- National Reference Centre, Dutch National Plant Protection Organization (NPPO), Wageningen, Netherlands
Ralstonia pseudosolanacearum (Ralstonia solanacearum phylotype I) isolates found in stunted, yellowing, and wilted ornamental rose (Rosa spp.) were assessed for their pathogenic ability in two rose cultivars (cv. “Armando” and cv. “Red Naomi”) and in four solanaceous crops: tomato (Solanum lycopersicum cv. “Money Maker”), tobacco (Nicotiana tabacum cv. “White Burley”), eggplant (Solanum melongena cv. “Black Beauty”) and sweet pepper (Capsicum annum cv. “Yolo Wonder”). Significant differences were observed in susceptibility between the two rose cultivars as well as between the two modes of inoculation performed. The cultivar “Armando” was significantly more susceptible than cultivar “Red Naomi,” exhibiting higher disease severity and incidence. Similarly, stem inoculation after wounding was found to be significantly more effective than soil drenching, resulting in higher disease severity. Additionally, a temperature dependency in susceptibility was observed for both cultivars irrespective of the mode of inoculation, however, this was significantly more pronounced upon soil drenching. The solanaceous crops all showed to be susceptible to the R. pseudosolanacearum isolates originated from the Rosa spp. plants. Furthermore, both rose cultivars were able to harbor symptomless infections with other R. pseudosolanacearum and R. solanacearum isolates than those isolated from rose. Our results clearly demonstrated that latent infections in a rose cultivar such as cv. “Red Naomi” do occur even at temperatures as low as 20°C. This latency poses high risks for the entire floricultural industry as latently infected Rosa spp. plants are propagated and distributed over various continents, including areas where climatic conditions are optimal for the pathogen.
Introduction
Ralstonia solanacearum (R. solanacearum) has been recently classified as the second most important plant pathogenic bacterium, in terms of scientific and economic relevance (Mansfield et al., 2012). Its significance is to be attributed to its persistence, systemic nature, wide host range, broad geographical distribution, and huge genome plasticity (Elphinstone, 2005; Denny, 2006; Castillo and Greenberg, 2007). More specifically, R. solanacearum genes involved in virulence evolve at a much faster rate than its genome as a whole (Remenant et al., 2010). The species R. solanacearum, as positioned taxonomically by Yabuuchi et al. (1995) appeared to be a heterogeneous species exhibiting considerable variation in virulence to diverse hosts (Prior and Steva, 1990; Marin and El-Nashaar, 1993; Jaunet and Wang, 1999). It was additionally proven to be a species complex (Palleroni and Doudoroff, 1971; Roberts et al., 1990; Gillings et al., 1993; Taghavi et al., 1996) that might represent more than one true species (Fegan and Prior, 2005). The complete group of strains belonging to this species complex is referred to as the “R. solanacearum species complex” (RSSC) (Fegan and Prior, 2005). Strains of the RSSC are traditionally divided into five races based on host range (Buddenhagen et al., 1962; He et al., 1983), and six biovars based on utilization of three disaccharides and three hexose alcohols (Hayward, 1964, 1994a). With the development of molecular tools for bacterial characterization, the classification of the RSSC has undergone major changes. As a result, a hierarchical classification system based on phylogenetic relationships was introduced by Fegan and Prior (2005). The four defined phylotypes showed to be correlated with the geographical origin of the strains. Phylotype I correlates with strains originating from Asia, phylotype II with those from America, phylotype III with those from Africa, and phylotype IV with those from Indonesia, Australia and Japan, including the strains of Ralstonia syzygii. More recently, a revision of the RSSC was proposed (Safni et al., 2014), introducing the division of the “species complex” into three genospecies: R. solanacearum, Ralstonia pseudosolanacearum and R. syzygii. The genospecies R. solanacearum consists of strains of R. solanacearum phylotype II only, including the type strain. R. pseudosolanacearum is composed of R. solanacearum strains that belong to phylotypes I and III. Finally, R. syzygii contains only R. solanacearum phylotype IV strains.
Although the RSSC has been studied intensively for decades, still new host species have been recently reported (Chandrashekara and Prasannakumar, 2010; Prieto Romo et al., 2012; Lin et al., 2015; Jiang et al., 2016; Tjou-Tam-Sin et al., 2017). The potential movement of RSSC representatives via propagative material between countries has probably been underestimated. The risk of bacterial dissemination might even be greater in cases where plants are considered non-hosts. When new undescribed RSSC pathotypes are introduced via propagative material into a new environment, the potential impact on local crops in this new environment is greatly unpredictable (Norman et al., 2009b). The increased trade in plant products around the world together with the rapid adaptation potential of RSSC (Genin and Boucher, 2004) can result in major outbreaks of this disease on known and on new host plants. Recently, isolates of R. pseudosolanacearum (phylotype I, race 1, biovar 3) were isolated from ornamental roses showing typical bacterial wilt symptoms and were confirmed to cause the disease symptoms (Tjou-Tam-Sin et al., 2017). Objective of this study was to assess the virulence of R. pseudosolanacearum isolates (phylotype I, race 1, biovar 3) acquired from symptomatic rose plants. The pathogenic ability of these isolates on two commercial rose cultivars, as well as on tomato, tobacco, eggplant and sweet pepper plants was investigated. Additionally, the pathogenic ability of a panel of R. pseudosolanacearum and R. solanacearum isolates originating from plants other than rose and belonging to phylotype I, II, and III has been evaluated on rose. The influence of temperature, botanical variation between rose cultivars and mode of inoculation (stem inoculation after wounding versus soil drenching) on susceptibility and disease conductivity was additionally evaluated.
