
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 September 2017
Sec. Plant Membrane Traffic and Transport
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01688
This article is part of the Research Topic From Structure to Signalosomes: New Perspectives About Membrane Receptors and Channels View all 16 articles
The ERECTA family genes, ERECTA (ER), ERECTA-LIKE1 (ERL1), and ERECTA-LIKE2 (ERL2), encode leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Knocking out these three genes can cause severe phenotypes, which indicates that they play significant roles in plant growth and development. However, the molecular mechanism within remains unclear. Here we show that the short hypocotyl phenotypes of er erl1 erl2 mutants are mainly due to the defects of cell elongation rather than the cell division. In contrast, in the ERECTA overexpression transgenic plants, the hypocotyl length is increased with elongated cells. Moreover, we show that the er erl1 erl2 triple mutant contains a low level of auxin, and the expression levels of the key auxin biosynthesis genes are significantly reduced. Consistent with this observation, increasing exogenous or endogenous auxin levels could partially rescue the cell elongation defects of the er erl1 erl2 triple mutant. Therefore, our results provide a molecular basis for auxin mediated ERECTA control of the hypocotyl length in Arabidopsis thaliana.
Plant cell size is one of the most important features in plant morphology, which is controlled strictly by the inheritance and influenced by the external environment. The cell size directly correlated with cell division and cell elongation. Schleiden and Schwann (1838-1839) established the “cell theory,” which regard that the cell division and cell elongation activities determine the growth and development of living organism. Cell elongation is very important for the process of plant differentiation and morphogenesis of plant organs. The growth of hypocotyls in Arabidopsis is the elongation and division of hypocotyl cells in essence. In particular, the cell elongation growth plays a decisive role in hypocotyl length (Stuart et al., 1977; Fenwick et al., 1999). The hypocotyl is an important structure connecting root, shoot tip and leaves, and also an important channel for transporting water, nutrients and signaling molecules in Arabidopsis. At the same time, hypocotyls are very sensitive to endogenous signal molecules and external environment signal (Gray et al., 1998; Nozue et al., 2007; de Lucas et al., 2008; Christie et al., 2011; Sun et al., 2012). The morphological structure of Arabidopsis thaliana hypocotyl is simple, consisting of more than 20 cells in longitudinal direction, which is below cotyledons to the above of radicle. Because most of these cells are formed in the embryonic stage, only a few are produced by cell division after germination, suggests that the hypocotyl growth is mainly caused by the cell elongation (Gendreau et al., 1997). Therefore, the hypocotyl of Arabidopsis has become an important model system for studying plant cell elongation due to its simple structure and important physiological functions.
Plant hormones are important regulatory signal molecules of hypocotyl elongation, and auxin is one of the most important types. For a single cell, the main physiological function of auxin is to regulate cell elongation, and the effect of auxin on acceleration of cell elongation is very rapid, and the lag time from treatment to effectiveness is only about 10 min. Previous studies have found that mutants with excessive synthesis of auxin in light exhibit a longer hypocotyl length, while mutants with deficient synthesis of auxin have shorter hypocotyl length (Romano et al., 1991; Jensen et al., 1998; Zhao et al., 2001; Cheng et al., 2006). This indicates that an appropriate concentration of auxin is essential for the growth and development of hypocotyls. It has now been shown that the effect of auxin on hypocotyl length is achieved by stimulating the degradation of IAA3, which in turn causes the ARF transcription factor to be released (Vernoux et al., 2011; Oh et al., 2014). ARF transcription factors, especially ARF6 and ARF8, combine the signals from endogenous development and exogenous environments to control the elongation of hypocotyl cells by activating the expression of cell elongation factors such as PRE, SAUR family genes (Bai et al., 2012; Oh et al., 2014; Challa et al., 2016).
The intercellular communication is very important for the formation of normal tissues, organs and organisms of plants. The cell surface receptor protein kinase located on the cytoplasmic membrane plays a vital role in the initiation of cell signal transmission. The ERECTA (ER) gene in Arabidopsis encodes a leucine-rich repeat receptor-like protein kinase, whose protein structure consists of a hydrophobic signal peptide in the extracellular domain, a leucine repeat sequence for protein interaction, a single transmembrane domain, and intracellular serine/threonine kinase domain (Torii et al., 1996). ERECTA is widely expressed in plant tissues and plays an important role in plant development and responses to external environmental signals (Yokoyama et al., 1998; Shpak, 2013; Jorda et al., 2016; Gao et al., 2017). ERECTA knockout mutants show obvious dysplasia phenotypes, for instance, stem node, siliques and petiole become short and inflorescences are densely clustered. Although Landsberg deficit in ERECTA gene is widely used as an Arabidopsis ecotype, there is no in-depth study on how ERECTA gene mutations have led to such a markedly intense phenotype. In addition, ERECTA, as a multi-functional major gene in the plant, has two homologous genes ERECTA LIKE 1 (ERL1) and ERECTA LIKE 2 (ERL2), both of which have partial functional redundancy with ERECTA. The triple mutant er erl1 erl2 intensifies ERECTA knockout mutant phenotype, and the plant is extremely dwarfed (Shpak et al., 2004). Previous studies have been shown that ERECTA, ERL1 and ERL2 play roles in the organ morphology and size determination in the reproductive stage of anthers and eggs by altering cell cycle progression (Shpak et al., 2004; Pillitteri et al., 2007). The mechanism of ERECTA gene family in cell-to-cell communication has been intensively studied in the stomatal development. ERECTA can recognize small peptide of the EPF/EPFL family, by forming a complex with TMM, affects stomatal development and differentiation by triggering MAPK cascade (Meng et al., 2012; Jewaria et al., 2013; Lin et al., 2017). However, the role of ERECTA family in the regulation hypocotyls remains unclear. Previous studies have shown that overexpression of YUCCA5, a member of YUCCA family of flavin monooxygenase in Arabidopsis auxin synthesis, inhibits mutant phenotype of er-103 (one of the ERECTA mutants) (Woodward et al., 2005). In addition, ERECTA family regulates the transport of auxin by regulating the expression of PIN1 at the initial stage of leaf primordia (Chen et al., 2013). However, the genetic interaction between the ERECTA family and auxin signaling in control of the hypocotyl development is still poorly understood.
In this study we found that, the hypocotyl lengths of single mutant er-105 and triple mutant er erl1 erl2 were significantly shorter than that of the wild type, whereas hypocotyls of ER-overexpressed plants was longer. We further showed that this change is due to a change in the length of a single cell in the hypocotyl rather than a change in the number of cells. In addition, we found that DR5::GFP signal of auxin reporter in the triple mutant er erl1 erl2 was significantly reduced than that of wild type. Moreover, the transcriptional levels of auxin early response gene and the major auxin synthetase genes were significantly repressed in the er erl1 erl2 triple mutant, suggesting that the auxin synthesis in the er erl1 erl2 triple mutant is sabotaged. Consistent with the observation, the exogenous auxin and endogenous auxin were able to rescue the shortened hypocotyl and cell length phenotype of the er erll erl2 triple mutant, indicating that these defects are due to the lack of auxin synthesis. Therefore we concluded that the ERECTA gene family controls the cell elongation in the hypocotyl by positively regulating the auxin biosynthesis in Arabidopsis thaliana.
All of the Arabidopsis lines used in this study were in the Col-0 background. The ER family mutants and overexpression plants have been described previously (Torii et al., 1996; Shpak et al., 2004; Shen et al., 2015). The seeds for er-105, er-/- erl1+/- erl2-/- mutant were kindly provided by Prof. Masao Tasaka (Nara Institute of Science and Technology). The seeds for 35S::ERECTA were kindly provided by Prof. Zuhua He (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences). The seeds for DR5::GFP-NLS in wild type were kindly provided by Prof. Yuling Jiao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences). Other Arabidopsis seeds were obtained from self-preserved, genetic hybridization or agrobacterium-mediated transformation. All seeds were sterilized with 70% ethanol and 0.5% Tween for 10 min, followed by washing two times with 95% ethanol and air drying. After sterilization, seeds were sown onto 1/2 Murashige and Skoog (MS) medium containing 1% sucrose and 0.8% agar, then incubated at 4°C in darkness for 2 days, finally grown vertically at 21°C under long-day condition (16 h of light and 8 h of darkness).
