
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 February 2018
Sec. Evolutionary and Population Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00142
 Wen-Bin Yu1,2*
Wen-Bin Yu1,2* Christopher P. Randle3
Christopher P. Randle3 Lu Lu4
Lu Lu4 Hong Wang4
Hong Wang4 Jun-Bo Yang5
Jun-Bo Yang5 Claude W. dePamphilis6
Claude W. dePamphilis6 Richard T. Corlett1
Richard T. Corlett1 De-Zhu Li5*
De-Zhu Li5*Phtheirospermum (Orobanchaceae), a hemiparasitic genus of Eastern Asia, is characterized by having long and viscous glandular hairs on stems and leaves. Despite this unifying character, previous phylogenetic analyses indicate that Phtheirospermum is polyphyletic, with Phtheirospermum japonicum allied with tribe Pedicularideae and members of the Ph. tenuisectum complex allied with members of tribe Rhinantheae. However, no analyses to date have included broad phylogenetic sampling necessary to test the monophyly of Phtheirospermum species, and to place these species into the existing subfamiliar taxonomic organization of Orobanchaceae. Two other genera of uncertain phylogenetic placement are Brandisia and Pterygiella, also both of Eastern Asia. In this study, broadly sampled phylogenetic analyses of nrITS and plastid DNA revealed hard incongruence between these datasets in the placement of Brandisia. However, both nrITS and the plastid datasets supported the placement of Ph. japonicum within tribe Pedicularideae, and a separate clade consisting of the Ph. tenuisectum complex and a monophyletic Pterygiella. Analyses were largely in agreement that Pterygiella, the Ptheirospermum complex, and Xizangia form a clade not nested within any of the monophyletic tribes of Orobanchaceae recognized to date. Ph. japonicum, a model species for parasitic plant research, is widely distributed in Eastern Asia. Despite this broad distribution, both nrITS and plastid DNA regions from a wide sampling of this species showed high genetic identity, suggesting that the wide species range is likely due to a recent population expansion. The Ph. tenuisectum complex is mainly distributed in the Hengduan Mountains region. Two cryptic species were identified by both phylogenetic analyses and morphological characters. Relationships among species of the Ph. tenuisectum complex and Pterygiella remain uncertain. Estimated divergence ages of the Ph. tenuisectum complex corresponding to the last two uplifts of the Qinghai–Tibet Plateau at around 8.0–7.0 Mya and 3.6–1.5 Mya indicated that the development of a hot-dry valley climate during these uplifts may have driven species diversification in the Ph. tenuisectum complex.
Mountain ranges often support a high diversity of plant life (Favre et al., 2015; Hughes and Atchison, 2015). Many of the 35 biodiversity hotspots are located in mountain ranges (Myers et al., 2000; Mittermeier et al., 2011). The Hengduan Mountains region is the core of the Mountains of Southwest China Biodiversity Hotspot (Zhang et al., 2009; Boufford, 2014). This region is characterized by extremely complex topography, with altitudes ranging from less than 2,000 meters in some valley floors to 7,558 meters at the summit of Gongga Mountain. Generally, the mountain ridges are oriented in the north–south direction, perpendicular to the main Himalayas. Several major river systems originated or were reorganized in this region during the uplift of the mountains, e.g., the Jinshajiang (Upper Yangtze) and its tributaries (Yalongjiang, Daduhe, Jialingjiang), as well as the Salween (Nujiang), and Mekong (Lancangjiang) (Zhang et al., 2011c). The Hengduan Mountains region harbors more than 9,000 species of vascular plants, around 32% of which are endemic (Li and Li, 1993; Wang and Wu, 1993, 1994; Zhang et al., 2009). More than 30 genera are endemic to this region and the adjacent Himalayas (Ying and Zhang, 1984; Wu et al., 2005; Boufford, 2014). Of these, several endemic genera have been subsumed within other widely distributed genera (Friesen et al., 2000; Pan et al., 2009; Tian et al., 2011; Zhang et al., 2011b; Nie et al., 2013). However, discovery of new genera is ongoing (Al-Shehbaz et al., 2004; Zhang et al., 2011a; Wang W. et al., 2013; Wang Y.-J. et al., 2013). Species richness and diversity in the Hengduan Mountains region have been ascribed to the accumulation of migrants and in situ diversification accelerated by the uplift of the mountains (Wen et al., 2014; Xing and Ree, 2017). This uplift event may have contributed to recent diversification of species in Phtheirospermum Bunge (Orobanchaceae), most of which can be found in the Hengduan Mountains region.
Phtheirospermum is a genus of hemiparasitic plants characterized by having long and viscous glandular hairs on stems and leaves, yet monophyly and species boundaries remain uncertain. Historically, this genus was divided into five species. Tao (1996) organized species in two sections in a full revision of the genus; section Phtheirospermum contains Ph. japonicum (Thunb.) Kanitz and Ph. tenuisectum Bureau & Franch. The former is an experimental model for the study of genetics and development of the haustorium in a generalist parasite (Cui et al., 2016; Ishida et al., 2016). Section Minutusepala (not validly published) includes the remaining species, Ph. glandulosum (Benth.) Hook. f., Ph. muliense C. Y. Wu & D. D. Tao and Ph. parishii Hook. f. Based on the Chinese material of Ph. glandulosum, Hong (1979) established Pseudobartsia, with one species Pseudobartsia yunnanensis Hong (Tao, 1993); because Ps. yunnanensis cannot be distinguished from Ph. glandulosum, Yu et al. (2015b) created the combination Ps. glandulosum. Molecular and morphological evidence supports the independent generic status of Pseudobartsia (Lu et al., 2007; Dong et al., 2013, 2015).
Of the remaining four species of Phtheirospermum, Ph. japonicum occurs throughout China (except Xinjiang), extending to eastern Russia, the Korean Peninsula, and Japan; Ph. tenuisectum is restricted to Western China (Guizhou, Qinghai, Sichuan, and Xizang); Ph. muliense is known by only the type collection from Muli in Sichuan, southwestern China; and Ph. parishii occurs in southern Myanmar and Thailand (Yamazaki, 1990). Tao (1996) reported three collections for Ph. parishii in Sichuan. Based on comparisons of type materials and herbarium specimens, however, the three gatherings, including seven sheets conserved at CDBI, KUN, and PE, are short plants of Ph. tenuisectum, and all of them were originally identified as Ph. tenuisectum. Phtheirospermum parishii was firstly collected from Tanintharyi region of southern Myanmar, then it was also found in northern Thailand (Yamazaki, 1990).
Even with Ph. glandulosum excluded from Phtheirospermum, monophyly of the remaining species is still not resolved. Phylogenetic analyses including Ph. japonicum or Ph. tenuisectum alone indicates that Ph. japonicum is close to tribe Pedicularideae (Bennett and Mathews, 2006; McNeal et al., 2013), whereas Ph. tenuisectum was excluded from tribe Pedicularideae as an independent lineage (Ree, 2005). Dong et al. (2013) were the first to sample three species of Phtheirospermum, Ph. japonicum, Ph. tenuisectum, and Ph. muliense. However, they included the sample J. S. Ying 4144 from Sichuan, which is one of three gatherings misidentified as Ph. parishii by Tao (1996). The phylogenetic analyses showed that Phtheirospermum spp. separated into two clades: one clade including Ph. japonicum as sister to Pedicularis spp., and another clade includes Ph. muliense and Ph. tenuisectum, as sister to Pterygiella. In the study of Yu et al. (2015a), Ph. tenuisectum and Ph. japonicum were included as outgroups; similar to previous studies, Ph. tenuisectum is resolved as sister to Pterygiella nigrescens, while Ph. japonicum is included in a clade of tribe Pedicularideae. Accompanied by a broad sampling of tribe Rhinantheae sensu lato, Pinto-Carrasco et al. (2017) used the data of the “Pterygiella complex” from Dong et al. (2013) to recover the polyphyly of Phtheirospermum. Although polyphyly of Phtheirospermum is suggested by these prior studies, complete species sampling and placement of species in a broad phylogenetic context of the family Orobanchaceae are needed. In the more broadly sampled study of McNeal et al. (2013), Pterygiella was placed at the base of tribe Rhinantheae sensu stricto using PHYA and PHYB datasets, but was weakly supported as sister to Brandisia using nrITS and the plastid datasets. In this study, Brandisia was close to tribe Cymbarieae, or sister to the clade including tribes Pedicularideae + Buchnereae + Rhinantheae (including Pterygiella) using PHYA and PHYB datasets, respectively. Therefore, instability of the placement of Brandisia and Pterygiella casts further into doubt the placement of Ph. tenuisectum and Ph. muliense, and fails to provide an indication as to which, if any, existing tribes these genera can be placed.
Currently, two species of Phtheirospermum, Ph. japonicum and Ph. tenuisectum, have been adopted in Chinese Floras (Tsoong and Yang, 1979; Hong et al., 1998). Morphologically, it is easy to distinguish Ph. tenuisectum from Ph. japonicum in that the former has dissected leaves with linear pinnae (vs. narrowly ovate to orbicular pinnae in the latter), smaller yellow corollas (vs. red/pink corollas), and smaller fruits and seeds. In herbaria, specimens having dissected leaves with linear pinnae, yellow flowers and small fruits, have almost all been labeled as Ph. tenuisectum. Based on examination of herbarium specimens, we found that Ph. tenuisectum varied extensively in habit, leaf morphology, calyx form, and corolla shape. With further observation of fresh materials in the field, we have recognized four distinct morphotypes (Figure 1). These four morphotypes appear to be of the same lineage, called the Ph. tenuisectum complex hereafter. One morphotype with dimorphic leaves (Figures 1J,K) has been described as Ph. muliense (Tao, 1996), which has been supported by molecular analyses (Dong et al., 2013). Except for Ph. tenuisectum (Figures 1A–D), the remaining two morphotypes are treated as two new taxa (species 1: Figures 1E,F; species 2: Figures 1G–I).

