- 1Guangdong Provincial Key Laboratory of Applied Botany, Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China
- 2College of Life Science, Institute of Plant Biology, National Taiwan University, Taipei, Taiwan
In eukaryotic cells, histone acetylation and deacetylation play an important role in the regulation of gene expression. Histone acetylation levels are modulated by histone acetyltransferases and histone deacetylases (HDACs). Recent studies indicate that HDACs play essential roles in the regulation of gene expression in plant response to environmental stress. In this review, we discussed the recent advance regarding the plant HDACs and their functions in the regulation of abiotic stress responses. The role of HDACs in autophagy was also discussed.
Introduction
In eukaryotes, DNA is packaged into chromatin with the core histone proteins including histone H2A, H2B, H3, and H4. Histone proteins are subjected to various post-translational modifications including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP ribosylation (Berger, 2007). Histone acetylation plays fundamental roles in the regulation of gene expression in many aspects of biological processes. Histone acetylation levels are reversibly regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs can acetylate the specific lysine residues on N-termini of histone H3 and H4, resulting in activation of gene transcription. On the contrary, HDACs link to transcriptional repression and gene silencing through deacetylation of lysine residues. Chromatin structure can be affected by altering the acetylation of the histone tails by HATs and HDACs through mechanism of the effect of so-called “open/closed DNA” for transcription. HDACs lack intrinsic DNA-binding activity and are recruited to target genes via their association with transcriptional factors as well as their incorporation into large multiprotein transcriptional complexes. Furthermore, removing acetyl groups from histone by HDACs causes the chromatin more tightly packed and leads to a reduced accessibility for transcription factors to bind to the DNA, resulting in transcriptional repression. Additionally, the removal of acetyl group from N- terminal tails of histone may also change the structure of chromatin and modulate the interaction of histone tails with its interacting partners. Multiple specific lysine sites of histone acetylation have been identified, including histone H2AK5, H2B (K5, K12, K15, K20), H3 (K4, K9, K14, K18, K23, K27), and H4 (K5, K8, K16, K12, K16) (Peterson and Laniel, 2004). In general, H2A and H2B acetylation is associated with gene activation. Similarly, the combination of H3K9 and H3K14 acetylation is also linked to transcriptional activation. In addition, H4K5 acetylation is connected with transcriptional activation, histone deposition and DNA repair (Peterson and Laniel, 2004).
HDACs in Plants
Histone deacetylases are highly conserved in many organisms, including fungi, animals, and plants (Ekwall, 2005). Multiple HDACs have been identified and characterized in plants (Hollender and Liu, 2008; Hu et al., 2009; Aquea et al., 2010; Liew et al., 2013; Zhao L. M. et al., 2015; Peng et al., 2017). Eighteen HDACs identified in Arabidopsis can be grouped into three main families including the RPD3/HDA1 family (homologous to yeast RPD3), Sir2 family (homologous to yeast Sir2), HD family (plant specific HDACs). The RPD3 family HDACs can be further divided into three classes by sequence similarity: class I (HDA6, HDA7, HDA9, and HDA19), class II (HDA5, HDA15, and HDA18), and class III (HDA8, HDA10, HDA14, and HDA17). HD2 proteins are plant specific HDACs, which was firstly identified in maize by Lusser et al. (1997). There are four HD2 type HDACs including HD2A, HD2B, HD2C, and HD2D in Arabidopsis. Sir2 family HDACs are nicotinamide adenine dinucleotide (NAD) dependent HDACs and there are two members of Sir2-like HDACs, SIR1 and SIR2. In Arabidopsis, HDACs play vital roles in plant development and in responses to various stresses (Hollender and Liu, 2008; Luo et al., 2012a,d, 2015; Han et al., 2016; Yu et al., 2017).
The rice genome also contains 18 HDACs, including 14 members of the RPD3/HDA1 family, two members of the Sir2 family and two members of the HD2 family (Hu et al., 2009). Several rice HDACs were reported to function in responses to various abiotic stress (Hu et al., 2009; Zhao J. H. et al., 2015; Zhao et al., 2016). Fifteen HDACs were characterized in maize (Zea mays), including 10 members of the RPD3/HDA1 family, one member of the SIR2 family, and four members of HD2 family (Hu et al., 2011). It was reported that ZmHDACs might participate in cold stress responses by selectively regulate the transcription of cold-responsive genes (Hu et al., 2011). Twenty-eight HDACs were identified in Soybean (Glycine max), including six members in the HD2 family, four members in the SIR2 family, and 14 members in the RPD3/HDA1 family. Genome-wide RNA-seq analysis indicated that GmHDACs might be involved in the gene regulation during flower initiation (Liew et al., 2013). The grape (Vitis vinifera) genome contains 13 HDAC genes (Aquea et al., 2010). Eleven HDACs were characterized in litchi (Litchi chinensis Sonn. cv. Feizixiao), which might play important roles in fruit abscission (Peng et al., 2017). In addition, 15 HDACs were identified in tomato (Solanum lycopersicum), which might be involved in gene regulation during reproductive development (Zhao L. M. et al., 2015).
