- State Key Laboratory of Genetic Engineering and Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China
Leaf senescence is an integral part of plant development, and the timing and progressing rate of senescence could substantially affect the yield and quality of crops. It has been known that a circadian rhythm synchronized with external environmental cues is critical for the optimal coordination of various physiological and metabolic processes. However, the reciprocal interactions between the circadian clock and leaf senescence in plants remain unknown. Here, through measuring the physiological and molecular senescence related markers of several circadian components mutants, we found that CIRCADIAN CLOCK-ASSOCIATED 1 inhibits leaf senescence. Further molecular and genetic studies revealed that CCA1 directly activates GLK2 and suppresses ORE1 expression to counteract leaf senescence. As plants age, the expression and periodic amplitude of CCA1 declines and thus weakens the inhibition of senescence. Our findings reveal an age-dependent circadian clock component of the process of leaf senescence.
Introduction
Leaf senescence is an integral part of plant development coupled with dramatic transcriptome reprogramming and the massive breakdown and recycling of macromolecules (Lim et al., 2007; Breeze et al., 2011; Wu et al., 2012). Senescence occurs autonomously in an age-dependent manner controlled by an innate genetic program and environmental stresses. Molecular genetic studies using Arabidopsis thaliana have identified many senescence-associated genes (SAGs) (leaf senescence database1) (Li et al., 2014). Among them, ORESARA 1 (ORE1, also named NAC2, NAC6, and NAC092) is an age- and ethylene-induced senescence regulator that positively regulates senescence initiation and chlorophyll degradation (Kim et al., 2009; Qiu et al., 2015). GLKs (Golden2-like transcription factors) act as critical maintainers of chloroplast activity and overexpressing GLK2 delays leaf senescence (Waters et al., 2009). A recent study showed that the protein interaction between ORE1 and GLKs (GLK1, GLK2) regulates the balance between growth and senescence. The progressive accumulation of ORE1 counteracts the function of GLKs and eventually shifts the balance toward senescence initiation (Rauf et al., 2013).
Circadian clocks orchestrate diverse developmental and physiological processes to enhance adaptability to diurnally changing environmental cues (Harmer, 2009). Loss of circadian regulation dampens fitness (Green et al., 2002). CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and its homologous protein LATE ELONGATED HYPOCOTYL (LHY), together with TIMING OF CAB EXPRESSION 1 (TOC1), form a core loop by repressing one another’s expression (Nagel and Kay, 2012). Circadian output regulates diverse processes such as photosynthesis, chlorophyll and starch metabolism (Harmer, 2000), hormone synthesis and signaling (Zheng et al., 2015) and the stress response (Wang et al., 2011; Takeuchi et al., 2014) to modulate their activity differentially at different time points of the day, which is known as the “gating” effect.
There are increasing lines of evidence that disruptions to the circadian system can substantially impact aging in animals (Banks et al., 2016), but the reciprocal interaction between circadian clock and senescence in plant systems has not been studied. Here, we investigated leaf senescence process in circadian clock-related mutants and revealed the possible mechanisms of inhibition of CCA1 in leaf senescence. With aging, the reduced expression of CCA1 attenuates the inhibition and leads to accelerated leaf senescence.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type in this study. The mutants used have been described previously: cca1-1 (Green and Tobin, 1999) (Ws background back-crossed with Col-0 three times), cca1-11 (Hall et al., 2003) (Ws background back-crossed with Col-0 three times), lhy (Salk_031092), cca1lhy, elf3-7 (Hicks et al., 2001), and toc1-2 (Strayer et al., 2000) (C24 background back-crossed with Col-0 three times). The mutant line ore1 (SALK_090154) was obtained from ABRC, and cca1-1ore1 was generated by crossing. The CDS (coding sequence) of GLK2 was cloned into pCMBIA1302 and then transformed into Agrobacterium strain GV3101 to generate line 35S:GLK2/cca1-1. Seeds were cold-treated at 4°C for 3 days. Seedlings were grown in soil under 100 μmol m-2 s-1 fluorescent light in a 22°C growth room with relative humidity of 65%.
Electrophoretic Mobility Shift Assay
The full-length CCA1 CDS was cloned into pMAL-c5g, and the empty vector pMAL-c5g was used for MBP expression. Plasmids were transformed into Escherichia coli strain Rosetta (DE3), and the proteins were induced by 0.5 mM isopropyl thio-β-D-galactoside (IPTG) at 18°C for 16 h in 200 mL LB medium. Amylose resin (New England Biolabs, United States) was used to purify MBP-CCA1 fusion protein and MBP following the manufacturer’s instructions. EMSA was conducted as described previously (Qiu et al., 2015).
Chromatin Immunoprecipitation
Detached leaves from 4-week-old Col-0 and CCA1OX (35S:CCA1-Flag) plants (Li et al., 2011) were collected at ZT1. Flag M2 antibody (Sigma) was used for immunoprecipitation. ChIP-qPCR results were first normalized with input sample as follows: cycle threshold (Ct) = CtChIP-CtInput. Relative enrichment was then calculated as the ratio of normalized results from 35S:CCA1-Flag relative to the WT control. Primers are listed in Supplementary Table 2.
Dual-Luciferase Transient Expression Assay
The promoters of ORE1 (2043 bp) and GLK2 (1589 bp) were amplified from Col-0 genomic DNA and cloned into the transient expression vector pGreenII0800-Luc to generate reporter constructs. The full-length CCA1 CDS was cloned into the pCHF3 binary vector to generate an effector construct. The constructs were transformed into Agrobacterium strain GV3101. A mixture of cells containing constructs with both effector and reporter and pSoup-P19 was infiltrated into young Nicotiana benthamiana leaves. The firefly and Renilla luciferase activities of Nicotiana benthamiana extracts were quantified using the Dual-Luciferase Reporter Assay System (Promega, United States) and detected with a Synergy 2 multi-mode microplate (Bio-Tek).
Physiological Measurements
Relative chlorophyll contents were measured using a Konica Minolta SPAD 502 dual-wavelength chlorophyll meter; we measured three leaf areas from the tip to base, and the average value was used for further analysis. Measurements of photochemical efficiency (Fv/Fm) were conducted as described previously (Song et al., 2014). DAB staining was conducted as in a previous report (Lai et al., 2012). Ethylene treatment was conducted as described previously (Liu and Wen, 2012).
Transcript Analysis
Total RNA of leaves was extracted using an RNAiso plus kit (Takara). A Reverse Transcription System (Takara) was used for first-strand cDNA synthesis. Quantitative PCR was performed with SYBR® premix Ex TaqTM II (Takara) on the MyiQ2 Real Time PCR Detection System (Bio-Rad, Hercules, CA, United States), and ACTIN2 was used as an internal control to normalize the gene expression level. Primers used in this study are listed in Supplementary Table 2.
