
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 November 2017
Sec. Plant Metabolism and Chemodiversity
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01974
MYB transcription factors hold vital roles in the regulation of plant secondary metabolic pathways. Laticifers in rubber trees (Hevea brasiliensis) are of primary importance in natural rubber production because natural rubber is formed and stored within these structures. To understand the role of MYB transcription factors in the specialized cells, we identified 44 MYB genes (named HblMYB1 to HblMYB44) by using our previously obtained transcriptome database of rubber tree laticifer cells and the public rubber tree genome database. Expression profiles showed that five MYB genes were highly expressed in the laticifers. HblMYB19 and HblMYB44 were selected for further study. HblMYB19 and HblMYB44 bound the promoters of HbFDPS1, HbSRPP, and HRT1 in yeast. Furthermore, the transient overexpression of HblMYB19 and HblMYB44 in tobacco plants significantly increased the activity of the promoters of HbFDPS1, HbSRPP, and HRT1. Basing on this information, we proposed that HblMYB19 and HblMYB44 are the regulators of HbFDPS1, HbSRPP, and HRT1, which are involved in the biosynthesis pathway of natural rubber.
MYB transcription factors (TFs) comprise a TF family highly rich in plants (Dubos et al., 2010). Plant MYB TFs contain highly conserved MYB domains involved in DNA binding (Rosinski and Atchley, 1998; Jin and Martin, 1999). On the basis of the number of MYB repeats present in their sequences, MYB TFs are divided into the following four groups: 1R-MYB, 2R-MYB, 3R-MYB, and 4R-MYB (Rosinski and Atchley, 1998; Jiang et al., 2004). Each MYB repeat contains approximately 52 amino-acid residues, which form three α-helices (Kanei-Ishii et al., 1990). Since the first plant MYB gene ZmC1 was characterized from Zea mays (Paz-Ares et al., 1987), numerous MYB genes have been identified and characterized from plants (Stracke et al., 2001; Dubos et al., 2010; Katiyar et al., 2012; Hou et al., 2014; Wang et al., 2015; Zhou et al., 2015; He et al., 2016). At least 155 and 197 MYB genes have been identified in rice and Arabidopsis, respectively (Katiyar et al., 2012). MYB TFs are involved in plant growth and development (Cominelli and Tonelli, 2009; Oh et al., 2011; Huang et al., 2013; Cai et al., 2015), hormone signal transduction (Shin et al., 2007; Zhao et al., 2014), secondary metabolism (Chezem and Clay, 2016; Chezem et al., 2016; Zhai et al., 2016), abiotic stress responses (Dai et al., 2007; Oh et al., 2011; Peng et al., 2011), and disease resistance (Liu et al., 2013; Zhang et al., 2015).
In rubber tree (Hevea brasiliensis), natural rubber (NR) is obtained from latex, which constitutes the cytoplasmic content of laticifer cells (Kush, 1994). Laticifers are specialized cells located inside the phloem tissue of rubber trees (Kush, 1994; Hao and Wu, 2000). NR is synthesized in the rubber particles of laticifers (Kush et al., 1990). Laticifers are of primary importance in NR production. However, the biological functions of the rubber molecule and latex remain unclear (Ko et al., 2003). The regulatory mechanisms of NR biosynthesis are also poorly understood (Li et al., 2016b; Tang et al., 2016; Yamashita et al., 2016). MYB TFs play vital roles in regulating plant secondary metabolic pathways, such as the general phenylpropanoid pathway and lignin, flavonoid, and glucosinolate pathways (Chezem et al., 2016). However, few MYB TF genes related to the NR biosynthesis pathway in rubber trees have been reported. To understand the MYB TFs in laticifers, we identified and characterized 44 MYB genes (named HblMYB1 to HblMYB44) in this study. We found that five MYB genes were more highly expressed in laticifers than in other tissues. Furthermore, HblMYB19 and HblMYB44 may be the regulators participating in the NR biosynthesis pathway.
Two-year-old trees of H. brasiliensis clones of Wenchang11 were grown in the Rubber Research Institute of Hainan Agricultural Reclamation, Wenchang, Hainan, China. Rubber tree shoots were treated by 0.07% methyl jasmonate (JA), 0.5% Ethrel (ET), 200 μm abscisic acid (ABA), and 200 μm salicylic acid (SA) as described previously (Hao and Wu, 2000). Then, seven groups of 10 trees were used in each treatment, in which the plant growth regulator was applied at 1, 3, 6, 9, 12, 24, and 48 h before tapping. One group served as an untreated control. After the treatments at all time points, latex samples from all the tested trees were collected and mixed together thoroughly. The resulting solution was then divided into five equal volumes for RNA extraction (Tang et al., 2007). The other tissues (leaves, barks, roots, and flowers) of the rubber tree were then collected and stored in liquid nitrogen for RNA extraction.
DNA was extracted from young leaves of H. brasiliensis through the cetyl trimethylammonium bromide method (Allen et al., 2006). The total RNA from latex was isolated in accordance with Tang’s method (Tang et al., 2007), whereas that from the other tissues was extracted as described previously (Li et al., 2011). Three biological replicates were used for RNA extraction.
The analytical software NCBI-Blast-2.2.28+-win32 and the genomic data of H. brasiliensis (Rahman et al., 2013; Tang et al., 2016) were downloaded from the National Center for Biotechnology Information (NCBI1). They were used to establish a local H. brasiliensis genome database. The MYB unigenes were obtained from the transcriptome database of the rubber tree latex in our previous study (Li et al., 2016a). The MYB unigenes were employed as query sequences for a BLAST search in the local rubber tree genome database. All candidate MYB genes were further analyzed for confirmation by using the NCBI Conserved Domain Search database2. The physical and chemical properties of HblMYB were analyzed with ExPASy3), and the CDS of HblMYBs were analyzed with GSDS4. The predicted amino-acid sequences of HblMYBs were aligned with ClustalX. The highly conservative domains of HblMYB proteins were illustrated with espript5, and the 3D structure was constructed with SWISS-MODEL6.
Arabidopsis MYB protein sequences were downloaded from the phytozome7. Arabidopsis MYB protein sequences and the deduced amino-acid sequences of HblMYBs were aligned using ClustalX. Then, using the neighbor-joining method and the MEGA6.0 program, we constructed the phylogenetic tree between HlMYBs and the known MYB from Arabidopsis, and bootstrap analysis was conducted with 1,000 replicates (Yang et al., 2015).
The cDNA synthesis for qRT-PCR was performed with a RevertAidTM First-Strand cDNA Synthesis Kit (Fermentas, Lithuania). qRT-PCR was performed using a SYBR Premix EX Taq Kit (TaKaRa, Japan). The primers for the HblMYBs were designed using the Primer Premier 5 software (Supplementary Table S1). HbACT7 was amplified through the following primers as standard control: 5′-TGTCAGCAACT GGGACGATATGG-3’ as primer 1 and 5′-GAGTCATCTTCTCTCTGTTGGC-3′ as primer 2 (Li et al., 2016a). qRT-PCR was performed as follows: 3 min at 95°C for denaturation, 40 cycles for 10 s at 95°C, 20 s at 58°C, and 25 s at 72°C. The quantitative value obtained from qRT-PCR is considered the cycle threshold (Ct). The normalized expression values for each gene were calculated through the following formula: 2-(Ct[gene]-Ct[HbACT7]), in which HbACT7 was used as a housekeeping gene for normalization. Three individual reactions were replicated. Data were analyzed by ANOVA to analyze the significant differences on the basis of Fischer’s LSD test (P < 0.05 and P < 0.01; Quirk et al., 2016).
The open reading frames (ORFs) of HblMYBs were amplified by PCR using primers (Supplementary Table S1). PCR products were ligated into the pCAMBIA1302 vector, generating pHblMYBs-GFP. pCAMBIA1302 and pHblMYBs-GFP were introduced into the onion epidermis by Agrobacterium-mediated transformation. The transformed onion epidermis was then cultured on an MS solid medium in the dark at 26°C for 5 h and then observed with a confocal microscope (Zeiss LSM510, Germany).
