
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 08 February 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00122
This article is part of the Research Topic Phenotyping at Plant and Cell Levels: The Quest for Tolerant Crop Development View all 39 articles
HIGHLIGHTS:
• Major environmental and genetic factors determining stress-related protein abundance are discussed.
• Major aspects of protein biological function including protein isoforms and PTMs, cellular localization and protein interactions are discussed.
• Functional diversity of protein isoforms and PTMs is discussed.
Abiotic stresses reveal profound impacts on plant proteomes including alterations in protein relative abundance, cellular localization, post-transcriptional and post-translational modifications (PTMs), protein interactions with other protein partners, and, finally, protein biological functions. The main aim of the present review is to discuss the major factors determining stress-related protein accumulation and their final biological functions. A dynamics of stress response including stress acclimation to altered ambient conditions and recovery after the stress treatment is discussed. The results of proteomic studies aimed at a comparison of stress response in plant genotypes differing in stress adaptability reveal constitutively enhanced levels of several stress-related proteins (protective proteins, chaperones, ROS scavenging- and detoxification-related enzymes) in the tolerant genotypes with respect to the susceptible ones. Tolerant genotypes can efficiently adjust energy metabolism to enhanced needs during stress acclimation. Stress tolerance vs. stress susceptibility are relative terms which can reflect different stress-coping strategies depending on the given stress treatment. The role of differential protein isoforms and PTMs with respect to their biological functions in different physiological constraints (cellular compartments and interacting partners) is discussed. The importance of protein functional studies following high-throughput proteome analyses is presented in a broader context of plant biology. In summary, the manuscript tries to provide an overview of the major factors which have to be considered when interpreting data from proteomic studies on stress-treated plants.
Stress can be defined as any environmental factor which adversely affects plant growth and development as well as crop quality and the final yield. Globally, the major abiotic stress factors—drought, high and low temperatures, and salinity—lead to the major reductions in crop yield. All plants, including crops, induce stress response leading either to stress escape, i.e., survival of stress treatment in metabolically inactive dormant stage such as seeds, or stress resistance, i.e., an active plant response to stress treatment. Stress resistance includes the strategies of stress avoidance, i.e., plant response aimed at maintenance of unstressed conditions at cellular and tissue levels, or stress tolerance, i.e., an active plant stress response to altered environment (Levitt, 1980).
The role of proteins in plant stress response is crucial since proteins are directly involved in shaping novel phenotype by adjustment of physiological traits to altered environment. The term “proteome” was introduced by Marc Wilkins in 1994 as protein complement of the genome, representing a whole of proteins in a given organism at a given time period. Unlike genome which is a static structure inherited from parents and defining plant genotype, changes in plant epigenome, transcriptome, proteome, and metabolome shape plant phenotype in response to both plant developmental and health stage as well as ambient environment. Proteins are directly involved in plant stress response both as structural proteins and also proteins involved in regulation of plant epigenome, transcriptome, and metabolome. Moreover, protein function is not dependent only on its molecular structure, but also on its cellular localization, post-translational modifications and interacting partners (Jorrín-Novo et al., 2009; Kosová et al., 2011).
Plant stress proteomics is a dynamic discipline aimed at the study of plant proteome and protein biological functions in plants exposed to stress. The number of publications on plant proteome under stress has risen geometrically in the last 15 years (Figure 1). In recent years, more than 150 original research and review papers are being published on this topic each year, and it is practically impossible to provide an extensive summary of the whole issue in a single review paper. A total number of publications for “plant proteome and stress” amounts 1,634 according to Web of Science; October 23rd, 2017. This burst of proteomic studies is enabled by the advancements in high-throughput instrumentation techniques aimed at separation of complex protein mixtures and identification of the individual protein species as well as by advancements in genomics leading to publication of reference genome sequences for important crop species in recent years (summarized in Kosová et al., 2015). Moreover, several review papers on plant stress proteomics were published in the recent years including summarizing reviews on plant abiotic stress proteomics (Kosová et al., 2011), proteomics of major abiotic stresses such as drought, salinity and extreme temperatures (Ahmad et al., 2016), proteomics of low temperature stress (Janmohammadi et al., 2015; Johnová et al., 2016), dehydration stress (Johnová et al., 2016), heavy metal stress (Ahsan et al., 2009; Hossain and Komatsu, 2013), plant biotic stresses (Sergeant and Renaut, 2010) with a special focus on fungal pathogens (Rampitsch and Bykova, 2012a) and Fusarium head blight disease (Yang et al., 2013a). Moreover, specialized reviews were published on plant root proteome response to abiotic stress (Ghosh and Xu, 2014), plant post-translational modifications under abiotic stress (Wu et al., 2016), plant phosphoproteomics (Rampitsch and Bykova, 2012b), redox proteomics (Rinalducci et al., 2008; Mock and Dietz, 2016), S-nitrosoproteomics (Romero-Puertas et al., 2013), subcellular proteomics under stress (Hossain et al., 2012), chloroplast proteome under abiotic stress (Ning and Wang, 2016), crop proteomics (Salekdeh and Komatsu, 2007), plant proteome responses to salinity (Zhang et al., 2012; Kosová et al., 2013a,b), stress responses of major crops (Tan et al., 2017) including rice (Agrawal et al., 2009), maize (Pechanova et al., 2013), wheat and barley (Komatsu et al., 2014; Kosová et al., 2014), soybean (Wang and Komatsu, 2016; Yin and Komatsu, 2017), common bean (Zargar et al., 2017), Solanaceae species (Ghatak et al., 2017), stress proteomics of crops grown in temperate climate (Kosová et al., 2015), and others.
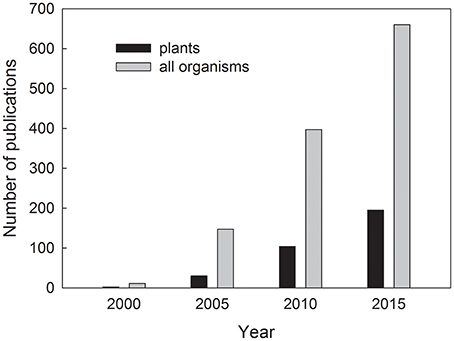
Figure 1. Number of publications found in Web of Science database as a reply to a query “plant proteome and stress” (black columns) for plants and “proteome and stress” (gray columns) for all organisms, respectively, for the years 2000, 2005, 2010, and 2015.
Protein biological functions reflect diversity and versatility of life. The aim of this review is to summarize major factors determining stress-related protein accumulation as well as their cellular localization and final biological function. The crucial stress-related factors determining protein accumulation include the dynamics of the given stress treatment, the joint effect of multiple stress factors as well as genotypic background underlying differential gene expression between stress-tolerant and stress-susceptible genotypes. The major factors determining protein biological functions include protein cellular localization, protein posttranscriptional and post-translational modifications (PTMs), and protein interactions with other protein and non-protein compounds. Finally, functional studies should complement high-throughput proteome analysis and can thus contribute to uncover protein role in plant stress response. All these factors represent the major aims of plant stress proteomics studies and are discussed in the manuscript (Figure 2).
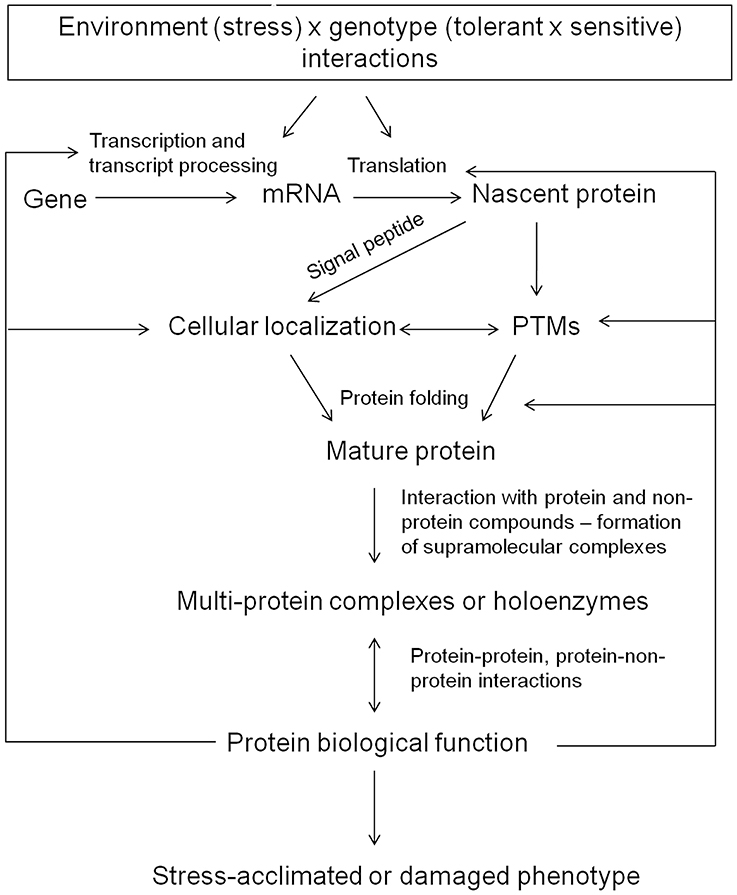
Figure 2. A schematic representation of the major factors determining protein accumulation and protein biological function.
Generally, stress factors represent environmental constraints which induce disturbances in cellular homeostasis including imbalances in plant water regime (reduced water uptake by roots; dehydration stress) as well as imbalances in cellular metabolic pathways, especially aerobic respiration and photosynthesis, leading to enhanced formation of reactive oxygen species (ROS; oxidative stress). However, there are also several effects of the individual stress factors which are specific for the different stress factors (Kosová et al., 2011).
Drought means a decreased soil water potential which causes reduced water uptake by roots. Plant response at molecular level lies in osmotic adjustment, i.e., a decrease in osmotic potential of cell cytoplasm due to an enhanced accumulation of several osmolytes and hydrophilic proteins (e.g., LEA proteins). In leaves, drought leads to stomatal closure associated with reduced CO2 uptake resulting in imbalances between photosynthetic electron transport processes and carbon assimilation. As a consequence, cellular dehydration is also associated with enhanced ROS formation resulting in an induction of several ROS scavenging enzymes such as several thioredoxin (Trx) isoforms in drought-treated sugar beet (Hajheidari et al., 2007). At proteome level, drought-induced imbalances in cellular metabolism including photosynthesis result in alterations of several photosynthesis-related proteins (RubisCO large subunit, FBP aldolase) leading to an establishment of new homeostasis (Perlikowski et al., 2014).
At molecular level, heat as enhanced temperature causes enhanced kinetics of biomolecules leading to an enhanced risk of protein misfolding. Thus, plant response includes an induction of several heat-shock transcription factors (HSFs) and downstream heat-shock proteins (HSPs). Moreover, heat causes enhanced water evaporation from soil surface as well as enhanced leaf transpiration thus usually resulting in water deficit under field conditions. Heat thus also causes dehydration stress and oxidative stress (Kosová et al., 2015).
In contrast, low temperatures (cold and frost) cause decreased kinetics of biomolecules leading to reduced cell membrane fluidity and a decreased rate of enzymatic reactions. Frost leads to formation of ice crystals in soil thus leading to reduced water uptake by roots resulting in cellular dehydration. As a response, plant induces an accumulation of several osmolytes and hydrophilic proteins such as dehydrins. Temperature stress (both heat and cold) also leads to an imbalance between photosynthetic electrontransport processes and carbon assimilation processes resulting in enhanced photoinhibition and thermal energy dissipation (Kosová et al., 2015).
Salinity stress represents enhanced levels of salt ions in soil water solution. As a consequence of enhanced ion levels, decreased soil water potential reveals a so-called osmotic effect on plant cells which leads to an accumulation of several osmolytes in the process of osmotic adjustment. In addition, enhanced ions in plant water solution induce a so-called ionic effect which progresses with stress duration and leads to penetration of toxic ions such as Na+ into plant cells. Plant response lies either in active ion exclusion or intracellular compartmentation into vacuoles leading to tissue tolerance. Both processes require energy and are thus associated with enhanced levels of several ATP-dependent ion transporters such as Na+/H+-ATPases, V-ATPases, and inorganic pyrophosphatases (PPi ases). Osmotic effect is rapid and common to all dehydration stresses while ionic effect is time-progressive and specific to salinity stress (reviewed in Munns, 2002; Munns and Tester, 2008).
Flooding stress leads to anaerobiosis in plant roots thus inducing fermentation processes. Fermentation leads to an enhanced accumulation of organic acids resulting in acidic pH of cell cytoplasm which adversely affects activity of several cellular enzymes. Recently, Komatsu and her colleagues published several proteomic studies on soybean root tip response to flooding including studies with a focus on several cellular organelles (nucleus, nucleolus, mitochondria, endoplasmic reticulum, plasma membrane, cell wall). For example, alterations in U3 snoRNP ribonucleoprotein and NOP1/NOP56 complex indicating changes in ribosome biosynthesis and thus novel protein biosynthesis were found in nuclear proteome of soybean root tips at initial phases of flooding (Yin and Komatsu, 2015, 2016).
Heavy metal stress also reveals toxic ionic effects due to metal ion penetration into the cells resulting in either ion exclusion or vacuolar compartmentation as utilized in plant hyperaccumulators for phytoremediation (Ahsan et al., 2009; Hossain and Komatsu, 2013).