For construction of pER::3 × VENUS-NLS, a 1.4-kb upstream sequence of ERECTA before ATG was used as a promoter. For the pERL1::3 × VENUS-NLS, a 4.1-kb upstream sequence of ERL1 before ATG was used as a promoter. For the pERL2::3 × VENUS-NLS, a 3.6-kb upstream sequence of ERL2 before ATG was used as a promoter. For construction of pER::ER-GFP, pERL1::ERL1-GFP and pERL2::ERL2-GFP, the genomic sequence of these three genes were amplified and used the same promoter we described above. The pER::ER-GFP construct was then transformed into the er-105 mutant to rescue its defects, while other constructs were transformed into the Col-0. For pER::iaaM, the 1.4-kb promoter of ERECTA was used to drive iaaM sequences that were obtained from Prof. Yunde Zhao (UC San Diego). This construct was then transformed into the er-/- erl1+/- erl2-/- mutant. The primer sequences used in the plasmid construction are listed in Supplementary Table S1.
Whole seedlings were dissected and immediately transferred to liquid nitrogen. The Tripure Isolation Reagent (Roche) was used to isolate total RNA from the plant samples. The PrimeScriptTM RT Reagent Kit (TaKaRa) was used for cDNA synthesis. Quantitative PCR was performed with the Thermo PIKO REAL96 Real-Time PCR system using the GoTaq® qPCR Master Mix (Promega) with the following PCR conditions: 95°C for 5 min and 40 cycles of 95°C for 10 s, 57°C for 30 s and 72°C for 30 s, followed by 72°C for 10 min and 20°C for 10 s. TUBULIN was used to normalize the mRNA levels. Primers used for qRT-PCR are listed in Supplementary Table S1.
Propidium Iodide (PI) powder (Sigma) was dissolved with PBS solution to 5 mg/ml as a stock solution, which was stored in dark. The working solution was diluted to 5 μg/ml. The whole seedling was put into the solution for staining 10 min, then washed with water three times and then observed under the confocal microscope (OLYMPUS LSM1200).
For the length of hypocotyls in the seedling stages, pictures were taken under the microscope; The Image J software was used for the measurement and statistical analysis. The cells in longitudinal direction from the top to the base of the hypocotyls epidermis were used for the measurement of each cell length, and 25 hypocotyls were used for biological repetitions, and the Image J software was used for the statistical analysis. The total cells number in the hypocotyls was counted directly under the microscope.
IAA and yucasin powder (Sigma) were dissolved in the DMSO and diluted to 50 μM for IAA and 5 mM for yucasin as the stock solutions. The stock solutions were diluted 1000x and added to the corresponding 1/2MS media. The control group was added with the same amount of DMSO in the media. The plants were grown vertically at 21°C under long-day condition (16 h of light and 8 h of darkness).
Where appropriate, statistical analyses were performed with analysis of variance (ANOVA) test. Otherwise, comparisons between two groups were conducted using Student’s t-test. The p-value level was set to 5%.
One of the most striking defects in the ERECTA family mutants (erf) mutants is the dwarf phenotype (Shpak et al., 2004). The er-105 single mutant showed reduced plant height comparing to that of wild type, whereas the plant height was increased in the ERECTA overexpressing plants (Supplementary Figure S1A) (Shen et al., 2015). In the er erl1 erl2 triple mutant, we observed even severe phenotypes in the reduction of the plant height (Supplementary Figure S1A), suggesting that ERECTA, ERL1 and ERL2 were functionally redundant in control of the plant height. To further support these observations, we measured the plant height of the 35S::ERECTA, er-105 and er erl1 erl2 plants. We observed a slight increase of the plant height in the 35S::ERECTA plants, however, a significant decrease in the er-105 and er erl1 erl2 mutants (Supplementary Figure S1B).
Consistent with the observation of plant height, in the 6-day-old seedling, we observed that the length of hypocotyl in the 35S::ERECTA plants was significantly increased comparing to that of wild type seedlings (Figures 1A,B). Conversely, the hypocotyl lengths in the er-105 and er er1 erl2 mutants were dramatically reduced (Figures 1A,B). We conclude that the ERECTA gene family are involved in the regulation of hypocotyl length in Arabidopsis.
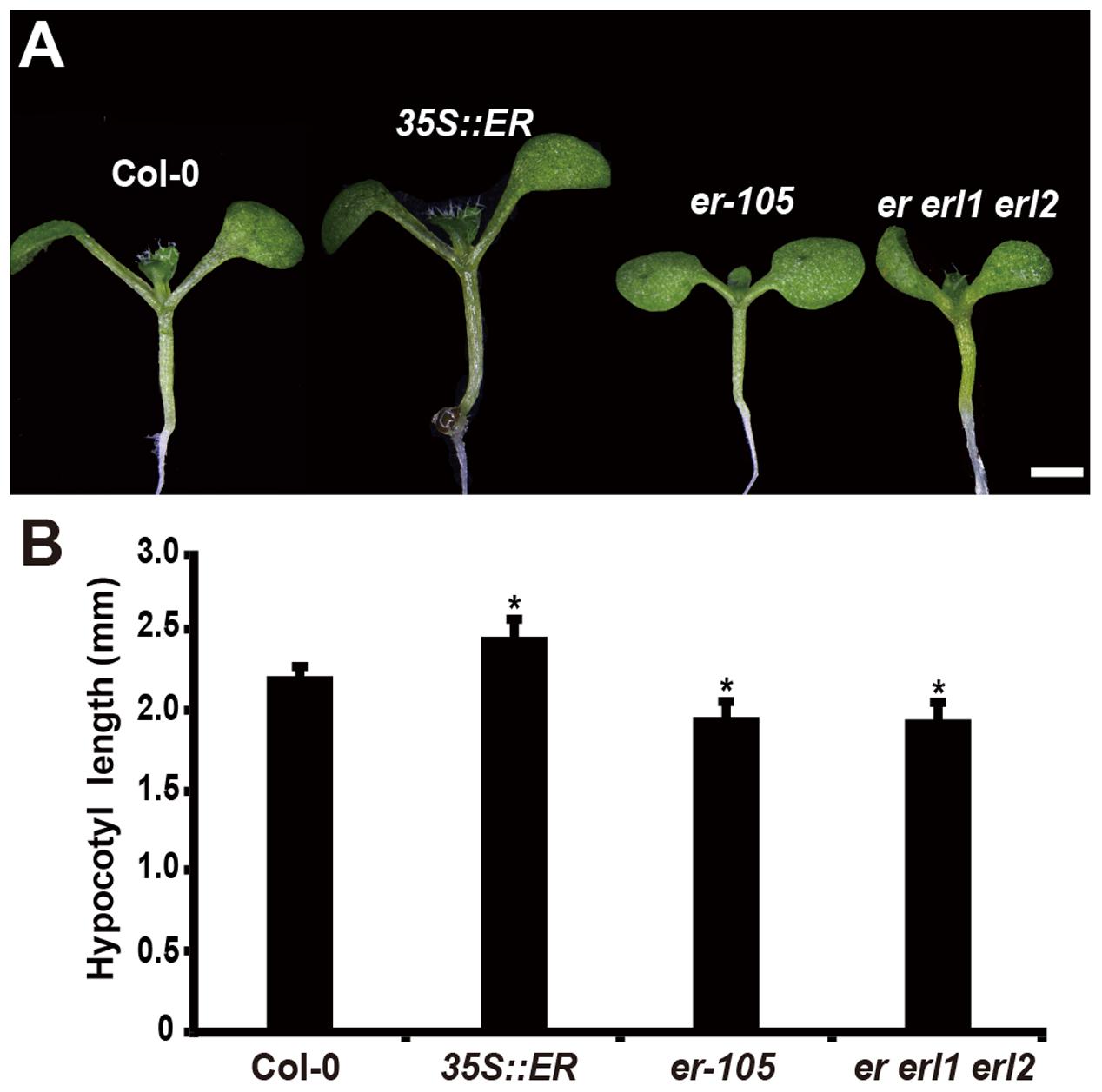
FIGURE 1. Hypocotyl length is shorten in the erf mutants. (A) Six-day-old seedlings of the wild-type plant, 35S::ERECTA transgenic plant, er-105 mutant and er erl1 erl2 mutant. (B) Average hypocotyl lengths of the 6-day-old seedlings in (A) (n = 40). Scale bar, 1 mm. ∗P < 0.05, Student’s t-test.
