
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 22 December 2017
Sec. Plant Nutrition
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.02197
This article is part of the Research TopicPlant Adaptations to Phosphate DeficiencyView all 13 articles
Arsenic (As) is toxic to organisms, and elevated As accumulation in rice (Oryza sativa) grain may pose a significant health risk to humans. The predominant form of As in soil under aerobic conditions is As(V), which has a chemical structure similar to that of PO43-. Rice roots take up As(V) by phosphate (Pi) transporters, such as OsPT1 and OsPT8. In the present study, we investigated the contribution of OsPT4, belonging to the Pht1 family, on rice As(V) uptake and transport. We determined the mRNA amounts of OsPTs in rice seedlings, and expressions of OsPT1, OsPT4, and OsPT8 were up-regulated under As(V) conditions. OsPT4-overexpressing plants were obtained to examine the As (V) transport activity of OsPT4 in rice. When transgenic rice grew in hydroponic culture with 25 and 50 μM As(V), the plants showed sensitivity to As(V) stress with aboveground parts showing delayed growth and the roots stunted. The OsPT4 CRISPR lines showed the opposite phenotype. When plants were grown in 5 μM As(V) solution for 7 days, the As accumulation of OsPT4-overexpressing plants increased up to twice in roots and shoots. Furthermore, the arsenate uptake rates of OsPT4-overexpressing lines were higher compared with wild type. The Vmax of As(V) uptake in OsPT4-overexpressing plants increased 23–45% compared with Nipponbare. In the flooded soil, the As accumulation of OsPT4-overexpressing plants increased 40–66% and 22–30% in straw and grain, respectively. While in OsPT4-cr plants As accumulation in roots decreased 17–30% compared with Nipponbare. Therefore, the present study indicates that OsPT4 is involved in As(V) uptake and transport and could be a good candidate gene to generate low As-accumulating rice.
Inorganic arsenic (As) is a highly toxic metalloid listed as a Class-1 carcinogen by the International Agency for Research on Cancer (Smith et al., 2002). Humans ingest As unintentionally in contaminated food and drinking water. Excessive ingestion of As causes a series of acute and chronic human health problems, including skin lesions, cancers and nervous exhaustion (Anawar et al., 2002; Abernathy et al., 2003; Meharg and Rahman, 2003; Das et al., 2004). Rice (Oryza sativa), the most important staple food for half of the world’s people especially in South and Southeast Asia (Meharg, 2004; Alamdar et al., 2017), is a major dietary source of inorganic As because of higher As accumulation in rice than in other cereal crops (Meharg and Rahman, 2003). The As contamination in soil is made worse by non-ferrous mining, which has elevated As accumulation in rice grain up to 723 ng g-1 – far in excess of the Chinese maximum concentration of 200 ng g-1 for inorganic As in rice (Zhu et al., 2008; Williams et al., 2009; Okkenhaug et al., 2012). It is necessary to understand the mechanism of rice As accumulation and to generate low-As rice to protect human health.
Inorganic As in soil is classified into two chemical species, depending on the redox status of the soil: arsenite [As(III)] and arsenate [As(V)] (Abedin et al., 2002). As(III) is the predominant form in anaerobic paddy soil and As(V) in soil under aerobic conditions. Plant roots take up different kinds of As by different pathways. As(III) can enter root cells through nodulin26-like intrinsic proteins. Previous studies suggested that nodulin26-like intrinsic proteins are involved in As(III) transport and determine the sensitivity to As(III) stress in Arabidopsis thaliana (Isayenkov and Maathuis, 2008; Kamiya et al., 2009; Xu et al., 2015). In rice, the silicon transporter OsNIP2;1 (Lsi1) is functional in As(III) uptake (Zhao et al., 2010), and As(III) efflux from rice root cells to the xylem is through OsLsi2 (Ma et al., 2008; Li et al., 2009). However, when rice is grown in non-flooded soil for a long time, the soils become aerobic and then the rice roots primarily absorb As(V). In addition, because rice roots release oxygen, As(III) can be oxidized to As(V) in the rice rhizosphere (Seyfferth et al., 2010). It was reported that the chemical structure of As(V) is similar to that of PO43- (Das et al., 2004). In various plant species including rice, phosphorus (Pi) competes with As(V) for uptake, suggesting that they both have the same transporters (De la Rosa et al., 2006; Catarecha et al., 2007). In Arabidopsis, As(V) absorption is closely related to the expression of Pi transporters Pht1;1 and Pht1;4 (Shin et al., 2004; Gonzalez et al., 2005). In rice, the Pi transport pathway genes also contribute to As uptake and transport. Overexpressing the gene for the transcription factor OsPHR2 (phosphate starvation response 2) led to doubling of the As concentration in root and shoot compared with wild-type, suggesting that this gene was involved in As(V) uptake and root–shoot translocation (Wu et al., 2011). In contrast, the Pi transporter OsPHF1 (phosphate transporter traffic facilitator 1) mutant lost more than half of its ability to take up As(V) (Wu et al., 2011). The Pht1 family genes (OsPT1–OsPT13) in the rice genome encode Pi transporters that localize in the plasma membrane. Kamiya et al. (2013) reported that the As accumulation in rice shoots is consistent with OsPT1 expression, indicating that OsPT1 is involved in As(V) uptake from soil to apoplast. In addition, OsPT8 was found to have a high affinity for As(V) and was a key transporter for As(V) uptake into rice roots (Wu et al., 2011; Kamiya et al., 2013). Overexpressing OsPT8 increased the maximum As(V) influx by fivefold and mutation of OsPT8 partially lost As(V) uptake ability (Wang et al., 2016).
After being absorbed by rice roots, As(V) is then transported into xylem vessels (Gilbert-Diamond et al., 2011). Because of its chemical structure being similar to Pi, As(V) can compete with Pi during Pi absorption and phosphorylation, forming As(V) esters and leading to imbalance in Pi metabolism (Finnegan and Chen, 2012). The As(V) esters are much less stable and hydrolyze faster than Pi esters. For example, As(V) competes with PO43- in ATP (adenosine triphosphate) synthesis and replaces it to form unstable adenosine diphosphate-As(V), resulting in disruption of energy flows in the cell (Hartley-Whitaker et al., 2001; Meharg and Hartley-Whitaker, 2002; Cozzolino et al., 2010). Besides, most As(V) should be reduced to As(III) inside plant cells (Su et al., 2010; Shi et al., 2016). OsACR2 has been suggested to be involved in As(V) reduction in rice (Duan et al., 2007). The latest study suggested that OsHAC1;1 and OsHAC1;2 function as As(V) reductase and were involved in the reduction of As(V) to As(III) in rice plants (Shi et al., 2016). Overexpression of OsHAC1;1 and OsHAC1;2 increased As(III) efflux and decreased As accumulation in rice shoots. The mode of action of As(III) differs from that of As(V), with As(III) acting as a cross-linking agent by binding to monothiol molecules, thiol-containing proteins and co-factors (Ha et al., 1999; Raab et al., 2005; Song et al., 2010). On the one hand, the binding of As(III) to proteins has negative effects on folding of these proteins, resulting in inactivation of many enzymes. On the other hand, As(III) complexation is the main detoxification pathway for both As(III) and As(V) (Xu et al., 2007).
Apart OsPT1 and OsPT8, it is unclear whether other Pht1 family genes are involved in As(V) uptake. The objective of the present study was to investigate the function of Pi transporter OsPT4 in rice As(V) uptake. The mRNA amount of OsPT4 in Nipponbare was measured and OsPT4 was induced by As(V) stress. Overexpressing OsPT4 significantly increased the As concentration in roots and shoots, and showed higher As(V) sensitivity at high As(V) levels. This study shows that OsPT4 plays an important role in As(V) absorption.
Five rice (O. sativa) lines, the wild type Nipponbare, OsPT2 overexpression line (OsPT2-ov), OsPT4 overexpression line (OsPT4-ov), OsPT4 RNA interference line (OsPT4-Ri), and OsPT4 CRISPR (Clustered regularly interspaced short palindromic repeats) line (OsPT4-cr), were used in this study. The generation of OsPT4-cr is described below. OsPT2-ov, OsPT4-ov, and OsPT4-Ri were characterized previously (Liu et al., 2010; Ye et al., 2015).
To generate the construct of OsPT4-CRISPR vector, we designed the DNA spacer in NEB cutter1. The amplified PCR product including U3 promoter, spacer of OsPT4 and sgRNA (small guide RNA) were cloned into vector PJE 45/pH-Ubi-cas9-7 (Miao et al., 2013). The primers were OsPT4-spcer-F:5′-AGCCGGGGCTCTTGGACGCCGTTTTAGAGCTATGCTGAAA-3′, spacer-sgRNA-R: 5′-AAAAAGCAGGCTTAAAAAAAAAGCACCGACTCG- 3′, OsPT4-spcer-F: 5′- GGCGTCCAAGAGCCCCGGCTTGCCACGGATCATCTGCAC-3′ and Pu3-spacer-F: 5′- AGAAAGCTGGGTAAAGGGATCTTTAAACATAC GAAC-3′. The construct was transformed into Nipponbare via Agrobacterium tumefaciens-mediated transformation (Wu et al., 2003).
Standard rice culture solution was used in hydroponic experiments. The composition of the culture solution follows: 1.44 mM NH4NO3, 0.5 mM K2SO4, 1.0 mM CaCl2, 1.6 mM MgSO4, 0.17 mM Na2SiO3, 0.3 mM NaH2PO4, 50 μM Fe-EDTA, 0.06 μM (NH4)6Mo7O24, 15 μM H3BO3, 8 μM MnCl2, 0.12 μM CuSO4, 0.12 μM ZnSO4, 29 μM FeCl3, and 40.5 μM citric acid at pH 5.5 (Yoshida et al., 1976). The transgenic lines were grown in solution containing different concentrations of As using Na3AsO4. The solution was renewed every 5 days. The Nipponare and transgenic lines were grown in a greenhouse under 16 h/8 h, 30/22°C, day/night conditions after germination, with c. 60% relative humidity.
