- 1State Key Laboratory of Crop Stress Biology for Arid Areas and College of Life Sciences, Northwest A&F University, Yangling, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas and College of Plant Protection, Northwest A&F University, Yangling, China
Highly conserved mitogen-activated protein kinase (MAPK) cascades regulate numerous plant processes, including hormonal responses, stress, and innate immunity. In this research, TaMAPK4 was predicted to be a target of tae-miR164. We verified the binding and suppression of TaMAPK4 by co-expression in Nicotiana benthamiana. Moreover, we found TaMAPK4 was localized in the cytoplasm and nucleus using transient expression analyses. TaMAPK4 transcripts increased following salicylic acid (SA) treatment and when host plants were infected with an avirulent race of the stripe-rust pathogen. Silencing of TaMAPK4 by virus-induced gene silencing permitted increased colonization by the avirulent pathogen race. Detailed histological results showed increased Puccinia striiformis (Pst) hyphal length, hyphal branches, and infection uredinial size compared to the non-silenced control. SA accumulation and the transcript levels of TaPR1, TaPR2, and TaPR5 were significantly down-regulated in TaMAPK4 knockdown plants. Overall, these results suggest that TaMAPK4 plays an important role in signaling during the wheat-Pst interaction. These results present new insights into MAPK signaling in wheat defense to rust pathogen.
Introduction
MicroRNAs (miRNAs) are negative post-transcriptional regulators of gene expression and play important roles in multiple biological processes, including plant immunity (Bartel, 2004; Navarro et al., 2006). The conserved plant miRNA miR164 regulates multiple target genes through cleavage to mRNA (Kim et al., 2009). miR164 participates in a range of physiological processes, including flower development, age-dependent cell death, and plant defense (Laufs et al., 2004; Kim et al., 2009; Li et al., 2014). Plants miR164 plays important roles in responding to biotic stresses; this molecule regulates the transcript levels of the genes to which it binds. For example, miR164 was induced in rice inoculated with Magnaporthe oryzae (Li et al., 2014). TaNAC21 in wheat is a target gene of miR164, leading to increase susceptibility to stripe-rust in wheat (Feng et al., 2014). In previous research, we predicted that miR164 binds to a mitogen-activated protein kinase (MAPK) gene in wheat (Wang et al., 2014). MAPK cascades are pivotal signal transduction modules in plants.
Mitogen-activated protein kinase pathways are evolutionarily conserved across the animal and plant kingdoms (Tena et al., 2001). Three kinase classes, including MAPKKK, MAPKK, and MAPK, compose a typical MPK cascade (Hao et al., 2015). Multiple MAPK genes have been described in plants, including Arabidopsis, tomato, and rice (Zhang and Klessig, 1998; Cardinale et al., 2002; Hamel et al., 2006).
Mitogen-activated protein kinase cascades play important roles in plant defense involving pathogen-associated molecular pattern (PAMPs)-triggered immunity (PTI), which was composed of plant-innate immunity (Jones and Dangl, 2006; Cristina et al., 2010). The best characterized MAPKs are MPK3 and MPK6 in Arabidopsis, which play positive roles in regulating of the defense response (Pitzschke et al., 2009). Pathogen effectors suppress MAPK activation to override plant defense responses. For example, MPK3 and MPK6 are inactivated by bacterial pathogen effector HopAI1 to suppress PAMP-induced genes (Hématy et al., 2009). Moreover, MAPK also can play negative regulatory roles in plant defense (Petersen et al., 2000). In tobacco (Nicotiana tabacum), the genes controlling salicylic acid-induced protein kinase (SIPK) are activated by various pathogen-related signals (Zhang and Klessig, 1998). An MAPK gene in rice, BWMK1, mediates the expression of many pathogenesis-related genes by the activation of the transcription factor OsEREBP1 (rice ethylene-responsive element-binding protein 1), resulting in increased resistance to pathogens (Cheong et al., 2003). MAPK cascades participate in various plant defense signaling processes such as biosynthesis and the subsequent signaling of salicylic acid (SA) (Petersen et al., 2000; Beckers et al., 2009). In the present study, we predicted one candidate target of miR164 in wheat was TaMAPK4 (not the homolog of Arabidopsis MPK4), which involved wheat responses to multiple stresses (Hao et al., 2015). TaMAPK4 shared high-sequence similarities to Arabidopsis MPK1, whose role in the plant defense reaction was still unknown.
Wheat stripe rust, which is caused by Puccinia striiformis f. sp. tritici (Pst), is among the most destructive wheat diseases worldwide, and wheat yield can be greatly reduced or even completely destroyed at an epidemic level (Wang et al., 2017). In the present study, we demonstrate TaMAPK4 could be regulated by miR164. TaMAPK4 localizes to the cytoplasm and the nucleus. Moreover, functional characterization supported that TaMAPK4 is a positive regulator of resistance to Pst in wheat. Our results suggest that miRNAs and MAPK signaling might be involved in regulating plant immunity and defense.
Materials and Methods
Plant Materials and Inoculation
Su11 is a Chinese wheat cultivar susceptible to Pst race CYR31 (compatible interaction) and resistant to CYR23 (incompatible interaction). The method of wheat seedling culture, inoculation, and incubation followed was previously described by Kang et al. (2002). Parallel mock inoculations were performed using tap water. After inoculation for 24 h, the seedlings were transferred to a 14°C growth chamber with a 16-h photoperiod. The control and inoculated wheat leaves were harvested at 0, 12, 24, 48, 72, and 120 h post-inoculation (hpi) and immediately frozen in liquid nitrogen. Nicotiana benthamiana was cultured in a growth chamber with a 16-h/8-h photoperiod at 25°C.
Hormone Treatments
For the hormone treatments, 2-week-old wheat seedlings at the second to third leaf-growth stages were separately sprayed with 2 mM SA, 100 μM methyl jasmonate (MeJA), a 100 μM ethephon (ET) solution, all of which were dissolved in 0.1% (v/v) ethanol (Wang et al., 2017). The control plants were sprayed with 0.1% (v/v) ethanol as for hormone treatments. Leaf samples were harvested at 0, 6, 12, and 24 hours post-treatment (hpt).
RNA Extraction and Quantitative Real-Time PCR Analysis
A PureLink RNA Mini Kit (Invitrogen, Beijing, China) was used to total RNA extract. A Revert Aid First-strand cDNA Synthesis Kit from Fermentas (Waltham, MA, United States) was used to synthesize cDNA from RNA. Quantitative real-time PCR (qRT-PCR) was performed on a CFX96 Real-Time System (Bio-Rad, Munich, Germany) using SYBR Green I (Invitrogen) for fluorescence. The total volume for PCR was 25 μl. The following PCR conditions were used: 95°C for 1 min, followed by 39 cycles at 95°C for 10 s, 55°C for 30 s, and 72°C for 1 min, with a final cycle at 72°C for 5 min. To standardize the data, the wheat-elongation factor TaEF-1α was used as an endogenous control for qRT-PCR analyses in wheat. The primers used in qRT-PCR are listed in Supplementary Table S2. Relative gene expression data were quantified using the comparative 2-ΔΔCT method (Livak and Schmittgen, 2001).
