
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 February 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00233
This article is part of the Research TopicBiosafety of Genetically Modified Organisms, Volume IIView all 23 articles
Transgenic glyphosate-tolerant plants overproducing EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) may exhibit enhanced fitness in glyphosate-free environments. If so, introgression of transgenes overexpressing EPSPS into wild relative species may lead to increased competitiveness of crop-wild hybrids, resulting in unpredicted environmental impact. Assessing fitness effects of transgenes overexpressing EPSPS in a model plant species can help address this question, while elucidating how overproducing EPSPS affects the fitness-related traits of plants. We produced segregating T2 and T3 Arabidopsis thaliana lineages with or without a transgene overexpressing EPSPS isolated from rice or Agrobacterium (CP4). For each of the three transgenes, we compared glyphosate tolerance, some fitness-related traits, and auxin (indole-3-acetic acid) content in transgene-present, transgene-absent, empty vector (EV), and parental lineages in a common-garden experiment. We detected substantially increased glyphosate tolerance in T2 plants of transgene-present lineages that overproduced EPSPS. We also documented significant increases in fecundity, which was associated with increased auxin content in T3 transgene-present lineages containing rice EPSPS genes, compared with their segregating transgene-absent lineages, EV, and parental controls. Our results from Arabidopsis with nine transgenic events provide a strong support to the hypothesis that transgenic plants overproducing EPSPS can benefit from a fecundity advantage in glyphosate-free environments. Stimulated biosynthesis of auxin, an important plant growth hormone, by overproducing EPSPS may play a role in enhanced fecundity of the transgenic Arabidopsis plants. The obtained knowledge is useful for assessing environmental impact caused by introgression of transgenes overproducing EPSPS from any GE crop into populations of its wild relatives.
Genetically engineered (GE) herbicide-tolerant crops are cultivated extensively over the world owing to their substantial agronomic, environmental, economic, health and social benefits (James, 2016). Herbicide-tolerant GE crops occupy ~76% of the total GE crop cultivation area, including herbicide-tolerant and stacked herbicide-tolerant/insect-resistant GE crops (James, 2016). Of these, glyphosate-tolerance represents the world's most widespread GE crop trait (Duke and Powles, 2008; Vats, 2015; James, 2016). The commercial cultivation of glyphosate-tolerant GE crops has greatly promoted the glyphosate application in agricultural ecosystems, consequently arousing global concerns over its potential environmental impact. Many weed species have evolved glyphosate tolerance under selective pressure after long-term glyphosate applications (Duke and Powles, 2008; Délye et al., 2013). Glyphosate-selective-pressure induced target-site mutations (Gaines et al., 2010, 2011; Chen et al., 2015) and amplification of the EPSPS genes (Nandula et al., 2013; Sammons and Gaines, 2014) have been found in those resistant weeds resulting new environmental problems as farmers shift to less environmentally friendly herbicides. In addition, transgene flow from glyphosate-tolerant GE plants to populations of wild or weedy relatives has been found, becoming an environmental biosafety concern (Reichman et al., 2006; Warwick et al., 2008; Wegier et al., 2011; Zapiola and Mallory-Smith, 2017). Yet, some researchers posit little or no environmental impact from introgression of glyphosate-tolerance transgenes into wild relative populations because they believe that such transgenes offer no fitness advantage in natural ecosystems in the absence of glyphosate (Cerdeira and Duke, 2006; Vila-Aiub et al., 2014).
The findings of significantly enhanced fecundity of crop-weed (Wang et al., 2014) and crop-wild (Yang et al., 2017b) rice hybrid progeny containing the glyphosate-tolerance transgene in a glyphosate-absent habitat suggests that introgression of such a glyphosate-tolerance transgene might result in transgene persistence and spread in populations of wild relatives, possibly causing environmental consequences (Qiu, 2013; Ryffel, 2014; Vila-Aiub et al., 2015; Martin et al., 2017). Glyphosate can competitively inhibit EPSPS (5-enolpyruvylshikimate-3-phosphate synthase, EC 2.5.1.19), resulting in weakness or even death of plants at the proper dosages (Roberts et al., 1998; Mueller et al., 2003; Duke and Powles, 2008). EPSPS is a key enzyme in the shikimate pathway, which is extremely important because ~35% or more plant biomass in the form of dry matter is represented by aromatic molecules derived directly from this pathway (Franz et al., 1997). In addition, EPSPS is essential for the production of aromatic amino acids (e.g., tryptophan, phenylalanine, and tyrosine) and other secondary metabolites (Weaver and Herrmann, 1997), suggesting that EPSPS is crucial for the survival, growth, and development of plants.
Overproduction of EPSPS, attempting to provide sufficient surplus of EPSPS binding to glyphosate to reduce its fatal toxicity, is one of the strategies to increase plants' tolerance to glyphosate. This strategy includes developing GE plants with multiple copies of the EPSPS gene (Rogers et al., 1983; Goldsbrough et al., 1990; Shyr et al., 1992) and increasing basal levels of the EPSPS enzyme by fusing an EPSPS gene with a strong promotor (Klee et al., 1987; Su et al., 2008). Notably, certain GE glyphosate-tolerant crops overexpressing EPSPS not only have increased glyphosate tolerance (Klee et al., 1987; Su et al., 2008; Vats, 2015; Yang et al., 2017a), but unexpectedly also showed increased yield (Zhou et al., 2003; Owen et al., 2010) and other fitness traits. The increased yield can also be manifest as increased fecundity in GE hybrid progeny with weedy (O. sativa f. spontanea, Wang et al., 2014) and wild (O. rufipogon, Yang et al., 2017b) rice overexpressing EPSPS, even under glyphosate-free conditions. Wang et al. (2014) also reported significantly increased Trp concentrations in crop-weed F2 transgene-present hybrid lineages of the GE rice (Oryza sativa) line overexpressing EPSPS and four weed rice populations in the glyphosate-free environment. It has been proven that Trp is associated with the biosynthesis of plant growth hormone auxin (Zhao, 2012). In addition, Yang et al. (2017b) reported considerably altered phenology of F2 EPSPS transgene-present hybrid lineages with two wild rice populations in the glyphosate-free environment. Altogether, these findings suggest that transgenes overproducing EPSPS will change fitness of crop-weed/wild hybrids.
Gressel et al. (2014) and Grunewald and Bury (2014) questioned whether the enhanced fecundity and metabolic traits in the glyphosate-tolerant crop-weed hybrids overexpressing EPSPS was due to the position effect of the gene insertion or the possible linkage with neighboring sequences because only a single transgenic event was involved in the study of Wang et al. (2014). It is therefore critical to confirm the observed increases in fecundity and metabolic traits are caused by the EPSPS transgene per-se, rather than the other actions of transgenes. In addition, it is necessary to address whether the observed changes in fecundity by overexpression of the EPSPS transgene is a general phenomenon? In other words, can the phenomenon observed in rice (Oryza) be also found in very distant plant species such as Arabidopsis? To answer the above question, we produced multiple independent events of transgenic Arabidopsis thaliana plants overexpressing exogenous EPSPS genes from different sources (rice and Agrobacterium), driven by a cauliflower mosaic virus 35S promotor (pCaMV35S). We also produced transgenic plants only containing the selective marker gene (nptII) driven by pCaMV35S as the empty vector (EV) control. The T2 and T3 segregating transgene-present and transgene-absent lineages, EV and parent controls were compared to estimate differences in glyphosate tolerance and fitness-related traits (Figure 1) to test whether the observed changes in rice would also occur in a phylogenetically distant plant species.