FIGURE 1. Plants of the Phtheirospermum tenuisectum complex. (A–D) Morphological variation in Ph. tenuisectum: (A) dissected leaves with wide lobes, and (B) flowers with spreading lower-lip and entire calyx lobes from Kangding (population T6, see Figure 2 and Supplementary Table S1, hereafter); (C) yellow flower with drooping lower-lip and branched calyx lobes from Lijiang (T12); and (D) yellow flower with spreading lower-lip and branched calyx lobes (T2). (E,F) Phtheirospermum sp. 1 from Lijiang (D1 and D2): (E) a whole plant and habitat; (F) flowering branch. (G–I) Morphological variation in Phtheirospermum sp. 2: (G) a flowering plant, and (H) leaves/bracts and flower with long and entire calyx lobes from Batang (H7); and (I) leaves with linear lobes from Zhongdian (H2). (J,K) Ph. muliense: (J) plants and habitat; (K) vegetative branch.
The main goals of this study were to: (1) investigate the suspected polyphyly of Phtheirospermum; (2) delimit morphology-based species using molecular and phylogenetic approaches, and (3) estimate the evolutionary histories of species in Phtheirospermum. To achieve our goals, we sampled the Ph. tenuisectum complex extensively. A population of Ph. muliense was newly discovered in Shangri-La, in northwestern Yunnan. Two individuals from this population were sequenced in this study. We reconstruct the phylogeny of the Ph. tenuisectum complex, and re-evaluated the phylogenetic relationship between the Ph. tenuisectum complex and Ph. japonicum on the base of large-scale sampling of Phtheirospermum, as well as selected genera from six recognized tribes (McNeal et al., 2013), and Brandisia, and Rehmannia (Stevens, 2001 onward; Xia et al., 2009; Zeng et al., 2017). To understand the evolutionary history of Phtheirospermum, divergence times were estimated using large-scale sampling of Lamiales and four fossil calibrations.
To reconstruct the phylogenetic relationship of Phtheirospermum, 68 accessions from Orobanchaceae (including Rehmannia) were sampled, representing all five recognized tribes, 22 genera and 30 taxa (Supplementary Table S1). For Ph. japonicum, we sampled nine individuals from nine populations in Yunnan, Sichuan, Henan, Zhejiang, and Liaoning Provinces in China, and from Kanagawa Prefecture in Japan. We sampled 13 individuals of Ph. tenuisectum, three individuals of Ph. muliense, and four and eight individuals of two undescribed taxa, respectively. Geographic sampling of the Ph. tenuisectum complex is presented in Figure 2. We sampled the three recognized species of Pterygiella including seven individuals to test the relationship between the Ph. tenuisectum complex and Pterygiella. We included sequences for Ps. glandulosa, and the samples T8 (J. S. Ying 4144), L1 (S. G. Wu 3582), and H1 (L. Lu LJ377) from the study of Dong et al. (2013). Forty-nine samples were sequenced for this study. Thirty-six sequences of four DNA barcoding loci (nrITS, matK, rbcL, and trnH-psbA) from two Pedicularis samples (Yu et al., 2011) and seven Pterygiella samples (Dong et al., 2011) have been published, as well as two trnL-F sequences from Pedicularis (Yu et al., 2015a), and one from Pterygiella (Dong et al., 2011). Forty-six sequences extracted from seven published plastomes [Bartsia inaequalis Benth., Castilleja paramensis F. González & Pabón-Mora, Cistanche phelypaea (L.) Cout., Lathraea squamaria L., Phelipanche purpurea Soják, Schwalbea americana L., and Striga hermonthica (Delile) Benth.], and the available nrITS sequences, were included for phylogenetic analyses. Voucher information or GenBank accessions are presented in Supplementary Table S1.
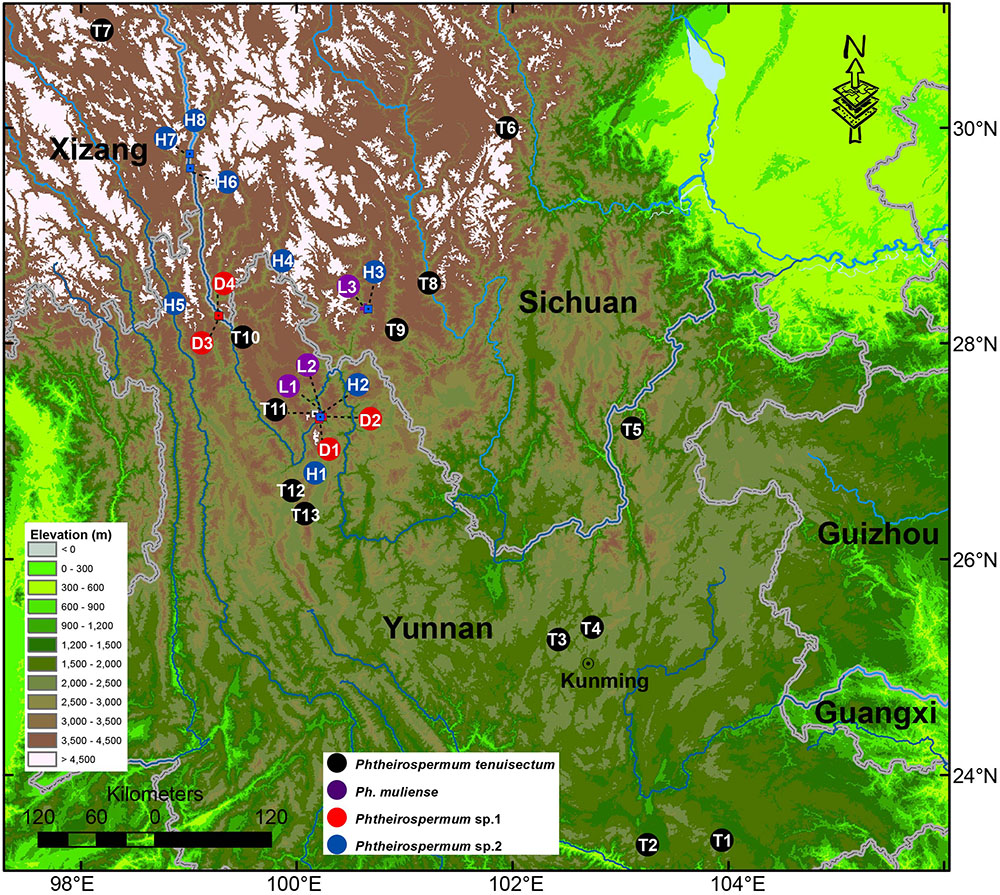
FIGURE 2. Geographical sampling of the Ph. tenuisectum complex in Southwest China. The river systems are highlighted in blue. The color of circles corresponds to taxa. Numbers and letters indicate populations; these abbreviations are also used in the Figures 3–6. More information regarding collection vouchers can be found in Supplementary Table S1.
Total genomic DNA was extracted from silica-gel-dried leaves using a modified CTAB protocol (Doyle and Doyle, 1987). We amplified and sequenced one nuclear (ribosomal internal transcribed spacer, nrITS) and seven plastid markers (matK, rbcL, rps2, rps16, trnK-matK, trnH-psbA, and trnL-F). There was no overlap between matK and the trnK-matK regions. Primer information is listed in Supplementary Table S2. Polymerase chain reaction (PCR) amplification conditions were: one cycle at 94°C for 3min; 35 cycles of 94°C for 45 s, 53–55°C for 60 s and 72°C for 60 s; followed by 72°C for 5 min (Yu et al., 2011). PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA, United States). Sequencing reactions were performed using the ABI Prism BigDye Terminator Kits (Applied Biosystems, Inc.), following a modified protocol (Yu et al., 2011). Automated sequencing was performed on ABI 3730xl DNA sequencer (Applied Biosystems) in Kunming Institute of Botany, Chinese Academy of Sciences.
The newly obtained raw sequences were assembled and edited using Geneious (Biomatters, Inc.) version 7.1 (Kearse et al., 2012). Assembled sequence consensuses were automatically aligned using MAFFT version 7.2 (Katoh and Toh, 2010), then adjusted manually in Geneious. The aligned matrices were concatenated using SequenceMatrix version 1.73 (Vaidya et al., 2011). Sequence characteristics were calculated using MEGA version 6.0 (Tamura et al., 2013).
Maximum Likelihood (ML) and Bayesian Inference (BI) methods were used to reconstruct phylogenetic trees. The nrITS and the concatenated plastid datasets were analyzed separately. No nucleotide positions were excluded from analyses. The ML tree searches and bootstrap estimation of clade support were conducted with RAxML version 8.2.10 (Stamatakis et al., 2008). These analyses used the GTR substitution model with gamma-distributed rate heterogeneity among sites and the proportion of invariable sites estimated from the data. The concatenated plastid dataset was partitioned by gene. Support values for the node and clade were estimated from 1000 bootstrap replicates. Bootstrap supports (BS) ≥ 70 were considered well supported (Hillis and Bull, 1993). Partitioned BI analyses were performed using MrBayes version 3.2.6 (Ronquist and Huelsenbeck, 2003), with DNA substitution models selected for each gene partition by the Bayesian information criterion (BIC) using jModeltest version 2.1.10 (Guindon and Gascuel, 2003; Darriba et al., 2012). Markov Chain Monte Carlo (MCMC) analyses were run in MrBayes for 10,000,000 generations for each dataset, with two simultaneous runs, and each run comprising four incrementally heated chains. The BI analyses started with a random tree, and sampled every 1000 generations. The number of generations for the datasets was considered sufficient when the average standard deviation of split frequencies was lower than 0.005, and Potential Scale Reduction Factor (PSRF) of Convergence Diagnostic (Gelman and Rubin, 1992) was 1.00. The first 25% of the trees was discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. Clades recovering posterior probability values (PP) ≥ 0.95 were considered as well supported (Alfaro et al., 2003; Erixon et al., 2003; Kolaczkowski and Thornton, 2007). Both ML and BI analyses, as well as jModeltest, were performed at the CIPRES Science Gateway1.
