- 1State Key Laboratory of Plant Genomics and National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
Strigolactones (SLs) are the latest confirmed phytohormones that regulate shoot branching by inhibiting bud outgrowth in higher plants. Perception of SLs depends on a novel mechanism employing an enzyme-receptor DWARF14 (D14) that hydrolyzes SLs and becomes covalently modified. This stimulates the interaction between D14 and D3, leading to the ubiquitination and degradation of the transcriptional repressor protein D53. However, the regulation of SL perception in rice remains elusive. In this study, we provide evidences that D14 is ubiquitinated after SL treatment and degraded through the 26S proteasome system. The Lys280 site of the D14 amino acid sequence was important for SL-induced D14 degradation, but did not change the subcellular localization of D14 nor disturbed the interaction between D14 and D3, nor D53 degradation. Biochemical and genetic analysis indicated that the key amino acids in the catalytic center of D14 were essential for D14 degradation. We further showed that D14 degradation is dependent on D3 and is tightly correlated with protein levels of D53. These findings revealed that D14 degradation takes place following D53 degradation and functions as an important feedback regulation mechanism of SL perception in rice.
Introduction
Strigolactones (SLs), a group of carotenoid-derived terpenoid lactones produced by plants, were initially characterized as signals that enable parasitic plants to detect their host (Cook et al., 1966), and also as signals recognized by arbuscular mycorrhizal (AM) fungi in the rhizosphere to build the symbiotic association with host plants (Akiyama et al., 2005). Besides the roles of SLs in the rhizosphere, SLs have been identified as endogenous phytohormones that are transported from roots to shoots and suppress shoot branching by inhibiting the outgrowth of axillary buds (Beveridge et al., 1996; Foo et al., 2001; Booker et al., 2005; Gomez-Roldan et al., 2008; Umehara et al., 2008). In addition, SLs have profound effects in many aspects of plant development, including internode elongation, leaf shape and senescence, shoot gravitropism, stem secondary thickening, root architecture, and the drought tolerance (Al-Babili and Bouwmeester, 2015; Waters et al., 2017).
The key components required for SL biosynthesis and signaling have been identified from genetic characterizations of highly branched mutants, including ramosus (rms) in pea (Pisum sativum), more axillary growth (max) in Arabidopsis thaliana, decreased apical dominance (dad) in petunia (Petunia hybrida), and dwarf (d) or high-tillering dwarf (htd) in rice (Oryza sativa). In the SL biosynthetic pathway, SLs are derived from all-trans-β-carotene, which is converted to 9-cis-β-carotene by the isomerase DWARF27 (D27) (Lin et al., 2009; Alder et al., 2012), and subsequently catalyzed into carlactone by carotenoid cleavage oxygenase 7 (CCD7) and CCD8 (Beveridge et al., 1996; Sorefan et al., 2003; Booker et al., 2004; Seto et al., 2014). The subsequent catalytic reactions are diverse in different species. In Arabidopsis, the cytochrome P450 enzyme MAX1 converts carlactone to carlactonoic acid (CLA), which is further converted to a methyl carlactonoate (MeCLA) by an unknown enzyme (Abe et al., 2014). Subsequently, LATERAL BRANCHING OXIDOREDUCTASE (LBO) converts MeCLA to an unknown SL-like product (MeCLA+16 Da) (Brewer et al., 2016). In rice, the MAX1 homolog Os01g0700900 is responsible for the oxidation of carlactone to 4-deoxyorobanchol (4DO), while another MAX1 homolog, Os01g0701400, functions as an orobanchol synthase that converts 4DO to orobanchol (Zhang et al., 2014). In sorghum, a newly identified sulfotransferase LOW GERMINATION STIMULANT1 (LGS1) is responsible for a change of the dominant SL in root exudates from 5-deoxystrigol to orobanchol via an unknown mechanism and regulates the Striga resistance (Gobena et al., 2017).
In SL signaling, three key components have been identified from genetic screen of SL-insensitive mutants in rice, Arabidopsis, pea, and petunia, which include the α/β-fold hydrolase D14/AtD14/RMS3/DAD2, the F-box protein D3/MAX2/RMS4/PhMAX2A, and the repressor proteins D53/D53-Like SMXLs (Stirnberg et al., 2002, 2007; Ishikawa et al., 2005; Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Hamiaux et al., 2012; Waters et al., 2012; Jiang et al., 2013; Nakamura et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Wang et al., 2015). Perception of SLs depends on a novel mechanism involving the formation of a covalently linked intermediate molecule (CLIM) from the binding of a SL molecule to the receptor D14 and being hydrolyzed by D14. This reaction promotes a conformational change of D14, leading to the interaction between D14 and D3 and triggering the SL signal transduction (Yao et al., 2016). S Ls can also induce the interaction between D14 and D53, leading to ubiquitination and degradation of D53 in a D3- and D14-dependent manner (Jiang et al., 2013; Zhou et al., 2013). D53 contains three ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs, which are essential to recruit the transcriptional co-repressor TOPLESS (TPL) and TPL-related proteins (TPRs) and could potentially repress the activities of target transcription factors (Jiang et al., 2013). Recently, Ideal Plant Architecture1 (IPA1), a key regulator of plant architecture in rice, has been identified as one of the long-speculated transcription factors involved in SL signaling. IPA1 can physically interact with D53 and plays an essential role in the feedback regulation of SL-induced D53 expression (Song et al., 2017).
Although the “de-repression activation” mechanism in SL signaling is similar to the signaling pathways of auxin, gibberellic acid (GA) and jasmonic acid (JA) (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Ueguchi-Tanaka et al., 2005; Chini et al., 2007; Thines et al., 2007; Jiang et al., 2013; Zhou et al., 2013), the SL receptor D14 functions as a non-canonical receptor (Yao et al., 2016). Therefore, the molecular mechanism underlying the inactivation of SL signaling triggered by the D14-SCFD3-D53 complex becomes an important open question. The degradation of signal receptors by the 26S proteasome system is critical for fine-tuning on signal transduction of plant hormones and environmental signals, such as JA, abscisic acid (ABA), ethylene and the blue light (Chen et al., 2007; Kevany et al., 2007; Yan et al., 2013; Kong et al., 2015; Tao et al., 2015; Liu et al., 2016). In Arabidopsis thaliana, AtD14 has been shown to undergo 26S proteasome-dependent degradation, but the molecular mechanism of AtD14 degradation remains elusive (Chevalier et al., 2014). In rice, whether D14 undergoes degradation and the regulatory mechanism underlies SL perception remain unknown. Due to the profound effects of SLs on tillering, a key agronomic trait in cereal crops, investigating the regulation of SL perception is important for improving plant architecture and grain yield of rice. In this study, we show that SLs stimulate the ubiquitination and degradation of D14 through 26S proteasome in rice. A point mutation at Lys280 of D14 could greatly impair D14 degradation, but has little effect on SL signal transduction. We also show that the hydrolase activity of D14 and the intact functions of D3 and D53 are both required for SL-induced D14 degradation. These discoveries have paved a way for elucidating of the inactivation mechanism of the SL perception in rice.
Materials and Methods
Plant Materials
Rice (Oryza sativa L. spp. japanica) mutants used in this study were d53 and d14 from Nipponbare and d3 from Zhonghua 11 (ZH11) as described previously (Jiang et al., 2013; Sang et al., 2014). Rice plants were cultivated in the experimental field of the Institute of Genetics and Developmental Biology at Beijing in the summer and Hainan in the winter. For quantitative PCR with reverse transcription and transient expression analysis, the seedlings of wild-type and mutants were grown in a growth chamber with a 16-h light at 28°C and 8-h dark at 25°C photoperiod with approximately 200 μM m-2 s-1 photon density and 70% humidity. For calli treatment assays, rice calli were cultured on selection medium (Gamborg et al., 1968; Chu et al., 1975) for 7 days at 28°C in a light-avoided environment in greenhouse.
Chemicals and Reagents
The synthetic SL analog GR24, a racemic mixture (rac-GR24) comprising amounts of GR245DS and GR24ent-5DS, was the product from Chiralix, MG132 from Calbiochem, the complete protease inhibitor cocktail (Cat#04693132001) and anti-GFP antibody (Cat#11814460001) from Roche, anti-actin antibody (Cat#M20009L) from Abmart, anti-Flag antibody (Cat#M20008) from Abmart, GFP-Trap®A beads (Cat#120716001A) from Chromotek, TRIzol kit (Cat#15596018) and Superscript III RT kit (Cat#18080-051) from Invitrogen, TURBO DNA-freeTM Kit (Cat#AM1907) from Ambion, SsoFast EvaGreen supermix (Cat#172-5201AP) from Bio-Rad, and Glutathione Sepharose 4 Fast Flow (Cat#17-5132-01) from GE healthcare. The anti-ubiquitin and anti-D53 antibodies are generated as described previously (Jiang et al., 2013; Tian et al., 2015).