The HbFDPS1 promoter (1,066 bp) and HbSRPP promoter (1,735 bp) were amplified by PCR with the primers as described previously (Guo et al., 2010, 2014). The 1,136 bp HRT1 promoter was amplified using the primers (Supplementary Table S1) based on the HRT1 sequence from the rubber tree genome database (GenBank accession: LVXX01000000; Tang et al., 2016). These promoters were cloned into the SpeI/MluI sites of the pHiS2.1 vectors (Clontech) to form the bait vectors pHiS-pHbSRPP, pHiS-HbFDPS1, and pHiS-HRT1. The ORFs of HbMYB19 and HbMYB44 were fused into the GAL4 domains of the pGAD7 vectors to generate the prey constructs pGAD-HbMYBs. The bait and prey vectors were then transformed into the yeast strain Y187 (Clontech). Afterward, the introduced yeast was cultivated on an SD/-Trp/-His/-Leu medium supplemented with 70 mM 3-amino-1,24-triazole (3-AT) at 30°C for 3 days.
The Dual-LUC assay was performed as described previously (Hellens et al., 2005). In brief, the promoters of HbSRPP, HbFDPS1, and HRT1 were cloned into pGreenII 0800 vectors, in which the expression of Renilla luciferase (REN-Luc) provided an internal control (Hellens et al., 2005). The ORFs of HbMYB19 and HbMYB44 were amplified with the primers (Supplementary Table S1) and then with the inserted pGreenII 62Sk vectors. All constructs were introduced into the Agrobacterium tumefaciens strain GV3103. The introduced GV3103 harboring pGreen-pHbSRPP, pGreen-pHbFDPS1, or pGreen-pHRT1 were mixed with the introduced GV3103 harboring pGreenII 62Sk-HblMYBs in a volume ratio of 1:5. The mixtures were injected into tobacco leaves. After culturing for 3 days, the infected areas of the leaves were obtained by puncher, and the protein was extracted. The activities of the luciferase and REN-Luc were measured through the Dual-LUC Reporter Assay System in accordance with the manufacturer’s manual (Promega). The binding ability of the HblMYBs to the promoters of HbSRPP, HbFDPS1, and HRT1 were represented by LUC/REN (ratio of LUC to REN-Luc). Three biological repeats were measured. The data were analyzed by ANOVA to determine the significant differences on the basis of the Fischer’s LSD test (P < 0.05 and P < 0.01) (Quirk et al., 2016).
The transcriptome database of the rubber tree latex was obtained in our previous study (Li et al., 2016a). A total of 76 MYB unigenes were obtained using our previously established latex transcriptome database. A total of 44 MYB genes were confirmed in the present study after the 76 MYB unigenes were searched in BLAST against those from the public rubber tree genome database. These MYB genes were named as HblMYB1 to HblMYB44. The sequences and properties of the identified 44 HblMYBs from the laticifer cells of the rubber tree are listed in Supplementary Table S2. The genomic structure of the 44 HblMYB genes was analyzed by GSDS. The 44 HblMYBs vary with respect to exon–intron gene structure. In particular, six HblMYBs contain only one exon, six HblMYBs contain two exons and one intron, 17 HblMYBs contain three exons and two introns, and the other HblMYBs containing more than five exons (Figure 1).
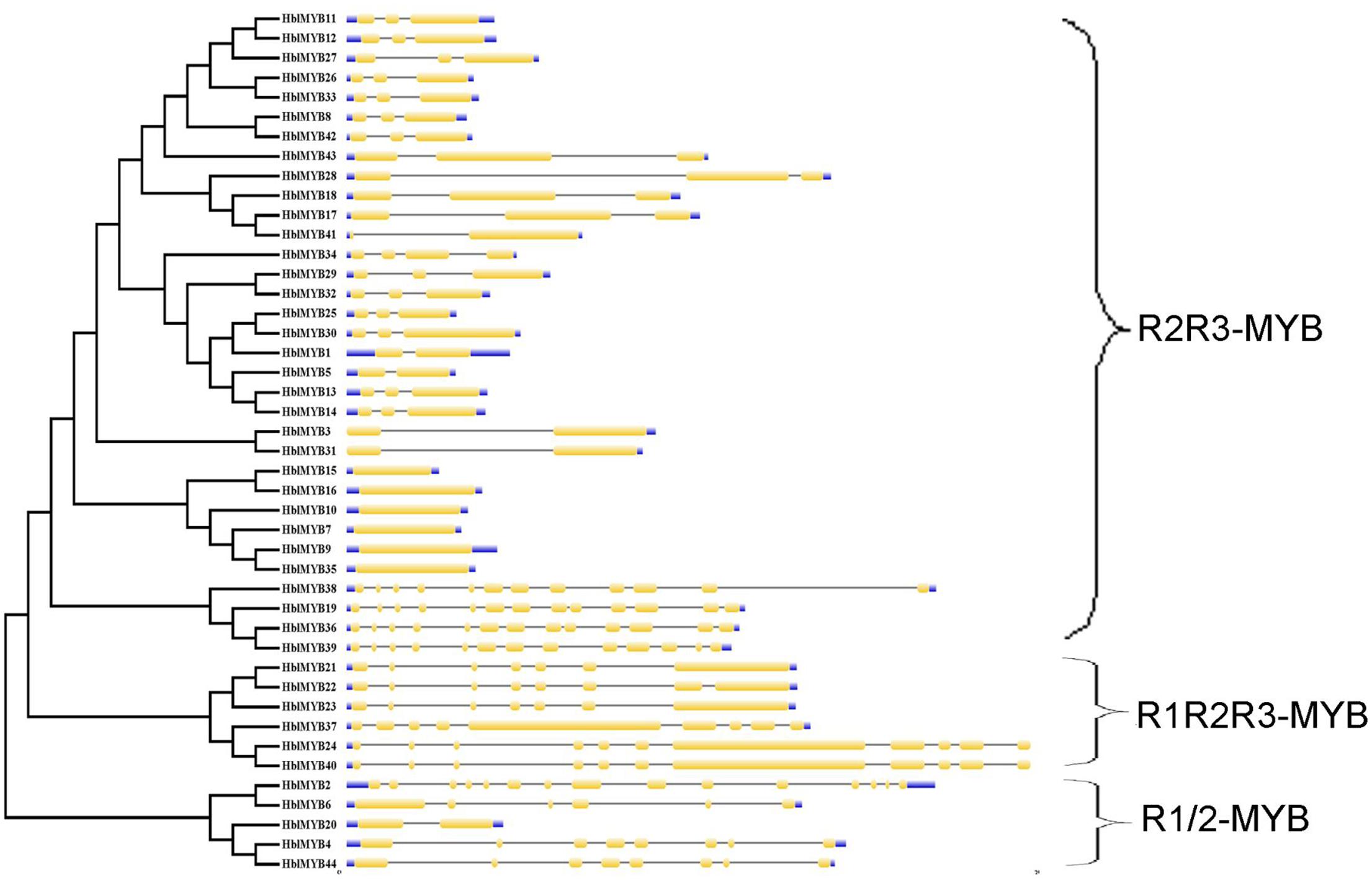
FIGURE 1. Exon–intron structure of HblMYBs based on their evolutionary relationships. The NJ evolutionary tree was generated with 1,000 bootstrap replicates based on the full-length sequences of the HblMYBs (left side). Exon–intron analyses of the HblMYBs were performed with GSDS. Meanwhile, the intron–exon structures of HblMYBs are described in the right portion. The exons and introns are indicated by yellow boxes and single lines, and the 5′-UTR or 3′-UTR are indicated by blue boxes. The lengths of the exons and introns for the corresponding HblMYBs are shown proportionally.
The number of amino acids in the HblMYB proteins ranges from 246 (HblMYB26, HblMYB33) to 1,043 (HblMYB24); protein molecular weights, from 27.24 (HblMYB10) to 115.12 (HblMYB24); and pI, from 5 (HblMYB37) to 9.44 (HblMYB3; Supplementary Table S2). Further analysis showed that the MYB domain (R unit) was highly conserved in the N-terminus. Among all the 44 HblMYBs, 5 HblMYBs contained one R unit, 33 HblMYBs had two R units, and 6 HblMYB contained three R units (Figure 1). The R2 repeats of R2R3-HblMYBs harbored three highly conserved tryptophan residues (W) at positions 6, 26, and 46 (Figure 2A). By contrast, the tryptophan residues were highly conserved at positions 25 and 44 of the R3 repeats (Figure 2B). Other highly conserved residues, except the tryptophan residues, were confirmed in the R2 and R3 domains. These residues included Gly (G4), Glu (E10), Asp (D11), Gly (G22), Arg (R37), Gly (G39), Lys (K40), Cys (C42), Arg (R43), Arg (R45), Asn (N48), Leu (L50), and Pro (P52) in the R2 repeat (Figure 2A), as well as Glu (E10), His (H18), Gly (G22), Asn (N23), Gly (G34), Arg (R35), Thr (T36), Asp (D37), Asn (N38), Lys (K41), and Asn (N42) in the R3 repeat (Figure 2B). Additionally, the 3D protein structure prediction showed that the R2 and R3 domains of the HblMYB proteins formed three α-helices (Figures 2C,D), which participate in transcriptional regulation (Dubos et al., 2010).
