
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 September 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01597
This article is part of the Research Topic Plant-Microbe-Insect Interaction: Source for Bio-fertilizers, Bio-medicines and Agent Research View all 43 articles
Herbivory defense systems in plants are largely regulated by jasmonate-(JA) and salicylate-(SA) signaling pathways. Such defense mechanisms may impact insect feeding dynamic, may also affect the transmission-acquisition relationship among virus, plants and vectoring insects. In the context of the tomato – whitefly – Tomato Yellow Leaf Curl Virus (TYLCV) biological model, we tested the impact of pre-infesting plants with a non-vector insect (aphid Myzus persicae) on feeding dynamics of a vector insect (whitefly Bemisia tabaci) as well as virus transmission-acquisition. We showed that an aphid herbivory period of 0–48 h led to a transient systemic increase of virus concentration in the host plant (root, stem, and leaf), with the same pattern observed in whiteflies feeding on aphid-infested plants. We used real-time quantitative PCR to study the expression of key genes of the SA- and JA-signaling pathways, as well as electrical penetration graph (EPG) to characterize the impact of aphid pre-infestation on whitefly feeding during TYLCV transmission (whitefly to tomato) and acquisition (tomato to whitefly). The impact of the duration of aphid pre-infestation (0, 24, or 48 h) on phloem feeding by whitefly (E2) during the transmission phase was similar to that of global whitefly feeding behavior (E1, E2 and probing duration) during the acquisition phase. In addition, we observed that a longer phase of aphid pre-infestation prior to virus transmission by whitefly led to the up-regulation and down-regulation of SA- and JA-signaling pathway genes, respectively. These results demonstrated a significant impact of aphid pre-infestation on the tomato – whitefly – TYLCV system. Transmission and acquisition of TYLCV was positively correlated with feeding activity of B. tabaci, and both were mediated by the SA- and JA-pathways. TYLCV concentration during the transmission phases was modulated by up- and down-regulation of SA- and JA-pathways, respectively. The two pathways were inconsistent during the acquisition phase; SA- related genes were up-regulated, whereas those up- and down-stream of the JA pathway showed a more complex relationship. These findings enhance our understanding of plant – herbivore – virus interactions, which are potentially important for development of ecologically sound pest and pathogen management programs.
In recent decades, there have been many studies on the ecological and physiological mechanisms underlying plant-mediated interactions among herbivore arthropods and plant pathogens, particularly regarding plant defense responses (Al-Bitar and Luisoni, 1995; Beale et al., 2006; Bleeker et al., 2011; Baldin et al., 2013; Mouttet et al., 2013). Although it is well established that infection by pathogenic microorganisms may indirectly influence the behavior of the subsequent herbivore insects of the plant (Beale et al., 2006; Goggin, 2007; Mouttet et al., 2011), there has been less work on the effect of insect herbivore infestation on plant pathogens (Czosnek et al., 1988; Mouttet et al., 2011, 2013). The defense responses induced in plants after incidence of herbivory is central to the longstanding co-evolutionary system of host plant, pathogenic microorganism and herbivore arthropod (including vectors). Furthermore, these responses often impact prevalence of disease and pests, as well as the degree of damage to plants (Mayer et al., 2002; Cory and Hoover, 2006; Huot et al., 2013). However, there are key gaps in understanding of host plant – vector insect – pathogen interaction systems. For example, it is unclear how phytohormone-mediated plant defenses which are induced by insect infestations or virus transmission may influence behaviors of virus-carrying insects. Thus, understanding the mechanisms by which virus transmission occurs in plants showing herbivore-induced defense may be helpful for development of novel pest management strategies.
There have been numerous studies describing various defense systems that are induced in plants in response to damage caused by pathogens and herbivore arthropods (Karban and Baldwin, 1997; Huot et al., 2013). Key signaling pathways identified as regulators of plant defense are Jamonate (JA) for insect infestations and Salicylate (SA) for pathogen infestations, although the relationship between gene expression and function can be complex (Thaler et al., 2010). Further, understanding signal interactions within a plant – insect – pathogen system requires consideration of ecological context and the temporal sequence of interactions, such as the effect that an initial herbivore attack might have on the system during subsequent infestations. These types of interactions are more representative of incidents in natural and agricultural settings, and thus must be studied to develop more effective solutions for pest/pathogen management (Agrawal et al., 2000; Howe and Jander, 2008; Taggar and Gill, 2016).
Tomato Yellow Leaf Curl Virus is a cosmopolitan geminivirus which can infect dicotyledons, including tomato (Goggin, 2007; Heil, 2008), and is usually transmitted by the globally invasive whitefly Bemisia tabaci (Green and Ryan, 1972; Rubinstein and Czosnek, 1997). The transmission of TYLCV by B. tabaci causes huge losses in tomato yield in many regions, including China (Pan et al., 2012). The acquisition of TYLCV by B. tabaci occurs when a non-infected whitefly feeds on the phloem of a TYLCV-infected plant, and transmission occurs when a virus infected B. tabaci feeds on a previously healthy plant (Czosnek et al., 2002). It has been shown that within this defense system, a prior attack by aphid Myzus persicae can influence the feeding strategy and population dynamics of subsequent herbivores (Tan et al., 2014). It is unclear whether TYLCV transmission and acquisition are also affected by the feeding behavior of whiteflies, or whether these are influenced by aphid pre-infestation.
Plants infected by TYLCV may attract more whiteflies, and generally create conditions suiting colonization and suppression of the virus (Stout et al., 2006; Luan et al., 2013). Previous research has shown that plant defense responses induced by aphids are similar to those induced by pathogens (Thompson and Goggin, 2006). Aphid attacks can activate the SA-signaling pathway and suppress the JA-signaling pathway of the plant, and consequentially attract more whiteflies (via JA) and suppress TYLCV (SA) (Zarate et al., 2007; Verhage et al., 2010). Increasing this attraction can lead to aggregation of whiteflies and increase their feeding, both of which may increase TYLCV transmission and acquisition. Therefore, evaluation of the SA- and JA-pathway under aphid pre-feeding may help us to understand the mechanism by which induced defense is regulated.
In order to better understand the interactions in the plant- herbivore-virus system under conditions of sequential herbivory, we tested the hypothesis that pre-infestation by aphid M. persicae affects the expression of hormone-related genes and the SA- and JA-signaling pathways, consequentially impacting feeding behavior of subsequent whiteflies and thus the transmission and acquisition of TYLCV. TaqMan Real-Time PCR has been used for rapid and efficient quantitative detection of organismal DNA. It has proven highly sensitive for detecting DNA viruses in insect vectors and plants (Lei et al., 1987; Kunkel and Brooks, 2002), and there have been reports on using this technology for detection of TYLCV in tomato (Liu et al., 2007). We use Real-Time PCR to quantify TYLCV in infected tissues of tomato plant as well as in B. tabaci. We tested if pre-infestation with M. persicae (i) affect the feeding behavior of B. tabaci, (ii) modulate TYLCV transmission and acquisition between host plant and B. tabaci, and (iii) impact the expression of SA- and JA-signaling pathway genes (and potential link with vector feeding behavior and virus transmission and acquisition).
Tomato plants, Solanum lycopersicum L. (var. Baofen-F1, 2008, Changfeng Institute of Vegetable, Lintong, Xi’an, Shaanxi, China) were cultivated in plastic trays (50.0 × 25.0 × 15.0 cm), with eight plants per tray. Seedlings, about 4–5 cm in height, were transplanted into plastic pots (20 cm in depth and 15 cm in diameter) and placed in clean cages (60 × 60 × 60 cm, plastic frame, screened with 120-mesh nylon yarn net). Plants used in all experiments were approximately 30 cm in height with 4–6 true, fully expanded leaves. The experiments were conducted in an environmental chamber at 25 ± 2°C, 65 ± 5% relative humidity and photoperiod of 16/8 h of light/dark with artificial lighting of 3500 lux. About 15,000 M. persicae individuals were collected from pepper plants (Capsicum annuum L. var. Jingyuan New Prince, provided by Qing County Modern Agricultural Technology Promotion Center, Hebei Province) in a greenhouse on the campus of Northwest A&F University, Yangling, Shaanxi (116°22′42″E and 39°59′58″N) in March 2011. The aphids were maintained on tomato plants under the laboratory conditions described above. All tested aphids were offspring (>five generations) of the original collected specimens.