Plasmid Construction
The plasmids of Actin:D14-GFP, Actin:D14S147A-GFP and Actin:D14 H297Y-GFP were generated in previous studies (Jiang et al., 2013). To construct the 35S:D14-GFP plasmid, the coding sequence of D14 was amplified with primers of pBI221-D14-F/R (Supplementary Table 1) and recombined into pBI221-GFP vector. To generate the 35S:D14K280E-GFP plasmid, the full-length of D14K280E coding sequences were derived from 35S:D14-GFP plasmid by site-directed mutagenesis using the primers of 35S-D14K280E-F/R (Supplementary Table 1), and subsequently cloned into the pBI221-GFP vector. To construct the plasmid of D14-GFP, the coding sequence of D14 was amplified with primers of Ubi-D14-F/R (Supplementary Table 1) and cloned into pTCK303. To generate the plasmids of D14K33E-GFP, D14K55E-GFP, D14K166E-GFP, D14K246E-GFP and D14K280E-GFP, the full-length of the coding sequences of D14K33E, D14K55E, D14K166E, D14K246E and D14K280E were derived from 35S:D14-GFP by site-directed mutagenesis using primers of D14K33E-F/R, D14K55E-F/R, D14K166E-F/R, D14K246E-F/R and D14K280E-F/R (Supplementary Table 1), respectively, and subsequently recombined into the binary vector pTCK303.
Chemical Treatment of Rice Calli
To examine whether SLs could induce the degradation of D14-GFP, D14K33E-GFP, D14K55E-GFP, D14K166E-GFP, D14K246E-GFP, and D14K280E-GFP, calli of these transgenic lines were cultured on the selection medium (Gamborg et al., 1968; Chu et al., 1975) for 7 days at 28°C and then transferred into a liquid medium. After the treatment with rac-GR24 at the indicated concentration for various hours, calli were collected and frozen at -80°C. Total proteins were extracted with the extraction buffer (50 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl, 10% (v/v) glycerol, 0.1% NP-40, and 1× complete protease inhibitor cocktail). The supernatant was boiled with 5× SDS buffer at 100°C for 10 min and immunoblotting was performed with anti-GFP and anti-Actin antibodies.
In Vivo Ubiquitination Assay
The seedlings of wild-type were cultured in greenhouse for 2 weeks. Protoplasts were prepared from shoot tissues and transformed with 35S:D14-GFP as described (Bart et al., 2006). After incubation at 28°C for 12 h in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES pH 5.7), the protoplasts were collected and pretreated with 50 μM MG132 for 1 h and then treated with 20 μM rac-GR24 or DMSO at 28°C for 3 h. Total proteins were extracted with the extraction buffer (50 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl, 10% (v/v) glycerol, 0.1% NP-40, 50 mM MG132, and 1× complete protease inhibitor cocktail). The lysates were centrifuged at 18,000 g for 20 min at 4°C. The supernatant was then taken for immunoprecipitation and subsequent immunoblot analysis using anti-ubiquitin and anti-GFP antibodies. Under the guidance of the supplier’s instruction, 20 μL of GFP-Trap®A beads were added into 1.2 mL totally extracted proteins and incubated at 4°C for 3 h with gentle rotation. The beads were washed three times with the washing buffer without NP-40 and then boiled with 50 μL SDS-PAGE sample buffer for protein blotting. Mouse anti-ubiquitin monoclonal antibody was used at a 1:2,000 dilution and mouse anti-GFP polyclonal antibodies at a 1:3,000 dilution.
Gene Expression Analysis
The seedlings of wild-type were hydroponic-cultured (pH 5.5) in greenhouse for 2 weeks. After 5 μM rac-GR24 treatment, the shoot base (0.5 cm) of the seedlings were harvested at different time points, total RNAs were extracted using a TRIzol kit according to the manufacturer’s manual and then treated with the TURBO DNA-freeTM Kit and used for complementary DNA synthesis with the Superscript III RT kit. About 12.5 μg total RNA was added into a 20-μL TURBO DNase mixture reaction system and incubated at 37°C for 30 min, and then added 2 μL DNase inactivation reagent and centrifuged at 12,000 g for 10 min, and finally 4 μL of the supernatants was used for complementary DNA synthesis. The quantitative PCR with reverse transcription experiments were performed with gene-specific primers of D14-RT-F/R and D53-RT-F/R (Supplementary Table 1) on a CFX 96 real-time PCR detection system (Bio-Rad). Each reaction volume was set as 10 μL, consisting of 5 μL SsoFast EvaGreen supermix, 0.5 μL sense primer (5 μM), 0.5 μL antisense primer (5 μM), 2.0 μL diluted cDNA, and 2 μL ddH2O. Rice UBIQUITIN (LOC_Os03g13170) gene was used as the internal control.
Co-IP Assay
The seedlings of the wild type were hydroponic-cultured (pH 5.5) in greenhouse for 2 weeks. Protoplasts generated from shoot tissues were transformed with 35S:D14-Flag, 35S:D3-GFP, or 35S-GFP (Bart et al., 2006). After incubation at 28°C for 12 h in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7), proteins were extracted from the collected protoplasts using the extraction buffer (50 mM sodium phosphate buffer, pH 7.0, 150 mM NaCl, 10% (v/v) glycerol, 0.1% NP-40, 50 μM MG132, 1× complete protease inhibitor cocktail) by centrifugation at 20,000 g for 20 min at 4°C and then the supernatant was taken out for Co-IP experiments. Then 20 μL of GFP-Trap®A beads was added into 1.2 mL extracted total proteins and incubated at 4°C for 3 h with gentle rotation in the presence or absence of 10 μM rac-GR24. The beads were washed three times with the washing buffer lack of NP-40 and then boiled with 50 μL SDS-PAGE sample buffer for protein blot. The proteins of D3-GFP and GFP were detected by mouse anti-GFP antibody at a 1:3,000 dilution, and the D14-Flag proteins by mouse anti-FLAG antibody at a 1:2,000 dilution.
Microscopy Analyses
Protoplasts were prepared from shoot tissues of 2-week-old seedlings and transformed with the plasmid of 35S:GFP, 35S:D14-GFP or 35S:D14K280E-GFP. The SV40NLS-mCherry plasmid, which bears a strong nucleus localization signal peptide, was co-transformed into the protoplasts to label the nucleus (Ye et al., 2012). After incubation for 14 h at 28°C in the dark, the protoplasts were collected to observe the GFP and mCherry signals with confocal microscope at the excitation wavelengths of 488 and 559 nm, respectively (FluoView FV1000; Olympus).
Results
SLs Stimulate the Ubiquitination and Degradation of D14
To explore whether the SL receptor undergoes a feedback regulation in rice, we first investigated the D14 protein levels after SL treatment. In Actin:D14-GFP transgenic calli treated with 20 μM rac-GR24, the D14-GFP fusion protein amount begun to decrease at 1 h, and dramatically reduced at 4 h (Figure 1A). We then examined the expression levels of D14 and D53 upon 5 μM rac-GR24 treatment in 2-week-old seedlings, and found that D14 transcripts were unaffected within 6 h treatment, while D53 transcripts were strongly induced after 2 h treatment (Figure 1B). These results indicate that SLs could induce the degradation of the D14 protein but have no effect on D14 transcription.