Materials and Methods
Bacterial Isolates and Culture Conditions
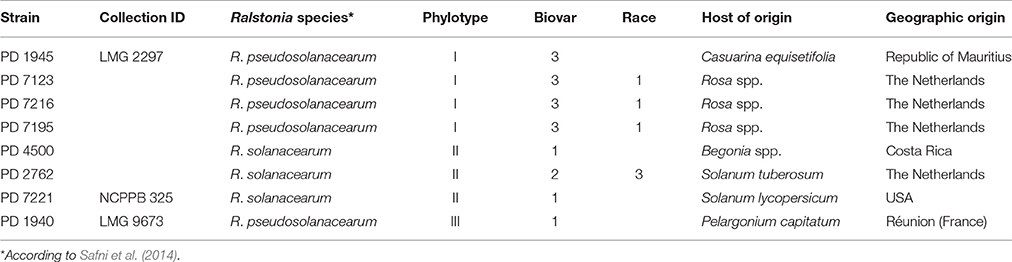
Eight representative R. pseudosolanacearum and R. solanacearum isolates selected according to their phylotype, geographic origin, and host of origin (Table 1) were assessed for their virulence on Rosa spp. and on Solanaceae. Isolates PD 7123, PD 7195, and PD 7216 were isolated in 2015 from naturally-infected Rosa spp., exhibiting typical symptoms and originating from glasshouse production systems in different locations in the Netherlands (Tjou-Tam-Sin et al., 2017). More specifically, PD 7123 was isolated from cv. “Red Naomi,” PD 7195 from cv. “Maritiem,” and PD 7216 from cv. “Armando.” Bacteria were isolated on YPG agar medium or on SMSA after incubation at 28°C for 2 or 6 days, respectively. Bacterial cultures were stored lyophilised as well as in sterile water at room temperature. Phylotype determination assigned the PD 7123, PD 7195, and PD 7216 isolates to phylotype I (Fegan and Prior, 2005).
Pathogenicity Assays
Two independent experiments were performed.
Design Experiment 1
In Experiment 1, eight R. pseudosolanacearum and R. solanacearum isolates were included for the inoculations (Table 1) in two rose cultivars (cv. “Armando” and cv. “Red Naomi”) and in four solanaceous crops: tomato (Solanum lycopersicum cv. “Money Maker”), tobacco (Nicotiana tabacum cv. “White Burley”), eggplant (Solanum melongena cv. “Black Beauty”), and sweet pepper (Capsicum annum cv. “Yolo Wonder”). The number of replicates per combination of plant species × isolate was: n = 3 for each of the rose cultivars inoculated and n = 10 for each of the inoculated solanaceous plant species. Suspensions of the bacterial isolates were prepared from 48 to 72 h old cultures by suspending them in phosphate buffer (PB 0.01M) to a final concentration of ca 108 CFU/mL. Sterile phosphate buffer (PB 0.01M) has been used as negative control. Rose plants were directly inoculated into the stem after pre-injuring their woody stems by wounding them with a scalpel. Thereafter, 20–50 μl bacterial suspension was injected per plant with a syringe and needle (21G × 2″; 0.8 × 50 mm) into the stem wounds (stem inoculation after wounding). For the four solanaceous crops, stem inoculations were performed by injuring them by stabbing with the needle (25G × 5/8″; 0.6 × 16 mm) of a syringe and leaving a drop of bacterial suspension behind on both entrances of the wound. Each stem was inoculated at two sites: at its lower part, just above the soil level and a few centimeters higher. Inoculated plants were kept at a 24/21°C diurnal temperature regime for up to 42 days post inoculation (dpi). Monitoring of disease was performed by assessing the disease incidence up to 42 dpi. Disease incidence was scored as the percentage (%) of symptomatic plants in a plant x isolate treatment, at 42 dpi, relatively to the total number of plants included in that treatment.
Design Experiment 2
In experiment 2, two R. pseudosolanacearum (PD 7123 and PD 7216) isolates (Table 1) were inoculated in Rosa sp. cv. “Armando” and cv. “Red Naomi”, and in tomato (S. lycopersicum cv. “Money Maker”). Experiment 2 was executed at two temperature levels (20 and 28°C) and inoculations were performed either by 1. Stem inoculation after wounding or by 2. Soil drenching. Preparation of bacterial suspensions and stem inoculation after wounding were both performed as described in Experiment 1. However, a final bacterial concentration of ca 107 CFU/mL was used for both, stem inoculation after wounding and soil drenching. For soil drenching, a total volume of 100 mL of bacterial suspension was poured onto the rooted soil of a plant potted in peaty soil (volume per pot, ca. 1.5 liter). Sterile phosphate buffer (PB 0.01M) has been used as negative control. The number of replicates per combination of plant species × isolate × temperature × mode of infection was: n = 10 for inoculations directly into the stem after wounding and n = 12 for soil drenching. Plants were kept at 20°C and at 28°C for up to 116 dpi. Disease progress was monitored initially at four dpi, thereafter, for the first 2 weeks, at a frequency of twice up to three times per week and thereafter once per week. At each observation point, disease severity was assessed only for the plant (per treatment) that was in the most advanced disease stage (Figure 1). At the end of experiment 2 (at 116 dpi), the disease incidence (%) and the disease severity were assessed for the total number of plants included.

