- Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, China
Sweet cherry (Prunus avium L.) is an important fruit crop in which fruit size is strongly associated with commercial value; few genes associated with fruit size have, however, been identified in sweet cherry. Members of the CYP78A subfamily, a group of important cytochrome P450s, have been found to be involved in controlling seed size and development in Arabidopsis thaliana, rice, soybean, and tomato. However, the influence of CYP78A members in controlling organ size and the underlying molecular mechanisms in sweet cherry and other fruit trees remains unclear. Here, we characterized a P. avium CYP78A gene PaCYP78A9 that is thought to be involved in the regulation of fruit size and organ development using overexpression and silencing approaches. PaCYP78A9 was significantly expressed in the flowers and fruit of sweet cherry. RNAi silencing of PaCYP78A9 produced small cherry fruits and PaCYP78A9 was found to affect fruit size by mediating mesocarp cell proliferation and expansion during fruit growth and development. Overexpression of PaCYP78A9 in Arabidopsis resulted in increased silique and seed size and PaCYP78A9 was found to be highly expressed in the inflorescences and siliques of transgenic plants. Genes related to cell cycling and proliferation were downregulated in fruit from sweet cherry TRV::PaCYP78A9-silencing lines, suggesting that PaCYP78A9 is likely to be an important upstream regulator of cell cycle processes. Together, our findings indicate that PaCYP78A9 plays an essential role in the regulation of cherry fruit size and provide insights into the molecular basis of the mechanisms regulating traits such as fruit size in P. avium.
Introduction
Sweet cherry (Prunus avium L.) is an economically valuable horticultural crop that is widely cultivated in temperate regions; its fleshy fruits are recognized as having nutraceutical properties and antioxidant activity (Li et al., 2010). As large fruit size in sweet cherry generates a premium market price (Whiting et al., 2006), increasing cherry fruit size has long been one of the most important goals for breeding selection during domestication and modern horticultural crop breeding (Zhang et al., 2010). Additional insight into the genetic and molecular mechanisms responsible for controlling sweet cherry fruit should, therefore, help to inform strategies to acquire larger fruit. While several previous studies have determined that fruit size is controlled by multiple genetic loci in sweet cherry and other horticultural fruit trees (Zhang et al., 2010; Rosyara et al., 2013; Campoy et al., 2015), only a few genes related to the molecular mechanisms regulating fruit size have thus far been identified. Thus, characterization of genes associated with fruit size and the molecular mechanisms that determine final fruit size is urgently required to assist in the development of strategies to increase fruit size.
Plant organ size is one of the most important agronomical traits targeted during domestication. Plant organ growth and development are genetically determined by both cell division and cell expansion (Horiguchi et al., 2006; Li and Li, 2015, 2016; Si et al., 2016). Several studies have suggested that organ size, including seed and fruit size, is ultimately controlled by multiple factors such as plant hormones, ubiquitin, microRNAs, and cytochrome P450s (CYPs) (Fang et al., 2012; Nitsch et al., 2012; Du et al., 2014; Ma et al., 2015, 2016; Yao et al., 2015). For instance, AUXIN RESPONSE FACTOR 2 (ARF2) and DA1 (DA means ‘large’ in Chinese, ubiquitin receptor) limit cell proliferation in the integuments of ovules and developing seeds, ultimately maternally affecting seed size (Schruff et al., 2006; Li et al., 2008). Abscisic acid (ABA)-biosynthesis related ABSCISIC ACID DEFICIENT2 (ABA2) and ABSCISIC ACID-INSENSITIVE5 (ABI5) regulate seed size by mediating embryonic cell proliferation and early endosperm cellularization in early seed development (Cheng et al., 2014). TRANSPARENT TESTA GLABRA2 (TTG2) and APETALA2 (AP2) play important roles in controlling seed size and growth by influencing cell elongation in the maternal integuments (Johnson, 2002; Jofuku et al., 2005). The RING-type E3 ubiquitin ligases, ENHANCER OF DA1 (EOD1) and DA2 regulate seed size by restricting cell proliferation in the maternal integuments (Li et al., 2008; Xia et al., 2013). MicroRNA172 (miRNA172) governs floral organ development and organ size by inhibiting translation of APETALA2 (AP2) (Yao et al., 2015).
Cytochrome P450s is one of the largest enzymatic protein families that is found in land plants from bryophytes to angiosperms; members of this family play vital roles in a variety of metabolic pathways by producing primary and secondary metabolites such as sterols, isoflavonoids, terpenoids, flavonoids, steroids, anthocyanins, phytohormones, and phytoalexin to promote plant growth and development and protect plants from stress (Ohkawa et al., 1998; Li et al., 2012; Xu J. et al., 2015). CYP78A, an important CYP subfamily, is a highly conserved plant-specific gene family (Nelson, 2006; Mizutani and Ohta, 2010). In Arabidopsis thaliana, six CYP78A genes, CYP78A5, CYP78A6, CYP78A7, CYP78A8, CYP78A9, and CYP78A10, have been found to be involved in the control of organ growth and development (Wang et al., 2008; Adamski et al., 2009; Bak et al., 2011; Fang et al., 2012; Sotelo-Silveira et al., 2013); several CYP78As have also been found to regulate organ size and development in other plants such as rice, wheat, tomato, and soybean (Chakrabarti et al., 2013; Nagasawa et al., 2013; Ma et al., 2015, 2016; Xu F. et al., 2015; Zhao et al., 2016). For example, KLUH/CYP78A5 regulated seed or fruit size by promoting integument cell proliferation in Arabidopsis, tomato, wheat, and soybean (Adamski et al., 2009; Chakrabarti et al., 2013; Ma et al., 2016; Zhao et al., 2016); TaCYP78A3 controls seed size by increasing cell proliferation in the integument in wheat (Ma et al., 2015); and OsCYP78A13 regulates grain size by mediating cell-cycle progression to balance the embryo/endosperm size in rice (Nagasawa et al., 2013). AtCYP78A9 acting redundantly with EOD3/CYP78A6 and CYP78A8 plays a critical role in regulating floral organ growth and integument development by promoting cell proliferation, thereby ultimately affecting seed size in A. thaliana (Fang et al., 2012; Sotelo-Silveira et al., 2013). However, the role of CYP78A members in the control of organ size and development has not yet been reported in sweet cherry or other fruit trees.
In this study, the role of P. avium PaCYP78A9, a CYP78A member, in the regulation of fruit size and organ development in sweet cherry was characterized using overexpression and silencing approaches. Fruit expression levels of PaCYP78A9 were detected in a landrace sweet cherry, ‘Longguan,’ and a wild sweet cherry, ‘Mazzard,’ respectively. PaCYP78A9 overexpression in Arabidopsis accelerated silique and seed development, resulting in enlarged seeds. Moreover, silencing of PaCYP78A9 during sweet cherry fruit development caused decreases in fruit mesocarp cell number and size, leading to a reduction in fruit size (Kawai et al., 2016; Cui and Wang, 2017). Together, our results provide direct evidence that PaCYP78A9 is involved in the regulation of fruit size; these results further contribute to an understanding of the cellular basis and genetic regulation of sweet cherry fruit size and development that may assist in the generation of new lines in the future with increased yield.
Materials and Methods
Plant Materials
Two P. avium varieties, a wild sweet cherry, ‘Mazzard’, and a landrace sweet cherry, ‘Longguan’, were grown in the resource garden of the National Fruit Tree Germplasm Repository, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (Zhengzhou, China). A single fruit of ‘Longguan’, a cultivated larger cherry, weighs more than 8 g, while ‘Mazzard’, a wild forest cherry of northern European origin, possesses very small fruit (∼2 g). The Arabidopsis cyp78a9 insertional mutant used in this study was provided by Prof. Huixian Zhao and Dr. Meng Ma from the College of Life Sciences, Northwest A (Agriculture) & F (Forestry) University in Yangling Shaanxi, China. A. thaliana used in this study was preserved in our laboratory. All Arabidopsis plants were grown at 20–22°C in a greenhouse with a 16 h light/8 h dark cycle and 60–75% relative humidity.
Pacyp78a9 Gene Isolation and Phylogenetic Analysis
Total RNA was extracted and purified from the leaves and fruit of sweet cherry using an EASYspin RNA Plant RNA rapid extraction kit (Yuanpinghao Bio, Tianjin, China) according to the manufacturer’s protocol. First-strand cDNA was synthesized from purified RNA samples using the FastQuant RT (With gDNase) kit (Tiangen Bio, Beijing, China) in accordance with the manufacturer’s instructions. Specific primers (PaCYP78A9-F and PaCYP78A9-R) were used to amplify the full-length PaCYP78A9 gene from the first-strand cDNA. MEGA 5.01 software was used to construct the phylogenetic tree of the PaCYP78A9 protein, other reported CYP78A family proteins from Triticum urartu, A. thaliana, Oryza sativa, and unreported CYP78A family proteins from the completed genome-sequencing fruit tree, including peach, apple, pear, grape; this was done using the neighbor-joining method with 500 bootstrap replicates (Tamura et al., 2011).
Vector Construction
The pBI121-PaCYP78A9 expression vector p35S::PaCYP78A9 was constructed by In-Fusion cloning as described previously (Frandsen et al., 2012). Full-length PaCYP78A9 was amplified using the primer pairs PaCYP78A9-e-F and PaCYP78A9-e-R that included 16-bp-long 5′ overhangs identical to the corresponding pBI121 sequence digested with XbaI or BamHI. The vector pBI121 was digested with XbaI and BamHI. Next, the full-length PaCYP78A9 fragment and the digested pBI121 were fused together with the In-FusionTM HD Cloning kit (Clontech, Mountain View, CA, United States) to obtain the p35S::PaCYP78A9 plasmid.
A 469-bp fragment of PaCYP78A9 was amplified (bases 511–980) with the primers PaCYP78A9-RNAi-F and PaCYP78A9-RNAi-R containing the EcoRI and KpnI restriction enzyme sites, respectively. The PaCYP78A9 fragment was inserted into a TRV2 vector digested with XbaI and BamHI using T4 ligase to construct the TRV:PaCYP78A9 plasmid.
Arabidopsis Transformation
The expression vector p35S::PaCYP78A9 was introduced into the Arabidopsis Columbia ecotype or the Arabidopsis cyp78a9 insertional mutant by the Agrobacterium tumefaciens-mediated floral-dip method when the Arabidopsis plants were 30-days-old. The seeds of transformants were collected and screened on MS medium supplemented with 50 mg/L kanamycin. Kanamycin-resistant plants were selected and transferred to growth soil chambers in a greenhouse at 20–22°C with a 16 h light/8 h dark cycle and 60–75% relative humidity where they were grown until maturity. The T3 generation of transgenic PaCYP78A9 overexpression and cyp78a9-complementation lines were used for further phenotypic analysis.
TRV-Mediated Pacyp78a9 Gene Silencing in Cherry Fruit
TRV-mediated PaCYP78A9 gene silencing was performed as described previously with Agrobacterium tumefaciens-mediated transformation with the following modifications for cherry fruit (Fu et al., 2005; Shen et al., 2014; Li et al., 2015). (i) Agrobacterium strain GV3101 were cultured overnight at 28°C to OD600 of 0.8, and were resuspended in the Agrobacterium infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, 100 μM acetosyringone) to a final OD600 of 0.8–1.0. (ii) Mixed Agrobacterium GV3101 strains containing the pTRV1, pTRV2, or pTRV:PaCYP78A9 vectors were infiltrated with a needle-less syringe into the basal pedicel of cherry fruit 7 days after full bloom (DAFB) until the whole fruit was permeated. (iii) The inoculated fruits were treated with bagging for 2 days. Fruits were evaluated at 10 days post-inoculation (dpi). The experiment was repeated three times with 30 transformed fruits per strain per replicate come from the same tree of 10 years old.