Plasmid Constructions
The primers used for plasmid construction in the present study are listed in Supplementary Table S2. TaMAPK4 was cloned from the cDNA of the wheat cultivar Su11 using FastPfu DNA Polymerase (TransGen Biotech, Beijing). We used TaMAPK4 in the vector pCAMBIA-1302 as a template to amplify the gene using specific primers with PstI and XbaI restriction enzyme sites.
The tae-miR164 precursor miR164 was digested using restriction endonucleases SacI and BamHI, and cloned into the pBI121 vector. The region containing the cleavage site in TaMAPK4 sequences was digested using the restriction endonucleases XbaI and BamHI and cloned into the pBI121 vector.
The plasmids used for virus-induced gene silencing (VIGS) were based on previously described constructs (Holzberg et al., 2002). RNA-derived clones (BSMV-TaMAPK4) were created using BSMV-TaPDS in which wheat phytoene desaturase (TaPDS) sequence fragments were replaced with specific TaMAPK4 sequences. The amplicon of TaMAPK4 was cut using NotI and PacI, and ligated into the sites of the BSMV: γ vector.
Subcellular Localization
A TaMAPK4-GFP fusion vector was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Strains carrying the different recombinant vectors [empty vector (EV) and pCAMBIA-1302-TaMAPK4] were cultured to an optical density of 0.8 at 600 nm (OD600) for injection into the leaves of N. benthamiana (4-week-old). Infiltrated seedlings were cultured in a growth chamber with a 16-h/8-h photoperiod at 25°C. Infiltrated leaf tissue samples were harvested at 2 days post-infiltration. DAPI was added to the cell suspensions at a concentration of 5 μg ml-1 to counter-stain the nuclei. GFP signals were detected using an Olympus BX-51 microscope (Olympus, Corp., Tokyo, Japan).
Next, 200 mg of N. benthamiana leaves transformed with A. tumefaciens strain containing pCAMBIA-1302-TaMAPK4-eGFP and pCAMBIA-TaMAPK4-eGFP were ground in liquid nitrogen and resuspended in extraction buffer (1 mM EDTA, 50 mM Tris pH 7.4, 1% sodium deoxycholate, 1% SDS, 150 mM NaCl, 5 mM NaF, 1% Triton X-100, 1 mM DTT, and 1 mM PMSF). The proteins were separated on 12% SDS-PAGE gels and transferred onto nitrocellulose blotting membranes (0.45 μm pore size; Bio-Rad, United States) by wet electroblotting. After blocking with 5% milk at room temperature for 2 h, the membranes were incubated with primary antibody (mouse anti-eGFP antibody, Sigma–Aldrich Chemie GmbH, Taufkirchen, Germany) at a 1:1,000 dilution to detect the eGFP-tagged fusion protein. An anti-mouse antibody was used as a secondary antibody at a 1:5,000 dilution (Sigma–Aldrich Chemie GmbH, Taufkirchen, Germany). Protein expression was detected using an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific, Waltham, MA, United States).
Prediction of miR164 RNA Target and Co-transformation of miR164 and TaMAPK4
The target gene of miR164 was predicted using the psRNATarget1 program at default settings. A maximum of two continuous mismatches were allowed, using a score cutoff ≤3.0.
Co-transformation was based on Feng et al. (2014). TaMAPK4 and miR164 were each integrated into the pBI121 vector containing the GUS reporter gene. A. tumefaciens strain GV3101 was used to introduce the recombinant vectors into the same N. benthamiana leaves. Strains carrying different recombinant vectors (EV, TaMAPK4, and miR164) were cultured to an optical density of 0.8 at OD600 prior to injection. The samples were diluted to OD600 = 0.5 using 10 mM of MgCl2. miR164+TaMAPK4 were mixed in equal volumes, and the mixture (OD600 = 1.0), was used to test for the cleavage activity of miR164. Similarly, the mix of TaMAPK4 and miR159 was used as a control. Liquid (1 mL) from each treatment was infiltrated into N. benthamiana leaves. Histochemical staining and quantification of GUS were performed as described (Jefferson et al., 1987).
BSMV-Mediated TaMAPK4 Gene Silencing
Wheat seedlings were infected with BSMV (barley-stripe mosaic virus) using the method described by Hein et al. (2005). The wild-type seedlings were used as a control check (CK). TaPDS (phytoene desaturase) was used as a positive control. The wheat seedlings were inoculated with each of the three viruses (BSMV: γ, BSMV: TaPDS, and BSMV: TaMAPK4). Ten days after virus inoculation, the fourth leaves were infected with urediniospores of races CYR23 (avirulent) or CYR31 (virulent). The fourth leaves were sampled at 0, 24, 48, and 120 hpi with Pst for cytological observation and RNA isolation.
The silencing efficiency of TaMAPK4 was calculated from the qRT-PCR data. To estimate changes in the fungal biomass, the single-copy target genes PstRTP1 (rust transferred protein 1 of Pst) and TaEF1 were used to analyze the slopes of the standard curves (Panwar et al., 2013).
For SA analysis, 200 mg of fresh leaf tissue per sample was ground in liquid nitrogen and used to extract SA for HPLC-MS (API 2000; AB SCIEX, Framingham, MA, United States) as previously described (Segarra et al., 2006).
Histological Determination of Fungal Growth
The harvested samples were decolorized as previously described (Wang et al., 2007). The staining procedure for wheat germ agglutinin (WGA) conjugated to the fluorophore Alexa 488 (Invitrogen, Carlsbad, CA, United States) was used to obtain high-quality images of the developing uredinia. The cleared wheat leaf segments were examined using an Olympus BX-51 microscope (Olympus, Corp., Tokyo, Japan). Five randomly selected leaf segments were examined to record fungal development and defense responses in infected host cells. The hyphal lengths and infection areas were measured using DP-BSW software (Olympus, Corp., Tokyo, Japan). One-way analysis of variance was performed using SPSS (IBM2). Three independent biological replicates were performed.
Results
Tae-miR164 Regulates Expression of TaMAPK4
In this study, we predicted target genes of miR164 in wheat, and found that one candidate target of miR164 in wheat is TaMAPK4 (Supplementary Table S1). To verify the suppression of TaMAPK4 by miR164, we co-expressed factors in N. benthamiana (Figure 1A). A recombinant pBI121 vector containing the β-glucuronidase (GUS) as a reporter was introduced into N. benthamiana leaves using the Agrobacterium-mediated transformation system. The pBI121 EV was used as a control. Leaves infiltrated with pBI121-TaMAPK4, in which the target sequence was fused upstream of the GUS gene, showed a similar phenotype to the EV. The pBI121-pre-miR164 construct produced no GUS phenotype in the leaf, in which the GUS gene was replaced by the precursor of miR164. However, GUS staining was markedly reduced in leaves co-expressing the mixture (pBI121-TaMAPK4 and pBI121-pre-miR164). Moreover, we used miR159 as a control, which shared no sequence similarity with miR164. Co-expression of the mixture (pBI121-TaMAPK4 and pBI121-pre-miR159) showed a similar phenotype to the EV.