Figure 1. Assessment of disease severity by making use of a scale categorizing the symptoms induced by Ralstonia pseudosolanacearum observed on Rosa spp. according to level 0–3 (corresponding with pictures A–D): (A) (0 = no symptoms), (B) (1 = starting symptoms), (C) (2 = clear, typical symptoms), (D) (3 = plant death).
Disease severity was assessed by using a scale as follows: 0 = healthy plants; 1 = first appearance of doubtful or mild symptoms of bacterial wilt; 2 = typical bacterial wilt expression with rolling and hanging leaves, leaf necrosis protruding from the leaf margins toward the leave veins, yellowing of leaves and early leaf drop, and possibly dark brown to black necrosis of stems dying back; 3 = irreversible total wilt and plant death, later followed by dark brown necrosis of the stems (Figure 1).
Observation data were analyzed in order to assess whether the isolate, the temperature, the botanical variation and the mode of inoculation significantly influenced disease severity. Analysis of variance (ANOVA) was performed using SPSS version 17.0. Statistical significance was accepted at α = 0.01.
Latency Testing and Identity Confirmation of Re-Isolates
In both experiments, re-isolations on SMSA medium were performed from at least one individual plant per treatment where typical symptoms were developed, in order to confirm the R. pseudosolanacearum and R. solanacearum infections. The identity of re-isolates exhibiting typical colony morphology for R. pseudosolanacearum and R. solanacearum on SMSA medium was confirmed by TaqMan PCR (Weller et al., 2000) with a minor grove binding unit (mgb)-adjusted probe as described in Vreeburg et al. (2016). Plants showing no symptoms till the end of the experiments were also included for the re-isolations. For this, extract from a ca. 2 cm section stem tissue per each stalk of a rose plant (avoiding the point of inoculation and at least 1 cm stem jointed to the inoculation point) was plated on SMSA medium, in order to confirm either the absence of R. pseudosolanacearum and R. solanacearum infections or the presence of latent infections. For experiment 1, all plants within one treatment combination were pooled for re-isolation at 42 dpi. For experiment 2, re-isolation was performed from each plant individually at 116 dpi.
Results
Experiment 1
Disease incidence (%) in Rosa spp. and in the four solanaceous plant species at 42 dpi when incubated at a 24°C, is presented in Table 2. Both, Rosa sp. cultivars and the four solanaceous plant species were highly susceptible for the isolates PD 7123, PD 7216, and PD 7195 originating from the naturally-infected rose. Disease incidence in both Rosa sp. cv. “Armando” and Rosa sp. cv. “Red Naomi” was 100% at 42 dpi, with plants exhibiting typical disease symptoms. Plant death at 42 dpi was observed only for cv. “Armando” plants. However, cv “Red Naomi” plants developed typical symptoms for R. pseudosoalanacearum in rose as in Figure 1C, but they did not exhibit plant death within the 42 dpi period. Remarkably, upon stem inoculation after wounding with a panel of R. pseudosolanacearum and R. solanacearum isolates belonging to phylotypes I, II, and III (Table 1) both cv. “Armando” and cv. “Red Naomi” were able to harbor infections, however, without showing symptoms at 42 dpi (Table 2). These symptomless infections on rose plants at 24°C were confirmed by the results obtained from latency testing.
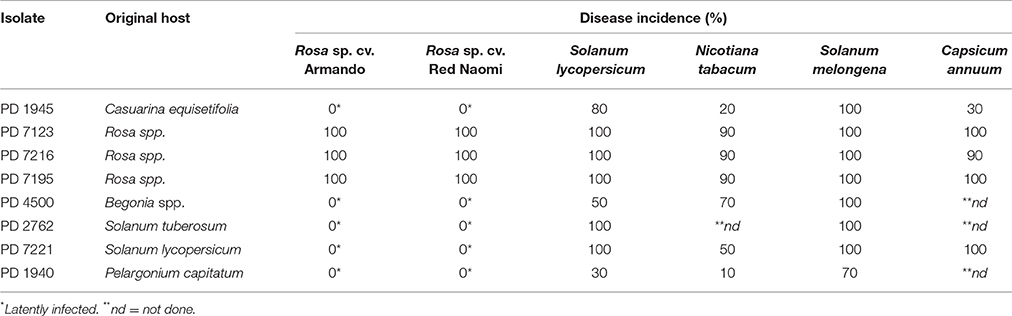
Table 2. Disease incidence (%) of bacterial wilt on two ornamental rose cultivars (n = 3) and four solanaceous plant species (n = 10) after stem inoculation with eight Ralstonia solanacearum and Ralstonia pseudosolanacearum isolates at 24/21°C diurnal temperature regime, 42 dpi, (Experiment 1).
Irrespective of the R. pseudosolanacearum (phylotype I) isolate (PD 7123, PD 7216, and PD 7195) inoculated on cv. “Armando”, the first symptomatic plant at 24°C appeared at 6–7 dpi, with isolate PD 7123 being slightly delayed in symptom development. However, the first symptomatic cv. “Red Naomi” plant appeared later; at 11 dpi (for PD 7216), at 13 dpi (for PD 7195), and at 15 dpi (for PD 7123). On the other hand, irrespective of the R. pseudosolanacearum (phylotype I) isolate (PD 7123, PD 7216, and PD 7195) used for inoculation, the first plant exhibiting plant death was observed at 21 dpi, only for Rosa sp. cv. “Armando”. From Rosa sp. cv. “Red Naomi” no plants exhibited plant death within 42 dpi. These results underline the difference in susceptibility for bacterial wilt caused by R. pseudosolanacearum (phylotype I) between rose cultivars.