Semi-Quantitative Reverse Transcription PCR (RT-PCR) and Quantitative Real-Time PCR (qRT-PCR) Analysis
Reverse transcription PCR and qRT-PCR assays were performed as previously described by Qi et al. (2015). Total RNA from different plant tissues (leaf, flower-bud, blossom, and fruit) were isolated as described above. A FastQuant RT (With gDNase) kit (Tiangen Bio) was used to synthesize the cDNA from RNA. The expression levels of target genes were quantified relative to the constitutively expressed Histone2 (Pav_sc0000671.1) gene using qRT-PCR in an ABI7500 PCR thermocycler (Applied Biosystems, Foster City, CA, United States) with the TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, Chain) containing SYBR Green fluorescent intercalating dye.
Equal amounts of RNA from sweet cherry fruit harvested from the TRV::PaCYP78A9- and TRV::00-silencing lines were used for reverse transcription of the first-strand cDNA via a FastQuant RT (With gDNase) kit (Tiangen Bio) according to the manufacturer’s protocol. RT-PCR was performed with equal amounts of cDNA as previously described, and using Histone2 as an internal control. The primers used in the RT-PCR and qRT-PCR assays are listed in Supplementary Table S1. The expression level was calculated from three replicates.
Morphological and Cytological Characterization
Morphological analysis of plant organs, including organ size and organ weight, was conducted according to the methods described previously (Disch et al., 2006; Adamski et al., 2009; Chakrabarti et al., 2013).
To assess cytologically the fruit cells from the exocarp and endocarp of sweet cherry fruit, morphological and cytological observations were performed as previously described with some modifications using pericarp tissue samples taken from sweet cherry fruit harvested from TRV::PaCYP78A9- and TRV::00-silencing lines at 40 DAFB (Adamski et al., 2009; Chakrabarti et al., 2013; Rojas-Gracia et al., 2017). Whole fresh cherry fruit were fixed in FAA solution (5% formaldehyde, 5% acetic acid, and 50% ethanol) at 4°C overnight after vacuuming. The samples were then dehydrated through a graded ethanol concentration series composed of 75% ethanol for 4 h, 85% ethanol for 2 h, 90% ethanol for 2 h, 95% ethanol for 1 h, and finally absolute ethanol for 1 h before being transitioned into histo-clear xylene for 20 min. The fresh tissue samples were embedded in paraffin using TKY-BMB (JB-T5, Wuhan Junjie Equipment Factory, Wuhan, China), and sectioned (4 μm in thickness) using a rotary microtome (Leica RM2016; Leica Microsystems, Germany). Subsequently, the paraffin sections were dewaxed with xylene and stained with hematoxylin and eosin, followed gradient alcohol dehydration, vitrification by xylene, and neutral gum sealing. Finally, the samples of stained cherry fruit epicarp and mesocarp sections were observed using light microscopy (Microscope Nikon Eclipse Ci, Nikon, Tokyo, Japan) with a Nikon DS-U3 digital camera (Nikon, Tokyo, Japan). Three replicates were performed for each sample.
Statistical Analysis
All data of results are provided as the mean ± standard deviation (SD). Statistically significant differences indicated in figures were based on t-test or one-way ANOVA (p ≤ 0.05) using SPSS 17.0 version (SPSS Inc., Chicago, IL, United States). Figures were made by Origin 9.1 (Microcal Software Inc., Northampton, MA, United States).
Results
Phylogenetic Analysis of CYP78A in Plants
To understand the cellular and molecular mechanism underlying fruit size in sweet cherry (P. avium), the peach genome v.2.1 sequence released by the International Peach Genome Initiative (GDR database1) was used to isolate the PaCYP78A9 gene from P. avium. An open reading frame 1653 bp was obtained for PaCYP78A9 with a start codon ATG and a stop codon TAA; this encoded a protein with 550 amino acid residues. To better understand the relationships between PaCYP78A9 and other members of the CYP78A family, multiple sequence alignment of the PaCYP78A9 protein and other CYP78A family members from various plant species was used to construct a phylogenetic tree. This showed that PaCYP78A9 was closely related to PmCYP78A9, PpCYP78A9, and MdCYP78A9, which have not been functionally characterized, and displayed high similarity to Arabidopsis CYP78A9 (67% identity), CYP78A6 (66% identity), CYP78A8 (65% identity), and to wheat CYP78A3 (59% identity), to tomato CYP78A5 (52% identity, Figure 1). These results demonstrated that PaCYP78A9 in P. avium is an ortholog of PpCYP78A9, PmCYP78A9, and MdCYP78A9, and might have the same function as AtCYP78A9 (Sotelo-Silveira et al., 2013) and TaCYP78A3 (Ma et al., 2015) that are known to play critical roles in regulating seed size.
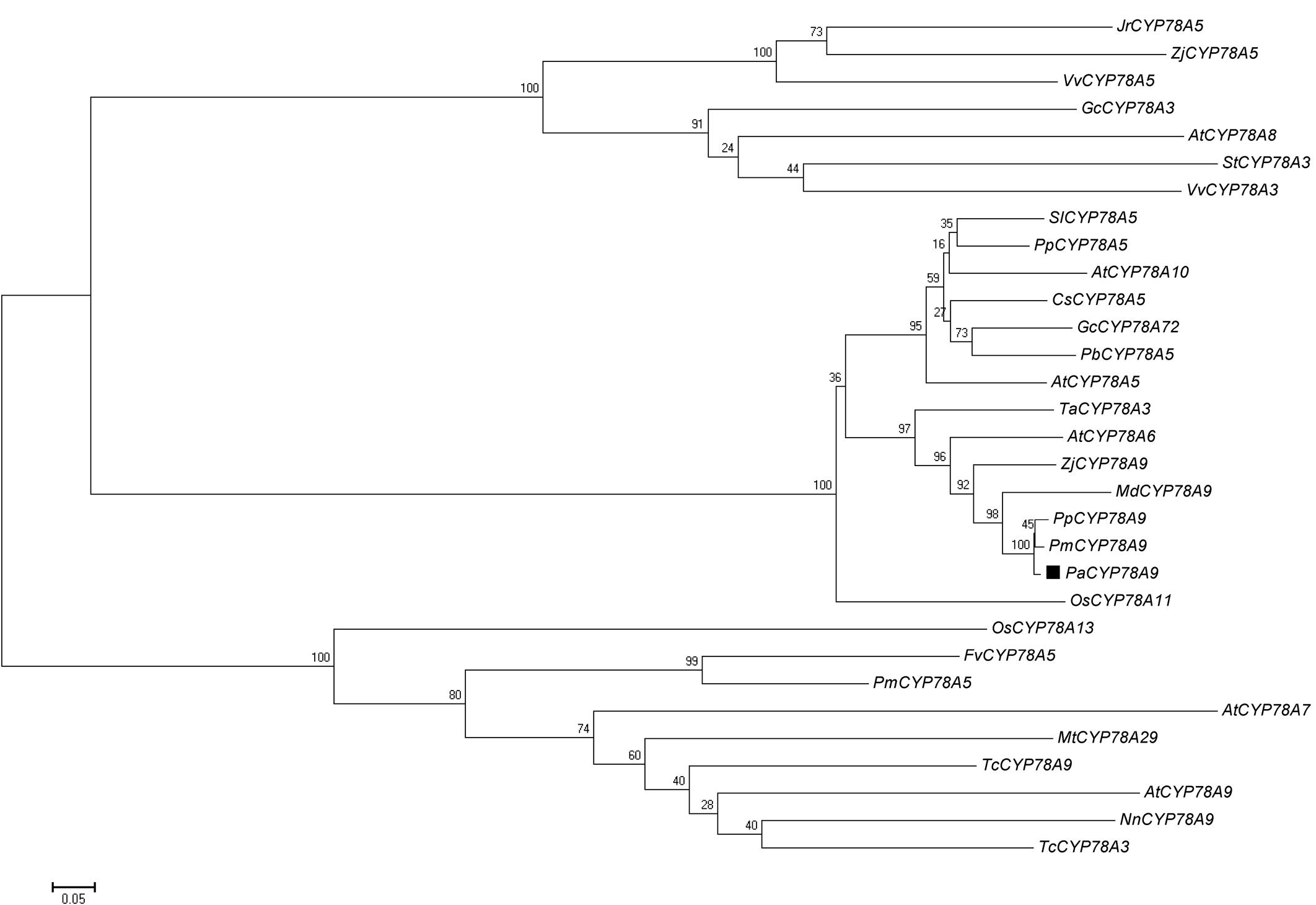
FIGURE 1. Phylogenetic analysis of the Prunus avium PaCYP78A protein and CYP78A orthologs in other plant species. Source organisms and accession numbers for homologous CYP78A proteins were as follows: JrCYP78A5 (Juglans regia, XM_018969352.1), ZjCYP78a5 (Ziziphus jujuba, XM_016027740.1), VvCYP78A5 (Vitis vinifera, XM_002265274.3), GcCYP78A3 (Glycine max, NM_001317595.1), AtCYP78A8 (Arabidopsis thaliana, NM_100001.2), StCYP78A3 (Solanum tuberosum, XM_006339692.2), VvCYP78A3 (V. vinifera, XM_002266457.2), SlCYP78A5 (S. lycopersicum, XM_004236016.3), PpCYP78A5 (Prunus persica, XM_007209835.2), AtCYP78A10 (A. thaliana, NM_106071.1), CsCYP78A5 (Citrus sinensis, XM_006477669.1), GcCYP78A72 (Glycine max, XM_003554632.3), PbCYP78A5 (Pyrus × bretschneideri, XM_009358148.2), AtCYP78A5 (A. thaliana, NM_101240.4), TaCYP78A3 (Triticum aestivum, KP768392.1), AtCYP78A6 (A. thaliana, NM_130231.4), ZjCYP78A9 (Ziziphus jujuba, XM_016033300.1), MdCYP78A9 (Malus × domestica, XM_008345221.2), PpCYP78A9 (P. persica, XM_020556145.1), PmCYP78A9 (P. mume, XM_008234862.2), PaCYP78A9 (P. avium, XM_021959332.1), OsCYP78A11 (Oryza sativa, XM_015757991.1), OSCYP78A13 (O. sativa, XM_015790749.1), FvCYP78A5 (Fragaria vesca, XM_004298317.2), PmCYP78A5 (P. mume, XM_008241890.2), AtCYP78A7 (A. thaliana, NM_121034.2), Mt CYP78A29 (Medicago truncatula, DQ335794.1), TcCYP78A9 (Theobroma cacao, XM_007051407.2), AtCYP78A9 (A. thaliana, NM_116053.3), NnCYP78A9 (Nelumbo nucifera, XM_010269146.1), TcCYP78A3 (T. cacao, XM_018117954.1). The tree was constructed using MEGA (version 5.01). A neighbor-joining evolutionary phylogeny test and 500 bootstrap replicates were selected for the analysis. The scale bar represents 0.2 substitutions per site.
