- 1Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
- 2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kongens Lyngby, Denmark
Malaria is still an eminent threat to major parts of the world population mainly in sub-Saharan Africa. Researchers around the world continuously seek novel solutions to either eliminate or treat the disease. Artemisinin, isolated from the Chinese medicinal herb Artemisia annua, is the active ingredient in artemisinin-based combination therapies used to treat the disease. However, naturally artemisinin is produced in small quantities, which leads to a shortage of global supply. Due to its complex structure, it is difficult chemically synthesize. Thus to date, A. annua remains as the main commercial source of artemisinin. Current advances in genetic and metabolic engineering drives to more diverse approaches and developments on improving in planta production of artemisinin, both in A. annua and in other plants. In this review, we describe efforts in bioengineering to obtain a higher production of artemisinin in A. annua and stable heterologous in planta systems. The current progress and advancements provides hope for significantly improved production in plants.
Introduction
Malaria is still a global concern with around 214 million annual cases and 430,000 annual deaths, mainly among of children younger than 5 (World Health Organization [WHO], 2016). This fatal disease is caused by Plasmodium sp. particularly Plasmodium falciparum that proliferate in female Anopheles mosquitoes (Cox, 2010). Since the 1940s there has been continuous attempts to halt the spread of the disease and this has succeeded in Europe, North America, and parts of Asia and Latin America (Carter and Mendis, 2002). However, not in Sub-Saharan Africa where 80% of the annual malaria patients are found. Besides measures such as vector control and insecticide-treated nets, research and development has led to new drugs and a vaccine. The current preferred therapy is artemisinin combination therapy (ACT) (Banek et al., 2014; Lalloo et al., 2016) that is based on artemisinin produced in the natural source Artemisia annua. Artemisinin can also be produced heterologously in the plants Nicotiana benthamiana and Physcomitrella patens (Han et al., 2016; Wang et al., 2016; Ikram et al., 2017). The vaccine toward Plasmodium is called PfSPZ and can be produced in N. benthamiana and P. patens plants (Rosales-Mendoza et al., 2014, 2017; Boes et al., 2016; Epstein et al., 2017).
Malaria drugs have contributed significantly to the reductions in malaria mortality and morbidity. The focus for many years has been to screen traditional medicine to find new antimalarial drugs (Simonsen et al., 2001; Adia et al., 2016; Nondo et al., 2017). The malaria drug artemisinin is an example of this and originates from A. annua, a Chinese medicinal plant (Qinghao), commonly known as sweet wormwood. It was discovered by the Chinese researcher You-You Tu and her team in 1972, and was named Qinghaosu (Klayman, 1985; Tu, 2011). Chemically, artemisinin is a sesquiterpene lactone with a unique endoperoxide structure, without the nitrogen containing heterocyclic ring like other antimalarial compounds (Luo and Shen, 1987). The in planta accumulation of artemisinin is 0.01–1.4% dry weight depending on the plant variety and artemisinin is stored in the glandular trichomes of A. annua (Duke et al., 1994; Van Agtmael et al., 1999; Bhakuni et al., 2001; Muangphrom et al., 2016). The current production using plants with a “low” content of artemisinin can only just cover the global need, which have led to an increase in price (Peplow, 2016). In 2006, World Health Organization (WHO) recommended artemisinin as the first-choice treatment for malaria. Rapid emergence of antimalarial drug resistance drew attention to formulation of artemisinin-based combination therapy (ACT) with artemisinin as the primary substance and is now the preferred treatment (World Health Organization [WHO], 2015).
To secure the global need of artemisinin, there are continuous and extensive efforts to enhance the production of artemisinin in the native plant A. annua. A. annua is currently the primary commercial source of artemisinin and significant breeding programs has contributed to higher artemisinin content in the plant (Ma et al., 2015; Pulice et al., 2016; Xie et al., 2016), including establishment of mutant libraries (Pandey et al., 2016). Several plant-breeding techniques have been applied to create superior cultivars of A. annua. For example, conventional breeding by crossing A. annua with high artemisinin content in wild population has led to hybrid lines with 2% artemisinin d.w (Delabays et al., 2001; Cockram et al., 2012). A detailed genetic map of A. annua comprising of genes and markers controlling artemisinin yield has been established to generate robust high yielding crops (Graham et al., 2010). Identification of A. annua superior parental lines with desired traits from these genetic maps has provided two high-yielding hybrids and diallel crossing of the parental lines and the hybrids has showed consistent results for the development of improved A. annua hybrids (Townsend et al., 2013). Doubling the number of chromosomes generated a new variety of tetraploid cultivar with higher artemisinin content and this might become a new elite line (Banyai et al., 2010b). The overall production of the new cultivars from various laboratories have increased the level of artemisinin to about 1 to 2% d.w. (Delabays et al., 1993; Ferreira et al., 2005; Graham et al., 2010; Brisibe et al., 2012), but not all the established plant lines are stable over generations (Delabays et al., 2001).
Efforts in plant breeding have been challenging due to the heterozygous nature of A. annua, which results in transgenic plants with varying degrees of artemisinin content even though they were generated in the same laboratory (Delabays et al., 2001; Graham et al., 2010; Larson et al., 2013). This variation is due to the segregation of the heterozygous wild type progeny leading to a different genetic background than the parent plant. Although several high content lines have been created, the unstable yield in the progeny of these cultivars were insufficient to increase the global supply of artemisinin (Shretta and Yadav, 2012; Paddon et al., 2013).
Accumulation of artemisinin in A. annua is limited to the small 10 cell glandular trichomes (GT) mostly on leaves and other aerial parts (Ferreira and Janick, 1995; Lommen et al., 2006; Ling et al., 2016). Low GT numbers are correlated to low artemisinin content (Graham et al., 2010; Kjær et al., 2012). Attempts to increase the number of GTs by physical and chemical stress have not been successful (Kjær et al., 2012). One study expressed the β glucosidase (bgl1) gene in A. annua through Agrobacterium-mediated transformation, which resulted in an increase of GT density by 20% on leaves and 66% on flowers and an increase in artemisinin content of 1.4% in leaves and 2.56% in flowers (d.w). Manipulating GT density together with biosynthetic pathway engineering may further increase artemisinin content in A. annua. In depth understanding of A. annua GT generation at the molecular level, will broaden the opportunities of increasing the artemisinin production. This approximately though require a greater acceptance of GMO crops in open fields to ensure the global supply.
Plant tissue culture has also been investigated to establish a production of artemisinin in A. annua hairy root or cell suspension cultures (Nair et al., 1986; Baldi and Dixit, 2008). Several manipulations of the growth conditions such as different sugar supply, light irradiation, UV-B radiation and chilling treatment have led to production of artemisinin in A. annua tissue cultures (Woerdenbag et al., 1993; Liu et al., 2002; Wang and Weathers, 2007; Baldi and Dixit, 2008; Yin et al., 2008; Pandey and Pandey-Rai, 2014). Generating somaclonal variants tolerant against salt stress through gamma-rays irradiation has resulted in 13 somaclonal variants (ASV1 to ASV13) of which one of the variants, ASV12 is a stable salt-tolerant line with a higher expression profile of artemisinin key genes (ADS, CYP71AV1, DBR2, and ALDH1) and a higher artemisinin content as compared to wild type. In addition, treatments with elicitors such as methyl jasmonate has significantly increased artemisinin production by up to 49% including up-regulating the expression of artemisinin biosynthesis genes as well as increased GT index (0.128) (Baldi and Dixit, 2008; Wang et al., 2010; Dangash et al., 2014; Xiang et al., 2015). Other elicitors such as chitosan, gibberellic acid, and salicylic acid also aid in the accumulation of artemisinin (Guo et al., 2010; Banyai et al., 2011). Combinations of various cultivation and elicitation methods are currently being geared for a mass production of artemisinin in A. annua hairy roots via bioreactors with 6.3 g/L dry weight (37.50 g fresh weight) biomass and 0.32 mg/g artemisinin content after 25 days (Patra and Srivastava, 2017).