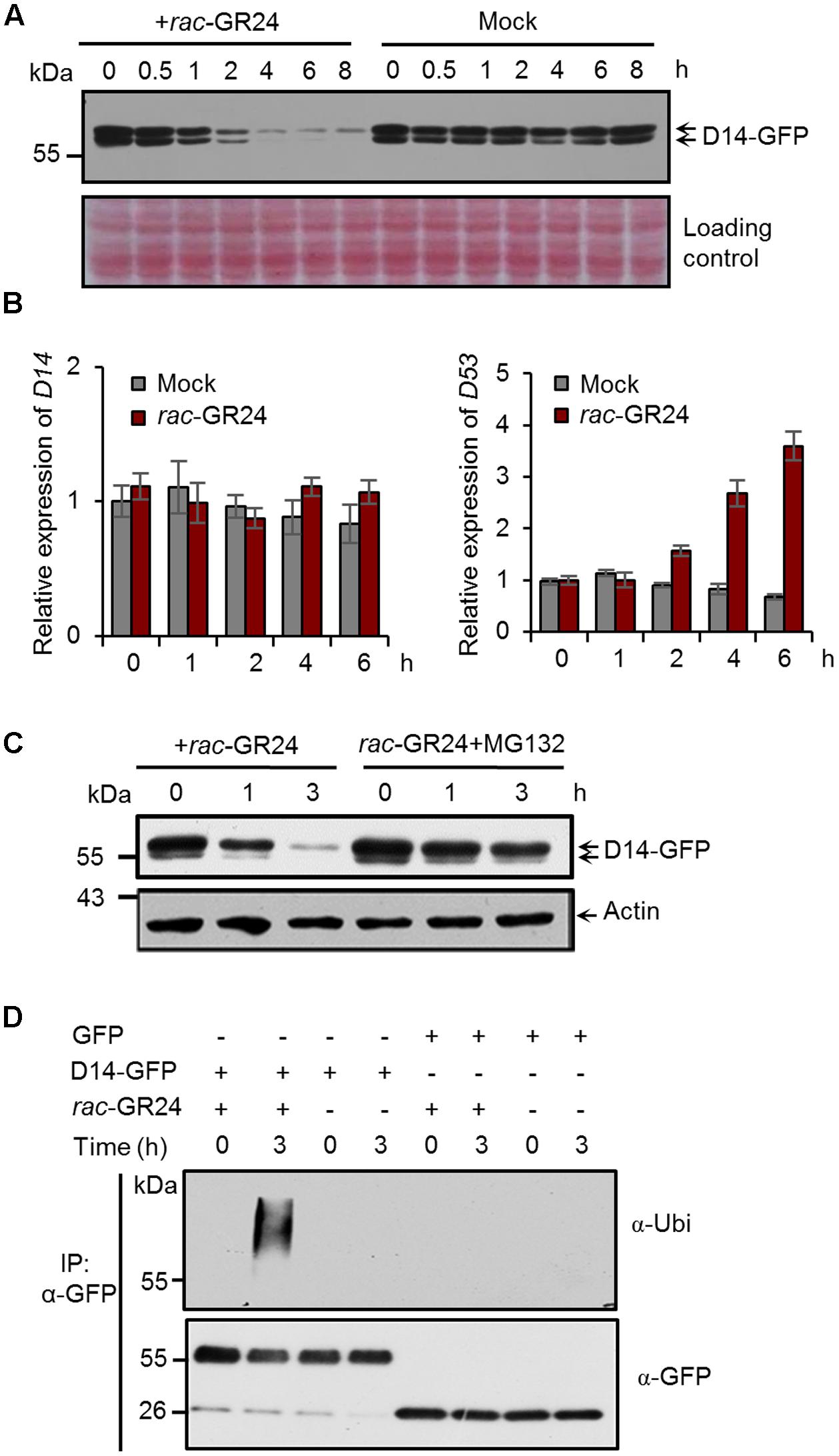
FIGURE 1. Strigolactones (SLs) promote ubiquitination and degradation of D14 in rice. (A) Protein levels of D14-GFP in calli of the Act:D14-GFP/d14 transgenic line at different time points after 20 μM rac-GR24 treatment with DMSO as a control. D14-GFP was detected by immunoblotting with an anti-GFP monoclonal antibody. Relative protein levels were determined by densitometry and normalized to loadings determined by Ponceau staining (red) in the immunoblotting analyses. (B) Relative expression levels of D14 and D53 in 2-week-old seedlings after 5 μM rac-GR24 treatment. Values represent means ± SEM, n = 3. (C) D14-GFP protein levels in calli of the D14-GFP/d14 transgenic line at different time points after rac-GR24 treatment in the presence or absence of MG132. Calli are pretreated with 50 μM MG132 for 1 h and then treated with 20 μM rac-GR24 or DMSO for 3 h. D14-GFP was detected by immunoblotting with an anti-GFP monoclonal antibody. Actin1 were used as loading control in the immunoblotting analyses. (D) Ubiquitination analysis of D14-GFP in rice protoplasts. Rice (Nipponbare) protoplasts were transformed with 35S:D14-GFP plasmids and incubated for 12 h, then pretreated with 50 μM MG132 for 1 h and immediately treated with 20 μM rac-GR24 or DMSO for 3 h. Proteins were extracted for affinity purification with an agarose-immobolized anti-GFP monoclonal antibody and followed by immunoblotting analysis with an anti-ubiquitin (upper panel) or anti-GFP (lower panel) monoclonal antibody.
To investigate whether D14 is degraded through the ubiquitin-26S proteasome system, we detected D14 protein levels in the Actin:D14-GFP transgenic calli treated with 20 μM rac-GR24 in the presence or absence of MG132, and found that the SL-induced D14 degradation was strongly inhibited by MG132, indicating that the 26S proteasome pathway is involved in the degradation of D14 (Figure 1C). We further examined the polyubiquitination of D14 upon rac-GR24 treatment in rice protoplasts transformed with 35S:D14-GFP. After 3 h treatment, the transiently expressed D14-GFP recombinant protein was polyubiquitinated, but no polyubiquitination signal was detected in any other negative control (Figure 1D). Collectively, these data demonstrate that SLs stimulate the ubiquitination and degradation of D14 via the ubiquitin-26S proteasome system in rice.
Identification of Key Amino Acids Responsible for SL-Induced D14 Degradation
To identify the potential ubiquitination sites of D14, we analyzed the protein sequence of D14 using BDM-PUB1. Five lysine sites, K33, K55, K166, K246, and K280, were predicted as potential candidate ubiquitination sites of D14, of which K280 has the highest score (Figure 2A). We then tested whether the degradation of D14 is affected when each ubiquitination site was mutated through creating the point-mutation constructs containing D14K33E-GFP, D14K55E-GFP, D14K166E-GFP, D14K246E-GFP or D14K280E-GFP and expressed them in rice protoplasts, respectively (Figure 2B). After treatment with 10 μM rac-GR24 for 2 h, these recombinant proteins showed degradation at different extents. Compared with the wild-type D14-GFP, the degradation of D14K280E-GFP was severely impaired and the degradation of D14K33E-GFP was moderately decreased upon SL treatment, but the degradation of D14K55E-GFP, D14K166E-GFP or D14K246E-GFP was relatively unaffected (Figure 2C), suggesting that K280 might be the key amino acid for SL-induced D14 degradation.
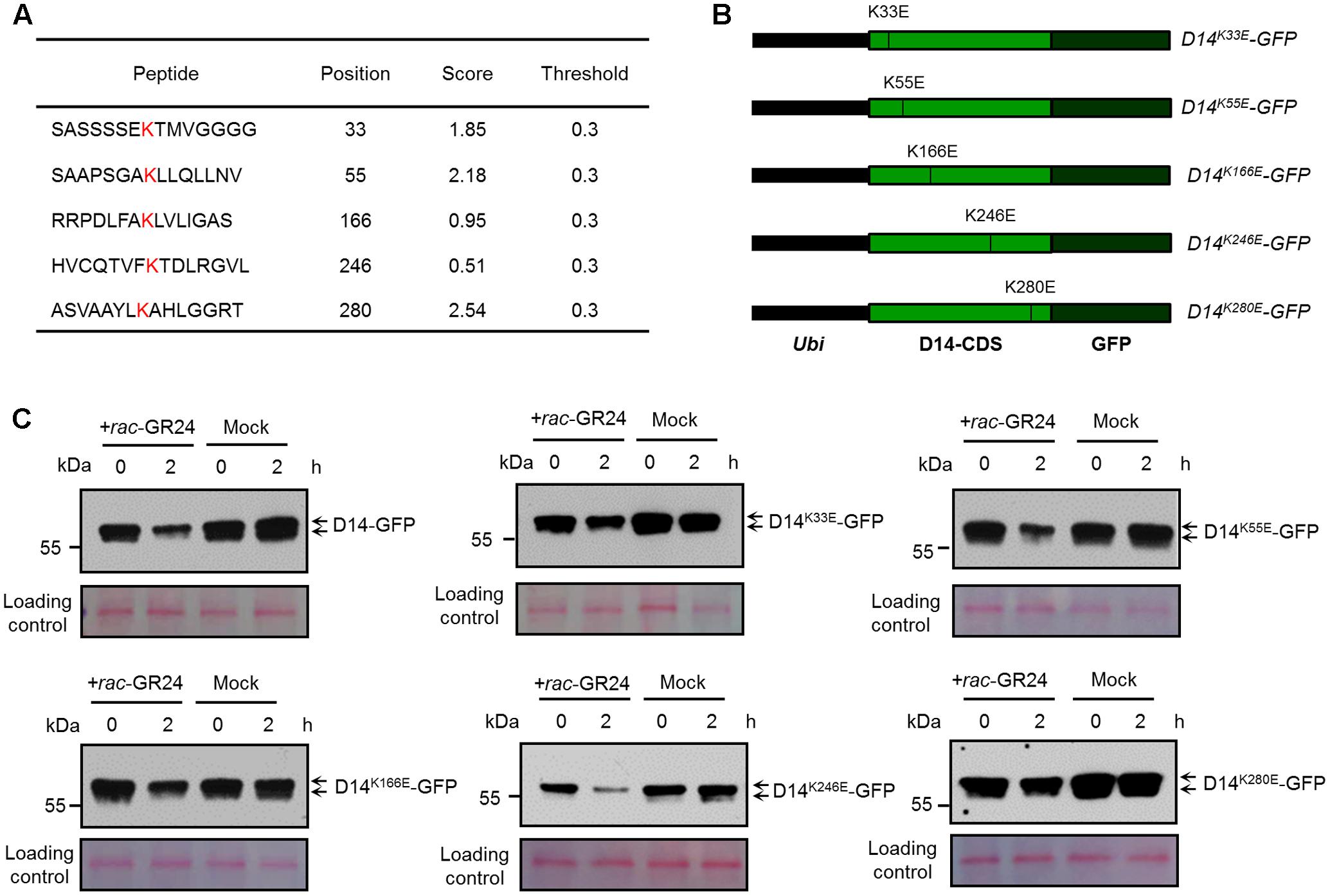
FIGURE 2. Characterization of key amino acid residues in D14 for its degradation. (A) The lysine sites in the D14 amino acid sequence and their predicted scores from the BDM-PUB website. (B) Schematic diagrams showing the constructs of D14 with point mutation of candidate ubiquitination sites. (C) Protein levels of D14-GFP, D14K33E-GFP, D14K55E-GFP, D14K166E-GFP, D14K246E-GFP and D14K280E-GFP in rice protoplasts transformed with plasmids shown in (B) after treatment with 10 μM rac-GR24 or DMSO for 2 h. Protein levels were detected by immunoblotting with an anti-GFP monoclonal antibody. Relative protein levels were normalized to the loading control determined by Ponceau staining (red).
