
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 25 October 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01806
 Araz S. Abdullah1*
Araz S. Abdullah1* Caroline S. Moffat1
Caroline S. Moffat1 Francisco J. Lopez-Ruiz1
Francisco J. Lopez-Ruiz1 Mark R. Gibberd1
Mark R. Gibberd1 John Hamblin2
John Hamblin2 Ayalsew Zerihun1
Ayalsew Zerihun1Studies of plant–pathogen interactions have historically focused on simple models of infection involving single host-single disease systems. However, plant infections often involve multiple species and/or genotypes and exhibit complexities not captured in single host-single disease systems. Here, we review recent insights into co-infection systems focusing on the dynamics of host-multi-pathogen interactions and the implications for host susceptibility/resistance. In co-infection systems, pathogen interactions include: (i) Competition, in which competing pathogens develop physical barriers or utilize toxins to exclude competitors from resource-dense niches; (ii) Cooperation, whereby pathogens beneficially interact, by providing mutual biochemical signals essential for pathogenesis, or through functional complementation via the exchange of resources necessary for survival; (iii) Coexistence, whereby pathogens can stably coexist through niche specialization. Furthermore, hosts are also able to, actively or passively, modulate niche competition through defense responses that target at least one pathogen. Typically, however, virulent pathogens subvert host defenses to facilitate infection, and responses elicited by one pathogen may be modified in the presence of another pathogen. Evidence also exists, albeit rare, of pathogens incorporating foreign genes that broaden niche adaptation and improve virulence. Throughout this review, we draw upon examples of co-infection systems from a range of pathogen types and identify outstanding questions for future innovation in disease control strategies.
Plant pathology has focused predominantly on single host-single disease interactions. Whilst this simplification has proved useful, plants in nature interact with multiple pathogen species/genotypes (Kozanitas et al., 2017; Tollenaere et al., 2017). This complex interaction, known as co-infection, is of particular interest since it tends to alter the course of the disease and the severity of expression (i.e., overall virulence) and has been the subject of a recent review in plant epidemiology (Tollenaere et al., 2016).
Three key interactions can cause damage in co-infected plants: host–pathogen, pathogen–pathogen, and host-multiple-pathogen complexes. Host–pathogen interactions are well-studied and are generally detrimental to the plant resulting in reduced fitness (Brown, 2015). In contrast, pathogen–pathogen and host-multiple-pathogen interactions are less studied. These interactions can lead to various results: antagonism, synergism, coexistence, mutualism, or cooperation (Table 1). The level of disease damage the plant experiences varies depending on the outcome of the interactions and the corresponding host responses. For example, several strains of Pseudomonas bacteria secrete antimicrobial compounds that are antagonistic to sensitive pathogens within the host (Walsh et al., 2001). Many such compounds are also phytotoxic and may exacerbate the level of disease damage (Maurhofer et al., 1992). Furthermore, some pathogens, such as the biotroph Blumeria graminis f. sp. tritici and the necrotroph Zymoseptoria tritici of wheat, do not interact directly to cause damage to the host as one pathogen can inhibit the development of the other (Orton and Brown, 2016). This inhibition can be so profound that the plant plays an active role in promoting the growth of disease suppressive pathogens (Smith et al., 1999). Therefore, moving beyond how heterogeneity of infection influences overall virulence requires a holistic understanding of how a host responds to co-infection and how pathogens interact and coexist.
Recent advances in genomic and molecular techniques have led to new insights into host–pathogen dynamics. For example, metagenomics and microbial tag sequencing have created novel opportunities for studying the wide range of pathogens associated with a single host (Petrosino et al., 2009; Tollenaere et al., 2012). These tools have provided insights into the prevalence of multiple infections in the field, and current knowledge indicates that the extent of co-infection can be significant in some pathosystems (Susi et al., 2015; Tollenaere et al., 2017). Progress in this area has revealed that co-infections can lead to several outcomes: (i) competitive exclusion where over time one pathogen excludes the other (Al-Naimi et al., 2005); (ii) mutualistic coexistence in which both pathogens receive benefits from the interaction (Mordecai et al., 2016); and (iii) emergence of new recombinant genomes where one pathogen incorporates a complementary gene set from another pathogen, leading to large-scale epidemics (Friesen et al., 2006). Indeed, populations of one pathogen may modify host environments to the advantage/disadvantage of other pathogens, affecting their frequencies and persistence within a pathogenic population (Perefarres et al., 2014). Hence, understanding within host–pathogen interaction is crucial for the prediction of long-term dynamics of multiple disease outcomes.
In this review, we discuss recent insights into within-host disease diversity and dynamics of pathogen interactions focusing on the current understanding of pathogen competition and cooperation, and the mechanisms that allow long-term coexistence to occur. We draw upon examples of co-infections from a range of pathogen types that provide useful insights for understanding the evolution of pathogen interactions and coexistence.
The overwhelming majority of studies on plant defense responses to pathogenic infections have been performed on single plant-disease pathosystems. However, under conducive conditions, plants frequently encounter multiple pathogens, often with various modes of action. Hence, a successful plant defense system will incorporate several resistance (R) genes that coordinate a response to multiple attacks. The genomes of plants encode a coordinated array of R-genes that permit recognition of the pathogen and a rapid defense response (Dangl and Jones, 2001). Prioritization of defense may occur, leading to larger investments in defense metabolites against certain pathogens depending on their modes of action (Hacquard et al., 2016; Castrillo et al., 2017). This raises the question: does an infection by one-pathogen influence a host defense to subsequent infection by other pathogens?
Some pathogenic infections can be detrimental to the defense systems predisposing the plant to subsequent secondary infections. For example, infection of Arabidopsis by the foliar bacterium Pseudomonas syringae renders plants vulnerable to invasion by the necrotrophic ascomycete Alternaria brassicicola (Spoel et al., 2007). Infection by the biotrophic oomycete Albugo candida suppresses Arabidopsis defenses, permitting subsequent infections by several otherwise avirulent pathogens (Cooper et al., 2008). Similarly, infection of maize by the phytopathogenic fungi Fusarium verticillioides facilitates infection by several related fungi, through the suppression of production of major secondary defense metabolites in the plant host (Saunders and Kohn, 2008). The mechanisms that lead to the suppression of defense have been defined in some cases. For example, the natriuretic peptide receptor NPA expressed by P. syringae permits subsequent infection by virulent A. brassicicola in Arabidopsis through downregulating a large range of defense-related genes (Spoel et al., 2003; Cooper et al., 2008). Similarly, fusaric acid secreted by F. oxysporum suppresses expression of genes that regulate the antimicrobial activity of 2,4-diacetylphloroglucinol and predisposes wheat to P. fluorescens infection (Notz et al., 2002).
Some pathogenic infections can enhance the defensive capacity of their hosts and activate responses against subsequent attacks. For example, infection by the foliar bacterium P. fluorescens suppresses flagellin-triggered defense in Arabidopsis via apoplastic secretion of low-molecular-weight defense metabolites (Millet et al., 2010). Upon exposure to the bacterium, the defense system of the plant is locally suppressed; though a defense-signaling cascade develops systematically and spreads across infected plant parts conferring resistance to subsequent attacks (Van Der Ent et al., 2009). Some root infections can confer resistance by forming rhizosphere networks that connect infected plants and signal-induced resistance to neighboring plants (Song et al., 2010). Similarly, induced resistance has been reported for co-infections by species of the same genus, where non-pathogenic F. oxysporum primed tomato plants to pathogenic F. oxysporum in a vaccine-like fashion (Aime et al., 2013). The molecular mechanisms involved in this priming have not been fully elucidated, although direct antagonism, detoxification of pathogen effectors and elevated expressions of plant defense-related genes have been recorded (Aime et al., 2013; Ravensdale et al., 2014; Conrath et al., 2015).