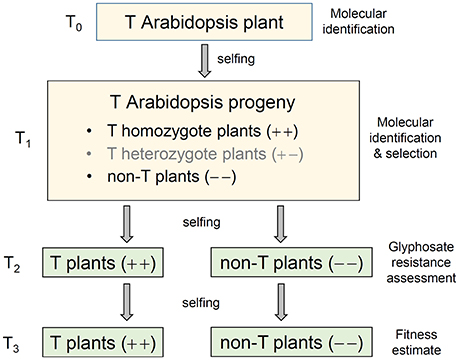
Figure 1. A schematic pedigree to illustrate the production of transgenic (T) Arabidopsis progeny containing genes overexpressing the 5-enolpyruvoylshikimate-3-phosphate synthase (EPSPS). A T0 transgenic Arabidopsis plant was self-pollinated to produce the T1 progeny that contained transgene-homozygote (+ +), transgene-heterozygote (+ –), and transgene-absent (– –) plants. The isogenic transgene-homozygote (+ +) and transgene-absent (– –) lineages generated from T1 plants through self-pollination (selfing) and molecular identification were retained for the experiments of glyphosate tolerance (T2), and EPSPS transgene expression, biomass and auxin (IAA) content, and fitness assessment (T3).
The objectives of this study are to address the following questions: (1) Does overproduction of EPSPS increase glyphosate tolerance of the Arabidopsis plants in transgenic lineages? (2) Does overexpression of exogenous EPSPS genes enhance fitness of transgenic Arabidopsis plants? (3) If so, is enhanced fitness of the transgenic plants caused by the position effect of an inserted gene? (4) Do transgene-present lineages that overproduce EPSPS synthesize more auxin than transgene-absent lineages? Answers to these questions will increase our understanding of the general effects of overexpressing EPSPS genes on the phenotypes of plant species.
An A. thaliana strain Columbia (Col 0, coded as P) was used as the parent to produce the comparative transgene-present and transgene-absent lineages in this study. Three EPSPS genes were used to develop transgenic constructs: two isolated from cultivated rice (Zhou et al., 2006; Su et al., 2008) and one (CP4, coded as C) from an Agrobacterium sp. strain (Padgette et al., 1996). The two genes from rice included one normal EPSPS (coded as E, Zhou et al., 2006) and another mutant EPSPS that was a C317-T mutation (coded as Em, Su et al., 2008). This Em gene was obtained by an error-prone PCR (polymerase chain reaction) technique and an Em transgenic rice line conferred high level of tolerance to glyphosate (Su et al., 2008; Lu et al., 2014a,b). The vector pCHF3 was used for overexpression transgenic constructs that contained one of the target EPSPS genes and a selective marker gene (nptII), both driven by pCaMV35S, respectively (Figure S1, upper panel). The nptII (neomycin phosphotransferase) marker gene conferred tolerance to kanamycin. An empty vector only including the selective marker gene (nptII) driven by pCaMV35S was also developed (Figure S1, lower panel) as a control.
The transgenic constructs were introduced into Agrobacterium tumefaciens first and then transformed into Arabidopsis plants by the floral-dip method (Clough and Bent, 1998) for genetic transformation. All seeds from the transformation-treated Arabidopsis plants were germinated on the 1/2 MS culture media, including 30 g sucrose, 2.2 g M519 (Murashige & Skoog Basal Medium with Vitamins), 50 ng kanamycin, and 8 g agar per liter (pH 5.7). All survived plants (T0) from the 1/2 MS culture media were subjected to molecular identification for the target and marker transgenes ($Appendix S2) to confirm their transgenic status. The primer sequences for the EPSPS and mutant EPSPS transgenes were 5′-acgaatgagggagagaccga-3′ and 5′-accatcagcgaagagtgcaa-3′; whereas those for the CP4 transgene were 5′-gcgtcgccgatgaaggtgctgt-3′ and 5′-cggtccttcatgttcggcggtctc-3′. Primer sequences for the nptII marker gene were 5′-taaagcacgaggaagcggtc-3′ and 5′-gatggattgcacgcaggttc-3′. All the primers were synthesized by the Shanghai Sangon Biotech Co., Ltd (Shanghai, China). To retain transgenic plants that are presumed to have a single copy of the transgenes (E, Em, or C), we selected T1 progeny generated from self-pollination of a T0 transgenic plant with a 3: 1 segregation ratio for kanamycin tolerant: sensitive, and confirmed by molecular identification (Figure S2) for further experiments.
To determine the transgene insertion or position effect causing the fitness change, we retained three transgenic events from each of the three transgenic constructs (E, Em, or C) with the above-average level of EPSPS expression for each transgenic construct in the T2 and T3 generations for further analyses (Appendix S3). The reason for measuring transgenic events in T2 and T3 was to examine the stability of EPSPS expression between generations. Transgenic events with the highest level of EPSPS expression in the T2 and T3 generations (Table S1) were excluded from the experiment to avoid the extreme cases. We only retained the isogenic lineages with transgenic homozygote (+ +) and transgene-absent plants (– –) from T1 segregating populations after molecular identification and selection for the transgenes in the T2 generation (Figure 1, Appendix S2). A total of 20 Arabidopsis lineages, including three transgene-present lineages (E+, Em+, and C+) for each of the three events (9), three segregated transgene-absent lineages (E–, Em–, and C–) for each of the three events each representing the three transgenes (9), one empty vector lineage (EV), and one parental strain (P), were used for further experiments.
Twenty Arabidopsis lineages in the T2 generation (Figure 1) were used to test glyphosate (Roundup®, glyphosate isopropyl amine salt aqueous solution: 41%) tolerance in a dose-response experiment. For each lineage (event, EV, or parent), three replicates (n = 3) each included three plants grown in a growth chamber (22°C), were included. A total of 180 plants were included in the test for glyphosate tolerance for the 20 lineages at each glyphosate concentration. Forty days after seeds were sown, all plants were sprayed with glyphosate aquatic solution at nine different concentrations: 0, 0.1, 0.2, 0.4, 0.8, 1.0, 1.2, 2.0, 5.0 mM, in which 0.4 mM equivalent to the concentration of 840 g/ha is the commonly used dosage for glyphosate application in fields for the agricultural weed control (Norsworthy et al., 2008). Therefore, a total of 1,620 plants was used in the glyphosate tolerance experiment. Survival ratios were determined as the number of surviving Arabidopsis plants as percent of the total number of plants used for analyses 15 days after glyphosate application. Plants that were partially green but with green meristems and central rosettes were scored as “surviving;” plants that had white meristems and central rosettes were scored as “dead” (Figure S3).
Twenty Arabidopsis lineages in the T3 generation (Figure 1) were used to determine EPSPS transgene expression and EPSPS protein content. Each Arabidopsis plant in the T3 generation was equally divided as two parts for measuring EPSPS transgene expression and the EPSPS protein content, respectively. For each of the 20 lineages (6 E+ and E–, 6 Em+ and Em–, 6 C+ and C–, 1 EV and 1 P), a pooled sample (each including 8 plants) with three replicates (n = 3) were used for these measurements 30 days after seeds were sown. Therefore, a total of 480 plants was used in this experiment.
