
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 05 March 2018
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00247
A growing body of evidence demonstrates a significant relationship between cellular redox state and circadian rhythms. Each day these two vital components of plant biology influence one another, dictating the pace for metabolism and physiology. Diverse environmental stressors can disrupt this condition and, although plant scientists have made significant progress in re-constructing functional networks of plant stress responses, stress impacts on the clock-redox crosstalk is poorly understood. Inter-connected phenomena such as redox state and metabolism, internal and external environments, cellular homeostasis and rhythms can impede predictive understanding of coordinated regulation of plant stress response. The integration of circadian clock effects into predictive network models is likely to increase final yield and better predict plant responses to stress. To achieve such integrated understanding, it is necessary to consider the internal clock not only as a gatekeeper of environmental responses but also as a target of stress syndromes. Using chlorophyll fluorescence as a reliable and high-throughput probe of stress coupled to functional genomics and metabolomics will provide insights on the crosstalk across a wide range of stress severity and duration, including potential insights into oxidative stress response and signaling. We suggest the efficiency of photosystem II in light conditions (Fv′/Fm′) to be the most dynamic of the fluorescence variables and therefore the most reliable parameter to follow the stress response from early sensing to mortality.
The link between the circadian clock and oxygenic metabolism is likely to have originated with the rise in oxygen concentration 3 billion years ago, when early photosynthetic bacteria started to use water as an electron donor. Given the lack of redox systems and the loss of an endogenous clock in archaeal taxa living in the absence of oxygen, the co-evolution of cellular clockwork and aerobic metabolism seems extremely probable (Schippers et al., 2013). It is thus reasonable to expect the Great Oxidation Event would cause selection for organisms capable of respiring and/or evolving molecular oxygen, with the most successful organisms acquiring Reactive Oxygen Species (ROS) removal systems to avoid relegation to anaerobic niches (Edgar et al., 2012).
Although the clock/redox state relationship lasts for the entire life of a plant, our current understanding of it is very limited. We present current advances in the study of the clock/redox state association in plants with particular attention to the influence of environmental stressors on this dynamic duo. We advocate for the use of chlorophyll a fluorescence, not only to monitor the plant physiological status during and after stress, but also to gain relevant information on possible clock alterations caused by this disturbance.
All living things on Earth encounter daily oscillations in environmental factors. A diverse range of organisms has evolved an endogenous clock, which permits anticipation of predictable fluctuations in environmental conditions arising from the daily rotation of our planet (McClung, 2006), enabling organisms to coordinate the timing of biological processes with the environmental conditions (Pittendrigh, 1960). The clock may be entrained by diverse external inputs, providing circadian (circa, about, and dian, a day) rhythms with a periodicity of approximately 24-h (DeCoursey et al., 2000; Green et al., 2002), which persist under constant conditions (de Mairan, 1729; Salome and McClung, 2004; Harmer, 2009; McClung, 2011; Guerriero et al., 2012). The resonance between endogenous clock and exogenous cycles affects performance (Todd et al., 2003; Dodd et al., 2005; de Montaigu et al., 2015; de Montaigu and Coupland, 2017), as does quantitative (naturally occurring) clock variation (Salmela et al., 2016; Rubin et al., 2017). The clock can be differentially affected by external cues over 24-h. Based on the time of day, the gating property of the clock causes different amplitude in the transcriptional responses of clock-regulated genes to the same environmental stimulus (Wilkins et al., 2009, 2010; Grundy et al., 2015).
The clock drives temporal gene expression with physiological consequences, such as gas exchange, from individual to ecosystem scales (Resco de Dios and Gessler, 2017) and many aspects of plant development and its interactions with the environment (Dodd et al., 2005; Gibon et al., 2006; Khan et al., 2010; Edwards et al., 2011, 2012; Kerwin et al., 2011; Wulund and Reddy, 2015; Resco de Dios, 2017; Hubbard et al., 2018). At dawn, the clock enhances the resistance to oxidative species produced during the light-harvesting processes (Doherty and Kay, 2010), it is responsible for part of the signaling that governs stomata opening (Hotta et al., 2007), and it controls the mobilization of carbohydrates at dusk (Graf and Smith, 2011). The clock also causes hormonal waves influencing life-history traits, such as plant size at reproduction (Hanano et al., 2006) and floral development (Somers et al., 1998; Doyle et al., 2002; Song et al., 2012). Clock modulation for several hormones is characterized in the model organism Arabidopsis thaliana, and some of these hormones have been recently shown to mediate a response to changes in light/dark cycles for controlled environments (Nitschke et al., 2016). In Arabidopsis, about 30% of the transcriptome is clock-regulated in day/night growing conditions (Covington et al., 2008). This percentage rises to 80% in free running settings (Barak et al., 2000; Haydon et al., 2013; Nagel et al., 2015). This crucial role for the clock has been confirmed for other dicots (Ramos et al., 2005; Wilkins et al., 2009; Yon et al., 2012; Marcolino-Gomes et al., 2014); while metabolic rhythmicity has been shown in monocot crops (Filichkin et al., 2011; Calixto et al., 2015).
While they were originally viewed as mere clock outputs, metabolic oscillations have now been shown to feed back to the clock (Harmer, 2009; Schippers et al., 2013), resulting in a complex and fine-tuned cellular network (Gallego and Virshup, 2007; Pruneda-Paz and Kay, 2010; Sanchez et al., 2011). For instance, the circadian clock tunes both timing and capacity of sugar production via photosynthesis but, at the same time, sugar signals entrain the clock (Haydon et al., 2013). Importantly, plants’ nutritional status seems to feed back to the circadian clock, with the most relevant impacts for nitrogen and iron assimilation (Gutiérrez et al., 2008; Chen et al., 2013). Indeed nitrogen transporters are up regulated at night in drought stressed plants to counter-act lower water uptake rates (Greenham et al., 2017). Currently, redox mechanisms seem to be the best candidate to set the rhythms for this biochemical oscillator.