Disease incidence in all four solanaceous plant species inoculated at 24°C with the R. pseudosolanacearum (phylotype I) isolates PD 7123, PD 7216, and PD 7195 was 90–100% at 42 dpi (Table 2). Irrespective of the isolate (PD 7123, PD 7216, and PD 7195) used for inoculation and irrespective of the solanaceous plant species used, the first symptomatic solanaceous plant always appeared within 4 dpi. However, the first plant exhibiting plant death in all four solanaceous plant species has been monitored between 6 and 21 dpi: up to 6–11 dpi for tomato, eggplant, and sweet pepper, and 21 days for tobacco.
Experiment 2
There were no significant differences (P = 0.078) in disease severity between the isolates PD 7123 and PD 7216 (Table 3). However, significant differences in disease severity have been indicated by ANOVA for the other main factors: temperature, mode of inoculation and botanical variation (Table 3). With the exception of the “temperature*botanical variation*mode of inoculation” interaction (P < 0.0005), all other interactions were not significantly different (Table 3). Disease severity was in general lower in treatment combinations including cv. “Red Naomi,” at 20°C, and after soil drenching application (Figure 2). However, it is worthwhile to mention that although cv. “Armando” plants produced typical symptoms upon stem inoculation after wounding at 20°C, they did not exhibit plant death within 116 dpi. Additionally, only one cv. “Red Naomi” plant, inoculated with isolate PD 7123, showed mild symptoms within 116 dpi upon stem inoculation after wounding at 20°C. Re-isolations from the symptomless cv. “Red Naomi” plants within the treatment combination of stem inoculation after wounding at 20°C revealed high levels of latent infections, namely 60 and 70% for isolates PD 7123 and PD 7216, respectively (Table 4).
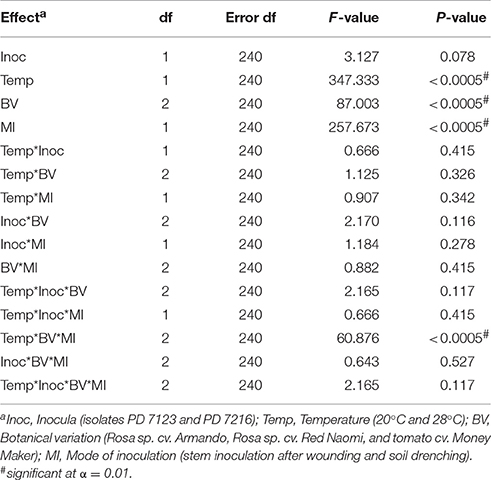
Table 3. ANOVA analysis of fixed effects and interactions on severity of bacterial wilt caused by Ralstonia pseudosolanacearum upon stem inoculation after wounding (n = 10) or soil dreching (n = 12), at 116 dpi (Experiment 2).
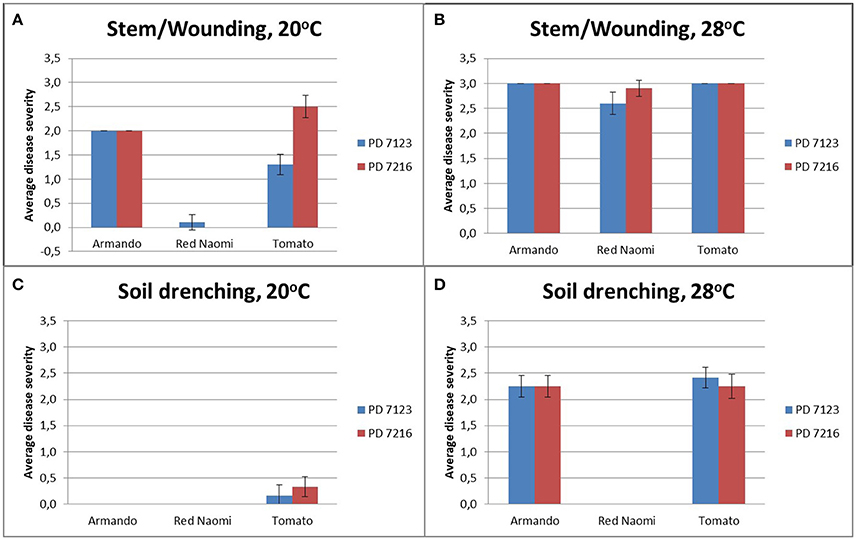
Figure 2. Average disease severity of bacterial wilt caused by Ralstonia pseudosolanacearum isolates PD 7123 and PD 7216 on plants of Rosa sp. cv. “Armando,” Rosa sp. cv. “Red Naomi” and Solanum lycopersicon, cv. “Money Maker,” at 20°C (A,C) and 28°C (B,D) at 116 dpi. Disease severity is given as the average of n = 10 plants for the stem inoculation after wounding (A,B) and n = 12 plants for the soil drenching treatment (C,D). Error bars indicate the standard error observed.