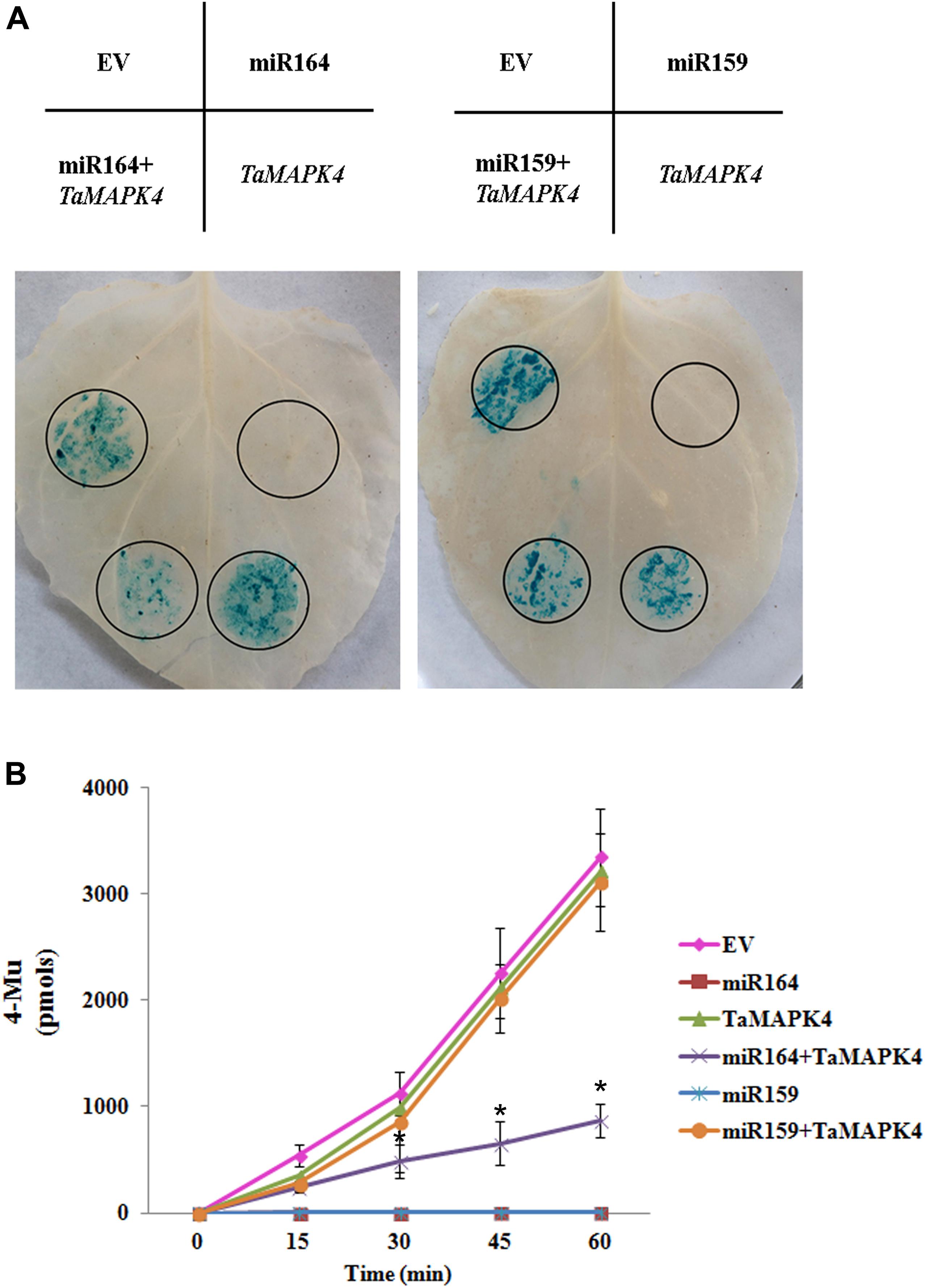
FIGURE 1. Co-expression of tae-miR164 and TaMAPK4 in Nicotiana benthamiana leaves. (A) β-glucuronidase (GUS) phenotypes observed by histochemical staining. (B) Quantitative detection of GUS activity in leaves inoculated with different recombinant vectors at different time points using a fluorospectrophotometer. EV, empty vector; 4-MU, 4-methyl-umbelliferyl- β-D-glucuronide. The results are presented as the means ± standard errors of three biological replications. Significant differences were determined using Student’s t-test: ∗P < 0.05.
To confirm the histochemical results, the levels of GUS in the leaves were measured using a fluorospectrophotometer (Figure 1B). Fluorescence in the EV, pBI121-TaMAPK4 and the mixture treatments (pBI121-TaMAPK4 and pBI121-miR159) in the inoculated leaf samples increased with reaction times. These results indicated that GUS was still present and functional. No fluorescence was detected in leaves injected with pBI121-miR164 or pBI121-miR159. Compared with the EV treatments, fluorescence with the mixture (pBI121-TaMAPK4 and pBI121-miR164) showed a slow increase, demonstrating that miR164 effectively regulated TaMAPK4.
TaMAPK4 Protein Is Localized in the Nucleus and Cytoplasm
Prediction analysis revealed that TaMPK4 is localized in the cytoplasm (Hao et al., 2015). We conducted subcellular localization experiments in N. benthamiana leaf cells to better characterize the biological function of TaMAPK4 and determine the subcellular localization of TaMAPK4. Interestingly, when N. benthamiana leaves were infused with the pCAMBIA-1302-TaMAPK4 constructs green fluorescent protein (GFP), fluorescence was transiently expressed in both the cytoplasm and the nucleus, and GFP was uniformly distributed throughout the cell in the control (Figure 2A). To confirm the subcellular localization results, we used western blotting to analyze the expression and stability of the TaMAPK4 fusion protein and found that both GFP and TaMAPK4 were successfully expressed (Figure 2B). Moreover, we expressed the TaMAPK4-GFP fusion protein in wheat protoplasts, found that the GFP fusion protein also expressed in both the cytoplasm and the nucleus (Supplementary Figure S1). We inferred that TaMAPK4 might have multiple roles in signaling either from the nucleus or its localization in the cytoplasm.