Real-time PCR was applied to determine the expression of EPSPS transgene relative to an ubiquitin reference gene (UBQ) from Arabidopsis. Total RNA was isolated from the 30-day-old plants using the RNAsimple Total RNA kit (TianGen, Beijing, China). DNA removal and RNA reverse-transcription were conducted using the PrimeScript® RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). The primers for real-time PCR were designed using the software Primer PREMIER ver. 5.0. The primer sequences for the EPSPS and mutant EPSPS transgenes were 5′-aaggatgcgaaagagg-3′ and 5′-caacccgacaaccaa-3′; whereas those for the CP4 transgene were 5′-tggattgcgatgaggg-3′ and 5′-tgatcgagatgggtggc-3′. The primer sequences for the reference gene (UBQ) were 5′-aatgtgaaggcgaagatccaagac-3′ and 5′-agacggaggacgagatgaagc-3′. The expression of the EPSPS genes in non-transgenic, EV, and parental lineages was measured using the Arabidopsis endogenous EPSPS gene. The primer sequences for the Arabidopsis endogenous EPSPS gene were 5′-aacgcaagttatgtcc-3′ and 5′-gcagttagtgccaag-3′. All the primers were synthesized by the Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). The real-time PCR reaction mixture kit (TaKaRa, Dalian, China) included 1 μl of template cDNA, 0.4 μl each of the forward and reverse 10 μM primers, and 1 × SYBR®Premix Ex TaqTM in a final volume of 20 μl. PCR reaction was conducted at 94°C 30 s; 94°C 15 s, 60°C 15 s, and 40 s at 72°C for 40 cycles.
The sandwich technique of ELISA (enzyme linked immunosorbent assay) was performed to determine the EPSPS protein content. The total proteins were extracted based on the method of phosphate-buffered saline (PBS, 28.7 g Na2HPO4-12H2O and 2.96 g NaH2PO4-2H2O, pH 7.4) with 10% (v/v) methanol in 0.1 M PBS to suspend the samples after ground into powder in liquid nitrogen. The samples were then incubated in ice bath for 40 min, followed by centrifuged at 8,000 × g, at 4°C for half an hour. The supernatant was collected for EPSPS protein content determination with the Quantiplate kit (Envirologix, Portland, OR, USA) for detecting EPSPS protein in plants and bacteria following the manufacturer's instructions. The Microplate Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to detect the optical density (OD) at the wavelength of 450 nm.
Twenty Arabidopsis lineages in the T3 generation were used to estimate fitness (Figure 1). Nine fitness-related traits were used for the measurement at different times: seed germination under normal and stressed conditions, leaf area, plant height and branching, the number of siliques per plant, number of seeds per silique and per plant (Table S2). To estimate seed germination ratios for each lineage, six replicate samples (n = 6) each with 50 seeds were germinated on 1/2 MS culture media under the normal, heat, and drought stress conditions, respectively (Table S2). To estimate the other fitness-related traits (Appendix S5), six replicates (n = 6) each with four plants were grown in each of the 20 lineages in a growth chamber (22°C). The layout of the total 120 replicates (pots) followed a completely randomized design.
Six Arabidopsis lineages with or without the EPSPS transgenes (E, Em, and C) in the T3 generation (Figure 1; Table S1) were used to measure biomass and auxin content. The auxin content was determined as the average total weight (ng) of auxin in plants at the same growth stage. One transgenic event (E2, Em3, or C2) each representing one of the three transgenic constructs (see Table S1) was randomly selected to measure plant biomasses and the auxin content, using fresh samples of the entire 30-day-old plants. From each event, five independent replicates (n = 5), each represented by five different plants of transgene-present or transgene-absent lineages, were measured for the biomass and auxin content (a pooled sample from five plants per replicate, Table S2), following the methods of Chen et al. (2012). A total of 150 plants were used for determining biomasses and the auxin content. The empty vector and parental lineages were not included for analyses.
One way ANOVA was conducted to examine the effect of the presence or absence of each EPSPS transgene, involving transgene-present (E+, Em+, or C+), transgene-absent (E–, Em–, or C–), EV, and parental lineages. One way ANOVA was also conducted to determine the effect of different events from each transgenic construct (E, Em, or C) on fitness-related traits. Duncan's multiple range test was conducted to determine significant differences in gene expression, protein content, and the fitness-related traits that showed the significant transgenic effect among transgene-present, transgene-absent, EV, and parental lineages based on one way ANOVA. The independent t-tests with Bonferroni corrections were conducted to test for significant differences in all measured fitness-related traits and auxin content between transgene-present and transgene-absent lineages, after the measured values were subject to the Levene's test for equality of variances. All statistical analyses were performed using the software SPSS ver. 19.0 (IBM Inc., New York, USA).
Substantially increased tolerance to glyphosate was detected in Arabidopsis plants of transgene-present lineages of all events representing the three transgenic constructs, compared to those of their transgene-absent, EV, and parental lineages in the T2 generation (Table 1; Figure S3a,b). The three types of transgenic Arabidopsis plants (E+, Em+, and C+) survived at different concentrations (0.1–1.2 mM) of glyphosate, although with some degrees of variation (~10–35%) among transgenic constructs at the same concentration. More than 50% transgenic plants of the three constructs survived when the concentration of glyphosate increased to 1.2 mM (Table 1). However, none of the transgene-absent Arabidopsis plants (E–, Em–, and C–) survived at the concentration of 0.4 mM glyphosate (Table 1), which was equivalent to the commonly applied glyphosate dosage (840 g/ha) in the field. Notably, ~7% plants in the transgenic CP4 lineages (C+) survived at the concentration of 2.0 mM glyphosate.
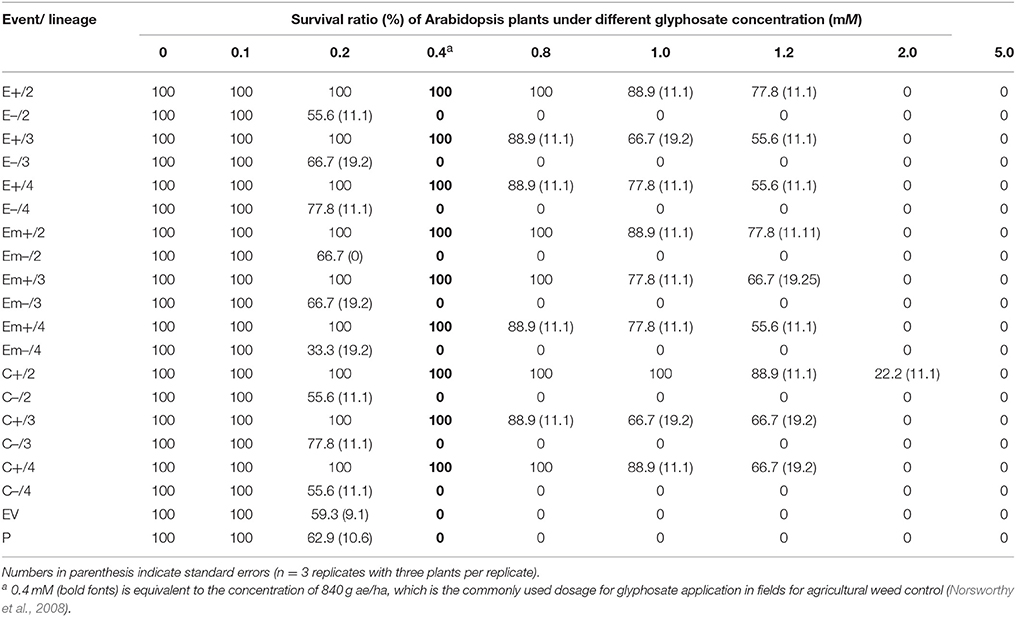
Table 1. Average survival ratios (%) of Arabidopsis thaliana plants in T2 transgene-present (E+, Em+, or C+) lineages overexpressing EPSPS and their segregating transgene-absent (E–, Em–, or C–), empty vector (EV), and parental (P) lineages under different glyphosate concentrations.