Recently, there has been growing interest in plant defense responses to co-infection at the hormonal level. This involves a comparative pathway analysis following infections by multiple organisms. Expression levels of genes responsive to jasmonic acid (JA), ethylene (Et), and salicylic acid (SA) are commonly measured in such analyses. Generally, JA and Et are considered to be mutual pathways linked to defense against necrotrophic pathogens such as Botrytis cinerea (Grant and Jones, 2009). SA, on the other hand, is often linked to defense against biotrophic pathogens such as P. syringae (Glazebrook, 2005). Antagonistic crosstalk between JA/ET and SA is well-documented (Pieterse et al., 2009; Robert-Seilaniantz et al., 2011), permitting the plant to mount the appropriate defense responses to the attacking pathogen. In Arabidopsis, elevated expression of JA/Et-responsive genes resulted in antagonistic effects on SA-responsive genes and increased plant resistance to B. cinerea (Moffat et al., 2012). Similarly, exogenously applied SA revealed antagonist effects on expressions of JA-responsive genes but simultaneously increased Arabidopsis resistance to P. syringae (Gazzarrini and Mccourt, 2003). Pathogens have evolved sophisticated mechanisms to exploit this antagonism and counter host defense responses. For example, the polyketide phytotoxin coronatine secreted by P. syringae is structurally analogous to jasmonoyl-isoleucine and can bind to JA receptors, hijacking the JA mediated defense and causing disease susceptibility to P. syringae (Katsir et al., 2008). Similarly, the avirulent effector AvrPtoB produced by P. syringae disrupts hormonal signaling components in Arabidopsis creating the potential for vulnerability to subsequent infections (De Torres-Zabala et al., 2007).
Although SA and JA signaling can be activated separately, recent studies have shown varying degrees of involvement of both pathways depending on plant–pathogen combinations. An elegant comparative transcriptomics study revealed significant overlap in Arabidopsis responses to a set of biotrophic and necrotrophic attackers (De Vos et al., 2005). Global gene expression analyses revealed that all Arabidopsis-attackers stimulated JA biosynthesis (De Vos et al., 2005). Similarly, infection by the non-necrotroph P. syringae induced a JA-mediated defense in Arabidopsis localized to infected regions (Cui et al., 2005). These results suggest the model of SA-mediated defense against biotrophs, and JA/Et-mediated defense against necrotrophs is too simplistic. The defense responses are likely to be fine-tuned to particular plant–pathogen combinations. There is much yet to be learned about mechanisms that allow for these differences, and this is an active area of research.
Many plant microbes can have latent pathogenic capacities and specialize in causing disease when host conditions are suitable. These microbes can be commensal, living benignly with limited or no damage to the plant, but may become pathogenic should host physiological conditions change (Hentschel et al., 2000). The foliar fungi Ramularia collo-cygni is a good example of a microbe that may be either commensal or pathogenic depending on the developmental stage of its barley host. The fungus becomes more aggressive during the late stages of barley development, and physiological events associated with flowering are thought to trigger the shift to necrotrophic lifestyle (Mcgrann et al., 2016). Even closely related members of a genus, notably species belonging to Colletotrichum, can be symbiotic under certain conditions and highly pathogenic under other conditions (Hiruma et al., 2016). Similarly, the switch to the pathogenic lifestyles for many microbes only occurs in defense-compromised plants following other aggressive disease encounters (Lahrmann et al., 2015; Fesel and Zuccaro, 2016). For instance, infection by highly virulent P. syringae induces systematic susceptibility in Arabidopsis stimulating the pathogenic capacity of an otherwise avirulent P. syringae isolate (Thomma et al., 2001).
The defense systems of the plant are as likely to recognize and respond to commensal microbes as they are to pathogenic ones (Dangl and Jones, 2001). Pathogenic microbes can suppress the defense systems of the plant to the extent that disease develops. Whilst commensal microbes can also suppress the defense systems of the plant; this is not usually to the extent that severe disease develops. Therefore, pathogenic interactions are those that outweigh the defense systems of the host resulting in disease, whereas commensal interactions are balanced and remain asymptomatic (reviewed in Schulz and Boyle, 2005). Whether a particular plant–microbe interaction leads to a disease depends on host factors that maintain the balance between host defense and pathogen virulence (Schulz et al., 1999). If this balance shifts to favor the microbe, commensal interactions can then become pathogenic. For example, suboptimal nutrition has been shown to weaken the defense systems of plants resulting in disease and growth reduction caused by an otherwise avirulent microbe (Kuldau and Yates, 2000). Similarly, highly pathogenic encounters with a necrotrophic P. syringae compromised the defense balance in Arabidopsis and rendered the host vulnerable to infection by avirulent P. syringae (Thomma et al., 2001). Hence, plant defense responses to microbes are highly unpredictable, and yet undiscovered mechanisms may determine the outcome of such interactions.
What factors the altered microbial metabolism between commensal and pathogenic is a promising area for future research. Key factors for such shifts are not well-studied; although, sudden changes in temperature and/or humidity are possible triggers (Mcgrann et al., 2016). Some changes in metabolism can be induced by pathogen signals that alter host defense responses. Moniliophthora perniciosa is a fungal species often found in cocoa shoots as an endophyte living benignly without causing apparent disease. Colonization of cocoa shoots by this fungus gradually increases SA production while decreasing JA production triggering a lifestyle shift and rendering the plant susceptible to the necrotrophic phase of the fungus (Chaves and Gianfagna, 2006). M. perniciosa is thought to have acquired the ability to produce SA to facilitate active competition with other plant microbes and initiate the transition to more aggressive lifestyles (Bezemer et al., 2006). Regardless of what the actual triggers for this shift are, it may indicate a decline in host available resources that are vital for pathogen survival and reproduction creating a potential for multi-pathogen competition. It remains unclear how plants discriminate and respond appropriately to closely related microbes with variable metabolism.
Recent advances in metagenomics have highlighted the vast diversity of the community in which plant pathogens reside (Schenk et al., 2012). Due to the complex nature of these communities and the limited host resources, pathogens often enter into fierce competition. Fundamentally, competition between coexisting pathogens occurs for growth- and fitness-limiting resources. Resource competition involves utilization of limited host nutrients by one pathogen that then restricts supply to other pathogens sharing the same host. Monod (1949) was the first to point out the relationship between nutrients and pathogen growth: within a defined space in which all nutrients are provided, pathogens may stably coexist. Suboptimal nutrition leads to competition whereby some species may dominate. The severity and type of competition is determined by the consumption of nutrients over time, and this principle has been applied to plant and animal populations as an explanation of the dynamics of competing individuals (Adee et al., 1990; Chesson, 2000).
In nutritionally defined niches, such as plant leaves, pathogens with similar nutritional requirements compete for finite resources (West et al., 2006). Competition under such conditions may lead to selection for the more virulent species, or conversely, all associated pathogens may suffer (Rankin et al., 2007). For example, when maize was co-infected by Ustilago maydis and F. verticillioides, initially both fungal species had increased growth followed by decreased growth over time due to the depletion of nutrient resources (Jonkers et al., 2012). Pathogen traits such as cell-wall adhesion can support greater nutrient acquisition and provide competitive advantage (Hibbing et al., 2010). Adherent cell walls can enable greater resource capture efficiency even when growth substrates are present at low concentrations (Figure 1A). Similarly, pathogens that can halt certain metabolic processes, such as toxin production, when the necessary nutrients are exhausted, may show a greater competitive advantage (Glenn et al., 2008). Improved nutrient acquisition can also be achieved by the release of cell-wall degrading enzymes or toxins to sequester host nutrients (Greenberg and Yao, 2004; Rohmer et al., 2011). However, these compounds may also provide a competitive advantage to other opportunistic pathogens that do not have to bear the energetic costs for their production (Hentzer et al., 2003; Cornelis and Dingemans, 2013; Ghoul et al., 2014).