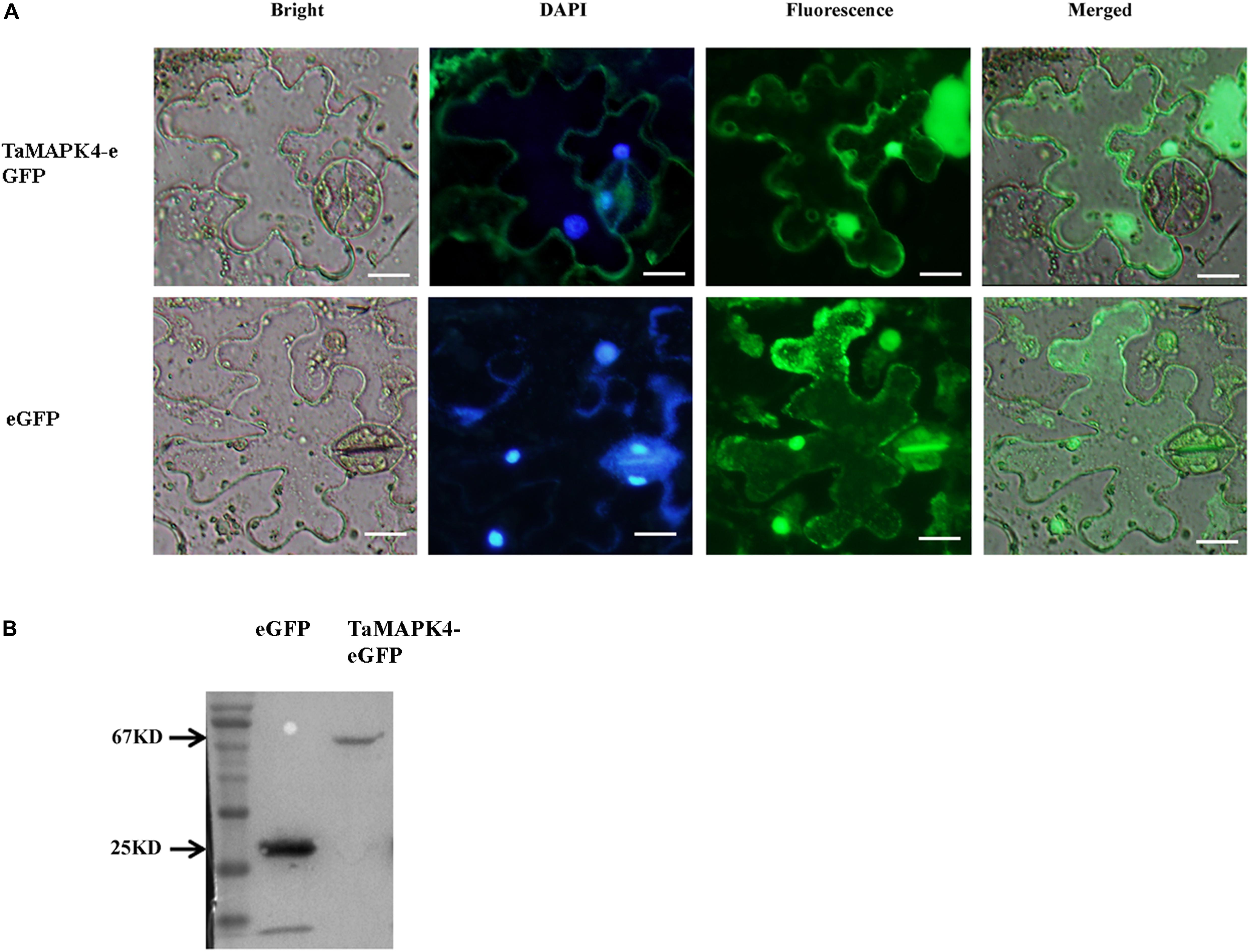
FIGURE 2. Subcellular localization of TaMAPK4 protein. (A) TaMAPK4-GFP fusion protein and green fluorescent protein (GFP) (control) were transiently expressed in N. benthamiana. (B) Western blot analysis of GFP and TaMAPK4-GFP fusion protein. Bar = 20 μm. Similar results were obtained from three biological replicates.
Transcriptional Responses of TaMAPK4 to Pst and Hormone Treatment
We examined the transcript levels of TaMAPK4 in Pst-infected wheat leaves using quantitative real-time PCR (qRT-PCR). TaMAPK4 transcript levels increased as early as 12 hpi and exhibited an approximately 2.8-fold peak response at 48 hpi in leaves inoculated with avirulent Pst CYR23 (Figure 3A). The transcript level of TaMAPK4 was only up-regulated at 12 hpi, and then showed no obvious change in leaves inoculated with virulent Pst CYR31. Therefore, the relative transcript level of TaMAPK4 in leaves inoculated with CYR23 was higher than in leaves inoculated with CYR31.
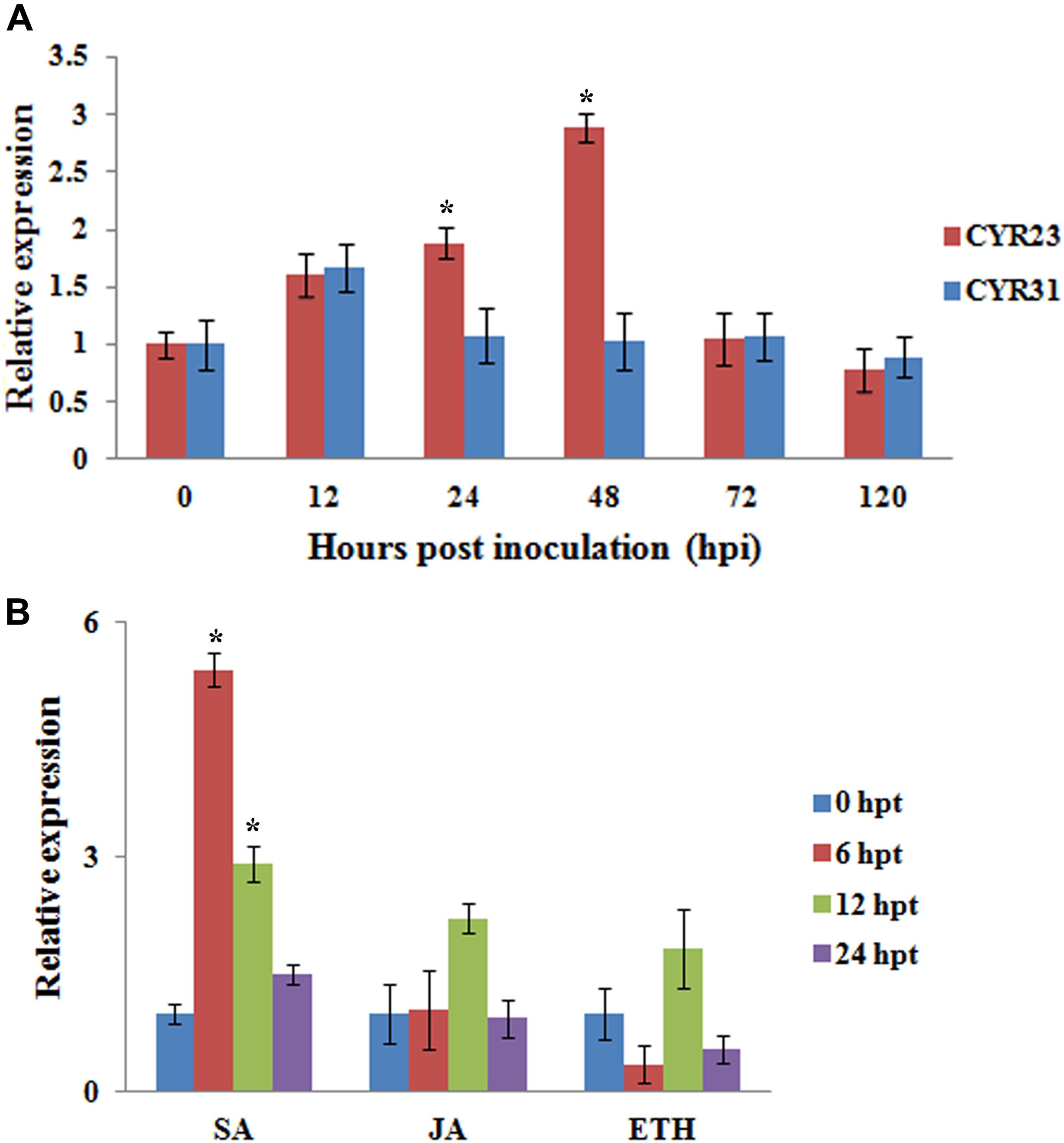
FIGURE 3. qRT-PCR analysis of relative transcript levels of TaMAPK4. (A) Transcript profiles of TaMAPK4 in wheat leaves (Su11) inoculated with Pst races CYR31 (virulent) and CYR23 (avirulent), respectively. (B) TaMAPK4 transcript profiles in wheat leaves treated with hormone elicitors. SA, salicylic acid; MeJA, methyl jasmonate; ET, ethylene. Mean expression values were calculated from three independent replicates. Significant differences were determined using Student’s t-test: ∗P < 0.05.