Stress treatment as well as stress response are dynamic processes leading to an establishment of novel cellular homeostasis. According to Levitt (1980) and further researchers (see reviewed in Kosová et al., 2011), the following phases of plant stress response can be distinguished: an alarm phase, an acclimation phase, a resistance phase, an exhaustion phase, and a recovery phase or death. Each phase of plant stress response can be characterized by its unique proteome composition (Table 1). Therefore, it is important to precisely define environmental conditions and stress treatments applied in proteomic experiments. Moreover, regarding phenotype-based differential plant stress tolerance, it is becoming obvious that the same ambient stress conditions can cause differential stress response at cellular level as indicated by different plant-related physiological and biochemical characteristics. For example, different plants grown under the same conditions can significantly differ in physiological and biochemical characteristics such as tissue relative water content (RWC) or tissue Na+ levels under drought and salinity stresses, respectively (Flowers and Colmer, 2008; Tardieu, 2012). Stress adaptability therefore results from a complex combination of traits that influence ratio between supply and demand in a given environmental scenario (Passioura, 2012; Vadez et al., 2013). In agriculture, the major attention is paid to the effects of environmental stress on crop yield which impact, however, depends on the type of crop and growing purpose. An example of yield-related characteristics reflecting the effects of environmental stress can be drought-tolerance indices in wheat (Ali and El-Sadek, 2016). Proteomic studies have revealed that relatively low differences in ambient environments can result in profound alterations at proteome level if stress timing is well estimated. For example, a comparison of spring barley crowns grown under two levels of soil water capacity (30 and 35% SWC) led to differential response in proteins related to energy metabolism and protein degradation indicating an active plant acclimation upon 35% SWC while profound imbalances in energy metabolism and enhanced protein damage upon 30% SWC (Vítámvás et al., 2015). Therefore, in proteomic experiments, it is important (however, very often not mentioned at all) to provide relevant characterization of plant physiological and biochemical status not only by a description of ambient conditions, but also by providing plant-related characteristics (e.g., plant growth-related characteristics, phenological stage, plant and soil water regime-related characteristics, cellular levels of salt ions, mineral nutrients, heavy metals, etc.) reflecting stress impact at cellular level and different plant strategies to cope with stress (reviewed in Passioura, 2006; Tardieu and Tuberosa, 2010; Tardieu et al., 2011; Poorter et al., 2012a,b; Claeys et al., 2014; Blum, 2016). This creates a significant problem in detailed comparisons of published data or using proteomic meta-data analysis, especially in research comparing adaptable vs. susceptible genotypes.
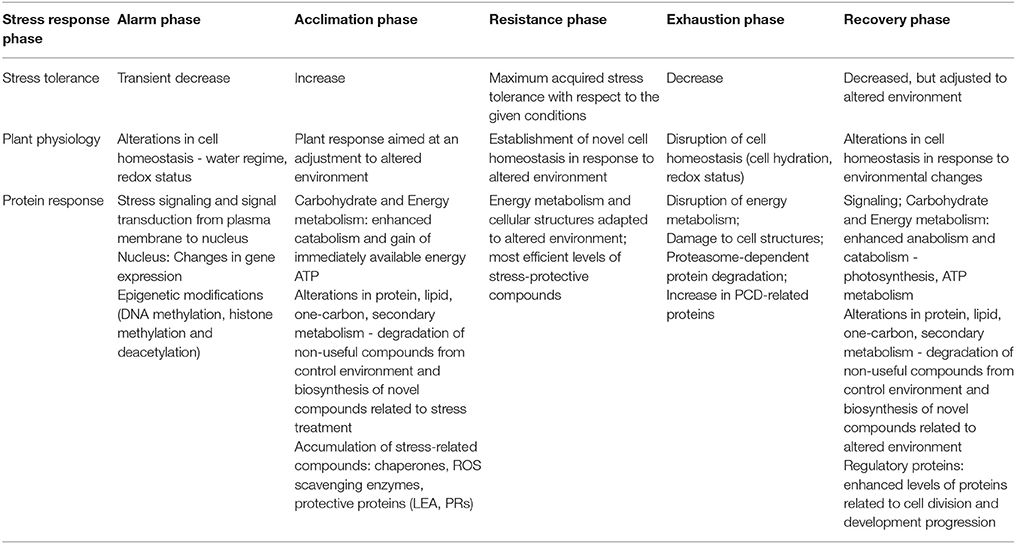
Table 1. Brief characteristics of the individual phases of plant stress response (alarm phase, acclimation phase, resistance phase, exhaustion phase, recovery phase) with respect to plant stress tolerance, physiological response, and proteome response.
In the following text, brief characteristics of the individual phases of plant stress response at proteome level are given. An alarm phase can be characterized by initial stress factor treatment leading to disturbances in cellular homeostasis and thus inducing stress signaling resulting in alterations in gene expression. Changes in ambient conditions are sensed by receptors at plasma membrane inducing signaling pathways transferring stress signal from plasma membrane to nucleus and leading to the changes in gene expression. Unlike transcriptomic studies, only few proteomic studies focused on the investigation of the initial phases of plant stress response since relatively few changes were found at the proteome level at that phase. For example, Arabidopsis chloroplast proteome was investigated 1 day after the beginning of cold treatment (5°C), but only minimum alterations were found (Goulas et al., 2006). A decreased abundance of Ca2+ sensing receptor was found in maize chloroplasts after 4 h of salt treatment (Zörb et al., 2009). In contrast, alterations in several photosynthesis-related proteins (OEE1, PSB28, PSI type III chlorophyll a/b binding protein, Trx m-type) were found in cold-treated forage grass Festuca pratensis at 26 h of cold treatment (Kosmala et al., 2009). The susceptibility/adaptability of individual cultivars is connected to alarm phase, as very susceptible cultivars respond extremely on each environmental stimulus. The tolerant cultivars respond more slowly and can show later signs of stress (delayed stress onset; Lawlor, 2013). This different responsiveness strategy can likely be universal across all other phases.
An acclimation phase can be characterized by profound changes in protein metabolism including both protein biosynthesis and protein degradation resulting in enhancement of plant stress tolerance. During stress acclimation phase, alterations in gene expression lead to profound changes in several metabolic pathways (protein metabolism, carbohydrate metabolism, lipid metabolism, energy metabolism, secondary metabolism, phytohormone metabolism, etc.,) as well as an accumulation of stress-related proteins (proteins with chaperone and protective functions such as LEA, dehydrins; PR proteins, ROS scavenging and detoxification enzymes). Metabolic alterations are aimed at a synthesis of novel stress-related compounds while a degradation of several other ones. Alterations in energy metabolism are aimed at an enhancement of energy production in an immediately available form such as ATP since processes associated with an enhanced biosynthesis of stress-related compounds reveal enhanced energy requirements. An accumulation of several protective proteins, detoxification-related and ROS scavenging enzymes mitigates harmful effects of stress on cellular microenvironment such as increased dehydration and oxidative stress as well as increased amounts of toxic byproducts of cellular metabolism as a consequence of imbalances in cellular homeostasis (Kosová et al., 2011, 2015).
Resistance phase is characterized by achieving a maximum acquired stress tolerance under given ambient conditions. At molecular level, resistance phase can be characterized by efficient adjustment of cellular metabolism to altered ambient conditions as well as by sufficient levels of stress-protective proteins minimizing harmful effects of stress on cellular structures and metabolisms (Kosová et al., 2011).
Exhaustion phase is characterized by a decline in acquired stress response and can be achieved when stress treatment lasts too long or is too severe leading to exploiting plant resources necessary for maintenance of enhanced stress tolerance. Exhaustion phase can be characterized by rapid imbalances in cellular homeostasis, imbalances in cellular metabolism, transition from more efficient aerobic processes to less efficient anaerobic processes, and emerging signs of cellular damage including programmed cell death (PCD)-related changes (Kosová et al., 2011; Maršálová et al., 2016).
Recovery phase follows cessation of ambient stress treatment and can be characterized by an establishment of novel cellular homeostasis under non-stress conditions. Recovery phase is often overlooked in plant stress proteomics studies; however, it is equally important as stress treatment since efficient recovery affects further plant growth and development. During recovery phase, several stress-related compounds are degraded and novel compounds are synthesized, and these processes are associated with enhanced energy requirements. Enhanced abundances of 8 out of 12 glycolysis enzymes as well as photosynthesis-related proteins were found in drought-tolerant Australian spring wheat Excalibur at 24 h after rewatering indicating efficient restoration of energy metabolism after stress (Ford et al., 2011). In some cases of recovery, physiological parameters at whole plant level indicate return to control conditions while proteomic analysis reveals profound alterations in plant cellular environment under recovery with respect to control conditions prior to stress treatment as it was observed in chloroplast proteome of tomato plants subjected to 19 days of water withholding (drought stress) followed by 6 days of watering (recovery) (Tamburino et al., 2017). Decreased levels of proteins associated with photosynthesis including photosystem components (plastocyanin, chlorophyll a-b binding protein 4), Calvin cycle enzymes (PGK, PRK) and ATP synthase complex subunits (ATP β,γ) after 6 days of recovery indicate an ongoing decline in cellular energy metabolism following severe stress and a long way to reestablishment of cellular processes (Tamburino et al., 2017).
Under field conditions, plants are usually exposed to combinations of multiple stress factors whose joint impact is not merely additive, i.e., it does not equal to the sum of the effects of the individual stress factors when acting separately (Mittler, 2006; Suzuki et al., 2014). Moreover, several stress factors also reveal secondary stress effects; for example, heat often leads to a dehydration stress since high temperatures enhance water evaporation and leaf transpiration. However, several different stress factors reveal some common features at cellular level. Under aerobic conditions, stress treatment leads to disturbances in cellular homeostasis resulting in enhanced ROS production (oxidative stress). Several abiotic stress factors also lead to reduced plant water uptake thus causing cell dehydration (dehydration stress). Drought can be accompanied by a strong oxidative stress due to photoinhibition of the components of photosynthetic electron transport chain. Flooding is associated with hypoxia (anoxia) leading to a shift in cellular metabolism from aerobic to anaerobic processes resulting in cytoplasm acidification due to an accumulation of organic acids as products of fermentation (Oh and Komatsu, 2015). Enhanced aluminum in soil is associated with soil acidity leading to changes in the soil mineral availability which results in an increased level of glutathione and other detoxification agents in rice roots (Yang et al., 2007).
At proteome level, the following stress factors comparisons either in different treatments or stress combinations within a single treatment were studied: a comparison of drought and salinity on salt-susceptible wheat cultivar Jinan 177 and a salt-tolerant T. aestivum × Thinopyrum ponticum hybrid Shanrong 3 (Peng et al., 2009), combined drought and heat treatments in Chinese arid shrub Carissa spinarum (Zhang et al., 2010), parallel and combined drought and heat treatments in a Syrian barley landrace Arta and an Australian cultivar Keel (Rollins et al., 2013), freezing combined with either drought or waterlogging in winter wheat (Li et al., 2014), a comparison of contrasting water stress treatments of drought and flooding on soybean seedlings (Oh and Komatsu, 2015).
Comparative proteome analysis of drought and salinity revealed larger changes in proteome composition induced by salinity than drought due to an ionic effect of salt stress (Peng et al., 2009). Similarly, heat caused larger alterations in proteome composition than drought in barley as well as in arid shrub Carissa spinarum due to the adverse effects of heat on plant photosynthetic apparatus (Zhang et al., 2010). In susceptible barley, heat led to a decrease in several photosynthesis-related proteins as well as an increase in thermostable RubisCO activase B isoform (Rollins et al., 2013). In relatively tolerant Carissa spinarum, heat induced several proteins involved in protection of photosynthetic apparatus such as thermostable RubisCO activase isoform, several sHSP and chaperonin proteins (Zhang et al., 2010). A comparison of the effects of two contrasting types of water stress—drought and waterlogging—on soybean root proteome revealed some similarities as well as differences in soybean root proteome response to these contrasting stresses (Oh and Komatsu, 2015). Both stresses induced an accumulation of glycolysis enzymes indicating an activation of carbohydrate catabolism and energy release; in addition, waterlogging induced fermentation enzymes and proteins related to cell wall modification while drought induced proteins involved in redox metabolism and alleviation of oxidative stress. Differential effects of these stresses can be found in S-adenosylmethionine synthetase (SAMS). In soybean roots, three isoforms of SAMS were found revealing differential dynamics in response to drought and waterlogging, respectively (Oh and Komatsu, 2015).
Moreover, a pretreatment with mild stress factor can help a plant to mitigate adverse effects of another stress. The corresponding effects deserve to be studied at proteome level. For example, mild drought or waterlogging applied prior to freezing can enhance freezing tolerance in winter wheat due to a positive effect of the mild stress on activation of cellular redox metabolism resulting in lower damage of photosynthetic apparatus under the subsequent freezing (Li et al., 2014). Similarly, a positive effect of an inoculation of barley plants with a mutualistic symbiont Piriformospora indica on mitigation of drought stress was found (Ghabooli et al., 2013). At proteome level, P. indica inoculation led to an increase in proteins involved in energy metabolism including photosynthesis as well as proteins involved in redox metabolism thus increasing barley tolerance to water stress (Ghabooli et al., 2013). Recently, a positive effect of arbuscular mycorrhizal fungus Glomus masseae on drought-treated wheat roots was reported leading to modulation of proteins related to sugar metabolism and cell wall rearrangement thus resulting in reduction of osmotic stress and maintenance of cellular integrity (Bernardo et al., 2017).
Summary: Abiotic stresses induce an active plant stress response at proteome level which is aimed at either maintenance of optimum cell environment (stress avoidance) or at an active acclimation to altered stress conditions (stress tolerance). Plant stress response is a dynamic process where several phases (alarm, acclimation, resistance, exhaustion, and recovery phase) with specific proteome composition can be distinguished. Relatively small differences in stress treatments can result in significant differences in plant proteome indicating either stress acclimation or stress damage. Combined stress treatments as occurring in the field reveal unique impacts on plant proteome which cannot be described as a mere sum of the distinct stress treatments.