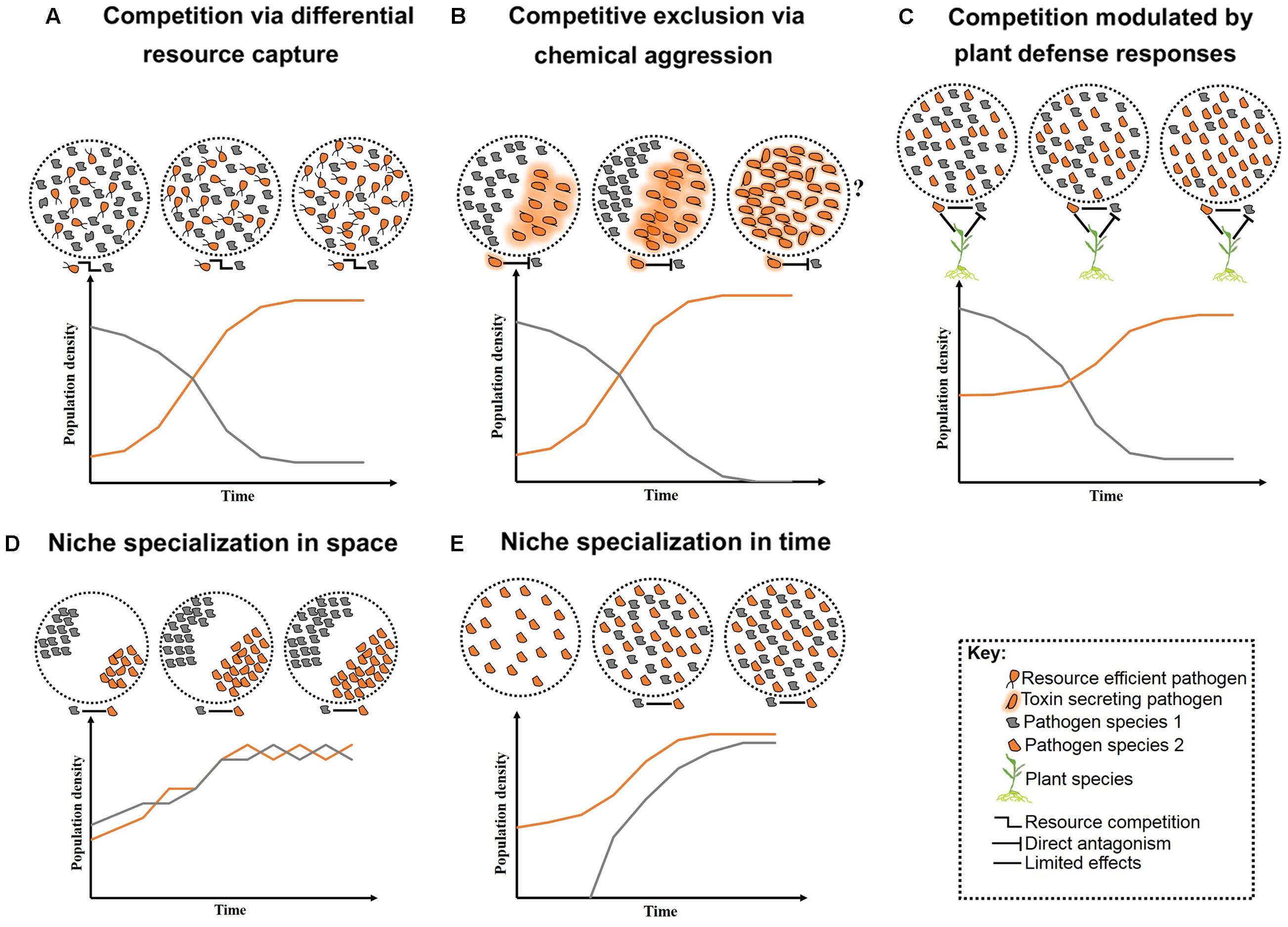
FIGURE 1. Schematic representation of long-term pathogen–pathogen interactions. Competition over growth limiting resources and/or host niches can initially create high diversity, but low stability. Overtime, competition restricts diversity and allows competitive species to outgrow less competitive ones (A–C). Alternatively, neutral interactions occur when pathogens co-reside distinct niches within their host (D) or arrive at various time intervals (E). Question marks represent less studied cases where toxin production may lead to reduce fitness in the absent of competitors.
More aggressive forms of competition between pathogens include direct chemical exclusion (Figure 1B). A classic example of chemical aggression includes tenuazonic acid secreted by the finger millet colonizing endophyte Phoma sp. which prevents growth of several pathogens including the toxigenic fungus F. graminearum (Mousa et al., 2015, 2016a). Mechanical aggression also occurs through disruption of cell membranes and formation of multilayer physical barriers limiting growth and infection success of competitors. For example, the root-inhabiting bacteria Enterobacter sp. forms specialized root-pathogen structures to prevent infection by F. graminearum in finger millet (Mousa et al., 2016b). A molecular mechanism involved in pathogen competition has been identified. The N-alkylated benzylamine secreted by the non-pathogenic bacterium P. putida acts as a lytic enzyme that inhibits fungal growth of B. cinerea in field bean (Ongena et al., 2005). Competition can also occur indirectly, mediated by the host through targeted defense responses against at least one pathogen (Figure 1C). However, the targeted defense can also provide a competitive advantage to pathogens that can counteract the recognition process of the host or that are capable of evading the plant defense system (Ren and Gijzen, 2016).
An expected outcome of competition is a localized reduction in diversity and concurrent specialization of pathogens to various tissues and/or host species – i.e., niche specialization. An individual plant may contain several niches in which pathogens can exist (Figure 1D), and heterogeneity in the biology and epidemiology of pathogens may permit niche specialization (Figure 1E). For example, differences in disease onset resulted in temporal separation and stable coexistence between two related fungal pathogens of canola, Leptosphaeria maculans and L. biglobosa (Toscano-Underwood et al., 2003). Niche specialization can reduce the severity of competition between pathogens permitting coexistence (Fitt et al., 2006), although pathogens occupying various niches within a plant may interact indirectly by stimulating a common host defense response (Aghnoum and Niks, 2012). Nevertheless, on an evolutionary timescale, competition may result in exclusion, enabling species to coexist when arriving at various times (Fitt et al., 2006; Pfennig and Pfennig, 2009; Perefarres et al., 2014). However, when displacement leads to the exclusion of the less competitive pathogen, the newly evolved highly competitive species may compete with other pathogens for their optimal niches (Perefarres et al., 2014). This situation may arise if a species integrates foreign genes allowing the invasion of novel niches (Friesen et al., 2006). Individuals of other species would then have fewer unoccupied niches in which to gain footholds, resulting in potentially large scale epidemics (Perefarres et al., 2014). Although the role of the integration of foreign genes in niche specialization and expansion is less clear, interspecies acquisition of the ToxA gene has allowed Pyrenophora tritici-repentis to infect ToxA-sensitive wheat varieties (Friesen et al., 2006).