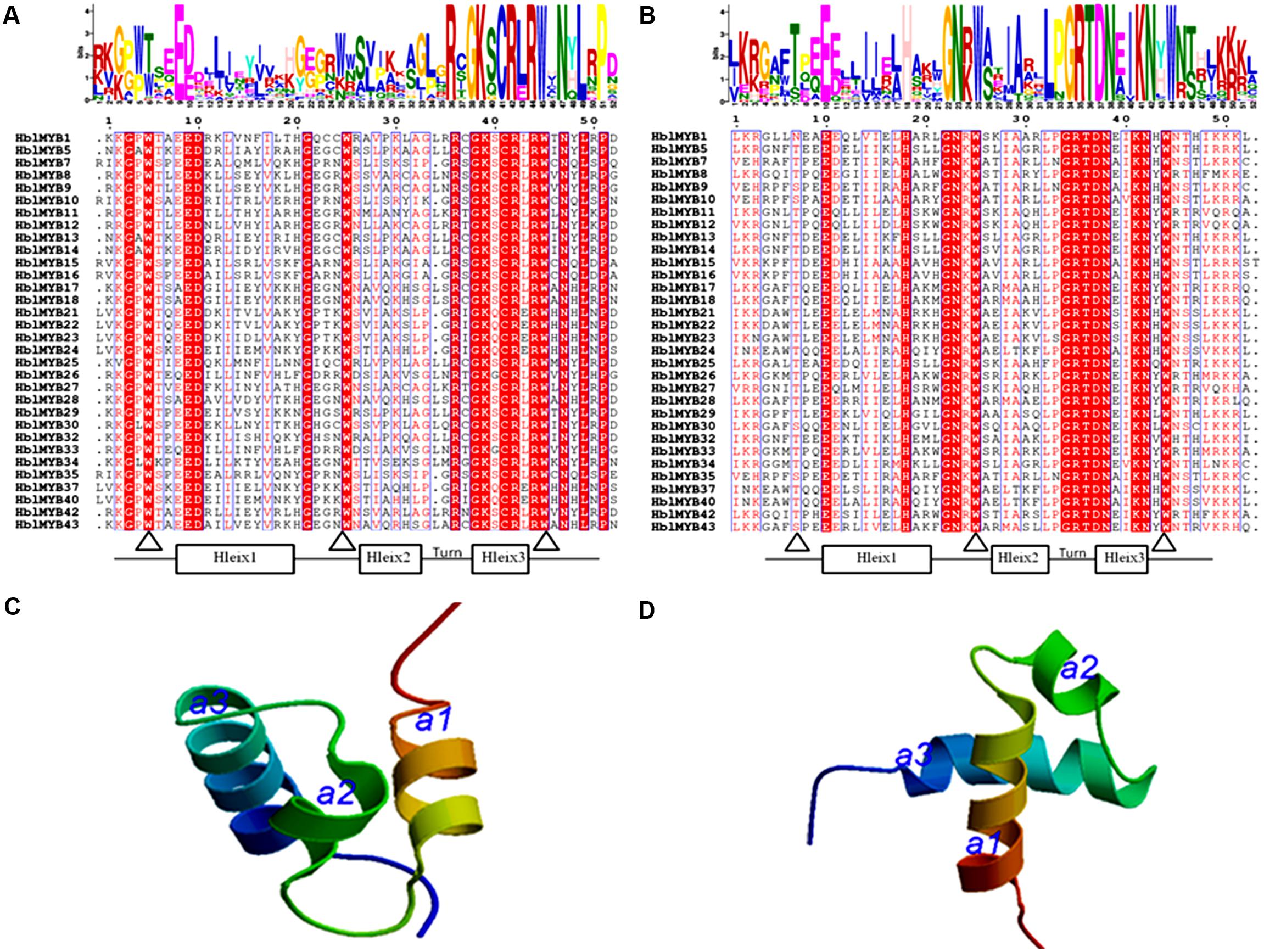
FIGURE 2. Predicted domains in the HblMYBs. The conserved domains were determined by MEME through the protein sequences of the HblMYBs. This online software was used to create the logo representations of the R2 (A) and R3 domains (B). The y-axis (measured in bits) depicts the overall height of the stack and indicates the sequence conservation at that position. Meanwhile, the symbol height within the stack indicates the relative frequency of each amino acid at that position. The predicted 3D structure models of the R2 (C) and the R3 domains are presented (D). The R2 and R3 domains of HblMYB1 were utilized to construct structural models.
To infer the evolutionary relationships between HlMYBs and the known MYB from Arabidopsis, we constructed a phylogenetic tree between the obtained 44 HblMYBs and 126 Arabidopsis MYB TFs (Figure 3). All the 170 MYB were classified into 29 subgroups. Meanwhile, 44 HblMYB proteins were divided into 17 subgroups (S2, 5, 8, 9, 13, 14, 15, 17, 18, 19, 20, 22, and 23 and G1, 2, 3, and 4). Their orthologous MYBs were from Arabidopsis. This result suggests the existence of few closely related orthologous MYBs between rubber trees and Arabidopsis. Of the 29 subgroups, 10 subgroups (S1, 4, 6, 7, 10, 11 12, 16, 21, and 25) did not exhibit any rubber tree ortholog, and two subgroups (G1 and G4) did not present any Arabidopsis ortholog. The phylogenetic tree indicated the existence of an ancestral set of MYB genes prior to the divergence of rubber tree and Arabidopsis.
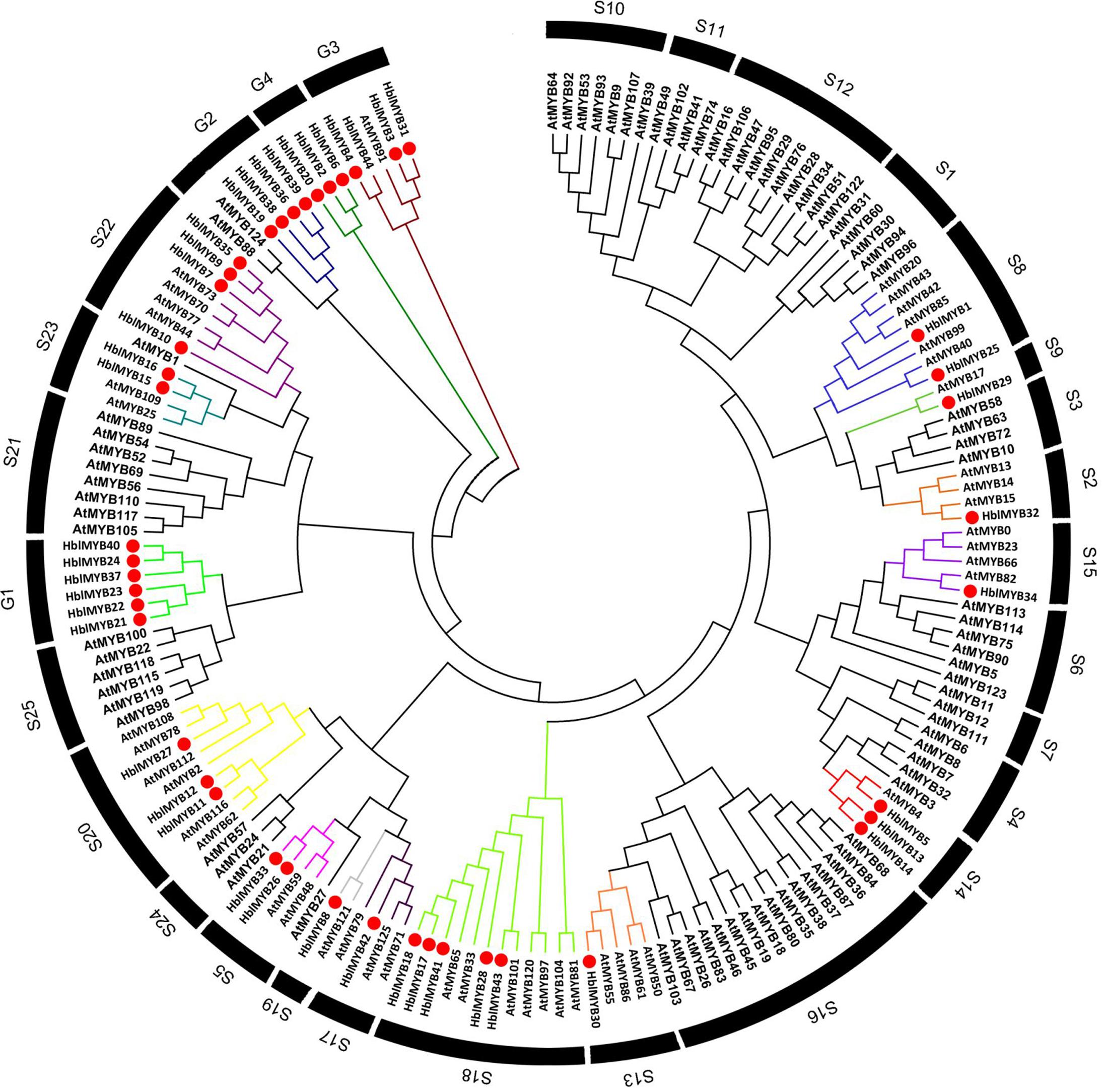
FIGURE 3. Phylogenetic analysis of HblMYBs with Arabidopsis MYB TFs by MEGA6.0 from CLUSTALW alignments. The Arabidopsis MYB TFs used in the evolutionary analysis were retrieved from the phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), and the internal branch support was estimated with 1,000 bootstrap replicates.