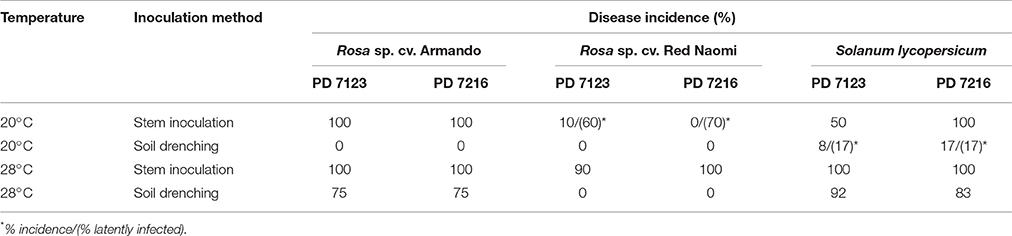
Table 4. Disease incidence (%) of bacterial wilt caused by Ralstonia pseudosolanacearum isolates PD 7123 and PD 7216 at 20 and 28°C on Rosa sp. cv. Armando, Rosa sp. cv. Red Naomi, and Solanum lycopersicum, cv. Money Maker, upon stem inoculation after wounding (n = 10) or soil drenching (n = 12), at 116 dpi (Experiment 2).
Stem inoculation after wounding in both cv. “Armando” and cv. “Red Naomi” at 28°C showed typical symptoms and exhibited plant death. The first plant exhibiting plant death at 28°C has been monitored already after 18 dpi for cv. “Armando” and after 43 dpi for cv. “Red Naomi” (Figure 3). These observations acquired by stem inoculation after wounding underline a statistically significant effect due to the interaction between the cultivar, the temperature and the mode of inoculation (Table 3), that is also supported by the disease progress curves (Figure 3). No significant differences (P = 0.116) were observed between cv. “Armando” and cv. “Red Naomi” upon inoculation with either isolate PD 7123 or PD 7216 in terms of disease severity (Table 3).
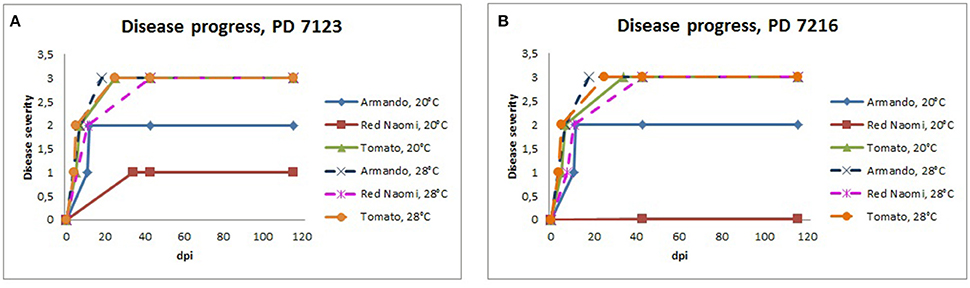
Figure 3. Severity of bacterial wilt caused by Ralstonia pseudosolanacearum isolates PD 7123 (A) and PD 7216 (B) on plants of Rosa sp. cv. “Armando,” Rosa sp. cv. “Red Naomi” and Solanum lycopersicum, cv. “Money Maker” in time, at 20 and 28°C up to 116 dpi. At each observation day, an assessment was done of disease severity of one plant (per treatment) that was in the most advanced disease stage.
The effect of temperature was statistically significant (P < 0.0005) after application of soil drenching in cv. “Armando” but also in S. lycopersicum cv. “Money Maker” plants (Table 3). After soil drenching at 20°C, both cv. “Armando” and cv. “Money Maker” presented no symptoms and only minor disease expression [<20% disease incidence (Table 4) and average severity <0.5% (Figure 2C)]. However, after soil drenching at 28°C higher values for disease incidence (Table 4) and disease severity (Figure 2D) were observed in both cv. “Armando” and cv. “Money Maker.” This effect on the disease severity was statistically supported (Table 3). Additionally, most cv. “Armando” plants exhibited plant death within the 116 dpi period of monitoring after soil drenching at 28°C. On the other hand, cv. “Red Naomi” plants showed no symptoms at all after soil drenching, irrespective of the incubation temperature, for up to 116 dpi (Figures 2C,D). Testing of the symptomless cv. “Red Naomi” plants after soil drenching at 28°C revealed no latent infections (Table 4). This highlights the need of mechanical injury to result in an assessable amount of disease severity (Figure 2) and disease incidence (Table 4) in cv. “Red Naomi” at either 20°C or 28°C and in cv. “Armando” at 20°C. Within the symptomless S. lycopersicum cv. “Money Maker” plants, upon soil drenching, re-isolations after 116 dpi at 20°C revealed the presence of latent infections at a level of 17% (Table 4). In the treatment combination of stem inoculation after wounding at 20°C, our results indicated that the disease severity (Figures 2A, 3) and disease incidence (Table 4) in both, cv “Armando” and S. lycopersicum cv. “Money Maker” were higher than those observed in cv “Red Naomi.”
Discussion
Bacterial wilt of ornamental and crop field plants caused by RSSC is frequently catastrophic, resulting in major economic losses in many countries (Hayward, 1991). Although, traditionally the most predominantly represented plant families harboring hosts of R. pseudosolanacearum and R. solanacearum are the Solanaceae and Musaceae (Buddenhagen, 1961; Hayward, 1994b; Elphinstone, 2005; Alvarez et al., 2010; Albuquerque et al., 2014), still new host species have been recently reported belonging to different plant families (Chandrashekara and Prasannakumar, 2010; Prieto Romo et al., 2012; Lin et al., 2015; Jiang et al., 2016). Recently, we reported, for the first time worldwide, the finding of R. pseudosolanacearum (phylotype I, race 1, biovar 3), causing bacterial wilt in Rosa spp. plants under greenhouse cultivation in the Netherlands (Tjou-Tam-Sin et al., 2017). The main aim of our study was to assess the virulence of R. pseudosolanacearum isolates from naturally-infected rose plants on two representative commercial cultivars of Rosa spp., namely cv. “Armando” and cv. “Red Naomi.”
