
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 December 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.02154
This article is part of the Research TopicStructure and Function of ChloroplastsView all 25 articles
The green alga Chlamydomonas reinhardtii is a key model organism for studying photosynthesis and oxidative stress in unicellular eukaryotes. Using a forward genetics approach, we have identified and characterized a mutant x32, which lacks a predicted protein named CGLD1 (Conserved in Green Lineage and Diatom 1) in GreenCut2, under normal and stress conditions. We show that loss of CGLD1 resulted in minimal photoautotrophic growth and PSII activity in the organism. We observed reduced amount of PSII complex and core subunits in the x32 mutant based on blue-native (BN)/PAGE and immunoblot analysis. Moreover, x32 exhibited increased sensitivity to high-light stress and altered tolerance to different reactive oxygenic species (ROS) stress treatments, i.e., decreased resistance to H2O2/or tert-Butyl hydroperoxide (t-BOOH) and increased tolerance to neutral red (NR) and rose bengal (RB) that induce the formation of singlet oxygen, respectively. Further analysis via quantitative real-time PCR (qRT-PCR) indicated that the increased singlet-oxygen tolerance of x32 was largely correlated with up-regulated gene expression of glutathione-S-transferases (GST). The phenotypical and physiological implications revealed from our experiments highlight the important roles of CGLD1 in maintaining structure and function of PSII as well as in protection of Chlamydomonas under photo-oxidative stress conditions.
Microalgae are major source of sustainable biofuel feedstock and CO2 sinks for the future (Brennan and Owende, 2010; Scranton et al., 2015). Compared to higher plants, microalgae have advantages in developing such kind of future because these organisms can be cultivated with CO2 supply toward reducing greenhouse gas emissions in non-arable areas (Pulz and Gross, 2004; Oey et al., 2013). However, there are limitations in available wild-type strains, including low efficiency of photosynthetic light utilization and high sensitivity to photo-oxidative stress, that impact the algal resource being economically competitive (Wijffels and Barbosa, 2010; Gimpel et al., 2013). To overcome these limitations, generation and characterization of novel mutant strains with desired phenotype is essential.
Chlamydomonas reinhardtii (henceforth referred to as Chlamydomonas) is one of the dominant model organisms for studying photosynthesis and oxidative stress in unicellular eukaryotes (Harris et al., 1989; Dent et al., 2015). This is virtually due to its advances in availability of genetic tools for transformation and selection, fully annotated genome (Merchant et al., 2007) that facilitates molecular genetics and ‘omics’ studies toward understanding of fundamental biological processes such as photosynthesis and stress responses (Fischer et al., 2005; Gonzalez-Ballester et al., 2010; Blaby et al., 2013, 2015; Heinnickel and Grossman, 2013; Wu et al., 2015), as well as the processes with biofuel significance including H2 photoproduction (Matthew et al., 2009; Chen et al., 2010; Toepel et al., 2013). Also, the organism is haploid capable of heterotrophic growth in the dark and highly sensitive to light, which makes Chlamydomonas extremely ideal for efficient generation and identification of photo-oxidative mutants such as sor1 and sak1 (Dent et al., 2001; Fischer et al., 2012; Wakao et al., 2014). The findings based on the studies of the mutants have improved our understanding of cell response in Chlamydomonas to oxidative stress substantially.
Recent advances in phylogenomics have established the GreenCut2 database which reveals 597 nucleus-encoded proteins conserved in plants and algae (Karpowicz et al., 2011). These proteins are presumed to be central to photosynthetic process (Dent et al., 2015). However, functional significance for more than half of these proteins including CGLD (Conserved in the Green Lineage and Diatom) remains to be experimentally determined (Karpowicz et al., 2011). Using forward genetic approach, we have isolated a number of Chlamydomonas mutants with reduced photosynthetic activity, including msf1 (Zhao et al., 2017) and x32 which lacks a predicted protein named CGLD1 encoded by Cre02.g084350. Most recent work from Arabidopsis thaliana and Synechocystis sp. Strain PCC 6803 showed that the homolog proteins of CGLD1 were presumably involved in efficient uptake of Mn2+ to thylakoids (Schneider et al., 2016) or homeostasis of Ca2+ or Mn2+ in Arabidopsis (Wang et al., 2016) or cyanobacterial cells (Brandenburg et al., 2017; Gandini et al., 2017). In contrast, phenotypic and physiological characterization of cgld1 mutant in Chlamydomonas, which is a key model system for photosynthesis research with great potential in synthetic biology and application (Scharff and Bock, 2014), is still limited (Dent et al., 2015; Schneider et al., 2016). No sufficient information is available regarding functional significance of CGLD1 in Chlamydomonas under various adverse conditions.
Here, we have investigated phenotypic and physiological characteristics of x32 mutant under normal and photo-oxidative stress conditions. We show that loss of CGLD1 resulted in minimal photoautotrophic growth and PSII activity in Chlamydomonas. We also show that the amount of PSII core proteins and PSII complex was drastically reduced in x32. Furthermore, we found that x32 mutant exhibits increased photoinhibition and altered tolerance to different reactive oxygen species (ROS) treatments. The increased tolerance to singlet oxygen could be largely attributed to up-regulated gene expression of glutathione-S-transferases (GST) in the mutant under such conditions.
Chlamydomonas reinhardtii wild-type strains CC400 (mt-), 137c (mt+), CC4051 (mt+) and cgld1 mutant (strain CAL029_02_05) was obtained from the Chlamydomonas Genetics Center1. The x32 mutant was isolated from an insertion mutant library constructed in our laboratory (Zhao et al., 2017) described in more detail below. The algal cells were cultured in TAP (Gorman and Levine, 1965) or in high-salt minimal (HSM) medium under continuous cool-white fluorescent light (60 μmol photons m-2 s-1) at 25°C. For all experiments, cells were grown to mid-exponential phase and harvested by centrifugation followed by adjusting the cell density to 2–4 × 106 cells ml-1. High-light stress treatment (1,300 μmol photons m-2 s-1) was performed according to (Zhao et al., 2013) except that the culture was transferred to flasks (25 ml) and the cell density was adjusted to 2 × 106 cells ml-1. ROS stress treatments were done as described (Chen et al., 2016). For mRNA and protein analysis, cells were harvested by centrifugation at 2500 g (4°C) for 5 min. After washing once with 0.01 M sodium phosphate buffer (pH 7.4), the cell pellets were stored at -70°C.
The insertion mutant library construction and photosynthetic mutants screen were described (Zhao et al., 2017). Briefly, the wild-type strain (CC400) was used and the mutant library was constructed by transforming this strain via the glass bead method with KpnI linearized plasmid pSI103 containing the aphVIII gene conferring paromomycin resistance (Kindle, 1990). Transformants that grew on TAP plates with 10 μg ml-1 paromomycin (Sigma) were isolated for photosynthetic mutants screening. Mutant screening was based on chlorophyll a fluorescence measurements. Sample preparation was done as previously (Zhao et al., 2013) and the measurements were taken with a chlorophyll fluorometer (Maxi-Imaging PAM; Walz, Effeltrich, Germany) by following the manufacturer’s instructions. Among the mutants with both the lowest Fv/Fm and Y(II) values, x32 was chosen for subsequent characterization.
Oxygen evolution rate of Chlamydomonas was measured as previously described (Sun et al., 2013) with a Chlorolab-2 oxygen electrode (Hansatech, Norfolk, United Kingdom). 77K fluorescence emission spectra were measured with a fluorescence spectrophotometer (F-2500; Hitachi, Japan) as described (Yang et al., 2014) with minor changes (Zhao et al., 2017). An excitation wavelength of 435 nm (5 nm bandwidth) was used to induce chlorophyll a fluorescence. The emission spectra were recorded (600–750 nm) and normalized at 716 nm. Light-induced redox changes of P700 were monitored by measuring absorbance at 820 nm using PAM101 fluorometer equipped with a dual-wavelength P700 unit (ED800T). Sample preparation was done according to Berry et al. (2011). A far-red light illumination (FR, 720 nm, 24 μmol photons m-2 s-1) was provided for 45 s to enable oxidation of P700 to a steady state then turned off to monitor the initial rate of P700+ dark reduction according to Klughammer and Schreiber (1998).