Redox state indicates the balance of oxidized versus reduced forms of electron donors and acceptors in a cell. When plants interact with the surrounding environment, the redox state is determined mainly by sudden production of highly reactive molecules, trigging signaling at a systemic level (Woodson, 2016). Reactive Oxygen Species (ROS) are inevitable by-products of aerobic metabolism and electron transport processes. Plants produce ROS because of the electron-transfer to and from molecular oxygen taking place in chloroplasts, mitochondria, and plasma membranes (Baxter et al., 2014). Peroxisomes are also involved in oxygen metabolism as signal mediators, regulating an array of oxidases-catalases to maintain H2O2 balance (Bonekamp et al., 2009). These reactive molecules are harmful when their generation exceeds their elimination. ROS can extensively damage all biomolecules – from lipids to proteins to nucleic acids – possibly leading to cell death (Ahmad et al., 2008). However, the perpetuation of a ROS signal, from the site of stress origin to the target of response, is crucial for coping with environmental stimuli (Bailey-Serres and Mittler, 2006). Therefore, a complex network of enzymatic and non-enzymatic antioxidants continuously scavenges excess ROS: the activity of these scavengers de facto translates into ROS homeostasis at the cellular level under favorable conditions (Doherty and Kay, 2010; Gilroy et al., 2014; Kangasjärvi and Kangasjärvi, 2014).
A number of studies have shown that the redox state is both regulated by and acts as a feedback on the endogenous clock under several environmental conditions (Krishnan and Pereira, 2008; Stangherlin and Reddy, 2013; Milev et al., 2015). The presence of ubiquitous sinks for H2O2 (peroxiredoxin protein family) supports the hypothesis of prevailing interlinks among ROS, metabolic pathways and the clock (Edgar et al., 2012; Yon et al., 2012; Hoyle and O’Neill, 2015). Rhythmic oscillations in ROS production seem to be altered under stress, as a consequence of redox feedbacks; yet, several components of redox-signaling pathways in plant cells remain undescribed (Suzuki et al., 2012; Kangasjärvi and Kangasjärvi, 2014).
The use of clock mutants has been critical in identifying circadian regulation of ROS homeostasis under several environmental conditions (Baxter et al., 2014; Greenham and McClung, 2015). ROS production and the activity of enzymatic scavengers have been shown to synchronously peak at specific times of the day (Lai et al., 2012). At the cellular level, a functioning clock directly regulates the redox state, coordinating the temporal activity of several scavengers. Fluctuations in one or multiple environmental factors affect the link between the clock and plant redox state eventually influencing growth, development and metabolism at the whole-plant level (Ahmad et al., 2008; Das and Roychoudhury, 2014; Gyöngyösi and Káldi, 2014).
Cellular redox state and circadian rhythms influence one another continuously and diverse environmental stressors will most likely impact both traits. When plant cells are healthy and completely functional, cellular homeostasis is actively maintained, and plants are in a dynamic equilibrium with the environment (Strasser, 1988). Under this equilibrium, the relation between redox state and circadian rhythms is effective, with a functional clock resonating with the environmental cycles (Figure 1A – working clock). Any significant change in the environmental conditions triggers what is commonly known as ‘oxidative stress,’ a sudden change in the redox state compromising cellular homeostasis (Cramer et al., 2011). This state of disequilibrium seems to affect antioxidant enzymes gated by the clock (Lai et al., 2012), slowing scavenging activity and leading to elevated ROS levels (Figure 1B – unknown clock activity, question mark). Through a phase of recovery (Figure 1C – unknown clock activity, question mark), plants frequently reach a new dynamic equilibrium after stress exposure (Figure 1D). During recovery, RNA metabolism and post-transcriptional gene silencing appear to play key roles in resetting both the epigenome and transcriptome, but it remains unclear how the circadian clock responds at this stage (Crisp et al., 2016). ROS dynamics in the recovery phase are likewise ambiguous in plants (Einset et al., 2007; Ahuja et al., 2010; Zhang and Kay, 2010; Obata and Fernie, 2012), primarily because these molecules are highly reactive and have a short half-life (Ahmad et al., 2008; Suzuki et al., 2012). Quantifying ROS and antioxidant pools is challenging due to measurement artifacts and to the occurrence of small to moderate changes in some component pools with stress (Queval et al., 2008; Noctor et al., 2016). Protein- and metabolomics allow for consistent quantification of lipid and protein peroxidation or glutathione accumulation to estimate ROS-dependent changes (Kranner et al., 2006; Noctor and Foyer, 2016; Abdelrahman et al., 2017). However, fine-resolved transcriptomic data may be the best means to characterize the mechanistic response to ROS production and scrubbing under abiotic stress (Noctor et al., 2012).
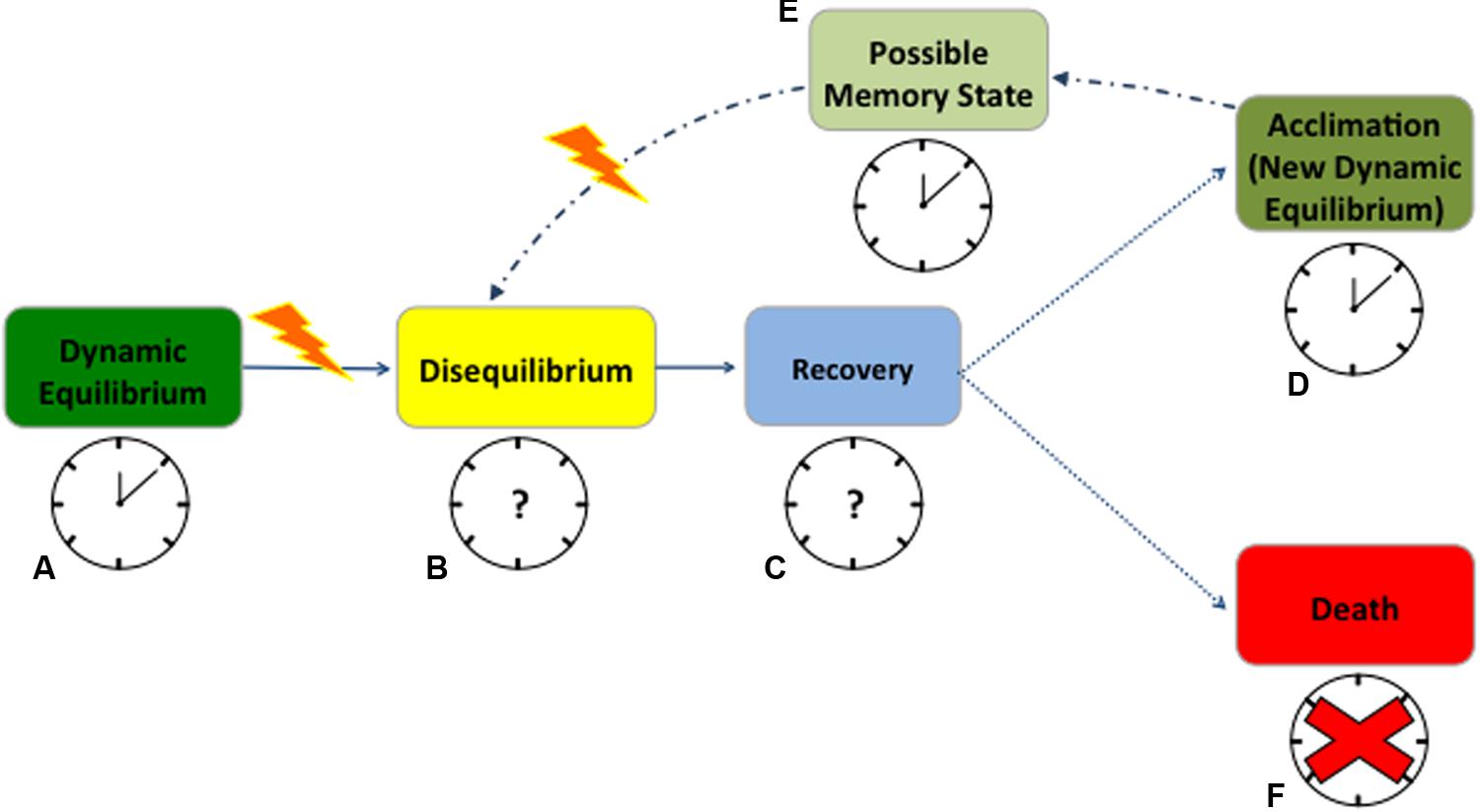