Significantly increased expression of the three EPSPS transgenic constructs (E, Em, and C) and content of the EPSPS proteins were detected in Arabidopsis plants of transgene-present lineages, compared to those of transgene-absent, EV, and the parental lineages (Table 2). The average values of relative expression of the three transgenes: EPSPS, mutant EPSPS, and CP4, measured by real-time PCR were significantly greater (P < 0.01) in the T3 transgene-present lineages (E+, Em+, and C+) than those in their corresponding transgene-absent (E–, Em–, and C–), the EV, and parental lineages based on the Duncan's multiple range test (Table 2). In addition, the average values of EPSPS protein content measured by ELISA were also significantly greater (P < 0.05) in the T3 transgene-present lineages (E+, Em+, and C+) than those in their corresponding transgene-absent (E–, Em–, and C–), the EV and parental lineages based on the Duncan's multiple range test (Table 2).
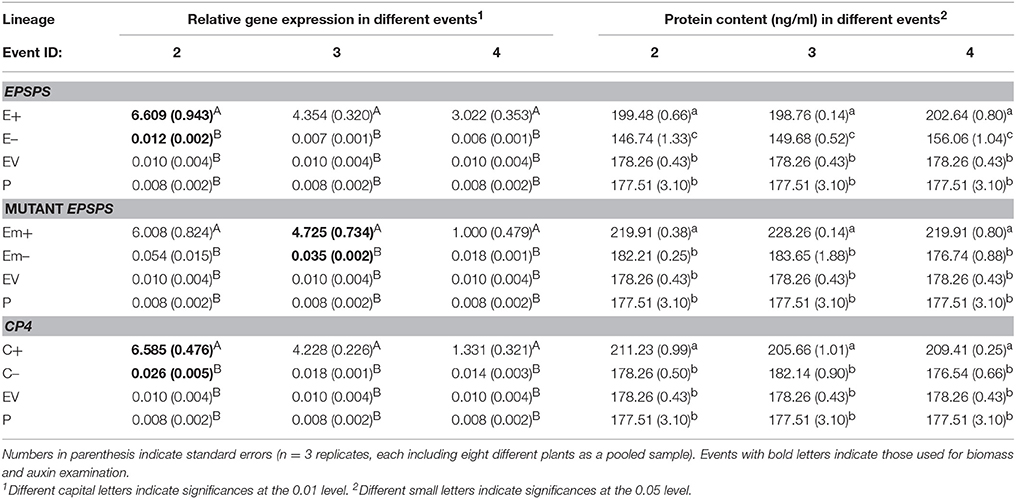
Table 2. Means of relative transgene expression and EPSPS protein content in T3 Arabidopsis thaliana plants of transgene-present (E+, Em+, and C+), transgene-absent (E–, Em–, and C–), empty vector (EV), and the parental lineages, measured by real-time PCR (polymerase chain reaction) and ELISA (enzyme linked immunosorbent assay), respectively.
We first tested differences in all included fitness-related traits among the three transgenic events based on each transgene-present or transgene-absent lineage using one-way ANOVA. Because none of these traits showed significant differences among the transgenic events (P < 0.05, Table S3), we grouped data from three events each with six replicates (n = 18) for each transgenic construct in subsequent analyses (Table S4). Thus, for seed germination, we analyzed data from 1,800 seeds per transgene for each treatment, while for other fitness-related traits, we analyzed data from 144 plants per transgene. One-way ANOVAs indicated significant effects of the presence versus absence of the three transgenes (E, Em, or C) on some fitness-related traits in the T3 generation (Table 3), including relative leaf area, plant height, and the numbers of siliques and seeds per plant (Table 3).
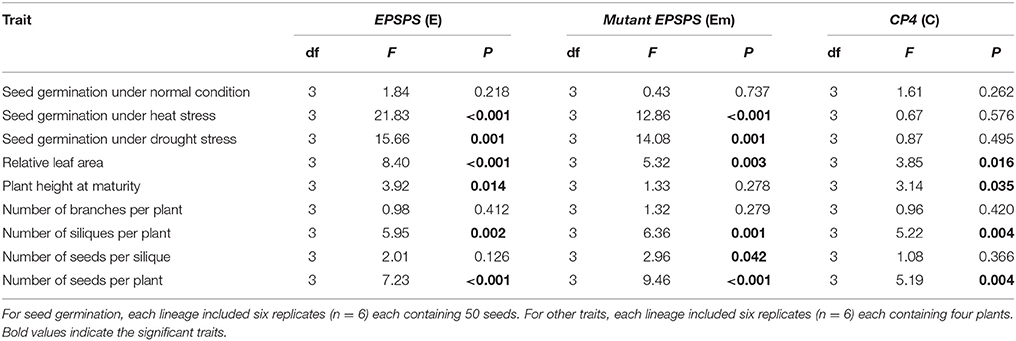
Table 3. One-way ANOVA to test the effects of each of the three epsps transgenes on fitness-related traits in T3 Arabidopsis thaliana plants, including the transgene-present (3), transgene-absent (3), empty vector (1), and parental (1) lineages.
All of these significant differences were in the direction of increased fitness for the transgene-present lineages relative to the controls. For example, overproduction of EPSPS was associated with increases of ~12–22% (n = 18) more seeds to germinate under the heat and drought stresses for the E and Em transgene-present lineages (Figures 2A–F; Table S4), although no significant differences in seed germination under the normal condition (22°C) (Table S4). Notably, the C+ transgene-present lineage showed a significant increase in seed germination under the drought stress (Figure 2F). In addition, overproduction of EPSPS resulted in 22–28% (n = 18) more siliques and 23–27% (n = 18) more seeds per plant (Figures 3A–F; Table S4). The number of branches per plant and seeds per silique was very similar across lineages (Table S4). Overall, the presence of the three transgenes was associated with larger plants (Figures 4A–F; Table S4) with enhanced fecundity (Figures 3A–F; Table S4) in the glyphosate-free environment. We did not detect evidence of fitness benefits or costs associated with the single EV lineage, compared to the parental and transgene-absent lineages.
