
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 08 September 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01514
This article is part of the Research TopicInduction of new cell types or organs in plants during biotic interactionsView all 16 articles
Root-knot nematodes (RKN), Meloidogyne spp., are distributed worldwide and impose severe economic damage to many agronomically important crops. The plant cell cycle machinery is considered one of the pivotal components for the formation of nematode feeding sites (NFSs) or galls. These feeding sites contain five to nine hypertrophied giant cells (GC) resulting from developmental reprogramming of host root cells by this pathogen. GC undergo synchronous waves of mitotic activity uncoupled from cytokinesis giving rise to large multinucleate cells. As development of the NFS progresses, multiple rounds of DNA synthesis occur in the nuclei of GC, coupled with nuclear and cellular expansion. These cells are highly metabolically active and provide the nematode with nutrients necessary for its development and completion of its life cycle. In Arabidopsis seven cyclin dependent kinase inhibitors (CKIs) belonging to the interactors/inhibitors of the cyclin dependent kinases (ICK) family, also referred as Kip-Related Proteins (KRPs) have been identified. Interactions of KRPs with CDK/Cyclin complexes decrease CDK activity, affecting both cell cycle progression and DNA content in a concentration-dependent manner. We performed the functional analysis of all Arabidopsis KRP gene members during RKN interaction in Arabidopsis to obtain more insight into their role during gall development. We demonstrated that three members of this family (KRP2, KRP5, and KRP6) were highly expressed in galls and were important for cell cycle regulation during NFS development as shown by their different modes of action. We also pointed out that cell cycle inhibition through overexpression of all members of the KRP family can affect NFS development and consequently compromise the nematode’s life cycle. In this review we summarized our recent understanding of the KRP family of genes, and their role in controlling cell cycle progression at the RKN feeding site.
A large number of crop species worldwide are harmed by plant-parasitic nematodes like root-knot nematodes (RKN), namely Meloidogyne spp. In the case of RKN, roots of the host plants are invaded by the motile, infective second-stage juveniles (J2), which induce dramatic changes in particular to the root vascular cells, ultimately producing a complex nematode feeding site (NFS) (Jones and Northcote, 1972). These sophisticated changes that occur in root cells during nematode infection, involve the formation of root swellings named galls, which contain the giant cells (GC) that are the nematode feeding cells. GC are the main source of nutrients that allow development and reproduction of this parasitic nematode. Parallel to GC development, a network of surrounding neighboring cells (NC) divides asymmetrically, ultimately supporting the transfer of nutrients into the GC (Hoth et al., 2008; Rodiuc et al., 2014). Together GC and NC constitute the complete NFS, which surrounded by cortical and epidermal cells, is apparent in the host root as the typical root-knot or gall.
The large number of host genes involved demonstrates the complexity of the plant–RKN interaction. Transcriptional data showed an extensive regulation of various host molecular pathways, most likely through the crosstalk between the nematode-secreted proteins and their host molecular targets (Gheysen and Mitchum, 2009). Although the molecular mechanisms behind the formation and development of the gall are still far from being completely understood, the activation and regulation of the host cell cycle machinery by RKN has been confirmed to be an essential process leading to the formation of multinucleated GC and gall expansion (de Almeida Engler et al., 2015).
The apparently balanced cell cycle gene expression occurring during the NFS development is characterized by two major cell cycle mechanisms. First, a recurring synchronized mitotic phase uncoupled from cytokinesis that lasts around 10 days in Arabidopsis, drives the formation of the multinucleate state of each giant cell followed by endoreduplication. Multiple rounds of DNA synthesis lacking mitosis accompany GC expansion and nuclei enlargement with a corresponding increase in ploidy level (de Almeida Engler et al., 1999, 2012). During both activation and progression of the cell cycle in NFS, the nematode most likely regulates the formation of the gall by its secreted proteins, and also acquires nutrients from the GC that are required for their development and maturation. RKN development comprises distinct juvenile developmental stages (J1 to J4) up to the formation of the typical egg laying, pear-shaped female, followed by offspring production. Distinct cellular features of these highly metabolically active GC are dense cytoplasm filled with profuse organelles, small vacuoles and cell wall ingrowths (Rodiuc et al., 2014).
The basic control mechanisms that regulate progression through the cell cycle are remarkably well conserved among eukaryotes. The cell cycle in plant cells is controlled by highly conserved complexes formed by cyclin-dependent kinases (CDKs) with their regulatory cyclin subunits (CYCs), which are needed for ensuring correct temporal and unidirectional progression of the cell cycle phases (Inzé and De Veylder, 2006). Expression of cell cycle genes in the plant host appears tightly regulated as the gall matures, implying a strict control of the cell cycle machinery and its molecular components during the plant–nematode interaction (de Almeida Engler et al., 2015). Cell cycle genes identified to date that participate in the ontogeny of RKN-induced galls have been extensively reviewed elsewhere (de Almeida Engler and Gheysen, 2013; de Almeida Engler et al., 2015). Herein, we focus on providing a summarized and global assessment of our recent understanding of a particular family of cyclin-dependent kinase inhibitor (CKI) genes that regulate cell cycle progression in the RKN feeding site using Arabidopsis as a model host.