Genetic analysis and complementation was done as described (Zhao et al., 2017) with minor modifications. For DNA blot analysis, genomic DNA was isolated from wild-type (CC400) and x32 mutant using the Plant Genomic DNA Kit by following the manufacturer’s instructions (Tiangen Biotech; Beijing, China). About 10 μg of genomic DNA was digested overnight with the restriction endonucleases KpnI and Hind III (New England Biolabs). The fragments resulting from the digestion were separated by 0.8% agarose gel electrophoresis followed by blotting onto nitrocellulose membranes and hybridizing with the DIG high prime DNA labeling and detection starter kit II from Roche (Catalog NO. 11585614910). The detection was achieved using a chemiluminescent substrate CSPD (Disodium 3-(4-methoxyspiro {l,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) phenyl phosphate) (Catalog NO. 11 755 633 001).
For gene mapping, genomic DNA flanking the aphVIII gene was isolated using high efficiency TAIL-PCR with the specific primers listed in Supplementary Table 1 according to (Liu and Chen, 2007) with slight modifications. The primary amplification reactions (20 μL) was composed of 2 μL of PCR buffer (Takara Bio Inc, Otsu, Shiga; Japan), 200 μM of dNTPs (TransGen Biotech; Beijing, China), 1 μM of any of the LAD primers, 0.3 μM of SP0, 0.5 μL of LA Taq (Takara Bio Inc, Otsu, Shiga; Japan) and 20–30 ng of DNA. Each 25-μL secondary reaction contained 2.5 μL of PCR buffer, 200 μM each of dNTPs, 0.3 μM of AC1 and SP1, 0.5 μl of LA Taq, and 1 μL of 50-fold diluted primary product. The amplified products from the secondary reactions were analyzed by agarose gel electrophoresis and were purified prior to sequencing. Sequencing reactions were performed by Sunbiotech (Beijing Sunbiotech; China) and the data were used to search the Chlamydomonas genome.
For tetrad analysis, genetic crosses between x32 and the wild-type (137c, mt+) and zygote dissection were performed accordingly to (Harris et al., 1989). The mutant was backcrossed twice and four progeny from distinct zygotes of the second generation (T2-x32) were obtained. To complement the x32 mutant, the gene was amplified from wild-type (CC400) with primers listed in Supplementary Table 1. The amplification product was digested with NdeI and EcoRI and subcloned into similarly treated vector pDble (Fischer and Rochaix, 2001). The constructed plasmids were introduced into x32 mutant by transformation. The transformants were then analyzed by chlorophyll fluorescence measurements. Colonies that displayed a wild-type phenotype were also analyzed for the integration of x32 by PCR with the specific primers listed in Supplementary Table 1.
Cloning and heterologous expression were performed as previously described (Chen et al., 2010). Total RNA isolation/purification and reverse transcription reactions were done as described (Sun et al., 2013). Briefly, the coding region of the CGLD1 gene without transmembrane sequence was amplified by PCR with PrimeSTAR HS DNA Polymerase (Takara, Ohtsu, Japan) using specific primers Pet28-CGLD1-F/R (Supplementary Table 1). The amplified fragment was cloned directly into pEasy-blunt vector (Beijing TransGen Biotech, China), which was then transformed into competent Escherichia coli DH5α cells. Positive clones containing the recombinant plasmid were selected and sequenced to ensure the authenticity of the ORFs (Beijing Sunbiotech, China). Extraction and purification of E. coli proteins were done as described (Zhou et al., 2008). A rabbit serum was produced by MBL (MBL, Nagoya, Japan) using the purified recombinant CGLD1 protein as immunogen. Specificity of the antibody was verified by immunoblotting using proteins extracted from the E. coli and Chlamydomonas cells, respectively.
Cells breakage, isolation of membrane proteins for blue-native (BN)/SDS-PAGE and SDS-PAGE were done as described (Chen et al., 2016). BN gel was prepared and electrophoresis was performed at 4°C with the running program previously reported (Yang et al., 2014) and immunoblot analysis were done according to (Zhao et al., 2013, 2017). The antibodies against D1, D2, CP43, CP47, Cytb6f, and AtpB were from Agrisera and those against PsaB, Lhcb4, and LhcII were from J.-D. Rochaix (University of Geneva). The dilutions for the specific antibodies used in this study were: anti-CGLD1 (1:1000), anti-CP43 and CP47 (1:3000), anti-AtpB (1:4000), anti-D1 and D2 (1:5000), anti-PsaB, Cytb6, LhcII and Lhcb4 (1:10000). The immuno-signal was detected using the Pro-light HRP ECL detection system (Tiangen Biotech; Beijing, China). The blots were scanned using a UMAX Power-Look 2100XL scanner (Willich, Germany). Protein content was determined according to Peterson (1977) using BSA as standard.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was done as described (Zhao et al., 2013) using CBLP as the internal control. Gene-specific PCR primer pairs for psbA and psbC were designed using the online program Primer32. The primer pairs previously reported for APX1, CAT1, GSTS1, GSTS2, and GPXH (Fischer et al., 2012; Pokora et al., 2017) were used and listed in Supplementary Table 1. Relative abundance was expressed as the fold change in expression level relative to the reference, calculated as 2-ΔΔCT.
For enzyme activity assay, crude extracts preparation and determination of SOD and CAT activity was done as described (Chen et al., 2016).
The x32 mutant was isolated during a genetic screen for Chlamydomonas insertion mutants with decreased photosynthetic activity as described previously (for details see Zhao et al., 2017). Figure 1 shows that, compared to wild-type, growth of x32 in TAP and high salt minimal (HSM) medium (Figure 1A) was severely repressed, suggesting impaired photosynthesis in the mutant. This was confirmed by the measurements of in vivo chlorophyll a fluorescence (Imaging PAM, Heinz Walz, Germany). The maximal and effective quantum yield of PSII, Fv/Fm and Y(II), was reduced to the minimal (6.6 and 3.0% of the wild-type level, respectively) (Figure 1B). In line with these, the rate of photosynthetic oxygen evolution in x32 mutant was undetectable (Figure 1C). To obtain further functional insights of photosystems in x32, low temperature fluorescence (77K) emission spectra of x32 and wild-type cells were then compared (Figure 1D). A differential reduction of the fluorescence emission peak at 686 nm, which is characteristic for PSII mutants (Össenbuhl et al., 2004), was observed in x32 (Figure 1D). This indicates the level of functional PSII in the mutant was significantly lower than wild-type. Since no difference in the light-induced redox kinetics of PSI was revealed between wild-type and the mutant (Figure 1E), we conclude that only PSII is affected in the Chlamydomonas mutant x32.
