- 1Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Guangdong Engineering Research Center for Litchi, College of Horticulture, South China Agricultural University, Guangzhou, China
- 2Department of Horticulture, Zhongkai University of Agriculture and Engineering, Guangzhou, China
- 3State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Institute of Microbiology, Guangzhou, China
Arbuscular mycorrhizal fungi (AMF) establish symbiosis with most terrestrial plants, and greatly regulate lateral root (LR) formation. Phosphorus (P), sugar, and plant hormones are proposed being involved in this regulation, however, no global evidence regarding these factors is available so far, especially in woody plants. In this study, we inoculated trifoliate orange seedlings (Poncirus trifoliata L. Raf) with an AMF isolate, Rhizophagus irregularis BGC JX04B. After 4 months of growth, LR formation was characterized, and sugar contents in roots were determined. RNA-Seq analysis was performed to obtain the transcriptomes of LR root tips from non-mycorrhizal and mycorrhizal seedlings. Quantitative real time PCR (qRT-PCR) of selected genes was also conducted for validation. The results showed that AMF significantly increased LR number, as well as plant biomass and shoot P concentration. The contents of glucose and fructose in primary root, and sucrose content in LR were also increased. A total of 909 differentially expressed genes (DEGs) were identified in response to AMF inoculation, and qRT-PCR validated the transcriptomic data. The numbers of DEGs related to P, sugar, and plant hormones were 31, 32, and 25, respectively. For P metabolism, the most up-regulated DEGs mainly encoded phosphate transporter, and the most down-regulated DEGs encoded acid phosphatase. For sugar metabolism, the most up-regulated DEGs encoded polygalacturonase and chitinase. For plant hormones, the most up-regulated DEGs were related to auxin signaling, and the most down-regulated DEGs were related to ethylene signaling. PLS-SEM analysis indicates that P metabolism was the most important pathway by which AMF regulates LR formation in this study. These data reveal the changes of genome-wide gene expression in responses to AMF inoculation in trifoliate orange and provide a solid basis for the future identification and characterization of key genes involved in LR formation induced by AMF.
Introduction
Root is essential for plant growth and development, serving a multiple functions including anchorage, absorption of mineral nutrients and water, and production of exudates with growth regulatory properties (Bailey et al., 2002; Chen et al., 2017). The characteristic of root system is mainly presented as root system architecture (RSA), namely the spatial configuration of root system such as root angle, root branching, root diameter and so on (Lynch, 1995). Among all the components of plant RSA, lateral root (LR), formed post-embryonically by LR primordium derived from specific cells of pericycle (Vilches-Barro and Maizel, 2015), is the most important (Nibau et al., 2008; Vilches-Barro and Maizel, 2015; Zhao et al., 2015). RSA is highly plastic, responding to the soil water and nutrient availability, soil matrix heterogeneity, and biotic interactions (Péret et al., 2009; Chen W. L. et al., 2016). As an example for the latter, a mutualistic association develops between roots and arbuscular mycorrhizal fungi (AMF) (Smith and Read, 2008), which comprehensively affect plant RSA, especially the LR formation (Gutjahr and Paszkowski, 2013; Jiang et al., 2015; Chen W. L. et al., 2016). Fusconi (2014) proposed a comprehensive model that AMF affect LR formation probably through three pathways: regulating phosphorus (P) metabolism (increased absorption), regulating carbohydrate metabolism (changed carbohydrate partitioning and pathway), and regulating plant hormone metabolism (modulation of concentration, transport and sensitivity) in host plants. This proposal has been partially verified (Jiang et al., 2015; Chen et al., 2017), however, a comprehensive verification is needed and the contribution of each pathway is also to be quantified.
Plant RSA is strictly controlled by genes, and recently, some experiments revealed the molecular mechanisms underlying the modified RSA with the RNA-Seq technique. Gutjahr et al. (2015) showed that genes expressed in crown root of rice (Oryza sativa L.) were related to plant hormone and secondary cell wall metabolism, while they were related to nutrient transportation in large LRs and fine LRs. After AMF inoculation, colonized crown roots adopted an expression profile more related to the mycorrhizal large LRs than to non-colonized crown roots, suggesting a reprogramming of crown root character by AMF. Another transcriptomic analysis of maize (Zea mays L.) root showed that, under local nitrate supply, different root types produced different gene expression models, resulting in various patterns of LR formation (Yu et al., 2016). Additionally, gene network was obtained to uncover the mechanism of the adventitious root formation of carnation (Dianthus caryophyllus L.) cutting in response to exogenous auxin (Villacorta-Martín et al., 2015). These results demonstrate the potential application of RNA-Seq technique to reveal the comprehensive pathway by which AMF regulates LR formation.
So far, most studies with RNA-Seq technique focused on herbaceous plant roots (Nawy et al., 2005; Brady et al., 2007; Zhang et al., 2014; Gao et al., 2015; Opitz et al., 2016; Song et al., 2016), and in contrast, few study shed light on the woody plants. Citrus, one of the most important woody fruit crops worldwide, strongly depends on AMF for acquiring nutrients because of lacking of root hairs (Wu et al., 2016). Previous studies indicated the significant regulation of LR formation in citrus plants by AMF (Yao et al., 2009; Chen et al., 2017). In this study, we chose trifoliate orange (P. trifoliata L. Raf), commonly used as the rootstock in the citrus-producing areas of China (Gao et al., 2016), as host plants to establish symbiosis with AMF. We aimed to dissect the pathways by which AMF regulates LR formation at molecular level with RNA-Seq technique, and to probe into the respective contribution of different pathway to the regulation.
Materials and Methods
Experimental Material and Experimental Design
To establish AM symbiont in pot culture, the commercially obtained trifoliate orange (P. trifoliata L. Raf) seeds were used as host plants, and AMF isolate was Rhizophagus irregularis BGC JX04B provided by Beijing Academy of Agriculture and Forestry Sciences (Chen et al., 2017). R. irregularis inocula were propagated with clover (Trifolium repense L.) and sorghum (Sorghum bicolor) as hosts for 4 months in the greenhouse. At harvest, the spore density was quantified to be ca. 49 spores per gram inoculum. The growth substrate was the mixture of autoclaved (121°C, 2 h) soils and peat (1:2, v:v). The soils were collected from the experimental orchard of South China Agricultural University, and the soil chemical properties were determined as follows: pH 4.60, organic matter content 1.58%, available N 65.0 mg·kg−1, available P 20.5 mg·kg−1, and available K 57.1 mg·kg−1. The substrate was additionally applied with 200 mg·kg−1 N (NH4NO3), 10 mg·kg−1 P (KH2PO4), and 100 mg·kg−1 K (KNO3).
We conducted two pot experiments to reveal the influence of AMF inoculation on LR formation of trifoliate seedlings. The first experiment was set up for RNA-Seq analysis with two treatments [non-mycorrhizal (C) and mycorrhizal (T)]. Each treatment comprised of 3 replicates. Each pot was filled with 450 g substrate and 50 g AMF inocula (T treatment) or sterilized inocula (C treatment). The second experiment was set up with the same treatments as in the first experiment, and the sugar contents in roots as affected by AMF inoculation were investigated.
Trifoliate orange seeds were surface-sterilized with 70% ethanol for 15 min, rinsed in distilled water for 5 times, and then germinated in autoclave-sterilized peat at 28°C in dark. Seedlings with five leaves and similar vigor were selected and transplanted to pots (two seedlings in each pot). All mycorrhizal and non-mycorrhizal seedlings grew in the greenhouse under natural light conditions with 22~30°C and 60%~80% relative humidity. Plants were harvested at 4 months after transplanting.
Harvest and Physio-Biochemical Parameters Measurement
At harvest, shoots and roots were separated and the fresh weights were recorded respectively, then the LR numbers of different orders were counted and LR forming capacity (LRC) was calculated as the value of (the number of N order LR)/(the number of N-1 order LR). LR tips (about 1 cm length) were randomly selected and cut, and preserved (about 0.2 g) at −80°C for RNA-Seq and qRT-PCR analysis. The leftover roots were cut into fragments (about 1 cm length) and 50 fragments were randomly picked up for the measurement of mycorrhizal colonization. Other root fragments were used for carbohydrate determination.