Other efforts to enhance artemisinin production have been attempted through genetic engineering of the artemisinin biosynthetic pathway genes in microbial heterologous hosts. Extensive work on the development of microbial production of artemisinin precursors led to semi-synthesis of artemisinin, but this is only partly commercially successful (Benjamin et al., 2016; Peplow, 2016; Singh et al., 2017). In this review, the progress and recent bioengineering advances in artemisinin production in stable heterologous in planta systems including genetic modifications of A. annua is summarized.
Artemisinin Biosynthesis in Artemisia annua
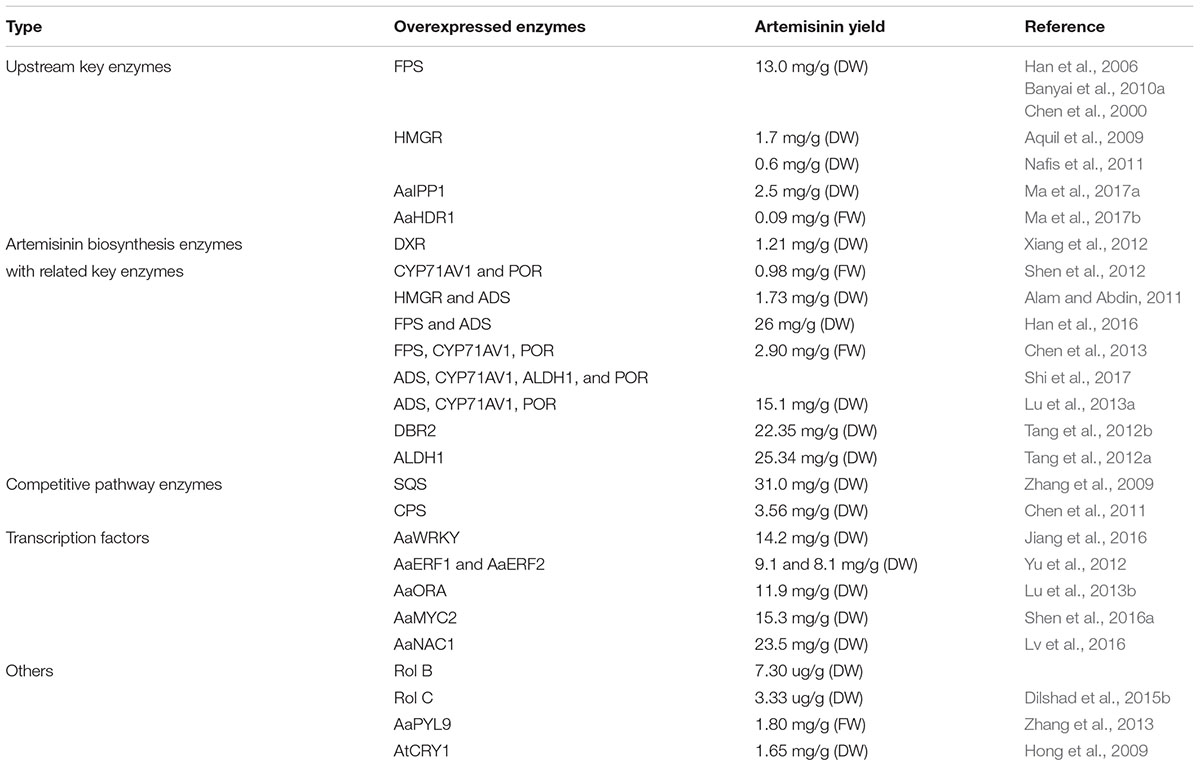
The biosynthesis of Artemisinin (Figure 1) has been explored for many years. However, not every detail about the regulation and biosynthesis is completely understood, but the discovery that the whole biosynthesis is located in the glandular trichomes of A. annua has facilitated in-depth regulatory studies (Olsson et al., 2009; Olofsson et al., 2011). Derived from the general terpenoid biosynthesis, two molecules of isopentenyl diphosphate (IPP) and one dimethylallyl diphosphate (DMAPP) are condensed by farnesyl diphosphate synthase (FPPS/FPS) into farnesyl diphosphate (FPP, farnesyl pyrophosphate), the C15 sesquiterpenoid precursor (Weathers et al., 2006; Brown, 2010; Wen and Yu, 2011). Overexpression of FPS in A. annua resulted in an increase of artemisinin production (Han et al., 2006; Banyai et al., 2010a), which confirms the role of FPS and availability of the substrates in the regulation of artemisinin biosynthesis similar to other sesquiterpene lactones (Simonsen et al., 2013).
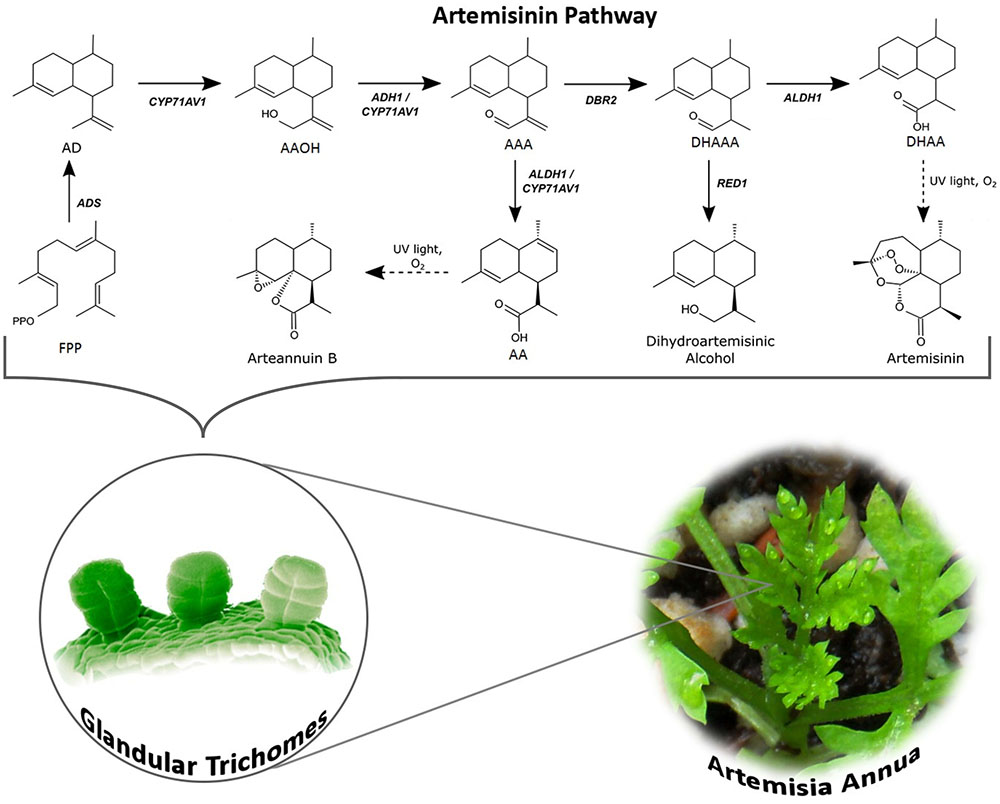
FIGURE 1. Artemisinin biosynthesis pathway occurs in the glandular trichomes of Artemisia annua. The pathway intermediates are defined as FPP, farnesyl diphosphate; AD, amorpha-4,11-diene; AAOH, artemisinic alcohol; AAA, artemisinic aldehyde; AA, artemisinic acid; DHAAA, dihydroartemisinic aldehyde; DHAA, dihydroartemisinic acid. The full name of the enzymes is stated in the text.
