- College of Plant Protection, Southwest University, Chongqing, China
Various classes of plant pathogenesis-related proteins have been identified in the past several decades. PR-Q, a member of the PR3 family encoding chitinases, has played an important role in regulating plant resistance and preventing pathogen infection. In this paper, we functionally characterized NtPR-Q in tobacco plants and found that the overexpression of NtPR-Q in tobacco Yunyan87 resulted in higher resistance to Ralstonia solanacearum inoculation. Surprisingly, overexpression of NtPR-Q led to the activation of many defense-related genes, such as salicylic acid (SA)-responsive genes NtPR1a/c, NtPR2 and NtCHN50, JA-responsive gene NtPR1b and ET production-associated genes NtACC Oxidase and NtEFE26. Consistent with the role of NtPR-Q in multiple stress responses, NtPR-Q transcripts were induced by the exogenous hormones SA, ethylene and methyl jasmonate, which could enhance the resistance of tobacco to R. solanacearum. Collectively, our results suggested that NtPR-Q overexpression led to the up-regulation of defense-related genes and enhanced plant resistance to R. solanacearum infection.
Introduction
Plants have a two-branched immune system when responding to the microbial infection. The first branch uses pattern recognition receptors (PRR) to recognize molecules common to many classes of microbes, generally known as PAMP-triggered immunity (PTI). The second layer of the immune response uses NB-LRR (nucleotide binding and leucine rich repeat) proteins which are encoded by most R genes to recognize pathogen effector, also known as effector-triggered immunity (ETI) (Jones and Dangl, 2006). Plant resistance refers to the ability of plants to overcome or inhibit the effect of a pathogen or other damaging environment factors to a certain extent (Agrios, 1988). Plants respond to external environmental stress through rapid changes in resistance gene expression, resulting in producing a number of specific proteins, including resistance-related proteins (RRP) and resistance-related metabolites (RRM), which are either only present in resistant genotypes or are found in higher accumulation in the resistant genotype than in the susceptible genotype (Kushalappa et al., 2016). It is a common strategy to improve plant resistance by constructing transgenic plants that overexpress transcription factors (Dang et al., 2013; Wang et al., 2013; Jia et al., 2015) or pathogenesis-related genes. Phytohormones, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), act as signal molecules during plant growth and stress response, and their content and activity in plant are highly susceptible to changes of external conditions and cell growth. It is found that SA-dependent responses act against biotrophs pathogens, and JA/ET-dependent responses act against necrotrophs pathogens (Glazebrook, 2005; Vlot et al., 2009).
The pathogenesis-related (PR) proteins are a class of proteins that suppress pathogens, detoxify toxins or virulence factors produced by pathogens and prevent pathogen advancement by enforcing cell walls (Kushalappa et al., 2016). Pathogenesis-related proteins have attracted an increasing research interest in view of their possible involvement in plant resistance to pathogens. There is a close relationship between PRs and SAR (Payne et al., 1990), and they have been considered as a marker of the induced state (Ward et al., 1991b; Uknes et al., 1992; Kessmann et al., 1994). Thaumatin-like protein (TLP-3) has been reported to enhance resistance to Alternaria alternata in transgenic tobacco plants (Safavi and Zareie, 2012). A pepper gene, CABPR1, which encodes PR1, has been reported to be strongly induced by wounding, ethephon treatment, and tobacco mosaic virus (TMV) infection (Sarowar et al., 2005). PRs have been classified into 17 families, and the PR-3 family has chitinase properties (Sels et al., 2008). Chitinases can be divided into two categories: exochitinases and endochitinases, and PR-Q belongs to the latter category (Payne et al., 1990). Many plant endochitinases exhibit an additional lysozyme or lysozyme-like activity. It is known that chitinases have significant antifungal activities against pathogenic fungi, such as Fusarium oxysporum and Cercospora nicotianae (Zhu et al., 1994; Jongedijk, 1995). PR-3 proteins can cleave the cell wall chitin polymers in situ, so that the cell wall weaken and enhance osmotic sensitivity to fungi. The expression of chitinases is regulated by wounding, methyl jasmonate (MeJA), ethylene, and gibberellin (Golshani et al., 2015). In tomato, the simultaneous expression of tobacco chitinase gene PR-3d and glucanase gene PR-2e lead to stronger resistance to F. oxysporum f.sp. lycopersici, whereas transgenic plants expressing either one of these genes are susceptible (Jongedijk, 1995). PR-Q, a member of the PR-3 family, is a class II chitinase. The acidic enzyme is found in the extracellular fluid from leaves, suggesting that it is localized in the apoplastic compartment (Payne et al., 1990; Ohme-Takagi et al., 1998). It is part of the systemic resistance and preventing pathogen infection.
As a complex species (Prior and Fegan, 2005), Ralstonia solanacearum has complex genetic diversity and strong adaptability to the environment (Horita et al., 2010; Liu et al., 2017b). R. solanacearum causes wilting symptoms to a variety of crops, including vegetables (Solanum tuberosum, S. lycopersicum, Capsicum spp.), tobacco (Nicotiana tabacum), peanuts (Arachis hypogaea), banana (Musa spp.), and so on (Ji et al., 2005). Since the first report of bacterial wilt appeared in 1864, the disease, resulting in serious economic losses, has been found in major countries and regions worldwide where Solanaceae crops are cultivated. At present, the control of bacterial wilt mainly depends on chemical pesticides (Yang et al., 2016), but based on the strong variability and soil-borne characteristics of R. solanacearum, the challenge for actual crop production remains considerable. It is important to study the functions of disease-resistance genes to improve plant resistance.
In this study, the NtPR-Q gene was overexpressed in tobacco, and its biological function in defending against R. solanacearum was analyzed. This study provides new information for the disease control network and a new strategy for disease resistance breeding.
Materials and Methods
Plant Materials and Growth Conditions
A N. tabacum cultivar, Yunyan87, was used in this study as the wild type (WT-Yunyan87). The surface of the seeds were disinfected with 75% alcohol for 30 s and 10% H2O2 for 10 min, then washed five times with sterile water and placed under greenhouse conditions for 2–3 weeks on MS agar medium (Murashige and Skoog, 1962; Dang et al., 2013). Regenerated shoots and healthy resistant shoots were grown on selective shooting medium and rooting medium, respectively, which both contained 500 mg⋅L-1 carbenicillin and 100 mg⋅L-1 kanamycin. The transgenic plants for this study were obtained by asexual reproduction. Well-developed rooted plants were transferred to matrix in plastic pots and then placed under greenhouse conditions at 25 ± 2°C with a relative humidity of 80% and 16/8 h light/dark cycle.
