
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 14 November 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01946
Survival of plants and nearly all organisms depends on the pterin based molybdenum cofactor (Moco) as well as its effective biosynthesis and insertion into apo-enzymes. To this end, both the central Moco biosynthesis enzymes are characterized and the conserved four-step reaction pathway for Moco biosynthesis is well-understood. However, protection mechanisms to prevent degradation during biosynthesis as well as transfer of the highly oxygen sensitive Moco and its intermediates are not fully enlightened. The formation of protein complexes involving transient protein-protein interactions is an efficient strategy for protected metabolic channelling of sensitive molecules. In this review, Moco biosynthesis and allocation network is presented and discussed. This network was intensively studied based on two in vivo interaction methods: bimolecular fluorescence complementation (BiFC) and split-luciferase. Whereas BiFC allows localisation of interacting partners, split-luciferase assay determines interaction strengths in vivo. Results demonstrate (i) interaction of Cnx2 and Cnx3 within the mitochondria and (ii) assembly of a biosynthesis complex including the cytosolic enzymes Cnx5, Cnx6, Cnx7, and Cnx1, which enables a protected transfer of intermediates. The whole complex is associated with actin filaments via Cnx1 as anchor protein. After biosynthesis, Moco needs to be handed over to the specific apo-enzymes. A potential pathway was discovered. Molybdenum-containing enzymes of the sulphite oxidase family interact directly with Cnx1. In contrast, the xanthine oxidoreductase family acquires Moco indirectly via a Moco binding protein (MoBP2) and Moco sulphurase ABA3. In summary, the uncovered interaction matrix enables an efficient transfer for intermediate and product protection via micro-compartmentation.
Molybdenum (Mo) belongs to the group of essential metals like iron, zinc, manganese, or copper, which are used as cofactors or in prosthetic groups. These metals ensure redox enzyme functions in all organisms (Hänsch and Mendel, 2009). The transition metal Mo is important due to its differing electron configurations (Holm et al., 2010). However, Mo is biologically inactive unless it is complexed by a specific prosthetic group. With the exception of the bacterial nitrogenase (Hu and Ribbe, 2013), all Mo-containing enzymes (Mo-enzyme) use Mo imbedded in a pterin based scaffold (Mendel, 2013). This molybdenum cofactor (Moco) is highly conserved during evolution and Moco containing enzymes can be found throughout all kingdoms of life (Schwarz and Mendel, 2006). Mo-enzymes are essential for the global carbon, sulphur, and nitrogen cycles (Hille, 2002).
The Moco biosynthesis four-step reaction pathway as well as the biosynthesis enzymes are highly conserved and can be found in nearly all prokaryotes and eukaryotes. These enzymes are designated in plants by an abbreviation for cofactor for nitrate reductase and xanthine dehydrogenase (Cnx1-3 and 5-7). As shown in Figure 1A, Cnx2 and Cnx3 catalyse the first step by converting 5'-GTP to cyclic pyranopterin monophosphate inside mitochondria (cPMP; Hänzelmann et al., 2002; Santamaria-Araujo et al., 2004). The resulting cPMP is exported into the cytosol by the ATM3 transporter (Teschner et al., 2010) for introducing a dithiolene group by transferring two sulphur atoms forming molybdopterin (MPT). This second reaction step is catalysed by the MPT synthase, a heterotetramer with two large Cnx6 and two small Cnx7 subunits. After sulphur transfer, the Cnx7 subunits need to be re-sulphurated via the MPT synthase sulphurase Cnx5 (Matthies et al., 2004). The Mo insertase Cnx1 catalyses the last two steps of biosynthesis. Cnx1 consists of a larger N-terminal Cnx1E domain and a smaller C-terminal Cnx1G domain. MPT is bound by the Cnx1G domain and is activated through adenylation, thus forming MPT-AMP in the third step (Kuper et al., 2003). This intermediate is transferred to the Cnx1E domain, which releases AMP and inserts Mo derived from molybdate (Llamas et al., 2006). This required molybdate is imported by specific molybdate transporters (Gasber et al., 2011; Tejada-Jiménez et al., 2013).
