
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 September 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01564
The sessile lifestyle of plants requires them to cope with stresses in situ. Plants overcome abiotic stresses by altering structure/morphology, and in some extreme conditions, by compressing the life cycle to survive the stresses in the form of seeds. Genetic and molecular studies have uncovered complex regulatory processes that coordinate stress adaptation and tolerance in plants, which are integrated at various levels. Investigating natural variation in stress responses has provided important insights into the evolutionary processes that shape the integrated regulation of adaptation and tolerance. This review primarily focuses on the current understanding of how transcriptional, post-transcriptional, post-translational, and epigenetic processes along with genetic variation orchestrate stress responses in plants. We also discuss the current and future development of computational tools to identify biologically meaningful factors from high dimensional, genome-scale data and construct the signaling networks consisting of these components.
Plants must mount appropriate responses to ever-changing environmental conditions, by altering growth and development through specialized metabolism, modifications in morphology, or changes in life history. Coordination of these responses is accomplished through multilevel regulatory processes, about which much is now known from investigations in Arabidopsis and emerging crop model systems. With the increasing availability of genomic tools, we can develop models of stress signaling networks across a broader range of species. Multilevel signal transduction processes are presented here that are not often viewed as an integrated system. This includes natural variation in regulatory processes that shape responses to major stressors, for example, drought, salt, and flooding. Regulatory mechanisms at the level of transcriptional regulation, alternative splicing and the rapid turnover/generation of regulatory proteins via ubiquitination and sumoylation shape complex networks that act to which in turn, modulate processes such as membrane transport to maintain cellular ion homeostasis, and chromatin remodeling. A consideration of data-driven modeling of stress signaling networks concludes our review. We suggest that the generation of massive datasets together with systematic analyses through careful database curation and integration of statistical and computational tools is necessary to enable accurate hypothesis building from these high-dimensional datasets.
Environmental variation has selected diverse responses among plant lineages, landraces, and wild crop relatives. This natural variation is an important tool for elucidating gene function without the confounding effects of expression outside the natural genomic context. Studies in natural variation have provided novel insights into evolutionary processes shaping stress responses as well as uncovering previously undescribed loci involved in stress responses. Predictably, whole genome duplications among Angiosperms and within lineage gene duplications have played an important role in shaping evolutionary patterns of stress response genes (van Veen et al., 2014). Altogether, exploring natural variation in stress response traits is uncovering important sources of genetic variation that improve our understanding of the coordinated regulation of these responses and are available to improve agronomic crops. While important insights have, and will continue to, come from studies in Arabidopsis, here we highlight insights gleaned from systems not traditionally considered models.
Natural genetic variation of drought responsive genes is found as both allelic variation at previously described loci as well as novel loci (Mickelbart et al., 2015; Zhang H. et al., 2016). For instance, in the wild tomato species Solanum pimpinellifolium, a screen of 94 genotypes uncovered 2 alleles for DREBA1 (drought-responsive element-binding A1) which together accounted for 25% of trait associated phenotypic variation (Rao et al., 2015). Further, patterns of allelic variation in transcription factors (TF)s and drought responsive genes have been shown to be important for selection and local adaptation (Muthamilarasan et al., 2015). Studying drought responsive Asr (ABSCISIC ACID STRESS RIPENING) gene family evolution in wild tomatoes, S. chilense and S. peruvianum, Fischer et al. (2011) found patterns of molecular evolution consistent with purifying selection at Asr1 and local adaptation for Asr4. Thus, natural populations provide a source of evolutionarily stable alternative alleles for adapting domesticated lines. While the tomato family is likely the most studied system for drought responses outside of Arabidopsis (Moyle and Muir, 2010), similar patterns are found across most crops (Zhang H. et al., 2016).
In addition to variation at known loci, wild crop relatives have enabled the identification of novel loci used to bolster crop plants (Mickelbart et al., 2015). For example, a new C2H2-type zinc-finger TF, GsZFP1, was identified from the soybean wild relative Glycine soja (Luo et al., 2011). Transgenic overexpression of GsZFP1 in alfalfa significantly increased the expression of drought responsive genes (Tang et al., 2013). Recently, quantitative trait locus (QTL) mapping in S. habrochaites, a drought-tolerant wild tomato revealed a QTL that co-localized to C2H2-type zinc-finger TFs on chromosome 9 of cultivated tomato (Arms et al., 2015). Additional analyses should reveal if these loci are homologous or represent convergent solutions to drought stress.
Salt stress is an important current and impending stress, evidenced by the estimated 18,000 patents granted which pertain to salt tolerance (Erskine et al., 2014). Natural variation in salt stress is mediated through common and novel genetic mechanisms which target sodium ion exclusion under saline growth conditions. For instance, a screen across wild wheat, Triticum monococcum, identified a locus Nax2 that, when introgressed into durum wheat (T. turgidum ssp. durum) increased yield by 25% on saline soils (James et al., 2006). Molecular characterization of this locus revealed a gene region TmHKT1;5-A encoding a Na+ transporter that is expressed in root tissue plasma membranes and thereby reduces xylem [Na+] (Munns et al., 2012). Further, the HKT (high-affinity potassium transporters) encoded in wild wheat relatives are orthologs of the HKT from Arabidopsis (AtHKT1;1) and rice OsHKT1;5 (Horie et al., 2009). Thus, variation in saline tolerance across monocot crops appears to be driven by ancestral polymorphism in the class 1 HKT.
Salt tolerance in soybean is, in part, governed by natural variation at a locus encoding a cation/H+ exchanger localized to the endoplasmic reticulum, GmSALT3 (Guan R. et al., 2014). While having a clearly defined sodium/proton exchanger domain, alignments revealed that this genic region shared ca. 59% identity with Arabidopsis AtCHX20 (Guan R. et al., 2014), underscoring the levels of gene duplication and reduced selection found for the CHX (cation:proton) antiporter family (Ye et al., 2013). Haplotypic analysis of GmSALT3 across diverse landraces and the wild soybean, G. soja, of nine haplotypes—two tolerant and seven sensitive—uncovered a single haplotype (H1) as the ancestral allele (Guan R. et al., 2014). Further, low nucleotide diversity and associated patterns of geographic occurrence revealed that salt-tolerant haplotypes are under strong selection in saline regions while alternative haplotypes were favored in lineages occupying lower saline environments (Guan R. et al., 2014). These molecular evolutionary patterns suggest that salt tolerance alleles are under strong selection and that variation results from relaxed selection on salt-sensitive alleles.
Natural variation in the cold acclimation associated C-repeat binding factor (CBF) gene family reveals complex patterns of evolution. In freezing tolerance, there are three important regulatory proteins CBF1, 2, and 3 that make up the so-called CBF regulon (Mckhann et al., 2008). In Arabidopsis CBF/DREB1 genes are arrayed in tandem as they are in wild tomato however, in a close relative, potato, there are additional copies (CBF4 and 5) which likely arose from a duplication of the ancestral cluster (Pennycooke et al., 2008). Investigating variation in CBF alleles across potato, the domesticated species (S. tuberosum) showed the physical linkage between CBF4 and CBF5, but CBF5 was missing in the wild species S. commersonii (Pennycooke et al., 2008). Further, comparing across five Solanum spp. lineages encompassing two groups (tomato and potato) only CBF3 and CBF5 formed distinct clades, independent of grouping, indicating that they are likely orthologs (Pennycooke et al., 2008). This suggests that substitutions in CBF1 and CBF2 are lineage specific and therefore obscuring orthologous relationships. This is supported by population level investigations within two species of wild tomato, S. peruvianum and S. chilense. Mboup et al. (2012) found that CBF3 showed significantly reduced nucleotide diversity across all populations/species consistent with the strong purifying selection at that locus. Interestingly, CBF2 showed patterns consistent with a trans-species polymorphism wherein two populations (one per species) revealed a haplotype structure with two diverged alleles, implying that balancing selection maintains polymorphism in CBF2 (Mboup et al., 2012). Finally, CBF1, as is the fate of most duplicated genes, was found to be a pseudogene in this group (Mboup et al., 2012). This complex evolutionary history for CBF genes shows the advantage of using natural variation to uncover gene function within the proper genomic context.
Flooding stress affects primary plant growth and development by disrupting, light interception, gas exchange, and therein reducing photosynthetic and aerobic respiration rates. These large-scale impacts have resulted in equally diverse plant responses that are centered on O2-sensing (Voesenek and Bailey-Serres, 2013). First discovered in rice (Oryza sativa), the group VII ERFs (ethylene response factor) are key regulators conserved across the roughly 130–200 million years divergence (Wolfe et al., 1989) between rice and Arabidopsis (Voesenek and Bailey-Serres, 2013). In fact, a recent phylogenetic analysis coupled with synteny across the highly conserved APETALA2 domain from whole genomes revealed that Angiosperm ERF-VIIs are derived from two ancestral loci, SBI and SBII which likely resulted from the duplication leading to Angiosperms (van Veen et al., 2014). Lineage-specific gene diversification in ERF-VII members, e.g., 15 members in rice and 5 in Arabidopsis, and high rates of nucleotide diversity suggest an important role for gene duplication and relaxed selection (outside of the highly conserved domains—APETALA2 and N-Terminus) have played an important role in the evolution of ERF-VII mediated flooding responses (van Veen et al., 2014).
Insights from domesticated and wild rice underscore the role of duplication in structuring regulatory elements of flooding responses. In rice (Oryza spp.), the locus Sub1 encodes one to three of the ERF-VII proteins (e.g., SUB1A, SUB1B, and SUB1C) which have diversified via duplication as indicated by orthology (Fukao et al., 2008). Patterns of allelic diversity further indicate lineage-specific evolution where Sub1 derived alleles phylogenetically cluster within lineage. For example, genome sequence analysis of nine Oryza species revealed that all of the rice genomes surveyed contain at least one SUB1-like gene, six of which possess both SUB1B- and SUB1C-like genes side by side on chromosome 9 as observed in domesticated rice (O. sativa; Dos Santos et al., 2017). SUB1A-like genes have been recognized only in limited accessions of O. sativa, O. rufipogon, and O. nivara; the presence of this gene was correlated with submergence tolerance in these species (Xu K. et al., 2006; Niroula et al., 2012). However, it appears that SUB1A is not essential for stress tolerance in some Oryza species because submergence-tolerant O. rhizomatis and O. eichingeri lack SUB1A (Niroula et al., 2012).
Exploring natural variation in stress responses has provided new insights into the multiple layers of regulatory processes coordinating stress responses across plant families. Further, the reciprocal insights from non-model and model systems are driving our understanding of the evolutionary processes that shape variation in stress responses. Characterizing regulatory processes shaping stress responses in non-model systems will continue to benefit from advances in high-throughput approaches. For instance, remote sensing of physiological status will allow screening of thousands as opposed to hundreds of individuals for tolerance traits. Further, novel sequencing approaches such as translating ribosome affinity purification (TRAP-seq; Reynoso et al., 2015), which captures the ‘translatome,’ will provide novel insights for post-translational regulation associated with abiotic stress responses.
Genetic and molecular studies have identified numerous TFs that are instrumental in the adaptation of plants to abiotic stresses. Functional characterization of key TFs that govern multiple signaling processes and directly regulate stress-responsive genes has contributed to dissecting intricate regulatory networks. In this section, we will focus on the representative TFs involved in drought, cold, heat, and flooding tolerance.
Abscisic acid (ABA) is a central signaling molecule activating adaptive responses to osmotic stress. Many ABA-responsive genes contain conserved ABA-responsive elements (ABREs) in their promoter regions (Fujita et al., 2013). ABRE-binding proteins/factors (AREBs/ABFs) are a subfamily of the basic leucine zipper (bZIP) family. These TFs activate the transcription of ABA-inducible genes through direct interaction with the ABRE motif. The Arabidopsis genome encodes nine AREBs/ABFs, four of which (AREB1/ABF2, AREB2/ABF4, ABF3, and ABF1) have been recognized as key TFs that regulate drought-responsive gene expression (Yoshida et al., 2014, 2015). Full activation of ABRE/ABF TFs requires multiple-site phosphorylation of their conserved region (Uno et al., 2000; Furihata et al., 2006; Fujii et al., 2007). Mutant and phosphoproteome studies suggested that ABRE/ABF proteins are the substrates of subclass III SNF1-related kinase 2 (SnRK2) such as SnRK2.2/SRK2D, SnRK2.3/SRK2I, and SnRK2.6/SRK2E, which are strongly activated by ABA (Yoshida et al., 2014).
NAC TFs are other players in ABA-dependent gene expression during drought. Arabidopsis ANAC019, ANAC055, ANAC072/RD26, and ANAC096 specifically bind to the NAC recognition sequence in the promoter region of stress-inducible genes to drive their expression (Tran et al., 2004; Xu et al., 2013). Overexpression of each of these NACs increases drought tolerance in Arabidopsis. In addition, ABA responsiveness and ABA-inducible gene expression are enhanced by constitutive expression of ANAC072/RD26 and ANAC096 (Fujita et al., 2004; Xu et al., 2013). Interestingly, ANAC096 directly interacts with ABF2 and ABF4, but not ABF3 (Xu et al., 2013). Moreover, anac096 abf2 abf4 triple knockout mutant plants exhibit reduced ABA sensitivity and osmotic stress tolerance compared with anac096 single mutant and abf2 abf4 double mutant plants. These results suggest that NAC and AREB/ABF TFs can cooperatively regulate expression of genes associated with ABA response and drought tolerance.
Drought-responsive element-binding proteins 2 (DREB2s) regulate drought-inducible gene expression in an ABA-independent manner (Yoshida et al., 2014). DREB2s physically interact with a conserved drought-responsive element (DRE) in the promoter region of drought-inducible genes. DREB2A is a key regulator for drought tolerance in Arabidopsis, but it also inhibits plant growth and reproduction. Therefore, the mRNA and protein accumulation of DREB2A are restricted via transcriptional and post-translational regulation under non-stress conditions. Growth-regulating factor 7 (GRF7) directly binds to the short promoter regions of DREB2A, suppressing its expression (Kim et al., 2012). In addition, DREB2A protein is ubiquitinated by ubiquitin E3 ligases, DREB2A-interacting proteins 1 and 2 (DRIP1 and 2) and subsequently degraded through proteasome-mediated proteolysis (Qin et al., 2008).
DREB1s/CBFs are major transcriptional regulators for cold acclimation (Lata and Prasad, 2011). In Arabidopsis, DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2 lie in tandem on chromosome 4. Although all the three DREB1s/CBFs are involved in acclimation responses to low temperature, they do not have fully overlapping functions. Time-course analysis of DREB1 expression revealed that DREB1C is induced later than DREB1A and DREB1B under cold (Novillo et al., 2004). This is consistent with the observation that DREB1C negatively regulates the expression of DREB1A and DREB1B under low-temperature stress (Figure 1A). Moreover, molecular analysis of DREB1A and DREB1B RNAi lines demonstrated that these two TFs are not involved in the regulation of other DREB1 genes in contrast to DREB1C (Novillo et al., 2007).
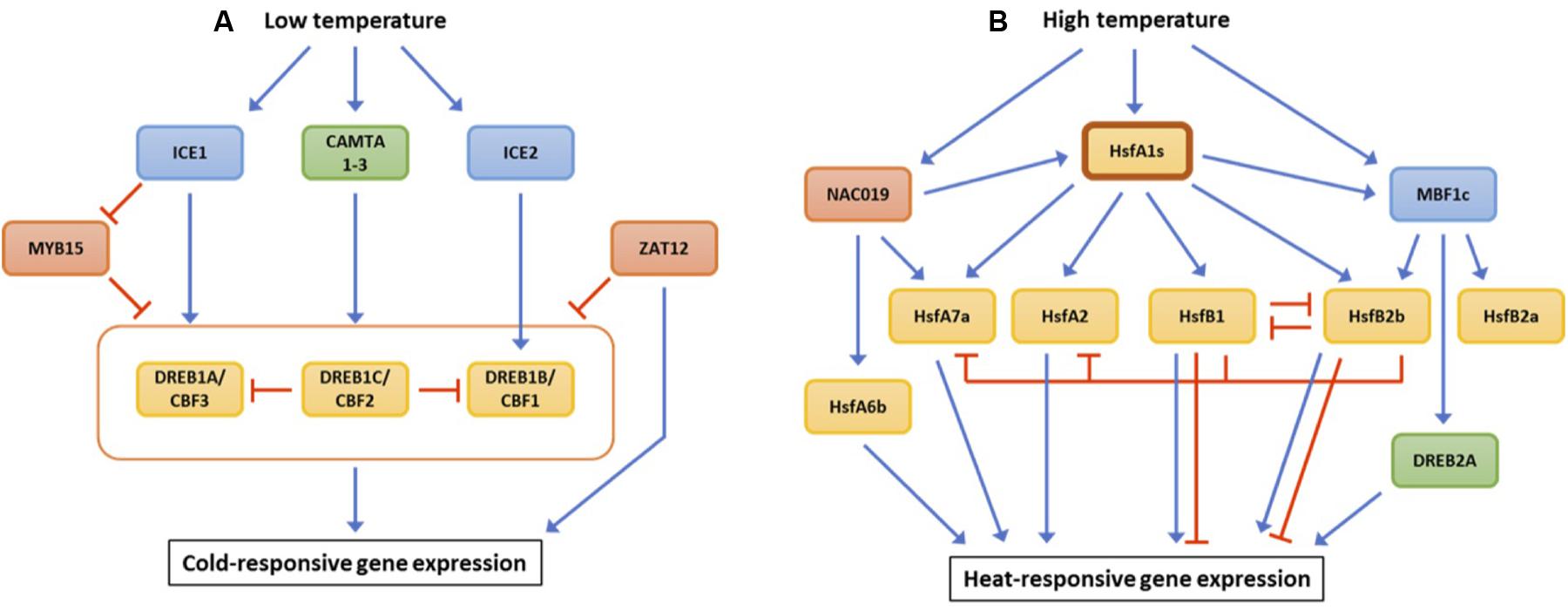
FIGURE 1. Transcriptional regulation of key transcription factors responsible for tolerance to low temperature (A) and high temperature (B). Blue and red lines represent up- and down-regulation of target gene expression, respectively.