AtD14 has been reported to be degraded after SL treatment in Arabidopsis thaliana (Chevalier et al., 2014). We then compared the amino acid sequences of D14 with its orthologues including AtD14 from Arabidopsis, RMS3 from pea and DAD2 from petunia, and found that the K280 site of D14 is conserved in pea and petunia, but changes to Arginine in Arabidopsis (Supplementary Figure 1), suggesting that the mechanism of D14 degradation might be different between rice and Arabidopsis. Further analysis on the destabilizing effects of the K280E mutation indicate that no obvious conformational change occurs in D14K280E based on structural prediction and comparison (Supplementary Figure 2). Taken together, these results indicate that the ubiquitination and degradation of D14 may involve multiple amino acids, of which K280 appears to play a major role.
D14K280E Mutation Does Not Affect SL-Induced D53 Degradation
We then evaluated whether the K280 mutation influence the function of D14 in SL signaling. It has been reported that AtD14 is localized in the nucleus and cytoplasm (Chevalier et al., 2014). We therefore investigated the subcellular localization of D14-GFP and D14K280E-GFP in a transient expression system using the SV40NLS-mCherry as a nuclear marker (Wang et al., 2015). As shown in Figure 3A, both D14-GFP and D14K280E-GFP proteins were localized in cytoplasm and nucleus, suggesting that K280 mutation does not affect the subcellular localization of D14. We then tested the interaction between D14K280E and D3 in wild-type protoplasts using the co-IP assay and found that D14K280E could interact with D3 after rac-GR24 treatment (Figure 3B). Furthermore, we compared the SL-induced degradation of D53 in calli of d14, D14-GFP/d14 and D14K280E-GFP/d14. The protein levels of D14 and D14K280E were comparable in different transgenic lines and displayed no obvious decrease after rac-GR24 treatment for 30 min (Supplementary Figure 3). Consistent with our previous study (Jiang et al., 2013), D53 degradation upon rac-GR24 treatment is dramatically impaired in d14 mutant. Importantly, both D14-GFP and D14K280E-GFP rescue the defect of D53 degradation in d14, indicating that D14K280E-GFP could trigger SL-induced D53 degradation and potential SL signaling (Figure 3C). Collectively, the D14K280E mutation doses not disturb the process of SL perception, which requires D14-D3 interaction and D53 degradation.
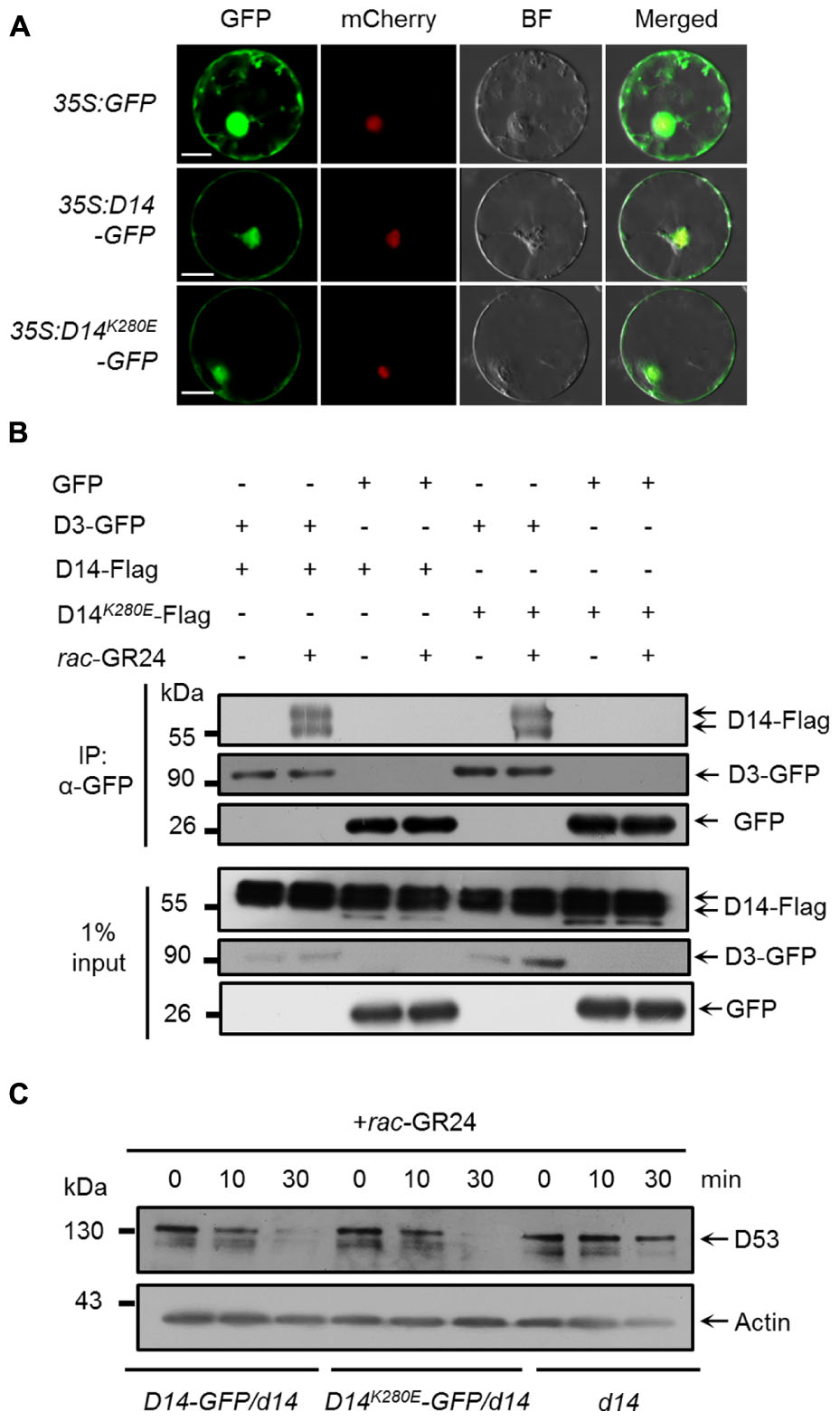
FIGURE 3. Effect of D14K280E mutation on SL signaling. (A) Subcellular localizations of GFP, D14-GFP and D14K280E-GFP in rice protoplasts. The 35S:SV40NLS-mCherry plasmid was cotransformed to label the nucleus. BF, bright field. Bar = 10 μm. (B) In vivo interaction between D14-Flag and D3-GFP revealed by co-IP assay in rice protoplasts. After transformation and incubation for 12 h, protoplasts were treated with 10 μM rac-GR24 for 1 h, and then the supernatant extracted from the protoplasts was incubated with an agarose-conjugated anti-GFP monoclonal antibody at 4°C for 3 h in the presence or absence of 10 μM rac-GR24, following which the D14 recombinant protein was detected with an anti-Flag monoclonal antibody, while D3-GFP and GFP were detected with an anti-GFP monoclonal antibody. Input refers to the total protein lysate before immunoprecipitation. (C) D53 protein levels in calli of Act:D14-GFP/d14, Act:D14K280E-GFP/d14 and d14 treated with 10 μM GR24. D53 was detected by immunoblotting with anti-D53 polyclonal antibodies. Rice Actin1 contents were used as loading controls in the immunoblotting analyses.
SL-Induced D14 Degradation Requires the Hydrolase Activity of D14
D14 encodes a member of the α/β hydrolase superfamily, which features the canonical triad catalytic center (Hamiaux et al., 2012; Kagiyama et al., 2013). The 147th serine and the 297th histidine sites are essential for the hydrolase activity and SL perception of D14 (Figure 4A) (Zhao et al., 2013; Yao et al., 2016). To address whether the hydrolase activity of D14 is required for its degradation, we mutated these key amino acids and generated the D14S147A-GFP and D14H297Y-GFP overexpression transgenic lines in the d14 background. Compared to the Actin:D14-GFP/d14 transgenic calli, the SL-stimulated D14 degradation was severely inhibited in Actin:D14S147A-GFP/d14 and Actin:D14H297Y-GFP/d14 calli (Figure 4B), indicating that the hydrolase activity of D14 is essential for the SL-induced D14 degradation. More importantly, the Actin:D14S147A-GFP/d14 and Actin:D14H297Y-GFP/d14 transgenic plants exhibited dwarf and high tillering phenotypes as the d14 mutant (Figure 4C), demonstrating that the S147 and H297 sites are both indispensable for the signal perception of SLs in vivo.
