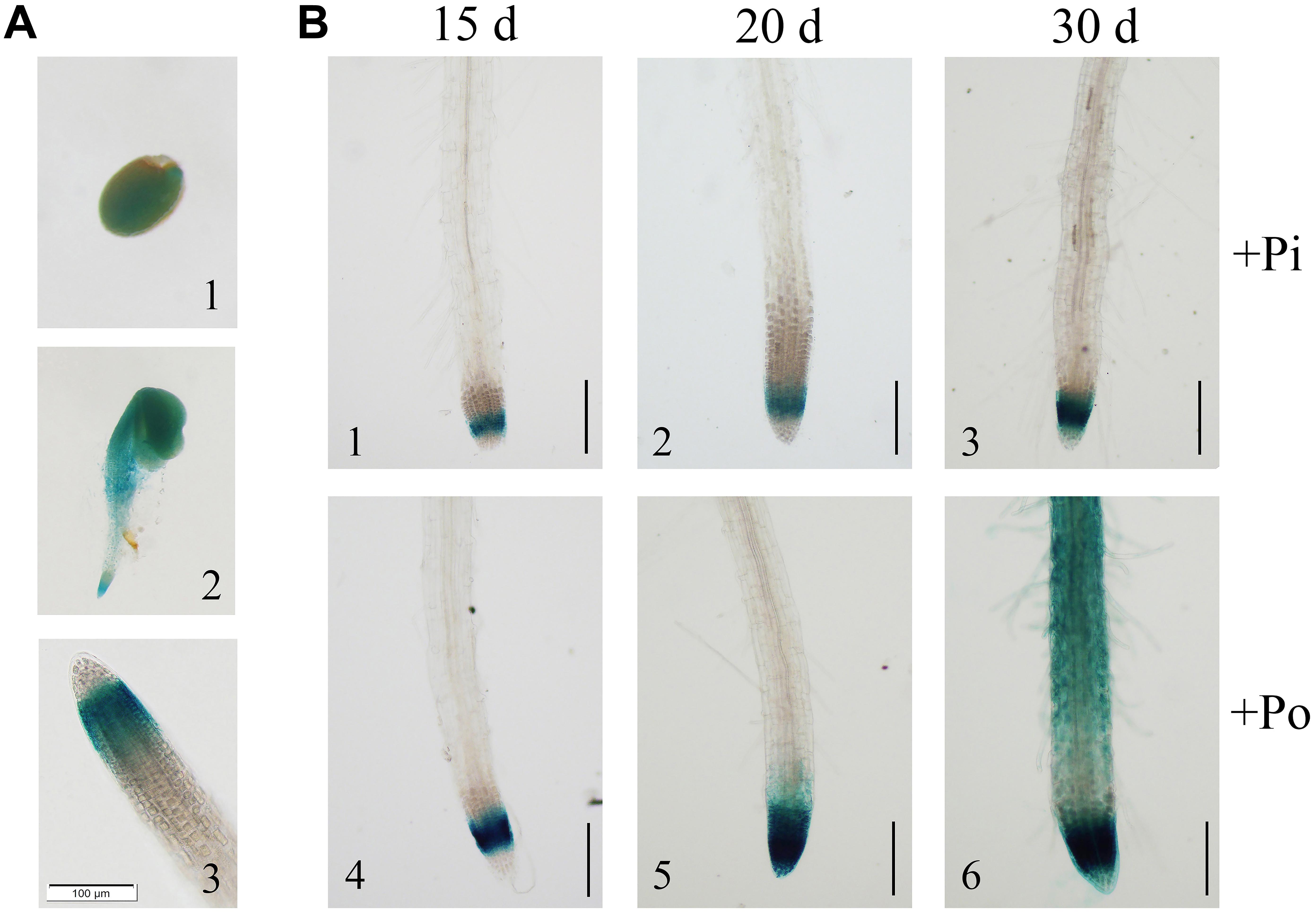
FIGURE 2. Analysis of the GmPAP14 promoter in transgenic plants containing the GmPAP14 promoter-GUS (PGmPAP14-GUS) reporter. (A) Histochemical localisation of GUS activity in transgenic plants (1) germinating seed. (2) 2-day-old seedling. (3) Root tip of 15-day-old seedling. The scale bar = 100 μm. (B) Analysis of the GmPAP14 promoter response to the Po condition. The plants were sown in vermiculite and treated with KH2PO4 (+Pi) or Phytate (+Po). After 15, 20, and 30 days, the samples were harvested for GUS staining. The scale bar = 50 μm.
Subsequently, we analyzed the response of the GmPAP14 promoter to low P at different developmental stages (Figure 2B). Stronger GUS staining was observed in the meristematic zone of the root 15 days after germination under the Po condition. As the plant developed, GUS staining was much stronger and expanded from the meristematic zone to the maturation zone and the root hairs under the Po condition. These results indicate that the GmPAP14 promoter is induced, and its expression expands under the Po condition.
GmPAP14 had all seven conserved amino acids required for its catalytic activity. To test its activity, recombinant 6 × His-GmPAP14 protein was purified by Ni-Agarose and detected on SDS–PAGE (Supplementary Figure S1A), which was further confirmed by immunoblotting with anti-His antibody (Supplementary Figure S1B). Enzymatic assays demonstrated that the pure GmPAP14 protein had APase activity with the highest activity at a pH of 7.5 (Supplementary Figure S1C). Additionally, GmPAP14 was found to be moderately thermostable with the highest activity at 35°C (Supplementary Figure S1D). An examination of its substrate specificity revealed that GmPAP14 had relatively high phytase activity (86% of APase activity, Supplementary Figure S1E).