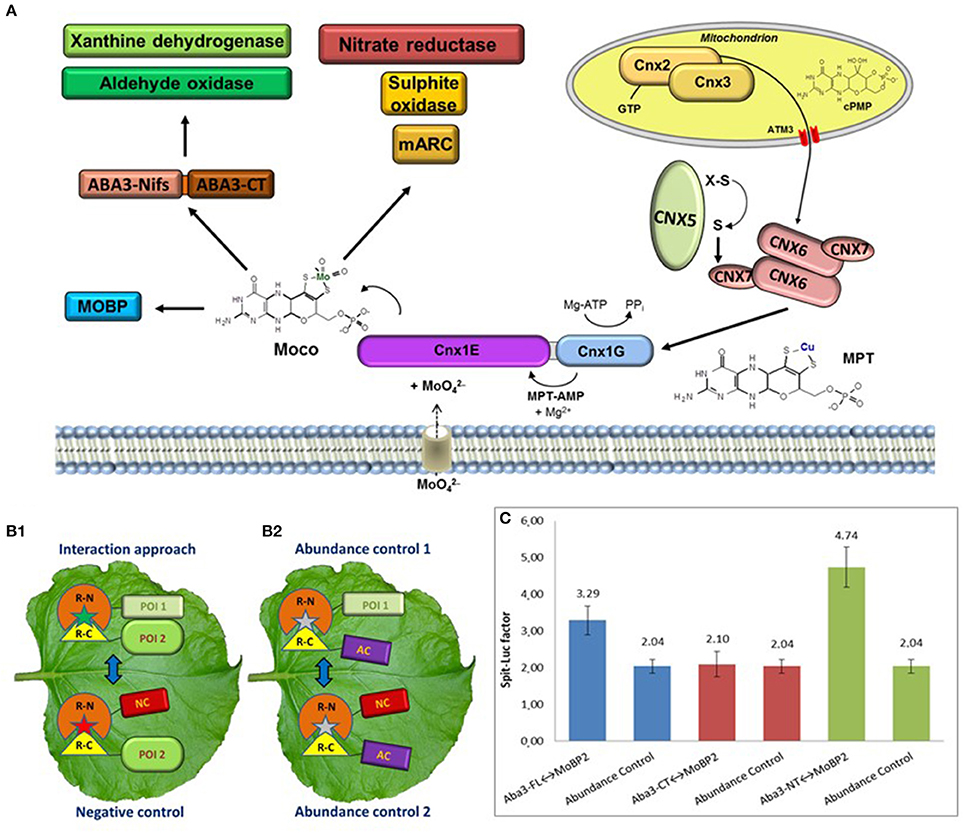
Figure 1. (A) The Moco enzyme pathway with the four-step reaction catalysed by Cnx2 and Cnx3 inside mitochondria as well as Cnx6, Cnx7, Cnx5, and Cnx1 in the cytosol. After biosynthesis, Moco is distributed to both families of Mo-enzymes and to Moco binding proteins. (B) Schematic presentation of an interaction approach and the necessary three controls for a split-protein assay. Depicted are the used constructs with the N-terminal (R-N) and the C-terminal (R-C) fragment of a reporter. (B1) The interaction approach needs two POI, while the negative control replace one POI for a non-interacting negative control protein (NC). (B2) In the abundance controls, a non-interacting abundance control protein (AC) is used. (C) FLuCI interaction study between ABA3 and MoBP2. Analysed was the full-length protein (FL) as well as the single N-terminal (NT) and C-terminal domain. Shown is the split-luciferase factor of the interaction approach in relation to the negative control. In addition, the split-luciferase factors of the two abundance controls are depicted, demonstrating the background factor without interaction. For interaction between the POI's, the split-luciferase factor has to be higher than the background factor.
The five Mo-enzymes found in plants are classified in two families based on the coordination chemistry of Mo in their active centre (Mendel and Schwarz, 2011). The directly produced di-oxo form of Moco is used by the first family, named sulphite oxidase family (SO-family). The SO-family consists of three members: sulphite oxidase (SO; Hänsch and Mendel, 2005), nitrate reductase (NR; Campbell, 1999), and mitochondrial amidoxine reducing component (mARC; Havemeyer et al., 2006). The second Mo-enzyme family, the xanthine oxidoreductase family (XOR-family), consists of aldehyde oxidase (AO; Seo et al., 2000) and xanthine dehydrogenase (XDH; Hille and Nishino, 1995). The XOR-family possesses the sulphurised mono-oxo Moco, which is produced by an additional biosynthesis step through the Moco sulphurase ABA3 in Arabidopsis (Wollers et al., 2008) and FLACCA in tomato (Sagi et al., 2002).
For plant survival, the most important Mo-enzyme is the cytosolic NR. It catalyses the first step of nitrate assimilation. The conversion of nitrate to nitrite is essential for plant growth and development (Campbell, 1996). The peroxisomal SO enables stress tolerance against sulphur dioxide (Brychkova et al., 2015; Baillie et al., 2016). XDH has a role in the catabolism of purines by catalysing the oxidation of hypoxanthine to xanthine and finally to uric acid (Brychkova et al., 2008) while producing superoxide anions (Zarepour et al., 2010). AO is involved in the biosynthesis of the phytohormone abscisic acid (Seo et al., 2000) as well as in detoxification of carbonyl aldehydes in stressed Arabidopsis siliques (Srivastava et al., 2017). The dependence of plants on functional Mo-enzymes highlights the necessity of efficient and reliable Moco production as well as its correct insertion into the apo-enzymes. However, both Moco and its intermediates are described as highly oxygen sensitive (Rajagopalan and Johnson, 1992). Therefore, a freely diffusible pool of molecules in the cytosol is rather unlikely due to the high degradation risk during biosynthesis and transfer. This problem can be circumvented by a protection mechanism via direct transfer of intermediates from one protein to the next, which has not been described for any organism so far. However, such a metabolic channelling in a protein complex is an efficient strategy to protect sensitive molecules (Miles et al., 1999; James and Viola, 2002) which can be studied using molecular-biological methods on cellular level. The presented study on the Moco-biosynthesis complex can function as a blueprint for investigations of other physiological protein networks in plants.