Given the fact that disturbing the functions of ERECTA gene family causes the severe defects in the hypocotyl length, we assumed that ERECTA gene family are expressed in the hypocotyl cells. To test whether the ERECTA family genes are transcribed in the hypocotyl cells, we first performed the promoter activation analysis. To this end, we constructed pER::3 × VENUS-NLS, pERL1::3 × VENUS-NLS, pERL2::3 × VENUS-NLS transgenic plants. We observed evenly distributed fluorescence signals in the whole hypocotyl of these three transgenic plants (Figures 2A–C), suggesting that ERECTA, ERL1 and ERL2 genes are expressed in the whole hypocotyl of Arabidopsis.
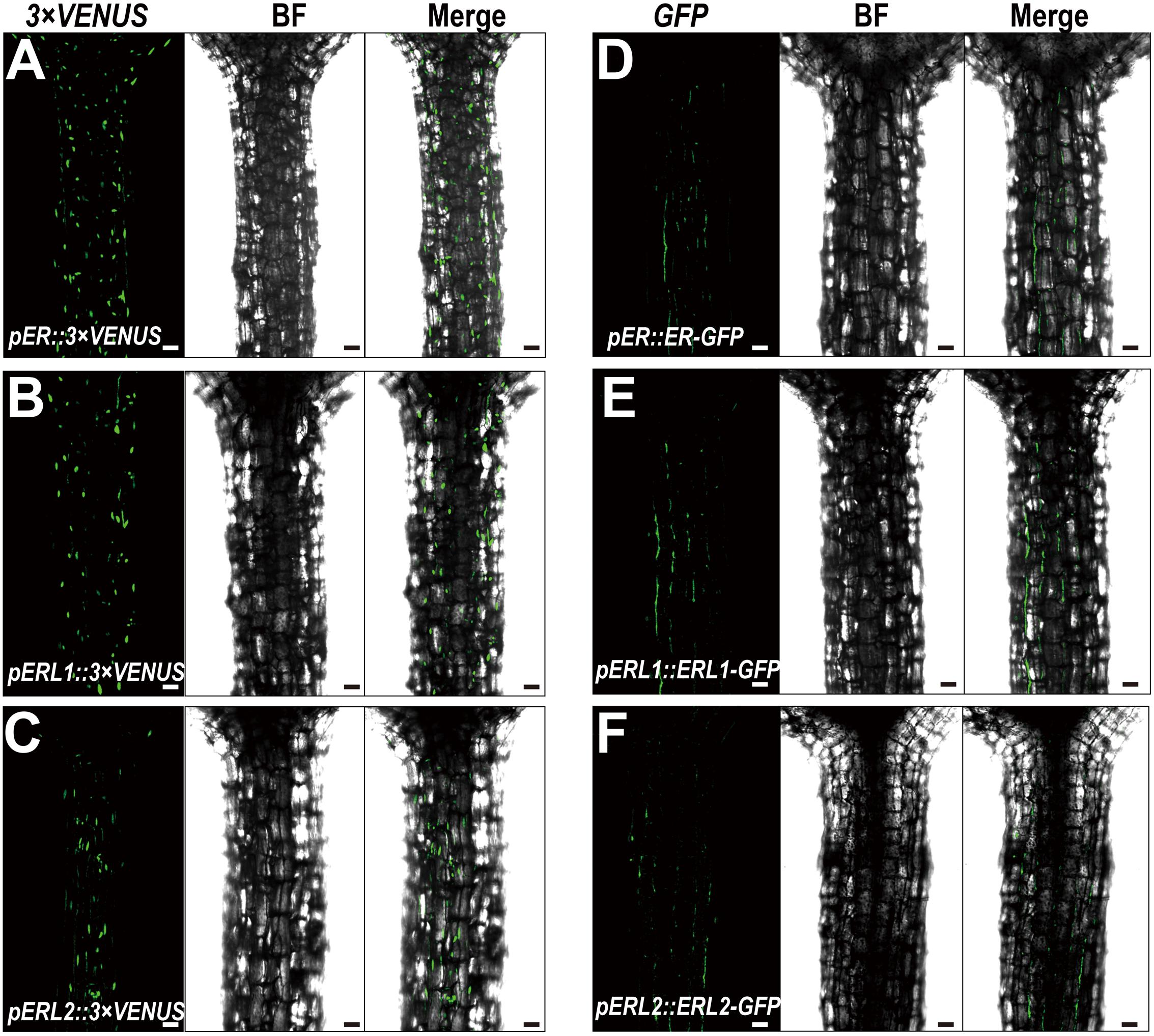
FIGURE 2. Expression patterns of the ERECTA family genes in hypocotyls. (A) The expression of the ERECTA promoter in hypocotyls determined by the fluorescent signals of pER::3 × VENNS-NLS. (B) The expression of the ERL1 promoter in hypocotyls determined by the fluorescent signals of pERL1::3 × VENNS-NLS. (C) The expression of the ERL2 promoter in hypocotyls determined by the fluorescent signals of pERL2::3 × VENNS-NLS. (D) The localization of ERECTA proteins in hypocotyls. (E) The localization of ERL1 proteins in hypocotyls. (F) The localization of ERL2 proteins in hypocotyls. Scale bars, 50 μm.
Because ERECTA family genes encode leucine-rich repeat receptor-like kinases, we then test whether the ERECTA, ERL1 and ERL2 proteins are localized in the hypocotyl cells. We transformed the pER::ER-GFP construct into the er-105 mutant and observed a fully rescue of the mutant defects. By the confocal microscopy, we observed that ERECTA proteins were strongly accumulated in the hypocotyls (Figure 2D).
To check the protein localization of two homologous genes of the ERECTA, the pERL1::ERL1-GFP, pERL2::ERL2-GFP transgenic plants were also analyzed under the confocal microscope. Consistent with the observation of ERECTA proteins, we found that the both of proteins, ERL1 and ERL2, were also localized in hypocotyls (Figures 2E,F).
The organ size and shape of multicellular organism is directly related to the cells division and the size of cells. Therefore, the individual cell length and the total number of cells could determine the hypocotyl length in Arabidopsis. Previous studies have shown that ERECTA family genes may regulate cell number in some organs, including stomata, ovule and stamen (Shpak et al., 2004; Woodward et al., 2005; Pillitteri et al., 2007; Hord et al., 2008). However, whether the ERECTA family genes control the cells division and cell elongation in the hypocotyl is still unclear. To test this, we performed Propidium Iodide (PI) staining on the hypocotyls of the wild-type plants, 35S::ERECTA plants, er-105 single mutant plants, and er erl1 erl2 triple mutant plants (Figures 3A–D). By the observation under the confocal microscope, we found that the individual cell size of hypocotyl in the 35S::ERECTA overexpression plants was large than that of wild type plants (Figure 3B). On the contrary, we observed size largely reduced cells in the hypocotyls of the er-105 single mutant and er erl1 erl2 triple mutant plants comparing to that of wild type plants (Figures 3C,D). By the quantitative analysis of the average cells length in the middle column of the hypocotyl epidermis, we found that the average cells length in the 35S::ERECTA overexpression plants was significantly increased, whereas it was remarkably decreased in the er-105 and er erl1 erl2 mutants (Figure 3E).