A soil experiment was performed in the experimental field in Huazhong Agricultural University, Wuhan, China. The experimental field was divided into two parts with or without Pi fertilizer. The Pi concentrations of these two fields were 15 mg kg-1 soil P (-P) and 30 mg kg-1 soil P (+P). Seedlings of Nipponbare and OsPT4-overexpressing plants (20-day-old) were transplanted into the soil and grown to maturity. Each treatment had 10 replicates.
Plant tissue samples (50–100 mg) were cut and ground with a mortar and pestle to a fine powder in liquid nitrogen. Afterward, total RNA was extracted using TRizol regent (Invitrogen, Carlsbad, CA, United States). Then, the resulted total RNAs were checked by gel electrophoresis (Supplementary Figure S1A). According to the manufacturer’s instructions, 3 μg of total RNA was used to synthesize the first-strand cDNA in 20 μL of reaction mixture using M-MLV reverse transcriptase (Invitrogen). Real-time PCR was performed using the SYBR Premix Ex TaqTM (TaKaRa, Shiga, Japan) with the following gene-specific primers (Table 1). The amplification reaction was performed on an Applied Biosystems (Foster City, CA, United States) 7500 PCR instrument. The rice Ubiquitin 5 gene was used as the internal control.
Rice seeds were soaked in deionized water overnight and germinated at 37°C in darkness for 3 days. Seedlings were transferred to 0.5 mM CaCl2 solution containing a gradient of As(V) concentrations: 0, 25, and 50 μM. Each treatment was replicated with 10 seedlings. Seedlings were grown in a controlled-environment room at 25°C constant temperature and 12-h day length. After 7 days, we photographed the growth phenotype and measured root length and shoot height.
Shoots and roots were harvested separately and roots washed with distilled water before sampling. After drying at 80°C for 3 days, all samples were digested in 65% nitric acid in a MARS6 microwave (CEM) at a temperature gradient of 120–180°C for 45 min, and then diluted in deionized water. The As content of samples was determined with inductively coupled plasma-mass spectrometry (Agilent 7700 series, CA, United States).
Fresh samples were milled in liquid nitrogen and kept at 4°C until samples thawed. The milled samples were homogenized in 10% (w/v) perchloric acid:5% (w/v) perchloric acid (1:9) and placed on ice for 30 min. Following centrifugation at 10 000 g for 10 min at 4°C, the supernatant was used for Pi measurement by molybdenum blue method. The working fluid was a 6:1 ratio 0.4% (w/v) ammonium molybdate dissolved in 0.5M H2SO4 mixed with 10% ascorbic acid. Of the working fluid, 2 mL was added to 1 mL of sample solution, incubated in a water bath at 42°C for 20 min, and then cooled on ice. Sample absorbance was measured at 820 nm, and Pi concentration was calculated by normalization to fresh-weight values.
Data were examined using one-way ANOVA, followed by comparisons of means using the LSD test (Fisher’s Least Significant Difference).
As(V) is a chemical analog of Pi and can be taken up by Pi transporters, so we assayed expression levels of the Pht1 family members except for OsPT11 and OsPT13, which are induced specifically during mycorrhizal symbiosis. The transcript levels of OsPT1, OsPT2, OsPT4, and OsPT8 were significantly higher than that of other Pht1 family members (Figure 1). When rice had been grown in hydroponic conditions with 5 μM As(V) for 7 days, the expression levels of OsPT1, OsPT4, and OsPT8 were significantly increased by 1.7, 2.7, and 5.0 times, respectively. Considering that OsPT1 and OsPT8 are involved in As(V) uptake and transport in rice, OsPT2 and OsPT4 may also have roles in As(V) uptake and transport. The OsPT2- and OsPT4-overexpressing plants were obtained via A. tumefaciens-mediated transformation, and expression levels of OsPT2 and OsPT4 were measured using real-time PCR (Supplementary Figures S1B,C). When transgenic plants had been grown in culture solution with 5 μM As(V) for 3 days, differences in As concentration in both shoots and roots were observed in OsPT4-overexpressing but not OsPT2-overexpressing plants (Supplementary Figures S1D,E). Thus, we focused on the role of OsPT4 in As(V) uptake and transport.
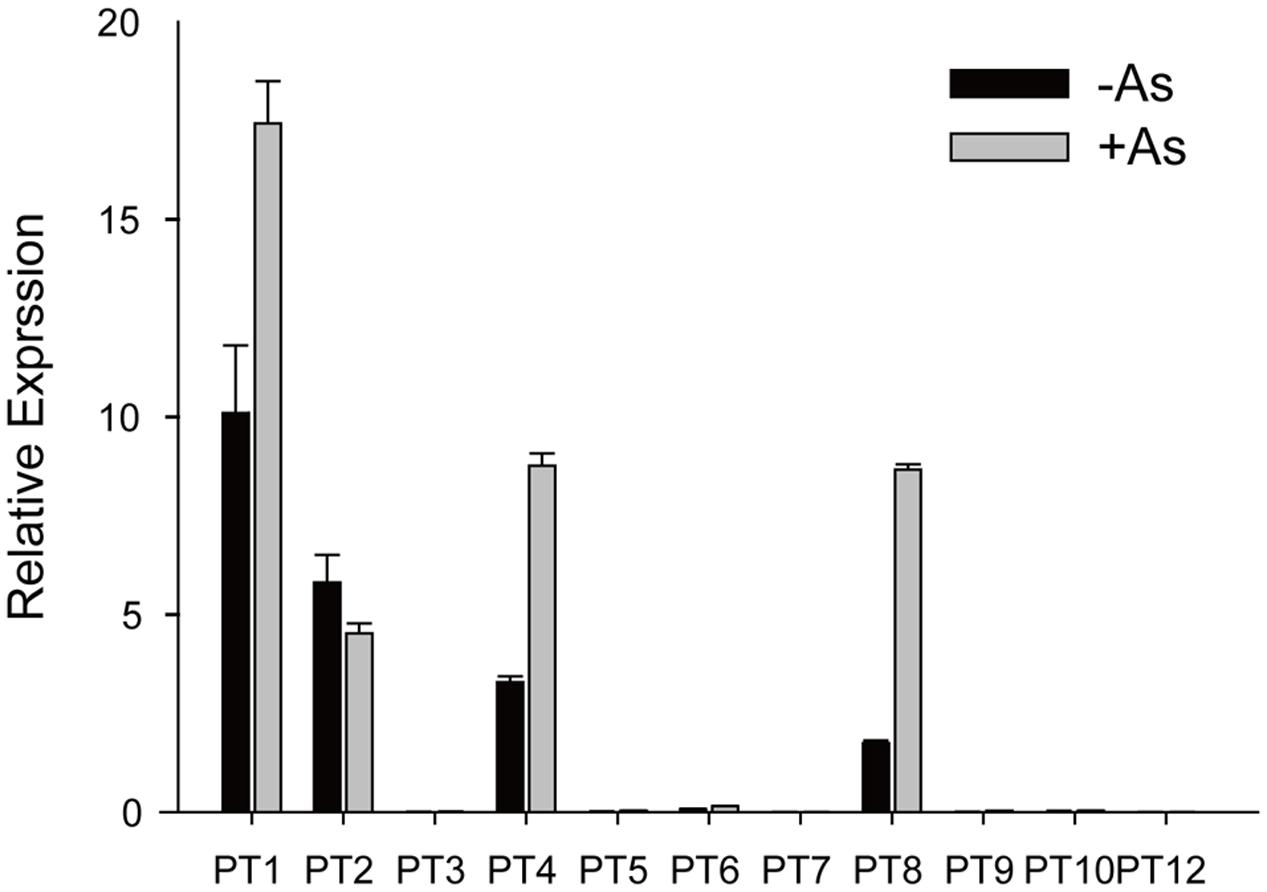
FIGURE 1. Relative expression levels of Pht1 family members in roots of Nipponbare grown in culture solution with or without 5 μM arsenate for 3 days.
To examine the hypothesis that OsPT4 was involved in As(V) absorption in rice plants, a phytotoxicity experiment was performed to observe growth of OsPT4-overexpressing rice. The Nipponbare and OsPT4-overexpressing lines were seeded in hydroponic conditions with 25 or 50 μM As(V). When exposed to the As(V) condition for 7 days, rice showed an As-toxicity phenotype in which growth of aboveground parts was delayed and the roots were stunted (Figures 2A–C). The OsPT4-overexpressing plants were more sensitive to As(V) and their root length and shoot height decreased 50 and 30% compared with wild-type, respectively (Figures 2D,E). Furthermore, As(V) uptake by OsPT4-overexpressing lines and wild-type roots were determined. At both 25 and 50 μM As(V), the As concentration in roots of OsPT4-overexpressing rice increased 10 and 33% compared with Nipponbare (Figure 2F).