FIGURE 1. Anticipated redox state and circadian rhythms response to abiotic stress in plants. When cells are healthy, plants are in a dynamic equilibrium with the environment and the clock promptly resonates with environmental cycles (A). At stress occurrence, changes in the redox state compromise cellular homeostasis, leading to a state of disequilibrium, with almost unknown consequences on the clock (B). During a phase of recovery or strain, plants try to cope with the stress but how the clock behaves during this time is still uncertain (C). A successful recovery will lead to a new dynamic equilibrium (acclimation) after stress exposure (D). When acclimated, plants maintain a state of memory for which they will possibly experience fewer disturbances from the next stress event (E). In both acclimation and memory state the clock will maintain its functions but very little is known on its possible phase shifts (D,E). On the contrary, if the stress becomes exceptionally destructive, the clock will succumb, plants will not recover and die (F). Solid arrows point to the most certain consequences of stress occurrence; dotted lines point to the possible consequences of recovery after stress; and dash/dotted lines point to the underlying mechanism of memory. Working clocks are coupled with identified physiological states of equilibrium while clock with the question marks represent the states where clock behavior is completely unknown. The red cross signifies the complete disruption of the clock when plants die. The yellow lightening bolt represents stress occurrence.
There are potentially beneficial aspects to stress, for instance stimulating improved resistance to future stress (Larcher, 1980). A stress can be harmless if a plant manages to rapidly alter its homeostasis, adjusting metabolism, structure, and function to acclimate to altered conditions (Figure 1D – working clock) (Tsimilli-Michael et al., 1996). In both humans and rodents, stressors have been shown to lead to hormesis, appropriately priming organisms for future stress response (Foster and Kreitzman, 2014; Fleta-Soriano and Munné-Bosch, 2016). In the same manner, plants seem to retain a memory of the stress, improving their ability to respond to future variations in environmental conditions (Fleta-Soriano and Munné-Bosch, 2016) (Figure 1E – working clock). Possibly, RNA turnover contributes to acclimation and stress memory, but there is no clear understanding of how this mechanism competes with the epigenetics in memory development (Crisp et al., 2016). While clock function is retained, phase shifts are commonly associated with proximal stress response (Figures 1D,E). However, changes in clock gene frequencies can be also part of an adaptive evolutionary response, as in crop plants undergoing selection for agronomically desirable traits (Kevers et al., 2004; Yarkhunova et al., 2016). In short, after an initial destabilization, plants acclimate to the stress or improve their resistance to it, to the extent that physiological systems can buffer the changes in its redox state (Figures 1A–E). However, if the limits of plant tolerance are exceeded, then the stress becomes destructive, leading to permanent damage, loss of productivity or death (Figure 1F – absence of the clock).
While in rodents the connection between the circadian clock and stress response is well characterized (Koch et al., 2016), plant clock responses from early stress sensing to death, and its potential for re-setting after stress occurrence is still unclear (Figures 1B,C) (Grundy et al., 2015; Wulund and Reddy, 2015). Several stochastic models have successfully predicted clock activity at the molecular level in response to predictable variation in environmental cues (Ruoff et al., 2007; Resco et al., 2009; Domjian and Rand, 2011; Kosová et al., 2011). However, circadian clocks also experience extrinsic noise, namely irregular fluctuations in the environment, which are mostly omitted from current process models (Pokhilko et al., 2013; Guerriero et al., 2014; De Caluwé et al., 2016). Yet, synchronized metabolic responses driven by the core oscillator seem to be fundamental in plant response to environmental stress (Sanchez et al., 2011). Based on studies in mammals and algae, it seems probable that acute oxidative stress can reset the clock, resulting in the concurrent activation of a network of circadian genes that will propagate an antioxidant, cell survival response (Tamaru et al., 2013). This hypothesis considers H2O2 as a signal transducer, relaying information about the external environment to the circadian pacemaker. Susceptibility to oxidative stress through disruption of the circadian oscillator is another proposed mechanism linking ROS and the expression of the circadian clock (Qian et al., 2010).
We suggest more detailed studies during the recovery phase from the stress (Figure 1C). Recovery is not merely a return to the pre-stress state, but is instead a regulated mechanism, and its resolution would help in predicting plant adjustments to changing environmental conditions. Future research should focus on determining if ROS levels after a stress event may reset the periodicity of scavenger activity and affect clock gene expression (Figure 1 – question marks). Moreover, it remains unclear how the clock behaves in extremely stressed plants close to mortality (Sanchez et al., 2011; Zhang et al., 2013; Resco de Dios and Gessler, 2017). In this scenario, a functional characterization of the effects of environmental noise on the core oscillator is key to integrating metabolic information, such as ROS dynamics, into current clock models (Einset et al., 2007; Miller et al., 2010; Zhang and Kay, 2010; Obata and Fernie, 2012; Haydon et al., 2013).
Chlorophyll a fluorescence is a fast, non-invasive method commonly used to assess plant performance (Baker, 2008; Croce and van Amerongen, 2014). The fluorescence signal (and its derived parameters) reliably mirror plant stress response under biotic and abiotic stress, for stress of different duration/intensity and across a variety of species, with higher responsiveness in light than dark conditions (Lichtenthaler et al., 1986; Baker and Rosenqvist, 2004; Woo et al., 2008; Papageorgiou and Govindjee, 2011). Although excessive ROS accumulation has been shown to occur together with changes in fluorescence parameters (Aldea et al., 2006; Moradi and Ismail, 2007) the direct mechanistic relation is still unclear (Gill and Tuteja, 2010). So far, no predictive understanding of the correlation between ROS and the fluorescence signal is possible and the association will depend on the stress type, intensity and duration.