The expression patterns of 44 HblMYBs were detected in roots, barks, leaves, flowers, and latex by qRT-PCR (Figure 4). The results showed that the expression profiles of the 44 HblMYBs differed across different tissues. Five HblMYBs (HblMYB19, 20, 25, 40, and 44) showed higher transcription levels in latex, whereas 36 HblMYBs in the leaves, and 22 HblMYBs in the flowers. By contrast, all HblMYBs clearly presented with lower expression in the barks and roots.
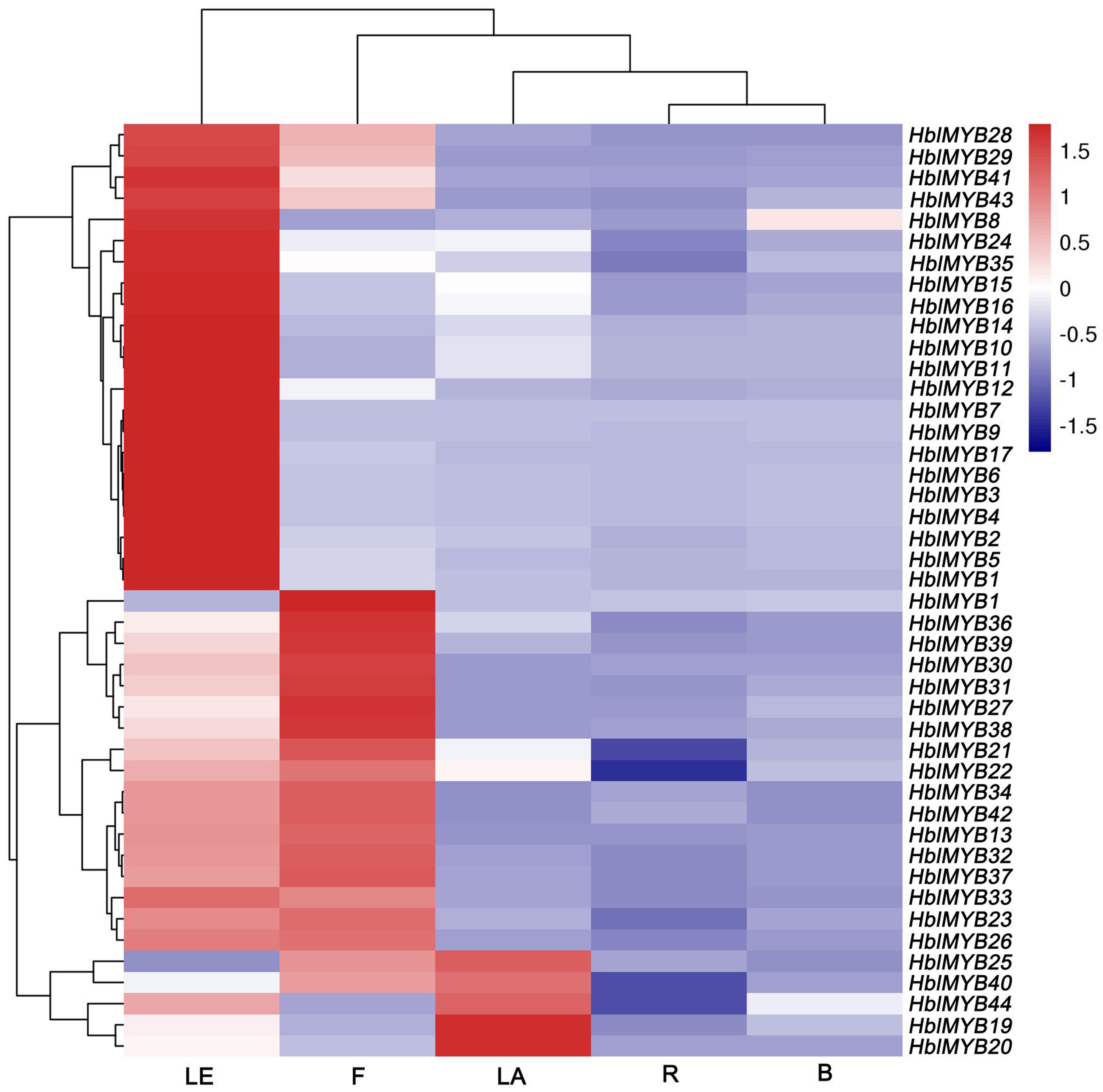
FIGURE 4. Expression patterns of HbMYBs in different tissues. The heatmap was created using log2-based values from three replicates of qRT-PCR data. The scale represents the relative signal intensity values. R, root; B, bark; LE, leaf; F, flower; LA, latex.
Given the expression of HblMYBs in different tissues, five HblMYBs (HblMYB19, 20, 25, 40, and 44, which showed high expression levels in latex), were selected for further analysis on their response to exogenous phytohormone. Overall, the five HblMYBs showed different expression patterns under MeJA, ET, ABA, and SA treatments (Figure 5). Results showed that JA induced the expression of HblMYB19, 20, 40, and 44 but down-regulated that of HblMYB25 at the 6 h time point. ET treatment upregulated the transcript abundance of HblMYB20, 25, and 40 but downregulated those of HblMYB19 and 44 at the 12 or 9 h time point. ABA stress induced the expression of HblMYB44 at 24 h time point. However, we repressed the HblMYB19, 20, 25, and 40 expression at 12 or 6 h time point. Lastly, SA treatment repressed the HblMYB20, 25, and 44 expression at 24, 9, or 3 h time point, but did not significantly affect the HblMYB19 and 44 expression.