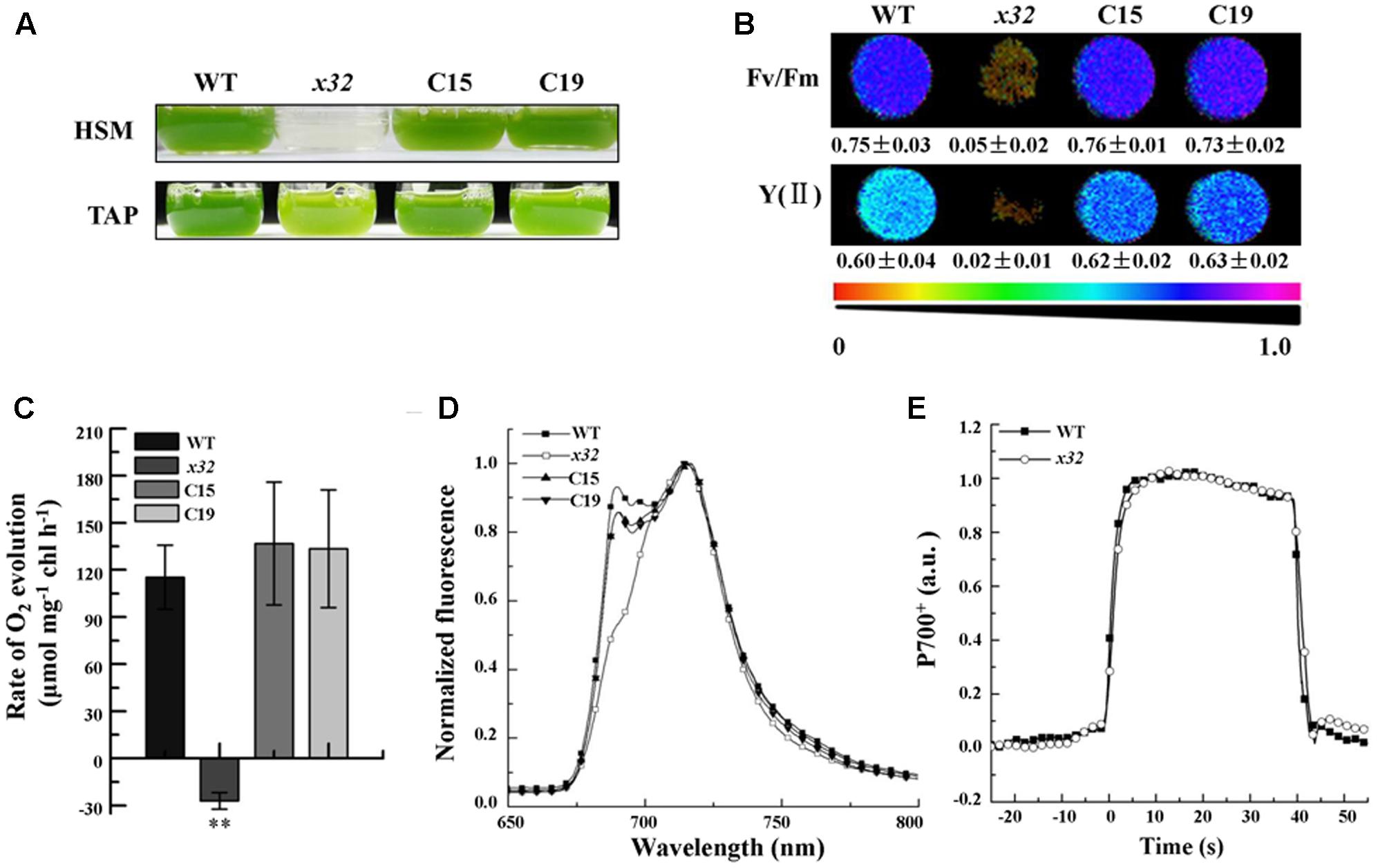
FIGURE 1. Growth and photosynthetic characters of wild-type (WT), x32 and the complemented strains (C15 and C19). (A) Growth in HSM and TAP medium with an irradiance of 60 μmol photons m–2 s–1 for 6 and 3 days, respectively. (B) Maximal (Fv/Fm) and effective quantum yield of PSII (YII) of the indicated strains. Standard deviations were estimated from three biological replicates. All experiments were repeated twice with similar results. (C) Rate of O2 evolution in different strains. Standard deviations were estimated from three biological replicates. All experiments were repeated twice with similar results. ∗∗p-value < 0.01 in Student’s t-test. (D) 77K fluorescence emission spectra of the indicated strains. The spectra were normalized at 716 nm. All experiments were repeated twice with similar results. (E) Light-induced redox kinetics of P700 in wild-type (WT) and x32 mutant. All experiments were repeated twice with similar results.
DNA blot analysis revealed a single insertion of the paromomycin resistance cassette in the genome of x32 (Figure 2A). The insertion site was mapped on the genome of Chlamydomonas via sequencing the flanking regions of the insert through thermal asymmetric interlaced PCR (Liu and Chen, 2007). Analysis of this genomic region in x32 revealed a DNA rearrangement that resulted in the deletion of five genes, i.e., Cre02.g084250, Cre02.g084300, Cre02.g084350, Cre02.g084400, and Cre02.g084450 (Figure 2B). Prediction by PredAlgo software3 shows that only the gene Cre02.g084350 encoding CGLD1 in the GreenCut2 (Karpowicz et al., 2011) is localized in the chloroplast. We therefore introduced the gene into x32 for complementation. The x32 mutant phenotype could be fully rescued in the complemented strains (C15, C19) based on growth, Fv/Fm and Y(II) values (Figure 1A). Analysis of genetic crosses between x32 mutant and wild-type was in agreement with these data (Supplementary Figures 1A,B). The photosynthetic defect of x32 co-segregated with paromomycin resistance in six complete tetrads analyzed, suggesting that the phenotype is linked to this genetic disruption.
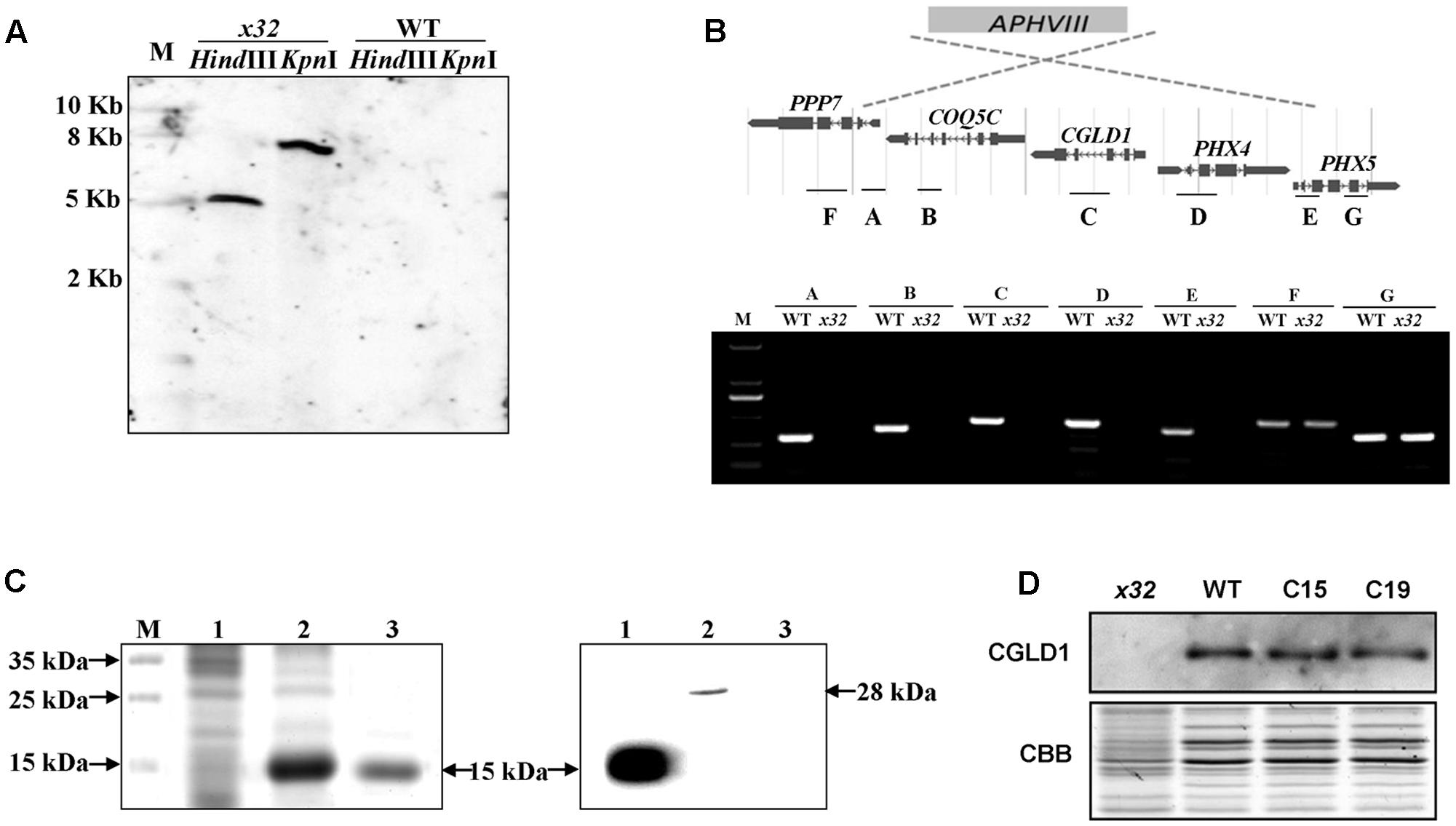
FIGURE 2. Genetic analysis of the x32 mutant. (A) DNA blot analysis of wild-type and x32. Genomic DNA was digested with HindIII and KpnI, fractionated by agarose gel electrophoresis and hybridized with a DNA probe of 407bp of APHVIII gene. (B) Mapping of the deletion in x32 caused by insertion of the paromomycin resistance cassette (APHVIII) by comparative analysis of wild-type and x32 DNA using PCR specific primer pairs. (C) Heterologous expression of recombinant CGLD1 protein and specificity of CGLD1 antibody. Coomassie-stained SDS-PAGE gel separating proteins extracted from Escherichia coli cells (left panel). M, protein molecular mass marker. 1 and 2, total proteins without or with isopropyl β-D-1-thiogalactopyranoside (IPTG) induction, respectively. 3, recombinant CGLD1 purified using Ni-NTA column. Specificity detection of the CGLD1 antibody by immunoblotting (right panel). Lane 1, purified recombinant CGLD1 protein (0.02 μg). Lanes 2 and 3, proteins (20 μg) extracted from wild-type and x32, respectively. (D) Immunoblot detection of CGLD1 in wild-type, x32 and the complemented strains C15 and C19. Membrane proteins (20 μg per lane) were separated by SDS-PAGE (16%) followed by immunoblotting with the CGLD1 antibody.