FPP is converted to amorpha-4,11-diene by amorpha-4,11-diene synthase (ADS) via carbocation formation and cyclization (Bouwmeester et al., 1999; Mercke et al., 2000; Picaud et al., 2005, 2006). In the following two oxidization steps, amorpha-4,11-diene is hydroxylated into artemisinic alcohol and oxidized to artemisinic aldehyde by amorphadiene monooxygenase (CYP71AV1), a cytochrome P450 enzyme (Teoh et al., 2006; Wang et al., 2011). The activity of the CYP71AV1 has also been confirmed through a knock-out of the endogenous gene in A. annua showing that these plants do not produce any downstream products of amorphadiene (Czechowski et al., 2016). It has later been discovered that the alcohol dehydrogenase (ADH1, a dehydrogenase/reductase enzyme) is specific toward artemisinin alcohol and oxidizes this to the aldehyde. This specificity and strong expression in A. annua glandular trichomes confirms that ADH1 is responsible for oxidation of artemisinic alcohol to artemisinic aldehyde (Olofsson et al., 2011; Paddon et al., 2013; He et al., 2017). Artemisinic aldehyde is further reduced to dihydroartemisinic aldehyde by artemisinic aldehyde Δ11 (13) reductase (DBR2) and subsequently oxidized to dihydroartemisinic acid by aldehyde dehydrogenase (ALDH1), which is also expressed in the trichomes (Zhang et al., 2008; Teoh et al., 2009; Rydén et al., 2010; Liu et al., 2016). Besides catalyzing the oxidation of dihydroartemisinic aldehyde to the acid, ALDH1 also catalyzes the oxidation of artemisinic aldehyde to artemisinic acid (a reaction that in yeast is catalyzed by CYP71AV1) (Teoh et al., 2006, 2009). Another enzyme, dihydroartemisinic aldehyde reductase (RED1) converts dihydroartemisinic aldehyde to dihydroartemisinic alcohol, a “dead end” substance, which affects the production yield of artemisinin (Rydén et al., 2010). The final step is a light-induced non-enzymatic spontaneous reaction converting dihydroartemisinic acid to artemisinin and artemisinic acid to arteannuin B (Sy and Brown, 2002; Teoh et al., 2006; Czechowski et al., 2016).
Bioengineering of Artemisinin Production in Green Plant Cells
Bioengineering of Biosynthetic Genes in Artemisia annua
Characterization of enzymes in the artemisinin biosynthetic pathway provides new tools and advances the possibility of engineering the production of artemisinin. This can be achieved by enhancing the general terpenoid metabolism and through overexpression of several genes involved in artemisinin biosynthesis in A. annua (Tang et al., 2014). Overexpression of key terpenoid genes encoding for the enzymes IDI, FPS, HMGR, the plastid targeted DXR and HDR have increased production significantly (some by 2 to 3 fold) in many different studies in A. annua (Han et al., 2006; Aquil et al., 2009; Banyai et al., 2010a; Nafis et al., 2011; Xiang et al., 2012; Ma et al., 2017a). Co-expression of FPS, CYP71AV1 and its redox partner, POR (cytochrome P450 reductase) increased production by 3.6 fold, whereas combining four genes ADS, CYP71AV1, ALDH1, and POR from A. annua yielded a 3.4 fold increase in the artemisinin levels (Chen et al., 2013; Shi et al., 2017). Additionally, the production also increased by overexpression of ADS, CYP71AV1, and HMGR (Ma et al., 2009; Alam et al., 2016). The expression of several genes in the pathway clearly have an effect on the artemisinin level and do increase the amount of biomass obtained (Shen et al., 2012; Alam et al., 2016). Thus, utilizing genetic engineering to target the expression of both upstream and specific artemisinin genes should be pursued.
The overexpression of DBR2 clearly showed that this is a key enzyme that regulates the production of artemisinin by guiding the metabolic flow from artemisinic acid toward dihydroartemisinic acid. Without the activity of DBR2 the plants solely make artemisinic acid and thereby arteannuin B (Zhang et al., 2008), thus showing that overexpression of this enzyme will enhance artemisinin production. Collectively, the overexpression studies have provided insights into the understanding of the pathway and how to upregulate it.
Another strategy in bioengineering is to block competing reactions such as the squalene synthase (SQS) and β-caryophyllene synthase, enzymes consuming FPP for sterol and β-caryophyllene biosynthesis. This has been proven to elevate artemisinin production by 3.14 and 5.49 fold, respectively (Zhang et al., 2009; Chen et al., 2011). Since RED1 competes with ALDH1 in artemisinin biosynthesis of A. annua, removing RED1 could also lead to the increase of artemisinin production in A. annua (Rydén et al., 2010).
FPS in general has a higher kcat value than sesquiterpene synthases and this is true for the FPS and ADS in A. annua. Thus, it has been investigated whether a fusion of the two enzymes would increase the turnover of FPP to amorphadiene (Han et al., 2016). The findings that such fusion can facilitate metabolite channeling through a biosynthesis pathway has recently been shown for other metabolites (Laursen et al., 2016). The metabolite channeling from FPS to ADS is supported by a 2–3 fold increase of amorphadiene in plants where these two genes are fused (Han et al., 2016). The dynamic artemisinin content in the transgenic and wild type plants is associated with the expression of these genes involved in the artemisinin pathway. Table 1 summarizes the work on genetic manipulation in A. annua to improve the production of artemisinin.
Bioengineering the Regulation of Artemisinin Biosynthesis
Over the last 20 years Agrobacterium rol A, B, and C genes have been shown to increase the biosynthesis of stress response metabolites in different plant families (Bulgakov, 2008). Rol genes are a potential activator of secondary metabolites which directly upregulate artemisinin production by induction of the gene expression, leading to higher amounts of enzymes and thus more products. Transformation of rol genes in other Artemisia sp. resulted not only in the overexpression of artemisinin pathway genes, but also artemisinin content in the plant (Dilshad et al., 2015a,b; Amanullah et al., 2016). Integration of individual and combined rol B and C genes in A. annua increases the production of artemisinin by up to ninefold (Ghosh et al., 1997; Dilshad et al., 2015a,b).
Identifying transcription factors involved in regulating artemisinin production has also contributed to a higher production of artemisinin and was recently reviewed (Shen et al., 2016b). Overexpression of the transcription factor AaWRKY1 shows a 4.4 fold increase of artemisinin compared to the control plant. Overexpression of another transcription factor jasmonate-responsive AP2/ERF-type; AaERF1 and AaERF2 increases the gene expression levels of ADS, CYP71AV1, and DBR2 resulting in a higher accumulation of artemisinin and artemisinic acid in A. annua (Shen et al., 2016b). What is clear from recent work is that there are parts of the artemisinin pathway, which have promoters that are specific for trichomes (Chen et al., 2017). Therefore changing these to a strong constitutive promoter might be a novel engineering target with CRISPR/Cas9 technology.
Metabolic Engineering in Nicotiana spp.
Introducing artemisinin pathway genes in heterologous plants has been successful in both stable and transient expression but the artemisinin yield is relatively low (Farhi et al., 2011; Zhang et al., 2011; Ting et al., 2013). Currently, only Nicotiana spp. has been used as the plant alternative in the artemisinin research as it is cheap, well-established with rapid growth and high biomass. The expression of ADS in Nicotiana tabacum resulted in an increased production of the first product amorpha-4,11-diene (Wallaart et al., 2001). The addition of CYP71AV1, DBR2, and ALDH1 produced 4 mg/g fresh weight of amorph-4,11-adiene in leaves followed by 0.01 mg/g dry weight of artemisinic alcohol (Zhang et al., 2011). Stable expression of five multiple genes from the MVA and artemisinin pathway constructed in a single vector into N. tabacum produces 0.48–6.8 μg/g dry weight of artemisinin (Farhi et al., 2011). However, transient expression combining ADS, FPS, HMGR, and CYP71AV1 in N. benthamiana produced artemisinic acid that was further modified by endogenous glycosyl transferase into artemisinic acid-12-β-diglucoside (Van Herpen et al., 2010). There is a high production of glycosylated artemisinin precursors with the expression of artemisinin genes in N. benthamiana (Ting et al., 2013).
Glycosylation is a problem in the Nicotiana spp. (Van Herpen et al., 2010; Ting et al., 2013). To overcome this, attempts were made to target the biosynthesis into different cellular compartments such as the chloroplast. Fuentes et al. (2016) introduced the artemisinin pathway into N. tabacum chloroplast via a stable plastid genome transformation followed by a combinatorial transformation resulting in a transformation of transplastomic recipient lines (COSTREL) that produces 120 μg/g artemisinic acid.