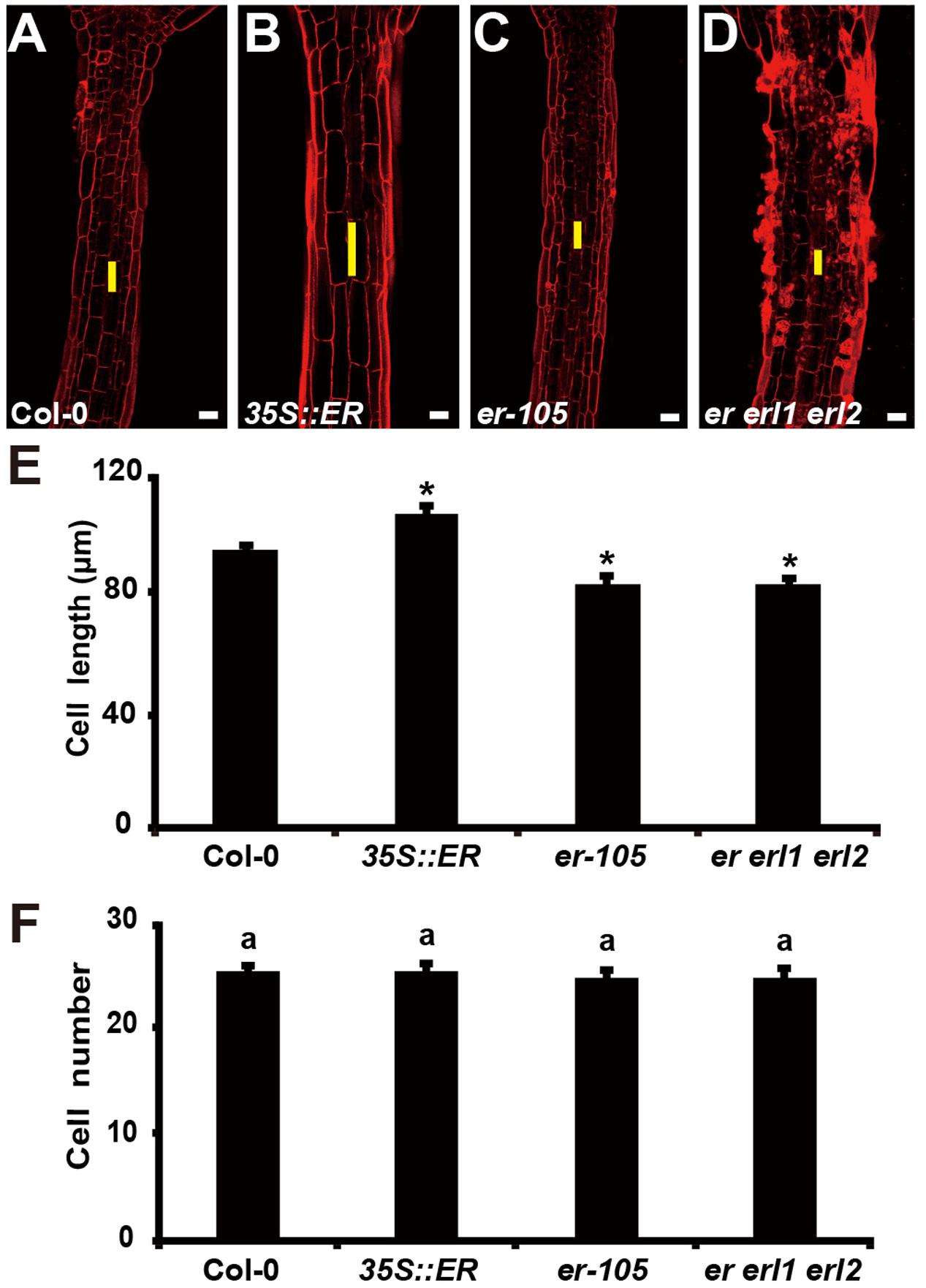
FIGURE 3. The ERECTA family genes controls cell elongation in the hypocotyl. (A–D) PI staining of hypocotyls in the 6-day-old wild-type (A), 35S::ERECTA plant (B), er-105 mutant (C) and er erl1 erl2 mutant (D). (E) Average individual cell lengths of the hypocotyls in the 6-day-old wild-type, 35S::ERECTA plant, er-105 mutant and er erl1 erl2 mutant (n = 25). (F) Total cell number in the hypocotyls of 6-day-old wild-type, 35S::ERECTA plant, er-105 mutant and er erl1 erl2 mutant (n = 25). The yellow line represents the length of a single cell. Scale bars, 50 μm. ANOVA-test in (F), same letters represent no statistically significant differences (P < 0.05). Student’s t-test in (E), ∗P < 0.05.
To test whether cells division is involved in the ERECTA family genes controlled hypocotyl elongation, we directly counted the total number of cells in the middle column of epidermis, and found that there were no significant difference in all the genotypes we test including the wild type, 35S::ERECTA, er-105 and er erl1 erl2 plants (Figure 3F). We concluded that the ERECTA gene family regulates the hypocotyl length by controlling the cell elongation rather than the cell division.
Given the fact that the cell elongation was significantly reduced in the erf mutants, we then asked by what means does the ERECTA gene family regulate individual cell length in the hypocotyl? Previous studies have shown that the auxin plays an important role in regulation of cell elongation (Rayle and Cleland, 1992; Galinha et al., 2007). The endogenous auxin increased mutants exhibit hypocotyls-elongated phenotypes, whereas auxin deficient mutants have the reduced size of cells (Romano et al., 1991; Zhao et al., 2001; Cheng et al., 2006). To examine whether the auxin is involved in the ERECTA gene family mediated controls of cell elongation, two different auxin sensors, DR5::GFP and DII-VENUS, were applied to detect the endogenous auxin levels in the er erl1 erl2 mutants. The DR5::GFP is a positive auxin sensor by which GFP transcriptions are activated by auxin (Ulmasov et al., 1997; Benkova et al., 2003). Under the confocal microscope, we observed evenly distributed fluorescence signals of DR5::GFP in the entire hypocotyl (Figure 4A), suggesting that the auxin was distributed in all the hypocotyl of Arabidopsis. However, we can barely detect the fluorescence signals in the er erl1 erl2 mutant hypocotyl (Figure 4B), suggesting that the endogenous auxin levels were dramatically reduced in the triple mutant hypocotyl. To confirm this observation, the transgenic plants that harbored the negative auxin sensor DII-VENUS by which fluorescence proteins were degradated by auxin (Brunoud et al., 2012) were further analyzed. Consistent with the observation of DR5::GFP, there were only extremely weak signals can be detected in the wild type hypocotyl (Figure 4C), but stronger DII-VENUS signals in the er erl1 erl2 triple mutants (Figure 4D). At the molecular level, we tested the transcriptional levels of auxin early response gene IAA19, which is commonly used to monitor the endogenous levels of auxin in Arabidopsis. We observed that the expression levels of IAA19 in the 35S::ERECTA transgenic plants were up regulated, whereas significantly down regulated in the er-105 and er erl1 erl2 triple mutants (Figure 4E). These data suggest that the ERECTA gene family is essential to maintain the endogenous auxin levels in the hypocotyl.
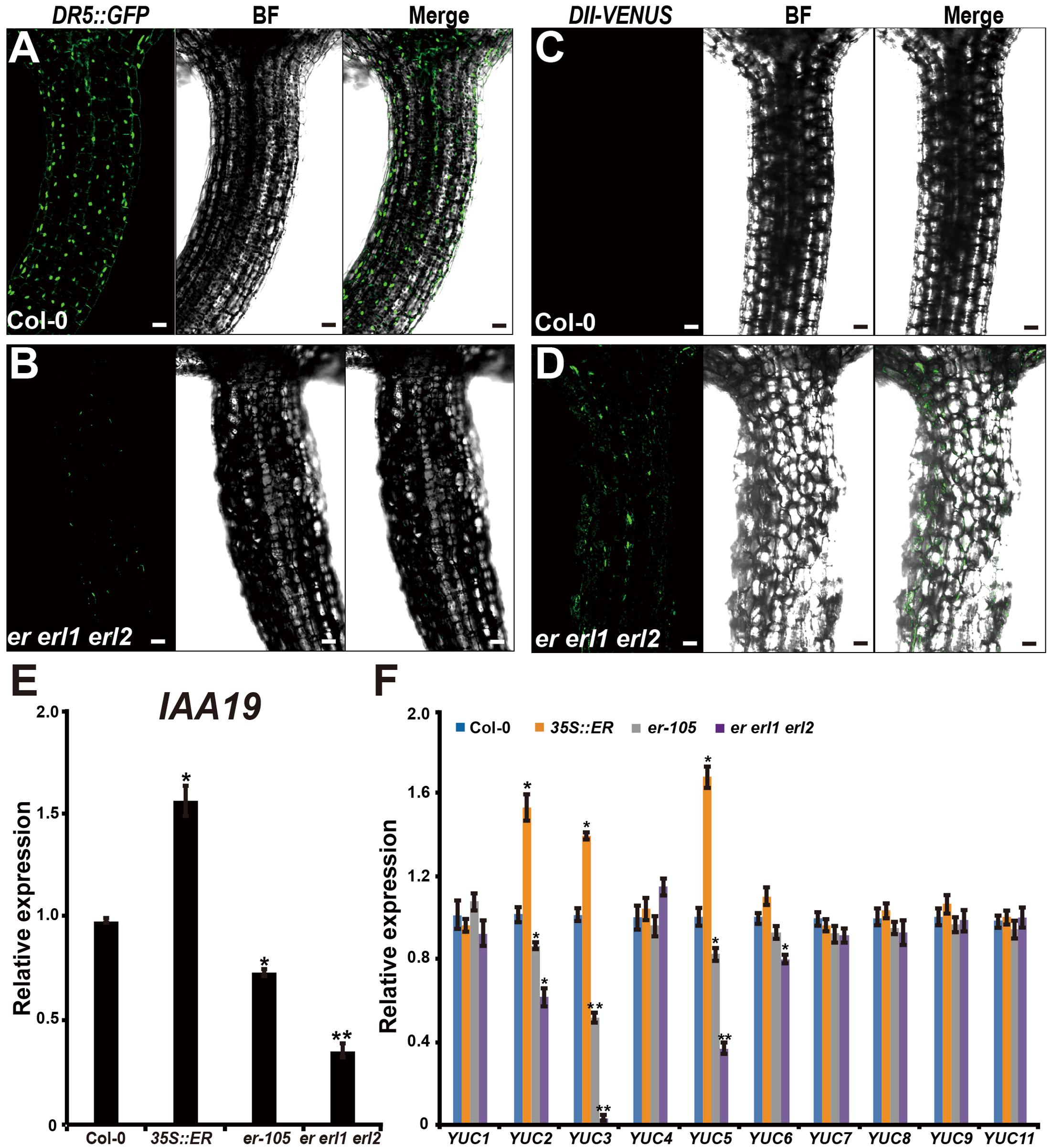
FIGURE 4. The ERECTA family genes controls auxin biosynthesis in the hypocotyl. (A) The fluorescent signals of DR5::GFP in the wild-type hypocotyl. (B) The fluorescent signals of DR5::GFP in the er erl1 erl2 mutant hypocotyl. (C) The fluorescent signals of DII-VENUS in the wild-type hypocotyl. (D) The fluorescent signals of DII-VENUS in the er erl1 erl2 mutant hypocotyl. (E) The expression levels of IAA19 in the wild-type, 35S::ERECTA, er-105 and er erl1 erl2 hypocotyls. The 6-day-old hypocotyls of each genotype were used for the RNA extraction. (F) The expression levels of YUC family genes in the wild-type, 35S::ERECTA, er-105 and er erl1 erl2 hypocotyls. The 6-day-old hypocotyls of each genotype were used for the RNA extraction. Scale bars, 50 μm. Error bars in the (E,F) indicate the SD of three biological repeats. ∗P < 0.05, ∗∗P < 0.01, Student’s t-test.
