- 1London Research and Development Centre, Agriculture and Agri-Food Canada, London, ON, Canada
- 2Department of Biology, University of Western Ontario, London, ON, Canada
Soybean (Glycine max [L.] Merr) is one of the main grain legumes worldwide. Soybean farmers lose billions of dollars’ worth of yield annually due to root and stem rot disease caused by the oomycete Phytophthora sojae. Many strategies have been developed to combat the disease, however, these methods have proven ineffective in the long term. A more cost effective and durable approach is to select a trait naturally found in soybean that can increase resistance. One such trait is the increased production of phytoalexin glyceollins in soybean. Glyceollins are isoflavonoids, synthesized via the legume-specific branch of general phenylpropanoid pathway. The first key enzyme exclusively involved in glyceollin synthesis is chalcone reductase (CHR) which coacts with chalcone synthase for the production of isoliquiritigenin, the precursor for glyceollin biosynthesis. Here we report the identification of 14 putative CHR genes in soybean where 11 of them are predicted to be functional. Our results show that GmCHRs display tissue-specific gene expression, and that only root-specific GmCHRs are induced upon P. sojae infection. Among 4 root-specific GmCHRs, GmCHR2A is located near a QTL that is linked to P. sojae resistance suggesting GmCHR2A as a novel locus for partial resistance that can be utilized for resistance breeding.
Introduction
Soybean (Glycine max [L.] Merr) is one of the most important grain legumes in the world. The seeds are an excellent source of protein, oil, micronutrients and specialized metabolites such as isoflavonoids and saponins, making it a profitable crop for human or livestock consumption and many industrial products. From this versatility, the United States Department of Agriculture has estimated that the Global Soybean Production in 2017/2018 is 348.04 million metric tons, which is an increase of 11.18% in soybean production worldwide1. In Canada, soybean was the 4th largest crop grown in 2016, producing 6.46 million metric tons and generating $2.4 billion in profits2. Though these numbers appear promising, soybean farmers encounter about $50 million of soybean yield loss annually in Canada, and $1–2 billion worldwide due to stem and root rot disease caused by Phytophthora sojae.
The traditional strategies to reduce the incidence of P. sojae infection such as soil treatment with calcium application (Sugimoto et al., 2010), fungicide application (Anderson and Buzzell, 1982) and improved soil drainage and tillage (Workneh et al., 1998) have not only proven to be ineffective, but also found to place selective pressures on P. sojae leading to resistance (Li et al., 2010). An alternative approach to this problem is selecting a cultivar of soybean with optimal resistance to P. sojae infection. Resistance to pathogen infection in soybeans can either be complete or partial. Complete resistance or race-specific resistance is conferred by single dominant resistance to P. sojae (Rps) genes, which counteract the virulence genes within P. sojae. This relationship is similar to effector-triggered immune response in other pathosystems (Sugimoto et al., 2012). To date, there are 27 Rps genes and more than 200 identified races of P. sojae (Sahoo et al., 2017). However, the rapid evolution of P. sojae races continues to diversify and as a result the new races become ineffective to the Rps genes leading to disease susceptibility. Partial resistance, referred to as field resistance, is more durable and a broad-spectrum non-race-specific trait conferred by several minor genes which involves various defense components. Cultivars with this type of resistance contain fewer damaged roots than completely susceptible cultivars, show delayed disease progression, and are effective against all races of P. sojae (Schmitthenner, 1985). Some key traits such as increased levels of soybean root suberin and increased production of phytoalexin glyceollins have been correlated with strong partial resistance in soybean (Thomas et al., 2007; Lygin et al., 2013). Isoflavonoids act as phytoalexins in soybean that play a role as plant’s basal and innate response to biotic and abiotic stress. When isoflavonoid production was compromised by down regulating the biosynthetic genes, chalcone reductase (CHR) and isoflavone synthase (IFS), it reduced the plants’ ability to fight off the pathogen attack (Subramanian et al., 2005; Graham et al., 2007; Lozovaya et al., 2007).
CHR (formally known as polyketide reductase) is the key enzyme for isoflavone aglycone daidzein biosynthesis which ultimately leads to the production of the phytoalexin glyceollins in soybean (Dakora and Phillips, 1996). The isoflavonoid pathway begins with the general phenylproponoid pathway that branches off into two sub-branches: (i) chalcone synthase (CHS) producing naringenin chalcone, a compound that subsequently synthesizes genistein, a core isoflavone aglycone and/or many other flavonoids; (ii) CHS coacting with CHR producing isoliquiritigenin, the building block of the other two core isoflavone aglycones, glycitein and daidzein (Figure 1).
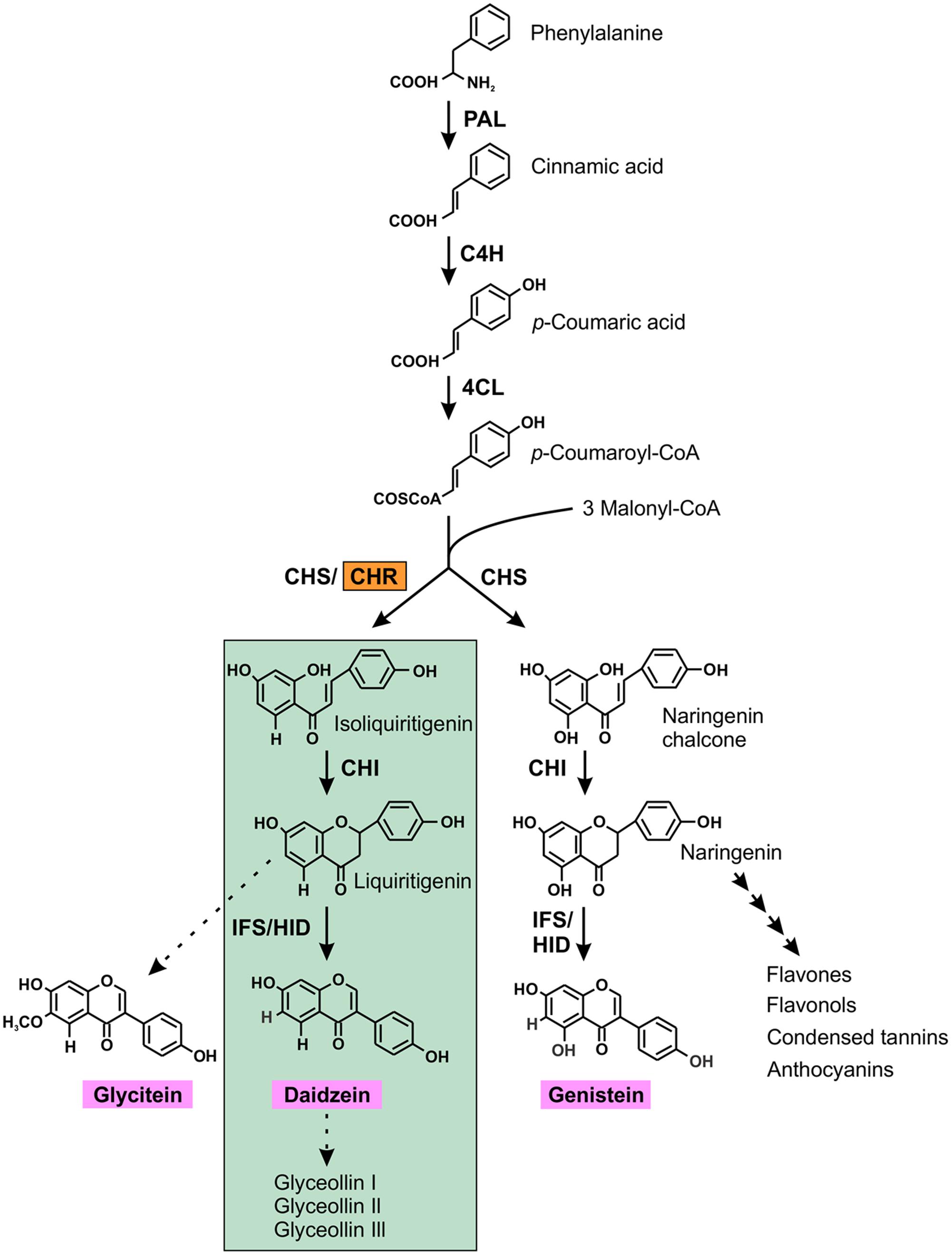
FIGURE 1. (Iso) flavonoid biosynthetic pathway in soybean. The first key legume-specific enzyme chalcone reductase (CHR) is highlighted in orange that leads into the synthesis of isoflavonoid phytoalexins (green highlights) in soybean. The three isoflavone aglycones glycitein, daidzein and genistein are shown in pink. The dotted arrow indicates speculative steps while multiple arrows indicate two or more steps in the pathway. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHR, chalcone reductase; CHS, chalcone synthase; CHI, chalcone isomerase; IFS, isoflavone synthase; 2HID, 2-hydroxyisoflavanone dehydratase.
Chalcone reductase belongs to the aldo-keto reductase (AKR) sub-family 4 in the large AKR superfamily (Jez et al., 1997). All members of this superfamily fold into a monomeric, (α/β)8 barrel structure and contain a catalytic tetrad of Asp-53, Tyr-58, Lys-87, and His-120 and a common NAD(P)(H) binding site that is located in a deep, large and hydrophobic pocket at the C-terminus end (Bomati et al., 2005). All CHRs predominantly contain hydrophobic and aromatic residues that line the unoccupied entrance to the active site cavity molded by Pro-29, Ala-57, Trp-89, Phe-130, and Phe-132. Largely polar residues define the base of this catalytic surface and include the catalytic tetrad, Trp-121 and Asn-167 (Bomati et al., 2005). Beside these facts, very little is known about CHR enzyme since it acts on intermediates for CHS, and identity of its substrate still remains unknown.
The first CHR activity was discovered in the crude extracts of Glycyrrhiza echinata (Ayabe et al., 1988). Up to now, CHR-like enzymes have been identified in a variety of leguminous plant species, including Medicago sativa (Ballance and Dixon, 1995), Sesbania rostrata (Goormachtig et al., 1999), Pueraria montana var. lobata (He et al., 2011), Glycyrrhiza glabra (Hayashi et al., 2013), and Lotus japonicus (Shimada et al., 2006). Graham et al. (2007) identified 4 soybean CHRs through the Expressed Sequence Tag (EST) database search3. RNAi silencing of these CHRs in soybean hairy roots resulted into reduced levels of isoflavonoids and increased susceptibility to P. sojae infection compared to the control (Graham et al., 2007).