To further confirm the candidate gene, we compared the phenotype of x32 and another cgld1 mutant obtained from the Chlamydomonas Resource Center4, which was derived from wild-type strain CC4051 with an insertion in exon2 of the CGLD1 gene (Dent et al., 2015; Schneider et al., 2016), in the presence of additional manganese (Mn2+) and calcium (Ca2+) (Supplementary Figure 1C). The similar results of growth profiles and Fv/Fm values of the two cgld1 mutants (Schneider et al., 2016; Supplementary Figure 1C) verify that disruption of CGLD1 is the cause of the phenotype observed in x32.
In the genome of Chlamydomonas, CGLD1 is annotated as a predicted protein (Phytozome v12.15). To determine the expression of CGLD1 protein in wild-type Chlamydomonas and verify the lack of CGLD1 in x32, we generated an antibody against recombinant CGLD1 protein and verified its specificity by immunoblotting using protein extracts from the wild-type Chlamydomonas (Figure 2C). Heterologous expression and purification of the recombinant CGLD1 protein was performed as previously reported (Zhou et al., 2008; Sun et al., 2013; Zhao et al., 2017). As shown in Figure 2C, the E. coli Transetta (DE3) cells produced a substantial amount of the expected recombinant protein possessing an estimated molecular mass of 15 kDa. This corresponds to the molecular mass calculated from the coding region of Chlamydomonas CGLD1 gene without the transmembrane sequences (Figure 2C, left panel). Total proteins extracted from the E. coli cells and Chlamydomonas cells were analyzed by immunoblotting with the antibody. A single band at 28 and 15 kDa was detected in wild-type Chlamydomonas and E. coli expressing the recombinant CGLD1 protein (Figure 2C, right panel), respectively. The former corresponds to the molecular mass that was calculated from the coding sequence of CGLD1 gene in Chlamydomonas. Since no band was detected in x32 mutant (Figure 2C, right panel), this antibody was therefore used for quantification of CGLD1 in wild-type, x32 and the complemented (C15, C19) strains (Figure 2D).
The decreased PSII activity of x32 could be due to reduced accumulation of thylakoid membrane proteins. To clarify this, we analyzed membrane protein complexes of x32 and wild-type cells by blue-native (BN)/SDS-PAGE using a slightly modified protocol (Yang et al., 2014) as described (Chen et al., 2016). As shown in Figure 3A, the overall BN-gel profile of thylakoid membranes isolated from wild-type was similar to earlier reported (Dewez et al., 2009; Chen et al., 2016), demonstrating high technical reproducibility of the BN-PAGE from different experiments. Comparision of the BN-PAGE gel pattern between wild-type and x32 revealed a clear reduction of the band corresponding to a PSII supercomplex, PII1 containing core subunits of PSII (D1, D2, CP43, CP47) (Rexroth et al., 2003; Dewez et al., 2009), in the mutant (Figure 3A). This was further confirmed by immunoblotting using the antibodies specific for key proteins of photosynthetic apparatus (Figure 3B). The levels of core subunits of PSII (D1, D2, CP43, and CP47) in x32 were 30% or less of that in wild-type whereas the levels of the key subunits of PSI (PsaB), Cytb6f (Cytb6), and ATP synthase (AtpB) as well as the LHCII proteins (LhcII and Lhcb4) remained unchanged in x32 (Figure 3B). To test whether the reduced levels of the core PSII proteins in x32 was due to limited transcription, we then compared the psbA and psbC mRNA levels between wild-type and x32 by quantitative real-time PCR (qRT-PCR) analysis. The transcript level of both genes was decreased 67.2% (psbA) and 68.3% (psbC) in relation to the wild-type (Figure 3C). Thus, these experimental data demonstrate a significant and specific impact of CGLD1 on PSII at both mRNA and protein level in Chlamydomonas.
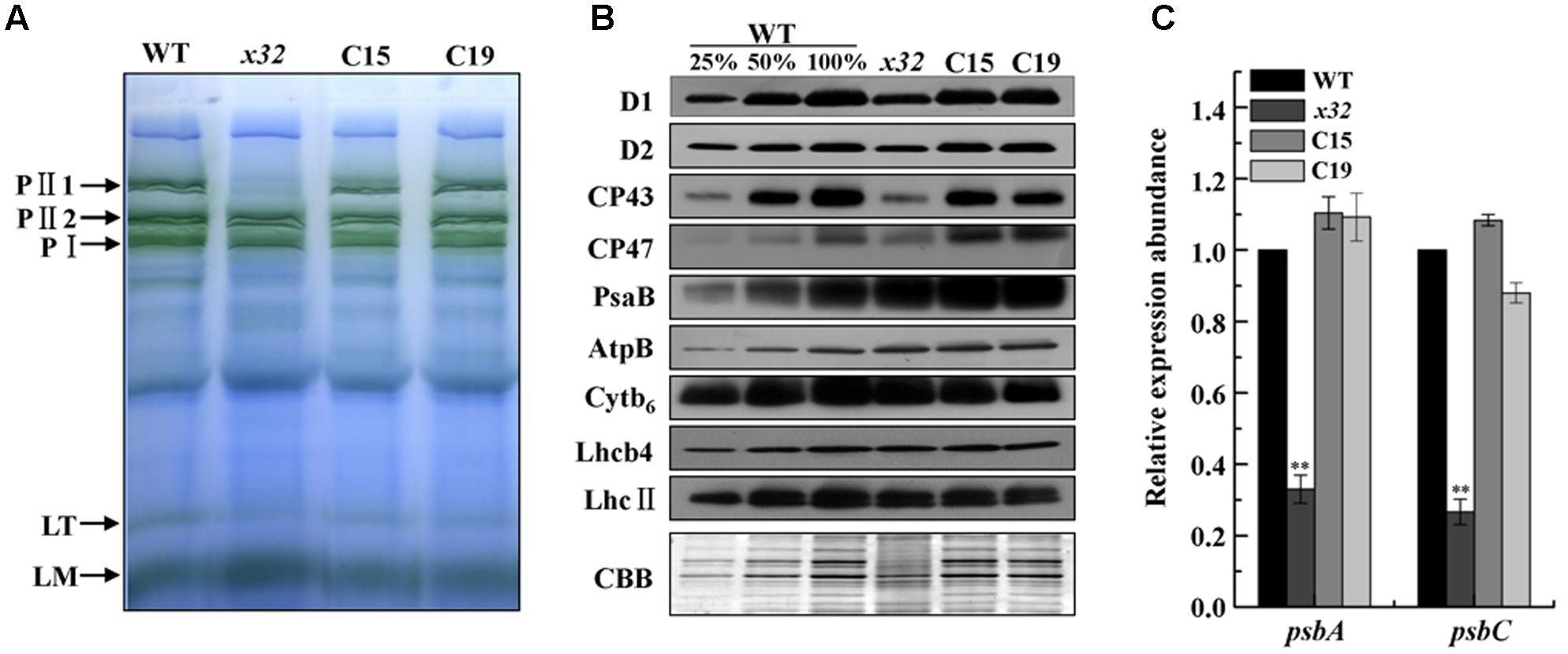
FIGURE 3. Abundance of photosynthetic proteins and complexes in thylakoid membranes of WT, x32 and the complemented strains (C15 and C19). (A) BN-PAGE of the thylakoid membranes. DM-solubilized membrane proteins (25 μg per lane) were separated by BN-PAGE. PII1, PII2, PSII supercomplexes; PI, PSI-LHCI complex; LT, LHCII trimer; LM, LHCII monomer. Similar results were obtained in two independent experiments. (B) Immunoblot detection of various photosynthetic proteins. Membrane proteins (10 μg per lane) were separated by 12% SDS-PAGE followed by immunoblot detection of indicated proteins. A dilution series of wild-type protein extract was used to quantify the amount of proteins in x32. Similar results were obtained in at least three independent experiments. (C) qRT-PCR analysis of mRNA levels of psbA and psbC in the indicated strains. Standard deviations were estimated from three biological replicates. Similar results were obtained in three independent experiments. CBLP gene was used as a control. ∗∗p-value < 0.01 in Student’s t-test.
