
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 January 2018
Sec. Plant Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00034
This article is part of the Research Topic Structure and Function of Chloroplasts View all 25 articles
 Min Chen1,2†
Min Chen1,2† Xiao Liu1,2†
Xiao Liu1,2† Shenghui Jiang1,2
Shenghui Jiang1,2 Binbin Wen1,2
Binbin Wen1,2 Chao Yang1,2
Chao Yang1,2 Wei Xiao1,2
Wei Xiao1,2 Xiling Fu1,2
Xiling Fu1,2 Dongmei Li1,2
Dongmei Li1,2 Xiude Chen1,2
Xiude Chen1,2 Dongsheng Gao1,2*
Dongsheng Gao1,2* Ling Li1,2*
Ling Li1,2*Peach is an ideal species for fruit tree research because of its small, fully sequenced genome. Chloroplast development is dependent on the tight cooperation between the nuclear and plastid genomes, and is regulated by GLK transcription factors. In this work, the pigment content was monitored and the chloroplast-to-chromoplast conversion during the fruit ripening was visualized by transmission electron microscopy. Localization and expression analyses showed that PpGLK1 was located in the nucleus and expressed mainly in the leaves and fruit skin. A transcriptome analysis showed that PpGLK1 and its target genes were significantly differentially expressed in ripening peach fruit skin. PpGLK1 silencing affected chlorophyll accumulation in peach leaves and fruits. Overexpression of PpGLK1 rescued the phenotypes of the Arabidopsis Atglk1Atglk2 double mutant and the tomato uniform ripening mutant. The results of a yeast two-hybrid analysis showed that PpGLK1 is autoactivated and that PpGLK1 (301-542 a.a.) interacted with PpARF5. Together, our results indicate that PpGLK1 regulates chloroplast development in green tissues in peach. Therefore, it may be a promising target gene for improving the production and quality of peach by genetic engineering and breeding approaches.
Peach (Prunus persica), an economically important diploid tree crop that originated in China, is one of the most highly genetically characterized deciduous trees in fruit and forest tree research (Verde et al., 2013). Photosynthesis converts light energy into electrochemical energy, and is the source of all organic molecules in plants (Alric and Johnson, 2017); therefore, photosynthesis is a target to increase crop productivity. Leaves are the main photosynthetic organ in plants, but most fruits have partially photosynthetic metabolism before they undergo the transition to truly heterotrophic metabolism. Pavel and Dejong (1993) showed that developing peach fruit provide 5–9% of the fruit photosynthate. Chloroplast development and chlorophyll content can affect the photosynthetic capacity of green tissues (Nadakuduti et al., 2014).
Many studies have been reported in peach. For example, PpYUC11, which participates in auxin biosynthesis, was identified as a candidate gene involved in the regulation of the stony hard phenotype in peach (Pan et al., 2015). Zhou et al. (2015) demonstrated that PpNAC1 can activate PpMYB10.1, a key gene in blood-flesh peach. Cao et al. (2016) described 12 key agronomic traits including flesh color and non-acid fruit in peach. Su et al. (2012) documented the distinctive physiological characteristics of red and green peel during fruit maturation, and Karagiannis et al. (2016) studied the effect of altitude on peach skin quality traits using comparative physiological and proteomic analyses. However, chloroplast development and photosynthesis in green tissues of peach are not fully understood.
Regulating chloroplast development and increasing the photosynthetic capacity are alternative strategies to improve the production and quality of plants. Chloroplasts are not only the sites for photosynthesis but also play important roles in the biosynthesis of many metabolites (Lopez-Juez and Pyke, 2005). In recent years, there has been increasing interest in the genetic analysis of photosynthetic processes and chloroplast development. In tomato, a ripening-related transcription factor ARABIDOPSIS PSEUDO RESPONSE REGULATOR 2-LIKE (SlAPRR2-LIKE), was shown to influence pigmentation, and the overexpression of SlAPPR2-Like produced a more intense green color (Pan et al., 2013). The down-regulation of SlARF4 causes a dark-green fruit phenotype at the pre-ripening stage (Sagar et al., 2013). The high-pigment mutants (hp1, hp2) had higher chlorophyll level in immature fruits than wild-type (Galpaz et al., 2008). The Curl (Cu) mutant, a dominant gain-of-function mutation of TKN2, showed elevated chlorophyll level and increased chloroplast numbers across the entire fruit surface (Nadakuduti et al., 2014). Cytokinin has been shown to affect chloroplast function via modulating photosynthetic performance (Cortleven and Schmülling, 2015) and activating chloroplast-related genes (Cortleven et al., 2016). In addition, inactivation of the Golden 2-like transcription factor SlGLK2 during breeding selection affected fruit chloroplast development, resulting in light green tomato fruit with reduced sugars (Powell et al., 2012).
The GLK transcription factors were first identified in maize, and were subsequently found in Arabidopsis, maize, rice, sorghum, and the moss Physcomitrella patens. To date, land plants have been found contain 1-4 GLKs but no GLK has been found in algal genomes (Wang et al., 2013). The GLK genes contain two conserved domains: a specific GLK/C-terminal box (GCT-box) only in the GLKs of land plants; and a DNA-binding domain (DBD) (Rossini et al., 2001). In Arabidopsis, the two GLKs act redundantly and the double mutant Atglk1Atglk2 exhibited a pale-green phenotype (Rossini et al., 2001; Fitter et al., 2002). In tomato, the two GLKs are functionally equivalent and only SlGLK2 is expressed in fruit, mainly in the green shoulder region. A truncated SlGLK2 caused the loss of the green shoulder (Powell et al., 2012). Overexpression of either SlGLK1 or SlGLK2 led to uniform ripening, resulting in darker green immature fruit, and improved quality of mature fruit (Cheng and Lai, 2013; Nguyen et al., 2014). In pepper, CaGLK2 was shown to regulate chloroplast development throughout the entire fruit (Brand et al., 2014). Kobayashi et al. (2012) demonstrated that the auxin signaling pathway can regulate chloroplast development via GLKs in fruit and roots. BZR1-1D transgenic lines produced fruits with a dark-green shoulder region and asynchronous ripening (the Uniform ripening (U) phenotype) and up-regulated expression of SlGLK2 (Liu et al., 2014). Using chromatin immunoprecipitation analyses, Waters et al. (2009) showed that nuclear photosynthetic genes including Lhcb3, PpLhcb5, Lhcb4.2, Lhcb6, Lhcb2.2, Lhca1, Lhca2, Lhca3, PORA, CHLM, CHLH, PsaK, PsaD, and GUN4 are potential target genes of GLK in Arabidopsis. However, little is known about the roles of GLK in peach.
In this study, the chloroplast-to-chromoplast conversion in ripening peach fruits was observed by transmission electron microscopy (TEM). A transcriptome analysis of the immature to mature epicarp showed that PpGLK1 and its target genes were significantly differentially expressed during fruit ripening. We tested the role of PpGLK1 by virus-induced gene silencing (VIGS) in peach and overexpression in Arabidopsis and tomato. Finally, we confirmed that PpGLK1 interacts with PpARF5 in peach. In conclusion, these results confirm that PpGLK1 regulates chloroplast development in peach. Thus, it is a candidate target gene for improving photosynthesis via genetic engineering or breeding strategies.
Peach trees (Prunus persica L.cv LuYou Tao1) were grown in a greenhouse at Shandong Agriculture University, Tai'an, China. Ten peach fruits from eight different trees were sampled from 19 to 68 DAFB (days after full bloom). They were sampled at approximately 7-day intervals at 10:00 to 11:00. The exocarp was collected, and immediately frozen in liquid N2, and then stored at −80°C. Part of the exocarp was directly used for TEM analysis.