Expression of DREB1A, DREB1B, and DREB1C are directly regulated by several upstream TFs (Figure 1A). Inducer of CBF expression 1 (ICE1) is a MYC-type bHLH TF that induces the transcription of all DREB1s (Chinnusamy et al., 2003). Another MYC-type bHLH TF, ICE2, activates the expression of DREB1B (Fursova et al., 2009). DREB1 genes are up-regulated in response to ABA although their induction levels are lower as compared to cold-induced gene expression (Knight et al., 2004). ICE1 may be responsible for the ABA-dependent expression of DREB1A. Indeed, loss-of-function mutation of ice1 repressed ABA-induced accumulation of DREB1A mRNA as compared to wild-type (Chinnusamy et al., 2006). There is an additional class of TFs, calmodulin-binding transcription activators (CAMTAs), which up-regulate the transcription of DREB1 genes. Studies of single and double camta mutants revealed that the three CAMTA TFs are required for the regulation of all three DREB1s (Doherty et al., 2009; Kim and Kim, 2013).
Some TFs negatively regulate the expression of the three DREB1 genes. MYB15 protein physically binds to the MYB recognition domains in the promoter regions of DREB1s, repressing their expression (Agarwal et al., 2006). Interestingly, expression of MYB15 is restricted by ICE1, suppressing MYB1-mediated down-regulation of DREB1s. ZAT12 is a C2H2 zinc-finger TF that regulates 24 cold-inducible genes, seven of which are members of the DREB1C regulon (Vogel et al., 2005). Despite its role in cold acclimation, Arabidopsis transgenic lines overexpressing ZAT12 showed reduced expression of all three DREB1s.
The expression of genes associated with heat tolerance is primarily regulated by heat shock transcription factors (HSFs). Of the HSFs identified to date, HsfA1s serve as central regulators that coordinate downstream TFs and other signaling components (Figure 1B). HsfA1 directly induces the expression of HsfA2 and HsfA7a, pivotal TFs activating heat-responsive genes in Arabidopsis (Charng et al., 2007; Yoshida et al., 2011). Members of another class of HSFs, HsfB1 and HsfB2b, are also up-regulated by HsfA1s. HsfB1 and HsfB2b negatively regulate the expression of HSF genes, HsfA2, HsfA7a, HsfB1, HsfB2b and several heat shock protein genes, suggesting the significance of HsfB1 and HsfB2b as signaling attenuators (Ikeda et al., 2011). HsfA1s directly enhance the expression of a non-HSF TF, MBF1c (Yoshida et al., 2011). MBF1c induces 36 different transcripts under heat stress, including HsfB2a, HsfB2b, and DREB2A (Suzuki et al., 2011). An Arabidopsis NAC TF, NAC019, physically interacts with the promoter regions of target genes, HsfA1b, HsfA6b, and HsfA7a, and up-regulates their expression (Guan Q. et al., 2014). In this manner, HsfA1s serve as regulatory hubs to orchestrate transcription factor networks consisting of HSF and other TF family genes.
Group VII of ethylene responsive factor (ERF)-type TFs (ERF-VIIs) are the best understood regulators of flooding and low oxygen tolerance (Fukao and Xiong, 2013). Xu K. et al. (2006) identified a highly submergence-inducible ERF-Vll gene, SUB1A, in a submergence-tolerant rice accession, FR13A. Introgression of SUB1A into intolerant genotypes significantly enhanced submergence tolerance. The major function of SUB1A under submergence is to limit carbohydrate consumption, amino acid metabolism, and elongation growth through hormonal regulation (Tamang and Fukao, 2015). A recent study revealed that SUB1A protein directly increases the expression of MAP kinase 3 (MPK3), whereas MPK3 phosphorylates SUB1A protein (a positive feedback loop) (Singh and Sinha, 2016). Mutant analysis of mpk6 suggested that phosphorylation of SUB1A is necessary for SUB1A-mediated submergence tolerance.
The Arabidopsis genome encodes five ERF-VII genes; HRE1, HRE2, RAP2.2, RAP2.3, and RAP2.12, all of which are involved in adaptation to submergence or low oxygen stress (Tamang and Fukao, 2015). Although these ERF-VII TFs up-regulate a similar set of hypoxia-responsive genes, transactivation studies suggest that RAP2.2, RAP2.3, and RAP2.12 are more powerful activators than HRE1 and HRE2 (Bui et al., 2015; Gasch et al., 2016). As observed in other stress-responsive TFs, the activity of RAP2.12 appears to be modulated by negative feedback loops. In fact, RAP2.12 increases the expression of a trihelix TF, hypoxia response attenuator 1 (HRA1), but HRA1 protein physically binds to RAP2.12 protein to inhibit its transactivation capacity (Giuntoli et al., 2014). In addition, HRA1 down-regulates the activation of its own promoter. Another layer of ERF-VII regulation is the N-end rule pathway of targeted proteolysis (Tamang and Fukao, 2015). This pathway consists of several protein-modifying enzymes, one of which requires molecular oxygen as a co-substrate (White et al., 2017). Under ambient oxygen concentrations, ERF-VII proteins are constitutively degraded through this pathway. However, low oxygen inhibits the oxygen-dependent reaction, leading to the escape of ERF-VII proteins from targeted proteolysis.
Molecular characterization of TF functions and interactions has advanced our understanding of how stress adaptation is orchestrated by transcriptional regulation. However, plant response and tolerance to abiotic stresses are coordinated through other processes such as epigenetic, alternative splicing, post-transcriptional, translational, and post-translational regulation. Thus, comprehensive analysis of TFs and other signaling components at various levels is crucial to uncover the integrated regulatory networks governing abiotic stress tolerance.
Alternative splicing (AS), the tissue, development and stress-dependent production of varying transcripts from a single gene, is a widespread phenomenon in plants (Reddy, 2007; Reddy et al., 2013). The process affects transcript stability, sequence, and subcellular localization of protein products. In this section, we will focus on the effect of splicing on plant abiotic stress responses.
Splicing events occur at the spliceosomal complex in the nucleus, which contains a variable population of RNA and protein molecules. They are regulated by specific splicing factors (SF) such as the serine-arginine (SR) and SR-like proteins (Carvalho et al., 2010), and supersensitive to abscisic acid and drought 1 (SAD1) (Cui et al., 2014), which channel signals to specific downstream pathways (Reddy, 2007), including, for example, plant responses to high light, heat and dehydration (Filichkin et al., 2010), and influences on circadian clock regulation of plant temperature responses (Seo et al., 2013; Filichkin et al., 2014).
Changes in the expression of SFs under specific conditions are determining factors in changes in AS patterns (Staiger and Brown, 2013), and subsequent phenotypic changes (Meyer et al., 2015). Current estimates are that up to 60% of multi-exon plant genes produce alternatively spliced variants (SVs) under different developmental or environmental conditions (Reddy et al., 2013). This number will increase as data accumulate for different experimental conditions, tissues, cell types (Efroni et al., 2016; Li et al., 2016), and plant species (Shen et al., 2014b; Thatcher et al., 2014). Many AS events in plants result in intron retention and the appearance of premature termination codons PTC (Reddy et al., 2013). Some of these truncated mRNA molecules are subjected to non-sense-mediated decay, (NMD), a mechanism that subjects targeted transcripts for degradation during the first round of translation (Reddy et al., 2013). NMD may regulate transcript abundance, however, it is important to note that other PTC-containing transcripts are not subject to NMD, suggesting a functional role for these non-coding transcripts, perhaps as dominant negative regulators. This has already been shown to be the case, for example, for an intron-containing JAZ10 SV, which acts as a negative regulator of jasmonic acid signaling (Chung et al., 2010). AS has been studied extensively with respect to responses to temperature, drought, and salt stress (Staiger and Brown, 2013). A key stress/ABA-related protein kinase, SnRK1, is regulated by a SF, SR45 (Carvalho et al., 2016). SR45 bound RNAs are enriched in stress and hormone related genes (Xing et al., 2015).
Alternative splicing also plays a role in temperature mediated effects on expression of the genes that control the circadian clock (Capovilla et al., 2015), a phenomenon known as a “molecular thermometer.” The splicing patterns of different SR pre-mRNAs are affected differentially by slight changes in ambient temperature, potentially affecting large populations of physiologically relevant SVs (Streitner et al., 2013). A study of the effects of thermal stress on the behavior of a key regulator of the circadian clock, CCA1, showed a change in transcripts with intron retention (Filichkin et al., 2014). The authors interpreted these results as the manifestation of a modulation of transcript abundance through changing ratios of SVs that are susceptible to NMD, and also to sequestration by the SR45 protein, compared to “functional” transcripts (Filichkin et al., 2014). In other work, SR45 has been implicated indirectly in AS of other circadian clock transcripts (Wang et al., 2012).
Data suggest that there is not a one-to-one correspondence between gene expression and alternative splicing events, and, therefore, it appears that this post-transcriptional process constitutes a distinct regulatory mechanism. In a study of the effects of drought imposition on AS in tissues of developing maize plants, it was found that drought imposition resulted in the appearance of a large number of novel SVs in ears, but a relatively small amount of gene expression changes (Thatcher et al., 2016). Furthermore, 77 SFs showed changes in gene expression over time, 46 showed differences in AS, but only 6 SFs showed both types of regulation, indicating that gene activation and alternative splicing are separate regulatory events. The expression level of a maize SF, PRP18, correlated well with drought-mediated AS across the different tissues studied, pointing to the central role of the specific expression of SF genes in AS-mediated cellular events.
Ding et al. (2014) conducted a transcriptomics study of the effects of varying concentrations of salt on AS in Arabidopsis. AS was enhanced by salt stress, with ca. 2000 AS events detected, compared with ca. 1300 such events detected in control plants. In contrast, Ding et al. (2014) reported that only 214 genes were differentially expressed (DE) under salt stress. There were differences among the over-represented categories between the DE and AS populations, with RNA processing related categories appearing as significantly enriched for AS events, while more general categories, such as “response to hormones” were enriched in the case of the DE genes. Included among the AS events were splicing of two SR pre-mRNAs, At-RSP41 and ATSCL33, where the SVs produced under salt stress did not retain introns that were present in the SVs present under control conditions. This result suggests that the alternative splicing induced by salt is an integral part of a salt response mechanism.
An intensive study of the effect of cold stress on the composition and cellular localization of SVs encoded by rice cyclophilin 19-4, (OsCYP19-4), suggests that isoforms lacking known functional domains may nonetheless have functional, and cell-specific, roles in stress responses (Lee et al., 2016). OsCYP19-4 had previously been shown by the authors to play a role in cold acclimation. In their 2016 study they showed that, under cold conditions, eight SVs encoded by OsCYP19-4 are present in rice seedlings, produced by combinations of intron retention and exon skipping. Only one of the SVs includes the protein phosphatase domain, which is associated with the known action of the encoded protein. The protein products of two of the SVs, which lacked the functional domain that confers protein phosphatase activity, were shown, nonetheless, to interact with a regulatory subunit of PP2A that is involved in the positive regulation of ABA signaling in guard cells. Proteins encoded by these two SVs were localized to guard cells and subsidiary cells, whereas the protein containing the known functional domain was detected at cell boundaries in all epidermal cells. This cellular “specialization” of the various protein products of the different SVs encoded by OsCYP19-4 strongly suggests unique functional roles for those transcripts that lack a known functional domain, in addition to the fully spliced SV.
Although much evidence has accumulated over the years, pointing to the importance of AS for abiotic stress responses, it has only recently been shown that a direct connection exists between specific splicing events and activation of the ABA signaling pathway (Ling et al., 2017). Using Pladienolide B, (PB), an inhibitor of the action of a SF in mammals, Ling et al. (2017), showed that PB treatment resulted in a mimicking of stress signals in Arabidopsis, as manifested in the appearance of 8000 genes with altered SVs, with a specific increase in intron retention, and a decrease in other forms of AS. Functional analysis of the responsive genes showed an enrichment in the categories of drought and salt stress, ABA responses, and RNA processing. Their data also showed that PB regulates the localization of SR45 within the nucleus. SR45 is a key SF protein, which acts as a negative regulator of ABA signaling (Carvalho et al., 2010). Furthermore, PB was shown not to bind to members of the ABA receptor protein family, eliminating the possibility that PB affects this phase of ABA signaling and strengthening the hypothesis of Ling et al. (2017) that the splicing mechanism itself is directly associated with abiotic stress responses, and ABA-related responses in particular.
Ubiquitin (Ub), a small protein with 76-amino acids that are conserved across all eukaryotic organisms, functions as a protein modifier to ubiquitylate a vast number of proteins – named ubiquitylation substrates. Quick switches of growth behavior during the plant stress responses rely on the removal of many preexisting regulatory proteins and the assembly of new ones. UPS is one of the primary mechanisms fulfilling this function—allowing plants to quickly respond, and adapt to ever-changing environmental cues. In this section, we will focus on the role of the UPS on plant abiotic stress tolerance.
Through ubiquitylation, the ubiquitylated proteins are often recognized by the 26S proteasome for degradation when they are modified by a chain of multi-Ubs that are linked together through one of 7 lysine (K) residues of Ub, primarily K48 and K11 (Kim et al., 2013). In addition to serving as a degradation signal, monoubiquitylation or polyubiquitination via other lysine residues of Ub, such as K63, can change the activity or localization of a ubiquitylation substrate (Komander and Rape, 2012).
The proteolytic function of the UPS can be sequentially separated into ubiquitylation and degradation stages. Ubiquitylation begins with the activation of Ub by an Ub activating enzyme (E1) through forming a high-energy thioester bond of its active Cys residue with the carboxyl Gly of Ub. The unstable Ub is easily transferred to a Cys residue on a Ub-conjugating enzyme (E2) by trans-esterification. Finally, the activated Ub on E2 is conjugated either directly using a Ub ligase (E3) or via an E3-Ub intermediate onto the 𝜀-amine group of a Lys residue on a ubiquitylation substrate or on another Ub that has been conjugated with the substrate (Hua and Vierstra, 2011). The specificity of this three-step enzymatic reaction is determined by the physical interactions between E3s and the ubiquitylation substrates. Consistent with the role of the UPS in plants in combating various stresses, the group of E3 members is extremely expanded in plants. Genomic studies estimated that ∼1,500 loci encode E3 proteins/subunits that are responsible for targeting an approximately equal number of ubiquitylation substrates (Kim et al., 2013). Based on the number of subunits, E3s are categorized as monosubunit and multisubunit enzymes. In Arabidopsis, there are 7 Homology to E6-ASSOCIATED CARBOXY-TERMINUS (HECT), 61 U-box, and 476 REALLY INTERESTING NEW GENE (RING) monosubunit E3s and ∼1,000 multisubunit E3s, many of which are CULLIN-RING (CRL) based. Based on the types of substrate receptors, there are three major groups of CRLs, SKP1-CULLIN1-F-BOX (SCF), BRIC-A-BRAC/TRAMTRACK/BROAD COMPLEX (BTB)-CULLIN3a/b, and DDB1-BINDING WD40 (DWD)-CULLIN4, which recognize their substrates through the protein products of 80 BTB (Gingerich et al., 2005), ∼900 F-box (Hua et al., 2011, 2013), and 85 DWD (Lee et al., 2008) loci, respectively, in Arabidopsis.
Since plants are constantly exposed to various stress conditions, it is not surprising that over the past decade many E3 ligase loci have been functionally characterized that are involved in responses/adaptations to many abiotic stresses (Supplementary Table 1). Of the characterized E3 ligase genes, 33 of 44 (75%) encode a monosubunit RING E3 ligase. It remains unknown if this result reflects the importance of RING E3 ligase genes in abiotic stress responses/adaptations. In the future, a study of the differential regulatory functions among different E3 ligase gene families is warranted in order to understand whether these differential functions are due to their different evolutionary mechanisms and/or evolutionary constraints.