Virulence of many plant pathogens depends on secretion of growth promoting factors that subvert host defenses and improve their own nutrient uptake. These growth-promoting factors are often deployed into the extracellular matrix of the host and may hence be indirectly accessible by other pathogens in a local community (Harrison et al., 2006). In such cases, producing species can be vulnerable to exploitation by opportunists that can utilize virulence factors of their neighbors without contributing to the production of these factors. P. syringae provides a good example where common virulence factors give rise to opportunistic exploitation. Virulence in this pathogen is facilitated by the type III secretion system (T3SS), a needle-like apparatus that enables injection of toxins into host leaf mesophyll (Hauck et al., 2003). Virulence factors required for host manipulation by this pathogen are expressed in a bistable fashion, leading to a slow-growing toxin-secreting (T3SS+) strain and a fast-growing toxin-lacking (T3SS-) strain (Barrett et al., 2011). Co-infection experiments initiated with wild-type Arabidopsis revealed a growth advantage to the less virulent T3SS- strain resulting from the opportunistic exploitation of the toxins secreted by T3SS+ (Barrett et al., 2011). Iron-scavenging siderophores provide an additional example whereby co-infection gives rise to opportunistic exploitation. Many opportunistic bacteria can take up heterologous siderophores, diverting iron away from siderophore producing strains and simultaneously passing the cost of production to co-infecting neighbors (Khan et al., 2006). Hence, siderophore-producing bacteria are vulnerable to opportunistic bacteria that are able to utilize heterologous iron-binding products (Khan et al., 2006). The bacteria are not subject to the energetic costs associated with siderophore production and can outgrow more virulent producing genotypes (Alizon and Lion, 2011). Indeed, during co-infection of Arabidopsis with several strains of P. aeruginosa, siderophore lacking bacteria evolved more rapidly and dominated siderophore producers in iron-limited conditions (Khan et al., 2006).
Besides competition, pathogens may also cooperate in associations that are essential for pathogenesis. These associations can be facilitated by biophysical and/or biochemical means. For example, hyphae of the fungal ascomycete Didymella bryoniae physically transported four bacterial species that co-infect Styrian oil pumpkin (Grube et al., 2011; Figure 2A). Samples including both the fungus and bacteria have been recovered from the field, indicating that mutualistic effects on pathogenesis may occur in nature (Grube et al., 2011). The association between Rhizopus microsporus and Burkholderia sp. is an example where a precise biochemical fungal-bacterial association has been identified. R. microsporus is a highly destructive zygomycetous fungus that causes blight in rice seedlings. The fungus is thought to secrete a phytotoxin known as rhizoxin; although no standard polyketide synthesis genes could be identified in the fungus genome (Partida-Martinez and Hertweck, 2005). The rhizoxin was, however, found to be secreted by the endosymbiotic bacteria that are harbored by the fungus. Thus, it appears that R. microsporus owes its pathogenicity to the presence of an endosymbiont bacteria belonging to the genus Burkholderia (Partida-Martinez and Hertweck, 2005). Intimate biochemical fungal-bacterial symbiosis occurs when both a recognition system and a molecular dialog bind the two. For instance, recognition of fusaric acid secreted by certain isolates of the fungus F. oxysporum stimulates growth in the bacterial pathogen P. fluorescens in tomato (De Weert et al., 2004; Kamilova et al., 2008).
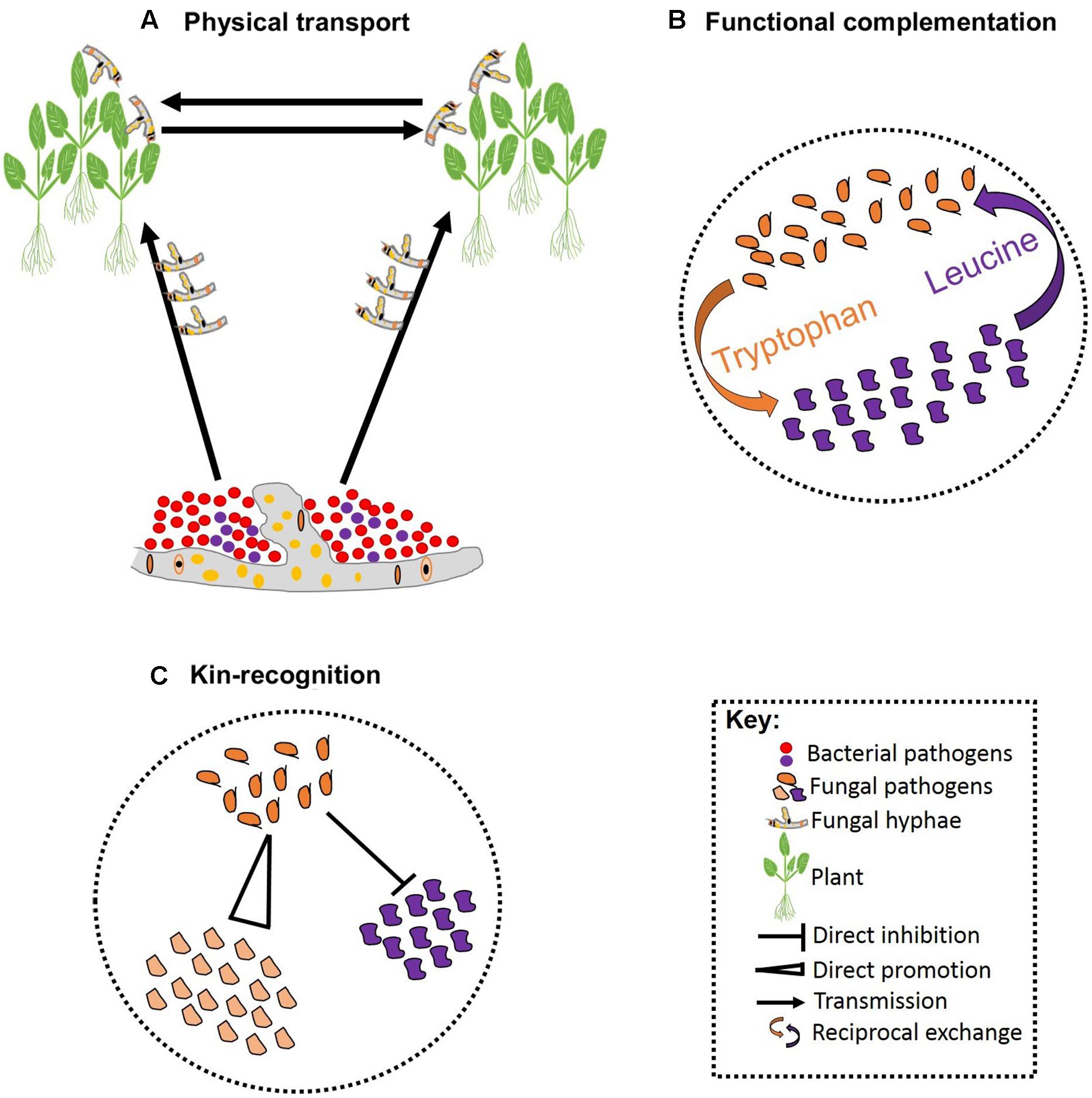
FIGURE 2. Schematic representation of pathogen–pathogen cooperation. Some bacteria can establish symbiotic associations with fungi that allowing them to exploit new niches and move across infected hosts (A). Cooperation can be vital for survival of both parties involved leading to reciprocal exchange of vital growth substrates (B). Cooperation can also involve kinship and some pathogens can facilitate fitness of their kin while restricting fitness of non-kin (C).
Endophytes, although less studied in the context of co-infection, has been considered as an additional component to the multi-pathogen symbiosis (Aime et al., 2013). Endophytes colonize internal plant tissues without causing visible disease and produce antimicrobial compounds that enhance general plant fitness and affect pathogen interactions directly and indirectly (for review, see Partida-Martínez and Heil, 2011). Among the most studied endophytes is the biocontrol agent F. oxysporum. This root-inhabiting fungus produces antimicrobial compounds that enhance plant resistance to pathogenic F. oxysporum (Aime et al., 2013). Recent microscopic and molecular investigations revealed the presence of a consortium of ectosymbiotic bacteria associated with F. oxysporum and crucial for the biocontrol properties of the fungus (Minerdi et al., 2008). However, when the fungus was cured of its associated bacteria, the biocontrol strain became pathogenic (Minerdi et al., 2008). This demonstrates that mutualistic cooperation that involves endophytes is not clear-cut, and complex dynamics involving other microbes may alter the properties of endophytic interactions.