Results
CCA1 Negatively Regulates Leaf Senescence
To investigate the effect of the circadian clock on leaf senescence, we examined the physiological parameters of senescence in several circadian clock component mutants (cca1-1, cca1-11, lhy, cca1lhy, and elf3-7) (Figure 1). After 6 weeks’ long day growth (16 h light and 8 h dark, 16L/8D), all detected circadian mutants displayed faster progression than Col-0 of senescence related indexes. The cca1-1, cca1-11, and lhy single mutants exhibited a significantly accelerated yellowing phenotype relative to Col-0, and the cca1lhy double mutant aggravated the early senescence phenotype. elf3-7 displayed a similar phenotype with that of cca1lhy (Figure 1A). Typical rosette leaves of 6-week old plants from the different genotypes showed the leaf longevity to evident the senescence process (Figure 1B). The chlorophyll content, Fv/Fm and ion leakage of the 5th and 6th true leaves was recorded from the third week to the sixth week after germination (Figures 1C–E). The single mutants of cca1 and lhy seemed normal at the third week but initiated accelerated changes on all the measured physiological parameters from the fourth week. The mutant of cca1lhy and elf3-7 had reduced chlorophyll content and Fv/Fm at the start of measurements. It has been known that single mutant of cca1-1 and lhy flowers at a similar time to wild type under long day, but earlier than wild type under short day (Mizoguchi et al., 2002). When photoperiod was changed to 12L/12D (12 h light and 12 h dark) growth condition, early senescence process in cca1-1 and lhy was even exaggerated (Supplementary Figure 1). Moreover, the changes of transcriptional level of senescence marker genes (CAB, SAG12, and SEN1) were also enhanced in circadian mutants after 6 weeks long day growth (Figure 1F). Taken together, these phenotypic and gene expression data suggest that circadian modulators, such as CCA1, LHY and ELF3, are negative regulators of natural leaf senescence.
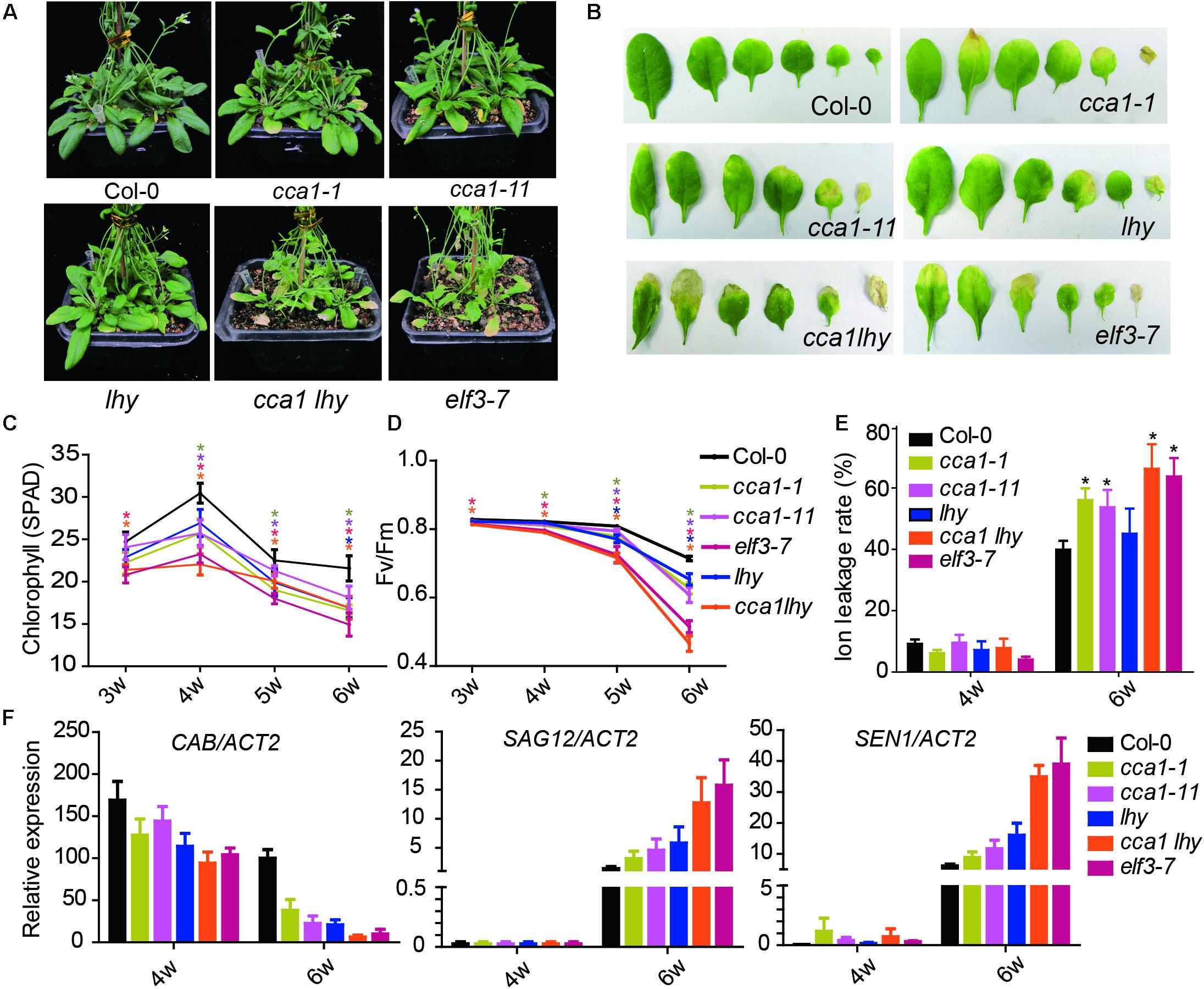
FIGURE 1. Circadian related mutants show altered physiological parameters of senescence. (A) Phenotypes of the Col-0 and circadian related mutants at 6 weeks under long day after germination. (B) Representative rosette leaves from different genotypes at 6 weeks under long day after germination. Chlorophyll contents (measured as SPAD) (C), Fv/Fm ratios (D) and ion leakage rates (E) of the 5th and 6th true leaves from different genotypes measured at 3, 4, 5, and 6 weeks after germination. Asterisks indicate where the difference between Col-0 and mutant is statistically significant (∗P < 0.05). (F) Relative expressions of CAB, SAG12, and SEN1 were detected in the 5th and 6th true leaves from 4- and 6-week-old long day grown plants. Three biological replicates were performed. ACTIN2 is an internal control for quantitative real-time PCR.