The chlorophyll contents in leaves were measured according to Chen Z. et al. (2016). After digested with HNO3, the shoot P contents were measured by a spectrophotometer according to Arshad et al. (2016). Mycorrhizal staining was performed according to Phillips and Hayman (1970) and mycorrhizal colonization was quantified according to Biermann and Linderman (1981). The carbohydrate contents (sucrose, glucose, and fructose) in primary roots and LR were colorimetrically determined according to Wu et al. (2015) with some modification. Briefly, 100 mg fresh roots were homogenized with 4 mL 80% ethanol, incubated for 30 min at 80°C, and then centrifuged at 8,000 rpm for 10 min. The residues were extracted twice as described above, and all the supernatants were combined for the analysis of carbohydrate content. Sucrose was assayed with a mixture of 0.4 mL supernatant and 0.2 mL 2 mol·L−1 NaOH at 100°C for 5 min, which were then mixed together with 2.8 mL 5 mol·L−1 HCl and 0.8 mL 0.1% resorcinol at 100°C for 10 min followed by measurement of absorbance at 480 nm. Glucose concentration was determined by mixing 1 mL of supernatant with 2 mL of prepared solution (1 mg·mL−1 o-dianisidine dihydrochloride, 1 mg·mL−1 horseradish peroxidase, and 1 U·mL−1 glucose oxidase) at 30°C for 5 min, then added 4 mL 10 mol·L−1 H2SO4 solution to terminate the reaction, and followed by measurement of absorbance at 460 nm. Fructose was assayed with a mixture of 0.4 mL supernatant, 0.8 mL 0.1% resorcinol and 0.4 mL 5 mol·L−1 HCl at 100°C for 10 min followed by measurement of absorbance at 480 nm. The carbohydrate contents were quantified according to the linear standard curves constructed with respective sugars.
RNA-Seq and Transcriptomic Analysis
Total RNA in the root tips from C and T treatments (3 independent replicates in each treatment) was isolated with RNA extraction kit (Huayueyang Biotechnology Co., LTD, Beijing) according to the manufacturer's protocol. The RNA integrity was checked with Agilent 2100 Bioanalyzer and Agilent RNA 6000 Nano Kit, and RNA concentration was determined using a Nanodrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). One aliquot was sent to Annoroad Gene Technology Co. Ltd. (Beijing, China) for RNA-seq. The libraries were sequenced on an Illumina Hiseq™ 4000 platform and 150 bp paired-end reads were generated.
After removing reads containing adapters or more than 50% N and low-quality reads from the raw reads, clean reads were obtained for all subsequent analyses. The Citrus sinensis reference genome (Xu et al., 2012), directly downloaded from the citrus genome website (http://citrus.hzau.edu.cn/orange/), was used for the mapping of clean reads by TopHat (Trapnell et al., 2009).
The RPKM value (Reads Per Kilobase per Million reads) and Fold Change value for each gene were measured by applying GFOLD algorithm (Feng et al., 2012). Differential expression analysis of two treatments was performed using the DESeq (Anders, 2010). The threshold for significantly differential expression was set as follows: adjusted P < 0.05 and |log2(fold change)| > 1 (Anders and Huber, 2012). The P-values were adjusted using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). The volcano plot of differentially expressed genes (DEGs) and cluster diagrams were made by R packages “gplots” and “ggplot2,” respectively.
Gene ontology (GO) enrichment analysis of DEGs was implemented by the Blast2GO software (Conesa et al., 2005). Based on Kyoto encyclopedia of genes and genomes (KEGG, http://www.genome.jp/kegg/), the KOBAS 2.0 software (http://kobas.cbi.pku.edu.cn/) was used to test the statistical enrichment of DEG in KEGG pathways (Xie et al., 2011).
Quantitative Real-Time PCR
Another aliquot of extracted RNA was used for the quantitative real-time PCR (qRT-PCR) according to previous protocol (Chen et al., 2017), with 3 independent replicates for each treatment. Briefly, cDNA was synthesized with the extracted RNA as template using iScript™ cDNA Synthesis Kit (Bio-Rad, USA). 18SrRNA gene was used as internal reference (Yan et al., 2012) in this study. All gene-specific primers designed with Batchprimer3 (https://probes.pw.usda.gov/batchprimer3/) were described in Supplementary Table 1 and the specificity was checked using Blasting Tool of NCBI. Primers were synthesized by Shanghai Sangon BioTech Co. PCR was conducted with iTaq™ Universal SYBR Green Supermix Kit (Bio-Rad, USA) using CFX96 Real-Time System. Each reaction mixture was 20 μL containing 5 μL diluted cDNA template, 250 nmol·L−1 of forward and backward primers, 10 μl SYBR Green supermix and 4 μL sterilized ddH2O. The two-step qRT-PCR was run as follows: 95°C for 30 s, 39 cycles of 95°C for 5 s, and 60°C for 30 s in 96-well plates (Bio-rad, USA). All samples were amplified in triplicate from the same RNA preparation and the mean value was considered. The relative expression of each target gene was calculated by using 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
All data were presented as mean ± standard error of three replicates. The t-test was performed and the correlations between the LR numbers and DEGs were calculated with bivariate correlations analysis using IBM SPSS v.21 statistical software (SPSS Inc., Chicago, IL).
To identify PT (encoding phosphate transporter) genes induced by AMF, phylogeny tree was constructed with the DEG data against AMF-induced PT genes in citrus reported previously (Shu et al., 2012) by using MEGA6 software. To discover the correlations between DEGs and LR formation, redundancy analysis (RDA) was performed using CANOCO 4.5 statistical package (Braak and Smilauer, 2002), and the results were visualized with the extension CanoDraw. To explore the relationships between AMF inoculation, LR formation, and metabolisms of P, sugar and plant hormones, a hypothetical model was specified and analyzed with Partial Least Squares Structural Equation Modeling (PLS-SEM) with the support of the SmartPLS 2.0 M3 software (Ringle et al., 2005). The standardized path coefficient values were generated with the PLS algorithm using Path weighting scheme, and a bootstrapping method was used to obtain the significance of path coefficients.
Results
Mycorrhizal Colonization and Plant Growth
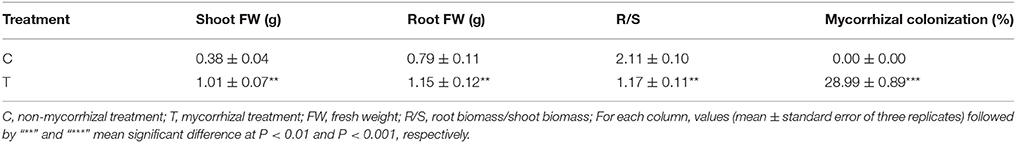
In the inoculated roots (T treatment), the mycorrhizal colonization was 28.99% (Table 1), while no mycorrhizal structures was observed in the non-inoculated roots (C treatment).
Compared with the non-mycorrhizal treatment (C), AMF inoculation significantly increased the growth of trifoliate orange seedlings, as revealed by the increase in either shoot FW or root FW, but decreased the R/S (Table 1, Figure 1A). It was clear that the root system were more vigorous and complex after AMF inoculation (Figure 1B).
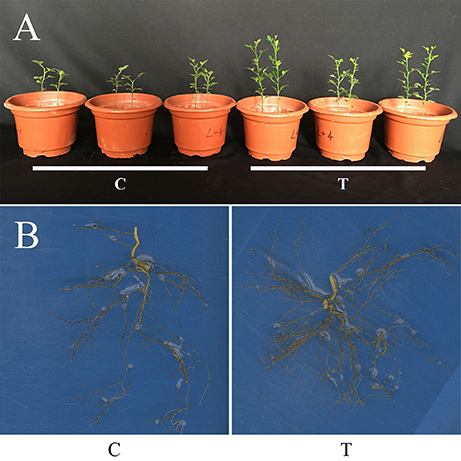
Figure 1. Plant growth of trifoliate orange seedlings as affected by AMF inoculation. (A) Photo showing the overall plant growth; (B) The root systems. C: non-mycorrhizal treatment, T: mycorrhizal treatment.