Bemisia tabaci whiteflies (507 males and 631 females) of Middle East-Asia Minor 1 (MEAM 1) species were collected from tomato plants in a greenhouse and their identity confirmed by mitochondrial DNA marker analysis as described by Chu et al. (2007). They were reared on cotton plants under the same conditions described for green peach aphids during March and April 2011. Newly emerged adult whiteflies were used in all experiments after they had occupied the tomato plants for more than five generations. TYLCV-infected tomato plants were provided by the Natural Enemy Application and Research Laboratory, Institute of Plant and Environment Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China. The plants were cultivated in the State Key Laboratory of Crop Stress Biology of Northwest A&F University using ventilated culturing cages (60 × 60 × 60 cm, plastic frame, screened with 120-mesh nylon yarn net) containing 100 pairs of whitefly adults. The whiteflies were moved to the cages containing 15 tomato plants with 4–6 leaves each. TYLCV-infected plants were tested after 20 days. Bemisia tabaci specimens were obtained from cotton leaves from which pseudo pupae had been cut off and the petiole had been inserted in a glass bottle filled with water and placed in a plastic cage (13 × 13 × 30 cm; 100-mesh nylon yarn net door on one side). Non-viruliferous whiteflies were maintained on cotton plants (Gossypium arboreum cv. SN407) that had been identified as non-infected hosts for TYLCV. Newly emerged Non-viruliferous adult whiteflies were used in experiments after they were moved from cotton plants and had occupied the tomato plants for more than five generations. Newly emerged whiteflies were collected and released onto the TYLCV-infected tomato plants in ventilated culturing cages for a duration of 48 h, after which the whiteflies were used in the experiments; previous studies indicate that the viral concentration in whitefly peaks after 12–48 h of feeding on tomato plants infected with TYLCV (Mason et al., 2008; Luo et al., 2010; Lucas-Barbosa et al., 2011).
Leaves of five TYLCV-infected tomato plants were ground into powder in a pre-chilled motor in liquid nitrogen. Genomic DNA was extracted from 40 mg powder using the WizardTM Genomic DNA purification kit (Promega Corporation, Madison, WI, United States) according to manufacturer’s instructions. Genomic DNA from healthy leaves of non-infected tomato plants were used as control. The extracted DNA was used as a template for PCR reaction or kept at -20°C for later use.
Based on the complete genomic sequence of TYLCV (GenBank accession number: NC_004005.1), two primer pairs: F1 (5′-CCAATAAGGCGTAAGCGTGTAGAC-3′) and R1 (5′-ACGCATGCCTCTAATCCAGTGTA-3′), F2 (5′-TCCCCTCTATTTCAAGATAACAGAACA-3′) and R2 (5′-TCTGAGGCTGTAATGTCGTCCA-3′), and TaqMan probe (5′-FAM- CCCAATGCCTTCCTG-MGB-3′) were designed by GeneCore Bio Technologies Co. Ltd. (Shanghai, China) to amplify 543 and 161 bp regions of the TYLCV gene. Actin (ACT) gene of B. tabaci (GenBank accession number: AF071908) and S. lycopersicum (GenBank accession number: AB199316) were used as internal controls, and 174 and 191 bp regions of BtACT and SlACT genes were amplified using F3 (5′-TCCTTCCAGCCATCCTTCTTG-3′) and R3 (5′-CGGTGATTTCCTTCTGCATT-3′) (Li et al., 2013) and F4 (5′-TGGTCGGAATGGGACAGAAG-3′) and R4 (5′-CTCAGTCAGGAGAACAGGGT-3′) (Chen et al., 2017) as primer pairs, respectively.
The PCR reactions were conducted in a C1000 Thermal Cycler (BioRad, Hercules, CA, United States) using F1 and R1 as primer pair in 30 μL of mixture containing 1 μL of template DNA, 1 μL of each primer (10 μM), 3 μL of 10 × PCR buffer (150 mM Tris–HCl, 500 mM Tris–HCl, pH 8.0 with 25 mM MgCl2), 1 μL of dNTP Mix (2.5 mM), 0.5 μL of Premix Ex Taq DNA polymerase (5 U/μL, Takara Biotechnology Co. Ltd., Dalian, China) and 22.5 μL of double distilled H2O. The PCR conditions were 10 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 57°C and 2 min at 72°C and a final extension at 72°C for 10 min. The PCR products were separated on a 2% agarose gel and purified using the Biospin Gel Extraction Kit (Bioer Technology Co. Ltd., Hangzhou, China) as described by the manufacturer. The recycled PCR products were ligated into the pMD-19 T vector (Takara) and transformed into DH5α cells. After blue-white selection, positive clones were sequenced by the Shanghai Sunny Biotech Co. Ltd., China. The sequence-verified clones were cultivated and plasmids were extracted. The concentration of extracted plasmid DNA was measured using a UV-Vis spectrophotometer (NanoDrop 2000 c). Copy number was determined according to the equation: copy number/mL = 6.02 × 1023 (cn/mol) × plasmid concentration (g/mL)/MW (g/mol). The standard curve of TYLCV coat protein gene was obtained by using serial 10-fold diluted plasmids (9.16 × 1010 to 9.16 × 10 copies) as templates, and by using F2 and R2 as primer pairs and TaqMan fluorescence probe (Figure 1). Amplifications were performed in 25 μl volumes containing 12.5 μL of Premix Ex Taq (Probe qPCR), 2 μL of template, 0.5 μL of each 10 μM F2 and R2 primers using thermocycler conditions as follows: 95°C for 10 s, followed by 40 cycles of 10 s at 95°C and 30 s at 58°C. Then, the linear equation of log concentration versus Ct curve was generated, where Ct values were plotted from 10-fold serial dilutions of the plasmid DNA. The estimation of copy number in samples was performed by computing the estimates of linear regression coefficients. The quantifications of DNA samples were calculated based on the fluorescence (ΔRn) values. All samples were run in duplicate by TaqMan Real-Time PCR assay for accuracy of results.
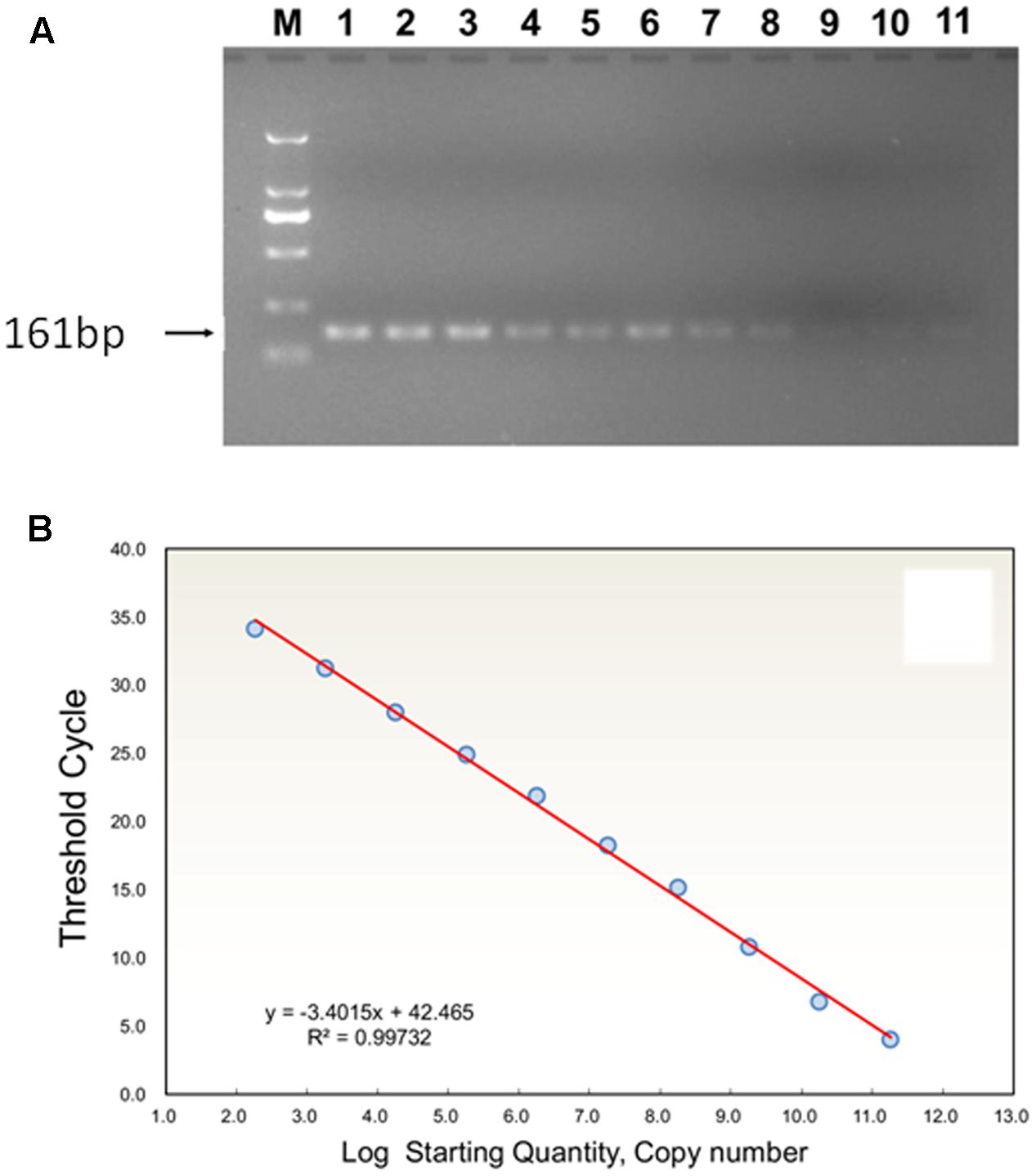
FIGURE 1. (A) Agarose gel analysis of TYLCV (TYLCV means tomato yellow leaf curl virus) using serial dilutions of plasmid standard product DNA, showing the amplification products obtained by PCR with TYLCV; M: 161-bp ladder DNA marker. Standards (copy numbers per μl of target DNA): 1, 9.16 × 1010; 2, 9.16 × 109; 3, 9.16 × 108; 4,9.16 × 107; 5, 9.16 × 106; 6, 9.16 × 105; 7, 9.16 × 104; 8, 9.16 × 103; 9, 9.16 × 102; 10, 9.16 × 101; 11, 9.16 × 100. (B) Standard curve analysis: the relationship between TYLCV concentration and corresponding Ct value is shown by linear regression equation of Ct (x) transformed into log (starting quantity).