Histone deacetylases can contribute to the establishment of epigenetic states and mediate the crosstalk of histone acetylation with other histone modifications. Our previous studies revealed that HDA6 interacts with the DNA methyltransfrase MET1 to regulate DNA methylation and histone deacetylation in Arabidopsis (Liu et al., 2012). In addition, HDA5 and HDA6 form a protein complex with the histone demethylase FLD, suggesting that regulatory crosstalk between histone demethylation and deacetylation through the direct interaction between HDA5/HDA6 and FLD (Yu et al., 2011; Luo et al., 2015). Furthermore, HDA6 also interacts with histone methyltransferases SUVH4/5/6 and they function collaboratively in transposon silencing by removing the acetyl group from histone H3 and adding the methyl group to histone H3K9 (Yu et al., 2017). Furthermore, a specific acetylation site, H3K14, which is associated with transcription activation, is identified to be propionylated and butyrylated in vivo, suggesting histone acetylation and other modifications may acts in combination to modulate chromatin condensation and transcription outputs (Kebede et al., 2017).
Functions of HDACs in Salt and Drought Stress Responses in Arabidopsis
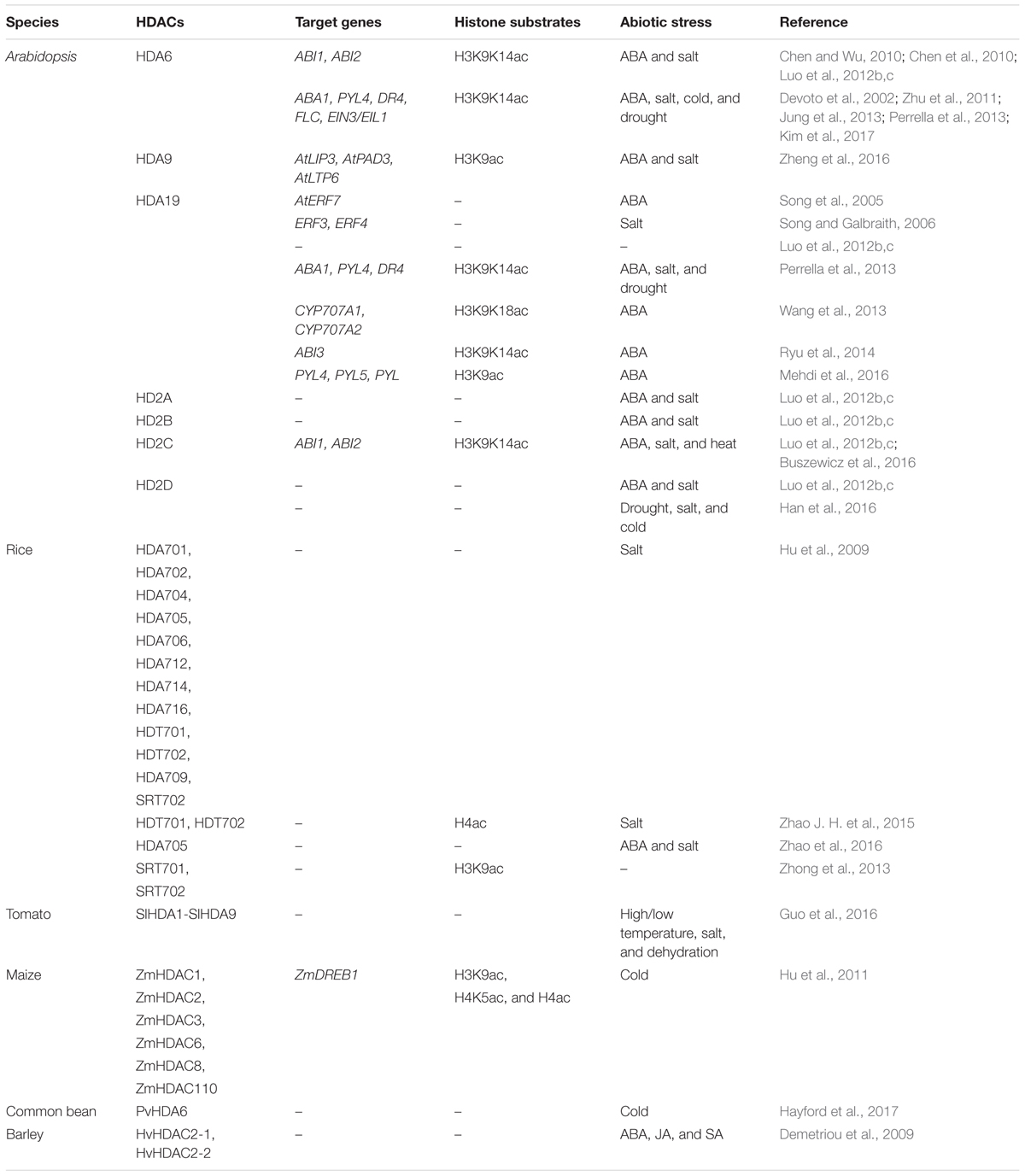