Here, we performed a genome-wide search of AKR gene family members and identified 14 CHR genes in soybean. The phylogenetic history of the GmCHRs, and their sequence analysis suggested 11 GmCHRs as catalytically active for its role. Transcript analysis revealed that 4 root-specific GmCHRs are induced upon stress. Some of the stress induced GmCHRs locate in the QTL regions that are linked to P. sojae resistance. These GmCHR genes together with other minor genes and Rps gene can be stacked in the same cultivar for future resistant breeding.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of soybean (Glycine max L. Merr.) cv Harosoy63, OX760-6 and Conrad, and Nicotiana benthamiana were grown in a growth room with 16 h light at 23°C and 8 h dark cycle at 18°C, with 60–70% relative humidity and a light intensity of 100–150 μmol m-2 s-1.
In Silico Analysis
To identify all CHR gene family members in the soybean genome, a search was conducted in the annotated G. max Wm82.a2.v1 genome of Phytozome4. The keywords “aldo-keto” and “aldo/keto” were used to find all the soybean aldo-keto reductases (GmAKRs). To ensure no GmAKRs were missed in the keyword search, each GmAKR was used as a query for a BLAST search again. For generating a phylogenetic tree, protein sequences were aligned using CLUSTALO5 and a Neighbor-joining tree based with 1000 bootstrap replications was created using MEGA7 (Kumar et al., 2016). The Poisson method was selected to calculate the evolutionary distance of the phylogenetic tree and pairwise deletion was selected for gaps/missing data treatment. To determine whether all candidate GmCHRs contain residues deemed important for catalytic activity, the protein sequences of the candidate GmCHR were aligned using CLUSTALO followed by BOXSHADE 3.216. Critical residues were manually spotted based on Bomati et al. (2005).
The QTLs and QTL markers from the year 2003 to 2016 corresponding to P. sojae resistance were extracted from the SoyBase and Soybean Breeder’s Toolbox7. To ensure no QTLs or QTL marker were missed in the search; a literature search was also conducted. Relative positions of GmCHRs, and QTL markers were mapped onto the chromosomes. QTLs regions in base pairs were noted from the G. max genome assembly on Soybase.org.
RNA Extraction, cDNA Synthesis, RT/qRT-PCR and Gene Expression Analysis
Total RNA was extracted from 100 mg of tissue using the RNeasy Plant Mini Kit (Qiagen) following manufacturer’s instruction with some modification. An on-column DNaseI (Promega) treatment was used to digest DNA. Thermoscript RT-PCR System (Life Technologies) was used to synthesize cDNA from total RNA. PCR amplification was performed using gene-specific primers (Supplementary Table S1). SsoFast EvaGreen Supermix Kit (BioRad) was used for qPCR reaction with gene-specific primers (Supplementary Table S1). CONS4 was used as a reference gene to normalize the expression (Libault et al., 2008).
A publicly available RNA-seq database containing transcriptome sequencing of soybean8 was mined for the tissue-specific expression profiles of GmCHR gene family members. The relative expression was normalized across the libraries corresponding to each tissue. A heatmap for GmCHR transcripts was generated in R.
Western Blot Analysis
Samples (0.5 g) were ground in liquid nitrogen and re-suspended in protein extraction buffer (25 mM Tris-HCL pH 8.0, 1 mM EDTA pH 8.0, 20 mM NaCl) supplemented with protease inhibitor cocktail (Sigma–Aldrich) for protein extraction. Total soluble proteins (30 μg) were separated on a SDS-PAGE, transferred onto PVDF membrane (Bio-Rad) using a Trans-Blot Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). The fusion proteins with YFP were detected using an anti-GFP (1:7000 dilution) mouse primary antibody and conjugated horseradish peroxidase goat anti-mouse (1:5000 dilution) secondary antibody, followed by treatment with ECL Prime Western Blot detection reagents (GE Health Care Life Sciences).
Stress Treatment
Seven-day old seedlings of soybean cv. L76-1988 were inoculated with P. sojae race 7. The stems of the infected plants were collected at 24, 48, and 72 h post-inoculation. For the AgNO3 treatment, soybean cv. Harosoy63 was grown in water-soaked vermiculite in the dark at 25°C for 6 days. Prior to the treatment, 10 etiolated seedlings per treatment were transferred into glass trays, after which, 5–10 drops of 10 μL of either water (control) or 1 mM of AgNO3 were placed onto the hypocotyl of each seedling (Supplementary Figure S1). The trays were transferred back in the dark to 25°C. Samples were collected at 6, 12, 24, 48, or 72 h after the treatment.
Plasmid Construction and Subcellular Localization
For subcellular localization study, GmCHRs were amplified by PCR using gene-specific primers (Supplementary Table S1) and cloned into the gateway entry vector pDONR-Zeo (Invitrogen) using the BP clonase reaction mix (Invitrogen), transformed into Escherichia coli DH5α. The recombinant plasmid pDONR-zeo-GmCHR was recombined with the destination vector pEarlyGate101 using the LR clonase reaction mix (Invitrogen). The sequence confirmed pEarlyGate101 was transformed into Agrobacterium tumefaciens GV3101 via electroporation.
The constructs in A. tumefaciens GV3101 were transformed into N. benthamiana leaves by infiltration (Sparkes et al., 2006), and transient expression was visualized through a Leica TCS SP2 inverted confocal microscope. To confirm the nuclear localization of the proteins, A. tumefaciens GV3101 with the construct containing nuclear localization signal fused to CFP (pEarleyGate100-NLS-CFP) and pEarleyGate101-GmCHR were mixed 1:1 and infiltrated into the leaves followed by confocal microscopy. For YFP visualization, an excitation wavelength of 514 nm was used and emissions were collected between 525 and 545 nm. For visualization of CFP, an excitation wavelength of 434 nm was used and emissions were collected between 460 and 490 nm.
Results
The Soybean Genome Contains 14 Putative GmCHRs
Since CHRs belong to the AKR superfamily, we performed a keyword search in the annotated G. max Wm82.a2.v1 genome on Phytozome to identify all the GmAKRs. Using the keywords “aldo-keto” and “aldo/keto”, protein databases KOG, Pfam and PANTHER identified 34 and 67 GmAKRs, respectively. Results from both keyword searches were compared and compiled to ensure no duplicates. Each GmAKR sequence was then used as a query for a BLAST search in G. max Wm82.a2.v1 genome, until no new GmAKRs were identified. This process identified a total of 67 unique AKR genes in soybean.
The AKR superfamily consists of 16 families; where CHRs from different plant species group into the AKR4 subfamily (Jez et al., 1997; Penning, 2015). To identify GmCHRs in soybean, a phylogenetic analysis was performed using the 67 candidate GmAKRs and previously characterized AKRs from Bomati et al. (2005) with the assumption that the GmCHRs would cluster together with the known CHRs from other plant species. As shown in Figure 2, 14 putative soybean CHRs clustered together with known CHRs from other plant species. GmCHR2A, GmCHR14, GmCHR18, GmCHR2B, GmCHR15 and GmCHR20 clustered closely with CHRs from M. sativa and P. montana var. lobata, POR from L. japonicus and G. glaba. Other inter-species clustering consisted of GmCHR9B, GmCHR16B, GmCHR9A, GmCHR16A, GmCHR9C, GmCHR16C, GmCHR9D, and GmCHR12 with CHR from S. rostrata. Codeinone reductase, a non-CHR, is also found in the same branch as CHR, however, none of the GmCHRs cluster together with this group.
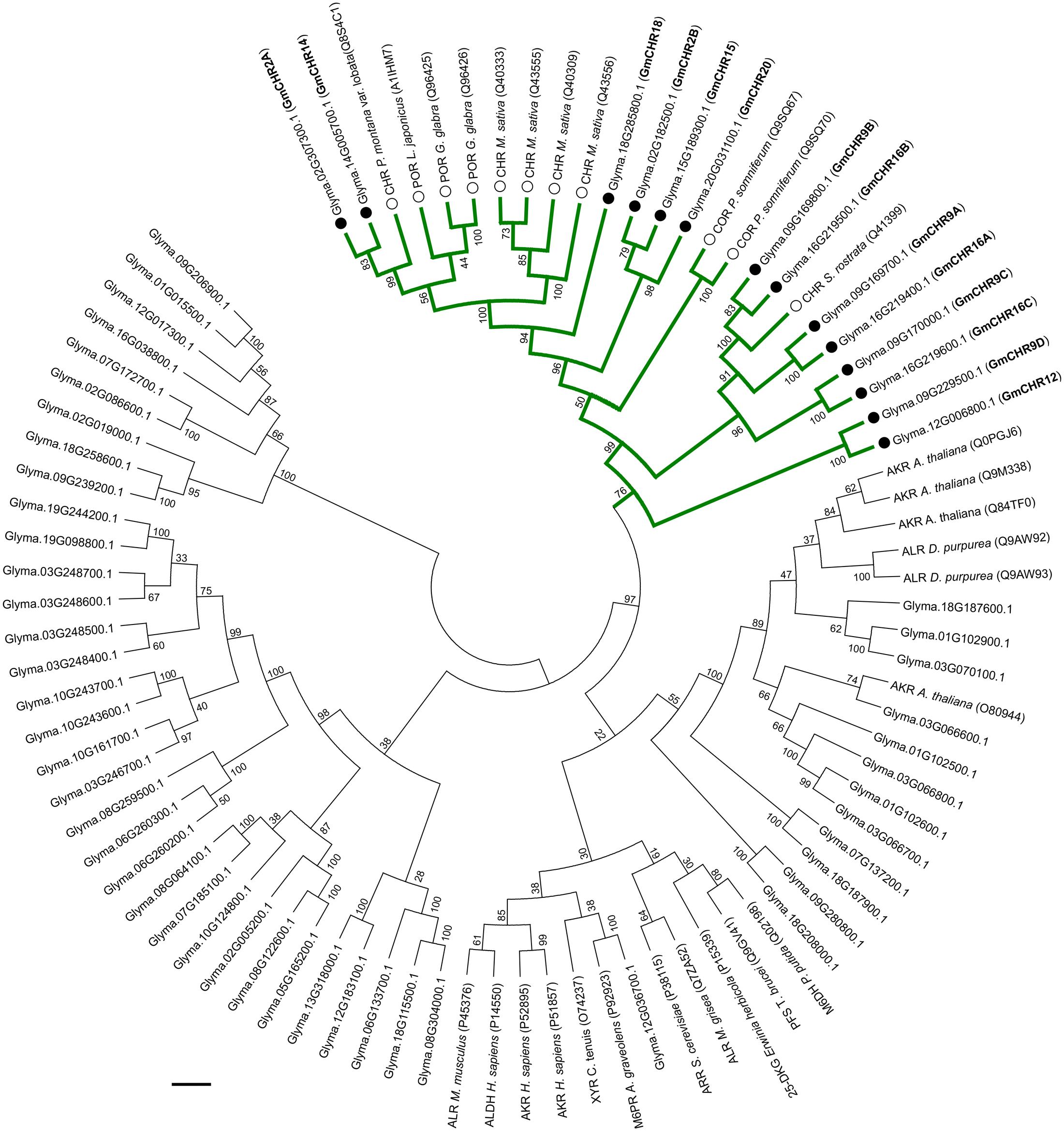
FIGURE 2. Phylogenetic analysis of GmAKRs and GmCHRs. The deduced amino acid sequences of putative GmAKRs and known AKRs from other plant species were aligned using CLUSTALO and the Neighbor-joining tree was constructed using MEGA7 software. The green branch indicates CHR-specific AKRs, black circles (●) and white circles (○) indicate putative GmCHRs and CHRs from other plants species, respectively. POR, polyketide reductase; COR, codeinone reductase; AKR, aldo-keto reductase; XYR, xylose reductase; M6PR, mannose-6-phosphate reductase; M6DH, morphine 6-dehydrogenase; ALHD, alcohol dehydrogenase; ALR, aldose reductase; ARR, arabinose reductase; 2,5 DKG, 2,5-diketo-D-gluconic acid reductase B; and PFS, prostaglandin F synthase. Common nomenclature for GmCHRs is also shown in parenthesis (bold).