In the aboveground part, the phosphorus content was significantly increased in the mycorrhizal plants, over two times higher than that in the non-mycorrhizal plants (Figure 2A). There was no obvious difference in the contents of chlorophyll a, b and total chlorophyll between the two treatments, although AMF inoculation slightly increased these parameters (Figure 2B).
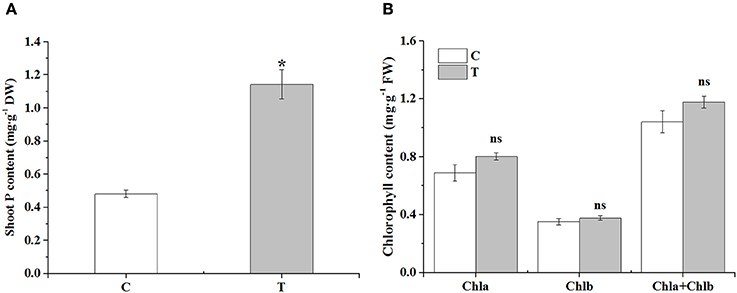
Figure 2. The effects of AMF on shoot P contents and leaf chlorophyll contents. (A) Shoot P contents; (B) Leaf chlorophyll contents. C: non-mycorrhizal treatment, T: mycorrhizal treatment; DW: dry weight, FW: fresh weight; “*” and “ns” mean significant difference at P < 0.05 and no significant difference, respectively.
Enhancement of Lateral Root Formation by AMF Inoculation
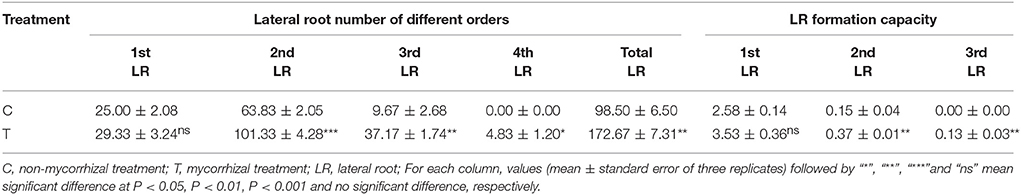
AMF inoculation greatly increased the LR formation except for the 1st order LR (Table 2). In detail, the 2nd order LR number was increased by nearly one time, and the 3rd order LR number was increased by over 3 times. The 4th order LR number was about 5 for the mycorrhizal plants, while no 4th order LR occurred for the non-mycorrhizal plants (Table 2). Moreover, AMF inoculation significantly elevated the formation capacity of the 2nd and 3rd LR (Table 2).
To verify the effect of AMF inoculation on the LR formation, we conducted a separate experiment, and got the similar results, where AMF inoculation also significantly increased numbers of the 1st order LR and the 3rd order LR (Supplementary Figure 1A). Meanwhile, the contents of glucose and fructose in the primary root were significantly increased (Supplementary Figure 1B) and the sucrose content in lateral roots was significantly increased by AMF inoculation (Supplementary Figure 1C).
Bioinformatics Analysis of RNA-Seq Data and Identification of DEGs in Roots
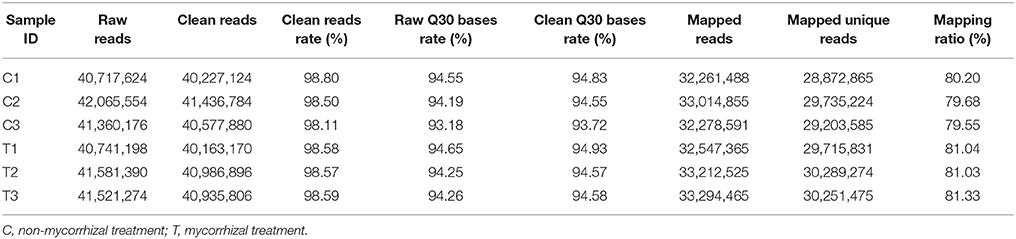
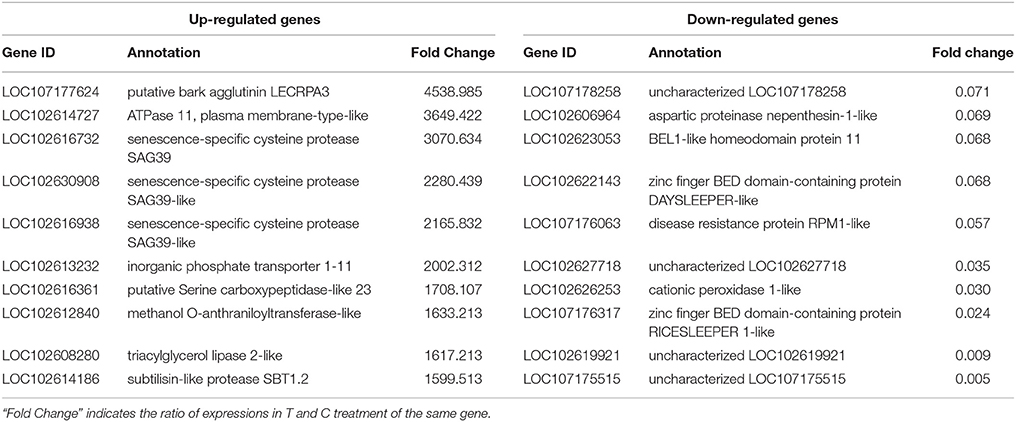
We performed RNA-Seq analysis to reveal the molecular mechanisms involved in the regulation of LR formation by AMF inoculation. An overview of the sequencing and assembly was outlined in Table 3. High-quality reads (clean reads) were obtained after trimming and applying quality filter, ranging from 40,163,170 to 41,436,784 reads for individually sequenced libraries. On the whole, about 80% of these reads (Table 3) mapped into the genome of Citrus sinensis were used to the subsequent analysis. The RNA-Seq dataset was deposited in NCBI Short Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under the accession number SRP119883. The published citrus genome includes 28,195 genes, and in present work, we detected the expressions of 16,383 genes in trifoliate orange root tips, including all experimental replicates. By comparing their expression levels under mycorrhizal and non-mycorrhizal treatment, 909 DEGs were identified (see in Supplementary Data 1), among which 647 were up regulated, and 262 down regulated by AMF inoculation (Figure 3A). Grouping by fold change, most DEGs distributed in the range of >10 (23.43%), 2~4 (24.31%), and 0.25~0.50 (22.66%) fold. 64.25% of DEGs were up regulated by AMF inoculation, not including the uniquely expressed genes (6.93%) in the mycorrhizal root tips (Figure 3B), while less than one third of DEGs (28.49%, Figure 3B) was down regulated in response to AMF inoculation. Additionally, only 3 genes were specifically expressed in the non-mycorrhizal plants, in contrast to 63 genes specifically expressed in mycorrhizal plants. Among all DEGs, 121 genes were uncharacterized according to the reference genome (Supplementary Data 1). The most 10 up-regulated and down-regulated genes were listed in Table 4. It is worthy to note that gene (LOC102613232) encoding inorganic phosphate transporter was up-regulated by almost 2000 times by AMF.