A bioinformatics analysis showed that GmPAP14 had a signal peptide sequence at its N terminal, indicating that it might be a secreted protein. To test this hypothesis and investigate the function of GmPAP14 in utilizing extracellular Po, the full-length cDNA of GmPAP14 driven by the CaMV 35S promoter was transferred into Arabidopsis. Three independent transgenic lines expressing GmPAP14 were obtained (Figure 3A). Immunoblotting also revealed that GmPAP14 proteins were highly and stably expressed in transgenic lines (Figure 3B). Subsequently, we measured the external enzyme activities of transgenic and WT plants. The results showed that the three independent transgenic lines had higher external APase activities than the WT plants (Figure 4A). Consistent with the higher APase activities, three GmPAP14-overexpressing lines also had higher external phytase activities than the WT plants (Figure 4B). These results suggested that GmPAP14 overexpression could significantly increase secreted APase and phytase activities in Arabidopsis.
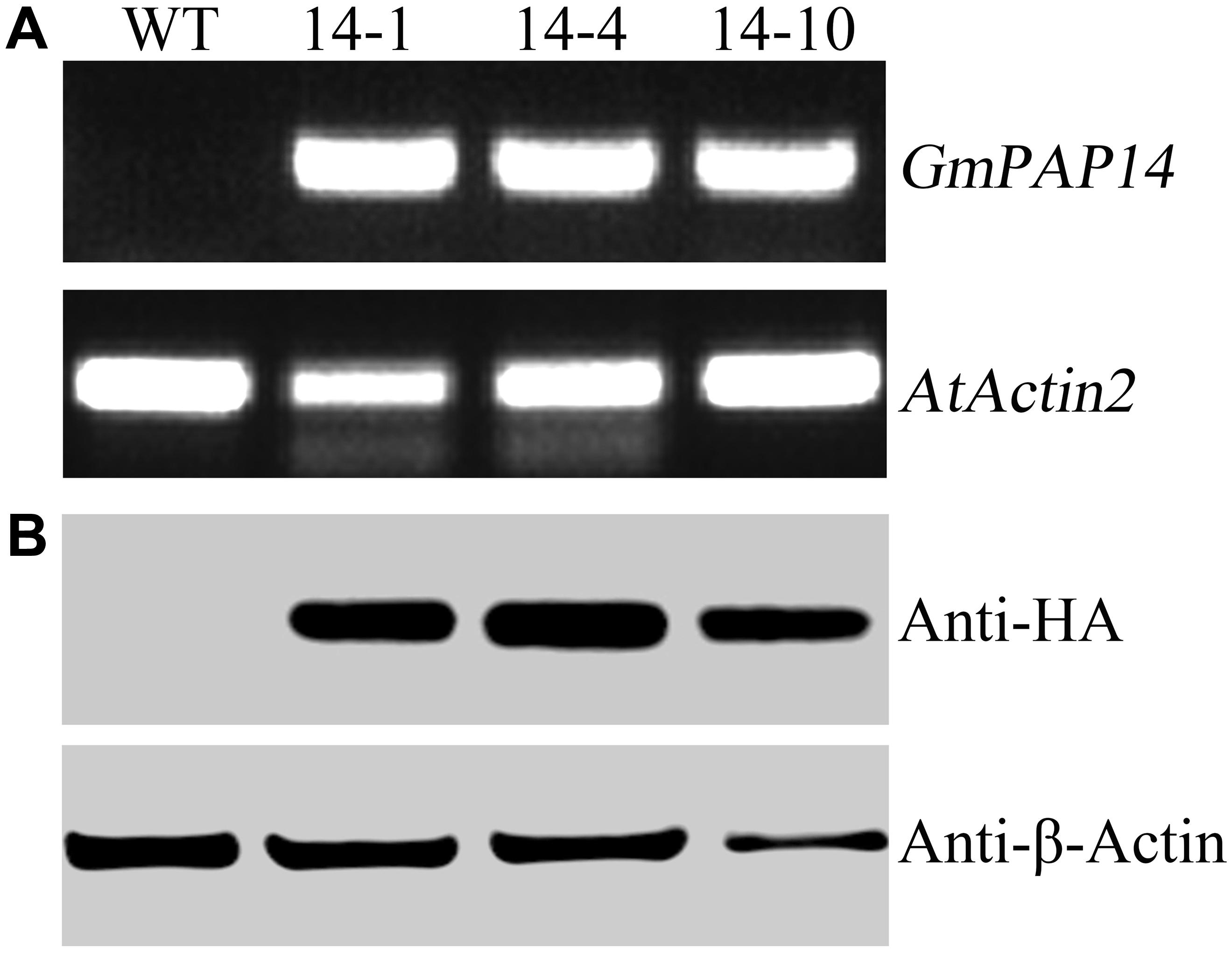
FIGURE 3. Analysis of GmPAP4 transcripts and protein levels in transgenic plants. (A) Analysis of RT-PCR in wild-type (WT) and transgenic plants. (B) Western blot detection of GmPAP14 in WT and transgenic plants. RNA and protein were extracted from 30-day-old seedlings of transgenic lines 14-1, 14-4, and 14-10. A total of 1 μg of RNA was used for RT-PCR. A total of 6 μg of protein was loaded onto an SDS–PAGE gel. Separated proteins were transferred to a nitrocellulose filter membrane and probed with anti-HA proteins.