High specificity was determined for primers (Figure 1). PCR amplification showed that the product of TYLCV DNA corresponded to a 161 bp band, and lacking primer dimers. The data for the estimation of a standard curve for absolute quantitative and TaqMan Real-Time PCR assays were obtained and analyzed by an iQTM5 Multicolor Real-Time PCR Detection System. Thus, sample Ct values were obtained automatically. Ct values and the logarithm of TYLCV DNA copy number (log) had a linear relationship. The logarithmic average of TYLCV DNA copy number (log) was calculated through a standard curve, giving y = -3.4015x + 42.465, slope -3.4015, R2 = 0.9973 from each sample of TYLCV, an amplification efficiency (E) of 96.8%, and showed a robust linear relationship (Figure 1B).
We assessed pre-infestation effects on viruses, separately for the processes of transmission and acquisition, and each by varying the duration of pre-infestation. We sequentially added aphids then whiteflies, then measured virus concentration. For transmission, Each tomato plant with 4–5 expanded leaves was infested separately with 60 fourth-instar aphid nymphs, then the aphids were removed using a soft brush after 12, 24, or 48 h. Aphids were randomly assigned to each plant. Sixty viruliferous adult whiteflies were introduced throughout the tomato plant (with sufficient space for flight in the mini cage) and fed for 24 h after removal of aphids. The whiteflies were removed using an aspirator. A group of five tomato plants (including roots) was collected, the roots washed with sterile distilled water, and stored at -80°C. Muniyappa et al. (2000) showed that the virus can be transmitted from whiteflies that have fed on TYLCV-infected plants for 24 h, to uninfected plants after exposure of only 20 min. Jiu et al. (2006) reported that the shortest time of transmission of TYLCV to plants by whiteflies was 15–30 min. Thus, in the present study, a 24 h feeding time was thought sufficient to transmit TYLCV to tomato plants. The experiment was repeated five times, each time with five tomato plants. Control tomato plants were not infested with aphids. Leaves of five TYLCV-infected tomato plants were ground into powder in a pre-chilled motor in liquid nitrogen. Virus concentration was assessed by DNA extraction and PCR; the TaqMan PCR amplification system followed the methods described above. For virus acquisition, we also measured TYLCV for B. tabaci via feeding on virus-infested tomato plants. Viruliferous whitefly were first used to transmit virus to the tomato plant, followed by aphid preinfestation and subsequent whitefly exposure. Tomato plants treated as above were individually placed in a square plastic cage (13 × 13 × 30 cm), and 60 viruliferous adult whitefly introduced using an aspirator. The whitefly adults were removed after feeding for 24 h, and then the plants were pre-infested with aphids as above. Fifty newly emerged non-viruliferous whitefly adult females were introduced into the cage to acquire virus from the infected tomato plant after removing the aphids. After feeding for 24 h, 40 whiteflies per plant from each group of five tomato plants were collected and stored at -80°C. Control tomato plants were not infested with aphids. The experiment was replicated five times, each time with five different tomato plants. For virus detection in whitefly, DNA was extracted using the WizardTM Genomic DNA purification kit (Promega, Madison, WI, United States) from 200 whiteflies of a group of five tomato plants, which were ground into homogenate in 600 μL of nuclear pyrolysis liquid. The extracted DNA was diluted 10 times for TaqMan PCR assay or saved at -28°C for later use. PCR amplification system and response procedures were as reported above.
Each tomato plant with 4–5 expanded leaves was infested separately with 60 fourth-instar aphid nymphs, then the aphids were removed using a soft brush after 24 or 48 h. Aphids were randomly assigned to each plant. Because the concentration of TYLCV was similar for aphid feeding of 12 and 24 h, this test used tomato plants infested with aphids for 24 or 48 h. The experiment was repeated three times, each time with eight whiteflies recorded.
For transmission, whitefly feeding during the virus transmission phase was recorded. Feeding behavior was assessed for newly emerged viruliferous whiteflies from TYCLV infected tomato plants. For acquisition, whitefly probing behavior during virus acquisition (from tomato) was recorded for newly emerged whiteflies from non-infected tomato.
Whitefly feeding was monitored using the electrical penetration graph (EPG) method (Liu et al., 2013). In EPG, the insect and its host plant creates an electrical circuit that is completed when the mouthparts of the phloem-feeding insect penetrate the plant. The resulting electrical signals are amplified and digitized. Fluctuations in voltage and electrical resistance are recorded on computer, and can be matched to specific feeding events. Prior to recording, whiteflies were immobilized by placing them in a freezer for several minutes, then waiting 1 h without feeding. The recording electrode of the EPG system (a thin gold wire of length 2 cm and diameter 10 μm), was connected to the whitefly’s dorsum with silver paint. Electrode adhesion was conducted under a microscope, and then the whitefly was put on the abaxial surface of the leaf and allowed to crawl freely. The wired whiteflies were then connected to the Giga-8 probe input, and another electrode was inserted into the soil at the base of the tomato plant. The whole system was placed into an electrically grounded Faraday cage to guard against external electrical noise. The EPG signals were digitized with a DI710-UL analog-to-digital converter, and the output was acquired and stored with PROBE3.4 software. The data were analyzed subsequently with STYLET 2.0 software. Phases of feeding behavior are described by 14 EPG parameters related to the pathway phase (C waveform, F waveform and potential drops), phloem phase (E waveform) and xylem phase (G waveform). Phloem subphase 1 (E1: salivary secretion into sieve elements) and phloem subphase 2 (E2: phloem ingestion) were extracted from each recording and compared among treatments. 12 h of EPGs were continuously recorded for each replicate. All experiments were carried out under artificial light (1,500 lx) with a 16 h light/8 h dark regime and at 25°C ± 2°C, at 70% relative humidity (RH).
Each treatment combination was replicated three times with eight plants for biological replicates, and each biological replicate contained three technical replicates. The expression of target genes involved in the JA-signaling [lipoxygenase D (LOXD), proteinase inhibitor II (PI-II)] and SA-signaling [phenylalanine ammonia lyase (PAL), and β-1,3-glucanase (PR2b)] pathways were determined.
The RNeasy Mini Kit (Qiagen, Valencia, CA, United States) was used to isolate total RNAs from tomato leaves (100-mg samples) before viruliferous whitefly and non-infected whitefly feeding, and 1 μg of RNAs were used to generate the cDNAs. The mRNA amounts of seven target genes were measured by real-time quantitative PCR.
Specific primers for each gene were designed from the tomato plant expressed sequence tag sequences using PRIMER5 software (Table 1). The reaction mixture contained the following reagents: 1 μL of first-strand cDNA, 2 μL of reaction buffer, 1 μL of MgCl2 (50 mM), 0.3 μL of SYBR Green, 0.2 μL of PLATINUM Taq polymerase, 0.5 μL of dNTPs (10 mM) and 10 μM of each primer in a total volume of 20 μL. Reactions were carried out on the Mx 3500P detection system (Stratagene), and PCR was performed under the following conditions: 2 min at 95°C; followed by 40 cycles of 30 s at 95°C and 30 s at 60°C. Following the real-time quantitative PCR, a melting curve was generated by gradually increasing the temperature to 95°C to test the homogeneity of PCR products. Relative standard curves for the transcripts of every gene were prepared with linear gradient (five-fold) cDNA as template and were included within each real-time quantitative PCR. The relative level of each target gene was standardized by comparing the copy numbers of target mRNA with SlACT and SlTUB CT values, which remained constant under different treatment conditions. The fold-changes of target genes were calculated using the 2ΔΔCT method. All PCR runs were performed with three technical replicates. The three independent biological replicates, each containing eight plants, were analyzed.