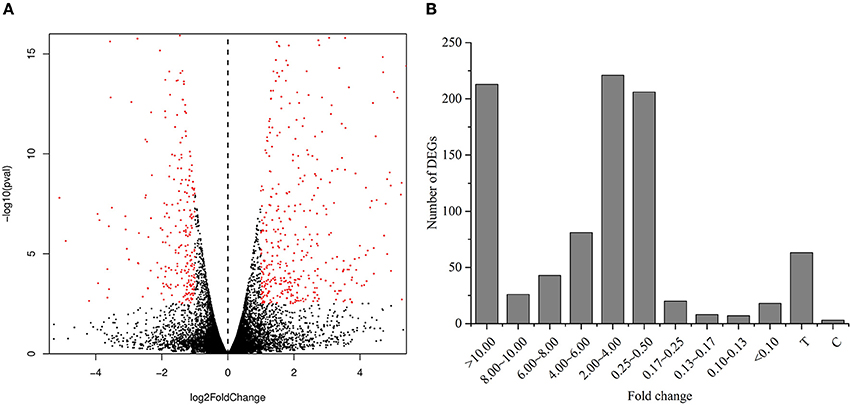
Figure 3. Analysis of differentially expressed genes between the mycorrihzal and non-mycorrhizal treatment. (A) Volcano plot showing differentially expressed genes. Red dots in left and right indicate the significantly down-regulated and up-regulated genes, respectively. (B) Number of DEGs grouped by range of fold change. C: non-mycorrhizal treatment, T: mycorrhizal treatment; “C” and “T” mean these genes only found to be expressed in C and T treatment, respectively.
Validation of Sequencing Results by qRT-PCR
To validate the RNA-Seq data, qRT-PCR of 14 randomly selected genes was performed. The expressions of these genes identified by qRT-PCR were as the same trend as observed in RNA-Seq (Supplementary Figure 2), and there was a significantly positive correlation (R2 = 0.7661, Supplementary Figure 3) between RPKM values and qRT-PCR data, indicating that the qRT-PCR results of these genes were consistent with their transcript abundance changes detected by RNA-Seq, which means the RNA-Seq data were credible for exploring the mechanism of phenotypic changes of root system induced by AMF inoculation.
Functional Classification of DEGs
All DEGs were assigned to GO categories, involved in 242 biological processes, 102 molecular functions, and 30 cellular components. Top 10 of each category were showed in Supplementary Figure 4. In detail, in the biological process category, oxidation-reduction process (72 DEGs), biological process (35 DEGs) and defense response to fungus (27 DEGs) were strongly represented. In the molecular functional category, 22, 21, and 12 DEGs were identified as belonging to kinase activity, metal ion binding, and oxygen binding subcategories, respectively. In the cellular component category, subcaterories of membrane (59 DEGs) and cytoplasmic membrane-bounded vesicle (46 DEGs) contained more differentially expressed genes.
Pathway enrichment analysis using KOBAS was implemented to analyze the biological functions of DEGs induced by AMF inoculation. Metabolic pathways (96 DEGs) and biosynthesis of secondary metabolites (70 DEGs) were the two enriched pathways comprising the most DEGs (Supplementary Figure 5). In addition, 14 and 7 DEGs were annotated with the starch and sucrose metabolism and plant hormone signal transduction pathways, respectively.
DEGs Related to Metabolisms of P, Sugar, and Plant Hormones
Due to the close relationship between LR formation and P status, sugar supply, and plant hormone, we selected the genes related to P metabolism, sugar metabolism, and plant hormone for further analysis (Figures 4A–C).
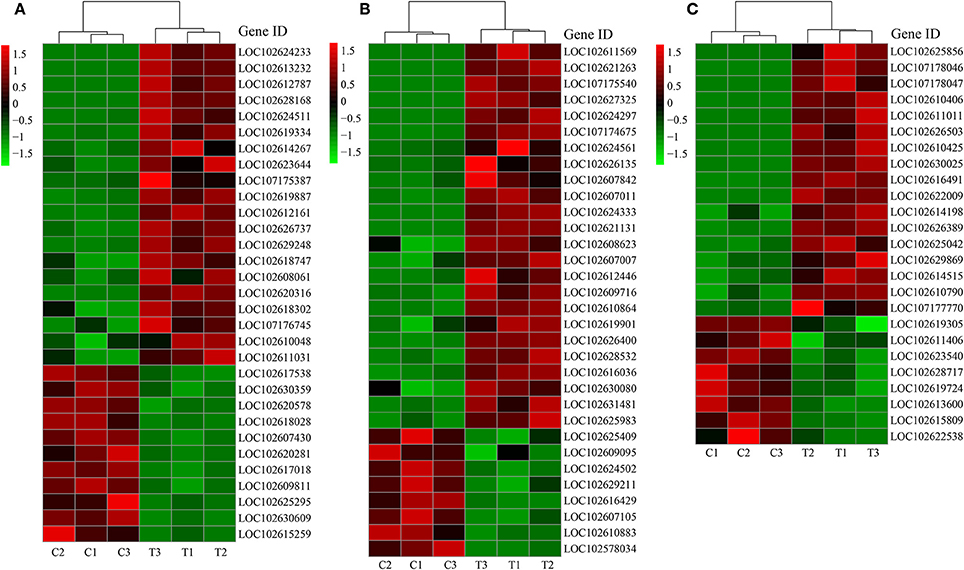
Figure 4. DEGs related to different categories. (A) DEGs related to P metabolism; (B) DEGs related to sugar metabolism; (C) DEGs related to plant hormone. C: non-mycorrhizal treatment, T: mycorrhizal treatment. The annotation of DEGs is listed in Supplementary Data 3.
About two-third of the P metabolism related DEGs were up-regulated by AMF inoculation (Figure 4A). Five PT genes (LOC102613232, LOC102628168, LOC102626737, LOC102608061, LOC102609811) were found as DEGs (DEGs related to phosphorus metabolism). Particularly, the expression of LOC102613232 and LOC102628168 in T treatment were over 1000 times more than those in C treatment (Figure 4A). Phylogeny tree indicates that both were AM-induced PT genes (Supplementary Figure 6). In contrast, most of the down-regulated genes encoded for acid phosphatase. The expression of 24 out of 32 DEGs related to sugar metabolism were higher in T treatment than in C treatment (Figure 4B). Moreover, 6 genes functioned with phosphorus involved. Interestingly, the most promoted DEGs were associated with polygalacturonase (PG), chitinase (CHI) and their analogs (PG-like protein, CHI-like protein). Three chitinase-related DEGs belong to endochitinase, among which LOC102607011 was a chitinase class III gene with over 9 times expression level in mycorrhizal root than that in the non-mycorrhizal root. For the genes involved in plant hormone metabolism, 17 were up-regulated and 8 were down-regulated after AMF inoculation (Figure 4C). In details, 10 DEGs were related to auxin, 7 DEGs to ethylene, 4 DEGs to gibberellic acid (GA), 2 DEGs to abscisic acid (ABA), and 2 DEGs to cytokinin, respectively. There were 8, 4, 3, 1, and 1 DEGs related to auxin, ethylene, GA, ABA, and cytokinin were up-regulated by AMF inoculation.
Correlation analysis (Supplementary Tables 2–4) showed that all genes were not significant correlated with the 1st LR number. Moreover, we performed RDA to investigate the correlation between the LR formation (LR number) and the main factors (P metabolism, sugar metabolism, and plant hormone metabolism). It is interesting that all the down-regulated genes were positively correlated with the 1st LR number, which was not affected by AMF inoculation; and in contrast, all the up-regulated genes were positively correlated with the 3rd LR number, which was significantly promoted by AMF inoculation (Figure 5). In details, for phosphorus metabolism, some genes encoding phosphatase (LOC102612161, LOC 102618747, LOC102620316, LOC102618302, LOC102611031) were closely correlated with 3rd LR number (Figure 5A, Supplementary Table 2); for carbohydrate metabolism, some genes encoding chitinase (LOC102627325, LOC102607011, LOC102621131) and glucose-6-phosphate 1-dehydrogenase (LOC102624333) were closely correlated with 3rd LR number (Figure 5B, Supplementary Table 3); and for plant hormones, some genes (LOC102622009, LOC102629869, LOC102610790) involved in auxin metabolism were closely correlated with 3rd LR number (Figure 5C, Supplementary Table 4).