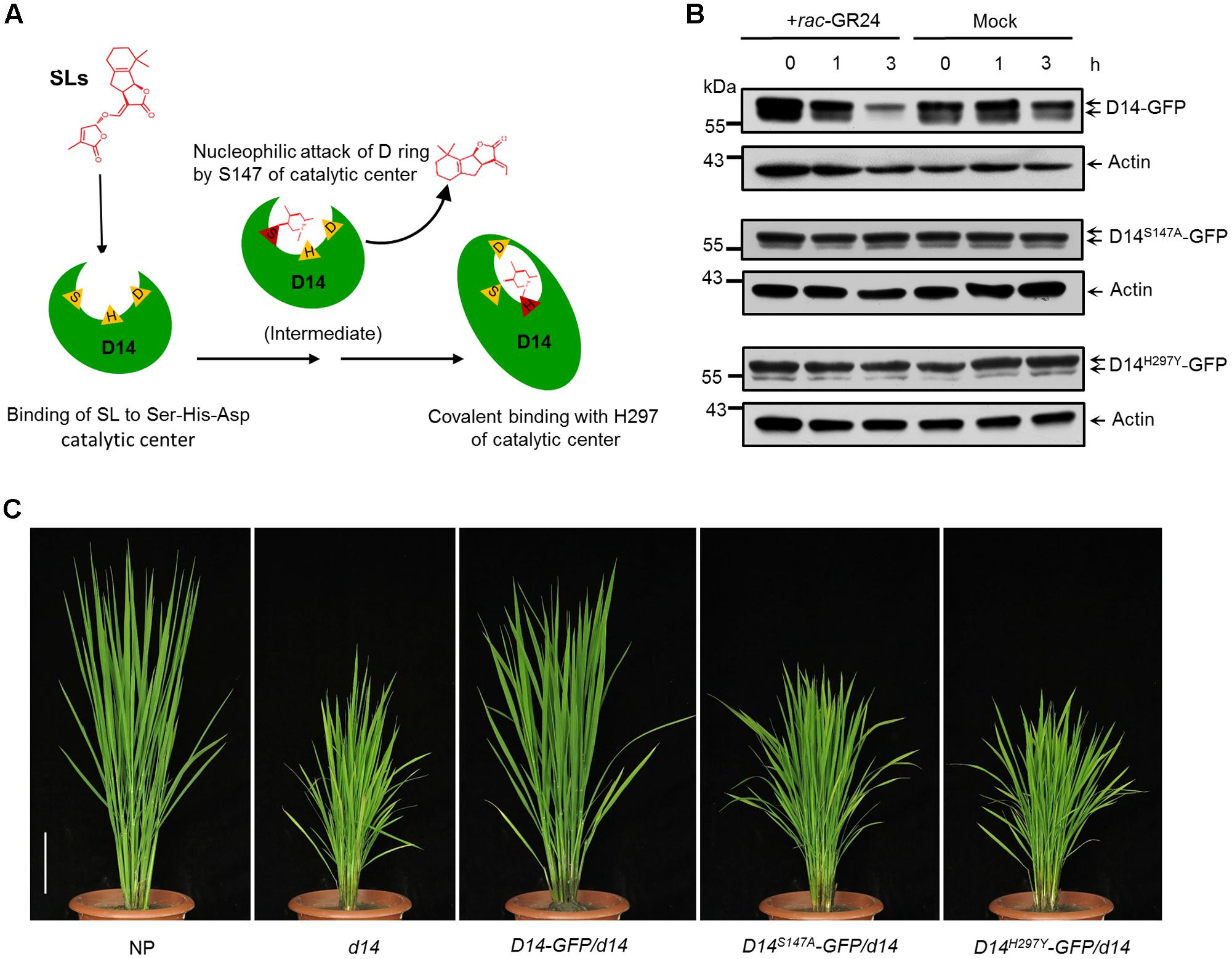
FIGURE 4. Requirement of hydrolase activity in SL-induced D14 degradation. (A) The schematic diagram of SL perception by D14. The open state D14 binds SLs with the Ser-His-Asp catalytic pocket (triangles). The Ser residue (S147) is involved in nucleophilic attack of the D-ring of SLs to initiate the catalytic reaction. Then the His residue (H297) covalently binds the aldehyde carbonyl of the S147-bound moiety to induce the conformational change of D14 from the open state to the close state. (B) Protein levels of D14-GFP, D14S147A-GFP and D14H297Y -GFP in transgenic calli after the 20 μM rac-GR24 or DMSO treatment. The D14-GFP fusion protein was detected with an anti-GFP monoclonal antibody. Actin1 contents were used as loading controls in the immunoblot analyses. (C) The complementation analyses of the d14 mutant with D14-GFP, D14S147A-GFP and D14H297Y-GFP. Bar = 10 cm.
SL-Induced D14 Degradation Is Dependent on D3 and Coupled to D53 Degradation
D3 encodes an F-box protein, which is a subunit of the Skp-Cullin-F-box (SCF) complex and is responsible for substrate recognition (Smith and Li, 2014). SLs trigger the interaction between D14 and D3, which in turn leads to the ubiquitination and degradation of D53, and probably relieves the repression on gene expression (Jiang et al., 2013; Zhou et al., 2013). To check whether the degradation of D14 is depended on D3, we first identified a loss-of-function mutant of d3 in the ZH11 background, which contains a premature translation mutation due to a 1-bp (base pair) deletion in the first exon and displayed dwarf and high-tillering phenotypes (Supplementary Figure 4). We then treated the Actin:D14-GFP transgenic calli in the wild-type or d3 background with 20 μM rac-GR24. Consistent with the role of D3 in targeting D53 for ubiquitination and degradation, we found that D53 degradation upon SL treatment was strongly inhibited in the d3 mutant. More importantly, D14 degradation was also blocked in the d3 mutant (Figure 5A), suggesting that D3 may directly involves in the proteolysis of D14 or D3 regulates the expression of unknown SL-responsive genes and consequently control D14 degradation.
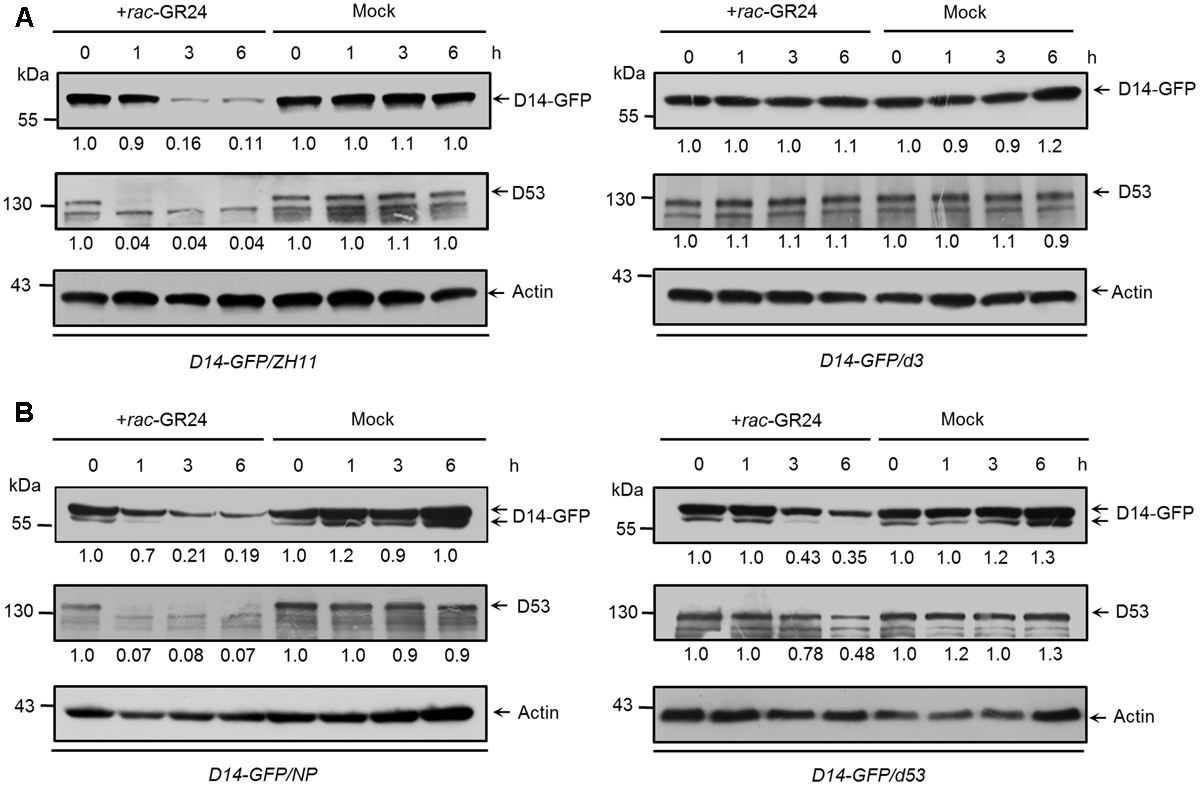
FIGURE 5. The SL-triggered D14 degradation in d3 and d53 mutants. (A) D14-GFP and D53 protein levels in Act:D14-GFP transgenic calli in the wild-type (ZH11) or d3 background after 20 μM rac-GR24 or DMSO treatment. (B) D14-GFP and D53 protein levels in transgenic calli of wild-type (Nipponbare) or d53 background after 20 μM rac-GR24 or DMSO treatment at different time points. D14-GFP and D53 levels were detected by immunoblotting with an anti-GFP monoclonal antibody and anti-D53 polyclonal antibodies, respectively. Relative amounts of proteins were determined by densitometry and normalized to actin1 and expressed relative to the value at zero time. Actin1 contents were used as loading controls in all the immunoblotting analyses.