In Arabidopsis seven CDK inhibitors belonging to the interactors/inhibitors of CDK (ICK) family, also referred to as Kip-Related Proteins (KRPs), have been identified (Wang et al., 1997; De Veylder et al., 2001). The KRPs are small proteins with a C-terminal domain required for CDK- or cyclin-binding, and they function as inhibitors (De Veylder et al., 2001; Inzé and De Veylder, 2006). They have different spatial expression and distinct temporal and functional patterns (De Veylder et al., 2001; Menges and Murray, 2002; Ormenese et al., 2004; Menges et al., 2005; Verkest et al., 2005a; Wang et al., 2006). This family of genes is known to bind to CDKA-CYCD complexes (Wang et al., 2006; Van Leene et al., 2010). The fine-tuning of KRP protein levels in plant cells is a key factor to maintain the balance between cell proliferation and cell differentiation (Verkest et al., 2005b; Inzé and De Veylder, 2006). Interactions of KRPs and CDK/CYCs complexes reduce CDK activity, and can influence both cell cycle progression and DNA replication in a concentration-dependent manner (Verkest et al., 2005b). Because RKN infection leads to the formation of multinucleate GC involving recurrent activity of the host cell cycle machinery, the specific involvement of the complete KRP gene family has been investigated. Our studies revealed that these cell cycle inhibitors exert distinct functions in a RKN-induced feeding site. Transcriptional activity and functional studies of KRP2, KRP5 and KRP6 revealed their participation in the regulation of the cell cycle machinery implicated in gall formation and expansion. Although KRP1, KRP3, KRP4 and KRP7 are not expressed in galls, their ectopic expression inhibited gall development and nematode maturation. Herein, we focused on major results obtained during our functional analyses (summarized in Table 1) of this gene family and briefly discuss their potential use for biotechnology applications.
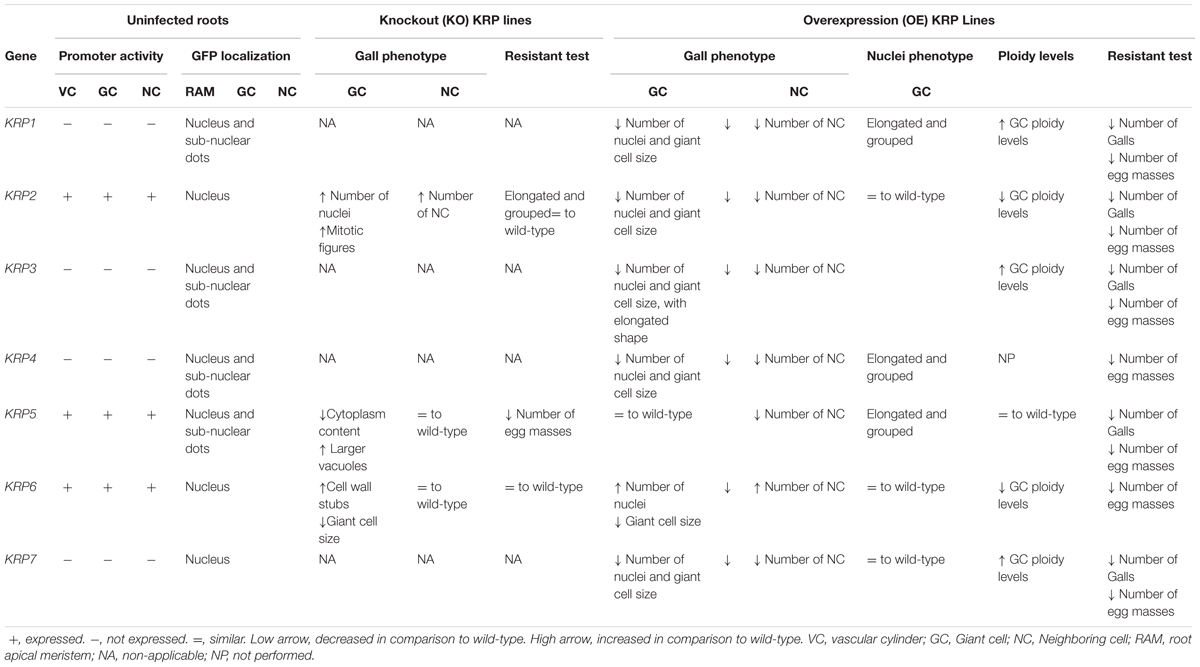
TABLE 1. Summarized overview of functional characterization studies of the Arabidopsis Kip-Related Protein (KRP) family in galls induced by the root-knot nematode Meloidogyne incognita.
Promoter activity as well as expression analysis of the seven Arabidopsis KRP genes (KRP1 to KRP7) have been examined in detail in uninfected roots and during several stages of gall development (Vieira et al., 2012, 2013, 2014; Coelho et al., 2017). Our data illustrated that three out of the seven members of the KRP gene family, namely KRP2, KRP5 and KRP6, are expressed in RKN induced galls and present during a specific time span of the NFS development (Table 1). KRP2, KRP5 and KRP6 transcripts were detected during early gall development [∼7 days after inoculation (DAI)] in agreement with the high mitotic activity occurring in the GC. This suggests that a level of cell cycle regulation must be triggered in order to control GC formation. During gall maturation (>14DAI), only KRP2 and KRP5 showed a significant transcription activity in GC implying their role in the endoreduplication cycle at this stage of gall development (Vieira et al., 2013, 2014; Coelho et al., 2017). Conversely, the remaining KRP1, KRP3, KRP4 and KRP7 genes were not expressed in the galls at any time point during nematode infection. KRP genes (KRP2, KRP5, and KRP6) present in the root vascular of non-infected tissues were also expressed in the galls. Likewise, no particular induction of the remaining KRP genes (KRP1, KRP3, KRP4, and KRP7) was seen in roots upon RKN infection. Therefore, for gall growth and development, KRP genes appeared to be regulated and followed their natural location in the root during nematode infection (Vieira et al., 2013).
Kip-Related Proteins are proteins that show a strict nuclear localization during interphase, with some members, namely KRP1, KRP3, KRP4 and KRP5, also displaying a subnuclear localization (Bird et al., 2007). This punctuated pattern has been associated with chromocenters (Jakoby et al., 2006; Bird et al., 2007), which are densely stained regions linked with chromatin (Jégu et al., 2013). Protein localization followed by time-lapse confocal microscopy of uninfected Arabidopsis root cells revealed that during mitosis KRP1-GFP, KRP3-GFP, GFP-KRP4, and GFP-KRP5 proteins co-localized with chromosomes (Table 1). This localization suggests their activity during interphase, as well as during different stages of mitosis. On the other hand, GFP-KRP2, GFP-KRP6, KRP7-GFP proteins became dispersed into the cytoplasm after nuclear envelope break down during mitosis presenting no apparent co-localization to the chromosomes. These proteins will later uniformly re-accumulate in the nuclei of the two newly formed daughter cells (Vieira et al., 2013, 2014; Coelho et al., 2017).