Pathogens and Inoculation Procedures
Ralstonia solanacearum CQPS-1 (phylotype I, race 1, biovar 3) (Liu et al., 2017a) was maintained in our laboratory and was isolated from Chongqing Municipality, China. R. solanacearum strains were cultured at 180 rpm, 30°C in NB medium (3 g⋅L-1 beef extract, 1 g⋅L-1 yeast extract, 5 g⋅L-1 peptone and 10 g⋅L-1 glucose), and bacterial suspension was cultured to about 1 × 109 CFU mL-1.
To study the relative expression levels of NtPR-Q in response to R. solanacearum infections, WT-Yunyan87 plants were inoculated with 10 mL of bacterial suspension (1 × 108 CFU mL-1). Leaf samples were harvested at 1, 3, 5, and 7 days post inoculation (dpi), frozen and stored at -80°C until further analysis. Control plants were inoculated with sterile water. Three individual plants were mixed to yield one sample.
For the pathogenicity assays, transgenic plants and WT-Yunyan87 plants were root inoculated with a 10 mL bacterial suspension (1 × 108 CFU mL-1). Assays were conducted with 10 plants and were repeated three times. The disease index of bacterial wilt were surveyed every day for 30 days using a 0 to 4 grading standards, where 0 = asymptomatic plant, 1 = less than 25% leaves wilted, 2 = less than 50% wilted leaves, 3 = more than 50% wilted leaves and 4 = completely wilted leaves (dead plant). The data presented with the average disease index (Wang et al., 2013).
For monitoring a bacterial growth assay in leaf tissues, bacterial suspension (5 × 107 CFU mL-1) was washed twice with sterile water and resuspended in sterile water. Transgenic plants and WT-Yunyan87 plants were inoculated by infiltrating 20 μL inoculum into the third leaves from the top using a blunt syringe. The inoculated leaves were harvested at the indicated time points (30 min, 1, 3, and 5 days) using a leaf punch 1 cm in diameter and were then grounded, serially diluted, and spread on TTC medium [3 g⋅L-1 beef extract, 1 g⋅L-1 yeast extract, 5 g⋅L-1 peptone, 10 g⋅L-1 glucose, 20 g⋅L-1 agar configured to NA medium, 100 mL NA medium supplemented with 500 μL triphenyl tetrazolium chloride (TTC) before use]. The number of colonies was counted after 2 days, and the number of R. solanacearum cells per square centimeter of leaves was calculated. The assay repeated three times.
To test the expression of defense-related genes in transgenic tobacco, we harvested the leaf of transgenic tobacco and WT-Yunyan87 tobacco which were grown on MS medium. To study the relative expression levels of defense-related genes in response to R. solanacearum infection, transgenic plants and WT-Yunyan87 plants were inoculated with 10 mL bacterial suspension (1 × 108 CFU mL-1). Leaf samples were harvested at 1, 3, and 5 dpi, frozen and stored at -80°C until further analysis. Control plants were inoculated with sterile water. Three individual plants were mixed to yield one sample.
Plant Treatment with Exogenous Hormones
WT-Yunyan87 tobacco plants at the four-leaf stage were sprayed with 0.1 mM MeJA, 2 mM SA, or 7 mM ethephon. SA was dissolved in 10% ethanol, and the others were dissolved in sterile water. Control plants were sprayed with 10% ethanol and sterile water. The disease index of bacterial wilt were surveyed every day for 26 days using 0 to 4 grading standards. Assays were conducted with 10 plants and were repeated three times. Plants treated with SA, MeJA, or ethephon were also harvested as samples at 3, 6, 12, and 24 h post treatment, frozen and stored at -80°C for the quantitative real-time PCR analysis. Three individual plants were mixed to yield one sample.
RNA Extraction, cDNA Synthesis
The leaves of plants were grounded into powder in liquid nitrogen using mortars. RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, DP432, Beijing, China). The RNA sample was then reverse transcribed with an iScriptTM cDNA Synthesis Kit (BIO-RAD, 1708891, Hercules, CA, United States) in a 20 μL reaction.
Quantitative Real-Time PCR
To analyze the relative transcript levels of various genes, quantitative real-time PCR (RT-qPCR) was performed with specific primers, NtUBI3 as the reference gene (Zhao et al., 2009) (see Table 1 for the gene-specific primers) using SsoFastTM EvaGreen® Supermix (BIO-RAD, 1725201, Hercules, CA, United States) and the CFX96 real-time system (BIO-RAD, United States). The 20 μL reaction mixtures contained 1 μL cDNA, 10 μL SsoFastTM EvaGreen® Supermix, 1 μL of each gene-specific primer (10 μM) and 7 μL ddH2O. The amplifications were performed at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, 55°C for 20 s and elongation at 56°C for 30 s. Melt curve analyses at 95°C were included at the end to ensure the consistency of the amplified products. The expression level and data analyses were performed by using Bio-Rad CFX Manager 3.0 software with the ΔΔCq method (Li et al., 2017). Each assay was performed with three independent technical repeats and the expression level of the genes was calculated from three biological replicates.
Construction of Plant Overexpression Vector and Agrobacterium Transformation
The full-length cDNA of NtPR-Q with a downstream in-frame His-Tag was cloned using BglII and BstEII restriction sites. The specific primers of NtPR-Q vector used in this study are listed in Table 1. The plant expression vector pVCT2024 was provided by Dr. Xingguo Zhang (College of Horticulture and Landscape Architecture, Southwest University). After digestion with BglII and BstEII, the NtPR-Q cDNA was inserted into pVCT2024 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The positive recombinant plasmid pVCT2024-NtPR-Q was transformed into Agrobacterium tumefaciens strain EHA 105, which was then transformed into WT-Yunyan87 plants using the leaf disk method described by Oh et al. (2005). Transgenic plants were further confirmed to have T-DNA insertions by standard PCR and RT-qPCR.
PCR Analysis of the T-DNA Insertion
To analyze the T-DNA insertion in transgenic tobacco, standard PCR was performed with the primers of NtPR-Q insertion and NtACTIN (see Table 1), WT-Yunyan87 tobacco plants as the blank control, NtACTIN served as an endogenous control (Dang et al., 2013). The 20 μL reaction mixtures contained 1 μL cDNA, 2 μL 10 × buffer (TaKaRa, Dalian, China), 1 μL of each gene-specific primer (10 μM), 1.6 μL dNTP (TIANGEN, Beijing, China), 0.1 μL Taq DNA polymerase (TIANGEN, Beijing, China), and 13.3 μL ddH2O. The amplifications were performed at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, 59°C for 30 s and elongation at 72°C for 2.5 min, 72°C for 5 min. PCR product was electrophoresed on 1% agarose gel and observed by Bio-Rad gel imager.