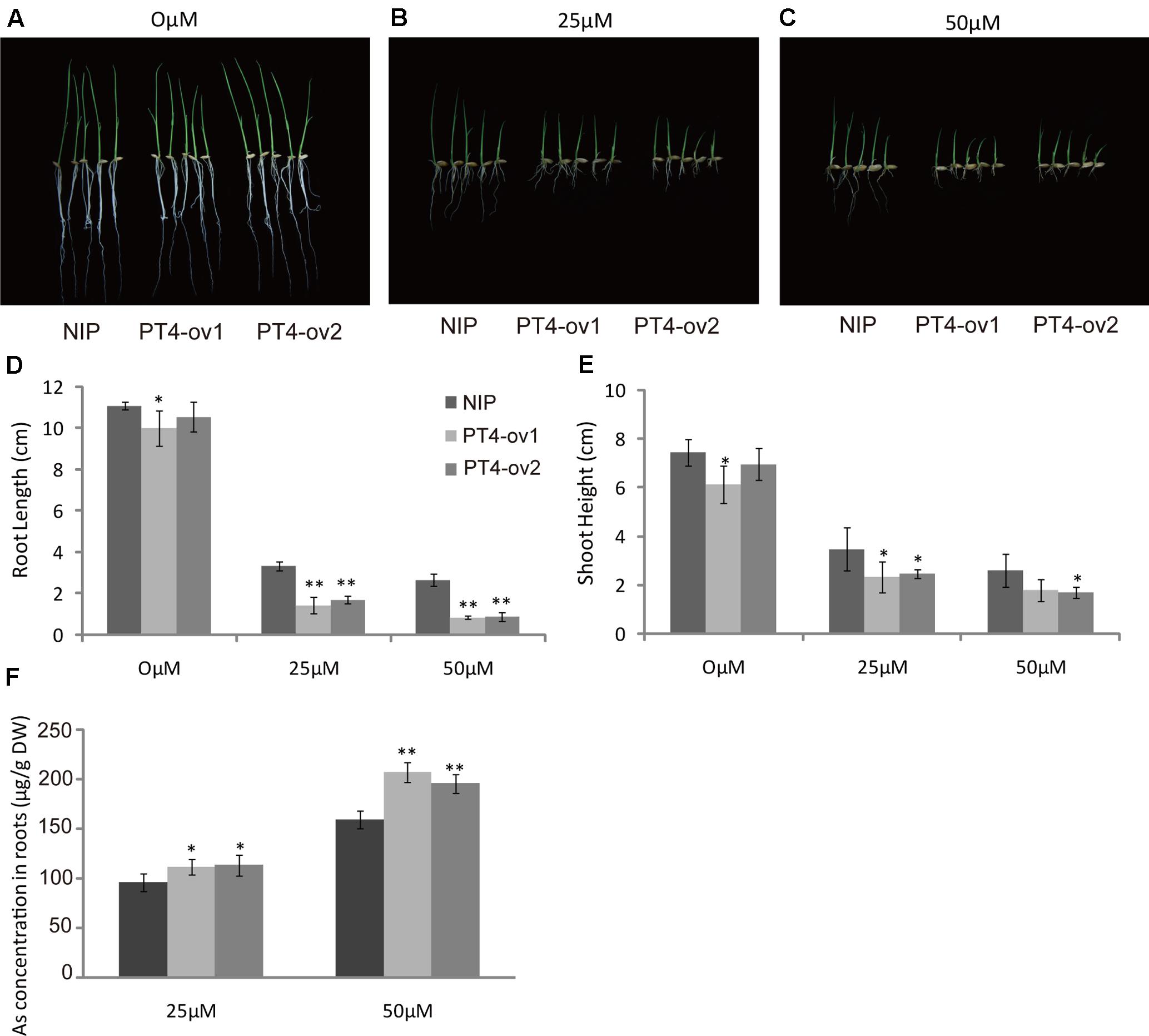
FIGURE 2. Characterization of two OsPT4-overexpressing plants in Nipponbare background. (A–C) The growth phenotype of OsPT4-overexpressing plants and wild-type. Plants were grown in nutrient solutions to which 0, 25, and 50 μM arsenate were added for 7 days. (D,E) Phenotypic analysis of OsPT4-overexpressing plants. The root length and shoot height were obtained from the 7-day-old wild-type and overexpressing plants grown in nutrient solution with different arsenate concentrations. Five plants per line were measured. (F) The As concentration of roots in wild-type and transgenic plants. Data are means ± SD of five biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW, dry weight.
A time-course experiment was used to investigate the ability of OsPT4 to take up As(V). The As accumulation was determined when plants were exposed to hydroponic culture with 5 μM As(V) for 2 h, 1 and 7 days. The concentration of As in rice roots and shoots increased significantly with the As(V) treatment time (Figure 3). When OsPT4-overexpressing lines grew in culture with 5 μM As(V) for 7 days, the As accumulation increased by twice in both roots and shoots.
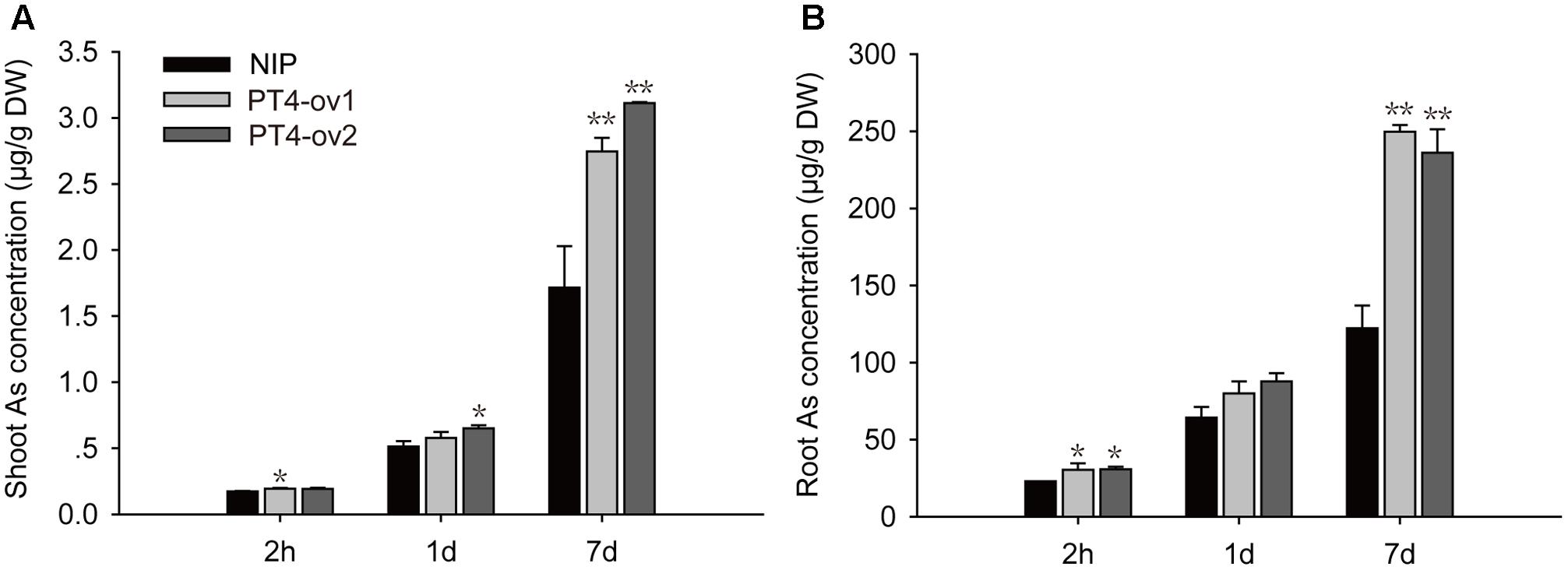
FIGURE 3. The As concentration in shoots and roots (A,B) of Nipponbare and OsPT4-overexpressing seedlings after 2 h, 1 and 7 days of exposure to 5 μM arsenate. Data are means ± SD of three biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW, dry weight.
Certainly, the overexpression lines of OsPT4 accumulated more As both in shoots and roots. It is important to determine the As uptake rate of Nipponbare and OsPT4-overexpressing rice. Wild type and overexpression lines were cultured in the hydroponic solution with 1–50 μM As(V) concentration under +P (100 μM) and -P (0 μM). According to the results (Figure 4), the As(V) uptake rate of OsPT4 overexpression lines was higher compared with wild type under the condition with or without Pi. In the absence of Pi, the As(V) uptake kinetics could be described by a Michaelis–Menten equation (Table 2). The Vmax (maximum influx velocity) of As(V) uptake in OsPT4-overexpressing plants increased 23–45% compared with Nipponbare. Additionally, the Km values of As(V) influx in overexpression lines were 8–28% higher than that in wild type. Under the +P condition, the As(V) uptake rates of rice were significantly lower compared with plants grown in the -P condition. Moreover, As(V) uptake rate was linear over the range of As(V) concentrations tested in the solution with Pi and the slopes of OsPT4 overexpressing plants were 1.4 to 2 times greater compared with Nipponbare. The data suggested that OsPT4 contributes to the As(V) uptake in rice root.