Expression Patterns of PaCYP78A9 during Fruit Growth and Development in Sweet Cherry
To analyze the function of PaCYP78A9 during fruit growth and development, qRT-PCR was used to examine the expression pattern of PaCYP78A9 in leaf, flower-bud, blossom, and fruit tissue obtained at different growth and developmental stages from both the wild sweet cherry ‘Mazzard’ and the landrace sweet cherry ‘Longguan.’ The expression level of PaCYP78A9 was low in the leaves of both ‘Mazzard’ and ‘Longguan’ but was relatively high in the flower-bud and flowers of both lines during the blossom stages (Figure 2). During early fruit growth and development (1–25 DAFB) of ‘Mazzard’ and ‘Longguan,’ PaCYP78A9 expression was more highly upregulated in fruit than in leaves, with expression peaking in fruit at 15 DAFB, suggesting that PaCYP78A9 plays an important role in fruit growth and development (Figure 2). Furthermore, the fruit transcript levels of PaCYP78A9 in the landrace sweet cherry ‘Longguan’ were significantly higher than in the wild sweet cherry ‘Mazzard’ (Figure 2). In Arabidopsis, AtCYP78A9 was expressed only in floral organs, not in vegetative tissue (Sotelo-Silveira et al., 2013). Similarly, the PaCYP78A9 gene was specifically expressed in sweet cherry floral organs that were related to the developing fruit. Together, these results indicate that the specific expression pattern of PaCYP78A9 could be involved in regulating fruit expansion and ultimately fruit size during the early stages of fruit growth and development.
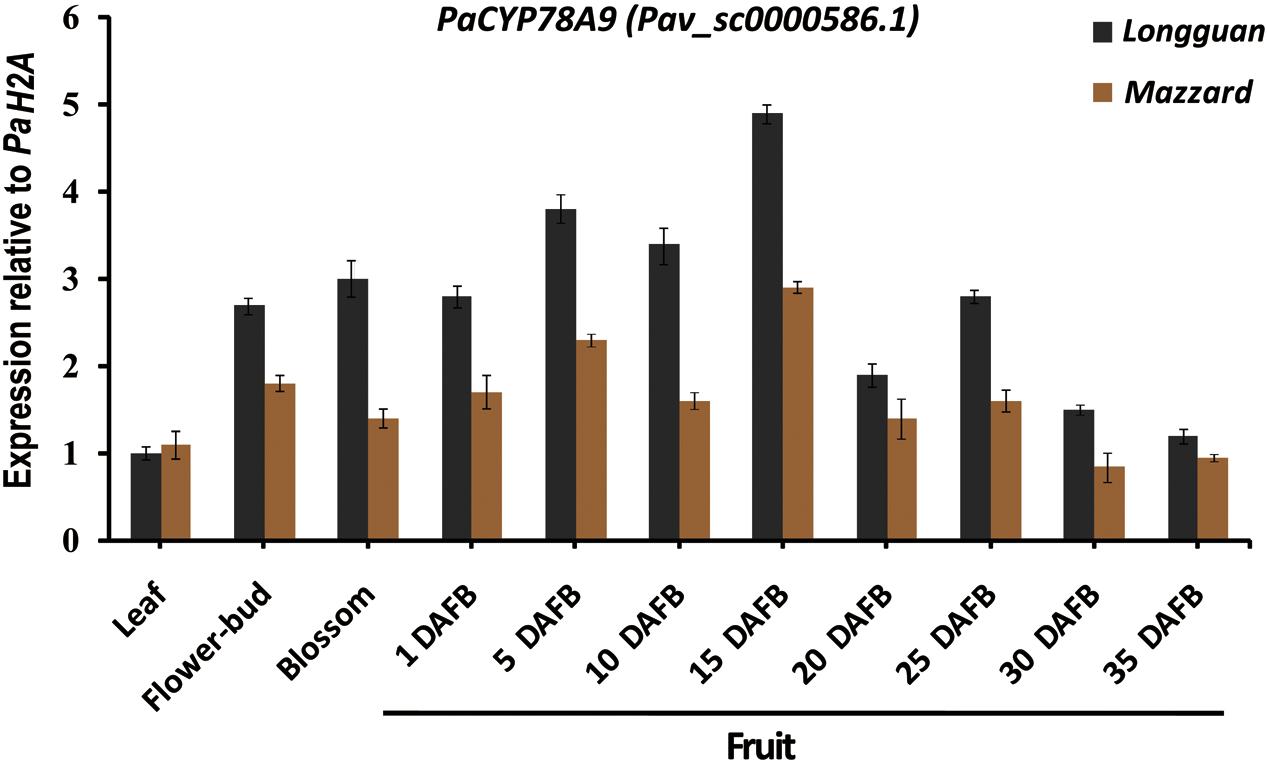
FIGURE 2. Expression profile of PaCYP78A9 in the ‘Longguan’ landrace sweet cherry and ‘Mazzard’ wild sweet cherry cultivars. The expression levels of PaCYP78A9 in leaves, flower-buds, blossoms, and fruit harvested at 1, 5, 10, 15, 20, and 25 days after full bloom (DAFB) were analyzed using qRT-PCR. Expression of PaCYP78A9 in the mature leaves of ‘Longguan’ was set to 1.0. P. avium Histone2A (Pav_sc0000671.1) was used as an internal control to calculate the relative expression levels of PaCYP78A9. The error bars represent the standard deviation of three independent experiments.
Silencing of PaCYP78A9 Decreases Fruit Size in Sweet Cherry
Previous studies have confirmed that Arabidopsis AtCYP78A9 control seed size by promoting cell proliferation (Sotelo-Silveira et al., 2013). To determine whether PaCYP78A9 influences fruit size during fruit growth and development, the tobacco rattle virus-induced gene silencing (TRV-VIGS) technique was used to knock down expression of the PaCYP78A9 gene in the sweet cherry landrace ‘Longguan.’. RT-PCR was performed using cDNA samples from infiltrated fruit at 15 dpi to verify that the PaCYP78A9 gene in ‘Longguan’ was effectively silenced. This showed that the expression of PaCYP78A9 was markedly reduced in the TRV::PaCYP78A9-infected fruit compared with the TRV::00-infected fruit (Figure 3B).
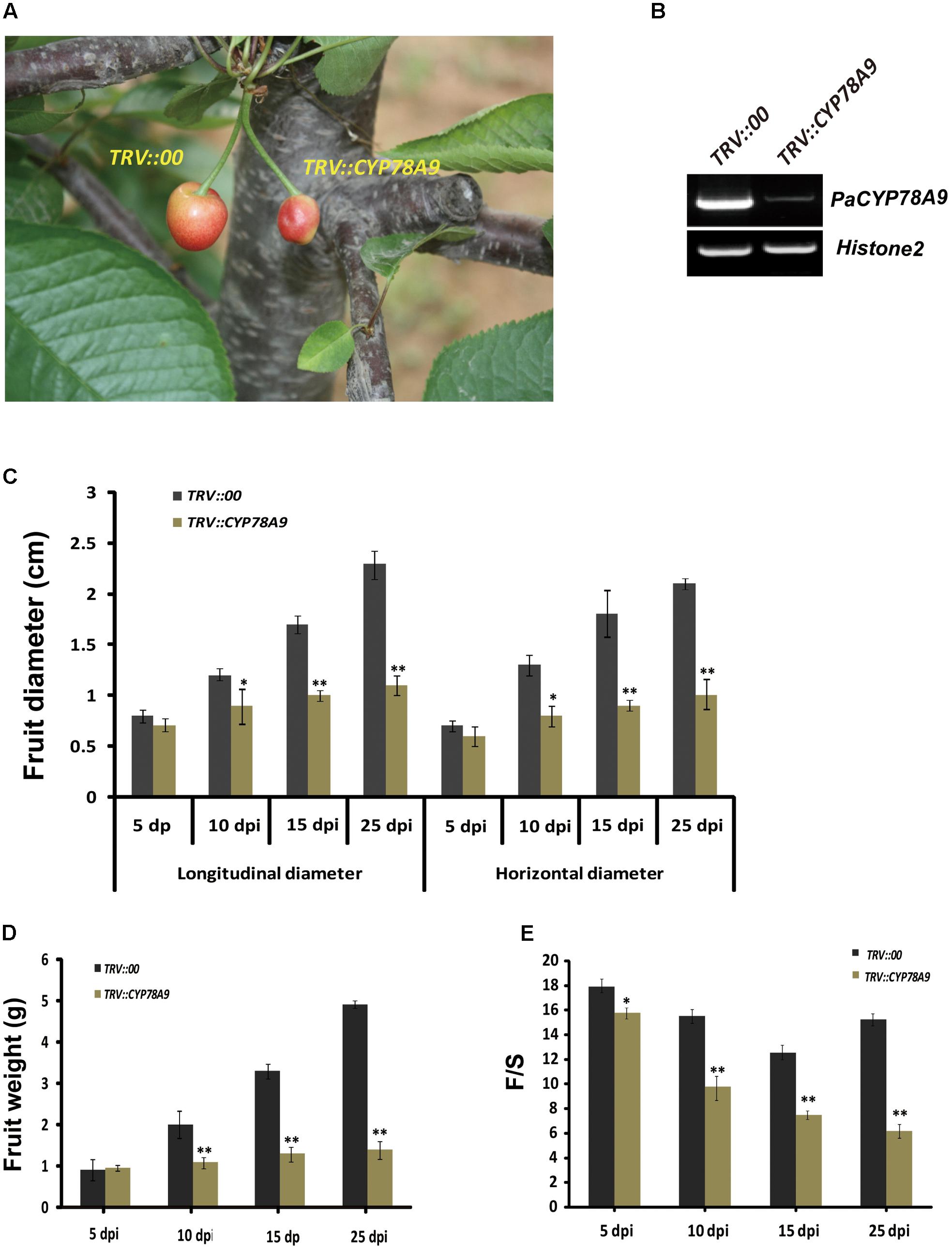
FIGURE 3. Silencing of PaCYP78A9 decreased fruit size in sweet cherry. (A) The phenotype of fruit infected with TRV::PaCYP78A9 or TRV::00 at 25 days post-inoculation (dpi). (B) PaCYP78A9 transcript levels in the fruit of sweet cherry TRV::PaCYP78A9- and TRV::00-silencing lines at 15 dpi by semi-quantitative RT-PCR. Histone2A was used as an internal control. (C–E) The fruit longitudinal diameter (C), horizontal diameter (D), and the weight ratio of fruit flesh and fruit stone (F/S, E) of the TRV::PaCYP78A9-infected fruit were analyzed at 5, 10, 15, and 25 dpi. Values represent the mean ± SD from three independent replicates. Statistical significance was determined by the Student’s t-test. ∗∗P-value < 0.01, ∗P-value < 0.05.
Investigation of plant phenotypes, measurement, and statistical analysis showed that the TRV::PaCYP78A9-infected fruit were dramatically smaller than the TRV::00-infected fruit at 5, 10, 15, and 25 dpi (Figure 3). The longitudinal diameter of the TRV::PaCYP78A9-infected fruit was reduced by 12, 38, 50, and 60%, respectively, when compared with the TRV::00-infected fruit at 5, 10, 15, and 25 dpi. Similarly, the horizontal diameter of TRV::PaCYP78A9-infected fruit also observably reduced (Figure 3C). Furthermore, the weight ratio of fruit flesh and fruit stone (F/S) of the TRV::PaCYP78A9-infected fruit were significantly reduced compared with the TRV::00-infected fruit (Figures 3D,E). However, there was no distinct change in the fruit stone size between TRV::PaCYP78A9-infected and TRV::00-infected sweet cherry fruits. These results suggest that the PaCYP78A9 gene is a significant positive regulator of sweet cherry fruit size during the stages of fruit growth and development.