Since disease development in plants only occurs when a virulent pathogen, a susceptible host and a favorable environment are simultaneously present, the influence of a number of abiotic parameters have been evaluated in our study; i.e., the of influence temperature and mode of inoculation (stem inoculation after wounding or soil drenching). Significant differences in disease severity among the 20°C and the 28°C treatments were found, irrespective of the mode of inoculation, with disease severity and disease incidence being consistently higher at the 28°C treatments. As strains of R. pseudosolanacearum (phylotype I, race 1, biovar 3) are generally known to require a relatively high optimum temperature (24–35°C) for symptom development (Kelman, 1953; Bocsanczy et al., 2014), the outcome of our study showing high disease expression at 28°C but only poor to moderate at 20°C is in accordance with the expectation. An additional explanation for the relatively poor disease development at 20°C, may be a virulence barrier that RSSC representatives exhibit under low temperature conditions, at the stage of host rhizosphere colonization (Bocsanczy et al., 2012).
In general, stem inoculation after wounding resulted in higher disease severity and disease incidence than soil drenching at both temperatures, however, this was more pronounced at 20°C than at 28°C. Up to 116 dpi, cv. “Red Naomi” showed no symptoms upon stem inoculation after wounding at 20°C, however, re-isolations resulted in high levels of latent infections (60–70%). This implicates a high risk for import of propagative plant material potentially harboring latent infections when kept at temperature as low as 20°C. The effectiveness of soil drenching on susceptibility to bacterial wilt showed to be significantly low for cv. “Red Naomi,” irrespective of the temperature, as no symptoms were monitored upon soil drenching for up to 116 dpi. However, no latent infections were found in this case, underlining a high dependency of mechanical stem injury in the infection process for effective disease development in cv. “Red Naomi.” On the other hand, highly susceptible cultivars such as cv. “Armando” can be effectively infected upon soil drenching via the root system. Based on our data, the use of stalks acquired from rose cut flowers as source of propagation material in nurseries poses a potentially high risk for the dissemination of R. pseudosolanacearum through systemic (latently) infected Rosa sp. plants and especially under greenhouse cultivation conditions, as previously highlighted (Norman and Yuen, 1998, 1999) for propagative plant material, in general.
The influence of botanical variation between cv. “Armando” and cv. “Red Naomi” on susceptibility to bacterial wilt is statistically supported in our experiments. This is in accordance with previous studies performed on other plant species (Jaunet and Wang, 1999; Norman et al., 2009a; Lebeau et al., 2011). On the other hand, no differences in virulence were observed among the isolates PD 7123, PD 7216, and PD 7195, irrespective of the temperature, mode of inoculation or botanical variation. These isolates are genetically closely related and formed a distinct monophyletic clade in phylotype I (unpublished data). Remarkably, both cv. “Armando” and cv. “Red Naomi” upon stem inoculation after wounding by a panel of RSSC isolates belonging to phylotypes I, II, and III were able to harbor latent infections at 42 dpi. Among these RSSC isolates inoculated in rose, an isolate of R. solanacearum phylotype IIB was included, formerly referred to as race 3 biovar 2 which is adapted to temperate climates and is known as the causal agent of potato brown rot. Our observations of harbored latent infections of this and other RSSC isolates in rose implicates an additional risk for import of latently infected rose plants by other RSSC isolates than the one isolated from rose. Our results highlight the urgency to study these latent RSSC infections upon exposure to different environmental conditions (i.e. temperature, water stress) and beyond the 6 week period (42 dpi).
The present work confirmed that R. pseudosolanacearum isolates PD 7123, PD 7216, and PD 7195 acquired from naturally-infected rose can cause severe disease to young plants of Solanaceae plants. The disease incidence within the Solanaceae plants was 90–100% at 42 dpi upon stem inoculation after wounding at 24°C. However, the susceptibility of potato to R. pseudosolanacearum (phylotype I) isolates from rose was not included in this study, as previously done for R. solanacearum phylotype II isolates from pelargonium (Williamson et al., 2002). In the current study we mainly concentrated on the risk of the R. pseudosolanacearum (phylotype I) isolates from rose toward glasshouse cultivated crops in temperate climatic regions. However, in warmer climatic zones R. pseudosolanacearum biovar 1 isolates are known to infect outdoor potato crops (Cruz et al., 2008). Our results highlight the urgency to study the pathogenic ability of R. pseudosolanacearum (phylotype I) isolates from rose on potato as well.
In conclusion, this study showed that temperature, botanical variation and mode of inoculation, alone or in combination, can significantly influence disease severity in rose upon inoculation with R. pseudosolanacearum (phylotype I, race 1, biovar 3) isolates from naturally-infected rose plants in greenhouse cultivation in the Netherlands. Additionally, it underlined the ability of these isolates to occur frequently as latent infections in the plant. This represents a serious challenge for the entire floricultural industry, worldwide. Although our results greatly contribute to the knowledge of the epidemiology of the R. pseudosolanacearum from rose, yet the limited host plants included here illustrate that the precise host range of this pathogen is still poorly characterized and greatly unknown.
Author Contributions
Conceived and designed the experiments: NT, MB. Performed the experiments NT, JvdB, PG, ML, MW. Analyzed the data: NT. Wrote the paper: NT, MB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by research grant OS 2016339, on the R. solanacearum species complex, of the Ministry of Economic Affairs in the Netherlands.