Several proteomic studies focused on a comparison of proteome response to a given stress between a tolerant and a susceptible plant genotype or related species (Table 2, Supplementary Table S1A). Unfortunately, some authors often use genotypes with not well-documented differences at physiological level or they use only one physiological trait to describe it, making the result less usable in further studies. Simply, lower water use or higher biomass upon stress do not mean superior stress tolerance because of interactions between genotype × environment × management (Vadez, 2014; Vadez et al., 2014). For example, a comparison of drought-treated inbred vs. hybrid maize plants revealed an adverse effect of higher plant biomass and larger leaf area on a balance between photosynthesis and leaf transpiration rates leading to lower water use efficiency (WUE) and drought tolerance in the hybrid with respect to its parents (Holá et al., 2017). Comparison of changes in proteome composition under stress with respect to control conditions reveals higher numbers of differentially abundant proteins between control and stress conditions in stress-susceptible materials than in stress-tolerant ones. This indicates more profound disturbances in cellular homeostasis in the susceptible plants. For example, a comparison of differentially abundant proteins in salt-treated Arabidopsis thaliana vs. salt-treated Thellungiella halophila with respect to control conditions revealed 88 differentially abundant proteins in glycophyte A. thaliana while only 37 differentially abundant proteins in halophyte T. halophila under the same conditions (Pang et al., 2010). The possible reason for these differences between tolerant and susceptible species may lie in the fact that the given stress conditions do not cause such large disturbances in cellular homeostasis in tolerant halophytes than in susceptible glycophytes. The explanation of these differences may also lie in the fact that proteomic studies in stress-tolerant plants often identified constitutively enhanced abundance of several protective proteins, e.g., LEA proteins such as dehydrins, germin-like proteins GLP, universal stress protein USP (Vítámvás et al., 2010; Benešová et al., 2012; Kosová et al., 2013c) as well as several detoxification enzymes which efficiently remove toxic byproducts of cellular metabolism (Askari et al., 2006; Maršálová et al., 2016). Enhanced levels of ROS scavenging enzymes APX, Cu/Zn-SOD, and Mn-SOD were found in drought-tolerant maize and sunflower genotypes with respect to the susceptible ones, respectively (Benešová et al., 2012; Ghaffari et al., 2013), in salt-tolerant T. aestivum × Lophopyrum elongatum amphiploid with respect to its parental common wheat cv. Chinese Spring (Jacoby et al., 2013) as well as in differentially cold-tolerant winter wheats (Xu et al., 2013). In contrast, enhanced levels of APX and MDAR were found in salt-susceptible barley roots with respect to salt-tolerant ones since salinity treatment caused larger disturbances in redox homeostasis (both ionic and toxic effects of salt) in the susceptible genotype than in the tolerant one (Witzel et al., 2009). Differential levels of lactoylglutathione lyase (glyoxalase) involved in detoxification of methylglyoxal as a byproduct during glycolysis and threonine biosynthesis were found in barley root (Witzel et al., 2009) and barley crown (Maršálová et al., 2016) under salinity and in oilseed rape leaves under drought (Urban et al., 2017). Enhanced levels of cyanate hydratase involved in degradation of cyanate, a toxic byproduct of ethylene biosynthesis, were found in a halophytic plant Suaeda aegyptiaca (Askari et al., 2006). Due to constitutively increased levels of several stress-protective proteins in tolerant genotypes, a higher stress-inducible increase of some stress-responsive proteins such as HSP70 and thioredoxin h (Trx h) was found in susceptible genotypes with respect to tolerant ones under stress treatment (Manaa et al., 2011). Correspondingly, enhanced levels of stress-responsive transcription factors (TFs) such as NACα (Maršálová et al., 2016), bHLH (Vincent et al., 2007), and MYB-like (Wendelboe-Nelson and Morris, 2012) were found in tolerant plants with respect to the susceptible ones. Stress treatment thus causes larger disturbances in cellular homeostasis in susceptible plants than in tolerant (or stress-adapted) ones. Studies of physiological characteristics have shown that tolerant plants usually reveal also lower impacts of environmental stress factors on plant-related characteristics such as RWC (drought) or tissue Na+ content (salinity) with respect to the susceptible plants when exposed to the same ambient environment (Flowers and Colmer, 2008, 2015).

Table 2. Examples of major proteins (protein groups) revealing differential abundance between stress-susceptible and stress-tolerant plant genotypes. More details on proteomic experiments dealing with stress response in differentially stress-responsive genotypes are given in Supplementary Table S1A.
The differences in the impact of stress on tolerant and susceptible plants are also reflected in cellular metabolism. Larger disturbances in cellular homeostasis represent a higher risk of oxidative stress (ROS formation) thus adversely affecting aerobic metabolism, especially photosynthesis and ATP biosynthesis. A decrease in proteins of photosynthetic electron transport chain such as components of oxygen evolving complex (OEC) and photosystem II reaction center (RC PSII) as well as in RubisCO subunits and Calvin cycle enzymes was found in susceptible plants (Caruso et al., 2008; Gharechahi et al., 2014; Holá et al., 2017). In contrast, tolerant plants were able to adjust to altered environment and thus they revealed increased levels of crucial photosynthetic proteins such as OEC components of PSII as well as RubisCO subunits and RubisCO-related proteins such as RubisCO activase and carbonic anhydrase (Askari et al., 2006; Ge et al., 2012; Guo et al., 2012; Budak et al., 2013; Kausar et al., 2013; Witzel et al., 2014). Following RubisCO carboxylation activity, enhanced abundance of Calvin cycle enzymes was also found in tolerant plants. Similarly, an enhanced level of mitochondrial malate dehydrogenase (mtMDH) catalyzing dehydrogenation of malate to oxaloacetate was found in highly frost-tolerant winter wheat Mironovskaya 808 with respect to less tolerant winter wheat Bezostaya 1 under long-term cold (Vítámvás et al., 2012) as well as in salt-tolerant T. aestivum × Lophopyrum elongatum amphiploid with respect to its parental common wheat cv. Chinese Spring (Jacoby et al., 2013) indicating more efficient mitochondrial respiration in the more tolerant genotypes. Due to efficient elimination of oxidative stress and enhancement of aerobic metabolism, tolerant genotypes can cover enhanced demands on energy during active stress acclimation process and/or likely use this in more efficient recovery. These differences are reflected in enhanced levels of proteins involved in ATP biosynthesis, especially the components of mitochondrial or plastid ATP synthases, in tolerant plants (Cheng et al., 2016). In contrast, in susceptible genotypes, an enhanced risk of oxidative stress leads to disturbances in energy metabolism indicated by opposite patterns of different isoforms of proteins involved in energy metabolism such as glycolytic enzymes (Rasoulnia et al., 2011; Vítámvás et al., 2015). Moreover, an enhanced risk of oxidative damage can lead to a shift from aerobic metabolism to anaerobic fermentation processes which are, however, less efficient in ATP production.
Other genotypic differences in stress-treated plants include differences in proteins involved in several metabolic pathways such as protein metabolism including both protein biosynthesis and proteasomal degradation (Maršálová et al., 2016), proteins involved in S-adenosylmethionine (SAM) metabolism (Faghani et al., 2015) as well as proteins involved in hormone biosynthesis such as gibberellin (GA) (Wang et al., 2008) and jasmonic acid (JA) (Pang et al., 2010).
Moreover, differences between tolerant and susceptible plants subjected to stress treatments were also observed in regulatory proteins. Tolerant plants which were able to acclimate to stress conditions reveal enhanced levels of those regulatory proteins which contribute to maintenance of fully acclimated state or are involved in stimulation of cell division indicating restoration of plant growth and development. Stress treatments lead to alterations in the levels of several small glycine-rich proteins (sGRPs) which are involved in regulation of transcription and transcript processing including alternative splicing of mRNAs leading to synthesis of different protein isoforms under stress with respect to control. An increased abundance of several sGRPs was found in plants exposed to several abiotic stress factors such as in transgenic Arabidopsis thaliana, poplar, and Nicotiana tabacum under salt and flooding stresses, respectively (Kwak et al., 2005; Durand et al., 2010; Wang et al., 2012). Another example of regulatory proteins revealing genotypic differences represents a pentatricopeptide repeat protein (PPR) in two drought- and salinity-treated grapevines (Vincent et al., 2007); PPR proteins are known to localize to organellar genomes where they are involved in posttranscriptional regulation of gene expression due to their RNA binding activities (Barkan and Small, 2014).
A lower damage of plant tissues and lower energy costs on stress acclimation in tolerant genotypes in comparison to susceptible ones could also result in a relatively more positive effect on novel protein biosynthesis, plant growth, and development in tolerant genotypes compared to susceptible ones when subjected to stress. An increase in eIF3 and mitochondrial EF-TuM was found in drought-tolerant maize genotype CE704 subjected to 6 days of dehydration while a decrease in eEF1D was found in drought-susceptible maize genotype 2023 under the same conditions (Benešová et al., 2012). Differences in eIF5A3 level between salt-susceptible common wheat cultivar Jinan 177 and salt-tolerant T. aestivum × Thinopyrum ponticum introgression hybrid Shanrong 3 indicate a higher antisenescence ability of the hybrid with respect to its parental cultivar under salinity (Wang et al., 2008). In contrast, susceptible plants can reveal enhanced levels of apoptosis-related proteins indicating regulated degradation of damaged cells. Translationally controlled tumor protein (TCTP) is known to be involved in negative regulation of p53-induced apoptosis due to its binding to p53 leading to a downregulation of its activity (Chen et al., 2013). Genotypic differences in TCTP levels were found in differentially tolerant drought-treated barley cultivars (Mostek et al., 2015) as well as in salt-susceptible H. vulgare and salt-tolerant H. marinum under high salinity of 300 mM NaCl (Maršálová et al., 2016). Environmental stresses also profoundly affect plant development leading to regulation of timing of vegetative-to-reproductive phase transition. In temperate climates where long-term periods of low temperatures occur during winter several plants had evolved a requirement of a long-term period of low temperatures prior to their transition to flowering, a phenomenon known as vernalization (Sung and Amasino, 2005; Kosová et al., 2008). At proteome level, differential response to 21 day-cold treatment was found between a winter and a spring cultivar of common wheat. Results showed an accumulation of several regulatory proteins such as germin E and lectin VER2, involved in maintenance of vegetative phase in the winter wheat while an induction of proteins involved in transition to flowering in the spring wheat including eIF5A-2, glycine-rich RNA-binding protein, and adenine phosphoribosyltransferase involved in cytokinin biosynthesis (Rinalducci et al., 2011a,b; Kosová et al., 2013d).
Last but not least, genotypic differences were also found in structural proteins including storage proteins such as legumin-like protein in cold-treated winter wheat crowns (Vítámvás et al., 2012), membrane transport-related proteins such as annexin (Mostek et al., 2015), porin (Alvarez et al., 2014), V-ATPase (Mostek et al., 2015), and cell wall modification-related proteins such as β-expansin (Alvarez et al., 2014), and lignin biosynthetic enzymes caffeoyl-coenzyme A O-methyltransferase CCOMT (Sugimoto and Takeda, 2009) and caffeic acid O-methyltransferase COMT (Riccardi et al., 2004; Vincent et al., 2005).
However, it has to be kept in mind that “stress tolerance” vs. “stress susceptibility” are relative terms depending on the given environmental conditions and the dynamics of stress treatment. Moreover, an eco-geographic origin of the individual genotype or cultivar plays a significant role in the aspect of its phenotypic response. In fact, stress-tolerant and stress-susceptible genotypes can differ in biological strategies applied when to cope with stress and can even show opposite trends in mild vs. severe stress or for different developmental stages. Thus, any stress treatment should be based on previous detailed review of target population of environment (Tardieu, 2012) and application supported by hypothesis-driven occurrence (Vadez et al., 2013). For example, in winter oilseed rapes, the different water-related strategy (water-savers, water-spenders) influenced significantly the rate of water uptake from the pot, and, consequently, the differential protein abundances (Urban et al., 2017). In the water-savers group, proteins related to nitrogen assimilation, ATP metabolism and redox homeostasis increased under stress, while in the water-spenders category, carbohydrate/energy, photosynthesis, stress related and rRNA processing proteins were increased upon stress. However, both groups contain drought-adaptable cultivars. Thus, if it has to be decided which cultivar or genotype is “tolerant,” the rate of stress onset, duration of stress, and actual plant developmental stage have to be specified.
Examples of proteins revealing differential abundances between stress-tolerant and stress-susceptible plants is given in Table 2. More detailed information on the proteomic experiments carried out in genotypes with differential stress response is provided in Supplementary Table S1A.
Summary: Different plant genotypes or crop cultivars within a given plant species reveal differential adaptability to environment including stress treatments which is reflected by their differential ability to survive and recover following stress. Differential adaptability can be described by the terms of “susceptibility” vs. “tolerance,” respectively, which are, however, relative terms depending on the given genotypes and stress treatments. Proteomics approaches could help the researchers to uncover differences in plant stress response at molecular level prior than they can be detected at the whole plant level by determination of physiological characteristics. Recent studies revealed that at proteome level, tolerant genotypes reveal constitutively enhanced levels of several detoxification-related and stress-protective proteins thus it can be hypothesized that tolerant plants can better cope with stress-induced imbalances in cellular homeostasis than susceptible ones. As a consequence, stress treatment leads to lower level of oxidative stress in tolerant plants with respect to susceptible ones thus enabling the tolerant plants to efficiently adjust energy metabolism to enhanced demands of stress acclimation process. As a result, tolerant plants are able to efficiently acclimate to stress conditions and to restore their growth and development while susceptible plants reveal signs of cellular damage caused by adverse effects of stress.
The major factors determining protein biological function include cellular localization, posttranscriptional and post-translational modifications, and protein interactions.
Cellular fractionation represents an efficient means how to reduce complexity of cellular proteome. Separation techniques working with total proteome extracts usually do not ensure clear separation of all individual proteins present in the mixture leading to the situation that relatively low-abundant organellar proteins are overlaid by relatively high-abundant cytosolic proteins. Plant cell is composed of cell wall (extracellular matrix, ECM) as a part of apoplast and cytosol as a part of symplast connected with other cells via plasmodesmata. Apoplast and symplast are separated by plasma membrane representing a dynamic interface between these environments. Symplast contains organelles with double membrane envelopes such as nucleus, plastids and mitochondria, and vesicular compartments surrounded only by a single membrane such as components of secretory pathway including endoplasmic reticulum (ER), Golgi complex and trans-Golgi network (TGN), vacuoles, peroxisomes, glyoxisomes, and other vesicular structures. Each organelle plays a specific role in plant stress acclimation: nucleus is a site of stress signal transformation to gene expression, and chloroplasts and mitochondria are sites of aerobic metabolism, which are crucial for energy supply during stress acclimation. Classical approaches to cellular fractionation via differential centrifugation result in separation of four major cell fractions - nuclear, plastidial, mitochondrial, and microsomal. However, proteomic studies focused on organellar response to stress are still relatively scarce when compared to total proteome studies. A summary of the major proteins revealing alterations in their relative abundance under stress in the individual compartments is provided in Figure 3 (Table 3). More detailed information on proteomic experiments focused on subcellular proteomics in stress-treated plants is provided in Supplementary Table S1B and in specialized reviews on organellar proteomics under stress (Hossain et al., 2012).