The chlorophyll and total carotenoid contents in peach fruit skin were measured each week during ripening and were calculated as described previously (Lichtenthaler, 1987). To measure the anthocyanin content, 0.5 g fruit skin was ground into a powder in liquid N2, mixed with 5 mL cold methanol containing 0.1% HCl, and then kept at 4°C for 24 h in the dark. After this, a two-buffer assay system was used to determine anthocyanin content (Jin et al., 2016).
Samples including fruit skin, leaves, and siliques were used for the microscopic analyses. All samples were fixed in 3.5% glutaraldehyde and washed with 0.1 M phosphate buffered saline (PBS). The samples were briefly post-fixed in 1% osmium tetroxide and dehydrated in an ascending ethanol series (10–70% ethanol). After this, the samples were subjected to endosmosis, and then imbedded and polymerized in Epon812 resin. Ultra-thin sections were cut using an LKB-V ultramicrotome and stained with uranium acetate and lead citrate. Finally, ultrastructure was examined under a JEOL-1200EX TEM (JEOL, Tokyo, Japan).
The full coding sequence of PpGLK1 without the stop codon was PCR-amplified and inserted into the pPZP211 vector. The primer was shown in Table S1. The plasmids harboring the PpGLK1-GFP and the control GFP were transformed into Agrobacterium tumefaciens strains GV3101 and LBA4404. A. tumefaciens LBA4404 was used to infect onion epidermal cells (Wang et al., 2017). Equal volumes of A. tumefaciens GV3101 and P19 were mixed together and used to infiltrate young tobacco leaves (Jiang et al., 2017). GFP fluorescence was detected under a Zeiss LSM880 microscope (Carl Zeiss, Oberkochen, Germany) and the pictures were analyzed using ZEN lite software (https://www.zeiss.de/corporate/home.html). The subcellular localization assays were repeated more than three times with similar results.
The P. persica genome annotations, downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html), were used as a reference. Bowtie2 v2.2.3 (Langmead and Salzberg, 2012) was used to build the genome index, and clean data were mapped to the reference genome using TopHat v2.0.12. The number of reads for each gene in the samples was counted by HTSeq v0.6.0. The expression levels of the genes in each sample were estimated as Reads Per Kilobase Million Mapped Reads. DEGseq v1.14.0 software was used to identify the DEGs between two biological replicate samples using a model based on negative binomial distribution (Wang et al., 2010). A P-value was assigned to each gene and adjusted by the Benjamini and Hochberg approach to control the false discovery rate. Genes with q ≤ 0.05 and |log2_ratio| ≥ 1 were identified as DEGs.
Total RNA was isolated from 1 g fruit skin using an RNeasy Plus Mini Kit (Qiagen) as the manufacturer's instructions and extracted three times for each sample. Three duplicates of three total RNA were reverse-transcribed for qRT-PCR first-strand cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen) in a total volume of 20 μL. The qRT-PCR mixture consisted of 10 μL SYBR Premix Ex Taq (TaKaRa Biotechnology, Dalian, China), 1 μL cDNA, and 1 μL each primer pair. The qRT-PCR conditions followed the manufacturer's instructions. The primers were designed using Beacon Designer 7 software, and produced 120–200 bp amplicons (see Table S1). The thermal cycling protocol was 10 min at 95°C, 40 cycles of 95°C for 15 s, and 1 min at 58°C for annealing and extension. The specificity was assessed by a melting curve analysis and size estimation of the amplified product. The relative expression levels of DEGs were calculated using the 2−ΔΔt method. All statistic analysis were performed using DPSv7.05.The Actin reference genes Prupe.6G163400 and Prupe. 3G205200 were used to normalize the assayed genes; Prupe.6G163400 was selected because of its stable expression during fruit development (Figure S1). The results shown are the average of three independent biological replicates repeated three times.
A specific cDNA fragment of PpGLK1 was amplified and inserted into pTRV2. pTRV2-PpGLK1, pTRV2, and pTRV1 were individually transformed into A. tumefaciens strain GV3101 for the VIGS experiments. The primer was shown in Table S1. The A. tumefaciens GV3101 lines containing pTRV2-PpGLK1, pTRV2, and pTRV1 were selected, incubated, resuspended to an OD600 of 0.8 in infiltration buffer (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone), and then kept at room temperature without shaking for 2 h. Considering the firmness of immature peach (63 DAFB), the pTRV2-PpGLK1 mixture (pTRV1:pTRV2-PpGLK1 = 1:1, v/v) and the control mixture (pTRV1:pTRV2 = 1:1, v/v) were gently vacuum-infiltrated into peach leaves and fruits for 30 min and 1 h, respectively. The infiltrated leaves were kept in the dark at 21°C overnight and then transferred to a light growth chamber. Peach leaves and fruits were sampled and photographed at 3 and 7 d after infiltration, respectively.
PpGLK1 was introduced into the pRI101-AN vector containing a GFP tag sequence to form 35S:: PpGLK1-GFP (Wang et al., 2017). The recombinant plasmid was transformed into A. tumefaciens LBA4404 and GV3101. The A. tumefaciens cells were then used to transform the vector into Arabidopsis (GV3101) and tomato cotyledons (LBA4404) (Fillatti et al., 1987). In Arabidopsis, T1 transgenic seedlings were isolated by selection on MS solid medium containing kanamycin and then grown in a light incubator. Seeds of the T3 generation were collected for later use. Transgenic tomato plants were selected on MS solid medium containing kanamycin. Tomato seedlings were grown in a light growth chamber (16-h light/8-h dark, 27°C/19°C). Three lines were verified as transgenic plants. Fruits of the selected plants were used for further analyses.
A Yeast Two-Hybrid Library was constructed using peach fruit skin. Cloning and testing bait for autoactivation and toxicity, and two-hybrid screening using yeast mating were performed according to the Matchmaker® Gold Yeast Two-Hybrid System User Manual (Clontech, Palo Alto, CA, USA). The domain-deleted form (301-542 a.a.) of PpGLK1, recombined into the pGBKT7 vector, was used for screening and the Y2H assay. The CDS of PpARF5 was cloned into the pGADT7 vector (Clontech). These two recombinant plasmids were co-transformed into Y2H Gold yeast cells, and then the cells cultured on selective medium lacking Trp and Leu (-T/-L) at 30°C. The putative transformants were transferred to selective medium lacking Trp, Leu, His, and adenine (-Leu/-Trp/-His/-Ade) with or without X-α-gal.
The CDS without stop codon of PpGLK1 was cloned to the 35S-pSPYNE-YFP vector and the CDS without stop codon of PpARF5 was cloned to the 35S-pSPYCE-YFP vector. These two recombinant plasmids were co-transformed into A. tumefaciens LBA4404. The mixture (35S-PpGLK1-pSPYNE:35S-PpARF5-pSPYCE = 1:1, v/v) and the control (35S-pSPYNE:35S-pSPYCE = 1:1, v/v) were used to infect onion epidermal cells for 25–30 min. Then, the onion epidermal cells were cultured on MS solid medium for 24–48 h at 28°C. Finally, YFP fluorescence was detected at an excitation wavelength of 488 nm under a confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany).
The pull-down assay was conducted as described by Wang et al. (2017). The PpGLK1 coding sequence was combined into the pET-32a (+) vector (Novagen, Madison, WI, USA) for His-tag fusion, and PpARF5 was cloned into the pGEX-4T-1 vector containing a GST-tag sequence. Then, both recombinant plasmids were individually transformed into Escherichia coli BL21 (DE3) to induce tag proteins. Subsequently, His-PpGLK1 was incubated with GST-PpARF5 or GST. After immunoprecipitation with an anti-His column, the pellet fraction was detected via immunoblotting using an anti-GST antibody.