Statistical Analysis
The SPSS version 17.0 software was used for statistical analysis. The data between two treatments were compared and statistically analyzed through the Student t-test. The data among multiple treatments were compared and statistically analyzed though one-way analysis of variance and Student–Newman–Keuls test.
Results
Response of NtPR-Q Transcript Levels to R. solanacearum Infection and Hormone Application
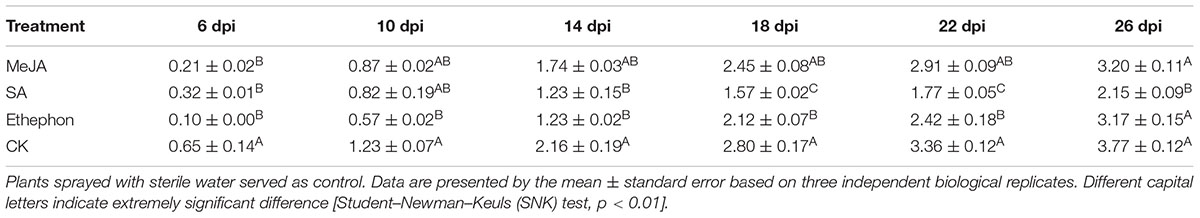
To test whether NtPR-Q is involved in the plant immune response against R. solanacearum, the transcriptional expression levels of NtPR-Q in WT-Yunyan87 was determined by RT-qPCR after inoculation with R. solanacearum strain CQPS-1 (Figure 1A). Compared with 0 hpi, NtPR-Q transcripts levels were enhanced in tobacco leaves starting at 1 dpi, and reached the maximum at 3 dpi. Subsequently, at 5 dpi, the NtPR-Q transcript levels decreased to the level of 1 dpi and maintained until 7 dpi. These results showed that transcripts of NtPR-Q accumulate in response to inoculation with R. solanacearum, suggesting NtPR-Q might be involved in defense reactions of tobacco to R. solanacearum.
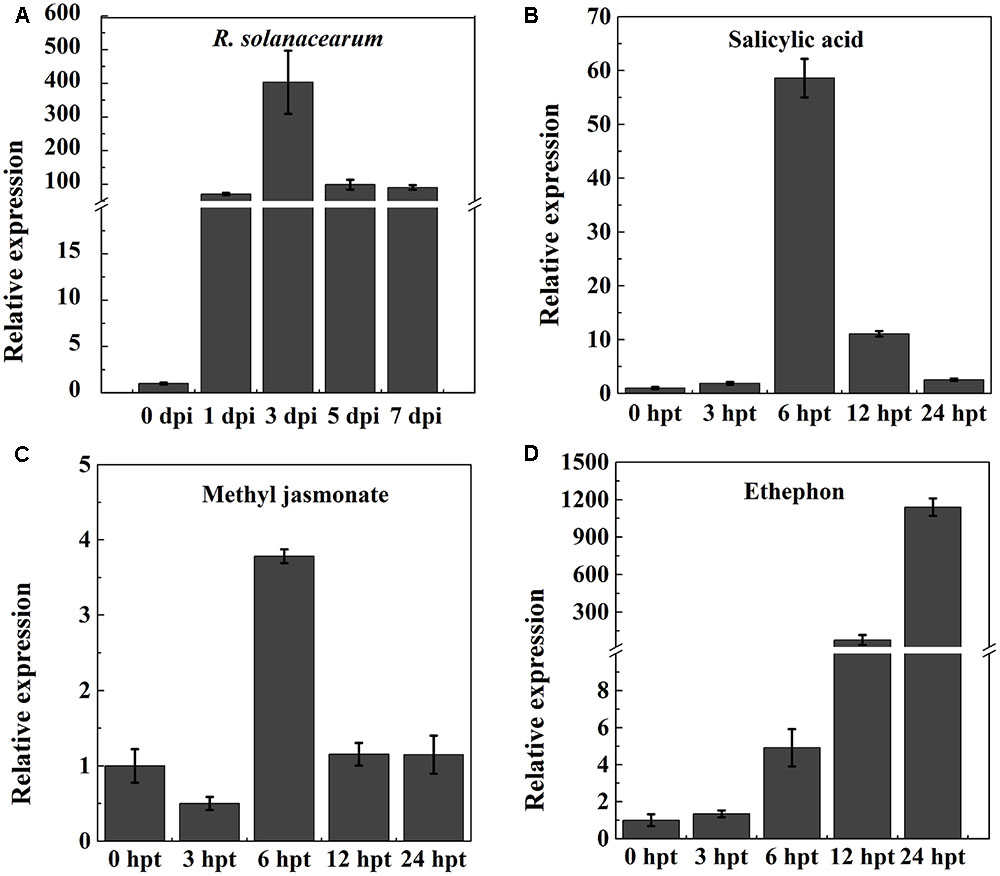
FIGURE 1. RT-qPCR analysis of the relative NtPR-Q transcript levels in WT-Yunyan87 tobacco plants exposed to stress. (A) Relative NtPR-Q transcript levels measured at different time points in WT-Yunyan87 tobacco after inoculation with Ralstonia solanacearum. Plants inoculated with sterile water served as control. (B–D) Relative NtPR-Q transcript levels measured at different time points in WT-Yunyan87 tobacco after treatment with salicylic acid, methyl jasmonate and ethephon. Control plants were sprayed with 10% ethanol or sterile water. NtUBI3 as the reference gene. The averages presented are based on three independent biological replicates for each set of plants.
Phytohormones are important signaling molecules that play an important role in the regulation of plant defense reactions against external stresses and mediating the expression of downstream defense genes (Fujita et al., 2006). To evaluate the possible involvement of NtPR-Q in the hormone signaling pathways, NtPR-Q transcript levels were determined by RT-qPCR in tobacco leaves after exogenous treatments with SA, MeJA, and ethephon. SA application enhanced the NtPR-Q transcript levels between 0 and 24 h, reaching the maximum at approximately 6 h (Figure 1B). In response to MeJA, the transcript levels of NtPR-Q was reduced at 3 h, but increased to the maximal level at 6 h and then returned to the basal level at 24 h (Figure 1C). After ethephon treatment, the NtPR-Q transcript levels increased between 0 and 24 h (Figure 1D). The transcript levels of NtPR-Q after the application of ethephon were the highest of the three treatments.