Phylogenetic incongruence between ML and BI analyses of each dataset was visually compared using TreeGraph version 2.12 (Stover and Muller, 2010). The incongruence length difference test (ILD) (Farris et al., 1995) was not used to assess topological conflict between the nuclear and the concatenated plastid datasets because this analysis has been shown to be misleading (Baker et al., 2011). We used a conservative PP ≥ 0.95 and BS ≥ 70 as thresholds for identifying strongly incongruent clades between nrITS and the plastid datasets. In addition, topological incongruence between nrITS and the plastid datasets was also investigated using the Shimodaira–Hasegawa (SH) test (Shimodaira and Hasegawa, 1999) and the approximately unbiased (AU) test (Shimodaira, 2002). Topologies were constrained using Mesquite version 3.2 (Maddison and Maddison, 2017). The SH and AU tests were performed using PAUP version 4.10 (Swofford, 2003).
The nrITS and the plastid datasets of the Ph. tenuisectum complex were combined to recover a phylogenetic network using SplitsTree version 4.14.3 (Huson and Bryant, 2006). Pterygiella samples were included as outgroups. The Neighbor-net model was chosen using the Kimura 2-parameter (K2P) distance and Ordinary Least Square Method. Bootstrap values for the respective splits were estimated from 1000 bootstrap replicates.
There is no reliable fossil in Orobanchaceae to calibrate the phylogeny; divergence times were indirectly estimated from calibrating fossils from other families of flowering plants (Bremer et al., 2004; Magallón et al., 2015; Wang et al., 2015; Wikström et al., 2015). Due to inconsistency in taxonomic sampling and marker choice in different studies, the mean crown ages of Orobanchaceae varied from 26.0 Mya (Wikström et al., 2015) to 74.54 Mya (Wang et al., 2015). To improve the molecular dating on Orobanchaceae and the Ph. tenuisectum complex, we selected 24 families and 102 genera from Lamiales, and Solanum tuberosum as the outgroup (Supplementary Table S3). Two chloroplast DNA regions, rps2, and trnK were concatenated for molecular dating. Dating analyses were conducted using Markov Chain Monte Carlo (MCMC) methods in BEAST version 2.4 (Bouckaert et al., 2014), which was performed at the CIPRES Science Gateway2. For setting parameters of BEAUti, we chose “BEAST model test” for “Site model,” “Relaxed Clock Log Normal” for “Clock model,” and “Yule Model” for speciation. Meanwhile, we selected four fossils to calibrate stem nodes of four families using CladeAge package (Matschiner et al., 2017). Of them, Acanthaceae (Acanthus rugatus Reid & Chandler), Bignoniaceae (Radermachera pulchra Reid & Chandler), Lamiaceae (Melissa parva Reid & Chandler) have reliable fossils from the Bembridge Flora (England) (Reid and Chandler, 1926) of the Late Eocene (Collinson et al., 2010), and Oleaceae (Fraxinus wilcoxiana Berry) from the Claiborne Formation of western Tennessee (United States) of the Middle Eocene (Call and Dilcher, 1992). The stem age of Acanthaceae (C4), Bignoniaceae (C3), and Lamiaceae (C5) was set as 33.9–37.8 Mya, and that of Oleaceae (C2) as 41.2–47.8 Mya. In addition, we used a normal model to constrain the crown age for Lamiales (C1) and Orobanchaceae (C6) as 84 ± 10 Mya and 56 ± 10 Mya, respectively. These ages were adopted from the TimeTree website3 (Hedges et al., 2006). Three independent MCMC runs with the same parameters were conducted. Each MCMC ran 100,000,000 generations, and was sampled every 10,000 generations. The first 5,000 generations were removed as “Pre Burnin.” Three Log outputs of the BEAST analyses were jointly evaluated using Tracer version 1.6. Effective sample sizes (ESS) of all parameters were more than 200, indicating that MCMC sampling was adequate (Lanfear et al., 2016). Trees of three runs were combined using LogCombiner by burning 20% of the initial trees. The Maximum Clade Credibility (MCC) tree was generated using TreeAnnotator by setting “Mean heights” for the “Node heights”. The MCC tree was visualized using FigTree version 1.4.24.
To evaluate sequence matrix characteristics, we classified the samples of Phtheirospermum spp. and Pterygiella spp. in several groups. First, four major groups were adopted, i.e., Phtheirospermum, Pterygiella, the Ph. tenuisectum complex and the Ph. tenuisectum complex + Pterygiella. Then, every species with two or more samples in Phtheirospermum was treated as an independent group. Five species groups in Phtheirospermum were recognized, i.e., four groups in the Ph. tenuisectum complex, and Ph. japonicum.
Sequence characteristics of nrITS, seven plastid DNA loci, and the concatenated plastid datasets are presented in Table 1. The nrITS dataset was the most informative marker (except in the Ph. muliense and Phtheirospermum sp. 1), followed by two chloroplast intergenic spacers (trnH-psbA and trnL-F) and two introns (matK-trnK, and rps16). Of the three coding genes, matK was the most informative. Sequences of Ph. japonicum had no informative variation across seven plastid markers throughout nine populations. Only four deletions/insertions were found in the Japanese sample (J9), i.e., indels of a single nucleotide for rps16 and trnK, and two nucleotides in trnL-F. In contrast, the sequences of Ph. tenuisectum showed the highest variation at the species level across all eight DNA regions. The remaining three taxa of the Ph. tenuisectum complex were restricted to the Hengduan Mountains region, and showed fewer variations. Though 4.40% of nrITS sites of Pterygiella were variable, only 0 to 0.29% of the sites across seven plastid loci were variable.
Phylogenetic trees using nrITS and the plastid datasets are presented in Figures 3, 4, respectively. Pterygiella + Phtheirospermum + Xizangia (hereafter referred to as the Pterygiella group) was well supported as monophyletic (nrITS: BS/PP = 61/0.99; plastid: BS/PP = 100/1.00), and not nested in any other formerly recognized tribe. The plastid dataset supported Brandisia spp. as sister to this group (BS/PP = 69/0.96). None of the analyses resolved Pterygiella group close to tribe Rhinantheae.
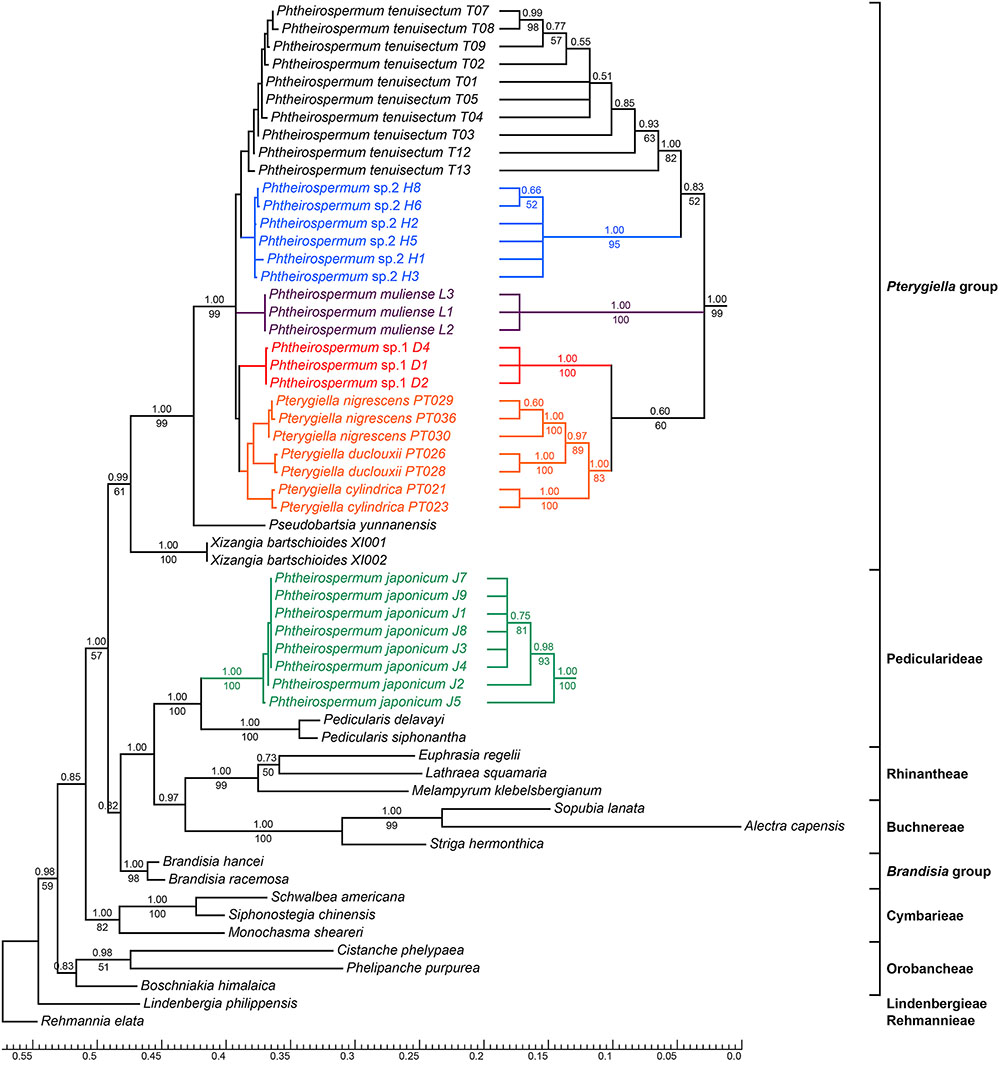
FIGURE 3. Bayesian inference (BI) tree inferred from the nrITS dataset. ML Bootstrap values are presented under branches, and BI posterior probabilities are shown above branches. The topology of some clades with short branch lengths appear on the right. The bottom scale bar represents the number of substitutions per site.