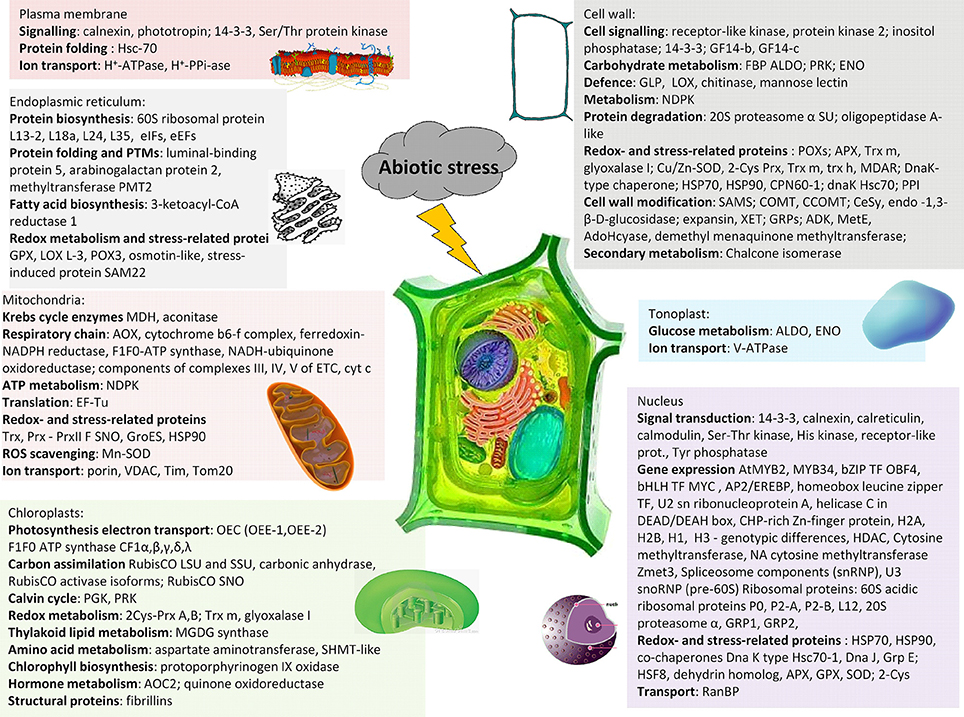
Figure 3. A schematic summary of the major stress-responsive proteins in the individual plant cell compartments. 2-Cys Prx, 2-cysteine peroxiredoxin; AOC, allene oxide cyclase; APX, ascorbate peroxidase; AOX, alternative oxidase; ENO, enolase; FBP ALDO, fructose-1,6-bisphosphate aldolase; GPX, glutathione peroxidase; GRP, glycine-rich protein; HSF, heat-shock (transcription) factor; HSP, heat-shock protein; LOX, lipoxygenase; MDAR, monodehydroascorbate reductase; MDH, malate dehydrogenase; OEC, oxygen-evolving center; PGK, phosphoglycerokinase; POX, peroxidase; PRK, phosphoribulokinase; SHMT, serine hydroxymethyltransferase; SNO, S-nitrosylation; SOD, superoxide dismutase; TF, transcription factor; Trx, thioredoxin.

Table 3. Examples of major proteins (protein groups) revealing differential abundance in cellular organelles (cell wall, plasma membrane, nucleus, chloroplast, mitochondria, tonoplast, endoplasmic reticulum) under stress.
Cell wall, also known as extracellular matrix (ECM), represents not only apoplastic mechanical envelope of the cell, but also a dynamic structure actively involved in stress sensing and signaling. Plant stress proteomic studies dealing with ECM fraction were mainly focused on two contrasting stresses—dehydration (drought) and flooding. Extracellular matrix proteome in rice shoots under dehydration stress revealed alterations in proteins involved in stress signaling (nucleoside diphosphate kinase, NDPK, involved in γ-phosphate transfers and G-protein signaling), proteins involved in detoxification and ROS scavenging (APX, thioredoxin, glyoxalase I, chitinase), chaperones (DnaK, CPN60, and HSP20) as well as proteins involved in cell wall modification such as enzymes of phenylpropanoid biosynthetic pathway and methyltransferases involved in methylation of lignin components (Pandey et al., 2010). Moreover, several cytosol-related proteins were also identified in ECM including proteins involved in carbohydrate metabolism as pentose phosphate pathway (phosphoribulokinase, transketolase). They are proposed to be involved in biosynthesis of sugars as a part of osmotic adjustment under dehydration stress or to be involved in ROS scavenging due to production of NADPH (Pandey et al., 2010).
Alterations in proteome composition of soybean root cell walls exposed to flooding indicate a reduced cell wall lignification which is probably associated with decreased levels of Cu/Zn-superoxide dismutase (Cu/Zn-SOD), four germin-like proteins, lipoxygenases and glycoprotein precursors. The results indicate a suppression of lignification due to a downregulation of ROS and jasmonate biosynthesis under flooding (Komatsu et al., 2010). In contrast, enhanced levels of proteins involved in production of apoplastic ROS, namely H2O2, such as oxalate oxidase, germins, APX, Cu/Zn-SOD, Trx m were found in elongation zone of maize root cell walls (Zhu et al., 2007) as well as chickpea seedlings shoot cell walls (Bhushan et al., 2007) subjected to dehydration stress, respectively. Apoplastic ROS are involved in enhanced cell wall loosening necessary for elongation root growth as well as for cross-linking of monolignols and other phenolics during cell wall lignification process. Cell wall lignification is associated with an enhanced abundance of β-D-glucosidases involved in release of monolignols essential for lignin biosynthesis (Bhushan et al., 2007).
Plasma membrane is the primary site of stress sensing and its transformation into signaling. Alterations in ambient conditions are sensed by receptors at plasma membrane; for example, temperature decrease leading to alterations in membrane fluidity results in conformational changes in plasma membrane-located two-component histidine-type kinases (Murata and Los, 1997; Suzuki et al., 2000); or alterations in Na+ levels induce signaling associated with an active Na+ efflux via SOS1/SOS2/SOS3 complex (Zhu, 2002). Recently, several proteomic studies dealt with plasma membrane fraction under stress. Nouri and Komatsu (2010) investigated proteomic response of soybean plasma membrane to osmotic stress using both gel-based (2-DE) and gel-free (LC-MS/MS) approaches. Three H+-ATPase isoforms involved in ion efflux and revealing an increase under hyperosmotic stress were identified. The study has also identified several proteins regulating H+-ATPase activity including calnexin, one protein phosphatase, phototropin, and three isoforms of protein kinases. Similarly, several isoforms of H+-ATPase and H+-pyrophosphatase (H+-PPi-ase) were found in plasma membranes in aleurone layers of germinating barley embryos (Hynek et al., 2009). A study on plasma membrane fraction isolated from flooding-treated soybean indicated a role of 14-3-3 proteins and Ser/Thr protein kinase in regulating activity of plasma membrane H+-ATPase and maintenance of ion homeostasis (Komatsu et al., 2009).
Nucleus represents the major organelle involved in plant phenotype remodeling in response to environmental stress since it is involved in stress signal transformation into changes in gene expression. The study on nuclear proteome in drought-treated chickpea revealed the major protein functional groups belonging to gene expression, signal transduction, chaperones, chromatin remodeling, ROS scavenging enzymes, proteins involved in nucleocytoplasmic transport, and other regulatory proteins (Pandey et al., 2008).
Proteins involved in signal transduction include 14-3-3 proteins as well as secondary messengers involved in Ca2+ signaling such as calreticulin and calnexin. Alterations were found in several signaling- and regulatory-related proteins such as receptor-activated protein kinase C1 (RACK1), epsilon2-COP, beta-catenin, and others, revealing interactions with other proteins such as 14-3-3ζ, cAMP signaling pathway, and others determining the final protein function (Komatsu et al., 2014). Proteins involved in stress-induced gene expression include several ABA-dependent and ABA-independent transcription factors such as bZIP, bHLH, MYB, MYC, NAC, etc., (Bae et al., 2003). Proteins involved in chromatin remodeling include proteins involved in DNA methylation such as cytosine methyltransferase as well as proteins involved in histone PTMs such as histone deacetylase (HDAC) revealed alterations under stress (Pandey et al., 2008; Subba et al., 2013). It is also known that differential expression of various histone isoforms such as H2A (a canonical isoform H2A vs. a cold-responsive isoform H2A.Z) affects chromatin remodeling under low-temperature stress leading to transcriptome reprogramming (Janská et al., 2011). Differential H2A isoforms and decreased H1 and H3 were also found in nuclear proteome of soybean after the onset of flooding indicating profound chromatin remodeling (Yin and Komatsu, 2016). Nucleus also represents a site of formation of ribosomal subunits; therefore, several ribosomal proteins were identified among proteins responding to cold in A. thaliana nuclei (Bae et al., 2003). Several snoU3 RNA-associated proteins and NOP1/NOP56 complex which are involved in 60S ribosomal subunit biogenesis were found declined in flooded soybean indicating suppressed novel protein biosynthesis (Yin and Komatsu, 2016). Furthermore, stress also induced an accumulation of chaperones from HSP70 and HSP90 families to protect other proteins from disassembly under stress (Bae et al., 2003; Pandey et al., 2010). In addition, alterations in proteins involved in nucleocytoplasmic transport via nuclear pores such as Ran-binding protein (RanBP) were found under drought stress (Pandey et al., 2008).
Stress treatments also lead to alterations in protein PTMs. An analysis of nuclear phosphoproteins in soybean root tip during flooding led to identification of 27 phosphoproteins including zinc-finger/BTB domain-containing protein 47, glycine-rich protein, and rRNA processing protein Rrp5 which were regulated by ABA and phosphorylated in response to flooding (Yin and Komatsu, 2015).
Chloroplasts as sites of photosynthetic machinery are profoundly affected by stress treatments. Photosynthetic apparatus is very susceptible to alterations in cellular redox homeostasis affected by stress. Oxidative stress, especially a severe one such as ozone treatment, leads to decreased levels of components of photosynthetic apparatus including both photosynthetic electron transport chain and carbon assimilation proteins. In contrast, proteins involved in ROS scavenging and carbohydrate catabolism increased under oxidative stress which corresponds to decreased starch and increased sucrose levels (Ahsan et al., 2010; Hüner et al., 2012). The most crucial components of photosynthetic electron transport chain revealing alterations in response to stress are proteins of OEC complex involved in electron release from water, components of cytochrome b6f complex as well as components of electron acceptor complexes ferredoxin-NADP reductase and NAD(P)H-quinone oxidoreductase. Following electron acceptors, components of CF1-CF0 ATP synthase also revealed alterations under stress, especially the subunits of CF1 component including the subunits α, β, γ, ε, and λ. Stress also leads to alterations in proteins associated with carbon fixation including RubisCO subunits, RubisCO activase A and carbonic anhydrase. Alterations in photosynthetic electrontransport chain lead to profound alterations in chloroplast redox homeostasis and detoxification metabolism which result in enhanced levels of redox-related enzymes such as 2-cysteine peroxiredoxins (2-Cys Prx), thioredoxin m (Trx m) isoform, and lactoylglutathione lyase (glyoxalase I) levels as found in Arabidopsis chloroplast proteome under cold (Goulas et al., 2006). Alterations in crucial components of photosynthetic electrontransport chain such as the final electron acceptor ferredoxin-NADPH oxidoreductase and in subunits of CF0-CF1 ATP synthase complex were found in maize chloroplasts at early stages of salt treatment (Zörb et al., 2009).
Furthermore, stress also affects other plastidial functions such as plastidial glycolysis and proteosynthesis. Alterations in plastidial isoforms of GAPDH as well as plastidial translation elongation factor EF-Tu were found in wheat chloroplast proteome under salinity (Kamal et al., 2012). An increase in enzymes such as monogalactosyl diacylglycerol (MGDG) synthase involved in the biosynthesis of thylakoid membrane glycolipids were found in maize chloroplasts at early phases of salt stress indicating alterations in thylakoid membrane composition in response to stress (Zörb et al., 2009). Structural proteins are also affected by stress; as an example, fibrillins as the major protein components of plastoglobuli reveal alterations in response to drought in Festuca arundinacea (Kosmala et al., 2012), however, in oilseed rapes, fibrillins were found significantly accumulated in all cultivars upon drought (Urban et al., 2017). Last, but not least, protoporhyrinogen IX oxidase was found enhanced in maize chloroplasts under salt stress (Zörb et al., 2009); the enzyme catalyzes a conversion of chlorophyll precursor protoporhyrinogen which is known to act as a photosensitizer involved in ROS (singlet oxygen 1O2) production under stress. Enhanced protoporhyrinogen IX oxidase thus reduces the amount of protoporhyrinogen decreasing a risk of ROS formation (Zörb et al., 2009).
An analysis of tomato chloroplast proteome under drought stress and subsequent recovery revealed the importance of chloroplast as an environmental sensor of stress signals leading to specific chloroplast-to-nucleus (retrograde) signaling pathways and a crosstalk with nucleus-based (anterograde) ABA-dependent signaling network (Tamburino et al., 2017).
Mitochondria as a major site of aerobic respiration face an enhanced risk of oxidative stress during stress acclimation. Therefore, enhanced levels of several ROS scavenging enzymes (Mn-SOD) were observed in mitochondrial proteome under stress (Taylor et al., 2005; Jacoby et al., 2010). Stress also led to enhanced levels of Krebs cycle enzymes such as malate dehydrogenase (MDH), components of F1Fo ATP synthase complex as well as alternative oxidase (AOX) which transfers electrons directly from ubiquinone pool to oxygen thus omitting cytochrome complexes of respiratory pathway (Jacoby et al., 2013). Unlike cytochromes, AOX is resistant to cyanide as a potential byproduct of plant metabolism under stress. Moreover, AOX activity prevents an overreduction of terminal electron acceptor thus preventing superoxide formation. Mitochondrial redox homeostasis is maintained by several thioredoxins (Trx) and peroxiredoxins (Prx). Besides ROS, NO can also modify several target proteins in mitochondria including ATP synthase CF1 β subunit, HSP90 and peroxiredoxins. It was found that S-nitrosylation of PrxII F in salt-treated pea resulted in a decrease of the protein biological activity (Camejo et al., 2013). Anoxia leads to increased levels of γ-aminobutyrate and tricarboxylic acid (TCA) cycle intermediates and, in contrast, to a decrease in components of mitochondrial electrontransport chain (ETC), especially complexes III, IV, and V, as found in soybean mitochondria under flooding (Komatsu et al., 2011). Stress also leads to altered levels of ion transporters such as voltage-dependent anion channel (VDAC), outer mitochondrial membrane porin (Jacoby et al., 2010), mitochondrial outer and inner membrane translocases Tom and Tim (Komatsu et al., 2011) as well as proteins with protective functions such as GroES chaperonin, chaperones and heat shock proteins in mitochondria thus preventing damage of protein complexes in respiratory chain (Taylor et al., 2005; Jacoby et al., 2013).