The fruit skin of peach fruits was analyzed at eight stages of growth to monitor the changes in pigment during fruit development. During fruit development, the chlorophyll and total carotenoid concentration decreased from 40 days after full bloom (DAFB) (Figures 1A,B). However, the anthocyanin content significantly increased, and was especially rapid from 61 DAFB onwards (Figure 1C). Then, we observed the plastids in peach fruit skin by transmission electron microscopy (TEM) at 19, 26, 33, 47, 61, and 68 DAFB. The chloroplasts in the fruit skin changed into chromoplasts as the fruit color changed (Figure 1D). Elongated chloroplasts with a clear inner structure were present in peach fruit skin at 19, 26, and 33 DAFB. Generally, these chloroplasts showed a typical arrangement of grana layers, starch grains, and parallel stromal thylakoids. Fruit coloration in peach occurred at 47 DAFB, and the chloroplast-to-chromoplast conversion caused ultrastructural changes because of the appearance of an elliptical plastid with disordered parallel stromal thylakoids and disrupted granal thylakoids. The plastoglobules were obsedved and the chloroplasts had changed into chromoplasts at 68 DAFB. Together, these results illustrated the conversion of chloroplasts into chromoplasts during peach fruit development.
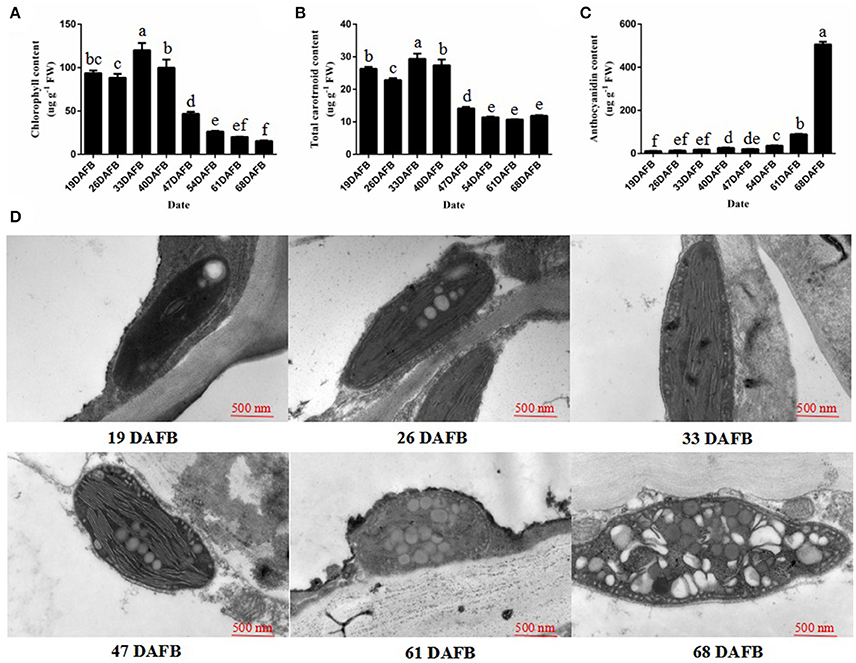
Figure 1. Chlorophyll, the total carotrnoid, anthocyanin contents, and chloroplast development during fruit ripening. (A) Chlorophyll content in peach skin during fruit development. (B) Total carotenoid content in peach skin during fruit development. (C) Anthocyanin content in peach skin during fruit development. (D) Transmission electron micrographs of chloroplast development in peach skin during fruit ripening. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
The peach genome contains only one GLK gene, so we named it PpGLK1. Sequence analysis showed that PpGLK1 contained 542 amino acids with a putative molecular weight of 59.17 kDa and a theoretical isoelectric point of 6.06. A multiple sequence alignment analysis demonstrated that PpGLK1 had a TEA DNA-binding domain and specific GCT-box at the C-terminal (Figure 2A).
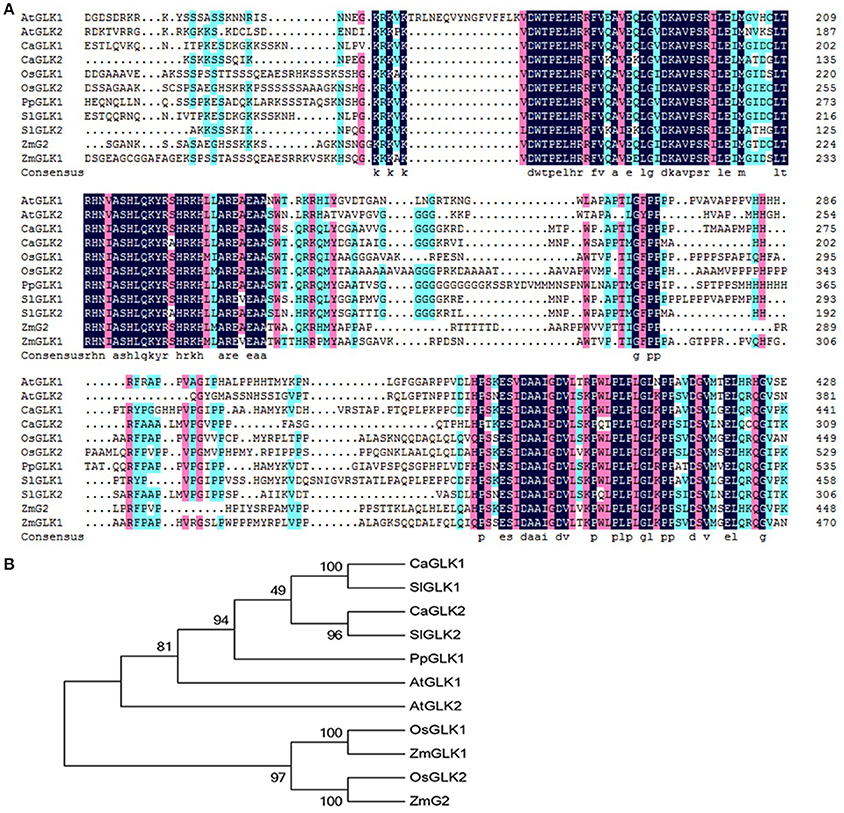
Figure 2. Analysis of GLKs in different species including Arabidopsis, tomato, pepper, rice, maize, and peach. (A) Multiple alignment of GLK amino acid sequences from Arabidopsis, tomato, pepper, rice, maize, and peach. (AtGLK1: AT2G20570; AtGLK2: AT5G44190; SlGLK1: Solyc07g053630.2.1; SlGLK2: Solyc10g008160.2.1; CaGLK1: LOC107878377; CaGLK2: LOC107845460; OsGLK1: LOC4340977; OsGLK2: LOC4326363; ZmGLK1: GRMZM2G026833; ZmG2: GRMZM2G087804; PpGLK1: Prupe.3G127700). (B) Phylogenetic tree analysis of GLKs from Arabidopsis, tomato, pepper, rice, maize, and peach. Phylogenetic tree was built using MEGA5.1 with the neighbor-joining method. Data of sequences used for phylogenetic analysis is provided in Table S4.
We constructed a phylogenetic tree using the GLK amino acid sequences of Arabidopsis thaliana, Oryza sativa, Capsicum annuum, Solanum lycopersicum, and Zea mays (Figure 2B). In the phylogenetic tree, PpGLK1 was most closely related to AtGLK1 and SlGLK2. Generally, orthologs that cluster together are likely to have similar functions (Tatusov et al., 1997).
The subcellular localization of PpGLK1 was predicted as the nucleus by WoLFPSORT (http://www.genscript.com/tools/wolf-psort). To verify the accuracy of this prediction, a fusion protein PpGLK1-GFP and a control protein GFP driven by the CaMV 35S promoter were generated and then transiently expressed in tobacco leaves and onion epidermal cells. Confocal imaging showed that the fluorescence of the control was diffuse throughout the whole cell in the tobacco leaves (Figure 3A) and the onion epidermal cells (Figure 3B). The fluorescence of PpGLK1-GFP was detected exclusively in the nucleus (Figures 3A,B), confirming the nuclear localization of PpGLK1. Next, the transcript levels of PpGLK1 were measured in various tissues (root, stem, flower, leaf, fruit skin, fruit flesh, and seed) by qRT-PCR. There were high transcript levels of PpGLK1 in the leaf and fruit skin, similar to the pattern of SlGLK1 expression (Figure 3C).