The specificity of the ubiquitylation regulatory process resides in at least two proteins, the E3 ligase and the cognate substrate. Although genomic studies have predicted a significant number of E3 ligase genes and genetic characterization has identified a number of abiotic stresses that are mediated by protein ubiquitylation processes, the pairwise relationship between E3s and the ubiquitylation substrates remains ambiguous. For example, only a few ubiquitylation substrates have been detected in abiotic stress responses and/or adaptations (Supplementary Table 1). However, of those known ubiquitylation substrates, some are important transcription factors (e.g., ABI5 by KEG), epigenetic regulators (e.g., PRMT4b by PQT3), and enzymes involved in the metabolism and signaling transduction of the central stress hormone, ABA (e.g., PYL8 by COP10, PP2CA by RGLG1, and RGLG5 in ABA signaling: Supplementary Table 1 and Section 3: Transcriptional Regulation of Stress Responses). In the future, as more ubiquitylation substrates are characterized and the development of computational modeling of plant abiotic stress responses (see Modeling of Plant Abiotic Stress Responses), it may be possible to build a more comprehensive understanding regarding the regulatory cascades of abiotic stress signaling mediated by the UPS across multiple levels.
In addition to Ub, all eukaryotic cells also express a number of small Ub-like proteins, termed Ub-like protein modifier family (Ubl). Interestingly, Ub and Ubl proteins modify their substrates through three enzymatic reactions that are sequentially catalyzed by activating, conjugating, and ligating enzymes (Vierstra, 2012). The substrates modified by the SMALL UB-LIKE MODIFIERs (SUMOs) are the second largest group of proteins that are targeted for post-translational modification by a short peptide in all eukaryotes. Like ubiquitylation, sumoylation substrates could be modified by one or multiple single SUMO moieties or by a chain of SUMOs. It is not known whether the topology of SUMO chains also determines the final destination of the substrates. However, it has been noticed that a polySUMO chain can serve as a degradation signal for substrate turnover (Geoffroy and Hay, 2009). Specifically, the polySUMO chain is recognized and polyubiquitylated by SUMO-TARGETED UBIQUITIN RING-E3 LIGASEs (STUBLs) via the lysine residues of one or more SUMO moieties. The polyUb chain eventually drags the ubiquitylated and sumoylated substrates into the 26S proteasome for degradation. In Arabidopsis, there are 6 STUBLs (Elrouby et al., 2013). It is yet unclear whether and how the STUBLs are involved in abiotic stress signaling. Based on the proteolytic function of this process, it would be worthwhile to investigate the proteome-wide substrates to characterize the specific regulatory functions in this field. Although sumoylation can cause protein degradation through STUBLs, the majority of sumoylation substrates are stable and result in conformational changes upon the covalent attachment of SUMOs.
Although a full set of SUMO E1, E2, and E3 is present in eukaryotic cells, biochemical reconstitution studies suggested that E1 and E2 are sufficient to drive the sumoylation process both in vitro and in Escherichia coli cells (Elrouby and Coupland, 2010; Augustine et al., 2016), indicating a more general regulatory function of SUMO than Ub. Consistent with these findings, several proteome-wide studies discovered that rather than some specific proteins being targeted for sumoylation upon abiotic stress treatments, a wide range of sumoylation substrates are induced in Arabidopsis seedlings after a short period of cold, heat, or oxidative stress (hydrogen peroxide) exposure (Miura et al., 2007; Saracco et al., 2007; Golebiowski et al., 2009; Miller et al., 2013; Augustine et al., 2016). Interestingly, this massive increase in the pool of sumoylated proteins was rapidly de-sumoylated when plants were recovered from the stress. Further proteomic analysis revealed that heat shock stress only significantly increased the abundance of pre-existing 172 SUMO conjugates rather than modifying new targets (Miller et al., 2013), suggesting that protein sumoylation regulates plant stress physiology in a different manner as does protein ubiquitylation. The significant enrichment of sumoylation substrates for nuclear proteins involved in chromatin remodeling/repair, transcription, RNA metabolism, and protein trafficking further suggests that sumoylation may leverage many regulatory directions either negatively or positively in response to abiotic stresses (Miller et al., 2010).
In mammals, it is recognized that sumoylation can also change the functional status of specific substrates (Elrouby, 2017). For example, once sumoylated, the mammalian thymine DNA glycosylase is released from its bound DNA region then deSUMOylated by SUMO isopeptidase, which in turn allows the deSUMOylated form to further bind to other DNA regions to remove thymine moieties from G/T mismatches (Hardeland et al., 2002; Takahashi et al., 2005). Consequently, unlike ubiquitylation that often results in the turnover of substrates, sumoylation and de-sumoylation are reversible processes that can serve as regulatory switches to alter the function of their substrates. To date, specific substrates of abiotic stress-induced sumoylation have not yet been reported in plants, although proteome-wide data are clearly connected with protein sumoylation induced by heat, salt, and/or oxidative stresses (Supplementary Table 1). In the future, the discovery of such specific proteins will benefit the development of abiotic stress-tolerant crops through manipulating sumoylation pathway.
Maintenance of the cellular ion homeostasis under stress requires the tight coordination of numerous transmembrane proteins responsible for the transport of ions and water across cellular membranes (Figure 2A). In this section, we present examples of how post-translational modifications and control of subcellular localization contribute to stress adaptation.
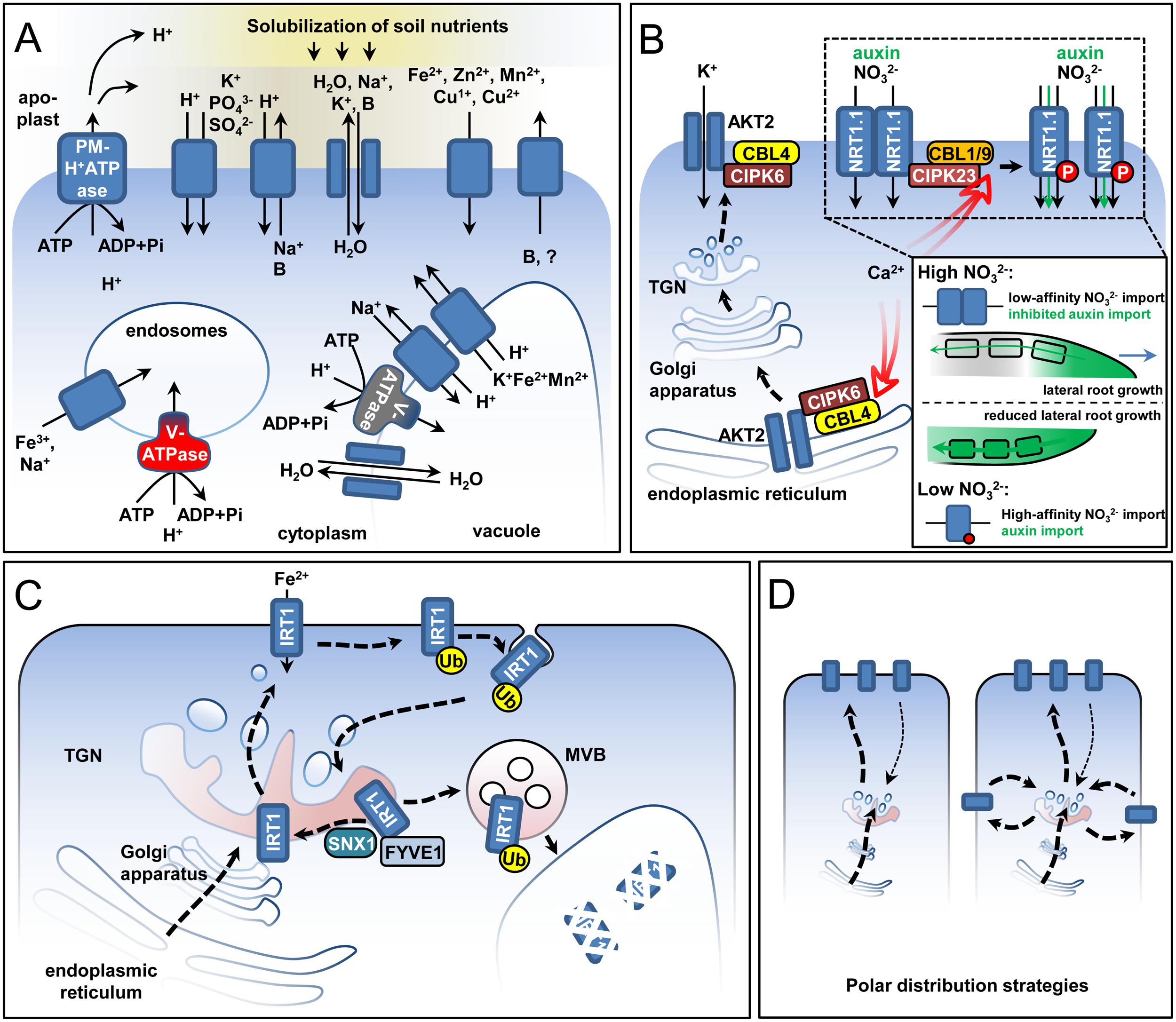
FIGURE 2. Post-transcriptional regulation of ion transport. Several principles of transport regulation are represented in idealized plant cells. (A) Ions and water are transported with the help of channels (drawn as adjacent shapes with arrows between them) and carriers. Certain carriers transport ions along the concentration gradient (shapes with a single arrow), while others are energized by functioning as proton symporters or antiporters (shapes with two arrows). The required proton gradients are generated by different membrane proton pumps with the help of ATP hydrolysis. (B) Ca-dependent kinases as activators of transport across the plasma membrane. Two examples are shown: the switch of NRT1.1 from low- to high-affinity state through phosphorylation and the translocation of the AKT2 channel from the endoplasmic reticulum to the plasma membrane with the help of the kinase-containing complex but in the absence of phosphorylation. The insert represents the developmental effect from the long-term activation of the NRT1.1 transporter. High-affinity NRT1.1 can transport auxin, promoting auxin depletion from the lateral root tip and inhibiting lateral root growth, thus directly affecting root architecture under nitrogen starvation. (C) Intracellular trafficking of the Fe2+ transporter IRT1. IRT1 is translocated to the plasma membrane, from where it can be ubiquitinated and endocytosed back to the trans-Golgi Network (TGN). In a sorting step at the TGN, the protein is either targeted for vacuolar degradation through the multivesicular body (MVB), or is recycled and retargeted toward the plasma membrane with the help of the SNX1 and FYVE1 proteins. (D) Polar localization of transporters can be achieved by either direct polar targeting (left image) or by the inhibition of transporter endocytosis at certain membrane regions causing depletion from other membrane domains (right image). An example for the former case is the polar targeting of IRT1 in the absence of its secondary substrates (Zn2+, Mn2+, and Co2+) (Barberon et al., 2014) and for the latter is the localization of the boron transporter NIP5;1 (Takano et al., 2010; Wang et al., 2017). In the case of IRT1, the polarization is environmentally driven as it depends on the external soluble ion concentrations.
Phosphorylation/dephosphorylation events are key for rebalancing cellular ion homeostasis in response to stress in plants. Multiple proteins involved in the translocation of ions have been identified as kinase/phosphatase targets. A classic example of signaling-affected transporter activity is the plasma membrane (PM) Na+/H+ antiporter SALT OVERLY SENSITIVE 1/Na+/H+ EXCHANGER 7 (SOS1/NHX7). Under high salinity conditions, an excess of cytoplasmic Na+ is potentially toxic for the plant. As a response, Na+ is either transported out to the rhizosphere or stored in the vacuole. SOS1 is kept inactive under unstressed conditions by a C-terminal autoinhibitory domain (Shi et al., 2000; Quintero et al., 2011). Under Na excess, a signaling-induced calcium transient is perceived by Calcineurin-B like 4 (CBL4/SOS3), which interacts and activates CBL-INTERACTING PROTEIN KINASE 24 (CIPK24/SOS2) (Qiu et al., 2002; Quintero et al., 2002). The CBL4-CIPK24 couple phosphorylates serine 1138 within the SOS1 autoinhibitory domain leading to the activation of Na efflux (Quintero et al., 2011). A similar regulatory principle, with Ca as a secondary messenger perceived by Ca-binding effector proteins, such as the CBL-CIPK system for example, was uncovered for other transport proteins under stress (Schulz et al., 2013; Steinhorst and Kudla, 2013). The CBL1-CIPK23 and CBL9-CIPK23 couples mediate the activation of the ARABIDOPSIS K+ TRANSPORTER 1 (AKT1), a channel for potassium uptake in plants (Li et al., 2006; Xu J. et al., 2006) in addition to controling the conversion of the NRT1.1 transporter from a low- to a high-affinity state under nitrogen limitation (Liu and Tsay, 2003; Ho et al., 2009). In the case of NRT1.1, phosphorylation of threonine 101 by CIPK23 switches the transporter from a low-affinity dimer to a high-affinity monomeric state (Liu and Tsay, 2003; Ho et al., 2009; Parker and Newstead, 2014; Sun J. et al., 2014) (Figure 2B). Mutant analysis has shown that NRT1.1 acts as both transporter and receptor for NO3-. Threonine 101 phosphorylation also affects the receptor function, as the two forms elicit different responses (Ho et al., 2009; Bouguyon et al., 2012). NRT1.1 plays an important role in long-term adaptation responses during N starvation, since it is able to import auxin, a process inhibited by high NO3- concentrations (Krouk et al., 2010; Mounier et al., 2014). Thus, NRT1.1 modulates auxin transport in lateral roots and its activity alters root architecture in response to external NO3- availability. Classical nitrogen deficiency root development effects are dependent on the phosphorylated NRT1.1 form (Bouguyon et al., 2015).
Phosphorylation can also affect the activity of transport proteins by changing their subcellular distribution. The exit of the phosphate transporter PHT1;1 from the endoplasmic reticulum (ER) depends on the phosphorylation status of its serine 514 residue. A phosphomimicking PHT1;1 mutant was retained in the ER, while a phospho-null form showed a predominant PM/endosome localization (Bayle et al., 2011). The potassium channel AKT2 requires the CBL4-CIPK6 couple for ER exit and PM localization (Figure 2B). Surprisingly, AKT2 is not phosphorylated in the process and the binding to CIPK6 is sufficient for the translocation (Held et al., 2011). Such pools of PM proteins at the ER have been observed in other cases as well and might represent a common mechanism assuring fast responses to external stimuli (Zelazny et al., 2007; Ivanov and Gaude, 2009; Bleckmann et al., 2010).
Metabolic enzymes might also influence PM transport through direct protein–protein interaction. In a particularly intriguing case, the high-affinity PM sulfate transporter SULTR1;2 was found to interact with the cytoplasmic enzyme O-acetylserine (thiol)lyase (OASTL), responsible for the incorporation of the sulfur into cysteine. The interaction inhibits the transporter but activates the OASTL activity, thus creating a module for the coordination of the intracellular sulfur levels balancing import and fixation under sulfur limitation (Shibagaki and Grossman, 2010). Interestingly, based on the analysis of new mutant alleles, SULTR2;1 has been proposed to function as a receptor, similar to NRT1.1 (Zhang et al., 2014).
A key element in balancing ion fluxes across the PM is the activity of the PM H+-ATPases. The proton pumping activity of these proteins is critical because it is used for energizing the transport process and because minerals, such as iron, can only be solubilized, and thus made available to the plant, by acidification of the rhizosphere (Brumbarova et al., 2015) (Figure 2A). Therefore, mutants with decreased PM H+-ATPase activity are not able to survive in alkaline soils (Fuglsang et al., 2007). In Arabidopsis there are 11 genes encoding PM H+-ATPases (Palmgren, 2001) and many of them respond to stress conditions (Colangelo and Guerinot, 2006; Janicka-Russak and Klobus, 2007; Janicka-Russak et al., 2008; Ivanov et al., 2012; Mlodzinska et al., 2015). Among these, AHA2 was found to be the dominant form in the root and, in addition to a response at the gene expression level, its activity is modulated post-translationally (Fuglsang et al., 2007). Activation of AHA2 depends on its interaction with a 14-3-3 protein. The ATPase can be inactivated by CBL2-CIPK11-mediated serine 931 phosphorylation, which inhibits this interaction. Consistent with this, CIPK11 loss-of-function plants display higher acidification capacity and perform better under alkaline conditions (Fuglsang et al., 2007). Multiple other sites within the AHA2 C-terminal cytoplasmic region were found to be phosphorylated in response to different environmental and developmental cues (Haruta et al., 2010; Fuglsang et al., 2014; Veshaguri et al., 2016).