To shed light on the molecular mechanism underlying the ability of the ERECTA gene family controlling auxin levels in the hypocotyl, we examined the expression levels of the YUCCA family genes, which encodes the major auxin biosynthesis genes (Cheng et al., 2006). We observed that the transcriptional levels of YUC2, YUC3 and YUC5 were significantly increased in the ERECTA overexpression plants compared with wild type (Figure 4F). Conversely, these three key auxin biosynthesis genes were markedly decreased in the er-105 and er erl1 erl2 triple mutants; especially the expression levels of YUC3 were drastically reduced in the er erl1 erl2 triple mutants (Figure 4F). We draw the conclusion that the ERECTA gene family positively regulates auxin biosynthesis via activating the expressions of YUC2, YUC3 and YUC5 genes.
Given the fact that the erf mutants contain a very low amount of auxin and the key auxin biosynthesis genes are under the positive control of ERECTA gene family, we hypothesized exogenous increase of auxin might rescue the defects of cell elongation and short hypocotyl phenotypes of the erf mutants. While, previous studies have shown that the high concentrations of auxin might inhibit the elongation of hypocotyls in wild-type Arabidopsis (Collett et al., 2000; Rashotte et al., 2003), which was also found in our experiments (data not shown). To exclude the possibility the high auxin inhibits the cell elongation in our test, we carefully select a very low concentration of IAA (50 nM), and performed our analysis. By applying this low concentration of IAA to the wild type seedlings, we did not observe any significant differences in term of the hypocotyl length comparing with the mock treatments (Figures 5A,B). However, this low concentration of IAA almost fully rescued the hypocotyl-shortened phenotypes of the er-105 single mutants and er erl1 erl2 triple mutants (Figure 5A). By quantifying the hypocotyl length, we observed significant increases of the hypocotyl length in the er-105 and er erl1 erl2 mutants upon the auxin treatments, which showed no differences to that of wild type seedlings (Figure 5B), suggesting that the hypocotyl-shortened defects in the erf mutants are due to the lacking of auxin, and the increase of exogenous auxin completely restores the mutant phenotypes. To further confirm this observation, we examine the cell elongation in the erf mutants after the auxin treatments. In the wild type hypocotyl, the 50 nM auxin treatments did not change the cell length comparing with the mock treatments (Figures 6A,B). However, in the er-105 and er erl1 erl2 mutants, the same amount of auxin effectively promoted the cell length to that of mock treatments (Figure 6A), and the cell length was significantly increased to the levels that comparable with the wild type seedlings (Figure 6B). To test the possibility whether the cell division was also involved in the rescue of erf mutants defects during the low auxin treatments, we directly counted the total number of cells in the middle column of epidermis with or without the low auxin treatments. We observed no significant difference among all the genotypes after the auxin treatments (Supplementary Figure S2). Therefore, we conclude that the hypocotyl-shortened phenotypes in the erf mutants are due to the cell elongation defects, which can be fully rescued by the treatments of exogenous auxin.
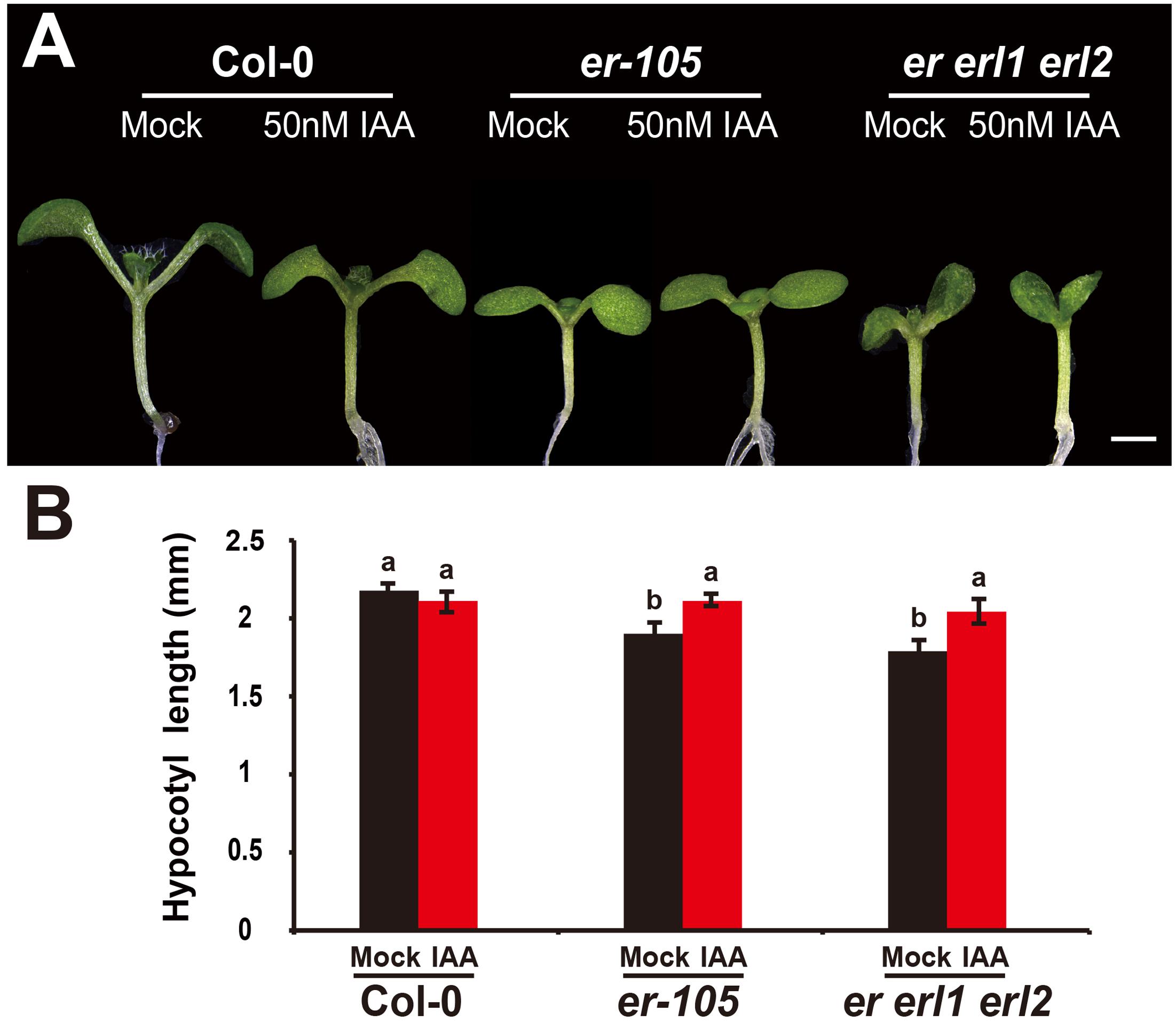
FIGURE 5. The low auxin rescues the erf hypocotyl-shortened phenotypes. (A) Six-day-old seedlings of the wild type, er-105 and er erl1 erl2 mutant grown in the 1/2MS media with or without 50 nM IAA. (B) The hypocotyl lengths of the seedlings in (A) (n = 40). Scale bar, 1 mm. Different letters represent statistically significant differences (P < 0.05), ANOVA-test.