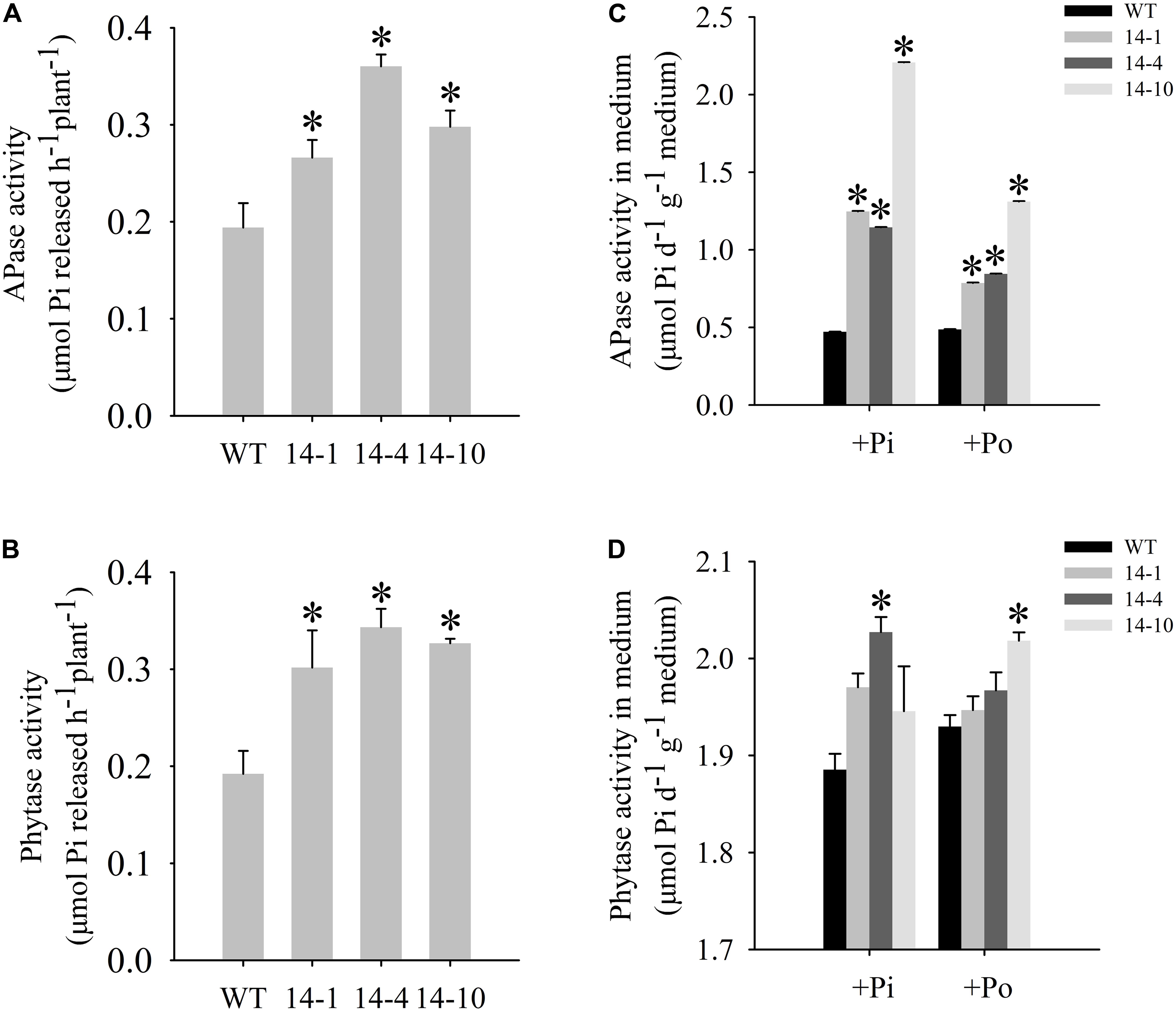
FIGURE 4. Measurement of secreted enzyme activities. (A,B) Secreted APase and phytase activities in the roots of transgenic plants (14-1, 14-4, and 14-10) and WT plants. 15-day-old seedlings from the Pi condition were transferred to 2-mL Eppendorf tubes containing 1.5 mL of a liquid medium supplemented with 1 mmol/L ρ-NPP or phytate. After being held for 1 day at 24°C, APase activity was measured at 410 nm. APase activity was expressed as ρ-NP released per hour per plant. Phytase activity was measured at 650 nm. Phytase activity was defined as Pi released per hour per plant. (C,D) APase and phytase activities in the medium of WT and transgenic plants. Transgenic and WT plants were sown on vermiculite under +Pi and +Po conditions. After 30 days of growth, the proteins were concentrated from the vermiculite, and enzyme activities were measured. The data are the mean ± SE (n = 5). Error bars represent the SE. Asterisks represent significant differences between transgenic and WT plants under the same condition (P < 0.05, t-test).
Next, we examined the APase and phytase activities of transgenic lines grown in vermiculite. The APase activity in vermiculite in which the transgenic plants were grown increased approximately 2–5 times under the Po condition compared to the activity in vermiculite in which WT plants were grown (Figure 4C). Phytase activity also increased in vermiculite in which the transgenic lines were grown (Figure 4D). These results add further support to GmPAP14 as a secreted protein and demonstrate its stability in vermiculite.
In this paper, the T3 transgenic and WT plants displayed different growth phenotypes under the Po condition. When Po was supplied, the three transgenic lines grew much better than the WT plants (Figures 5A,B) with shoot weight increases of 225.8, 54.3, and 52.8% (Figure 5C), and root weight increases of 183.9, 68.2, and 60.0% (Figure 5D). These results show that GmPAP14 overexpression significantly increases plant biomass under the Po condition.