We also tested the expression of TaMAPK4 under methyl jasmonate (MeJA), SA, and ethephon (ET) stresses. TaMAPK4 transcript levels were strongly elevated, reaching a 5.4-fold peak at 6 hpt in response to SA (Figure 3B), but there was no significant change in the transcript levels of TaMAPK4 following JA treatment. These results suggest that SA signaling was involved in TaMAPK4 regulation.
Knockdown of TaMAPK4-Decreased Resistance to Pst
We used a VIGS system to characterize the function of TaMAPK4 in the stripe rust-resistant wheat cultivar Su11. The plant displayed mild chlorotic mosaic symptoms and no other obvious defects at 10 days post-virus inoculation (dpi). As a control to confirm that the VIGS system was functioning correctly, we silenced the wheat phytoene desaturase (PDS) gene through inoculation with the recombinant virus BSMV: TaPDS, causing severe symptoms of chlorophyll photobleaching at 10 dpi. The fourth leaves of Su11 plants were inoculated with avirulent Pst CYR23 and virulent CYR31 at 10 dpi with the virus. Conspicuous hypersensitive responses (HR) were elicited by CYR23-infected control seedlings (CK) and seedlings that were previously infected with BSMV: γ. However, there was increased uredinial development and limited urediniospore production in leaves infected with BSMV: TaMAPK4 at 14 dpi (Figure 4A). There was no phenotypic change in the compatible interaction compared with the CK, which exhibited normal disease development and profuse sporulation on all leaves inoculated with CYR31.
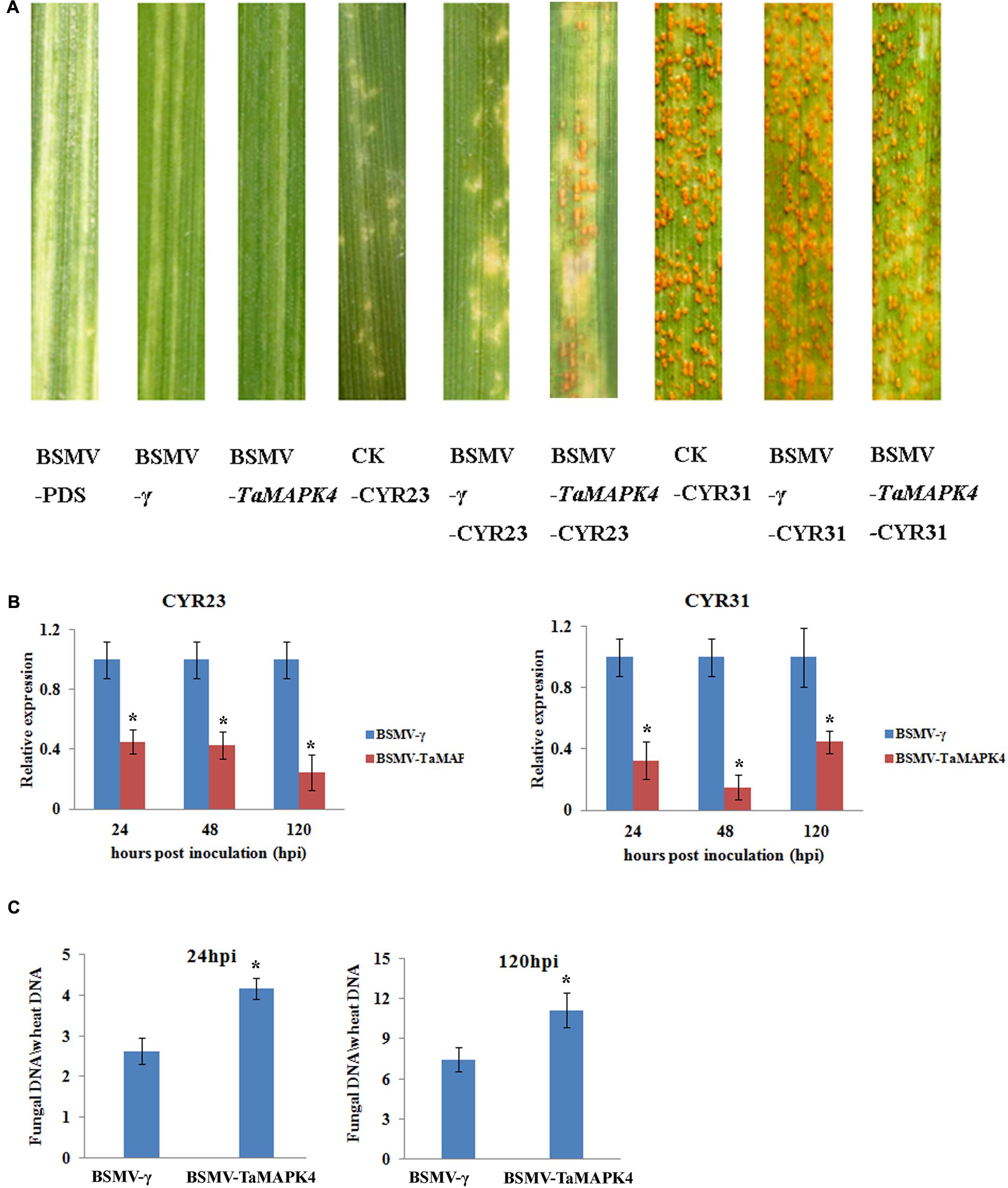
FIGURE 4. Functional characterization of TaMAPK4 after virus-induced gene silencing. (A) Phenotypic changes in the fourth leaves of plants pre-inoculated with positive control vector (BSMV-PDS), FES buffer (CK), or empty BSMV vector (BSMV-γ) at 14 days post-virus treatment. Phenotypes for the fourth leaves inoculated with Puccinia striiformis f. sp. tritici (Pst) races CYR23, or CYR31 at 14 dpi. (B) Relative transcript levels of TaMAPK4 in TaMAPK4 knockdown leaves. RNA samples were isolated from TaMAPK4 knockdown leaves infected with Pst races CYR23 (left), or CYR31 (right). BSMV-γ leaves infected with Pst races CYR23, or CYR31 were used as control, respectively. (C) Biomass of Pst (CYR23) measured at 24 and 120 hpi. Means and standard deviations were calculated from three independent replicates. Significant differences were determined using Student’s t-test: ∗P < 0.05.