Recent studies indicated that HDACs play important roles in plant abiotic stress responses (Table 1). In Arabidopsis, members of the RPD3/HDA1 family were found to interact with various proteins involved in plant stress responses and regulate gene expression through histone deacetylation (Figure 1). HDA6 is involved in drought stress tolerance by regulating gene expression in the acetate biosynthesis pathway (Kim et al., 2017). HDA6 also regulates the jasmonate (JA) associated stress response by interacting with COI1 and JAZ1, the key regulators of JA signaling (Devoto et al., 2002; Zhu et al., 2011). hda9 mutant plants showed an enhanced tolerance to salt and drought stress. A large number of stress response genes, especially water deprivation stress-related genes, were up-regulated and hyper-deacetylated in hda9 mutants, indicating that HDA9 may act as a negative regulator in modulating stress responsive gene expression through histone deacetylation (Chen et al., 2016; Kim et al., 2016; Zheng et al., 2016).
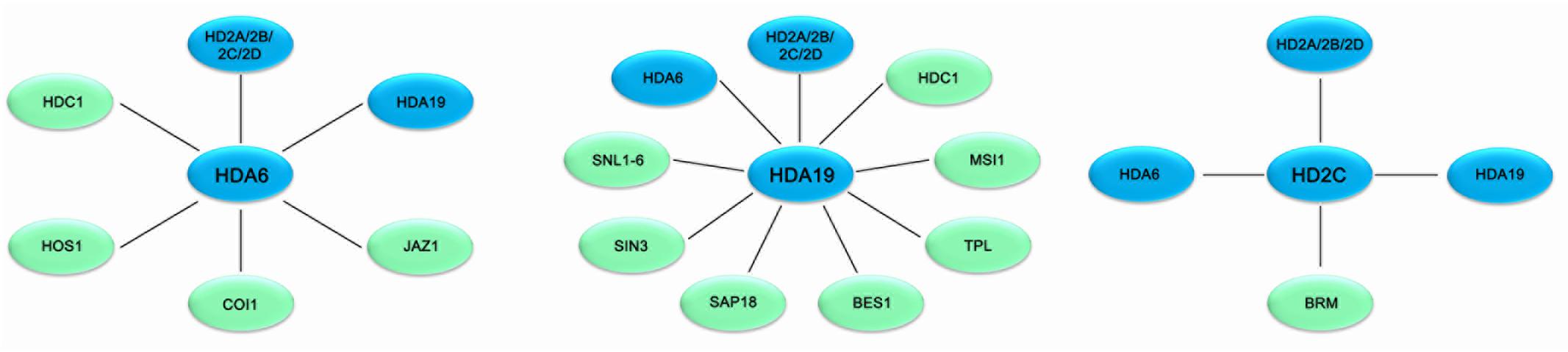
FIGURE 1. Association networks of HDACs and interacting proteins in plant stress response in Arabidopsis. HDACs are associated with various protein involved in stress response in Arabidopsis. Solid bars indicate physical interaction between HDACs (blue) and other protein (green).