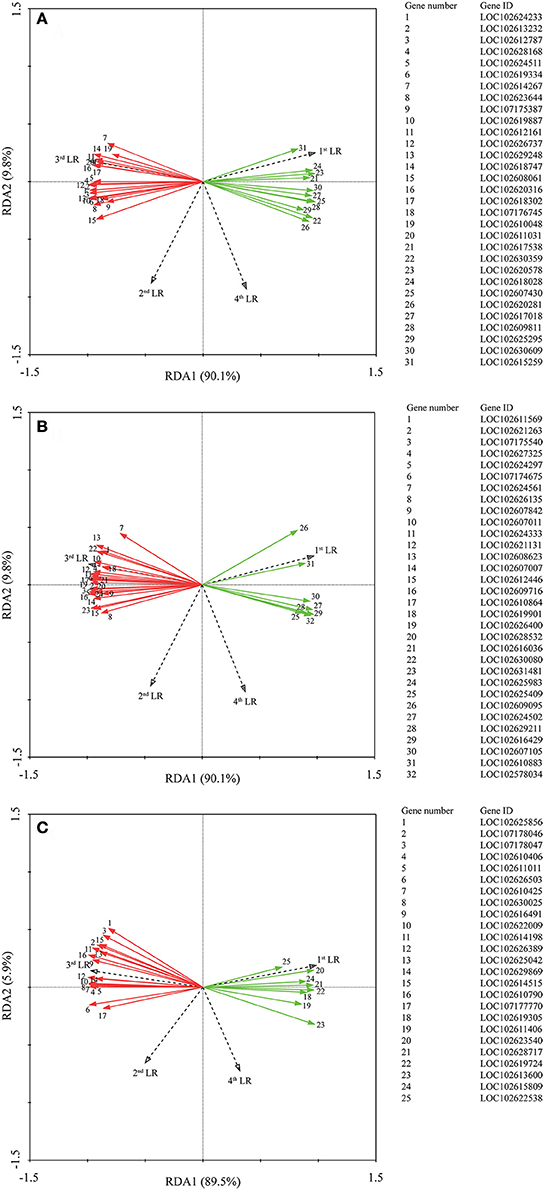
Figure 5. Redundancy analysis (RDA) indicating the correlations between DEGs related to different categories and LR numbers. (A) DEGs related to P metabolism. (B) DEGs related to sugar metabolism; (C) DEGs related to plant hormone. Red and green arrows indicated the up-regulated and down-regulated DEGs due to AMF inoculation, respectively. The annotation of DEGs is listed in Supplementary Data 3.
DEGs Related to Metabolisms of Lipid Biosynthesis and Metabolism
Totally, 28 DEGs were related to lipid biosynthesis and metabolism, with 11 down-regulated and 17 up-regulated (Supplementary Table 5). Among these DEGs, 7 DEGs were involved in fatty acid metabolism, with 4 up-regulated by AMF, and 8 DEGs were related to steroid biosynthesis, with all strongly up-regulated by AMF. The up-regulated and fatty acid metabolism related genes participated in lipid synthesis (LOC102626302, LOC102618665, LOC102612001) and the desaturation reactions (LOC102611163).
Probing Into Pathways by Which AMF Inoculation Affects LR Formation
To reveal how AMF inoculation affected the LR formation via the metabolisms of phosphorus, carbohydrate, and plant hormones, and to gain a comprehensive view of these pathways, we performed PLS-SEM analysis. In PLS-SEM, all latent variables were significant (Supplementary Table 6) and goodness-of-fit measures, such as average variance extracted (AVE; indicator for converge validity) and composite reliability (indicator for internal consistency reliability) were higher than 0.5 and 0.7, respectively (Hair et al., 2011; Supplementary Table 7), indicating this model was reliable. As showed in Figure 6, AMF inoculation promoted the metabolisms of P (0.991), sugar (0.995), auxin (0.990), and ethylene (0.987). The total effect of AMF on LR formation (0.999) was significantly positive (Supplementary Table 6), in which the indirect effects via P metabolism (1.854) and auxin (0.746) were dominant (Figure 6). Besides, the indirect effects via sugar metabolism (−1.084) and ethylene metabolism (−0.383) were negative, and the effect of other unknown pathway (−0.151) was negative.
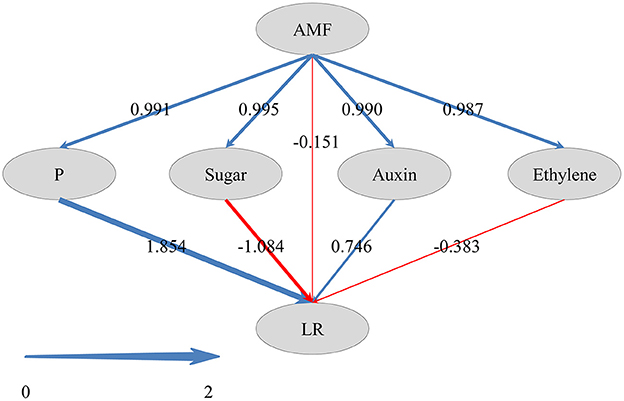
Figure 6. Directed graph of the Partial Least Squares Structural Equation Modeling (PLS-SEM). Larger path coefficients were reflected in the width of the arrows; and blue and red arrows indicate positive and negative effects, respectively.
Disscussion
In this study, the mycorrhizal colonization of trifoliate orange seedlings colonized by R. irregularis was 28.99%, similar to that colonized by Acaulospora scrobiculata (15.8%) and Funneliformis mosseae (38.9%) in the same plant species (Wu et al., 2016). DEGs related to Ser/Thr receptor kinases (51 DEGs), chitinases (11), lectins (5), and glutathione S-transferases (4) were observed (Supplementary Data 1), which are mycorrhiza-induced genes according to Guether et al. (2009), highlighting the evolutionary conservation of genetic processes supporting such symbiosis.
Promotion of LR formation by AMF has been demonstrated both in annual plants (Ronco et al., 2008; Gutjahr et al., 2009) and perennial woody species (Padilla and Encina, 2005; Yao et al., 2009; Orfanoudakis et al., 2010). In this study, LR numbers were significantly increased in the mycorrhizal seedlings, in line with our previous results (Chen et al., 2017), evidencing the important role of AMF in regulating LR formation. RNA-Seq represents a powerful tool to disentangle plant-AMF interaction at molecular level. With this technique, Shu et al. (2016) demonstrated that girdling regulated litchi mycorrhizal root development by altering carbohydrate metabolism (glyoxylate and dicarboxylate metabolism, citrate cycle, and pentose phosphate pathway). In grapevine roots, AMF significantly altered the expression of genes involved in nutrient transport, transcription factors, and cell wall-related genes (Balestrini et al., 2017). However, these work did not address how AMF regulated LR formation in woody plants. To gain insight into the regulatory network of LR formation induced by AMF, we performed RNA-Seq and analyzed differences in gene expression between the roots of non-mycorrhizal and mycorrhizal trifoliate orange seedlings. 909 DEGs were obtained, and according to Gene Ontology categories, 15 DEGs in the root tip were associated with cell wall reorganization (Supplementary Data 2), leading to continued root growth in mycorrhizal trifoliate orange (Opitz et al., 2016). Besides, DEGs related to oxidation-reduction process, transmembrane transport, kinase activity and membrane also possibly played roles in root development in response to AMF inoculation.
It is well accepted that AMF regulates LR formation via three pathways (see review by Fusconi, 2014), e.g., P metabolism, carbohydrate metabolism, and plant hormone metabolism. P is one of the most important constraints for plant growth and development in terrestrial ecosystems, especially in acid soils with low P availability (Xu et al., 2003; Kochian et al., 2004). In this study, shoot P content in the mycorrhizal plants was ~2 times that in the non-mycorrhizal plants, indicating the involvement of P metabolism in the regulation of LR formation by AMF. Actually, the 31 DEGs related to P metabolism demonstrate the active but complex P metabolism in roots following AMF inoculation. Phylogenetic analysis indicated that LOC102613232 and LOC102628168 encoded the mycorrhizal- inducible P transporter, positively responding to AMF colonization in trifoliate orange plants. In the same species, Shu et al. (2012) found that AMF increased the expression of PtaPT4 and PtaPT5 by up to 800-fold, indicating the homology of LOC102613232 and LOC102628168 with PtaPT4 and PtaPT. In contrast, the down-regulated DEGs related to acid phosphatase are probably indicative of alleviated P starvation in mycorrhizal plants, because P deficiency induced the PtPAP expression and acid phsophatase activity in trifoliate orange plants (Shu et al., 2014). PLS-SEM analysis showed that P metabolism was the most influential pathway by which AMF regulated the LR formation in trifoliate orange.