Furthermore, we compared the amounts of D14-GFP and D53 in Actin:D14-GFP transgenic calli in the wild-type and d53 background (Figure 5B). The d53 mutant shows typical SL-deficient phenotypes including dwarf and high tillering, and is insensitive to exogenous SL treatment. These developmental defects are caused by a significant attenuation of D53 degradation due to an in-frame deletion of amino acid 813-817 and an amino acid substitution at 812 of D53 protein (Jiang et al., 2013). Thus the SL signaling is impaired in the d53 mutant. After rac-GR24 treatment for 1 h, endogenous D53 protein almost disappeared in wild type (left) but remained stable in the d53 mutant (right), consistent with results in Figure 3C and our previous observation that D53 was almost disappeared within 30 min but remained stable in d53 (Jiang et al., 2013). Meanwhile, the protein levels of D14-GFP in wild type began to decrease (left) but remained stable in d53 (right). With extended SL treatment for 3 and 6 h, endogenous D53 in the d53 mutant began to degrade and reached about half of the value at zero time. The D14-GFP also degraded in d53 (right) but the degradation degree was significantly impaired compared to that of wild type (left) (Figure 5B). These results indicate that D14 degradation is coupled to D53 degradation, which is closely related to the status of SL signaling.
Discussion
The degradation of receptors exists as a fine-tune mechanism in the signaling of hormones, such as ABA, JA, brassinolide and ethylene (Kevany et al., 2007; Wu et al., 2011; Yan et al., 2013; Kong et al., 2015). In this study, we found that D14 degradation was observed from 1 h and reached maximum level at about 3–4 h (Figures 1A,C, 4B, 5), while D53 protein is degraded at 10 min and almost disappeared at 30 min after GR24 treatment (Figure 3C), indicating that the SL receptor D14 is degraded through the 26S proteasome following the degradation of D53 in rice. SLs are recognized and hydrolyzed into a D-ring-derived molecule by D14 and form a covalently link bridge with the catalytic sites of D14 (Yao et al., 2016). This process stimulates a conformational change of D14 and the formation of a SCFD3-D14-D53 complex, leading to the ubiquitination and degradation of D53 (Jiang et al., 2013; Zhou et al., 2013). Subsequently, the repression of D53 on SL signaling is relieved, and the downstream components are activated to regulate plant development. Meanwhile, SLs could in turn trigger the degradation of D14 through the 26S proteasome pathway, leading to the inactivation of SL perception (Figure 6).
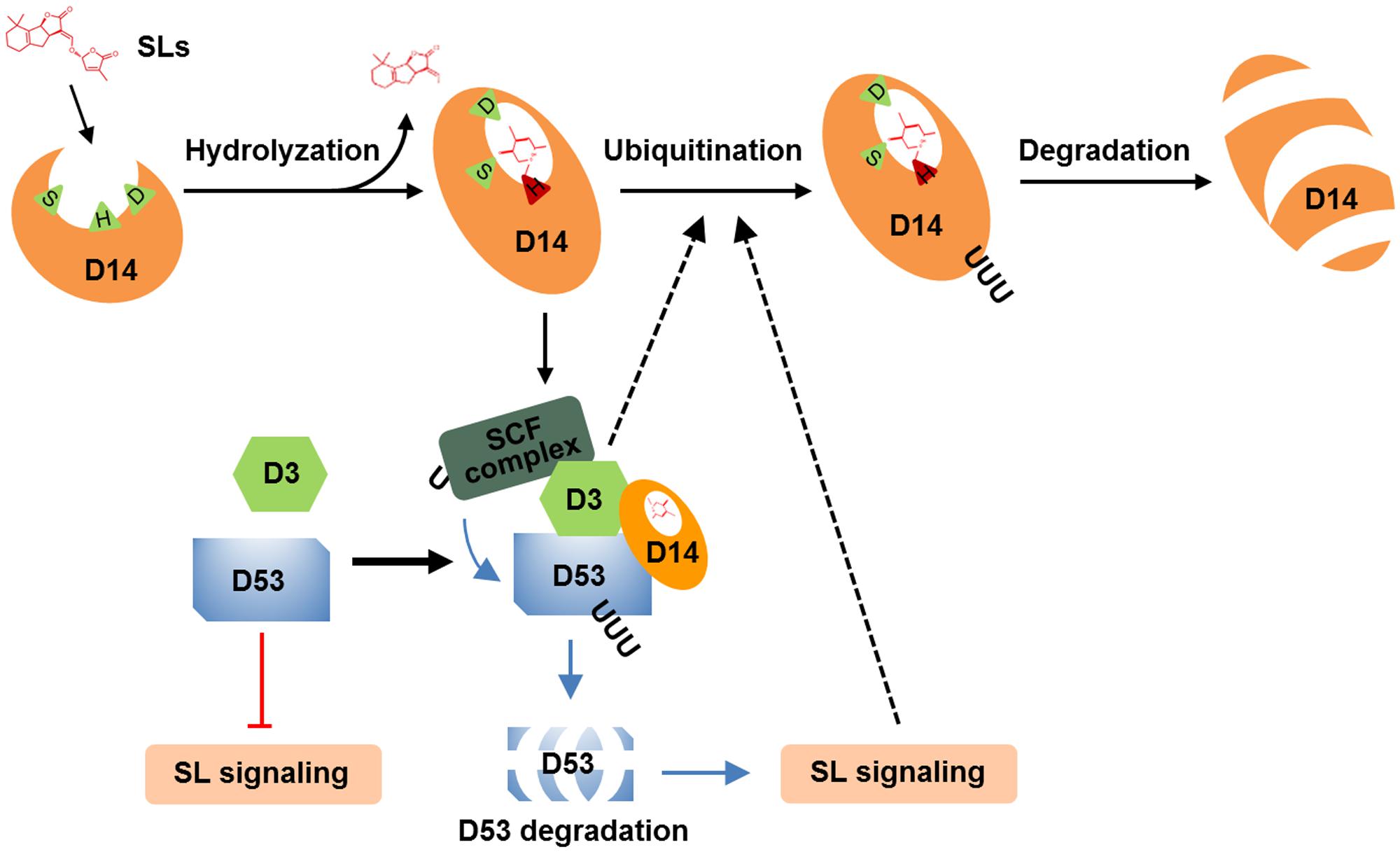
FIGURE 6. A proposed mechanism of D14 degradation in rice. D14 binds, attacks and hydrolyses SLs into ABC-ring and D-ring-derived molecules, covalently binds the D-ring-derived molecule, and forms a complex with D3 and D53. D53 undergoes ubiquitination and degradation, which relieve the repression on SL signal transduction. Subsequently, D14 is ubiquitinated and degraded through unknown mechanism to down regulate or shut down the SL signal transduction. D3 or the downstream targets of SL signaling may involve in this important feedback regulation.
The molecular mechanisms by which D14 was ubiquitinated and degraded are still open questions in higher plants (Chevalier et al., 2014; Wang et al., 2017). We found that D14 degradation was blocked in the loss-of-function mutant of d3 that displays completed obstructed D53 degradation, and was impaired in the dominant mutant d53 that displays significantly attenuated D53 degradation (Figure 5). These phenomena raised two possibilities for D14 degradation in vivo. The F-box protein D3 may target D53 and then D14 for ubiquitination and degradation. In this case, the over-accumulated D53 protein in the d53 mutant may hold more D3 protein and disturb the ubiquitination of D14. But another scenario also fit the available information. An unknown E3 ligase downstream of SL signaling may involve in the ubiquitination and degradation of D14. When SL perception is blocked (as in d3 mutant) or attenuated (as in d53 mutant), the expression or modification of this E3 ligase may change accordingly and thus influence D14 degradation (Figure 6). Crucially, further analysis of D14 ubiquitination and degradation based on in vitro reconstitution of SCFD3 E3 ligase activity and the structural analysis of D14-D3-D53 complex will give direct evidence of whether D3 direct target D14 for ubiquitination and degradation.