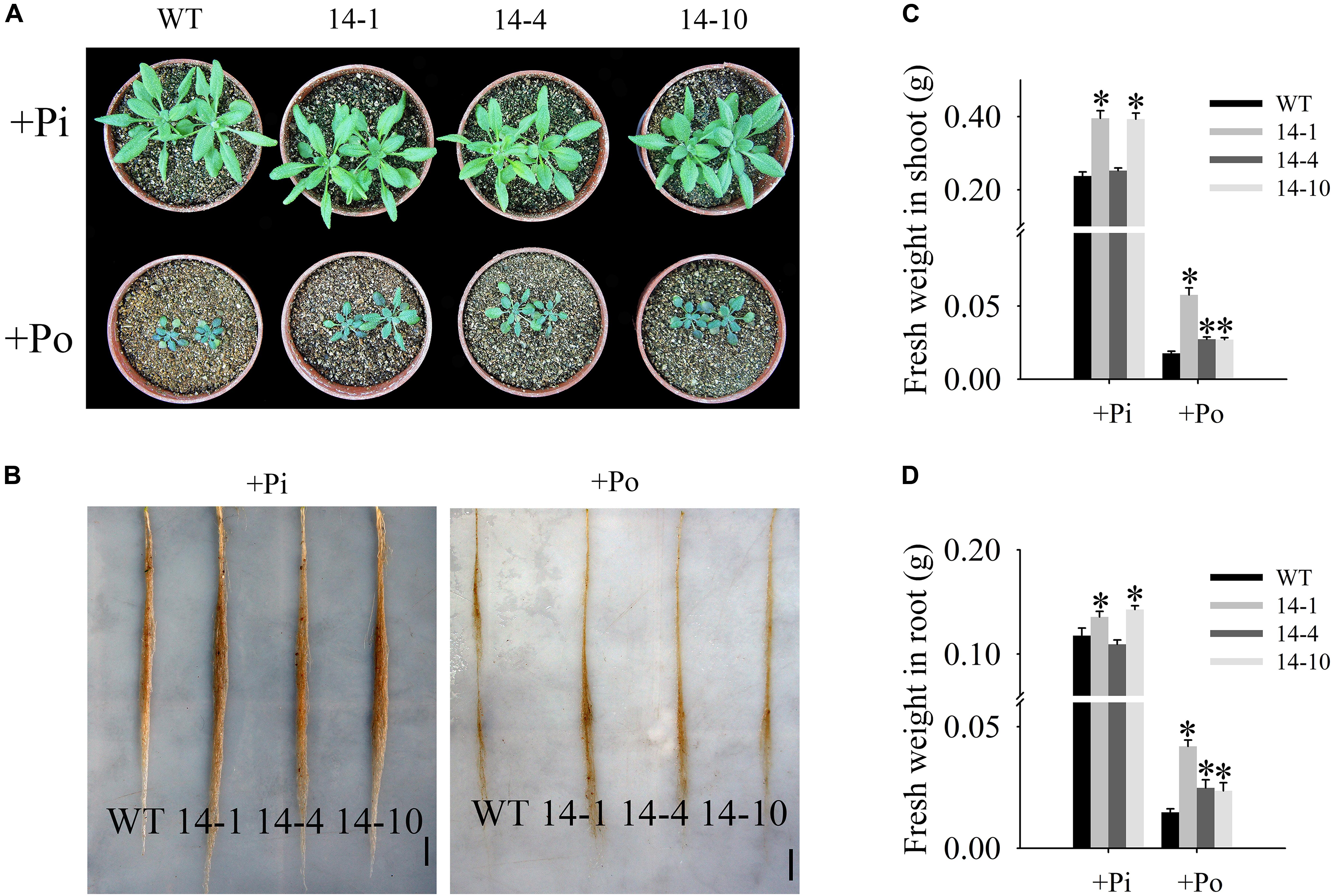
FIGURE 5. Measurement of transgenic and WT plant biomass under different phosphorus conditions. (A,B) Appearance of 30-day-old seedlings after treatment with KH2PO4 (+Pi) and Phytate (+Po). (C,D) Fresh weights of shoots and roots of transgenic and WT plants under +Pi and +Po conditions. The data are the mean ± SE (n = 5). Error bars represent the SE. Asterisks represent significant differences between transgenic and WT plants under the same condition (P < 0.05, t-test).
An analysis of Po utilization revealed that P content increased in shoots and roots of GmPAP14-overexpressing plants under the Po condition compared to the WT plants (Figures 6A,B). Additionally, P content in vermiculite in which the transgenic plants were grown significantly decreased under the Po condition (Figure 6C).
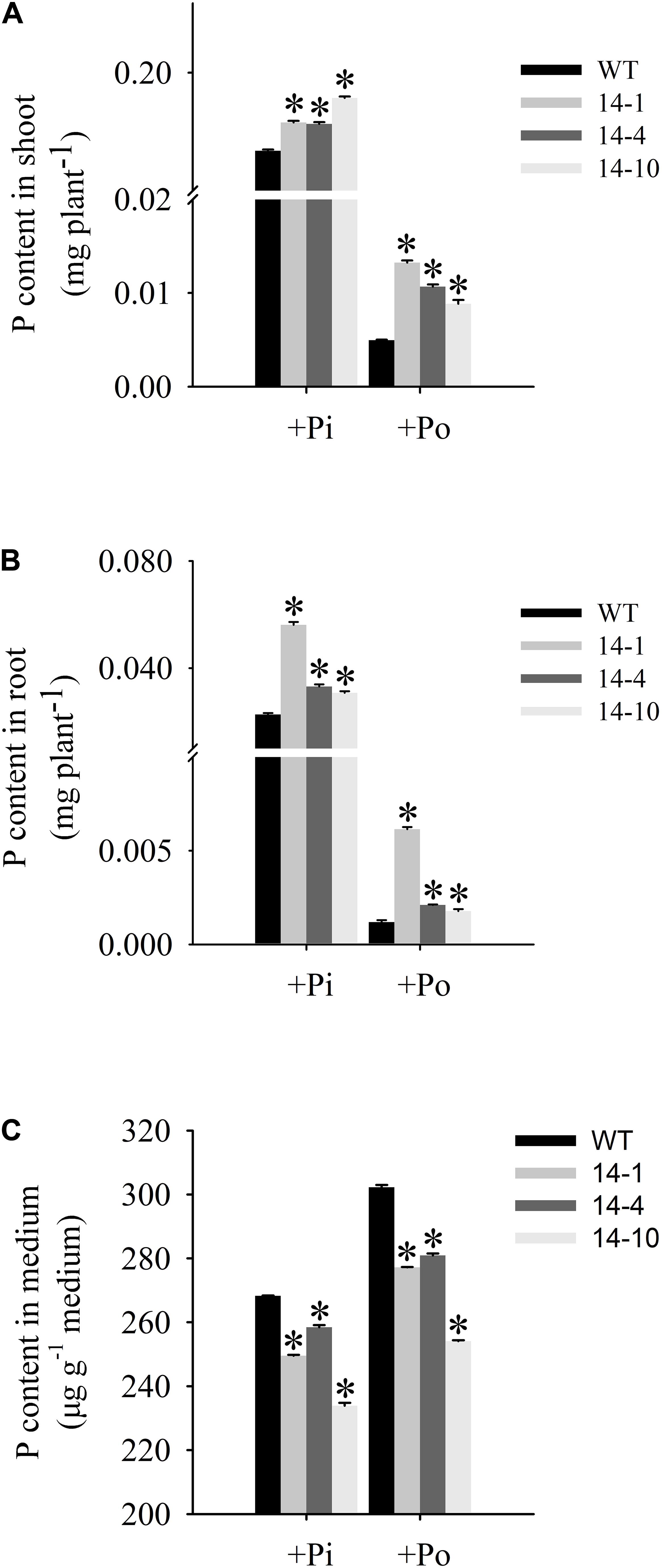
FIGURE 6. Organic phosphorus utilization efficiency of transgenic and WT plants under different phosphorus conditions. (A) P content in shoot. (B) P content in root. (C) P content in medium. After 30 days of growth, shoots, roots, and growth mediums were harvested separately for P content measurements. WT, wild-type plant. 14-1, 14-4, and 14-10: the three transgenic lines used. The data are the mean ± SE (n = 5). Error bars represent the SE. Asterisks represent significant differences between transgenic and WT plants under the same condition (P < 0.05, t-test).