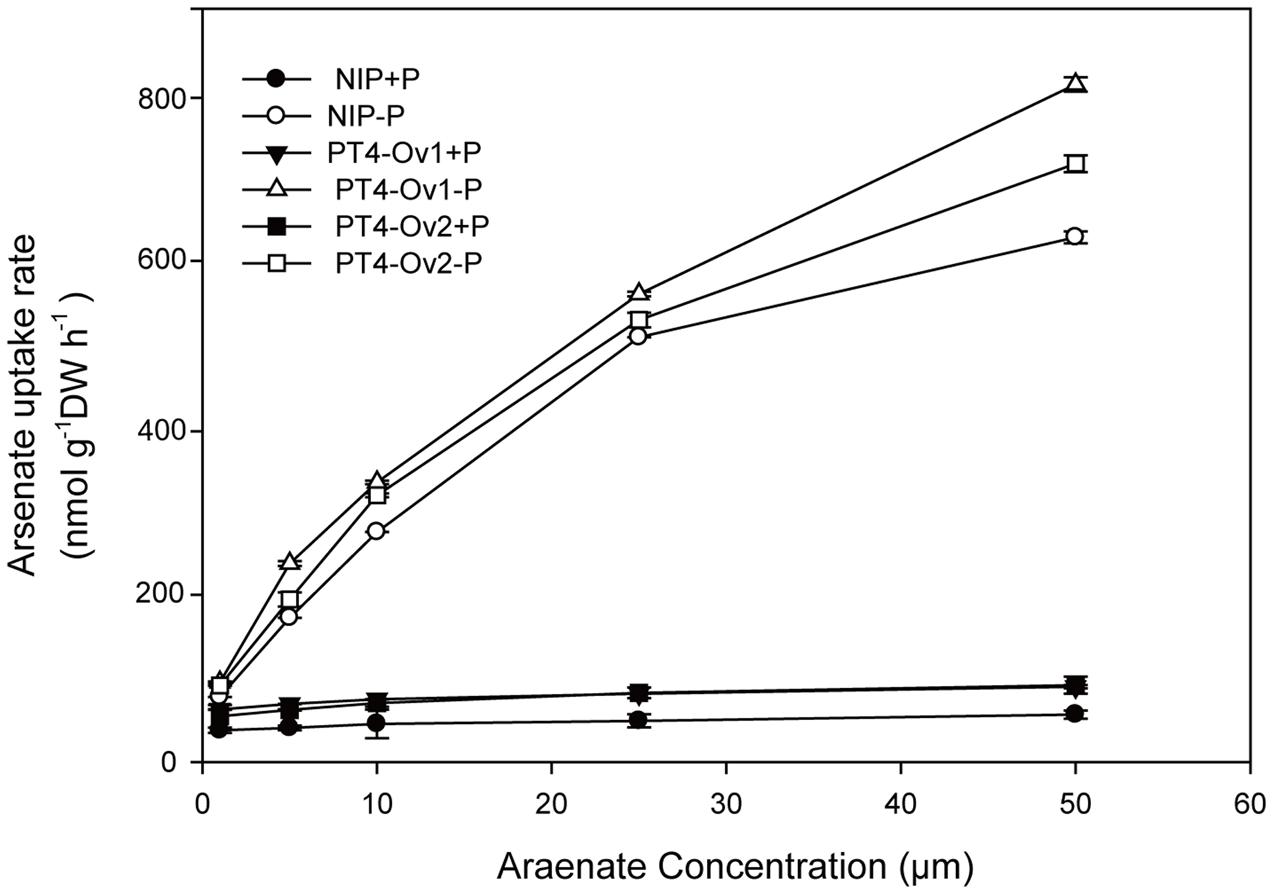
FIGURE 4. Arsenate uptake kinetics of wild type and OsPT4 overexpressing plants. Data are means ± SD of four biological replicates. DW, dry weight.
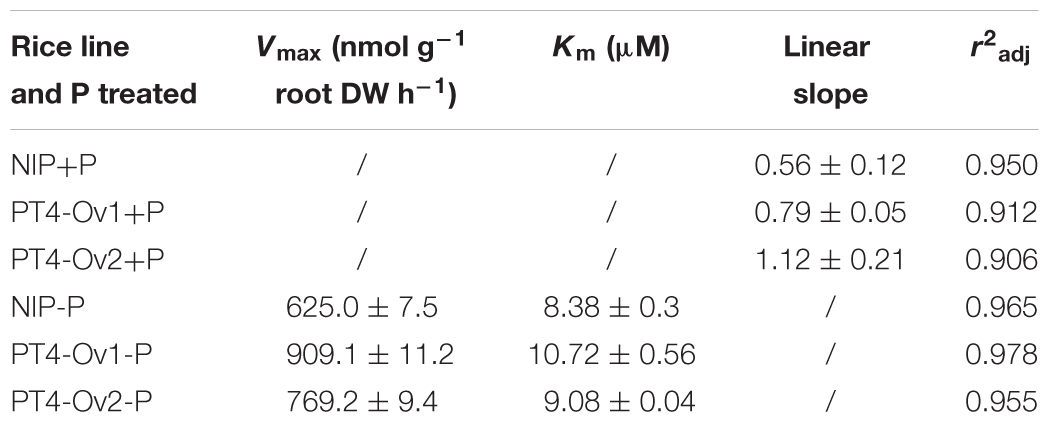
TABLE 2. Fitted parameters of arsenate uptake kinetics of Nipponbare and the PT4 overexpression line of rice.
To further understand the role of OsPT4 in As concentration and distribution in rice, the Nipponbare and OsPT4-overexpressing plants were grown to heading stage in hydroponic culture with 25 μM As(V). In this study, As accumulated mainly in the roots and to a lesser degree in aboveground organs (Figure 5A). In shoots, As was mainly in the nodes, which is the most important storage location for various metallic elements. The sum of As content in nodes accounted for 60% of total As in aboveground parts (Figure 5B). The As content in OsPT4-overexpressing lines increased in both roots and shoots by 22 and 47%, respectively. In addition, the As content of all organs in aboveground parts increased significantly, especially in nodes. The As content of the first node in OsPT4-overexpressing lines increased by twice that for the wild-type, while the second and third node only increased by 55 and 39% compared with wild-type, respectively. Furthermore, the As accumulation in flag leaf and flag sheath of OsPT4-overexpressing lines increased by 40 and 32%. The total As distribution of Nipponbare and OsPT4-overexpressing lines grown in the field soil was similar to that in the hydroponic experiment (Supplementary Figure S2A). This result was consistent with the expression pattern in different organs of rice, in which OsPT4 was mainly expressed in root and flag leaf (Supplementary Figure S2B).
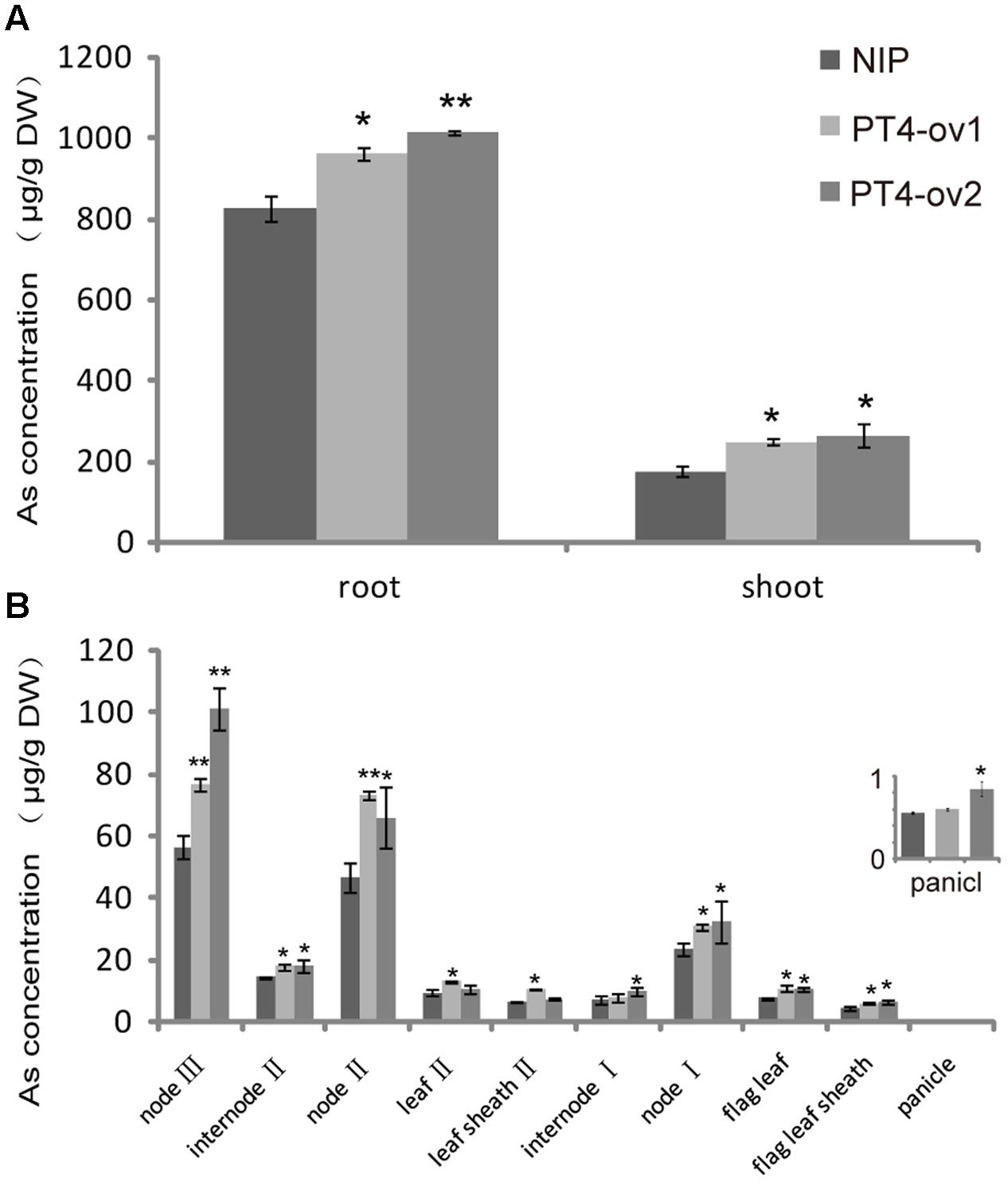
FIGURE 5. The As concentration and distribution in Nipponbare and OsPT4-overexpressing lines grown in hydroponic culture with 5 μM arsenate. (A) The As concentration of shoots and roots of Nipponbare and OsPT4-overexpressing lines. (B) The As concentration of Nipponbare and OsPT4-overexpressing lines in different organs of shoots. Data are means ± SD of three biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW: dry weight.
Previous studies suggested that OsPT1 and OsPT8 were involved in As(V) uptake. The transcript levels of OsPT1, OsPT4, and OsPT8 in rice grown in 5 μM As(V) for 2 h, 1 and 7 days were measured to determine any interaction between these genes. In roots, OsPT8 expression rapidly increased by 10 times, then decreased and remained at a high level; however, OsPT1 and OsPT4 expression increased gradually and maintained a constant level until 7 days (Figure 6A). In the shoot, Real-time PCR analysis showed that expressions of these three genes were enhanced by As(V) (Figure 6B). OsPT1 and OsPT8 were significantly induced by 30 and 8 times, respectively, within a short period and then quickly returned to their original state. The transcript level of OsPT4 increased gradually with treatment time, and finally increased 11 times at 7-days treatment. The induction of OsPT1, OsPT4, and OsPT8 by As(V) raised the question of whether there was functional redundancy across the three genes. To determine this, the relative expression levels of OsPT1 and OsPT8 were evaluated in Nipponbare and OsPT4-overexpressing rice grown in hydroponic culture with or without As(V) (Figures 6C,D). Interestingly, expressions of OsPT1 and OsPT8 had no change in OsPT4-overexpressing rice cultured under normal conditions. The expression levels of OsPT1 both in roots and shoots of OsPT4-overexpressing rice significantly increased in the As(V) condition. However, expression of OsPT8 in OsPT4-overexpressing rice decreased over twofold in roots and was almost unchanged in shoots. The results suggest a lack of functional redundancy among OsPT1, OsPT4 and OsPT8.