Overexpression of NtPR-Q Enhances the Resistance of Tobacco to R. solanacearum
Differences in expression of NtPR-Q in response to R. solanacearum inoculation and the treatment of hormones suggested a role for this gene in plant immunity. To test this possibility, we established transgenic tobacco NtPR-Q-OE under the control of the 35S promoter in the pVCT2024 vector (Figure 2A). PCR analysis was used to verify the T-DNA insertion in transgenic tobacco lines (Figure 2B). The NtPR-Q transcript levels in NtPR-Q-OE and WT-Yunyan87 plants were determined in 10 independent transgenic T1 tobacco lines via RT-qPCR. The transgenic lines 8 and 9 exhibited higher levels of NtPR-Q mRNA than other NtPR-Q-OE lines and were therefore used for subsequent experiments (Figure 2C). We did not observe any differences in the growth phenotype between the NtPR-Q-OE transgenic plants and WT-Yunyan87 plants (Figure 2D).
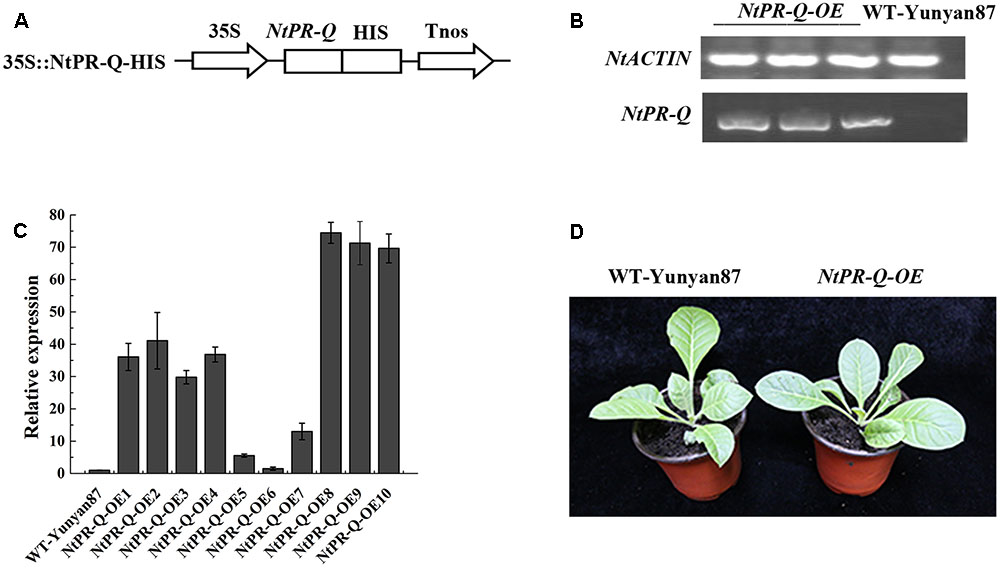
FIGURE 2. Generation of NtPR-Q-overexpressing tobacco plants. (A) Schematic diagram of the 35S::NtPR-Q-HIS fusion protein construct. (B) PCR analysis of the T-DNA insertion in representative transgenic tobacco plants. WT-Yunyan87 tobacco plants as the blank control, and NtACTIN served as an endogenous control. (C) Relative transcripts of NtPR-Q in NtPR-Q-OE lines and WT-Yunyan87 plants were tested by RT-qPCR. The relative transcript levels in WT-Yunyan87 plants as the control, NtUBI3 as the reference gene. The averages presented are based on three independent biological replicates for each set of plants. Error bars indicate the standard error. (D) Growth phenotypes of NtPR-Q-OE plants and WT-Yunyan87 plants.
NtPR-Q-OE and WT-Yunyan87 tobacco plants were infected by highly virulent R. solanacearum strain CQPS-1 via root irrigation. At 15 dpi, we observed clear wilting symptoms on a majority of WT-Yunyan87 plants, whereas only a few NtPR-Q-OE tobacco plants exhibited slight disease symptom (Figure 3A). At 10, 14, 18, and 22 dpi, the disease indexes of NtPR-Q-OE tobacco plants were approximately 0.06, 0.28, 0.55, and 1.11, respectively, but more intense wilting symptoms were observed in the WT-Yunyan87 plants, the disease indexes were approximately 1.56, 2.61, 3.33, and 3.89, respectively (Figure 3B). To measure the bacterial growth, WT-Yunyan87 plants and NtPR-Q-OE tobacco were inoculated by 20 μL inoculum (1 × 108 CFU mL-1) into the third leaves. The results showed that the amount of R. solanacearum in the leaves of WT-Yunyan87 plants was significantly higher than NtPR-Q-OE tobacco plants at 0, 1, 3, and 5 dpi (Figure 3C).