In non-limiting light conditions, stress will differentially affect the variable fluorescence signal (Fv) and fluorescence derived variables, such as the efficiency of photosystem II in light (Fv′/Fm′) and the Non-Photochemical Quenching (NPQ) (Figure 2). Each parameter reflects a specific aspect of photosynthetic activity, and in sum they depict a picture of the state of the photosynthetic light harvesting machinery (Roháček, 2002; Maxwell and Johnson, 2004). For the entire spectrum of stress response, Fv remains fairly constant (Figure 2 – dotted line) until membrane failure at the cellular scale takes place, proximally leading to death of the plant after a distal cause such as severe drought (Guadagno et al., 2017). NPQ (Figure 2 – solid line) is known to have a tight correlation with increasing stress (both for duration and/or intensity) (Müller et al., 2001; Demmig-Adams et al., 2014) until complete cellular failure at which point Fv will decline to zero and the plant is considered dead (Guadagno et al., 2017). Fv′/Fm′ seems to be the most reliable parameter to follow stress response dynamics from early sensing to mortality (Figure 2 – dashed line). For instance, Fv′/Fm′ is able to capture the onset of the stress (early sensing) and increases to compensate for limited gas exchange due to stomatal closure with drought (Greenham et al., 2017). This is an informative outcome because in the past the theoretical maximum efficiency of PSII (Fv/Fm) has always been considered the main indicator of plant stress response (Murchie and Lawson, 2013). Typically, Fv/Fm values decrease most significantly only under marked stress, when the survival of plants may have already been irretrievably reduced (Chen et al., 2015). On the other hand, Fv′/Fm′ dynamics more closely follow stress dynamics and the relative changes in redox state from early sensing to death in both conifers and herbaceous plants (Guadagno et al., 2017) (Figure 2 – dashed line).
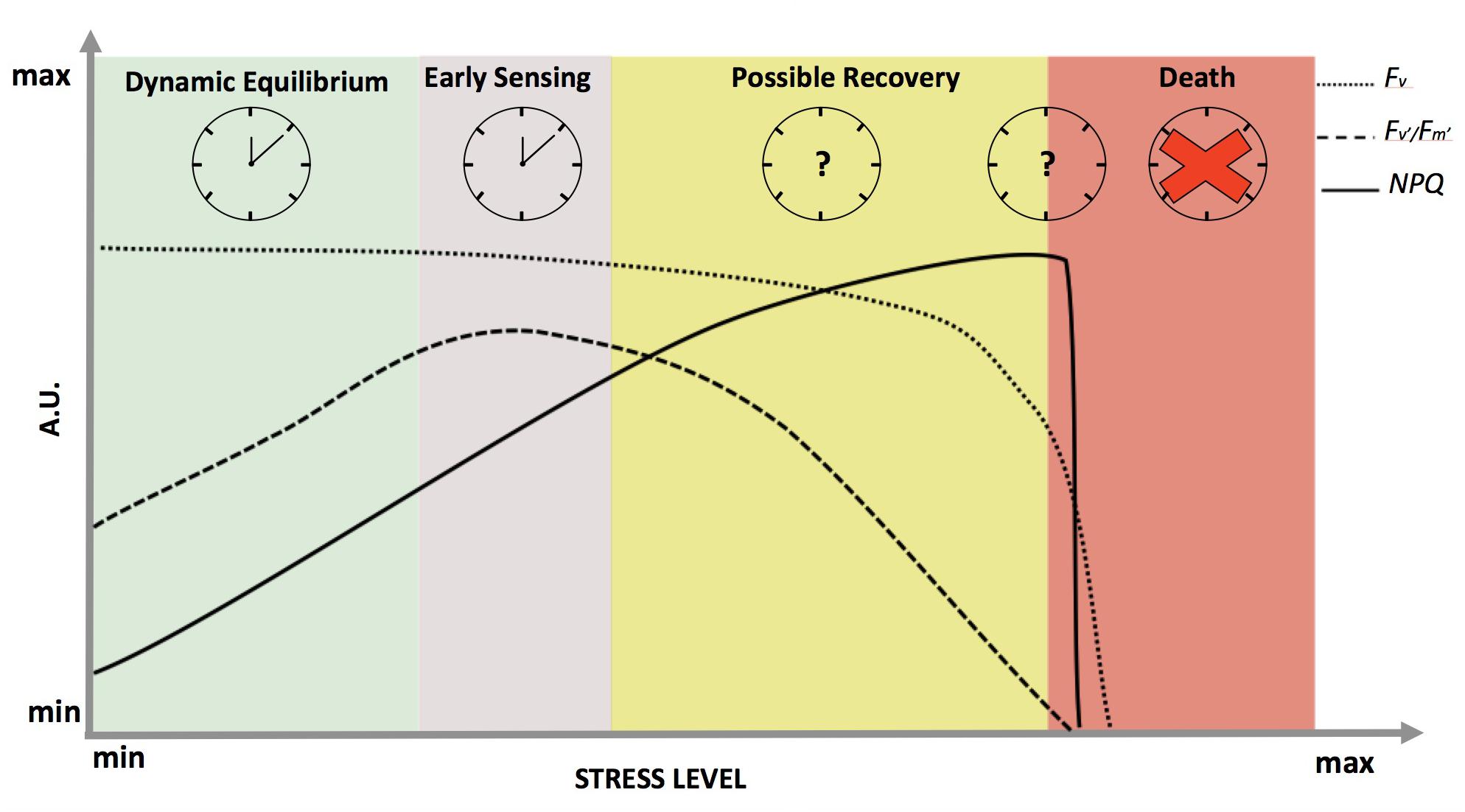
FIGURE 2. Dynamics of chlorophyll a fluorescence and derived parameters in response to abiotic stress in optimal light conditions. On the x-axis, the stress level is reported as a continuum between a minimum value at physiological equilibrium (green shadow), passing through early sensing (purple shadow) and possible recovery (yellow shadow), till death (red shadow). The y-axis represents fluorescence in arbitrary units: dotted, dashed and solid lines representing Fv (variable fluorescence signal), Fv′/Fm′ (maximum efficiency of photosystem II in light) and NPQ (Non-Photochemical Quenching), respectively. On the top part of the panel, working clocks are reported for the most studied physiological states while clock with the question marks represent the states where clock behavior is completely unknown. The red cross signifies the complete disruption of the clock when plants die.