A protected Moco transfer requires tight protein-protein interaction between involved proteins within the plant cell. In vivo protein-protein interaction studies were performed inside living cells thus providing natural conditions for protein-protein interaction. In contrast, in vitro experiments just give indications for the physiological processes but cannot mimic e.g., compartmentation and physiological micro-environment inside the living cell and thus lack the complexity of in vivo experiments. Conditions of in vivo experiments have to be monitored very accurately. The used concentration of proteins of interest (POI) in the cells depends on transformation and expression/degradation levels, respectively. Furthermore, results will be influenced by both reaction conditions as well as reaction partners depending on age, growing stage or vitality of the plant cells. These prerequisites have to be equalised by many experimental replications and the usage of appropriate negative and abundance controls. However, only in vivo interaction studies are able to verify the preconditions of a potential substrate channelling with so many partners in one protein complex.
The majority of developed in vivo interaction studies are based on fusion proteins consisting of the POI's and a split reporter protein (Stynen et al., 2012). There are only a few alternative in vivo methods like yeast-two-hybrid assay (Y2H; Causier and Davies, 2002) or Foerster resonance energy transfer (FRET; Bhat et al., 2006). The Y2H is often used for screenings for new interaction partners, however, the heterologous system counteract measuring in the natural environment. On the other hand, FRET studies are performed inside plant cells, but measurement and interpretation are much more sophisticated than for split reporter systems. Both the bimolecular fluorescence complementation (BiFC; Waadt et al., 2008; Gehl et al., 2009) and the floated leaf luciferase complementation imaging assay (FLuCI; Gehl et al., 2011), use the ability of a split reporter to reconstitute and to resume activity after bringing the split parts into close proximity. Interaction of the fused POI's increases the reconstitution of the reporter termini, which increases the measureable fluorescence or luminescence, respectively, of the analysed cells compared to a negative control with reconstitution only by chance (Figure 1B1). It has to be considered, that a fused reporter protein could mask an interaction site by steric hindrance. Therefore, all possible orientations of reporter fusions to both POI's have to be tested for reliable results.
All split-protein assays need additional controls to validate the results of the interaction approach (Figure 1B2). Random reconstitution depends on the concentrations of both fusion partners. However, the abundance of constructs with the POI and the negative control construct may be variable due to altered protein expression or degradation, which could yield false positive or negative results. Moreover, the behaviour of random reconstitution under in vivo conditions is multifaceted. This fact demanded the measurement of the background level of random reconstitution inside a living cell, called abundance control, which additionally has to be performed when measuring in vivo protein-protein interactions (Kaufholdt et al., 2013).
Both assays have differences in the nature of reconstitution of the used reporter proteins, which makes the combination of these assays reasonable. Fluorescence reporter termini in the BiFCs assay reconstitute irreversibly to full reporter functionality (Rose et al., 2010), which allows semi-quantitative measurement of interactions, even if these interactions are weak or only once occurring in a protein life cycle (Kaufholdt et al., 2016b). In contrast, the reconstituted luciferase termini of the FluCI assay are reversibly bound and able to disjoin from each other after the contact between the POI's is finished. This dynamic reporter system allows for the measurement of interaction strengths (Chen et al., 2008).
The basis for a direct transfer of metabolites via protein-protein interaction is the localisation of all involved enzymes in the same cell compartment. Therefore, localisation of all Moco pathway proteins was investigated via fluorescence reporter studies. With exception of the enzymes of the first Moco biosynthesis step Cnx2 and Cnx3 that are localised inside mitochondria, the enzymes Cnx1, Cnx5, Cnx6, Cnx7, and ABA3 were all localised within the cytosol (Kaufholdt et al., 2013). The Mo-enzymes NR, XDH, and AO have also been localised within the cytosol (Dalling et al., 1972; Koiwai et al., 2004; Kaufholdt et al., 2016b). A tonoplast association is discussed for XDH (Ma et al., 2016). The exceptions are the peroxisomal SO (Nowak et al., 2004) and mARC, which was localised in the outer membrane of mitochondria in mammals (Havemeyer et al., 2006).
Starting at reaction step one of the Moco biosynthesis, Kaufholdt et al. (2013) tested the mitochondrial protein pair Cnx2/Cnx3 using BiFC and FLuCI assays. Interaction approaches in organelles demand organelle-specific negative and abundance control proteins. Such a set of mitochondria-specific control vectors enabled interaction studies between the proteins of the first biosynthesis step Cnx2 and Cnx3. BiFC revealed an interaction between these two proteins and showed a punctual fluorescence distributed within the cytosol representing mitochondria. This interaction could be verified by the FLuCI assay.