Actually, it is not surprising of the earlier senescence in these circadian mutants because some of them show earlier flowering phenotypes (Harmer and Kay, 2005). Although the molecular link between flowering and senescence is not built yet and cca1-1 displayed the similar flowering time with wild type under long day, the same chronological age but different physiological age/stage may confuse the consequence of phenotypes. Plant hormones, such as abscisic acid (ABA) and jasmonic acid, can rapidly induce detached leaf senescence (Zhu et al., 2015; Gao et al., 2016). To further verify the involvement of CCA1 in senescence, we investigated ABA and MeJA-induced leaf senescence in detached leaf of cca1-1. Before the treatment, the physiological stage (Chlorophyll contents and Fv/Fm) of leaves from Col-0 and cca1-1 is similar. After 3 days ABA and MeJA treatments, accelerated senescence was observed in cca1-1 compared with that in Col-0 (Supplementary Figure 2). CCA1 might play a part in these hormone responses, but CCA1 is involved in the regulation of leaf senescence process.
Bioinformatics Screen for Targets of CCA1
The negative regulations of CCA1 on leaf senescence could be caused by (1) the overall physiological state due to shorter period (Dodd et al., 2005; Ni et al., 2009; Graf et al., 2010; Nagel and Kay, 2012); (2) the role on hormone responses (Mizuno and Yamashino, 2008; Atamian and Harmer, 2016); (3) the suppression effect of CCA1 on senescence associated genes (SAGs), and so on. Among these possibilities, we investigated the direct regulation of CCA1 on SAGs firstly.
Two cis-elements related to circadian clocks have been well-documented: CCA1-Binding Site (CBS, AAMAATCT) and Evening Element (EE, AAAATATCT) (Harmer and Kay, 2005). To examine whether circadian regulated cis-elements are enriched in the promoter regions of senescence-associated genes (SAGs), we used the Patmatch tool on the TAIR website2 to search for CBS or EE in gene promoter regions (1000 bp sequence upstream of translation start site). According to the Tair 10 genome background, 2330 genes (6.93%) with one or more EEs and 9134 genes (27.18%) with one or more CBSs were identified among 33,602 gene promoter regions. When we examined 1961 senescence induced genes (SIGs) and 1554 senescence repressed genes (SRGs) during natural senescence (van der Graaff, 2006), we found that 30.04% (589/1961) of the SIGs and 28.44% (442/1554) of the SRGs had one or more CBSs, whereas 8.77% (172/1961) of the SIGs and 9.07% (141/1554) of the SRGs had one or more EEs in their promoters. Genes with promoter bearing either CBSs or EEs showed significant enrichment in SIGs relative to the whole genome background, and those bearing EE showed enrichment in SRGs (Fisher’s exact test, Figure 2A), indicating that senescence associated genes might be widely under the circadian regulation.
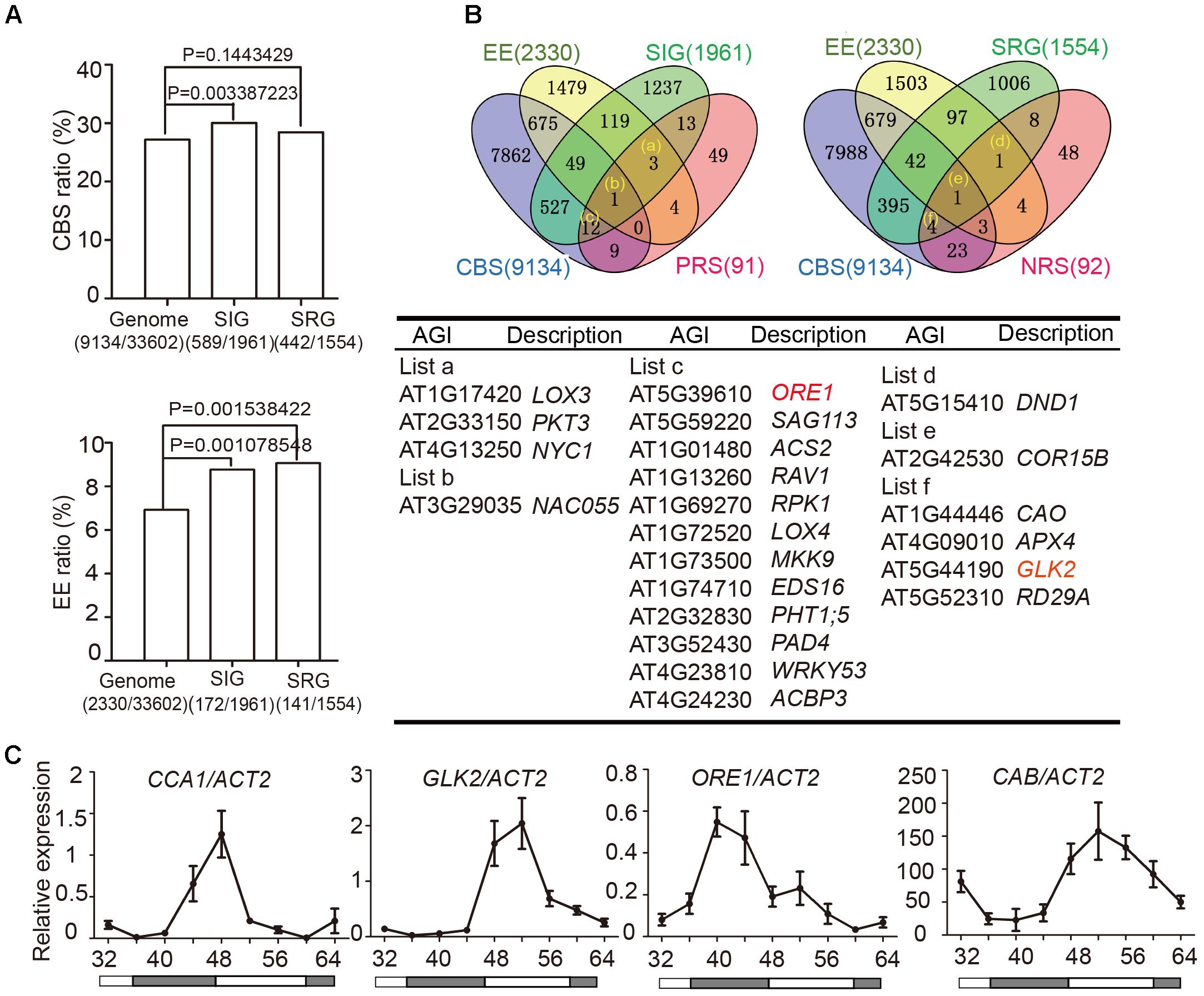
FIGURE 2. Identification of circadian-associated senescence regulators. (A) The ratios of CBS and EE in genome background, senescence induced genes (SIGs) and senescence repressed genes (SRGs), Fisher’s exact test was used in statistical analysis. (B) Venn diagrams show the intersections among EE (Evening Element genes), CBS (CCA1 Binding Site genes), SIG (Senescence Induced Genes), SRG (Senescence Repressed Genes), PRS (Positive Regulate natural Senescence genes), and NRS (Negative Regulate natural Senescence genes). The genes belong to indicated groups are listed in the table below. (C) Rhythmic expressions of CCA1, ORE1, GLK2, and CAB in the 5th and 6th true leave from 12-h light/12-h dark and then transferred to constant light. Shaded areas indicate subjective night periods. The leaves were harvested at indicated time points. Three biological replicates were performed. ACTIN2 is an internal control for quantitative real-time PCR.