We emphasize the diurnal timing of plant response to abiotic stress can be critical, as we recently showed in Brassica rapa under mild drought stress (Des Mairas, 2017; Greenham et al., 2017). During the day, a phase shift in expression pattern for genes related to photosystem efficiency and light response pathways (e.g., Light Harvesting Chlorophyll a/b Binding-LHCB2.2, Photosystem II Manganese Binding-PSBY) was observed, consistent with the increase in Fv′/Fm′ (Greenham et al., 2017). Diel changes in chlorophyll a fluorescence were first shown in algae and phytoplankton (Prézelin and Ley, 1980; Sorek et al., 2013). Later, the same fluorescence parameters were observed to have rhythmicity in Arabidopsis mutants and barley under constant blue and white light conditions (Litthauer et al., 2015; Hussien et al., 2017), suggesting fluorescence as a possible high-throughput marker for circadian rhythms in plants as well as for changes in clock phase resulting from stress.
During the last few years, several studies have confirmed circadian rhythms in redox state across species, suggesting the existence of a strong clock/redox interconnection. Although it seems clear that redox state and the circadian clock are interlocked in stress response, it remains unknown if the clock is reset by stress and if any type of protective acclimation is triggered at the cell level. In this perspective, we propose that a timely avenue of research lies in investigating the details of the recovery phase from the stress. We suggest a more intensive use of chlorophyll a fluorescence to assess variation in circadian rhythms, and summarized the importance of fluorescence dynamics at different stress levels. Fluorescence data as a high-throughput screen, coupled with ROS analysis, proteomic, metabolomics and gene expression, will inform and improve existing process models: acquiring realistic predictions for plant responses to a changing environment and ultimately improving breeding strategies.
CRG conceptualized, reviewed, validated formerly collected the data, and wrote the original draft of the manuscript. CRG and BEE worked on the visualization of the perspective. BEE and CW acquired the funding and reviewed and edited the manuscript.
NSF funding IOS 1025965, IOS 1547796 and IOS-1444571 supported this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Stanley DeVore for data collection on ROS activity that helped conceptualize this Perspective. We also thank Heather N. Speckman, Daniel Beverly, Mallory Lai, and Dr. Dave Millar for critically reading the manuscript.
Abdelrahman, M., Burritt, D. J., and Phan Tran, L.-S. (2017). The use of metabolomic quantitative trait locus mapping and osmotic adjustment traits for the improvement of crop yields under environmental stresses. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2017.06.020 [Epub ahead of print].
Ahmad, P., Sarwat, M., and Sharma, S. (2008). Reactive oxygen species, antioxidants and signaling in plants. J. Plant Biol. 51, 167–173. doi: 10.1007/BF03030694
Ahuja, I., de Vos, R. C. H., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Aldea, M., Frank, T. D., and DeLucia, E. H. (2006). A method for quantitative analysis of spatially variable physiological processes across leaf surfaces. Photosynth. Res. 90, 161–172. doi: 10.1007/s11120-006-9119-z
Bailey-Serres, J., and Mittler, R. (2006). The roles of reactive oxygen species in plant cells. Plant Physiol. 141:311. doi: 10.1104/pp.104.900191
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Baker, N. R., and Rosenqvist, E. (2004). Applications of chlorophyll fluorescence can improve crop production strategies: an examination of future possibilities. J. Exp. Bot. 55, 1607–1621. doi: 10.1093/jxb/erh196
Barak, S., Tobin, E. M., Andronis, C., Sugano, S., and Green, R. M. (2000). All in good time: the Arabidopsis circadian clock. Trends Plant Sci. 5, 517–522. doi: 10.1016/S1360-1385(00)01785-4
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bonekamp, N. A., Völkl, A., Fahimi, H. D., and Schrader, M. (2009). Reactive oxygen species and peroxisomes: struggling for balance. Biofactors 35, 346–355. doi: 10.1002/biof.48
Calixto, C. P., Waugh, R., and Brown, J. W. (2015). Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes. J. Mol. Evol. 80, 108–119. doi: 10.1007/s00239-015-9665-0
Chen, D., Wang, S., Xiong, B., Cao, B., and Deng, X. (2015). Carbon/Nitrogen imbalance associated with drought-induced leaf senescence in Sorghum bicolor. PLoS One 10:e0137026. doi: 10.1371/journal.pone.0137026
Chen, Y.-Y., Wang, Y., Shin, L.-J., Wu, J.-F., Shanmugam, V., Tsednee, M., et al. (2013). Iron is involved in the maintenance of circadian period length in Arabidopsis. Plant Physiol. 161, 1409–1420. doi: 10.1104/pp.112.212068
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-r130
Cramer, G. R., Urano, K., Deirot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. doi: 10.1186/1471-2229-11-163
Crisp, P. A., Ganguly, D., Eichten, S. R., Borevitz, J. O., and Pogson, B. J. (2016). Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2:e1501240. doi: 10.1126/sciadv.1501340
Croce, R., and van Amerongen, H. (2014). Natural strategies for photosynthetic light harvesting. Nat. Chem. Biol. 10, 492–501. doi: 10.1038/nchembio.1555
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
De Caluwé, J., Xiao, Q., Hermans, C., Verbruggen, N., Leloup, J.-C., and Gonze, D. (2016). A compact model for the complex plant circadian clock. Front. Plant Sci. 7:74. doi: 10.3389/fpls.2016.00074
DeCoursey, P. J., Walker, J. K., and Smith, S. A. (2000). A circadian pacemaker in free-living chipmunks: essential for survival? J. Comp. Physiol. A 186, 169–180. doi: 10.1007/s003590050017
de Montaigu, A., and Coupland, G. (2017). The timing of GIGANTEA expression during day/night cycles varies with the geographical origin of Arabidopsis accessions. Plant Signal. Behav. 12:e1342026. doi: 10.1080/15592324.2017.1342026