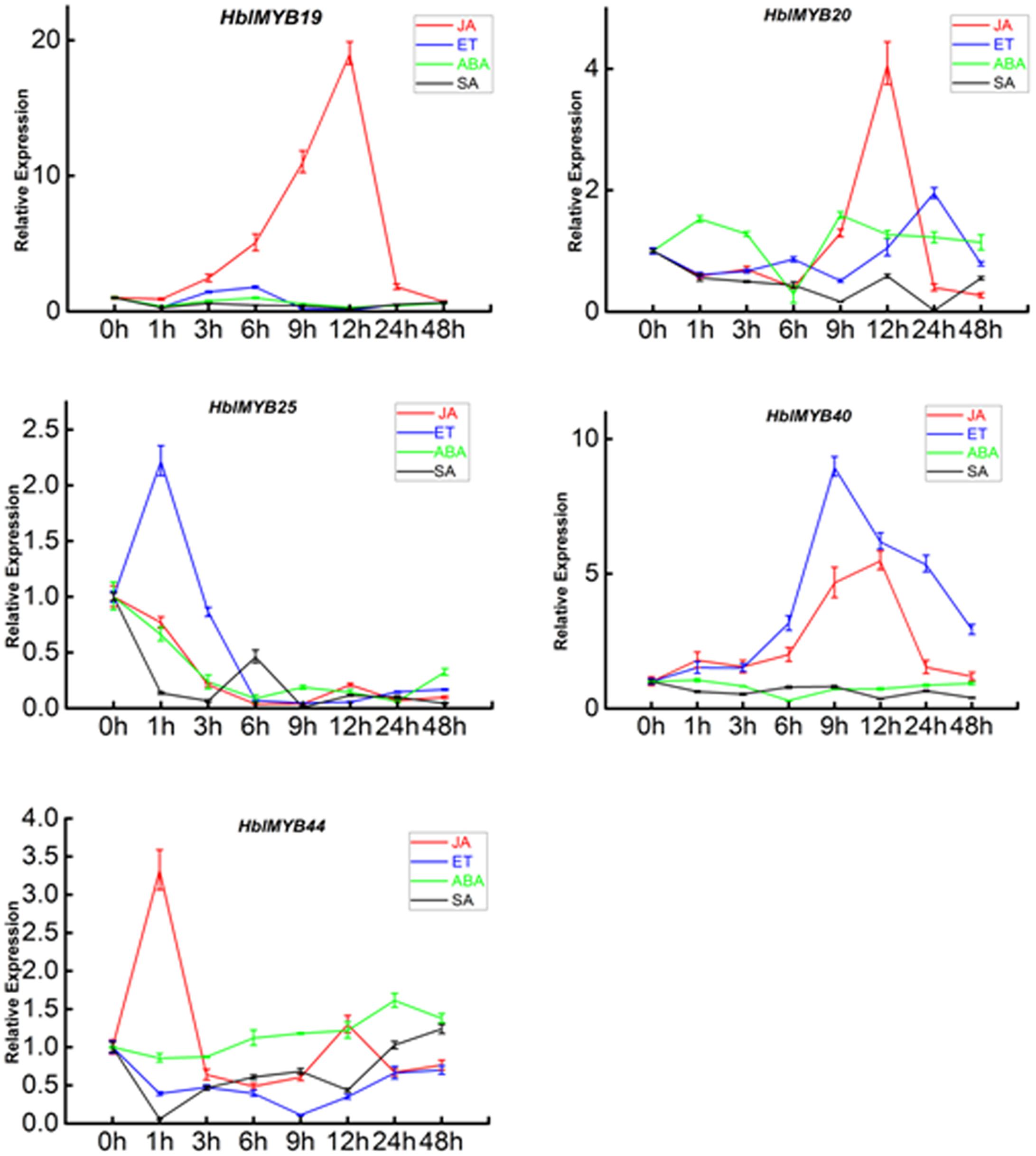
FIGURE 5. Expression patterns of the five HbMYBs responding to phytohormone treatment. The relative transcript abundances of HbMYBs were examined via qRT-PCR. The y-axis is the scale of the relative transcript abundance level. The x-axis shows the time course of the phytohormone treatment. The average of three independent biological replicates was computed at each time. Data are presented as mean ± standard error (SE) (n = 3). The statistical significance of the differences was assessed by ANOVA (one or two stars correspond to P < 0.05 and P < 0.01, respectively).
Given the expression of HblMYBs and response to JA in laticifers, HblMYB19 and HblMYB44 were selected for further analysis. First, subcellular localization analysis was performed on HblMYB19 and HblMYB44. We found that the GFP signals expressed the fusion proteins of HblMYB19, and 44 were present only in the nucleus of onion epidermal cells. By contrast, the GFP signals expressing the GFP protein were present obviously both in the nuclei and cytosol (Figure 6).
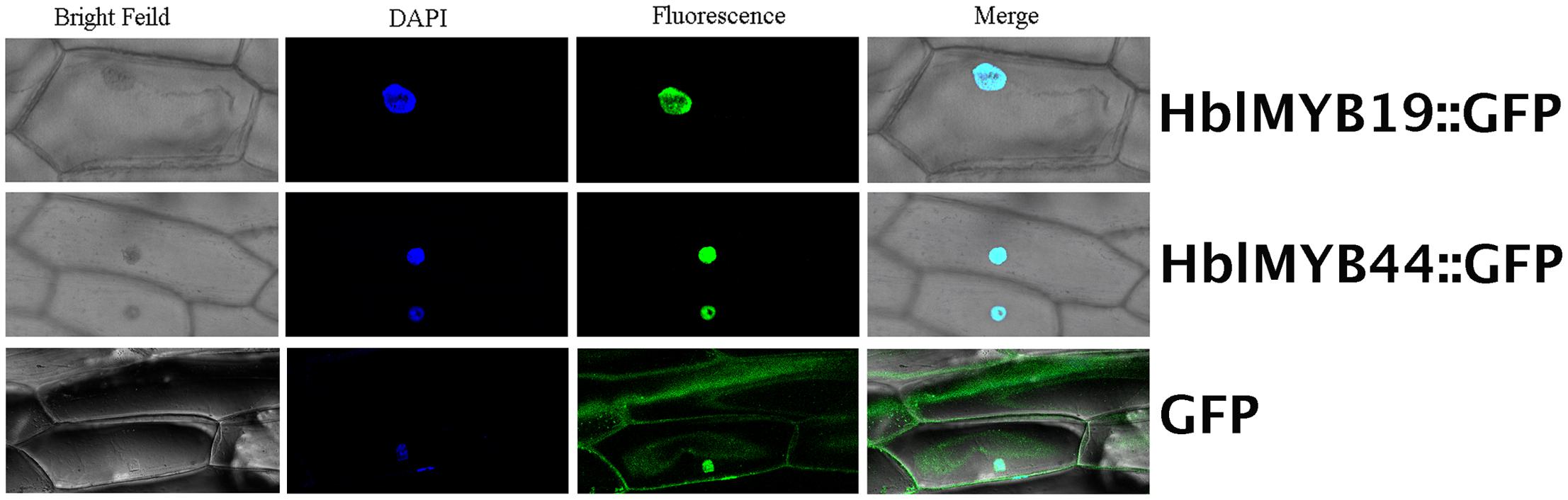
FIGURE 6. Nuclear localization of HbMYB19 and HbMYB44. The upper panel shows the corresponding bright-field image, DAPI image, fluorescence image, and merged image of HbMYB19-GFP. The middle panel displays the corresponding bright-field image, DAPI image, fluorescence image, and merged image of HbMYB44-GFP. The lower panel shows the corresponding bright-field image, DAPI image, fluorescence image, and merged image of GFP.
Farnesyl diphosphate synthase (FDPS), small rubber particle protein (SRPP), and Hevea cis-prenyltransferases or rubber transferase (HRT) from H. brasiliensis are responsible for the cis-1,4-polymerization of isoprene units from isopentenyl diphosphate (IPP) and implicated in NR yield (Light et al., 1989; Oh et al., 1999; Asawatreratanakul et al., 2003). To determine whether HblMybs bind the promoters of HbFDPS1, HbSRPP, and HRT1, we performed a yeast one-hybrid analysis. The yeast clones harboring pHblMyb19+pHIS2-pHbSRPP, pHblMyb19+pHIS2-pHRT1, pHblMyb19+pHIS2-pHbFDPS1, pHblMyb44+pHIS2-HbSRPP, pHblMyb44+pHIS2-pHRT1, and pHblMyb44+pHIS2-pHbFDPS1 can grow on SD/-Trp/-His/-Leu selective medium added with 70 mM 3-AT (Figure 7). This result indicated that HblMyb19 and HblMyb44 bound the promoters of HbSRPP, HRT1, and HbFDPS1 in yeast.