We obtained 589+442 CBS-containing SIG/SRG genes and 172+141 EE-containing SIG/SRG genes. We further looked for the overlap between these candidates with 91 Positive Regulate natural Senescence genes (PRS) and 92 Negative Regulate natural Senescence genes (NRS) (Li et al., 2014) (Figure 2B and Supplementary Table 1). The overlapped genes could be the putative senescence associated targets of CCA1. Taking advantage of Diurnal database3, the diurnal transcriptional expression of some candidates were displayed in Supplementary Figure 3.
The positive senescence regulators ORE1 (Kim et al., 2009; Qiu et al., 2015) and chloroplast activity maintainer gene GLK2 (Waters et al., 2009; Rauf et al., 2013) appeared in the circadian regulated gene list and their expressional patterns are rhythmic (Figure 2B and Supplementary Figure 3). Then we conducted a 48-h time-course experiment to test the expression of CCA1, ORE1, GLK2, and CAB in 4-week old Col-0 plants grown under 12L/12D and following by constant light. The expression levels of ORE1 and GLK2 exhibited a diurnal rhythm, indicating that CCA1 might regulate the expression of these two genes (Figure 2C).
CCA1 Negatively Regulates Leaf Senescence Through Repressing ORE1 and Activating GLK2
To address the direct or indirect regulation of CCA1 on ORE1 and GLK2, we searched the CBS motif in promoter region of ORE1 and GLK2. There are two putative CBSs [-38 to -44, -457 to -465 bp upstream of TSS (translation start site)] in ORE1’s promoter and one CBS at 224 bp from the TSS in GLK2’s promoter. Electrophoretic mobility shift assay (EMSA) showed that an MBP-CCA1 fusion protein purified from E. coli binds to those CBSs in the promoters of both ORE1 and GLK2 (Figure 3A). The binding of CCA1 with ORE1-Probe1 appears to be stronger than that with ORE1-Probe2, probably due to 1 bp difference on conserved binding motif. Non-labeled DNA competitor successfully eliminated the binding ability in a dosage-dependent manner, suggesting that the binding of CCA1 to those probes is specific.
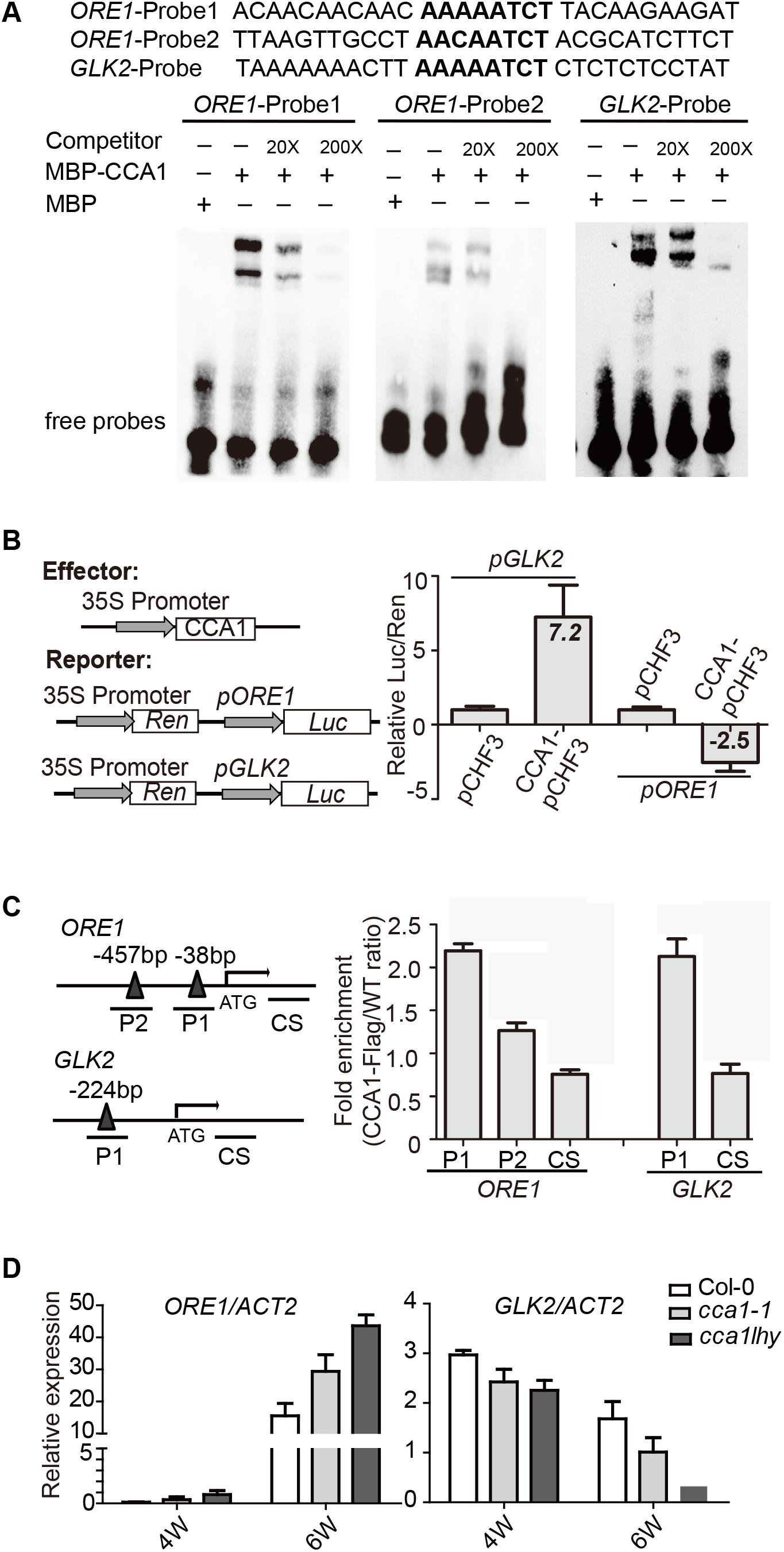
FIGURE 3. CCA1 binds to the promoters and regulates gene expression of ORE1 and GLK2. (A) CCA1 protein physically interacts with GLK2/ORE1’s promoters in EMSA assay. MBP protein was used as a negative control. The probes are listed on the top. (B) Effects of CCA1 on the activity of GLK2/ORE1 promoters in tobacco leaves. Effector construct contains CCA1 driven by CaMV 35S promoter. Reporter construct contains the Luc reporter gene driven by the promoter of GLK2/ORE1. Effects of CCA1 on GLK2/ORE1’s promoter were expressed as a ratio of Luc to Ren. Relative Luc/Ren was normalized to control (pCHF3). (C) Binding of CCA1 with the promoter of GLK2/ORE1 in vivo. ChIP assays were quantified by real-time PCR after normalizing with wild type samples. The fragments of GLK2/ORE1 coding sequences (CS) were used as negative control. Bars represent SEM, two independent experiments. Black triangles represent CBS cis-element. (D) Relative expressions of GLK2/ORE1 in 4- and 6-week-old Col-0, cca1-1, and cca1lhy double mutants. Three biological replicates were performed. ACTIN2 is an internal control for quantitative real-time PCR.