Regardless, mutualistic cooperation between pathogens can have major epidemiological implications, and certain plant pathogens are only host destructive when they cooperate with other independent pathogens. For example, obligate mutualism between maize dwarf mosaic virus and wheat streak mosaic virus causes lethal maize necrosis – neither of these pathogens is known to cause lethal necrosis alone (Uyemoto, 1983). Similarly, co-infection of tobacco mosaic virus and potato virus cause defoliation streak and a high rate of mortality in young tomato leaves (Hull, 2014). Cooperation can also enhance pathogen persistence by supporting greater reproduction rates, increasing the chance of the host being a source of inoculum in the next season (Fondong et al., 2000). Co-infection of Nicotiana benthamiana plants with two strains of the cassava mosaic virus showed symptoms covering all leaves, while single-strain infection exhibited partial coverage and some leaves remained asymptomatic (Fondong et al., 2000).
Reciprocal exchange of growth substrates is a common strategy through which pathogens establish stable cooperation. Such cooperative associations are prominent when resources are deficient, and one species produces the growth substrates required by the other (Figure 2B). Hence, cooperation in such cases is vital for the success of all species involved (Hoek et al., 2016). In addition, there are a few well-characterized studies of altruistic cooperation between closely related species. Altruistic cooperation occurs when co-infection leads to the reduced availability of resources per species, such that the less competitive species experience reduced reproduction rates preserving resources for the reproduction of the other species (West et al., 2002; Kummerli et al., 2009). Hence, the reproductive advantage of the competitive species is dependent on the other species (Figure 2C). Hamilton’s kin-selection theory provides a plausible explanation for the altruistic association between relatives: by helping a close relative, a pathogen is indirectly passing its genes to the next generation (Hamilton, 1963). This can occur when the degree of relatedness between the benefactor and the beneficiary is high, and the benefit outweighs the cost of the cooperation (Eberhard, 1975; Chao et al., 2000). Coexisting pathogens that exhibit altruism may display rapid evolution to overcome pesticides as a result of the reproductive advantage of the more competitive species. Furthermore, initiating defense responses against various pathogens acting together may prove costly to host resources and defense systems, although evidence for such interactions in plant pathogens is currently lacking.
Recent research into pathogen interactions has considered the genetic adaptation that close proximity facilitates. It has been suggested that genomes of some species display reduced mutation rates as an adaptation strategy to the presence of other coexisting species (Cordero and Polz, 2014). Fungi isolated from tree-hole rainwater pools exhibited similar resource acquisition strategies that should lead to severe competition. However, culturing these isolates on media that resembled their natural habitat resulted in a reciprocal exchange of growth substrates (Madsen et al., 2016). When multiple species are in close proximity, genetic recombination may occur through the fusion of haploid cells. For instance, reshuffling of alleles between genetically distant fungi resulted in novel genetic diversity in Z. pseudotritici (Stukenbrock et al., 2012). Novel hybrids often display new characteristics that enable colonization of previously unexploited niches. For example, the horizontal transfer of the host-specific gene ToxA from P. nodorum to P. tritici-repentis allows P. tritici-repentis to invade ToxA sensitive wheat cultivars on a large scale (Friesen et al., 2006).
Co-infection can also provide a selective advantage for the adaptation of low-frequency pathogen populations. For example, the persistence of non-virulent strains of P. syringae was partially linked to their growth advantage through coexistence with virulent strains (Barrett et al., 2011). Similarly, laboratory-based co-infection studies demonstrated a fitness advantage to less competent pathogens conditional on their coexistence with fully-adapted pathogens (Bashey, 2015). Perhaps the simplest explanation for such fitness advantage would be a mass-action mechanism – i.e., more disease load within the plant implies greater disease transmission rates and greater infection opportunities for both species (Perefarres et al., 2014). A less direct explanation, especially applicable for viruses, may involve heteroencapsidation. This may occur when two co-infecting species display sufficient genetic variability in which case the more competent species may complement the less competent species providing benefits in accumulation and transmission within and among plants (Perefarres et al., 2014). Direct testing of mechanisms by which co-infection contributes to the maintenance of pathogen diversity is an exciting research area.
Current knowledge suggests that virulence is an inevitable requirement for host exploitation. Evolution of virulence can be constrained by the reproduction rate of a pathogen (Alizon et al., 2013). Hence, increased virulence may initially be advantageous, but subsequent consequences resulting from increased host mortality are not (Chao et al., 2000; Ebert and Bull, 2003). This is particularly true in pathosystems where both the host and pathogen have comparable generation times, in which case the costs of increased virulence are spread among all species sharing the host – the so-called tragedy of the commons (Hardin, 1968). Under co-infection conditions, pathogens are thought to utilize host-limited resources more efficiently with natural selection favoring the coexistence of pathogens that are less harmful to their hosts (Van Baalen and Sabelis, 1995; Brown et al., 2002). Early insights into the evolution of virulence were provided by the classical three-way model of Levin and Pimentel (1981) which included a host and two pathogens differing in virulence. Based on this model, when two pathogens invade the same host, virulence of one species is always considered relative to the virulence of the other species (Levin and Pimentel, 1981; Brown et al., 2002).
Early during co-infection, the more virulent pathogen may quickly take over. Nevertheless, both more and less virulent pathogens can coexist (Van Baalen and Sabelis, 1995). In vitro experiments suggest that coexistence between two pathogen species varying in virulence can occur. In one system, the population of the more virulent pathogen U. maydis was able to reach higher frequencies over the less virulent species F. verticillioides, though both species coexisted stably (Jonkers et al., 2012). However, when the same system was conducted in planta using maize, the less virulent species experienced lower resistance from the host and hence gained a competitive advantage over the more virulent pathogen (Estrada et al., 2012). As the less virulent species dominate, local competition of some degree takes place, and the balance may shift to favor the more virulent species (Macho et al., 2007). Once again, as the more virulent pathogen becomes abundant, plant defense systems prioritize responses against the more virulent pathogen, thereby indirectly allowing the less virulent pathogen to regain lost ground (De Roode et al., 2005; Thieme, 2007; Cobey and Lipsitch, 2013). This cycle is key for pathogen coexistence in space and time and implies, from an evolutionary perspective, that neither too high nor too low levels of virulence are advantageous.
If we are to make significant progress in plant disease management, research efforts should embrace field representative systems including multiple-pathogen infections. The role of pathogen–pathogen interactions and their impact on plant defense systems should increasingly be recognized as a priority of equal importance to studying single plant–pathogen interactions. Although rare, interaction between pathogens potentially allows the exchange of genes encoding virulence factors broadening pathogen infection strategies and allowing them to exploit new niches (Friesen et al., 2006). However, some interactions can alert the defense system of the plant making subsequent infections less likely. Progress in utilizing pathogen–pathogen interactions for developing holistic disease management strategies has been underwhelming, indicating an area that requires attention (Martinez-Medina et al., 2016).
In co-infection, pathogens can produce antimicrobial compounds toxic to other pathogens sharing the same host. However, whether production of such compounds truly represents an adaptation strategy to competition is unclear. It would be debilitating for a pathogen to produce compounds toward future threats if these threats are not realized. Such pathogens would be selected against under a reasonable assumption that the pathogen must target direct enemies or risk wasting vital resources. Nevertheless, pathogens can coexist and share a host, mainly due to conditions favoring the occurrence of multiple pathogens. An outstanding question is how changes in natural (e.g., climatic) and man-made (e.g., new varieties with polygenic or major gene resistances) conditions alter coexistence on the long-term. Changes to conditions may favor one pathogen over another, leading to potential invasion by large-scale aggressive pathogens. Other challenges that require future attention is cases of below- and above-ground co-infections. Pathogens colonizing various plant parts can interact via systemic host defenses, making studying these interactions particularly intriguing (Filgueiras et al., 2016). Other challenging questions that may have significant epidemiological implications are cases where pathogens invade novel niches/species. The metabolic changes that are required for a pathogen to optimize nutrient acquisition from novel niches remain unclear. High-throughput multi-species transcriptomics and metabolomics should help to unravel some of the mechanisms. Such methods will provide a better understanding of pathogen interactions allowing the development of disease control measures against multiple pathogens.