qRT-PCR was used to confirm that the RNAi system was functional by determining the silencing efficiency of TaMAPK4. Compared to leaves inoculated with BSMV: γ, the transcript levels of TaMAPK4 were reduced by 55, 58, and 76% at 24, 48, and 120 hpi, respectively, in BSMV: TaMAPK4 leaves inoculated with CYR23 (Figure 4B). In the compatible interaction, the expression of TaMAPK4 was reduced by 56–86%. The Pst biomass was also increased in the leaves of plants with TaMAPK4 knocked-down (Figure 4C). Moreover, we found miR164 was up-regulated in the TaMAPK4 knocked-down leaves inoculated with CYR23 (Supplementary Figure S2).
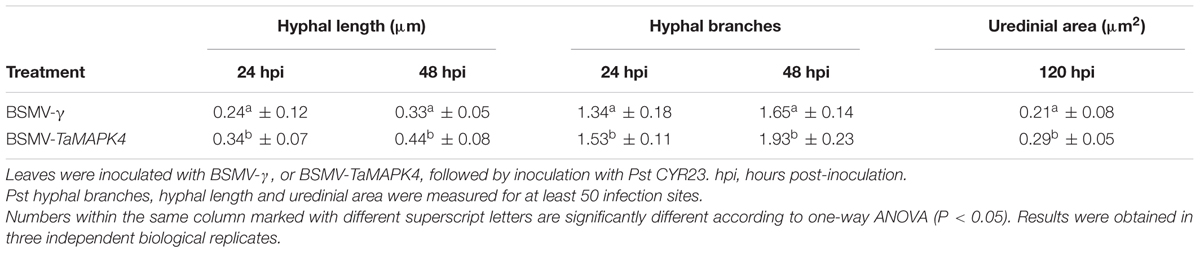
Su11 leaves inoculated with BSMV: TaMAPK4 showed increased uredinial development in response to CYR23. To further verify these phenotypes, detailed histological changes were compared to the control (Table 1). Pst hyphal length and hyphal branches in BSMV: TaMAPK4 knockdown leaves were obviously longer (P < 0.05) than those in the BSMV: γ-treated leaves at 24 and 48 hpi. Moreover, the uredinial area in wheat leaves pre-inoculated with BSMV: TaMAPK4 was significantly (P < 0.05) increased at 120 hpi.
SA Accumulation and Defense-Related Genes in Tamapk4 Knockdown Wheat Leaves
TaMAPK4 transcript levels were strongly elevated in response to SA. To determine whether the knockdown of TaMAPK4 affects SA accumulation in wheat leaves, we assessed the SA accumulation in TaMAPK4 knockdown wheat leaves after infection with Pst. SA accumulation was decreased at 24 and 48 hpi (Figure 5A).
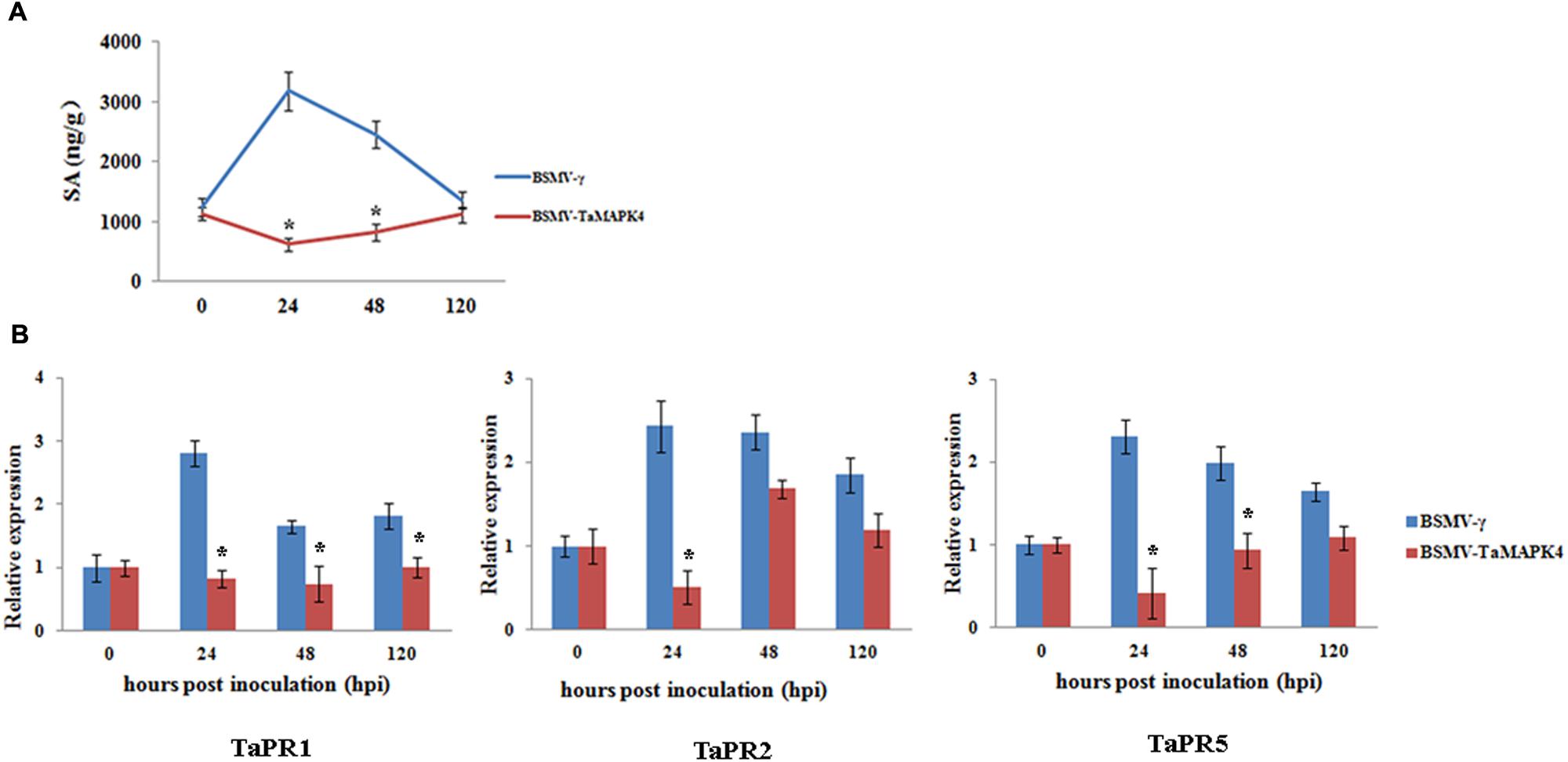
FIGURE 5. Salicylic acid accumulation and transcript levels of pathogenesis-related (PR) genes in TaMAPK4 knockdown leaves. (A) SA accumulation in TaMAPK4 knockdown leaves inoculated with Pst race CYR23. SA, salicylic acid; ng/g, SA accumulation (ng) per fresh leaves weight (g); (B) Transcript levels of TaPR1, TaPR2, and TaPR5 genes in TaMAPK4 knockdown leaves challenged with Pst race CYR23. TaPR1, pathogenesis-related protein 1; TaPR2, β-1,3-glucanase; TaPR5, thaumatin-like protein. BSMV-γ leaves infected with Pst race CYR23 were used as control. Means and standard deviations were calculated from three independent replicates. Significant differences were determined using Student’s t-test: ∗P < 0.05.