Our protein localization studies suggest that the dynamic post-translational regulation of the KRP members (KRP2, KRP5 and KRP6) implicated in gall formation might also occur during the RKN–plant interaction. It is still unknown as to whether the post-translational regulation is targeted directly or indirectly by the RKN secreted proteins. Although transcript analyses illustrated continuous detection of KRP2, the protein expression profile exposed a weak GFP-KRP2 accumulation in nuclei of young GC (2-14DAI), while in maturing galls (>14DAI) the enhanced accumulation of GFP-KRP2 in the GC nuclei suggested its involvement in endoreduplication control (Vieira et al., 2013). Previously, we had shown a strong CDKB1;1 promoter activity in young galls (de Almeida Engler et al., 1999). CDKA functions in coordination with B-type CDKs to promote the transition from G2 to mitosis, and KRP2 is considered an important regulator of the G2-M transition in Arabidopsis (Inzé and De Veylder, 2006). As KRP2 operates in a dosage-dependent manner, B-type CDKs phosphorylate KRP2 triggering their destruction and allowing cell proliferation. B-type CDK activity ceases in cells triggered to enter endoreduplication resulting in stabilization of KRPs, which bind and inhibit CDK-CYC complexes that have a role in mitosis (Verkest et al., 2005b). Accordingly, the reduced expression levels of CDKB1;1 at intermediate to advanced GC developmental stages (14–21 DAI) (de Almeida Engler et al., 1999), overlap with the increased labeling of GFP-KRP2 within the GC nuclei (Vieira et al., 2013). Protein profiles suggest that the fluctuations of KRP2 in the GC nuclei may reflect its regulation by CDKB;1 at early time points, allowing mitotic divisions to take place within the GC. At later time points KRP2 accumulation could mediate the switch from a mitotic cell cycle toward the endoreduplication status of the GC.
In the case of KRP5, in vivo examination of RKN-infected roots treated with the proteasome inhibitor MG132, showed enhanced GFP-KRP5 accumulation in the GC nuclei suggesting that KRP5 protein levels are potentially controlled by degradation of the 26S proteasome protein (Coelho et al., 2017). Our observations indicated that KRP5 accompanied the process of GC maturation, and this might be important for cell cycle regulation during the NFS expansion (Coelho et al., 2017). KRP5 has been found to be preferentially expressed in endoreduplicating cells during etiolation and is considered a positive regulator of endoreduplication (Jégu et al., 2013). It has also been shown that KRP5 binds to various loci on chromatin in nuclear localization studies (Jégu et al., 2013). In addition to its CDK/CYC inhibitory capacity, KRP5 has been also implicated in the regulation of transcription, suggesting multifaceted functions for KRP5 that include cell elongation and endoreduplication (Jégu et al., 2013). The fact that KRP5 can target heterochromatin regions suggests that KRP5 is also involved in the modulation of chromatin organization, as its overexpression resulted in an increase of chromocenter decondensation (Jégu et al., 2013). Similar to uninfected plants (Jégu et al., 2013), KRP5 expression could be linked to the expansion of the GC (Coelho et al., 2017).
Detection of GFP-KRP6 within the gall tissues was restricted to the mitotic nuclei of both GC and NC at early stages of gall development (2–10 DAI). As GC development progressed, a decline to absence of GFP-KRP6 expression took place within the GC nuclei, while expression continued in nuclei of the dividing NC (Vieira et al., 2014). This implies that the mitotic phase of GC appears to rely on the presence of proteins like KRP6 that promote mitosis and possibly inhibit cytokinesis (Vieira et al., 2014). The combination of these studies supports the incipient connection between cell cycle activity and the importance of KRP genes as regulatory agents with distinct substrates (e.g., different CDK/CYC complexes) and phase-specific functions. Therefore, KRPs might be acting and contributing to the sequential progression of the cell cycle in galls.
To determine the potential role of KRP genes normally expressed in NFS, loss of function mutants of KRP2, KRP5 and KRP6 were challenged with RKN. For krp2 and krp2-/-/krp6-/- mutants, morphological analyses of galls revealed an intensification of the mitotic activity leading to an increased number of nuclei within the GC and proliferation of the surrounding NC (Vieira et al., 2013). This accelerated mitotic activity in the gall was supported by data showing that loss of function of krp2 increased the rate of lateral root formation (Sanz et al., 2011). In addition, downregulation of multiple KRPs (Anzola et al., 2010) and a combination of multiple knockout KRP genes also contributed to an increase of CDK activity in Arabidopsis that stimulated cell proliferation. This misregulation of the cell cycle in the mutant krp galls most likely contributed to the accumulation of CDKA;1 in roots (Cheng et al., 2013), causing the super-activation of CDK/CYC complexes in the absence of single (Boruc et al., 2010), or multiple KRP members, and thus allowing a faster entry into the mitotic phase during RKN interaction. Despite the enhanced mitotic activity observed in the krp2 and krp2-/-/krp6-/- mutants, GC matured normally, without compromising nematode viability (Vieira et al., 2013).