Mesocarp Cell Number Is Reduced in PaCYP78A9-Silenced Sweet Cherry Fruit
Some CYP78A family members, such as TaCYP78A3, TaCYP78A5, AtCYP78A9, and AtCYP78A5, influenced ultimate seed size by regulating cell proliferation in the integument, which is the future seed coat, in Arabidopsis and wheat (Adamski et al., 2009; Sotelo-Silveira et al., 2013; Ma et al., 2015, 2016). To determine whether the decreased fruit size observed in sweet cherry PaCYP78A9-silencing lines was caused by reduced cell volume in the ovary wall, the mesocarp cell volume of fruit from the TRV::PaCYP78A9- and TRV::00-silencing sweet cherry lines was examined. Mesocarp cell volumes were significantly reduced in fruit from sweet cherry PaCYP78A9-silencing lines compared with fruit from the TRV::00-silencing lines at 25 dpi (Figures 4, 5). Furthermore, the mesocarp cell length and fruit size were smaller for the PaCYP78A9-silencing lines than the TRV::00-silencing lines (Figures 4C,D). Therefore, the decreased fruit size could be caused by a reduction in the mesocarp cell volume and expansion.
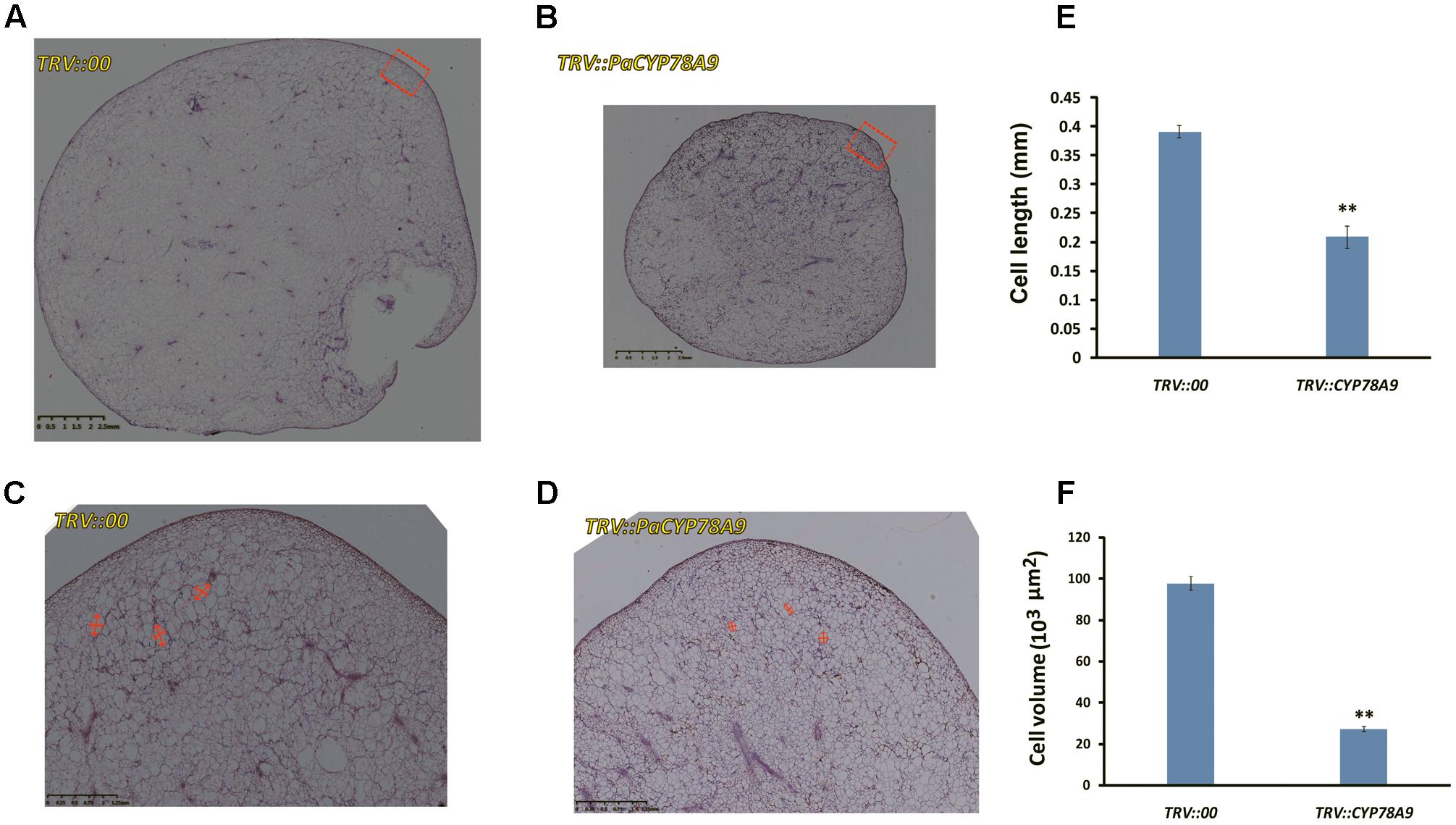
FIGURE 4. The tissue morphology of flesh taken from mesocarp longitudinal sections of sweet cherry fruit. (A,B) Mesocarp longitudinal sections from the fresh fruit of TRV::00-silencing (A) and TRV::PaCYP78A9-silencing (B) lines at 25 dpi stained with hematoxylin (Scale bars = 2.5 mm). (C,D) Magnified views of the mesocarp longitudinal sections (the red virtual box area) corresponding to (A,B) (Scale bars = 1.25 mm). (E,F) Quantification of cell length and Cell volume of the sections from the fresh fruit of TRV::00-silencing and TRV::PaCYP78A9-silencing lines at 25 dpi. Values represent the mean ± SD from three independent replicates. ∗∗Significant differences as calculated using the Student’s t-test at p < 0.01.
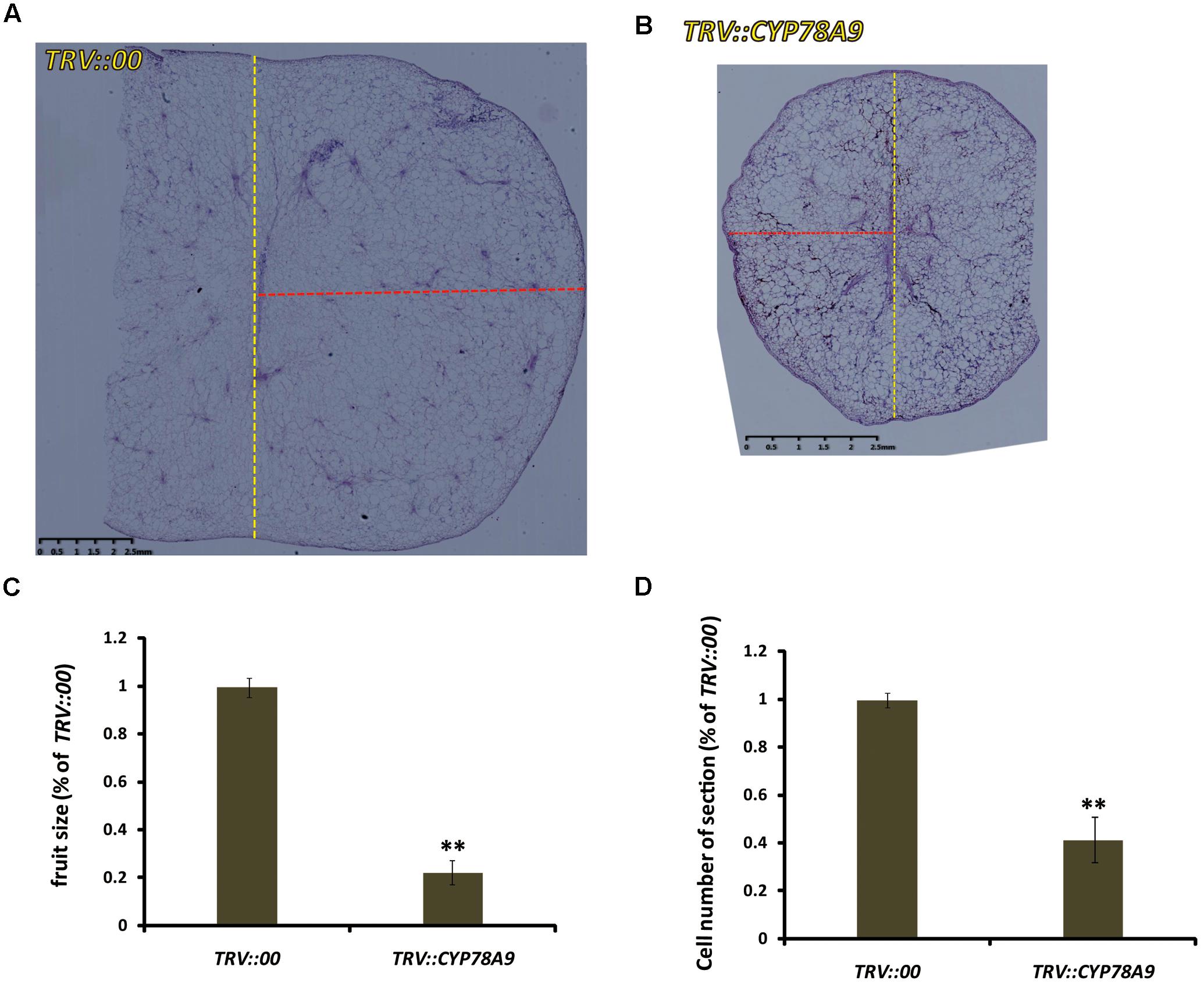
FIGURE 5. The tissue morphology of mesocarp equatorial sections taken from fresh sweet cherry fresh fruit. (A,B) Mesocarp equatorial sections from the fruit of TRV::00-silencing (A) and TRV::PaCYP78A9-silencing (B) lines at 25 dpi stained with hematoxylin (Scale bars = 2.5 mm). The yellow dashed lines showed the meridian of the fruit, the red dashed lines showed the half of equator of the fruit. (C,D) Quantification of the fruit size (C) and cell number of the sections (D) from the fresh fruit of TRV::PaCYP78A9-silencing lines compared with those TRV::00-silencing lines at 25 dpi. ∗∗Indicate significant differences from TRV::00-silencing fruit at P < 0.01 (t-test).
Downregulation of Cell Cycle Genes in Sweet Cherry Fruit from PaCYP78A9-Silencing Lines
Previous studies have shown that cell division activity was restricted by decreases in the transcript levels of cell proliferation-related marker genes and cell cycle genes, including cyclin-dependent kinases (CDKA1, CDKB1, CDKB2, and CDKD3), cyclins (CycA1, CycB2, and CycD3), transcription factors (E2Fa-like and E2Fb-like) encoding promoters of the cell cycle, and other cell division regulation factors such as the cell number regulator 1-like factor (fw2.2), FASCIATED (FAS), and LOCULE NUMBER (LC); downregulation of these genes ultimately resulted in small fruit (Inzé and De Veylder, 2006; Rodríguez et al., 2011; Azzi et al., 2015; Okello et al., 2015). To further analyze the function of PaCYP78A9, the transcript levels of the cell cycle genes and cell proliferation-related marker genes were detected in the fruit of sweet cherry TRV::PaCYP78A9- and TRV::00-silencing lines using qRT-PCR. Notably, in TRV::PaCYP78A9-silencing fruit, the expression level of CDKA1 (Pav_sc0001111.1), CDKD3 (Pav_sc0000103.1), CycA1 (Pav_sc0000195.1), CycB2 (Pav_sc0001288.1), E2Fa-like (Pav_sc0000667.1), E2Fb-like (Pav_sc0000271.1), FAS (Pav_sc0001640.1), and LC (Pav_sc0000044.1) were significantly lower than in the fruit of TRV::00-silencing lines (Figures 6, 7). Conversely, the transcript levels of CDKB1 (Pav_sc0001699.1), CDKB2 (Pav_sc0000600.1), CycD3 (Pav_sc0000136.1), and fw2.2 (Pav_sc0002451.1) were higher in the fruit of TRV::PaCYP78A9-silencing lines compared with the fruit of TRV::00-silencing lines (Figures 6, 7). Previous studies have shown that overexpression of CycD3 reduced cell proliferation in Arabidopsis and that FW2.2 and CDKB1/2 negatively regulated fruit size by controlling cell proliferation during early fruit development in tomato (Frary et al., 2000; Czerednik et al., 2012). Similarly, these genes appear to show analogous functions in the regulation of fruit size in cherry. The downregulation of most cell cycle and proliferation genes in the fruit of TRV::PaCYP78A9-silencing lines suggests that PaCYP78A9 is likely to an important upstream protein factor of cell cycle processes.