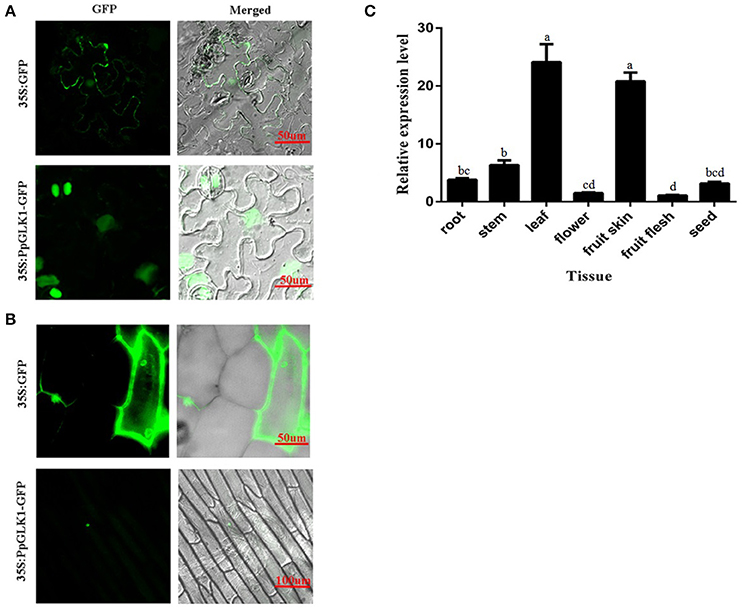
Figure 3. Subcellular localization and tissue-specific expression of PpGLK1. (A) Subcellular localization of PpGLK1 in tobacco lower epidermal cells. Fluorescence, and merged images are shown (left to right). (B) Subcellular localization of PpGLK1 in onion epidermal cells. (C) Expression patterns of PpGLK1 in different tissues including root, stem, leaf, flower, fruit skin, flesh, seed. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
To study the molecular basis of chloroplast development in peach fruit skin, we performed a transcriptome analysis of the epicarp at immature to mature stages. Six cDNA libraries were constructed using mRNA from 19 DAFB and 68 DAFB fruit skin samples, with three biological replicates (GenBank Accession number: SRX3469636 and SRX3469635). Significant differentially expressed genes (DEGs) were identified based on their | log2Ratio | ≥ 1 and q < 0.05 threshold. A Gene Ontology (GO) term analysis showed that large proportions DEGs were related to photosynthesis, light harvesting, chloroplast, and chlorophyll binding (Table S2). Importantly, the GLK transcription factor and its target genes such as PpLhcb1.3, PpLhcb2.2, PpLhca3, PpHEMA, PpGUN4 all showed significant differences in expression during ripening, consistent with the changes in chlorophyll content. When gene transcript levels were determined using an alternative methodology, qRT-PCR, the results were consistent with those of the transcriptome analysis (Figure 4). The transcript levels of PpGLK1 and its target genes decreased during fruit development. Based on these results, we concluded that PpGLK1 may play an important role in chlorophyll biosynthesis and chloroplast development.
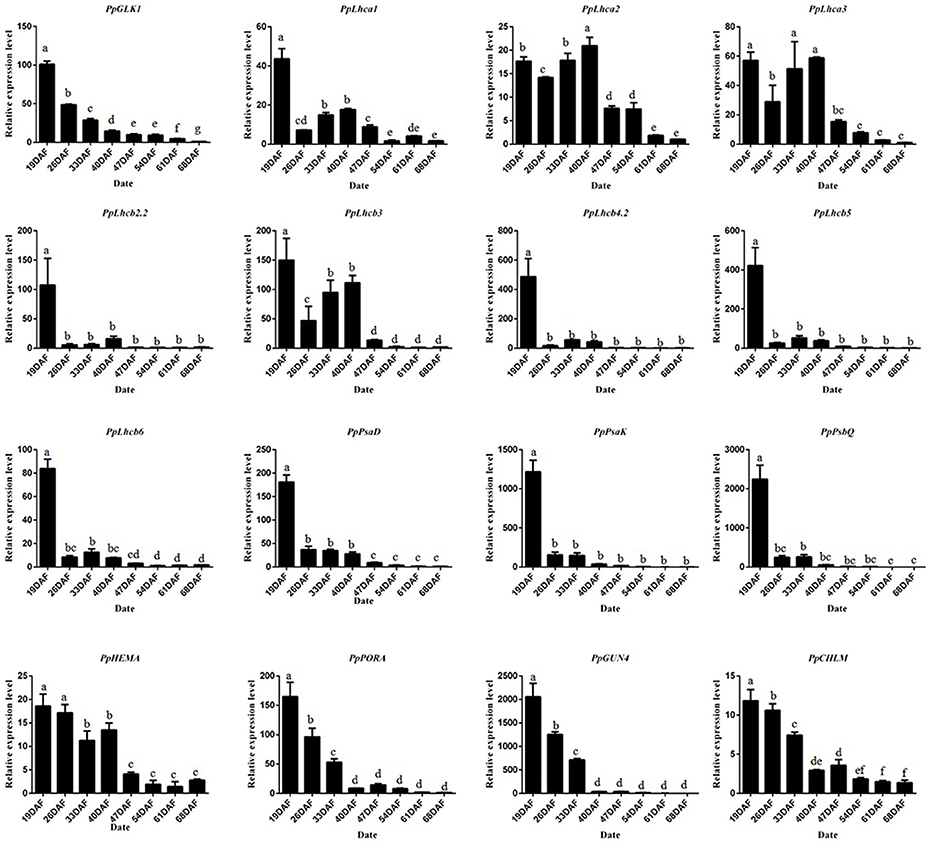
Figure 4. Expression analysis of PpGLK1 and its target genes in fruit skin of 19–68 DAFB. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
Next, we used VIGS to eliminate PpGLK1 expression in peach. We selected a specific cDNA fragment of the PpGLK1 gene and introduced it into the pTRV2 vector to produce pTRV2-PpGLK1. pTRV2 was used as the control. Both vectors were infiltrated into peach fruit and leaves along with pTRV1 for VIGS. Mature leaves and immature fruit of peach were selected to validate the effect of PpGLK1 silencing. In mature leaves, the infiltration sites on the leaves became pale green by 3 days after transformation with pTRV1 and pTRV2-PpGLK1, whereas no clear phenotype was observed after transformation with the control vector (Figure 5A). Further analyses showed that chlorophyll content at the infiltration sites of pTRV1 and pTRV2-PpGLK1 was 0.73 mg g−1, much lower than that at sites infiltrated with the control vector (1.41 mg g−1; Figure 5B). The chlorophyll content was 1.52 mg g−1 at non-infiltrated sites. At 3 days after transformation, the transcript level of PpGLK1 at sites infiltrated with pTRV1 and pTRV2-PpGLK1 was 91.20% lower than that at non-infiltrated sites (Figure 5C).