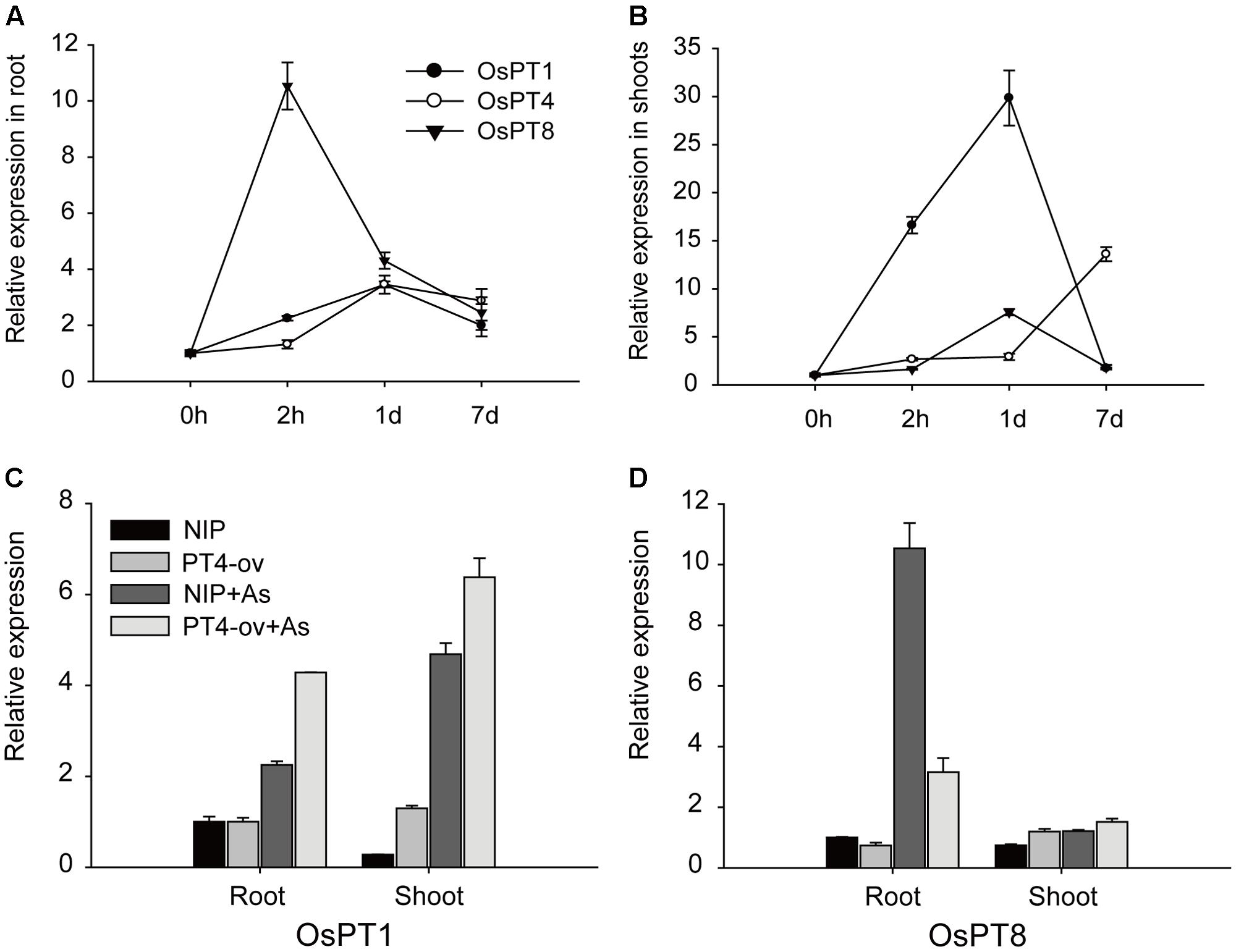
FIGURE 6. Transcript levels of OsPT1, OsPT4, and OsPT8 in Nipponbare under arsenate stress and the effect of altered expression of OsPT4 on OsPT1, OsPT8. (A,B) Expression of OsPT1, OsPT4, and OsPT8 in roots (A,B) of Nipponbare seedlings grown in nutrient solutions with 5 μM arsenate after 2 h, 1 and 7 days. (C,D) Expression of OsPT1 and OsPT8 in wild type and OsPT4-overexpressing lines grown in solution with or without arsenate. Error bars indicate ±SD (n = 3).
A long-term experiment was used to investigate transgenic and wild-type plants grown to maturity in flooded soil conditions with two levels of P: -P (15 mg P kg-1 soil) and +P (30 mg P kg-1 soil). Overexpression of OsPT4 significantly increased total Pi concentrations in both grain and straw both in -P field and +P field (Figures 7A,B). In -P field, OsPT4 overexpression significantly enhanced grain and straw As accumulation by 22–30% and 40–66%, respectively, but no significant difference was shown in +P field (Figures 7C,D).
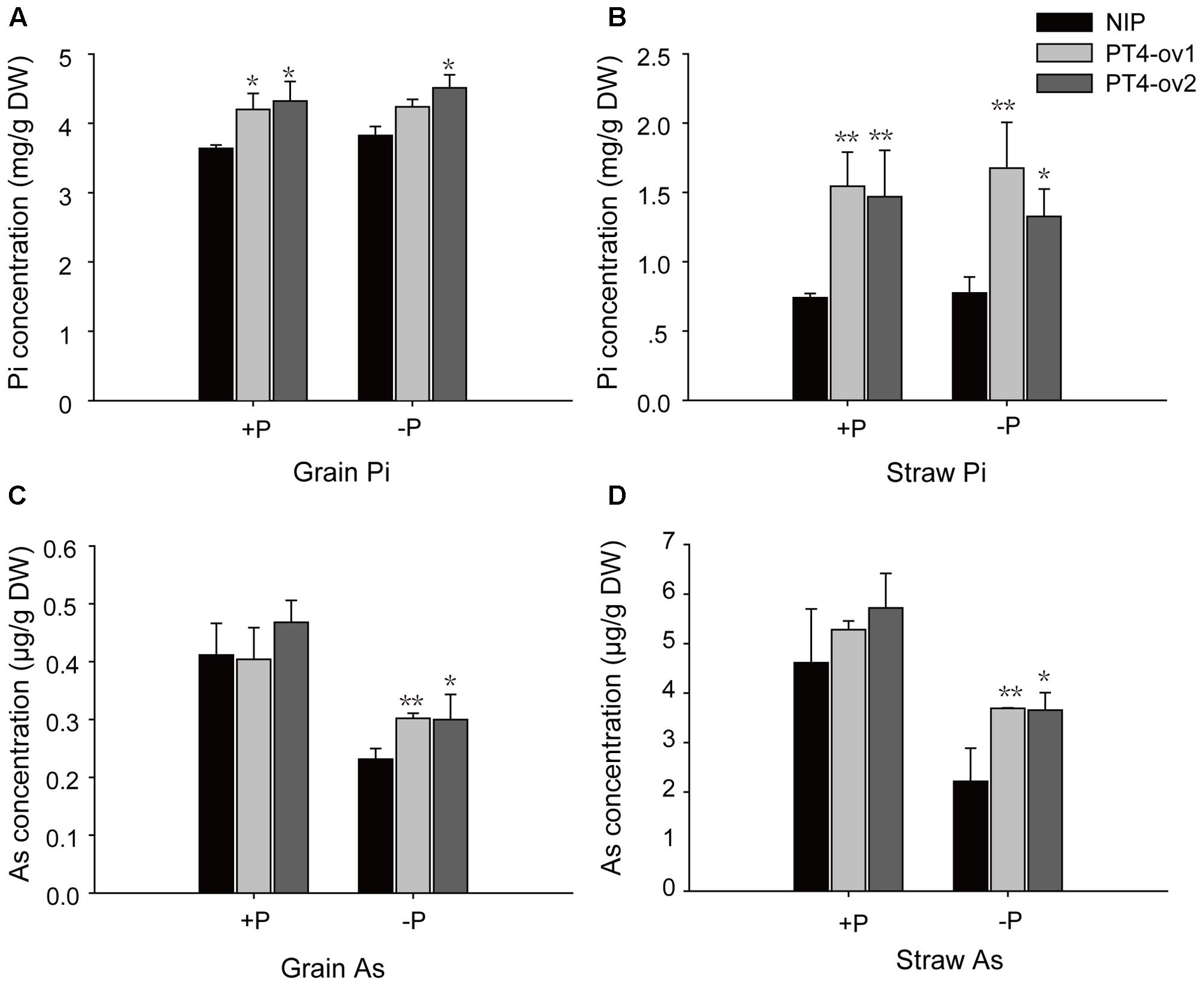
FIGURE 7. The Pi and As concentrations of wild-type and OsPT4-overexpressing lines in the field. The rice was grown to maturity in soil under flooded conditions including two levels of P: –P (15 mg P kg-1 soil) and +P (30 mg P kg-1 soil). (A,B) The Pi concentration of grain (A) and straw (B) in wild-type and OsPT4-overexpressing lines. (C,D) The As concentration of grain (C) and straw (D) in wild-type and OsPT4-overexpressing lines. Data are means ± SD of three biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW: dry weight.