One of the obvious symptoms of plant Pi stress is anthocyanin accumulation in shoots, which is believed to protect chloroplasts from photoinhibition (Vance et al., 2003). Under the Po condition, anthocyanin concentrations in three independent transgenic lines were significantly lower than those in WT plants (Figure 7A). Compared to WT plants under the Po condition, measurements of carbon assimilation rates showed that GmPAP14 overexpressing improved plant photosynthetic potential with 84.2, 100.0, and 126.3% increases in assimilation rates in the three independent transgenic lines (Figure 7B).
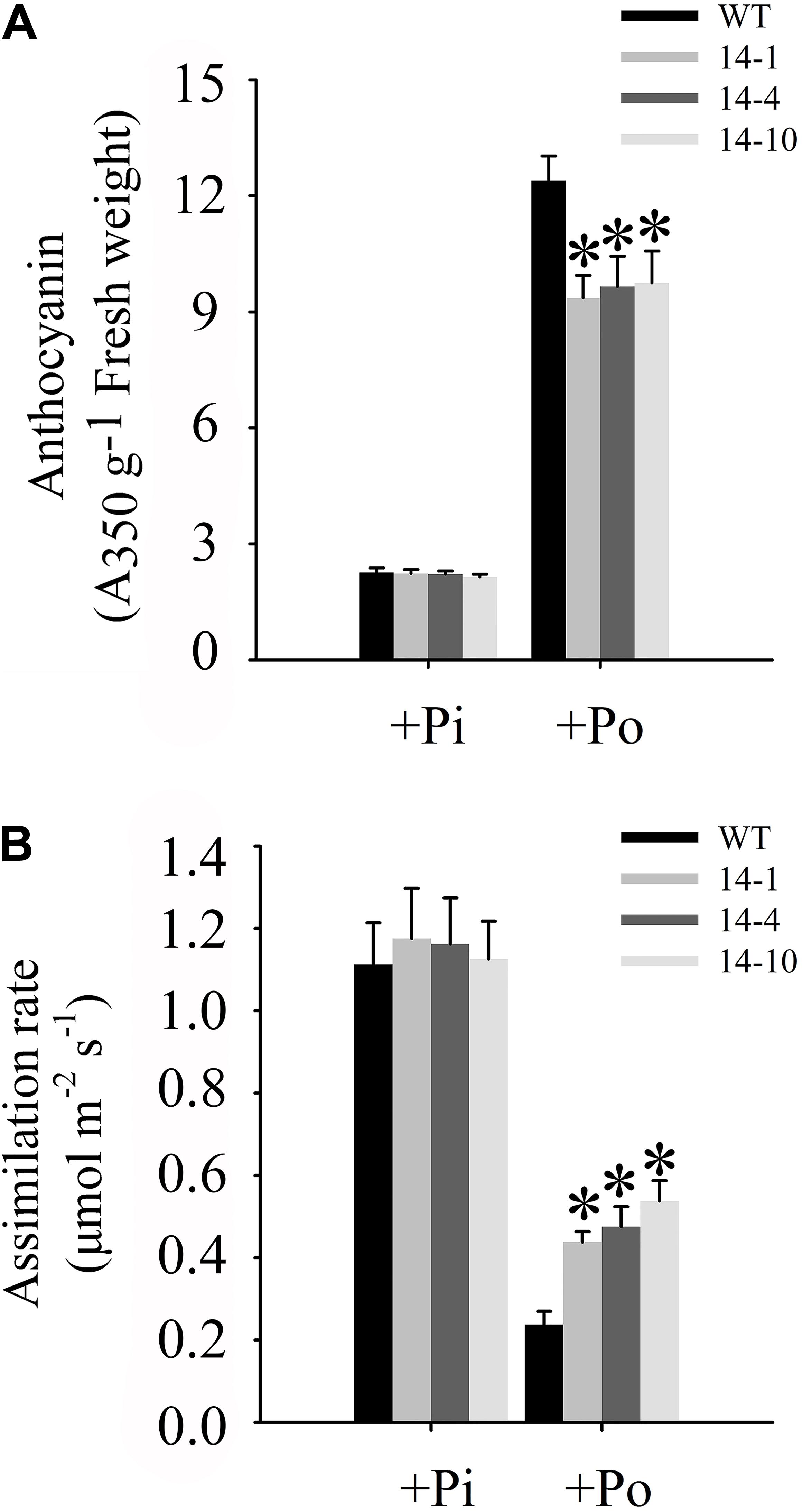
FIGURE 7. Measurements of photosynthetic efficiency in transgenic and WT plants under different phosphorus conditions. (A) Measurement of anthocyanin in transgenic lines (14-1, 14-4, and 14-10) and WT plants. (B) Assimilation rates of transgenic and WT plants were measured under +Pi and Po conditions. Transgenic and WT plants were sown on vermiculite under +Pi and +Po conditions. After 30 days of growth, leaves were harvested to measure anthocyanin and assimilation rates were measured using CIRAS-3. The data are the mean ± SE (n = 5 for A; n = 30 for B). The error bars represent the SE. Asterisks represent significant differences between transgenic and WT plants under the same condition (P < 0.05, t-test).