AMF can enhance shoot photosynthesis in citrus via the activation of the light-harvesting complex family of proteins in photosystem II (Gao et al., 2016), and regulate the sucrose metabolism in shoots (Wu et al., 2015). AMF colonizing roots represent a strong sink for photosynthates, and drive the transport of photosynthates (mainly as sucrose) from shoots to roots. These shoot-derived carbohydrates fulfill a dual role, acting as metabolism substrates and as signaling molecules (Smeekens et al., 2010). In this study, the increased glucose and fructose contents in primary roots (Supplementary Figure 1B) by AMF probably contribute to metabolisms essential to LR formation. Moreover, 14 DEGs were enriched in “starch and sucrose metabolism” (Supplementary Figure 5), indicating the involvement of sugar metabolism in the regulation. However, we did not identify any DEG related to sugar signaling, which was proposed by Hammond and White (2011). Although Zhou et al. (2008) demonstrated that sugar signaling mediated the cluster root formation induced by P starvation in white lupin, it seems that sugar signaling was not involved in the LR formation in trifoliate orange. In contrast, DEGs related to polygalacturonase (PG), chitinase (CHI) and their analogs (PG-like protein, CHI-like protein) were drastically up-regulated in inoculated seedlings. It is reported that PG activity increased during the LR emergence, and played an important role in the LR outgrowth (Peretto et al., 1992). CHI catalyzes the hydrolysis of 1,4-β-linkages inside chitin, and thus degrade the fungal cell wall to limit pathogen invasion (Eckardt, 2008). In mycorrhizal root, class III chitinase genes were strongly induced (Krajinski and Frenzel, 2007), in accordance with our result that LOC102607011 was up-regulated by AMF. According to Salzer et al. (2000), expression of class III chitinase genes were mycorrhiza-specific, while expression of defense-related chitinase genes (class I, II, and IV) were kept at a low and basal level in mycorrhiza. Recently, increasingly accumulated evidence indicate that CHI-like proteins may be also involved in the growth and developmental processes of plants (Kesari et al., 2015), including root morphogenesis (Hermans et al., 2010, 2011). It is noteworthy that sugar metabolism pathway negatively affect the LR formation as revealed by PLS-SEM analysis, probably indicating the competition for carbohydrate between AMF and LR formation. In overall, these data probably suggested that sugar metabolites were involved in the AMF regulated LR formation, but sugar signaling was not.
Plant hormones, especially auxin, play vital roles in LR development and RSA construction (Qu et al., 2016; Chen et al., 2017). In this study, among 25 DEGs related to plant hormones, 10 were associated with auxin (Figure 4C), demonstrating the primary role of auxin in LR formation regulated by AMF. It is evidenced that the biosynthesis of auxin in plant was promoted following AMF inoculation (Kaldorf and Ludwig-Müller, 2000; Alenazi et al., 2015), leading to increased LR formation. Another important plant hormone in this study was ethylene, involved in reducing LR formation (He et al., 2005; Singh et al., 2015), and seven DEGs related to ethylene were identified, four of which were ethylene-response transcription factors (ERF) (Figure 4C). In Arabidopsis, RNA interference-mediated suppression of AtERF070 (Arabidopsis thaliana Ethylene Response Factor070), a Pi starvation induced TF belonging to the APETALA2/ETHYLENE RESPONSE FACTOR family, led to augmented LR development (Ramaiah et al., 2014). Three of 4 ERFs were down-regulated, consistent with the increasing LR number in inoculated plants. The positive effect of auxin and negative effect of ethylene were also confirmed in the PLS-SEM analysis. Despite the contribution of gibberellin (Gou et al., 2010), abscisic acid (Soucek et al., 2007; Hong et al., 2013), and cytokinin (Aloni et al., 2006; Laplaze et al., 2007) on LR development, there were few DEGs related to them in this study, especially the latter two plant hormones, indicating that they might be not involved in AMF's modification on LR formation.
Our data demonstrated that AMF increased LR formation in trifoliate orange. Interestingly, however, AMF only colonized large LR but did not colonize fine LR in rice (Fiorilli et al., 2015). With RNA-Seq technique, they found the down regulation of genes involved in AM establishment in fine LR compared with large LR, indicating some mechanisms suppressing AM symbiosis in fine LR. These findings highlight the complexity in the interaction between AMF and LR formation, and thus call for more intensive work in this aspect.
Recently, several studies demonstrated that in addition to sugars, lipids are another major C source delivered to AMF in mycorrhiza (Jiang et al., 2017; Luginbuehl et al., 2017). In this study, we identified 28 DEGs related to lipid biosynthesis and metabolism. Particularly, 4 DEGs related to fatty acid metabolism were up-regulated by AMF. Similarly, some DEGs related to fatty acid biosynthetic process and long-chain fatty acid metabolic process were up-regulated upon AMF colonization (Fiorilli et al., 2015). These indicated that AMF can alter fatty acid metabolism of the host plant by their requirement of lipids. Moreover, steroids act as early AM signals and play an important role in many aspects of AM symbiosis, e.g., hyphal penetration and arbuscule formation (Bucher, 2010). This may explain the up-regulation of 8 DEGs AMF in this study.
Conclusion
To gain insights into the roles of P metabolism, sugar metabolism and plant hormones in the regulation of LR formation by AMF, we inoculated trifoliate orange seedlings with Rhizophagus irregularis. AMF inoculation strongly promoted the LR formation, as well as plant biomass and P concentration. RNA-Seq analysis revealed 909 DEGs in response to AMF inoculation, with 31, 32, and 23 DEGs related to P metabolism, sugar metabolism and plant hormones. These DEGs mainly encoded phosphate transporter, acid phosphatase, polygalacturonase, chitinase, auxin signaling, and ethylene signaling. PLS-SEM analysis indicates that AMF regulated LR formation mainly via the pathway of P metabolism in this study. In general, this transcriptomic data provide a global view of the pathways by which AMF regulates LR formation.
Author Contributions
WC, HZ, and QY designed the experiments. WC, JL, and PX performed the experiments. WC and PX carried out the bioinformatics analyses. WC, HZ, JC, and QY analyzed the data. WC and QY wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was financially supported by the National Natural Science Foundation of China [31270448], the Guangdong Higher Education Talents Project [(2013)246], and the Guangdong Science and Technology Foundation [2015TX01N036, 2015A02029087, 2016B090918090].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02039/full#supplementary-material
References
Alenazi, M. M., Egamberdieva, D., and Ahmad, P. (2015). Arbuscular mycorrhizal fungi mitigates NaCl induced adverse effects on Solanum lycopersicum L. Pak. J. Bot. 47, 327–340.
Aloni, R., Aloni, E., Langhans, M., and Ullrich, C. I. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97, 883–893. doi: 10.1093/aob/mcl027
Anders, S. (2010). Analysing RNA-Seq data with the DESeq package. Available online at: http://www.bioconductor.org/help/course-materials/2011/BioC2011/LabStuff/DESeq.pdf
Anders, S., and Huber, W. (2012). Differential Expression of RNA-Seq Data at the Gene Level–the DESeq Package. Heidelberg: European Molecular Biology Laboratory (EMBL).