HDA19 is recruited by the AP2/EREBP transcription factor AtERF7 and forms a repressor complex with AtSin3 to repress transcription of stress response genes in ABA and drought stress response (Song et al., 2005). Furthermore, HDA19 interacts with SNL1 and forms a repressor complex with SNL2 to regulate ABA synthesis through histone deacetylation of the specific target genes (Wang et al., 2013). In addition, HDA19 is associated with BES1 and TPL and the BES1-TPL-HDA19 repressor complex modulates ABI3 expression through histone deacetylation (Ryu et al., 2014). HDA19 was also found to act in a protein complex with the WD40-repeat protein MSI1 and the SIN3-like proteins to fine-tune ABA signaling in Arabidopsis (Mehdi et al., 2016). Both MSI1 antisense and HDA19 RNAi plants showed enhanced tolerance to salt stress treatment. The ABA sensitivity of gene expression is modulated through the MSI1-HDA19-SIN3 complex via modulating the transcription of ABA receptor genes (Mehdi et al., 2016). In addition, both HDA6 and HDA19 form a protein complex with HDC1 required for deacetylation of H3K9K14 in the plant abiotic stress responses (Perrella et al., 2013).
HD2-type proteins are plant specific HDACs and have been characterized in Arabidopsis, rice, soybean, barley tomato and maize (Demetriou et al., 2009; Luo et al., 2012b; Kim et al., 2015). The expression of Arabidopsis HD2A, HD2B, HD2C, and HD2D is repressed by ABA and high salt treatment (Luo et al., 2012b). Overexpressing HD2D in transgenic Arabidopsis resulted in increased tolerance to drought and salt stresses, suggesting that HD2D is involved in environmental stress responses (Han et al., 2016). Furthermore, overexpression of HD2C enhances salt and drought tolerance by modulating ABA-responsive genes (Sridha and Wu, 2006). HD2C functionally associates with HDA6 and regulates ABA-responsive gene expression through histone deacetylation (Luo et al., 2012b).
Functions of HDACs in Temperature Stress Responses in Arabidopsis
HDA6 plays a critical role in cold tolerance by regulating the expression of cold stress responsive genes (To et al., 2011; Kim et al., 2012; Jung et al., 2013). Moreover, the cold signaling attenuator, HOS1, interacts with HDA6 and inhibits the binding of HDA6 to FLC chromatin, resulting in a delayed flowering under short-term cold stress (Jung et al., 2013).
Overexpressing HD2D in transgenic Arabidopsis resulted in increased tolerance to cold stresses (Han et al., 2016). HD2C acts as a negative regulator of heat-activated genes in plants exposed to heat treatment through interacting with the chromatin remodeling factor BRAHMA (BRM) (Buszewicz et al., 2016). HD2A, HD2C, and HD2D can interact with both HDA6 and HDA19 (Luo et al., 2012c), suggesting that HD2-type HDACs functionally associate with RPD3-type HDACs in the multiprotein complex to regulate stress response genes in plants.
Additionally, the HDAC inhibitor trichostatin A (TSA) treatment can lead to an increased ROS level in animal cells (Sun et al., 2014). Similarly, an increased ROS level and enhanced antioxidant activity were detected by TSA treatment in Arabidopsis, suggesting that HDACs may be involved in the regulation of the ROS content under stress conditions in Arabidopsis (Jadko, 2015). Moreover, total HDAC activity was reduced by GSNO and S-nitroso-N-acetyl-DL-penicillamine treatment in plants (Mengel et al., 2017). ChIP-seq analysis revealed that the global H3K9/14ac was affected by NO treatment, resulting in the hyperacetylation of stress-related genes (Mengel et al., 2017).
Functions of HDACs in Abiotic Stress Responses in Rice and Other Plant Species
In rice, microarray analysis revealed that the expression of OsHDA703 and OsHDA710 was induced, whereas the expression of several other HDAC genes was repressed under high salt and drought treatment (Hu et al., 2009). In addition, the expression of OsHDT701 and OsHDT702 was affected under ABA, salt, and PEG stresses (Zhao J. H. et al., 2015). Interestingly, OsHDA709 and OsSRT702 were induced by drought treatment, but repressed by high salt. Genome-wide acetylation and binding analysis indicated that OsSRT701 may directly regulate the expression of stress-related genes by H3K9 deacetylation (Zhong et al., 2013). Overexpressing of OsHDT701 in transgenic rice resulted in increasing tolerance to salt and drought, and enhanced resistance to both Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae (Xoo) (Ding et al., 2012; Zhao J. H. et al., 2015). In contrast, overexpression of OsHDA705 in transgenic rice resulted in a decreased salt and ABA stress resistance during seed germination (Zhao et al., 2016). In addition, transcription of ABA biosynthetic genes is increased in the HDA705 overexpressing transgenic plants, suggesting that HDA705 may play vital roles in response to abiotic stresses in rice (Zhao et al., 2016).