de Montaigu, A., Giakountis, A., Rubin, M., Tóth, R., Cremer, F., Sokolova, V., et al. (2015). Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc. Natl. Acad. Sci. U.S.A. 112, 905–910. doi: 10.1073/pnas.1422242112
Demmig-Adams, B., Koh, S. C., Cohu, C. M., Muller, O., Stewart, J. J., and Adams, W. W. III (2014). “Non-photochemical quenching in contrasting plant species and environments,” in Non-Photochemical Fluorescence Quenching and Energy Dissipation in Plants, Algae, and Cyanobacteria. Advances in Photosynthesis and Respiration, eds B. Demmig-Adams, W. W. III Adams, G. Garab, and Govindjee (Dordrecht: Springer). doi: 10.1007/978-94-017-9032-1_24
Des Mairas, D. L. (2017). Into the fourth dimension - The influence of time on the drought response of Brassica rapa, an agriculturally important species of plant, has been clarified. eLife 6:e31328. doi: 10.7554/eLife.31328
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Doherty, C. J., and Kay, S. A. (2010). Circadian control of global gene expression patterns. Annu. Rev. Genet. 44, 419–444. doi: 10.1146/annurev-genet-102209-163432
Domjian, M., and Rand, D. A. (2011). Balance equations can buffer noisy and sustained environmental perturbations of circadian clocks. Interface Focus 1, 177–186. doi: 10.1098/rsfs.2010.0007
Doyle, M. R., Davis, S. J., Bastow, R. M., McWatters, H. G., Kozma-Bognar, L., Nagy, F., et al. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. doi: 10.1038/nature00954
Edgar, R. S., Green, E. W., Zhao, Y., van Ooijen, G., Olmedo, M., Qin, X., et al. (2012). Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. doi: 10.1038/nature11088
Edwards, C. E., Ewers, B. E., McClung, C. R., and Weinig, C. (2012). Quantitative variation in water-use efficiency across watering regimes and its relationship with circadian, vegetative, reproductive, and leaf gas-exchange traits. Mol. Plant 5, 653–658. doi: 10.1093/mp/sss004
Edwards, C. E., Ewers, B. E., Williams, D. G., Xie, Q., Lou, P., Xu, X., et al. (2011). The genetic architecture of ecophysiological and circadian traits in Brassica rapa. Genetics 189, 375–390. doi: 10.1534/genetics.110.125112
Einset, J., Winge, P., and Bones, A. (2007). ROS signaling pathways in chilling stress. Plant Signal. Behav. 2, 365–367. doi: 10.4161/psb.2.5.4461
Filichkin, S. A., Breton, G., Priest, H. D., Dharmawardhana, P., Jaiswal, P., Fox, S. E., et al. (2011). Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One 6:e16907. doi: 10.1371/journal.pone.0016907
Fleta-Soriano, E., and Munné-Bosch, S. (2016). Stress memory and the inevitable effects of drought: a physiological perspective. Front. Plant Sci. 7:143. doi: 10.3389/fpls.2016.00143
Foster, R. G., and Kreitzman, L. (2014). The rhythms of life: what your body clock means to you! Exp. Physiol. 99, 599–606. doi: 10.1113/expphysiol.2012.071118
Gallego, M., and Virshup, D. M. (2007). Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148. doi: 10.1038/nrm2106
Gibon, Y., Usadel, B., Blaesing, O. E., Kamlage, B., Hoehne, M., Trethewey, R., et al. (2006). Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7:R76. doi: 10.1186/gb-2006-7-8-r76
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gilroy, S., Suzuki, N., Miller, G., Choi, W. G., Toyota, M., Devireddy, A. R., et al. (2014). A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19, 623–630. doi: 10.1016/j.tplants.2014.06.013
Graf, A., and Smith, A. M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16, 169–175. doi: 10.1016/j.tplants.2010.12.003
Green, R. M., Tingay, S., Wang, Z.-Y., and Tobin, E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584. doi: 10.1104/pp.004374
Greenham, K., Guadagno, C. R., Gehan, M. A., Mockler, T. C., Weinig, C., Ewers, B. E., et al. (2017). Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. eLife 6:e29655. doi: 10.7554/eLife.29655
Greenham, K., and McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610. doi: 10.1038/nrg3976
Grundy, J., Stoker, C., and Carré, I. A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6:648. doi: 10.3389/fpls.2015.00648
Guadagno, C. R., Ewers, B. E., Speckman, H. N., Aston, T. L., Huhn, B. J., DeVore, S. B., et al. (2017). Dead or alive? Using membrane failure and chlorophyll a fluorescence to predict plant mortality from drought. Plant Physiol. 175, 223–234. doi: 10.1104/pp.16.00581
Guerriero, M. L., Akman, O. E., and van Ooijen, G. (2014). Stochastic models of cellular circadian rhythms in plants help to understand the impact of noise on robustness and clock structure. Front. Plant Sci. 5:564. doi: 10.3389/fpls.2014.00564
Guerriero, M. L., Pokhilko, A., Fernandez, A. P., Halliday, K. J., Millar, A. J., and Hillston, J. (2012). Stochastic properties of the plant circadian clock. J. R. Soc. Interface 9, 744–756. doi: 10.1098/rsif.2011.0378
Gutiérrez, R. A., Stokes, T. L., Thum, K., Xu, X., Obertello, M., Katari, M. S., et al. (2008). Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. U.S.A. 105, 4939–4944. doi: 10.1073/pnas.0800211105
Gyöngyösi, N., and Káldi, K. (2014). Interconnections of reactive oxygen species homeostasis and circadian rhythm in Neurospora crassa. Antioxid. Redox Signal. 20, 3007–3023. doi: 10.1089/ars.2013.5558
Hanano, S., Domagalska, M. A., Nagy, F., and Davis, S. J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11, 1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E., and Webb, A. A. R. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. doi: 10.1038/nature12603
Hotta, C. T., Gardner, M. J., Hubbard, K. E., Baek, S. J., Dalchau, N., Suhita, D., et al. (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 30, 333–349. doi: 10.1111/j.1365-3040.2006.01627.x
Hoyle, N. P., and O’Neill, J. S. (2015). Oxidation-reduction cycles of peroxiredoxin proteins and non-transcriptional aspects of timekeeping. Biochemistry 54, 184–193. doi: 10.1021/bi5008386