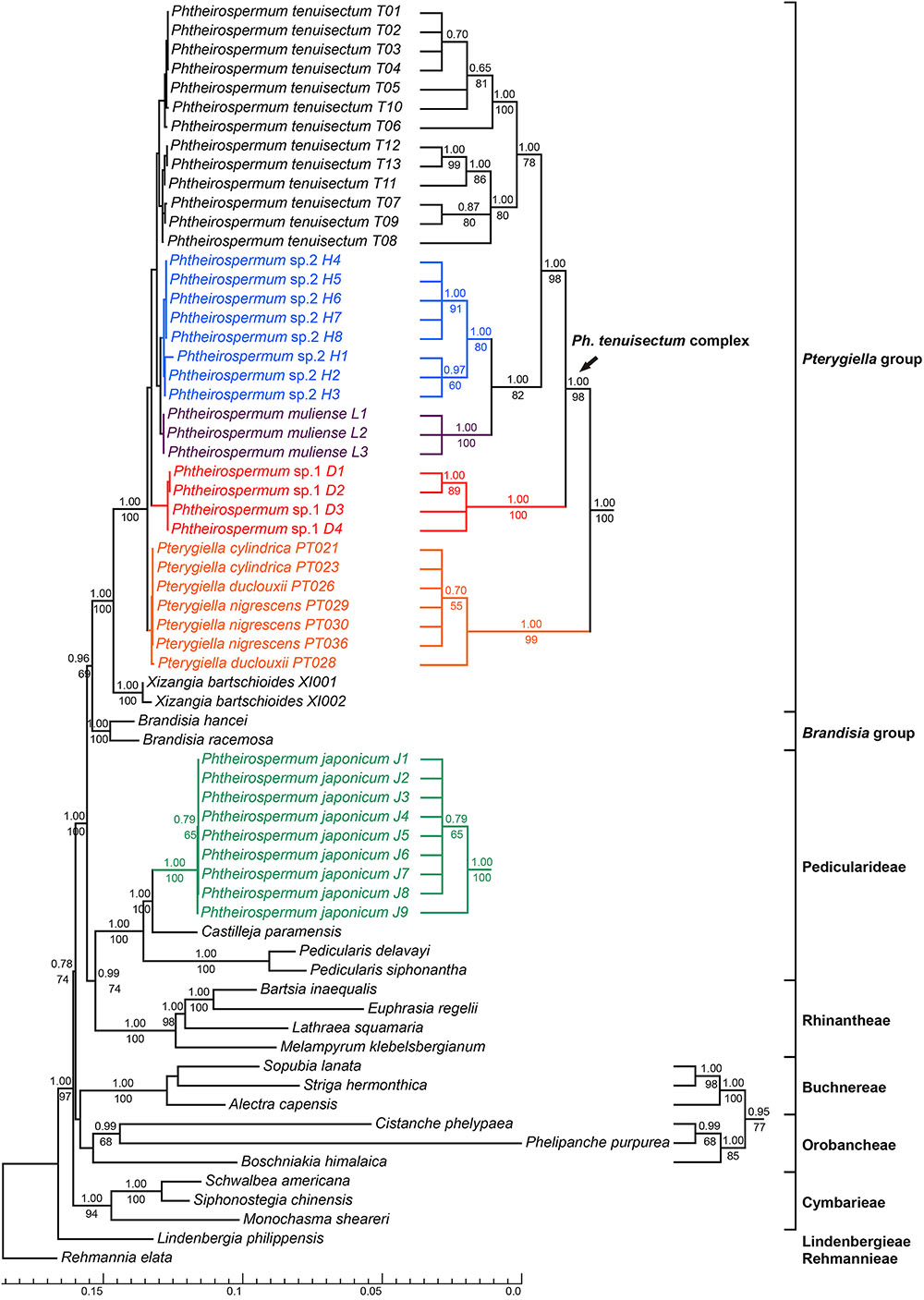
FIGURE 4. Bayesian inference tree inferred from the combined plastid dataset. The seven plastid markers include matK, rbcL, rps2, rps16, trnK-matK, trnH-psbA and trnL-F. ML Bootstrap values are presented under branches, and BI posterior probabilities are shown above branches. The topology of some clades with short branch lengths appear on the right. The bottom scale bar represents the number of substitutions per site.
Both nrITS and the plastid datasets recovered Ph. japonicum nested within Pedicularideae (nrITS and plastid: BS/PP = 100/1.00). The Ph. tenuisectum complex and Pterygiella spp. also formed a well-supported clade in both analyses (nrITS: BS/PP = 99/1.00; plastid: BS/PP = 100/1.00). The four morphotypes of the Ph. tenuisectum complex were recovered as reciprocally monophyletic, well-supported lineages. The Ph. tenuisectum complex was strongly supported as monophyletic by the plastid dataset (BS/PP = 99/1.00), and phylogenetic relationships for the four morphotypes were fully resolved (Figure 4). In contrast, Pterygiella spp. were nested within the Ph. tenuisectum complex in nrITS analyses (BS/PP = 100/1.00). In this analysis, Ph. tenuisectum was supported as sister to Phtheirospermum sp. 2 (BS/PP = 59/0.92), and Phtheirospermum sp. 1 and Pterygiella spp. formed a clade (BS/PP = 60/0.60).
Based on support values, we found hard topological conflicts between nrITS and the plastid datasets in the placement of tribes Orobancheae and Buchnereae. Additionally, these placements disagree with those recovered by McNeal et al. (2013). Further topological comparisons using SH and UA tests are presented in Table 2. In constrained analyses using nrITS dataset, both SH and UA tests indicated that the monophyly of the Ph. tenuisectum complex and Brandisia spp. sister to Pterygiella group were supported (P > 0.2). On the other hand, sister relationship between tribes Orobancheae and Buchnereae, and tribes Rhinantheae and Pedicularideae were rejected (P < 0.05). The constrained analyses using the plastid data rejected the paraphyly of the Ph. tenuisectum complex by the AU test (P < 0.05) and the sister relationship between tribes Buchnereae and Rhinantheae by both SH and UA tests. Therefore, nrITS and the plastid datasets should be analyzed separately.
To further explore phylogenetic incongruence within the Ph. tenuisectum complex, a phylogenetic network was inferred from the total dataset (Figure 5). A well supported split separated the Ph. tenuisectum complex from Pterygiella spp. (BS = 99). Each of the three species of Pterygiella were well resolved as monophyletic (BS = 100). The four morphotypes of the Ph. tenuisectum complex were also recovered as separate clusters. This pattern of relationships is similar to the plastid phylogeny, in which that Ph. tenuisectum was sister to Phtheirospermum sp. 2 + Ph. muliense (BS = 96). In addition, Phtheirospermum sp. 1 had connections with Pt. duclouxii + Pt. nigrescens (BS = 73), likely based on signal from the nrITS dataset.
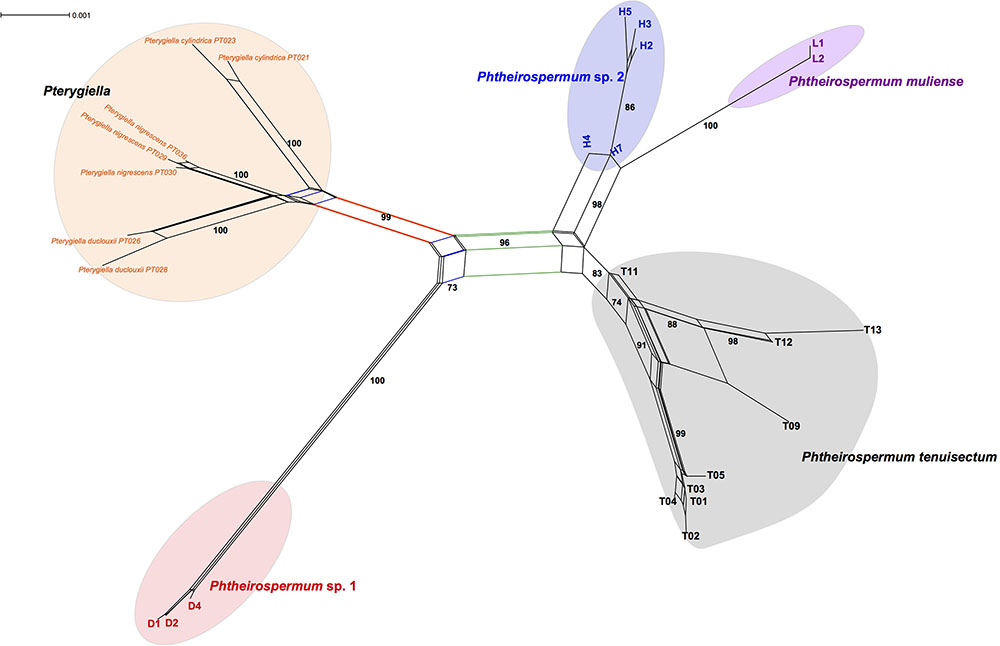
FIGURE 5. Phylogenetic network of the Ph. tenuisectum complex and Pterygiella spp. using all loci. The scale bar on the top left represents the number of substitutions per site.
The MCC tree is shown in Figure 6 and Supplementary Figure S1, including mean ages and 95% HPD interval bars. The exact values for six calibrated and 13 annotated nodes are summarized in Table 3. The mean substitution rate was 1.10 × 10-3 per site per million years (95% HPD: 9.04 × 10-4 - 1.29 × 10-3). The Yule speciation rate was 4.65 × 10-2 (95% HPD: 3.63 × 10-2 - 5.67 × 10-2). The coefficient of variation was 1.08 (95% HPD: 0.91 - 1.27), indicating that a high degree of rate heterogeneity was observed across the tree and that a relaxed-clock model was suitable for this dataset (Drummond et al., 2006).