In order to further study the role of OsPT4 in rice As(V) uptake and transport, we studied the phenotype of OsPT4-Ri plants in solution with 0, 25, and 50 μM arsenate. The growth of OsPT4-Ri plants was similar to the wild type. No obvious difference in As concentration between NIP and OsPT4-Ri plants was observed (Supplementary Figure S3). Meanwhile, we obtained two different OsPT4 CRISPR lines that were treated with different As concentrations. As we expected, the OsPT4-cr lines showed stronger resistance to As(V) compared to wild type (Figure 8). The root length and shoot height of OsPT4-cr plants were significantly longer than wild type. Furthermore, the As accumulation of roots in OsPT4-cr plants decreased 17–30% compared with Nipponbare. The results obtained from OsPT4-cr plants confirmed that OsPT4 is involved in As(V) uptake and transport.
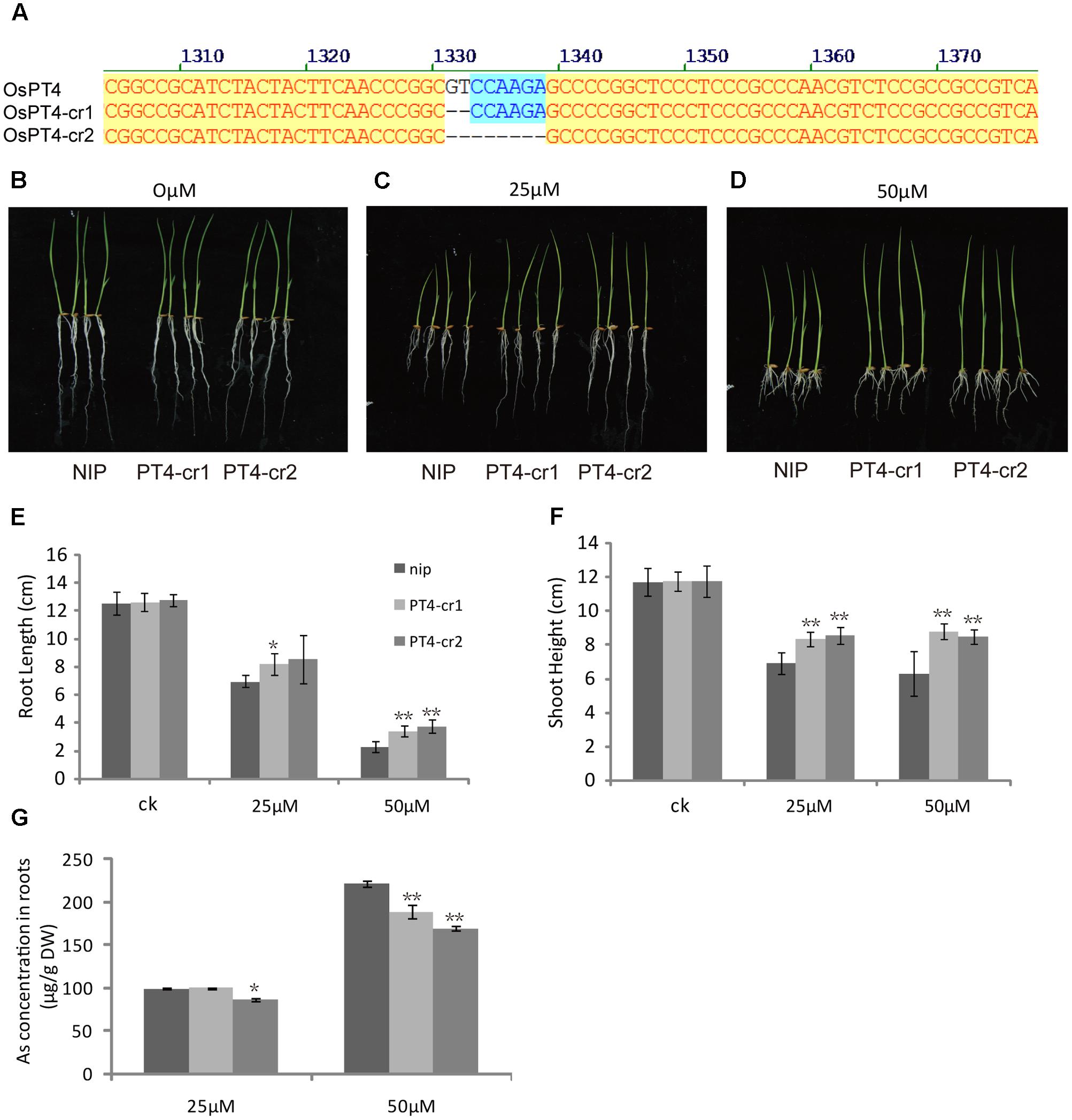
FIGURE 8. Characterization of two OsPT4 CRISPR plants in Nipponbare background. (A) Detection of mutations in OsPT4. The position of target site on the gene structure is indicated. (B–D) The growth phenotype of OsPT4-cr plants and wild-type. Plants were grown in nutrient solution to which 0, 25, and 50 μM arsenate were added for 7 days. (E,F) Phenotypic analysis of OsPT4 CRISPR plants. The root lengths and shoot heights were obtained from the 7-day-old wild type and OsPT4-cr plants grown in nutrient solution with different arsenate concentrations. Five plants per line were measured. (G) As concentrations of roots in wild-type and transgenic plants. Data are means ± SD of five biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW: dry weight.
As(V) is absorbed in roots and transported from vegetative tissues to rice grain. Elevated As accumulation in rice grain may pose a significant health risk to humans. It is important to determine how Pi transport genes contribute to As accumulation in rice. In the present study, we identified OsPT4 as an important component of As(V) homeostasis and As tolerance in rice.
As(V) is a toxic analog of Pi, so it can be absorbed and transported via Pi transport in plant (Xu et al., 2007). In Arabidopsis, previous studies suggested that AtPht1;1 and AtPht1;4 mediated a significant proportion of the As(V) uptake and a AtPHF1 mutant was more resistant to As(V) than wild-type (Shin et al., 2004; Gonzalez et al., 2005). In rice, research has shown that Pht1 family genes participate in As(V) uptake – OsPT1 was involved in As(V) transport from soil to apoplast and OsPT8 functioned in As(V) uptake and resulted in a high affinity for As (Wu et al., 2011; Kamiya et al., 2013).
It was reported that the expression of OsPT4 significantly increased in root of BRRT51 under As stress (Begum et al., 2016). In our study, the transcript levels of Pht1 family genes in Nipponbare grown in normal and As(V) conditions were measured. The expression level of OsPT4 significantly increased in hydroponic culture containing As(V). The up-regulated expression of OsPT4 hinted at a role in As(V) uptake. Furthermore, the ability of OsPT4 to absorb As was clearly demonstrated in the As phytotoxicity, hydroponic and field experiments. Root length and shoot height of OsPT4-overexpressing lines decreased 50 and 30%, respectively, compared with Nipponbare, and the As accumulation in roots of transgenic lines increased 33%. Meanwhile, the OsPT4-cr plants produced the opposite phenotype. Furthermore, the As(V) uptake rates of OsPT4-overexpressing plants were significantly higher than that in wild type under the growth condition with or without Pi. Differences in As concentration were also observed in grain and straw of OsPT4-overexpressing plants compared with wild-type in the flooded soil. All these results suggested that OsPT4 was a functional transporter in As(V) uptake.
Previous studies showed that OsPT4, a Pi-influx transporter involved in Pi acquisition and mobilization in rice, facilitates embryo development (Ye et al., 2015; Zhang et al., 2015). OsPT4 was highly expressed in roots, specifically in the exodermis cells and cortex. Although the sclerenchymatous cells at the exodermis in rice is the first apoplastic barrier to entry of toxic As, it does not completely stop entry of As because it shares transporters with essential Pi. The strong expression of OsPT4 in the root exodermis cells and cortex (Ye et al., 2015) may explain the high As accumulation in roots and the heightened effect of As(V) in the As phytotoxicity experiment. Overall, the results suggested that OsPT4 was a Pi transporter sharing an ion channel with As(V) and playing an important role in As(V) uptake.
In rice, OsPT4 is constitutively expressed in roots and shoots (Ye et al., 2015), with the highest expression in flag leaves (Supplementary Figure S2). However, the As concentration in roots was much higher than that in shoots (Figure 5). The reason for this discrepancy is likely to be due to the mitigation strategies in rice As uptake and transport. The As(V) was absorbed by OsPT4 and the other transporters, and then quickly reduced into As(III). OsHAC1;1 and OsHAC1;2 have been reported to act as arsenate reductases in rice (Shi et al., 2016). Most of As(III) was mainly fastened in root cortex and stele, forming complex with thiol (Su et al., 2010). The uncomplexed As(III) is transported to shoots and even to grain. The formation of As(III)-thiol complex in rice roots helps to explain the reason why the As concentration in roots is four times higher than that in shoots.
The flag leaf is an essential tissue for the growth and development of rice panicles, and plays a key role in remobilization of many mineral elements from leaves to developing grain. In addition, Zhang and his team reported that OsPT4 was involved in Pi mobilization that facilitates embryo development in rice (Zhang et al., 2015). In our work, overexpressing OsPT4 resulted in higher As concentration than wild-type in various organs of rice shoots, such as nodes, flag leaves and panicles Thus, we deduced that OsPT4 was probably involved in As mobilization from flag leaf to panicles and immobilization in grain.
Many researches have reported that nodes are critical hubs in controlling the distribution of mineral elements including As. Nodes have a markedly larger concentration of As than the other tissues of rice shoots (Moore et al., 2014). This was confirmed in present study, showing that the As concentration of nodes represents 60% of the total As in shoot. The possible explanation is that a large portion of the node tissues are vascular bundles and that As accumulates strongly in the phloem (Chen et al., 2015). The nodes that produce or are near crown roots may accumulate higher concentrations of As accumulation. This may explain the observation that OsPT4-overexpressing rice accumulated more As in node II and node III where generates crown roots. In the last few years, many mineral element transporters have been reported to function in rice nodes. A member of the rice C-type ATP-binding cassette (ABC) transporter family, OsABCC1, was reported as a As(III)-phytochelatin transporter. Knockout of OsABCC1 in rice resulted in less As accumulation in the nodes and more As accumulation in the grain (Song et al., 2014). A strategy to prevent As accumulating in the grain is accumulate As in the nodes, especially those close to crown roots.