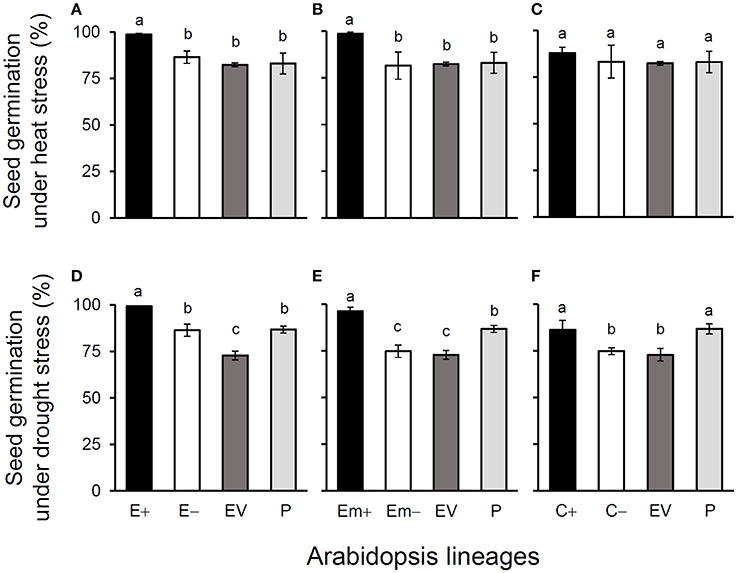
Figure 2. Average % seed germination of three Arabidopsis transgenic events under heat (A–C) and drought (D–F) stresses in T3 transgene-present, transgene-absent, empty vector (EV), and parent (P) lineages in the glyphosate-free environment. Different letters above the columns indicate significances at P < 0.05 based on Duncan's multiple range test (n = 18). E+: EPSPS transgene-present lineages, E–: EPSPS transgene-absent lineages, Em+: mutant EPSPS transgene-present lineages, Em–: mutant EPSPS transgene-absent lineages; C+: CP4 transgene-present lineages, C–: CP4 transgene-absent lineages. Bars represent standard errors.
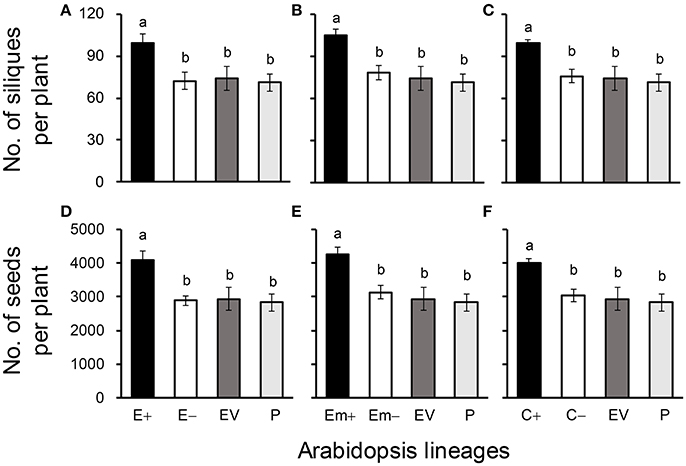
Figure 3. Average silique (A–C) and seed production (D–F) of three Arabidopsis transgenic events in T3 transgene-present, transgene-absent, empty vector (EV), and parent (P) lineages in the glyphosate-free environment. Different letters above the columns indicate significances at P < 0.05 based on Duncan's multiple range test (n = 18). E+: EPSPS transgene-present lineages, E–: EPSPS transgene-absent lineages, Em+: mutant EPSPS transgene-present lineages, Em–: mutant EPSPS transgene-absent lineages; C+: CP4 transgene-present lineages, C–: CP4 transgene-absent lineages. Bars represent standard errors.
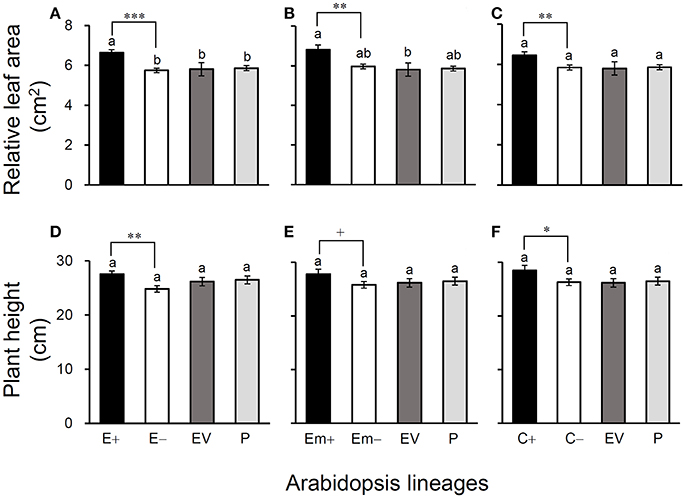
Figure 4. Average relative leaf-area (A–C) and plant height (D–F) of three Arabidopsis transgenic events in T3 transgene-present, transgene-absent, empty vector (EV), and parent (P) lineages in the glyphosate-free environment. Different letters above the columns indicate significances at P < 0.05 based on Duncan's multiple range test (n = 18). Differences between transgene-present and transgene-absent lineages were compared based on the independent t-test after Bonferroni correction (n = 18). +P <0.1, *P < 0.05, **P < 0.01, ***P < 0.001. E+: EPSPS transgene-present lineages, E–: EPSPS transgene-absent lineages, Em+: mutant EPSPS transgene-present lineages, Em–: mutant EPSPS transgene-absent lineages; C+: CP4 transgene-present lineages, C–: CP4 transgene-absent lineages. Bars represent standard errors.
Significant increases in biomass and auxin content were detected in 30-day-old plants in transgene-present E+ and Em+ lineages in the T3 generation (Figures 5A, B). However, no significant differences in biomass or auxin content were observed between transgene-present C+ and transgene-absent C– lineages, although the values were slight higher for the C+ lineage. About 28–33% increases in biomass were observed in the transgene-present E+ and Em+ lineages, compared with their transgene-absent counterparts (Figure 5A). Increases in auxin content of around 25–33% were detected in transgene-present E+ and Em+ lineages, compared with their segregating transgene-absent counterparts (Figure 5B).
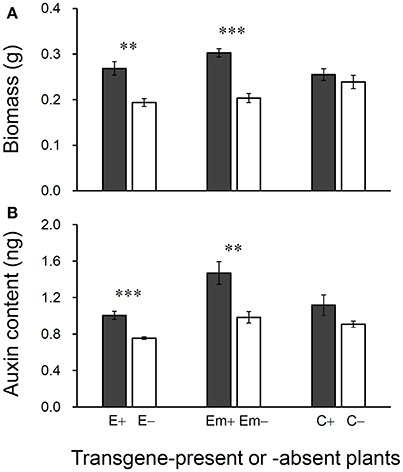
Figure 5. Differences in biomass (A) and auxin (IAA) content (B) of 30-day plants between T3 transgene-present and transgene-absent Arabidopsis lineages based on the independent t-test (n = 5). E+: EPSPS transgene-present lineage, E–: EPSPS transgene-absent lineages (event E/2); Em+: mutant EPSPS transgene-present lineages, Em–: mutant EPSPS transgene-absent lineages (event Em/3); C+: CP4 transgene-present lineages, C–: CP4 transgene-absent lineages (event C/2). **P < 0.01, ***P < 0.001. Bars represent standard errors.