The coordination of nutrient partitioning within the cell is a critical factor for stress adaptation. The endosomal localization of different NHX-family transporters suggests that upon uptake, cytoplasmic ions are sequestered in the endomembrane system, preventing toxicity (Bassil et al., 2011). Indeed, expression of the trans-Golgi network (TGN)-localized NHX5 transporter improved salt tolerance in both mono and dicotyledonous species (Shi et al., 2008; Li et al., 2011a,b). While the mechanism behind this effect is not yet clear, it is proposed that the TGN-localized NHX proteins function in TGN-to-vacuole protein trafficking (Ashnest et al., 2015). There are indications that correct TGN-to-vacuole trafficking of material is crucial for maintaining cellular homeostasis under salt stress, however, the strategies of plants might differ. In Arabidopsis, salt tolerance involves the inhibition of vacuolar trafficking through the depletion of the tonoplast-localized v-SNARE protein VAMP711 (Leshem et al., 2006, 2010), while in rice (O. sativa) this trafficking step is enhanced through the expression of the genes encoding the RAB GTPase OsRAB11 and the GTPase-activating protein OsGAP1 (Son et al., 2013; Pizarro and Norambuena, 2014). The vacuole is a major storage compartment for ions. Multiple transport systems exist to partition ions and water on both sides of the tonoplast (Figure 2A). Several studies have shown that the activation of such transporters under stress involves Ca signaling. Similarly, the calcium-dependent kinase CPK3 phosphorylates the K+ channel TPK1, thus promoting its activation by the 14-3-3 protein GRF6 (Latz et al., 2013). An interesting example is the CIPK24/SOS2 protein kinase responsible for the activation of, among others, the vacuolar Na-/H+ antiporter NHX1 (Qiu et al., 2004) and Ca2+/H+ antiporter CAX1 (Cheng et al., 2004). CIPK24 thus represents an example of coordination of PM and tonoplast transport as it also regulates Na+ import at the PM (discussed above). Another example is the ABA-activated SnRK2 kinase OST1, which has several targets including the PM-localized AtSLAC1 and the vacuolar AtCLCa anion transporters to coordinate stomata closure in response to environmental change (Vahisalu et al., 2010; Wege et al., 2014).
Regulation of proton gradients by the proton pumping activity of vacuolar pyrophosphatases (V-PPase), primarily, the vacuolar ATPases (V-ATPase), is key for the function of many transporters at the endomembranes and the tonoplast. V-ATPase is a multisubunit complex localized throughout the endomembrane system with the exact complex composition varying among compartments (Neuhaus and Trentmann, 2014). It was shown that cold acclimation resulted in the increased abundance of V-ATPase subunits at the tonoplast, consistent with increased proton pumping activity (Schulze et al., 2012). CIPK24 was shown to interact strongly and phosphorylate the V-ATPase, thus activating it under salt stress (Batelli et al., 2007). Interestingly, however, the loss of the V-ATPase activity at the TGN, but not at the tonoplast, makes Arabidopsis hypersensitive to salt stress, indicating that the TGN has an important role for reaction and adaptation to stress (Krebs et al., 2010).
Plasma membrane-localized proteins undergo constant cycles of endocytosis and PM retargeting. At the TGN, endocytosed transporters can be either recycled, or sent for degradation. Recycling was shown to be crucial for the survival of Arabidopsis under iron limitation (Figure 2C). In the absence of the endosomal sorting protein SORTING NEXIN 1 (SNX1), the principal iron transporter IRT1 fails to recycle and is instead degraded, leading to failure of snx1 mutant plants to cope with iron deficiency (Blum et al., 2014; Ivanov et al., 2014). SNX1 and its interaction partners are regulated in response to different environmental cues, suggesting that protein sorting at the TGN is stress-sensitive (Brumbarova and Ivanov, 2016). The plant-unique ESCRT subunit FYVE1/FREE1 was shown to interact with IRT1 and promote its recycling (Barberon et al., 2014). In contrast, transporters which have been marked by ubiquitination are targeted via the multivesicular bodies (MVB) for vacuolar degradation (Figure 2C). Ubiquitination has been shown to affect the availability of IRT1 and the boron transporter BOR1 (Barberon et al., 2011; Kasai et al., 2011; Shin et al., 2013). A ubiquitination-defective form of IRT1 remains at the PM and plants expressing it suffer due to metal hyper accumulation (Barberon et al., 2011). Endomembrane trafficking, in particular clathrin-mediated endocytosis, is an important way of coordinating the directional ion transport (Takano et al., 2010; Barberon et al., 2014; Wang et al., 2017) (Figure 2D). The boron importer NIP5;1 and exporter BOR1 are polarly distributed on opposite domains of the epidermis cell thus ensuring that imported boron will be transported to inner root tissues (Takano et al., 2010).
Understanding the mechanistic complexity of transporter regulation in response to environmental stress is of great importance since common regulatory principles exist. However, investigating case-specific differences will allow us to understand how adaptation to stress is achieved. A widely-used strategy of improving plant capacity to respond to abiotic stress in laboratory conditions is the overexpression of transporters. However, many transporters were found to a have broad substrate range (Korshunova et al., 1999; Corratge-Faillie and Lacombe, 2017). Thus, in field conditions overexpression might cause unwanted accumulation of additional compounds to potentially toxic levels. Therefore, steps, such as the modulation of regulatory and signaling components, might be necessary to rebalance the intracellular partitioning of imported compounds. Alternatively, it was shown that transporter substrate specificity could be manipulated by exchanging key amino acids (Rogers et al., 2000). While the underlying mechanism remains unclear, increasing transporter selectivity might be a strategy to circumvent some of the crop improvement problems and create targeted solutions for specific abiotic stresses. It remains to be seen, however, whether such strategies will be successful in the dynamic and complex environment outside the laboratory.
Chromatin, which is defined as DNA wrapped around histone proteins, plays a major role in allowing or blocking transcriptional response to abiotic stress. Histone-modifying enzymes have been shown to directly regulate transcription by modulating the histone marks of stress responsive genes. In this section, we will assemble the existing information on chromatin and highlight the possible role of histone variants and histone modifications in stress responses.
The basic organization of DNA wrapped around protein units is called the nucleosome (Luger et al., 1997). The nucleosome is an octamer formed by two copies of each of the histone subunits, H2A, H2B, H3, and H4, and is associated with 146 bp of DNA (Arya and Schlick, 2009; Zhou et al., 2013; Asensi-Fabado et al., 2017). At the edge of the nucleosome, the histone H1 is linked to the DNA region responsible for connecting nucleosomes (Kim et al., 2015; Asensi-Fabado et al., 2017). Specific histone variants such as H2A.Z, H3.3, and CenH3 can be recruited to the nucleosome during specific developmental stages or in response to environmental stimuli (Deal et al., 2007; Zhang K. et al., 2007; Coleman-Derr and Zilberman, 2012).
In eukaryotes, basic amino acids such as lysine and arginine distributed on the N-terminal tail can be reversibly modified by the addition of different chemical groups that ultimately alter chromatin compaction and DNA accessibility (Jenuwein and Allis, 2001). Based on DNA accessibility, chromatin is classified as euchromatin (lightly packed) or heterochromatin (heavily packed). Altogether the structure of the chromatin is also referred as the “histone code” and includes a wide range of chemical modifications, such as methylation, acetylation, phosphorylation, and ubiquitination (Kouzarides, 2007). Each modification has been linked to various biological processes such as, DNA replication, transcription, repair and chromosome condensation (Kouzarides, 2007).
In addition to histone modifications, DNA can be chemically modified by the addition of methyl groups to cytosines (C) in a symmetric or asymmetric context (CG, CHG, and CHH). Enrichment in DNA methylation occurs in the centromeric and pericentromeric regions of the chromosome where many transposable elements (TEs) are located and their transcription is prevented (Finnegan et al., 1998; Tariq and Paszkowski, 2004). Silencing mechanisms involving DNA methylation include synthesis of short interfering RNAs (small RNAs) as well as histone modifications such as H3K9me2 (Mathieu et al., 2005; Zhou et al., 2010). Exposure of plants to high temperature leads to the activation of transposable elements (Pecinka et al., 2010; Ito et al., 2011). Such activation is not due to loss in DNA methylation but is caused by heterochromatin de-condensation (Pecinka et al., 2010). Furthermore, the activity of retrotransposons after heat exposure can be trans-generationally inherited when production of sRNAs is compromised (Ito et al., 2011), suggesting the importance of sRNAs biogenesis during the resetting process in the germ line.
Plant histone family contains a number of variants with small differences in amino acid sequence and structure, resulting in changes in affinities for DNA or histone binding proteins. The most characterized histone variants belong to the H3 and H2A families (Probst and Mittelsten Scheid, 2015). In Arabidopsis histone H3 is present in two variants: H3.1 and H3.3 which differ by four amino acids (Shi et al., 2011). Histone H2A variants are instead H2AX, H2AZ, and H2AW, with H2AW playing a major role in silencing heterochromatin (Talbert and Henikoff, 2010; Yelagandula et al., 2014). H3.3 and H2AZ have been shown to be involved in active transcription. ChIP-seq experiments indicated that H2AZ deposition occurs at the first nucleosome after the Transcriptional Start Sites (TSS) and in regions that are low in DNA methylation (Zilberman et al., 2008). H3.3 containing nucleosomes are enriched in gene bodies as well as in a subset of promoter regions (Deal and Henikoff, 2011; Shu et al., 2014).
In plants, histone variants can be stress-inducible, suggesting that environmental stress signals can alter chromatin structure by replacing H3 and H2A with one of their variants (Zhou et al., 2013). For example, deposition of H2A.Z across gene bodies was positively correlated with gene responsiveness, either among different tissues or in response to different biotic or abiotic stimuli (Coleman-Derr and Zilberman, 2012). Further, recent studies have shown that H2AZ can positively or negatively regulate transcription based on its accumulation in gene bodies or on TSS (Sura et al., 2017). Concordantly, the results of Kumar and Wigge (2010) revealed that H2A.Z is important in regulating responses to heat and cold stress (Kumar and Wigge, 2010). Using a forward genetic screen approach, nucleosomes containing the H2A.Z variant were found to be essential for temperature perception (Kumar and Wigge, 2010). Transcriptome analysis of plants without correct H2A.Z incorporation into chromatin displayed a constitutive up-regulation of genes induced by warm temperature (27°C), when the plants were grown at 12°C (Figure 3) (Kumar and Wigge, 2010). A ChIP profile of H2A.Z on the HSP70 gene showed eviction of H2A.Z during exposure to high temperatures at transcriptional start sites. Lack of H2A.Z allows RNA Polymerase (POL II) to initiate transcription. Therefore, failure of H2A.Z incorporation leads to a constitutively high expression of genes induced by heat (Figure 3).
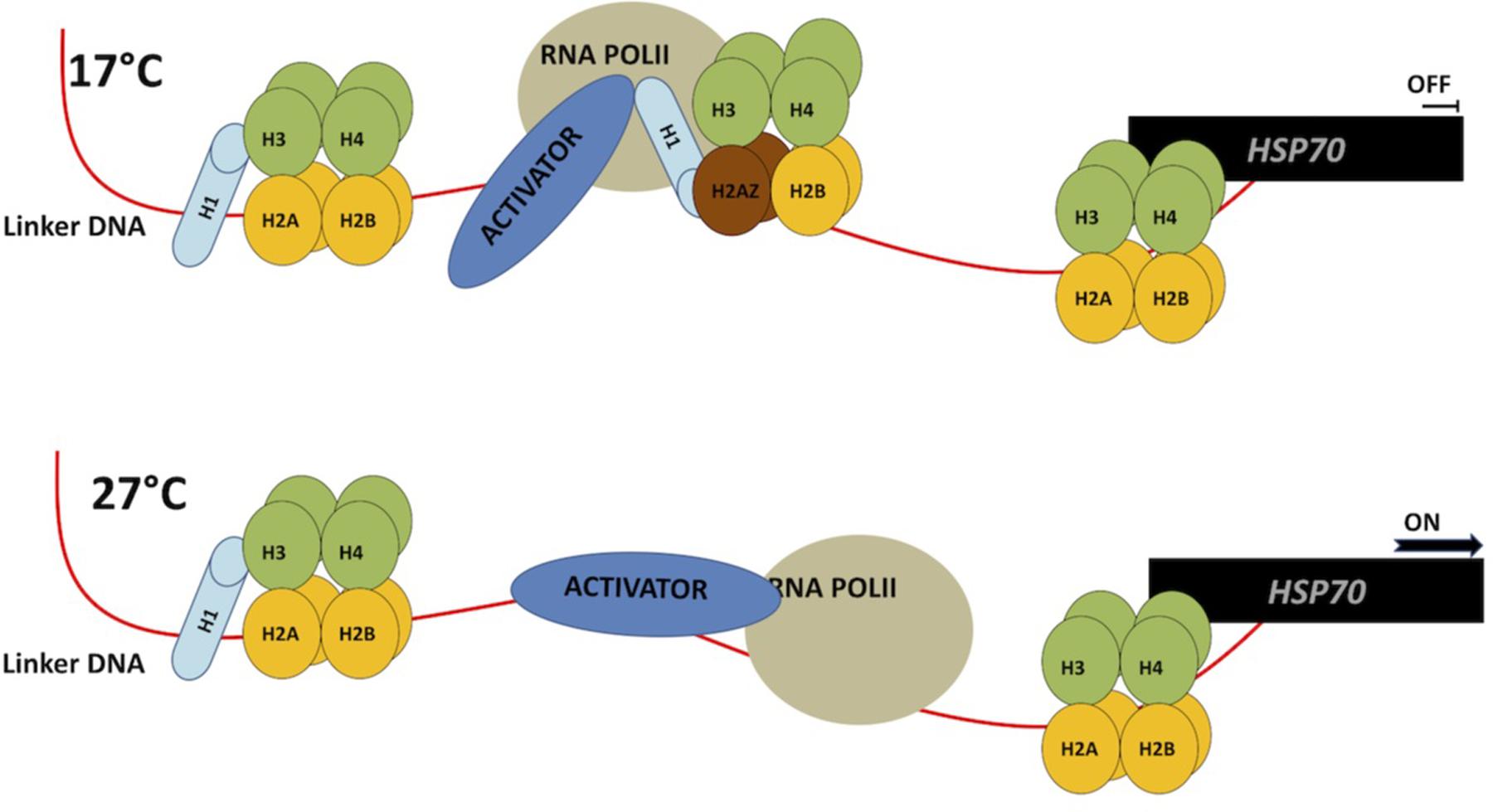
FIGURE 3. H2AZ modulates gene expression during temperature perception. At low temperature (17C) H2AZ-containing nucleosomes have high occupancy and prevent HSP70 transcription by blocking RNA PolII and transcriptional activator progression (top). At high temperature H2AZ occupancy is reduced therefore allowing increase in HSP70 gene expression (bottom). This model in based on the data shown in Kumar and Wigge (2010).
In Arabidopsis, histone H1 has three histone variants: H1.1, H1.2, and H1.3 (Wierzbicki and Jerzmanowski, 2005). The first two are present under unstressed conditions whereas H1.3 is induced by ABA and water stress (Ascenzi and Gantt, 1997). Further studies showed that H1.3 expression is also regulated by combination of low light and ABA (Rutowicz et al., 2015). Under low light intensity, H1.3 protein was predominantly localized to guard cells. Bisulfite sequencing revealed an increase in total DNA methylation in h1.3 mutant plants compared to wild type, mostly under low light conditions (Rutowicz et al., 2015). When low light was combined with drought, h1.3 plants showed a higher leaf number and fresh/dry weight than wild type (Rutowicz et al., 2015).
Plant histone modification sites have been identified by mass spectrometry and biochemical assays (Earley et al., 2007; Zhang X.Y. et al., 2007). Histone acetylation is often considered a positive regulator of transcription by allowing access to the RNA polymerase and transcription factors (Kuo et al., 1996; Zhang et al., 1998; Shahbazian and Grunstein, 2007). Conversely, de-acetylating histones increase the affinity between DNA and histones, thereby reducing gene expression. (Kadosh and Struhl, 1998; Rundlett et al., 1998; Chen et al., 2010; To et al., 2011). Histone acetylation is modulated by histone modifying enzymes such as histone acetyltransferases (HATs) and histone deacetylases (HDACs). The majority of them have been identified in different plant species (Chen et al., 2010; Papaefthimiou et al., 2010; Pontvianne et al., 2010; Aquea et al., 2011; Huang et al., 2011; Aiese Cigliano et al., 2013). Among the different HATs and HDACs found to be able to alter acetylation within the H3 and H4 tails, some have been indicated as key players by modulating gene expression in response to abiotic stress.
In Arabidopsis, the histone acetyltransferase GCN5 forms a complex with transcriptional co-activators ADA and Spt-Ada-Gcn5 acetyltransferase (SAGA) (Vlachonasios et al., 2003). Ada2b and sgf29 mutants displayed hyposensitivity to salt, with reduced expression of salt-responsive genes such as RESPONSIVE TO ABA18 (RAB18), COLD-RESPONSIVE 6.6 (COR6.6) and RESPONSIVE TO DESSICATION29B (RD29B). ChIP-PCR experiments indicated a drastic overall reduction in histone acetylation for RAB18 and COR6.6, whereas only H3K9/K14Ac residues was affected on RD29B (Kaldis et al., 2011). Overall, the data indicates that ADA2b is a putative positive regulator of stress response by acetylating salt-induced genes (Kaldis et al., 2011).