Both Cnx2 and Cnx3 are unable to interact with the other Moco biosynthesis enzymes for direct cPMP transfer because of the localisation in different compartments (Figure 1). In addition, an interaction of these enzymes with the transporter ATM3 to transfer their product cPMP is still unknown. However, an unprotected export is also feasible because cPMP is the least oxygen sensitive Moco biosynthesis intermediate. Furthermore, cPMP is stable enough for cPMP-mediated therapy of human Moco deficiency, which is unique for Moco intermediates (Santamaria-Araujo et al., 2012).
As a next step, the cytosolic enzymes were analysed for interaction (Kaufholdt et al., 2013). MPT-synthase consists of two Cnx6 and two Cnx7 subunits. Interactions were found between the protein pairs Cnx6/Cnx6 and Cnx6/Cnx7 via BiFC and FLuCI assays. The interaction strength of both pairs is very high and the strongest of all protein pairs tested, which indicates a permanent contact of these proteins. Therefore, a stable heterotetrameric complex of the four subunits was concluded, which verified the results of the crystal structure analysis of Escherichia coli MPT-synthase complex by Rudolph et al. (2001).
The C-terminal region of the small MPT synthase subunit Cnx7 is highly conserved in all Moco depending organisms. The terminal double glycine motif carries the sulphur atom for transfer to cPMP and inserts into a pocket of each large subunit forming two probably independent active sites (Rudolph et al., 2001). Mutations of the glycine motif in E. coli (Schmitz et al., 2007) and human (Hänzelmann et al., 2002) homologs decreased the sulphur transfer activity dramatically. In the FLuCI-studies, Kaufholdt et al. (2013) investigated whether this functional impairment is caused by a disturbance of protein-protein interaction between the MPT subunits by point mutations of the penultimate glycine. The interaction strength of Cnx6 to wildtype and mutant Cnx7 were directly compared to each other. Both a mutation of glycine to a larger phenylalanine as well as to the charged glutamate decreased the interaction strength by a third and by half, respectively. This indicates that mutations in the last two glycines lead to a loss of function of the MPT-synthase due to a disturbance of the heterotetramer formation.
MPT-synthase sulphurase Cnx5 as well as the Moco insertase Cnx1 were tested each against both subunits of the MPT-synthase. Interactions were detected for the protein pairs Cnx5/Cnx7 and Cnx1/Cnx6, while the protein pair Cnx5/Cnx6 as well as Cnx1/Cnx7 showed no interaction. Therefore, a direct contact was concluded for the sulphur transfer from Cnx5 to Cnx7 as well as for the MPT transfer from Cnx6 to Cnx1. In addition, the E-domain of Cnx1 was identified as the interaction domain with Cnx6, which was also verified by additional crosslinking experiments (Kaufholdt et al., 2013). However, in contrast to the permanent interactions within the MPT-synthase, interaction strengths of the protein pairs Cnx5/Cnx7 and Cnx1/Cnx6 were distinct but less intensive, so they seem to be of transient nature.
Micro-compartmentation of biosynthesis complexes at the cytoskeleton is frequently observed in plant cells (Gutierrez et al., 2009; Marek et al., 2011). First indications for a cytoskeleton anchoring were also seen for the Moco biosynthesis complex. The plant Mo insertase Cnx1 has an animal homologue named gephyrin, which was shown to anchor neuronal postsynaptic inhibitory glycine receptors to polymerised tubulin (Kirsch et al., 1993). In addition, in vitro studies depicted that plant Cnx1 was co-sedimented with rabbit filamentous actin (Schwarz et al., 2000). To verify these indications inside the plant cell, BiFC assays were used by Kaufholdt et al. (2016a) for analyses of possible interactions of the Moco biosynthetic complex with both actin filaments and microtubules. A non-invasive indirect labelling of the cytoskeleton was used via cytoskeleton binding proteins fused to the reporter fragments. Direct fusion of cytoskeleton proteins disturbs cytoskeleton multimerisation or can mask binding sites. Two binding proteins were used for both cytoskeleton types: (i) Lifeact from Saccharomyces cerevisiae (Riedl et al., 2008; Era et al., 2009) and the actin binding domain 2 of fimbrin from Arabidopsis thaliana (Sano et al., 2005) for actin filaments as well as (ii) microtubule binding domains of the protein Casein-Kinase-1-Like-6 (Ben-Nissan et al., 2008) and the Microtubule Associated Protein 65-1 (Smertenko et al., 2004) from A. thaliana for microtubules.
Cnx1 was the only cytosolic Moco biosynthesis protein that showed an interaction with actin filaments. Beside a stronger fluorescence compared to the negative control, a specific nucleus-concentrated fluorescence pattern—forming a star-like pattern on the nuclear basket—also indicates interaction of the Cnx1 to the cytoskeleton in this kind of BiFC approach. A more detailed analysis of the two Cnx1 domains identified Cnx1G as the main interaction site of Cnx1 to filamentous actin. However, microtubule interaction was not detectable for any of the Moco proteins.