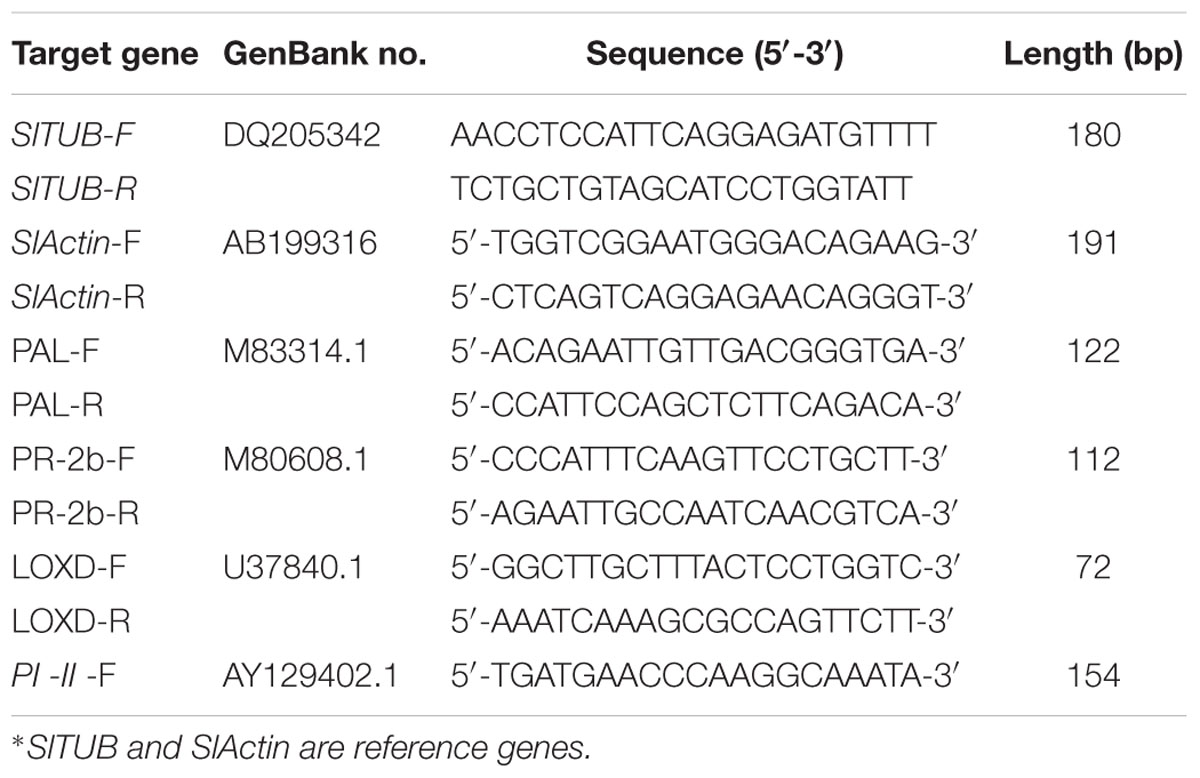
TABLE 1. Overview of the target genes used in this study, showing their GenBank accession numbers and the primer pair used for qRT-PCR.
One-way analysis of variance (ANOVA) was used to analyze the data of log copy number of TYLCV DNA for the effects of duration of infestation by M. persicae on the transmission and acquisition of TYLCV, whitefly probing profiles (EPG) and defense gene expressions. ANOVA was performed with IBM SPSS statistics version 20.0 (SPSS, Chicago, IL, United States). An independent t-test was used to test the effects of aphid feeding duration on defense gene expression during transmission and acquisition. All data were checked for normality and equality of residual error variances, and were transformed (log or square root) where appropriate to satisfy assumptions of ANOVA. General Linear Mixed Model (GLMM) was used to analyze the effects of the duration of aphid pre-infestation, plant position and their interaction on TYLCV. In both cases, when a significant effect was found, Duncan’s test was used as post hoc test to compare the log copy number of TYLCV DNA (P < 0.05).
The GLMM tests highlighted significant differences in TYLCV DNA concentrations for different durations of aphid pre-infestation (F = 12.622, df = 3176, P < 0.05), and among different tomato plant tissues (F = 4.107, df = 2177, P < 0.05). Further, there was a significant interaction between duration of aphid pre-infestation, tomato plant tissues, and concentration of TYLCV DNA (F = 7.208, df = 6, P < 0.05) (Figure 2). The concentration of TYLCV DNA in B. tabaci after feeding on virus-infected tomato was influenced by duration of aphid pre-infestation (F = 3.769, df = 3176, P < 0.05; Figure 3).
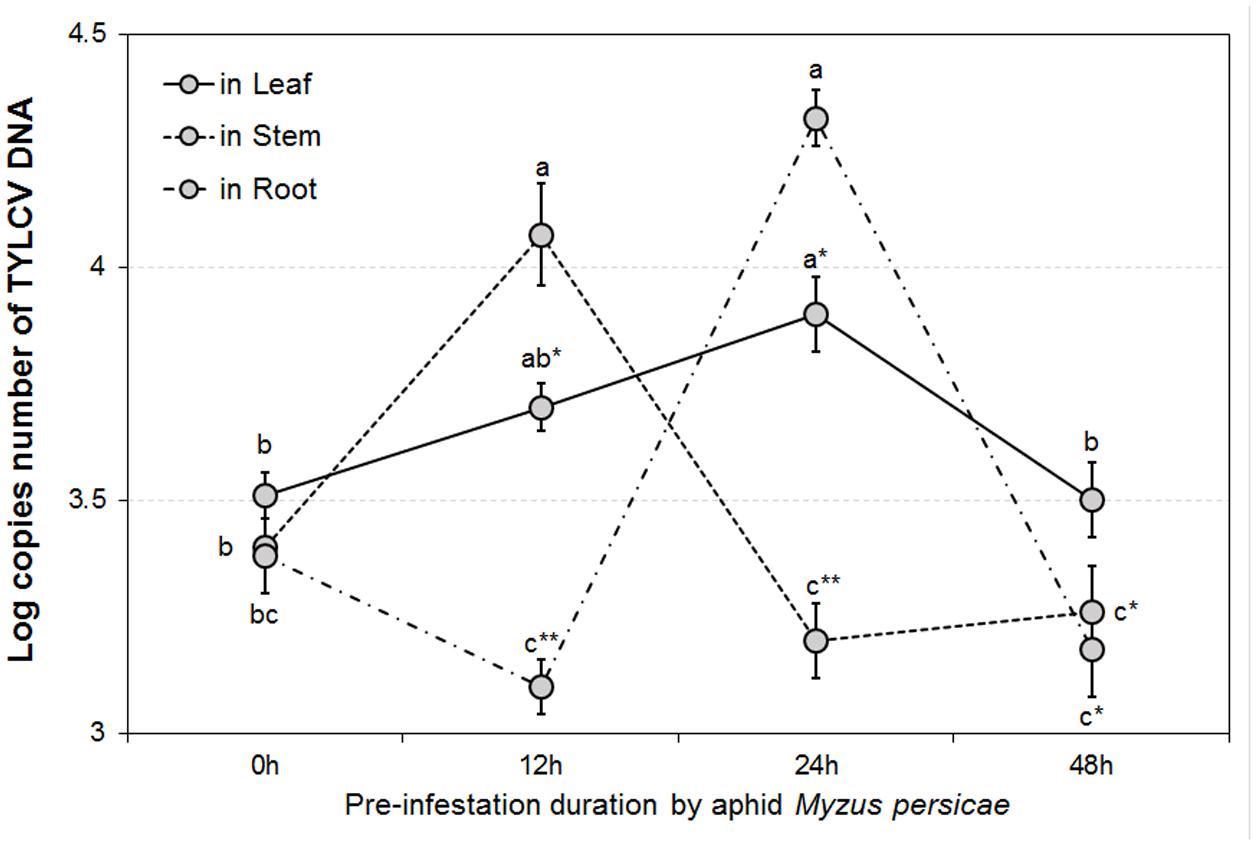
FIGURE 2. TYLCV DNA concentrations in different tomato plant parts after feeding by TYLCV-infested Bemisia tabaci according to various Myzus persicae pre-infestation duration treatments. The error bars are standard errors of the mean. Colored letters indicate significant differences of DNA concentration among different aphid pre-infestation durations in each tomato plant part, and ‘∗’ indicates significant differences among three plant parts in the same pre-infestation duration according to Duncan’s test at P = 0.05.
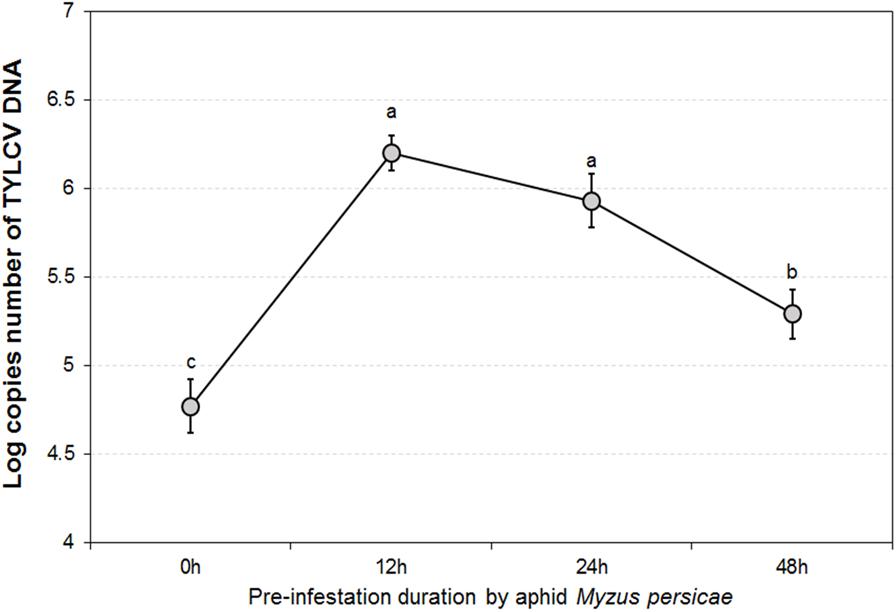
FIGURE 3. TYLCV DNA concentrations in Bemisia tabaci adults after feeding on TYLCV-infested tomato plants for 24 h according to various Myzus persicae pre-infestation. The error bars are standard errors of the mean. Letters indicate significant differences of DNA concentration among different aphid pre-infestation durations according to Duncan’s test at P = 0.05.