By fusing the ORE1 and GLK2 promoters with the luciferase gene, their transcriptional activation by overexpressed CCA1 can be tested in tobacco leaves. We found that CCA1 had a 7.2-fold activation effect on GLK2’s promoter and an approximately 2.5-fold inhibitory effect on ORE1’s promoter (Figure 3B).
To verify whether CCA1 interacts with ORE1/GLK2’s promoters in vivo, a 4-week-old transgenic line harboring 35S:CCA1-Flag in the Col-0 background was used for ChIP-qPCR. ChIP assays revealed that CCA1 was able to bind to the promoters of ORE1 and GLK2, including both the P1 and P2 regions, which contain CBS elements (Figure 3C).
Moreover, higher expression of ORE1 and lower expression of GLK2 were found in 6-week-old cca1-1 and cca1lhy mutants compared to that in Col-0 by qRT-PCR (Figure 3D).
These results collectively indicate that CCA1 directly binds to the promoters of ORE1 and GLK2 and regulates their expression.
ORE1 and GLK2 Act Downstream of CCA1
To discover the genetic relationship between CCA1 and its two target genes ORE1 and GLK2, we generated a cca1-1ore1 double mutant and ectopically expressed 35S:GLK2 in the cca1-1 background. The expression levels of CCA1, ORE1, and GLK2 were confirmed by qRT-PCR and western blotting in the above lines (Supplementary Figure 4). As expected, the cca1-1ore1 double mutant and cca1-1/G2OX (35S:GLK2/cca1-1) showed similar senescence process with ore1 and Col-0/G2OX (35S:GLK2/Col-0) (16L/8D), respectively. The earlier senescence phenotype of cca1-1 was reversed by knockdown of ORE1 and overexpression of GLK2 based on the measurement of chlorophyll content, Fv/Fm and ion leakage (Figures 4A–D). These data demonstrate that CCA1 negatively regulates senescence through repressing the expression of ORE1 and activating the expression of GLK2.
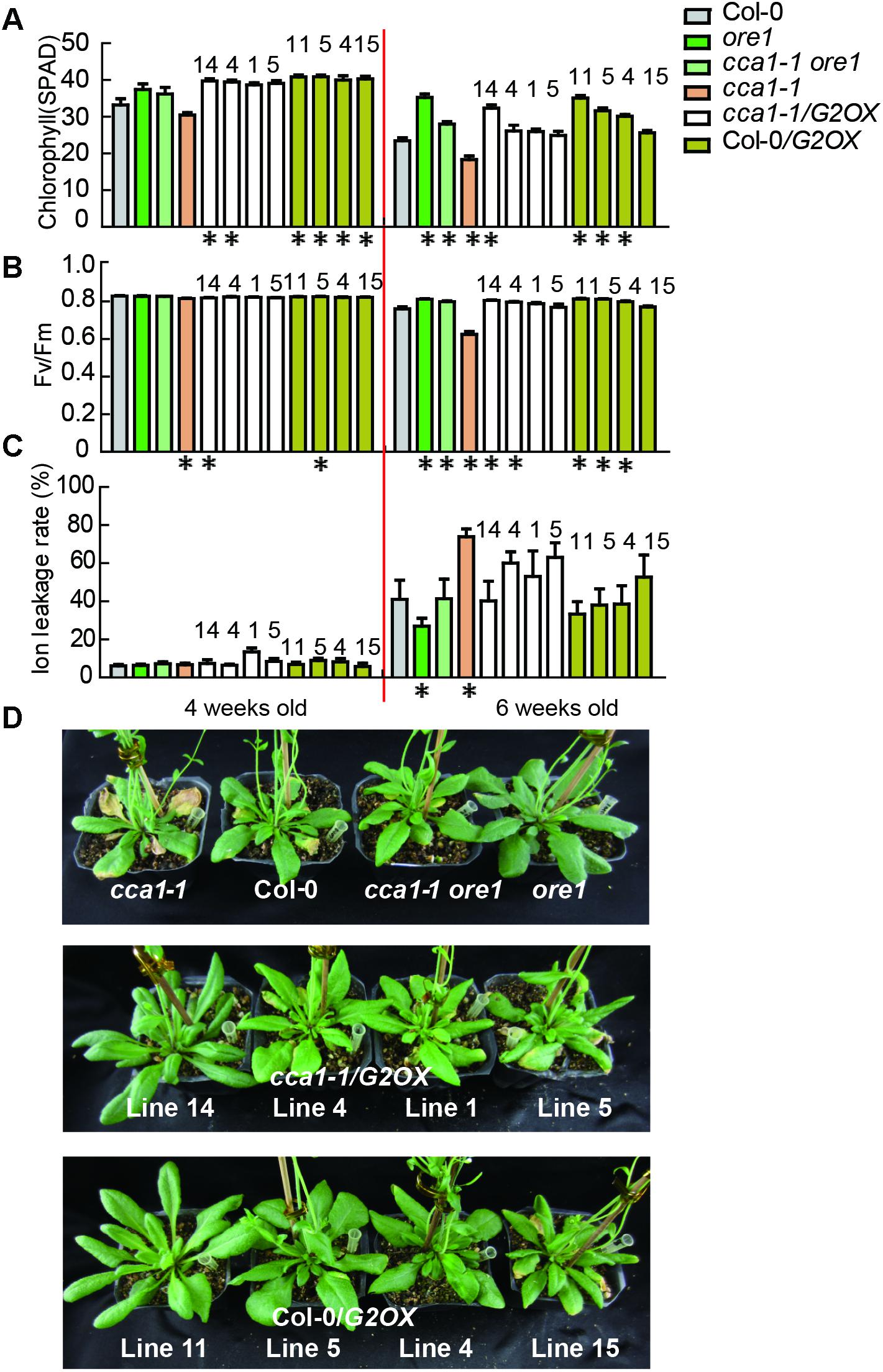
FIGURE 4. ORE1 and GLK2 act genetically downstream of CCA1. Chlorophyll contents (measured as SPAD) (A) Fv/Fm ratios (B) and ion leakage rates (C) of the 5th and 6th true leaves from different genotypes measured at 3, 4, 5, and 6 weeks under long day after germination. Four independent GLK2 overexpressed lines in Col-0 (Col-0/G2OX) or cca1-1 (cca1-1/G2OX) were shown. Asterisks indicate where the difference between Col-0 and mutant is statistically significant (∗P < 0.05). Numbers indicate different independent lines. (D) Phenotypes of different genotypes at 6 weeks under long day after germination.