After identification of Cnx1 as an actin binding protein, anchoring of the entire Moco biosynthesis complex to actin filaments was hypothesized and therefore studied by a BiFC approach including labelled Cnx6 and actin binding proteins with fluorescence protein fragments. This approach with additionally expressed unlabelled Cnx1 formed the star-like pattern characteristic for an actin filament interaction. Therefore, Cnx1 acts as bridge protein binding to filamentous actin with its Cnx1G domain as well as to proteins of the Moco biosynthesis with its Cnx1E domain.
After biosynthesis, Moco has to be transferred to apo-Mo-enzymes in a protected way. Cnx1 delivers di-oxo Moco, which is prepared for incorporation into the apo-Mo-enzymes of the SO-family. BiFC assays indicate direct interaction in the cytosol of Cnx1 with NR and SO, respectively. This leads to the assumption of a Moco insertion before importing holo-SO into peroxisomes.
No interaction of Cnx1 neither with the mono-oxo Moco using Mo-enzymes XDH/AO nor with ABA3 was detectable, which excludes direct metabolite transfer. However, an indirect transfer is plausible, because the Moco binding protein MoBP2 might act as a potential bridge protein between Cnx1 and ABA3. An interaction with both enzymes was identified in a screening using the FLuCI assay. As example, the selected interaction study of ABA3 and MoBP2 is depicted in Figure 1C to demonstrate the necessity of abundance controls for identification of the N-terminal domain of ABA3 as exclusive interaction domain with MoBP2. In addition, a one-time-only interaction of XDH1 with the C-terminal domain of ABA3 was identified. Therefore, after processing of Moco a direct transfer from ABA3 to the apo-XOR-enzymes is suggested.
In vivo protein-protein interaction assays helped to explain the multifaceted Moco biosynthesis protein network in subcellular compartments inside the plant cell. The irreversible complementation of the BiFC assay enabled to enlighten and to localise every type of interaction, permanent interactions of complexes as well as transient interactions. In contrast, the reversible and dynamic complementation of the luciferase reporter in the FLuCI assay enabled characterizing the interaction strength between protein pairs.
The presented data allow for generating an interaction matrix as well as defining interaction strengths for proteins of the Moco biosynthesis proteins in higher plants. This micro-compartmentation inside the cytosol is a precondition for metabolic channelling. During the passage through the three cytosolic reaction steps, the oxygen sensitive intermediates of Moco can be channelled from one protein to the next inside a biosynthesis complex. This protein complex structure permits an evolutionary physiological advantage by economising with the trace element molybdate via efficient resource and energy management.
Cnx1 anchors this whole complex on actin filaments as a bridge protein by mediating an indirect interaction of the MPT-synthase to the cytoskeleton. During sulphuration of Cnx7, Cnx5 is also part of this micro-compartmentation. Different hypotheses are possible to explain a spatial anchoring of Moco biosynthesis complex on the cytoskeleton for increasing biosynthesis efficiency. (i) Actin binding could increase the stability of the complex. Cnx1 first makes contact to actin followed by recruitment of the other components of the complex. A free diffusion in the cytosol could disturb these interactions and consequently the reaction efficiency. (ii) The anchoring could be important for positioning the whole complex near specific transporters to get the required substrates directly after import. One of these transporters is the mitochondrial ATM3 exporter, which transfers cPMP out of mitochondria. (iii) Another group of transporters provides molybdate for insertion into MPT by Cnx1. An efficient supply of molybdate is important for an uninterrupted reaction cascade inside the Moco biosynthesis complex. The animal Cnx1 homologue gephyrin clusters neuronal receptors at the postsynaptic membrane via microtubule binding. In an analogous manner, Cnx1 could cluster molybdate transporters on actin filaments for direct substrate channelling via protein interaction.
The interaction network between the Moco biosynthesis complex and the Mo-enzymes forms a potential allocation pathway reducing the risk of oxygen damage of Moco. Mo-enzymes using di-oxo Moco have a direct interaction with the biosynthesis complex while Mo-enzymes using mono-oxo Moco receive the prosthetic group via an indirect pathway. Furthermore, the bypass of Moco via a binding protein and ABA3 leads to the hypothesis that di-oxo Moco is processed prior to insertion into the members of the XOR-family.
For the first time, the complete Moco pathway interaction network has been identified in plants (Figure 2) which might help to understand the closely related biosynthesis in animal and human cells. Although a direct tracking of Moco intermediates through this complex is not practicable with available methods so far, this uncovered protein structure permits the conclusion that micro-compartmentation is a central mechanism inside plant cells to regulate and protect the substrate flow on physiological level.