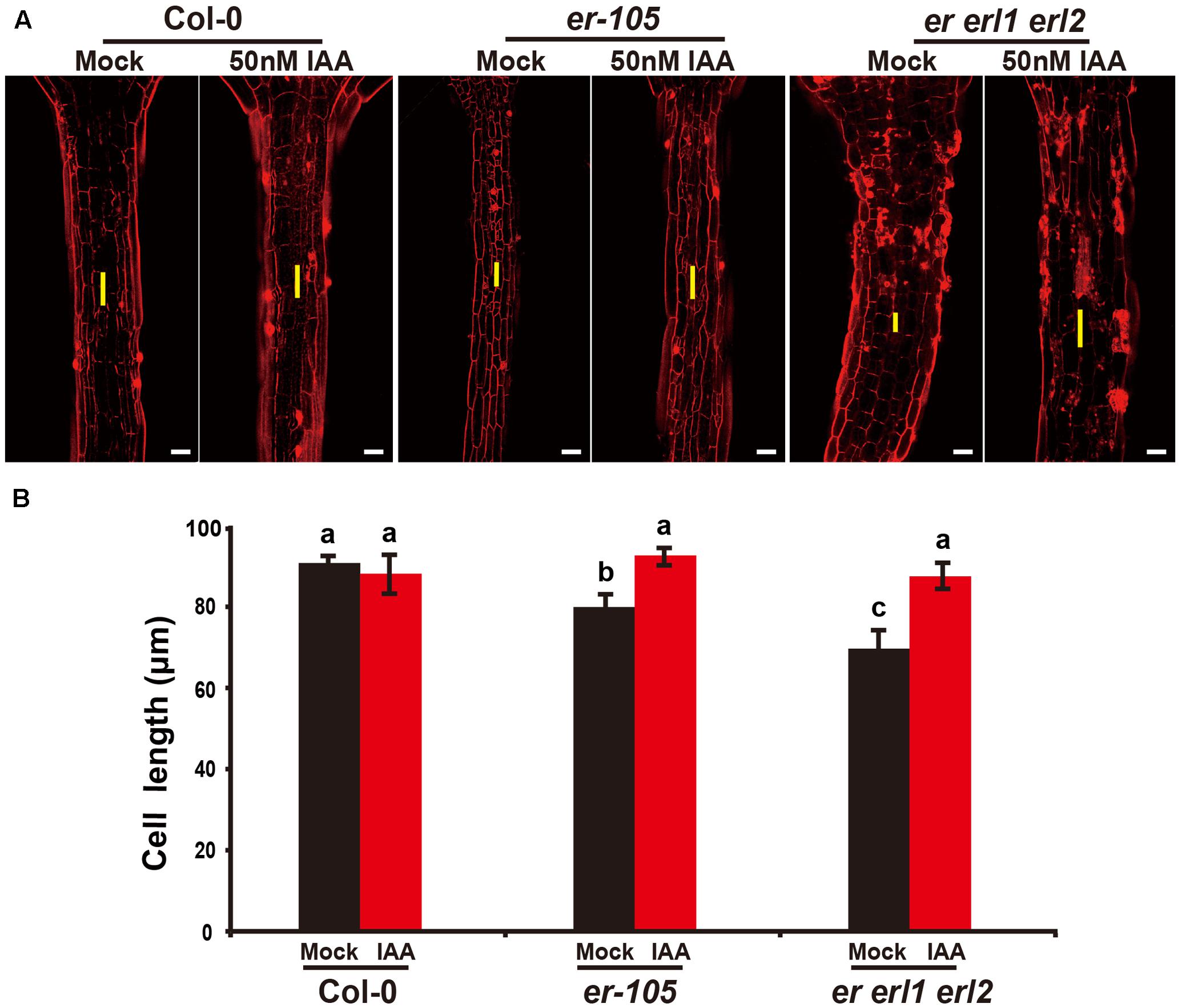
FIGURE 6. Exogenous auxin rescues the cell elongation defects in the erf mutants. (A) PI staining of hypocotyls in the wild type, er-105 and er erl1 erl2 mutant grown in the 1/2MS media with or without 50 nM IAA. (B) The average cell lengths of the hypocotyls in the wild type, er-105 and er erl1 erl2 mutant grown in the 1/2MS media with or without 50 nM IAA. (n = 25). The yellow line represents the length of a single cell. Scale bars, 50 μm. Different letters represent statistically significant differences (P < 0.05), ANOVA-test.
Given the fact that the ERECTA gene family positively regulates auxin biosynthesis in the hypocotyl by activating the expressions of several YUCCA family genes, and the overexpression of ERECTA causes the elevated expressions of YUCs (Figure 4F), we then tested this interaction genetically. We treated the 35S::ERECTA seedlings with the chemical yucasin, which have been shown to reduce the exogenous auxin levels by inhibiting the expression of YUCCA genes (Nishimura et al., 2014). We carefully select a low concentration of yucasin (5 μM) to perform the analysis, which showed no effect on the hypocotyl length in the wild type seedlings (Supplementary Figures S3A,B). While in the 35S::ERECTA seedlings, the hypocotyl length was significantly decreased (Supplementary Figures S3A,B). Likely, the hypocotyl cell length of the wild type plants did not respond the low yucasin treatments. However, the cell length of the 35S::ERECTA seedlings with increased YUCCA expressions, reduced dramatically upon the low yucasin treatments (Supplementary Figure S4), demonstrating that the ERECTA gene family controls the cell elongation in hypocotyls by positively regulating the auxin biosynthesis.
The bacterial auxin biosynthesis gene iaaM, which was firstly identified in the T-DNA of agrobacterium, is commonly used in plants to increase the endogenous auxin levels (Comai and Kosuge, 1982). To further investigate the mechanism by which the ERECTA gene family mediated auxin synthesis controls the cell elongation in hypocotyls, we expressed the iaaM gene under the control of the ERECTA promoter, and transformed the pER:iaaM construct into the er-/- erl1+/- erl2-/- mutants. By analyzing the expression levels of the iaaM and the IAA19 genes in eight independent transgenic lines, we observed a correlation between these two genes, suggesting that the ectopically expressed iaaM increased the endogenous auxin levels in the er erl1 erl2 triple mutant background (Supplementary Figure S5A). We measured the hypocotyl length of eight independent transgenic lines and selected three independent T1 transgenic lines with high expression of iaaM for the further analysis. (Supplementary Figure S5B), and observed that hypocotyls and individual cell lengths of pER:iaaM transgenic plants were higher than those of er erl1 erl2 triple mutants (Figures 7A–D). Thus we conclude that the ERECTA gene family mediated endogenous auxin biosynthesis controls the cell elongation in hypocotyls.