Acid phosphatases (EC 3.1.3.2) are considered important for plant responses to Pi starvation. Extracellular APases belong to a group of Pi starvation-inducible (PSI) phosphohydrolases secreted by roots (Tran et al., 2010). Secretion of PAPs, a specific group of PSI phosphohydrolases, has been shown to play an important role in external Po utilization (Wang et al., 2014). In this paper, GmPAP14 was mainly expressed and induced in soybean roots under the Po condition (Figure 1), indicating that GmPAP14 is a PSI gene that plays a crucial part in Po utilization. Moreover, GUS staining of transgenic plants with PGmPAP14-GUS indicated that the GmPAP14 promoter not only was strongly induced but also extended its expression region from meristematic to maturation regions and root hairs (Figure 2). In our previous study, GmPAP14 signal peptides and the prediction of its subcellular localisation suggested that it was a secreted protein (Kong et al., 2012). In our current study, increases in the secreted APase activities of transgenic plants and APase activities in vermiculite further demonstrated that GmPAP14 could be produced in roots and secreted to facilitate the utilization of more Po around the roots (Figures 4A,C).
Purple acid phosphatases hydrolyse a wide range of organic P to produce Pi with an acidic pH optimum (Tran et al., 2010). Phytate is the main contributor of Po, accounting for approximately 60% of total soil Po (Wang et al., 2013). However, only a few studies have reported on the ability of PAPs to hydrolyse phytate. AtPAP15 overexpression enhanced soybean plant growth on sand culture containing phytate (Kuang et al., 2009; Wang et al., 2009). Wang et al. (2011) reported that AtPAP10 had only moderate phytase activity (29% of APase activity), as determined by in vitro enzyme assays. In our previous work, we identified a soybean PAP gene, GmPAP4, which has phytase activity (45% of APase activity) and is involved in phytate hydrolysis and utilization (Kong et al., 2014). In this paper, we also found that GmPAP14 had characteristics of a phytase, and higher phytase activity (86% of APase activity). Evidence of GmPAP14’s phytase activity includes the following: (1) The significant induction of GmPAP14 under the phytate condition (Figure 1) and its strong expression in germinating seeds (Figure 2A) and cotyledons (Figure 2B) suggested that it might scavenge phytate. (2) In vitro enzyme assays showed that GmPAP14 possessed a higher phytase activity (86%) relative to APase activity (Supplementary Figure S1E). (3) GmPAP14 overexpression significantly improved secreted phytase activities of plants and phytase activities in vermiculite (Figures 4B,D). (4) Transgenic plants with GmPAP14 overexpression obtained more P under the phytate condition compared to WT plants (Figure 6). Cumulatively, this evidence supports the conclusion that GmPAP14 is a phytase-like PAP, playing an important role in phytate utilization.
PAPs were suggested to act as multifunctional enzymes with roles in plant growth and development and peroxidase activity, but not in Po hydrolysis (Del Pozo et al., 1999; Zhu et al., 2005). For instance, histochemical staining of transgenic plants expressing an AtPAP23 promoter-GUS reporter construct revealed that the transcription of AtPAP23 was much stronger in flowers, suggesting that AtPAP23 may play an important role in flower development (Zhu et al., 2005). In our previous work, we found that transgenic plants with 35S-GmPAP4 had more lateral roots than WT plants (Kong et al., 2014). Lateral roots are initiated from pericycle founder cells (Tang et al., 2017). Later work with GmPAP4 promoter-GUS plants also showed that GUS staining was detected in the stele (data not shown). This suggested that GmPAP4 played a role in the formation of lateral roots. However, GmPAP14, another member of the PAP family in soybean, was mainly detected in the meristematic region of the root (Figure 2). Additionally, no significant differences in lateral root development were found in transgenic plants overexpressing GmPAP14 (data not shown). Interestingly, GUS expression driven by a GmPAP14 promoter was also detected in stomatal guard cells (Supplementary Figure S2), providing novel evidence of a role for GmPAP14 in stomatal function. Although no direct support for our observations was found in the existing literature describing PAPs, several reports on AtPHO1 described it as an Pi-induced gene that was primarily expressed in the root vascular tissues and played a role in the transfer of Pi (Hamburger et al., 2002; Rouached et al., 2011). Additionally, AtPHO1 expression in guard cells influences stomatal closure in response to abscisic acid (ABA) in Arabidopsis (Zimmerli et al., 2012).
In summary, we have characterized the functions of GmPAP14. All our data clearly indicate that when plants suffer from low Pi stress, more GmPAP14 is mobilized and secreted from the root to utilize external Po. GmPAP14 expression in stomatal guard cells may also influence their response to P starvation; however, additional experiments are needed to demonstrate this. Therefore, we infer that GmPAP14 functions to improve efficiency of P-utilization and may also coordinate stomatal changes to promote photosynthesis under low-Pi conditions.
CZ, YK, and XL conceived and designed the research. YK and XL conducted the experiments and analyzed the data. BW, WL, and HD participated in parts of the experiments. YK drafted the manuscript. CZ revised the manuscript.