References
Albuquerque, G. M., Santos, L. A., Felix, K. C., Rollemberg, C. L., Silva, A. M., Souza, E. B., et al. (2014). Moko disease-causing strains of Ralstonia solanacearum from Brazil extend known diversity in paraphyletic phylotype II. Phytopathology 104, 1175–1182. doi: 10.1094/PHYTO-12-13-0334-R
Alvarez, B., Biosca, E. G., and López, M. M. (2010). “On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen,” in Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, Vol. 1, ed A Mendez-Vilas (Badajoz: Formatex), 267–279.
Bocsanczy, A. M., Achenbach, U. C., Mangravita-Novo, A., Chow, M., and Norman, D. J. (2014). Proteomic comparison of Ralstonia solanacearum strains reveals temperature dependent virulence factors. BMC Genomics 15:280. doi: 10.1186/1471-2164-15-280
Bocsanczy, A. M., Achenbach, U. C., Mangravita-Novo, A., Yuen, J. M., and Norman, D. J. (2012). Comparative effect of low temperature on virulence and twitching motility of Ralstonia solanacearum strains present in Florida. Phytopathology 15, 185–194. doi: 10.1094/PHYTO-05-11-0145
Buddenhagen, I., Sequeira, L., and Kelman, A. (1962). Designation of races in Pseudomonas solanacearum. Phytopathology 52, 726–1962.
Buddenhagen, I. W. (1961). Bacterial wilt of bananas: history and known distribution. Trop. Agric. 38, 107–121.
Castillo, J. A., and Greenberg, J. T. (2007). Evolutionary dynamics of Ralstonia solanacearum. Appl. Environ. Microbiol. 73, 1225–1238. doi: 10.1128/AEM.01253-06
Chandrashekara, K. N., and Prasannakumar, M. K. (2010). New host plants for Ralstonia solanacearum from India. Plant Pathol. 59, 11–78. doi: 10.1111/j.1365-3059.2010.02358.x
Cruz, L., Eloy, M., Quirino, F., and Carrinho, H. (2008). Ralstonia solanacearum biovar 1 associated with a new outbreak of potato brown rot in Portugal. Phytopathol. Mediterr. 47, 87–91. doi: 10.14601/Phytopathol_Mediterr-2610
Denny, T. P (2006). “Plant pathogenic Ralstonia species,” in Plant-Associated Bacteria, ed S. S. Gnanamanickam (Dordrecht: Springer), 573–644.
Elphinstone, J. G. (2005). “The current bacterial wilt situation: a global overview,” in Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex, eds C. Allen, P. Prior, and A. C. Hayward (St. Paul, MN: The American Phytopathological Society), 9–28.
Fegan, M., and Prior, P. (2005). “How complex is the Ralstonia solanacearum Species Complex,” in Bacterial Wilt Disease and the Ralstonia Solanaceaum Species Complex, eds C. Allen, P. Prior, and A. C. Hayward (St. Paul, MN: The American Phytopathological Society), 449–461.
Genin, S., and Boucher, C. (2004). Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42, 107–134. doi: 10.1146/annurev.phyto.42.011204.104301
Gillings, M., Fahy, P., and Davies, C. (1993). Restriction analysis of an amplified polygalacturonase gene fragment differentiates strains of the phytopathogenic bacterium Pseudomonas solanacearum. Lett. Appl. Microbiol. 17, 44–48. doi: 10.1111/j.1472-765X.1993.tb01432.x
Hayward, A. C. (1964). Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27, 265–277. doi: 10.1111/j.1365-2672.1964.tb04912.x
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
Hayward, A. C. (1994a). “Systematics and phylogeny of Pseudomonas solanacearum and related bacteria,” in Bacterial Wilt: The Disease and its Causative Agent, Pseudomonas solanacearum, eds A. C. Hayward and G. L. Hartman (Wallingford: CAB International), 123–135.
Hayward, A. C. (1994b). “The hosts of Pseudomonas solanacearum,” in Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas solanacearum, eds A. C. Hayward and G. L. Hartman (Wallingford: CAB International), 9–24.
He, L. Y., Sequeira, L., and Kelman, A. (1983). Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 67, 1357–1361. doi: 10.1094/PD-67-1357
Jaunet, T. X., and Wang, J. F. (1999). Variation in genotype and aggressiveness of Ralstonia solanacearum race 1 isolated from tomato in Taiwan. Phytopathology 89, 320–327. doi: 10.1094/PHYTO.1999.89.4.320
Jiang, Y., Li, B., Liu, P., Liao, F., Weng, Q., and Chen, Q. (2016). First report of bacterial wilt caused by Ralstonia solanacearum on fig trees in China. For. Pathol. 46, 256–258. doi: 10.1111/efp.12267
Kelman, A. (1953). The Bacterial Wilt Caused by Pseudomonas solanacearum: A Literature Review and Bibliography. Raleigh, NC: North Carolina Agricultural Experiment Station.
Lebeau, A., Daunay, M.-C., Frary, A., Palloix, A., Wang, J.-F., Dintinger, J., et al. (2011). Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101, 154–165. doi: 10.1094/PHYTO-02-10-0048
Lin, C. H., Chuang, M. H., and Wang, J. F. (2015). First report of bacterial wilt caused by Ralstonia solanacearum on chard in Taiwan. Plant Dis. 99:282. doi: 10.1094/PDIS-07-14-0715-PDN
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Marin, J. E., and El-Nashaar, H. M. (1993). “Pathogenicity of the new phenotypes of Pseudomonas solanacearum from Peru,” in Bacterial Wilt, eds G. L. Hartman and A. C. Hayward (Canberra, ACT: ACIAR Proceedings), 78–84.