Arshad, M., Ali, S., Noman, A., Ali, Q., Rizwan, M., Farid, M., et al. (2016). Phosphorus amendment decreased cadmium (Cd) uptake and ameliorates chlorophyll contents, gas exchange attributes, antioxidants, and mineral nutrients in wheat (Triticum aestivum L.) under Cd stress. Arch. Agron. Soil Sci. 62, 533–546. doi: 10.1080/03650340.2015.1064903
Bailey, P. H., Currey, J. D., and Fitter, A. H. (2002). The role of root system architecture and root hairs in promoting anchorage against uprooting forces in Allium cepa and root mutants of Arabidopsis thaliana. J. Exp. Bot. 53, 333–340. doi: 10.1093/jexbot/53.367.333
Balestrini, R., Salvioli, A., Dal Molin, A., Novero, M., Gabelli, G., Paparelli, E., et al. (2017). Impact of an arbuscular mycorrhizal fungus versus a mixed microbial inoculum on the transcriptome reprogramming of grapevine roots. Mycorrhiza 27, 417–430. doi: 10.1007/s00572-016-0754-8
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
Biermann, B., and Linderman, R. G. (1981). Quantifying vesicular-arbuscular mycorrhizas: a proposed method towards standardization. New Phytol. 87, 63–67. doi: 10.1111/j.1469-8137.1981.tb01690.x
Braak, C. J. F. T., and Smilauer, P. (2002). CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5). Ithaca, NY. Available online at: http://www.canoco.com
Brady, S. M., Orlando, D. A., Lee, J. Y., Wang, J. Y., Koch, J., Dinneny, J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. doi: 10.1126/science.1146265
Bucher, M. (2010). A novel lipid signal in the arbuscular mycorrhizal symbiosis within eyesight? New Phytol. 85, 593–595. doi: 10.1111/j.1469-8137.2009.03156.x
Chen, W. L., Li, J., Zhu, H. H., Chen, J. Z., and Yao, Q. (2016). A review of the regulation of plant root system architecture by rhizosphere microorganisms. Acta Ecol. Sin. 36, 5285–5297. doi: 10.5846/stxb201502260390
Chen, W. L., Li, J., Zhu, H. H., Xu, P. Y., Chen, J. Z., and Yao, Q. (2017). The differential and interactive effects of arbuscular mycorrhizal fungus and phosphorus on the lateral root formation in Poncirus trifoliata (L.). Sci. Hortic. 217, 258–265. doi: 10.1016/j.scienta.2017.02.008
Chen, Z., Zhao, J. T., Qin, Y. H., and Hu, G. B. (2016). Study on the graft compatibility between ‘Jingganghongnuo’ and other litchi cultivars. Sci. Hortic. 199, 56–62. doi: 10.1016/j.scienta.2015.12.020
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Eckardt, N. A. (2008). Chitin signaling in plants: insights into the perception of fungal pathogens and rhizobacterial symbionts. Plant Cell 20, 241–243. doi: 10.1105/tpc.108.058784
Feng, J. X., Meyer, C. A., Wang, Q., Liu, J. S., Liu, X. S., and Zhang, Y. (2012). GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28, 2782–2788. doi: 10.1093/bioinformatics/bts515
Fiorilli, V., Vallino, M., Biselli, C., Faccio, A., Bagnaresi, P., and Bonfante, P. (2015). Host and non-host roots in rice: cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 6:636. doi: 10.3389/fpls.2015.00636
Fusconi, A. (2014). Regulation of root morphogenesis in arbuscular mycorrhizae: what role do fungal exudates, phosphate, sugars and hormones play in lateral root formation? Ann. Bot. 113, 19–33. doi: 10.1093/aob/mct258
Gao, J., Zhang, Y. Z., Lu, C. L., Peng, H., Luo, M., Li, G., et al. (2015). The development dynamics of the maize root transcriptome responsive to heavy metal Pb pollution. Biochem. Biophys. Res. Commun. 458, 287–293. doi: 10.1016/j.bbrc.2015.01.101
Gao, X., Zhao, S., Xu, Q. L., and Xiao, J. X. (2016). Transcriptome responses of grafted Citrus sinensis plants to inoculation with the arbuscular mycorrhizal fungus Glomus versiforme. Trees 30, 1073–1082. doi: 10.1007/s00468-015-1345-6
Gou, J. Q., Strauss, S. H., Tsai, C. J., Fang, K., Chen, Y. R., Jiang, X. N., et al. (2010). Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22, 623–639. doi: 10.1105/tpc.109.073239
Guether, M., Balestrini, R., Hannah, M., He, J., Udvardi, M. K., and Bonfante, P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 182, 200–212. doi: 10.1111/j.1469-8137.2008.02725.x
Gutjahr, C., Casieri, L., and Paszkowski, U. (2009). Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 182, 829–837. doi: 10.1111/j.1469-8137.2009.02839.x
Gutjahr, C., and Paszkowski, U. (2013). Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 4:204. doi: 10.3389/fpls.2013.00204
Gutjahr, C., Sawers, R. J. H., Marti, G., Andrés-Hernández, L., Yang, S. Y., Casieri, L., et al. (2015). Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. U.S.A. 112, 6754–6759. doi: 10.1073/pnas.1504142112
Hair, J. F., Ringle, C. M., and Sarstedt, M. (2011). PLS-SEM: indeed a silver bullet. J. Mark. Theory Pract. 19, 139–152. doi: 10.2753/MTP1069-6679190202
Hammond, J. P., and White, P. J. (2011). Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 156, 1033–1040. doi: 10.1104/pp.111.175380
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., and Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. doi: 10.1111/j.1365-313X.2005.02575.x
Hermans, C., Porco, S., Vandenbussche, F., Gille, S., De Pessemier, J., Van Der Straeten, D., et al. (2011). Dissecting the role of CHITINASE-LIKE 1 in nitrate-dependent changes in root architecture. Plant Physiol. 157, 1313–1326. doi: 10.1104/pp.111.181461
Hermans, C., Porco, S., Verbruggen, N., and Bush, D. R. (2010). Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 152, 904–917. doi: 10.1104/pp.109.149849
Hong, J. H., Seah, S. W., and Xu, J. (2013). The root of ABA action in environmental stress response. Plant Cell Rep. 32, 1–13. doi: 10.1007/s00299-013-1439-9
Jiang, X., Chen, W. L., Xu, C. X., Zhu, H. H., and Yao, Q. (2015). Influences of arbuscular mycorrhizal fungus and phosphorus level on the lateral root formation of tomato seedlings. Chin. J. Appl. Ecol. 26, 1186–1192. doi: 10.13287/j.1001-9332.2015.0031
Jiang, Y., Wang, W. X., Xie, Q. J., Liu, N., Liu, L. X., Wang, D. P., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Kaldorf, M., and Ludwig-Müller, J. (2000). AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiol. Plantarum 109, 58–67. doi: 10.1034/j.1399-3054.2000.100109.x
Kesari, P., Patil, D. N., Kumar, P., Tomar, S., Sharma, A. K., and Kumar, P. (2015). Structural and functional evolution of chitinase-like proteins from plants. Proteomics 15, 1693–1705. doi: 10.1002/pmic.201400421
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soils? mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Krajinski, F., and Frenzel, A. (2007). Towards the elucidation of AM-specific transcription in Medicago truncatula. Phytochemistry 68, 75–81. doi: 10.1016/j.phytochem.2006.09.035
Laplaze, L., Benkova, E., Casimiro, I., Maes, L., Vanneste, S., Swarup, R., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900. doi: 10.1105/tpc.107.055863
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luginbuehl, L. H., Menard, G. N., Kurup, S., Van Erp, H., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
Lynch, J. (1995). Root architecture and plant productivity. Plant Physiol. 109, 7–13. doi: 10.1104/pp.109.1.7
Nawy, T., Lee, J., Colinas, J., Wang, J. Y., Thongrod, S. C., Malamy, J. E., et al. (2005). Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17, 1908–1925. doi: 10.1105/tpc.105.031724