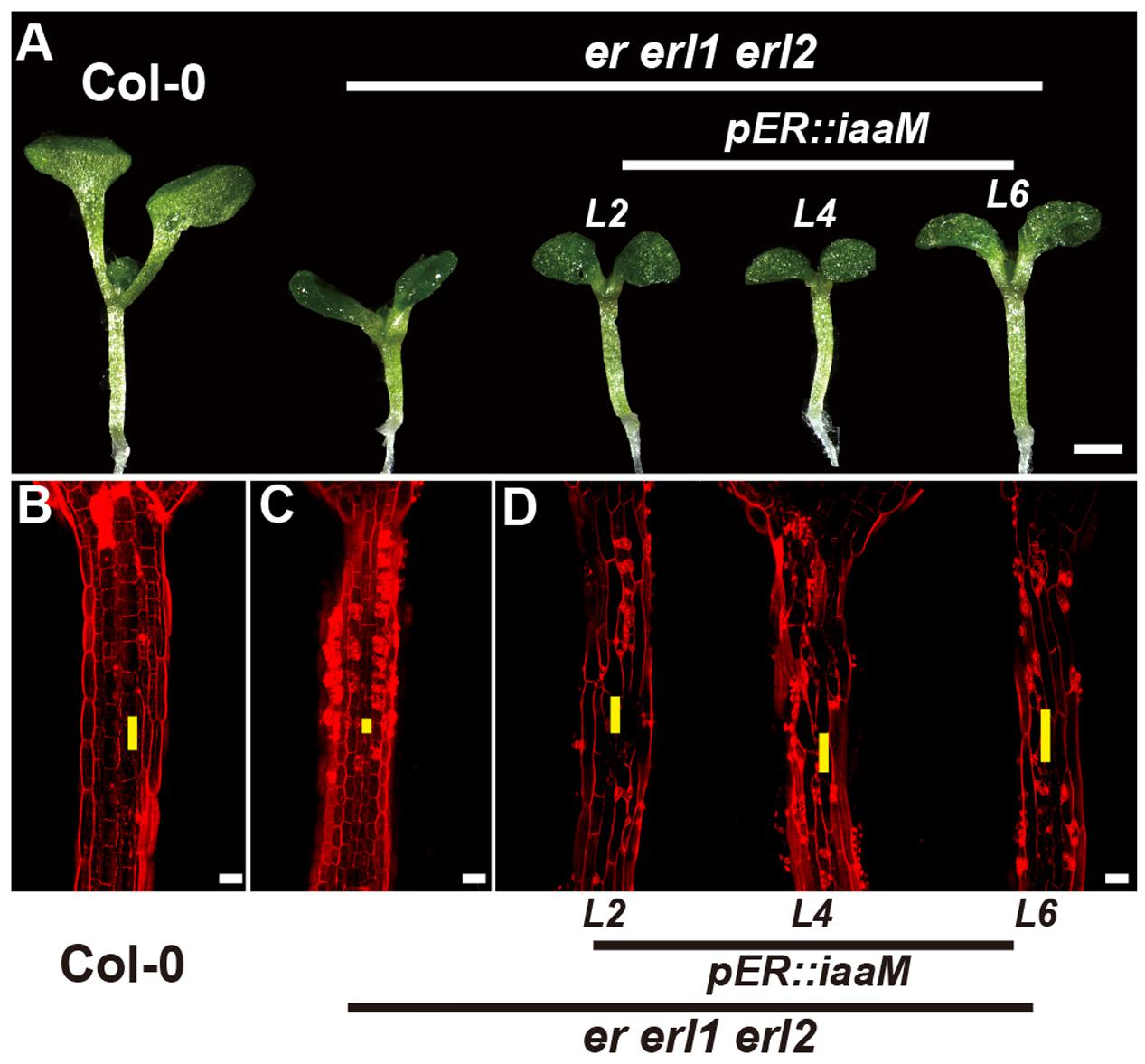
FIGURE 7. The endogenous auxin rescues the erf mutant phenotypes in the hypocotyl. (A) Six-day-old seedlings of the wild type, the er erl1 erl2 mutant and three independent pER:iaaM transgenic lines in the er erl1 erl2 mutant background (L2, L4 and L6). (B–D) PI staining of hypocotyls in the wild type (B), the er erl1 erl2 mutant (C) and three independent pER:iaaM transgenic lines in the er erl1 erl2 mutant background (D). The yellow line represents the length of a single cell. Scale bar in (A), 1 mm; scale bars in (B–D), 50 μm.
The ERECTA family genes, which encode leucine-rich repeat receptor-like kinases, have been shown to regulate multiple developmental processes including stomatal formation, inflorescence architecture, and ovule development (Shpak et al., 2004, 2005; Lee et al., 2012). Woodward et al have previously shown that a dominant mutant, super1-D, which causes the overexpression of one of the YUCCA family genes YUC5, is epistatic to er mutants (Woodward et al., 2005). However, little is known about the genetic interaction between the ERECTA and auxin signaling in control of the hypocotyl development. In this study, we analyzed the endogenous auxin levels in the erf mutants by using two different auxin sensors, DR5::GFP and DII-VENUS. Our results show that the endogenous auxin levels were dramatically reduced in the hypocotyl of the er erl1 erl2 triple mutant (Figures 2A–D). To further support this observation, we examined the expression levels of IAA19 in the hypocotyl. We found that the IAA19 transcripts are significantly reduced in the er-105 and er erl1 erl2 mutants, and increased in the 35S::ERECTA transgenic plants (Figure 4E). To further investigate the mechanism by which the ERECTA family genes regulate auxin levels in the hypocotyl, we examined the key auxin biosynthesis genes. We observed that the transcriptional levels of YUC2, YUC3 and YUC5 were significantly decreased in the er-105 and er erl1 erl2 triple mutants, whereas increased in the 35S::ERECTA transgenic plants (Figure 4F). Our data suggest that the ERECTA gene family positively regulates auxin biosynthesis by activating the key auxin biosynthesis genes.
Previous study has shown that the overexpression of YUC5 in the wild type, er-103 and er-105 cause the same extent of increase of hypocotyls regardless of their genotypes (Woodward et al., 2005), which raises the question whether the ERECTA family genes regulate the cell elongation in the hypocotyl via auxin biosynthesis. By careful selection of a very low concentration, 50 nM of IAA fully rescued the short hypocotyl and cell elongation defects in the er-105 and er erl1 erl2 mutants, but have no significant effects in the wild type (Figures 5, 6), suggesting that these defects of the erf mutants are caused by the auxin deficiency in the hypocotyl. Further support for this idea came from the observation that the elongated hypocotyl cells in the 35S::ERECTA seedlings are completely suppressed by using the 5 μM yucasin treatments to inhibit YUCCA genes (Supplementary Figure S4). Our data demonstrate that the ERECTA gene family controls cell elongation by positively regulating auxin biosynthesis. Likely, during leaf margin morphogenesis, the ligand-receptor pair of EPFL2-ER has been shown to be essential for maintaining an appropriate auxin concentration (Tameshige et al., 2016).
The small-secreted peptide PSY1, together with its LRR receptor, positively regulates cell elongation in hypocotyls by triggering a signaling cascade (Mahmood et al., 2014), suggesting a crucial role of peptide ligand-receptor pair in the hypocotyl regulation. ERECTA with its ligands EPF/EPFL peptides family, participate multiple functions during plant development, for example, EPF1, EPF2 and EPFL9 in stomatal regulation, EPFL4, EPFL5 and EPFL6 in inflorescence development and EPFL2 in tooth development (Abrash et al., 2011; Uchida and Tasaka, 2013; Tameshige et al., 2016; Lin et al., 2017). Moreover, in the secondary growth of Arabidopsis hypocotyls, ERECTA and ERL1 are very important in the xylem and phloem to prevent premature during sequential events in the secondary growth, suggesting that ERECTA family plays an important role in the morphogenesis of hypocotyls (Ikematsu et al., 2017). Therefore, we speculate that ERECTA family might regulate the cell elongation in hypocotyls with some members of the EPF/EPFL family or other ligands.
During the reproductive stage, the short internodes and pediceels phenotypes of the erf mutants in the inflorescences were mainly due to reduced cell proliferation (Shpak et al., 2003, 2004; Woodward et al., 2005). However, in the hypocotyl, mutations of the ERECTA gene family result in the defects specifically in the cell elongation rather than the cell division (Figure 3). This idea is further support by the early observation that, in Arabidopsis, hypocotyl elongation is mainly due to the cell elongation, while the number of cells is rather stable (Gendreau et al., 1997). These data suggest that the functions of ERECTA gene family in controlling of cell proliferation and cell elongation is highly tissue-specific, however, it remains to be shown how the functions of ERECTA gene family are specified in different tissues to fulfill their biological functions.
ZT, XQ, and ZZ designed the experiments, analyzed the data and wrote the paper. XQ performed the experiments.