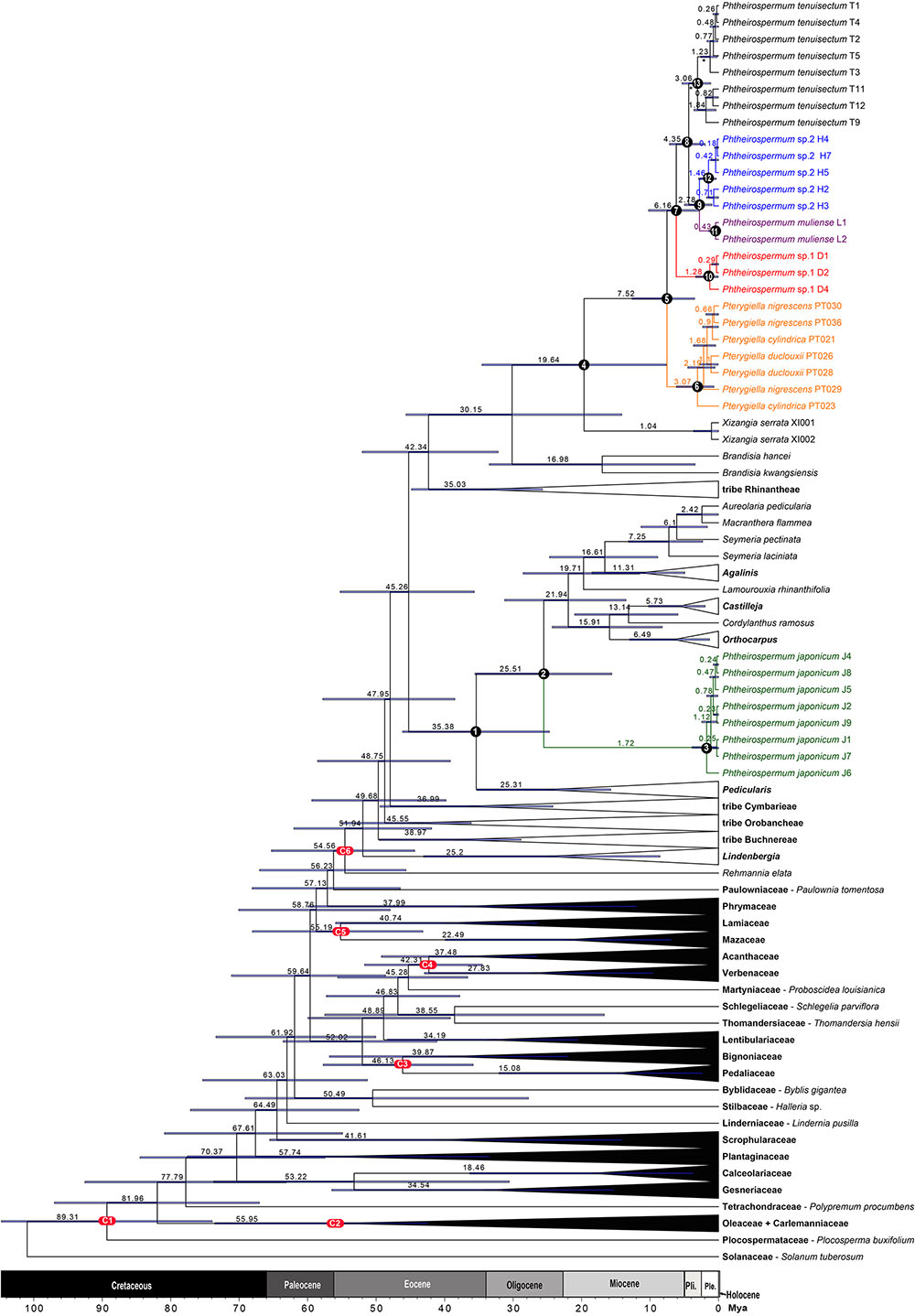
FIGURE 6. A simplified maximum clade credibility tree of Lamiales from BEAST divergence time analysis. The estimated age of nodes is presented above the branch. Node bars represent the 95% highest posterior density (HPD) interval. Six calibrated (red) and 13 key stem/crown nodes (black) were annotated by letters and/or numbers.
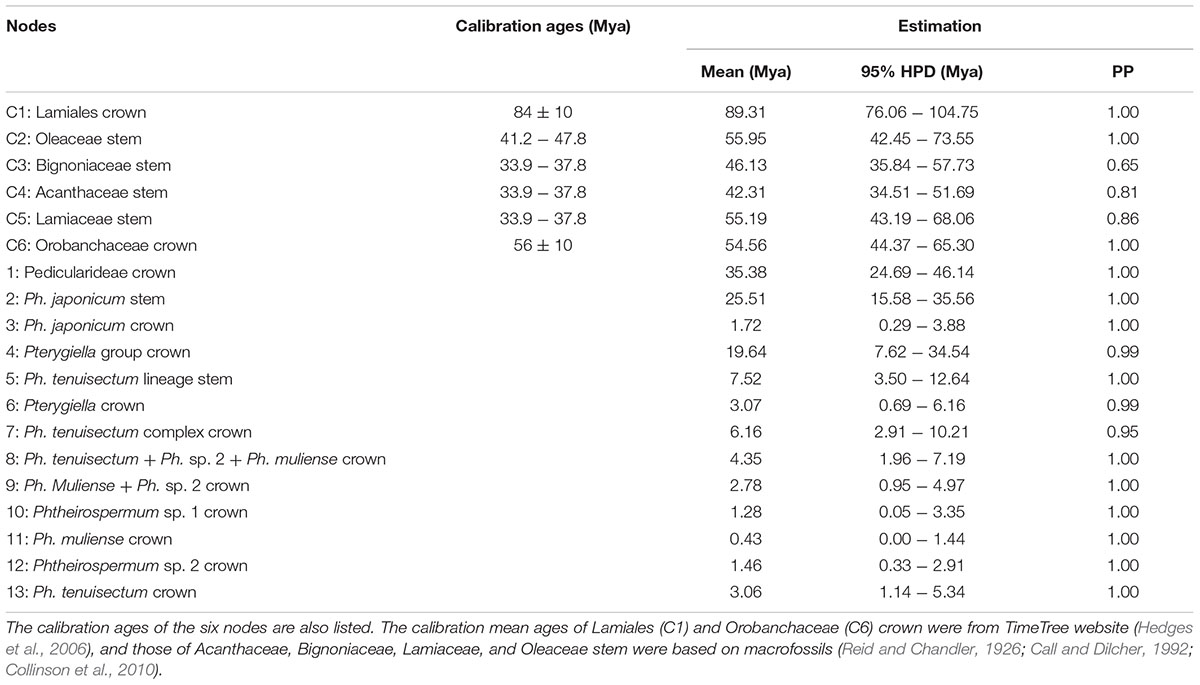
TABLE 3. Estimated age, 95% highest posterior density (HPD) interval, and posterior probability (PP) value of six calibrated and 13 key stem/crown nodes.
Familial relationships in Lamiales were not fully resolved, and only some nodes were well supported (PP ≥ 0.95). Paulowniaceae and Phrymaceae were strongly supported as sister to Orobanchaceae (PP = 1.00). In Orobanchaceae, nine clades were well resolved (PP ≥ 0.99). Of these, seven clades have been recognized as tribes, and Brandisia as an independent lineage (Stevens, 2001 onward; McNeal et al., 2013). The Pterygiella group was recovered as monophyletic. Tribe Rehmannieae was the most basal clade, followed by splits marking the divergences of tribes Lindenbergieae and Buchnereae from the remainder of the lineage. The following splits mark the divergences of tribes Orobancheae and Cymbarieae successively, followed by Pedicularideae and Rhinantheae, leaving Brandisia and the Pterygiella group as a strongly supported clade (PP = 1.00). Brandisia was weakly supported as sister to the Pterygiella group. Within the Pterygiella group, the Ph. tenuisectum complex was strongly supported as monophyletic, and phylogenetic relationships of four morphotypes were well resolved.
The divergence time estimate for Orobanchaceae was in the early Eocene (54.56 Mya). Tribe Pedicularideae separated from tribe Rhinantheae – the Pterygiella group at the mid-Eocene (45.26 Mya), then Ph. japonicum diverged from the new world genera of tribe Pedicularideae at the late Oligocene (25.51 Mya), and its subsequent diversification occurred at the mid-Pleistocene (1.72 Mya). Diversification of the Pterygiella group was estimated to be around the early Miocene (19.64 Mya). The Ph. tenuisectum complex diverged from Pterygiella spp. during the later Miocene (7.52 Mya), with the four morphotype lineages fully established by the early Pleistocene (2.78 Mya).
The current delimitation of Orobanchaceae includes Rehmannia and Trianeophora as the basal tribe, i.e., Rehmannieae (Stevens, 2001 onward), which was strongly supported by phylogenetic analyses (Xia et al., 2009; Zeng et al., 2017). To date, seven tribes and Brandisia have been recognized in Orobanchaceae (Stevens, 2001 onward; McNeal et al., 2013). Although PHYA and PHYB nuclear data strongly supported Pterygiella as sister to the remaining genera of tribe Rhinantheae; both nrITS and the combined rps2 and matK datasets poorly supported Pterygiella as sister to Brandisia, and only the plastid dataset weakly supported the clade Pterygiella + Brandisia as sister to the remaining genera of tribe Rhinantheae (McNeal et al., 2013). In this study, both nrITS and the plastid datasets strongly resolved the Pterygiella group as monophyletic. The plastid dataset strongly supported Brandisia spp. as sister to the Pterygiella group; however, nrITS weakly resolved Brandisia as sister to a clade comprising tribes Buchnereae, Pedicularideae, and Rhinantheae. Therefore, Brandisia and the Pterygiella group should be treated as two separate groups or tribes in Orobanchaceae.
All phylogenetic analyses recovered polyphyly of Phtheirospermum, e.g., Ph. japonicum grouped with tribe Pedicularideae and the Ph. tenuisectum complex was close to Pterygiella spp. in the clade including Pseudobartsia and Xizangia (Figures 3, 4). This is not surprising, given that previous analyses place Ph. japonicum within Pedicularideae (Bennett and Mathews, 2006; McNeal et al., 2013; Yu et al., 2015a; Pinto-Carrasco et al., 2017). In this tribe, Pedicularis is the most basal group, followed by Ph. japonicum. The remaining genera are mainly distributed in the New World (Bennett and Mathews, 2006; McNeal et al., 2013), including Castilleja, Orthocarpus, and Triphysaria, another important model for parasitic plant genetics, genomics, and evolution (Tomilov et al., 2005; Westwood et al., 2010; Bandaranayake et al., 2012; Yang et al., 2015). The Ph. tenuisectum complex is clearly excluded from tribe Pedicularideae, forming a lineage including Pterygiella, Xizangia, and Pseudobartsia. The taxonomic confusion of Phtheirospermum can be ascribed to J. D. Hooker who described the second species Ph. parishii in Phtheirospermum, and transferred Euphrasia glandulosa Benth. to this genus (Hooker and Thomson, 1884). Based on the ellipsoid seeds and the minutely reticulated seed surface, C. B. Clarke suggested treating Ph. parishii as a separate genus (i.e., “Emmenospermum”), but Hooker did not adopt this treatment, because seed morphology of Scrophulariaceae was high variable within and among genera (Hooker and Thomson, 1884). Based on Hooker’s delimitation of Phtheirospermum, Ph. tenuisectum and Ph. muliense were placed in this genus (Bureau and Franchet, 1891; Tao, 1996). However, the phylogenetic analyses demonstrate that the Ph. tenuisectum complex should be separated from Ph. japonicum. Furthermore, this separation is justified by morphology; compared to Ph. japonicum, members of the Ph. tenuisectum complex bear linear rather than ovate to orbicular pinnae, smaller flowers with yellow rather than red corollas, and smaller fruits and seeds.