Twenty-four hour aphid infestation significantly increased the time that viruliferous whiteflies spent in the salivation phase [E1] (F = 3.586, P = 0.033, df = 2, 69) and duration of phloem feeding [E2] (F = 4.296, P = 0.017, df = 2, 69; Figure 4A). By contrast there was no significant effect on the total duration of probing by whiteflies (F = 2.658, P = 0.077, df = 2, 69; Figure 4A). For 48-h aphid infestation, the duration of E2 was similar to plants not infested with aphids; whereas significant differences are observed at 48 h for the duration of E1 and total duration of probing, compared to 0 h.
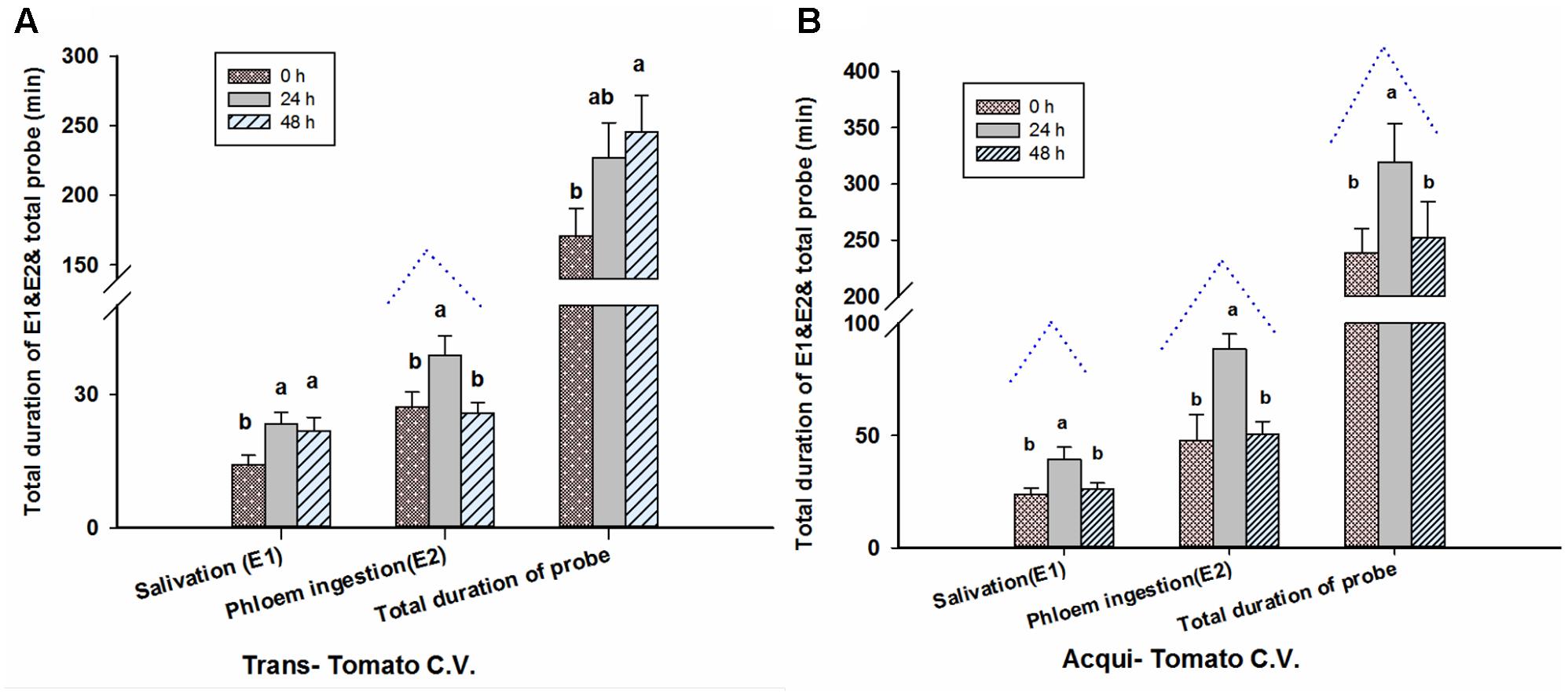
FIGURE 4. Aphid pre-infestation duration effects on feeding behavior (EPG) of transmission (A) of whiteflies on virus non-infected tomato plants and acquisition (B) of whiteflies on virus infected tomato plants. Letters indicate significant differences of feeding behavior among different aphid pre-infestation durations.
Pre-infestation with M. persicae significantly impacted several aspects of whitefly feeding on virus-infected leaves (E1: F = 4.836, P = 0.011; E2: F = 6.173, P = 0.003; and total duration of probing: F = 3.669, P = 0.031; all df = 2, 69; Figure 4B). 24 h of aphid infestation significantly increased E1, E2 and the total duration of probing by whiteflies. For 48-h aphid infestation, the durations of E1, E2 and of probing were similar to plants not infested with aphids. This trend was similar to the virus concentration in whiteflies with increasing aphid feeding duration on viruliferous tomato plants (Figure 4B).
The relative mRNA expressions of PAL (before transmission: F = 9.213, P = 0.0001, df = 2, 69; and before acquisition: F = 4.727, P = 0.012, df = 2, 69) and PR2b (before transmission: F = 19.881, P = 0.0001, df = 2, 69; and before acquisition: F = 2.553, P = 0.085, df = 2, 69) positively correlated with aphid feeding time prior to transmission by whiteflies (Figure 5). Similarly, prior to acquisition by whiteflies, expressions of PAL and PR2b in plants with 24 h of aphid feeding were higher than for those without aphids, but similar to those with 48 h of aphid feeding. Moreover, expressions of both genes were higher prior to acquisition by whiteflies than prior to transmission by whiteflies: PAL for 0 h, t = -2.924, df = 33.022, P = 0.006; for 24 h, t = -4.709, df = 35.63, P = 0.0001; and for 48 h, t = -2.132, df = 35.117, P = 0.04; and PR2b for 0 h, t = -6.654, df = 23.494, P = 0.0001; for 24 h, t = -4.547, df = 26.518, P = 0.0001; and for 48 h, t = -3.111, df = 35.117, P = 0.003 (Figure 5).
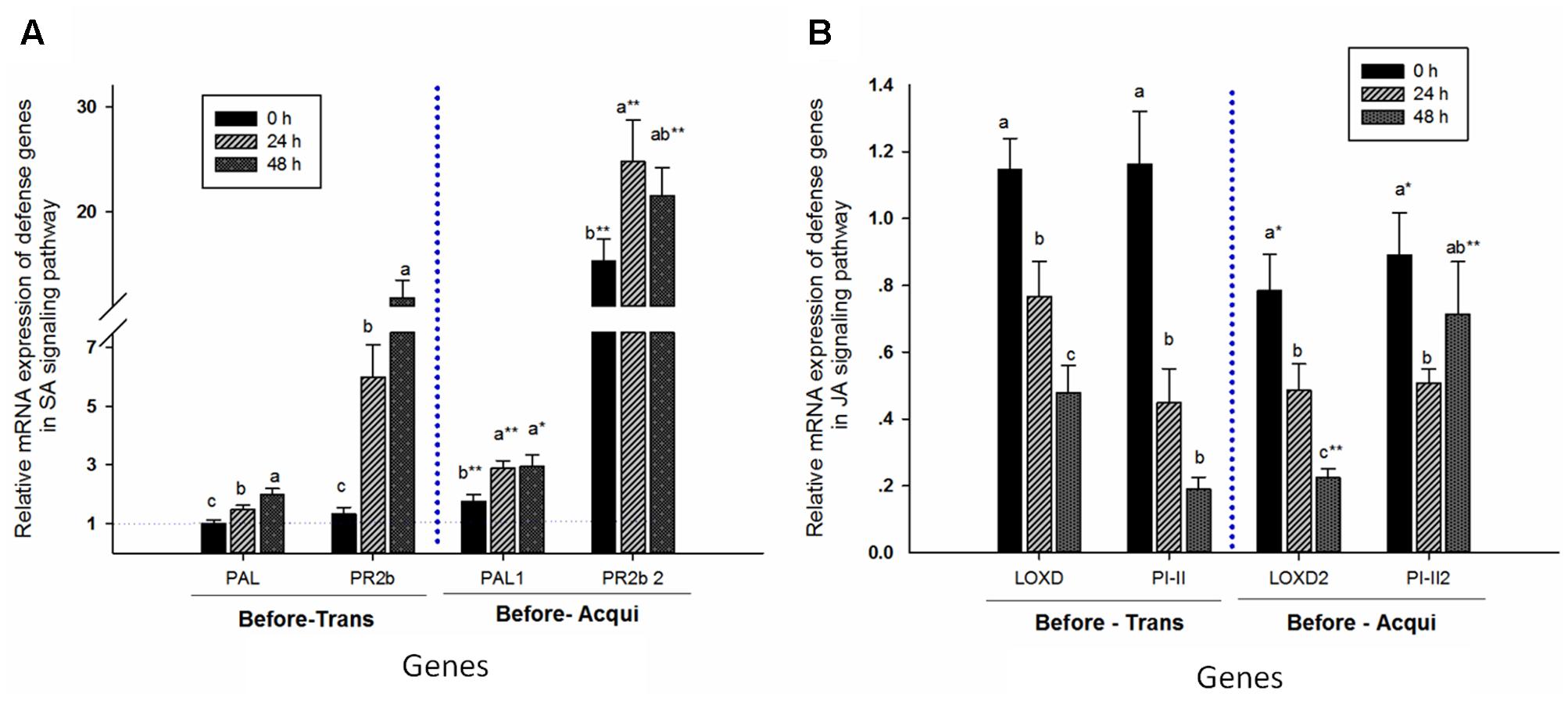
FIGURE 5. Aphid pre-infestation duration effects on related defense genes in the SA- (A) and JA- (B) signaling pathways in tomato plants. The relative mRNA expression was calculated according to the 2-ΔΔCT method, and the y axis represents the level of change. The expression level of each target gene was first normalized to the quantity of SlActin and SlTub. The error bar represents the standard deviation calculated from three replicates. Letters indicate significant differences in relative expression among different aphid pre-infestation durations, and ‘∗’ indicates significant differences in the same aphid pre-infestation duration according to Duncan’s test at P = 0.05.