This work was supported by the grant to ZT from the National Natural Science Foundation of China (31300248), and the grant to ZZ from the Ministry of Science and Technology of China (2013CB967300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Masao Tasaka, Prof. Zuhua He, Prof. Yuling Jiao, and Prof. Yunde Zhao for sharing seeds or plasmid.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01688/full#supplementary-material
Abrash, E. B., Davies, K. A., and Bergmann, D. C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23, 2864–2879. doi: 10.1105/tpc.111.086637
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, U810–U878. doi: 10.1038/ncb2546
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602.
Brunoud, G., Wells, D. M., Oliva, M., Larrieu, A., Mirabet, V., Burrow, A. H., et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. doi: 10.1038/nature10791
Challa, K. R., Aggarwal, P., and Nath, U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28, 2117–2130. doi: 10.1105/tpc.16.00360
Chen, M. K., Wilson, R. L., Palme, K., Ditengou, F. A., and Shpak, E. D. (2013). ERECTA family genes regulate auxin transport in the shoot apical meristem and forming leaf primordia. Plant Physiol. 162, 1978–1991. doi: 10.1104/pp.113.218198
Cheng, Y. F., Dai, X. H., and Zhao, Y. D. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Christie, J. M., Yang, H. B., Richter, G. L., Sullivan, S., Thomson, C. E., Lin, J. S., et al. (2011). phot1 Inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLOS Biol. 9:1001076. doi: 10.1371/journal.pbio.1001076
Collett, C. E., Harberd, N. P., and Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124, 553–561. doi: 10.1104/pp.124.2.553
Comai, L., and Kosuge, T. (1982). Cloning and characterization of iaaM, a virulence determinant of Pseudomonas-savastanoi. J. Bacteriol. 149, 40–46.
de Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Fenwick, K. M., Apperley, D. C., Cosgrove, D. J., and Jarvis, M. C. (1999). Polymer mobility in cell walls of cucumber hypocotyls. Phytochemistry 51, 17–22.
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. doi: 10.1038/nature06206
Gao, Y., Wu, Y. J., Du, J. B., Zhan, Y. Y., Sun, D. D., Zhao, J. X., et al. (2017). Both light-induced SA accumulation and ETI mediators contribute to the cell death regulated by BAK1 and BKK1. Front. Plant Sci. 8:622. doi: 10.3389/fpls.2017.00622
Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. doi: 10.1104/pp.114.1.295
Gray, W. M., Ostin, A., Sandberg, G., Romano, C. P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7197–7202. doi: 10.1073/pnas.95.12.7197
Hord, C. L. H., Suna, Y. J., Pillitteri, L. J., Torii, K. U., Wang, H. C., Zhang, S. Q., et al. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1, 645–658. doi: 10.1093/mp/ssn029
Ikematsu, S., Tasaka, M., Torii, K. U., and Uchida, N. (2017). ERECTA-family receptor kinase genes redundantly prevent premature progression of secondary growth in the Arabidopsis hypocotyl. New Phytol. 213, 1697–1709. doi: 10.1111/nph.14335
Jensen, P. J., Hangarter, R. P., and Estelle, M. (1998). Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116, 455–462. doi: 10.1104/pp.116.2.455
Jewaria, P. K., Hara, T., Tanaka, H., Kondo, T., Betsuyaku, S., Sawa, S., et al. (2013). Differential effects of the peptides stomagen, EPF1 and EPF2 on activation of MAP Kinase MPK6 and the SPCH protein level. Plant Cell Physiol. 54, 1253–1262. doi: 10.1093/pcp/pct076
Jorda, L., Sopena-Torres, S., Escudero, V., Nunez-Corcuera, B., Delgado-Cerezo, M., Torii, K. U., et al. (2016). ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Front. Plant Sci. 7:897. doi: 10.3389/fpls.2016.00897
Lee, J. S., Kuroha, T., Hnilova, M., Khatayevich, D., Kanaoka, M. M., McAbee, J. M., et al. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126–136. doi: 10.1101/gad.179895.111
Lin, G. Z., Zhang, L., Han, Z. F., Yang, X. R., Liu, W. J., Li, E. T., et al. (2017). A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31, 927–938. doi: 10.1101/gad.297580.117
Mahmood, K., Kannangara, R., Jorgensen, K., and Fuglsang, A. T. (2014). Analysis of peptide PSY1 responding transcripts in the two Arabidopsis plant lines: wild type and psy1r receptor mutant. BMC Genomics 15:441. doi: 10.1186/1471-2164-15-441
Meng, X. Z., Wang, H. C., He, Y. X., Liu, Y. D., Walker, J. C., Torii, K. U., et al. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960. doi: 10.1105/tpc.112.104695
Nishimura, T., Hayashi, K., Suzuki, H., Gyohda, A., Takaoka, C., Sakaguchi, Y., et al. (2014). Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 77, 352–366. doi: 10.1111/tpj.12399
Nozue, K., Covington, M. F., Duek, P. D., Lorrain, S., Fankhauser, C., Harmer, S. L., et al. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361. doi: 10.1038/nature05946
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., and Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. doi: 10.7554/eLife.03031
Pillitteri, L. J., Bemis, S. M., Shpak, E. D., and Torii, K. U. (2007). Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134, 3099–3109. doi: 10.1242/dev.004788
Rashotte, A. M., Poupart, J., Waddell, C. S., and Muday, G. K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133, 761–772. doi: 10.1104/pp.103.022582
Rayle, D. L., and Cleland, R. E. (1992). The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99, 1271–1274. doi: 10.1104/pp.99.4.1271
Romano, C. P., Hein, M. B., and Klee, H. J. (1991). Inactivation of auxin in tobacco transformed with the indoleacetic-acid lysine synthetase gene of Pseudomonas-savastanoi. Genes Dev. 5, 438–446. doi: 10.1101/gad.5.3.438
Shen, H., Zhong, X. B., Zhao, F. F., Wang, Y. M., Yan, B. X., Li, Q., et al. (2015). Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. doi: 10.1038/nbt.3321
Shpak, E. D. (2013). Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 55, 1238–1250. doi: 10.1111/jipb.12108
Shpak, E. D., Berthiaume, C. T., Hill, E. J., and Torii, K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491–1501. doi: 10.1242/dev.01028
Shpak, E. D., Lakeman, M. B., and Torii, K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. doi: 10.1105/tpc.010413
Shpak, E. D., McAbee, J. M., Pillitteri, L. J., and Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293. doi: 10.1126/science.1109710
Stuart, D. A., Durnam, D. J., and Jones, R. L. (1977). Cell elongation and cell division in elongating lettuce hypocotyl sections. Planta 135, 249–255. doi: 10.1007/BF00384897
Sun, J. Q., Qi, L. L., Li, Y. N., Chu, J. F., and Li, C. Y. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLOS Genet. 8:e1002594. doi: 10.1371/journal.pgen.1002594
Tameshige, T., Okamoto, S., Lee, J. S., Aida, M., Tasaka, M., Torii, K. U., et al. (2016). A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr. Biol. 26, 2478–2485. doi: 10.1016/j.cub.2016.07.014
Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F., et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. doi: 10.1105/tpc.8.4.735
Uchida, N., and Tasaka, M. (2013). Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 64, 5335–5343. doi: 10.1093/jxb/ert196
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971.
Vernoux, T., Brunoud, G., Farcot, E., Morin, V., Van den Daele, H., Legrand, J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7:508. doi: 10.1038/msb.2011.39
Woodward, C., Bemis, S. M., Hill, E. J., Sawa, S., Koshiba, T., and Torii, K. U. (2005). Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 139, 192–203. doi: 10.1104/pp.105.063495
Yokoyama, R., Takahashi, T., Kato, A., Torii, K. U., and Komeda, Y. (1998). The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 15, 301–310. doi: 10.1046/j.1365-313X.1998.00203.x
Keywords: Arabidopsis, ERECTA, auxin, hypocotyl length, cell elongation
Citation: Qu X, Zhao Z and Tian Z (2017) ERECTA Regulates Cell Elongation by Activating Auxin Biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 8:1688. doi: 10.3389/fpls.2017.01688
Received: 27 July 2017; Accepted: 14 September 2017;
Published: 27 September 2017.
Edited by:
Kai He, Lanzhou University, ChinaReviewed by:
Anja Thoe Fuglsang, University of Copenhagen, DenmarkCopyright © 2017 Qu, Zhao and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Zhao, emh6aGFvQHVzdGMuZWR1LmNu Zhaoxia Tian, enh0aWFuQHVzdGMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.