Analysis of mutants for histone deacetylase 6 (HDA6) showed hyposensitivity to ABA and NaCl during germination (Chen et al., 2010). ChIP-PCR on stress-induced DREB2A and RESPONSIVE TO DESSICATION29A (RD29A) genes indicated loss of hyper-acetylation in hda6 correlated with a lower expression than wild type (Chen et al., 2010). HDA6 has been shown to interact with another type of HDACs: HD2C (Luo et al., 2012b). Similarly to HDA6, loss of function for HD2C also resulted in a hypersensitive response to ABA and NaCl. This response was correlated with an increase in gene expression as well as in histone acetylation for ABA responsive genes ABA INSENSITIVE1 and 2(ABI1 and ABI2) (Luo et al., 2012a). Interestingly, over expression of HD2C led to a hyposensitive response to ABA and NaCl during germination (Sridha and Wu, 2006), postulating that such a response could depend on ABI1 and ABI2 expression levels. Recently, using pull-down approach, HD2C has been found to interact directly with several members of chromatin remodeling complexes (CRCs), including SWITCH SUBUNIT3 (SWI3B) (Buszewicz et al., 2016). HD2C expression was found to be up-regulated after heat stress and mutants for H2DC and for BRAHMA (BRM), another protein found to be part of HD2C complex, showed similar phenotypes when subjected to high temperature (Buszewicz et al., 2016). Transcriptional analysis of hd2c-3 and brm1 after heat treatment shows a subset of co-regulated genes, including HSP101 and ROTAMASE FKBP1 (ROF1) (Buszewicz et al., 2016).
Unlike HDA6, hda9 mutants showed hyposensitivity to PEG or salt during germination when compared to wild type (Zheng et al., 2016). This phenotype was correlated with an increased expression and histone hyper-acetylation for genes involved in water stress (Zheng et al., 2016). In many organisms, histone deacetylases form multi-protein complexes (Carrozza et al., 2005). The yeast histone deacetylase REDUCED POTASSIUM DEPENDENCY (RPD3) complex includes co-repressors SWI-INDEPENDENT3-like (SIN3-like) and different histone binding proteins. Some other proteins of unknown functions such as REGULATOR of TRANSCRIPTION 2 and 3 (RXT2 and RXT3) co-eluted within the complex. The Arabidopsis homolog of RXT3, named HISTONE DEACETYLASE COMPLEX1 (HDC1) was found to interact directly with histone deacetylases HDA6 and HDA19 (Perrella et al., 2013). Similarly to hda6 and hda19 mutants (Chen et al., 2011), hdc1-1 seedlings showed hypersensitivity to ABA and NaCl (Perrella et al., 2013). Overexpression of HDC1 led to a reduction in ABA and NaCl sensitivity, and to an increase in biomass (Perrella et al., 2013) (Figure 4). At the transcriptional level, when treated with salt, hdc1-1 plants showed a greater induction of stress-responsive genes like ABA DEFICIENT3 (ABA3), RD29A and RD29B. Genes like PYR1-LIKE4 (PYL4) and DROUGHT-REPRESSED4 (DR4) which are usually down-regulated in response to osmotic stress were up-regulated in hdc1-1 under control conditions. Because no phenotypes have been reported when histone deacetylases are over-expressed, it is likely that HDC1 can function as a rate-limiting component of the HDA complex (Figure 4).
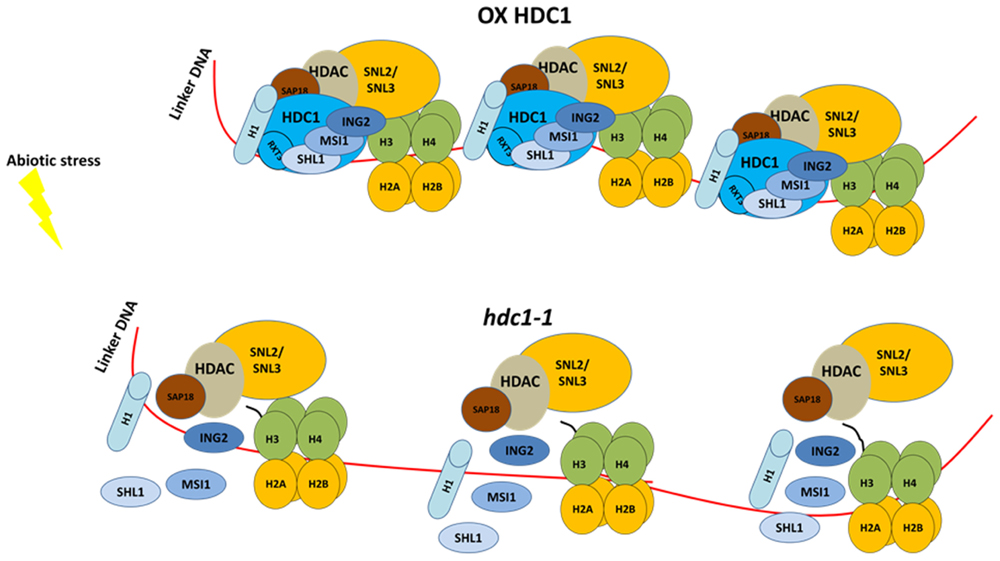
FIGURE 4. HDC1 is rate limiting component of HDAC complexes. During stress response, HDC1 over expression stabilizes the complex by maintaining a tighter association with DNA and chromatin and therefore enhancing the HDAC activity. Plants overexpressing HDC1 are less sensitive to salt and ABA during germination and have an increased growth in stressed conditions (top). Lack of HDC1 de-stabilizes the complex and allows increase of transcription of stress responsive genes (see text). Hdc1-1 loss of function phenocopies mutants for HDAC like hda6 and hda19. Hdc1-1 plants are hypersensitive to NaCl and ABA during germination and display a reduced growth under salt conditions (bottom). This model is based on the data shown in Perrella et al. (2013, 2016).
Consistent with this hypothesis, hdc1 mutants also showed an increase in histone acetylation levels with respect to wild type at the whole chromatin level, as well for single genes such as ABA1, DR4, and PYL4. Altogether the data showed that HDC1 is important for the fine-tuning of histone deacetylase activity during stress responses. Further studies also revealed that HDC1 is able to interact with histone binding proteins as well as H1 variants via RXT3 domain (Perrella et al., 2016). Arabidopsis plants over-expressing the HDC1 RXT3-like domain showed that the domain is sufficient to modulate some HDC1 responses, including germination and growth (Perrella et al., 2016) (Figure 4). In a different study, under control conditions, HDC1 was confirmed as the HDAC member in a protein complex including HDA19, H3-binding protein MULTICOPY SUPRESSOR of IRA1 (MSI1) and co-repressors SIN3 like (Derkacheva et al., 2013; Mehdi et al., 2015). Like hdc1-1, hda19 and msi1 mutants also displayed increased transcripts for RD29B, NAC DOMAIN CONTANING PROTEIN19 (ANAC019), COLD REGULATED 15A(COR15A) and PYL receptors 4, 5, and 6 in response to ABA. ChIP experiments showed that MSI1 is able to associate with PYL promoters (Mehdi et al., 2015).
Methylation of histone tails occurs primarily on lysine and arginine residues. Unlike acetylation, the position of the residues and the number of methylation groups are correlated with either active or repressed transcription. For instance, H3K4me2/3 is usually linked to transcriptional activation, whereas H3K9me2 is abundant in regions with low transcription (Liu et al., 2010; Asensi-Fabado et al., 2017). Methylation marks are applied by histone methyltransferases, whereas the removal is brought about by demethylases (Liu et al., 2010; Lu et al., 2011).
For most of the stress-responsive genes, increased expression is positively correlated with the addition of H3K4me3 marks (Zhang et al., 2009; Kim et al., 2012). Such transcriptional and chromatin changes are almost abolished in plants lacking histone methyltransferase HOMOLOG of TRITHORAX1 (ATX1) (Ding et al., 2011). Altogether this indicates a positive relationship between stress responses and histone methylation. Conversely, gain of function mutants for histone demethylase JUMONJI DOMAIN-CONTAINING PROTEIN 15 JMJ15 showed down-regulation of stress-related genes as well as the removal of the H3K4me3 marks (Shen et al., 2014a). However, down-regulation associated with histone demethylase activity was not constitutive. In fact stress-responsive genes such as RD29A and RD29B were instead up-regulated in jmj15 compared to the wild type (Shen et al., 2014a). This suggests that RD29A and RD29B are not targeted by JMJ15 demethylation.
In a time course study where Arabidopsis seedlings were exposed to 24 h cycles of dehydration stress, followed by recovery under control conditions, two groups of genes were identified as “not trainable” and “trainable” (Ding et al., 2012). Trainable genes such as RD29B and RAB18 showed an increase in gene expression dependent on the number of cycles whereas the “untrainable” RD29A and COR15A transcripts were similar after each stress. During the recovery period, RD29A and COR15A had H3K4me3 levels similar to those in control conditions, whereas the trainable genes showed an additive increase of H3K4me3 after each cycle. Such increases were accompanied by an accumulation of Pol II on RD29B and RAB18 (Ding et al., 2012). The trained plants wilted much slower than non-trained plants and lost less water when subjected to a dehydration/rehydration cycle (Ding et al., 2012).
The histone mark H3K27me3 is usually considered a repressive mark and it is mostly known for its role in repressing flowering locus C during vernalization (Angel et al., 2011; Hepworth and Dean, 2015). Recent studies have also shown a primary role for H3K27me3 during stress responses. Priming treatment of Arabidopsis plants with low salt showed changes at the genome-wide level mostly for H3K27me3 (Sani et al., 2013). More importantly, priming led to shortening and separation of the methylation levels. This chromatin feature was defined as “etching” effect which was still apparent after 10 days from the first treatment. At the expression level, some genes showed a long-lasting decrease (HKT1, PLASMA MEMBRANE INTRINSIC PROTEIN2E PIP2E) or increase (GH3.1, GH3.3) even after a second stronger treatment (Sani et al., 2013). Overall, these experiments show the existence of a long-term somatic memory whose information is a combination of chromatin and transcriptional changes.
In a heat stress study, H3K27me3 were found to be down-regulated at the FLOWERING LOCUS C (FLC) gene body when plants were treated at 29°C which then resulted in up-regulation of FLC expression (Gan et al., 2014). However, with loss of function for two histone demethylases (JUMONJI DOMAIN CONTANING PROTEIN 30 and 32,JMJ30,JMJ32), the levels of H3K27me3 did not decrease therefore FLC was no longer upregulated when the plants were exposed to high temperatures, suggesting that JMJ30/32 are required to demethylate FLC upon heat stress and that removal of H3K27me3 from FLC gene body is important for gene activation (Gan et al., 2014).
The data collected so far have shown the importance of changes in chromatin during stress responses. Studies of mutants for histone acetylation and methylation have further proven how chromatin modifications can be considered as a regulatory check-point for transcription. However, research is still limited for correlating transcription and histone modification at single loci. The approach in studying the role of few responsive genes during abiotic stress is, unfortunately, not exhaustive. More genome-wide approaches are needed and studies on early-time points will further distinguish stress response from adaptation.
Computational modeling has become an indispensable tool for research in plant abiotic stress responses. Two major branches of computational modeling are (1) genomic data driven modeling and (2) quantitative, dynamic modeling. Recently, tremendous amounts of data have been generated in the form of genomic sequences, chromatin modifications, and transcript, protein, and metabolite abundances. The goal of data-driven modeling is to identify biologically meaningful signals from genome-scale data. Data-driven modeling includes identification of causal SNPs in genome-wide association analysis (Li et al., 2010; Thoen et al., 2017), identification of DE genes (Geng et al., 2013; Bechtold et al., 2016), proteins (Lumba et al., 2014; Mostafa et al., 2016), metabolites (Töpfer and Niokoloski, 2013; Töpfer et al., 2014; Zhang S. et al., 2016), and the reflection of those changes in gene regulatory networks (Zaag et al., 2015; Landeghem et al., 2016). To validate modeling results, wet-bench experiments should be performed for candidate loci. In many cases, data-driven modeling can be validated using existing biological knowledge. Only proper validation can provide confidence to novel genes identified by data modeling approaches. Although genomic data have become increasingly available for various plants, most computational methodologies have been developed in the model plant Arabidopsis. Therefore, in this section, we will focus on recent progress in computational modeling in Arabidopsis.
Transcriptome profiling is the most widely used approach in genomic-scale studies of plant stress responses. One commonly performed analysis is to understand the functions of individual genes in a gene family. For example, mining of published expression profiles identified two CRF genes that are related to cold stress responses (Jeon et al., 2016). For species with reference genomes such as Arabidopsis, analysis of gene expression can be combined with analysis of TF binding sites in promoters. For example, RNA-seq analysis was used to construct transcription networks under proteotoxic stress (Gladman et al., 2016) and identified two NAC transcription factors that mediate proteasomal stress responses (Gladman et al., 2016). Co-expression and promoter analysis were also used to define combinatorial regulation of transcription factors in Arabidopsis stress response in single and combined stresses (Barah et al., 2016). In a comparative genomics study, promoter analysis of 30 angiosperm genomes showed conserved binding sites of ABRE and CE3-like motifs in the promoter regions of a stress regulated BAM1 gene (Thalmann et al., 2016). Increasingly, data are generated at high spatial and temporal resolution and the results are integrated with other large-scale data to provide detailed predictions. One example is combining live imaging and cell type-specific transcriptome profiling (Geng et al., 2013). In this study, ABA was shown to be a key hormone that connects growth recovery under salt stress in specific cell layers in Arabidopsis roots (Geng et al., 2013).
Reverse engineering of gene regulatory networks is the process of inferring transcription regulation using expression data. For example, more than one thousand microarray samples in Arabidopsis (Carrera et al., 2009) were used to infer regulatory network and the results show that regulatory interactions are more densely connected to genes responsive to environmental changes than other genes (Carrera et al., 2009). In anther example, Bayesian network modeling of transcriptome data has revealed hub TFs involved in drought responses in Arabidopsis (Bechtold et al., 2016).
Machine learning was applied to preselect informative genes from expression patterns and to integrate features from network analysis to predict functional genes that are related to stress responses (Ma et al., 2014). Any single machine learning method is often based on specific assumptions about the distribution of the underlying data. For example, linear support vector machines (SVM) assume samples are linearly separable in the feature space (Ni et al., 2016). Therefore, no single method can always out-perform other methods. Ensemble methods aggregate results from multiple inference approaches, and has been shown to improved performance in learning gene regulatory networks (Vermeirssen et al., 2014). The input of machine learning methods is typically a collection of data sets from multiple experiments. Wet-bench measurements are always carried out to validate the function of candidate genes (Ma et al., 2014; Vermeirssen et al., 2014).
Unlike networks inferred from transcriptomic data, ChIP-seq and DAP-seq (O’Malley et al., 2016) provide direct evidence of transcription factor and target gene interactions, furnishing the backbone of gene regulatory network. In Arabidopsis seedling, large-scale ChIP-seq and RNA-seq analyses showed that ABA increases TF binding at many promoter regions, but it reduces TF binding at other loci. Using this network data, previously un-annotated hub genes have been shown to be key regulators of ABA signaling (Song et al., 2016). Such techniques coupled with modeling may provide new insight into other stress responses such as heat stress (Ohama et al., 2015, 2017). Splicing regulation is another key step that determines the final sequence and concentration of many plant genes. In the same spirit of constructing TF-gene regulatory networks, computational modeling can help to identify sequence motifs and characterize splicing regulatory networks (Aghamirzaie et al., 2016).
In Arabidopsis, genome-wide association studies (GWAS) have been performed to identify SNPs related to adaptation to climate changes (Baxter et al., 2010; Li et al., 2010; Lasky et al., 2012, 2014) as well as other traits (Atwell et al., 2010). In recent years, a number of GWAS studies have been performed in Arabidopsis to identify candidate loci that are associated with abiotic stress tolerance, including various drought and low water potential conditions (Verslues et al., 2014; El-Soda et al., 2015; Bac-Molenaar et al., 2016), heat stress (Bac-Molenaar et al., 2015), and salt stress (Julkowska et al., 2016). In addition to identifying loci associated with abiotic stress condition, a large-scale GWAS was performed to associate SNPs for dozens of other stresses and combined stresses in Arabidopsis (Thoen et al., 2017). Many SNPs were identified in this study and the significant SNPs can be categorized as associated with multiple stresses or only associated with specific stressors (Thoen et al., 2017). A growing trend is to combine GWAS analysis with the underlying molecular networks. For example, by combining GWAS with co-expression networks, regulators of glucosinolates have been identified in Arabidopsis (Chan et al., 2011). In recent years, GWAS has been combined with co-expression networks to identify regulators of core-metabolic pathways (Wu et al., 2016), and new candidate genes involved in regulating amino acid metabolism (Angelovici et al., 2017).
To perform quantitative, dynamic modeling of stress responses, commonly used approaches require extensive existing knowledge of a given biological process. One example of dynamic modeling is multi-level modeling of guard cell signaling pathways, which revealed the interactions between ABA and red light responses in guard cells (Sun Z. et al., 2014). However, dynamics modeling is not feasible for many pathways due to a lack of detailed knowledge of interactions among signaling molecules. One way to systematically define the signaling network is through detection of protein–protein interactions. Recently, yeast-2-hybrid has been used to study the interaction between ABA-responsive genes and greatly expanded the interactions in the ABA signaling network (Lumba et al., 2014). However, integrating molecular interactions into network modeling is still a challenge, because the interactions between proteins do not provide the direction of information flow under stress conditions.