Age-Declined CCA1 Attenuates the Inhibition of Leaf Senescence
Even as the circadian rhythm efficiently restricts leaf senescence during the juvenile stage, senescence occurs eventually as the leaf ages. One explanation is a reduction of the expression of circadian-related genes during aging. To test this idea, we performed time-course qRT-PCR to monitor the expression of CCA1 at both dawn and dusk at 24, 30, 36, and 42 days. We collected samples at ZT0 and ZT12 since the mRNA level of CCA1 peaked at dawn (ZT0) and decreased to a low level at dusk (ZT12) (Wang et al., 2011). Interestingly, although the expression of CCA1 oscillated between dawn and dusk, the expression and the rhythmically amplitude were significantly decreased with aging (Figure 5A). Moreover, besides CCA1, the expressions of circadian component ELF3 and CHE were also decreased and the rhythmically amplitudes of CO and JMJD5 dramatically reduced with aging (Supplementary Figure 5). With transcriptional decline of CCA1, the expression levels of ORE1 and GLK2 followed their regulation by CCA1 (Figures 5B,C and Supplementary Figure 5). Further, the expression of PIF3, PIF4 and PIF5 increased with aging (Figures 5D–F and Supplementary Figure 5). Thus, age-declined CCA1 and GLK2 attenuate the inhibition of leaf senescence and accumulated PIFs and ORE1 promote senescence initiation.
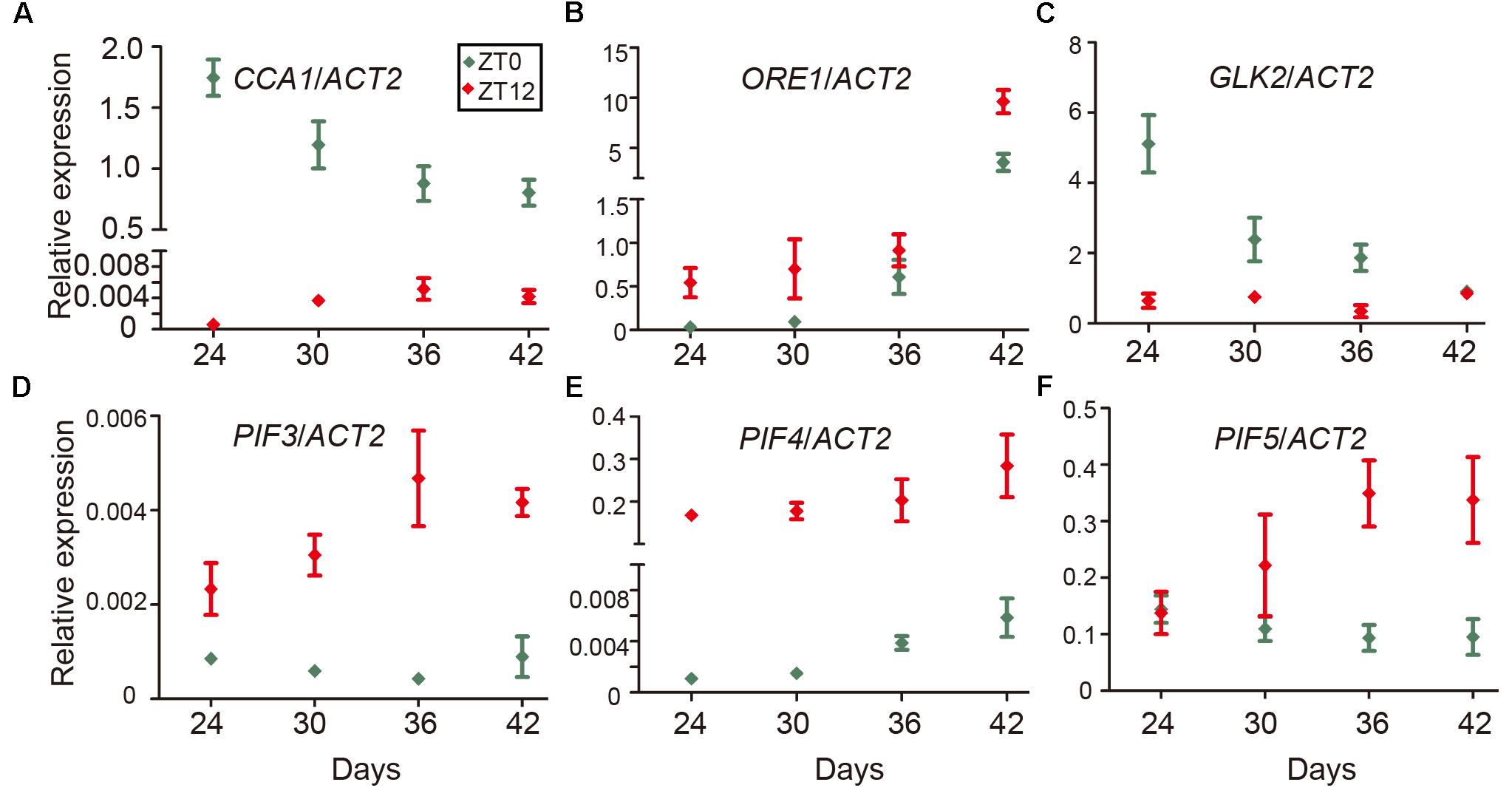
FIGURE 5. Expression changes of CCA1, ORE1, GLK2, and PIFs with aging. Relative expression levels of CCA1 (A), ORE1 (B), GLK2 (C), PIF3 (D), PIF4 (E), and PIF5 (F) in the 5th and 6th true leaves of wild type from the indicated days (Y-axis) after germination. Relative expression of genes at ZT0 and ZT12 were measured for each time point. Bars represent the mean with SEM, n = 3.