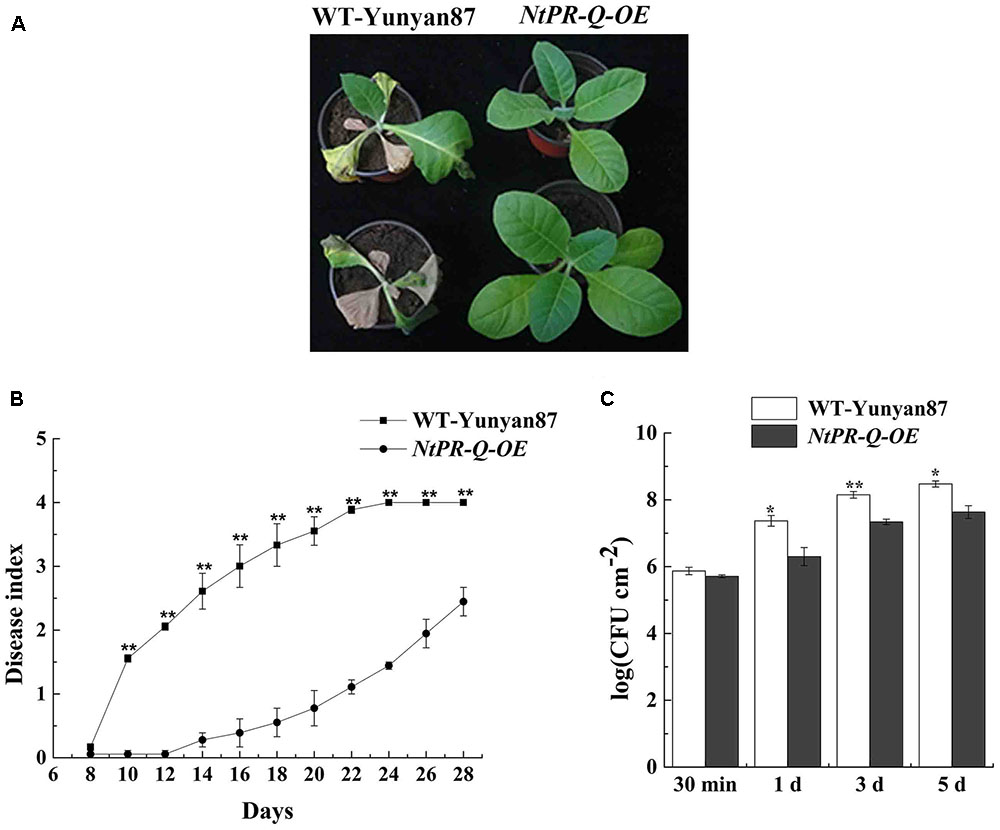
FIGURE 3. Overexpression of NtPR-Q enhances tobacco resistance to R. solanacearum. (A) Four-leaf-stage plants of the representative NtPR-Q-OE line and its parental WT-Yunyan87 15 days after irrigation with a 10 mL suspension (1 × 108 CFU mL-1). (B) R. solanacearum-inoculated plants were scored using a disease index ranging from 0 to 4: 0 = asymptomatic plant, 1 = less than 25% leaves wilted, 2 = less than 50% wilted leaves, 3 = more than 50% wilted leaves and 4 = completely wilted leaves (dead plant). The averages presented are based on three independent biological replicates for each set of plants. (C) R. solanacearum growth assay. The amount of R. solanacearum in the leaves of WT-Yunyan87 and NtPR-Q-OE tobacco plants at 30 min, 1, 3, and 5 dpi. The leaves were inoculated with a 20 μL suspension (1 × 108 CFU mL-1) of the highly virulent R. solanacearum strain CQPS-1. The averages presented are based on three independent biological replicates, each comprising three plants. Error bars indicate the standard error, Asterisks indicates a significant difference (t-test, ∗p < 0.05, ∗∗p < 0.01).
To further affirm the role of NtPR-Q in disease resistance and explore the possible mechanism that how NtPR-Q overexpression enhanced tobacco resistance to R. solanacearum, the transcriptional expression of defense-related genes in NtPR-Q-OE plants were investigated by RT-qPCR. We examined the transcript levels of the SA-responsive genes NtPR1a/c, NtPR2, and NtCHN50 (Brogue et al., 1991; Ward et al., 1991a), the JA-responsive gene NtPR1b (Sohn et al., 2007), ET production-associated genes NtACC Oxidase and NtEFE26 (Chen et al., 2003), the HR-associated genes NtHSR201 and NtHIN1 (Sohn et al., 2007) and the reactive oxygen species (ROS) detoxification-associated genes NtGST1 and NtCAT1 (Takahashi and Nagata, 1992; Takahashi et al., 1997). All the genes were shown to be up-regulated in response to pathogen infection (Rizhsky et al., 2002; Sohn et al., 2007; Chen et al., 2012). The results of RT-qPCR showed that the transcript levels of these genes were significantly increased in NtPR-Q-OE plants except for NtCAT1 (Figure 4).
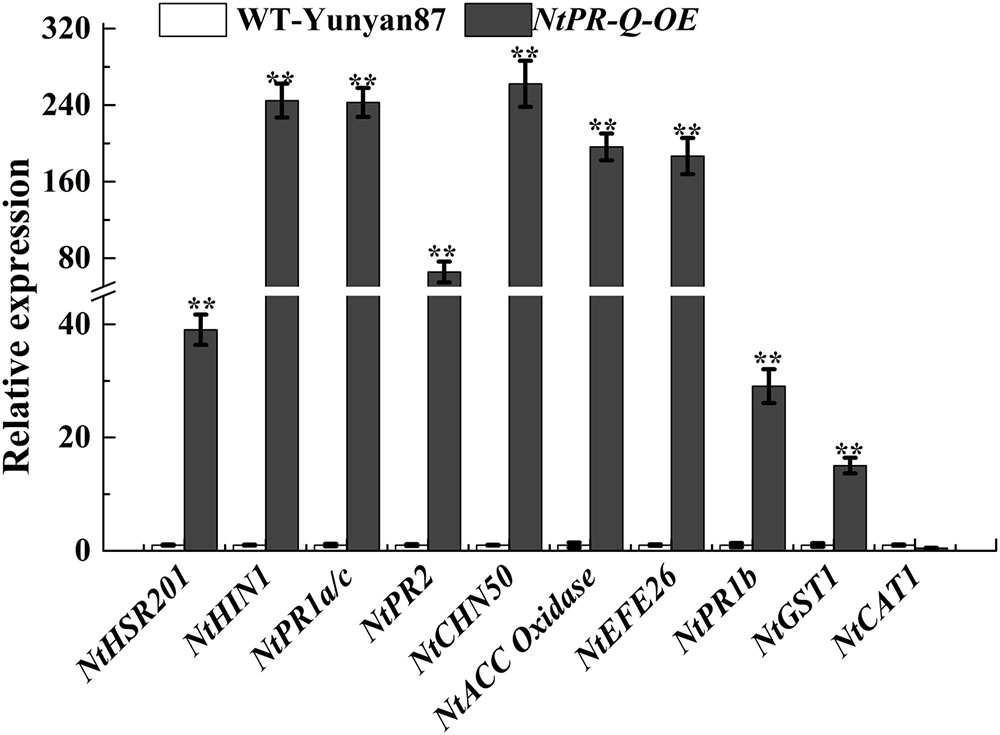
FIGURE 4. RT-qPCR analysis of relative expression of defense marker genes in NtPR-Q-OE plants compared with WT-Yunyan87 plants. Defense-related gene transcript levels in WT-Yunyan87 plants were used as controls, NtUBI3 as the reference gene. Error bars indicate the standard error. The averages presented are based on three independent replicates. Asterisks indicate a significant difference (t-test, ∗p < 0.05, ∗∗p < 0.01).
The transcriptional profiles of defense-related genes in R. solanacearum-infected tobacco NtPR-Q-OE8, NtPR-Q-OE9, and WT-Yunyan87 were investigated by RT-qPCR (Figure 5). After R. solanacearum infection, the transcript levels of HR- (NtHIN1), SA- (NtPR1a/c, NtPR2 and NtCHN50), ET- (NtEFE26, NtACC Oxidase), JA- (NtPR1b) and ROS- (NtGST1) associated genes in NtPR-Q-OE plants were higher than in WT-Yunyan87 plants at least at three of the four tested timepoints. While, expression levels of NtHIN1 in WT-Yunyan87 and NtPR-Q-OE plants were similar.