The active site of CHR is primarily defined by the AKR family “catalytic tetrad” (Asp-53, Tyr-58, Lys-87, and His-120) with the additional polar residues Trp-121, and Asn-167 (Bomati et al., 2005). To identify whether all the putative GmCHRs contain critical amino acid residues that are required for CHR function, the amino acid sequences of the 14 putative GmCHRs were aligned with the known CHRs from other plant species, and a selection of non-CHR AKRs and CHR-specific active site residues were searched manually. This process identified the following alterations in 3 candidate GmCHRs: (1) in GmCHR2B, Tyr-58 was missing; (2) in GmCHR9B, Asp-53, Tyr-58, Lys-87, His-120, and Trp-121 were missing; and (3) Asn-167 was not present in GmCHR16C (Figure 3). Based on these results, GmCHR2B, GmCHR9B, and GmCHR16C were eliminated for further study as they lack one or more critical residues for their function.
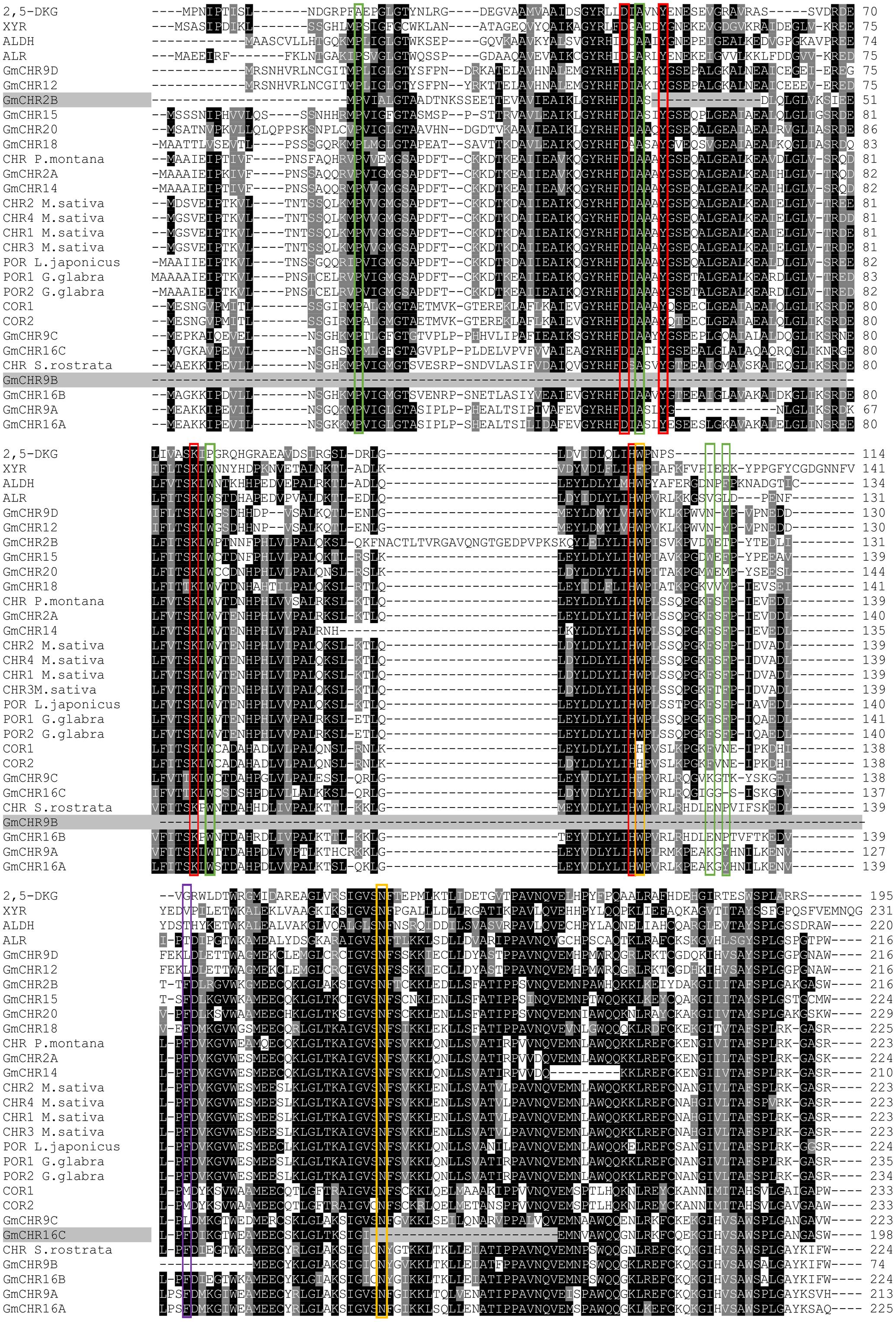
FIGURE 3. Identification of important amino acid residues in putative GmCHRs. Candidate GmCHRs obtained from the phylogenetic analysis were aligned using CLUSTALO with other known AKRs and CHRs from other plant species. Critical and other residues were noted: entrance of the catalytic site ( ), AKR catalytic tetrad (
), AKR catalytic tetrad ( ), unique amino acids from COR (
), unique amino acids from COR ( ), CHR active site with AKR catalytic tetrad (
), CHR active site with AKR catalytic tetrad ( ). GmCHRs which are missing critical residue are indicated through gray shading. Only an abridged version of the alignment is shown.
). GmCHRs which are missing critical residue are indicated through gray shading. Only an abridged version of the alignment is shown.
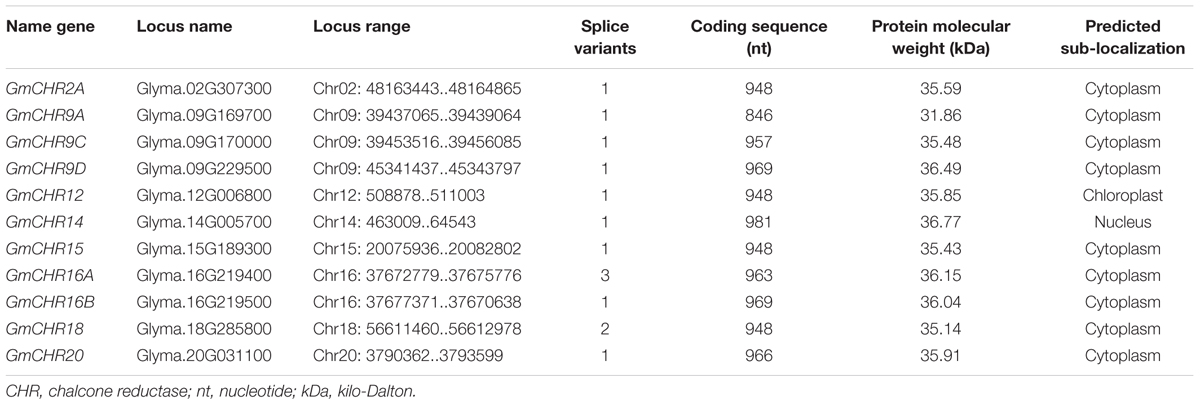
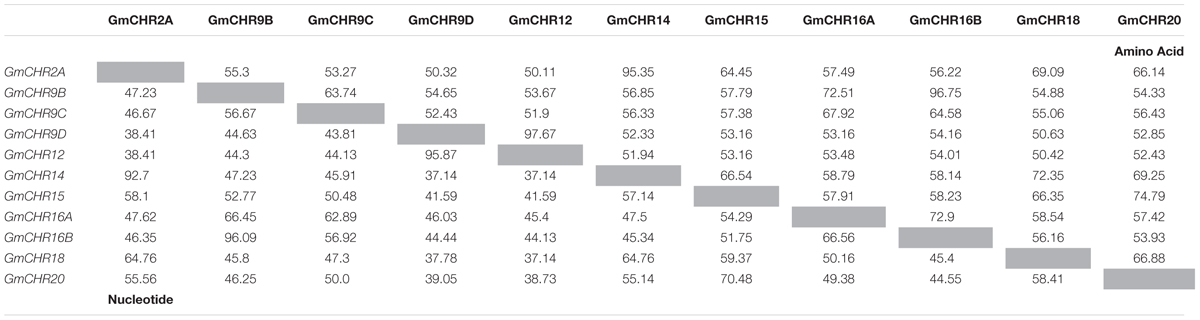
Detailed information on the remaining 11 putative GmCHRs, including gene location, coding sequence length, protein molecular mass and predicted subcellular localization is shown in Table 1. Except for GmCHR12 and GmCHR14, all other GmCHRs were predicted to localize in the cytoplasm. Pairwise amino acid sequence identity comparison of the 11 GmCHR gene family members ranged from 50 to 97%. However, pairwise nucleotide sequence identity comparison ranged from 37 to 96% (Table 2).