Nibau, C., Gibbs, D. J., and Coates, J. C. (2008). Branching out in new directions: the control of root architecture by lateral root formation. New Phytol. 179, 595–614. doi: 10.1111/j.1469-8137.2008.02472.x
Opitz, N., Marcon, C., Paschold, A., Malik, W. A., Lithio, A., Brandt, R., et al. (2016). Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. J. Exp. Bot. 67, 1095–1107. doi: 10.1093/jxb/erv453
Orfanoudakis, M., Wheeler, C. T., and Hooker, J. E. (2010). Both the arbuscular mycorrhizal fungus Gigaspora rosea and Frankia increase root system branching and reduce root hair frequency in Alnus glutinosa. Mycorrhiza 20, 117–126. doi: 10.1007/s00572-009-0271-0
Padilla, I. M. G., and Encina, C. L. (2005). Changes in root morphology accompanying mycorrhizal alleviation of phosphorus deficiency in micropropagated Annona cherimola Mill. plants. Sci. Hortic. 106, 360–369. doi: 10.1016/j.scienta.2005.05.001
Péret, B., Larrieu, A., and Bennett, M. J. (2009). Lateral root emergence: a difficult birth. J. Exp. Bot. 60, 3637–3643. doi: 10.1093/jxb/erp232
Peretto, R., Favaron, F., Bettini, V., De Lorenzo, G., Marini, S., Alghisi, P., et al. (1992). Expression and localization of polygalacturonase during the outgrowth of lateral roots in Allium porrum L. Planta 188, 164–172. doi: 10.1007/BF00216810
Phillips, J. M., and Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 118–158. doi: 10.1016/S0007-1536(70)80110-3
Qu, C. P., Xu, Z. R., Hu, Y. B., Lu, Y., Yang, C. J., Sun, G. Y., et al. (2016). RNA-SEQ reveals transcriptional level changes of poplar roots in different forms of nitrogen treatments. Front. Plant Sci. 7:51. doi: 10.3389/fpls.2016.00051
Ramaiah, M., Jain, A., and Raghothama, K. G. (2014). ETHYLENE RESPONSE FACTOR070 regulates root development and phosphate starvation-mediated responses. Plant Physiol. 164, 1484–1498. doi: 10.1104/pp.113.231183
Ringle, C. M., Wende, S., and Will, S. (2005). Smart PLS 2.0 (M3) Beta. Hamburg. Available online at: http://www.smartpls.de
Ronco, M. G., Ruscitti, M. E., Arango, M. C., and Beltrano, J. (2008). Glyphosate and mycorrhization induce changes in plant growth and in root morphology and architecture in pepper plants (Capsicum annuum L.). J. Hortic. Sci. Biotech. 83, 497–505. doi: 10.1080/14620316.2008.11512413
Salzer, P., Bonanomi, A., Beyer, K., Vögeli-Lange, R., Aeschbacher, R. A., Lange, J., et al. (2000). Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection. Mol. Plant Microbe Interact. 13, 763–777. doi: 10.1094/MPMI.2000.13.7.763
Shu, B., Li, W. C., Liu, L. Q., Wei, Y. Z., and Shi, S. Y. (2016). Transcriptomes of arbuscular mycorrhizal fungi and litchi host interaction after tree girdling. Front. Microbiol. 7:408. doi: 10.3389/fmicb.2016.00408
Shu, B., Wang, P., and Xia, R. X. (2014). Effects of mycorrhizal fungi on phytate-phosphorus utilization in trifoliate orange (Poncirus trifoliata L. Raf) seedlings. Acta Physiol. Plant 36, 1023–1032. doi: 10.1007/s11738-013-1480-x
Shu, B., Xia, R. X., and Wang, P. (2012). Differential regulation of Pht1 phosphate transporters from trifoliate orange (Poncirus trifoliata L. Raf) seedlings. Sci. Hortic. 146, 115–123. doi: 10.1016/j.scienta.2012.08.014
Singh, M., Gupta, A., and Laxmi, A. (2015). Ethylene acts as a negative regulator of glucose induced lateral root emergence in Arabidopsis. Plant Signal. Behav. 10:e1058460. doi: 10.1080/15592324.2015.1058460
Smeekens, S., Ma, J. K., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 273–278. doi: 10.1016/j.pbi.2009.12.002
Song, L., Prince, S., Valliyodan, B., Joshi, T., Maldonado Dos Santos, J. V., Wang, J. J., et al. (2016). Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genomics 17, 57. doi: 10.1186/s12864-016-2378-y
Soucek, P., Klíma, P., Reková, A., and Brzobohatý, B. (2007). Involvement of hormones and KNOXI genes in early Arabidopsis seedling development. J. Exp. Bot. 58, 3797–3810. doi: 10.1093/jxb/erm236
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Vilches-Barro, A., and Maizel, A. (2015). Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23, 31–38. doi: 10.1016/j.pbi.2014.10.005
Villacorta-Martín, C., Sánchez-García, A. B., Villanova, J., Cano, A., van de Rhee, M., de Haan, J., et al. (2015). Gene expression profiling during adventitious root formation in carnation stem cuttings. BMC Genomics 16, 789. doi: 10.1186/s12864-015-2003-5
Wu, Q. S., Cao, M. Q., Zou, Y. N., Wu, C., and He, X. H. (2016). Mycorrhizal colonization represents functional equilibrium on root morphology and carbon distribution of trifoliate orange grown in a split-root system. Sci. Hortic. 199, 95–102. doi: 10.1016/j.scienta.2015.12.039
Wu, Q. S., Srivastava, A. K., and Li, Y. (2015). Effects of mycorrhizal symbiosis on growth behavior and carbohydrate metabolism of trifoliate orange under different substrate P levels. J. Plant Growth Regul. 34, 499–508. doi: 10.1007/s00344-015-9485-x
Xie, C., Mao, X. Z., Huang, J. J., Ding, Y., Wu, J. M., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, Q., Chen, L. L., Ruan, X. A., Chen, D. J., Zhu, A. D., Chen, C. L., et al. (2012). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66. doi: 10.1038/ng.2472
Xu, R. K., Zhao, A. Z., Li, Q. M., Kong, X. L., and Ji, G. L. (2003). Acidity regime of the Red Soils in a subtropical region of southern China under field conditions. Geoderma 115, 75–84. doi: 10.1016/S0016-7061(03)00077-6
Yan, J. W., Yuan, F. R., Long, G. Y., Qin, L., and Deng, Z. N. (2012). Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol. Biol. Rep. 39, 1831–1838. doi: 10.1007/s11033-011-0925-9
Yao, Q., Wang, L. R., Zhu, H. H., and Chen, J. Z. (2009). Effect of arbuscular mycorrhizal fungal inoculation on root system architecture of trifoliate orange (Poncirus trifoliata L. Raf.) seedlings. Sci. Hortic. 121, 458–461. doi: 10.1016/j.scienta.2009.03.013
Yu, P., Baldauf, J., Lithio, A., Marcon, C., Nettleton, D., Li, C. J., et al. (2016). Root type specific reprogramming of maize pericycle transcriptomes by local high nitrate results in disparate lateral root branching patterns. Plant Physiol. 170, 1885–2015. doi: 10.1104/pp.15.01885
Zhang, N., Zhang, H. J., Zhao, B., Sun, Q. Q., Cao, Y. Y., Li, R., et al. (2014). The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 56, 39–50. doi: 10.1111/jpi.12095
Zhao, H. M., Ma, T. F., Wang, X., Deng, Y. T., Ma, H. L., Zhang, R. S., et al. (2015). OsAUX1 controls lateral root initiation in rice (Oryza sativa L.). Plant Cell Environ. 38, 2208–2222. doi: 10.1111/pce.12467
Keywords: arbuscular mycorrhizal fungus, trifoliate orange, lateral root formation, RNA-Seq, P metabolism, sugar metabolism, plant hormone
Citation: Chen W, Li J, Zhu H, Xu P, Chen J and Yao Q (2017) Arbuscular Mycorrhizal Fungus Enhances Lateral Root Formation in Poncirus trifoliata (L.) as Revealed by RNA-Seq Analysis. Front. Plant Sci. 8:2039. doi: 10.3389/fpls.2017.02039
Received: 02 August 2017; Accepted: 14 November 2017;
Published: 29 November 2017.
Edited by:
Andrea Genre, Università degli Studi di Torino, ItalyReviewed by:
Raffaella Balestrini, Consiglio Nazionale Delle Ricerche (CNR), ItalyOswaldo Valdes-Lopez, Universidad Nacional Autónoma de México, Mexico
Copyright © 2017 Chen, Li, Zhu, Xu, Chen and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Yao, eWFvcXNjYXVAc2NhdS5lZHUuY24=
 Weili Chen1
Weili Chen1 Qing Yao
Qing Yao