AA conceived the idea, revised the literature and drafted the manuscript. CM, FL-R, AZ, MG, and JH supervised the work and provided critical suggestions on the article. All authors read and approved the final version of the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge the support of the Grains Research and Development Corporation (CUR00023), and the Research Training Program (RTP) funded by the Australian government. The authors would also like to thank Leon Hodgson, Lincoln Harper, and Wesley Mair (Centre for Crop and Disease Management/Curtin University) for critically reading the manuscript.
Adee, S., Pfender, W., and Hartnett, D. (1990). Competition between Pyrenophora tritici-repentis and Septoria nodorum in the wheat leaf as measured with de wit replacement series. Phytopathology 80, 1177–1182. doi: 10.1094/Phyto-80-1177
Aghnoum, R., and Niks, R. E. (2012). Compatible Puccinia hordei infection in barley induces basal defense to subsequent infection by Blumeria graminis. Physiol. Mol. Plant Pathol. 77, 17–22. doi: 10.1016/j.pmpp.2011.10.003
Aime, S., Alabouvette, C., Steinberg, C., and Olivain, C. (2013). The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol. Plant Microbe Interact. 26, 918–926. doi: 10.1094/MPMI-12-12-0290-R
Alizon, S., De Roode, J. C., and Michalakis, Y. (2013). Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567. doi: 10.1111/ele.12076
Alizon, S., and Lion, S. (2011). Within-host parasite cooperation and the evolution of virulence. Proc. R. Soc. Lond. B Biol. Sci. 278, 3738–3747. doi: 10.1098/rspb.2011.0471
Al-Naimi, F., Garrett, K., and Bockus, W. W. (2005). Competition, facilitation, and niche differentiation in two foliar pathogens. Oecologia 143, 449–457. doi: 10.1007/s00442-004-1814-x
Barrett, L. G., Bell, T., Dwyer, G., and Bergelson, J. (2011). Cheating, trade-offs and the evolution of aggressiveness in a natural pathogen population. Ecol. Lett. 14, 1149–1157. doi: 10.1111/j.1461-0248.2011.01687.x
Bashey, F. (2015). Within-host competitive interactions as a mechanism for the maintenance of parasite diversity. Philos. Trans. R. Soc. B 370, 1–8. doi: 10.1098/rstb.2014.0301
Bezemer, T. M., Harvey, J. A., Kowalchuk, G. A., Korpershoek, H., and Van Der Putten, W. H. (2006). Interplay between Senecio jacobaea and plant, soil, and aboveground insect community composition. Ecology 87, 2002–2013. doi: 10.1890/0012-9658(2006)87[2002:IBSJAP]2.0.CO;2
Brown, J. K. (2015). Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. doi: 10.1146/annurev-phyto-102313-045914
Brown, S. P., Hochberg, M. E., and Grenfell, B. T. (2002). Does multiple infection select for raised virulence? Trends Microbiol. 10, 401–405.
Castrillo, G., Teixeira, P. J. P. L., Paredes, S. H., Law, T. F., De Lorenzo, L., Feltcher, M. E., et al. (2017). Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518. doi: 10.1038/nature21417
Chao, L., Hanley, K. A., Burch, C. L., Dahlberg, C., and Turner, P. E. (2000). Kin selection and parasite evolution: higher and lower virulence with hard and soft selection. Q. Rev. Biol. 75, 261–275. doi: 10.1086/393499
Chaves, F. C., and Gianfagna, T. J. (2006). Necrotrophic phase of Moniliophthora perniciosa causes salicylic acid accumulation in infected stems of cacao. Physiol. Mol. Plant Pathol. 69, 104–108. doi: 10.1016/j.pmpp.2007.02.003
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Cobey, S., and Lipsitch, M. (2013). Pathogen diversity and hidden regimes of apparent competition. Am. Nat. 181, 12–24. doi: 10.1086/668598
Conrath, U., Beckers, G. J., Langenbach, C. J., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Cooper, A., Latunde-Dada, A., Woods-Tör, A., Lynn, J., Lucas, J., Crute, I., et al. (2008). Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Mol. Plant Microbe Interact. 21, 745–756. doi: 10.1094/MPMI-21-6-0745
Cordero, O. X., and Polz, M. F. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273. doi: 10.1038/nrmicro3218
Cornelis, P., and Dingemans, J. (2013). Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol. 3:75. doi: 10.3389/fcimb.2013.00075
Cui, J., Bahrami, A. K., Pringle, E. G., Hernandez-Guzman, G., Bender, C. L., Pierce, N. E., et al. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. U.S.A. 102, 1791–1796. doi: 10.1073/pnas.0409450102
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
De Roode, J. C., Pansini, R., Cheesman, S. J., Helinski, M. E., Huijben, S., Wargo, A. R., et al. (2005). Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl. Acad. Sci. U.S.A. 102, 7624–7628. doi: 10.1073/pnas.0500078102
De Torres-Zabala, M., Truman, W., Bennett, M. H., Lafforgue, G., Mansfield, J. W., Egea, P. R., et al. (2007). Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26, 1434–1443. doi: 10.1038/sj.emboj.7601575
De Vos, M., Van Oosten, V. R., Van Poecke, R. M., Van Pelt, J. A., Pozo, M. J., Mueller, M. J., et al. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18, 923–937. doi: 10.1094/MPMI-18-0923
De Weert, S., Kuiper, I., Lagendijk, E. L., Lamers, G. E., and Lugtenberg, B. J. (2004). Role of chemotaxis toward fusaric acid in colonization of hyphae of Fusarium oxysporum f. sp. radicis-lycopersici by Pseudomonas fluorescens WCS365. Mol. Plant Microbe Interact. 17, 1185–1191. doi: 10.1094/MPMI.2004.17.11.1185
Eberhard, M. J. W. (1975). The evolution of social behavior by kin selection. Q. Rev. Biol. 50, 1–33. doi: 10.1086/408298
Ebert, D., and Bull, J. J. (2003). Challenging the trade-off model for the evolution of virulence: is virulence management feasible? Trends Microbiol. 11, 15–20.
Estrada, A. E. R., Jonkers, W., Kistler, H. C., and May, G. (2012). Interactions between Fusarium verticillioides, Ustilago maydis, and Zea mays: an endophyte, a pathogen, and their shared plant host. Fungal Genet. Biol. 49, 578–587. doi: 10.1016/j.fgb.2012.05.001
Fesel, P. H., and Zuccaro, A. (2016). Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis. Curr. Opin. Microbiol. 32, 103–112. doi: 10.1016/j.mib.2016.05.008
Filgueiras, C. C., Willett, D. S., Junior, A. M., Pareja, M., El Borai, F., Dickson, D. W., et al. (2016). Stimulation of the salicylic acid pathway aboveground recruits entomopathogenic nematodes belowground. PLOS ONE 11:e0154712. doi: 10.1371/journal.pone.0154712
Fitt, B. D., Huang, Y.-J., Van Den Bosch, F., and West, J. S. (2006). Coexistence of related pathogen species on arable crops in space and time. Annu. Rev. Phytopathol. 44, 163–182. doi: 10.1146/annurev.phyto.44.070505.143417
Fondong, V., Pita, J., Rey, M., De Kochko, A., Beachy, R., and Fauquet, C. (2000). Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81, 287–297. doi: 10.1099/0022-1317-81-1-287
Friesen, T. L., Stukenbrock, E. H., Liu, Z., Meinhardt, S., Ling, H., Faris, J. D., et al. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. doi: 10.1038/ng1839
Gazzarrini, S., and Mccourt, P. (2003). Cross-talk in plant hormone signalling: what Arabidopsis mutants are telling us. Ann. Bot. 91, 605–612. doi: 10.1093/aob/mcg064
Ghoul, M., West, S., Diggle, S., and Griffin, A. (2014). An experimental test of whether cheating is context dependent. J. Evol. Biol. 27, 551–556. doi: 10.1111/jeb.12319
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Glenn, A. E., Zitomer, N. C., Zimeri, A. M., Williams, L. D., Riley, R. T., and Proctor, R. H. (2008). Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant Microbe Interact. 21, 87–97. doi: 10.1094/MPMI-21-1-0087
Grant, M. R., and Jones, J. D. (2009). Hormone (dis) harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
Greenberg, J. T., and Yao, N. (2004). The role and regulation of programmed cell death in plant–pathogen interactions. Cell Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Grube, M., Fürnkranz, M., Zitzenbacher, S., Huss, H., and Berg, G. (2011). Emerging multi-pathogen disease caused by Didymella bryoniae and pathogenic bacteria on Styrian oil pumpkin. Eur. J. Plant Pathol. 131, 539–548. doi: 10.1007/s10658-011-9829-8
Hacquard, S., Kracher, B., Hiruma, K., Munch, P. C., Garrido-Oter, R., Thon, M. R., et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7, 1–12.