GmCHRs Family Members Display Tissue-Specific Gene Expression
To determine tissue-specific expression pattern of GmCHRs, publicly accessible RNA-seq data derived from high throughput sequencing using total RNA isolated from various soybean tissues at different developmental stages such as developing embryos, mature seed, and a number of vegetative tissues9 was retrieved. The relative expression was normalized across the libraries corresponding to each tissue and a heatmap was produced based on the expression level of each GmCHR for each tissue. Out of 11 putative GmCHRs, GmCHR2A, GmCHR14, GmCHR18, GmCHR12 and GmCHR20 were expressed at higher level in roots as compared to other tissues (Figure 4). Majority of the GmCHRs were expressed in the seedlings except for GmCHR9C. Among the 5 root-specific GmCHRs, GmCHR14 and GmCHR20 transcript also accumulated in stem and seedling tissues. Transcript levels of GmCHR15 and GmCHR9C were abundant in early embryo development indicating their role in embryo development. Tissue-specific transcript accumulation data for GmCHR9A and GmCHR9D could not be obtained from the RNAseq database8, therefore, they are not included in Figure 4. It is possible that these genes may not be expressed in whole seeds at various stages of development (globular, heart, cotyledon, early-, mid- and late-maturation, dry), leaf, root, stem, floral bud and seedling included in the study (Danzer et al., 2015).
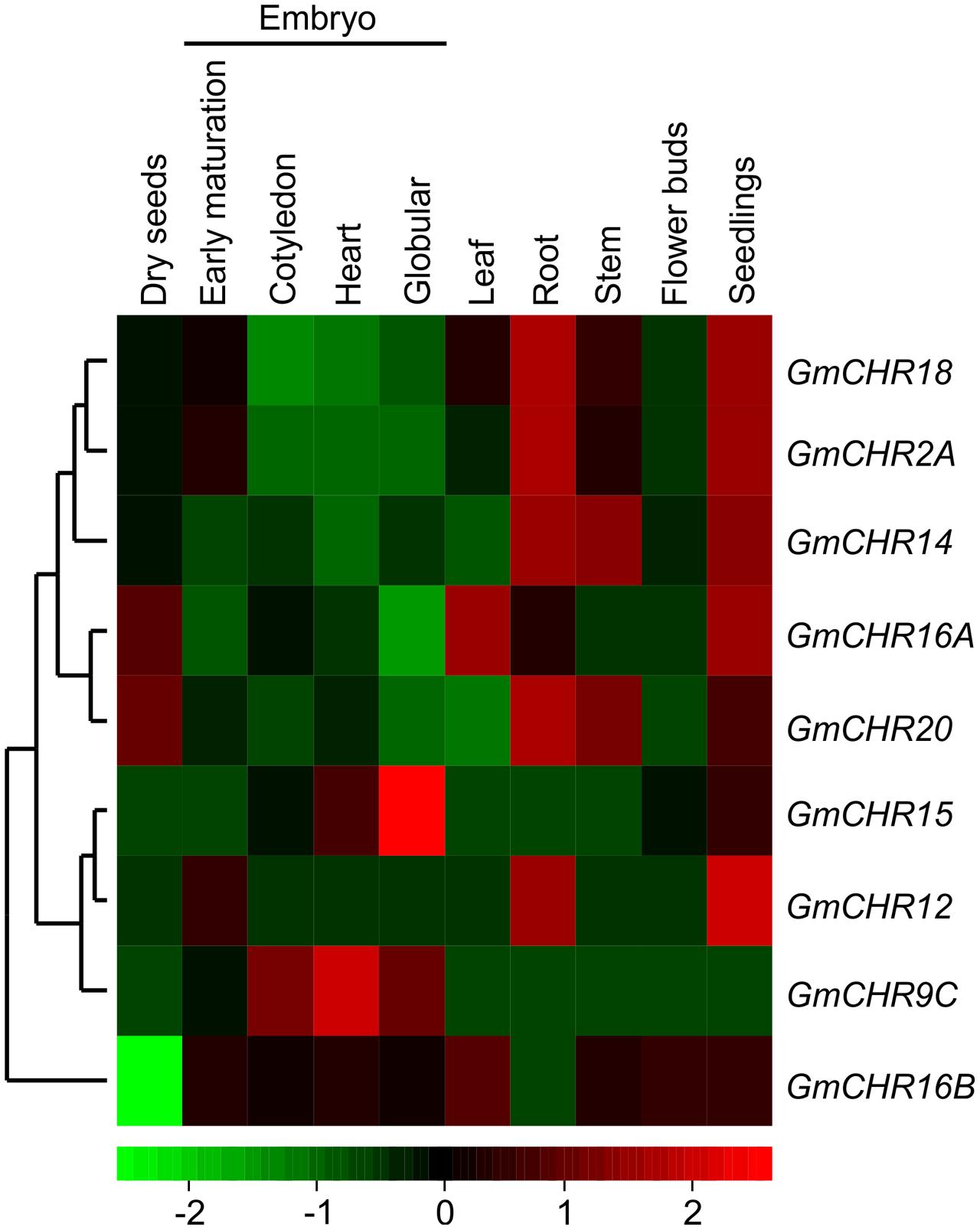
FIGURE 4. Tissue-specific expression profile of CHR genes in soybean. Soybean RNA-seq data across different tissues were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29163) for heat map construction. Transcript abundance is indicated by the color scale below the heat map with a gradient from red (high) to green (low).
Root-Specific GmCHRs Are Induced upon Stress
To identify GmCHR genes that are induced upon pathogen attack, stems of soybean cv. L76-1988 were infected with P. sojae agar culture. Stem samples were collected at 24, 48, and 72 h post-infection, and expression analysis of 11 putative GmCHRs was performed using RT-PCR with gene-specific primers. As shown in Figure 5A, expression of 4 root-specific GmCHR genes, GmCHR2A, GmCHR14, GmCHR18 and GmCHR20, were induced after 24 h and remained induced until 72 h post-infection except for GmCHR14 where no transcript accumulation was observed at 48 hr in treated tissue. Even though GmCHR12 was expressed in root (Figure 4), it did not show induced expression upon P. sojae infection. Expressions of GmCHR9C, GmCHR12 and GmCHR16B were also undetectable in both control and infected samples. However, accumulation of GmCHR16A remained unchanged in both infected and control samples.
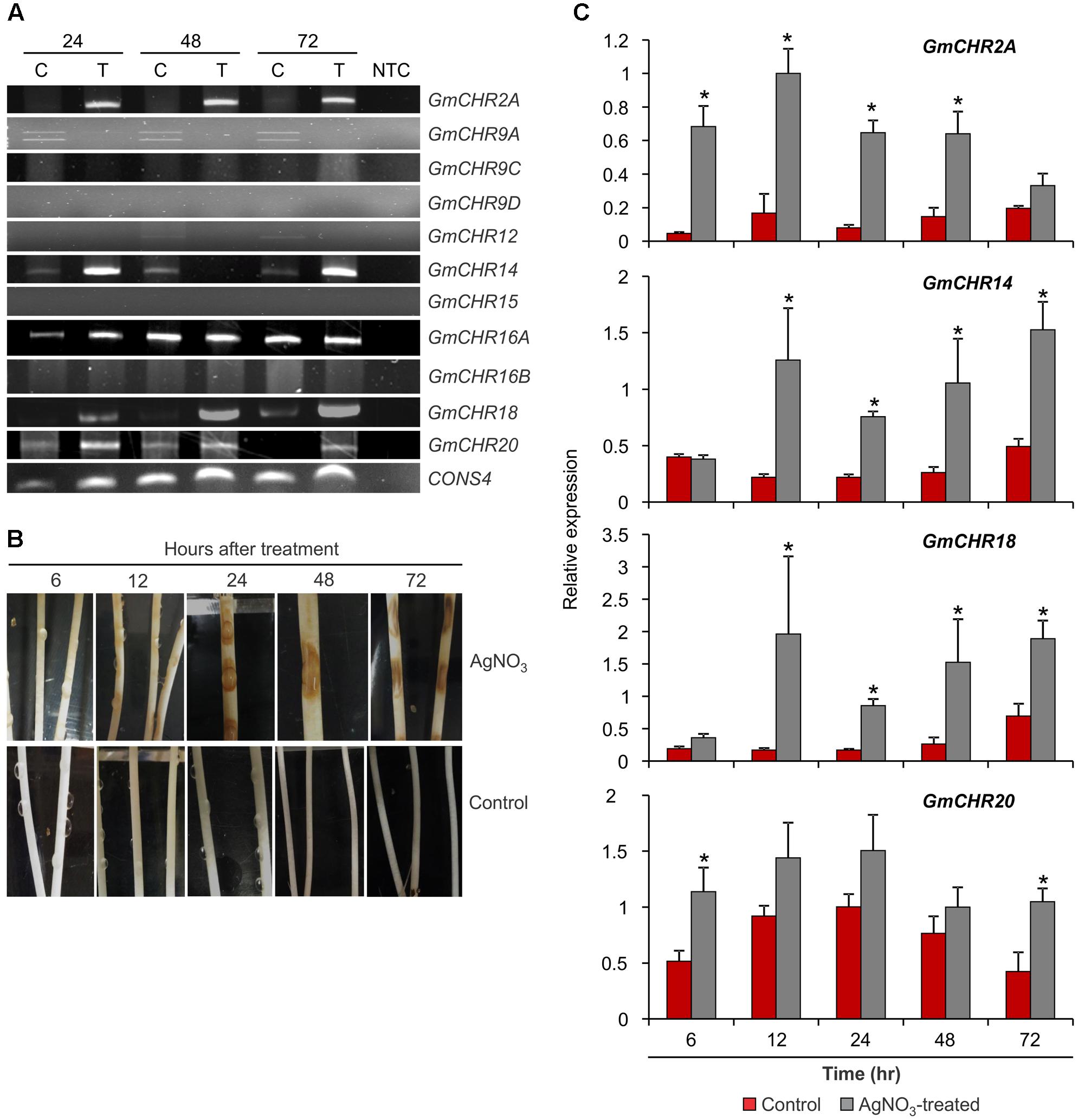
FIGURE 5. Expression of root-specific GmCHRs in response to stress. (A) Expression of GmCHRs in response to P. sojae infection. Total RNA (1 μg) from P. sojae-infected stems of soybean cv. L76-1988 (T) and control (C) for 24, 48 or 72 hr was used to synthesize cDNA. Expression analysis was conducted by RT-PCR with GmCHR gene-specific primers. NTC indicates no template control. CONS4 was used as a loading control. (B) Effects of AgNO3 on etiolated soybean cv. Harosoy63 hypocotyls. Soybean seeds were grown in dark at 24°C for 6 days in water-soaked vermiculite. The seedlings were then placed onto a tray and inoculated with either water (control) or 1 mM AgNO3 (treated). Photographs were taken at the time points as indicated. (C) Detail expression analysis of 4 root-specific GmCHRs. Total RNA (1 μg) of soybean cv. Harosoy63 was used to synthesized cDNA from untreated and AgNO3-treated hypocotyls. Expression analysis was conducted by qPCR with GmCHR gene-specific primers. Error bars indicates standard error of the mean (SEM) of two biological and three technical replicates per biological replicates. CONS4 was used as a reference gene. The asterisks (∗) denotes significant expression as determined by Student’s t-test (p < 0.05).