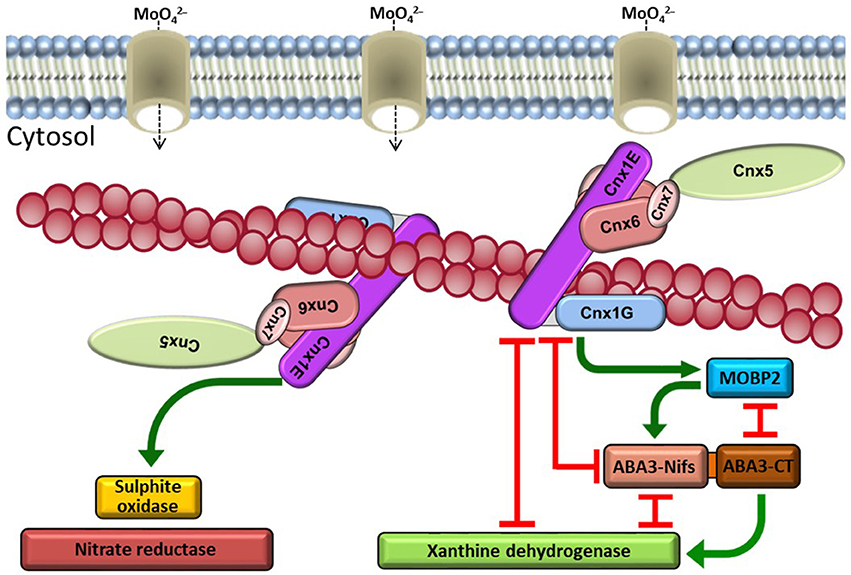
Figure 2. Schematic presentation of the Moco biosynthesis interaction network. The cytosolic Moco biosynthesis enzymes Cnx5, Cnx6, Cnx7, and Cnx1 form a multi-enzyme complex on actin filaments. Molybdate as substrate is provided by molybdate transporters. An assembly of the Moco biosynthesis complex near these transporters at the cytoskeleton is hypothesised. After insertion of Mo from molybdate, di-oxo Moco is inserted into enzymes of the SO family via interaction with Cnx1. The enzymes of the XOR-family receive the mono-oxo form of Moco from ABA3, which generates this form of Moco from di-oxo Moco supplied by Cnx1 via MoBP2 as mediating protein.
DK and RH were primarily involved in drafting the manuscript and DK produced the figures. C-KB and RM critically read the manuscript and improved the text. RRM and RH conceived the study and coordinated the work. All of the authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge the contributions of all bachelor and master students from our research institution that aided the authors in collecting the data for analyses.
Baillie, C. K., Kaufholdt, D., Karpinski, L. H., Schreiber, V., Hänsch, S., Evers, C., et al. (2016). Detoxification of volcanic sulfur surplus in planta: three different strategies of survival. Environ. Exp. Bot. 126, 44–54. doi: 10.1016/j.envexpbot.2016.02.007
Ben-Nissan, G., Cui, W., Kim, D., Yang, Y., Yoo, B., and Lee, J. (2008). Arabidopsis Casein Kinase 1-Like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 148, 1897–1907. doi: 10.1104/pp.108.129346
Bhat, R. A., Lahaye, T., and Panstruga, R. (2006). The visible touch: in planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2:12. doi: 10.1186/1746-4811-2-12
Brychkova, G., Alikulov, Z., Fluhr, R., and Sagi, M. (2008). A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 54, 496–509. doi: 10.1111/j.1365-313X.2008.03440.x
Brychkova, G., Yarmolinsky, D., Batushansky, A., Grishkevich, V., Khozin-Goldberg, I., Fait, A., et al. (2015). Sulfite oxidase activity is essential for normal sulfur, nitrogen and carbon metabolism in tomato leaves. Plants 4, 573–605. doi: 10.3390/plants4030573
Campbell, W. H. (1996). Nitrate reductase biochemistry comes of age. Plant Physiol. 111, 355–361. doi: 10.1104/pp.111.2.355
Campbell, W. H. (1999). Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 277–303. doi: 10.1146/annurev.arplant.50.1.277
Causier, B., and Davies, B. (2002). Analysing protein-protein interactions with the yeast two-hybrid system. Plant Mol. Biol. 50, 855–870. doi: 10.1023/A:1021214007897
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 46, 368–376. doi: 10.1104/pp.107.111740
Dalling, M. J., Tolbert, N. E., and Hageman, R. H. (1972). Intracellular location of nitrate reductase and nitrite reductase. I. Spinach and tobacco leaves. Biochim. Biophys. Acta 283, 505–512. doi: 10.1016/0005-2728(72)90266-6
Era, A., Tominaga, M., Ebine, K., Awai, C., Saito, C., Ishizaki, K., et al. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50, 1041–1048. doi: 10.1093/pcp/pcp055
Gasber, A., Klaumann, S., Trentmann, O., Trampczynska, A., Clemens, S., Schneider, S., et al. (2011). Identification of an Arabidopsis solute carrier critical for intracellular transport and inter organ allocation of molybdate. Plant Biol. 13, 710–718. doi: 10.1111/j.1438-8677.2011.00448.x
Gehl, C., Kaufholdt, D., Hamisch, D., Bikker, R., Kudla, J., Mendel, R. R., et al. (2011). Quantitative analysis of dynamic protein-protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. Plant J. 67, 542–553. doi: 10.1111/j.1365-313X.2011.04607.x
Gehl, C., Waadt, R., Kudla, J., Mendel, R. R., and Hänsch, R. (2009). New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Mol. Plant. 2, 1051–1058. doi: 10.1093/mp/ssp040
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M. C., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Hänsch, R., and Mendel, R. R. (2005). Sulfite oxidation in plant peroxisomes. Photosyn. Res. 86, 337–343. doi: 10.1007/s11120-005-5221-x
Hänsch, R., and Mendel, R. R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12, 259–266. doi: 10.1016/j.pbi.2009.05.006
Hänzelmann, P., Schwarz, G., and Mendel, R. R. (2002). Functionality of alternative splice forms of the first enzymes involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 277, 18303–18312. doi: 10.1074/jbc.M200947200
Havemeyer, A., Bittner, F., Wollers, S., Mendel, R. R., Kunze, T., and Clement, B. (2006). Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281, 34796–34802. doi: 10.1074/jbc.M607697200
Hille, R. (2002). Molybdenum and tungsten in biology. Trends Biochem. Sci. 27, 360–367. doi: 10.1016/S0968-0004(02)02107-2
Hille, R., and Nishino, T. (1995). Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. FASEB J. 9, 995–1003.