In contrast, the relative mRNA expressions of JA-signaling genes LOXD (before transmission: F = 12.418, P = 0.00001, df = 2, 69; and before acquisition: F = 12.566, P = 0.00001, df = 2, 69) and PI-II (before transmission: F = 20.617, P = 0.00001, df = 2, 69; and before acquisition: F = 2.611, P = 0.081, df = 2, 69) were significantly reduced with increase in aphid feeding time, except for PI-II in plant prior to acquisition by whiteflies. The expression of LOXD in plants was lower prior to acquisition than prior to transmission by whiteflies, except for during an aphid infestation period of 24 h (LOXD for 0 h, t = 2.497, df = 46, P = 0.016; for 24 h, t = 1.666, df = 45.831, P = 0.103; and for 48 h, t = -2.741, df = 28.652, P = 0.01). However, expression of PI-II was lower in the non-infested plant prior to acquisition but not prior to transmission (PI-II, 0 h: t = 2.304, df = 46, P = 0.026; 24 h: t = -0.532, df = 30.834, P = 0.598; 48 h: t = -4.889, df = 27.284, P = 0.00001) (Figure 5).
This study is the first investigation on impact of herbivore pre-infestation on virus transmission and acquisition by subsequent herbivores, in terms of key interactions such as feeding behavior and plant defenses. We demonstrated that virus transmission and acquisition between tomatoes and vector whiteflies under aphid pre-infestation follow two different mechanisms. The SA- and JA-signaling pathways had different roles and the feeding behavior of whiteflies dominated the acquisition process. We believe that this overlooked issue will be of wide ecological interest, since knowledge of the effects of herbivorous feeding activity on the acquisition and transmission of viruses can shed light on complex plant-mediated relationships among herbivores, arthropod vectors and pathogens (Mehta et al., 1994; Messina et al., 2002; Beale et al., 2006; Mithöfer and Boland, 2012). Feeding behavior (salivation/phloem ingestion/duration of feeding) of viruliferous and non-infected B. tabaci was also influenced by different aphid pre-infestation times, in contexts of both transmission and acquisition. Furthermore, we found that the expression of two SA-signaling pathway genes may increase when aphid pre-infestation period is prolonged, and expression of the related JA-signaling pathway is reduced, especially during the transmission phase. Previous studies have shown that attack by aphids can inhibit the JA-signaling pathways and consequentially reduce negative impacts of herbivory (Papayiannis et al., 2009, 2010; Giordanengo et al., 2010). Plant defense responses induced by sap-sucking herbivores or pathogens are often similar, since both responses can act to resist both insects and pathogens (Thaler et al., 2002; Murugan and Dhandapani, 2007). The defense signaling pathways relying on SA and JA may vary with injury time (Preston et al., 1999; Nombela et al., 2009). Tobacco plants infected with TMV developed SAR, while damage response via JA was not normally activated, possibly because infection by TMV increased SA levels, inhibiting the JA pathway (Ohgushi, 2005). However, this might reduce JA/ET-associated insect resistance (Salzman et al., 2005).
The pattern in transmission of virus from viruliferous B. tabaci to non-infected tomato was consistent among different plant organs tested (attacked leaves, stems, and roots), as well as under different durations of aphid pre-infestation. When measured directly on the aphid-attacked leaf, the amount of TYLCV initially increased, and was followed by a decline, with extension of period of pre-infestation. The stem and the root showed similar tendencies; the pre-infestation was found to stimulate systemic resistance, resulting in a more effective response to subsequent attacks. Generally, herbivore feeding action leads to a diverse and synergistic systemic defense, performed across different tissues of the plant (Pico et al., 1999; Schilmiller and Howe, 2005; Jung et al., 2009). The defense pathways may be induced in tissues away from the position of attack, although this reaction is somewhat delayed (Kessmann et al., 1994). In this study, TYLCV DNA concentration in roots showed a belated increase, which demonstrated that a period of time is required for systemic transmission of TYLCV within tomato plants. Thus, a threshold period is needed for transmission of injury messages within the plant and for producing the defense compounds responsible for defense (Sauge et al., 2002). The acquisition of TYLCV by whitefly from TYLCV-infected tomato plants also displayed a tendency to rise and then decline. When increasing duration of aphid infestation beyond the 12 h threshold, the TYLCV concentration in whiteflies began to decrease. Aphids feeding on tomato plants for sufficient time can induce a plant defense response which inhibits whiteflies from acquiring TYLCV. Our result indicated that extension of the pre-infestation phase may increase virus transmission and acquisition by whitefly in the short term. This result might be explained by feeding efficacy, as influenced by the plant induced defense response.
As our results show, the feeding behavior strategies of either TYLCV infected B. tabaci on non-infected tomato (virus transmission) or non-infected whitefly on viruliferous plants (virus acquisition) were both influenced by duration of pre-infestation by M. persicae. We found that the duration of phloem ingestion (E2) of B. tabaci may increase at 24 h and decrease at 48 h, with increasing of duration of pre-infestation. However, the total probing duration for whitefly was inconsistent between transmission/acquisition treatments. The transmission and acquisition of virus between whitefly and tomato plant may be due to different feeding strategies; for transmission of virus from whitefly to plant, net virus infestation is associated with salivation (E1). Thus, as indicated by the amount of TYLCV DNA in the host plant (Figure 2), virus infection in tomato may have a corresponding increase with salivation by viruliferous B. tabaci, although unaffected by total duration of probing. In contrast, the acquisition of virus by whitefly from infected plants was mainly related to phloem ingestion (E2). Clearly, our results showed similar responses to variation in aphid pre-infestation between the amount of TYLCV in whitefly and the duration of their phloem ingestion (Figures 3, 4). It seems that virus transmission/acquisition is influenced by the whitefly’s different feeding strategies. The feeding of whiteflies may be inhibited by the plant defense response triggered by aphid pre-infestation. Empirical reports indicate that defensive materials (like JA, SA and its derivants) may suppress phloem ingestion (Kehr, 2006). Thus, the non-viruliferous whitefly showed a reduced ability to combat this response, and struggle to maintain normal phloem ingestion feeding time. Interestingly, the virus infection may be quite specific in the way it regulates feeding behavior, with the whitefly enhancing only key processes, particularly salivation, which is vital for virus transmission.
Defense response to herbivore or plant pathogens occurs mostly in the Salicylic Acid (SA) and Jasmonic Acid (JA) signaling pathways (Lamb et al., 1989). Feeding by aphids induce plant responses similar to those induced by pathogens (Czosnek et al., 2002; Thompson and Goggin, 2006). Aphid infestation may activate the SA-signaling pathway and suppress the JA-signaling pathway, which will attract whiteflies and suppress TYLCV (Verhage et al., 2010). SA activation and JA suppression both inhibit viruses and improve feeding behavior of herbivore insects, respectively (Zarate et al., 2007). Our results showed that extending the period of aphid pre-infestation may reduce expression of the genes involved in the JA-signaling pathway, and increase expression of the genes involved in the SA-signaling pathway. According to our results, it seems that upregulating the SA pathway and suppressing the JA pathway with aphid pre-infestation may directly impact the feeding strategies of the whitefly, and consequentially, the transmission and acquisition of TYLCV between whitefly and tomato plant. Similarly, there have been several reports showing that induced plant defense responses are stronger against pathogens compared to insect herbivores (Ghanim et al., 2001; Tan and Liu, 2014). Ohgushi (2005) reported that infestation by tobacco mosaic virus can induce systemic acquired resistance, while not activating the JA signaling pathway. Generally, the SA pathway is activated by pathogens, and the JA pathway is induced by insects (Salzman et al., 2005). Previous studies have shown that attack by sap-sucking herbivores can induce plant responses similar to those induced by pathogen infestations, inhibiting the JA-signaling pathways and consequentially reducing negative impacts on herbivores (Papayiannis et al., 2009, 2010; Giordanengo et al., 2010). Plant defense responses induced by herbivores or pathogens are often similar, since both responses can act to resist both insects and pathogens, or to minimize interference between herbivores and pathogen defenses (Thaler et al., 2002; Murugan and Dhandapani, 2007). Tomato plants infested by phloem-feeding insects (e.g., aphids) evoked JA- and SA-signaling pathways to co-regulate the expression of plant defense genes (Muniyappa et al., 2000). The antagonism between SA and JA impacts feeding behavior of whiteflies and viral propagation. In the transmission test, the SA and JA was dominant during the whole process. However, during the acquisition test, the JA-signaling pathway was the key factor affecting virus acquisition, and the feeding behavior of whiteflies dominated the whole process. SA activity may deter the transmission of pathogens, but inhibition of JA might attract herbivorous insects (Chu et al., 2007). The results showed that a decrease in JA combined with an increase in SA led to a net positive effect of aphid infestation on increased feeding of whiteflies, outweighing inhibition of TYLCV transmission.