Endoplasmic reticulum (ER) is a major site of protein biosynthesis as well as folding and PTMs of nascent proteins. Investigation of ER-enriched fraction isolated from soybean root tips exposed to flooding revealed alterations in proteins involved in protein biosynthesis and PTMs, namely glycosylation, such as luminal-binding protein 5, arabinogalactan protein 2, and methyltransferase PMT2 (Komatsu et al., 2012). Moreover, an increase in 3-ketoacylCoA reductase 1 involved in fatty acid biosynthesis as well as in stress- and defense-related proteins such as stress-induced protein SAM22 (Starvation-associated message 22) and osmotin-like proteins were found. In addition, enhanced levels of proteins involved in anaerobic metabolism such as glycolysis (GAPDH, ENO) and fermentation (ADH1) as well as proteins involved in PCD induction (apoptosis-inducing factor homolog A) were found in ER-enriched fraction under flooding which is typical for anaerobiosis and aerenchyma formation (Komatsu et al., 2012).
Vacuole plays an important role in plant salinity tolerance due to Na+ intracellular compartmentation. The rate of Na+ vacuolar sequestration is affected by the activity of tonoplast-located V-ATPase, which functions as an ATP-dependent Na+/H+ antiporter. In their study on Arabidopsis los2 mutant lacking functional cytoplasmic enolase, Barkla et al. (2009) have shown a role of glycolytic enzymes aldolase (ALDO) and enolase (ENO) associated with tonoplast fraction for the activity of V-ATPase due to ATP supply.
Summary: Each cellular compartment reveals specific biological functions which correspond with specificities of their proteomes. Different protein isoforms located in different cellular compartements can reveal either analogous functions (e.g., cytosolic Cu/Zn-SOD, mitochondrial Mn-SOD, chloroplast Fe-SOD) or entirely different functions (e.g., cytosolic vs. nuclear enolase) depending on their interactions.
Protein biological function is also determined by protein isoforms and post-translational modifications (PTMs). Protein isoforms differ in their sequence (primary structure) while PTMs represent chemical modifications of amino acid residues which can range from small molecules (NO) to whole peptides (ubiquitin, SUMO). Protein isoforms arise either from a single gene as products of posttranscriptional modifications such as alternative splicing, RNA editing, and others, or they can be products of orthologous or paralogous genes. Protein PTMs are reversible and can alter throughout protein life cycle, e.g., reversible phosphorylation of protein kinases or ubiquitination of proteins targeted to degradation. Protein isoforms and PTMs often differ in their molecular weight or isoelectric point making them distinguishable by 1-DE and 2-DE. Currently, over than 300 kinds of protein PTMs were described (Wu et al., 2016); however, only few of them were studied with respect to plant stress treatments. Stress-related PTMs mostly include phosphorylation, glycosylation, ubiquitination, sumoylation, and modifications induced by reactive molecular species (RMS) such as protein carbonylation and nitrosylation, etc., which result in significant alterations of protein function. Analysis of Web of Science publication database revealed that currently, the most studied protein PTMs upon stress represent phosphoproteomic studies (73 studies found as a reply for a query “plant phosphoproteome and stress” in October, 2017), followed by redox proteomics, glycoproteomics, and recently also by protein S-nitrosylation while other PTMs remain largely untouched in stress-treated plants.
Phosphoproteome analysis of drought-treated wheat seedling leaves revealed phosphorylated signaling proteins belonging to ABA-induced SnRK and PP2C signaling pathways, calcium-dependent protein kinase (CDPK), mitogen-activated protein kinase (MAPK) pathway, and calcium-dependent signaling proteins such as phosphatidylinositol-4,5-diphosphate. Other phosphoproteins identified include phosphorylated transcription factors such as ABA-dependent transcription factors ABI5, MYB1R1, and bHLH as well as ABA-independent transcription factors such as zinc finger CCCH domain containing proteins. Other phosphoproteins include proteins involved in transport such as aquaporins (AQPs) and proton ion pumps (H+-ATPases), proteins involved in stress defense such as zinc finger ABI5, three E3 ubiquitin ligases isoforms, ROS scavenging-related proteins glutamate decarboxylase 1 and glutathione peroxidase, and LEA proteins such as WCOR615, WCOR719 and WRAB17 (Zhang et al., 2014). Phosphorylation-regulated proteins involved in signaling and signal transduction also play important roles in plant-pathogen interactions affecting the extent of infection as revealed by a phosphoproteomic study on wheat infected with Septoria tritici (Yang et al., 2013b). Pathogen is sensed by plant receptor-like kinases and G proteins on plasma membrane. The initial signal is then transduced by MAPK and CDPK signaling cascades to nucleus where transcription factors such as WRKY involved in expression of genes associated with defense response are activated by phosphorylation. Similarly, alterations in phosphorylation patterns were also found in several signaling proteins produced by fungal pathogen such as protein kinase A, 14-3-3 protein, G protein subunit α, Ras GTPase, several MAPKs and an ABC transporter indicating that phosphorylation might play an important role in plant-pathogen interactions. Differential phosphorylation of dehydrin-5 protein and LEAIII proteins (WRAB17) was found in differentially-tolerant drought-treated common wheat and durum wheat cultivars, respectively (Brini et al., 2007; Zhang et al., 2014) which might indicate differential protein cellular localization between cytoplasm and nucleus (Rorat, 2006).
Redox proteomics: Possibilities of PTM modifications by reactive species of oxygen (ROS), nitrogen (RNS), carbonyl (RCS) and sulfur (RSS) are given in recent review by Mock and Dietz (2016). A proteomic study on three Iranian drought-treated wheat cultivars led to an identification of three different isoforms of thioredoxin h (Trx h) revealing different abundance patterns in response to drought stress as well as several other proteins known as potential targets of Trx h activity indicating an importance of plant redox response for protein protection under drought (Hajheidari et al., 2007). Apart from highly studied ROS, the effects of nitric oxide (NO) as a stress signaling molecule and the resulting S-nitrosylation of cysteine residues which affects protein biological activity becomes recently studied. For example, S-nitrosylation of RubisCO subunits in Brassica juncea exposed to cold leads to a decrease in RubisCO carboxylation activity (Abat and Deswal, 2009).
Protein glycosylation plays an important role in plant response to pathogens. Differential patterns of non-glycosylated vs. glycosylated isoforms of xylanase inhibitors with non-glycosylated forms increased more strongly than their glycosylated counterparts were found in wheat treated with F. graminearum ΔTri mutant (Dornez et al., 2010). In wheat, DNA-damage inducible protein was found to undergo O-glycosylation under F. graminearum infection (Zhou et al., 2006). O-glycosylation may play an important role in response to low-temperature stress as reported for some dehydrin proteins isolated from cold-treated floral buds of blueberry (Levi et al., 1999) and pistachio (Yakubov et al., 2005); however, biological function of these glycosylated proteins remains unknown.
Examples of protein isoforms differential functions: Cellular redox metabolism, especially in chloroplasts and mitochondria, represents a finely tuned network of redox reactions, which are aimed at minimizing harmful damage by RMS, especially under stress. Cellular redox homeostasis thus represents a result of a coordinated action of several isoforms of redox proteins with specific, but overlapping functions enabling fine tuning of redox metabolism. For example, more than 20 Trx and Trx-like protein isoforms (Trx-f1, Trx-f2, Trx-m1-4, Trx-x, Trx-y1, Trx-y2, Trx-z, atypical Cys-His rich Trx), NTRC (NADPH-dependent Trx reductase C), protein disulfide isomerase-like (APR-1-3), thiol-based peroxidases and ascorbate peroxidases were identified in A. thaliana chloroplasts (Meyer et al., 2005; Dietz, 2016). The individual redox proteins are involved in reactions with thiol groups of target proteins in photosynthetic electron transport chain thus revealing specific, but overlapping functions which are still far from being fully understood (König et al., 2012). Similarly, several small HSP proteins (sHSPs) with overlapping chaperone functions were identified in heat-treated wheat grains (Skylas et al., 2002; Majoul et al., 2004) indicating an enhanced need of protein protection against heat-induced damage.
Other protein isoforms may differ in their tissue localization or induction by different stresses. Three isoforms of SAMS were identified in soybean revealing differential patterns in relative abundance both at transcript and protein levels in different seedling parts (root, cotyledon, hypocotyl) under drought and flooding, respectively (Oh and Komatsu, 2015; Wang et al., 2016). SAMS is known to be involved in methylation of lignin components; thus, the different abundances of different SAMS isoforms may underlie differences in plant cell growth and lignification of cell walls.
It is known that protein enzymatic activity depends on temperature. Therefore, alterations in protein isoforms of several enzymes differing in their thermostability were found on 2-DE (2D-DIGE) gels under temperature stress including both high and low temperatures. In Arabidopsis chloroplast fractions under cold, alterations in relative abundance of RubisCO large and small subunits were found under cold with respect to control indicating alterations in isoform composition of the whole RubisCO holoenzyme thus affecting its final activity under altered temperature (Goulas et al., 2006). Alterations in protein isoform composition become obvious, especially under heat. For example, cereals from the tribe Triticeae encode two RubisCO activase isoforms - a conventional RubisCO activase A and a thermostable RubisCO activase B. In heat-treated barley, thermostable RubisCO activase B revealed an increase while thermo-unstable RubisCO activase A revealed a decrease upon the same treatment (Rollins et al., 2013). A study in Chenopodium album revealed a higher thermostability of chloroplast APX and Cu/Zn-SOD isoforms than mitochondrial APX and Mn-SOD isoforms (Khanna-Chopra et al., 2011).
Protein isoforms can reveal either similar or entirely different biological functions. A few examples of distinct biological functions of protein isoforms are given below. Eukaryotic translation initiation factor eIF5A in cytosol is known to act as a translation initiation factor involved in regulation of protein biosynthesis. However, its nuclear isoforms eIF5A1 and eIF5A2, differing in hypusination level, were reported to differentially regulate cell cycle leading to either apoptosis (PCD) or cell division (Thompson et al., 2004). In a comparative proteomic study on salt-treated glycophytic H. vulgare and halophytic H. marinum, different eIF5A isoforms were found with eIF5A1 isoform in H. vulgare while eIF5A2 isoform in H. marinum (Maršálová et al., 2016). The presence of eIF5A1 in H. vulgare corresponds with enhanced levels of several apoptosis-related proteins found in H. vulgare under high salinity. Flooding stress-related translocation of cytochrome c from mitochondria into cytoplasm via VDAC leads to cytochrome c interaction with caspases and induction of PCD (Komatsu et al., 2011).
Similarly, nuclear isoforms of cytosolic proteins FBP aldolase, enolase, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) known as glycolytic enzymes are involved in important regulatory processes in nucleus. Nuclear isoform of fructose-bisphosphate aldolase (FBP aldolase) is known to act as a DNA-binding protein involved in regulation of expression of its own gene as well as other genes (Ronai et al., 1992). Nuclear isoform of GAPDH is known to act as a tRNA-binding protein involved in tRNA export (Singh and Green, 1993). It has also been reported that FBP aldolase may be involved in an integration of intra- and extracellular signals associated with growth, development, and sugar biosynthesis (Li et al., 2012) while non-phosphorylating isoform of GAPDH may be involved in regulation of ROS levels (Bustos et al., 2008). Nuclear isoform of enolase encoded by Los2 locus in A. thaliana functions as a transcriptional repressor of STZ/ZAT10 which is a repressor of cold-inducible CBF pathway; nuclear isoform of enolase thus acts as an indirect positive regulator of CBF pathway and CBF-regulated COR gene expression (Lee et al., 2002). Tonoplast-associated isoforms of aldolase and enolase are involved in activation of V-ATPase in salt-treated Mesembryanthemum crystallinum plants leading to Na+ vacuolar accumulation (Barkla et al., 2009). It can thus be concluded that biological function of a given protein is determined by PTM as well as cellular localization.
Proteomic analysis enables the researchers to identify proteins, which reveal quantitative or qualitative differences between control and stress treatments. These proteins might play a crucial role in plant stress response. Biological function of a given protein is dependent on its PTM, cellular localization as well as interaction partners. Therefore, functional studies of these proteins are required to characterize their role in plant stress response. Functional studies using knock-out mutants, enzyme assays, tools for protein-protein interaction study, etc., provide valuable data on protein biological functions which can complement the data on protein relative abundance obtained by high-throughput proteomic analysis.
In winter cereals, transition to flowering is regulated by a period of low temperatures, a phenomenon known as vernalization. An interactomics study in winter wheat using two independent methodical approaches, yeast two-hybrid GAL4 system and split-GFP fluorescent complementation in Nicotiana benthamiana, has confirmed physical interactions between proteins involved in flowering regulation (TaVRT1/VRN1, TaVRT2, VRN2, TaFT1/VRN3, TaHd) as well as between other protein groups such as proteins involved in signaling (two phospholipases C, a receptor-like protein kinase, G protein) and microtubule remodeling (α-tubulin and TaTil, a lipocalin known to be involved in microtubule polymerization) which opens new ways to study their functions under cold (Tardif et al., 2007).
Protein interactions play also crucial roles in plant-pathogen interactions. For example, wheat treatment with Fusarium graminearum ΔTri mutants led to changes in wheat xylanase inhibitors (Dornez et al., 2010). Fusarium graminearum ΔTri mutants are unable to produce mycotoxin deoxynivalenol (DON) thus they are less infectious than wild type pathogen. Differences in the level of plant xylanase inhibitors induced by pathogen were found. Another example is a study on triticale infected by F. culmorum where significant increases in β-amylase and α-amylase inhibitor from triticale were found. Data on alterations in protein relative abundance are accompanied by determination of α-amylase and β-amylase enzymatic activities leading to the hypothesis that the activity of plant β-amylase as well as high levels of plant α-amylase inhibitor involved in an inhibition of pathogen α-amylase activity represent important components of plant-pathogen interaction system at molecular level (Perlikowski et al., 2016).
Summary: Protein isoforms and PTMs differ in their primary sequence or amino acid residues modifications, respectively, making them distinguishable on 2DE gels. Different protein isoforms and PTMs derived from a single gene can reveal same, similar, or entirely different biological functions depending on their cellular localizations and protein-protein interactions. Interactomics studies are necessary to understand protein biological functions in the given molecular context.
Proteins play a crucial role in plant stress response leading to stress-adapted phenotype. However, it is becoming evident that protein role in plant stress response is determined by several factors which include the impacts of environmental stress treatments, genotypic differences affecting stress impacts on plant cellular homeostasis as well as differential protein biological functions determined by protein isoforms, PTMs, cellular localizations, and protein-protein interactions. The major points on the stress-related factors determining protein accumulation and biological function which have to be considered when interpreting proteomic data are summarized below as well as in Figure 4.