Since AgNO3 treatment induces defense responses and phytoalexin production in soybean, it has been used in the past to mimic the effect of pathogen attack (Ward et al., 1979; Bhattacharyya and Ward, 1986; Moy et al., 2004; Kubeš et al., 2014). For quantitative analysis of stress-induced GmCHR expression in soybean, we treated etiolated soybean (cv. Harosoy63) hypocotyls with 1 mM AgNO3 or water (control). Tissue samples were collected at 6, 12, 24, 48, or 72 hr after the treatment. Upon AgNO3 treatment, soybean hypocotyls displayed brown lesions at 12 h which became darker and bigger in the later time points. No such lesions were observed in the control hypocotyls at any time points (Figure 5B). RNA was isolated from these samples and expression profiles of 4 root-specific P. sojae induced GmCHRs were investigated in detail. Our qPCR results revealed that the expression of GmCHR2A was significantly higher at 6, 12, 24, and 48 h in AgNO3-treated soybean hypocotyls compared to the controls. Accumulation of GmCHR14 and GmCHR18 transcripts were significantly higher at 12, 24, 48, and 72 h post AgNO3 treatment compared to the control samples whereas increased accumulation of GmCHR20 was only observed at 6 and 72 h in AgNO3-treated hypocotyls compared to the controls (Figure 5C).
GmCHRs Localize in the Nucleus and Cytoplasm
To determine the subcellular localization of the stress-induced GmCHR genes, each member was translationally fused with YFP, and the fusion protein was transiently expressed in leaf epidermal cells of N. benthamiana followed by confocal microscopy. All 4 GmCHRs (GmCHR2A, GmCHR14, GmCHR18 and GmCHR20) were found in 2 cellular compartments, cytoplasm and nucleus. Confirmation of the nuclear localization of GmCHRs was carried out by co-expressing GmCHR-YFP and nuclear localization signal (NLS) containing control (NLS-CFP), and observing the overlap between the NLS-CFP and GmCHR-YFP signals (Figure 6A). To demonstrate that the nuclear localization of GmCHRs are not due to passive diffusion into the nucleus, we made a translation fusions of two tandem YFP proteins with GmCHR2A (GmCHR2A-YFP-YFP) and determined its subcellular location. Despite of its larger molecular mass (88.39 kDa), the fusion protein GmCHR2A-YFP-YFP was found in both nucleus and cytoplasm (Figure 6B) suggesting that presence of GmCHR2A in the nucleus was not due to diffusion. Western blot analysis of transiently expressed GmCHR2A-YFP-YFP in N. benthamiana suggested that the fluorescence observed in the nucleus was due to the intact GmCHR2A-YFP-YFP and not by the cleaved YFP fragments as there was only a single band of estimated size and that the recombinant protein was not cleaved in planta by plant proteases (Figure 6C).
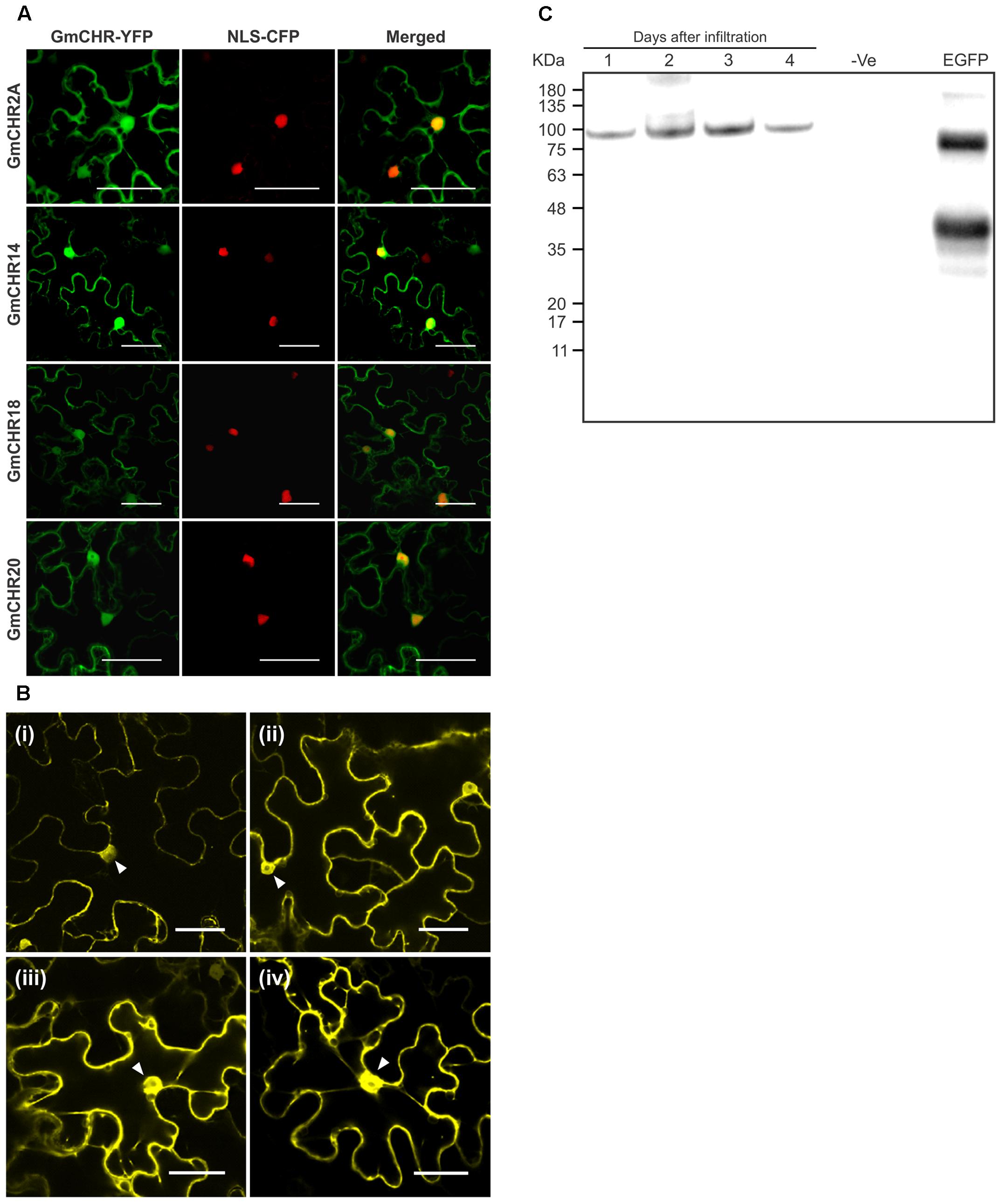
FIGURE 6. Subcellular localization of stress induced GmCHRs. (A) Four root-specific GmCHRs that show induced expression upon P. sojae infection were translationally fused with the YFP, and transiently expressed in N. benthamiana leaves by A. tumefaciens-mediated transformation followed by confocal microscopy. A nuclear localization signal fused with the CFP (NLS-CFP) was used to confirm nuclear localization. The scale bar indicates 50 μm. (B) Transient expression of GmCHR2A-YFP-YFP, in the epidermal cells of N. benthamiana as described in (A). Pictures were taken as indicated (i) Day 1, (ii) Day 2, (iii) Day 3 and (iv) Day 4 post-infiltration. Triangles indicate the nucleus. (C) Western blot analysis of transiently expressed GmCHR2A-YFP-YFP in N. benthamiana leaves. Total proteins were separate on SDS-PAGE and transferred into a PVDF membrane, followed by Western blot analysis. The blot was incubated with anti-GFP primary antibody (1:7000 dilution) and HRP conjugated goat anti-mouse secondary antibody (1:5000 dilution), followed by chemiluminescence detection. Proteins extracted from an uninfiltrated N. benthamiana leaf sample was used as a negative control (–ve), and eGFP with hydrophobin was used as a positive control (∼37 kDa).
QTLs and QTL Markers Linked to P. sojae Resistance Contain GmCHR Loci
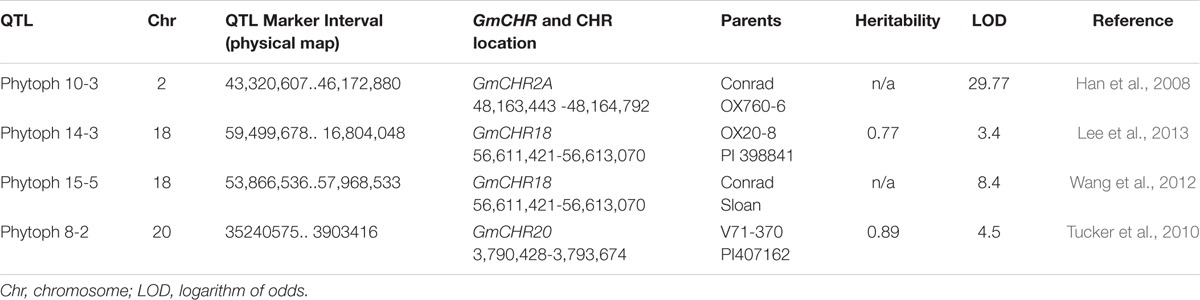
To determine QTLs and QTL markers that are linked to P. sojae resistance, a survey of soybean database and literature search were conducted. A search in the ‘SoyBase and Soybean Breeder’s Toolbox’ from the year 2003 to 2016 identified 77 QTLs that are linked to P. sojae resistance in soybean (Supplementary Table S2). These 77 QTLs were extensively researched for GmCHR loci, parental lines and LOD scores. Four QTLs, Phytoph 10-3 (Han et al., 2008), Phytoph 14-3 (Lee et al., 2013), Phytoph 8-2 (Tucker et al., 2010) and Phytoph 15-5 (Wang et al., 2012) were found which contain GmCHR loci. Highlights of the QTLs included: (1) Phytoph 10-3 flanks GmCHR2A locus 2 megabase pairs and has an LOD score of 29.7; (2) Phytoph 14-3 and Phytoph 15-5 contain GmCHR18 locus within the marker interval with lower LOD score; (3) Phytoph 8-2 contains GmCHR20 locus that stretches over 31 megabase pairs with the LOD score of 4.5 (Table 3). These details suggest that Phytoph 10-3 containing GmCHR2A is involved in P. sojae resistance in soybean.