Though the Ph. tenuisectum complex was strongly supported as monophyletic by the plastid dataset (Figure 4), nrITS resolved Ph. tenuisectum complex as paraphyletic, with Pterygiella nested within it (Figure 3). Pinto-Carrasco et al. (2017) have combined the Ph. tenuisectum complex with Pterygiella in a single taxon, creating new combinations Pt. muliensis (C.Y.Wu & D.D.Tao) Pinto-Carrasco, E.Rico & M.M.Mart.Ort., Pt. parishii (Hook. f.) Pinto-Carrasco, E.Rico & M.M.Mart.Ort., and Pt. tenuisecta (Bureau and Franch.) Pinto-Carrasco, E.Rico & M.M.Mart.Ort.. This taxonomic decision was justified on the basis of phylogenetic results, and morphological evidence including five-toothed calyces and pollen characters. However, we suggest that the Ph. tenuisectum complex and Pterygiella should be retained as separated groups. First, in the constrained analyses, monophyly of the Ph. tenuisectum complex was not rejected (Table 2). Moreover, the Ph. tenuisectum complex is morphologically quite distinct from Pterygiella. Pterygiella spp. are characterized by winged stems (except Pt. cylindrica P. C. Tsoong), lanceolate and entire leaves, and by a calyx that is broadly campanulate and 5-veined (Supplementary Figure S2). Conversely, the Ph. tenuisectum complex has cylindrical stems, pinnatisect leaves, and a tubular calyx. In addition, seed morphology and capsule indumentum also differ between Pterygiella and the Ph. tenuisectum complex (Dong et al., 2013, 2015). Therefore, we suggest treating the Ph. tenuisectum complex as an independent genus. So far, there is no available name for this lineage. The name “Emmenospermum” cannot be selected because it would be a later homonym of Emmenosperma F. Mueller (Rhamnaceae). A new genus name needs to be published in accordance with the International Code of Nomenclature for algae, fungi, and plants (McNeill et al., 2012), including comprehensive morphological comparison with Pterygiella spp. and other relatives.
The genus Phtheirospermum was established on the basis of Ph. chinense Bunge, which was collected in north China (Fischer and Meyer, 1835). Both the genus and species names are ascribed to A.A. Bunge in F.E.L. Fischer’s and C.A. Meyer’s edited monograph, Index Seminum [St. Petersburg]. However, an early name Gerardia japonica Thunberg had been described based on a Japanese specimen. Therefore, Ph. japonicum is the correct name to replace Ph. chinense (Kanitz and Weiss, 1878). Our phylogenetic results showed the polyphyly of Phtheirospermum, the current delimitation of Phtheirospermum include Ph. japonicum alone, and Ph. tenuisectum complex needs to be transferred to a new genus. Currently, Ph. japonicum has been established as the experimental model for the study of genetics and development of the haustorium (Cui et al., 2016; Ishida et al., 2016). Maintaining the name Ph. japonicum will therefore benefit molecular biologists using this model in publications or communications with public audiences and plant specialists. This taxonomic decision would forestall confusion as has arisen in the nomenclature of the monkeyflower (Mimulus guttatus DC.), a model organism for studies of evolution and ecology that was shown to be polyphyletic. The correct name of the monkeyflower was changed to Erythranthe guttata (DC.) G. L. Nesom (Barker et al., 2012). However, some recent publications still use the old name M. guttatus (Lee et al., 2016; Ferris et al., 2017; Puzey et al., 2017).
Phtheirospermum japonicum is a widely distributed species in Eastern Asia (Hong et al., 1998). In this study, we sampled nine individuals from southwestern (Yunnan: J2, J3, J7, and J8; Sichuan: J6), through central China (Henan: J5), to eastern (Zhejiang: J1) and northeastern China (Liaoning: J4), and extending to Japan (Kanagawa: J9). Both nrITS and the plastid evidence indicated that genetic variation in Ph. japonicum was low, and current distribution range might be a result of a recent population expansion. The results of molecular dating showed that a recent radiation occurred at around 1.72 Mya (95% HPD: 0.29–3.88 Mya), and the Japanese sample was derived from a Yunnan sample (J2) at around 0.23 Mya (95% HPD: 0.0–0.73 Mya).
The Ph. tenuisectum complex can be classified as four species lineages, each strongly supported by both nrITS and the plastid datasets (Figures 3, 4), and diagnosable by morphological characters (Figure 1). Phylogenetic relationships among the four lineages were inconsistent between nrITS and the plastid datasets. It is likely that because resolution of the nrITS tree was low, constrained analyses using nrITS dataset did not reject the constrained plastid topology (Table 2). The plastid dataset strongly supported that Phtheirospermum sp. 1 was the sister to the three other lineages, in which Phtheirospermum sp. 2 was sister to Ph. muliense. As an ancestral lineage in the Ph. tenuisectum complex, Phtheirospermum sp. 1 might share plesiomorphic characters with Pterygiella spp. (i.e., incomplete lineage sorting), so that the nrITS dataset weakly supported Pterygiella spp. as sister to Phtheirospermum sp. 1 (Figure 3), and phylogenetic network showed a well-supported genetic connection between Phtheirospermum sp. 1 and Pt. duclouxii + Pt. nigrescens (Figure 5). Meanwhile, ancient hybridization/introgression cannot be excluded between Pterygiella spp. and Phtheirospermum sp. 1. In addition, ancient hybridization/introgression and incomplete lineage sorting may also have occurred between Phtheirospermum sp. 2 and Ph. tenuisectum.
The last two uplifts of the Qinghai–Tibet Plateau happened at around 8.0–7.0 Mya and 3.6–1.5 Mya (Zheng et al., 2000; Garzione et al., 2003; Molnar, 2005; Ge et al., 2006; Li et al., 2015; but see Renner, 2016). Many species groups diversified rapidly with these uplifts in the Hengduan Mountains (Wen et al., 2014; Xing and Ree, 2017). In this study, the estimated ages for the origin of Ph. tenuisectum complex and its subsequent divergences fell into the range of the two uplifts (Table 3 and Figure 6). The uplift during 8.0–7.0 Mya may have driven divergence of the Ph. tenuisectum complex from Pterygiella. The center of diversity for both the Ph. tenuisectum complex and Pterygiella is in northwestern Yunnan and southwestern Sichuan, perhaps the place of origin of the Ph. tenuisectum complex. Overlapping species distributions among these genera out of the Hengduan Mountains may be the results of later population expansion. Of the four lineages in the Ph. tenuisectum complex, Ph. muliense, Phtheirospermum sp. 1, and Phtheirospermum sp. 2 grow along/near the dry valleys of Jinsha River and its tributaries, with only the sample H5 of Phtheirospermum sp. 2 extend to Lancang River (Figure 2). In contrast, Ph. tenuisectum mainly occurs in meadows and has a wider distribution. The valleys of Jinshan and Lancang Rivers and their tributaries in the Hengduan Mountains region experience a specialized dry-hot valley climate (Yang et al., 2016), which may have contributed to divergence of Ph. muliense, Phtheirospermum sp. 1, and Phtheirospermum sp. 2. To address the evolutionary history of Ph. tenuisectum complex in the future, a phylogeographic approach may be applied using more dense population samplings and multiple individuals per population.
W-BY, HW, and D-ZL conceived the study. W-BY, CR, LL, HW, and RC collected the data. W-BY analyzed the data. W-BY, CR, LL, CdP, and D-ZL interpreted the results. All authors wrote and revised the paper, and approved the final version.
This study was supported by grants from the National Natural Science Foundation of China (Grants 31470323 and 31200185), the National Basic Research Program of China (2014CB954100), the Major International Joint Research Project of National Natural Science Foundation of China (31320103919), the 1000 Talents Program (WQ20110491035), US National Science Foundation (DEB1119746), the Chinese Academy of Sciences (Y7ZK251B01), and the China Scholarship Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to J. Cai, L.-N. Dong, H.-J. He, G.-W. Hu, W. Jiang, M.-L. Liu, Y. Luo, J. Morawetz, B. Pan, H. Tang, X.-L. Yang, Z.-K. Wu, and S.-D. Zhang for their kind help in the field and/or providing samples for this study; to M.-L. Liu and J. Yang for their assistance in molecular experiments; and to two reviewers for their valuable comments and suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00142/full#supplementary-material
FIGURE S1 | A detailed maximum clade credibility tree of Lamiales from BEAST divergence time analysis. The estimated age of nodes was presented above the branch. Node bars represents 95% highest posterior density (HPD) interval. PP ≥ 95% is indicated by one asterisk (∗), and 95% ≥ PP ≥ 75% is indicated by two asterisks under the branch. Six calibrated (red) and 13 key stem/crown nodes (black) were annotated by letters and/or numbers.
FIGURE S2 | Photographs of Pterygiella. (A–C) Pterygiella cylindrica P. C. Tsoong; (D–F) Pt. duclouxii Franchet; and (G–I) Pt. nigrescens Oliver.
TABLE S1 | Voucher information for phylogeny of Phtheirospermum and Pterygiella, including 20 other genera of Orobanchaceae.
TABLE S2 | Primer information for PCR amplification and sequencing in this study.
TABLE S3 | Voucher information for molecular dating of Orobanchaceae, including selected genera and families from Lamiales.
Alfaro, M. E., Zoller, S., and Lutzoni, F. (2003). Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 20, 255–266. doi: 10.1093/molbev/msg028
Al-Shehbaz, I. A., Yue, J., and Sun, H. (2004). Shangrilaia (Brassicaceae), a new henus from China. Novon 14, 271–274.