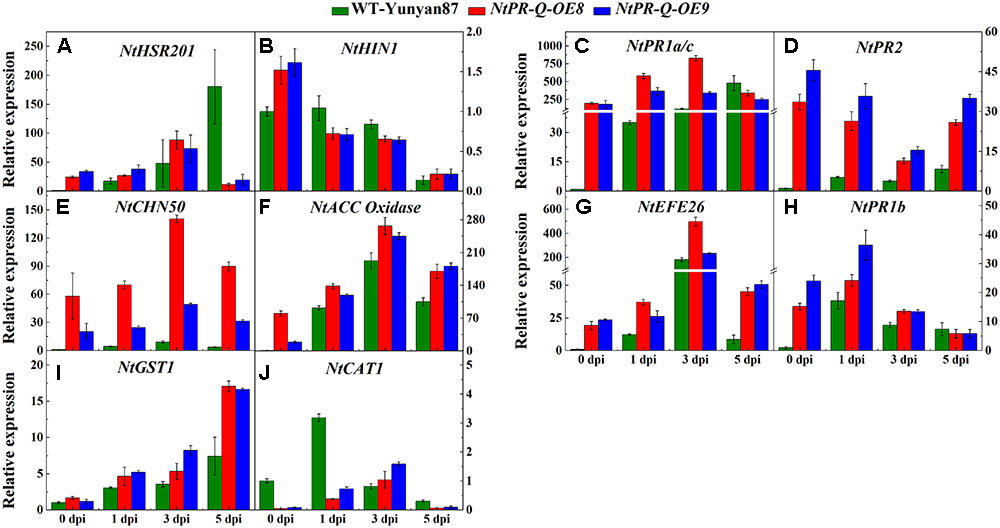
FIGURE 5. Relative expression of defense-related marker genes in NtPR-Q-OE8, NtPR-Q-OE9 and WT-Yunyan87 tobacco plants after R. solanacearum inoculation. RT-qPCR analysis of relative transcript levels of defense-related genes in NtPR-Q-OE and WT-Yunyan87 plants at 0, 1, 3, and 5 dpi. (A,B) Relative transcript levels of the hypersensitive response (HR)-maker genes NtHSR201 and NtHIN1. (C–E) Relative transcript levels of the salicylic acid (SA)-responsive genes NtPR1a/c, NtPR2 and NtCHN50. (F,G) Relative transcript levels of the ethylene biosynthesis-associated genes NtACC Oxidase and NtEFE26. (H) Relative transcript levels of the jasmonic acid-responsive gene NtPR1b. (I,J) Relative transcript levels of the reactive oxygen species detoxification-associated genes NtCAT1 and NtGST1. Plants inoculated with sterile water as control, NtUBI3 as the reference gene. The averages presented are based on three independent biological replicates for each set of plants. Error bars indicate the standard error.
Effect of Exogenous SA, MeJA, and Ethephon Application on Disease Control
Salicylic acid, MeJA, and ethephon application led to an increase in NtPR-Q transcript levels (Figure 1). To assess the capacity of them in the resistance of tobacco to R. solanacearum, the disease index was surveyed after treatment with 2 mM SA, 0.1 mM MeJA, and 7 mM ethephon. The results showed that the disease index after MeJA treatment was lower than control but the difference was not significant. Tobacco plants treated with SA were significantly more resistant than the control. Ethephon application also obvious enhanced the plants resistance at 10–22 days but had no significant effect at final stage. We found that the application of these hormones enhanced the resistance of tobacco to R. solanacearum, with the effect of SA application being the best (Table 2).
Discussion
Numerous studies have shown that the members of PR genes are involved in the resistance of plants to various biotic and abiotic stresses (Schultheiss et al., 2003). The expression levels of many PR genes are induced by many factors (Datta and Muthukrishnan, 1999), such as fungi, bacteria, and nematodes (Fujibe et al., 2000; Hamamouch et al., 2011), as well as SA, β-aminobutyric acid (Li et al., 2014), cold and drought (Gao et al., 2013; Chen et al., 2015), light (Sessa et al., 1995). Gao et al. (2013) found that the expression of PR3 was up-regulated under salt stress. Similarly, Xanthomonas campestris pv. pruni (Xcp) led to the induction of peach PR transcripts, including Pp-PR1b, three PR5 genes Pp-TLPA, Pp-TLP2, and Pp-TLP3, to a significantly higher degree in the resistant cultivar (Sherif et al., 2012). In addition, overexpression of defense-related proteins, such as PR protein and WRKY transcription factor lead to the up-regulation of PR-Q expression level. A pepper basic PR1 gene overexpression in tobacco enhances resistance against heavy metals and pathogens Phytophthora nicotianae, R. solanacearum and Pseudomonas syringae pv. tabaci and increases the expression of NtPR-Q and glutathione S-transferase genes (Sarowar et al., 2005). The expression of NtPR-Q in CaWRKY40-overexpressing tobacco was higher than that in wild-type plants (Dang et al., 2013). We found that the expression of NtPR-Q was markedly induced by R. solanacearum attack and a variety of plant hormone treatments. Overexpression NtPR-Q in tobacco enhanced the resistance to R. solanacearum attack. PR-Q, a member of the PR-3 family, is a class II chitinase. Plant chitinase were previously shown to be involved in the resistance of plant to fungi and harmful insects (Zhu et al., 1994; Golshani et al., 2015). In this paper, NtPR-Q as a tobacco chitinase was found to be functional in plant resistance to R. solanacearum infection.
The transcript accumulation of HR-associated genes and PR genes is typically observed during defense responses and serves as a marker for this type of biological condition. The enhanced resistance of NtPR-Q-OE plants was associated with the enhanced expression of defense-associated genes, such as the HR-associated genes NtHSR201 and NtHIN1, the SA-responsive genes NtPR1a/c, NtPR2, and NtCHN50, ET production-associated genes such as NtACC Oxidase and NtEFE26, the JA-responsive gene NtPR1b and the reactive oxygen species (ROS) detoxification-associated gene NtGST1. Thus, NtPR-Q, a PR protein, promotes the transcription levels of a range of HR- and defense-associated genes, resulting in enhancing tobacco resistance against R. solanacearum.