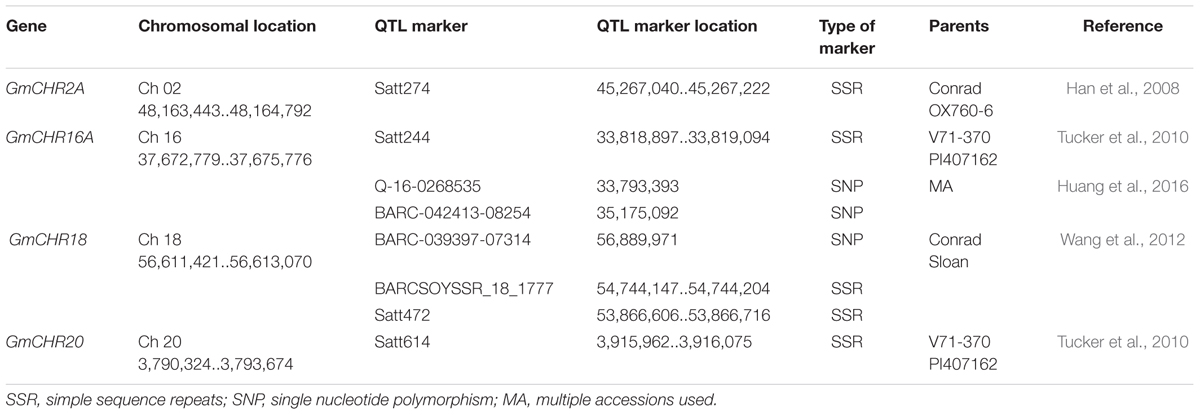
An additional literature search was conducted for QTL markers linked to P. sojae resistance in soybean. This process identified over 500 QTL markers (Supplementary Table S3). The markers that share the same chromosome as GmCHR were separated, and exact locations of the QTL markers were noted. A total of 8 QTL markers were found to flank GmCHR2A, GmCHR16A, GmCHR18 and GmCHR20 loci (Han et al., 2008; Tucker et al., 2010; Wang et al., 2010). The details on the QTL marker are summarized in Table 4.
P. sojae Resistant Soybean Cultivar Accumulates Higher Levels of Root-Specific GmCHR
Since several QTL markers and QTLs linked to P. sojae resistance contained GmCHR gene family members, we evaluated the parental cultivars of QTL Phytoph 10-3 for root-specific GmCHR gene expression. Root tissues of 2-week old seedlings of soybean cv. Conrad and OX760-6 were used for gene expression analysis. As shown in Figure 7A, expression of GmCHR2A, GmCHR14 and GmCHR18 were significantly higher in roots of Conrad (P. sojae resistant cultivar) compared to the OX760-6 (P. sojae susceptible cultivar). No difference in the expression of GmCHR20 was observed between Conrad and OX760-6. When the etiolated hypocotyls of Conrad and OX760-6 were treated with AgNO3, and expression of root-specific GmCHRs were monitored, only the expression of GmCHR2A was induced in response to AgNO3 treatment and its transcript accumulation was higher in Conrad compared to OX760-6 at later time points (Figure 7B). No difference in the level of other root-specific GmCHRs were observed in Conrad and OX760-6 both in control and AgNO3-treated samples. Furthermore, expression of GmCHR14 and GmCHR18 were not induced in response to AgNO3 treatment (not shown).
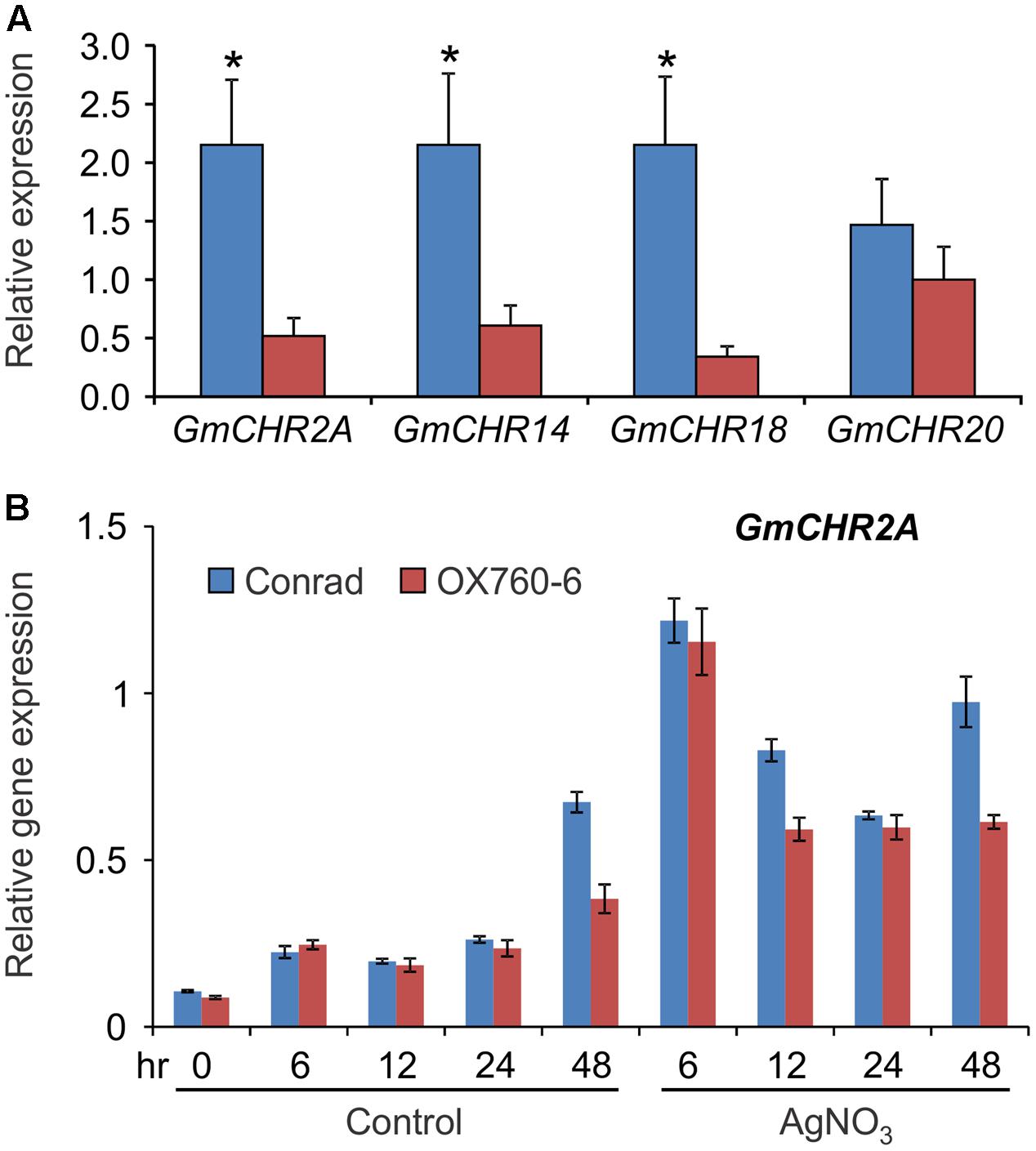
FIGURE 7. Accumulation of root-specific GmCHRs in Conrad and OX760-6. Total RNA (1 μg) was used to synthesize cDNA from (A) roots, and (B) untreated and AgNO3-treated etiolated hypocotyls of soybean cv. Conrad and OX760-6. Expression analysis was conducted by qPCR using gene-specific primers. Error bars indicates SEM of three technical replicates per biological replicates where 2 biological replicates for roots and 4 biological replicates for etiolated hypocotyls were used. CONS4 was used as a reference gene. The asterisks (∗) denotes significant difference in expression as determined by Student’s t-test (p < 0.05).
Discussion
The legume-specific enzyme CHR together with CHS, converts 1 molecule of p-coumaroyl-CoA and 3 molecules of malonyl-CoA to isoliquiritigenin, the building block of two core isoflavone aglycones, glycitein and daidzein, where daidzein serves as a precursor for the production of phytoalexin glyceollins in soybean. Many studies have reported the expression patterns of genes during infection and the heritability of resistance, however, little is known about the importance of CHR, the first key enzyme, which directs the flux to the production of phytoalexin glyceollins in soybean. Here we report the genome-wide identification of GmCHRs in soybean, investigate their subcellular location, and tissue-specific and pathogen induced gene expression. Our results demonstrate that the root-specific GmCHRs are induced upon pathogen infection, and are located near QTLs and QTL markers linked to P. sojae resistance.
Many CHR-like enzymes have been reported in a variety of leguminous plants (Ballance and Dixon, 1995; Goormachtig et al., 1999; Shimada et al., 2006; He et al., 2011; Hayashi et al., 2013). A recent study identified only 2 CHRs (GmCHR2A and GmCHR14) in soybean where only one sequence from GenBank (accession EU921437) was used as the query to search the soybean genome database (Chu et al., 2014). Here we identified 14 CHR gene family members in soybean. Our approach is more robust and provides confidence as it first identified all the 67 GmAKRs based on the current database annotation and other search tools, and then segregated the GmCHRs gene family members using their phylogenetic relationship with known CHRs (Figure 2). We also confirmed that the 4 GmCHRs previously identified (Subramanian et al., 2006) are also included in our list. Soybean is a paleopolyploid with a genome size of approximately 1 gigabase pairs that has undergone at least two whole genome duplications (Schmutz et al., 2010). As a result of the genome duplications, nearly 75% of soybean genes are present in multiple copies. The large number of GmCHRs could be the result of whole genome duplication events in soybean. Phylogenetic analysis of GmCHRs illustrated that most GmCHRs cluster in pairs, further supporting ancient genome duplication events (Figure 2).
Out of 14 putative GmCHRs, 11 were found to contain conserved critical residues (Figure 3). Since CHRs are a part of the AKR family, these enzymes must contain the catalytic sites (Bomati et al., 2005). The 3 GmCHRs: GmCHR2B, GmCHR9B and GmCHR16C lack one or more catalytic site residues, therefore, were eliminated from our study. However, it is possible that they may possess weak enzymatic activity or may be evolving new catalytic features.