Feedback regulation is an important and universal mechanism to maintain the homeostasis of a signaling pathway (Schwechheimer, 2008). In plants, the biosynthesis and signal transduction of hormones are adjusted by several transcriptional and post-translational feedback regulation (Benjamins and Scheres, 2008; Kendrick and Chang, 2008; To and Kieber, 2008; Vlot et al., 2009; Jaillais and Chory, 2010; Jaillais and Vert, 2016; Yu et al., 2016; Carvalhais et al., 2017). For examples, auxins trigger the degradation of repressor proteins AXIN/INDOLE-3-ACETIC ACIDs (Aux/IAAs) and subsequently release the AUXIN RESPONSE FACTOR (ARF) transcription factors to regulate signal transduction. However, ARF could also promote the expression of Aux/IAA genes to repress the auxin signaling (Benjamins and Scheres, 2008). In the SL biosynthetic pathway, SLs repress the transcription of MAX3 and MAX4, which are key biosynthetic genes in Arabidopsis (Mashiguchi et al., 2009; Wang et al., 2015). In addition, in max and d mutants, the expression levels of MAX4 and its rice orthologous gene D10 were up-regulated (Jiang et al., 2013; Zhou et al., 2013; Wang et al., 2015). In the SL signaling pathway, SLs trigger rapid degradation of D53 to promote SL signaling in rice, but in turn activate the transcription of the D53 gene to limit SL signaling (Jiang et al., 2013; Zhou et al., 2013). This feedback regulation is mediated by the transcription factor IPA1, which could interact with D53 and activate D53 expression through directly binding the D53 promoter (Song et al., 2017). In Arabidopsis, AtD14 has been reported to undergo SL-induced and MAX2-dependent degradation through the proteasome system (Chevalier et al., 2014). In this work, we showed that SLs also promote the D14 degradation in rice, which would form a negative feedback loop of SL signaling. Intriguingly, SLs could trigger D53 degradation from 5 min, then downregulate MAX3 and MAX4 expression levels while upregulate the D53 expression from about 1–2 h. The SL-induced D14 degradation was observed from 1 h and reached maximum level at about 3–4 h, suggesting that the precise feedback regulation loops in different time dimension would effectively modulate the duration and intensity of SL signaling.
Parasitic plants are the largest biotic cause of reduced crop yields throughout the Africa (Rodenburg et al., 2016; Gobena et al., 2017). The germination of parasitic plants, mainly including Striga and Orobanche, is triggered by SLs secreted from crop roots (Waters et al., 2017). Currently, several strategies have been proposed to control parasitic plants, including induction of Striga germination through treating the unplanted fields with SL-like compounds and kill the parasitic plants before planting crops, and breeding of new crop varieties with altered SL biosynthesis or secretion without harming their normal growth (Nakamura and Asami, 2014; Toh et al., 2015; Fernández-Aparicio et al., 2016; Holbrook-Smith et al., 2016). Recent studies on highly Striga-resistant sorghum lines indicated that the SL profiles of root exudates from the lgs1 variants displayed reduced 5-deoxystrigol, a highly active Striga germination stimulant, but enhanced orobanchol, an SL required for normal growth. This change did not bring disadvantageous effect on plant development, but could inhibit the stimulation of Striga germination effectively (Gobena et al., 2017). In this study, we found that the mutation of D14K280E did not affect the normal function of D14 and SL signaling, but inhibited the D14 ubiquitination and degradation following SL treatment (Figures 2, 3). It is speculated that overexpressing the D14K280E may lead to the hypersensitivity to SLs but this may also need other amino acids working cooperatively with K280, for example K33, which moderately regulate D14 degradation (Figure 2C). Furthermore, when the mutated D14 protein resistant to ubiquitination and degradation is overexpressed, tiller number in rice is speculated to decrease due to an enhanced SL perception. Overexpression of this kind of D14 protein may suppress the high tillering phenotypes d27 and d17 that display impaired SL biosynthesis. This may present an alternative approach to cultivate rice varieties that perform decreased SL biosynthesis and reduced germination of Striga without harming the internal SL signaling pathway and regular plant development. It is rational that genetic modifications on the biosynthesis and perception of SLs are applicable to control the parasitic plant growth in crops in the future.
Author Contributions
QH, YH, BW, and JL conceived this project and designed all experiments. QH, YH, LW, SL, XM, GL, YJ, MC, XS, and LJ performed some of the experiments. BW, QH, YH, HY, and JL analyzed the data and wrote the paper. All authors commented on the article.
Funding
This work was supported by grants from the National Key Research and Development Program of China (Grant 2016YFD0100901), National Natural Science Foundation of China (Grant 91635301), and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” (Grant XDPB0401).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Yonghong Wang and Linzhou Huang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the rice d3 mutant, Prof. Yijun Qi (Tsinghua University) for providing the plasmid of SV40NLS-mCherry, Prof. Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for technical assistance in ubiquitination analysis, and lab members (Dr. Jingbo Duan, Huihui Liu) for their suggestions and technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01935/full#supplementary-material
Footnotes
References
Abe, S., Sado, A., Tanaka, K., Kisugi, T., Asami, K., Ota, S., et al. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. U.S.A. 111, 18084–18089. doi: 10.1073/pnas.1410801111
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Al-Babili, S., and Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. doi: 10.1146/annurev-arplant-043014-114759
Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M., Bigler, P., et al. (2012). The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. doi: 10.1126/science.1218094
Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., et al. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. doi: 10.1093/pcp/pcp091
Bart, R., Chern, M., Park, C. J., Bartley, L., and Ronald, P. C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2:13. doi: 10.1186/1746-4811-2-13
Benjamins, R., and Scheres, B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443–465. doi: 10.1146/annurev.arplant.58.032806.103805
Beveridge, C. A., Ross, J. J., and Murfet, I. C. (1996). Branching in pea (action of genes Rms3 and Rms4). Plant Physiol. 110, 859–865. doi: 10.1104/pp.110.3.859
Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H., and Leyser, O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14, 1232–1238. doi: 10.1016/j.cub.2004.06.061
Booker, J., Sieberer, T., Wright, W., Williamson, L., Willett, B., Stirnberg, P., et al. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8, 443–449. doi: 10.1016/j.devcel.2005.01.009
Brewer, P. B., Yoneyama, K., Filardo, F., Meyers, E., Scaffidi, A., Frickey, T., et al. (2016). LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 6301–6306. doi: 10.1073/pnas.1601729113
Carvalhais, L. C., Schenk, P. M., and Dennis, P. G. (2017). Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 21, 42–47. doi: 10.1016/j.mib.2017.03.009
Chen, Y. F., Shakeel, S. N., Bowers, J., Zhao, X. C., Etheridge, N., and Schaller, G. E. (2007). Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. Cell Res. 282, 24752–24758.
Chevalier, F., Nieminen, K., Sanchez-Ferrero, J. C., Rodriguez, M. L., Chagoyen, M., Hardtke, C. S., et al. (2014). Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26, 1134–1150. doi: 10.1105/tpc.114.122903
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chu, C., Wang, C., Sun, C., Chen, H., Yin, K., Chu, C., et al. (1975). Establishment of an efficient medium for anther culture of rice through comparative experiments on nitrogen-sources. Sci. China 18, 659–668.