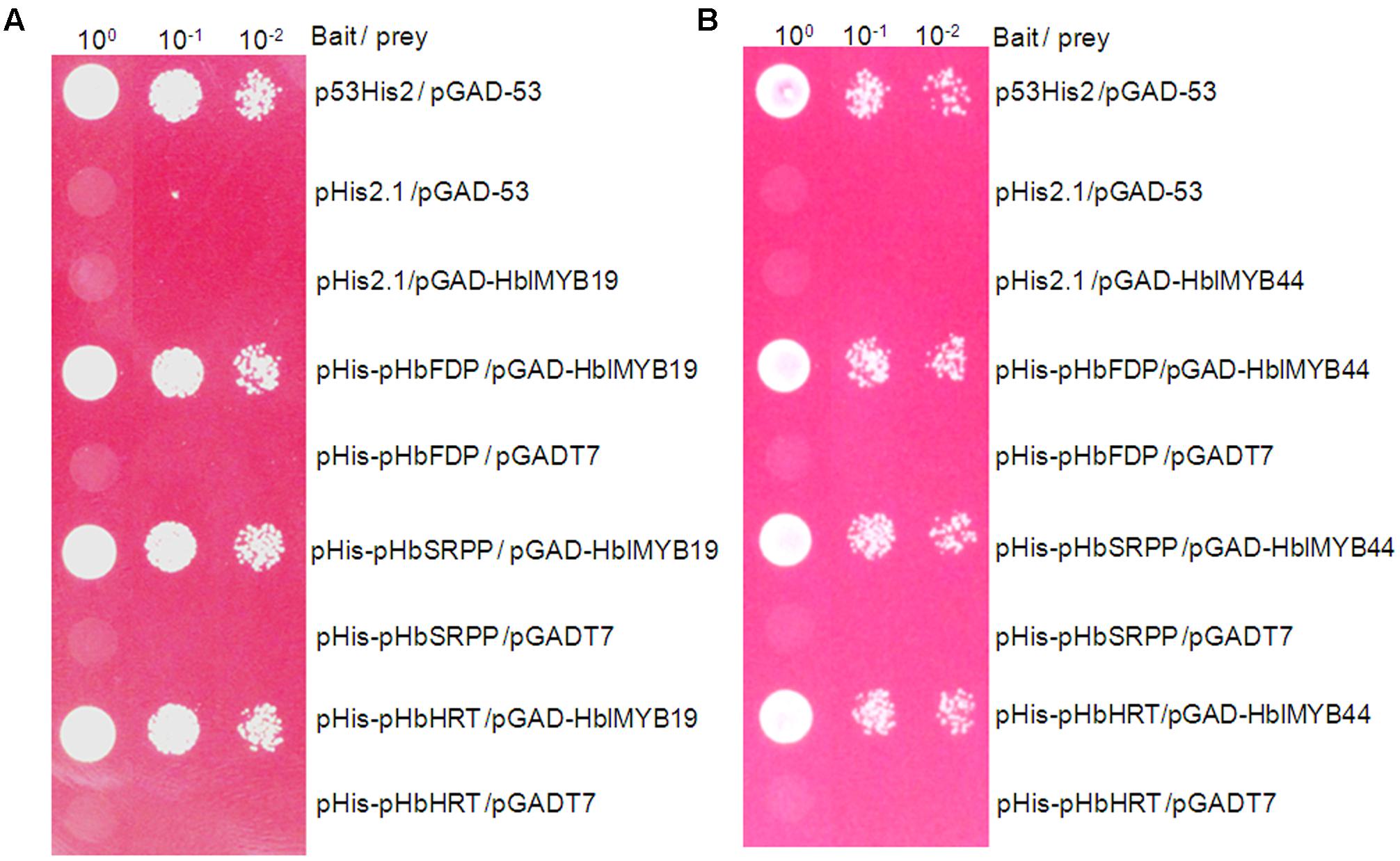
FIGURE 7. Activation of the promoters of HbFDPS1, HbSRPP, and HRT1 by HbMYB19 (A) and HbMYB44 (B) in yeast. Yeast cells carrying the bait vector and prey vector were grown in SD/-Leu selective medium containing 470 mM AbA at 30°C for 3 days.
Given the interaction between yeast HblMyb19 and HblMyb44 and the promoters of HRT1, HbSRPP, and HbFDPS1, we investigated whether HblMyb19 and HblMyb44 participate in the regulation of the promoters of HbHRT, HbSRPP, and HbFDPS1 in plants. For this purpose, HblMyb19 and HblMyb44 were transiently expressed in tobacco by Agrobacterium-mediated transformation (Figure 8A). The luciferase activity controlled by the HblMyb19 or HblMyb44 binding of the promoters of HRT1, HbSRPP, and HbFDPS1 was elevated (Figure 8B). Moreover, the expression of HblMyb19 or HblMyb44 resulted in a significant increase in luciferase activity. The data showed that the transient expression of HblMyb19 and HblMyb44 activated the promoters of HRT1, HbSRPP, and HbFDPS1.
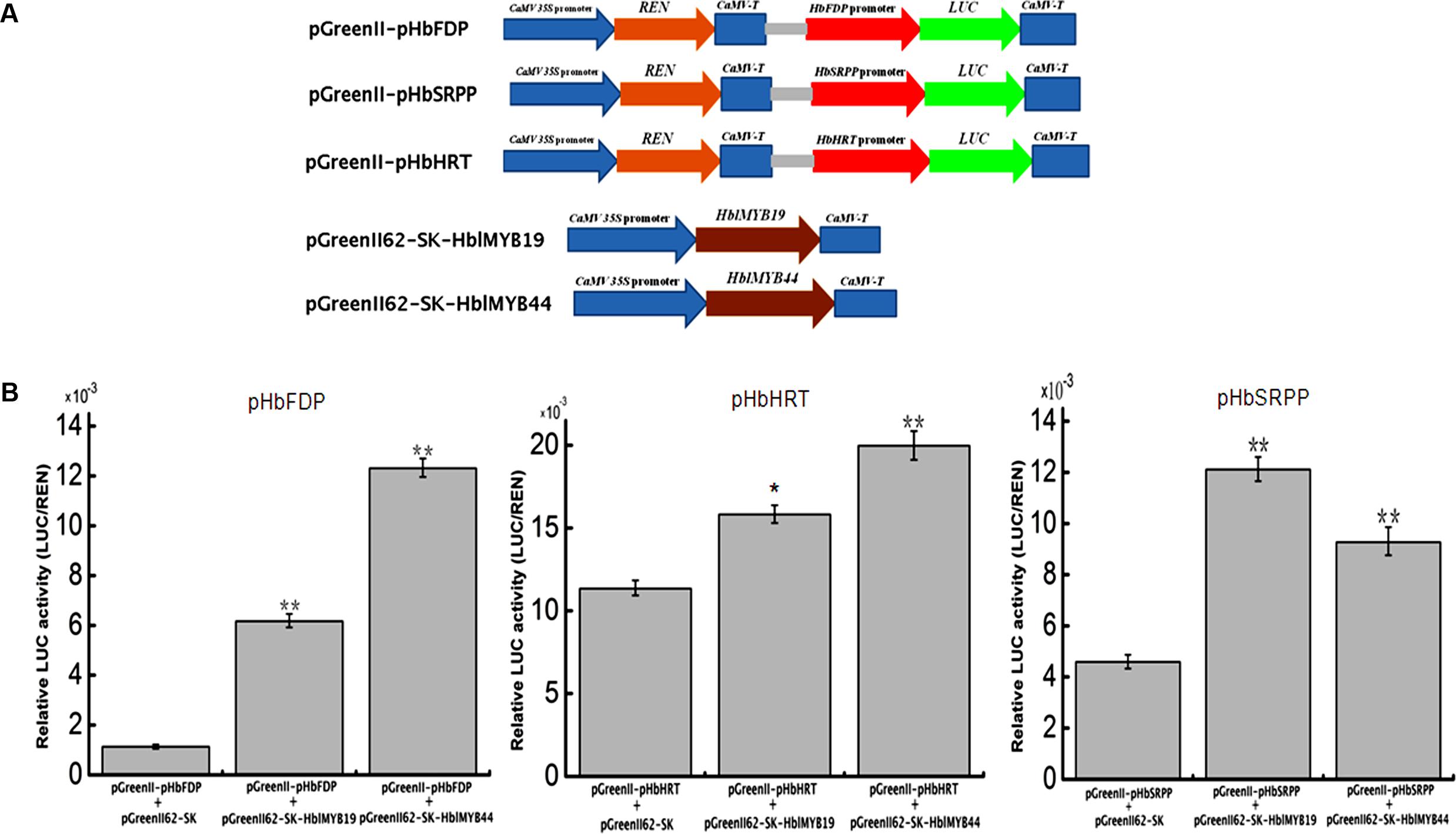
FIGURE 8. Activation of the promoters of HbFDPS1, HbSRPP, and HRT1 by HblMyb19 and HblMyb44 in a transient expression system. (A) Schematics of the transient expression vectors used in the transient expression analysis. (B) Relative LUC activity from the transient expression analysis of the promoters of HbSRPP, HRT1, and HbFDPS1 co-infiltrated with a plasmid containing the genes for HblMyb19 and HblMyb44 fused with the 35S promoter. Three replicates were included for each sample. Data are presented as mean ± SE (n = 3), and the statistical significance of the differences was assessed by ANOVA (one or two stars correspond to P < 0.05 and P < 0.01, respectively).