Norman, D. J., Huang, Q., Yuen, J. M. F., Mangravita-Novo, A., and Byrne, D. (2009a). Susceptibility of geranium cultivars to Ralstonia solanacearum. Hortscience 44, 1504–1508. Available online at: http://hortsci.ashspublications.org/content/44/5/1504.full
Norman, D. J., and Yuen, J. M. F. (1998). A distinct pathotype of Ralstonia (Pseudomonas) solanacearum race 1, biovar 1 entering Florida on infected pothos (Epipremnum aureum) cuttings. Can. J. Plant Pathol. 20, 171–175. doi: 10.1080/07060669809500423
Norman, D. J., and Yuen, J. M. F. (1999). First report of Ralstonia (Pseudomonas) solanacearum infecting pot anthurium production in Florida. Plant Dis. 83:300. doi: 10.1094/PDIS.1999.83.3.300A
Norman, D. J., Zapata, M., Gabriel, D. W., Duan, Y. P., Yuen, J. M. F., Mangravita-Novo, A., et al. (2009b). Genetic diversity and host range variation of Ralstonia solanacearum strains entering North America. Phytopathology 99, 1070–1077. doi: 10.1094/PHYTO-99-9-1070
Palleroni, N. J., and Doudoroff, M. (1971). Phenotypic charcterization and deoxyribonucleic acid homologies of Pseudomonas solanacearum. J. Bacteriol. 107, 690–696.
Prieto Romo, J., Morales Osorio, J. G., and Salazar Yepes, M. (2012). Identification of new hosts for Ralstonia solanacearum (Smith) race 2 from Colombia. Rev. Prot. Veg. 27, 151–161. Available online at: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S1010-27522012000300003
Prior, P., and Steva, H. (1990). Characteristics of strains of Pseudomonas solanacearum from the French West Indies. Plant Dis. 74, 13–17.
Remenant, B., Coupat-Goutaland, B., Guidot, A., Cellier, G., Wicker, E., Allen, C., et al. (2010). Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:379. doi: 10.1186/1471-2164-11-379
Roberts, S. J., Eden-Green, S. J., Joness, P., and Amber, D. J. (1990). Pseudomonas syzygii, sp. nov., the cause of Sumatra disease of cloves. Syst. Appl. Microbiol. 13, 34–43. doi: 10.1016/S0723-2020(11)80178-5
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Taghavi, M., Hayward, C., Sly, I., and Fegan, M. (1996). Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 46, 10–15. doi: 10.1099/00207713-46-1-10
Tjou-Tam-Sin, N. N. A., van de Bilt, J. L. J., Westenberg, M., Korpershoek, H. J., Vermunt, A. M. W., Meekes, E. T. M., et al. (2017). First report of bacterial wilt caused by Ralstonia solanacearum in ornamental Rosa sp. Plant Dis. 101:378. doi: 10.1094/PDIS-02-16-0250-PDN
Vreeburg, R. A. M., Bergsma-Vlami, M., Bollema, R. M., de Haan, E. G., Kooman-Gersmann, M., Smits-Mastebroek, L., et al. (2016). Performance of real-time PCR and immunofluorescence for the detection of Clavibacter michiganensis subsp. sepedonicus and Ralstonia solanacearum in potato tubers in routine testing. EPPO Bull. 46, 112–121. doi: 10.1111/epp.12278
Weller, S. A., Elphinstone, J. G., Smith, N. C., Boonham, N., and Stead, D. E. (2000). Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fuorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66, 2853–2858. doi: 10.1128/AEM.66.7.2853-2858.2000
Williamson, L., Nakaho, K., Hudelson, B., and Allen, C. (2002). Ralstonia solanacearum race 3, biovar 2 strains isolated from geranium are pathogenic on potato. Plant Dis. 86, 987–991. doi: 10.1094/PDIS.2002.86.9.987
Yabuuchi, E., Kosako, Y., Yano, I., Hotta, H., and Nishiuchi, Y. (1995). Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen.nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Douderoff 1973) comb.nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39, 897–904.
Keywords: Ralstonia pseudosolanacearum, Rosa spp., disease severity, disease incidence, temperature dependency, stem inoculation, soil drenching
Citation: Tjou-Tam-Sin NNA, van de Bilt JLJ, Westenberg M, Gorkink-Smits PPMA, Landman NM and Bergsma-Vlami M (2017) Assessing the Pathogenic Ability of Ralstonia pseudosolanacearum (Ralstonia solanacearum Phylotype I) from Ornamental Rosa spp. Plants. Front. Plant Sci. 8:1895. doi: 10.3389/fpls.2017.01895
Received: 30 June 2017; Accepted: 19 October 2017;
Published: 02 November 2017.
Edited by:
Carlos Alberto Lopes, Brazilian Agricultural Research Corporation, BrazilReviewed by:
Teresa Ann Coutinho, University of Pretoria, South AfricaMassimiliano Morelli, Istituto per la Protezione Sostenibile delle Piante (CNR), Italy
Copyright © 2017 Tjou-Tam-Sin, van de Bilt, Westenberg, Gorkink-Smits, Landman and Bergsma-Vlami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Napoleon N. A. Tjou-Tam-Sin, bi50am91LXRhbS1zaW5AbnZ3YS5ubA==
 Napoleon N. A. Tjou-Tam-Sin
Napoleon N. A. Tjou-Tam-Sin Jeroen L. J. van de Bilt
Jeroen L. J. van de Bilt Maria Bergsma-Vlami
Maria Bergsma-Vlami