Hamilton, W. D. (1963). The evolution of altruistic behavior. Am. Nat. 97, 354–356. doi: 10.1086/497114
Harrison, F., Browning, L. E., Vos, M., and Buckling, A. (2006). Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4:21. doi: 10.1186/1741-7007-4-21
Hauck, P., Thilmony, R., and He, S. Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. U.S.A. 100, 8577–8582. doi: 10.1073/pnas.1431173100
Hentschel, U., Steinert, M., and Hacker, J. (2000). Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8, 226–231. doi: 10.1016/S0966-842X(00)01758-3
Hentzer, M., Wu, H., Andersen, J. B., Riedel, K., Rasmussen, T. B., Bagge, N., et al. (2003). Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815. doi: 10.1093/emboj/cdg366
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hiruma, K., Gerlach, N., Sacristán, S., Nakano, R. T., Hacquard, S., Kracher, B., et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028
Hoek, T. A., Axelrod, K., Biancalani, T., Yurtsev, E. A., Liu, J., and Gore, J. (2016). Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLOS Biol. 14:e1002540. doi: 10.1371/journal.pbio.1002540
Jonkers, W., Estrada, A. E. R., Lee, K., Breakspear, A., May, G., and Kistler, H. C. (2012). Metabolome and transcriptome of the interaction between Ustilago maydis and Fusarium verticillioides in vitro. Appl. Environ. Microbiol. 78, 3656–3667. doi: 10.1128/AEM.07841-11
Kamilova, F., Lamers, G., and Lugtenberg, B. (2008). Biocontrol strain Pseudomonas fluorescens WCS365 inhibits germination of Fusarium oxysporum spores in tomato root exudate as well as subsequent formation of new spores. Environ. Microbiol. 10, 2455–2461. doi: 10.1111/j.1462-2920.2008.01638.x
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105. doi: 10.1073/pnas.0802332105
Khan, A., Geetha, R., Akolkar, A., Pandya, A., Archana, G., and Desai, A. J. (2006). Differential cross-utilization of heterologous siderophores by nodule bacteria of Cajanus cajan and its possible role in growth under iron-limited conditions. Appl. Soil Ecol. 34, 19–26. doi: 10.1016/j.apsoil.2005.12.001
Kozanitas, M., Osmundson, T. W., Linzer, R., and Garbelotto, M. (2017). Interspecific interactions between the Sudden Oak Death pathogen Phytophthora ramorum and two sympatric Phytophthora species in varying ecological conditions. Fungal Ecol. 28, 86–96. doi: 10.1016/j.funeco.2017.04.006
Kuldau, G. A., and Yates, I. E. (2000). “Evidence for Fusarium endophytes in cultivated and wild plants,” in Microbial Endophytes, eds C. W. Bacon and J. F. White (New York, NY: Marcel Dekker), 85–117.
Kummerli, R., Griffin, A. S., West, S. A., Buckling, A., and Harrison, F. (2009). Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. R. Soc. B 276, 3531–3538. doi: 10.1098/rspb.2009.0861
Lahrmann, U., Strehmel, N., Langen, G., Frerigmann, H., Leson, L., Ding, Y., et al. (2015). Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol. 207, 841–857. doi: 10.1111/nph.13411
Levin, S., and Pimentel, D. (1981). Selection of intermediate rates of increase in parasite-host systems. Am. Nat. 117, 308–315. doi: 10.1086/283708
Macho, A. P., Zumaquero, A., Ortiz-Martín, I., and Beuzon, C. R. (2007). Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae–plant interactions. Mol. Plant Pathol. 8, 437–450. doi: 10.1111/j.1364-3703.2007.00404.x
Madsen, J. S., Roder, H. L., Russel, J., Sorensen, H., Burmolle, M., and Sorensen, S. J. (2016). Co-existence facilitates interspecific biofilm formation in complex microbial communities. Environ. Microbiol. 18, 2565–2574. doi: 10.1111/1462-2920.13335
Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C. M., Pozo, M. J., et al. (2016). Recognizing plant defense priming. Trends Plant Sci. 21, 818–822. doi: 10.1016/j.tplants.2016.07.009
Maurhofer, M., Keel, C., Schnider, U., Voisard, C., Haas, D., and Défago, G. (1992). Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology 82, 190–195. doi: 10.1094/Phyto-82-190
Mcgrann, G. R., Andongabo, A., Sjökvist, E., Trivedi, U., Dussart, F., Kaczmarek, M., et al. (2016). The genome of the emerging barley pathogen Ramularia collo-cygni. BMC Genomics 17:584. doi: 10.1186/s12864-016-2928-3
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Minerdi, D., Moretti, M., Gilardi, G., Barberio, C., Gullino, M. L., and Garibaldi, A. (2008). Bacterial ectosymbionts and virulence silencing in a Fusarium oxysporum strain. Environ. Microbiol. 10, 1725–1741. doi: 10.1111/j.1462-2920.2008.01594.x
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLOS ONE 7:e35995. doi: 10.1371/journal.pone.0035995
Monod, J. (1949). The growth of bacterial cultures. Annu. Rev. Microbiol. 3, 371–394. doi: 10.1146/annurev.mi.03.100149.002103
Mordecai, E. A., Gross, K., and Mitchell, C. E. (2016). Within-host niche differences and fitness trade-offs promote coexistence of plant viruses. Am. Nat. 187, E13–E26. doi: 10.1086/684114
Mousa, W. K., Schwan, A., Davidson, J., Strange, P., Liu, H., Zhou, T., et al. (2015). An endophytic fungus isolated from finger millet (Eleusine coracana) produces anti-fungal natural products. Front. Microbiol. 6:1157. doi: 10.3389/fmicb.2015.01157
Mousa, W. K., Schwan, A. L., and Raizada, M. N. (2016a). Characterization of antifungal natural products isolated from endophytic fungi of finger millet (Eleusine coracana). Molecules 21:1171. doi: 10.3390/molecules21091171
Mousa, W. K., Shearer, C., Limay-Rios, V., Ettinger, C. L., Eisen, J. A., and Raizada, M. N. (2016b). Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat. Microbiol. 1, 16167. doi: 10.1038/nmicrobiol.2016.167
Notz, R., Maurhofer, M., Dubach, H., Haas, D., and Défago, G. (2002). Fusaric acid-producing strains of Fusarium oxysporum alter 2, 4-diacetylphloroglucinol biosynthetic gene expression in Pseudomonas fluorescens CHA0 in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 68, 2229–2235. doi: 10.1128/AEM.68.5.2229-2235.2002
Ongena, M., Jourdan, E., Schafer, M., Kech, C., Budzikiewicz, H., Luxen, A., et al. (2005). Isolation of an N-alkylated benzylamine derivative from Pseudomonas putida BTP1 as elicitor of induced systemic resistance in bean. Mol. Plant Microbe Interact. 18, 562–569. doi: 10.1094/MPMI-18-0562
Orton, E. S., and Brown, J. K. (2016). Reduction of growth and reproduction of the biotrophic fungus Blumeria graminis in the presence of a necrotrophic pathogen. Front. Plant Sci. 7:742. doi: 10.3389/fpls.2016.00742