Plant MYBs regulate secondary metabolism (Liu et al., 2015; Chezem et al., 2016). Several flavonoid-related MYB TFs were already identified in plants. TT2, the first identified proanthocyanidin (PA)-related MYB TF, induces the biosynthesis of PAs in seed coats of A. thaliana by activating DFR, ANS, and ANR (Nesi et al., 2001). In grapevines, VvMYBA1 and VvMYBA2 are specific regulators, which activate the UFGT of the anthocyanin pathway (Kobayashi et al., 2002). MdMYB10 alleles are the key regulatory factors during the coloration of apple fruits (Takos et al., 2006; Espley et al., 2007). VvMYBPA2 was identified as a direct regulator of several structural flavonoid pathway genes in grapevines (Terrier et al., 2009). MdMYB3 activates some flavonoid-biosynthesis-related genes, such as CHI, CHS, FLS, and UFGT, in apple fruits (Vimolmangkang et al., 2013). In strawberry, FaMYB10 regulates the anthocyanin-pathway-related genes, including most of the EBGs and LBGs in ripened fruit receptacles during ripening (Medina-Puche et al., 2014). Few MYB genes from H. brasiliensis have been reported. Overexpressed HbMyb1 in tobacco suppresses stress-induced cell death (Peng et al., 2011). Another MYB gene down-regulated in trees with tapping panel dryness was identified from the SSH library (Venkatachalam et al., 2007). The MYB gene was significantly induced by ET, ABA, JA, SA, and wounding treatments (Qin et al., 2014). To date, whether MYB TFs help regulate the NR synthesis pathway in rubber trees remains unknown. NR is synthesized from the precursor IPP (Archer and Audley, 1987; Madhavan et al., 1989; Cornish and Backhaus, 1990; Chow et al., 2007, 2012; Sando et al., 2008). During NR biosynthesis, FDPS and HRT, along with SRPP, are critical to NR biosynthesis and frequently used to determine the efficiency of a process (Light et al., 1989; Oh et al., 1999; Asawatreratanakul et al., 2003). However, the regulatory mechanism of rubber biosynthesis is incompletely understood (Yamashita et al., 2016; Tang et al., 2016). HbWRKY1 was identified as a negative transcription regulator of HbSRPP (Wang et al., 2013). Other WRKY proteins may participate in the regulation of NR biosynthesis (Li et al., 2014). HbMADS4 was also found as a negative transcriptional regulator of HbSRPP (Li et al., 2016b). HbCZF1, a CCCH-type zinc-finger protein, highly activates the hmg1 promoter, and HbCZF1 may help regulate NR biosynthesis (Guo et al., 2015). In the present study, HblMyb19 and HblMyb44 bind the promoters of HRT1, HbSRPP, and HbFDPS1 in yeast. Moreover, HblMyb19 and HblMyb44 activated the promoters of HRT1, HbSRPP, and HbFDPS1 in plants. These results strongly indicated that HRT1, HbSRPP, and HbFDPS1 are the target genes of HblMyb19 and HblMyb44, and HblMyb19 and HblMyb44 are transcriptional activators of HRT1, HbSRPP, and HbFDPS1. Moreover, JA signaling may regulate NR biosynthesis in laticifers (Zeng et al., 2009; Pirrello et al., 2014). The induction of the expression of HblMyb19 and HblMyb44 by MeJA showed that HblMyb19 and HblMyb44 may play a role in the JA signaling pathway. Additionally, HRT1, HbSRPP, and HbFDPS1 can be utilized to increase the NR content in rubber tree. As a result, upregulating HRT1, HbSRPP, and HbFDPS1 can improve NR productivity in the transgenic plants. The identification and characterization of the NR-biosynthesis-related MYB TFs would greatly help increase the understanding of the molecular mechanism of NR metabolism.
In the present study, 44 MYB genes (named HblMYB1 to HblMYB44) were identified form rubber tree laticifer cells, and we found that five genes were highly expressed in laticifers. HblMYB19 and HblMYB44 bind the promoter of HbSRPP, HRT1, and HbFDPS1 in yeast. Furthermore, the transient over-expression of HblMYB19 and HblMYB44 in tobacco plants significantly increased the activity of the promoters of HbSRPP, HRT1, and HbFDPS. Basing on all the above-mentioned information, we propose that HblMYB19 and HblMYB44 are regulators of HbSRPP, HRT1, and HbFDPS1, all of which participate in NR biosynthesis.
S-QP and YW designed the research, YW, D-FZ, H-LL, DG, and J-HZ performed the research, and YW and S-QP wrote the paper. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by National Natural Science Foundation of China (No. 31670611), Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630052016003).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01974/full#supplementary-material
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Archer, B. L., and Audley, B. G. (1987). New aspects of rubber biosynthesis. Bot. J. Linn. Soc. 94, 181–196. doi: 10.1111/j.1095-8339.1987.tb01045.x
Asawatreratanakul, K., Zhang, Y. W., Wititsuwannaku, D., Wititsuwannaku, R., Takahashi, S., Rattanapittayaporn, A., et al. (2003). Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. Eur. J. Biochem. 270, 4671–4680. doi: 10.1046/j.1432-1033.2003.03863.x
Cai, H., Tian, S., Dong, H., and Guo, C. (2015). Pleiotropic effects of TaMYB3R1 on plant development and response to osmotic stress in transgenic Arabidopsis. Gene 558, 227–234. doi: 10.1016/j.gene.2014.12.066
Chezem, W. R., and Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Chezem, W. R., Nicole, K., and Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Chow, K. S., Mat-Isa, M. N., Bahari, A., Ghazali, A. K., Alias, H., Mohd-Zainuddin, Z., et al. (2012). Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J. Exp. Bot. 63, 1863–1871. doi: 10.1093/jxb/err363
Chow, K. S., Wan, K. L., Isa, M. N., Bahari, A., Tan, S. H., Harikrishna, K., et al. (2007). Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 58, 2429–2440. doi: 10.1093/jxb/erm093
Cominelli, E., and Tonelli, C. (2009). A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 19, 1231–1232. doi: 10.1038/cr.2009.123
Cornish, K., and Backhaus, R. A. (1990). Rubber transferase activity in rubber particles of guayule. Phytochemistry 29, 3809–3813. doi: 10.1016/0031-9422(90)85337-F
Dai, X., Xu, Y., Ma, Q., Xu, W., Wang, T., Xue, Y., et al. (2007). Overexpression of an R1R2R3MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751. doi: 10.1104/pp.106.094532
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Guo, D., Li, H. L., and Peng, S. Q. (2010). Cloning and function analysis of 5′ regulatory region for HbFPS from Hevea brasiliensis. Trop. Agric. Eng. 34, 33–36.
Guo, D., Li, H. L., Tang, X., and Peng, S. Q. (2014). Molecular and functional characterization of the HbSRPP promoter in response to hormones and abiotic stresses. Transgenic Res. 23, 331–340. doi: 10.1007/s11248-013-9753-0
Guo, D., Yi, H. Y., Li, H. L., Liu, C., Yang, Z. P., and Peng, S. Q. (2015). Molecular characterization of HbCZF1, a Hevea brasiliensis CCCH-type zinc finger protein that regulates hmg1. Plant Cell Rep. 34, 1569–1578. doi: 10.1007/s00299-015-1809-6
Hao, B. Z., and Wu, J. L. (2000). Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. doi: 10.1006/anbo.1999.0995
He, Q., Jones, D. C., Li, W., Xie, F., Ma, J., Sun, R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 6:22980. doi: 10.1038/srep22980
Hellens, R. G., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Hou, X. J., Li, S. B., Liu, S. R., Hu, C. G., and Zhang, J. Z. (2014). Genome-wide classification and evolutionary and expression analyses of citrus MYB transcription factor families in sweet orange. PLOS ONE 9:e112375. doi: 10.1371/journal.pone.0112375
Huang, C., Hu, G., Li, F., Li, Y., Wu, J., and Zhou, X. (2013). NbPHAN, a MYB transcriptional factor, regulates leaf development and affects drought tolerance in Nicotiana benthamiana. Physiol. Plant. 149, 297–309. doi: 10.1111/ppl.12031
Jiang, C., Gu, J., Chopra, S., Gu, X., and Peterson, T. (2004). Ordered origin of the typical two- and three-repeat Myb genes. Gene 326, 13–22. doi: 10.1016/j.gene.2003.09.049
Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. doi: 10.1023/A:1006319732410
Kanei-Ishii, C., Sarai, A., Sawazaki, T., Nakagoshi, H., He, D. N., Ogata, K., et al. (1990). The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 265, 19990–19995.