Hubbard, C. J., Brock, M. T., van Diepen, L. T., Ewers, B. E., Maignien, L., and Weinig, C. (2018). The plant circadian clock influences rhizosphere community structure and function. ISME J. 12, 400–410. doi: 10.1038/ismej.2017.172
Hussien, D., Dakhiya, Y., Fridman, E., Kiflawi, M., and Green, R. (2017). Correlations between circadian rhythms and growth in challenging environments. Plant Physiol. 173, 1–11. doi: 10.1104/pp.17.00057
Kangasjärvi, S., and Kangasjärvi, J. (2014). Towards understanding extracellular ROS sensory and signaling systems in plants. Adv. Bot. 2014:538946. doi: 10.1155/2014/538946
Kerwin, R. E., Jimenez-Gomez, J. M., Fulop, D., Harmer, S. L., Maloof, J. N., and Kliebenstein, D. J. (2011). Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23, 471–485. doi: 10.1105/tpc.110.082065
Kevers, C., Franck, T., Strasser, R. J., Dommes, J., and Gaspar, T. (2004). Hyperhydricity of micropropagated shoots: a typically stress-induced change of physiological state. Plant Cell Tissue Organ Cult. 77, 181–199. doi: 10.1023/B:TICU.0000016825.18930.e4
Khan, S., Rowe, S. C., and Harmon, F. G. (2010). Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol. 10:126. doi: 10.1186/1471-2229-10-126
Koch, C. E., Leinweber, B., Drengberg, B. C., Blaum, C., and Oster, H. (2016). Interaction between circadian rhythms and stress. Neurobiol. Stress 6, 57–67. doi: 10.1016/j.ynstr.2016.09.001
Kosová, K., Vítámvás, P., Prášil, I. T., and Renaut, J. (2011). Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteomics 74, 1301–1322. doi: 10.1016/j.jprot.2011.02.006
Kranner, I., Birtić, S., Anderson, K. M., and Pritchard, H. W. (2006). Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death? Free Radic. Biol. Med. 40, 2155–2165. doi: 10.1016/j.freeradbiomed.2006.02.013
Krishnan, A., and Pereira, A. (2008). Integrative approaches for mining transcriptional regulatory programs in Arabidopsis. Brief. Funct. Genomics 7, 264–274. doi: 10.1093/bfgp/eln035
Lai, A. G., Doherty, C. J., Mueller-Roeber, B., Kay, S. A., Shippers, J. H. M., and Dijkwell, P. P. (2012). Circadian clock-associated 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134. doi: 10.1073/pnas.1209148109
Larcher, W. (1980). Physiological Plant Ecology. New York, NY: Springer. doi: 10.1007/978-3-642-96545-6
Lichtenthaler, H. K., Buschmann, C., Rinderle, U., and Schmuck, G. (1986). Application of chlorophyll fluorescence in ecophysiology. Radiat. Environ. Biophys. 25, 297–308. doi: 10.1007/BF01214643
Litthauer, S., Battle, M. W., Lawson, T., and Jones, M. A. (2015). Phototropins maintain robust circadian oscillation of PSII operating efficiency under blue light. Plant J. 83, 1034–1045. doi: 10.1111/tpj.12947
Marcolino-Gomes, J., Rodrigues, F. A., Fuganti-Pagliarini, R., Bendix, C., Nakayama, T. J., Celaya, B., et al. (2014). Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One 9:e86402. doi: 10.1371/journal.pone.0086402
Maxwell, K., and Johnson, G. N. (2004). Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 345, 659–668.
McClung, C. R. (2011). The genetics of plant clocks. Adv. Genet. 74, 105–138. doi: 10.1016/B978-0-12-387690-4.00004-0
Milev, N. B., Rey, G., Valekunja, U. K., Edgar, R. S., O’Neill, J. S., and Reddy, A. B. (2015). “Analysis of the redox oscillations in the circadian clockwork,” in Methods in Enzymology, Circadian Rhythms and Biological Clock, Vol. 552, ed. A. Sehgal (Amsterdam: Elsevier Inc.).
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Moradi, F., and Ismail, A. M. (2007). Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann. Bot. 99, 1161–1173. doi: 10.1093/aob/mcm052
Müller, P., Li, X.-P., and Niyogi, K. K. (2001). Non-photochemical quenching. a response to excess light energy. Plant Physiol. 125, 1558–1566. doi: 10.1104/pp.125.4.1558
Murchie, E. H., and Lawson, T. (2013). Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J. Exp. Bot. 64, 3983–3998. doi: 10.1093/jxb/ert208
Nagel, D. H., Doherty, C. J., Pruneds-Paz, J. L., Schmitz, R. J., Ecker, J. R., and Kay, S. A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E4802–E4810. doi: 10.1073/pnas.1513609112
Nitschke, S., Cortleven, A., Iven, T., Feussner, I., Havaux, M., Riefler, M., et al. (2016). Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28, 1616–1639. doi: 10.1105/tpc.16.00016
Noctor, G., and Foyer, C. H. (2016). Intracellular redox compartmentation and ROS related communication in regulation and signaling. Plant Physiol. 171, 1581–1592. doi: 10.1104/pp.16.00346
Noctor, G., Mhamdi, A., Chaouch, S., Han, Y. I., Neukermans, J., Marquez-Garcia, B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35, 454–484. doi: 10.1111/j.1365-3040.2011.02400.x
Noctor, G., Mhamdi, A., and Foyer, C. H. (2016). Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ. 39, 1140–1160. doi: 10.1111/pce.12726
Obata, T., and Fernie, A. R. (2012). The use of metabolomics to dissect plant responses to abiotic stress. Cell. Mol. Life Sci. 69, 3225–3243. doi: 10.1007/s00018-012-1091-5
Papageorgiou, G. C., and Govindjee, G. (2011). Photosystem II fluorescence: slow changes–scaling from the past. J. Photochem. Photobiol. 104, 258–270. doi: 10.1016/j.jphotobiol.2011.03.008
Pittendrigh, C. S. (1960). Circadian rhythms and the circadian organization of living systems. Cold Spring Harb. Symp. Quant. Biol. 25, 159–184. doi: 10.1101/SQB.1960.025.01.015
Pokhilko, A., Mas, P., and Millar, A. J. (2013). Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 7:23. doi: 10.1186/1752-0509-7-23
Prézelin, B. B., and Ley, A. C. (1980). Photosynthesis and chlorophyll a fluorescence rhythms of marine phytoplankton. Mar. Biol. 55, 295–307. doi: 10.1007/BF00393782
Pruneda-Paz, J. L., and Kay, S. A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15, 259–265. doi: 10.1016/j.tplants.2010.03.003
Qian, H., Yu, S., Sun, Z., Xie, X., Liu, W., and Fu, Z. (2010). Effects of copper sulfate, hydrogen peroxide and N-phenyl-2-naphthylamine on oxidative stress and the expression of genes involved photosynthesis and microcystin disposition in Microcystis aeruginosa. Aquat. Toxicol. 99, 405–412. doi: 10.1016/j.aquatox.2010.05.018