There are a reported 13 members of the rice Pht1 family and most share a similar protein structure and the same protein destination (localized to the plasma membrane). Among their encoding genes, OsPT1, OsPT4, and OsPT8 were induced by As(V) stress. Individually overexpressing these three genes led to higher As accumulation than their background. These results raised the question of whether there is a network among these three Pi transporters playing specific and/or overlapping roles in As accumulation in rice.
The sensitivity to As(V) stress of OsPT4-overexpressing plants and OsPT8 mutants, assessed in As phytotoxicity experiments, showed that both OsPT4 and OsPT8 were involved in As(V) uptake from roots (Wang et al., 2016) (Figure 2). Analyses of the GUS reporter gene driven by the promoters of OsPT4 and OsPT8 indicated a partial overlap in their spatial expression patterns, because the strongest expression of OsPT4 and OsPT8 were in the epidermis and cortex (Jia et al., 2011; Ye et al., 2015). Additionally, real-time PCR analysis revealed the same expression pattern of OsPT4 and OsPT8 in BRRI33 and BRRI51 under As stress along with lower regulation in BRRI33 and higher regulation in BRRI51 (Begum et al., 2016). And both OsPT4 and OsPT8 were up-regulated in overexpression lines of OsPAP21b and OsHAD1 under Pi-deficient conditions (Mehra et al., 2017; Pandey et al., 2017). Moreover, the transcript level of OsPT8 in roots was dramatically attenuated in OsPT4-overexpressing plants grown in As(V) conditions (Figure 6D). These results indicated that there may be a functional overlap of OsPT4 with OsPT8. However, contrary to this assumption, ospt8 mutants lost almost half of their As(V) uptake ability when seedlings were exposed to 1–2 μM As(V)(Wang et al., 2016). In our study, OsPT4-cr lines displayed significantly lower As concentrations in roots. The similar phenomenon has been observed in ospt4 mutant (Cao et al., 2017). The attenuation of function of OsPT8 was not compensated for by OsPT4, and so OsPT4 and OsPT8 were non-redundant in As(V) uptake. Furthermore, the expression variance of OsPT4 and OsPT8 in the time-course experiment also differed. In the present study, OsPT8 expression in rice roots quickly increased by 10 times when plants were treated with As(V) for 2 h and then dropped (Figure 6A). Over the same time periods, the transcript level of OsPT4 increased gradually and remained at a high level. In flooded soil, overexpressing OsPT8 did not change the As accumulation in straw and grain (Wu et al., 2011), but the As accumulation in OsPT4-overexpressing plants increased in Pi-replete conditions (Figures 7C,D). These results strongly suggest that OsPT4 and OsPT8 had similar expression patterns but different regulation pathways in As uptake.
In Arabidopsis, there was a possible functional overlap of Pht1;1 with Pht1;4. In the As(V) condition, the double mutant showed a more resistant phenotype compared with background and the mutants for single genes (Shin et al., 2004). In rice, the promoters of OsPT1 and OsPT4, unlike OsPT8, did not contain the P1BS element. The expression level of OsPT1 and OsPT4 was not effect by Pi supply conditions (Wu et al., 2013). Kamiya et al. (2013) reported that OsPT1 was involved in As accumulation in shoots, a conclusion consistent with our result that the expression of OsPT1 in shoots rapidly increased by 30 times (Figure 6B). The As accumulation in shoots of ospt1 mutant was not significantly different to its background in Pi-replete condition (Kamiya et al., 2013), indicating that there may be a functional overlap between OsPT1 and OsPT4. We also found that altered expression of OsPT4 did affect expression of OsPT1. The transcript level of OsPT1 in rice roots and shoots dramatically increased in OsPT4-overexpressing plants grown in normal and As(V) conditions (Figure 6C). Therefore, it was logical to assume that OsPT1 and OsPT4 shared a similar or the same pathway in rice As(V) uptake and translocation. Studies utilizing double mutants of these two genes are needed to test this hypothesis.
Although there were significant increases of As concentration in OsPT4-overexpressing plants grown in hydroponic solution with Pi supplement, differences in As concentration in grain and straw were observed under -P but not +P flooded soil (Figure 7). The latest study showed that knockout of OsPT4 could significantly decrease the inorganic As in rice grain (Cao et al., 2017). Since inorganic As is classified as Class-1 carcinogen, decreasing the concentration of inorganic As in rice grain is an important rice breeding target to protect human health. Under these circumstances, OsPT4 which is involved in Pi and As uptake is a candidate gene to generate a high Pi-efficiency and low As-accumulating rice.
Experimental design: XL and YY. Experiments: YY, PL, TX, LZ, JL, and DC. Data analysis: YY and MY. Manuscript preparation: XL and YY. Supervision, funding and reagents: XL.
This work was supported by grants from the National High Technology Research and Development Program of China (2014AA10A603), Special Fund for Agro-Scientific Research in the Public Interest (201403015), the National Natural Science Foundation of China (31520103914 and 31471932).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was conducted at the National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan. We thank Professor Lijia Qu for kindly providing the PJE 45/pH-Ubi-cas9-7 vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02197/full#supplementary-material
FIGURE S1 | The characteristics of OsPT2- and OsPT4-overexpressing plants. The expression levels and As contents of OsPT2 and OsPT4 were determined with real-time polymerase chain reaction and inductively coupled plasma mass spectrometry (ICP-MS), respectively. (A) Total RNA of wild type, OsPT2- and OsPT4-overexpressing plants. (B) Relative expression levels of OsPT2 in OsPT2-overexpressing plants compared with background. (C) Relative expression levels of OsPT4 in OsPT4-overexpressing plants compared with background. (D,E) The As accumulation in shoot (D) and root (E) of wild-type and transgenic plants after exposure to 5 μM arsenate for 7 days. Data are means ± SD of three biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW: dry weight.
FIGURE S2 | The As concentration in OsPT4-overexpressing plants and the expression pattern of OsPT4 in Nipponbare. (A) The As concentration of wild-type and OsPT4-overexpressing plants grown to heading stage in flooded soil. Data are means ± SD of three biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW: dry weight. (B) The relative expression levels of OsPT4 in different organs of Nipponbare. Error bars indicate ± SD (n = 3).
FIGURE S3 | Phenotypes of OsPT4 RNA interference plants. (A–C) The growth phenotype of OsPT4-Ri plants and wild type. Plants were grown in nutrient solution to which 0, 25, and 50 μM arsenate were added for 7 days. (D) As concentrations of roots in wild-type and OsPT4-Ri plants. Data are means ± SD of five biological replicates. Values are significantly different from those of wild-type: ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). DW, dry weight.
Abedin, M. J., Cotter-Howells, J., and Meharg, A. A. (2002). Arsenic uptake and accumulation in rice (Oryza sativa L.) irrigated with contaminated water. Plant Soil 240, 311–319. doi: 10.1023/A:1015792723288
Abernathy, C. O., Thomas, D. J., and Calderon, R. L. (2003). Health effects and risk assessment of arsenic. J. Nutr. 133(5 Suppl. 1), 1536S–1538S.