Discussion
Leaf senescence is a complex degenerative process under tight genetic control and also affected by environmental signal. Circadian resonance is likely to provide advantageous adaptation in all kingdoms, including plants. It has been reported that plant circadian clocks are involved in the regulation of photosynthesis, growth, survival, and competitive advantage (Dodd et al., 2005). Whether circadian clocks prevent leaf senescence in plants is not clear. Several senescence-associated genes have been reported to be regulated by circadian clocks. For example, the abundance of NPR1 (Non-expressor of Pathogenesis-Related gene 1) is circadian gated (Zheng et al., 2015; Zhou et al., 2015) and promotes leaf senescence (Morris et al., 2000; Fitzgerald et al., 2004). PIFs, the positive regulators of leaf senescence (Sakuraba et al., 2014; Song et al., 2014), diurnally oscillate at the transcriptional/translational levels (Soy et al., 2016). Circadian evening complex component ELF3 regulates dark-induced leaf senescence (Sakuraba et al., 2014). These findings raised the question of whether the circadian clock regulates plant senescence.
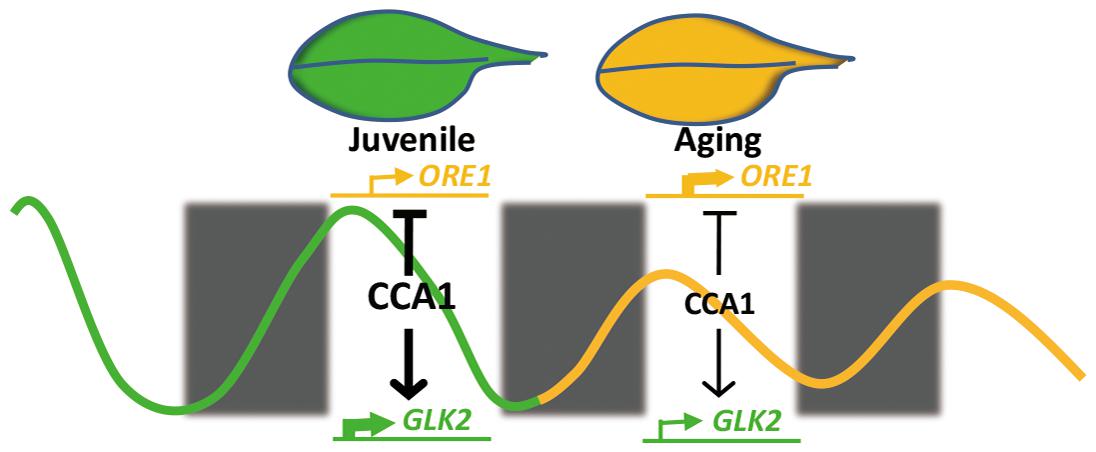
Our current study demonstrates that central circadian clock component CCA1 appears to regulate the expression of ORE1 and GLK2 to inhibit leaf senescence at the juvenile stage. With aging, declined CCA1 releases the inhibition and promotes the senescence initiation (Figure 6). This proposed mechanistic model is based on several evidences: (i) cca1 displays the accelerated yellowing phenotype; (ii) ORE1 and GLK2 belong to targets of CCA1; (iii) CCA1 directly binds to the promoter of ORE1 and GLK2; (iv) the expression of ORE1 and GLK2 is altered in cca1; (v) knocking down the expression of ORE1 and overexpressing of GLK2 rescued the earlier senescence phenotype of cca1; (vi) the transcriptional level of CCA1 decreased with aging.
As a central circadian rhythm regulator, CCA1 occupied more than 1000 genomic regions identified in the seedling stage according to ChIP sequencing data (Nagel et al., 2015). Among the targets of CCA1, although many of the target genes are repressed by CCA1, some of them are still activated, such as LHCB1.1 (CAB2) (Wang et al., 1997; Wang and Tobin, 1998). The flanking regions of CBS or EE and different interacting partners of CCA1 might modulate the expression of target genes and lead to the different action of CCA1 (Harmer and Kay, 2005). The confirmation of target genes of CCA1 emphasizes the broad role of the plant clock in regulating multiple biological functions. Based on our bioinformatics comparative analysis, ORE1 and GLK2 were identified and experimentally confirmed in ChIP-PCR assay using pre-senescence leaves. However, they are not shown in ChIP-Sequencing dataset using young seedlings, probably because co-partners of CCA1 are different during distinct developmental stages. Several interacting partners of CCA1 in early developmental stage have been published (Sugano et al., 1998; Andronis et al., 2008; Lau et al., 2011). Future work to identify CCA1 co-partners during senescence stage will be desired. Besides ORE1 and GLK2, more targets of CCA1 probably also contribute to leaf senescence, such as photosynthesis related genes including PORB (Ni et al., 2009), reactive oxygen species (ROS) homeostasis related genes (Lai et al., 2012).
Interestingly, CCA1 positively regulates GLK2 and negatively regulates ORE1 expression, which was reminiscent of a similar model that stress related senescence regulator ATAF1 executes its physiological role by simultaneously affecting both GLK1 and ORE1 (Garapati et al., 2015). Decreased expression of GLKs and enhanced accumulation of ORE1 constitutes a molecular transition mechanism from growth to senescence. These two key molecules of leaf senescence were regulated not only by CCA1, also by several modulators, such as EIN3 (Qiu et al., 2015), PIF4/5 (Sakuraba et al., 2014; Song et al., 2014). With aging, the dramatically transcriptional changes of these two genes were co-regulated by multiple signal pathways in senescence initiation.
In addition to the role of CCA1 on downstream gene regulation, the misalignment of the internal circadian clock and the external influence of zeitgebers in cca1 leads to complicated effects on plant metabolism that probably also contribute to reduced longevity in our detected condition. To assess the effect of circadian period on leaf senescence, we conducted a preliminary experiment to detect the senescence phenotype of plants grown in two distinct photoperiods (Photoperiod 1: 16 h light and 8 h dark; Photoperiod 2: 8 h light and 4 h dark). Plants grown in 16L/8D and 8L/4D showed similar chlorophyll content and Fv/Fm ratio at 4 weeks after sowing; however, plants in 8L/4D showed a more severe leaf yellowing phenotype relative to plants in 16L/8D at 6 weeks (Supplementary Figure 6), which indicates that accelerated circadian rhythm promotes leaf senescence in Arabidopsis. Besides the effect of photoperiod on leaf senescence, future work will be also required to figure out whether the circadian clock components regulate leaf senescence though modulating the hormone responses.
Taken together, we provided insights to how a circadian clock component regulates leaf senescence and how the expression of CCA1 is changed during aging. Our work suggests that circadian system acts as a timekeeper of natural senescence and enhances survival by timing initiation of leaf senescence.