GmCHRs displayed tissue-specific gene expression. The majority of the GmCHRs were either expressed in the seedlings, roots or dry seeds (Figure 4). Previously, it was found that CHR in soybean were moderately expressed in the flowers and weakly expressed in leaves, stems, roots, endosperms and embryos (Liu, 2009). Differential expression of CHRs have also been studied in other plant species such as Astragalus membranaceus (Xu et al., 2012) and P. montana var. lobata (He et al., 2011). It was found that CHRs from A. membranaceus and P. montana var. lobata were highly expressed in roots and stem, respectively. Out of 11 GmCHRs identified, GmCHR2A, GmCHR14, GmCHR12, GmCHR18 and GmCHR20 transcript accumulation was much higher in root tissue compared to other tissues under study. Studies have shown that CHS7, CHS8 (Yi et al., 2010), IFS1, IFS2 (Dhaubhadel et al., 2003) and CHI (Dastmalchi and Dhaubhadel, 2015) are also expressed in the roots which infers that they possibly assist in the role of root-specific phytoalexins production.
To evaluate if GmCHR family members respond differently upon pathogen infection, their expression levels were studied by RT-PCR at various time points after P. sojae infection. Interestingly, the expression levels of only 4 root-specific GmCHRs, GmCHR2A, GmCHR14, GmCHR18 and GmCHR20, were induced upon infection suggesting that they have a role in defense against P. sojae infection (Figure 5A). Upon infection, P. sojae releases elicitors which stimulate the plant defense response (Jones and Dangl, 2006). As a result, the plant induces the expression of resistance and defense related genes to counteract infection. Studies have shown the upregulation or induction of CHRs at the infection site during Fusarium attack in soybeans (Iqbal et al., 2005), cadmium treatment in Medicago truncatula (Aloui et al., 2012), and Colletotrichum falcatum infection in sugarcane (Selvaraj et al., 2014). Several studies have used the AgNO3 treatment to mimic pathogen infection and induce phytoalexin production in soybeans (Moy et al., 2004; Kubeš et al., 2014). The mechanism of this “elicitor effect” has not been identified yet. The quantitative analysis of root-specific GmCHRs in response to AgNO3 demonstrated a significant increase in transcript accumulation of GmCHR2A, GmCHR14 and GmCHR18 (Figure 5C). These root-specific GmCHRs respond to the AgNO3 treatment as early as 12 h which coincides with findings from (Alkharouf et al., 2006). Changes in gene expression within roots upon Heterodera glycines (the soybean cyst nematode) attack was investigated using a 6000 gene microarray (Alkharouf et al., 2006). It was found that CHR (Genbank BM108162) was induced as soon as 6 and 12 h upon infection.
All members of the GmCHR family displayed nuclear and cytoplasmic localization in N. benthamiana leaf epidermal cells (Figure 6). These findings are consistent with the localization of GmCHS which works together with GmCHR to produce isoliquiritigenin. Evidence has shown that other enzymes involved in the isoflavonoid biosynthesis such as GmCHI (Dastmalchi and Dhaubhadel, 2015), glycosyltransferase (UGT73F2) and malonyltransferase (GmMT7) (Dhaubhadel et al., 2008) are also localized to the nucleus and the cytoplasm. Similarly, nuclear localization of flavonoid enzymes have also been reported in Arabidopsis (Saslowsky et al., 2005). Flavanols and flavonols have been found in the nuclei of some tree species (Feucht et al., 2012) such as conifers Taxus baccata L., Tsuga canadensis L. (Feucht et al., 2014), however, a simple diffusion process was speculated for the presence of these metabolites in the nucleus. Since isoflavonoid biosynthesis requires multiple cytochrome P450 monooxygenases that are localized in the endoplasmic reticulum, synthesis of these metabolites in the nucleus can be completely ruled out. This could imply that nuclear localized GmCHRs might have an alternate function other than their catalytic role in the isoflavonoid biosynthesis. It is possible that the nuclear localized isoflavonoid enzymes may function as moonlighting proteins with two or more different functions at different locations within the cell (Copley, 2012; Jeffery, 2014).
Several studies have suggested that Rps genes (Dorrance and Schmitthenner, 2000; Sandhu et al., 2005), isoflavonoid biosynthetic genes (Subramanian et al., 2005; Graham et al., 2007), and genes involved in suberin production (Ranathunge et al., 2008) contribute to P. sojae resistance in soybean. Furthermore, molecular markers linked to Rps genes or QTLs and QTL markers linked to P. sojae resistance have been identified in soybean. However, little has been reported linking the candidate genes with the QTLs and the phenotype. Here, we have identified a total of 8 QTL markers and 4 QTLs which flank or are approximate to GmCHR2A, GmCHR18 and GmCHR20. Among the QTLs, Phytoph 14-3 covers most of the chromosome 18 (Table 3). Even though QTL regions can generally span several megabase pairs, and can contain several hundreds to thousands of genes (Dupuis and Siegmund, 1999), further fine-mapping experiments are required to pinpoint the spanning regions (Touzet et al., 1995; Holtan and Hake, 2003). Statistical association or validation studies can confirm the co-segregation of genes with a QTL. Based on these parameters, QTL Phytoph 10-3 was selected as the most reliable QTL found in this study as it is confined to a specific region on the chromosome and contains the highest LOD score. Even though the expression of root-specific GmCHRs, GmCHR2A, GmCHR14 and GmCHR18 were significantly higher in the roots of the parental cultivars Conrad (resistant cultivar) as compared to OX760-6 (susceptible cultivar) used in the QTL study by Han et al. (2008), only expression of GmCHR2A was induced upon stress and was higher in the cultivar Conrad as compared to OX760-6 (Figure 7B). This result together with the expression of root-specific GmCHRs (Figure 4), suggests that in addition to GmCHRs, there are other yet unidentified factors that play an important role in partial resistance in soybean. Identification of each component is crucial to understand the resistance mechanism so that genes encoding those factors could be stacked together into a single cultivar for resistance breeding.
Author Contributions
SD conceived and designed the experiments. CS and JY performed the experiments and analyzed the data. SD and CS wrote the article; all the authors read and commented on the article.
Funding
This work was supported by Agriculture and Agri-Food Canada’s Genomics Research and Development Initiative and Natural Sciences and Engineering Research Council of Canada grants to SD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Alex Molnar for his help with the preparation of the figures and Hong Mo Kim, Chelsea Ismael and Patrick Chapman for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02073/full#supplementary-material
FIGURE S1 | Set-up of AgNO3 stress treatment on soybean hypocotyl.
Footnotes
- ^ www.globalsoybeanproduction.com
- ^ soycanada.ca/statistics/at-a-glance/
- ^ http://compbio.dfci.harvard.edu/tgi/
- ^ http://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ http://www.ch.embnet.org/software/BOX_form.html
- ^ http://soybase.org/
- ^ http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29163
- ^ http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29163
References
Alkharouf, N. W., Klink, V. P., Chouikha, I. B., Beard, H. S., MacDonald, M. H., Meyer, S., et al. (2006). Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224, 838–852. doi: 10.1007/s00425-006-0270-8
Aloui, A., Dumas-Gaudot, E., Daher, Z., van Tuinen, D., Aschi-Smit, S., and Morandi, D. (2012). Influence of arbuscular mycorrhizal colonisation on cadmium induced Medicago truncatula root isoflavonoid accumulation. Plant Physiol. Biochem. 60, 233–239. doi: 10.1016/j.plaphy.2012.08.014
Anderson, T. R., and Buzzell, R. I. (1982). Efficacy of metalaxyl in controlling Phytophthora root and stalk rot of soybean cultivars differing in field tolerance. Plant Dis. 66, 1144–1145. doi: 10.1094/PD-66-1144
Ayabe, S.-I., Udagawa, A., and Furuya, T. (1988). NAD(P)H-dependent 6′-deoxychalcone synthase activity in Glycyrrhiza echinata cells induced by yeast extract. Arch. Biochem. Biophys. 261, 458–462. doi: 10.1016/0003-9861(88)90362-1
Ballance, G. M., and Dixon, R. A. (1995). Medicago sativa cDNAs encoding chalcone reductase. Plant Physiol. 107, 1027–1028. doi: 10.1104/pp.107.3.1027
Bhattacharyya, M. K., and Ward, E. W. B. (1986). Resistance, susceptibility and accumulation of glyceollins I–III in soybean organs inoculated with Phytophthora megasperma f. sp. glycinea. Physiol. Mol. Plant Pathol. 29, 227–237. doi: 10.1016/S0048-4059(86)80023-6
Bomati, E. K., Austin, M. B., Bowman, M. E., Dixon, R. A., and Noel, J. P. (2005). Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J. Biol. Chem. 280, 30496–30503. doi: 10.1074/jbc.M502239200
Chu, S., Wang, J., Cheng, H., Yang, Q., and Yu, D. (2014). Evolutionary study of the isoflavonoid pathway based on multiple copies analysis in soybean. BMC Genet. 15:76. doi: 10.1186/1471-2156-15-76
Copley, S. D. (2012). Moonlighting is mainstream: paradigm adjustment required. Bioessays 34, 578–588. doi: 10.1002/bies.201100191
Dakora, F. D., and Phillips, D. A. (1996). Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 49, 1–20. doi: 10.1006/pmpp.1996.0035
Danzer, J., Mellott, E., Bui, A. Q., Le, B. H., Martin, P., Hashimoto, M., et al. (2015). Down-regulating the expression of 53 soybean transcription factor genes uncovers a role for SPEECHLESS in initiating stomatal cell lineages during embryo development. Plant Physiol. 168, 1025–1035. doi: 10.1104/pp.15.00432
Dastmalchi, M., and Dhaubhadel, S. (2015). Soybean chalcone isomerase: evolution of the fold, and the differential expression and localization of the gene family. Planta 241, 507–523. doi: 10.1007/s00425-014-2200-5
Dhaubhadel, S., Farhangkhoee, M., and Chapman, R. (2008). Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 59, 981–994. doi: 10.1093/jxb/ern046
Dhaubhadel, S., McGarvey, B. D., Williams, R., and Gijzen, M. (2003). Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Biol. 53, 733–743. doi: 10.1023/B:PLAN.0000023666.30358.ae
Dorrance, A., and Schmitthenner, A. (2000). New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis. 84, 1303–1308. doi: 10.1094/PHYTO-97-0106
Dupuis, J., and Siegmund, D. (1999). Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151, 373–386.