For both wet-bench scientists and computational biologists, online databases with user-friendly interfaces are necessary tools to explore the vast amount of data and networks generated by computational modeling. Fortunately, we can now perform comparative network analyses to identify abiotic stress related genes in Arabidopsis (Landeghem et al., 2016), explore uniformly re-processed expression data to query abiotic stress related gene co-expression networks (Zaag et al., 2015), and to search curated databases for published proteins that are related to plant stress response (Mousavi et al., 2016).
We illustrated multilevel regulatory processes coordinating responses and tolerance to abiotic stresses in plants. Genomic, transcriptomic, and proteomic analyses revealed the involvement of chromatin modification, transcriptional regulation, alternative splicing, protein phosphorylation, and ubiquitination/sumoylation in controlling adaptive responses to abiotic stresses. Using the outcomes of these studies, models of stress signaling networks have been constructed. Most of these models were developed based on Arabidopsis data. However, non-Arabidopsis studies have also identified other regulatory processes in stress signaling models. For example, DREB2 mRNAs are regulated by alternative splicing in maize, wheat, and barley, and only functional forms induce the expression of drought-responsive genes (Xue and Loveridge, 2004; Egawa et al., 2006; Qin et al., 2007) (Figure 5). In pearl millet, DREB2A was shown to be a phosphoprotein; the phosphorylated DREB2A lacked the binding ability to the DRE motif (Agarwal et al., 2007). The involvement of these regulatory processes in the DREB2 pathway has not been reported in Arabidopsis and other dicot species. Similarly, the stability of DREB2A protein is controlled by ubiquitin-dependent proteasomal degradation in Arabidopsis (Qin et al., 2008), but it is unknown whether this targeted proteolysis takes place in other plants. It is possible that some signaling components and regulatory processes are added or removed during evolutionary processes such as gene duplication and deletion.
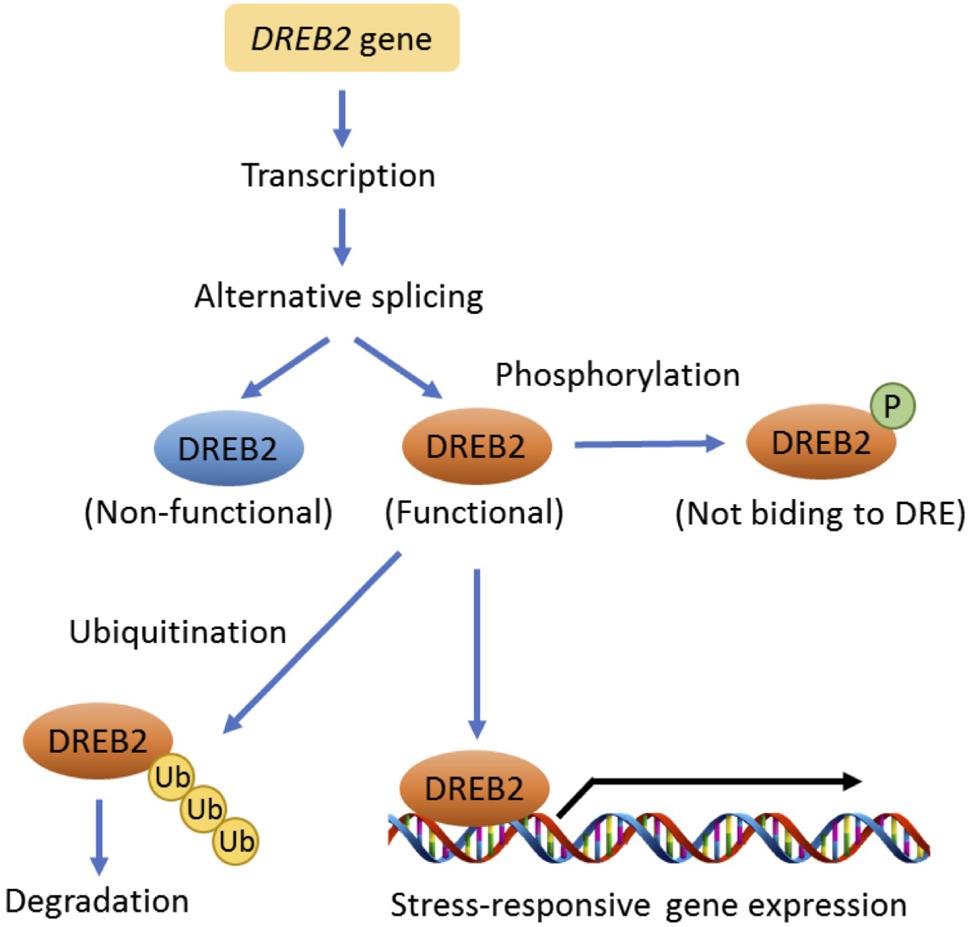
FIGURE 5. Model of the regulatory processes for DREB2 gene/protein under drought. In maize, wheat, and barley, DREB2 transcripts are regulated by alternative splicing, and only functional forms activate the expression of drought-responsive genes. In Arabidopsis, DREB2A protein is ubiquitinated by E3 ubiquitin ligases, DRIP1 and 2, promoting targeted proteolysis of DREB2A. Pearl millet DREB2A is a phosphoprotein and its phosphorylation inhibits the physical interaction with the DRE motif. It is unclear that these three processes regulating DREB2 gene/protein are conserved among higher plants. Evolutionary (speciation) processes might lead to addition or removal of some signaling components and processes as consequences of gene duplication and deletion.
To summarize, we anticipate several major future research trends in plant abiotic stress response research. First, emerging sequencing platforms and techniques will enable more detailed studies of individual regulatory components, which in turn, will drive the identification of new interactions and coordinated regulation among these components. For example, Iso-seq on the Pacific Biosciences sequencing platform is enabling the precise characterization of genome wide alternative splicing events, yielding new insights into the regulatory processes shaping this stress response. Second, computational modeling coupled with genomic-scale experimental data will continue to be a major source of discovery in stress response regulation. Multi-level models that integrate data from, genetic, epigenetic, and transcriptional studies with data from splicing and post-transcriptional regulation can provide novel insights into the global coordination of stress responses. Finally, evolutionary analysis of orthologous gene sequences and biochemical validation of signaling processes in diverse species will facilitate the identification of conserved and specific processes that coordinate stress adaptation and tolerance in plants.
All authors listed have made a substantial, direct and equal contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Funding from the following sources is acknowledged:
Thomas F. and Kate Miller Jeffress Memorial Trust, Bank of America, Trustee, Virginia Soybean Board, Virginia Corn Board (TF); NSF grants DBI-1062472 and MCB-1052145 and the Genomics, Bioinformatics, and Computational Biology doctoral program at Virginia Tech (RG); NSF grant DEB-1136707 (DH); Ohio University Grant SU1007172 (ZH); Strategic Research Fund, Heinrich-Heine University, Düsseldorf, Germany (SFF-F2014/730-15 Ivanov) and the German Research Foundation, through Collaborative Research Center 1208 (project B05) (RI); Industrial Partnership Award The Biotechnology and Biological Sciences Research Council BBSRC grant BB/K008218/1 (GP); The Virginia Agricultural Experiment Station and the Hatch Program of NIFA, USDA (TF, RG, DH, and SL).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01564/full#supplementary-material
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Agarwal, P., Agarwal, P. K., Nair, S., Sopory, S. K., and Reddy, M. K. (2007). Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol. Genet. Genomics 277, 189–198. doi: 10.1007/s00438-006-0183-z
Aghamirzaie, D., Collakova, E., Li, S., and Grene, R. (2016). CoSpliceNet: a framework for co-splicing network inference from transcriptomics data. BMC Genomics 17:845. doi: 10.1186/s12864-016-3172-6
Aiese Cigliano, R., Sanseverino, W., Cremona, G., Ercolano, M. R., Conicella, C., and Consiglio, F. M. (2013). Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14:57. doi: 10.1186/1471-2164-14-57
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–108. doi: 10.1038/nature10241
Angelovici, R., Batushansky, A., Deason, N., Gonzalez-Jorge, S., Gore, M. A., Fait, A., et al. (2017). Network-guided GWAS improves identification of genes affecting free amino acids. Plant Physiol. 173, 872–886. doi: 10.1104/pp.16.01287
Aquea, F., Vega, A., Timmermann, T., Poupin, M. J., and Arce-Johnson, P. (2011). Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep. 30, 1087–1097. doi: 10.1007/s00299-011-1015-0
Arms, E. M., Bloom, A. J., and Clair, D. A. S. (2015). High-resolution mapping of a major effect QTL from wild tomato Solanum habrochaites that influences water relations under root chilling. Theor. Appl. Genet. 128, 1713–1724. doi: 10.1007/s00122-015-2540-y
Arya, G., and Schlick, T. (2009). A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A 113, 4045–4059. doi: 10.1021/jp810375d
Ascenzi, R., and Gantt, J. S. (1997). A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34, 629–641. doi: 10.1023/A:1005886011722
Asensi-Fabado, M.-A., Amtmann, A., and Perrella, G. (2017). Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochim. Biophys. Acta 1860, 106–122. doi: 10.1016/j.bbagrm.2016.07.015
Ashnest, J. R., Huynh, D. L., Dragwidge, J. M., Ford, B. A., and Gendall, A. R. (2015). Arabidopsis intracellular NHX-type sodium-proton antiporters are required for seed storage protein processing. Plant Cell Physiol. 56, 2220–2233. doi: 10.1093/pcp/pcv138
Atwell, S., Huang, Y. S., Vilhjálmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Augustine, R. C., York, S. L., Rytz, T. C., and Vierstra, R. D. (2016). Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol. 171, 2191–2210. doi: 10.1104/pp.16.00353
Bac-Molenaar, J. A., Fradin, E. F., Becker, F. F., Rienstra, J. A., van der Schoot, J., Vreugdenhil, D., et al. (2015). Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27, 1857–1874. doi: 10.1105/tpc.15.00248
Bac-Molenaar, J. A., Granier, C., Keurentjes, J. J., and Vreugdenhil, D. (2016). Genome-wide association mapping of time-dependent growth responses to moderate drought stress in Arabidopsis. Plant Cell Environ. 39, 88–102. doi: 10.1111/pce.12595
Barah, P., B N, M. N., Jayavelu, N. D., Sowdhamini, R., Shameer, K., and Bones, A. M. (2016). Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucleic Acids Res. 44, 3147–3164. doi: 10.1093/nar/gkv1463
Barberon, M., Dubeaux, G., Kolb, C., Isono, E., Zelazny, E., and Vert, G. (2014). Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, 8293–8298. doi: 10.1073/pnas.1402262111
Barberon, M., Zelazny, E., Robert, S., Conejero, G., Curie, C., Friml, J., et al. (2011). Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, 450–458. doi: 10.1073/pnas.1100659108
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239. doi: 10.1105/tpc.110.079426
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q., Fujii, H., Pan, S., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27, 7781–7790. doi: 10.1128/MCB.00430-07
Baxter, I., Brazelton, J. N., Yu, D., Huang, Y. S., Lahner, B., Yakubova, E., et al. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1; 1. PLOS Genet. 6:e1001193. doi: 10.1371/journal.pgen.1001193
Bayle, V., Arrighi, J. F., Creff, A., Nespoulous, C., Vialaret, J., Rossignol, M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23, 1523–1535. doi: 10.1105/tpc.110.081067
Bechtold, U., Penfold, C. A., Jenkins, D. J., Legaie, R., Moore, J. D., Lawson, T., et al. (2016). Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. Plant Cell 28, 345–366. doi: 10.1105/tpc.15.00910
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A., and Simon, R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Blum, A., Brumbarova, T., Bauer, P., and Ivanov, R. (2014). Hormone influence on the spatial regulation of IRT1 expression in iron-deficient Arabidopsis thaliana roots. Plant Signal. Behav. 9:e28787. doi: 10.4161/psb.28787
Bouguyon, E., Brun, F., Meynard, D., Kubes, M., Pervent, M., Leran, S., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1:15015. doi: 10.1038/nplants.2015.15
Bouguyon, E., Gojon, A., and Nacry, P. (2012). Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 23, 648–654. doi: 10.1016/j.semcdb.2012.01.004
Brumbarova, T., Bauer, P., and Ivanov, R. (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 20, 124–133. doi: 10.1016/j.tplants.2014.11.004
Brumbarova, T., and Ivanov, R. (2016). Differential gene expression and protein phosphorylation as factors regulating the state of the Arabidopsis SNX1 protein complexes in response to environmental stimuli. Front. Plant Sci. 7:1456. doi: 10.3389/fpls.2016.01456
Bui, L. T., Giuntoli, B., Kosmacz, M., Parlanti, S., and Licausi, F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236, 37–43. doi: 10.1016/j.plantsci.2015.03.008
Buszewicz, D., Archacki, R., Palusiński, A., Kotliński, M., Fogtman, A., Iwanicka-Nowicka, R., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 39, 2108–2122. doi: 10.1111/pce.12756
Capovilla, G., Pajoro, A., Immink, R. G., and Schmid, M. (2015). Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 27, 97–103. doi: 10.1016/j.pbi.2015.06.016
Carrera, J., Rodrigo, G., Jaramillo, A., and Elena, S. F. (2009). Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 10:R96. doi: 10.1186/gb-2009-10-9-r96
Carrozza, M. J., Florens, L., Swanson, S. K., Shia, W.-J., Anderson, S., Yates, J., et al. (2005). Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731, 77–87. doi: 10.1016/j.bbaexp.2005.09.005
Carvalho, R. F., Carvalho, S. D., and Duque, P. (2010). The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 154, 772–783. doi: 10.1104/pp.110.155523
Carvalho, R. F., Szakonyi, D., Simpson, C. G., Barbosa, I. C., Brown, J. W., Baena-González, E., et al. (2016). The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 28, 1910–1925. doi: 10.1105/tpc.16.00301
Chan, E. K., Rowe, H. C., Corwin, J. A., Joseph, B., and Kliebenstein, D. J. (2011). Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLOS Biol. 9:e1001125. doi: 10.1371/journal.pbio.1001125
Charng, Y. Y., Liu, H. C., Liu, N. Y., Chi, W. T., Wang, C. N., Chang, S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chen, L.-T., Luo, M., Wang, Y.-Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, X., Hu, Y., and Zhou, D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426. doi: 10.1016/j.bbagrm.2011.03.004
Cheng, N. H., Pittman, J. K., Zhu, J. K., and Hirschi, K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H(+)/Ca(2+) antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279, 2922–2926. doi: 10.1074/jbc.M309084200
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Deve 17, 1043–1054. doi: 10.1101/gad.1077503
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2006). Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. 27, 141–177. doi: 10.1007/0-387-25856-6_9
Chung, H. S., Cooke, T. F., Depew, C. L., Patel, L. C., Ogawa, N., Kobayashi, Y., et al. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622. doi: 10.1111/j.1365-313X.2010.04265.x
Colangelo, E. P., and Guerinot, M. L. (2006). Put the metal to the petal: metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 9, 322–330. doi: 10.1016/j.pbi.2006.03.015
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLOS Genet. 8:e1002988. doi: 10.1371/journal.pgen.1002988
Corratge-Faillie, C., and Lacombe, B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. doi: 10.1093/jxb/erw499 [Epub ahead of print].
Cui, P., Zhang, S., Ding, F., Ali, S., and Xiong, L. (2014). Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 15:R1. doi: 10.1186/gb-2014-15-1-r1
Deal, R. B., and Henikoff, S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14, 116–122. doi: 10.1016/j.pbi.2010.11.005
Deal, R. B., Topp, C. N., Mckinney, E. C., and Meagher, R. B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A. Z. Plant Cell 19, 74–83. doi: 10.1105/tpc.106.048447
Derkacheva, M., Steinbach, Y., Wildhaber, T., Mozgová, I., Mahrez, W., Nanni, P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085. doi: 10.1038/emboj.2013.145
Ding, F., Cui, P., Wang, Z., Zhang, S., Ali, S., and Xiong, L. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15:431. doi: 10.1186/1471-2164-15-431
Ding, Y., Avramova, Z., and Fromm, M. (2011). The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought ’train’ transcriptional responses in Arabidopsis. Nat. Commun. 3:740. doi: 10.1038/ncomms1732
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dos Santos, R. S., Farias, D. D. R., Pegoraro, C., Rombaldi, C. V., Fukao, T., Wing, R. A., et al. (2017). Evolutionary analysis of the SUB1 locus across the Oryza genomes. Rice 10:4. doi: 10.1186/s12284-016-0140-3
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., and Pikaard, C. S. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 52, k615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Efroni, I., Mello, A., Nawy, T., Ip, P.-L., Rahni, R., Delrose, N., et al. (2016). Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165, 1721–1733. doi: 10.1016/j.cell.2016.04.046
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., and Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81, 77–91. doi: 10.1266/ggs.81.77
Elrouby, N. (2017). Regulation of plant cellular and organismal development by SUMO. Adv. Exp. Med. Biol. 963, 227–247. doi: 10.1007/978-3-319-50044-7_14
Elrouby, N., Bonequi, M. V., Porri, A., and Coupland, G. (2013). Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc. Natl. Acad. Sci. U.S.A. 110, 19956–19961. doi: 10.1073/pnas.1319985110
Elrouby, N., and Coupland, G. (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. U.S.A. 107, 17415–17420. doi: 10.1073/pnas.1005452107
El-Soda, M., Kruijer, W., Malosetti, M., Koornneef, M., and Aarts, M. G. (2015). Quantitative trait loci and candidate genes underlying genotype by environment interaction in the response of Arabidopsis thaliana to drought. Plant Cell Environ. 38, 585–599. doi: 10.1111/pce.12418
Erskine, W., Upadhyaya, H. D., and Malik, A. I. (2014). “Salinity,” in Plant Genetic Resources and Climate Change, eds M. Jackson, B. Ford-Lloyd, and M. Parry (Oxfordshire: CAB International), 236–250.