Holm, R. H., Solomon, E. I., Majumdar, A., and Tenderholt, A. (2010). Comparative molecular chemistry of molybdenum and tungsten and its relation to hydroxylase and oxotransferase enzymes. Coord. Chem. Rev. 255, 993–1015. doi: 10.1016/j.ccr.2010.10.017
Hu, Y., and Ribbe, M. W. (2013). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. J. Biol. Chem. 288, 13173–13177. doi: 10.1074/jbc.R113.454041
James, C. L., and Viola, R. E. (2002). Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry 41, 3726–3731. doi: 10.1021/bi0159074
Kaufholdt, D., Baillie, C. K., Bikker, R., Burkart, V., Dudek, C. A., von Pein, L., et al. (2016a). The molybdenum cofactor biosynthesis complex interacts with actin filaments via molybdenum insertase Cnx1 as anchor protein in Arabidopsis thaliana. Plant Sci. 244, 8–18. doi: 10.1016/j.plantsci.2015.12.011
Kaufholdt, D., Baillie, C. K., Meyer, M. H., Schwich, O. D., Timmerer, U. L., Tobias, L., et al. (2016b). Identification of a protein-protein interaction network downstream of molybdenum cofactor biosynthesis in Arabidopsis thaliana. J. Plant Physiol. 207, 42–50. doi: 10.1016/j.jplph.2016.10.002
Kaufholdt, D., Gehl, C., Geisler, M., Jeske, O., Voedisch, S., Ratke, C., et al. (2013). Visualization and quantification of protein interactions in the biosynthetic pathway of molybdenum cofactor in Arabidopsis thaliana. J. Exp. Bot. 64, 2005–2016. doi: 10.1093/jxb/ert064
Kirsch, J., Wolters, I., Triller, A., and Betz, H. (1993). Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature 366, 745–748. doi: 10.1038/366745a0
Koiwai, H., Nakaminami, K., Seo, M., Mitsuhashi, W., Toyomasu, T., and Koshiba, T. (2004). Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 134, 1697–1707. doi: 10.1104/pp.103.036970
Kuper, J., Winking, J., Hecht, H. J., Mendel, R. R., and Schwarz, G. (2003). The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G. Arch. Biochem. Biophys. 411, 36–46. doi: 10.1016/S0003-9861(02)00714-2
Llamas, A., Otte, T., Multhaup, G., Mendel, R. R., and Schwarz, G. (2006). The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J. Biol. Chem. 281, 18343–18350. doi: 10.1074/jbc.M601415200
Ma, X., Wang, W., Bittner, F., Schmidt, N., Berkey, R., Zhang, L., et al. (2016). Dual and opposing roles of xanthine dehydrogenase in defense-associated reactive oxygen species metabolism in Arabidopsis. Plant Cell 28, 1108–1126. doi: 10.1105/tpc.15.00880
Marek, M., Merten, O. W., Galibert, L., Vlak, J. M., and van Oers, M. M. (2011). Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J. Virol. 85, 5350–5362. doi: 10.1128/JVI.00035-11
Matthies, A., Rajagopalan, K. V., Mendel, R. R., and Leimkühler, S. (2004). Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 5946–5951. doi: 10.1073/pnas.0308191101
Mendel, R. R. (2013). The molybdenum cofactor. J. Biol. Chem. 288, 13165–13172. doi: 10.1074/jbc.R113.455311
Mendel, R. R., and Schwarz, G. (2011). Molybdenum cofactor biosynthesis in plants and humans. Coord. Chem. Rev. 255, 1145–1158. doi: 10.1016/j.ccr.2011.01.054
Miles, E. W., Rhee, S., and Davies, D. R. (1999). The molecular basis of substrate channeling. J. Biol. Chem. 274, 12193–12196. doi: 10.1074/jbc.274.18.12193
Nowak, K., Luniak, N., Witt, C., Wüstefeld, Y., Wachter, A., Mendel, R. R., et al. (2004). Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol. 45, 1889–1894. doi: 10.1093/pcp/pch212
Rajagopalan, K. V., and Johnson, J. L. (1992). The pterin molybdenum cofactors. J. Biol. Chem. 267, 10199–10202.
Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607. doi: 10.1038/nmeth.1220
Rose, R. H., Briddon, S. J., and Holliday, N. D. (2010). Bimolecular fluorescence complementation: lighting up seven transmembrane domain receptor signalling networks. Br. J. Pharmacol. 159, 738–750. doi: 10.1111/j.1476-5381.2009.00480.x
Rudolph, M. J., Wuebbens, M. M., Rajagopalan, K. V., and Schindelin, H. (2001). Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat. Struct. Mol. Biol. 8, 42–46. doi: 10.1038/83034
Sagi, M., Scazzocchio, C., and Fluhr, R. (2002). The absence of molybdenum cofactor sulfuration is the primary cause of the flacca phenotype in tomato plants. Plant J. 31, 305–317. doi: 10.1046/j.1365-313X.2002.01363.x
Sano, T., Higaki, T., Oda, Y., Hayashi, T., and Hasezawa, S. (2005). Appearance of actin microfilament ‘twin peaks’ in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP–fimbrin. Plant J. 44, 595–605. doi: 10.1111/j.1365-313X.2005.02558.x
Santamaria-Araujo, J. A., Fischer, B., Otte, T., Nimtz, M., Mendel, R. R., Wray, V., et al. (2004). The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 279, 15994–15999. doi: 10.1074/jbc.M311815200
Santamaria-Araujo, J. A., Wray, V., and Schwarz, G. (2012). Structure and stability of the molybdenum cofactor intermediate cyclic pyranopterin monophosphate. J. Biol. Inorgan. Chem. 17, 113–122. doi: 10.1007/s00775-011-0835-2
Schmitz, J., Wuebbens, M. M., Rajagopalan, K. V., and Leimkühler, S. (2007). Role of the C-terminal Gly-Gly motif of Escherichia coli MoaD, a molybdenum cofactor biosynthesis protein with a ubiquitin fold. Biochemistry 46, 909–916. doi: 10.1021/bi062011w
Schwarz, G., and Mendel, R. R. (2006). Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647. doi: 10.1146/annurev.arplant.57.032905.105437
Schwarz, G., Schulze, J., Bittner, F., Eilers, T., Kuper, J., Bollmann, G., et al. (2000). The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12, 2455–2471. doi: 10.1105/tpc.12.12.2455
Seo, M., Peeters, A. J., Koiwai, H., Oritani, T., Marion-Poll, A., Zeevaart, J. A., et al. (2000). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. U.S.A. 97, 12908–12913. doi: 10.1073/pnas.220426197
Smertenko, A. P., Chang, H., Wagner, V., Kaloriti, D., Fenyk, S., Sonobe, S., et al. (2004). The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell 16, 2035–2047. doi: 10.1105/tpc.104.023937
Srivastava, S., Brychkova, G., Yarmolinsky, D., Soltabayeva, A., Samani, T., and Sagi, M. (2017). Aldehyde Oxidase 4 plays a critical role in delaying silique senescence by catalyzing aldehyde detoxification. Plant Physiol. 173, 1977–1997. doi: 10.1104/pp.16.01939
Stynen, B., Tournu, H., Tavernier, J., and Van Dijck, P. (2012). Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 76, 331–382. doi: 10.1128/MMBR.05021-11
Tejada-Jiménez, M., Chamizo-Ampudia, A., Galván, A., Fernández, E., and Llamas, Á. (2013). Molybdenum metabolism in plants. Metallomics 5, 1191–1203. doi: 10.1039/c3mt00078h
Teschner, J., Lachmann, N., Schulze, J., Geisler, M., Selbach, K., Santamaria-Araujo, J., et al. (2010). A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 22, 468–480. doi: 10.1105/tpc.109.068478
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Wollers, S., Heidenreich, T., Zarepour, M., Zachmann, D., Kraft, C., Zhao, Y., et al. (2008). Binding of sulfurated molybdenum cofactor to the C-terminal domain of ABA3 from Arabidopsis thaliana provides insight into the mechanism of molybdenum cofactor sulfuration. J. Biol. Chem. 283, 9642–9650. doi: 10.1074/jbc.M708549200
Keywords: protein-protein interaction network, bimolecular fluorescent complementation (BiFC), split-luciferase, molybdenum cofactor, cytoskeleton, metabolic channelling
Citation: Kaufholdt D, Baillie C-K, Meinen R, Mendel RR and Hänsch R (2017) The Molybdenum Cofactor Biosynthesis Network: In vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 8:1946. doi: 10.3389/fpls.2017.01946
Received: 01 August 2017; Accepted: 30 October 2017;
Published: 14 November 2017.
Edited by:
Michael James Considine, University of Western Australia, AustraliaReviewed by:
Moshe Sagi, Ben-Gurion University of the Negev, Beersheba, IsraelCopyright © 2017 Kaufholdt, Baillie, Meinen, Mendel and Hänsch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert Hänsch, ci5oYWVuc2NoQHR1LWJzLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.