Cook, C. E., Whichard, L. P., Turner, B., and Wall, M. E. (1966). Germination of witchweed (Striga lutea Lour.) isolation and properties of a potent stimulant. Science 154, 1189–1190. doi: 10.1126/science.154.3753.1189
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Fernández-Aparicio, M., Reboud, X., and Gibot-Leclerc, S. (2016). Broomrape weeds. Underground mechanisms of parasitism and associated strategies for their control: a review. Front. Plant Sci. 7:135. doi: 10.3389/fpls.2016.00135
Foo, E., Turnbull, C. G., and Beveridge, C. A. (2001). Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol. 126, 203–209. doi: 10.1104/pp.126.1.203
Gamborg, O. L., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Gao, Z., Qian, Q., Liu, X., Yan, M., Feng, Q., Dong, G., et al. (2009). Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol. Biol. 71, 265–276. doi: 10.1007/s11103-009-9522-x
Gobena, D., Shimels, M., Rich, P. J., Ruyter-Spira, C., Bouwmeester, H., Kanuganti, S., et al. (2017). Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. U.S.A. 114, 4471–4476. doi: 10.1073/pnas.1618965114
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pages, V., Dun, E. A., Pillot, J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–U122. doi: 10.1038/nature07271
Hamiaux, C., Drummond, R. S., Janssen, B. J., Ledger, S. E., Cooney, J. M., Newcomb, R. D., et al. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036. doi: 10.1016/j.cub.2012.08.007
Holbrook-Smith, D., Toh, S., Tsuchiya, Y., and McCourt, P. (2016). Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat. Chem. Biol. 12, 724–729. doi: 10.1038/nchembio.2129
Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I., and Kyozuka, J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46, 79–86. doi: 10.1093/pcp/pci022
Jaillais, Y., and Chory, J. (2010). Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 17, 642–645. doi: 10.1038/nsmb0610-642
Jaillais, Y., and Vert, G. (2016). Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol. 33, 92–100. doi: 10.1016/j.pbi.2016.06.014
Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F., Wang, L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. doi: 10.1038/nature12870
Kagiyama, M., Hirano, Y., Mori, T., Kim, S. Y., Kyozuka, J., Seto, Y., et al. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18, 147–160. doi: 10.1111/gtc.12025
Kendrick, M. D., and Chang, C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485. doi: 10.1016/j.pbi.2008.06.011
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/nature03542
Kevany, B. M., Tieman, D. M., Taylor, M. G., Cin, V. D., and Klee, H. J. (2007). Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467. doi: 10.1111/j.1365-313X.2007.03170.x
Kong, L., Cheng, J., Zhu, Y., Ding, Y., Meng, J., Chen, Z., et al. (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 20:8630. doi: 10.1038/ncomms9630
Lin, H., Wang, R., Qian, Q., Yan, M., Meng, X., Fu, Z., et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525. doi: 10.1105/tpc.109.065987
Liu, Q., Wang, Q., Liu, B., Wang, W., Wang, X., Park, J., et al. (2016). The blue light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome2 requires multiple E3 ubiquitin ligases. Plant Cell Physiol. 57, 2175–2186. doi: 10.1093/pcp/pcw134
Liu, W., Wu, C., Fu, Y., Hu, G., Si, H., Zhu, L., et al. (2009). Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649–658. doi: 10.1007/s00425-009-0975-6
Mashiguchi, K., Sasaki, E., Shimada, Y., Nagae, M., Ueno, K., Nakani, T., et al. (2009). Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73, 2460–2465. doi: 10.1271/bbb.90443
Nakamura, H., and Asami, T. (2014). Target sites for chemical regulation of strigolactone signaling. Front. Plant Sci. 5:623. doi: 10.3389/fpls.2014.00623
Nakamura, H., Xue, Y. L., Miyakawa, T., Hou, F., Qin, H. M., Fukui, K., et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4:2613. doi: 10.1038/ncomms3613
Rodenburg, J., Demont, M., Zwart, S. J., and Bastiaans, L. (2016). Parasitic weed incidence and related economic losses in rice in Africa. Agric. Ecosyst. Environ. 235, 306–317. doi: 10.1016/j.agee.2016.10.020
Sang, D., Chen, D., Liu, G., Liang, Y., Huang, L., Meng, X., et al. (2014). Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 11199–11204. doi: 10.1073/pnas.1411859111
Schwechheimer, C. (2008). Understanding gibberellic acid signaling - are we there yet? Curr. Opin. Plant Biol. 11, 9–15. doi: 10.1016/j.pbi.2007.10.011
Seto, Y., Sado, A., Asami, K., Hanada, A., Umehara, M., Akiyama, K., et al. (2014). Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U.S.A. 111, 1640–1645. doi: 10.1073/pnas.1314805111
Smith, S. M., and Li, J. (2014). Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21, 23–29. doi: 10.1016/j.pbi.2014.06.003
Song, X., Lu, Z., Yu, H., Shao, G., Xiong, J., Meng, X., et al. (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27, 1128–1141. doi: 10.1038/cr.2017.102
Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., et al. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes. Dev. 17, 1469–1474. doi: 10.1101/gad.256603
Soundappan, I., Bennett, T., Morffy, N., Liang, Y., Stanga, J. P., Abbas, A., et al. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159. doi: 10.1105/tpc.15.00562
Stirnberg, P., Furner, I. J., and Leyser, O. (2007) . MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 8094. doi: 10.1111/j.1365-313X.2007.03032.x
Stirnberg, P., van de Sande, K., and Leyser, H. M. O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141.
Tao, J. J., Cao, Y. R., Chen, H. W., Wei, W., Li, Q. T., Ma, B., et al. (2015). Tobacco translationally controlled tumor protein interacts with ethylene receptor tobacco histidine kinase1 and enhances plant growth through promotion of cell proliferation. Plant Physiol. 169, 96–114. doi: 10.1104/pp.15.00355
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tian, M., Lou, L., Liu, L., Yu, F., Zhao, Q., Zhang, H., et al. (2015). The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 82, 81–92. doi: 10.1111/tpj.12797
To, J. P., and Kieber, J. J. (2008). Cytokinin signaling: two-components and more. Trends Plant Sci. 13, 85–92. doi: 10.1016/j.tplants.2007.11.005
Toh, S., Holbrook-Smith, D., Stogios, P. J., Onopriyenko, O., Lumba, S., Tsuchiya, Y., et al. (2015). Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207. doi: 10.1126/science.aac9476
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, B., Wang, Y., and Li, J. (2017). “Strigolactones,” in Hormone Metabolism and Signaling in Plants, eds J. Li, C. Li, and S. M. Smith (London: Academic Press Elsevier), 327–359. doi: 10.1016/B978-0-12-811562-6.00010-4
Wang, L., Wang, B., Jiang, L., Liu, X., Li, X., Lu, Z., et al. (2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142. doi: 10.1105/tpc.15.00605
Waters, M. T., Gutjahr, C., Bennett, T., and Nelson, D. C. (2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322. doi: 10.1146/annurev-arplant-042916-040925
Waters, M. T., Nelson, D. C., Scaffidi, A., Flematti, G. R., Sun, Y. K., Dixon, K. W., et al. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. doi: 10.1242/dev.074567
Wu, G., Wang, X., Li, X., Kamiya, Y., Otegui, M. S., and Chory, J. (2011). Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 4, 29. doi: 10.1126/scisignal.2001258
Yan, J., Li, H., Li, S., Yao, R., Deng, H., Xie, Q., et al. (2013). The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell 25, 486–498. doi: 10.1105/tpc.112.105486
Yao, R., Ming, Z., Yan, L., Li, S., Wang, F., Ma, S., et al. (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. doi: 10.1038/nature19073
Ye, R. Q., Wang, W., Iki, T., Liu, C., Wu, Y., Ishikawa, M., et al. (2012). Cytoplasmic assembly and selective nuclear import of Arabidopsis ARGONAUTE4/siRNA complexes. Mol. Cell 46, 859–870. doi: 10.1016/j.molcel.2012.04.013
Yu, F., Wu, Y., and Xie, Q. (2016). Ubiquitin-proteasome system in ABA signaling: from perception to action. Mol. Plant 9, 21–33. doi: 10.1016/j.molp.2015.09.015
Zhang, Y. X., van Dijk, A. D. J., Scaffidi, A., Flematti, G. R., Hofmann, M., Charnikhova, T., et al. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10, 1028–1033. doi: 10.1038/nchembio.1660
Zhao, L. H., Zhou, X. E., Wu, Z. S., Yi, W., Xu, Y., Li, S. L., et al. (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439. doi: 10.1038/cr.2013.19
Keywords: Oryza sativa, plant hormones, strigolactones, hydrolase, DWARF14, proteolysis, signal transduction
Citation: Hu Q, He Y, Wang L, Liu S, Meng X, Liu G, Jing Y, Chen M, Song X, Jiang L, Yu H, Wang B and Li J (2017) DWARF14, A Receptor Covalently Linked with the Active Form of Strigolactones, Undergoes Strigolactone-Dependent Degradation in Rice. Front. Plant Sci. 8:1935. doi: 10.3389/fpls.2017.01935
Received: 10 August 2017; Accepted: 26 October 2017;
Published: 09 November 2017.
Edited by:
Zuhua He, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Haiyang Wang, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, ChinaXuelu Wang, Huazhong Agricultural University, China
Copyright © 2017 Hu, He, Wang, Liu, Meng, Liu, Jing, Chen, Song, Jiang, Yu, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiayang Li, anlsaUBnZW5ldGljcy5hYy5jbg== Bing Wang, YmluZ3dhbmdAZ2VuZXRpY3MuYWMuY24=
†Present address: Lei Wang, Agricultural Genome Institute, Chinese Academy of Agricultural Sciences, Shenzhen, China; Liang Jiang, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany
‡These authors have contributed equally to this work.
 Qingliang Hu
Qingliang Hu Yajun He
Yajun He Lei Wang
Lei Wang Simiao Liu
Simiao Liu Xiangbing Meng
Xiangbing Meng Guifu Liu
Guifu Liu Yanhui Jing
Yanhui Jing Mingjiang Chen
Mingjiang Chen Xiaoguang Song
Xiaoguang Song Liang Jiang
Liang Jiang Hong Yu
Hong Yu Bing Wang
Bing Wang Jiayang Li
Jiayang Li