Furthermore, the defense responses of these two pathways may vary with duration of injury (Preston et al., 1999; Nombela et al., 2009). Our results not only showed a temporal component to variation in associated gene expression, but also showed that indirect systemic responses (variation in amount of TYLCV across plant organs) also vary according to duration of pre-infestation. Generally, herbivore feeding leads to systemic defense in the plant, enacted in a concerted manner across tissues and organs (Polston et al., 1990; Pico et al., 1999). Thus, further evaluation of defense-related gene expression induced by insect feeding on different plant parts is required, and how this is impacted by pre-infestation.
Generally, we have confirmed the hypothesis that aphid pre-infestation impacts transmission and acquisition of TYLCV between tomato and B. tabaci. Pre-infestation showed different respective influences to the feeding strategies during the transmission or acquisition processes, while associated with opposing trends in expression of the SA- and JA- pathways. Based on current understanding of the interactions between the SA- and JA- pathways, particularly cross-inhibition, we believe that the mechanism delineating the complex and precise interaction among these organisms is worthy of further research. Notwithstanding, due to the economic importance of tomato crop, our study might have implications in the development of novel strategies for the regulation of plant pathogens and insect vectors.
XT, JC, TL, and FG designed the study. All authors drafted the manuscript. XT, GB, ND, and XY implemented the analyses. All authors gave final approval for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. T. Giordani (University of Pisa, Italy) for reading and commenting on an earlier version of the manuscript, and thanks Dr S. Wang and Dr. H. X. Xu for providing part of the material and methods. This work was supported by the National Basic Research Program of China (973 Program) (grant No. 2013CB127605), the Special Fund for Agroscientific Research in the Public Interest (grant Nos. 201303024 and 201303108), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11050400), the National Nature Science Fund of China (grant No. 31370438), the R&D Special Fund for Public Welfare Industry (Agriculture 201303019) and the State Key Laboratory of Integrated Management of Pest Insects and Rodents (Grant No. ChineseIPM1611). We are grateful for the assistance of all staff and students in the Key Laboratory of Applied Entomology, Northwest A&F University at Yangling, Shaanxi, China.
Agrawal, A. A., Karban, R., and Colfer, R. G. (2000). How leaf domatia and induced plant resistance affect herbivores. natural enemies and plant performance. Oikos 89, 70–80. doi: 10.1034/j.1600-0706.2000.890108.x
Al-Bitar, L., and Luisoni, E. (1995). Tomato yellow leaf curl geminivirus: serological evaluation of an improved purification method 1. EPPO Bull. 25, 269–276. doi: 10.1111/j.1365-2338.1995.tb01467.x
Baldin, E. L. L., Crotti, A. E. M., Wakabayashi, K. A. L., Silva, J. P. G. F., Aguiar, G. P., Souza, E. S., et al. (2013). Plant-derived essential oils affecting settlement and oviposition of Bemisia tabaci (Genn.) biotype B on tomato. J. Pest Sci. 86, 301–308. doi: 10.1007/s13744-015-0356-8
Beale, M. H., Birkett, M. A., Bruce, T. J., Chamberlain, K., Field, L. M., Huttly, A. K., et al. (2006). Aphid alarm pheromone produced by transgenic plants affects aphid and parasitoid behavior. Proc. Natl. Acad. Sci. U.S.A. 103, 10509–10513. doi: 10.1073/pnas.0603998103
Bleeker, P. M., Spyropoulou, E. A., Diergaarde, P. J., Volpin, H., De Both, M. T., Zerbe, P., et al. (2011). RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol. Biol. 77, 323–336. doi: 10.1007/s11103-011-9813-x
Chen, S., Zhao, H., Wang, M., Li, J., Wang, Z., Wang, F., et al. (2017). Overexpression of E3 ubiquitin ligase gene AdBiL contributes to resistance against chilling stress and leaf mold disease in Tomato. Front. Plant Sci. 8:1109. doi: 10.3389/fpls.2017.01109
Chu, D., Jiang, T., Liu, G., Jiang, D., Tao, Y., Fan, Z., et al. (2007). Biotype status and distribution of Bemisia tabaci (Hemiptera: Aleyrodidae) in Shandong province of China based on Mitochondrial DNA markers. Environ. Entomol. 36, 1290–1295. doi: 10.1093/ee/36.5.1290
Cory, J. S., and Hoover, K. (2006). Plant-mediated effects in insect–pathogen interactions. Trends Ecol. Evol. 21, 278–286. doi: 10.1016/j.tree.2006.02.005
Czosnek, H., Ber, R., Navot, N., Zamir, D., Antignus, Y., and Cohen, S. (1988). Detection of tomato yellow leaf curl virus in lysates of plants and insects by hybridization with a viral DNA probe. Plant Dis. 72, 949–951. doi: 10.1094/PD-72-0949
Czosnek, H., Ghanim, M., and Ghanim, M. (2002). The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with Tomato yellow leaf curl virus. Annu. Appl. Biol. 140, 215–231. doi: 10.1111/j.1744-7348.2002.tb00175.x
Ghanim, M., Morin, S., and Czosnek, H. (2001). Rate of Tomato yellow leaf curl virus translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology 91, 188–196. doi: 10.1094/PHYTO.2001.91.2.188
Giordanengo, P., Brunissen, L., Rusterucci, C., Vincent, C., Van Bel, A., Dinant, S., et al. (2010). Compatible plant-aphid interactions: how aphids manipulate plant responses. C R Biol. 333, 516–523. doi: 10.1016/j.crvi.2010.03.007
Goggin, F. L. (2007). Plant–aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant Biol. 10, 399–408. doi: 10.1016/j.pbi.2007.06.004
Green, T., and Ryan, C. (1972). Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175, 776–777. doi: 10.1126/science.175.4023.776
Heil, M. (2008). Indirect defence via tritrophic interactions. New Phytol. 178, 41–61. doi: 10.1111/j.1469-8137.2007.02330.x
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Huot, O. B., Nachappa, P., and Tamborindeguy, C. (2013). The evolutionary strategies of plant defenses have a dynamic impact on the adaptations and interactions of vectors and pathogens. Insect Sci. 20, 297–306. doi: 10.1111/1744-7917.12010
Jiu, M., Zhou, X. P., and Liu, S. S. (2006). Acquisition and transmission of two Begomoviruses by the B and a non-B biotype of Bemisia tabaci from Zhejiang, China. J. Phytopathol. 154, 587–591. doi: 10.1111/j.1439-0434.2006.01151.x
Jung, H. W., Tschaplinski, T. J., Glazebrook, J., and Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
Karban, R., and Baldwin, I. T. (1997). Induced Responses to Herbivory. Chicago: University of Chicago Press. doi: 10.7208/chicago/9780226424972.001.0001
Kehr, J. (2006). Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 57, 767–774. doi: 10.1093/jxb/erj087
Kessmann, H., Staub, T., Hofman, C., Maetzke, T., and Herzog, J. (1994). Induction of systemic acquired disease resistance in plants by chemicals. Annu. Rev. Phytopathol. 32, 439–459. doi: 10.1146/annurev.py.32.090194.002255
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Lamb, C. J., Lawton, M. A., Dron, M., and Dixon, R. A. (1989). Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell 56, 215–224. doi: 10.1016/0092-8674(89)90894-5
Lei, X. Y., Li, H. F., and Chiu, W. F. (1987). Effect of induced resistance on virus infection of some plants and the mechanisms of the resistance inducer NS-83. Sci. Agric. 20, 1–6.