Another group aimed to engineer two mega-metabolic pathways separately into two different cellular compartments. They first elevated the IPP pools by introducing six genes from MVA pathway into N. tabacum chloroplast followed by the artemisinin pathway genes into the nuclear genome with subcellular targeting at DBR2, CPR, and CYP71AV1 via chloroplast transit peptide. The lines produced ∼0.8 mg/g dry weight of artemisinin (Malhotra et al., 2016). While various methods were explored in order to enhance artemisinin production in Nicotiana, the production levels remain minimal due to the complex nature of the gene expression and regulation in artemisinin biosynthesis pathway as well as the complex glycosylation response in Nicotiana.
Metabolic Engineering in Physcomitrella Patens
A new production platform is being established in a non-vascular plant, the moss P. patens (Simonsen et al., 2009; Buttner-Mainik et al., 2011; Ikram et al., 2015; Reski et al., 2015). Having unique molecular tools of highly efficient homologous recombination and a fully sequenced genome, P. patens is an attractive production system when compared to other plant production hosts (Schaefer and Zrÿd, 1997; Reski, 1998; Frank et al., 2005). Additionally, a novel transformation technology involving in vivo assembly of multiple DNA fragments in P. patens has been established, further increasing its attractiveness as a promising photosynthetic chassis for synthetic biology (King et al., 2016). This is also supported by several works utilizing P. patens as a “green factory,” for example, the expression of taxadiene synthase in P. patens produces taxadiene without any phenotypic change, making it a capable host for the production of paclitaxel (Anterola et al., 2009). In addition, three important sesquiterpenoids in the fragrance industry, patchoulol, β-santalene, and sclareol was also successfully produced in P. patens with productivity up to 1.3, 0.039, and 2.84 mg/g dry weight respectively (Zhan et al., 2014; Pan et al., 2015). We recently reported the successful production of artemisinin in P. patens (Ikram et al., 2017). All five artemisinin pathway genes were introduced into P. patens via the in vivo assembly of multiple DNA fragments method and the transgenic P. patens lines produces 0.21 mg/g DW of artemisinin, a significant level at only 3 days of culturing. A considerable advantage of P. patens as an artemisinin production platform is the absence of pathway intermediates (glycosylation and glutathione conjugation). P. patens has less glycosyltransferases as compared to higher plants that may lead to the possibility of lower risk of endogenous modifications to xenobiotic metabolites. Further research in bioengineering of P. patens for a higher artemisinin production is ongoing and could potentially help stabilize the supply of artemisinin and aid in containing malaria. Bioengineering of artemisinin biosynthesis pathway in heterologous in planta host with successful production of artemisinic acid and artemisinin is summarized in Table 2.
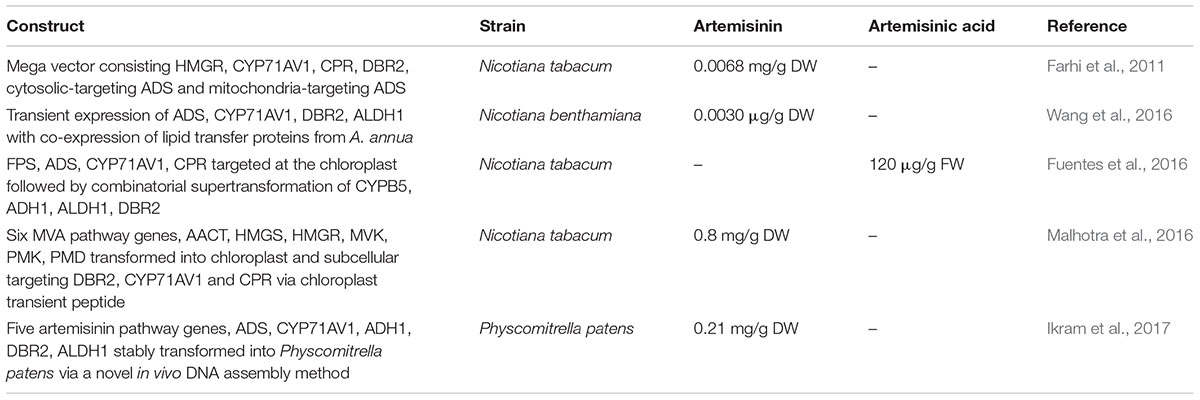
TABLE 2. Compilation of bioengineering works in heterologous in planta host producing artemisinin and artemisinic acid.
Perspectives
Current advances in genetic and metabolic engineering drive a more diverse research and development approach on improving in planta production of artemisinin. The successes achieved in heterologous plant hosts and engineering of A. annua remains are of great importance. Microbial engineering of artemisinic acid shows some potential, but the added costs for later chemical synthesis of artemisinin is a detracting factor for replacing A. Annua as the main artemisinin source. Progress in plant engineering and synthetic biology has significantly improved the awareness of using plant as production hosts leading to great efforts in the implementation and enhancement of artemisinin production in both in vivo and in vitro production. Furthermore, heterologous in planta production seems to be more cost effective and environmentally friendly than other current biotechnological platforms. Advances in multigene transformation, transcription factors along with targeting of cellular compartment techniques will enable elevation of production levels in future engineered plants bringing us closer to industrial scale plant factories for artemisinin production. Perhaps the continuous production of artemisinin and other valuable plant metabolites in suspended bioreactor cultures with in situ extraction to avoid cell toxicity is not too far in the future. This will avoid the regulatory restrictions on in field GMO plants, and allow for stable continues production of drugs.
Author Contributions
NI and HS collectively wrote the manuscript and initiated the work behind it. NI contributed with major parts of the literature research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
NI was supported by a grant from the Ministry of Higher Education, Malaysia and the University of Malaya.
References
Adia, M. M., Emami, S. N., Byamukama, R., Faye, I., and Borg-Karlson, A.-K. (2016). Antiplasmodial activity and phytochemical analysis of extracts from selected Ugandan medicinal plants. J. Ethnopharmacol. 186, 14–19. doi: 10.1016/j.jep.2016.03.047
Alam, P., and Abdin, M. Z. (2011). Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep. 30, 1919–1928. doi: 10.1007/s00299-011-1099-6
Alam, P., Kamaluddin, Sharaf-Eldin, M. A., Elkholy, S. F., and Abdin, M. Z. (2016). The effect of over-expression of rate limiting enzymes on the yield of artemisinin in Artemisia annua. Rendiconti Lincei 27, 311–319. doi: 10.1007/s12210-015-0481-7
Amanullah, B. M., Rizvi, Z. F., and Zia, M. (2016). Production of artemisinin and its derivatives in hairy roots of Artemisia dubia induced by rola gene transformation. Pak. J. Bot. 48, 699–706.
Anterola, A., Shanle, E., Perroud, P.-F., and Quatrano, R. (2009). Production of taxa-4(5),11(12)-diene by transgenic Physcomitrella patens. Transgenic Res. 18, 655–660. doi: 10.1007/s11248-009-9252-5
Aquil, S., Husaini, A. M., Abdin, M. Z., and Rather, G. M. (2009). Overexpression of the HMG-CoA reductase gene leads to enhanced artemisinin biosynthesis in transgenic Artemisia annua plants. Planta Med. 75, 1453–1458. doi: 10.1055/s-0029-1185775
Baldi, A., and Dixit, V. K. (2008). Yield enhancement strategies for artemisinin production by suspension cultures of Artemisia annua. Bioresour. Technol. 99, 4609–4614. doi: 10.1016/j.biortech.2007.06.061
Banek, K., Lalani, M., Staedke, S. G., and Chandramohan, D. (2014). Adherence to artemisinin-based combination therapy for the treatment of malaria: a systematic review of the evidence. Malar. J. 13:7. doi: 10.1186/1475-2875-13-7
Banyai, W., Kirdmanee, C., Mii, M., and Supaibulwatana, K. (2010a). Overexpression of farnesyl pyrophosphate synthase (FPS) gene affected artemisinin content and growth of Artemisia annua L. Plant Cell Tissue Organ Cult. 103, 255–265. doi: 10.1007/s11240-010-9775-8
Banyai, W., Sangthong, R., Karaket, N., Inthima, P., Mii, M., and Supaibulwatana, K. (2010b). Overproduction of artemisinin in tetraploid Artemisia annua L. Plant Biotechnol. J. 27, 427–433. doi: 10.5511/plantbiotechnology.10.0726a
Banyai, W., Mii, M., and Supaibulwatana, K. (2011). Enhancement of artemisinin content and biomass in Artemisia annua by exogenous GA3 treatment. Plant Growth Regul. 63, 45–54. doi: 10.1007/s10725-010-9510-9
Benjamin, K. R., Silva, I. R., Cherubim, J. O. P., Mcphee, D., and Paddon, C. J. (2016). Developing commercial production of semi-synthetic artemisinin, and of 2-farnesene, an isoprenoid produced by fermentation of Brazilian sugar. J. Braz. Chem. Soc. 27, 1339–1345. doi: 10.5935/0103-5053.20160119
Bhakuni, R., Jain, D., Sharma, R., and Kumar, S. (2001). Secondary metabolites of Artemisia annua and their biological activity. Curr. Sci. 80, 35–48.