Filichkin, S. A., Cumbie, J. S., Dharmawardhana, P., Jaiswal, P., Chang, J. H., Palusa, S. G., et al. (2014). Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 8, 207–227. doi: 10.1016/j.molp.2014.10.011
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. doi: 10.1101/gr.093302.109
Finnegan, E. J., Genger, R. K., Peacock, W. J., and Dennis, E. S. (1998). DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 223–247. doi: 10.1146/annurev.arplant.49.1.223
Fischer, I., Camus-Kulandaivelu, L., Allal, F., and Stephan, W. (2011). Adaptation to drought in two wild tomato species: the evolution of the Asr gene family. New Phytol. 190, 1032–1044. doi: 10.1111/j.1469-8137.2011.03648.x
Fuglsang, A. T., Guo, Y., Cuin, T. A., Qiu, Q., Song, C., Kristiansen, K. A., et al. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+ -ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634. doi: 10.1105/tpc.105.035626
Fuglsang, A. T., Kristensen, A., Cuin, T. A., Schulze, W. X., Persson, J., Thuesen, K. H., et al. (2014). Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J. 80, 951–964. doi: 10.1111/tpj.12680
Fujii, H., Verslues, P. E., and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19, 485–494. doi: 10.1105/tpc.106.048538
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takgi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Fujita, Y., Yoshida, T., and Yamaguchi-Shinozaki, K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147, 15–27. doi: 10.1111/j.1399-3054.2012.01635.x
Fukao, T., Harris, T., and Bailey-Serres, J. (2008). Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann. Bot. 103, 143–150. doi: 10.1093/aob/mcn172
Fukao, T., and Xiong, L. (2013). Genetic mechanisms conferring adaptation to submergence and drought in rice: simple or complex? Curr. Opin. Plant Biol. 16, 196–204. doi: 10.1016/j.pbi.2013.02.003
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Fursova, O. V., Pogorelko, G. V., and Tarasov, V. A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429, 98–103. doi: 10.1016/j.gene.2008.10.016
Gan, E.-S., Xu, Y., Wong, J.-Y., Geraldine Goh, J., Sun, B., Wee, W.-Y., et al. (2014). Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 5:5098. doi: 10.1038/ncomms6098
Gasch, P., Fundinger, M., Muller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. M. Y., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25, 2132–2154. doi: 10.1105/tpc.113.112896
Geoffroy, M. C., and Hay, R. T. (2009). An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564–568. doi: 10.1038/nrm2707
Gingerich, D. J., Gagne, J. M., Salter, D. W., Hellmann, H., Estelle, M., Ma, L., et al. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280, 18810–18821. doi: 10.1074/jbc.M413247200
Giuntoli, B., Lee, S. C., Licausi, F., Kosmacz, M., Oosumi, T., Van Dongen, J. T., et al. (2014). A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLOS Biol. 12:e1001950. doi: 10.1371/journal.pbio.1001950
Gladman, N. P., Marshall, R. S., Lee, K.-H., and Vierstra, R. D. (2016). The proteasome stress regulon is controlled by a Pair of NAC transcription factors in Arabidopsis. Plant Cell 28, 1279–1296. doi: 10.1105/tpc.15.01022
Golebiowski, F., Matic, I., Tatham, M. H., Cole, C., Yin, Y., Nakamura, A., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24. doi: 10.1126/scisignal.2000282
Guan, Q., Yue, X., Zeng, H., and Zhu, J. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26, 438–453. doi: 10.1105/tpc.113.118927
Guan, R., Qu, Y., Guo, Y., Yu, L., Liu, Y., Jiang, J., et al. (2014). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80, 937–950. doi: 10.1111/tpj.12695
Hardeland, U., Steinacher, R., Jiricny, J., and Schar, P. (2002). Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456–1464. doi: 10.1093/emboj/21.6.1456
Haruta, M., Burch, H. L., Nelson, R. B., Barrett-Wilt, G., Kline, K. G., Mohsin, S. B., et al. (2010). Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285, 17918–17929. doi: 10.1074/jbc.M110.101733
Held, K., Pascaud, F., Eckert, C., Gajdanowicz, P., Hashimoto, K., Corratge-Faillie, C., et al. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21, 1116–1130. doi: 10.1038/cr.2011.50
Hepworth, J., and Dean, C. (2015). Flowering locus C’s lessons: conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol. 168, 1237–1245. doi: 10.1104/pp.15.00496
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Horie, T., Hauser, F., and Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14, 660–668. doi: 10.1016/j.tplants.2009.08.009
Hua, Z., Pool, J. E., Schmitz, R. J., Schultz, M. D., Shiu, S. H., Ecker, J. R., et al. (2013). Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 16927–16932. doi: 10.1073/pnas.1316009110
Hua, Z., and Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Hua, Z., Zou, C., Shiu, S. H., and Vierstra, R. D. (2011). Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLOS ONE 6:e16219. doi: 10.1371/journal.pone.0016219
Huang, Y., Liu, C., Shen, W.-H., and Ruan, Y. (2011). Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol. 11:175. doi: 10.1186/1471-2229-11-175
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. doi: 10.1104/pp.111.179036
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Ivanov, R., Brumbarova, T., and Bauer, P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42. doi: 10.1093/mp/ssr065
Ivanov, R., Brumbarova, T., Blum, A., Jantke, A. M., Fink-Straube, C., and Bauer, P. (2014). SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26, 1294–1307. doi: 10.1105/tpc.113.116244
Ivanov, R., and Gaude, T. (2009). Endocytosis and endosomal regulation of the S-receptor kinase during the self-incompatibility response in Brassica oleracea. Plant Cell 21, 2107–2117. doi: 10.1105/tpc.108.063479
James, R. A., Davenport, R. J., and Munns, R. (2006). Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142, 1537–1547. doi: 10.1104/pp.106.086538
Janicka-Russak, M., Kabala, K., Burzynski, M., and Klobus, G. (2008). Response of plasma membrane H+-ATPase to heavy metal stress in Cucumis sativus roots. J. Exp. Bot. 59, 3721–3728. doi: 10.1093/jxb/ern219
Janicka-Russak, M., and Klobus, G. (2007). Modification of plasma membrane and vacuolar H+ -ATPases in response to NaCL and ABA. J. Plant Physiol. 164, 295–302. doi: 10.1016/j.jplph.2006.01.014
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074–1080. doi: 10.1126/science.1063127
Jeon, J., Cho, C., Lee, M. R., Van Binh, N., and Kim, J. (2016). CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell 28, 1828–1843. doi: 10.1105/tpc.15.00909
Julkowska, M. M., Klei, K., Fokkens, L., Haring, M. A., Schranz, M. E., and Testerink, C. (2016). Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exper. Bot. 67, 2127–2138. doi: 10.1093/jxb/erw015
Kadosh, D., and Struhl, K. (1998). Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18, 5121–5127. doi: 10.1128/MCB.18.9.5121
Kaldis, A., Tsementzi, D., Tanriverdi, O., and Vlachonasios, K. E. (2011). Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 233, 749–762. doi: 10.1007/s00425-010-1337-0
Kasai, K., Takano, J., Miwa, K., Toyoda, A., and Fujiwara, T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286, 6175–6183. doi: 10.1074/jbc.M110.184929
Kim, D. Y., Scalf, M., Smith, L. M., and Vierstra, R. D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25, 1523–1540. doi: 10.1105/tpc.112.108613
Kim, J. H., and Kim, W. T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162, 1733–1749. doi: 10.1104/pp.113.220103
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, J.-M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Knight, H., Zarka, D. G., Okamoto, H., Thomashow, M. F., and Knight, M. R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Phys. 135, k1710–1717. doi: 10.1104/pp.104.043562
Komander, D., and Rape, M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81, 203–229. doi: 10.1146/annurev-biochem-060310-170328
Korshunova, Y. O., Eide, D., Clark, W. G., Guerinot, M. L., and Pakrasi, H. B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40, 37–44. doi: 10.1023/A:1026438615520
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Krebs, M., Beyhl, D., Gorlich, E., Al-Rasheid, K. A., Marten, I., Stierhof, Y. D., et al. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 107, 3251–3256. doi: 10.1073/pnas.0913035107
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Kuo, M.-H., Brownell, J. E., Sobel, R. E., Ranalli, T. A., Cook, R. G., Edmondson, D. G., et al. (1996). Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272. doi: 10.1038/383269a0
Landeghem, S. V., Parys, T. V., Dubois, M., Inzé, D., and De Peer, Y. V. (2016). Diffany: an ontology-driven framework to infer, visualise and analyse differential molecular networks. BMC Bioinformat. 17:18. doi: 10.1186/s12859-015-0863-y
Lasky, J. R., Des Marais, D. L., Lowry, D. B., Povolotskaya, I., McKay, J. K., Richards, J. H., et al. (2014). Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol. Biol. Evolut. 31, 2283–2296. doi: 10.1093/molbev/msu170
Lasky, J. R., Des Marais, D. L., McKay, J., Richards, J. H., Juenger, T. E., and Keitt, T. H. (2012). Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol. Ecol. 21, 5512–5529. doi: 10.1111/j.1365-294X.2012.05709.x
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Lee, A., Lee, S. S., Jung, W. Y., Park, H. J., Lim, B. R., Kim, H. S., et al. (2016). The OsCYP19-4 gene is expressed as multiple alternatively spliced transcripts encoding isoforms with distinct cellular localizations and PPIase activities under cold stress. Int. J. Mol. Sci. 17, E1154. doi: 10.3390/ijms17071154
Lee, J. H., Terzaghi, W., Gusmaroli, G., Charron, J. B., Yoon, H. J., Chen, H., et al. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20, 152–167. doi: 10.1105/tpc.107.055418
Leshem, Y., Golani, Y., Kaye, Y., and Levine, A. (2010). Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 61, 2615–2622. doi: 10.1093/jxb/erq099
Leshem, Y., Melamed-Book, N., Cagnac, O., Ronen, G., Nishri, Y., Solomon, M., et al. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 103, 18008–18013. doi: 10.1073/pnas.0604421103
Li, L., Kim, B. G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Li, M., Li, Y., Li, H., and Wu, G. (2011a). Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiol. 31, 349–357. doi: 10.1093/treephys/tpr003
Li, M., Lin, X., Li, H., Pan, X., and Wu, G. (2011b). Overexpression of AtNHX5 improves tolerance to both salt and water stress in rice (Oryza sativa L.). Plant Cell Tiss. Organ. Cult. 107, 283–293. doi: 10.1007/s11240-011-9979-6
Li, S., Yamada, M., Han, X., Ohler, U., and Benfey, P. N. (2016). High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 39, 508–522. doi: 10.1016/j.devcel.2016.10.012
Li, Y., Huang, Y., Bergelson, J., Nordborg, M., and Borevitz, J. O. (2010). Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 21199–21204. doi: 10.1073/pnas.1007431107
Ling, Y., Alshareef, S., Butt, H., Lozano-Juste, J., Li, L., Galal, A. A., et al. (2017). Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 89, 291–309. doi: 10.1111/tpj.13383
Liu, C. Y., Lu, F. L., Cui, X., and Cao, X. F. (2010). “Histone methylation in higher plants,” in Annual Review of Plant Biology, eds S. Merchant, W. R. Briggs, and D. Ort (Palo Alto, CA: Annual Reviews).