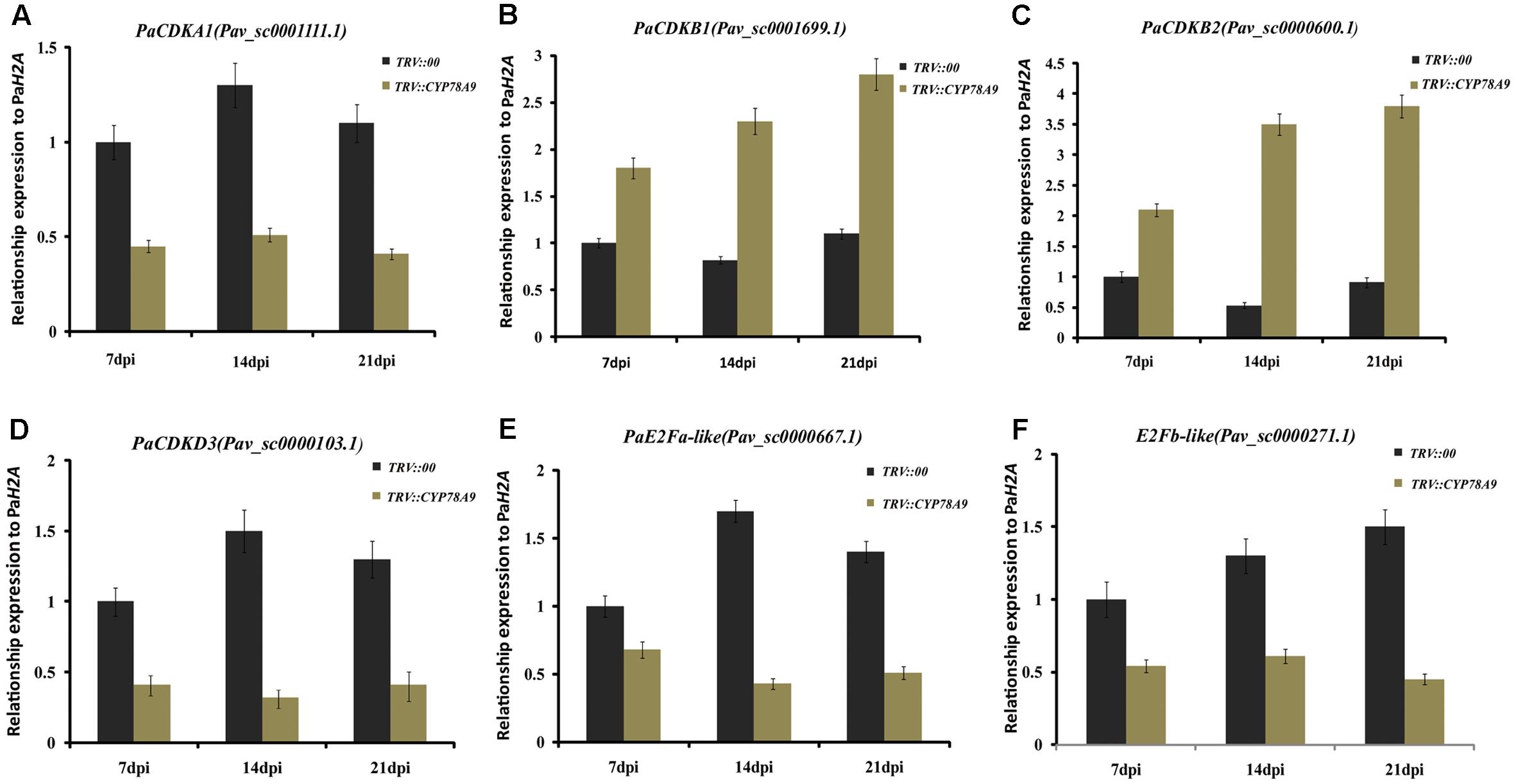
FIGURE 6. Quantitative real-time PCR analysis showing the expression patterns of cyclin-dependent kinase (CDKA1, CDKB1, CDKB2, and CDKD3) and transcription factors encoding promoters of the cell cycle (E2Fa-like and E2Fb-like) in fruit from the sweet cherry TRV::PaCYP78A9- and TRV::00-silencing lines at 7, 14, and 21 dpi. The expression of CDKA1 (A), CDKD3 (D), E2Fa-like (E), and E2Fb-like (F) in TRV::PaCYP78A9-silenced fruit at 7, 14, and 21 dpi was downregulated compared with their expression in the fruit of TRV::00-silencing lines. Conversely, the transcript levels of CDKB1 (B) and CDKB2 (C) increased in TRV::PaCYP78A9-silenced fruit compared with TRV::00-silenced fruit. The gene expression in TRV::00-silenced sweet cherry fruit at 7 dpi was set to 1.0 and Histone2A (Pav_sc0000671.1) was used as an internal standard. Values are means ± SD of three independent experiments performed in duplicate.
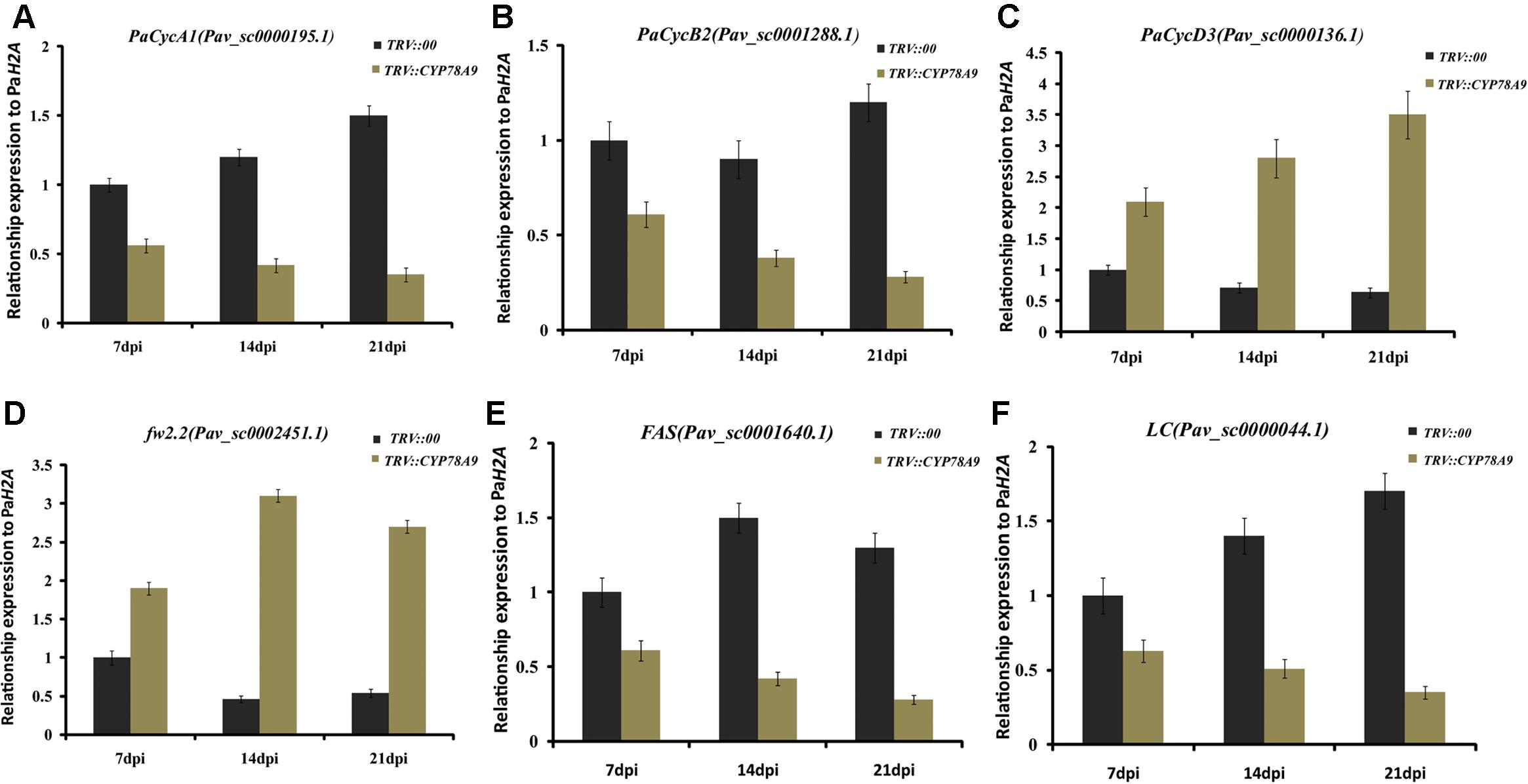
FIGURE 7. Quantitative real-time PCR analysis of the expression levels of cyclins (CycA1, CycB2, and CycD3) and other cell division regulation factors (fw2.2, FAS, and LC) in the fruit for sweet cherry TRV::PaCYP78A9- and TRV::00-silencing lines at 7, 14, and 21 dpi. The expression of CycA1 (A), CycB2 (B), FAS (E), and LC (F) at 7, 14, and 21 dpi in TRV::PaCYP78A9-silenced fruit was downregulated when compared with their expression in fruit of the TRV::00-silencing lines. Conversely, CycD3 (C) and fw2.2 (D) were upregulated in TRV::PaCYP78A9-silenced fruit compared with TRV::00-silenced fruit. Gene expression in TRV::00-silenced sweet cherry fruit at 7 dpi was set to 1.0 and Histone2A (Pav_sc0000671.1) was used as an internal standard. Values are means ± SD of three independent experiments performed in duplicate.
Overexpression of PaCYP78A9 Leads to Increased Seed Size in Arabidopsis
In Arabidopsis, overexpression of CYP78A9 previously resulted in large and seedless siliques and in larger seeds. Moreover, CYP78A9 was highly expressed in the floral organs, especially in the inner carpel wall, suggesting that CYP78A9 plays a role in fruit development (Sotelo-Silveira et al., 2013). To further characterize the function of PaCYP78A9 in this study, the overexpression vector p35S::PaCYP78A9 was assembled and introduced into wild-type (WT) Columbia Arabidopsis plants via Agrobacterium-mediated transformation. We observed that 20 T2 transgenic lines had larger siliques and seeds when compared with WT Columbia Arabidopsis; this increase in silique size was positively correlated with the expression level of PaCYP78A9 in the different transgenic lines (Supplementary Figure S1). Three independent T3 transgenic lines with the highest constitutive PaCYP78A9 expression, PaCYP78A9-3, PaCYP78A9-5, and PaCYP78A9-11 were selected for further analysis.