Baker, W. J., Norup, M. V., Clarkson, J. J., Couvreur, T. L. P., Dowe, J. L., Lewis, C. E., et al. (2011). Phylogenetic relationships among arecoid palms (Arecaceae: Arecoideae). Ann. Bot. 108, 1417–1432. doi: 10.1093/aob/mcr020
Bandaranayake, P. C. G., Tomilov, A., Tomilova, N. B., Ngo, Q. A., Wickett, N., Depamphilis, C. W., et al. (2012). The TvPirin gene is necessary for haustorium development in the parasitic plant Triphysaria versicolor. Plant Physiol. 158, 1046–1053. doi: 10.1104/pp.111.186858
Barker, W. R., Nesom, G. L., Beardsley, P. M., and Fraga, N. S. (2012). A taxonomic conspectus of Phrymaceae: a narrowed circumscription for Mimulus, new and resurrected genera, and new names and combinations. Phytoneuron 201, 1–60.
Bennett, J. R., and Mathews, S. (2006). Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93, 1039–1051. doi: 10.3732/ajb.93.7.1039
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2: a software patform for bayesian evolutionary analysis. PLOS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Bremer, K., Friis, E., and Bremer, B. (2004). Molecular phylogenetic dating of asterid flowering plants shows early cretaceous diversification. Syst. Biol. 53, 496–505. doi: 10.1080/10635150490445913
Bureau, E., and Franchet, A. (1891). Plantes nouvelles du Thibet et de la Chine occidentale recueillies pendant le voyage de M. Bonvalot et du Prince Henri d’Orleìans en 1890. J. Bot. 5, 128–130.
Call, V. B., and Dilcher, D. L. (1992). Investigations of angiosperms from the Eocene of southeastern North America: samaras of Fraxinus wilcoxiana Berry. Rev. Palaeobot. Palynol. 74, 249–266. doi: 10.1016/0034-6667(92)90010-E
Collinson, M., Manchester, S. R., Wilde, V., and Hayes, P. (2010). Fruit and seed floras from exceptionally preserved biotas in the European Paleogene. Bull. Geosci. 85, 155–162. doi: 10.3140/bull.geosci.1155
Cui, S., Wakatake, T., Hashimoto, K., Saucet, S. B., Toyooka, K., Yoshida, S., et al. (2016). Haustorial hairs are specialized root hairs that support parasitism in the facultative parasitic plant Phtheirospermum japonicum. Plant Physiol. 170, 1492–1503. doi: 10.1104/pp.15.01786
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dong, L.-N., Wang, H., Wortley, A. H., Li, D.-Z., and Lu, L. (2015). Fruit and seed morphology in some representative genera of tribe Rhinantheae sensu lato (Orobanchaceae) and related taxa. Plant Syst. Evol. 301, 479–500. doi: 10.1007/s00606-014-1087-8
Dong, L.-N., Wang, H., Wortley, A. H., Lu, L., and Li, D.-Z. (2013). Phylogenetic relationships in the Pterygiella complex (Orobanchaceae) inferred from molecular and morphological evidence. Bot. J. Linn. Soc. 171, 491–507. doi: 10.1111/j.1095-8339.2012.01326.x
Dong, L.-N., Wortley, A. H., Wang, H., Li, D.-Z., and Lu, L. (2011). Efficiency of DNA barcodes for species delimitation: a case in Pterygiella Oliv. (Orobanchaceae). J. Syst. Evol. 49, 189–202. doi: 10.1111/j.1759-6831.2011.00124.x
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 19, 11–15.
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., and Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLOS Biol. 4:e88. doi: 10.1371/journal.pbio.0040088
Erixon, P., Svennblad, B., Britton, T., and Oxelman, B. (2003). Reliability of bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst. Biol. 52, 665–673. doi: 10.1080/10635150390235485
Farris, J. S., Källersjö, M., Kluge, A. G., and Bult, C. (1995). Constructing a significance test for incongruence. Syst. Biol. 44, 570–572. doi: 10.1093/sysbio/44.4.570
Favre, A., Päckert, M., Pauls, S. U., Jähnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253. doi: 10.1111/brv.12107
Ferris, K. G., Barnett, L. L., Blackman, B. K., and Willis, J. H. (2017). The genetic architecture of local adaptation and reproductive isolation in sympatry within the Mimulus guttatus species complex. Mol. Ecol. 26, 208–224. doi: 10.1111/mec.13763
Fischer, F. E. L., and Meyer, C. A. (eds) (1835). Index Seminum, quae Hortus Botanicus Imperialis Petropolitanus pro Mutua Commutatione Offert. Saint Petersburg: Accedunt Animadversiones Botanicae Nonnullae.
Friesen, N., Fritsch, R. M., Pollner, S., and Blattner, F. R. (2000). Molecular and morphological evidence for an origin of the aberrant genus Milula within himalayan species of Allium (Alliacae). Mol. Phylogenet. Evol. 17, 209–218. doi: 10.1006/mpev.2000.0844
Garzione, C. N., Decelles, P. G., Hodkinson, D. G., Ojha, T. P., and Upreti, B. N. (2003). East-west extension and Miocene environmental change in the southern Tibetan plateau: Thakkhola graben, central Nepal. Geol. Soc. Am. Bull. 115, 3–20. doi: 10.1130/0016-7606(2003)115<0003:EWEAME>2.0.CO;2
Ge, X.-H., Ren, S.-M., Ma, L.-X., Wu, G.-D., Liu, Y.-J., and Yuan, S.-H. (2006). Multi-stage uplifts of the Qinghai-Tibet plateau and their environmental effects. Earth Sci. Front. 16, 118–130.
Gelman, A., and Rubin, D. B. (1992). Inference from iterative simulation using multiple sequences. Statistical. Sci. 7, 457–472. doi: 10.1214/ss/1177011136
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hedges, S. B., Dudley, J., and Kumar, S. (2006). TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972. doi: 10.1093/bioinformatics/btl505
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Hong, D.-Y. (1979). “Pseudobartsia D.Y.Hong,” in Flora Reipublicae Popularis Sinacae, eds P.-C. Tsoong and H.-B. Yang (Beijing: Science Press), 388–389.
Hong, D.-Y., Yang, H.-B., Jin, C.-L., Fischer, M. A., Holmgren, N. H., and Mill, R. R. (1998). Scrophulariaceae. Saint Louis: Missouri Botanical Garden Press.
Hughes, C. E., and Atchison, G. W. (2015). The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytol. 207, 275–282. doi: 10.1111/nph.13230
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Ishida, J. K., Wakatake, T., Yoshida, S., Takebayashi, Y., Kasahara, H., Wafula, E., et al. (2016). Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 28, 1795–1814. doi: 10.1105/tpc.16.00310
Kanitz, A., and Weiss, E. (eds) (1878). Expeditio Austro-Hungarica ad oras Asiae Orientalis: Anthophyta quae in Japonia legit Beat. Budapest: Typis Societ.
Katoh, K., and Toh, H. (2010). Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26, 1899–1900. doi: 10.1093/bioinformatics/btq224
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kolaczkowski, B., and Thornton, J. W. (2007). Effects of branch length uncertainty on Bayesian posterior probabilities for phylogenetic hypotheses. Mol. Biol. Evol. 24, 2108–2118. doi: 10.1093/molbev/msm141
Lanfear, R., Hua, X., and Warren, D. L. (2016). Estimating the effective sample size of tree topologies from Bayesian phylogenetic analyses. Genome Biol. Evol. 8, 2319–2332. doi: 10.1093/gbe/evw171
Lee, Y. W., Fishman, L., Kelly, J. K., and Willis, J. H. (2016). Fitness variation is generated by a segregating inversion in yellow monkeyflower (Mimulus guttatus). Genetics 202, 1473–1484. doi: 10.1534/genetics.115.183566
Li, J., Zhou, S., Zhao, Z., and Zhang, J. (2015). The Qingzang movement: the major uplift of the Qinghai-Tibetan Plateau. Sci. Chin. Earth Sci. 58, 2113–2122. doi: 10.1007/s11430-015-5124-4
Li, X.-W., and Li, J. (1993). A preliminary floristic study on the seed plants from the region of Hengduan Mountains. Acta Bot. Yunnanica 15, 217–231.
Lu, L., Wang, H., Blackmore, S., Li, D. Z., and Dong, L. N. (2007). Pollen morphology of the tribe Rhinantheae (Orobanchaceae) and its systematic significances. Plant Syst. Evol. 268, 177–198. doi: 10.1007/s00606-007-0562-x
Maddison, W. P., and Maddison, D. R. (2017). Mesquite: a Modular System for Evolutionary Analysis. Version 3.2. Available at: http://mesquiteproject.wikispaces.com/home [accessed February 10, 2017].
Magallón, S., Gómez-Acevedo, S., Sánchez-Reyes, L. L., and Hernández-Hernández, T. (2015). A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453. doi: 10.1111/nph.13264
Matschiner, M., Musilová, Z., Barth, J. M. I., Starostová, Z., Salzburger, W., Steel, M., et al. (2017). Bayesian phylogenetic estimation of clade ages supports trans-Atlantic dispersal of cichlid fishes. Syst. Biol. 66, 3–22. doi: 10.1093/sysbio/syw076
McNeal, J. R., Bennett, J. R., Wolfe, A. D., and Mathews, S. (2013). Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 100, 971–983. doi: 10.3732/ajb.1200448
McNeill, J., Barrie, F. R., Buck, W. R., Demoulin, V., Greuter, W., Hawksworth, D. L., et al. (eds) (2012). “International Code of Nomenclature for algae, fungi, and plants (Melbourne Code),” in Proceedings of the Eighteenth International Botanical Congress Melbourne, (Ruggell: A.R.G. Gantner Verlag).
Mittermeier, R. A., Turner, W. R., Larsen, F. W., Brooks, T. M., and Gascon, C. (2011). “Global biodiversity conservation: the critical role of hotspots,” in Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas, eds F. E. Zachos and J. C. Habel (Berlin: Springe), 3–22.