To determine potential physiological roles of CGLD1 protein in Chlamydomonas under adverse condition, we performed various stress experiments. Because high-light is the most common stress condition occurring to photosynthetic organisms and Chlamydomonas is a key model system for studying photo-oxidative stress in unicellular eukaryotes, we compared CGLD1 levels of wild-type cells after treatments with high-light (1,300 μmol photons m-2 s-1) and different ROS, i.e., H2O2, neutral red (NR) and rose bengal (RB). The stress treatments were done as described (Zhao et al., 2013; Chen et al., 2016) and the CGLD1 protein was monitored using immunoblot analyses. As shown in Figure 4A, the amount of CGLD1 was in most cases increased after these stress treatments, indicating that this protein is also involved in photoinhibition/photoprotection against photo-oxidative stress in the organism. To confirm these, we then investigated the sensitivity of x32 to high-light stress treatment (Figure 4B). As expected, cell growth of x32 was significantly repressed during high-light stress treatment (1,300 μmol photons m-2 s-1) compared to wild-type (Figure 4B). The remarkable reduction of cell growth was observed upon exposure of x32 to high light for 40 min. This was more apparent based on Fv/Fm values (Figure 4B, right panel), suggesting increased photoinhibition in the mutant.
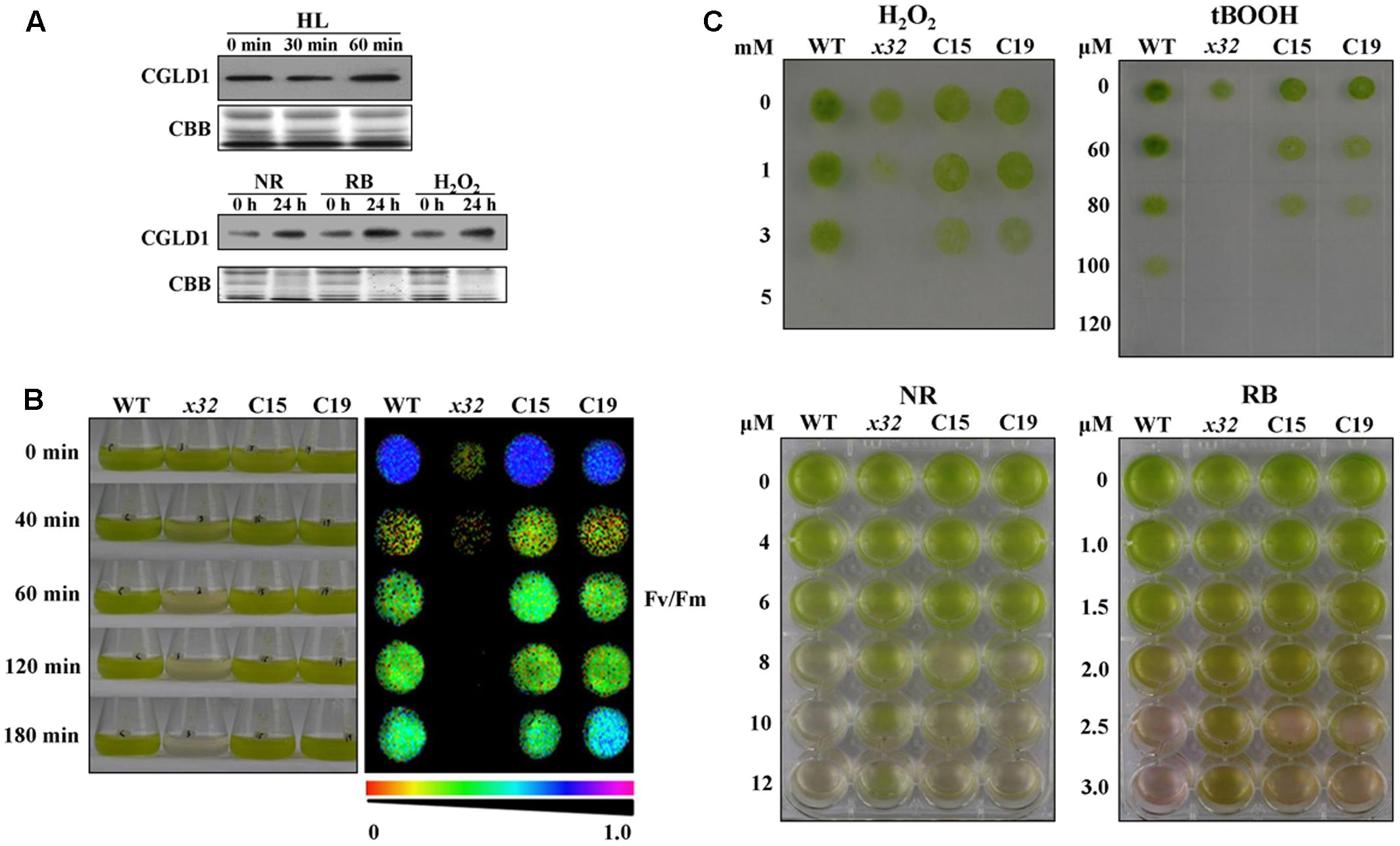
FIGURE 4. High-light and ROS-stress tolerance of WT, x32 and the complemented strains (C15 and C19). (A) Immunoblot analyses of CGLD1 amounts in Chlamydomonas under high-light (HL; 1,300 μmol photons m–2 s–1) and ROS-stress conditions. Membrane proteins (10 μg per lane) were separated by SDS-PAGE (16%) followed by immunoblotting with the CGLD1 antibody. Similar results were obtained in three independent experiments. (B) Growth and quantum yield of PSII (Fv/Fm) of the indicated strains under high-light treatment (1,300 μmol photons m–2 s–1). Similar results were obtained in at least three independent experiments. (C) ROS tolerance of the indicated strains. Growth recovery after treatment with H2O2, t-BOOH (tert-Butyl hydroperoxide) followed by spotting on TAP plates for 3 days (upper). Growth after treatment with NR (neutral red) and RB (rose bengal). Similar results were obtained in at least three independent experiments.