Boes, A., Reimann, A., Twyman, R. M., Fischer, R., Schillberg, S., and Spiegel, H. (2016). “A plant-based transient expression system for the rapid production of malaria vaccine candidates,” in Vaccine Design: Methods and Protocols: Vaccines for Veterinary Diseases, Vol. 2, ed. S. Thomas (New York, NY: Springer), 597–619.
Bouwmeester, H. J., Wallaart, T. E., Janssen, M. H., Van Loo, B., Jansen, B. J., Posthumus, M. A., et al. (1999). Amorpha-4, 11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 52, 843–854. doi: 10.1016/S0031-9422(99)00206-X
Brisibe, E. A., Udensi, O., Chukwurah, P. N., De Magalhäes, P. M., Figueira, G. M., and Ferreira, J. F. (2012). Adaptation and agronomic performance of Artemisia annua L. under lowland humid tropical conditions. Ind. Crops Prod. 39, 190–197. doi: 10.1016/j.indcrop.2012.02.018
Brown, G. D. (2010). The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao). Molecules 15, 7603–7698. doi: 10.3390/molecules15117603
Bulgakov, V. P. (2008). Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 26, 318–324. doi: 10.1016/j.biotechadv.2008.03.001
Buttner-Mainik, A., Parsons, J., Jerome, H., Hartmann, A., Lamer, S., Schaaf, A., et al. (2011). Production of biologically active recombinant human factor H in Physcomitrella. Plant Biotechnol. J. 9, 373–383. doi: 10.1111/j.1467-7652.2010.00552.x
Carter, R., and Mendis, K. N. (2002). Evolutionary and historical aspects of the burden of malaria. Clin. Microbiol. Rev. 15, 564–594. doi: 10.1128/CMR.15.4.564-594.2002
Chen, D., Ye, H., and Li, G. (2000). Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 155, 179–185. doi: 10.1016/S0168-9452(00)00217-X
Chen, J.-L., Fang, H.-M., Ji, Y.-P., Pu, G.-B., Guo, Y.-W., Huang, L.-L., et al. (2011). Artemisinin biosynthesis enhancement in transgenic Artemisia annua plants by downregulation of the β-caryophyllene synthase gene. Planta Med. 77, 1759–1765. doi: 10.1055/s-0030-1271038
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Chen, Y., Shen, Q., Wang, Y., Wang, T., Wu, S., Zhang, L., et al. (2013). The stacked over-expression of FPS, CYP71AV1 and CPR genes leads to the increase of artemisinin level in Artemisia annua L. Plant Biotechnol. Rep. 7, 287–295. doi: 10.1007/s11816-012-0262-z
Cockram, J., Hill, C., Burns, C., Arroo, R. R., Woolley, J. G., Flockart, I., et al. (2012). Screening a diverse collection of Artemisia annua germplasm accessions for the antimalarial compound, artemisinin. Plant Genet. Resour. 10, 152–154. doi: 10.1017/S1479262112000159
Cox, F. (2010). History of the discovery of the malaria parasites and their vectors. Parasit. Vectors 3:5. doi: 10.1186/1756-3305-3-5
Czechowski, T., Larson, T. R., Catania, T. M., Harvey, D., Brown, G. D., and Graham, I. A. (2016). Artemisia annua mutant impaired in artemisinin synthesis demonstrates importance of nonenzymatic conversion in terpenoid metabolism. Proc. Natl. Acad. Sci. U.S.A. 113, 15150–15155. doi: 10.1073/pnas.1611567113
Dangash, A., Pandya, N., Bharillya, A., Jhala, A., and Jain, D. (2014). Impact of exogenous elicitors on artemisinin production and trichome density in Artemisia annua L. under subtropical conditions. Not. Sci. Biol. 6, 349–353. doi: 10.15835/nsb.6.3.9109
Delabays, N., Collet, G., and Benakis, A. (1993). Selection and breeding for high artemisinin (Qinghaosu) yielding strains of Artemisia annua. Acta Hortic. 330, 203–208. doi: 10.17660/ActaHortic.1993.330.24
Delabays, N., Simonnet, X., and Gaudin, M. (2001). The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr. Med. Chem. 8, 1795–1801. doi: 10.2174/0929867013371635
Dilshad, E., Cusido, R. M., Estrada, K. R., Bonfill, M., and Mirza, B. (2015a). Genetic transformation of Artemisia carvifolia buch with rol genes enhances artemisinin accumulation. PLOS ONE 10:e0140266. doi: 10.1371/journal.pone.0140266
Dilshad, E., Cusido, R. M., Palazon, J., Estrada, K. R., Bonfill, M., and Mirza, B. (2015b). Enhanced artemisinin yield by expression of rol genes in Artemisia annua. Malar. J. 14, 424. doi: 10.1186/s12936-015-0951-5
Duke, M. V., Paul, R. N., Elsohly, H. N., Sturtz, G., and Duke, S. O. (1994). Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int. J. Plant Sci. 155, 365–372. doi: 10.1086/297173
Epstein, J. E., Paolino, K. M., Richie, T. L., Sedegah, M., Singer, A., Ruben, A. J., et al. (2017). Protection against Plasmodium falciparum malaria by PfSPZ vaccine. JCI Insight 2:e89154. doi: 10.1172/jci.insight.89154
Farhi, M., Marhevka, E., Ben-Ari, J., Algamas-Dimantov, A., Liang, Z., Zeevi, V., et al. (2011). Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol. 29, 1072–1074. doi: 10.1038/nbt.2054
Ferreira, J. F., and Janick, J. (1995). Floral morphology of Artemisia annua with special reference to trichomes. Int. J. Plant Sci. 156, 807–815. doi: 10.1086/297304
Ferreira, J. F., Laughlin, J., Delabays, N., and De Magalhães, P. M. (2005). Cultivation and genetics of Artemisia annua L. for increased production of the antimalarial artemisinin. Plant Genet. Resour. 3, 206–229. doi: 10.1079/PGR200585
Frank, W., Decker, E., and Reski, R. (2005). Molecular tools to study Physcomitrella patens. Plant Biol. 7, 220–227. doi: 10.1055/s-2005-865645
Fuentes, P., Zhou, F., Erban, A., Karcher, D., Kopka, J., and Bock, R. (2016). A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 5:e13664. doi: 10.7554/eLife.13664
Ghosh, B., Mukherjee, S., and Jha, S. (1997). Genetic transformation of Artemisia annua by Agrobacterium tumefaciens and artemisinin synthesis in transformed cultures. Plant Sci. 122, 193–199. doi: 10.4103/0973-1296.127372
Graham, I. A., Besser, K., Blumer, S., Branigan, C. A., Czechowski, T., Elias, L., et al. (2010). The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327, 328–331. doi: 10.1126/science.1182612
Guo, X.-X., Yang, X.-Q., Yang, R.-Y., and Zeng, Q.-P. (2010). Salicylic acid and methyl jasmonate but not Rose Bengal enhance artemisinin production through invoking burst of endogenous singlet oxygen. Plant Sci. 178, 390–397. doi: 10.1016/j.plantsci.2010.01.014
Han, J., Wang, H., Kanagarajan, S., Hao, M., Lundgren, A., and Brodelius, P. E. (2016). Promoting artemisinin biosynthesis in Artemisia annua plants by substrate channeling. Mol. Plant 9, 946–948. doi: 10.1016/j.molp.2016.03.004
Han, J. L., Liu, B. Y., Ye, H. C., Wang, H., Li, Z. Q., and Li, G. F. (2006). Effects of overexpression of the endogenous farnesyl diphosphate synthase on the artemisinin content in Artemisia annua L. J. Integr. Plant Biol. 48, 482–487. doi: 10.1111/j.1744-7909.2006.00208.x