While none of the transgenic overexpression lines displayed morphological or growth differences in the vegetative stage when compared with WT Arabidopsis plants, the flowers of these overexpression lines were larger, and they produced more siliques than the WT plants (Figures 8A,B,D,G). The seeds of all T3 PaCYP78A9 overexpression transgenic lines were significantly larger than those of WT plants, with both increased seed widths and lengths (Figures 8F,H). The shape of these seeds did not, however, observably change when compared with seed from the WT plants.
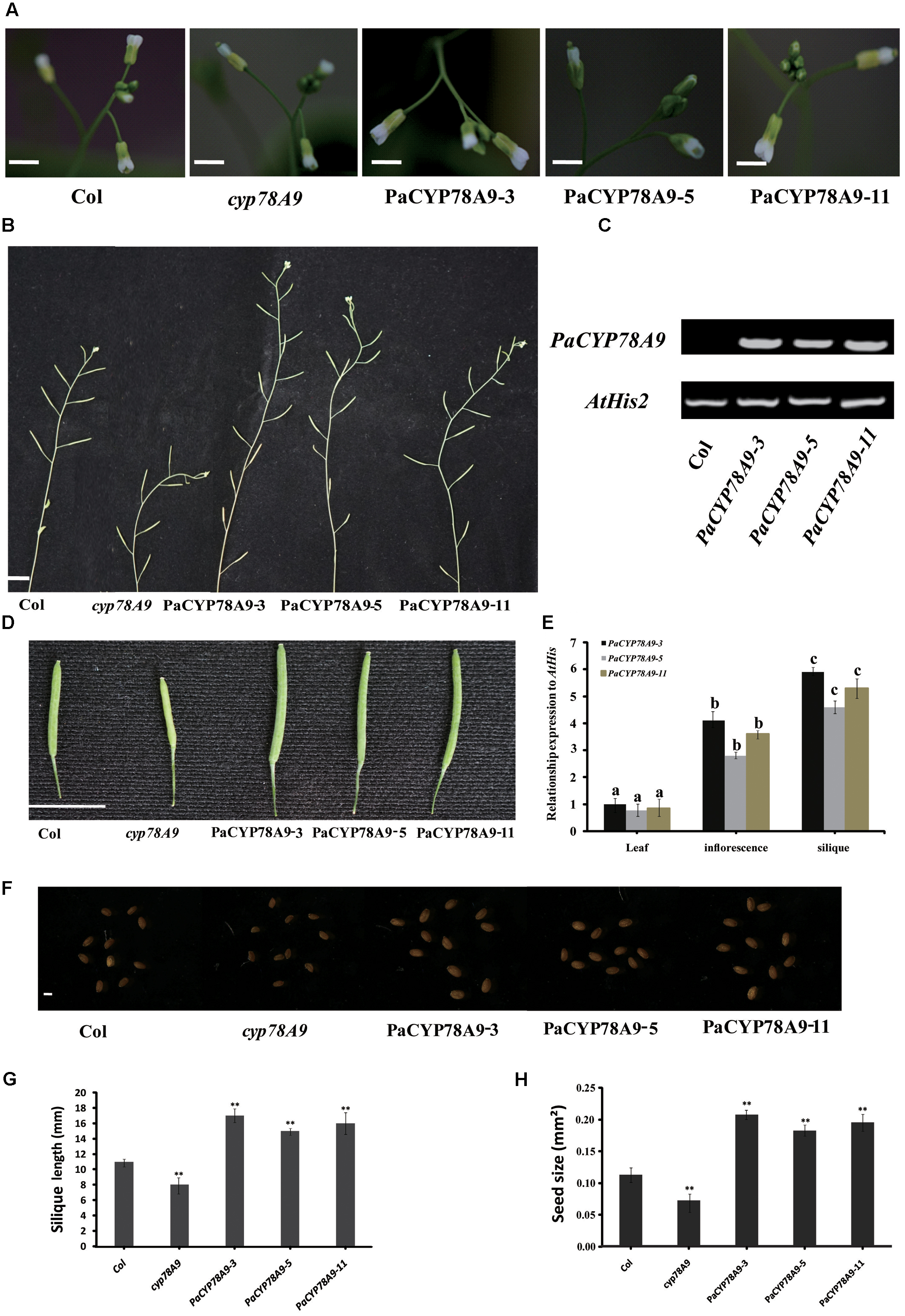
FIGURE 8. Overexpression PaCYP78A9 increased silique and seed size in Arabidopsis. (A) Inflorescence phenotypes of transgenic PaCYP78A9 overexpression lines, cyp78A9 mutant lines, and wild-type (WT) Arabidopsis (scale bars = 2.00 mm). (B) The phenotypes of the main inflorescence stem at stage 6.00 (The stage is in accordance with those described by Boyes et al., 2001) in PaCYP78A9 overexpression lines, cyp78A9 mutant lines, and WT (scale bar = 10.00 mm). (C) RT-PCR analysis of PaCYP78A9 in Arabidopsis transgenic lines. Total RNA was isolated from a mixture of inflorescences and siliques from 10 plants per line. (D) Silique phenotypes (scale bar = 10.00 mm). (E) Expression levels of PaCYP78A9 in different tissues from T3 transgenic lines. (F) Seed phenotypes (scale bar = 0.1 mm). (G,H) All siliques (G) and seeds (H) were measured for 10 transgenic Arabidopsis plants, cyp78A9 mutants, and WT plants. (H) Seed size as represented by the projected area of a seed. Values represent the mean ± SD from three independent replicates. Statistical significance (G,H) was determined by the Student’s t-test, ∗∗significant differences as calculated using the Student’s t-test at p < 0.01; and (E) by one-way ANOVA, letters indicate significant differences (P < 0.05, Duncan test).
We also examined the expression patterns of PaCYP78A9 in the developing flowers and seeds of three independent transgenic T3 overexpression lines and the WT control. Using different tissues from these transgenic lines, qRT-PCR indicated that PaCYP78A9 was largely detected in the inflorescences and siliques of transgenic T3 plants (Figure 8E). Moreover, seed size was correlated with the expression level of PaCYP78A9 in the different transgenic lines (Supplementary Figure S1 and Figure 8). These results indicate that overexpression of PaCYP78A9 promoted large seeds; combined with evidence that silencing of PaCYP78A9 decreased cherry fruit size, this suggests that PaCYP78A9 plays an essential role in the growth and development of reproductive organs in sweet cherry.
PaCYP78A9 Can Rescue the Phenotype of the Arabidopsis Cyp78a9 Deletion Mutant
To further demonstrate the function of PaCYP78A9 in promoting fruit growth and expansion in sweet cherry, a functional copy of PaCYP78A9 containing the promoter CaMV 35S and terminator NOS was transformed into homozygous cyp78a9 mutants (Salk_066588C, obtained from the ABRC) in the Columbia-0 background that display a small seed phenotype. The seed size of cyp78a9 mutants could be rescued to normal WT Arabidopsis levels through the expression of PaCYP78A9 in these lines (Figure 9). Furthermore, overexpression of PaCYP78A9 in the cyp78a9 mutant resulted in seeds that were larger than those of WT Arabidopsis (Figure 9C). These results indicate that PaCYP78A9 can rescue the defective phenotype of the Arabidopsis cyp78a9 insertional mutant, suggesting a conserved function for these two genes in promoting organ size.
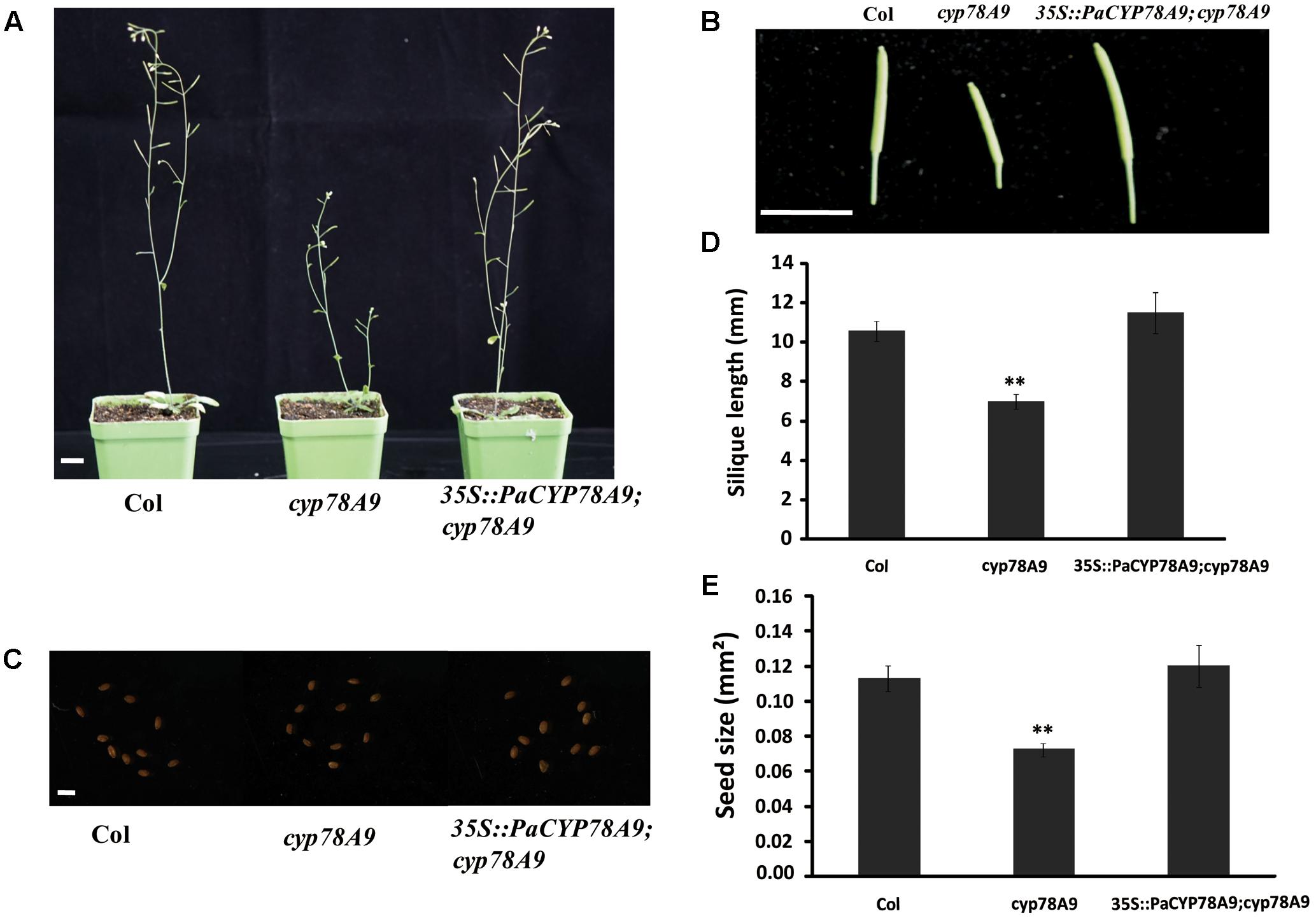
FIGURE 9. PaCYP78A9 can rescue the phenotype of the Arabidopsis cyp78a9 deletion mutant. (A) The phenotype of Arabidopsis plants at stage 6.00 (scale bar = 10.00 mm). (B) Silique phenotypes (scale bar = 10.00 mm). (C) Seed phenotypes (scale bar = 0.1 mm). (D,E) All siliques (D) and seeds (E) were measured for 10 plants of each genotype. Values represent the mean ± SD from three independent replicates. ∗∗Significant differences as calculated using the Student’s t-test at p < 0.01.