Alamdar, A., Eqani, S. A., Hanif, N., Ali, S. M., Fasola, M., Bokhari, H., et al. (2017). Human exposure to trace metals and arsenic via consumption of fish from river Chenab, Pakistan and associated health risks. Chemosphere 168, 1004–1012. doi: 10.1016/j.chemosphere.2016.10.110
Anawar, H. M., Akai, J., Mostofa, K. M. G., Safiullah, S., and Tareq, S. M. (2002). Arsenic poisoning in groundwater - Health risk and geochemical sources in Bangladesh. Environ. Int. 27, 597–604. doi: 10.1016/S0160-4120(01)00116-7
Begum, M. C., Islam, M. S., Islam, M., Amin, R., Parvez, M. S., and Kabir, A. H. (2016). Biochemical and molecular responses underlying differential arsenic tolerance in rice (Oryza sativa L.). Plant Physiol. Biochen. 104, 266–277. doi: 10.1016/j.plaphy.2016.03.034
Cao, Y., Ai, H., Mei, H., Liu, X., Sun, S., Xu, G., et al. (2017). Knocking out OsPT4 gene decreases arsenate uptake by rice plants and inorganic arsenic accumulation in rice grains. Environ. Sci. Technol. 51, 12131–12138. doi: 10.1021/acs.est.7b03028
Catarecha, P., Segura, M. D., Franco-Zorrilla, J. M., Garcia-Ponce, B., Lanza, M., Solano, R., et al. (2007). A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19, 1123–1133. doi: 10.1105/tpc.106.041871
Chen, Y., Moore, K. L., Miller, A. J., McGrath, S. P., Ma, J. F., and Zhao, F.-J. (2015). The role of nodes in arsenic storage and distribution in rice. J. Exp. Bot. 66, 3717–3724. doi: 10.1093/jxb/erv164
Cozzolino, V., Pigna, M., Di Meo, V., Caporale, A. G., and Violante, A. (2010). Effects of arbuscular mycorrhizal inoculation and phosphorus supply on the growth of Lactuca sativa L. and arsenic and phosphorus availability in an arsenic polluted soil under non-sterile conditions. Appl. Soil Ecol. 45, 262–268. doi: 10.1016/j.apsoil.2010.05.001
Das, H. K., Mitra, A. K., Sengupta, P. K., Hossain, A., Islam, F., and Rabbani, G. H. (2004). Arsenic concentrations in rice, vegetables, a fish in Bangladesh: a preliminary study. Environ. Int. 30, 383–387. doi: 10.1016/j.envint.2003.09.005
De la Rosa, G., Parsons, J. G., Martinez-Martinez, A., Peralta-Videa, J. R., and Gardea-Torresdey, J. L. (2006). Spectroscopic study of the impact of arsenic speciation on arsenic/phosphorus uptake and plant growth in tumbleweed (Salsola kali). Environ. Sci. Technol. 40, 1991–1996. doi: 10.1021/es051526s
Duan, G. L., Zhou, Y., Tong, Y. P., Mukhopadhyay, R., Rosen, B. P., and Zhu, Y. G. (2007). A CDC25 homologue from rice functions as an arsenate reductase. New Phytol. 174, 311–321. doi: 10.1111/j.1469-8137.2007.02009
Finnegan, P. M., and Chen, W. H. (2012). Arsenic toxicity: the effects on plant metabolism. Front. Physiol. 3:182. doi: 10.3389/Fphys.2012.00182
Gilbert-Diamond, D., Cottingham, K. L., Gruber, J. F., Punshon, T., Sayarath, V., Gandolfi, A. J., et al. (2011). Rice consumption contributes to arsenic exposure in US women. Proc. Natl. Acad. Sci. U.S.A. 108, 20656–20660. doi: 10.1073/pnas.1109127108
Gonzalez, E., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512. doi: 10.1105/tpc.105.036640
Ha, S. B., Smith, A. P., Howden, R., Dietrich, W. M., Bugg, S., O’Connell, M. J., et al. (1999). Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11, 1153–1164. doi: 10.2307/3870806
Hartley-Whitaker, J., Ainsworth, G., Vooijs, R., Ten Bookum, W., Schat, H., and Meharg, A. A. (2001). Phytochelatins are involved in differential arsenate tolerance in Holcus lanatus. Plant Physiol. 126, 299–306. doi: 10.1104/pp.126.1.299
Isayenkov, S. V., and Maathuis, F. J. M. (2008). The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Lett. 582, 1625–1628. doi: 10.1016/j.febslet.2008.04.022
Jia, H., Ren, H., Gu, M., Zhao, J., Sun, S., Zhang, X., et al. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175. doi: 10.1104/pp.111.175240
Kamiya, T., Islam, M. R., Duan, G. L., Uraguchi, S., and Fujiwara, T. (2013). Phosphate deficiency signaling pathway is a target of arsenate and phosphate transporter OsPT1 is involved in As accumulation in shoots of rice. Soil Sci. Plant Nutr. 59, 580–590. doi: 10.1080/00380768.2013.804390
Kamiya, T., Tanaka, M., Mitani, N., Ma, J. F., Maeshima, M., and Fujiwara, T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284, 2114–2120. doi: 10.1074/jbc.M806881200
Li, R. Y., Ago, Y., Liu, W. J., Mitani, N., Feldmann, J., McGrath, S. P., et al. (2009). The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol. 150, 2071–2080. doi: 10.1104/pp.109.140350
Liu, F., Wang, Z., Ren, H., Shen, C., Li, Y., Ling, H.-Q., et al. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. doi: 10.1111/j.1365-313X.2010.04170.x
Ma, J. F., Yamaji, N., Mitani, N., Xu, X. Y., Su, Y. H., McGrath, S. P., et al. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U.S.A. 105, 9931–9935. doi: 10.1073/pnas.0802361105
Meharg, A. A. (2004). Arsenic in rice - understanding a new disaster for South-East Asia. Trends Plant Sci. 9, 415–417. doi: 10.1016/j.tplants.2004.07.002
Meharg, A. A., and Hartley-Whitaker, J. (2002). Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154, 29–43. doi: 10.1046/j.1469-8137.2002.00363.x
Meharg, A. A., and Rahman, M. (2003). Arsenic contamination of Bangladesh paddy field soils: implications for rice contribution to arsenic consumption. Environ. Sci. Technol. 37, 229–234. doi: 10.1021/es0259842
Mehra, P., Pandey, B. K., and Giri, J. (2017). Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol. J. 15, 1054–1067. doi: 10.1111/pbi.12699
Miao, J., Guo, D. S., Zhang, J. Z., Huang, Q. P., Qin, G. J., and Zhang, X. (2013). Targered mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Moore, K. L., Chen, Y., Meene, A. M., Hughes, L., Geraki, T., Mosselmans, S. P., et al. (2014). Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 201, 104–115. doi: 10.1111/nph.12497
Okkenhaug, G., Zhu, Y. G., He, J. W., Li, X., Luo, L., and Mulder, J. (2012). Antimony (Sb) and arsenic (As) in Sb mining impacted paddy soil from Xikuangshan, China: differences in mechanisms controlling soil sequestration and uptake in rice. Environ. Sci. Technol. 46, 3155–3162. doi: 10.1021/es2022472
Pandey, B. K., Mehra, P., Verma, L., Bhadouria, J., and Giri, J. (2017). OsHAD1, a haloacid dehaligenase-like APase, enhances phosphate accumulation. Plant Physiol. 174, 2316–2332. doi: 10.1104/pp.17.00571
Raab, A., Schat, H., Meharg, A. A., and Feldmann, J. (2005). Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): formation of arsenic-phytochelatin complexes during exposure to high arsenic concentrations. New Phytol. 168, 551–558. doi: 10.1111/j.1469-8137.2005.01519.x
Seyfferth, A. L., Webb, S. M., Andrews, J. C., and Fendorf, S. (2010). Arsenic localization, speciation, and co-occurrence with iron on rice (Oryza sativa L.) roots having variable Fe coatings. Environ. Sci. Technol. 44, 8108–8113. doi: 10.1021/es101139z
Shi, S. L., Wang, T., Chen, Z., Tang, Z., Wu, Z. C., Salt, D. E., et al. (2016). OsHAC1;1 and OsHAC1;2 function as arsenate reductases and regulate arsenic accumulation. Plant Physiol. 172, 1708–1719. doi: 10.1104/pp.16.01332
Shin, H., Shin, H. S., Dewbre, G. R., and Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Smith, A. H., Lopipero, P. A., Bates, M. N., and Steinmaus, C. M. (2002). Public health - arsenic epidemiology and drinking water standards. Science 296, 2145–2146. doi: 10.1126/science.1072896
Song, W.-Y., Park, J., Mendoza-Cozatl, D. G., Suter-Grotemeyer, M., Shim, D., Hortensteiner, S., et al. (2010). Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. U.S.A. 107, 21187–21192. doi: 10.1073/pnas.1013964107
Song, W.-Y., Yamaki, T., Yamaji, N., Ko, D., Jung, K.-H., Fujii-Kashino, M., et al. (2014). A rice ABC transporter, OsABCC1, reduce arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U.S.A. 111, 15699–15704. doi: 10.1073/pnas.1414968111
Su, Y. H., McGrath, S. P., and Zhao, F. J. (2010). Rice is more efficient in arsenite uptake and translocation than wheat and barley. Plant Soil 328, 27–34. doi: 10.1007/s11104-009-0074-2
Wang, P., Zhang, W., Mao, C., Xu, G., and Zhao, F. J. (2016). The role of OsPT8 in arsenate uptake and varietal difference in arsenate tolerance in rice. J. Exp. Bot. 67, 6051–6059. doi: 10.1093/jxb/erw362
Williams, P. N., Lei, M., Sun, G. X., Huang, Q., Lu, Y., Deacon, C., et al. (2009). Occurrence and partitioning of cadmium, arsenic and lead in mine impacted paddy rice: Hunan, China. Environ. Sci. Technol. 43, 637–642. doi: 10.1021/es802412r
Wu, C., Yuan, W., Chen, G., Kilian, A., and Li, J. (2003). Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35, 418–427. doi: 10.1046/j.1365-313X.2003.01808.x
Wu, P., Shou, H., Xu, G., and Lian, X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16, 205–212. doi: 10.1016/j.pbi.2013.03.002
Wu, Z., Ren, H., McGrath, S. P., Wu, P., and Zhao, F. J. (2011). Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157, 498–508. doi: 10.1104/pp.111.178921
Xu, W., Dai, W., Yan, H., Li, S., Shen, H., Chen, Y., et al. (2015). Arabidopsis NIP3;1 plays an important role in arsenic uptake and root-to-shoot translocation under arsenite stress conditions. Mol. Plant 8, 722–733. doi: 10.1016/j.molp.2015.01.005
Xu, X. Y., McGrath, S. P., and Zhao, F. J. (2007). Rapid reduction of arsenate in the medium mediated by plant roots. New Phytol. 176, 590–599. doi: 10.1111/j.1469-8137.2007.02195.x
Ye, Y., Yuan, J., Chang, X., Yang, M., Zhang, L., Lu, K., et al. (2015). The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLOS ONE 10:e0126186. doi: 10.1371/journal.pone.0126186
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). Laboratory Manual for Physiological Studies of Rice. Manila: International Rice Research Institute.
Zhang, F., Sun, Y., Pei, W., Jain, A., Sun, R., Cao, Y., et al. (2015). Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82, 556–569. doi: 10.1111/tpj.12804
Zhao, F. J., Ago, Y., Mitani, N., Li, R. Y., Su, Y. H., Yamaji, N., et al. (2010). The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 186, 392–399. doi: 10.1111/j.1469-8137.2010.03192.x
Keywords: OsPT4, rice, arsenate, phosphate transporter, uptake
Citation: Ye Y, Li P, Xu T, Zeng L, Cheng D, Yang M, Luo J and Lian X (2017) OsPT4 Contributes to Arsenate Uptake and Transport in Rice. Front. Plant Sci. 8:2197. doi: 10.3389/fpls.2017.02197
Received: 21 September 2017; Accepted: 13 December 2017;
Published: 22 December 2017.
Edited by:
Vagner A. Benedito, West Virginia University, United StatesReviewed by:
Ahmad H. Kabir, University of Rajshahi, BangladeshCopyright © 2017 Ye, Li, Xu, Zeng, Cheng, Yang, Luo and Lian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingming Lian, eG1saWFuQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.