Salicylic acid, JA, and ET have been shown to activate the PR genes expression of plants in varying degrees and act either synergistically (Devadas et al., 2002; Mur et al., 2006) or antagonistically (Kunkel and Brooks, 2002) during defense signaling, dependent on their concentrations. In peach, Pp-PR1a was induced by SA, whereas the Pp-PR1b, Pp-TLPA, Pp-TLP2, and Pp-TLP3 genes were induced mainly by ET or MeJA treatments. The induction of the same set of PR genes in response to pathogen infection, MeJA or ethephon treatment suggests the involvement of JA/ethylene-signaling pathways in mediating resistance against Xcp (Sherif et al., 2012). In this paper, NtPR-Q was induced by SA, MeJA and ethephon application, and the overexpression of NtPR-Q enhanced the expression of SA-dependent NtPR1a/c, NtPR2, and NtCHN50, JA-responsive NtPR1b and ET associated genes NtACC Oxidase and NtEFE26 by R. solanacearum infection. These results suggest that NtPR-Q might influence SA, JA and ET-mediated defense signaling during R. solanacearum infection. Although there were a lot of studies to explore the role of chitinase in plant disease resistance, it only tested the plant resistance to fungi pathogens and the expression of other PR genes was not analyzed (Zhu et al., 1994; Jongedijk, 1995). Our RT-qPCR analysis showed that the NtPR-Q overexpression enhanced the expression of a whole range of defense-related genes, which seems to support the conclusion that NtPR-Q induces plant SAR. We speculate that the constitutive expression of PR-Q results in the accumulation of unfolded proteins in the endoplasmic reticulum (ER), which leads to ER stress and activates defense signal pathways, thereby enhances the resistance of plants to R. solanacearum. This phenomenon is referred as unfolded protein response (UPR) (Chakraborty et al., 2016; Wan and Jiang, 2016). However, to verify this hypothesis, we still need further evidence.
Conclusion
It was concluded that tobacco plants exhibited resistance to bacterial wilt induced by R. solanacearum when NtPR-Q was constitutively expressed, which indicates potency of NtPR-Q in plant resistance breeding. However, it is still needs further study on the specific biological functions of NtPR-Q and its involvement in the resistance to other environmental stresses.
Author Contributions
The experiment was designed by WD and QL. YT, QL, and YL accomplished the experiments. YT analyzed the data. YT and WD wrote the paper, and the paper was revised by YT, QL, and LZ.
Funding
The experiment was sponsored by the Key Projects of the China National Tobacco Corporation (110201502019, 110201601025 LS-05).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank Dr. Xingguo Zhang (College of Horticulture and Landscape Architecture, Southwest University) for providing the plant expression vector pVCT2024.
References
Agrios, G. N. (1988). “How pathogens attack plants,” in Plant Pathology (Elsevier: Academic Press), 63–86.
Brogue, K., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., et al. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. doi: 10.1126/science.254.5035.1194
Chakraborty, R., Ji, H. B., Bae, E. Y., Kim, W.-Y., Lee, S. Y., Kim, M. G., et al. (2016). Comparison and contrast of plant, yeast, and mammalian ER stress and UPR. Appl. Biol. Chem. 59, 337–347. doi: 10.1007/s13765-016-0167-6
Chen, L., Yu, S., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, L. J., Ren, H., Deng, X. G., Li, Y. N., Cha, W. Q., Lin, H. H., et al. (2015). Effects of light intensity on the susceptibility of Nicotiana tabacum to cucumber mosaic virus. J. Gen. Plant Pathol. 81, 399–408. doi: 10.1007/s10327-015-0602-2
Chen, N., Goodwin, P. H., and Hsiang, T. (2003). The role of ethylene during the infection of Nicotiana tabacum by Colletotrichum destructivum. J. Exp. Bot. 54, 2449–2456. doi: 10.1093/jxb/erg289
Dang, F. F., Wang, Y. N., Lu, Y. U., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Datta, S. K., and Muthukrishnan, S. (1999). Pathogenesis-Related Proteins of Plants. Boca Raton, FL: CRC Press, 21–47.
Devadas, S. K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1, mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. doi: 10.1046/j.1365-313X.2002.01300.x
Fujibe, T., Watanabe, K., Nakajima, N., Ohashi, Y., Mitsuhara, I., Yamamoto, K. T., et al. (2000). Accumulation of pathogenesis-related proteins in tobacco leaves irradiated with UV-B. J. Plant Res. 113, 387–394. doi: 10.1007/PL00013946
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Gao, Q., Zeng, X., Jia, L., Niu, D., Li, X., Guan, M., et al. (2013). The expression profiling of rice pathogenesis-related proteins in seedling stage under environmental stresses. Prog. Biochem. Biophys. 40, 1140. doi: 10.3724/SP.J.1206.2013.00014
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–207. doi: 10.1146/annurev.phyto.43.040204.135923
Golshani, F., Fakheri, B. A., Behshad, E., and Vashvaei, R. M. (2015). PRs proteins and their mechanism in plants. Biol. Forum Int. J. 7, 477–495.