Li, R., Xie, W., Wang, S., Wu, Q., Yang, N., Yang, X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLOS ONE 8:e53006. doi: 10.1371/journal.pone.0053006
Liu, B. M., Preisser, E. L., Chu, D., Pan, H., Xie, W., Wang, S., et al. (2013). Multiple forms of vector manipulation by a plant-infecting virus: Bemisia tabaci and tomato yellow leaf curl virus. J. Virol. 87, 4929–4937. doi: 10.1128/JVI.03571-12
Liu, S. S., De Barro, P., Xu, J., Luan, J.-B., Zang, L. S., Ruan, Y. M., et al. (2007). Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318, 1769–1772. doi: 10.1126/science.1149887
Luan, J. B., Yao, D. M., Zhang, T., Walling, L. L., Yang, M., Wang, Y. J., et al. (2013). Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol. Lett. 16, 390–398. doi: 10.1111/ele.12055
Lucas-Barbosa, D., van Loon, J. J., and Dicke, M. (2011). The effects of herbivore-induced plant volatiles on interactions between plants and flower-visiting insects. Phytochemistry 72, 1647–1654. doi: 10.1016/j.phytochem.2011.03.013
Luo, C., Jones, C., Devine, G., Zhang, F., Denholm, I., and Gorman, K. (2010). Insecticide resistance in Bemisia tabaci biotype Q (Hemiptera: Aleyrodidae) from China. Crop Prot. 29, 429–434. doi: 10.1016/j.cropro.2009.10.001
Mason, G., Caciagli, P., Accotto, G. P., and Noris, E. (2008). Real-time PCR for the quantitation of Tomato yellow leaf curl Sardinia virus in tomato plants and in Bemisia tabaci. J. Virol. Methods 147, 282–289. doi: 10.1016/j.jviromet.2007.09.015
Mayer, R. T., Inbar, M., McKenzie, C., Shatters, R., Borowicz, V., Albrecht, U., et al. (2002). Multitrophic interactions of the silverleaf whitefly, host plants, competing herbivores, and phytopathogens. Arch. Insect Biochem. Physiol. 51, 151–169. doi: 10.1002/arch.10065
Mehta, P., Wyman, J. A., Nakhla, M. K., and Maxwell, D. P. (1994). Polymerase chain reaction detection of viruliferous Bemisia tabaci (Homoptera: Aleyrodidae) two tomato-infecting geminiviruses. J. Econ. Entomol. 87, 1285–1290.
Messina, F. J., Durham, S. L., Richards, J. H., and McArthur, D. E. (2002). Trade-off between plant growth and defense? A comparison of sagebrush populations. Oecologia 131, 43–51. doi: 10.1007/s00442-001-0859-3
Mithöfer, A., and Boland, W. (2012). Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63, 431–450. doi: 10.1146/annurev-arplant-042110-103854
Mouttet, R., Bearez, P., Thomas, C., and Desneux, N. (2011). Phytophagous arthropods and a pathogen sharing a host plant: evidence for indirect plant-mediated interactions. PLOS ONE 6:e18840. doi: 10.1371/journal.pone.0018840
Mouttet, R., Kaplan, I., Bearez, P., Amiens-Desneux, E., and Desneux, N. (2013). Spatiotemporal patterns of induced resistance and susceptibility linking diverse plant parasites. Oecologia 173, 1379–1386. doi: 10.1007/s00442-013-2716-6
Muniyappa, V., Venkatesh, H., Ramappa, H., Kulkarni, R., Zeidan, M., Tarba, C. Y., et al. (2000). Tomato leaf curl virus from Bangalore (ToLCV-Ban4): sequence comparison with Indian ToLCV isolates, detection in plants and insects, and vector relationships. Arch. Virol. 145, 1583–1598. doi: 10.1007/s007050070078
Murugan, M., and Dhandapani, N. (2007). Induced systemic resistance activates defense responses to interspecific insect infestations on tomato. J. Veg. Sci. 12, 43–62. doi: 10.1300/J484v12n03_05
Nombela, G., Garzo, E., Duque, M., and Muñiz, M. (2009). Preinfestations of tomato plants by whiteflies (Bemisia tabaci) or aphids (Macrosiphum euphorbiae) induce variable resistance or susceptibility responses. Bull. Entomol. Res. 99, 183–191. doi: 10.1017/S0007485308006214
Ohgushi, T. (2005). Indirect interaction webs: herbivore-induced effects through trait change in plants. Annu. Rev. Ecol. Evol. Syst. 36, 81–105. doi: 10.1146/annurev.ecolsys.36.091704.175523
Pan, H. P., Chu, D., Yan, W. Q., Su, Q., Liu, B. M., Wang, S. L., et al. (2012). Rapid spread of tomato yellow leaf curl virus in china is aided differentially by two invasive whiteflies. PLOS ONE 7:e34817. doi: 10.1371/journal.pone.0034817
Papayiannis, L., Brown, J., Seraphides, N., Hadjistylli, M., Ioannou, N., and Katis, N. (2009). A real-time PCR assay to differentiate the B and Q biotypes of the Bemisia tabaci complex in Cyprus. Bull. Entomol. Res. 99, 573–582. doi: 10.1017/S0007485308006603
Papayiannis, L., Iacovides, T., Katis, N., and Brown, J. (2010). Differentiation of tomato yellow leaf curl virus and tomato yellow leaf curl Sardinia virus using real-time TaqMan® PCR. J. Virol. Methods 165, 238–245. doi: 10.1016/j.jviromet.2010.02.003
Pico, B., Díez, M. J., and Nuez, F. (1999). Improved diagnostic techniques for tomato yellow leaf curl virus in tomato breeding programs. Plant Dis. 83, 1006–1012. doi: 10.1094/PDIS.1999.83.11.1006
Polston, J., Al-Musa, A., Perring, T. M., and Dodds, J. (1990). Association of the nucleic acid of squash leaf curl geminivirus with the whitefly Bemisia tabaci. Phytopathology 80, 850–856. doi: 10.1094/Phyto-80-850
Preston, C. A., Lewandowski, C., Enyedi, A. J., and Baldwin, I. T. (1999). Tobacco mosaic virus inoculation inhibits wound-induced jasmonic acid-mediated responses within but not between plants. Planta 209, 87–95. doi: 10.1007/s004250050609
Rubinstein, G., and Czosnek, H. (1997). Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 78, 2683–2689. doi: 10.1099/0022-1317-78-10-2683
Salzman, R. A., Brady, J. A., Finlayson, S. A., Buchanan, C. D., Summer, E. J., Sun, F., et al. (2005). Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 138, 352–368. doi: 10.1104/pp.104.058206
Sauge, M. H., Lacroze, J. P., Poëssel, J. L., Pascal, T., and Kervella, J. (2002). Induced resistance by Myzus persicae in the peach cultivar ‘Rubira’. Entomol. Exp. Appl. 102, 29–37. doi: 10.1046/j.1570-7458.2002.00922.x
Schilmiller, A. L., and Howe, G. A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8, 369–377. doi: 10.1016/j.pbi.2005.05.008
Stout, M. J., Thaler, J. S., and Thomma, B. P. (2006). Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 51, 663–689. doi: 10.1146/annurev.ento.51.110104.151117
Taggar, G. K., and Gill, R. S. (2016). Host plant resistance in Vigna sp towards whitefly, Bemisia tabaci (Gennadius): a review. Entomol. Gen. 36, 1–24. doi: 10.1127/entomologia/2016/0184
Tan, X. L., and Liu, T. X. (2014). Aphid-induced plant volatiles affect the attractiveness of tomato plants to Bemisia tabaci (Hemiptera: Aleyrodidae) and associated natural enemies. Entomol. Exp. Appl. 151, 259–269. doi: 10.1111/eea.12190
Tan, X. L., Wang, S., Ridsdill-Smith, J., and Liu, T. X. (2014). Direct and indirect impacts of infestation of Myzus persicae (Hemiptera: Aphididae) on Bemisia tabaci (Hemiptera: Aleyrodidae). PLOS ONE 9:e94310. doi: 10.1371/journal.pone.0094310
Thaler, J. S., Agrawal, A. A., and Halitschke, R. (2010). Salicylate-mediated interactions between pathogens and herbivores. Ecology 91, 1075–1082. doi: 10.1890/08-2347.1
Thaler, J. S., Fidantsef, A. L., and Bostock, R. M. (2002). Antagonism between jasmonate-and salicylate-mediated induced plant resistance: effects of concentration and timing of elicitation on defense-related proteins, herbivore, and pathogen performance in tomato. J. Chem. Ecol. 28, 1131–1159. doi: 10.1023/A:1016225515936
Thompson, G. A., and Goggin, F. L. (2006). Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 57, 755–766. doi: 10.1093/jxb/erj135
Verhage, A., van Wees, S. C. M., and Pieterse, C. M. J. (2010). Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol. 154, 536–540. doi: 10.1104/pp.110.161570
Keywords: TaqMan Real-Time PCR, systemic induced defense, Bemisia tabaci, Myzus persicae, electrical penetration graph, jasmonic acid, salicylic acid, signaling pathways
Citation: Tan XL, Chen JL, Benelli G, Desneux N, Yang XQ, Liu TX and Ge F (2017) Pre-infestation of Tomato Plants by Aphids Modulates Transmission-Acquisition Relationship among Whiteflies, Tomato Yellow Leaf Curl Virus (TYLCV) and Plants. Front. Plant Sci. 8:1597. doi: 10.3389/fpls.2017.01597
Received: 05 December 2016; Accepted: 31 August 2017;
Published: 22 September 2017.
Edited by:
Gero Benckiser, Justus-Liebig-Universität Gießen, GermanyReviewed by:
Yuanyuan Song, Fujian Agriculture and Forestry University, ChinaCopyright © 2017 Tan, Chen, Benelli, Desneux, Yang, Liu and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tong X. Liu, dHhsaXVAbndzdWFmLmVkdS5jbg== Feng Ge, Z2VmQGlvei5hYy5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.