This research was funded by the National Natural Science Foundation of China (No. 31401405), the National Project from the Ministry of Agriculture of China (No. 2014ZX0800404B), and the Fund for Team Building of Crop Science, Hebei Agricultural University (TD2016C320).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Professor Zhiying Ma for critically reading the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00292/full#supplementary-material
FIGURE S1 | Biochemical characterization of GmPAP14. (A) SDS–PAGE analysis of purified GmPAP14. (1) protein molecular marker; (2) crude protein; (3) protein purified with Ni-agarose resin. (B) Immunoblot analysis of purified GmPAP14-His. (4) protein molecular marker; (5) GmPAP14-His hybridisation signal. (C) Thermal stability of GmPAP14. (D) pH response of GmPAP14 activity. (E) APase and phytase activities of purified GmPAP14. The data are the mean of three replicates with SE.
FIGURE S2 | Analysis of GmPAP14 promoter activity in leaves of transgenic plants. (A) Histochemical localisation of GUS activity in transgenic plants under the Pi condition. (B) Histochemical localisation of GUS activity in transgenic plants under the Po condition. The scale bar = 50 μm.
Bozzo, G. G., Raghothama, K. G., and Plaxton, W. C. (2004). Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Biochem. J. 377, 419–428. doi: 10.1042/bj20030947
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Del Pozo, J. C., Allona, I., Rubio, V., Leyva, A., De La Peña, A., Aragoncillo, C., et al. (1999). A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 19, 579–589. doi: 10.1046/j.1365-313X.1999.00562.x
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Fiorilli, V., Lanfranco, L., and Bonfante, P. (2013). The expression of GintPT, the phosphate transporter of Rhizophagus irregularis, depends on the symbiotic status and phosphate availability. Planta 237, 1267–1277. doi: 10.1007/s00425-013-1842-z
George, T. S., and Richardson, A. E. (2008). “Potential and limitations to improving crops for enhanced phosphorus utilization,” in The Ecophysiology of Plant-Phosphorus Interactions, eds J. P. Hammond and P. J. White (Dordrecht: Springer), 247–270. doi: 10.1007/978-1-4020-8435-5_11
Gilbert, N. (2009). Environment: the disappearing nutrient. Nat. News 461, 716–718. doi: 10.1038/461716a
Gonzalez-Munoz, E., Avendano-Vazquez, A. O., Montes, R. A., de Folter, S., Andres-Hernandez, L., Abreu-Goodger, C., et al. (2015). The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Front. Plant Sci. 6:341. doi: 10.3389/fpls.2015.00341
Hamburger, D., Rezzonico, E., Petétot, M. D. C., Somerville, C., and Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the Xylem. Plant Cell 14, 889–902. doi: 10.1105/tpc.000745
Holme, I. B., Dionisio, G., Madsen, C. K., and Brinch-Pedersen, H. (2016). Barley HvPAPhy_a as transgene provides high and stable phytase activities in mature barley straw and in grains. Plant Biotechnol. J. 15, 415–422. doi: 10.1111/pbi.12636
Hur, Y. J., Jin, B. R., Nam, J., Chung, Y. S., Lee, J. H., Choi, H. K., et al. (2010). Molecular characterization of OsPAP2: transgenic expression of a purple acid phosphatase up-regulated in phosphate-deprived rice suspension cells. Biotechnol. Lett. 32, 163–170. doi: 10.1007/s10529-009-0131-1
Hurley, B. A., Tran, H. T., Marty, N. J., Park, J., Snedden, W. A., Mullen, R. T., et al. (2010). The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 153, 1112–1122. doi: 10.1104/pp.110.153270
Kaida, R., Serada, S., Norioka, N., Norioka, S., Neumetzler, L., Pauly, M., et al. (2010). Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol. 153, 603–610. doi: 10.1104/pp.110.154138
Kong, Y., Li, X., Ma, J., Li, W., Yan, G., and Zhang, C. (2014). GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep. 33, 655–667. doi: 10.1007/s00299-014-1588-5
Kong, Y. B., Ning, Y. D., Liu, Y., Li, X. H., Chang, W. S., and Zhang, C. Y. (2012). Cloning and bioinformatics analysis of GmPAP14 gene in soybean. J. Agric. Univ. Hebei 35, 7–24.
Kuang, R., Chan, K. H., Yeung, E., and Lim, B. L. (2009). Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 151, 199–209. doi: 10.1104/pp.109.143180
Li, C., Gui, S., Yang, T., Walk, T., Wang, X., and Liao, H. (2012). Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 109, 275–285. doi: 10.1093/aob/mcr246
Li, C., Li, C., Zhang, H., Liao, H., and Wang, X. (2017). The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol. Plant. 159, 215–227. doi: 10.1111/ppl.12524
Li, D., Zhu, H., Liu, K., Liu, X., Leggewie, G., Udvardi, M., et al. (2002). Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 277, 27772–27781. doi: 10.1074/jbc.M204183200
Li, W. Y., Shao, G., and Lam, H. M. (2008). Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 178, 80–91. doi: 10.1111/j.1469-8137.2007.02356.x
Liang, C., Tian, J., Lam, H. M., Lim, B. L., Yan, X., and Liao, H. (2010). Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol. 152, 854–865. doi: 10.1104/pp.109.147918
Liao, H., Wong, F.-L., Phang, T.-H., Cheung, M.-Y., Li, W.-Y. F., Shao, G., et al. (2003). GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 318, 103–111. doi: 10.1016/S0378-1119(03)00764-9
Liu, X., Zhao, X., Zhang, L., Lu, W., Li, X., and Xiao, K. (2013). TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. 40, 329–341. doi: 10.1071/fp12242
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2--ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, L., Qiu, W., Gao, W., Tyerman, S. D., Shou, H., and Wang, C. (2016). OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ. 39, 2247–2259. doi: 10.1111/pce.12794
Marschner, H., and Rimmington, G. (1988). Mineral nutrition of higher plants. Plant Cell Environ. 11, 147–148. doi: 10.1111/1365-3040.ep11604921
Olczak, M., Morawiecka, B., and Watorek, W. (2003). Plant purple acid phosphatases-genes, structures and biological function. Acta Biochim. Pol. 50, 1245–1256.