To determine whether the tolerance of x32 mutant to oxidative stress was changed due to loss of CGLD1, we also compared the sensitivity of x32 and wild- type cells to different ROS treatments. Similar results were obtained with wild-type strain in the present (Figure 4B) and earlier investigations (Chen et al., 2016). However, the x32 mutant was more sensitive to H2O2 and tert-Butyl hydroperoxide (t-BOOH) than wild-type (Figure 4C, upper panel). Correlated with this, both gene expression of ascorbate peroxidase 1 (APX1) and catalase 1 (CAT1) (Supplementary Figure 2), which are known to be H2O2-responsive marker genes (Wakao et al., 2014), as well as the increase of activity of superoxide dismutases (SOD) and catalases (CAT), which are the dominant ROS scavenging enzymes known to be present in the chloroplasts, was less pronounced in x32 mutant than in wild-type (Supplementary Figure 3). Interestingly, our experimental data shows that x32 was more resistant to neutral red (NR) and rose bengal (RB) than wild-type (Figure 4B, lower panel). These observations raise the possibility that anti-singlet oxygen response is enhanced in x32 mutant under such stress conditions.
To test the possibility mentioned above, we measured the mRNA levels of several key genes encoding glutathione-S-transferases and thioredoxin peroxidase, i.e., GSTS and GPXH, that are known to be involved in singlet oxygen-induced acclimation process in Chlamydomonas (Leisinger et al., 2001; Fischer et al., 2005, 2009; Ledford et al., 2007) using qRT-PCR. Total mRNA isolated from different strains treated with NR or RB was subjected for the analysis. Figure 5 shows that expression of these genes was in most cases increased in all strains. The fold-increase of expression in wild-type was similar to the earlier report (Fischer et al., 2005). In x32, a differential increase pattern of expression was observed compared to that in wild-type (Figures 5A–C). Higher expression of GSTS1 and GSTS2 in x32 was most remarkable under NR (2.3-fold) and RB (8.9-fold) treatment, respectively. Based on these experimental results, we conclude that the singlet-oxygen resistance observed in the mutant could be largely attributed to the up-regulated expression of GST genes.
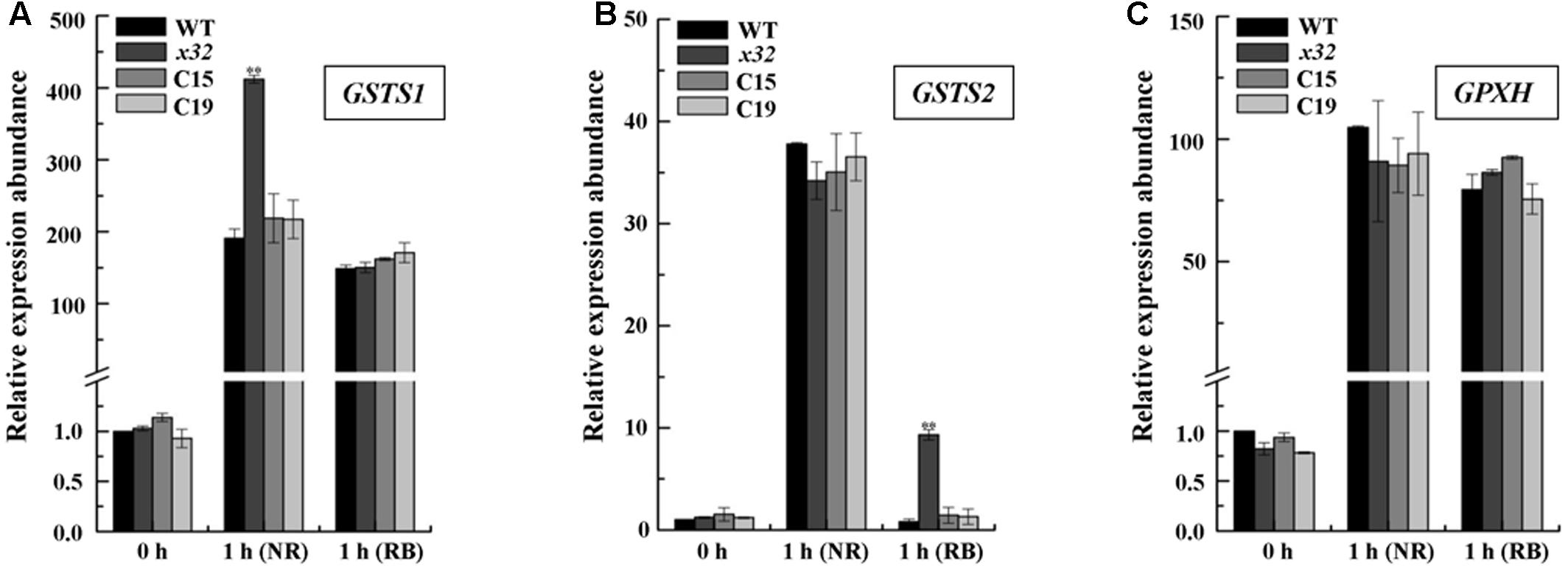
FIGURE 5. Up-regulated GSTS expression in x32 mutant under singlet oxygen stress. qRT-PCR analysis of expression of GSTS1 (A), GSTS2 (B), and GPXH (C) in WT, x32 and the complemented strains (C15 and C19) treated with NR and RB, respectively. Standard deviations were estimated from three biological replicates. Similar results were obtained in at least three independent experiments. CBLP gene was used as a control. ∗p-values < 0.05, ∗∗p < 0.01 in Student’s t-test, respectively.
In this work, we have isolated and characterized x32 mutant lacking CGLD1, a predicted protein belongs to the GreenCut2 superfamily, in Chlamydomonas. We have provided direct experimental evidence of CGLD1 in maintaining structure and function of PSII as well as in protecting Chlamydomonas against photo-oxidative stress. New insights revealed in this work are discussed below.
In the genome of Chlamydomonas. CGLD1 is annotated as a predicted protein with unknown function (Phytozome v12.16). Using a forward genetics approach, we have determined its important and specific role in PSII functionality of this organism (Figures 1, 3). This finding is somewhat different from that reported in Arabidopsis and Synechocystis, showing that not only PSII supercomplex but also PSI complex was significantly reduced (Schneider et al., 2016; Gandini et al., 2017). Indeed, functional and structural analysis of the photosystems, i.e., low temperature fluorescence (77K) emission spectra and light-induced redox kinetics of P700, BN-PAGE and immunoblot quantifications (Figures 1, 3), allows us to propose a crucial role of CGLD1 for maintaining PSII in Chlamydomonas. Also, based on the genomic information, we have generated a specific antibody against CGLD1 and verified the expression of CGLD1 in wild-type Chlamydomonas (Figure 2). These are novel and direct experimental evidence of presence and major function of CGLD1 protein in Chlamydomonas.
Similar to the observations in Arabidopsis and the original cgld1 mutant (Schneider et al., 2016), which was derived from a different wild-type Chlamydomonas strain (Dent et al., 2015), supplementation of excess Mn2+ rather than Ca2+ could largely restored the photosynthetic activity (Fv/Fm) in x32 mutant (Supplementary Figure 1). These consistent results obtained from different Chlamydomonas strains as well as the cyanobacterium Synechocystis (Brandenburg et al., 2017; Gandini et al., 2017), strongly support the suggestion that CGLD1 is involved in uptake/maintenance of Mn2+ homeostasis in photosynthetic organisms. Nevertheless, how this is exactly elicited in chloroplast and cyanobacteria remains an open question. Although direct experimental evidences are currently lacking it is presumed that in Arabidopsis the homolog of CGLD1 (PAM71) functions in Mn2+ uptake into thylakoids for optimal PSII performance (Schneider et al., 2016). In Chlamydomonas, it is not fully understood how deletion of CGLD1 impaires PSII under normal growth condition. Based on the similar photosynthetic phenotype of Chlamydomonas and Arabidopsis in response to excess Mn2+ (Supplementary Figure 1; Schneider et al., 2016), it could be postulated that CGLD1 protein in Chlamydomonas functions in the similar way that proposed for its homolog in Arabidopsis (Schneider et al., 2016) under normal growth condition.