Results from this study demonstrated that the transfer of overexpressing EPSPS genes isolated from different sources, namely rice (E or Em) and Agrobacterium (CP4), into A. thaliana plants substantially increased their tolerance to the glyphosate (Roundup®) herbicide although with considerable variation among the three EPSPS transgenes. This finding is based on the dose-response comparisons of >1,600 Arabidopsis plants among transgene-present lineages, their isogenic transgene-absent counterparts, the empty vector (EV), and parent controls in a climate chamber. In the dose-response experiment, all transgene-present Arabidopsis plants survived at the 0.4 mM glyphosate dosage, which is equivalent to the commonly used concentration of 840 g/ha for glyphosate application for agricultural weed control (Norsworthy et al., 2008). In contrast, none of the Arabidopsis plants in the corresponding transgene-absent lineages, EV, and parent controls survived at this dosage (0.4 mM). Increased glyphosate tolerance has also been observed in other GE plants overexpressing EPSPS transgenes, such as Petunia hybrida (Shah et al., 1986), rice (Su et al., 2008; Lu et al., 2014a,b), tobacco plants (Jones et al., 1996), and A. thaliana (Klee et al., 1987; Yang et al., 2017a), regardless of the origins (endogenous or exogenous) of EPSPS genes. All these results indicate that the transfer of an overexpressing EPSPS gene into plants, including the model plant Arabidopsis in this study and other studies (Klee et al., 1987; Yang et al., 2017a), can increase glyphosate tolerance of the transgenic plants due to overproduction of EPSPS.
Increased tolerance to glyphosate in our transgenic plants overexpressing an EPSPS gene is presumably due to the sufficiently surplus EPSPS that can bind glyphosate (Rogers et al., 1983), as reported in P. hybrida (Shah et al., 1986), tobacco plants (Jones et al., 1996), and Arabidopsis (Klee et al., 1987; Yang et al., 2017a). Thus, these results confirm the strategy of overproducing EPSPS driven by a strong promoter (e.g., pCaMV35S for dicots and pUbiquitin for monocots) to be effective in increase GE crops' tolerance to glyphosate, regardless of exogenous (as in this study) or endogenous (Klee et al., 1987; Su et al., 2008; Yang et al., 2017a). The application of an endogenous transgene overproducing EPSPS at a proper level to develop GE glyphosate-tolerant crops may have particular commercial values. That is, a glyphosate-tolerance transgene originating from the crop species rather than a bacterium or other sources may reduce consumers' concerns over the food safety issues (Kuiper et al., 2001; Qaim and Zilberman, 2003). In addition, the application would be particularly useful for GE crops such as sugar beets, potatoes, and vegetables, of which the consumed parts are vegetative organs, because the transgenic plants appear to have increased vigor, but with less opportunities of crop-to-wild/weed gene flow mediated by pollination.
As often happens in studies of transgenic plants, we also observed considerable variation in overexpression of the EPSPS genes among different transgenic events in our experiment, based on the real-time PCR analysis. Therefore, we only included transgenic events with a relatively high level of EPSPS expression because the main objective of this study was to determine whether overexpression of EPSPS genes would enhance fecundity of transgene-present plants. To address the question about the likelihood of insertion or position effect by a single transgenic event with enhanced fecundity of transgenic plants (Gressel et al., 2014; Grunewald and Bury, 2014), we included GE Arabidopsis plants of three transgenic events representing each of the three transgenic constructs with the above-average level of EPSPS overexpressing in our common-garden experiment for fitness comparisons. Our results did not show significant differences in fitness-related traits among the three included transgenic events of each transgene in transgene-present Arabidopsis lineages. In addition, we did not find significant differences in fitness-related traits between the EV and parental lineages. These results support previous findings that enhanced fitness/fecundity (as shown in (Su et al., 2008; Wang et al., 2014; Yang et al., 2017a),b) is not the consequence of an insertion or position effect (process of transgenesis), but due to the action of the transgene itself. We therefore confirm that overexpression of EPSPS in GE plants with a proper level can result in overproduction of their EPSPS and increased glyphosate tolerance.
In the common-garden experiment in a growth chamber, we observed significantly increased seed germination ratios in E+ and Em+ transgene-present lineages when seeds were exposed to the heat (28°C) and drought [200 mM D-mannitol (C6H14O6)] stresses, although no differences were found in seed germination among different lines when seeds were exposed to the normal temperature (22°C, ideal for Arabidopsis, Xiong et al., 1999). The CP4+ transgene-present lineages only showed significant increases in seed germination under the drought stress. Considering that auxin (IAA) can promote seed germination and plant growth under abiotic stresses (Woodward and Bartel, 2005; Liu et al., 2014; Naser and Shani, 2016), our explanation for enhanced seed germination under stresses can be attributed to the increased auxin content in transgenic Arabidopsis plants. The report of Leadem (1987) in which auxin stimulated seed germination under special conditions including heat and cold stresses supports our explanation. However, we propose more studies to test this hypothesis because very limited examples are found in the scientific literature.
In addition, our results indicated significantly greater values of a few major fitness-related traits, including the number of siliques and seeds per plant in transgene-present Arabidopsis lineages in glyphosate-free environment. Similar findings of increased fitness were also reported in transgene-present Arabidopsis plants containing a native gene overproducing EPSPS (Beres et al., in press). All these findings support our previous observation of the significantly enhanced fecundity in transgene-present crop-weed (Wang et al., 2014) and crop-wild (Yang et al., 2017b) hybrid lineages containing an EPSPS transgene from rice. It is apparent that the presence of a transgene overproducing EPSPS, regardless of its origin (endogenous or exogenous), may significantly enhance the fecundity of a plant. Altogether, findings from wild/weedy rice (monocot) and Arabidopsis (eudicot) indicate that overexpression of an EPSPS gene to a proper level with increased fecundity of GE plants in the glyphosate-free environments may be a general feature of angiosperms. Therefore, environmental impact caused by introgression of a transgene overexpressing EPSPS from GE glyphosate-tolerant crops into their wild/weedy relatives should be thoroughly assessed, even in the glyphosate-free environment. Further studies including hybrid descendants of transgenic crops overexpressing EPSPS with their wild relatives should be conducted to provide more evidence for the potential ecological impact.
What could be the underlying mechanisms for enhanced fecundity of GE plants containing an EPSPS overexpressing transgene in glyphosate-free environment? In this study, we detected increased auxin, an important plant growth hormone (Woodward and Bartel, 2005; Zhao, 2012; Liu et al., 2014), in transgene-present Arabidopsis lineages (E+ and Em+). We therefore hypothesize that increased total endogenous auxin may play a role in promoting the growth and development (probably also stress tolerance) of transgene-present Arabidopsis plants, eventually leading to increases in their fitness-related traits, although other factors such as enhanced photosynthetic rates by overproducing EPSPS (see Wang et al., 2014) can also promote the growth of transgene-present plants. As indicated in our previous study, significantly increased tryptophan (Trp) content was detected in four independent transgene-present crop-weed hybrid lineages overproducing EPSPS (Wang et al., 2014). Trp is an aromatic amino acid synthesized in the downstream of EPSPS in the shikimate pathway (Herrmann and Weaver, 1999; Maeda and Dudareva, 2012). Recent studies have revealed that auxin biosynthesis is a simple two-step pathway converting Trp to auxin in plants (Mashiguchi et al., 2011; Won et al., 2011). This suggests that overproduction of EPSPS may lead to increases in auxin through increased Trp (Zhao, 2012; Wang et al., 2014). Thus, the complete discovery of the precise biosynthesis pathway from EPSPS to auxin will provide deeper insight into mechanisms associated with fitness effect and environmental impact of transgenic plants that overproduce EPSPS in agricultural and natural habitats.