To determine whether the expression of defense-related genes was affected by TaMAPK4 silencing, we selected PR genes for qRT-PCR analysis. The transcript levels of TaPR1, TaPR2, and TaPR5 were decreased in TaMAPK4 knockdown plants infected with the Pst pathotype CYR23 (Figure 5B). These results indicated that TaMAPK4 might affect the transcript levels of defense-related genes in the wheat-Pst interaction.
Discussion
Mitogen-activated protein kinases are regulated by the sequential phosphorylation of MAPK kinase (Zhang and Klessig, 2001). In the present study, we verified that TaMAPK4 is a candidate target of miR164 by co-expression studies in N. benthamiana. A single miRNA could regulate multiple target genes with different roles in plant immunity. For example, miR863-3p, which targets two atypical receptor-like pseudokinases (ARLPKs) and SERRATE (SE), is induced by the bacterial pathogen Pseudomonas syringae. During an early stage of infection, miR863-3p silences the ARLPKs that act as negative regulators of plant defense. During infection, miR863-3p silences SE to positively regulate plant defense (Niu et al., 2016). miR164 also regulates multiple target genes, such as NAC genes and ethylene insensitive 3 (EIN3) (Kim et al., 2009). In a previous study, we observed that TaNAC21, a target of miR164, played a negative role in Pst resistance in wheat. The expression of TaNAC21 was induced at 24 and 48 hpi in the compatible interaction (Feng et al., 2014). However, in the present study, the transcript accumulation of TaMAPK4 was increased at 12, 24, and 48 hpi in the incompatible interaction. Thus, these results suggested that miR164 might regulate target genes in the wheat-Pst interaction. TaMAPK4 and TaNAC21 played different roles in the regulation of defense in different wheat-Pst interactions. However, the detailed mechanisms of the interplay between TaMAPK4 and miR164 in plant defense need to be further explored.
The characterization of where a gene exerts its function is needed to understand the biological function of the gene. As previously described, there are few reports on the subcellular localization of plant MAPKs (Zaïdi et al., 2010). The mechanism of MAPK localization is complex. It is well-established that MAPKs shuttle between the cytoplasm and nucleus in yeast and mammalian cells in response to different types of stimulation (Cyert, 2001). In plants, Arabidopsis MKP1 was primarily cytoplasmic and interacted with and dephosphorylated MPK6, and MPK1 might be involved in regulating basal MPK activity (Bartels et al., 2009). Moreover, TMPK1 (Triticum turgidum L. subsp. Durum) was found in the nucleus, and this protein may travel from the cytoplasm to the nucleus to regulate phosphatase activity and interact with TMPK3 and TMPK6 by associating with NLS-containing proteins, suggesting a regulatory function by interacting with specific nuclear components (Zaïdi et al., 2010). Our localization experiments determined that TaMAPK4 localized to both the cytoplasm and nucleus. Thus, we inferred that TaMAPK4 might have multiple roles in signaling either from the nucleus or its localization in the cytoplasm.
Mitogen-activated protein kinase cascades play central roles in multiple stress responses, including host immunity (Innes, 2001). In the present study, TaMAPK4 was up-regulated in the incompatible interaction. The knockdown of TaMAPK4 by VIGS-induced silencing increased uredinial development in the incompatible race CYR23: Su11 response. The histological results were consistent with the TaMAPK4 gene expression profiles. Taken together, these results indicated that TaMAPK4 might contribute to the wheat response to Pst.
Salicylic acid is an important component of plant defense signaling (Vlot et al., 2009). BWMK1 (the rice MAPK gene) was activated by SA signal and could phosphorylate another transcription factor(s) exclusively associated with SA-mediated gene expression (Cheong et al., 2003). In the present study, we found that TaMAPK4 was more highly expressed after SA treatment. These data inferred that TaMAPK4 might be activated by SA. Moreover, MAPK cascades have been implicated in both the regulation of defense hormone biosynthesis and signaling events downstream of hormone sensing. Arabidopsis MPK3 and, to a lesser extent, MPK6 play pivotal roles in the SA-mediated priming of plants for protection against pathogens (Beckers et al., 2009; Bethke et al., 2012). In contrast, Arabidopsis mpk4 (not the homolog of TaMAPK4) mutant plants exhibited constitutively elevated levels of SA, and increased pathogen resistance (Petersen et al., 2000). Moreover, the constitutive SA responses in the mpk4 mutant were triggered by the SUMM2 (an R protein)-mediated signaling pathway (Zhang et al., 2012). In another experiment, the SA levels in TaMAPK4 knockdown seedlings in the incompatible interaction were significantly lower than those in the compatible interaction, suggesting that TaMAPK4 is a positive regulator of SA signaling. Therefore, these results suggested that TaMAPK4 plays important roles in SA-mediated defense signaling pathways.
The accumulation of PR proteins in the plant is related to plant resistance responses (Van Loon and Van Strien, 2002). MAPK genes are involved in regulating the expression of PR genes. For example, the ectopic overexpression of BWMK1 in transgenic tobacco plants led to increased resistance to pathogens associated with elevated levels of PR gene expression (Cheong et al., 2003). We therefore assessed the expression of PR genes in TaMAPK4 knockdown seedlings and found that transcript levels of TaPR1, TaPR2, and TaPR5 were significantly reduced relative to non-knockdown controls. Based on these results, we concluded that the reduced resistance to Pst in TaMAPK4 knockdown wheat plants might be due to decreased defense responses.