Discussion
Cytochrome P450 is one of the largest families of plant proteins; the involvement of its members in the regulation of various metabolic pathways affecting plant growth and development has been studied systematically in many organisms (Ohkawa et al., 1998; Xu J. et al., 2015). Seven plant CYP78A family members, such as CYP78A5, CYP78A6, CYP78A9, CYP78A11, CYP78A13, and CYP78A3 have been identified in Arabidopsis, rice, wheat, tomato, Zea mays, and Physcomitrella patents where they were found to be involved in promoting and enhancing growth and development of the female reproductive organs (Imaishi et al., 2000; Adamski et al., 2009; Katsumata et al., 2011; Fang et al., 2012; Chakrabarti et al., 2013; Nagasawa et al., 2013; Sotelo-Silveira et al., 2013; Ma et al., 2015, 2016; Xu F. et al., 2015). However, the role of CYP78A members in controlling organ size and development in fruit trees was less well characterized.
In this study, we characterized the biological function of the P. avium PaCYP78A9 gene. Overexpression of PaCYP78A9 in Arabidopsis resulted in increased seed size, with the PaCYP78A9 gene found to be largely expressed in the inflorescences and siliques of transgenic plants, and in the flowers and fruit of sweet cherry. Sweet cherry lines silenced for PaCYP78A9 using RNAi produced smaller fruit than the control plants. Moreover, PaCYP78A9 influenced fruit size by affecting mesocarp cell proliferation and expansion during fruit growth and development. Our findings indicate that PaCYP78A9 plays an essential role in regulating cherry fruit size, thereby providing insights into the molecular basis underlying fruit traits such as fruit size and other horticultural characteristics in P. avium.
Although the development of seeds is a complex process, recent studies have shown that final seed size is influenced primarily by three major programs; the bi-parentally derived fertilization products, namely the embryo and endosperm, and the seed coat that is derived from the ovule integument in Arabidopsis, wheat, and rice (Ohto et al., 2005; Sundaresan, 2005; Berger et al., 2006). Some CYP78As and transcription factors such as DA1, TTG2, ARF2, AP2, AtCYP78A9/TaCYP78A3, EOD3/CYP78A6, CYP78A5/KLU, and a KLU homolog affected seed size by promoting cell proliferation of the ovule integuments (seed coat) (Adamski et al., 2009; Sotelo-Silveira et al., 2013). Likewise, mature stone fruit (e.g., peach, cherry, and apricot) derived from ovary tissues are composed of pericarp (exocarp, mesocarp, and endocarp) and seed. Cell expansion and division are important factors regulating fruit size (Gillaspy et al., 1993). In cherry, previous research suggested that fruit size within the same genotype was controlled through mesocarp cell size (Olmstead et al., 2007; Zhang and Whiting, 2011). Conversely, our results indicate that final fruit size was regulated by accelerating mesocarp cell proliferation and expansion during fruit growth and development. Interestingly, overexpression of PaCYP78A9 in Arabidopsis resulted in both enlarged siliques and seeds but also lengthened the reproduction phase. In Arabidopsis, CYP78A9 is involved in controlling floral organ size and also functions during reproductive development by participating in the conversion of dihydrokaempferol to dihydroquercetin in the flavonoid pathway (Sotelo-Silveira et al., 2013). In view of the above findings, we speculate that PaCYP78A9 maybe function by producing a signal of some sort, or by participating in the pathway regulating reproductive organ size through the proliferation of mesocarp cells during fruit growth and development. Taken together, our findings indicate that PaCYP78A9, like CYP78A family members in other plants, regulated reproductive organ development.
While we have characterization the function of CYP78A9 in the regulation of organ size, the regulation mechanism and genetic network mediating CYP78A9 remains unclear. Moreover, CYP78A subfamily genes are generally expressed in floral organs or specific meristems where they move within an inflorescence, not between inflorescences (Zondlo and Irish, 1999; Eriksson et al., 2010; Rolland-Lagan, 2010). For example, CYP78A9 expression in Arabidopsis was detected only in floral organs and not in vegetative tissue (Sotelo-Silveira et al., 2013). Furthermore, CYP78A5/KLU positively regulated a mobile signal downstream from KLU within an inflorescence that affected flower size by influencing cell proliferation (Rolland-Lagan, 2010). To uncover the genetic regulatory network of CYP78A9, the transcript level of cyclin-dependent kinases, cyclins, transcription factors that encode promoters of the cell cycle, and other cell division regulation factors involved in cell proliferation and the regulation of cell cycle genes were analyzed. Our results showed that, with the exclusion of CDKB1, CDKB2, CycD3, and fw2.2, cell cycle genes and transcription factors were most downregulated when PaCYP78A9 was silenced in sweet cherry fruit. During organ growth and development, plant cell proliferation was regulated by the cell cycle; this endoreduplication is largely controlled by Cyc and CDK dimers while cell expansion is related to endoreduplication (Joubes and Chevalier, 2000; Sugimoto-Shirasu and Roberts, 2003; Renaudin et al., 2017). Moreover, the size and number of fruit pericarp cells that were associated with endoreduplication in tomato influenced final tomato fruit size (Cheniclet et al., 2005; Gonzalez et al., 2007; Su et al., 2014; Musseau et al., 2017). Knock-down of CDKA1 and overexpression of either CDKB1 or CDKB2 resulted in smaller and irregularly shaped fruits by reducing the number of exocarp cell layers (Czerednik et al., 2012, 2015). Overexpression of cyclins CycA1, CycB2, and CycD3;3 enhanced growth rate by promoting cell proliferation (Doerner et al., 1996; Dewitte et al., 2007), and the transcription factors E2Fa and E2Fb, acting as activators of cell proliferation, played an important role in cell cycle progression and development (Sozzani et al., 2006). Furthermore, both FAS and LC were ultimately responsible for the growth of larger fruit by increasing floral meristem size and cell division (Cong et al., 2008; Muños et al., 2011). In this study, the expression of positive cell proliferation and cell expansion regulators, including two cyclin-dependent kinases (CDKA1 and CDKD3), three cyclins (CycA1, CycB2, and CycD3;3), two transcription factors (E2Fa and E2Fb) and two other cell division regulation factors (FAS and LC) was suppressed in cherry fruit with reduced PaCYP78A9 expression. We, therefore, hypothesize that PaCYP78A9 is an upstream regulator of during fruit growth and development. Previous studies have indicated that many CYPs are involved in the metabolism of different phytohormones and that CYP78A might be a novel mobile factor regulating organ growth and development in a manner similar to these phytohormones (Adamski et al., 2009; Xu J. et al., 2015). However, the mechanisms underlying the PaCYP78A9-mediated regulation of cell proliferation and cell expansion require further investigation, with more efforts needed in the future to characterize the mechanism of CYP78A9.
In summary, PaCYP78A9, an ortholog of Arabidopsis CYP78A9 in sweet cherry, was functionally characterized and found to affect fruit size by influencing mesocarp cell proliferation and expansion during fruit growth and development. The specific transcription pattern of PaCYP78A9 was positively and closely associated with final fruit size and we further used transgenic lines over-expressing or silenced for PaCYP78A9 to show that it is involved in regulating fruit size. These findings provide novel insights into understanding the molecular mechanisms underlying fruit size determination during fruit growth and development in fruit trees.
Author Contributions
ML, XQ, and CL conceived the research. XQ designed the experiments. XQ, CL, LS, and YL performed the experiments. XQ analyzed the results, and wrote the manuscript. ML provided scientific suggestions, and revised the manuscript. All authors approved the final manuscript, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by a grant from The Central Public-interest Scientific Institution Basal Research Fund (1610192017707) and The Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2017-ZFRI).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank Emma Tacken, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02076/full#supplementary-material
FIGURE S1 | Quantitative real-time PCR (qRT-PCR) analysis of the expression level (A) of PaCYP78A9 and seed size (B) of PaCYP78A9 over-expression T2 Arabidopsis transgenic lines compared with wild Arabidopsis. The endogenous ATCYP78A9 expression in WT Arabidopsis was set to 1.0. (t-test: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
Footnotes
References
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. doi: 10.1073/pnas.0907024106
Azzi, L., Deluche, C., Gévaudant, F., Frangne, N., Delmas, F., Hernould, M., et al. (2015). Fruit growth-related genes in tomato. J. Exp. Bot. 66, 1075–1086. doi: 10.1093/jxb/eru527
Bak, S., Beisson, F., Bishop, G., Hamberger, B., Höfer, R., Paquette, S., et al. (2011). Cytochromes P450. The Arabidopsis Book 9:e0144. doi: 10.1199/tab.0144
Berger, F., Grini, P. E., and Schnittger, A. (2006). Endosperm: an integrator of seed growth and development. Curr. Opin. Plant Biol. 9, 664–670. doi: 10.1016/j.pbi.2006.09.015
Boyes, D. C., Zayed, A. M., Ascenzi, R., McCaskill, A. J., Hoffman, N. E., Davis, K. R., et al. (2001). Growth stage–based phenotypic analysis of Arabidopsis a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510. doi: 10.1105/TPC.010011
Campoy, J. A., Le Dantec, L., Barreneche, T., Dirlewanger, E., and Quero-García, J. (2015). New insights into fruit firmness and weight control in sweet cherry. Plant Mol. Biol. Rep. 33, 783–796. doi: 10.1007/s11105-014-0773-6
Chakrabarti, M., Zhang, N., Sauvage, C., Muños, S., Blanca, J., Cañizares, J., et al. (2013). A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. U.S.A. 110, 17125–17130. doi: 10.1073/pnas.1307313110
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Cheniclet, C., Rong, W. Y., Causse, M., Frangne, N., Bolling, L., Carde, J. P., et al. (2005). Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 139, 1984–1994. doi: 10.1104/pp.105.068767
Cong, B., Barrero, L. S., and Tanksley, S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40, 800–804. doi: 10.1038/ng.144
Cui, H., and Wang, A. (2017). An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum pox virus via silencing of a host factor gene. Plant Biotechnol. J. 15, 344–356. doi: 10.1111/pbi.12629
Czerednik, A., Busscher, M., Angenent, G. C., and de Maagd, R. A. (2015). The cell size distribution of tomato fruit can be changed by overexpression of CDKA1. Plant Biotechnol. J. 13, 259–268. doi: 10.1111/pbi.12268
Czerednik, A., Busscher, M., Bielen, B. A., Wolters-Arts, M., de Maagd, R. A., and Angenent, G. C. (2012). Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. J. Exp. Bot. 63, 2605–2617. doi: 10.1093/jxb/err451
Dewitte, W., Scofield, S., Alcasabas, A. A., Maughan, S. C., Menges, M., Braun, N., et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542. doi: 10.1073/pnas.0704166104
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
Doerner, P., Jorgensen, J. E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. doi: 10.1038/380520a0
Du, L., Li, N., Chen, L., Xu, Y., Li, Y., Zhang, Y., et al. (2014). The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26, 665–677. doi: 10.1105/tpc.114.122663
Eriksson, S., Stransfeld, L., Adamski, N. M., Breuninger, H., and Lenhard, M. (2010). KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Curr. Biol. 20, 527–532. doi: 10.1016/j.cub.2010.01.039
Fang, W. J., Wang, Z. B., Cui, R. F., Li, J., and Li, Y. H. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70, 929–939. doi: 10.1111/j.1365-313X.2012.04907.x
Frandsen, R. J., Frandsen, M., and Giese, H. (2012). Targeted gene replacement in fungal pathogens via Agrobacterium tumefaciens-mediated transformation. Methods Protoc. 835, 17–45. doi: 10.1007/978-1-61779-501-5_2
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E., Cong, B., Liu, J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi: 10.1126/science.289.5476.85
Fu, D. Q., Zhu, B. Z., Zhu, H. L., Jiang, W. B., and Luo, Y. B. (2005). Virus-induced gene silencing in tomato fruit. Plant J. 43, 299–308. doi: 10.1111/j.1365-313X.2005.02441.x
Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: a developmental perspective. Plant Cell 5, 1439–1451. doi: 10.1105/tpc.5.10.1439
Gonzalez, N., Gévaudant, F., Hernould, M., Chevalier, C., and Mouras, A. (2007). The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J. 51, 642–655. doi: 10.1111/j.1365-313X.2007.03167.x
Horiguchi, G., Ferjani, A., Fujikura, U., and Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119, 37–42. doi: 10.1007/s10265-005-0232-4
Imaishi, H., Matsuo, S., Swai, E., and Ohkawa, H. (2000). CYP78A1 preferentially expressed in developing inflorescences of Zea mays encoded a cytochrome P450-dependent lauric acid 12-monooxygenase. Biosci. Biotechnol. Biochem. 64, 1696–1701. doi: 10.1271/bbb.64.1696
Inzé, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3117–3122. doi: 10.1073/pnas.0409893102
Johnson, C. S. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Joubes, J., and Chevalier, C. (2000). Endoreduplication in higher plants. Plant Mol. Biol. 43, 735–745. doi: 10.1023/A:1006446417196
Katsumata, T., Fukazawa, J., Magome, H., Jikumaru, Y., Kamiya, Y., Natsume, M., et al. (2011). Involvement of theCYP78A subfamily of cytochrome P450 monooxygenases in protonema growth and gametophore formation in the moss Physcomitrella patens. Biosci. Biotechnol. Biochem. 75, 331–336. doi: 10.1271/bbb.100759
Kawai, T., Gonoi, A., Nitta, M., Yamagishi, N., Yoshikawa, N., and Tao, R. (2016). Virus-induced gene silencing in various Prunus species with the Apple latent spherical virus vector. Sci. Hortic. 199, 103–113. doi: 10.1016/j.scienta.2015.12.031
Li, B., Xie, Z., Zhang, A., Xu, W., Zhang, C., Liu, C., et al. (2010). Tree growth characteristics and flower bud differentiation of sweet cherry (Prunus avium L.) under different climate conditions in China. Hortic. Sci. 37, 6–13.