JF: Conducted the experiment, analyzed data, and wrote the paper; PN, ZG, XG, and Y-QF: Conducted some part of the experiment; B-RL: Conceived and designed the experiment, analyzed data, and wrote the paper. All authors reviewed the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (31330014) and the National Program of Development of Transgenic New Species of China (2016ZX08011-006). A. A. Snow and N. C. Ellstrand provided critical suggestions on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00233/full#supplementary-material
Beres, Z. T., Yang, X., Jin, L., Zhao, W., Mackey, D. M., and Snow, A. A. (in press). Over-expression of a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) may enhance fecundity in Arabidopsis thaliana in the absence of glyphosate. Int. J. Plant. Sci.
Cerdeira, A. L., and Duke, S. O. (2006). The current status and environmental impacts of glyphosate-tolerant crops: a review. J. Environ. Qual. 35, 1633–1658. doi: 10.2134/jeq2005.0378
Chen, J. C., Huang, H. J., Zhang, C. X., Wei, S., Huang, Z., Chen, J., et al. (2015). Mutations and amplification of EPSPS gene confer tolerance to glyphosate in goosegrass (Eleusine indica). Planta 242, 859–868. doi: 10.1007/s00425-015-2324-2
Chen, M. L., Fu, X. M., Liu, J. Q., Ye, T. T., Hou, S. Y., Huang, Y. Q., et al. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 905, 67–74. doi: 10.1016/j.jchromb.2012.08.005
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Délye, C., Jasieniuk, M., and Le Corre, V. (2013). Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 29, 649–658. doi: 10.1016/j.tig.2013.06.001
Duke, S. O., and Powles, S. B. (2008). Glyphosate: a once-in-a-century herbicide. Pest Manag. Sci. 64, 319–325. doi: 10.1002/ps.1518
Franz, J. E., Mao, M. K., and Sikorski, J. A. (1997). Glyphosate: A Unique Global Herbicide. Washington, DC: American Chemistry Society.
Gaines, T. A., Shaner, D. L., Ward, S. M., Leach, J. E., Preston, C., and Westra, P. (2011). Mechanism of tolerance of evolved glyphosate-tolerant Palmer amaranth (Amaranthus palmeri). J. Agric. Food Chem. 59, 5886–5889. doi: 10.1021/jf104719k
Gaines, T. A., Zhang, W., Wang, D., Bukun, B., Chisholm, S. T., Shaner, D. L., et al. (2010). Proc. Natl. Acad. Sci. U.S.A. 107, 1029–1034. doi: 10.1073/pnas.0906649107
Goldsbrough, P. B., Hatch, E. M., and Huang, B. (1990). Gene amplification in glyphosate tolerant tobacco cells. Plant Sci. 72, 53–62. doi: 10.1016/0168-9452(90)90186-R
Gressel, J., Neal Stewart, C., Giddings, L. V., Fischer, A. J., Streibig, J. C., Burgos, N. R., et al. (2014). Overexpression of epsps transgene in weedy rice: insufficient evidence to support speculations about biosafety. New Phytol. 202, 360–362. doi: 10.1111/nph.12615
Grunewald, W., and Bury, J. (2014). Comment on ‘A novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) synthase transgene for glyphosate tolerance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide' by Wang et al. (2014). New Phytol. 202, 367–369. doi: 10.1111/nph.12683
Herrmann, K. M., and Weaver, L. M. (1999). The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503. doi: 10.1146/annurev.arplant.50.1.473
James, C. (2016). Global Status of Commercialized Biotech/GM Crops: 2016. ISAAA Brief No. 52. Ithaca, NY: ISAAA. Available online at: http://www.isaaa.org/resources/publications/briefs/52/default.asp (Accessed December 10, 2017).
Jones, J. D., Goldsbrough, P. B., and Weller, S. C. (1996). Stability and expression of amplified EPSPS genes in glyphosate tolerant tobacco cells and plantlets. Plant Cell Rep. 15, 431–436. doi: 10.1007/BF00232070
Klee, H. J., Muskopf, Y. M., and Gasser, C. S. (1987). Cloning of an Arabidopsis thaliana gene encoding 5-enolpyruvylshikimate-3-phosphate synthase: sequence analysis and manipulation to obtain glyphosate-tolerant plants. Mol. Gen. Genet. 210, 437–442. doi: 10.1007/BF00327194
Kuiper, H. A., Kleter, G. A., Noteborn, H. P., and Kok, E. J. (2001). Assessment of the food safety issues related to genetically modified foods. Plant J. 27, 503–528. doi: 10.1046/j.1365-313X.2001.01119.x
Leadem, C. L. (1987). The role of plant growth regulators in the germination of forest tree seeds. Plant Growth Regul. 6, 61–93. doi: 10.1007/BF00043950
Liu, L., Guo, G., Wang, Z., Ji, H., Mu, F., and Li, X. (2014). “Auxin in plant growth and stress responses,” in Phytohormones: A Window To Metabolism, Signaling And Biotechnological Applications, eds P. T. Lam-Son and P. Sikander (New York, NY: Springer), 1–35.
Lu, B. R., Snow, A. A., Yang, X., and Wang, W. (2014a). Using a single transgenic event to infer fitness effects in crop-weed hybrids: a reply to the Letter by Grunewald & Bury (2014). New Phytol. 202, 370–372. doi: 10.1111/nph.12748
Lu, B. R., Snow, A. A., Yang, X., and Wang, W. (2014b). Scientific data published by a peer-reviewed journal should be properly interpreted: a reply to the letter by Gressel et al. (2014). New Phytol. 202, 363–367. doi: 10.1111/nph.12684
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Martin, S. L., Benedict, L., Sauder, C. A., Wei, W., da Costa, L. O., Hall, L. M., et al. (2017). Glyphosate tolerance reduces kochia fitness: comparison of segregating tolerant and susceptible F2 populations. Plant Sci. 261, 69–79. doi: 10.1016/j.plantsci.2017.04.010
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Mueller, T. C., Massey, J. H., Hayes, R. M., Main, C. L., and Stewart, C. N. Jr. (2003). Shikimate accumulates in both glyphosate-sensitive and glyphosate-tolerant horseweed (Conyza canadensis L. Cronq.). J. Agric. Food Chem. 51, 680–684. doi: 10.1021/jf026006k
Nandula, V. K., Ray, J. D., Ribeiro, D. N., Pan, Z., and Reddy, K. N. (2013). Glyphosate tolerance in tall waterhemp (Amaranthus tuberculatus) from Mississippi is due to both altered target-site and nontarget-site mechanisms. Weed Sci. 61, 374–383. doi: 10.1614/WS-D-12-00155.1
Naser, V., and Shani, E. (2016). Auxin response under osmotic stress. Plant Mol. Biol. 91, 66–1672. doi: 10.1007/s11103-016-0476-5
Norsworthy, J. K., Griffith, G. M., Scott, R. C., Smith, K. L., and Oliver, L. R. (2008). Confirmation and control of glyphosate-resistant Palmer Amaranth (Amaranthus palmeri) in Arkansas. Weed Technol. 22, 108–113. doi: 10.1614/WT-07-128.1
Owen, M. D. K., Pedersen, P., De Bruin, J. L., and Stuart, J. (2010). Comparisons of genetically modified and non-genetically modified soybean cultivars and weed management systems. Crop Sci. 50, 2597–2604. doi: 10.2135/cropsci2010.01.0035
Padgette, S. R., Taylor, N. B., Nida, D. L., Bailey, M. R., MacDonald, J., Holden, L. R., et al. (1996). The composition of glyphosate-tolerant soybean seeds is equivalent to that of conventional soybeans. J. Nutr. 126, 702–716.