Mild defects were observed in krp5 knockout lines, mainly at later time points (14 and 21 DAI), as GC induced on these mutants contained less cytoplasm and larger vacuoles in comparison to their respective wild-type (Coelho et al., 2017). Likewise, KRP5 deficiency revealed only very mild defects in Arabidopsis plants, yet, showed some negative effect on the cortical cell size of etiolated hypocotyls (Jégu et al., 2013). Lack of KRP6 led to some extent to the inhibition of the mitotic activity within the GC, as these were significantly smaller than in the wild-type (Vieira et al., 2014). Interestingly, lack of KRP6 resulted in the presence of more cell wall stubs between the nuclei in a fraction of these GC, indicating the potential role of KRP6 in cytokinesis (Vieira et al., 2014). Single mutation in the krp genes caused minimal effects on plant phenotype, probably due to their partial functional redundancy (Cheng et al., 2013). Our results showed that the absence of KRP5 and KRP6 in cells with an amplified cell cycle status like the GC can have subtle deficiencies in their phenotype during gall development, as nematodes associated with krp5 and krp6 lines showed a delayed development and reduced offspring (Vieira et al., 2014; Coelho et al., 2017).
Several studies have demonstrated that KRPs are crucial negative regulators of the cell cycle (Wang et al., 1997; De Veylder et al., 2001; Liu et al., 2008; Jun et al., 2013). However, in vitro inhibition of kinase activity of the CDKA complex by KRPs is variable (Wang et al., 1997), and recent findings suggest that they might be implicated in a broader range of biological roles (Jégu et al., 2013; Vieira et al., 2014). In order to study the ectopic effects of each KRP gene (hereby named as KRP1OE to KRP7OE) in RKN-induced galls, we used a set of microscopy techniques, involving both semi-thin (Figure 1) and thick sectioning to whole-mount analyses of galls at different time points after nematode infection. One of the most prominent features of all KRPOE lines (with exception of KRP6OE) was a visual reduction of the NC asymmetrically surrounding GC, and the reduced size of the GC (Figure 1). Although inhibition levels were variable among the KRP genes, most galls displayed an overall reduced size compared to their wild-type counterpart. Another common feature of ectopic KRP levels (with exception of KRP6OE) was the significant reduction in the number of the GC nuclei associated with a severe blockage of mitotic activity (Vieira et al., 2012, 2013; Coelho et al., 2017).
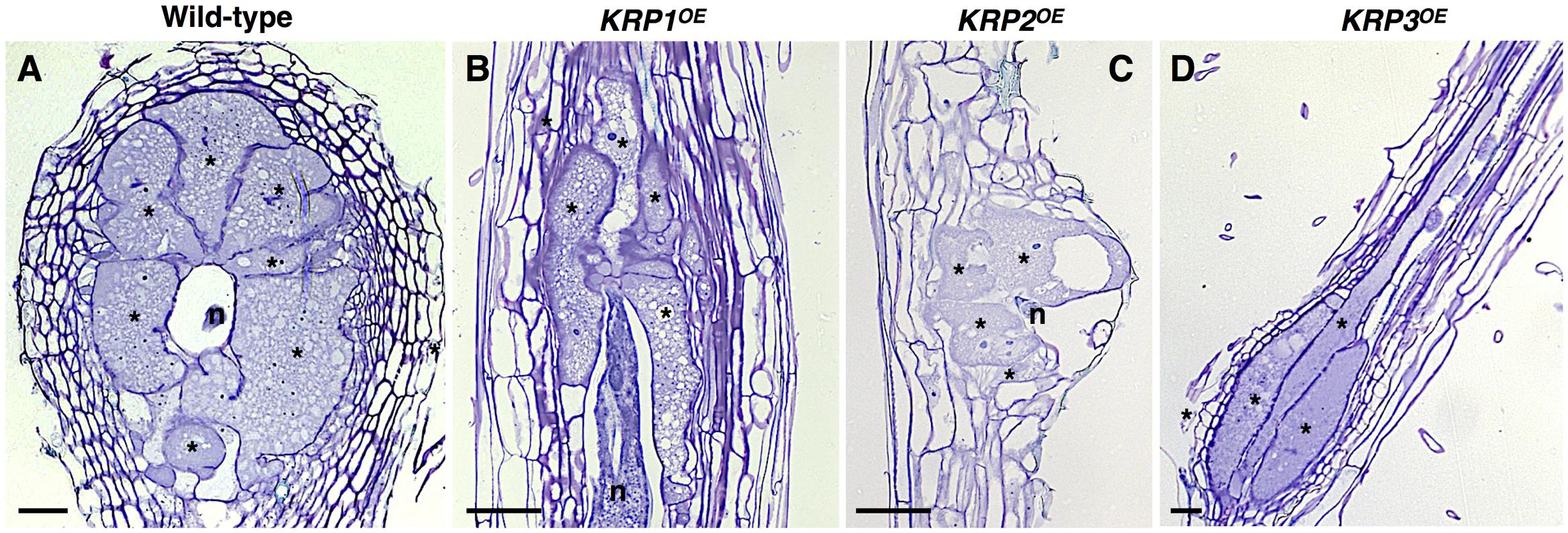
FIGURE 1. Functional analyses of the Arabidopsis Kip-Related Protein (KRP) gene family in galls induced by the root-knot nematode Meloidogyne incognita show a strong inhibition of mitosis in feeding sites leading to a drastic decrease in gall size. (A–D) Examples of the morphological changes observed throughout the ectopic expression of KRP1 (B), KRP2 (C) and KRP3 (D) compared to wild-type (A). An overall reduction of gall size was observed for the KRPOE lines as a consequence of blockage of mitotic activity in giant cells as well as in neighboring cells. A noteworthy change in giant cell morphology was observed for KRP3OE line, resulting in elongated giant cells with reduced size. Asterisk, giant cell; n, nematode. Scale bars = 50 μm.