He, Q., Fu, X., Shi, P., Liu, M., Shen, Q., and Tang, K. (2017). Glandular trichome-specific expression of alcohol dehydrogenase 1 (ADH1) using a promoter-GUS fusion in Artemisia annua L. Plant Cell Tissue Organ Cult. 130, 61–72. doi: 10.1007/s11240-017-1204-9
Hong, G.-J., Hu, W.-L., Li, J.-X., Chen, X.-Y., and Wang, L.-J. (2009). Increased accumulation of artemisinin and anthocyanins in Artemisia annua expressing the Arabidopsis blue light receptor CRY1. Plant Mol. Biol. Rep. 27, 334–341. doi: 10.1007/s11105-008-0088-6
Ikram, N. K. B., Beyraghdar Kashkooli, A., Peramuna, A. V., Van Der Krol, A. R., Bouwmeester, H., and Simonsen, H. T. (2017). Stable production of the antimalarial drug artemisinin in the moss Physcomitrella patens. Front. Bioeng. Biotechnol. 5:47. doi: 10.3389/fbioe.2017.00047
Ikram, N. K., Zhan, X., Pan, X., King, B. C., and Simonsen, H. T. (2015). Stable heterologous expression of biologically active terpenoids in green plant cells. Front. Plant Sci. 6:129. doi: 10.3389/fpls.2015.00129
Jiang, W., Fu, X., Pan, Q., Tang, Y., Shen, Q., Lv, Z., et al. (2016). Overexpression of AaWRKY1 leads to an enhanced content of artemisinin in Artemisia annua. Biomed Res. Int. 2016:7314971. doi: 10.1155/2016/7314971
King, B. C., Vavitsas, K., Ikram, N. K. B. K., Schrøder, J., Scharff, L. B., Hamberger, B., et al. (2016). In vivo assembly of DNA-fragments in the moss, Physcomitrella patens. Sci. Rep. 6:25030. doi: 10.1038/srep25030
Kjær, A., Grevsen, K., and Jensen, M. (2012). Effect of external stress on density and size of glandular trichomes in full-grown Artemisia annua, the source of anti-malarial artemisinin. AoB Plants 2012:ls018. doi: 10.1093/aobpla/pls018
Klayman, D. L. (1985). Qinghaosu (artemisinin): an antimalarial drug from China. Science 228, 1049–1055. doi: 10.1126/science.3887571
Lalloo, D. G., Shingadia, D., Bell, D. J., Beeching, N. J., Whitty, C. J. M., and Chiodini, P. L. (2016). UK malaria treatment guidelines 2016. J. Infect. 72, 635–649. doi: 10.1016/j.jinf.2016.02.001
Larson, T. R., Branigan, C., Harvey, D., Penfield, T., Bowles, D., and Graham, I. A. (2013). A survey of artemisinic and dihydroartemisinic acid contents in glasshouse and global field-grown populations of the artemisinin-producing plant Artemisia annua L. Ind. Crops Prod. 45, 1–6. doi: 10.1016/j.indcrop.2012.12.004
Laursen, T., Borch, J., Knudsen, C., Bavishi, K., Torta, F., Martens, H. J., et al. (2016). Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354, 890–893. doi: 10.1126/science.aag2347
Ling, X., Hexin, T., and Lei, Z. (2016). Artemisia annua glandular secretory trichomes: the biofactory of antimalarial agent artemisinin. Sci. Bull. 61, 26–36. doi: 10.1007/s11434-015-0980-z
Liu, C.-Z., Guo, C., Wang, Y.-C., and Ouyang, F. (2002). Effect of light irradiation on hairy root growth and artemisinin biosynthesis of Artemisia annua L. Proc. Biochem. 38, 581–585. doi: 10.1016/S0032-9592(02)00165-6
Liu, M., Shi, P., Fu, X., Brodelius, P. E., Shen, Q., Jiang, W., et al. (2016). Characterization of a trichome-specific promoter of the aldehyde dehydrogenase 1 (ALDH1) gene in Artemisia annua. Plant Cell Tissue Organ Cult. 126, 469–480. doi: 10.1007/s11240-016-1015-4
Lommen, W., Schenk, E., Bouwmeester, H., and Verstappen, F. (2006). Trichome dynamics and artemisinin accumulation during development and senescence of Artemisia annua leaves. Planta Med. 72, 336–345. doi: 10.1055/s-2005-916202
Lu, X., Shen, Q., Zhang, L., Zhang, F., Jiang, W., Lv, Z., et al. (2013a). Promotion of artemisinin biosynthesis in transgenic Artemisia annua by overexpressing ADS, CYP71AV1 and CPR genes. Ind. Crops Prod. 49, 380–385. doi: 10.1016/j.indcrop.2013.04.045
Lu, X., Zhang, L., Zhang, F., Jiang, W., Shen, Q., Zhang, L., et al. (2013b). AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198, 1191–1202. doi: 10.1111/nph.12207
Luo, X. D., and Shen, C. C. (1987). The chemistry, pharmacology, and clinical applications of qinghaosu (artemisinin) and its derivatives. Med. Res. Rev. 7, 29–52. doi: 10.1002/med.2610070103
Lv, Z., Wang, S., Zhang, F., Chen, L., Hao, X., Pan, Q., et al. (2016). Overexpression of a Novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 57, 1961–1971. doi: 10.1093/pcp/pcw118
Ma, C., Wang, H., Lu, X., Wang, H., Xu, G., and Liu, B. (2009). Terpenoid metabolic profiling analysis of transgenic Artemisia annua L. by comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. Metabolomics 5, 497–506. doi: 10.1007/s11306-009-0170-6
Ma, D., Li, G., Alejos-Gonzalez, F., Zhu, Y., Xue, Z., Wang, A., et al. (2017a). Overexpression of a type I isopentenyl pyrophosphate isomerase of Artemisia annua in the cytosol leads to high arteannuin B production and artemisinin increase. Plant J. 91, 466–479. doi: 10.1111/tpj.13583
Ma, D., Li, G., Zhu, Y., and Xie, D.-Y. (2017b). Overexpression and suppression of Artemisia annua 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 1 gene (AaHDR1) differentially regulate artemisinin and terpenoid biosynthesis. Front. Plant Sci. 8:77. doi: 10.3389/fpls.2017.00077
Ma, D.-M., Wang, Z., Wang, L., Alejos-Gonzales, F., Sun, M.-A., and Xie, D.-Y. (2015). A genome-wide scenario of terpene pathways in self-pollinated Artemisia annua. Mol. Plant 8, 1580–1598. doi: 10.1016/j.molp.2015.07.004
Malhotra, K., Subramaniyan, M., Rawat, K., Kalamuddin, M., Qureshi, M. I., Malhotra, P., et al. (2016). Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant Cells. Mol. Plant 9, 1464–1477. doi: 10.1016/j.molp.2016.09.013
Mercke, P., Bengtsson, M., Bouwmeester, H. J., Posthumus, M. A., and Brodelius, P. E. (2000). Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 381, 173–180. doi: 10.1006/abbi.2000.1962
Muangphrom, P., Seki, H., Fukushima, E. O., and Muranaka, T. (2016). Artemisinin-based antimalarial research: application of biotechnology to the production of artemisinin, its mode of action, and the mechanism of resistance of Plasmodium parasites. J. Nat. Med. 70, 318–334. doi: 10.1007/s11418-016-1008-y
Nafis, T., Akmal, M., Ram, M., Alam, P., Ahlawat, S., Mohd, A., et al. (2011). Enhancement of artemisinin content by constitutive expression of the HMG-CoA reductase gene in high-yielding strain of Artemisia annua L. Plant Biotechnol. Rep. 5, 53–60. doi: 10.1007/s11816-010-0156-x
Nair, M. S. R., Acton, N., Klayman, D. L., Kendrick, K., Basile, D. V., and Mante, S. (1986). Production of artemisinin in tissue cultures of Artemisia Annua. J. Nat. Prod. 49, 504–507. doi: 10.1021/np50045a021