Conclusion
TaMAPK4 was induced during the wheat-Pst incompatible interaction and likely contributed to the actual expression of incompatibility between the wheat host and Pst pathogen, referred to as the infection type. Our results suggest that TaMAPK4 is regulated by miR164, and these data provide further insights into how MAPK genes might be involved in plant defense.
Author Contributions
BW carried out most of the experiments; BW and ZK wrote the manuscript; NS performed the quantitative RT-PCR and analyzed the data; QZ and NW grew the plant samples; NW collected all the phenotypic data; and ZK revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31620103913), and the 111 Project from the Ministry of Education of China (No. B07049).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00152/full#supplementary-material
FIGURE S1 | Subcellular localization of the TaMAPK4 protein. GFP and TaMAPK4-GFP fusion proteins were expressed in wheat protoplasts. Bar = 20 μm.
FIGURE S2 | Relative transcript levels of miR164 in TaMAPK4 knockdown leaves infected with Pst race CYR23. BSMV-γ leaves infected with Pst race CYR23 were used as control. Means and standard deviations were from three independent replicates. Significant differences were determined using Student’s t-test: ∗P < 0.05.
TABLE S1 | Prediction of tae-miR164 target in wheat.
TABLE S2 | Primers designed for TaMAPK4 research.
Footnotes
- ^ http://plantgrn.noble.org/psRNATarget/
- ^ http://www.ibm.com, version 16.0
References
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartels, S., Anderson, J. C., Besteiro, M. A. G., Carreri, A., Hirt, H., Buchala, A., et al. (2009). MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21, 2884–2897. doi: 10.1105/tpc.109.067678
Beckers, G. J., Jaskiewicz, M., Liu, Y., Underwood, W. R., He, S. Y., Zhang, S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. doi: 10.1105/tpc.108.062158
Bethke, G., Pecher, P., Eschen-Lippold, L., Tsuda, K., Katagiri, F., Glazebrook, J., et al. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25, 471–480. doi: 10.1094/MPMI-11-11-0281
Cardinale, F., Meskiene, I., Ouaked, F., and Hirt, H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14, 703–711. doi: 10.1105/tpc.010256
Cheong, Y. H., Moon, B. C., Kim, J. K., Kim, C. Y., Kim, M. C., Kim, I. H., et al. (2003). BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972. doi: 10.1104/pp.103.023176
Cristina, M. S., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252
Cyert, M. S. (2001). Regulation of nuclear localization during signaling. J. Biol. Chem. 276, 20805–20808. doi: 10.1074/jbc.R100012200
Feng, H., Duan, X. Y., Zhang, Q., Li, X. R., Wang, B., Huang, L. L., et al. (2014). The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 15, 284–296. doi: 10.1111/mpp.12089
Hamel, L. P., Nicole, M. C., Sritubtim, S., Morency, M. J., Ellis, M., Ehlting, J., et al. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. doi: 10.1016/j.tplants.2006.02.007
Hao, L., Wen, Y., Zhao, Y., Lu, W., and Xiao, K. (2015). Wheat mitogen-activated protein kinase gene TaMPK4 improves plant tolerance to multiple stresses through modifying root growth, ROS metabolism, and nutrient acquisitions. Plant Cell Rep. 34, 2081–2097. doi: 10.1007/s00299-015-1853-2
Hein, I., Barciszewska-Pacak, M., Hrubikova, K., Williamson, S., Dinesen, M., Soenderby, I. E., et al. (2005). Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138, 2155–2164. doi: 10.1104/pp.105.062810
Hématy, K., Cherk, C., and Somerville, S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12, 406–413. doi: 10.1016/j.pbi.2009.06.007
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Innes, R. W. (2001). Mapping out the roles of MAP kinases in plant defense. Trends Plant Sci. 6, 392–394. doi: 10.1016/S1360-1385(01)02058-1
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, Z. S., Huang, L. L., and Buchenauer, H. (2002). Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis. J. Plant Dis. Protect. 109, 25–37.
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. doi: 10.1242/dev.01320
Li, Y., Lu, Y. G., Shi, Y., Wu, L., Xu, Y. J., Huang, F., et al. (2014). Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164, 1077–1092. doi: 10.1104/pp.113.230052
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Niu, D., Lii, Y. E., Chellappan, P., Lei, L., Peralta, K., Jiang, C., et al. (2016). miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 7:11324. doi: 10.1038/ncomms11324
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73, 521–532. doi: 10.1111/tpj.12047
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. doi: 10.1016/S0092-8674(00)00213-0
Pitzschke, A., Schikora, A., and Hirt, H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12, 421–426. doi: 10.1016/j.pbi.2009.06.008
Segarra, G., Jáuregui, O., Casanova, E., and Trillas, I. (2006). Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry. 67, 395–401. doi: 10.1016/j.phytochem.2005.11.017
Tena, G., Asai, T., Chiu, W. L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. doi: 10.1016/S1369-5266(00)00191-6
Van Loon, L. C., and Van Strien, E. A. (2002). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, B., Sun, Y. F., Song, N., Wei, J. P., Wang, X. J., Feng, H., et al. (2014). MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol. Biochem. 80, 90–96. doi: 10.1016/j.plaphy.2014.03.020
Wang, B., Sun, Y. F., Song, N., Zhao, M., Liu, R., Feng, H., et al. (2017). Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 215, 338–350. doi: 10.1111/nph.14577
Wang, C. F., Huang, L. L., Buchenauer, H., Han, Q. M., Zhang, H. C., and Kang, Z. S. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2- and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Zaïdi, I., Ebel, C., Touzri, M., Herzog, E., Evrard, J., Schmit, A. C., et al. (2010). TMKP1 is a novel wheat stress responsive MAP kinase phosphatase localized in the nucleus. Plant Mol. Biol. 73, 325–338. doi: 10.1007/s11103-010-9617-4
Zhang, S., and Klessig, D. F. (1998). Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 95, 7433–7438. doi: 10.1073/pnas.95.13.7433
Zhang, S., and Klessig, D. F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. doi: 10.1016/S1360-1385(01)02103-3
Keywords: host defense mechanisms, mitogen-activated protein kinase, Puccinia striiformis, wheat, virus-induced gene silencing
Citation: Wang B, Song N, Zhang Q, Wang N and Kang Z (2018) TaMAPK4 Acts as a Positive Regulator in Defense of Wheat Stripe-Rust Infection. Front. Plant Sci. 9:152. doi: 10.3389/fpls.2018.00152
Received: 14 November 2017; Accepted: 29 January 2018;
Published: 15 February 2018.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Steven Whitham, Iowa State University, United StatesXiaoquan Qi, Institute of Botany (CAS), China
Copyright © 2018 Wang, Song, Zhang, Wang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=
 Bing Wang1
Bing Wang1 Qiong Zhang
Qiong Zhang Zhensheng Kang
Zhensheng Kang