The importance of polyploidization for gall expansion and maturation has been also demonstrated in plants with impaired endocycle machinery (de Almeida Engler et al., 2012). Endoreduplication occurs in a myriad of organisms and constitutes an effective strategy for cell growth. It is often found in differentiated cells with high metabolic activity (Edgar and Orr-Weaver, 2001) and is used extensively in tissues reserved for nutrient uptake and storage (Lee et al., 2009). In addition, the association between DNA content and cell expansion is regarded as a key process for understanding the mechanism of cell expansion (Kondorosi and Kondorosi, 2004; Breuer et al., 2010). After the mitotic to endocycle transition, progression through the endocycle is modulated by a subset of the same molecular components that control progression through the mitotic cell cycle. These molecular components form a complex regulatory network that produce oscillations in the activity of CDKs responsible for control DNA synthesis, resulting in alternating S and G phases leading to polyploidy (Lee et al., 2009; De Veylder et al., 2011). Therefore, it is important to point out that when ectopically expressed, KRPs inhibit the endoreduplication cycle, as shown by the decreased ploidy levels in Arabidopsis leaves (e.g., KRP2, KRP6) (De Veylder et al., 2001; Liu et al., 2008). The ectopic expression of KRPs also negatively affected the endoreduplication in gall cells, and it was perceived by the reduced ploidy levels for the KRP2OE and KRP6OE infected roots (Vieira et al., 2014).
Among all members of the KRP family, ectopic expression of KRP2 revealed the most conspicuous reduction in size and structure of galls (Vieira et al., 2013). KRP2 has been implicated in the modulation of both mitosis and endoreduplication mechanisms, consistent with its ability to inhibit S-phase CDK activity (Verkest et al., 2005a). Similarly, in RKN-induced galls the effects caused by the ectopic KRP2 expression can be conceptually divided in two phases: a strong KRP2OE expression at early stages of gall development that correlated with decreased nuclear divisions in GC and reduced mitosis in NC. At later stages KRP2 accumulation above their normal threshold levels will arrest endoreduplication of the GC nuclei (Vieira et al., 2013). Based on its regulatory action, KRP2 might have a stronger affinity to enable blocking the CDK/CYC complexes in galls that are necessary for driving the characteristic feeding site induced by RKN.
In a similar trend, artificial KRP1, KRP3, KRP4 and KRP7 expression prompted inhibition of mitosis, and this led to reduced gall sizes, particularly in the overexpressing KRP1OE and KRP7OE lines (Vieira et al., 2012, 2013; Coelho et al., 2017). A noteworthy change in GC morphology was observed in KRP3OE galls, as an ectopic KRP3 expression caused an elongated GC phenotype with reduced cell size (Coelho et al., 2017). Increased levels of KRP3 have been correlated with an alteration in the architecture of the shoot apical meristem, including a reduced dome size and slightly changed cell morphology (Jun et al., 2013). The regulatory mechanisms that control gall organization and GC shape involving the cell cycle have not been reported, but the results obtained after ectopic expression of KRP3 could imply an effect of this gene on cell morphogenesis as well.
In the case of KRP5OE a less severe effect on the gall structure was observed in comparison to KRP1, KRP3, KRP4, and KRP7 overexpression lines. Cell cycle inhibition in the KRP5OE line was sufficient to cause an overall reduction of the gall size, even though KRP5 had less of an effect on cell proliferation when compared to the other KRPs. Although KRP5 is known as a positive regulator of endoreduplication in particular plant tissues (e.g., etiolated seedlings) (Jégu et al., 2013; Wen et al., 2013), stimulation of the endocycle was not observed in GCs recorded by flow cytometry analyses (Coelho et al., 2017).
The most unexpected results were obtained upon ectopic expression of KRP6. In contrast to the other Arabidopsis KRP members, mitotic activity was triggered rather than inhibited in gall tissues overexpressing KRP6. This unexpected effect was caused by the formation of large galls filled with proliferating NC and GC of reduced size filled with a large number of nuclei (Vieira et al., 2014). These results were strengthened by equivalent observations using Arabidopsis cell cultures, where overexpression of KRP6 was correlated with an accelerated G1-to-S and G2-to-M transitions, driving cells to enter mitosis earlier than wild-type. Likewise, phenotypic analysis of cultured Arabidopsis cells revealed a failure in cytokinesis, with the appearance of multinucleated cells as a consequence of ectopic KRP6 expression (Vieira et al., 2014). This outcome could be explained if KRP6 functioned through activation of CDK/CYCD complexes by promoting their assembly or stability, or their targeting to the nucleus, resulting in more efficient phosphorylation of target proteins (Vieira et al., 2014). Overall, KRP6 expression upon RKN infection and the phenotypic resemblance between KRP6OE cell cultures and GC morphology point toward the involvement of KRP6 in the multinucleate and acytokinetic state of GC.
Another intriguing morphological change within mature GC was the presence of aberrant elongated and apparently connected nuclei in the KRP1, KRP3, KRP4 and KRP5, overexpressing lines (Figure 2). In comparison, KRP2OE, KRP6OE and KRP7OE lines displayed the typical amoeboid-shaped nuclei similar to the observed phenotype in wild-type GC. Remarkably, KRP1, KRP3, KRP4 and KRP5 presented a conserved nuclear localization signal motif (YLQLRSRRL) that is responsible for the spots observed in the nuclei (Jakoby et al., 2006; Zhou et al., 2006), and these KRPs co-localized with chromosomes during different phases of mitosis. The specific role of these KRPs during mitosis was unclear, but depletion of these proteins in a precise timing associated with mitosis seemed to be imperative. For example, KRP4 degradation during sister chromatid separation appeared to be a prerequisite for normal mitosis (Boruc et al., 2010). It is conceivable that the progressive cumulative action of these KRPs during GC nuclear division leads to the aberrant, extended nuclei phenotype. The possible consequences of such KRP accumulation could involve defective DNA replication, erroneous chromosome segregation, or defects on chromosome organization within the nuclear envelope. Recent nuclear analysis of the aberrant nuclei seen in mature GC upon ectopic KRP3 and KRP5 expression revealed that the increased nuclear volume may not only be linked to increased ploidy levels, but also to be the result of accumulation of mitotic defects (Antonino de Souza et al., 2017).