Feucht, W., Schmid, M., and Treutter, D. (2014). Flavanols and flavonols in the nuclei of conifer genotypes with different growth. Forests 5, 2122–2135. doi: 10.3390/f5092122
Feucht, W., Treutter, D., and Polster, J. (2012). Flavanols in nuclei of tree species: facts and possible functions. Trees 26, 1413–1425. doi: 10.1007/s00468-012-0725-4
Goormachtig, S., Lievens, S., Herman, S., Van Montagu, M., and Holsters, M. (1999). Chalcone reductase-homologous transcripts accumulate during development of stem-borne nodules on the tropical legume Sesbania rostrata. Planta 209, 45–52. doi: 10.1007/s004250050605
Graham, T. L., Graham, M. Y., Subramanian, S., and Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144, 728–740. doi: 10.1104/pp.107.097865
Han, Y., Teng, W., Yu, K., Poysa, V., Anderson, T., Qiu, L., et al. (2008). Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 162, 231–239. doi: 10.1007/s10681-007-9558-4
Hayashi, H., Fujii, I., Iinuma, M., Shibano, M., Ozaki, K., and Watanabe, H. (2013). Characterization of a glycyrrhizin-deficient strain of Glycyrrhiza uralensis. Biol. Pharm. Bull. 36, 1448–1453. doi: 10.1248/bpb.b13-00164
He, X., Blount, J., Ge, S., Tang, Y., and Dixon, R. (2011). A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233, 843–855. doi: 10.1007/s00425-010-1344-1
Holtan, H. E. E., and Hake, S. (2003). Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165, 1541–1550.
Huang, J., Guo, N., Li, Y., Sun, J., Hu, G., Zhang, H., et al. (2016). Phenotypic evaluation and genetic dissection of resistance to Phytophthora sojae in the Chinese soybean mini core collection. BMC Genet. 17:85. doi: 10.1186/s12863-016-0383-4
Iqbal, M. J., Yaegashi, S., Ahsan, R., Shopinski, K. L., and Lightfoot, D. A. (2005). Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 110, 1429–1438. doi: 10.1007/s00122-005-1969-9
Jeffery, C. J. (2014). An introduction to protein moonlighting. Biochem. Soc. Trans. 42, 1679–1683. doi: 10.1042/BST20140226
Jez, J. M., Bennett, M. J., Schlegel, B. P., Lewis, M., and Penning, T. M. (1997). Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 326(Pt 3), 625–636. doi: 10.1042/bj3260625
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kubeš, J., Tùmová, L., Martin, J., Vildová, A., Hendrychová, H., and Sojková, K. (2014). The production of isoflavonoids in Genista tinctoria L. cell suspension culture after abiotic stressors treatment. Nat. Prod. Res. 28, 2253–2263. doi: 10.1080/14786419.2014.938336
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, S., Mian, M. A. R., McHale, L. K., Wang, H., Wijeratne, A. J., Sneller, C. H., et al. (2013). Novel quantitative trait loci for partial resistance to Phytophthora sojae in soybean PI 398841. Theor. Appl. Genet. 126, 1121–1132. doi: 10.1007/s00122-013-2040-x
Li, X. P., Han, Y. P., Teng, W. L., Zhang, S. Z., Yu, K. F., Poysa, V., et al. (2010). Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’ and ‘Hefeng 25’. Theor. Appl. Genet. 121, 651–658. doi: 10.1007/s00122-010-1337-2
Libault, M., Thibivilliers, S., Bilgin, D. D., Radwan, O., Benitez, M., Clough, S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome 1, 44–54. doi: 10.3835/plantgenome2008.02.0091
Liu, G. Y. (2009). Isolation, sequence identification and tissue expression profile of two novel soybean (Glycine max) genes-vestitone reductase and chalcone reductase. Mol. Biol. Rep. 36, 1991–1994. doi: 10.1007/s11033-008-9409-y
Lozovaya, V. V., Lygin, A. V., Zernova, O. V., Ulanov, A. V., Li, S., Hartman, G. L., et al. (2007). Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 225, 665–679. doi: 10.1007/s00425-006-0368-z
Lygin, A. V., Zernova, O. V., Hill, C. B., Kholina, N. A., Widholm, J. M., Hartman, G. L., et al. (2013). Glyceollin is an important component of soybean plant defense against Phytophthora sojae and Macrophomina phaseolina. Phytopathology 103, 984–994. doi: 10.1094/PHYTO-12-12-0328-R
Moy, P., Qutob, D., Chapman, P., Atkinson, I., and Gijzen, M. (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant Microbe Interact. 17, 1051–1062. doi: 10.1094/MPMI.2004.17.10.1051
Penning, T. M. (2015). The aldo-keto reductases (AKRs): overview. Chem. Biol. Interact. 234, 236–246. doi: 10.1016/j.cbi.2014.09.024
Ranathunge, K., Thomas, R. H., Fang, X., Peterson, C. A., Gijzen, M., and Bernards, M. A. (2008). Soybean root suberin and partial resistance to root rot caused by Phytophthora sojae. Phytopathology 98, 1179–1189. doi: 10.1094/PHYTO-98-11-1179
Sahoo, D. K., Abeysekara, N. S., Cianzio, S. R., Robertson, A. E., and Bhattacharyya, M. K. (2017). A novel Phytophthora sojae resistance Rps12 gene mapped to a genomic region that contains several Rps genes. PLOS ONE 12:e0169950. doi: 10.1371/journal.pone.0169950
Sandhu, D., Schallock, K., Rivera-Velez, N., Lundeen, P., Cianzio, S., and Bhattacharyya, M. (2005). Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J. Hered. 96, 536–541. doi: 10.1093/jhered/esi081
Saslowsky, D. E., Warek, U., and Winkel, B. S. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280, 23735–23740. doi: 10.1074/jbc.M413506200
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368. doi: 10.1094/PD-69-362
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Selvaraj, N., Ramadass, A., Amalraj, R., Palaniyandi, M., and Rasappa, V. (2014). Molecular profiling of systemic acquired resistance (SAR)-responsive transcripts in sugarcane challenged with Colletotrichum falcatum. Appl. Biochem. Biotechnol. 174, 2839–2850. doi: 10.1007/s12010-014-1230-6
Shimada, N., Nakatsuka, T., Nishihara, M., Yamamura, S., Ayabe, S.-I., and Aoki, T. (2006). Isolation and characterization of a cDNA encoding polyketide reductase in Lotus japonicus. Plant Biotechnol. 23, 509–513. doi: 10.5511/plantbiotechnology.23.509
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Prot. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Subramanian, S., Graham, M. A., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353. doi: 10.1104/pp.104.057257
Subramanian, S., Stacey, G., and Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273. doi: 10.1111/j.1365-313X.2006.02874.x
Sugimoto, T., Kato, M., Yoshida, S., Matsumoto, I., Kobayashi, T., Kaga, A., et al. (2012). Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 61, 511–522. doi: 10.1270/jsbbs.61.511
Sugimoto, T., Watanabe, K., Yoshida, S., Aino, M., Furiki, M., Shiono, M., et al. (2010). Field application of calcium to reduce phytophthora stem rot of soybean, and calcium distribution in plants. Plant Dis. 94, 812–819. doi: 10.1094/PDIS-94-7-0812
Thomas, R., Fang, X., Ranathunge, K., Anderson, T. R., Peterson, C. A., and Bernards, M. A. (2007). Soybean root suberin: anatomical distribution, chemical composition, and relationship to partial resistance to Phytophthora sojae. Plant Physiol. 144, 299–311. doi: 10.1104/pp.106.091090
Touzet, P., Winkler, R., and Helentjaris, T. (1995). Combined genetic and physiological analysis of a locus contributing to quantitative variation. Theor. Appl. Genet. 91, 200–205. doi: 10.1007/BF00220878
Tucker, D. M., Saghai Maroof, M. A., Mideros, S., Skoneczka, J. A., Nabati, D. A., Buss, G. R., et al. (2010). Mapping quantitative trait loci for partial resistance to in a soybean interspecific cross. Crop Sci. 50, 628–635. doi: 10.2135/cropsci2009.03.0161
Wang, H., Waller, L., Tripathy, S., St. Martin, S., Zhou, L., Krampis, K., et al. (2010). Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome 3, 23–40. doi: 10.1186/1471-2164-13-428
Wang, H., Wijeratne, A., Wijeratne, S., Lee, S., Taylor, C. G., St Martin, S. K., et al. (2012). Dissection of two soybean QTL conferring partial resistance to Phytophthora sojae through sequence and gene expression analysis. BMC Genomics 13:428. doi: 10.1186/1471-2164-13-428
Ward, E., Lazarovits, G., Unwin, C., and Buzzell, R. (1979). Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology 69, 951–955. doi: 10.1094/Phyto-69-951
Workneh, F., Yang, X. B., and Tylka, G. L. (1998). Effect of tillage practices on vertical distribution of Phytophthora sojae. Plant Dis. 82, 1258–1263. doi: 10.1094/PDIS.1998.82.11.1258
Xu, R.-Y., Nan, P., Pan, H., Zhou, T., and Chen, J. (2012). Molecular cloning, characterization and expression of a chalcone reductase gene from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao. Mol. Biol. Rep. 39, 2275–2283. doi: 10.1007/s11033-011-0977-x
Keywords: soybean, chalcone reductase, gene family, Phytophthora sojae, root and stem rot disease, resistance, quantitative trait loci, plant stress
Citation: Sepiol CJ, Yu J and Dhaubhadel S (2017) Genome-Wide Identification of Chalcone Reductase Gene Family in Soybean: Insight into Root-Specific GmCHRs and Phytophthora sojae Resistance. Front. Plant Sci. 8:2073. doi: 10.3389/fpls.2017.02073
Received: 11 July 2017; Accepted: 20 November 2017;
Published: 07 December 2017.
Edited by:
Stefan Martens, Fondazione Edmund Mach, ItalyReviewed by:
Li Tian, University of California, Davis, United StatesLorenzo Caputi, John Innes Centre (BBSRC), United Kingdom
Heidi Halbwirth, Vienna University of Technology, Austria
Copyright © 2017 Sepiol, Yu and Dhaubhadel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangeeta Dhaubhadel, c2FuZ2VldGEuZGhhdWJoYWRlbEBjYW5hZGEuY2E=
 Caroline J. Sepiol
Caroline J. Sepiol Jaeju Yu
Jaeju Yu Sangeeta Dhaubhadel
Sangeeta Dhaubhadel