Liu, K. H., and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. doi: 10.1038/38444
Lumba, S., Toh, S., Handfield, L.-F., Swan, M., Liu, R., Youn, J.-Y., et al. (2014). A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev. Cell 29, 360–372. doi: 10.1016/j.devcel.2014.04.004
Luo, M., Liu, X. C., Singh, P., Cui, Y. H., Zimmerli, L., and Wu, K. Q. (2012a). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136. doi: 10.1016/j.bbagrm.2011.06.008
Luo, M., Wang, Y.-Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012b). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Luo, X., Bai, X., Zhu, D., Li, Y., Ji, W., Cai, H., et al. (2011). GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta 235, 1141–1155. doi: 10.1007/s00425-011-1563-0
Ma, C., Xin, M., Feldmann, K. A., and Wang, X. (2014). Machine learning-based differential network analysis: a study of stress-responsive transcriptomes in Arabidopsis. Plant Cell 26, 520–537. doi: 10.1105/tpc.113.121913
Mathieu, O., Probst, A. V., and Paszkowski, J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24, 2783–2791. doi: 10.1038/sj.emboj.7600743
Mboup, M., Fischer, I., Lainer, H., and Stephan, W. (2012). Trans-species polymorphism and allele-specific expression in the CBF gene family of wild tomatoes. Mol. Biol. Evol. 29, 3641–3652. doi: 10.1093/molbev/mss176
Mckhann, H. I., Gery, C., Berard, A., Leveque, S., Zuther, E., Hincha, D. K., et al. (2008). Natural variation in CBF gene sequence, gene expression and freezing tolerance in the Versailles core collection of Arabidopsis thaliana. BMC Plant Biol. 8:105. doi: 10.1186/1471-2229-8-105
Mehdi, S., Derkacheva, M., Ramström, M., Kralemann, L., Bergquist, J., and Hennig, L. (2015). MSI1 functions in a HDAC complex to fine-tune ABA signaling. Plant Cell. 28, 42–54. doi: 10.1105/tpc.15.00763
Meyer, K., Koester, T., and Staiger, D. (2015). Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules 5, 1717–1740. doi: 10.3390/biom5031717
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Miller, M. J., Barrett-Wilt, G. A., Hua, Z., and Vierstra, R. D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517. doi: 10.1073/pnas.1004181107
Miller, M. J., Scalf, M., Rytz, T. C., Hubler, S. L., Smith, L. M., and Vierstra, R. D. (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol. Cell. Proteomics 12, 449–463. doi: 10.1074/mcp.M112.025056
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Mlodzinska, E., Klobus, G., Christensen, M. D., and Fuglsang, A. T. (2015). The plasma membrane H(+) -ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plant. 154, 270–282. doi: 10.1111/ppl.12305
Mostafa, I., Zhu, N., Yoo, M.-J., Balmant, K. M., Misra, B. B., Dufresne, C., et al. (2016). New nodes and edges in the glucosinolate molecular network revealed by proteomics and metabolomics of Arabidopsis myb28/29 and cyp79B2/B3 glucosinolate mutants. J. Proteomics 138, 1–19. doi: 10.1016/j.jprot.2016.02.012
Mounier, E., Pervent, M., Ljung, K., Gojon, A., and Nacry, P. (2014). Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ 37, 162–174. doi: 10.1111/pce.12143
Mousavi, S. A., Pouya, F. M., Ghaffari, M. R., Mirzaei, M., Ghaffari, A., Alikhani, M., et al. (2016). PlantPReS: a database for plant proteome response to stress. J. Proteomics 143, 69–72. doi: 10.1016/j.jprot.2016.03.009
Moyle, L. C., and Muir, C. D. (2010). Reciprocal insights into adaptation from agricultural and evolutionary studies in tomato. Evolut. Appl. 3, 409–421. doi: 10.1111/j.1752-4571.2010.00143.x
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Muthamilarasan, M., Bonthala, V. S., Khandelwal, R., Jaishankar, J., Shweta, S., Nawaz, K., et al. (2015). Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front. Plant Sci. 6:910. doi: 10.3389/fpls.2015.00910
Neuhaus, H. E., and Trentmann, O. (2014). Regulation of transport processes across the tonoplast. Front. Plant Sci. 5:460. doi: 10.3389/fpls.2014.00460
Ni, Y., Aghamirzaie, D., Elmarakeby, H., Collakova, E., Li, S., Grene, R., et al. (2016). A machine learning approach to predict gene regulatory networks in seed development in Arabidopsis. Front. Plant Sci. 7:1936. doi: 10.3389/fpls.2016.01936
Niroula, R. K., Pucciariello, C., Ho, V. T., Novi, G., Fukao, T., and Perata, P. (2012). SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J. 72, 282–293. doi: 10.1111/j.1365-313X.2012.05078.x
Novillo, F., Alonso, J. M., Ecker, J. R., and Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 3985–3990. doi: 10.1073/pnas.0303029101
Novillo, F., Medina, J., and Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. U.S.A. 104, 21002–21007. doi: 10.1073/pnas.0705639105
Ohama, N., Kusakabe, K., Mizoi, J., Zhao, H., Kidokoro, S., Koizumi, S., et al. (2015). The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell. 28, 181–201. doi: 10.1105/tpc.15.00435
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
O’Malley, R. C., Huang, S.-S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Palmgren, M. G. (2001). PLANT PLASMA MEMBRANE H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. doi: 10.1146/annurev.arplant.52.1.817
Papaefthimiou, D., Likotrafiti, E., Kapazoglou, A., Bladenopoulos, K., and Tsaftaris, A. (2010). Epigenetic chromatin modifiers in barley: III. Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA. Plant Physiol. Biochem. 48, 98–107. doi: 10.1016/j.plaphy.2010.01.002
Parker, J. L., and Newstead, S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. doi: 10.1038/nature13116
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., and Scheid, O. M. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Pennycooke, J. C., Cheng, H., Roberts, S. M., Yang, Q., Rhee, S. Y., and Stockinger, E. J. (2008). The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol. Biol. 67, 483–497. doi: 10.1007/s11103-008-9333-5
Perrella, G., Carr, C., Asensi-Fabado, M. A., Donald, N. A., Páldi, K., Hannah, M. A., et al. (2016). The histone deacetylase complex 1 protein of Arabidopsis has the capacity to interact with multiple proteins including histone 3-binding proteins and histone 1 variants. Plant Physiol. 171, 62–70. doi: 10.1104/pp.15.01760
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Gosselé, V., Verduyn, C., et al. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Pizarro, L., and Norambuena, L. (2014). Regulation of protein trafficking: posttranslational mechanisms and the unexplored transcriptional control. Plant Sci. 225, 24–33. doi: 10.1016/j.plantsci.2014.05.004
Pontvianne, F., Blevins, T., and Pikaard, C. S. (2010). Arabidopsis histone lysine methyltransferases. Adv. Bot. Res. 53, 1–22. doi: 10.1016/S0065-2296(10)53001-5
Probst, A. V., and Mittelsten Scheid, O. (2015). Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27, 8–16. doi: 10.1016/j.pbi.2015.05.011
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S. P., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Qiu, Q. S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., and Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279, 207–215. doi: 10.1074/jbc.M307982200
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108, 2611–2616. doi: 10.1073/pnas.1018921108
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., and Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99, 9061–9066. doi: 10.1073/pnas.132092099
Rao, E. S., Kadirvel, P., Symonds, R. C., Geethanjali, S., Thontadarya, R. N., and Ebert, A. W. (2015). Variations in DREB1A and VP1. 1 genes show association with salt tolerance traits in wild tomato (Solanum pimpinellifolium). PLOS ONE 10:e0132535. doi: 10.1371/journal.pone.0132535
Reddy, A. S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58, 267–294. doi: 10.1146/annurev.arplant.58.032806.103754
Reddy, A. S., Marquez, Y., Kalyna, M., and Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683. doi: 10.1105/tpc.113.117523
Reynoso, M. A., Juntawong, P., Lancia, M., Blanco, F. A., Bailey-Serres, J., and Zanetti, M. E. (2015). Translating Ribosome Affinity Purification (TRAP) followed by RNA sequencing technology (TRAP-SEQ) for quantitative assessment of plant translatomes. Methods Mol. Biol. 1284, 185–207. doi: 10.1007/978-1-4939-2444-8_9
Rogers, E. E., Eide, D. J., and Guerinot, M. L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. U.S.A. 97, 12356–12360. doi: 10.1073/pnas.210214197
Rundlett, S. E., Carmen, A. A., Suka, N., Turner, B. M., and Grunstein, M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835. doi: 10.1038/33952
Rutowicz, K., Puzio, M., Halibart-Puzio, J., Lirski, M., Kroteń, M. A., Kotliński, M., et al. (2015). A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol. 169, 2080–2101. doi: 10.1104/pp.15.00493
Sani, E., Herzyk, P., Perrella, G., Colot, V., and Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14:R59. doi: 10.1186/gb-2013-14-6-r59
Saracco, S. A., Miller, M. J., Kurepa, J., and Vierstra, R. D. (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145, 119–134. doi: 10.1104/pp.107.102285
Schulz, P., Herde, M., and Romeis, T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163, 523–530. doi: 10.1104/pp.113.222539
Schulze, W. X., Schneider, T., Starck, S., Martinoia, E., and Trentmann, O. (2012). Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 69, 529–541. doi: 10.1111/j.1365-313X.2011.04812.x
Seo, P. J., Park, M. J., and Park, C. M. (2013). Alternative splicing of transcription factors in plant responses to low temperature stress: mechanisms and functions. Planta 237, 1415–1424. doi: 10.1007/s00425-013-1882-4
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Shen, Y., Conde, E., Silva, N., Audonnet, L., Servet, C., Wei, W., et al. (2014a). Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 5:290. doi: 10.3389/fpls.2014.00290
Shen, Y., Zhou, Z., Wang, Z., Li, W., Fang, C., Wu, M., et al. (2014b). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. doi: 10.1105/tpc.114.122739
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, L., Wang, J., Hong, F., Spector, D. L., and Fang, Y. (2011). Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes. Proc. Natl. Acad. Sci. 108, 10574–10578. doi: 10.1073/pnas.1017882108
Shi, L. Y., Li, H. Q., Pan, X. P., Wu, G. J., and Li, M. R. (2008). Improvement of Torenia fournieri salinity tolerance by expression of Arabidopsis AtNHX5. Funct. Plant Biol. 35, 185–192. doi: 10.1071/FP07269
Shibagaki, N., and Grossman, A. R. (2010). Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J. Biol. Chem. 285, 25094–25102. doi: 10.1074/jbc.M110.126888
Shin, L. J., Lo, J. C., Chen, G. H., Callis, J., Fu, H., and Yeh, K. C. (2013). IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25, 3039–3051. doi: 10.1105/tpc.113.115212
Shu, H., Nakamura, M., Siretskiy, A., Borghi, L., Moraes, I., Wildhaber, T., et al. (2014). Arabidopsis replacement histone variant H3. 3 occupies promoters of regulated genes. Geno. Biol. 15:R62. doi: 10.1186/gb-2014-15-4-r62
Singh, P., and Sinha, A. K. (2016). A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 imparts submergence tolerance in rice. Plant Cell 28, 1127–1143. doi: 10.1105/tpc.15.01001
Son, Y. S., Im, C. H., Kim, D. W., and Bahk, J. D. (2013). OsRab11 and OsGAP1 are essential for the vesicle trafficking of the vacuolar H(+)-ATPase OsVHA-a1 under high salinity conditions. Plant Sci. 198, 58–71. doi: 10.1016/j.plantsci.2012.09.003
Song, L., Huang, S. S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354:aag1550. doi: 10.1126/science.aag1550
Sridha, S., and Wu, K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46, 124–133. doi: 10.1111/j.1365-313X.2006.02678.x
Staiger, D., and Brown, J. W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656. doi: 10.1105/tpc.113.113803
Steinhorst, L., and Kudla, J. (2013). Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol. 163, 471–485. doi: 10.1104/pp.113.222950
Streitner, C., Simpson, C. G., Shaw, P., Danisman, S., Brown, J. W., and Staiger, D. (2013). Small changes in ambient temperature affect alternative splicing in Arabidopsis thaliana. Plant Signal. Behav. 8:e24638. doi: 10.4161/psb.24638
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., and Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Sun, Z., Jin, X., Albert, R., and Assmann, S. M. (2014). Multi-level modeling of light-induced stomatal opening offers new insights into its regulation by drought. PLOS Comput. Biol. 10:e1003930. doi: 10.1371/journal.pcbi.1003930
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kuś-Slowinska, M., Pawełoszek, Ł., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29, 791–807. doi: 10.1105/tpc.16.00573
Suzuki, N., Sejima, H., Tam, R., Schlauch, K., and Mittler, R. (2011). Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 66, k844–851. doi: 10.1111/j.1365-313X.2011.04550.x
Takahashi, H., Hatakeyama, S., Saitoh, H., and Nakayama, K. I. (2005). Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 280, 5611–5621. doi: 10.1074/jbc.M408130200
Takano, J., Tanaka, M., Toyoda, A., Miwa, K., Kasai, K., Fuji, K., et al. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 5220–5225. doi: 10.1073/pnas.0910744107
Talbert, P. B., and Henikoff, S. (2010). Histone variants [mdash] ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275. doi: 10.1038/nrm2861
Tamang, B. G., and Fukao, T. (2015). Plant adaptation to multiple stresses during submergence and following desubmergence. Int. J. Mol. Sci. 16, 30164–30180. doi: 10.3390/ijms161226226
Tang, L., Cai, H., Ji, W., Luo, X., Wang, Z., Wu, J., et al. (2013). Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 71, 22–30. doi: 10.1016/j.plaphy.2013.06.024
Tariq, M., and Paszkowski, J. (2004). DNA and histone methylation in plants. Trends Genet. 20, 244–251. doi: 10.1016/j.tig.2004.04.005
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28, 1860–1878. doi: 10.1105/tpc.16.00143
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170, 586–599. doi: 10.1104/pp.15.01267
Thatcher, S. R., Zhou, W., Leonard, A., Wang, B. B., Beatty, M., Zastrow-Hayes, G., et al. (2014). Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell 26, 3472–3487. doi: 10.1105/tpc.114.130773
Thoen, M. P. M., Davila Olivas, N. H., Kloth, K. J., Coolen, S., Huang, P.-P., Aarts, M. G. M., et al. (2017). Genetic architecture of plant stress resistance: multi-trait genome-wide association mapping. New Phytol. 213, 1346–1362. doi: 10.1111/nph.14220
To, T. K., Kim, J.-M., Matsui, A., Kurihara, Y., Morosawa, T., Ishida, J., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLOS Genet. 7:e1002055. doi: 10.1371/journal.pgen.1002055
Töpfer, N., and Niokoloski, Z. (2013). Large-scale modeling provides insights into Arabidopsis’ acclimation to changing light and temperature conditions. Plant Signal. Behav. 8:e25480. doi: 10.4161/psb.25480
Töpfer, N., Scossa, F., Fernie, A., and Nikoloski, Z. (2014). Variability of metabolite levels is linked to differential metabolic pathways in Arabidopsis’s responses to abiotic stresses. PLOS Comput. Biol. 10:e1003656. doi: 10.1371/journal.pcbi.1003656
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analyis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early respnsie to dehydration sterss 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, k11632–11637. doi: 10.1073/pnas.190309197
Vahisalu, T., Puzorjova, I., Brosche, M., Valk, E., Lepiku, M., Moldau, H., et al. (2010). Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 62, 442–453. doi: 10.1111/j.1365-313X.2010.04159.x
van Veen, H., Akman, M., Jamar, D. C. L., Vreugdenhil, D., Kooiker, M., Van Tienderen, P., et al. (2014). Group VII Ethylene Response Factor diversification and regulation in four species from flood-prone environments. Plant Cell Environ. 37, 2421–2432. doi: 10.1111/pce.12302
Vermeirssen, V., De Clercq, I., Van Parys, T., Van Breusegem, F., and Van De Peer, Y. (2014). Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell 26, 4656–4679. doi: 10.1105/tpc.114.131417
Verslues, P. E., Lasky, J. R., Juenger, T. E., Liu, T. W., and Kumar, M. N. (2014). Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Phys. 164, 144–159. doi: 10.1104/pp.113.224014
Veshaguri, S., Christensen, S. M., Kemmer, G. C., Ghale, G., Moller, M. P., Lohr, C., et al. (2016). Direct observation of proton pumping by a eukaryotic P-type ATPase. Science 351, 1469–1473. doi: 10.1126/science.aad6429
Vierstra, R. D. (2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160, 2–14. doi: 10.1104/pp.112.200667
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption Mutations of ADA2b and GCN5 Transcriptional Adaptor Genes Dramatically Affect Arabidopsis Growth, Development, and Gene Expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Voesenek, L., and Bailey-Serres, J. (2013). Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol 16, 647–653. doi: 10.1016/j.pbi.2013.06.008
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Wang, S., Yoshinari, A., Shimada, T., Hara-Nishimura, I., Mitani-Ueno, N., Feng Ma, J., et al. (2017). Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. Plant Cell 29, 824–842. doi: 10.1105/tpc.16.00825
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295. doi: 10.1105/tpc.112.100081
Wege, S., De Angeli, A., Droillard, M. J., Kroniewicz, L., Merlot, S., Cornu, D., et al. (2014). Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci. Signal. 7:ra65. doi: 10.1126/scisignal.2005140
White, M. D., Klecker, M., Hopkinson, R. J., Weits, D. A., Mueller, C., Naumann, C., et al. (2017). Plant cysteine oxidase are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 8:14690. doi: 10.1038/ncomms14690
Wierzbicki, A. T., and Jerzmanowski, A. (2005). Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics 169, 997–1008. doi: 10.1534/genetics.104.031997
Wolfe, K. H., Gouy, M., Yang, Y. W., Sharp, P. M., and Li, W. H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. U.S.A. 86, 6201–6205. doi: 10.1073/pnas.86.16.6201
Wu, S., Alseekh, S., Cuadros-Inostroza,Á., Fusari, C. M., Mutwil, M., Kooke, R., et al. (2016). Combined use of genome-wide association data and correlation networks unravels key regulators of primary metabolism in Arabidopsis thaliana. PLOS Genet. 12:e1006363. doi: 10.1371/journal.pgen.1006363
Xing, D., Wang, Y., Hamilton, M., Ben-Hur, A., and Reddy, A. S. N. (2015). Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27, 3294–3308. doi: 10.1105/tpc.15.00641
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. doi: 10.1038/nature04920
Xu, Z. Y., Kim, S. Y., Hyeon, D. Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Xue, G. P., and Loveridge, C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37, 326–339. doi: 10.1046/j.1365-313X.2003.01963.x
Ye, C.-Y., Yang, X., Xia, X., and Yin, W. (2013). Comparative analysis of cation/proton antiporter superfamily in plants. Gene 521, 245–251. doi: 10.1016/j.gene.2013.03.104
Yelagandula, R., Stroud, H., Holec, S., Zhou, K., Feng, S., Zhong, X., et al. (2014). The Histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158, 98–109. doi: 10.1016/j.cell.2014.06.006
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, S., Mizoi, J., Nakashima, K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286, 321–332. doi: 10.1007/s00438-011-0647-7
Zaag, R., Tamby, J. P., Guichard, C., Tariq, Z., Rigaill, G., Delannoy, E., et al. (2015). GEM2Net: from gene expression modeling to -omics networks, a new CATdb module to investigate Arabidopsis thaliana genes involved in stress response. Nucleic Acids Res. 43, D1010–D1017. doi: 10.1093/nar/gku1155
Zelazny, E., Borst, J. W., Muylaert, M., Batoko, H., Hemminga, M. A., and Chaumont, F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. U.S.A. 104, 12359–12364. doi: 10.1073/pnas.0701180104
Zhang, B., Pasini, R., Dan, H., Joshi, N., Zhao, Y., Leustek, T., et al. (2014). Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J. 77, 185–197. doi: 10.1111/tpj.12376
Zhang, H., Mittal, N., Leamy, L. J., Barazani, O., and Song, B.-H. (2016). Back into the wild-Apply untapped genetic diversity of wild relatives for crop improvement. Evolut. Appl. 10, 5–24. doi: 10.1111/eva.12434
Zhang, K., Sridhar, V. V., Zhu, J., Kapoor, A., and Zhu, J.-K. (2007). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLOS ONE 2:e1210. doi: 10.1371/journal.pone.0001210
Zhang, S., Yang, W., Zhao, Q., Zhou, X., Jiang, L., Ma, S., et al. (2016). Analysis of weighted co-regulatory networks in maize provides insights into new genes and regulatory mechanisms related to inositol phosphate metabolism. BMC Genomics 17:129. doi: 10.1186/s12864-016-2476-x
Zhang, X. Y., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLOS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, W., Bone, J. R., Edmondson, D. G., Turner, B. M., and Roth, S. Y. (1998). Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17, 3155–3167. doi: 10.1093/emboj/17.11.3155
Zhang, X. Y., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Zhou, B.-R., Feng, H., Kato, H., Dai, L., Yang, Y., Zhou, Y., et al. (2013). Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 110, 19390–19395. doi: 10.1073/pnas.1314905110
Zhou, J., Wang, X., He, K., Charron, J.-B. F., Elling, A. A., and Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Keywords: abiotic stress, alternative splicing, protein modification, ion transport, chromatin modification, transcriptional and post-transcriptional regulation, data-driven modeling, network modeling
Citation: Haak DC, Fukao T, Grene R, Hua Z, Ivanov R, Perrella G and Li S (2017) Multilevel Regulation of Abiotic Stress Responses in Plants. Front. Plant Sci. 8:1564. doi: 10.3389/fpls.2017.01564
Received: 05 April 2017; Accepted: 28 August 2017;
Published: 20 September 2017.
Edited by:
Paul E. Verslues, Academia Sinica, TaiwanReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences, JapanCopyright © 2017 Haak, Fukao, Grene, Hua, Ivanov, Perrella and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruth Grene, Z3JlbmVAdnQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.