Molnar, P. (2005). Mio-pliocene growth of the Tibetan Plateau and evolution of East Asian climate. Palaeontol. Electron. 8, 1–23.
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Nie, Z.-L., Funk, V., Sun, H., Deng, T., Meng, Y., and Wen, J. (2013). Molecular phylogeny of Anaphalis (Asteraceae, Gnaphalieae) with biogeographic implications in the Northern Hemisphere. J. Plant Res. 126, 17–32. doi: 10.1007/s10265-012-0506-6
Pan, Y.-Z., Fang, L.-Q., Hao, G., Cai, J., and Gong, X. (2009). Systematic positions of Lamiophlomis and Paraphlomis (Lamiaceae) based on nuclear and chloroplast sequences. J. Syst. Evol. 47, 535–542. doi: 10.1111/j.1759-6831.2009.00050.x
Pinto-Carrasco, D., Scheunert, A., Heubl, G., Rico, E., and Martínez-Ortega, M. M. (2017). Unravelling the phylogeny of the root-hemiparasitic genus Odontites (tribe Rhinantheae, Orobanchaceae): evidence for five main lineages. Taxon 66, 886–908. doi: 10.12705/664.6
Puzey, J. R., Willis, J. H., and Kelly, J. K. (2017). Population structure and local selection yield high genomic variation in Mimulus guttatus. Mol. Ecol. 26, 519–535. doi: 10.1111/mec.13922
Ree, R. H. (2005). Phylogeny and the evolution of floral diversity in Pedicularis (Orobanchaceae). Int. J. Plant Sci. 166, 595–613. doi: 10.1086/430191
Reid, E. M., and Chandler, M. E. J. (1926). Catalogue of Cainozoic Plants in the Department of Geology. London: British Museum. doi: 10.5962/bhl.title.110151
Renner, S. S. (2016). Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. J. Biogeogr. 43, 1479–1487. doi: 10.1111/jbi.12755
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508. doi: 10.1080/10635150290069913
Shimodaira, H., and Hasegawa, M. (1999). Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stevens, P. F. (2001). Angiosperm Phylogeny Website. Version 12. Available at: http://www.mobot.org/MOBOT/research/APweb/ [accessed December 26, 2015].
Stover, B. C., and Muller, K. F. (2010). TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi: 10.1186/1471-2105-11-7
Swofford, D. (2003). PAUP∗. Phylogenetic analysis using parsimony (∗and other methods). version 4. Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tao, D.-D. (1993). Phtheirospermum glandulosum – a new recorded species of scrophulariaceae from Yunnan, China. Acta Bot. Yunnanica 15:232.
Tao, D.-D. (1996). A taxonomic study on the genus Phtheirospermum (Scrophulariaceae). Acta Bot. Yunnanica 18, 301–307.
Tian, X., Luo, J., Wang, A., Mao, K., and Liu, J. (2011). On the origin of the woody buckwheat Fagopyrum tibeticum ( = Parapteropyrum tibeticum) in the Qinghai–Tibetan Plateau. Mol. Phylogenet. Evol. 61, 515–520. doi: 10.1016/j.ympev.2011.07.001
Tomilov, A. A., Tomilova, N. B., Abdallah, I., and Yoder, J. I. (2005). Localized hormone fluxes and early haustorium development in the hemiparasitic plant Triphysaria versicolor. Plant Physiol. 138, 1469–1480. doi: 10.1104/pp.104.057836
Tsoong, P.-C., and Yang, H.-P. (eds) (1979). Flora Reipublicae Popularis Sinacae, Vol. 67. Beijing: Science Press.
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Wang, H.-J., Li, W.-T., Liu, Y.-N., Yang, F.-S., and Wang, X.-Q. (2015). Range-wide multilocus phylogenetic analyses of Pedicularis sect. Cyathophora (Orobanchaceae): implications for species delimitation and speciation. Taxon 64, 959–974. doi: 10.12705/645.6
Wang, W., Liu, Y., Yu, S.-X., Gao, T.-G., and Chen, Z.-D. (2013). Gymnaconitum, a new genus of Ranunculaceae endemic to the Qinghai-Tibetan Plateau. Taxon 62, 713–722. doi: 10.12705/624.10
Wang, Y.-J., Von Raab-Straube, E., Susanna, A., and Liu, J.-Q. (2013). Shangwua (Compositae), a new genus from the Qinghai-Tibetan Plateau and Himalayas. Taxon 62, 984–996. doi: 10.12705/625.19
Wang, W.-T., and Wu, S.-G. (eds) (1993). Vascular Plants of the Hengduan Mountains (Part I). Beijing: Science Press.
Wang, W.-T., and Wu, S.-G. (eds) (1994). Vascular Plants of the Hengduan Mountains (Part II). Beijing: Science Press.
Wen, J., Zhang, J., Nie, Z.-L., Zhong, Y., and Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5:4. doi: 10.3389/fgene.2014.00004
Westwood, J. H., Yoder, J. I., Timko, M. P., and Depamphilis, C. W. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235. doi: 10.1016/j.tplants.2010.01.004
Wikström, N., Kainulainen, K., Razafimandimbison, S. G., Smedmark, J. E. E., and Bremer, B. (2015). A revised time tree of the Asterids: establishing a temporal framework for evolutionary studies of the coffee family (Rubiaceae). PLOS ONE 10:e0126690. doi: 10.1371/journal.pone.0126690
Wu, Z.-Y., Sun, H., Zhou, Z.-K., Peng, H., and Li, D.-Z. (2005). Origin and differentiation of endemism in the Flora of China. Acta Bot. Yunnanica 27, 577–604.
Xia, Z., Wang, Y.-Z., and Smith, J. F. (2009). Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.l.) inferred from five gene regions. Am. J. Bot. 96, 519–530. doi: 10.3732/ajb.0800195
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yamazaki, T. (1990). “Scrophulariaceae,” in Flora of Thailand, Vol. 5, Part 2, eds T. Smitinand and K. Larsen (Bangkok: Royal Forest Department), 139–238.
Yang, J., Zhang, Z., Shen, Z., Ou, X., Geng, Y., and Yang, M. (2016). Review of research on the vegetation and environment of dry-hot valleys in Yunnan. Biodivers. Sci. 24, 462–474. doi: 10.17520/biods.2015251
Yang, Z., Wafula, E. K., Honaas, L. A., Zhang, H., Das, M., Fernandez-Aparicio, M., et al. (2015). Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 32, 767–790. doi: 10.1093/molbev/msu343
Ying, T.-S., and Zhang, Z.-S. (1984). Endemism in the flora of China - studies on the endemic genera. Acta Phytotaxon. Sin. 22, 259–268. doi: 10.1371/journal.pone.0151941
Yu, W.-B., Huang, P.-H., Ree, R. H., Liu, M.-L., Li, D.-Z., and Wang, H. (2011). DNA barcoding of Pedicularis L. (Orobanchaceae): evaluating four universal DNA barcoding loci in a large and hemiparasitic genus. J. Syst. Evol. 49, 425–437. doi: 10.1111/j.1759-6831.2011.00154.x
Yu, W.-B., Liu, M.-L., Wang, H., Mill, R. R., Ree, R. H., Yang, J.-B., et al. (2015a). Towards a comprehensive phylogeny of the large temperate genus Pedicularis (Orobanchaceae), with an emphasis on species from the Himalaya-Hengduan Mountains. BMC Plant Biol. 15:176. doi: 10.1186/s12870-015-0547-9
Yu, W.-B., Wang, H., and Li, D. (2015b). Pseudobartsia glandulosa, a new combination to replace Pseudobartsia yunnanensis (Orobanchaceae). Phytotaxa 217, 197–199. doi: 10.11646/phytotaxa.217.2.10
Zeng, S., Zhou, T., Han, K., Yang, Y., Zhao, J., and Liu, Z.-L. (2017). The complete chloroplast genome sequences of six Rehmannia species. Genes 8:103. doi: 10.3390/genes8030103
Zhang, D.-C., Boufford, D. E., Ree, R. H., and Sun, H. (2009). The 29 degrees N latitudinal line: an important division in the Hengduan Mountains, a biodiversity hotspot in southwest China. Nord. J. Bot. 27, 405–412. doi: 10.1111/j.1756-1051.2008.00235.x
Zhang, J.-W., Boufford, D. E., and Sun, H. (2011a). Parasyncalathium J.W. Zhang, Boufford & H. Sun (Asteraceae, Cichorieae): a new genus endemic to the Himalaya-Hengduan Mountains. Taxon 60, 1678–1684.
Zhang, J. W., Nie, Z. L., Wen, J., and Sun, H. (2011b). Molecular phylogeny and biogeography of three closely related genera, Soroseris, Stebbinsia, and Syncalathium (Asteraceae, Cichorieae), endemic to the Tibetan Plateau, SW China. Taxon 60, 15–26.
Zhang, T.-C., Comes, H. P., and Sun, H. (2011c). Chloroplast phylogeography of Terminalia franchetii (Combretaceae) from the eastern Sino-Himalayan region and its correlation with historical river capture events. Mol. Phylogenet. Evol. 60, 1–12. doi: 10.1016/j.ympev.2011.04.009
Keywords: cryptic species, Hengduan Mountains, Orobanchaceae, Phtheirospermum, phylogenetic incongruence, Pterygiella
Citation: Yu W-B, Randle CP, Lu L, Wang H, Yang J-B, dePamphilis CW, Corlett RT and Li D-Z (2018) The Hemiparasitic Plant Phtheirospermum (Orobanchaceae) Is Polyphyletic and Contains Cryptic Species in the Hengduan Mountains of Southwest China. Front. Plant Sci. 9:142. doi: 10.3389/fpls.2018.00142
Received: 06 September 2017; Accepted: 25 January 2018;
Published: 09 February 2018.
Edited by:
Renchao Zhou, Sun Yat-sen University, ChinaReviewed by:
Andi Wolfe, The Ohio State University, United StatesCopyright © 2018 Yu, Randle, Lu, Wang, Yang, dePamphilis, Corlett and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Bin Yu, eXV3ZW5iaW5AeHRiZy5hYy5jbg== De-Zhu Li, ZHpsQG1haWwua2liLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.