Queval, G., Hager, J., Gakiere, B., and Noctor, G. (2008). Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J. Exp. Bot. 59, 135–146. doi: 10.1093/jxb/erm193
Ramos, A., Pérez-Solís, E., Ibáñez, C., Casado, R., Collada, C., Gómez, L., et al. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. U.S.A. 102, 7037–7042. doi: 10.1073/pnas.0408549102
Resco, V., Hartwell, J., and Hall, A. (2009). Ecological implications of plants ability to tell the time. Ecol. Lett. 12, 583–592. doi: 10.1111/j.1461-0248.2009.01295.x
Resco de Dios, V. (2017). Circadian regulation and diurnal variation in gas exchange. Plant Physiol. 175, 34. doi: 10.1104/pp.17.00984
Resco de Dios, V., and Gessler, A. (2017). Circadian regulation of photosynthesis and transpiration from genes to ecosystems. Environ. Exp. Bot. (in press). doi: 10.1016/j.envexpbot.2017.09.010
Roháček, K. (2002). Chlorophyll fluorescence parameters: the definitions, photosynthetic meaning, and mutual relationships. Photosynthetica 40, 13–29. doi: 10.1023/A:1020125719386
Rubin, M. J., Brock, M. T., Knapp, M., Welch, S. M., Harmer, S. L., Maloof, J. N., et al. (2017). Circadian phenotypes vary across seasonal environments and correlate with fitness components. Mol. Ecol. 26, 5528–5540. doi: 10.1111/mec.14287
Ruoff, P., Zakhartsev, M., and Westerhoff, H. V. (2007). Temperature compensation through system biology. FEBS J. 274, 940–950. doi: 10.1111/j.1742-4658.2007.05641.x
Salmela, M. J., Greenham, K., Lou, P., McClung, C. R., Ewers, B. E., and Weinig, C. (2016). Variation in circadian rhythms is maintained among and within populations in Boechera stricta. Plant Cell Environ. 39, 1293–1303. doi: 10.1111/pce.12670
Salome, P. A., and McClung, C. R. (2004). The Arabidopsis thaliana clock. J. Biol. Rhythms 19, 425–435. doi: 10.1177/0748730404268112
Sanchez, A., Shin, J., and Davis, S. J. (2011). Abiotic stress and plant circadian clock. Plant Signal. Behav. 6, 223–231. doi: 10.4161/psb.6.2.14893
Schippers, J. H. M., Lai, A. G., Mueller-Roeber, B., and Dijkwel, P. P. (2013). Could ROS signals drive tissue-specific clocks? Transcription 4, 206–208. doi: 10.4161/trns.26362
Somers, D. E., Webb, A. A., Pearson, M., and Kay, S. A. (1998). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494.
Song, Y., Gao, Z., and Luan, W. (2012). Interaction between temperature and photoperiod in regulation of flowering time in rice. Sci. China Life Sci. 55, 241–249. doi: 10.1007/s11427-012-4300-4
Sorek, M., Yacobi, Y. Z., Roopin, M., Berman-Frank, I., and Levy, O. (2013). Photosynthetic circadian rhythmicity patterns of Symbiodium, the coral endosymbiotic algae. Proc. R. Soc. B 280:20122942. doi: 10.1098/rspb.2012.2942
Stangherlin, A., and Reddy, A. B. (2013). Regulation of circadian clocks by redox homeostasis. J. Biol. Chem. 288, 26505–26511. doi: 10.1074/jbc.R113.457564
Strasser, R. J. (1988). “A concept for stress and its application in remote sensing,” in Applications of Chlorophyll Fluorescence, ed. H. K. Lichtenthaler (Dordrecht: Kluwer Academic Publishers), 333–337.
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Tamaru, T., Hattori, M., Ninomiya, Y., Kawamura, G., Varès, G., Honda, K., et al. (2013). ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS One 8:e82006. doi: 10.1371/journal.pone.0082006
Todd, P. M., Salomé, P. A., Yu, H. J., Spencer, T. R., Sharp, E. L., McPeek, M. A., et al. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302, 1049–1053. doi: 10.1126/science.1082971
Tsimilli-Michael, M., Krüger, G. H. J., and Strasser, R. J. (1996). About the perpetual state changes in plants approaching harmony with their environment. Arch. Sci. GeneÌve 49, 173–203.
Wilkins, O., Bräutigam, K., and Campbell, M. M. (2010). Time of day shapes Arabidopsis drought transcriptomes. Plant J. 63, 715–727. doi: 10.1111/j.1365-313X.2010.04274.x
Wilkins, O., Waldron, L., Nahal, H., Provart, N. J., and Campbell, M. M. (2009). Genotype and time of day shape the Populus drought response. Plant J. 60, 703–715. doi: 10.1111/j.1365-313X.2009.03993.x
Woo, N. S., Badger, M. R., and Pogson, B. J. (2008). A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods 4:27. doi: 10.1186/1746-4811-4-27
Woodson, J. D. (2016). Chloroplast quality control—balancing energy production and stress. New Phytol. 212, 36–41. doi: 10.1111/nph.14134
Wulund, L., and Reddy, A. B. (2015). A brief history of circadian time: the emergence of redox oscillations as a novel component of biological rhythms. Perspect. Sci. 6, 26–37. doi: 10.1016/j.pisc.2015.08.002
Yarkhunova, Y., Edwards, C. E., Ewers, B. E., Baker, R. L., Aston, T. L., McClung, C. R., et al. (2016). Selection during crop diversification involves correlated evolution of the circadian clock and ecophysiological traits in Brassica rapa. New Phytol. 210, 133–144. doi: 10.1111/nph.13758
Yon, F., Seo, P.-J., Ryu, J. Y., Park, C.-H., Baldwin, I. T., and Kim, S. G. (2012). Identification and characterization of circadian clock genes in a native tobacco, Nicotiana attenuata. BMC Plant Biol. 12:172. doi: 10.1186/1471-2229-12-172
Zhang, C., Xie, Q., Anderson, R. G., Ng, G., Seitz, N. C., Peterson, T., et al. (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9:e1003370. doi: 10.1371/journal.ppat.1003370
Keywords: circadian rhythms, ROS, redox state, plant stress response, chlorophyll a fluorescence
Citation: Guadagno CR, Ewers BE and Weinig C (2018) Circadian Rhythms and Redox State in Plants: Till Stress Do Us Part. Front. Plant Sci. 9:247. doi: 10.3389/fpls.2018.00247
Received: 05 December 2017; Accepted: 12 February 2018;
Published: 05 March 2018.
Edited by:
Lam-Son Tran, RIKEN, JapanReviewed by:
Mostafa Abdelwahed Abdelrahman, Tohoku University, JapanCopyright © 2018 Guadagno, Ewers and Weinig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmela R. Guadagno, Y2d1YWRhZ25AdXd5by5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.