In common bean (Phaseolus vulgaris Linn.), the expression of PvHDA6 was increased during cold treatment, indicating PvHDA6 is a cold response gene involved in regulation of plant abiotic stress tolerance (Hayford et al., 2017). In tomato (Solanum lycopersicum), SlHDACs were induced in various degrees under high salinity, dehydration, and different high/low temperature treatments, suggesting that SlHDACs might function in different stress responses (Guo et al., 2016). Treatment with the HDAC inhibitor, suberoylanilide hydroxamic acid (SAHA), enhanced plant salinity stress tolerance, indicating that HDACs might function in salt stress tolerance in cassava (Patanun et al., 2017). In barley (Hordeum vulgare), the transcription of HvHD2 genes were affected by multiple plant hormones, such as ABA, salicylic acid, and JA (Demetriou et al., 2009). In maize (Zea mays), the transcriptional patterns of ZmHDACs, including ZmHDAC1, ZmHDAC2, ZmHDAC3, ZmHDAC6, ZmHDAC8, and ZmHDAC110 was altered in response to low temperature. Levels of histone H3K9ac, H4K5ac, and H4ac were decreased after cold treatment. In addition, the expression of ZmDREB1 and ZmCOR413 was repressed by trichostatin A (TSA) treatment under cold stress conditions. Chromatin immunoprecipitation assays suggested that ZmHDACs may directly activate the expression of ZmDREB1 through histone deacetylation (Hu et al., 2011).
HDACs and Autophagy
Autophagy is a tightly regulated pathway involving the lysosomal degradation of cytoplasmic organelles or cytosolic components. This pathway can be stimulated by multiple forms of cellular stress. Recent studies indicated that HDA9 may repress the expression of a number of autophagy related genes including APG9, ATG2, ATG13, and ATG8e in Arabidopsis (Chen et al., 2016). However, the molecular mechanism how HDACs participate in autophagy remains largely unknown in plants.
Involvement of protein acetylation in autophagy was widely studied in mammalian and yeast cells (Yi et al., 2012; Fullgrabe et al., 2013; Huang et al., 2015; Su et al., 2017). In Saccharomyces cerevisiae, Rpd3 is required for K9 and K18 acetylation/deacetylation of Autophagy-related 3 (Atg3) during autophagy (Yi and Yu, 2012; Yi et al., 2012). In mammals, Atg8 is deacetylated by Sirt1 in response to starvation, and overexpression of Sirt1 can stimulate autophagosome formation (Lee et al., 2008). Deacetylation of LC3, a key initiator of autophagy, at K49, K51 by Sirt1 is essential for starvation-induced autophagosome formation in human cells (Huang et al., 2015). The acetylation or deacetylation of LC3 mediated by Sirt1 is linked to its nucleocytoplasmic transport. Under normal conditions, both nuclear and cytoplasmic forms of LC3 are acetylated (Huang and Liu, 2015). However, in response to starvation, LC3 is redistributed from the nucleus to the cytoplasm. In addition, deacetylation levels of LC3 is required for the interaction between LC3 and Atg7 (Huang and Liu, 2015; Huang et al., 2015). Taken together, these data suggested that HDACs act as regulators of autophagy in mammalian and yeast cells. Further research is required to determine how plant HDAC functions in this process.
Conclusion and Future Prospects
Histone deacetylases are recruited by diverse DNA-binding transcriptional factors forming multiple protein complexes to fine-tune the chromatin structure and modulate the gene expression in plant responses to environment stresses. Identifying the key components of HDAC complexes though yeast two-hybrid screening and in vivo immunoprecipitation in combination with mass spectrometry (IP-MS) is indispensable for understanding the functional organization of protein-protein interaction networks in the regulation of abiotic stress responses. To further understand how HDACs are involved in plant responses to abiotic stress, it is also important to identify the transcriptional regulatory network and the genome-wide binding site of HDACs by using RNA-seq and ChIP-seq approaches.
Recent studies indicated that non-histone proteins can also be acetylated or deacetylated by HATs or HDACs (Kovacs et al., 2005; Tran et al., 2012; Hao et al., 2016). For instance, Arabidopsis HDA6 can enhance brassinosteroid (BR) signaling by directly deacetylating the GSK3-like kinase BR-INSENSITIVE 2 (BIN2) and inhibits its kinase activity in the BR signaling pathway (Hao et al., 2016). In addition, the NAD+-dependent HDAC SRT1 could also remove the lysine acetylation from Arabidopsis c-Myc-Binding Protein-1 (AtMBP-1) and significantly enhance its stability in regulating primary metabolism and stress response (Liu et al., 2017). Further research is required to investigate acetylation and deacetylation of non-histone proteins in plant abiotic stress response. Environmental stresses cause significant crop losses on an annual basis. Uncovering the function of HDACs in plant responses to abiotic stress will contribute to our understanding of how plants adapt to environmental changes, which will be applicable in improvement of agricultural productivity.