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Ko, J. H., Chow, K. S., and Han, K. H. (2003). Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree). Plant Mol. Biol. 5, 479–492. doi: 10.1023/B:PLAN.0000019119.66643.5d
Kobayashi, S., Ishimaru, M., Hiraoka, K., and Honda, C. (2002). Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215, 924–933. doi: 10.1007/s00425-002-0830-5
Kush, A., Goyvaerts, E., Chye, M. L., and Chua, N. H. (1990). Laticifer-specific gene expression in Hevea brasiliensis (rubber tree). Proc. Natl. Acad. Sci. U.S.A. 87, 1787–1790. doi: 10.1073/pnas.87.5.1787
Li, H. L., Guo, D., Yang, Z. P., Tang, X., and Peng, S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 104, 14–23. doi: 10.1016/j.ygeno.2014.04.004
Li, H. L., Guo, D., Zhu, J. H., Wang, Y., Chen, X. T., and Peng, S. Q. (2016a). Comparative transcriptome analysis of latex reveals molecular mechanisms underlying higher rubber yield in Hevea brasiliensis self-rooting juvenile clones. Front. Plant Sci. 7:1204. doi: 10.3389/fpls.2016.01204
Li, H. L., Wang, Y., Guo, D., Tian, W. M., and Peng, S. Q. (2011). Three MADS-box genes of Hevea brasiliensis expressed during somatic embryogenesis and in the laticifer cells. Mol. Biol. Rep. 38, 4045–4052. doi: 10.1007/s11033-010-0523-2
Li, H. L., Wei, L. R., Guo, D., Wang, Y., Zhu, J. H., Chen, X. T., et al. (2016b). HbMADS4, a MADS-box transcription factor from Hevea brasiliensis, negatively regulates HbSRPP. Front. Plant Sci. 7:1709. doi: 10.3389/fpls.2016.01709
Light, D. R., Lazarus, R. A., and Dennis, M. S. (1989). Rubber elongation by farnesyl pyrophosphate synthases involves a novel switch in enzyme stereospecificity. J. Biol. Chem. 264, 18598–18607.
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Liu, X., Yang, L., Zhou, X., Zhou, M., Lu, Y., Ma, L., et al. (2013). Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J. Exp. Bot. 64, 2243–2253. doi: 10.1093/jxb/ert084
Madhavan, S., Greenblatt, G. A., Foster, M. A., and Benedict, C. R. (1989). Stimulation of isopentenyl pyrophosphate incorporation into polyisoprene in extracts from guayule plants (Parthenium argentatum Gray) by low temperature and 2-(3,4-dichlorophenoxy) triethylamine. Plant Physiol. 89, 506–511. doi: 10.1104/pp.89.2.506
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114.
Oh, J. E., Kwon, Y., Kim, J. H., Noh, H., Hong, S. W., and Lee, H. (2011). A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 77, 91–103. doi: 10.1007/s11103-011-9796-7
Oh, S. K., Kang, H., Shin, D. H., Yang, J., Chow, K. S., Yeang, H. Y., et al. (1999). Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J. Biol. Chem. 274, 17132–17138. doi: 10.1074/jbc.274.24.17132
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P., and Saedler, H. (1987). The regulatory c1locus of Zea mays encodes a protein with homology to MYB oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558.
Peng, S. Q., Wu, K. X., Huang, G. X., and Chen, S. C. (2011). HbMyb1, a Myb transcription factor from Hevea brasiliensis, suppresses stress induced cell death in transgenic tobacco. Plant Physiol. Biochem. 49, 1429–1435. doi: 10.1016/j.plaphy.2011.09.007
Pirrello, J., Leclercq, J., Dessailly, F., Rio, M., Piyatrakul, P., Kuswanhadi, K., et al. (2014). Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 14:341. doi: 10.1186/s12870-014-0341-0
Qin, B., Zhang, Y., and Wang, M. (2014). Molecular cloning and expression of a novel MYB transcription factor gene in rubber tree. Mol. Biol. Rep. 41, 8169–8176. doi: 10.1007/s11033-014-3717-1
Quirk, T. J., Quirk, M. H., and Horton, H. F. (2016). “One-way analysis of variance (ANOVA),” in Excel 2016 for Physical Sciences Statistics, eds T. J. Quirk, M. H. Quirk, and H. F. Horton (Berlin: Springer), 171–187. doi: 10.1007/978-3-319-40075-4_8
Rahman, A. Y. A., Usharraj, A. O., Misra, B. B., Thottathil, G. P., Jayasekaran, K., Feng, Y., et al. (2013). Draft genome sequence of the rubber tree (Hevea brasiliensis). BMC Genomics 14:75. doi: 10.1186/1471-2164-14-75
Rosinski, J. A., and Atchley, W. R. (1998). Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J. Mol. Evol. 46, 74–83. doi: 10.1007/PL00006285
Sando, T., Takaoka, C., Mukai, Y., Yamashita, A., Hattori, M., Ogasawara, N., et al. (2008). Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem 72, 2049–2060. doi: 10.1271/bbb.80165
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi: 10.1105/tpc.107.050963
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tang, C., Qi, G., Li, H., Zhang, C., and Wang, Y. (2007). A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (para rubber tree). J. Biochem. Biophys. Methods 70, 749–754. doi: 10.1016/j.jbbm.2007.04.002
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2, 16073. doi: 10.1038/nplants.2016.73
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verriès, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Venkatachalam, P., Thulaseedharan, A., and Raghothama, K. (2007). Identification of expression profiles of tapping panel dryness (TPD) associated genes from the latex of rubber tree (Hevea brasiliensis Muell. Arg.). Planta 226, 499–515. doi: 10.1007/s00425-007-0500-8
Vimolmangkang, S., Han, Y., Wei, G., and Korban, S. S. (2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13:176. doi: 10.1186/1471-2229-13-176
Wang, Y., Guo, D., Li, H. L., and Peng, S. Q. (2013). Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 71, 283–289. doi: 10.1016/j.plaphy.2013.07.020
Wang, Z., Tang, J., Hu, R., Wu, P., Hou, X. L., Song, X. M., et al. (2015). Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genomics 16:17. doi: 10.1186/s12864-015-1216-y
Yamashita, S., Yamaguchi, H., Waki, T., Aoki, Y., Mizuno, M., Yanbe, F., et al. (2016). Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. eLife 5:e19022. doi: 10.7554/eLife.19022
Yang, Z. P., Li, H. L., Guo, D., and Peng, S. Q. (2015). Identification and characterization of the EIN3/EIL gene family in Hevea brasiliensis. Tree Genet. Genomes 11, 51. doi: 10.1007/s11295-015-0877-7
Zeng, R. Z., Duan, C. F., Li, X. Y., Tian, W. M., and Nie, Z. Y. (2009). Vacuolar-type inorganic pyrophosphatase located on the rubber particle in the latex is an essential enzyme in regulation of the rubber biosynthesis in Hevea brasiliensis. Plant Sci. 176, 602–607. doi: 10.1016/j.plantsci.2009.01.009
Zhai, R., Wang, Z., Zhang, S., Meng, G., Song, L., Wang, Z., et al. (2016). Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.). J. Exp. Bot. 67, 1275–1284. doi: 10.1093/jxb/erv524
Zhang, Z., Chen, J., Su, Y., Liu, H., Chen, Y., Luo, P., et al. (2015). TaLHY, a 1R-MYB transcription factor, plays an important role in disease resistance against stripe rust fungus and ear heading in wheat. PLOS ONE 10:e0127723. doi: 10.1371/journal.pone.0127723
Zhao, Y., Xing, L., Wang, X., Hou, Y. J., Gao, J., Wang, P., et al. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7, ra53. doi: 10.1126/scisignal.2005051
Keywords: Hevea brasiliensis, MYB-type transcription factor, laticifer, natural rubber, biosynthesis
Citation: Wang Y, Zhan D-F, Li H-L, Guo D, Zhu J-H and Peng S-Q (2017) Transcriptome-Wide Identification and Characterization of MYB Transcription Factor Genes in the Laticifer Cells of Hevea brasiliensis. Front. Plant Sci. 8:1974. doi: 10.3389/fpls.2017.01974
Received: 22 May 2017; Accepted: 01 November 2017;
Published: 15 November 2017.
Edited by:
Wanchai De-Eknamkul, Chulalongkorn University, ThailandCopyright © 2017 Wang, Zhan, Li, Guo, Zhu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Qing Peng, c2hxcGVuZ0AxNjMuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.