No information is available so far characterizing the expression of CGLD1 protein under adverse conditions including high-light irradiation and oxidative stress. Our finding of elevated level of CGLD1 protein in wild-type Chlamydomonas cells after these treatments (Figure 4) implicates its putative function in photo-oxidative responses. Physiological significance of CGLD1 under these stress conditions was demonstrated by the distinct phenotype of wild-type and x32 (Figure 4). Based on the increased sensitivity of the mutant to high-light and peroxide stress, we propose that CGLD1 is also involved in photoinhibition or photodamage/repair of PSII. Since D1 protein is widely accepted as the primary target damaged during photoinhibition in vivo (Mulo et al., 2012) and a repair mechanism which involves an intricate and multi-step process operates in all photosynthetic organisms (Nixon et al., 2010; Nickelsen and Rengstl, 2013), we could speculate that deletion of CGLD1 may impact efficient PSII repair (including D1 synthesis) in the organism. Further research is directed toward in-depth understanding of the underlying molecular mechanisms.
Interestingly, we found different response patterns of x32 to photo-oxidative stress in comparison with wild-type Chlamydomonas. While higher sensitivity of the mutant to high-light and peroxide stress, as frequently reported for PSII mutants as well as the reduced SOD and CAT activity in x32 treated with H2O2 (Supplementary Figure 3), was expected, the increased tolerance to singlet-oxygen stress was for the first time revealed in a Chlamydomonas mutant lacking CGLD1 (Figure 4). The reason for this novel phenotype of x32 is currently unclear. In sor1 mutant, which lacks a basic leucine zipper transcription factor, the singlet-oxygen resistance has been mainly attributed to up-regulated expression of the stress responsive genes GSTS and GPXH in Chlamydomonas (Fischer et al., 2012). Our finding of up-regulated GSTS expression in x32 under singlet oxygen stress (Figure 5) is of indication that the detoxifying system described in sor1 is also reinforced in this mutant under such stress conditions. Moreover, it has been previously reported that non-photochemical quenching (NPQ) was increased compared to the wild-type (Dent et al., 2015). Considering that NPQ is one of the efficient modules in photoprotection, we would presume that the NPQ mechanism is also enhanced in x32 mutant lacking CGLD1 and contributes at least partially to its increased resistance to singlet oxygen stress.
In summary, the present work determined expression and functional significance of CGLD1 in Chlamydomonas. Loss of CGLD1 leads to minimal photoautotrophic growth and PSII activity in x32 mutant. Biochemical analysis of x32 revealed that the steady levels of PSII supercomplex and core proteins was dramatically reduced compared to wild-type. Furthermore, we found that x32 was, due to loss of CGLD1, more tolerant to singlet oxygen stress than wild-type. Correlated with this, up-regulated GSTS expression was found increased more in x32. The phenotypical and physiological implications revealed from this study provide important information for in-depth studies toward understanding structure and function of CGLD1 in Chlamydomonas as well as a valuable alga strain with increased resistance to singlet oxygen stress for potential applications.
JX and FH conceived the research and designed the experiments. JX, PL, and LZ performed the experiments. JX and FH analyzed data and wrote the manuscript.
This study was supported by grants from the National Natural Science Foundation of China (Nos. 31670239, 31470340), National Basic Research Program of China (973 Program, No. 2015CB150100) and the funding from the Strategic Priority Research Program (Nos. XDB17000000; KGCX2-YW-373) of Chinese Academy of Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02154/full#supplementary-material
Berry, L. L., Brzezowski, P., and Wilson, K. E. (2011). Inactivation of the STT7 gene protects PsaF-deficient Chlamydomonas reinhardtii cells from oxidative stress under high light. Physiol. Plant. 141, 188–196. doi: 10.1111/j.1399-3054.2010.01421.x
Blaby, I. K., Blaby-Haas, C. E., Perez-Perez, M. E., Schmollinger, S., Fitz-Gibbon, S., Lemaire, S. D., et al. (2015). Genome-wide analysis on Chlamydomonas reinhardtii reveals the impact of hydrogen peroxide on protein stress responses and overlap with other stress transcriptomes. Plant J. 84, 974–988. doi: 10.1111/tpj.13053
Blaby, I. K., Glaesener, A. G., Mettler, T., Fitz-Gibbon, S. T., Gallaher, S. D., Liu, B., et al. (2013). Systems-level analysis of nitrogen starvation–induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25, 4305–4323. doi: 10.1105/tpc.113.117580
Brandenburg, F., Schoffman, H., Kurz, S., Kramer, U., Keren, N., Weber, A. P. M., et al. (2017). The Synechocystis manganese exporter Mnx is essential for manganese homeostasis in Cyanobacteria. Plant Physiol. 173, 1798–1810. doi: 10.1104/pp.16.01895
Brennan, L., and Owende, P. (2010). Biofuels from microalgae-a review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 14, 557–577. doi: 10.1016/j.rser.2009.10.009
Chen, M., Zhang, J., Zhao, L., Xing, J., Peng, L., Kuang, T., et al. (2016). Loss of algal proton gradient regulation 5 increases reactive oxygen species scavenging and H2 evolution. J. Integr. Plant Biol. 58, 943–946. doi: 10.1111/jipb.12502
Chen, M., Zhao, K., Sun, Y. L., Cui, S. X., Zhang, L. F., Yang, B., et al. (2010). Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J. Proteome Res. 9, 3854–3866. doi: 10.1021/pr100076c
Dent, R. M., Han, M., and Niyogi, K. K. (2001). Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii. Trends Plant Sci. 6, 364–371. doi: 10.1016/S1360-1385(01)02018-0
Dent, R. M., Sharifi, M. N., Malnoe, A., Haglund, C., Calderon, R. H., Wakao, S., et al. (2015). Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 82, 337–351. doi: 10.1111/tpj.12806
Dewez, D., Park, S., Garcia-Cerdan, J. G., Lindberg, P., and Melis, A. (2009). Mechanism of REP27 protein action in the D1 protein turnover and photosystem II repair from photodamage. Plant Physiol. 151, 88–99. doi: 10.1104/pp.109.140798
Fischer, B. B., Dayer, R., Schwarzenbach, Y., Lemaire, S. D., Behra, R., Liedtke, A., et al. (2009). Function and regulation of the glutathione peroxidase homologous gene GPXH/GPX5 in Chlamydomonas reinhardtii. Plant Mol. Biol. 71, 569–583. doi: 10.1007/s11103-009-9540-8
Fischer, B. B., Krieger-Liszkay, A., and Eggen, R. I. L. (2005). Oxidative stress induced by the photosensitizers neutral red (type I) or rose Bengal (type II) in the light causes different molecular responses in Chlamydomonas reinhardtii. Plant Sci. 168, 747–759. doi: 10.1016/j.plantsci.2004.10.008
Fischer, B. B., Ledford, H. K., Wakao, S., Huang, S. G., Casero, D., Pellegrini, M., et al. (2012). SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 109, E1302–E1311. doi: 10.1073/pnas.1116843109
Fischer, N., and Rochaix, J. D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894. doi: 10.1007/s004380100485
Gandini, C., Schmidt, S. B., Husted, S., Schneider, A., and Leister, D. (2017). The transporter SynPAM71 is located in the plasma membrane and thylakoids, and mediates manganese tolerance in Synechocystis PCC6803. New Phytol. 215, 256–268. doi: 10.1111/nph.14526
Gimpel, J. A., Specht, E. A., Georgianna, D. R., and Mayfield, S. P. (2013). Advances in microalgae engineering and synthetic biology applications for biofuel production. Curr. Opin. Chem. Biol. 17, 489–495. doi: 10.1016/j.cbpa.2013.03.038
Gonzalez-Ballester, D., Casero, D., Cokus, S., Pellegrini, M., Merchant, S. S., and Grossman, A. R. (2010). RNA-Seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22, 2058–2084. doi: 10.1105/tpc.109.071167
Gorman, D. S., and Levine, R. P. (1965). Cytochrome F and plastocyanin - their sequence in photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669. doi: 10.1073/pnas.54.6.1665
Harris, E. H., Burkhart, B. D., Gillham, N. W., and Boynton, J. E. (1989). Antibiotic-resistance mutations in the chloroplast 16s and 23s rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123, 281–292.