Author Contributions
ML and KW conceived the idea. ML and KC wrote the manuscript. KW revised the manuscript. ML, KC, YX, SY, and KW critically evaluated the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the Hong Kong Scholars Program (XJ2013011) and Youth Innovation Promotion Association, Chinese Academy of Sciences (2017399). This work was also supported by the National Natural Science Foundation of China (31371308 and 31771423) and the Ministry of Science and Technology of Taiwan (105-2311-B-002-012-MY3 and 106-2313-B-002-003).
References
Aquea, F., Timmermann, T., and Arce-Johnson, P. (2010). Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 48, 194–199. doi: 10.1016/j.plaphy.2009.12.009
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Buszewicz, D., Archacki, R., Palusiński, A., Kotliński, M., Fogtman, A., Iwanicka-Nowicka, R., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 39, 2108–2122. doi: 10.1111/pce.12756
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, L. T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Chen, X. S., Lu, L., Mayer, K. S., Scalf, M., Qian, S. M., Lomax, A., et al. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5:e17214. doi: 10.7554/eLife.17214
Demetriou, K., Kapazoglou, A., Tondelli, A., Francia, E., Stanca, M. A., Bladenopoulos, K., et al. (2009). Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol. Plant. 136, 358–368. doi: 10.1111/j.1399-3054.2009.01236.x
Devoto, A., Nieto-Rostro, M., Xie, D. X., Ellis, C., Harmston, R., Patrick, E., et al. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466. doi: 10.1046/j.1365-313X.2002.01432.x
Ding, B., Bellizzi, M. D., Ning, Y. S., Meyers, B. C., and Wang, G. L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24, 3783–3794. doi: 10.1105/tpc.112.101972
Ekwall, K. (2005). Genome-wide analysis of HDAC function. Trends Genet. 21, 608–615. doi: 10.1016/j.tig.2005.08.009
Fullgrabe, J., Lynch-Day, M. A., Heldring, N., Li, W. B., Struijk, R. B., Ma, Q., et al. (2013). The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468–471. doi: 10.1038/nature12313
Guo, J.-E., Hu, Z., Guo, X., Zhang, L., Yu, X., Zhou, S., et al. (2016). Molecular characterization of nine tissue-specific or stress-responsive genes of histone deacetylase in tomato. J. Plant Growth Regul. 36, 566–577. doi: 10.1007/s00344-016-9660-8
Han, Z. F., Yu, H. M., Zhao, Z., Hunter, D., Luo, X. J., Duan, J., et al. (2016). AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 7:310. doi: 10.3389/fpls.2016.00310
Hao, Y. H., Wang, H. J., Qiao, S. L., Leng, L. N., and Wang, X. L. (2016). Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. U.S.A. 113, 10418–10423. doi: 10.1073/pnas.1521363113
Hayford, R. K. A., Ligaba-Osena, A., Subramani, M., Brown, A., Melmaiee, K., Hossain, K., et al. (2017). Characterization and expression analysis of common bean histone deacetylase 6 during development and cold stress response. Int. J. Genomics. 2017:2502691. doi: 10.1155/2017/2502691
Hollender, C., and Liu, Z. C. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Hu, Y., Zhang, L., Zhao, L., Li, J., He, S. B., Zhou, K., et al. (2011). Trichostatin a selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLOS ONE 6:e22132. doi: 10.1371/journal.pone.0022132
Hu, Y. F., Qin, F. J., Huang, L. M., Sun, Q. W., Li, C., Zhao, Y., et al. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388, 266–271. doi: 10.1016/j.bbrc.2009.07.162
Huang, R., and Liu, W. (2015). Identifying an essential role of nuclear LC3 for autophagy. Autophagy 11, 852–853. doi: 10.1080/15548627.2015.1038016
Huang, R., Xu, Y., Wan, W., Shou, X., Qian, J., You, Z., et al. (2015). Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466. doi: 10.1016/j.molcel.2014.12.013
Jadko, S. I. (2015). Histone deacetylase activity and reactive oxygen species content in the tissue culture of Arabidopsis thaliana under normal conditions and development of acute osmotic stress. Ukr. Biochem. J. 87, 57–62. doi: 10.15407/ubj87.03.057
Jung, J. H., Park, J. H., Lee, S., To, T. K., Kim, J. M., Seki, M., et al. (2013). The cold signaling attenuator high expression of osmotically responsive gene1 activates flowering locus C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25, 4378–4390. doi: 10.1105/tpc.113.118364
Kebede, A. F., Nieborak, A., Shahidian, L. Z., Le Gras, S., Richter, F., Gómez, D. A., et al. (2017). Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol. doi: 10.1038/nsmb.3490 [Epub ahead of print].