Hamamouch, N., Li, C., Seo, P. J., Park, C. M., and Davis, E. L. (2011). Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 12, 355–364. doi: 10.1111/j.1364-3703.2010.00675.x
Horita, M., Suga, Y., Ooshiro, A., and Tsuchiya, K. (2010). Analysis of genetic and biological characters of Japanese potato strains of Ralstonia solanacearum. J. Gen. Plant Pathol. 76, 196–207. doi: 10.1007/s10327-010-0229-2
Ji, P., Momol, M. T., Olson, S. M., Pradhanang, P. M., and Jones, J. B. (2005). Evaluation of thymol as biofumigant for control of bacterial wilt of tomato under field conditions. Plant Dis. 89, 497–500. doi: 10.1094/PD-89-0497
Jia, H., Wang, C., Wang, F., Liu, S., Li, G., and Guo, X. (2015). GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLOS ONE 10:e0120646. doi: 10.1371/journal.pone.0120646
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jongedijk, E. (1995). Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica 85, 173–180. doi: 10.1007/BF00023946
Kessmann, H., Staub, T., Ligon, J., Oostendoro, M., and Ryals, J. (1994). Activation of systemic acquired disease resistance in plants. Eur. J. Plant Pathol. 100, 359–369. doi: 10.1007/BF01874804
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Kushalappa, A. C., Yogendra, K. N., and Karre, S. (2016). Plant innate immune response: qualitative and quantitative resistance. Crit. Rev. Plant Sci. 35, 38–55. doi: 10.1080/07352689.2016.1148980
Li, S., Xu, C., Wang, J., Guo, B., Yang, L., Chen, J., et al. (2017). Cinnamic, myristic and fumaric acids in tobacco root exudates induce the infection of plants by Ralstonia solanacearum. Plant Soil 412, 381–395. doi: 10.1007/s11104-016-3060-5
Li, X., Wei, F., Niu, D., Guan, M., Miao, L., Shi, J., et al. (2014). Expression analysis of pathogenesis-related 1 proteins in normal growth of rice leaves and interactions between rice and Xanthomonas oryzae pv.oryzae. Chin. Bull. Bot. 49, 127–138. doi: 10.3724/SP.J.1259.2014.00127
Liu, Y., Tang, Y., Qin, X., Yang, L., Jiang, G., Li, S., et al. (2017a). Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front. Microbiol. 8:974. doi: 10.3389/fmicb.2017.00974
Liu, Y., Wu, D., Liu, Q., Zhang, S., Tang, Y., Jiang, G., et al. (2017b). The sequevar distribution of Ralstonia solanacearum, in tobacco-growing zones of China is structured by elevation. Eur. J. Plant Pathol. 147, 541–551. doi: 10.1007/s10658-016-1023-6
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Oh, S. K., Park, J. M., Joung, Y. H., Lee, S., Chung, E., Kim, S. Y., et al. (2005). A plant EPF-type zinc-finger protein, CaPIF1, involved in defence against pathogens. Mol. Plant Pathol. 6, 269–285. doi: 10.1111/j.1364-3703.2005.00284.x
Ohme-Takagi, M. Jr., Meins, F., and Shinshi, H. (1998). A tobacco gene encoding a novel basic class II chitinase: a putative ancestor of basic class I and acidic class II chitinase genes. Mol. Genet. Genomics 259, 511–515. doi: 10.1007/s004380050842
Payne, G., Ahl, P., Moyer, M., Harper, A., Beck, J., Meins, F., et al. (1990). Isolation of complementary DNA clones encoding pathogenesis-related proteins P and Q, two acidic chitinases from tobacco. Proc. Natl. Acad. Sci. U.S.A. 87, 98–102. doi: 10.1073/pnas.87.1.98
Prior, P., and Fegan, M. (2005). Recent development in the phylogeny and classification of Ralstonia solanacearum. Acta Hortic. 695, 127–136. doi: 10.17660/ActaHortic.2005.695.14
Rizhsky, L., Liang, H., and Mittler, R. (2002). The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151. doi: 10.1104/pp.006858
Safavi, K., and Zareie, R. (2012). Constitutive expression of thaumatin-like protein (TLP-3) in transgenic tobacco plants leads to enhance resistance to Alternaria alternata. Arch. Phytopathol. Plant Prot. 45, 161–169. doi: 10.1080/03235408.2010.507947
Sarowar, S., Kim, Y. J., Kim, E. N., Kim, K. D., Hwang, B. K., Islam, R., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 24, 216–224. doi: 10.1007/s00299-005-0928-x
Schultheiss, H., Dechert, C., Király, L., Fodor, J., Michel, K., and Kogel, K. H. (2003). Functional assessment of the pathogenesis-related protein PR-1b in barley. Plant Sci. 165, 1275–1280. doi: 10.1016/S0168-9452(03)00336-4
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., and De, Bolle MF (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Sessa, G., Yang, X. Q., Raz, V., Eyal, Y., and Fluhr, R. (1995). Dark induction and subcellular localization of the pathogenesis-related PRB-1b protein. Plant Mol. Biol. 28, 537–547. doi: 10.1007/BF00020400
Sherif, S., Paliyath, G., and Jayasankar, S. (2012). Molecular characterization of peach PR genes and their induction kinetics in response to bacterial infection and signaling molecules. Plant Cell Rep. 31, 697–711. doi: 10.1007/s00299-011-1188-6
Sohn, S. I., Kim, Y. H., Kim, B. R., Lee, S. Y., Lim, C. K., Hur, J. H., et al. (2007). Transgenic tobacco expressing the hrpN(EP) gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea. Mol. Cells 24, 232–239.
Takahashi, H., Chen, Z., Du, H., Liu, Y., and Klessig, D. F. (1997). Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11, 993–1005. doi: 10.1046/j.1365-313X.1997.11050993.x
Takahashi, Y., and Nagata, T. (1992). parB: an auxin-regulated gene encoding glutathione S-transferase. Proc. Natl. Acad. Sci. U.S.A. 89, 56–59. doi: 10.1073/pnas.89.1.56
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., et al. (1992). Acquired resistance in arabidopsis. Plant Cell 4, 645–656. doi: 10.2307/3869523
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wan, S., and Jiang, L. (2016). Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253, 753–764. doi: 10.1007/s00709-015-0852-z
Wang, Y., Dang, F., Liu, Z., Wang, X., Eulgem, T., Lai, Y., et al. (2013). CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 14, 131–144. doi: 10.1111/j.1364-3703.2012.00836.x
Ward, E. R., Payne, G. B., Moyer, M. B., Williams, S. C., Dincher, S. S., Sharkey, K. C., et al. (1991a). Differential regulation of beta-1, 3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 96, 390–397. doi: 10.1104/pp.96.2.390
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., et al. (1991b). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. doi: 10.2307/3869297
Yang, L., Ding, W., Xu, Y., Wu, D., Li, S., Chen, J., et al. (2016). New insights into the antibacterial activity of hydroxycoumarins against Ralstonia solanacearum. Molecules 21:468. doi: 10.3390/molecules21040468
Zhao, D. Y., Shen, L., Fan, B., Yu, M., Zheng, Y., and Lv, S. (2009). Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits. FEBS Lett. 583, 3329–3334. doi: 10.1016/j.febslet.2009.09.029
Keywords: NtPR-Q, Nicotiana tabacum, Ralstonia solanacearum, plant resistance, pathogenesis-related proteins
Citation: Tang Y, Liu Q, Liu Y, Zhang L and Ding W (2017) Overexpression of NtPR-Q Up-Regulates Multiple Defense-Related Genes in Nicotiana tabacum and Enhances Plant Resistance to Ralstonia solanacearum. Front. Plant Sci. 8:1963. doi: 10.3389/fpls.2017.01963
Received: 15 February 2017; Accepted: 31 October 2017;
Published: 16 November 2017.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Felix Mauch, University of Fribourg, SwitzerlandMassimiliano Morelli, Istituto per la Protezione Sostenibile delle Piante (CNR), Italy
Copyright © 2017 Tang, Liu, Liu, Zhang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ding, ZHdpbmc4MThAMTYzLmNvbQ==
 Yuanman Tang
Yuanman Tang Qiuping Liu
Qiuping Liu Ying Liu
Ying Liu Wei Ding
Wei Ding