Partida-Martínez, L. P., and Heil, M. (2011). The microbe-free plant: fact or artifact? Front. Plant Sci. 2:100. doi: 10.3389/fpls.2011.00100
Partida-Martinez, L. P., and Hertweck, C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888. doi: 10.1038/nature03997
Perefarres, F., Thebaud, G., Lefeuvre, P., Chiroleu, F., Rimbaud, L., Hoareau, M., et al. (2014). Frequency-dependent assistance as a way out of competitive exclusion between two strains of an emerging virus. Proc. R. Soc. Lond. B Biol. Sci. 281, 1–9. doi: 10.1098/rspb.2013.3374
Petrosino, J. F., Highlander, S., Luna, R. A., Gibbs, R. A., and Versalovic, J. (2009). Metagenomic pyrosequencing and microbial identification. Clin. Chem. 55, 856–866. doi: 10.1373/clinchem.2008.107565
Pfennig, K. S., and Pfennig, D. W. (2009). Character displacement: ecological and reproductive responses to a common evolutionary problem. Q. Rev. Biol. 84, 253–276. doi: 10.1086/605079
Pieterse, C. M., Leon-Reyes, A., Van Der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Rankin, D. J., Bargum, K., and Kokko, H. (2007). The tragedy of the commons in evolutionary biology. Trends Ecol. Evol. 22, 643–651. doi: 10.1016/j.tree.2007.07.009
Ravensdale, M., Rocheleau, H., Wang, L., Nasmith, C., Ouellet, T., and Subramaniam, R. (2014). Components of priming-induced resistance to Fusarium head blight in wheat revealed by two distinct mutants of Fusarium graminearum. Mol. Plant Pathol. 15, 948–956. doi: 10.1111/mpp.12145
Ren, N., and Gijzen, M. (2016). Escaping host immunity: new tricks for plant pathogens. PLOS Pathol. 12:e1005631. doi: 10.1371/journal.ppat.1005631
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rohmer, L., Hocquet, D., and Miller, S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. doi: 10.1016/j.tim.2011.04.003
Saunders, M., and Kohn, L. M. (2008). Host-synthesized secondary compounds influence the in vitro interactions between fungal endophytes of maize. Appl. Environ. Microbiol. 74, 136–142. doi: 10.1128/AEM.01538-07
Schenk, P. M., Carvalhais, L. C., and Kazan, K. (2012). Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trends Biotechnol. 30, 177–184. doi: 10.1016/j.tibtech.2011.11.002
Schulz, B., and Boyle, C. (2005). The endophytic continuum. Mycol. Res. 109, 661–686. doi: 10.1017/S095375620500273X
Schulz, B., Römmert, A.-K., Dammann, U., Aust, H.-J., and Strack, D. (1999). The endophyte-host interaction: a balanced antagonism? Mycol. Res. 103, 1275–1283. doi: 10.1017/S0953756299008540
Smith, K. P., Handelsman, J., and Goodman, R. M. (1999). Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. U.S.A. 96, 4786–4790. doi: 10.1073/pnas.96.9.4786
Song, Y. Y., Zeng, R. S., Xu, J. F., Li, J., Shen, X., and Yihdego, W. G. (2010). Interplant communication of tomato plants through underground common mycorrhizal networks. PLOS ONE 5:e13324. doi: 10.1371/journal.pone.0013324
Spoel, S. H., Johnson, J. S., and Dong, X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. U.S.A. 104, 18842–18847. doi: 10.1073/pnas.0708139104
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate-and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Stukenbrock, E. H., Christiansen, F. B., Hansen, T. T., Dutheil, J. Y., and Schierup, M. H. (2012). Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc. Natl. Acad. Sci. U.S.A. 109, 10954–10959. doi: 10.1073/pnas.1201403109
Susi, H., Barres, B., Vale, P. F., and Laine, A.-L. (2015). Co-infection alters population dynamics of infectious disease. Nat. Commun. 6, 1–8. doi: 10.1038/ncomms6975
Thieme, H. R. (2007). “Pathogen competition and coexistence and the evolution of virulence,” in Mathematics for Life Science and Medicine, eds Y. Takeuchi, Y. Iwasa, and K. Sato (Berlin: Springer).
Thomma, B. P., Penninckx, I. A., Cammue, B. P., and Broekaert, W. F. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/S0952-7915(00)00183-7
Tollenaere, C., Lacombe, S., Wonni, I., Barro, M., Ndougonna, C., Gnacko, F., et al. (2017). Virus-bacteria rice co-Infection in Africa: field estimation, reciprocal effects, molecular mechanisms, and evolutionary implications. Front. Plant Sci. 8:645. doi: 10.3389/fpls.2017.00645
Tollenaere, C., Susi, H., and Laine, A.-L. (2016). Evolutionary and epidemiological implications of multiple infection in plants. Trends Plant Sci. 21, 80–90. doi: 10.1016/j.tplants.2015.10.014
Tollenaere, C., Susi, H., Nokso-Koivisto, J., Koskinen, P., Tack, A., Auvinen, P., et al. (2012). SNP design from 454 sequencing of Podosphaera plantaginis transcriptome reveals a genetically diverse pathogen metapopulation with high levels of mixed-genotype infection. PLOS ONE 7:e52492. doi: 10.1371/journal.pone.0052492
Toscano-Underwood, C., Huang, Y., Fitt, B. D., and Hall, A. (2003). Effects of temperature on maturation of pseudothecia of Leptosphaeria maculans and L. biglobosa on oilseed rape stem debris. Plant Pathol. 52, 726–736. doi: 10.1111/j.1365-3059.2003.00930.x
Uyemoto, J. (1983). Biology and control of maize chlorotic mottle virus. Plant Dis. 67, 7–10. doi: 10.1094/PD-67-7
Van Baalen, M., and Sabelis, M. W. (1995). The scope for virulence management: a comment on Ewald’s view on the evolution of virulence. Trends Microbiol. 3, 414–416. doi: 10.1016/S0966-842X(00)88991-X
Van Der Ent, S., Van Wees, S. C., and Pieterse, C. M. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70, 1581–1588. doi: 10.1016/j.phytochem.2009.06.009
Walsh, U. F., Morrissey, J. P., and O’gara, F. (2001). Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 12, 289–295. doi: 10.1016/S0958-1669(00)00212-3
West, S. A., Griffin, A. S., Gardner, A., and Diggle, S. P. (2006). Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607. doi: 10.1038/nrmicro1461
Keywords: multiple infections, plant defense to co-infection, pathogen competition, pathogen cooperation, pathogen coexistence, niche heterogeneity
Citation: Abdullah AS, Moffat CS, Lopez-Ruiz FJ, Gibberd MR, Hamblin J and Zerihun A (2017) Host–Multi-Pathogen Warfare: Pathogen Interactions in Co-infected Plants. Front. Plant Sci. 8:1806. doi: 10.3389/fpls.2017.01806
Received: 07 July 2017; Accepted: 04 October 2017;
Published: 25 October 2017.
Edited by:
Vincenzo Lionetti, Sapienza Università di Roma, ItalyReviewed by:
Walaa Kamel Mousa, University of Guelph, CanadaCopyright © 2017 Abdullah, Moffat, Lopez-Ruiz, Gibberd, Hamblin and Zerihun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Araz S. Abdullah, YXJhei5hYmR1bGxhaEBjdXJ0aW4uZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.