Kim, J. M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, J. M., To, T. K., Matsui, A., Tanoi, K., Kobayashi, N. I., Matsuda, F., et al. (2017). Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 3:17097. doi: 10.1038/nplants.2017.97
Kim, J. M., To, T. K., and Seki, M. (2012). An epigenetic integrator: new insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol. 53, 794–800. doi: 10.1093/pcp/pcs004
Kim, Y. J., Wang, R. Z., Gao, L., Li, D. M., Xu, C., Mang, H. G., et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863. doi: 10.1073/pnas.1618618114
Kovacs, J. J., Murphy, P. J. M., Gaillard, S., Zhao, X. A., Wu, J. T., Nicchitta, C. V., et al. (2005). HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18, 601–607. doi: 10.1016/j.molcel.2005.04.021
Lee, I. H., Cao, L., Mostoslavsky, R., Lombard, D. B., Liu, J., Bruns, N. E., et al. (2008). A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 105, 3374–3379. doi: 10.1073/pnas.0712145105
Liew, L. C., Singh, M. B., and Bhalla, P. L. (2013). An RNA-Seq transcriptome analysis of histone modifiers and RNA silencing genes in soybean during floral initiation process. PLOS ONE 8:e77502. doi: 10.1371/journal.pone.0077502
Liu, X., Wei, W., Zhu, W., Su, L., Xiong, Z., Zhou, M., et al. (2017). Histone deacetylase AtSRT1 rregulates metabolic flux and stress response in Arabidopsis. Mol. Plant 10, 1510–1522. doi: 10.1016/j.molp.2017.10.010
Liu, X., Yu, C. W., Duan, J., Luo, M., Wang, K., Tian, G., et al. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158, 119–129. doi: 10.1104/pp.111.184275
Luo, M., Liu, X., Singh, P., Cui, Y., Zimmerli, L., and Wu, K. (2012a). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136. doi: 10.1016/j.bbagrm.2011.06.008
Luo, M., Tai, R., Yu, C. W., Yang, S., Chen, C. Y., Lin, W. D., et al. (2015). Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J. 82, 925–936. doi: 10.1111/tpj.12868
Luo, M., Wang, Y. Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012b). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Luo, M., Wang, Y. Y., Liu, X., Yang, S., and Wu, K. (2012c). HD2 proteins interact with RPD3-type histone deacetylases. Plant Signal. Behav. 7, 608–610. doi: 10.4161/psb.20044
Luo, M., Yu, C. W., Chen, F. F., Zhao, L., Tian, G., Liu, X., et al. (2012d). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLOS Genet. 8:e1003114. doi: 10.1371/journal.pgen.1003114
Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. doi: 10.1126/science.277.5322.88
Mehdi, S., Derkacheva, M., Ramstrom, M., Kralemann, L., Bergquist, J., and Hennig, L. (2016). The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28, 42–54. doi: 10.1105/tpc.15.00763
Mengel, A., Ageeva, A., Georgii, E., Bernhardt, J., Wu, K. Q., Durner, J., et al. (2017). Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 173, 1434–1452. doi: 10.1104/pp.16.01734
Patanun, O., Ueda, M., Itouga, M., Kato, Y., Utsumi, Y., Matsui, A., et al. (2017). The histone deacetylase inhibitor suberoylanilide hydroxamic acid alleviates salinity stress in cassava. Front. Plant Sci. 7:2039. doi: 10.3389/fpls.2016.02039
Peng, M. J., Ying, P. Y., Liu, X. C., Li, C. Q., Xia, R., Li, J. G., et al. (2017). Genome-wide identification of histone modifiers and their expression patterns during fruit abscission in litchi. Front. Plant Sci. 8:639. doi: 10.3389/fpls.2017.00639
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Gossele, V., Verduyn, C., et al. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Peterson, C. L., and Laniel, M. A. (2004). Histones and histone modifications. Curr. Biol. 14, R546–R551. doi: 10.1016/j.cub.2004.07.007
Ryu, H., Cho, H., Bae, W., and Hwang, I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5:4138. doi: 10.1038/ncomms5138