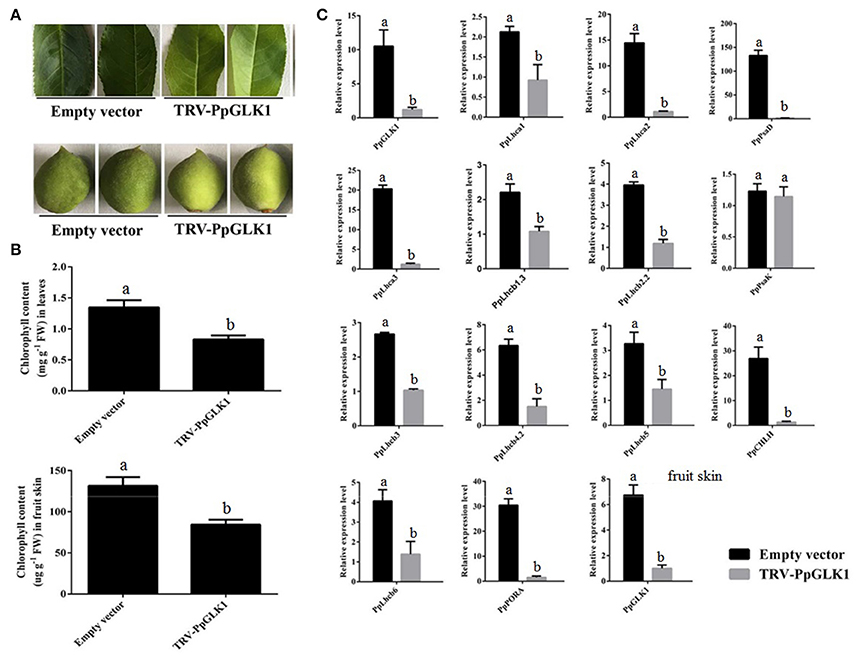
Figure 5. Pale-green phenotype of peach after PpGLK1 silencing. (A) Phenotypes of leaves and fruits of PpGLK1-silenced peach. (B) Chlorophyll contents in leaves and fruits of PpGLK1-silenced and control peach. (C) Transcript levels of PpGLK1 and its target genes in leaves and fruits of PpGLK1-silenced and control peach. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
Waters et al. (2009) demonstrated that GLK can bind to the promoter of many nuclear photosynthetic genes including Lhcb3, Lhcb5, Lhcb4.2, PpLhcb6, Lhcb2.2, Lhcb1.3, Lhca1, Lhca2, Lhca3, PORA, CHLM, CHLH, PsaK, PsaD, and GUN4. The transcript levels of these target genes were clearly decreased at sites infiltrated with pTRV1 and pTRV2-PpGLK1 (Figure 5C). There were no significant differences in the transcript levels of each of these genes between sites infiltrated with the control vector and uninfected leaves. In the immature fruit, infiltration sites were paler than the control by 7 days after transformation with pTRV1 and pTRV2-PpGLK1 (Figure 5A) and the expression of PpGLK1 and its target genes were down-regulated at the infiltrated sites (Figure S2). Measurements of chlorophyll content in peach fruit peel confirmed a decrease in chlorophyll content around the infiltrated sites (Figure 5B). Thus, silencing of PpGLK1 affected the expression of its target genes and chlorophyll content in peach. Together, these results showed that PpGLK1 can positively regulate chlorophyll accumulation in peach fruit peel.
The GLK transcription factors have been shown to regulate chloroplast development and formation of the photosynthetic apparatus in Arabidopsis (Waters et al., 2009). To further verify the function of PpGLK1, a CaMV-35S promoter (35S::PpGLK1) vector was constructed and heterologously transformed into the Arabidopsis glk1glk2 double mutant. Transgenic plants were identified by western blotting with GFP antibody (Figure 6A). Introduction of 35S::PpGLK1 recovered the glk1glk2 double mutant pale-green phenotype (Figure 6B); the chlorophyll content in 35S::PpGLK1 Arabidopsis was approximately 3.5 times that in glk1glk2 (Figure 6C). The TEM analyses showed that there was a general increase in the number of granal thylakoids (8.9) in PpGLK1-OE lines, compared with glk1glk2 leaves (1.5). The starch grains in the chloroplasts of PpGLK1-OE siliques were significantly larger than those of Arabidopsis glk1glk2 mutant (Figure 6D). These results proved that PpGLK1 could complement the chlorophyll biosynthesis phenotype of the glk1glk2 double mutant. This complementary analysis showed that PpGLK1 maintains its function to control chloroplast development in Arabidopsis, like in peach.
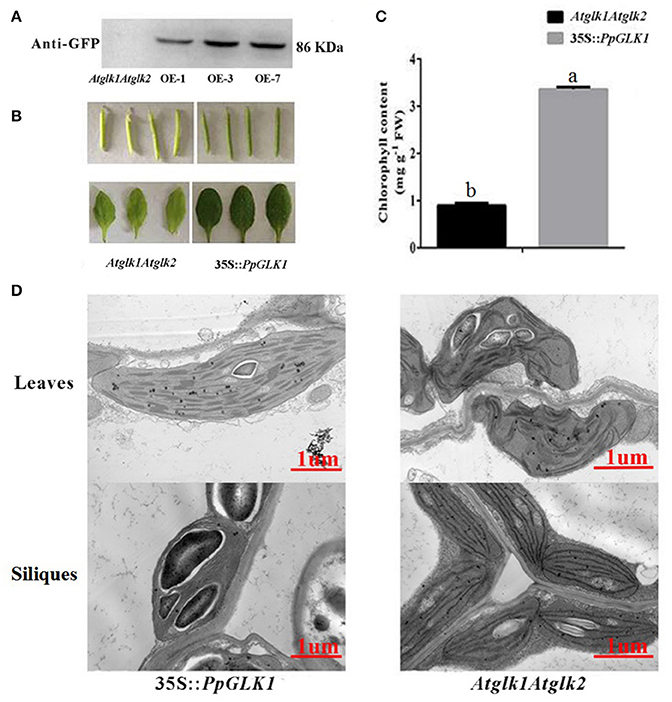
Figure 6. Overexpression of PpGLK1 in Arabidopsis double mutant Atglk1Atglk2. (A) Western blot analysis confirming PpGLK1 overexpression in Atglk1Atglk2. (B) Phenotype of PpGLK1-overexpressing Atglk1Atglk2. (C) Chlorophyll contents in Atglk1Atglk2 overexpressing PpGLK1 and Atglk1Atglk2. (D) Chloroplast development in siliques and leaves observed by TEM in Atglk1Atglk2 overexpressing PpGLK1 and Atglk1Atglk2. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
As reported by Powell et al. (2012), uniform ripening (u) encodes a truncated Slglk2 that affects fruit chloroplast development in tomato. Micro-Tom, which is homozygous for the u mutation (u/u), was selected for further experiments (Nadakuduti et al., 2014). Similar to PpGLK1-OE in Arabidopsis, Micro-Tom lines overexpressing PpGLK1-OE were selected by qRT-PCR and western analysis (Figure 7A). The Micro-Tom PpGLK1-OE lines produced uniform dark-green unripe fruit (Figure 7B). Chlorophyll content was elevated by 135-fold (Figure 7C) compared with that in untransformed Micro-Tom (u/u). The number and size of chloroplasts were increased and the number of thylakoids per granum was also greater in Micro-Tom PpGLK1-OE fruit than in u/u fruit (Figure 7D). Interestingly, the transcript levels of GLK target genes were also up-regulated in Micro-Tom PpGLK1-OE (Figure 8). Thus, the phenotypic characterization of PpGLK1-OE in u/u tomato revealed that PpGLK1 is associated with chloroplast development and chlorophyll biosynthesis in tomato, like in peach.