Heinnickel, M. L., and Grossman, A. R. (2013). The GreenCut: re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth. Res. 116, 427–436. doi: 10.1007/s11120-013-9882-6
Karpowicz, S. J., Prochnik, S. E., Grossman, A. R., and Merchant, S. S. (2011). The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286, 21427–21439. doi: 10.1074/jbc.M111.233734
Kindle, K. L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232. doi: 10.1073/pnas.87.3.1228
Klughammer, C., and Schreiber, U. (1998). Measuring P700 absorbance changes in the near infrared spectral region with a dual wavelength pulse modulation system. Photosynth. Mech. Effects I–V, 4357–4360. doi: 10.1007/978-94-011-3953-3_1008
Ledford, H. K., Chin, B. L., and Niyogi, K. K. (2007). Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot. Cell 6, 919–930. doi: 10.1128/Ec.00207-06
Leisinger, U., Rufenacht, K., Fischer, B., Pesaro, M., Spengler, A., Zehnder, A. J. B., et al. (2001). The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Mol. Biol. 46, 395–408. doi: 10.1023/A:1010601424452
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–656. doi: 10.2144/000112601
Matthew, T., Zhou, W. X., Rupprecht, J., Lim, L., Thomas-Hall, S. R., Doebbe, A., et al. (2009). The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 284, 23415–23425. doi: 10.1074/jbc.M109.003541
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–251. doi: 10.1126/science.1143609
Mulo, P., Sakurai, I., and Aro, E.-M. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim. Biophys. Acta 1817, 247–257. doi: 10.1016/j.bbabio.2011.04.011
Nickelsen, J., and Rengstl, B. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635. doi: 10.1146/annurev-arplant-050312-120124
Nixon, P. J., Michoux, F., Yu, J. F., Boehm, M., and Komenda, J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16. doi: 10.1093/aob/mcq059
Oey, M., Ross, I. L., Stephens, E., Steinbeck, J., Wolf, J., Radzun, K. A., et al. (2013). RNAi Knock-Down of LHCBM1, 2 and 3 increases photosynthetic H2 production efficiency of the green alga Chlamydomonas reinhardtii. PLOS ONE 8:e61375. doi: 10.1371/journal.pone.0061375
Össenbuhl, F., Gohre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J. D., and Eichacker, L. A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16, 1790–1800. doi: 10.1105/tpc.023226
Peterson, G. L. (1977). A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83, 346–356. doi: 10.1016/0003-2697(77)90043-4
Pokora, W., Aksmann, A., Bascik-Remisiewicz, A., Dettlaff-Pokora, A., Rykaczewski, M., Gappa, M., et al. (2017). Changes in nitric oxide/hydrogen peroxide content and cell cycle progression: study with synchronized cultures of green alga Chlamydomonas reinhardtii. J. Plant Physiol. 208, 84–93. doi: 10.1016/j.jplph.2016.10.008
Pulz, O., and Gross, W. (2004). Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 65, 635–648. doi: 10.1007/s00253-004-1647-x
Rexroth, S., Meyer, J. M. W., Tittingdorf, Z., Frank Krause, F., Dencher, N. A., and Seelert, H. (2003). Thylakoid membrane at altered metabolic state: challenging the forgotten realms of the proteome. Electrophoresis 24, 2814–2823. doi: 10.1002/elps.200305543
Scharff, L. B., and Bock, R. (2014). Synthetic biology in plastids. Plant J. 78, 783–798. doi: 10.1111/tpj.12356
Schneider, A., Steinberger, I., Herdean, A., Gandini, C., Eisenhut, M., Kurz, S., et al. (2016). The evolutionarily conserved protein PHOTOSYNTHESIS AFFECTED MUTANT71 is required for efficient manganese uptake at the thylakoid membrane in Arabidopsis. Plant Cell 28, 892–910. doi: 10.1105/tpc.15.00812
Scranton, M. A., Ostrand, J. T., Fields, F. J., and Mayfield, S. P. (2015). Chlamydomonas as a model for biofuels and bio-products production. Plant J. 82, 523–531. doi: 10.1111/tpj.12780
Sun, Y. L., Chen, M., Yang, H. M., Zhang, J., Kuang, T. Y., and Huang, F. (2013). Enhanced H2 photoproduction by down-regulation of ferredoxin-NADP+ reductase (FNR) in the green alga Chlamydomonas reinhardtii. Int. J. Hydrogen Energy 38, 16029–16037. doi: 10.1016/j.ijhydene.2013.10.011
Toepel, J., Illmer-Kephalides, M., Jaenicke, S., Straube, J., May, P., Goesmann, A., et al. (2013). New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol. J. 11, 717–733. doi: 10.1111/pbi.12062
Wakao, S., Chin, B. L., Ledford, H. K., Dent, R. M., Casero, D., Pellegrini, M., et al. (2014). Phosphoprotein SAK1 is a regulator of acclimation to singlet oxygen in Chlamydomonas reinhardtii. eLife 3:e02286. doi: 10.7554/eLife.02286
Wang, C., Xu, W. T., Jin, H. L., Zhang, T. J., Lai, J. B., Zhou, X., et al. (2016). A putative chloroplast-localized Ca2+/H+ antiporter CCHA1 is involved in calcium and pH homeostasis and required for PSII function in Arabidopsis. Mol. Plant 9, 1183–1196. doi: 10.1016/j.molp.2016.05.015
Wijffels, R. H., and Barbosa, M. J. (2010). An outlook on microalgal biofuels. Science 330, 796–799. doi: 10.1126/science.1189003
Wu, G. X., Hufnagel, D. E., Denton, A. K., and Shiu, S. H. (2015). Retained duplicate genes in green alga Chlamydomonas reinhardtii tend to be stress responsive and experience frequent response gains. BMC Genomics 16:149. doi: 10.1186/S12864-015-1335-5
Yang, H. M., Liao, L. B., Bo, T. T., Zhao, L., Sun, X. W., Lu, X. F., et al. (2014). Slr0151 in Synechocystis sp PCC 6803 is required for efficient repair of photosystem II under high-light condition. J. Integr. Plant Biol. 56, 1136–1150. doi: 10.1111/jipb.12275
Zhao, L., Chen, M., Cheng, D. M., Yang, H. M., Sun, Y. L., Zhou, H. Y., et al. (2013). Different B-type methionine sulfoxide reductases in Chlamydomonas may protect the alga against high-light, sulfur-depletion, or oxidative stress. J. Integr. Plant Biol. 55, 1054–1068. doi: 10.1111/jipb.12104
Zhao, L., Cheng, D. M., Huang, X. H., Chen, M., Dall’Osto, L., Xing, J. L., et al. (2017). A light harvesting complex- like protein in maintenance of photosynthetic components in Chlamydomonas. Plant Physiol. 174, 2419–2433. doi: 10.1104/pp.16.01465
Keywords: C. reinhardtii, x32 mutant, PSII, photo-oxidative stress, singlet oxygen
Citation: Xing J, Liu P, Zhao L and Huang F (2017) Deletion of CGLD1 Impairs PSII and Increases Singlet Oxygen Tolerance of Green Alga Chlamydomonas reinhardtii. Front. Plant Sci. 8:2154. doi: 10.3389/fpls.2017.02154
Received: 28 September 2017; Accepted: 05 December 2017;
Published: 15 December 2017.
Edited by:
Hongbo Gao, Beijing Forestry University, ChinaReviewed by:
Deqiang Duanmu, Huazhong Agricultural University, ChinaCopyright © 2017 Xing, Liu, Zhao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Huang, Zmh1YW5nQGliY2FzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.