• Abiotic stresses reveal both common and specific effects on plants. Several abiotic stresses lead to oxidative stress, several stresses (frost, heat, drought, salinity) lead to cellular dehydration.
• Plant stress response is a dynamic process where several phases (alarm, acclimation, resistance, exhaustion, recovery) with specific proteome composition can be distinguished.
• The impacts of combined stress treatments do not equal to the sum of the effects of the individual stress treatments and thus they need to be studied.
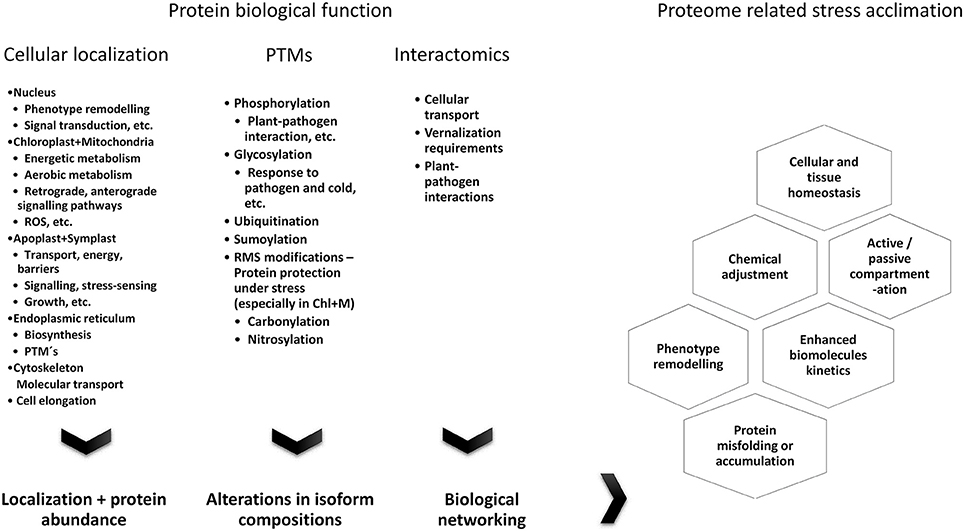
Figure 4. Summary of the major factors determining proteome composition and protein biological functions in stress-treated plants.
Comparison of stress-susceptible vs. stress-tolerant plants:
• The impacts of stress on plants have to be assessed using plant-related characteristics (e.g., water content in plant tissues) rather than environment-related characteristics (e.g., water content in soil).
• Tolerant plants reveal constitutively enhanced levels of several stress- and detoxification-related proteins thus the stress treatment causes weaker disturbances in their cellular homeostasis.
• Tolerant plants can adjust energy metabolism to enhanced demands of stress acclimation process due to activation of photosynthesis and aerobic respiration.
• Susceptible and tolerant plants also reveal differences in development-related proteins under stress: at molecular level, susceptible plants reveal cellular damage leading to activation of PCD-related processes while tolerant plants reveal restoration of active growth and development.
Protein isoforms and PTMs:
• Protein isoforms reveal differences in their primary sequence while protein PTMs represent differential modifications of amino acid residues.
• Protein isoforms can reveal same, similar or completely different biological functions which depend on their cell localization, PTMs or protein-protein interactions.
• Interactomics studies are necessary to understand protein biological functions in the given molecular context.
KK outlined the manuscript area and prepared preliminary manuscript text. The other authors PV, MU, IP, and JR contributed actively to the preliminary manuscript text, and designed figures and tables.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by an institutional project from Czech Ministry of Agriculture (MZe CZ) RO017, QJ1310055, and QK1710302 and by projects LD14087 and LD15167 supported by Czech Ministry of Education, Youth and Sports (MEYS CZ) as a part of international COST actions FA1208 “Pathogen-informed Strategies for Sustainable Crop Resistance” and FA1306 “The Quest for Tolerant Varieties - Phenotyping at Plant and Cellular Level,” respectively. This work was also supported by project LO1417 of Czech Ministry of Education, Youth and Sports (MEYS CZ) and from the National Fund for Research in Luxembourg, project SMARTWALL, C15/SR/10240550.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00122/full#supplementary-material
ADH, alcohol dehydrogenase; AOX, alternative oxidase; APX, ascorbate peroxidase; AQP, aquaporin; CCOMT, caffeoyl-coenzyme A O-methyltransferase; CBF, C-repeat binding factor; CDPK, calcium-dependent protein kinase; COMT, caffeic acid O-metyltransferase; COR, cold-regulated (protein); ECM, extracellular matrix; eIF, eukaryotic translation initiation factor; ENO, enolase; ETC, electrontransport chain; FBP ALDO, fructose 1,6-bisphosphate aldolase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GLP, germin-like protein; GRP, glycine-rich protein; HDAC, histone deacetylase; HSF, heat-shock transcription factor; HSP, heat-shock protein; LEA, late embryogenesis abundant (protein); MAPK, mitogen-activated protein kinase; MDAR, monodehydroascorbate reductase; MDH, malate dehydrogenase; MGDG, monogalactosyldiacylglycerol; NDPK, nucleoside diphosphate kinase; OEC, oxygen evolving complex; PCD, programmed cell death; PGK, phosphoglycerokinase; PPR, pentatricopeptide repeat protein; PRK, phosphoribulokinase; Prx, peroxiredoxin; PTM, posttranslational modification; RCS, reactive carbonyl species; RMS, reactive molecular species; RNS, reactive nitrogen species; ROS, reactive oxygen species; RSS, reactive sulfur species; RubisCO, ribulose-1,5-bisphosphate carboxylase; SAM, S-adenosylmethionine; SAMS, S-adenosylmethionine synthetase; SAM22, starvation-associated message 22; SOD, superoxide dismutase; SOS, salt overly sensitive (gene); TCA, tricarboxylic acid (cycle); TCTP, translationally-controlled tumor protein; TPI, triose phosphate isomerase; Trx, thioredoxin; USP, universal stress protein; VDAC, voltage-dependent anion channel.
Abat, J. K., and Deswal, R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380. doi: 10.1002/pmic.200800985
Agrawal, G. K., Jwa, N. S., and Rakwal, R. (2009). Rice proteomics: ending phase I and the beginning of phase II. Proteomics 9, 935–963. doi: 10.1002/pmic.200800594
Ahmad, P., Abdel Latef, A. A. H., Rasool, S., Akram, N. A., Ashraf, M., and Gucel, S. (2016). Role of proteomics in crop stress tolerance. Front. Plant Sci. 7:1336. doi: 10.3389/fpls.2016.01336
Ahsan, N., Nanjo, Y., Sawada, H., Kohno, Y., and Komatsu, S. (2010). Ozone stress-induced proteomic changes in leaf total soluble and chloroplast proteins of soybean reveal that carbon allocation is involved in adaptation in the early developmental stage. Proteomics 10, 2605–2619. doi: 10.1002/pmic.201000180
Ahsan, N., Renaut, J., and Komatsu, S. (2009). Recent developments in the application of proteomics to the analysis of plant responses to heavy metals. Proteomics 9, 2602–2621. doi: 10.1002/pmic.200800935
Ali, M. B., and El-Sadek, A. N. (2016). Evaluation of drought tolerance indices for wheat (Triticum aestivum L.) under irrigated and rainfed conditions. Commun. Biometry Crop Sci. 11, 77–89.
Alvarez, S., Roy Choudhury, S., and Pandey, S. (2014). Comparative quantitative proteomics analysis of the ABA response of roots of drought-sensitive and drought-tolerant wheat varieties identifies proteomic signatures of drought adaptability. J. Proteome Res. 13, 1688–1701. doi: 10.1021/pr401165b
Askari, H., Edqvist, J., Hajheidari, M., Kafi, M., and Salekdeh, G. H. (2006). Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 6, 2542–2554. doi: 10.1002/pmic.200500328
Bae, M. S., Cho, E. J., Choi, E. Y., and Park, O. K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36, 652–663. doi: 10.1046/j.1365-313X.2003.01907.x
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Barkla, B. J., Vera-Estrella, R., Hernández-Coronado, M., and Pantoja, O. (2009). Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21, 4044–4058. doi: 10.1105/tpc.109.069211
Benešová, M., Holá, D., Fischer, L., Jedelský, P. L., Hnilička, F., Wilhelmová, N., et al. (2012). The physiology and proteomics of drought tolerance in maize: early stomatal closure as a cause of lower tolerance to short-term dehydration? PLoS ONE 7:e38017. doi: 10.1371/journal.pone.0038017
Bernardo, L., Morcia, C., Carletti, P., Ghizzoni, R., Badeck, F. W., Rizza, F., et al. (2017). Proteomic insight into the mitigation of wheat root drought stress by arbuscular mycorrhizae. J. Proteomics 169, 21–32. doi: 10.1016/j.jprot.2017.03.024
Bhushan, D., Pandey, A., Choudhary, M. K., Datta, A., Chakraborty, S., and Chakraborty, N. (2007). Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteomics 6, 1868–1884. doi: 10.1074/mcp.M700015-MCP200
Blum, A. (2016). Stress, strain, signalling, and adaptation - not just a matter of definition. J. Exp. Bot. 67, 562–565. doi: 10.1093/jxb/erv497
Brini, F., Hanin, M., Lumbreras, V., Irar, S., Pagès, M., and Masmoudi, K. (2007). Functional characterization of DHN-5, a dehydrin showing a differential phosphorylation pattern in two Tunisian durum wheat (Triticum durum Desf.) varieties with marked differences in salt and drought tolerance. Plant Sci. 172, 20–28. doi: 10.1016/j.plantsci.2006.07.011
Budak, H., Akpinar, B. A., Unver, T., and Turktas, M. (2013). Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI-MS/MS. Plant Mol. Biol. 83, 89–103. doi: 10.1007/s11103-013-0024-5
Bustos, D. M., Bustamante, C. A., and Iglesias, A. A. (2008). Involvement of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase in response to oxidative stress. J. Plant Physiol. 165, 456–461. doi: 10.1016/j.jplph.2007.06.005
Camejo, D., Romero-Puertas, M. D., Rodríguez-Serrano, M., Sandalio, L. M., Lázaro, J. J., Jiménez, A., et al. (2013). Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteomics 79, 87–99. doi: 10.1016/j.jprot.2012.12.003
Caruso, G., Cavaliere, C., Guarino, C., Gubbiotti, R., Foglia, P., and Laganà, A. (2008). Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal. Bioanal. Chem. 391, 381–390. doi: 10.1007/s00216-008-2008-x
Chen, W., Wang, H., Tao, S., Zheng, Y., Wu, W., Lian, F., et al. (2013). Tumor protein translationally controlled 1 is a p53 target gene that promotes cell survival. Cell Cycle 12, 2321–2328. doi: 10.4161/cc.25404
Cheng, L., Wang, Y., He, Q., Li, H., Zhang, X., and Zhang, F. (2016). Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 16:188. doi: 10.1186/s12870-016-0871-8
Claeys, H., Van Landeghem, S., Dubois, M., Maleux, K., and Inzé, D. (2014). What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol. 165, 519–527. doi: 10.1104/pp.113.234641
Dietz, K. J. (2016). Thiol-based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 39, 20–25. doi: 10.14348/molcells.2016.2324
Dornez, E., Croes, E., Gebruers, K., Carpentier, S., Swennen, R., Laukens, K., et al. (2010). 2-D DIGE reveals changes in wheat xylanase inhibitor protein families due to Fusarium graminearum Delta Tri5 infection and grain development. Proteomics 10, 2303–2319. doi: 10.1002/pmic.200900493
Durand, T. C., Sergeant, K., Planchon, S., Carpin, S., Label, P., Morabito, D., et al. (2010). Acute metal stress in Populus tremula x P. alba (717-1B4 genotype): Leaf and cambial proteome changes induced by cadmium(2+). Proteomics 10, 349–368. doi: 10.1002/pmic.200900484
Faghani, E., Gharechahi, J., Komatsu, S., Mirzaei, M., Khavarinejad, R. A., Najafi, F., et al. (2015). Comparative physiology and proteomic analysis of two wheat genotypes contrasting in drought tolerance. J. Proteomics 114, 1–15. doi: 10.1016/j.jprot.2014.10.018
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Flowers, T. J., and Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Ford, K. L., Cassin, A., and Bacic, A. (2011). Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2:44. doi: 10.3389/fpls.2011.00044
Ge, P., Ma, C., Wang, S., Gao, L., Li, X., Guo, G., et al. (2012). Comparative proteomic analysis of grain development in two spring wheat varieties under drought stress. Anal. Bioanal. Chem. 402, 1297–1313. doi: 10.1007/s00216-011-5532-z
Ghabooli, M., Khatabi, B., Ahmadi, F. S., Sepehri, M., Mirzaei, M., Amirkhani, A., et al. (2013). Proteomics study reveals the molecular mechanisms underlying water stress tolerance induced by Piriformospora indica in barley. J. Proteomics 94, 289–301. doi: 10.1016/j.jprot.2013.09.017
Ghaffari, M., Toorchi, M., Valizadeh, M., and Komatsu, S. (2013). Differential response of root proteome to drought stress in drought sensitive and tolerant sunflower inbred lines. Funct. Plant Biol. 40, 609–617. doi: 10.1071/FP12251