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P. C., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Song, C. P., and Galbraith, D. W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257. doi: 10.1007/s11103-005-3880-9
Sridha, S., and Wu, K. Q. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. doi: 10.1111/j.1365-313X.2006.02678.x
Su, H., Yang, F., Wang, Q. T., Shen, Q. H., Huang, J. T., Peng, C., et al. (2017). VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell 67, 907–921. doi: 10.1016/j.molcel.2017.07.024
Sun, S. J., Han, Y. Y., Liu, J., Fang, Y., Tian, Y., Zhou, J. F., et al. (2014). Trichostatin a targets the mitochondrial respiratory chain, increasing mitochondrial reactive oxygen species production to trigger apoptosis in human breast cancer cells. PLOS ONE 9:e91610. doi: 10.1371/journal.pone.0091610
To, T. K., Nakaminami, K., Kim, J. M., Morosawa, T., Ishida, J., Tanaka, M., et al. (2011). Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 406, 414–419. doi: 10.1016/j.bbrc.2011.02.058
Tran, H. T., Nimick, M., Uhrig, R. G., Templeton, G., Morrice, N., Gourlay, R., et al. (2012). Arabidopsis thaliana histone deacetylase 14 (HDA14) is an alpha-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J. 71, 263–272. doi: 10.1111/j.1365-313X.2012.04984.x
Wang, Z., Cao, H., Sun, Y. Z., Li, X. Y., Chen, F. Y., Carles, A., et al. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25, 149–166. doi: 10.1105/tpc.112.108191
Yi, C., Ma, M. S., Ran, L. L., Zheng, J. X., Tong, J. J., Zhu, J., et al. (2012). Function and molecular mechanism of acetylation in autophagy regulation. Science 336, 474–477. doi: 10.1126/science.1216990
Yi, C., and Yu, L. (2012). How does acetylation regulate autophagy? Autophagy 8, 1529–1530. doi: 10.4161/auto.21156
Yu, C. W., Liu, X., Luo, M., Chen, C., Lin, X., Tian, G., et al. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156, 173–184. doi: 10.1104/pp.111.174417
Yu, C. W., Tai, R., Wang, S. C., Yang, P., Luo, M., Yang, S., et al. (2017). HISTONE DEACETYLASE6 acts in concert with histone Methyltransferases SUVH4, SUVH5, and SUVH6 to regulate transposon silencing. Plant Cell 29, 1970–1983. doi: 10.1105/tpc.16.00570
Zhao, J., Li, M., Gu, D., Liu, X., Zhang, J., Wu, K., et al. (2016). Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 470, 439–444. doi: 10.1016/j.bbrc.2016.01.016
Zhao, J. H., Zhang, J. X., Zhang, W., Wu, K. L., Zheng, F., Tian, L. N., et al. (2015). Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front. Plant Sci. 5:764. doi: 10.3389/fpls.2014.00764
Zhao, L. M., Lu, J. X., Zhang, J. X., Wu, P. Y., Yang, S. G., and Wu, K. Q. (2015). Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front. Plant Sci. 5:760. doi: 10.3389/fpls.2014.00760
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Zhong, X., Zhang, H., Zhao, Y., Sun, Q., Hu, Y., Peng, H., et al. (2013). The rice NAD(+)-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLOS ONE 8:e66807. doi: 10.1371/journal.pone.0066807
Keywords: histone deacetylation, HDACs, abiotic stress, autophagy, protein complexes
Citation: Luo M, Cheng K, Xu Y, Yang S and Wu K (2017) Plant Responses to Abiotic Stress Regulated by Histone Deacetylases. Front. Plant Sci. 8:2147. doi: 10.3389/fpls.2017.02147
Received: 29 September 2017; Accepted: 04 December 2017;
Published: 15 December 2017.
Edited by:
Stéphane Bourque, Université de Bourgogne, FranceReviewed by:
Giorgio Perrella, University of Glasgow, United KingdomSerena Varotto, Università degli Studi di Padova, Italy
Copyright © 2017 Luo, Cheng, Xu, Yang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Luo, bHVvbWluZ0BzY2JnLmFjLmNu Keqiang Wu, a2V3dUBudHUuZWR1LnR3
†These authors have contributed equally to this work.
 Ming Luo
Ming Luo Kai Cheng
Kai Cheng Yingchao Xu
Yingchao Xu Songguang Yang
Songguang Yang Keqiang Wu
Keqiang Wu