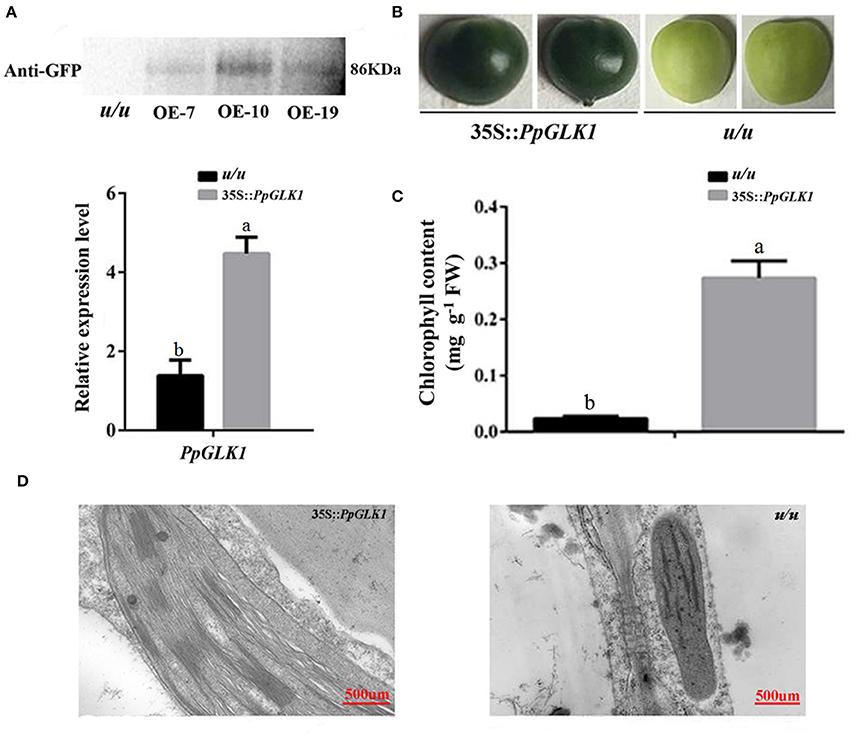
Figure 7. Overexpression of PpGLK1 in u/u tomato. (A) qRT-PCR and western blot analysis confirming PpGLK1 overexpression in u/u tomato. (B) Phenotype of PpGLK1-overexpressing u/u tomato. (C) Chlorophyll contents in u/u tomato and PpGLK1-overexpressing u/u tomato. (D) Chloroplast development in fruit observed by TEM in PpGLK1-overexpressing u/u tomato fruit and u/u tomato fruit. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
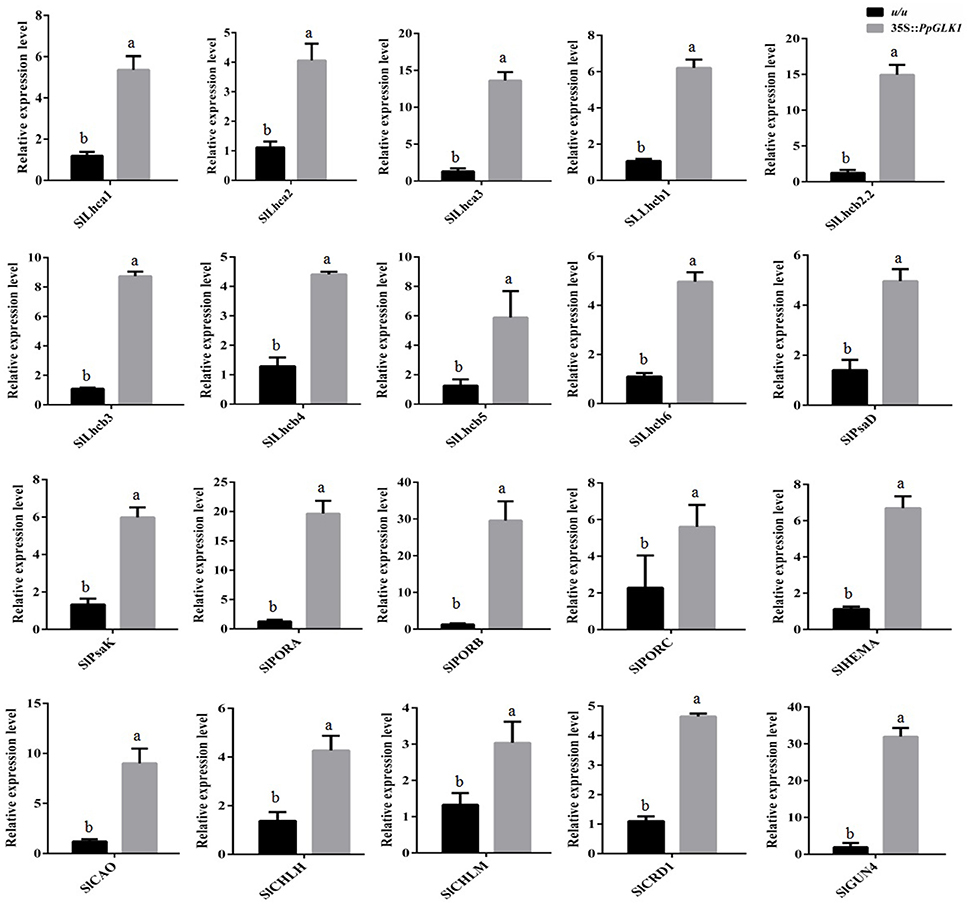
Figure 8. Expression patterns of PpGLK1 target genes in PpGLK1-overexpressing u/u tomato fruit. Data is presented as the means ± SD, n = 3. Different letters above bars denote statistical significance according to one-way ANOVA and Duncan's test (P < 0.05).
To test bait for autoactivation and toxicity, the full-length PpGLK1 coding region fused to pGBKT7 was generated and transformed into competent Y2HGold cells; pGBKT7-53 served as the positive control. After selection on SD/-Trp, SD/-Trp/ X-a-Gal and SD/-Trp/X-a-Gal/AbA medium, we found that the expression of our bait protein was not toxic in yeast and that the PpGLK1 protein autoactivated (Figure 9A and Figure S3). To select the fragment for yeast two-hybrid (Y2H) screening, the PpGLK1 protein was cut into two fragments: 1-300 a.a. and 301-542 a.a. The 301-542 a.a. fragment did not autoactivate and was used as bait for Y2H screening (Figure 9A). In the Y2H screening, several positive colonies harbored the PpARF5 protein, which is homolog of AtARF5. In a Y2H assay, PpGLK1 (301–542) interacted with the full-length PpARF5 protein (Figure 9B and Figure S3). When this interaction was tested in a bimolecular fluorescence complementation (BiFC) analysis, PpGLK1 interacted with the full-length PpARF5 protein in vivo (Figure 9C). A pull-down assay confirmed the interaction between PpGLK1 and PpARF5 in vitro (Figure 9D).
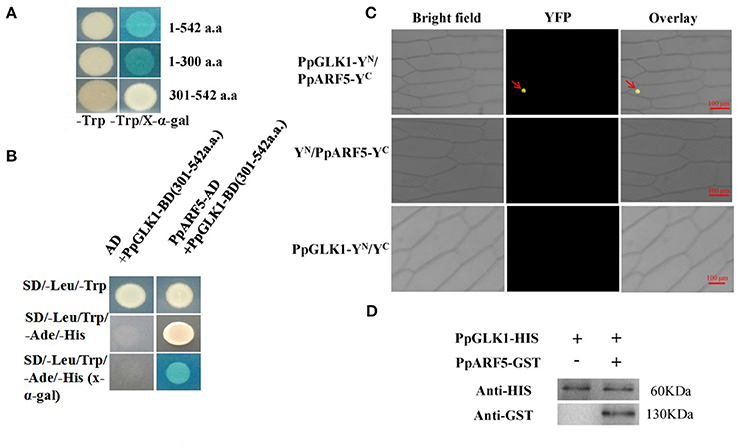
Figure 9. Interaction between PpGLK1 and PpARF5. (A) Autoactivation of PpGLK1 in yeast cells. (B) Interaction between PpGLK1 and PpARF5 in Y2H assay. (C) Interaction between PpGLK1 and PpARF5 in BiFC assay. (D) Interaction between PpGLK1 and PpARF5 in Vitro in pull-down assay.