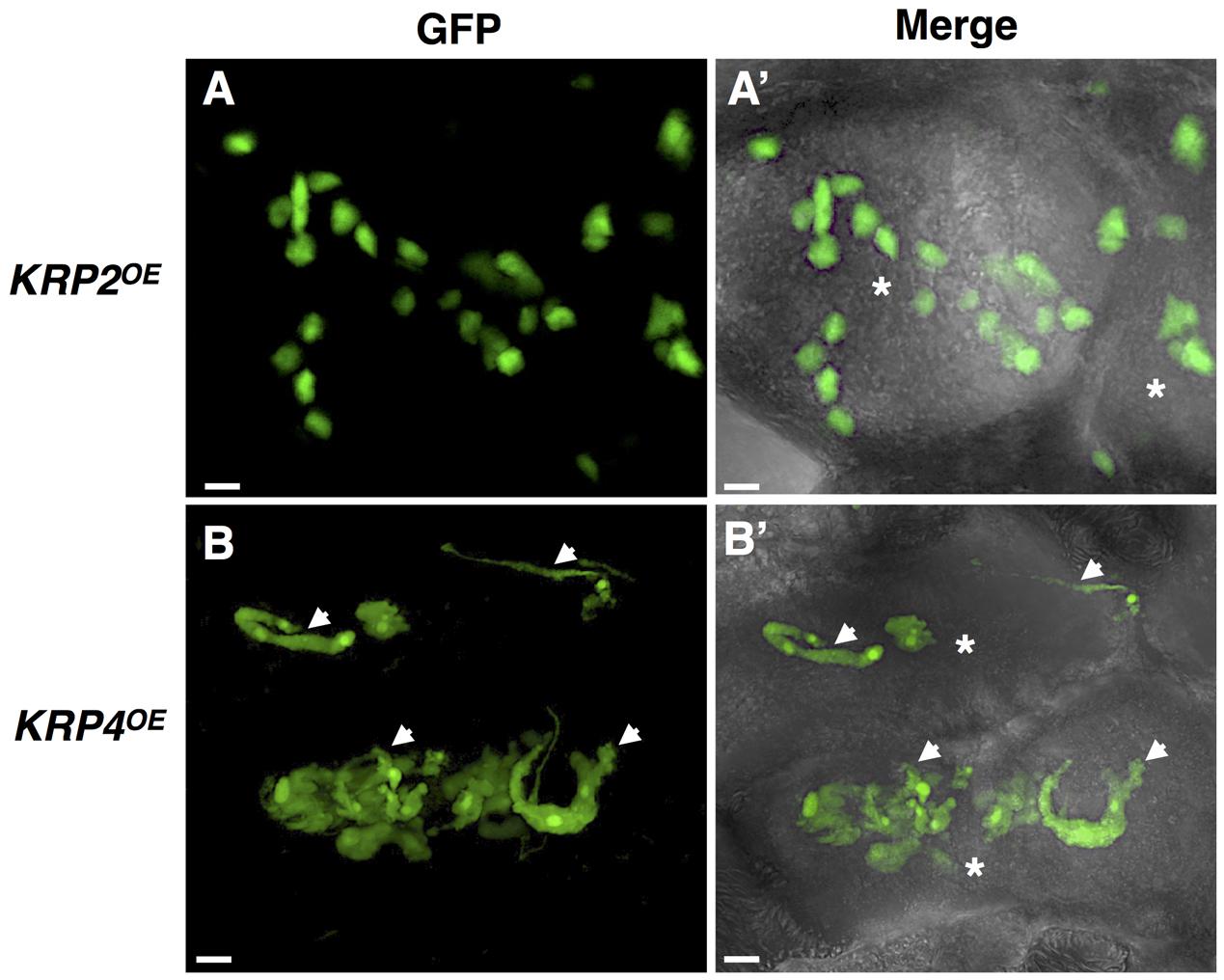
FIGURE 2. In vivo nuclear localization of GFP-KRP2 and GFP-KRP4 in induced galls of Meloidogyne incognita. Images are 3D confocal projections. (Left) Correspond to GFP fluorescence images, and (Right) Overlays of GFP fluorescence with differential interference contrast. While KRP2OE lines (A,A’) displayed the typical amoeboid shaped nuclei similar to the observed phenotype in wild-type giant cell, the KRP4OE lines (B,B’) presented aberrant elongated and apparently connected nuclei (white arrows). Asterisk, giant cell. Bars = 5 μm.
The RKN NFS is considered a resilient metabolic sink, showing features of specialized transfer cells with high metabolic activity and an increased cytoplasmic density. The last feature implicates the replacement of a large central vacuole by several smaller ones, an increased number and size of nuclei, proliferation of organelles and thickened cell walls with finger-like protuberance allowing for an increase in membrane surface for solute uptake (Berg et al., 2008; Rodiuc et al., 2014). As mentioned above, KRP overexpression caused a decrease in GC size and structure, thus reducing the capacity of acting as a sink-like cell and apparently slowing down the metabolic activity needed for gall functioning. In parallel, the reduced network of NC that surrounds the GC may exert a significant impact by causing a reduction of nutrient transfer into the GC needed for nematode development.
It is remarkable that RKN were able to induce a feeding site in all KRPOE lines, reinforcing the robust capability of the RKN to maneuver the plant cell cycle machinery in their favor. Despite the different levels of decreased gall size observed for the different KRPOE lines, these lines were able to limit the nutrient demands for RKN development and affect offspring production. Restrained nematode development was more pronounced in the KRP1OE, KRP2OE and KRP7OE lines and to a lesser extent in the remaining KRP members (Vieira et al., 2012, 2013, 2014; Coelho et al., 2017).