Nondo, R. S. O., Moshi, M. J., Erasto, P., Masimba, P. J., Machumi, F., Kidukuli, A. W., et al. (2017). Anti-plasmodial activity of norcaesalpin D and extracts of four medicinal plants used traditionally for treatment of malaria. BMC Complement. Altern. Med. 17:167. doi: 10.1186/s12906-017-1673-8
Olofsson, L., Engstrom, A., Lundgren, A., and Brodelius, P. (2011). Relative expression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol. 11:45. doi: 10.1186/1471-2229-11-45
Olsson, M. E., Olofsson, L. M., Lindahl, A.-L., Lundgren, A., Brodelius, M., and Brodelius, P. E. (2009). Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70, 1123–1128. doi: 10.1016/j.phytochem.2009.07.009
Paddon, C. J., Westfall, P., Pitera, D., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Pan, X.-W., Han, L., Zhang, Y.-H., Chen, D.-F., and Simonsen, H. T. (2015). Sclareol production in the moss Physcomitrella patens and observations on growth and terpenoid biosynthesis. Plant Biotechnol. Rep. 9, 149–159. doi: 10.1007/s11816-015-0353-8
Pandey, N., Meena, R. P., Rai, S. K., and Pandey-Rai, S. (2016). In vitro generation of high artemisinin yielding salt tolerant somaclonal variant and development of SCAR marker in Artemisia annua L. Plant Cell Tissue Organ Cult. 127, 301–314. doi: 10.1007/s11240-016-1050-1
Pandey, N., and Pandey-Rai, S. (2014). Short term UV-B radiation-mediated transcriptional responses and altered secondary metabolism of in vitro propagated plantlets of Artemisia annua L. Plant Cell Tissue Organ Cult. 116, 371–385. doi: 10.1007/s11240-013-0413-0
Patra, N., and Srivastava, A. K. (2017). “Mass production of artemisinin using hairy root cultivation of Artemisia annua in bioreactor,” in Bioprocessing of Plant in Vitro Systems, eds A. Pavlov and T. Bleys (Cham: Springer International Publishing), 1–17.
Peplow, M. (2016). Synthetic malaria drug meets market resistance: first commercial deployment of synthetic biology for medicine has modest impact. Nature 530, 389–391. doi: 10.1038/530390a
Picaud, S., Mercke, P., He, X., Sterner, O., Brodelius, M., Cane, D. E., et al. (2006). Amorpha-4,11-diene synthase: mechanism and stereochemistry of the enzymatic cyclization of farnesyl diphosphate. Arch. Biochem. Biophys. 448, 150–155. doi: 10.1016/j.abb.2005.07.015
Picaud, S., Olofsson, L., Brodelius, M., and Brodelius, P. E. (2005). Expression, purification, and characterization of recombinant amorpha-4,11-diene synthase from Artemisia annua L. Arch. Biochem. Biophys. 436, 215–226. doi: 10.1016/j.abb.2005.02.012
Pulice, G., Pelaz, S., and Matías-Hernández, L. (2016). Molecular Farming in Artemisia annua, a promising approach to improve anti-malarial drug production. Front. Plant Sci. 7:329. doi: 10.3389/fpls.2016.00329
Reski, R. (1998). Development, genetics and molecular biology of mosses. Bot. Acta 111, 1–15. doi: 10.1111/j.1438-8677.1998.tb00670.x
Reski, R., Parsons, J., and Decker, E. L. (2015). Moss-made pharmaceuticals: from bench to bedside. Plant Biotechnol. J. 13, 1191–1198. doi: 10.1111/pbi.12401
Rosales-Mendoza, S., Nieto-Gómez, R., and Angulo, C. (2017). A perspective on the development of plant-made vaccines in the fight against Ebola virus. Front. Immunol. 8:252. doi: 10.3389/fimmu.2017.00252
Rosales-Mendoza, S., Orellana-Escobedo, L., Romero-Maldonado, A., Decker, E. L., and Reski, R. (2014). The potential of Physcomitrella patens as a platform for the production of plant-based vaccines. Expert Rev. Vaccines 13, 203–212. doi: 10.1586/14760584.2014.872987
Rydén, A.-M., Ruyter-Spira, C., Quax, W. J., Osada, H., Muranaka, T., Kayser, O., et al. (2010). The molecular cloning of dihydroartemisinic aldehyde reductase and its implication in artemisinin biosynthesis in Artemisia annua. Planta Med. 76, 1778–1783. doi: 10.1055/s-0030-1249930
Schaefer, D. G., and Zrÿd, J. P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11, 1195–1206. doi: 10.1046/j.1365-313X.1997.11061195.x
Shen, Q., Chen, Y., Wang, T., Wu, S., Lu, X., Zhang, L., et al. (2012). Overexpression of the cytochrome P450 monooxygenase (cyp71av1) and cytochrome P450 reductase (cpr) genes increased artemisinin content in Artemisia annua (Asteraceae). Genet. Mol. Res. 11, 3298–3309. doi: 10.4238/2012.September.12.13
Shen, Q., Lu, X., Yan, T., Fu, X., Lv, Z., Zhang, F., et al. (2016a). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210, 1269–1281. doi: 10.1111/nph.13874
Shen, Q., Yan, T., Fu, X., and Tang, K. (2016b). Transcriptional regulation of artemisinin biosynthesis in Artemisia annua L. Sci. Bull. 61, 18–25. doi: 10.1007/s11434-015-0983-9
Shi, P., Fu, X., Liu, M., Shen, Q., Jiang, W., Li, L., et al. (2017). Promotion of artemisinin content in Artemisia annua by overexpression of multiple artemisinin biosynthetic pathway genes. Plant Cell Tissue Organ Cult. 129, 251–259. doi: 10.1007/s11240-017-1173-z
Shretta, R., and Yadav, P. (2012). Stabilizing supply of artemisinin and artemisinin-based combination therapy in an era of wide-spread scale-up. Malar. J. 11:399. doi: 10.1186/1475-2875-11-399
Simonsen, H. T., Drew, D. P., and Lunde, C. (2009). Perspectives on using Physcomitrella patens as an alternative production platform for thapsigargin and other terpenoid drug candidates. Perspect. Medicin. Chem. 3, 1–6. doi: 10.4137/PMC.S2220
Simonsen, H. T., Nordskjold, J. B., Smitt, U. W., Nyman, U., Palpu, P., Joshi, P., et al. (2001). In vitro screening of Indian medicinal plants for antiplasmodial activity. J. Ethnopharmacol. 74, 195–204. doi: 10.1016/S0378-8741(00)00369-X
Simonsen, H. T., Weitzel, C., and Christensen, S. B. (2013). “Guaianolide sesquiterpenoids - Their pharmacology and biosynthesis,” in Handbook of Natural Products - Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes, eds K. G. Ramawat and J. M. Merillon (Berlin: Springer-Verlag), 3069–3098.
Singh, D., Mcphee, D., Paddon, C. J., Cherry, J., Maurya, G., Mahale, G., et al. (2017). Amalgamation of synthetic biology and chemistry for high-throughput nonconventional synthesis of the antimalarial drug artemisinin. Org. Process Res. Dev. 21, 551–558. doi: 10.1021/acs.oprd.6b00414
Sy, L.-K., and Brown, G. D. (2002). The mechanism of the spontaneous autoxidation of dihydroartemisinic acid. Tetrahedron 58, 897–908. doi: 10.1016/S0040-4020(01)01193-0
Tang, K., Chen, Y., Shen, Q., Wang, T., Wu, S., and Wang, G. (2012a). Overexpression ALDH1 gene increased artemisinin content in Artemisia annua L. China Patent Application CN201210014242.