Although many genes have been reported to regulate chloroplast development, only GLKs control a large number of photosynthesis-related genes (Waters et al., 2009). Thus, it is critical to elucidate the function and effects of PpGLK1 in peach. In this study, our results showed that PpGLK1 regulates chloroplast development in peach, leading to chlorophyll accumulation. These results indicate that PpGLK1 is a good target for improvements in fruit quality and production via breeding or transgenic approaches.
The transition from chloroplasts into chromoplasts has been documented for ripening goji berries (Hempel et al., 2017), and was shown to be induced by fruit shading in grapefruit (Lado et al., 2015). Yang et al. (2015) demonstrated that chlorophyll content was significantly lower in a rubescent leaf color mutant of Anthurium andraeanum than in the wild type, and analyses of the micro- and ultra-structures in the chloroplasts showed that a chloroplast-to-chromoplast transition occurred in the leaf color mutants (Cheung et al., 1993; Yang et al., 2015). A comparative proteomic analysis showed that the proteins involved in the light reactions of photosynthesis decreased in abundance during the chloroplast-to-chromoplast transition in tomato (Barsan et al., 2012). In our study, the chlorophyll and the total carotrnoid content decreased during fruit development and analyses of ultra-structural changes in chloroplasts revealed the details of the chloroplast-to-chromoplast transition. This transition has also been documented in tomato (Harris and Spurr, 1969) and bell pepper (Spurr and Harris, 1968).
Transcriptomic profiling can reveal the patterns of gene expression over time and in different parts of the plant (Wang et al., 2009). In the present study, we added a transcriptome analysis to reveal the molecular foundation of the chloroplast-to-chromoplast transition. Interestingly, the transcript levels of PpGLK1 and its target genes decreased during the fruit ripening in peach. In Arabidopsis and tomato, homologs of PpGLK1 have been shown play an important role in chloroplast development (Waters et al., 2009; Powell et al., 2012). In addition, mutants of GLK target genes (including CAO and CHLM) exhibited pale-green phenotypes. However, whether PpGLK1 also functions in chloroplast development in peach was yet to be confirmed. The GLK genes are members of the GARP family (Riechmann et al., 2000) which contains 35 genes in peach. Rossini et al. (2001) demonstrated that GLK genes contain two highly conserved domains. Based on these two conserved domains, we searched for and found only one GLK gene in peach. One explanation for this result may be that ancestral triplicated blocks are fragmentary and a recent whole-genome duplication event has not occurred in peach (Verde et al., 2013). In the phylogenetic analysis, PpGLK1 was most closely related to AtGLK1 and SlGLK2 (Figure 2B) and the multiple sequence alignment showed that they have the same functional domains (Figure 2A). All these results suggested that PpGLK1 may have the same function in peach, tomato, and Arabidopsis. Overexpression of PpGLK1 complemented the double mutant Atglk1Atglk2 (Arabidopsis) and u/u (tomato) (Figures 6B, 7B). In addition, the PpGLK1-complemented mutants showed increased expression of chlorophyll-related genes and increased chlorophyll content. These results provided further evidence PpGLK1 has the same function as AtGLK1 and SlGLK2, which are known to be involved in chloroplast development and chlorophyll biosynthesis. Improving the photosynthetic capacity can lead to increased carbohydrate and carotenoids contents in mature fruit (Powell et al., 2012).
The VIGS system, which is composed of TRV1 and TRV2, is a powerful tool for gene functional characterization in vivo (Sun et al., 2016). It has been successfully used to investigate gene function in the leaves and fruits of Pyrus betulaefolia (Li et al., 2017), apple (Hu et al., 2016; Jiang et al., 2017), pear (Zhai et al., 2016), and peach (Zhou et al., 2015). The Arabidopsis double mutant Atglk1Atglk2 exhibited a pale-green phenotype and the tomato double mutant u/u produced uniformly light-green unripe fruit. In peach, silencing of PpGLK1 decreased the chlorophyll content and led to pale-green fruit and leaves (Figures 5A,B) suggesting that GLK regulates chloroplast development in peach leaves and fruits. On the basis of our results, we concluded that PpGLK1 is positive regulator of chloroplast development like GLKs in other species.
Complex hormonal signals activate the transcription factors that regulate chloroplast development-related genes (Nakamura et al., 2009). Brassinosteroids regulate plant growth and development through BES1 and BZR1 transcription factors. The bes1-D mutant exhibited a pale-green color and exhibited a reduced chlorophyll content. Another BES1 mutant with repressed GLK1 and GLK2 expression showed inhibited chloroplast development (Yu et al., 2011). Chloroplast biogenesis in greening roots was shown to depend on the combination of HY5 and GLKs, which function downstream of light and auxin/cytokinin signaling pathways (Kobayashi et al., 2012). GLK proteins need partners to bind DNA (Waters et al., 2009). Tamai et al. (2002) demonstrated that AtGLK1 and AtGLK2 transactivate transcription in yeast and can interact with GBF1. In addition, AtGLK1 was shown to interact with GBF3. Our results showed that PpGLK1 can autoactivate, like AtGLK1 and AtGLK2 (Figure 9A). The results of the Y2H, pull-down, and BiFC assays all confirmed the interaction between PpGLK1 and PpARF5 (Figures 9B–D). The coordinated actions of ARF and Aux/IAA transcriptional repressors play a foremost role in auxin action. Sagar et al. (2013) showed that auxin can repress the expression of GLK2, and that down-regulation of SlARF4 resulted in a dark-green fruit phenotype. It is noteworthy that the promoter of PpGLK1 contained an auxin-responsive element, the AACGAC box (Table S3). The coordinated action of ARF and Aux/IAA transcriptional repressors play a major role in auxin action. ARF5/MP broadly regulates the expression of Aux/IAA genes in distinct subclades and is under negative feedback control by Aux/IAA genes (Krogan et al., 2014). Together, the results of this study provide further evidence for a link between auxin signaling and chloroplast development.
MC, DG, and LL designed the research. MC and XL performed the experiments. MC, SJ, and XL analyzed the data. LL, DL, XC, DG, and XF contributed new models. MC, SJ wrote the manuscript. LL, CY revised the intellectual content of this manuscript. All authors read and approved the final manuscript.
This study was funded by the National Natural Science Foundation of China (Grant number 31372050 and 31672137) and Funds of Shandong “Double Tops” Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00034/full#supplementary-material
Figure S1. The Cq of the Actin genes (Prupe.6G163400 and Prupe. 3G205200).
Figure S2. Transcript levels of PpGLK1 target genes in fruit skin of PpGLK1-silenced and control peach.
Figure S3. The original figures of Figures 9A,B.
Table S1. The primers used in this article.
Table S2. GO term enrichment analysis of DEGs.
Table S3. The analysis of PpGLK1 promoter.
Table S4. Data of sequences used for phylogenetic analysis.
Alric, J., and Johnson, X. (2017). Alternative electron transport pathways in photosynthesis: a confluence of regulation. Curr. Opin. Plant Biol. 37, 78–86. doi: 10.1016/j.pbi.2017.03.014
Barsan, C., Zouine, M., Maza, E., Bian, W., Egea, I., Rossignol, M., et al. (2012). Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol. 160, 708–725. doi: 10.1104/pp.112.203679
Brand, A., Borovsky, Y., Hill, T., Rahman, K. A. A., Bellalou, A., Deynze, A. V., et al. (2014). CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 127, 2139–2148. doi: 10.1007/s00122-014-2367-y
Cao, K., Zhou, Z., Wang, Q., Guo, J., Zhao, P., Zhu, G., et al. (2016). Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 7:13246. doi: 10.1038/ncomms13246
Cheng, L., and Lai, K. (2013). Golden2- Like (GLK2) Transcription Factor: Developmental Control of Tomato Fruit Photosynthesis and Its Contribution to Ripe Fruit Characteristics. M.S. thesis, University of California, Davis, CA.