Qin, L., Zhao, J., Tian, J., Chen, L., Sun, Z., Guo, Y., et al. (2012). The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159, 1634–1643. doi: 10.1104/pp.112.199786
Ravichandran, S., Stone, S. L., Benkel, B., and Prithiviraj, B. (2013). Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol. 13:107. doi: 10.1186/1471-2229-13-107
Ravichandran, S., Stone, S. L., Benkel, B., Zhang, J., Berrue, F., and Prithiviraj, B. (2015). Optimal level of purple acid phosphatase5 is required for maintaining complete resistance to Pseudomonas syringae. Front. Plant Sci. 6:568. doi: 10.3389/fpls.2015.00568
Rouached, H., Stefanovic, A., Secco, D., Bulak, A. A., Gout, E., Bligny, R., et al. (2011). Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 65, 557–570. doi: 10.1111/j.1365-313X.2010.04442.x
Schenk, G., Ge, Y., Carrington, L. E., Wynne, C. J., Searle, I. R., Carroll, B. J., et al. (1999). Binuclear metal centers in plant purple acid phosphatases: Fe–Mn in sweet potato and Fe–Zn in soybean. Arch. Biochem. Biophys. 370, 183–189. doi: 10.1006/abbi.1999.1407
Song, H., Yin, Z., Chao, M., Ning, L., Zhang, D., and Yu, D. (2014). Functional properties and expression quantitative trait loci for phosphate transporter GmPT1 in soybean. Plant Cell Environ. 37, 462–472. doi: 10.1111/pce.12170
Sun, F., Liang, C., Whelan, J., Yang, J., Zhang, P., and Lim, B. L. (2013). Global transcriptome analysis of AtPAP2–overexpressing Arabidopsis thaliana with elevated ATP. BMC Genomics 14:752. doi: 10.1186/1471-2164-14-752
Tang, L. P., Zhou, C., Wang, S. S., Yuan, J., Zhang, X. S., and Su, Y. H. (2017). FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 213, 1740–1754. doi: 10.1111/nph.14313
Tian, J., Wang, C., Zhang, Q., He, X., Whelan, J., and Shou, H. (2012). Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. J. Integr. Plant Biol. 54, 631–639. doi: 10.1111/j.1744-7909.2012.01143.x
Tran, H. T., Hurley, B. A., and Plaxton, W. C. (2010). Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci. 179, 14–27. doi: 10.1016/j.plantsci.2010.04.005
Vance, C. P. (2001). Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 127, 390–397. doi: 10.1104/pp.010331
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Veljanovski, V., Vanderbeld, B., Knowles, V. L., Snedden, W. A., and Plaxton, W. C. (2006). Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 142, 1282–1293. doi: 10.1104/pp.106.087171
Wang, F., Duan, Y., Chen, L., Wang, Y., Zhu, X., and Li, W. (2012). Glutathione peroxidase cDNA cloning and expression in soybean roots under heterodera glycines infection. J. Agric. Sci. 4, 163–171. doi: 10.5539/jas.v4n7p163
Wang, L., Li, Z., Qian, W., Guo, W., Gao, X., Huang, L., et al. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 157, 1283–1299. doi: 10.1104/pp.111.183723
Wang, L., Lu, S., Zhang, Y., Li, Z., Du, X., and Liu, D. (2014). Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 56, 299–314. doi: 10.1111/jipb.12184
Wang, X., Wang, Y., Tian, J., Lim, B. L., Yan, X., and Liao, H. (2009). Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol. 151, 233–240. doi: 10.1104/pp.109.138891
Wang, Y., Ye, X., Ding, G., and Xu, F. (2013). Overexpression of phyA and appA genes improves soil organic phosphorus utilisation and seed phytase activity in Brassica napus. PLoS One 8:e60801. doi: 10.1371/journal.pone.0060801
Wu, P., Shou, H., Xu, G., and Lian, X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16, 205–212. doi: 10.1016/j.pbi.2013.03.002
Xiao, K., Katagi, H., Harrison, M., and Wang, Z. Y. (2006). Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci. 170, 191–202. doi: 10.1016/j.plantsci.2005.08.001
Zhang, Q., Wang, C., Tian, J., Li, K., and Shou, H. (2011). Identification of rice purple acid phosphatases related to posphate starvation signalling. Plant Biol. 13, 7–15. doi: 10.1111/j.1438-8677.2010.00346.x
Zhang, W., Gruszewski, H. A., Chevone, B. I., and Nessler, C. L. (2008). An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 146, 431–440. doi: 10.1104/pp.107.109934
Zhu, H., Qian, W., Lu, X., Li, D., Liu, X., Liu, K., et al. (2005). Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 59, 581–594. doi: 10.1007/s11103-005-0183-0
Keywords: purple acid phosphatase, acid phosphatase, phytate, phosphorus utilization efficiency, soybean
Citation: Kong Y, Li X, Wang B, Li W, Du H and Zhang C (2018) The Soybean Purple Acid Phosphatase GmPAP14 Predominantly Enhances External Phytate Utilization in Plants. Front. Plant Sci. 9:292. doi: 10.3389/fpls.2018.00292
Received: 23 November 2017; Accepted: 19 February 2018;
Published: 12 March 2018.
Edited by:
Hatem Rouached, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Ajay Kumar Pandey, National Agri-Food Biotechnology Institute, IndiaCopyright © 2018 Kong, Li, Wang, Li, Du and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiying Zhang, emhhbmdjYWl5aW5nQGhlYmF1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.