Author Contributions
YS, YJ, BK, and LL designed the research; YS and YJ performed the experiments; YS, YJ, and LL analyzed the data; LL wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Key R&D Program of China (Grant No. 2017YFA0503800) and the National Natural Science Foundation of China (Grant Nos. 31470374, 31500973).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Guodong Ren (Fudan University, China) and Dr. Chi-Kuang Wen (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, China) for critically reading the manuscript. We thank Dr. Hongtao Liu (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, China) for providing seeds of cca1-1, cca1-11, cca1lhy, lhy, elf3-7, and toc1 and Dr. Gang Li (Shandong Agricultural University, China) for sharing seeds of 35S:CCA1-Flag (Li et al., 2011).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00280/full#supplementary-material
Footnotes
References
Andronis, C., Barak, S., Knowles, S. M., Sugano, S., and Tobin, E. M. (2008). The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 1, 58–67. doi: 10.1093/mp/ssm005
Atamian, H. S., and Harmer, S. L. (2016). Circadian regulation of hormone signaling and plant physiology. Plant Mol. Biol. 91, 691–702. doi: 10.1007/s11103-016-0477-4
Banks, G., Nolan, P. M., and Peirson, S. N. (2016). Reciprocal interactions between circadian clocks and aging. Mamm. Genome 27, 332–340. doi: 10.1007/s00335-016-9639-6
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Fitzgerald, H. A., Chern, M. S., Navarre, R., and Ronald, P. C. (2004). Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 17, 140–151. doi: 10.1094/mpmi.2004.17.2.140
Gao, S., Gao, J., Zhu, X., Song, Y., Li, Z., Ren, G., et al. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9, 1272–1285. doi: 10.1016/j.molp.2016.06.006
Garapati, P., Xue, G. P., Munne-Bosch, S., and Balazadeh, S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. doi: 10.1104/pp.15.00567
Graf, A., Schlereth, A., Stitt, M., and Smith, A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. U.S.A. 107, 9458–9463. doi: 10.1073/pnas.0914299107
Green, R. M., Tingay, S., Wang, Z. Y., and Tobin, E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584. doi: 10.1104/pp.004374
Green, R. M., and Tobin, E. M. (1999). Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. U.S.A. 96, 4176–4179. doi: 10.1073/pnas.96.7.4176
Hall, A., Bastow, R. M., Davis, S. J., Hanano, S., McWatters, H. G., Hibberd, V., et al. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15, 2719–2729. doi: 10.1105/tpc.013730
Harmer, S. L. (2000). Orchestrated transcription of key pathways in Arabidopsis by the Circadian Clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Harmer, S. L., and Kay, S. A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17, 1926–1940. doi: 10.1105/tpc.105.033035
Hicks, K. A., Albertson, T. M., and Wagner, D. R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13, 1281–1292. doi: 10.1105/tpc.13.6.1281
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Lai, A. G., Doherty, C. J., Mueller-Roeber, B., Kay, S. A., Schippers, J. H., and Dijkwel, P. P. (2012). CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134. doi: 10.1073/pnas.1209148109
Lau, O. S., Huang, X., Charron, J. B., Lee, J. H., Li, G., and Deng, X. W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43, 703–712. doi: 10.1016/j.molcel.2011.07.013
Li, G., Siddiqui, H., Teng, Y., Lin, R., Wan, X. Y., Li, J., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13, 616–622. doi: 10.1038/ncb2219
Li, Z., Zhao, Y., Liu, X., Peng, J., Guo, H., and Luo, J. (2014). LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res. 42, D1200–D1205. doi: 10.1093/nar/gkt1061
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu, Q., and Wen, C. K. (2012). Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol. 158, 1193–1207. doi: 10.1104/pp.111.187757
Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H. R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. doi: 10.1016/S1534-5807(02)00170-3
Mizuno, T., and Yamashino, T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49, 481–487. doi: 10.1093/pcp/pcn008
Morris, K., MacKerness, S. A., Page, T., John, C. F., Murphy, A. M., Carr, J. P., et al. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23, 677–685. doi: 10.1046/j.1365-313x.2000.00836.x
Nagel, D. H., Doherty, C. J., Pruneda-Paz, J. L., Schmitz, R. J., Ecker, J. R., and Kay, S. A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E4802–E4810. doi: 10.1073/pnas.1513609112
Nagel, D. H., and Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Ni, Z., Kim, E. D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Qiu, K., Li, Z., Yang, Z., Chen, J., Wu, S., Zhu, X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11:e1005399. doi: 10.1371/journal.pgen.1005399
Rauf, M., Arif, M., Dortay, H., Matallana-Ramirez, L. P., Waters, M. T., Gil Nam, H., et al. (2013). ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 14, 382–388. doi: 10.1038/embor.2013.24
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., and Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4 and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Soy, J., Leivar, P., González-Schain, N., Martín, G., Diaz, C., Sentandreu, M., et al. (2016). Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113, 4870–4875. doi: 10.1073/pnas.1603745113
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Sugano, S., Andronis, C., Green, R. M., Wang, Z. Y., and Tobin, E. M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. U.S.A. 95, 11020–11025. doi: 10.1073/pnas.95.18.11020
Takeuchi, T., Newton, L., Burkhardt, A., Mason, S., and Farre, E. M. (2014). Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles. J. Exp. Bot. 65, 6003–6012. doi: 10.1093/jxb/eru339
van der Graaff, E. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. doi: 10.1104/pp.106.079293
Wang, W., Barnaby, J. Y., Tada, Y., Li, H., Tor, M., Caldelari, D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. doi: 10.1038/nature09766
Wang, Z. Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M. S., and Tobin, E. M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. doi: 10.1105/tpc.9.4.491
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. doi: 10.1016/S0092-8674(00)81464-6
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Wu, X. Y., Kuai, B. K., Jia, J. Z., and Jing, H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54, 936–952. doi: 10.1111/jipb.12005
Zheng, X.-Y., Zhou, M., Yoo, H., Pruneda-Paz, J. L., Spivey, N. W., Kay, S. A., et al. (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. U.S.A. 112, 9166–9173. doi: 10.1073/pnas.1511182112
Zhou, M., Wang, W., Karapetyan, S., Mwimba, M., Marqués, J., Buchler, N. E., et al. (2015). Redox rhythm reinforces the circadian clock to gate immune response. Nature 523, 472–476. doi: 10.1038/nature14449
Keywords: circadian clock, leaf senescence, CCA1, ORE1, GLK2, aging
Citation: Song Y, Jiang Y, Kuai B and Li L (2018) CIRCADIAN CLOCK-ASSOCIATED 1 Inhibits Leaf Senescence in Arabidopsis. Front. Plant Sci. 9:280. doi: 10.3389/fpls.2018.00280
Received: 22 November 2017; Accepted: 19 February 2018;
Published: 06 March 2018.
Edited by:
Hong Qiao, University of Texas at Austin, United StatesReviewed by:
Shangwei Zhong, Peking University, ChinaLei Wang, Institute of Botany, Chinese Academy of Sciences, China
Copyright © 2018 Song, Jiang, Kuai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, bGlubGlAZnVkYW4uZWR1LmNu
† These authors have contributed equally to this work.
 Yi Song
Yi Song Yupei Jiang
Yupei Jiang Benke Kuai
Benke Kuai Lin Li
Lin Li