Former efforts have been conducted toward understanding the molecular mechanisms driving the formation and development of RKN feeding sites (Gheysen and Mitchum, 2009). The importance of a balanced cell cycle during gall formation and development has been supported by several studies (de Almeida Engler et al., 1999, 2012; Vieira et al., 2013). Functional analyses of the seven KRP inhibitor genes of Arabidopsis revealed that different family members might exert distinct functions during RKN feeding site development, although possessing an inhibitory effect in their cell cycle machinery. During gall development a level of regulation of the cell cycle by KRP2, KRP5 and KRP6 most likely takes place in order to prevent irreversible damage to the host plant root. These three KRPs naturally expressed in galls might fine-tune CDK activity and control the transition of the mitotic cycle to the endocycle. The control of the gall-related cell cycle machinery does not seem to involve the activation of the four remaining KRPs (KRP1, KRP3, KRP4 and KRP7). Nevertheless, our functional studies ectopically expressing KRPs have shown that inhibition of the cell cycle CDK/CYC complexes compromised nematode development leading to decreases in their offspring.
A detailed analysis of the diverse members of the KRP multi-gene family highlighted the extent of the functional regulation of this family of CKIs during gall formation and uncovered potential novel functions for this family. The occurrence of an aberrant cell cycle in GC makes them a useful model for dissecting cell cycle gene function in galls, and as well as in plant cells. These studies provide an example of how artificial and continuous expression of KRP inhibitors in GCs can be deleterious for gall development. Future studies focused on engineering expression of KRP genes using promoters preferentially active in galls are in progress and can be considered an attractive route for RKN control. For this approach, within this family of cell cycle inhibitors, KRP2 followed by KRP1, KRP7, KRP3 and KRP4 seem to be the best candidates to be considered for management of the cell cycle in galls. An analogous strategy might be envisaged for controlling the infection of other plant pathogens that exploit the host cell cycle machinery.
PV and JAE wrote the review and thank all the authors who contributed with data for all KRP manuscripts mentioned herein.
PV is funded by National Funds through FCT – Foundation for Science and Technology under the Project UID/AGR/00115/2013.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are very grateful to all authors of the manuscripts related to this work, to Dr. Gilbert Engler for critically reading this manuscript, and Dr. Kathryn Kamo for editing the final version.
Antonino de Souza, J. D. Jr., Pierre, O., Coelho, R. R., Grossi-de-Sa, M. F., Engler, G., and de Almeida Engler, J. (2017). Application of nuclear volume measurements to comprehend the cell cycle in root-knot nematode-induced giant cells. Front. Plant Sci. 8:e961. doi: 10.3389/fpls.2017.00961
Anzola, J. M., Sieberer, T., Ortbauer, M., Butt, H., Korbei, B., Weinhofer, I., et al. (2010). Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. U.S.A. 107, 10308–10313. doi: 10.1073/pnas.0913918107
Berg, R. H., Fester, T., and Taylor, C. G. (2008). “Development of the root-knot nematode feeding cell,” in Plant Cell Monographs, Cell Biology of Plant Nematode Parasitism, eds R. H. Berg and C. G. Taylor (Berlin: Springer), 115–152.
Bird, D. A., Buruiana, M. M., Zhou, Y., Fowke, L. C., and Wang, H. (2007). Arabidopsis cyclin-dependent kinase inhibitors are nuclear-localized and show?different localization patterns within the nucleoplasm. Plant Cell Rep. 26, 861–872. doi: 10.1007/s00299-006-0294-3
Boruc, J., Van den Daele, H., Hollunder, J., Rombauts, S., Mylle, E., Hilson, P., et al. (2010). Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22, 1264–1280. doi: 10.1105/tpc.109.073635
Breuer, C., Ishida, T., and Sugimoto, K. (2010). Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 13, 654–660. doi: 10.1016/j.pbi.2010.10.006
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., and Liu, H. et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Coelho, R. R., Vieira, P., Sousa Junior, J. D. A., Martin-Jimenez, C., De Veylder, L., Cazareth, J., et al. (2017). Exploiting cell cycle inhibitor genes of the KRP family to control root-knot nematode induced feeding sites in plants. Plant Cell Environ. 40, 1174–1188. doi: 10.1111/pce.12912
de Almeida Engler, J., De Vleesschauwer, V., Burssens, S., Celenza, J. L., Inzé, D., Van Montagu, M., et al. (1999). Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11, 793–808. doi: 10.1105/tpc.11.5.793
de Almeida Engler, J., and Gheysen, G. (2013). Nematode-induced endoreduplication in plant host cells: why and how? Mol. Plant Microbe Interact. 26, 17–24. doi: 10.1094/MPMI-05-12-0128-CR
de Almeida Engler, J., Kyndt, T., Vieira, P., Van Cappelle, E., Bouldolf, V., Sanchez, V., et al. (2012). CCS52 and DEL1 genes are key components of the endocycle in nematode induced feeding sites. Plant J. 72, 185–198. doi: 10.1111/j.1365-313X.2012.05054.x
de Almeida Engler, J., Vieira, P., Rodiuc, N., Grossi de Sa, M. F., and Engler, G. (2015). The plant cell cycle machinery: usurped and modulated by plant parasitic nematodes. Adv. Bot. Res. 73, 91–118. doi: 10.1016/bs.abr.2014.12.003
De Veylder, L., Beeckman, T., Beemster, G. T. S., Krols, L., Terras F., Landrieu I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. doi: 10.1105/tpc.13.7.1653
De Veylder, L., Larkin, J. C., and Schnittger, A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634. doi: 10.1016/j.tplants.2011.07.001