Gharechahi, J., Alizadeh, H., Naghavi, M. R., and Sharifi, G. (2014). A proteomic analysis to identify cold acclimation associated proteins in wild wheat (Triticum urartu L.). Mol. Biol. Rep. 41, 3897–3905. doi: 10.1007/s11033-014-3257-8
Ghatak, A., Chaturvedi, P., Paul, P., Agrawal, G. K., Rakwal, R., Kim, S. T., et al. (2017). Proteomics survey of Solanaceae family: current status and challenges ahead. J. Proteomics 169, 41–57. doi: 10.1016/j.prot.2017.05
Ghosh, D., and Xu, J. (2014). Abiotic stress responses in plant roots: a proteomics perspective. Front. Plant Sci. 5:6. doi: 10.3389/fpls.2014.00006
Goulas, E., Schubert, M., Kieselbach, T., Kleczkowski, L. A., Gardeström, P., Schröder, W., et al. (2006). The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 47, 720–734. doi: 10.1111/j.1365-313X.2006.02821.x
Guo, G., Ge, P., Ma, C., Li, X., Lv, D., Wang, S., et al. (2012). Comparative proteomic analysis of salt response proteins in seedling roots of two wheat varieties. J. Proteomics 75, 1867–1885. doi: 10.1016/j.jprot.2011.12.032
Hajheidari, M., Eivazi, A., Buchanan, B. B., Wong, J. H., Majidi, I., and Salekdeh, G. H. (2007). Proteomics uncovers a role for redox in drought tolerance in wheat. J. Proteome Res. 6, 1451–1460. doi: 10.1021/pr060570j
Holá, D., Benešová, M., Fischer, L., Haisel, D., Hnilička, F., Hniličková, H., et al. (2017). The disadvantages of being a hybrid during drought: a combined analysis of plant morphology, physiology and leaf proteome in maize. PLoS ONE 12:e0176121. doi: 10.1371/journal.pone.0176121
Hossain, Z., and Komatsu, S. (2013). Contribution of proteomic studies towards understanding plant heavy metal stress response. Front. Plant Sci. 3:310. doi: 10.3389/fpls.2012.00310
Hossain, Z., Nouri, M. Z., and Komatsu, S. (2012). Plant cell organelle proteomics in response to abiotic stress. J. Proteome Res. 11, 37–48. doi: 10.1021/pr200863r
Hüner, N. P., Bode, R., Dahal, K., Hollis, L., Rosso, D., Krol, M., et al. (2012). Chloroplast redox imbalance governs phenotypic plasticity: the “grand design of photosynthesis” revisited. Front. Plant Sci. 3:255. doi: 10.3389/fpls.2012.00255
Hynek, R., Svensson, B., Jensen, O. N., Barkholt, V., and Finnie, C. (2009). The plasma membrane proteome of germinating barley embryos. Proteomics 9, 3787–3794. doi: 10.1002/pmic.200800745
Jacoby, R. P., Millar, A. H., and Taylor, N. L. (2010). Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. J. Proteome Res. 9, 6595–6604. doi: 10.1021/pr1007834
Jacoby, R. P., Millar, A. H., and Taylor, N. L. (2013). Investigating the role of respiration in plant salinity tolerance by analyzing mitochondrial proteomes from wheat and a salinity-tolerant amphiploid (wheat x Lophopyrum elongatum). J. Proteome Res. 12, 4807–4829. doi: 10.1021/pr400504a
Janmohammadi, M., Zolla, L., and Rinalducci, S. (2015). Low temperature tolerance in plants: changes at the protein level. Phytochemistry 117, 76–89. doi: 10.1016/j.phytochem.2015.06.003
Janská, A., Aprile, A., Zámečník, J., Cattivelli, L., and Ovesná, J. (2011). Transcriptional responses of winter barley to cold indicate nucleosome remodelling as a specific feature of crown tissues. Funct. Integr. Genomics 11, 307–325. doi: 10.1007/s10142-011-0213-8
Johnová, P., Skalák, J., Saiz-Fernández, I., and Brzobohatý, B. (2016). Plant responses to ambient temperature fluctuations and water-limiting conditions: a proteome-wide perspective. Biochim. Biophys. Acta Proteins Proteomics 1864, 916–931. doi: 10.1016/j.bbapap.2016.02.007
Jorrín-Novo, J. V., Maldonado, A. M., Echevarría-Zomeno, S., Valledor, L., Castillejo, M. A., Curto, M., et al. (2009). Plant proteomics update (2007-2008): Second-generation proteomic techniques, an appropriate experiemntal design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage, and expand biological knowledge. J. Proteomics 72, 285–314. doi: 10.1016/j.jprot.2009.01.026
Kamal, A. H., Cho, K., Kim, D. E., Uozumi, N., Chung, K. Y., Lee, S. Y., et al. (2012). Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol. Biol. Rep. 39, 9059–9074. doi: 10.1007/s11033-012-1777-7
Kausar, R., Arshad, M., Shahzad, A., and Komatsu, S. (2013). Proteomics analysis of sensitive and tolerant barley genotypes under drought stress. Amino Acids 44, 345–359. doi: 10.1007/s00726-012-1338-3
Khanna-Chopra, R., Jajoo, A., and Semwal, V. K. (2011). Chloroplasts and mitochondria have multiple heat tolerant isozymes of SOD and APX in leaf and inflorescence in Chenopodium album. Biochem. Biophys. Res. Commun. 412, 522–525. doi: 10.1016/j.bbrc.2011.06.179
Komatsu, S., Kamal, A. H., and Hossain, Z. (2014). Wheat proteomics: proteome modulation and abiotic stress acclimation. Front. Plant Sci. 5:684. doi: 10.3389/fpls.2014.00684
Komatsu, S., Kobayashi, Y., Nishizawa, K., Nanjo, Y., and Furukawa, K. (2010). Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 39, 1435–1449. doi: 10.1007/s00726-010-0608-1
Komatsu, S., Kuji, R., Nanjo, Y., Hiraga, S., and Furukawa, K. (2012). Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteomics 77, 531–560. doi: 10.1016/j.jprot.2012.09.032
Komatsu, S., Wada, T., Abaléa, Y., Nouri, M. Z., Nanjo, Y., Nakayama, N., et al. (2009). Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Proteome Res. 8, 4487–4499. doi: 10.1021/pr9002883
Komatsu, S., Yamamoto, A., Nakamura, T., Nouri, M. Z., Nanjo, Y., Nishizawa, K., et al. (2011). Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 10, 3993–4004. doi: 10.1021/pr2001918
König, J., Muthuramalingam, M., and Dietz, K. J. (2012). Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Curr. Opin. Plant Biol. 15, 261–268. doi: 10.1016/j.pbi.2011.12.002
Kosmala, A., Bocian, A., Rapacz, M., Jurczyk, B., and Zwierzykowski, Z. (2009). Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J. Exp. Bot. 60, 3595–3609. doi: 10.1093/jxb/erp205
Kosmala, A., Perlikowski, D., Pawłowicz, I., and Rapacz, M. (2012). Changes in the chloroplast proteome following water deficit and subsequent watering in a high- and a low-drought-tolerant genotype of Festuca arundinacea. J. Exp. Bot. 63, 6161–6172. doi: 10.1093/jxb/ers265
Kosová, K., Prášil, I. T., and Vítámvás, P. (2008). The relationship between vernalization-and photoperiodically-regulated genes and the development of frost tolerance in wheat and barley. Biol. Plant. 52, 601–615. doi: 10.1007/s10535-008-0120-6
Kosová, K., Prášil, I. T., and Vítámvás, P. (2013b). Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 14, 6757–6789. doi: 10.3390/ijms14046757
Kosová, K., Vítámvás, P., Planchon, S., Renaut, J., Vanková, R., and Prášil, I. T. (2013d). Proteome analysis of cold response in spring and winter wheat (Triticum aestivum) crowns reveals similarities in stress adaptation and differences in regulatory processes between the growth habits. J. Proteome Res. 12, 4830–4845. doi: 10.1021/pr400600g
Kosová, K., Vítámvás, P., and Prášil, I. T. (2014). Proteomics of stress responses in wheat and barley-search for potential protein markers of stress tolerance. Front. Plant Sci. 5:711. doi: 10.3389/fpls.2014.00711
Kosová, K., Vítámvás, P., Prášil, I. T., and Renaut, J. (2011). Plant proteome changes under abiotic stress - Contribution of proteomics studies to understanding plant stress response. J. Proteomics 74, 1301–1322. doi: 10.1016/j.jprot.2011.02.006
Kosová, K., Vítámvás, P., Prášilová, P., and Prášil, I. T. (2013c). Accumulation of WCS120 and DHN5 proteins in differently frost-tolerant wheat and barley cultivars grown under a broad temperature scale. Biol. Plant. 57, 105–112. doi: 10.1007/s10535-012-0237-5
Kosová, K., Vítámvás, P., Urban, M. O., Klíma, M., Roy, A., and Prášil, I. T. (2015). Biological networks underlying abiotic stress tolerance in temperate crops - a proteomic perspective. Int. J. Mol. Sci. 16, 20913–20942. doi: 10.3390/ijms160920913
Kosová, K., Vítámvás, P., Urban, M. O., and Prášil, I. T. (2013a). Plant proteome responses to salinity stress - comparison of glycophytes and halophytes. Funct. Plant Biol. 40, 775–786. doi: 10.1071/FP12375
Kwak, K. J., Kim, Y. O., and Kang, H. S. (2005). Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 56, 3007–3016. doi: 10.1093/jxb/eri298
Lawlor, D. W. (2013). Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J. Exp. Bot. 64, 83–108. doi: 10.1093/jxb/ers326
Lee, H., Guo, Y., Ohta, M., Xiong, L. M., Stevenson, B., and Zhu, J. K. (2002). LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21, 2692–2702. doi: 10.1093/emboj/21.11.2692
Levi, A., Panta, G. R., Parmentier, C. M., Muthalif, M. M., Arora, R., Shanker, S., et al. (1999). Complementary DNA cloning, sequencing and expression of an unusual dehydrin from blueberry floral buds. Physiol. Plant. 107, 98–109.
Levitt, J. (1980). Responses of Plants to Environmental Stresses: Chilling, Freezing and High Temperature Stresses. New York, NY: Academic Publisher.
Li, G., Zhang, Z. S., Gao, H. Y., Liu, P., Dong, S. T., Zhang, J. W., et al. (2012). Effects of nitrogen on photosynthetic characteristics of leaves from two different stay-green corn (Zea mays L.) varieties at the grain-filling stage. Can. J. Plant Sci. 92, 671–680. doi: 10.4141/CJPS2012-039
Li, X., Cai, J., Liu, F., Dai, T., Cao, W., and Jiang, D. (2014). Physiological, proteomic and transcriptional responses of wheat to combination of drought or waterlogging with late spring low temperature. Funct. Plant Biol. 41, 690–703. doi: 10.1071/FP13306
Majoul, T., Bancel, E., Triboï, E., Ben Hamida, J., and Branlard, G. (2004). Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from non-prolamins fraction. Proteomics 4, 505–513. doi: 10.1002/pmic.200300570
Manaa, A., Ben Ahmed, H., Valot, B., Bouchet, J.-P., Aschi-Smiti, S., Causse, M., et al. (2011). Salt and genotype impact on plant physiology and root proteome variations in tomato. J. Exp. Bot. 62, 2797–2813. doi: 10.1093/jxb/erq460
Maršálová, L., Vítámvás, P., Hynek, R., Prášil, I. T., and Kosová, K. (2016). Proteomic response of Hordeum vulgare cv. Tadmor and Hordeum marinum to salinity stress: similarities and differences between a glycophyte and a halophyte. Front. Plant Sci. 7:1154. doi: 10.3389/fpls.2016.01154
Meyer, Y., Reichheld, J. P., and Vignols, F. (2005). Thioredoxins in Arabidopsis and other plants. Photosyn. Res. 86, 419–433.doi: 10.1007/s11120-005-5220-y
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Mock, H. P., and Dietz, K. J. (2016). Redox proteomics for the assessment of redox-related posttranslational regulation in plants. Biochim. Biophys. Acta Proteins Proteomics 1864, 967–973. doi: 10.1016/j.bbapap.2016.01.005
Mostek, A., Börner, A., Badowiec, A., and Weidner, S. (2015). Alterations in root proteome of salt-sensitive and tolerant barley lines under salt stress conditions. J. Plant Physiol. 174, 166–176. doi: 10.1016/j.jplph.2014.08.020
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Murata, N., and Los, D. A. (1997). Membrane fluidity and temperature perception. Plant Physiol. 115, 875–879. doi: 10.1104/pp.115.3.875
Ning, F., and Wang, W. (2016). “The response of chloroplast proteome to abiotic stress,” in Drought Stress Tolerance in Plants, eds M. A. Hossain, S. H. Wani, S. Bhattacharjee, D.J. Burritt, and L. S. P. Tran (Verlag: Springer), 232–249.