Cheung, A. Y., McNellis, T., and Piekos, B. (1993). Maintenance of chloroplast components during chromoplast differentiation in the tomato mutant Green Flesh. Plant Physiol. 101, 1223–1229. doi: 10.1104/pp.101.4.1223
Cortleven, A., Marg, I., Yamburenko, M. V., Schlicke, H., Hill, K., Grimm, B., et al. (2016). Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 172, 464–478. doi: 10.1104/pp.16.00640
Cortleven, A., and Schmülling, T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. doi: 10.1093/jxb/erv132
Fillatti, J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. doi: 10.1038/nbt0787-726
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., and Langdale, J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1046/j.1365-313X.2002.01390.x
Galpaz, N., Wang, Q., Menda, N., Zamir, D., and Hirschberg, J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730. doi: 10.1111/j.1365-313X.2007.03362.x
Harris, W. M., and Spurr, A. R. (1969). Chromoplasts of tomato fruit. II. the red tomato. Am. J. Bot. 56, 380–389. doi: 10.2307/2440813
Hempel, J., Schädle, C. N., Sprenger, J., Heller, A., Carle, R., and Schweiggert, R. M. (2017). Ultrastructural deposition forms and bioaccessibility of carotenoids and carotenoid esters from goji berries (Lycium barbarum L.). Food Chem. 218, 525–533. doi: 10.1016/j.foodchem.2016.09.065
Hu, D. G., Sun, C. H., Zhang, Q. Y., An, J. P., You, C. X., and Hao, Y. J. (2016). Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 12:e1006273. doi: 10.1371/journal.pgen.1006273
Jiang, Y., Liu, C., Yan, D., Wen, X., Liu, Y., Wang, H., et al. (2017). MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 68, 1055–1069. doi: 10.1093/jxb/erx029
Jin, W., Wang, H., Li, M., Wang, J., Yang, Y., Zhang, X., et al. (2016). The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. doi: 10.1111/pbi.12568
Karagiannis, E., Tanou, G., Samiotaki, M., Michailidis, M., Diamantidis, G. S., Minas, I., et al. (2016). Comparative physiological and proteomic analysis reveal distinct regulation of peach skin quality traits by altitude. Front. Plant Sci. 7:1689. doi: 10.3389/fpls.2016.01689
Kobayashi, K., Baba, S., Obayashi, T., Sato, M., Toyooka, K., Keränen, M., et al. (2012). Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24, 1081–1095. doi: 10.1105/tpc.111.092254
Krogan, N. T., Yin, X., Ckurshumova, W., and Berleth, T. (2014). Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. New Phytol. 204, 474–483. doi: 10.1111/nph.12994
Lado, J., Cronje, P., Alquézar, B., Page, A., Manzi, M., Gómez-Cadenas, A., et al. (2015). Fruit shading enhances peel color, carotenes accumulation and chromoplast differentiation in red grapefruit. Physiol. Plant. 154, 469–484. doi: 10.1111/ppl.12332
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, K., Xing, C., Yao, Z., and Huang, X. (2017). PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 15, 1186–1203. doi: 10.1111/pbi.12708
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Liu, L., Jia, C., Zhang, M., Chen, D., Chen, S., Guo, R., et al. (2014). Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol. J. 12, 105–115. doi: 10.1111/pbi.12121
Lopez-Juez, E., and Pyke, K. A. (2005). Plastids unleashed: their development and their integration in plant development. Int. J. Dev. Biol. 49, 557–577. doi: 10.1387/ijdb.051997el
Nadakuduti, S. S., Holdsworth, W. L., Klein, C. L., and Barry, C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78, 1022–1033. doi: 10.1111/tpj.12529
Nakamura, H., Hakata, M., Ueno, O., Nagamura, Y., Hirochika, H., Takano, M., et al. (2009). Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 50, 1933–1949. doi: 10.1093/pcp/pcp138
Nguyen, C. V., Vrebalov, J. T., Gapper, N. E., Zheng, Y., Zhong, S., Fei, Z., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26, 585–601. doi: 10.1105/tpc.113.118794
Pan, L., Zeng, W., Niu, L., Lu, Z., Lu, H., Cui, G., et al. (2015). PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 66, 7031–7044. doi: 10.1093/jxb/erv400
Pan, Y., Bradley, G., Pyke, K., Ball, G., Lu, C., Fray, R., et al. (2013). Network inference analysis identifies an APRR2-Like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161, 1476–1485. doi: 10.1104/pp.112.212654
Pavel, E. W., and Dejong, T. M. (1993). Source-and sink-limited growth periods of developing peach fruits indicated by relative growth rate analysis. J. Am. Soc. Hortic. Sci. 6, 820–824.
Powell, A. L., Nguyen, C. V., Hill, T., Cheng, K. L., Figueroa-Balderas, R., Aktas, H., et al. (2012). Uniform ripening encodes a Golden2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715. doi: 10.1126/science.1222218
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rossini, L., Cribb, L., Martin, D. J., and Langdale, J. A. (2001). The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13, 1231–1244. doi: 10.1105/tpc.13.5.1231
Sagar, M., Chervin, C., Mila, I., Hao, Y., Roustan, J. P., Benichou, M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374. doi: 10.1104/pp.113.213843
Spurr, A. R., and Harris, W. M. (1968). Ultrastructure of chloroplasts and chromoplasts in Capsicum annuum. I. thylakoid membrane changes during fruit ripening. Am. J. Bot. 55, 1210–1224. doi: 10.2307/2440743
Su, M., Chen, K., Ye, Z., Zhang, B., Guo, J., Xu, C., et al. (2012). Physical changes and physiological characteristics of red and green peel during nectarine (cv. Hu018) maturation. J. Sci. Food Agric. 92, 1448–1454. doi: 10.1002/jsfa.4724
Sun, D., Nandety, R. S., Zhang, Y., Reid, M. S., Niu, L., and Jiang, C. Z. (2016). A petunia ethylene-responsive element binding factor, PhERF2, plays an important role in antiviral RNA silencing. J. Exp. Bot. 67, 3353–3365. doi: 10.1093/jxb/erw155
Tamai, H., Iwabuchi, M., and Meshi, T. (2002). Arabidopsis GARP transcriptional activators interact with the pro-rich activation domain shared by G-box- binding bZIP factors. Plant Cell Physiol. 43, 99–107. doi: 10.1093/pcp/pcf011
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637.
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., Marroni, F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. doi: 10.1038/ng.2586
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, P., Fouracre, J., Kelly, S., Karki, S., Gowik, U., Aubry, S., et al. (2013). Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 237, 481–495. doi: 10.1007/s00425-012-1754-3
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Yang, Y., Chen, X., Xu, B., Li, Y., Ma, Y., and Wang, G. (2015). Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 6:139. doi: 10.3389/fpls.2015.00139
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Zhai, R., Wang, Z., Zhang, S., Meng, G., Song, L., Wang, Z., et al. (2016). Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 67, 1275–1284. doi: 10.1093/jxb/erv524
Keywords: chloroplast development, chlorophyll, GLK transcription factor, peach, PpARF5
Citation: Chen M, Liu X, Jiang S, Wen B, Yang C, Xiao W, Fu X, Li D, Chen X, Gao D and Li L (2018) Transcriptomic and Functional Analyses Reveal That PpGLK1 Regulates Chloroplast Development in Peach (Prunus persica). Front. Plant Sci. 9:34. doi: 10.3389/fpls.2018.00034
Received: 19 October 2017; Accepted: 09 January 2018;
Published: 26 January 2018.
Edited by:
Rebecca L. Roston, University of Nebraska-Lincoln, United StatesReviewed by:
Shan Lu, Nanjing University, ChinaCopyright © 2018 Chen, Liu, Jiang, Wen, Yang, Xiao, Fu, Li, Chen, Gao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Gao, ZHNnYW9Ac2RhdS5lZHUuY24=
Ling Li, bGlsaW5nc2RhdUAxNjMuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.