Edgar, B. A., and Orr-Weaver, T. L. (2001). Endoreplication cell cycles: more for less. Cell 105, 297–306. doi: 10.1016/S0092-8674(01)00334-8
Gheysen, G., and Mitchum, M. G. (2009). “Molecular insights in the susceptible plant response to nematode infection”, in Plant Cell Monographs: Cell Biology of Plant Nematode Interactions, eds R. H. Berg and C. G. Taylor (Berlin: Springer), 45–81. doi: 10.1007/978-3-540-85215-5_3
Hoth, S., Stadler, R., Sauer, N., and Hammes, U. Z. (2008). Differential vascularization of nematode-induced feeding sites. Proc. Natl. Acad. Sci. U.S.A. 105, 12617–12622. doi: 10.1073/pnas.0803835105
Inzé, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
Jakoby, M. J., Weinl, C., Pusch, S., Kuijt, S. J., Merkle, T., Dissmeyer, N., et al. (2006). Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 141, 1293–1305. doi: 10.1104/pp.106.081406
Jégu, T., Latrasse, D., Delarue, M., Mazubert, C., Bourge, M., and Hudik, E. et al. (2013). Multiple functions of Kip-related protein5 connect endoreduplication and cell elongation. Plant Physiol. 161, 1694–1705. doi: 10.1104/pp.112.212357
Jones, M. G. K., and Northcote, D. H. (1972). Multinucleate transfer cells induced in coleus roots by the root-knot nematode, Meloidogyne arenaria. Protoplasma 75, 381–395. doi: 10.1007/BF01282117
Jun, S. E., Okushima, Y., Nam, J., Umeda, M., and Kim, G.-T. (2013). Kip-related protein 3 is required for control of endoreduplication in the shoot apical meristem and leaves of Arabidopsis. Mol. Cells 35, 47–53. doi: 10.1007/s10059-013-2270-4
Kondorosi, E., and Kondorosi, A. (2004). Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 567, 152–157. doi: 10.1016/j.febslet.2004.04.075
Lee, H. O., Davidson, J. M., and Duronio, R. J. (2009). Endoreduplication: polyploidy with purpose. Genes Dev. 23, 2461–2477. doi: 10.1101/gad.1829209
Liu, J., Zhang, Y., Qin, G., Tsuge, T., Sakaguchi, N., Luo, G., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING- type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20, 1538–1554. doi: 10.1105/tpc.108.059741
Menges, M., de Jager, S. M., Gruissem, W., and Murray, J. A. H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41, 546–566. doi: 10.1111/j.1365-313X.2004.02319.x
Menges, M., and Murray, J. A. H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. doi: 10.1046/j.1365-313X.2002.01274.x
Ormenese, S., de Almeida Engler, J., De Groodt, R., De Veylder, L., Inzé, D., and Jacqmard, A. (2004). Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann. Bot. 93, 575–580. doi: 10.1093/aob/mch077
Rodiuc, N., Vieira, P., Banora, M., and de Almeida Engler, J. (2014). On the track of transfer cells formation by specialized plant-parasitic nematodes. Front. Plant Sci. 5:160. doi: 10.3389/fpls.2014.00160
Sanz, L., Dewitte, W., Forzani, C., Patell, F., Nieuwland, J., Wen, B. et al. (2011). The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23, 641–660. doi: 10.1105/tpc.110.080002
Van Leene, J., Hollunder, J., Eeckhout, D., Persiau, G., Van De Slijke, E., Stals, H., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397. doi: 10.1038/msb.2010.53
Verkest, A., Manes, C.-L., Vercruysse, M., Maes, S., Van Der Schueren, E., and Beeckman, T. et al. (2005a). The cyclin-dependent kinase inhibitor KRP2 controls the mitosis-to-endocycle transition during Arabidopsis leaf development through a specific inhibition of the mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736. doi: 10.1105/tpc.105.032383
Verkest, A., Weinl, C., Inzé, D., De Veylder, L., and Schnittger, A. (2005b). Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol. 139, 1099–1106. doi: 10.1104/pp.105.069906
Vieira, P., De Clercq, A., Stals, H., Van Leene, J., Van De Slijke, E., Van Isterdael, G., et al. (2014). The cyclin-depedent kinase inhibitor KRP6 induces mitosis and impairs cytokinesis in giant cells induced by plant-parasitic nematodes in Arabidopsis. Plant Cell 26, 2633–2647. doi: 10.1105/tpc.114.126425
Vieira, P., Engler, G., and de Almeida Engler, J. (2012). Whole-mount confocal imaging of nuclei in giant feeding-cells induced by root-knot nematodes in Arabidopsis. New Phytol. 195, 488–496. doi: 10.1111/j.1469-8137.2012.04175.x
Vieira, P., Escudero, C., Rodiuc, N., Boruc, J., Russinova, E., Glab, N., et al. (2013). Ectopic expression of Kip-related proteins restrains root-knot nematode-feeding site expansion. New Phytol. 199, 505–519. doi: 10.1111/nph.12255
Wang, H., Fowke, L. C., and Crosby, W. L. (1997). A plant cyclin-dependent kinase inhibitor gene. Nature 386, 451–452. doi: 10.1038/386451a0
Wang, H., Zhou, Y., and Fowke, L. C. (2006). The emerging importance of cyclin-dependent kinase inhibitors in the regulation of the plant cell cycle and related processes. Can. J. Bot. 84, 640–650. doi: 10.1139/b06-043
Wen, B., Nieuwland, J., and Murray J. A. (2013). The Arabidopsis CDK inhibitor ICK3/KRP5 is rate limiting for primary root growth and promotes growth through cell elongation and endoreduplication. J. Exp. Bot. 64, 1135–1144. doi: 10.1093/jxb/ert009
Keywords: cyclin dependent kinase, giant cells, kip-related proteins, Meloidogyne incognita, Arabidopsis thaliana
Citation: Vieira P and de Almeida Engler J (2017) Plant Cyclin-Dependent Kinase Inhibitors of the KRP Family: Potent Inhibitors of Root-Knot Nematode Feeding Sites in Plant Roots. Front. Plant Sci. 8:1514. doi: 10.3389/fpls.2017.01514
Received: 24 May 2017; Accepted: 17 August 2017;
Published: 08 September 2017.
Edited by:
Mary C. Wildermuth, University of California, Berkeley, United StatesReviewed by:
Lei Zhang, Washington State University, United StatesCopyright © 2017 Vieira and de Almeida Engler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janice de Almeida Engler, amFuaWNlLmRlLWFsbWVpZGFAaW5yYS5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.