Qaim, M., and Zilberman, D. (2003). Yield effects of genetically modified crops in developing countries. Science 299, 900–902. doi: 10.1126/science.1080609
Qiu, J. (2013). Genetically modified crops pass benefits to weeds: Herbicide tolerance could confer an advantage on plants in the wild. Nature. 500, 389–390. doi: 10.1038/nature.2013.13517
Reichman, J. R., Watrud, L. S., Lee, E. H., Burdick, C. A., Bollman, M. A., Storm, M. J., et al. (2006). Establishment of transgenic herbicide-tolerant creeping bentgrass (Agrostis stolonifera L.) in nonagronomic habitats. Mol. Ecol. 15, 4243–4255. doi: 10.1111/j.1365-294X.2006.03072.x
Roberts, F., Roberts, C. W., Johnson, J. J., Kyle, D. E., Krell, T., Coggins, J. R., et al. (1998). Evidence for the shikimate pathway in apicomplexan parasites. Nature 393, 801–805. doi: 10.1038/31723
Rogers, S. G., Brand, L. A., Holder, S. B., Sharps, E. S., and Brackin, M. J. (1983). Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl. Environ. Microbiol. 46, 37–43.
Ryffel, G. U. (2014). Transgene flow: facts, speculations and possible countermeasures. GM Crops Food 5, 249–258. doi: 10.4161/21645698.2014.945883
Sammons, R. D., and Gaines, T. A. (2014). Glyphosate tolerance: state of knowledge. Pest Manag. Sci. 70, 1367–1377. doi: 10.1002/ps.3743
Shah, D. M., Horsch, R. B., Klee, H. J., Kishore, G. M., Winter, J. A., Tumer, N. E., et al. (1986). Engineering herbicide tolerance in plants. Science 233, 478–481. doi: 10.1126/science.233.4762.478
Shyr, Y. Y., Hepburn, A. G., and Widholm, J. M. (1992). Glyphosate selected amplification of the 5-enolpyruvylshikimate-3-phosphate synthase gene in cultured carrot cells. Mol. Gen. Genet. 232, 377–382. doi: 10.1007/BF00266240
Su, J., Chen, G. M., Tian, D. G., Zhu, Z., and Wang, F. A. (2008). A gene encodes 5-enolpyruvylshikimate-3-phosphate mutagenized by error-prone PCR conferred rice with high glyphosate-tolerance. Mol. Plant Breed. 7, 830–836. doi: 10.3969/j.issn.1672-416X.2008.05.002
Vats, S. (2015). Herbicides: history, classification and genetic manipulation of plants for herbicide tolerance. Sustain. Agric. Rev. 15, 153–192. doi: 10.1007/978-3-319-09132-7_3
Vila-Aiub, M. M., Goh, S. S., Gaines, T. A., Han, H., Busi, R., Yu, Q., et al. (2014). No fitness cost of glyphosate tolerance endowed by massive EPSPS gene amplification in Amaranthus palmeri. Planta 239, 793–801. doi: 10.1007/s00425-013-2022-x
Vila-Aiub, M. M., Gundel, P. E., and Preston, C. (2015). Experimental methods for estimation of plant fitness costs associated with herbicide-tolerance genes. Weed Sci. 63, 203–216. doi: 10.1614/WS-D-14-00062.1
Wang, W., Xia, H., Yang, X., Xu, T., Si, H. J., Cai, X. X., et al. (2014). A novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) synthase transgene for glyphosate tolerance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide. New Phytol. 202, 679–688. doi: 10.1111/nph.12428
Warwick, S., Legere, A., Simard, M., and James, T. (2008). Do escaped transgenes persist in nature? The case of an herbicide tolerance transgene in a weedy Brassica rapa population. Mol Ecol. 17, 1387–1396. doi: 10.1111/j.1365-294X.2007.03567.x
Weaver, L. M., and Herrmann, K. M. (1997). Dynamics of the shikimate pathway in plants. Trends Plant Sci. 2, 346–351. doi: 10.1016/S1360-1385(97)84622-5
Wegier, A., Piñeyro-Nelson, A., Alarcón, J., Gálvez-Mariscal, A., Alvarez-Buylla, E. R., and Piñero, D. (2011). Recent long-distance transgene flow into wild populations conforms to historical patterns of gene flow in cotton (Gossypium hirsutum) at its centre of origin. Mol. Ecol. 20, 4182–4194. doi: 10.1111/j.1365-294X.2011.05258.x
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 185–191. doi: 10.1073/pnas.1108436108
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Annu. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Xiong, L., Ishitani, M., and Zhu, J. K. (1999). Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 119, 205–212. doi: 10.1104/pp.119.1.205
Yang, X., Beres, Z. T., Jin, L., Parrish, J. T., Zhao, W., Mackey, D., et al. (2017a). Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate tolerance in Arabidopsis thaliana. PLoS ONE 12:e0175820. doi: 10.1371/journal.pone.0175820
Yang, X., Li, L., Jiang, X., Wang, W., Cai, X., Su, J., et al. (2017b). Genetically engineered rice endogenous 5-enolpyruvoylshikimate-3-phosphate synthase (epsps) transgene alters phenology and fitness of crop-wild hybrid offspring. Sci. Rep. 7:6834. doi: 10.1038/s41598-017-07089-9
Zapiola, M. L., and Mallory-Smith, C. A. (2017). Pollen-mediated gene flow from transgenic perennial creeping bentgrass and hybridization at the landscape level. PLoS ONE 12:e0173308. doi: 10.1371/journal.pone.0173308
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zhou, H., Berg, J. D., Blank, S. E., Chay, C. A., Chen, G., Eskelsen, S. R., et al. (2003). Field efficacy assessment of transgenic roundup ready wheat. Crop Sci. 43, 1072–1075. doi: 10.2135/cropsci2003.1072
Keywords: abiotic stress, Arabidopsis thaliana, fitness, glyphosate-tolerance, growth hormone, indole-3-acetic acid, seed germination, transgenic plant
Citation: Fang J, Nan P, Gu Z, Ge X, Feng Y-Q and Lu B-R (2018) Overexpressing Exogenous 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) Genes Increases Fecundity and Auxin Content of Transgenic Arabidopsis Plants. Front. Plant Sci. 9:233. doi: 10.3389/fpls.2018.00233
Received: 15 January 2018; Accepted: 09 February 2018;
Published: 27 February 2018.
Edited by:
Andrew F. Roberts, International Life Sciences Institute, United StatesReviewed by:
Alan John Gray, Centre for Ecology & Hydrology, United KingdomCopyright © 2018 Fang, Nan, Gu, Ge, Feng and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Rong Lu, YnJsdUBmdWRhbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.