Li, D. M., Wang, Y., and Han, K. L. (2012). Recent density functional theory model calculations of drug metabolism by cytochrome P450. Coord. Chem. Rev. 256, 1137–1150. doi: 10.1016/j.ccr.2012.01.016
Li, N., and Li, Y. (2015). Maternal control of seed size in plants. J. Exp. Bot. 66, 1087–1097. doi: 10.1093/jxb/eru549
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, Q., Chen, P., Dai, S., Sun, Y., Yuan, B., Kai, W., et al. (2015). PacCYP707A2 negatively regulates cherry fruit ripening while PacCYP707A1 mediates drought tolerance. J. Exp. Bot. 66, 3765–3774. doi: 10.1093/jxb/erv169
Li, Y. H., Zheng, L. Y., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Ma, M., Wang, Q., Li, Z., Cheng, H., Li, Z., Liu, X., et al. (2015). Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.) affects seed size. Plant J. 83, 312–325. doi: 10.1111/tpj.12896
Ma, M., Zhao, H., Li, Z., Hu, S., Song, W., and Liu, X. (2016). TaCYP78A5 regulates seed size in wheat (Triticum aestivum). J. Exp. Bot. 67, 1397–1410. doi: 10.1093/jxb/erv542
Mizutani, M., and Ohta, D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315. doi: 10.1146/annurev-arplant-042809-112305
Muños, S., Ranc, N., Botton, E., Bérard, A., Rolland, S., Duffé, P., et al. (2011). Increase in tomato locule number is controlled by two key SNP located near Wuschel. Plant Physiol. 156, 2244–2254. doi: 10.1104/pp.111.173997
Musseau, C., Just, D., Jorly, J., Gévaudant, F., Moing, A., Chevalier, C., et al. (2017). Identification of two new mechanisms that regulate fruit growth by cell expansion in tomato. Front. Plant Sci. 8:988. doi: 10.3389/fpls.2017.00988
Nagasawa, N., Hibara, K., Heppard, E. P., Vander Velden, K. A., Luck, S., Beatty, M., et al. (2013). GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 75, 592–605. doi: 10.1111/tpj.12223
Nelson, D. R. (2006). Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 5, 193–204. doi: 10.1007/s11101-006-9015-3
Nitsch, L., Kohlen, W., Oplaat, C., Charnikhova, T., Cristescu, S., Michieli, P., et al. (2012). ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 169, 878–883. doi: 10.1016/j.jplph.2012.02.004
Ohkawa, H., Imaishi, H., Shiota, N., Yamada, T., Inui, H., and Ohkawa, Y. (1998). Molecular mechanisms of herbicide resistance with special emphasis on cytochrome P450 monooxygenases. Plant Biotechnol. 15, 173–176. doi: 10.5511/plantbiotechnology.15.173
Ohto, M. A., Fischer, R. L., Goldberg, R. B., Nakamura, K., and Harada, J. J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3123–3128. doi: 10.1073/pnas.0409858102
Okello, R. C., Heuvelink, E., de Visser, P. H., Lammers, M., de Maagd, R. A., Marcelis, L. F., et al. (2015). Fruit illumination stimulates cell division but has no detectable effect on fruit size in tomato (Solanum lycopersicum). Physiol. Plant. 154, 114–127. doi: 10.1111/ppl.12283
Olmstead, J. W., Iezzoni, A. F., and Whiting, M. D. (2007). Genotypic differences in sweet cherry fruit size are primarily a function of cell number. J. Am. Soc. Hortic. Sci. 132, 697–703.
Qi, X. L., Su, X. F., Guo, H. M., Qi, J. C., and Cheng, H. M. (2015). A ku70 null mutant improves gene targeting frequency in the fungal pathogen Verticillium dahliae. World J. Microbiol. Biotechnol. 31, 1889–1897. doi: 10.1007/s11274-015-1907-1
Renaudin, J. P., Deluche, C., Cheniclet, C., Chevalier, C., and Frangne, N. (2017). Cell layer-specific patterns of cell division and cell expansion during fruit set and fruit growth in tomato pericarp. J. Exp. Bot. 68, 1613–1623. doi: 10.1093/jxb/erx058
Rodríguez, G. R., Muños, S., Anderson, C., Sim, S. C., Michel, A., Causse, M., et al. (2011). Distribution of SUN, OVATE, LC, and FAS alleles in tomato germplasm and their effect on fruit morphology. Plant Physiol. 156, 275–285. doi: 10.1104/pp.110.167577
Rojas-Gracia, P., Roque, E., Medina, M., Rochina, M., Hamza, R., Angarita-Díaz, M. P., et al. (2017). The parthenocarpic hydra mutant reveals a new function for a SPOROCYTELESS-like gene in the control of fruit set in tomato. New Phytol. 214, 1198–1212. doi: 10.1111/nph.14433
Rolland-Lagan, A. G. (2010). Organ size: the role of mobile signals. Curr. Biol. 20, R268–R269. doi: 10.1016/j.cub.2010.02.017
Rosyara, U. R., Bink, M. C., van de Weg, E., Zhang, G., Wang, D., Sebolt, A., et al. (2013). Fruit size QTL identification and the prediction of parental QTL genotypes and breeding values in multiple pedigreed populations of sweet cherry. Mol. Breed. 32, 875–887. doi: 10.1007/s11032-013-9916-y
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., and Fenby, N. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Shen, X., Zhao, K., Liu, L., Zhang, K., Yuan, H., Liao, X., et al. (2014). A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 55, 862–880. doi: 10.1093/pcp/pcu013
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sotelo-Silveira, M., Cucinotta, M., Chauvin, A. L., Chávez, Montes R. A., Colombo, L., Marsch-Martínez, N., et al. (2013). Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiol. 162, 779–799. doi: 10.1104/pp.113.218214
Sozzani, R., Maggio, C., Varotto, S., Canova, S., Bergounioux, C., Albani, D., et al. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140, 1355–1366. doi: 10.1104/pp.106.077990
Su, L., Bassa, C., Audran, C., Mila, I., Cheniclet, C., Chevalier, C., et al. (2014). The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion. Plant Cell Physiol. 55, 1969–1976. doi: 10.1093/pcp/pcu124
Sugimoto-Shirasu, K., and Roberts, K. (2003). ‘Big it up’: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553. doi: 10.1016/j.pbi.2003.09.009
Sundaresan, V. (2005). Control of seed size in plants. Proc. Natl. Acad. Sci. U.S.A. 102, 17887–17888. doi: 10.1073/pnas.0509021102
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, J. W., Schwab, R., Czech, B., Mica, E., and Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. doi: 10.1105/tpc.108.058180
Whiting, M. D., Ophardt, D., and McFerson, J. R. (2006). Chemical blossom thinners vary in their effect on sweet cherry fruit set, yield, fruit quality, and crop value. HortTechnology 16, 66–70.
Xia, T., Li, N., Dumenil, J., Li, J., Kamenski, A., Bevan, M. W., et al. (2013). The Ubiquitin receptor DA1 Interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25, 3347–3359. doi: 10.1105/tpc.113.115063
Xu, F., Fang, J., Ou, S., Gao, S., Zhang, F., Du, L., et al. (2015). Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 38, 800–811. doi: 10.1111/pce.12452
Xu, J., Wang, X. Y., and Guo, W. Z. (2015). The cytochrome P450 superfamily: key players in plant development and defense. J. Integr. Agric. 14, 1673–1686. doi: 10.1016/S2095-3119(14)60980-1
Yao, J. L., Xu, J., Cornille, A., Tomes, S., Karunairetnam, S., Luo, Z., et al. (2015). A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 84, 417–427. doi: 10.1111/tpj.13021
Zhang, C., and Whiting, M. D. (2011). Improving ‘Bing’ sweet cherry fruit quality with plant growth regulators. Sci. Hortic. 127, 341–346. doi: 10.1016/j.scienta.2010.11.006
Zhang, G. R., Sebolt, A. M., Sooriyapathirana, S. S., Wang, D. C., Bink, M. C., Olmstead, J., et al. (2010). Fruit size QTL analysis of an F1 population derived from a cross between a domesticated sweet cherry cultivar and a wild forest sweet cherry. Tree Genet. Genomes 6, 25–36. doi: 10.1007/s11295-009-0225-x
Zhao, B., Dai, A., Wei, H., Yang, S., Wang, B., Jiang, N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. doi: 10.1007/s11103-015-0392-0
Keywords: Prunus avium L, CYP78A, fruit size, VIGS, PaCYP78A9
Citation: Qi X, Liu C, Song L, Li Y and Li M (2017) PaCYP78A9, a Cytochrome P450, Regulates Fruit Size in Sweet Cherry (Prunus avium L.). Front. Plant Sci. 8:2076. doi: 10.3389/fpls.2017.02076
Received: 17 August 2017; Accepted: 20 November 2017;
Published: 05 December 2017.
Edited by:
Rafael Lozano, University of Almería, SpainReviewed by:
Muriel Quinet, Université catholique de Louvain, BelgiumYong Xu, National Engineering Research Center for Vegetables, China
Maria Luisa Badenes, El Instituto Valenciano de Investigaciones Agrarias, Spain
Copyright © 2017 Qi, Liu, Song, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Li, bGltaW5nMDZAY2Fhcy5jbg==
 Xiliang Qi
Xiliang Qi Congli Liu
Congli Liu Lulu Song
Lulu Song