Tang, K., Shen, Q., Chen, Y., Wang, T., Wu, S., and Lu, X. (2012b). Overexpression DBR2 gene increased artemisinin content in Artemisia annua L. China Patent Application CN201210014227.
Tang, K., Shen, Q., Yan, T., and Fu, X. (2014). Transgenic approach to increase artemisinin content in Artemisia annua L. Plant Cell Rep. 33, 605–615. doi: 10.1007/s00299-014-1566-y
Teoh, K. H., Polichuk, D. R., Reed, D. W., and Covello, P. S. (2009). Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87, 635–642. doi: 10.1139/B09-032
Teoh, K. H., Polichuk, D. R., Reed, D. W., Nowak, G., and Covello, P. S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416. doi: 10.1016/j.febslet.2006.01.065
Ting, H. M., Wang, B., Rydén, A. M., Woittiez, L., Herpen, T., Verstappen, F. W., et al. (2013). The metabolite chemotype of Nicotiana benthamiana transiently expressing artemisinin biosynthetic pathway genes is a function of CYP71AV1 type and relative gene dosage. New Phytol. 199, 352–366. doi: 10.1111/nph.12274
Townsend, T., Segura, V., Chigeza, G., Penfield, T., Rae, A., Harvey, D., et al. (2013). The use of combining ability analysis to identify elite parents for Artemisia annua F1 hybrid production. PLOS ONE 8:e61989. doi: 10.1371/journal.pone.0061989
Tu, Y. (2011). The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 17, 1217–1220. doi: 10.1038/nm.2471
Van Agtmael, M. A., Eggelte, T. A., and Van Boxtel, C. J. (1999). Artemisinin drugs in the treatment of malaria: from medicinal herb to registered medication. Trends Pharmacol. Sci. 20, 199–205. doi: 10.1016/S0165-6147(99)01302-4
Van Herpen, T. W., Cankar, K., Nogueira, M., Bosch, D., Bouwmeester, H. J., and Beekwilder, J. (2010). Nicotiana benthamiana as a production platform for artemisinin precursors. PLOS ONE 5:e14222. doi: 10.1371/journal.pone.0014222
Wallaart, T. E., Bouwmeester, H. J., Hille, J., Poppinga, L., and Maijers, N. C. (2001). Amorpha-4,11-diene synthase: cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin. Planta 212, 460–465. doi: 10.1007/s004250000428
Wang, B., Kashkooli, A. B., Sallets, A., Ting, H.-M., De Ruijter, N. C., Olofsson, L., et al. (2016). Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua. Metab. Eng. 38, 159–169. doi: 10.1016/j.ymben.2016.07.004
Wang, H., Ma, C., Li, Z., Ma, L., Wang, H., Ye, H., et al. (2010). Effects of exogenous methyl jasmonate on artemisinin biosynthesis and secondary metabolites in Artemisia annua L. Ind. Crops Prod. 31, 214–218. doi: 10.1016/j.indcrop.2009.10.008
Wang, Y., and Weathers, P. (2007). Sugars proportionately affect artemisinin production. Plant Cell Rep. 26, 1073–1081. doi: 10.1007/s00299-006-0295-2
Wang, Y., Yang, K., Jing, F., Li, M., Deng, T., Huang, R., et al. (2011). Cloning and characterization of trichome-specific promoter of cpr71av1 gene involved in artemisinin biosynthesis in Artemisia annua L. Mol. Biol. 45, 751. doi: 10.1134/S0026893311040145
Weathers, P. J., Elkholy, S., and Wobbe, K. K. (2006). Artemisinin: the biosynthetic pathway and Its regulation in Artemisia annua, a terpenoid-rich species. In Vitro Cell Dev. Biol. Plant 42, 309–317. doi: 10.1079/IVP2006782
Wen, W., and Yu, R. (2011). Artemisinin biosynthesis and its regulatory enzymes: progress and perspective. Pharmacogn. Rev. 5, 189–194. doi: 10.4103/0973-7847.91118
Woerdenbag, H., Lüers, J. J., Van Uden, W., Pras, N., Malingré, T., and Alfermann, A. W. (1993). Production of the new antimalarial drug artemisinin in shoot cultures of Artemisia annua L. Plant Cell Tissue Organ Cult. 32, 247–257. doi: 10.1007/BF00029850
World Health Organization [WHO] (2015). Guidelines for the Treatment of Malaria, 3rd Edn. Geneva: WHO.
Xiang, L., Zeng, L. X., Yuan, Y., Chen, M., Wang, F., Liu, X. Q., et al. (2012). Enhancement of artemisinin biosynthesis by overexpressing dxr, cyp71av1 and cpr in the plants of Artemisia annua L. Plant Omics 5, 503–507.
Xiang, L., Zhu, S., Zhao, T., Zhang, M., Liu, W., Chen, M., et al. (2015). Enhancement of artemisinin content and relative expression of genes of artemisinin biosynthesis in Artemisia annua by exogenous MeJA treatment. Plant Growth Regul. 75, 435–441. doi: 10.1007/s10725-014-0004-z
Xie, D.-Y., Ma, D.-M., Judd, R., and Jones, A. L. (2016). Artemisinin biosynthesis in Artemisia annua and metabolic engineering: questions, challenges, and perspectives. Phytochem. Rev. 15, 1093–1114. doi: 10.1007/s11101-016-9480-2
Yin, L., Zhao, C., Huang, Y., Yang, R.-Y., and Zeng, Q.-P. (2008). Abiotic stress-induced expression of artemisinin biosynthesis genes in Artemisia annua L. Chin. J. Appl. Environ. Biol. 14, 1–15.
Yu, Z.-X., Li, J.-X., Yang, C.-Q., Hu, W.-L., Wang, L.-J., and Chen, X.-Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zhan, X., Zhang, Y.-H., Chen, D.-F., and Simonsen, H. T. (2014). Metabolic engineering of the moss Physcomitrella patens to produce the sesquiterpenoids patchoulol and α/β-santalene. Front. Plant Sci. 5:636. doi: 10.3389/fpls.2014.00636
Zhang, F., Lu, X., Lv, Z., Zhang, L., Zhu, M., Jiang, W., et al. (2013). Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin content in Artemisia annua L. PLOS ONE 8:e56697. doi: 10.1371/journal.pone.0056697
Zhang, L., Jing, F., Li, F., Li, M., Wang, Y., Wang, G., et al. (2009). Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Biochem. 52, 199–207. doi: 10.1042/BA20080068
Zhang, Y., Nowak, G., Reed, D. W., and Covello, P. S. (2011). The production of artemisinin precursors in tobacco. Plant Biotechnol. J. 9, 445–454. doi: 10.1111/j.1467-7652.2010.00556.x
Zhang, Y., Teoh, K. H., Reed, D. W., Maes, L., Goossens, A., Olson, D. J., et al. (2008). The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J. Biol. Chem. 283, 21501–21508. doi: 10.1074/jbc.M803090200
Keywords: plant biotechnology, malaria, artemisinin, Artemisia annua, bioengineering
Citation: Ikram NKBK and Simonsen HT (2017) A Review of Biotechnological Artemisinin Production in Plants. Front. Plant Sci. 8:1966. doi: 10.3389/fpls.2017.01966
Received: 15 August 2017; Accepted: 31 October 2017;
Published: 15 November 2017.
Edited by:
Tomasz Czechowski, University of York, United KingdomReviewed by:
Patrick Smithers Covello, National Research Council Plant Biotechnology Institute, CanadaDeyu Xie, North Carolina State University, United States
Copyright © 2017 Ikram and Simonsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henrik T. Simonsen, aGV0c0BkdHUuZGs=
 Nur K. B. K. Ikram
Nur K. B. K. Ikram Henrik T. Simonsen
Henrik T. Simonsen