Nouri, M. Z., and Komatsu, S. (2010). Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 10, 1930–1945. doi: 10.1002/pmic.200900632
Oh, M., and Komatsu, S. (2015). Characterization of proteins in soybean roots under flooding and drought stresses. J. Proteomics 114, 161–181. doi: 10.1016/j.jprot.2014.11.008
Pandey, A., Chakraborty, S., Datta, A., and Chakraborty, N. (2008). Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell. Proteomics 7, 88–107. doi: 10.1074/mcp.M700314-MCP200
Pandey, A., Rajamani, U., Verma, J., Subba, P., Chakraborty, N., Datta, A., et al. (2010). Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: a proteomic approach. J. Proteome Res. 9, 3443–3464. doi: 10.1021/pr901098p
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Passioura, J. B. (2006). The perils of pot experiments. Funct. Plant Biol. 33, 1075–1079. doi: 10.1071/FP06223
Passioura, J. B. (2012). Phenotyping for drought tolerance in grain crops: when is it useful to breeders? Funct. Plant Biol. 39, 851–859. doi: 10.1071/FP12079
Pechanova, O., Takác, T., Samaj, J., and Pechan, T. (2013). Maize proteomics: an insight into the biology of an important cereal crop. Proteomics 13, 637–662. doi: 10.1002/pmic.201200275
Peng, Z., Wang, M., Li, F., Lv, H., Li, C., and Xia, G. (2009). A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol. Cell. Proteomics 8, 2676–2686. doi: 10.1074/mcp.M900052-MCP200
Perlikowski, D., Kosmala, A., Rapacz, M., Kościelniak, J., Pawłowicz, I., and Zwierzykowski, Z. (2014). Influence of short-term drought conditions and subsequent re-watering on the physiology and proteome of Lolium multiflorum/Festuca arundinacea introgression forms, with contrasting levels of tolerance to long-term drought. Plant Biol. 16, 385–394. doi: 10.1111/plb.12074
Perlikowski, D., Wiśniewska, H., Kaczmarek, J., Góral, T., Ochodzki, P., Kwiatek, M., et al. (2016). Alterations in kernel proteome after infection with Fusarium culmorum in two triticale cultivars with contrasting resistance to Fusarium head blight. Front. Plant Sci. 7:1217. doi: 10.3389/fpls.2016.01217
Poorter, H., Buhler, J., van Dusschoten, D., Climent, J., and Postma, J. A. (2012a). Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct. Plant Biol. 39, 839–850. doi: 10.1071/FP12049
Poorter, H., Fiorani, F., Stitt, M., Schurr, U., Finck, A., Gibon, Y., et al. (2012b). The art of growing plants for experimental purposes: a practical guide for the plant biologist Review. Funct. Plant Biol. 39, 821–838. doi: 10.1071/FP12028
Rampitsch, C., and Bykova, N. V. (2012a). Proteomics and plant disease: advances in combating a major threat to the global food supply. Proteomics 12, 673–690. doi: 10.1002/pmic.201100359
Rampitsch, C., and Bykova, N. V. (2012b). The beginnings of crop phosphoproteomics: exploring early warning systems of stress. Front. Plant Sci. 3:144. doi: 10.3389/fpls.2012.00144
Rasoulnia, A., Bihamta, M. R., Peyghambari, S. A., Alizadeh, H., and Rahnama, A. (2011). Proteomic response of barley leaves to salinity. Mol. Biol. Rep. 38, 5055–5063. doi: 10.1007/s11033-010-0651-8
Riccardi, F., Gazeau, P., Jacquemot, M. P., Vincent, D., and Zivy, M. (2004). Deciphering genetic variations of proteome responses to water deficit in maize leaves. Plant Physiol. Biochem. 42, 1003–1011. doi: 10.1016/j.plaphy.2004.09.009
Rinalducci, S., Egidi, M. G., Karimzadeh, G., Jazii, F. R., and Zolla, L. (2011a). Proteomic analysis of a spring wheat cultivar in response to prolonged cold stress. Electrophoresis 32, 1807–1818. doi: 10.1002/elps.201000663
Rinalducci, S., Egidi, M. G., Mahfoozi, S., Godehkahriz, S. J., and Zolla, L. (2011b). The influence of temperature on plant development in a vernalization-requiring winter wheat: a 2-DE based proteomic investigation. J. Proteomics 74, 643–659. doi: 10.1016/j.jprot.2011.02.005
Rinalducci, S., Murgiano, L., and Zolla, L. (2008). Redox proteomics: basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 59, 3781–3801. doi: 10.1093/jxb/ern252
Rollins, J. A., Habte, E., Templer, S. E., Colby, T., Schmidt, J., and von Korff, M. (2013). Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 64, 3201–3212. doi: 10.1093/jxb/ert158
Romero-Puertas, M. C., Rodríguez-Serrano, M., and Sandalio, L. M. (2013). Protein S-nitrosylation in plants under abiotic stress: an overview. Front. Plant Sci. 4:373. doi: 10.3389/fpls.2013.00373
Ronai, Z., Robinson, R., Rutberg, S., Lazarus, P., and Sardana, M. (1992). Aldolase-DNA interactions in a SEWA cell system. Biochim. Biophys. Acta 1130, 20–28. doi: 10.1016/0167-4781(92)90456-A
Rorat, T. (2006). Plant dehydrins - tissue location, structure and function. Cell. Mol. Biol. Letters 11, 536–556. doi: 10.2478/s11658-006-0044-0
Salekdeh, G. H., and Komatsu, S. (2007). Crop proteomics: aim at sustainable agriculture of tomorrow. Proteomics 7, 2976–2996. doi: 10.1002/pmic.200700181
Sergeant, K., and Renaut, J. (2010). Plant biotic stress and proteomics. Curr. Proteomics 7, 275–297. doi: 10.2174/157016410793611765
Singh, R., and Green, M. R. (1993). Sequence-specific binding of transfer-RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259, 365–368. doi: 10.1126/science.8420004
Skylas, D. J., Cordwell, S. J., Hains, P. G., Larsen, M. R., Basseal, D. J., Walsh, B. J., et al. (2002). Heat shock of wheat during grain filling: proteins associated with heat-tolerance. J. Cereal Sci. 35, 175–188. doi: 10.1006/jcrs.2001.0410
Subba, P., Kumar, R., Gayali, S., Shekhar, S., Parveen, S., Pandey, A., et al. (2013). Characterisation of the nuclear proteome of a dehydration-sensitive cultivar of chickpea and comparative proteomic analysis with a tolerant cultivar. Proteomics 13, 1973–1992. doi: 10.1002/pmic.201200380
Sugimoto, M., and Takeda, K. (2009). Proteomic Analysis of specific proteins in the root of salt-tolerant barley. Biosci. Biotechnol. Biochem. 73, 2762–2765. doi: 10.1271/bbb.90456
Sung, S., and Amasino, R. M. (2005). Remembering winter: toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56, 491–508. doi: 10.1146/annurev.arplant.56.032604.144307
Suzuki, I., Los, D. A., Kanesaki, Y., Mikami, K., and Murata, N. (2000). The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19, 1327–1334. doi: 10.1093/emboj/19.6.1327
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tamburino, R., Vitale, M., Ruggiero, A., Sassi, M., Sannino, L., Arena, S., et al. (2017). Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicum L.). BMC Plant Biol. 17:40. doi: 10.1186/s12870-017-0971-0
Tan, B. C., Lim, Y. S., and Lau, S. E. (2017). Proteomics in commercial crops: an overview. J. Proteomics 169, 176–188. doi: 10.1016/j.jprot.2017.05.018
Tardieu, F. (2012). Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. J. Exp. Bot. 63, 25–31. doi: 10.1093/jxb/err269
Tardieu, F., Granier, C., and Muller, B. (2011). Water deficit and growth. Co-ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 14, 283–289. doi: 10.1016/j.pbi.2011.02.002
Tardieu, F., and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Tardif, G., Kane, N. A., Adam, H., Labrie, L., Major, G., Gulick, P., et al. (2007). Interaction network of proteins associated with abiotic stress response and development in wheat. Plant Mol. Biol. 63, 703–718. doi: 10.1007/s11103-006-9119-6
Taylor, N. L., Heazlewood, J. L., Day, D. A., and Millar, A. H. (2005). Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteomics 4, 1122–1133. doi: 10.1074/mcp.M400210-MCP200
Thompson, J. E., Hopkins, M. T., Taylor, C., and Wang, T. W. (2004). Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 9, 174–179. doi: 10.1016/j.tplants.2004.02.008
Urban, M. O., Vašek, J., Klíma, M., Krtková, J., Kosová, K., Prášil, I. T., et al. (2017). Proteomic and physiological approach reveals drought-induced changes in rapeseeds: water-saver and water-spender strategy. J. Proteomics 152, 188–205. doi: 10.1016/j.jprot.2016.11.004
Vadez, V. (2014). Root hydraulics: the forgotten side of roots in drought adaptation. Field Crops Res. 165, 15–24. doi: 10.1016/j.fcr.2014.03.017
Vadez, V., Kholova, J., Medina, S., Kakkera, A., and Anderberg, H. (2014). Transpiration efficiency: new insights into an old story. J. Exp. Bot. 65, 6141–6153. doi: 10.1093/jxb/eru040
Vadez, V., Kholová, J., Zaman-Allah, M., and Belko, N. (2013). Water: the most important 'molecular' component of water stress tolerance research. Funct. Plant Biol. 40, 1310–1322. doi: 10.1071/FP13149
Vincent, D., Ergül, A., Bohlman, M. C., Tattersall, E. A. R., Tillett, R. L., Wheatley, M. D., et al. (2007). Proteomic analysis reveals differences between Vitis vinifera L. cv. Chardonnay and cv. Cabernet Sauvignon and their responses to water deficit and salinity. J. Exp. Bot. 58, 1873–1892. doi: 10.1093/jxb/erm012
Vincent, D., Lapierre, C., Pollet, B., Cornic, G., Negroni, L., and Zivy, M. (2005). Water deficits affect caffeate O-methyltransferase, lignification, and related enzymes in maize leaves. A proteomic investigation. Plant Physiol. 137, 949–960. doi: 10.1104/pp.104.050815
Vítámvás, P., Kosová, K., Prášilová, P., and Prášil, I. T. (2010). Accumulation of WCS120 protein in wheat cultivars grown at 9 degrees C or 17 degrees C in relation to their winter survival. Plant Breed. 129, 611–616. doi: 10.1111/j.1439-0523.2010.01783.x
Vítámvás, P., Prášil, I. T., Kosová, K., Planchon, S., and Renaut, J. (2012). Analysis of proteome and frost tolerance in chromosome 5A and 5B reciprocal substitution lines between two winter wheats during long-term cold acclimation. Proteomics 12, 68–85. doi: 10.1002/pmic.201000779
Vítámvás, P., Urban, M. O., Škodáček, Z., Kosová, K., Pitelková, I., Vítámvás, J., et al. (2015). Quantitative analysis of proteome extracted from barley crowns grown under different drought conditions. Front. Plant Sci. 6:479. doi: 10.3389/fpls.2015.00479
Wang, C., Zhang, D. W., Wang, Y. C., Zheng, L., and Yang, C. P. (2012). A glycine-rich RNA-binding protein can mediate physiological responses in transgenic plants under salt stress. Mol. Biol. Rep. 39, 1047–1053. doi: 10.1007/s11033-011-0830-2
Wang, M. C., Peng, Z. Y., Li, C. L., Li, F., Liu, C., and Xia, G. M. (2008). Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8, 1470–1489. doi: 10.1002/pmic.200700569
Wang, X., and Komatsu, S. (2016). Plant subcellular proteomics: application for exploring optimal cell function in soybean. J. Proteomics 143, 45–56. doi: 10.1016/j.jprot.2016.01.011
Wang, X., Oh, M. W., and Komatsu, S. (2016). Characterization of S-adenosylmethionine synthetases in soybean under flooding and drought stresses. Biol. Plant. 60, 269–278. doi: 10.1007/s10535-016-0586-6
Wendelboe-Nelson, C., and Morris, P. C. (2012). Proteins linked to drought tolerance revealed by DIGE analysis of drought resistant and susceptible barley varieties. Proteomics 12, 3374–3385. doi: 10.1002/pmic.201200154
Witzel, K., Matros, A., Strickert, M., Kaspar, S., Peukert, M., Mühling, K. H., et al. (2014). Salinity stress in roots of contrasting barley genotypes reveals time-distinct and genotype-specific patterns for defined proteins. Mol. Plant 7, 336–355. doi: 10.1093/mp/sst063
Witzel, K., Weidner, A., Surabhi, G. K., Börner, A., and Mock, H. P. (2009). Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J. Exp. Bot. 60, 3545–3557. doi: 10.1093/jxb/erp198
Wu, X., Gong, F., Cao, D., Hu, X., and Wang, W. (2016). Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics 16, 847–865. doi: 10.1002/pmic.201500301
Xu, J., Li, Y., Sun, J., Du, L., Zhang, Y., Yu, Q., et al. (2013). Comparative physiological and proteomic response to abrupt low temperature stress between two winter wheat cultivars differing in low temperature tolerance. Plant Biol. 15, 292–303. doi: 10.1111/j.1438-8677.2012.00639.x
Yakubov, B., Barazani, O., Shachack, A., Rowland, L. J., Shoseyov, O., and Golan-Goldhirsh, A. (2005). Cloning and expression of a dehydrin-like protein from Pistacia vera L. Trees 19, 224–230. doi: 10.1007/s00468-004-0385-0
Yang, F., Jacobsen, S., Jørgensen, H. J. L., Collinge, D. B., Svensson, B., and Finnie, C. (2013a). Fusarium graminearum and its interactions with cereal heads: studies in the proteomics era. Front. Plant Sci. 4:37. doi: 10.3389/fpls.2013.00037
Yang, F., Melo-Braga, M. N., Larsen, M. R., Jørgensen, H. J. L., and Palmisano, G. (2013b). Battle through signalling between wheat and the fungal pathogen Septoria tritici revealed by proteomics and phosphoproteomics. Mol. Cell. Proteomics 12, 2497–2508. doi: 10.1074/mcp.M113.027532
Yang, Q., Wang, Y., Zhang, J., Shi, W., Qian, C., and Peng, X. (2007). Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7, 737–749. doi: 10.1002/pmic.200600703
Yin, X., and Komatsu, S. (2015). Quantitative proteomics of nuclear phosphoproteins in the root tip of soybean during the initial stages of flooding stress. J. Proteomics 119, 183–195. doi: 10.1016/j.jprot.2015.02.004
Yin, X., and Komatsu, S. (2016). Nuclear proteomics reveals the role of protein synthesis and chromatin structure in root tip of soybean during the initial stage of flooding stress. J. Proteome Res. 15, 2283–2298. doi: 10.1021/acs.jproteome.6b00330
Yin, X., and Komatsu, S. (2017). Comprehensive analysis of response and tolerant mechanisms in early stage soybean at initial-flooding stress. J. Proteomics 169, 225–232. doi: 10.1016/j.jprot.2017.01.014
Zargar, S. M., Mahajan, R., Nazir, M., Nagar, P., Kim, S. T., Rai, V., et al. (2017). Common bean proteomics: Present status and future strategies. J. Proteomics 169, 239–248. doi: 10.1016/j.jprot.2017.03.019
Zhang, H., Han, B., Wang, T., Chen, S., Li, H., Zhang, Y., et al. (2012). Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 11, 49–67. doi: 10.1021/pr200861w
Zhang, M., Li, G., Huang, W., Bi, T., Chen, G., Tang, Z., et al. (2010). Proteomic study of Carissa spinarum in response to combined heat and drought stress. Proteomics 10, 3117–3129. doi: 10.1002/pmic.200900637
Zhang, M., Lv, D., Ge, P., Bian, Y., Chen, G., Zhu, G., et al. (2014). Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteomics 109, 290–308. doi: 10.1016/j.jprot.2014.07.010
Zhou, W., Eudes, F., and Laroche, A. (2006). Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 6, 4599–4609. doi: 10.1002/pmic.200600052
Zhu, J., Alvarez, S., Marsh, E. L., LeNoble, M. E., Cho, I.-J., Sivaguru, M., et al. (2007). Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol. 145, 1533–1548. doi: 10.1104/pp.107.107250
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: stress dynamics, multiple stress treatments, stress-susceptible genotypes, stress-tolerant genotypes, protein isoforms and PTMs, functional studies
Citation: Kosová K, Vítámvás P, Urban MO, Prášil IT and Renaut J (2018) Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 9:122. doi: 10.3389/fpls.2018.00122
Received: 30 October 2017; Accepted: 23 January 2018;
Published: 08 February 2018.
Edited by:
Sebastien Christian Carpentier, Bioversity International (Belgium), BelgiumReviewed by:
Arkadiusz Kosmala, Institute of Plant Genetics (PAN), PolandCopyright © 2018 Kosová, Vítámvás, Urban, Prášil and Renaut. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klára Kosová, a29zb3ZhQHZ1cnYuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.